Abstract
The phosphorylated histone variant, γ-H2AX, is known to play a key role in DNA damage repair. However, the clinical significance of H2AX mRNA expression in breast cancer remains unclear. Utilizing a bioinformatical approach, a total of 3594 breast cancer patients with clinical and transcriptomic data were investigated. Bioinformatical analysis showed that high expression of H2AX is associated with worse disease-free, disease-specific, and overall survival consistently in two independent cohorts. High H2AX expressing tumors were associated with upregulated DNA repair gene sets. Although H2AX was not predictive of chemotherapy response, it was significantly downregulated after effective chemotherapy or radio-chemotherapy. Notably, tumors with high H2AX expression were enriched for DNA replication and MYC targets gene sets, and associated with increased MKI67 expression, suggesting alterations in cell proliferation machinery. H2AX knockdown cells showed decreased cell proliferation as compared to the control cells. Finally, H2AX mRNA expression was higher in the metastatic clones as compared to the parental cells and in the metastatic tumors as compared to the primary tumors in patients, with higher H2AX mRNA expression found in advanced stage cancer patients. In conclusion, high H2AX mRNA expression is associated with increased DNA repair, cell proliferation, metastasis, and worse survival in breast cancer patients.
Keywords: H2AX, breast cancer, mRNA, prognosis, cancer, DNA repair
Introduction
Breast cancer is the second leading cause of cancer-related death among women both in the United States and worldwide [1]. Despite a 5-year survival rate of 90%, 40,000 women still die from breast cancer every year in the US [2,3]. These facts tell us that further investigation of cancer recurrence and metastasis is the key to improve breast cancer survival. In this regard, remarkable progress in genomic analyses during the last decade now allows researchers to link cancer biology with its clinical relevance utilizing bioinformatical analyses [4]. In addition to experimental approach which is essential for deeper understanding in mechanisms of cancer biology, investigation of the clinical relevance of these mechanisms is important to apply the knowledge to clinical practice that may have a direct impact on patient outcome.
H2AX is a variant type of one of a core histone H2A. H2AX is phosphorylated to form γ-H2AX, which plays a key role in DNA damage repair when DNA double-strand breaks occur [5,6], thus it is often used as a marker of DNA damage and repair. A prior study has shown that upregulation of H2AX is associated with generation of reactive oxygen species (ROS) [7-10]. It was also reported that increased cell proliferation by activated MYC signaling-induced DNA damage correlated with increased levels of ROS [11]. High expression of γ-H2AX was observed in various types of cancer as compared to normal tissue [12]. High expression of γ-H2AX, which accounts for 5-20% of the each cohort, are associated with patient prognosis and treatment response in some types of cancer [13], where it is associated with early metastasis in breast cancer [12]. However, the clinical relevance of H2AX mRNA expression in breast cancer remains unclear. Compared to immunohistochemical detection of γ-H2AX expression, assessment of H2AX mRNA expression is less subjective and cost effective with less proficiency bias. In this study, we hypothesized that extremely high H2AX mRNA expression is associated with DNA repair, cancer aggressiveness and patient poor prognosis similar to γ-H2AX in breast cancer.
Materials and methods
Data acquisition and pre-processing
We used publicly available cohort, The Cancer Genome Atlas (TCGA) [14], as a discovery cohort. Out of 1098, 1093 patients have both mRNA expression from RNA sequence and clinical data. Among them, 1090 and 999 patients have overall survival (OS) and disease-free survival (DFS) data, respectively. As a validation cohort, we obtained Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort [15]. Out of 1904 patients which have both clinical and gene expression data, 481 patients who died of other cause were excluded from disease-specific survival (DSS) analysis. The clinical data and gene expression level quantification data (mRNA expression Z-score from RNA sequence) were downloaded through cBioPortal [16,17] for both TCGA and METABRIC cohorts. As chemotherapy cohorts, we obtained datasets from the Gene Expression Omnibus (GEO) database and gene expression data was analyzed after log2 transformed. As a chemotherapy cohort, we used GSE28844 [18], in which there are 28 breast cancer patients who treated with anthracycline based regimen with tumor gene expression data of before and after chemotherapy. Out of 28 patients, 5, 10 and 13 patients were diagnosed pathologically good (Miller & Payne grade 4 or 5), mid (Miller & Payne grade 3), and bad response (Miller & Payne grade 1 or 2) to the chemotherapy [18]. We analyzed GSE25066 [19] as another neoadjuvant cohort. Out of 488 patients treated by neoadjuvant taxane-anthracycline chemotherapy, 99 and 389 patients were diagnosed as pathological complete response (pCR) and residual disease (RD). As a neoadjuvant radio-chemotherapy cohort, GSE15781 [20] was analyzed. Among 21 resectable rectal cancer patients, 10 and 11 patients were undertaken surgery with or without neoadjuvant radio-chemotherapy. As a neoadjuvant radio-chemotherapy breast cancer cohort, GSE22513 [21] was analyzed. Out of 14 patients treated with paclitaxel followed by concurrent paclitaxel and radiation therapy, 4 and 10 patients were diagnosed as pCR or non-pCR. As a metastatic breast cancer cohort, GSE110590 [22] was analyzed. There were 13, 14, 12 and 10 tumors from primary, liver, lung and brain metastatic lesions of 16 metastatic breast cancer patients.
Cell culture and reagents
Human breast cancer cell lines, MCF7 and MDA-MB-231 were obtained from ATCC (Manassas, VA). The lung metastatic clone of MDA-MB-231, LM2-4 was kindly provided by Prof. Robert S. Kerbel of Sunnybrook Research Institute, University of Toronto [23]. Murine breast cancer cell line, 4T1 was obtained from Caliper Life Science (Hopkinton, MA) and its bone metastatic clone, 4T1.2 was kindly provided by Prof. Cheryl L. Jorcyk of Boise State University. All cell lines were cultured in RPMI-1640 (Gibco, Gaithersburg, MD) with 10% fetal bovine serum (FBS) (Gibco) in a humidified incubator at 37°C in 5% CO2. All cell lines were used within 20 passages after revival and were shown to be mycoplasma free using the PlasmoTest kit (InVivoGen, San Diego, CA). For H2AX knockdown experiments, MCF7 and 4T1 cells were transduced with shNT (non-targeting) or shH2AX and selected under either 2 µg/ml of puromycin (Gibco) for MCF7 or 200 µg/ml of G418 (Sigma) for 4T1 to establish stable knockdown cells. shRNA sequences are shown in Table S1.
Western blot
Cells were lysed with NP40 cell lysis buffer (Invitrogen, Carlsbad, CA), and lysates were separated by electrophoresis and transferred onto a nitrocellulose membrane. Membranes were blocked with 5% milk for 1 h at room temperature, and then incubated with primary antibody (H2AX; 1:500, from Cell Signaling Technology (Danvers, MA), Actin; 1:40,000 from Millipore Sigma (Burlington, MA)) at 4°C overnight. Blots were developed with HRP-labelled secondary antibodies, followed by Clarity Western ECL detection system (BioRad, Hercules, CA). Chemiluminescence signals were acquired using a ChemiDoc MP imager (BioRad).
Cell proliferation
3,000 cells were seeded per well of a 96-well plate and incubated overnight. Cell proliferation was measured at the indicated time point using either Cell Counting Kit-8 (Dojindo, Japan) or CellTiter 96 AQueous One Solution Cell Proliferation Assay Kit (Promega, Madison, WI), according to the manufacturer’s protocols.
Gene expression analysis of cell lines with RNA sequence
Gene expression of human breast cancer cell line MDA-MB-231 comparing with its highly lung metastatic clone LM2-4 (n=3, each) [24], and murine mammary adenocarcinoma cell line 4T1 comparing with its highly bone metastatic clone 4T1.2 (n=3, each) [25] were analyzed by RNA sequence using the TruSeq Stranded Total RNA kit (Illumina). Data was normalized as Reads Per Kilobase of transcript, per Million mapped reads (RPKM) and then log2 transformed.
Mice experiment
Approval from the Roswell Park Comprehensive Cancer Center Animal Care and Use Committee was obtained for all experiments. Female BALB/c mice were obtained from Jackson Laboratory. 1×105 of either control or H2AX knockdown 4T1 cells were inoculated into the #2 mammary fat pad. Radical mastectomy was performed 8 days after cell inoculation as described before [26,27]. Total bioluminescence was measured at 5 minutes intervals up to 40 minutes by IVIS spectrum (PerkinElmer), then determined by the peak value over this time frame.
Statistical analysis
For gene expression comparison, statistical analyses were performed using Student t-tests. The prognostic differences between H2AX high and low expression tumors, including OS, DFS and DSS, were analyzed using Kaplan-Meier curve with log-rank test. The patients were dichotomized based on H2AX expression and cutoff points were determined as top 5 percent in TCGA and METABRIC cohorts. Gene set enrichment analysis (GSEA) was performed by comparing H2AX high and low expressing tumors using software provided by the Broad Institute. False discovery rate (FDR) <0.25 was considered as statistically significant. All statistical analyses were performed using R software (http:///www.r-project.org/) together with Bioconductor (http://bioconductor.org/) and GraphPad Prism (GraphPad).
Results
High H2AX expression is associated with worse survival in breast cancer patients
Based on previous reports that γ-H2AX expression is elevated in 5-20% of tumors in a given cohort [13], we defined the top 5% as H2AX high expression group in the each cohort. Patients with high expression of H2AX tumor showed significantly worse DFS (Figure 1A, 5-year DFS rates; 65.0% vs 82.4%, P=0.010) and OS (Figure 1B, 5-year OS rates; 68.6% vs 82.7%, P=0.032) in TCGA cohort. These results were validated in the METABRIC cohort, where high expression of H2AX was associated with significantly worse DSS (Figure 1C, 5-year DSS rates; 70.8% vs 85.6%, P=0.002). These findings suggest that extremely high expression of H2AX is associated with poor survival of breast cancer patients.
Figure 1.
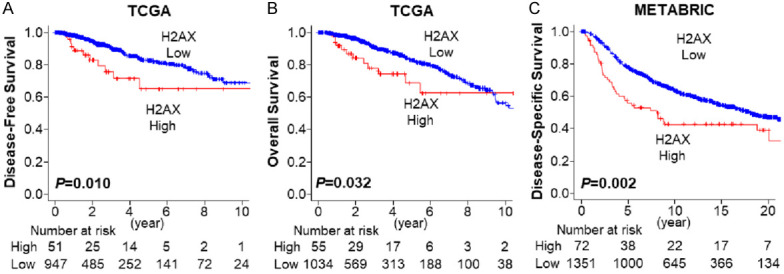
Breast cancer patient survival by H2AX expression. A. Disease-free survival by H2AX expression in TCGA breast cancer cohort. H2AX high: n=51, H2AX low: n=947. B. Overall survival by H2AX expression in TCGA breast cancer cohort. H2AX high: n=55, H2AX low: n=1034. C. Disease-specific survival by H2AX in METABTIC breast cancer cohort. H2AX high: n=72, H2AX low: n=1351. Red and blue lines represent H2AX high and low expression patients.
When we stratified breast cancer patients into subtypes, H2AX expression was higher in the ER-negative tumors as compared to the ER-positive tumors in both TCGA (Figure S1A, P<0.001) and METABRIC (Figure S1D, P<0.001) cohorts. However, the survival difference was not consistent in TCGA and METABRIC cohorts (Figure S1B, S1C, S1E, S1F). Similar findings were seen in triple-negative breast cancers (TNBCs), in which H2AX expression was significantly higher in TNBC as compared to non-TNBC in both TCGA (P<0.001) and METABRIC (P<0.001) cohorts (Figure S2A, S2D), but there was no consistent survival difference in H2AX high expressing tumor between TCGA and METABRIC cohorts (Figure S2B, S2C, S2E, S2F). H2AX expression was significantly higher in the HER2-positive tumors as compared to HER2-negative tumors in both TCGA (P=0.048) and METABRIC (P<0.001) cohorts (Figure S3A, S3D). There was no significant difference in survival by H2AX expression in HER2-positive or negative tumors in both TCGA and METABRIC cohorts (Figure S3B, S3C, S3E, S3F). These findings imply that the impact of H2AX expression on patient survival is not affected by subtype.
High H2AX expressing tumor is associated with DNA repair
Since γ-H2AX indicates an early cellular response to DNA double-strand breaks [28], we hypothesized that tumors that express extremely high levels of H2AX represent high γ-H2AX to promote DNA repair. As we expected, GSEA revealed that H2AX high breast cancer patients show significant enrichment in DNA repair and base excision repair gene sets consistently in both TCGA (DNA repair; normalized enrichment score (NES) =2.00, FDR=0.009, Base excision repair; NES=1.87, FDR=0.024) and METABRIC cohorts (DNA repair; NES=1.44, FDR=0.130, Base excision repair; NES=1.82, FDR=0.025) (Figure 2A). Although high γ-H2AX was reported to be associated with increased reaction oxygen species (ROS) in cancer [11], H2AX high expressing tumor showed enrichment of ROS gene set only in TCGA (NES=1.81, FDR=0.038), but not in METABRIC (NES=0.84, FDR=0.823) (Figure 2B). These findings suggest that H2AX high expressing tumor is associated with DNA repair and base excision repair, which is in agreement with elevated γ-H2AX.
Figure 2.
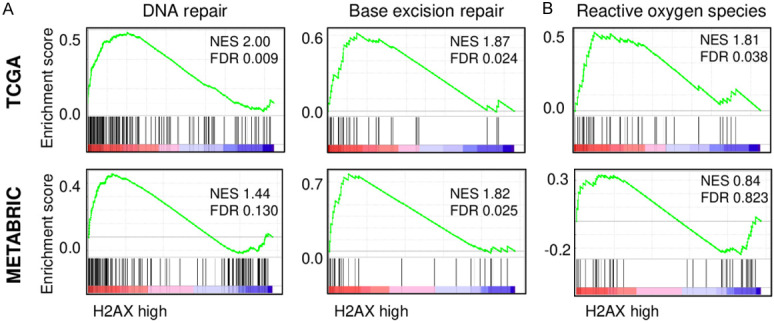
The association of H2AX with DNA repair and reactive oxygen species pathway in breast cancer. A. Gene set enrichment analysis (GSEA) of DNA repair and base excision repair pathway with respect to high and low H2AX levels in breast cancers in TCGA (upper) and METABRIC (lower) cohort. B. GSEA of reactive oxygen species pathway with respect to high and low H2AX levels in breast cancer in TCGA (upper) and METABRIC (lower) cohort. H2AX high: n=55, H2AX low: n=1034 for TCGA, H2AX high: n=72, H2AX low: n=1351 for METABRIC.
H2AX expression decreases with effective chemotherapy
Since H2AX high expressing tumors were associated with activation of DNA repair pathway, we investigated changes in H2AX mRNA expression with DNA damaging chemotherapy treatments. H2AX expression levels in breast cancer before the treatment did not predict response to epirubicin or doxorubicin in GSE28844 cohort (Figure 3A). In another neoadjuvant cohort (GSE25066), the expression levels of H2AX were trend to be higher in pCR patients as compared to RD patients, however, it did not reach the statistical difference (P=0.065) (Figure 3B). Interestingly, we found that H2AX expression level was significantly downregulated after neoadjuvant chemotherapy as compared to the pre-treatment tumor (Figure 3C, P=0.004), particularly in the tumor with good response to chemotherapy (P<0.001), but not in the mid (P=0.384) or bad (P=0.151) response (Figure 3D). On the other hand, the expression levels of other H2A coding genes, HIST1H2AB, HIST1H2AC and HIST1H2AE did not show the difference before and after treatment (Figure S4). These findings suggest that H2AX level does not predict chemotherapy response, however, its decrease was associated with good response to chemotherapy independent of H2A.
Figure 3.
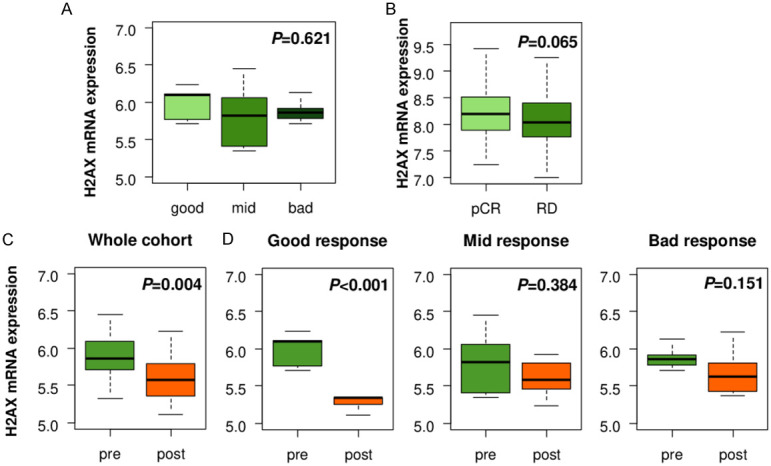
Association between H2AX levels and chemotherapy in breast cancer. (A) H2AX expression assessment in pre-chemotherapy tumors by treatment response in GSE28844 breast cancer cohort. Good response: n=5, mid response: n=10, bad response: n=13. (B) H2AX expression comparison between pathological complete response (pCR) and residual disease (RD) groups in GSE25066 breast cancer cohort. pCR: n=99, RD: n=389. H2AX expression comparison between pre- and post-chemotherapy tumors in (C) whole cohort (n=28), (D) good (n=5), mid (n=10), and bad (n=13) response group in GSE28844 breast cancer cohort.
H2AX expression decrease with radio-chemotherapy
It is reported that high γ-H2AX expression tumor shows resistance to radiation therapy in colorectal cancer [29]. Thus, we investigated if H2AX mRNA expression is also associated with radiotherapy response. We found that H2AX expression in the tumor after radio-chemotherapy (RCT) was significantly lower as compared to the non-treated tumors in rectal cancer cohort, GSE15781 (Figure 4A, P<0.001). This change in H2AX expression by RCT was not seen in the normal tissue (Figure 4A, P=0.293). Interestingly, the H2AX level trended to be higher in the tumors achieved pCR after RCT as compared to the non-pCR tissues among the breast cancers treated by paclitaxel chemotherapy followed by concurrent paclitaxel/radiation. However, it did not reach a statistically significant difference (Figure 4B, P=0.188).
Figure 4.
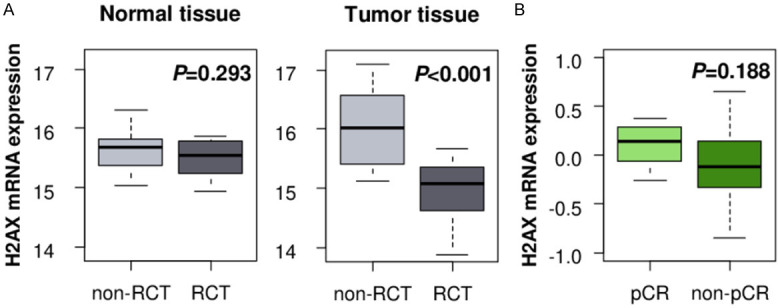
Association between H2AX and radiation therapy. A. H2AX expression analysis of post-treatment normal and tumor tissues with and without neoadjuvant radio-chemotherapy (RCT) in rectal cancer cohort GSE15781. Non-RCT normal: n=10, RCT normal: n=10, non-RCT tumor: n=13, RCT tumor: n=9. B. H2AX expression comparison in pre-RCT treatment tumors between pathological complete response (pCR, n=8) and non-pCR (n=20) tumors in breast cancer cohort GSE22513.
H2AX high expressing tumor is associated with increased cell proliferation
Given that increased cancer cell proliferation induce DNA damage [11], we hypothesized that high H2AX expressing tumor with enhanced DNA repair reflects enhanced cell proliferation. As expected, H2AX high expressing tumor enriched DNA replication gene sets consistently in both TCGA (NES=1.84, FDR=0.030) and METABRIC (NES=1.77, FDR=0.037) cohorts (Figure 5A). As shown in Figure 5B, H2AX high expressing tumor significantly enriched MYC targets gene sets consistently in both TCGA (targets v1; NES=2.06, FDR=0.007, target v2; NES=2.07, FDR=0.008) and METABRIC (targets v1; NES=1.34, FDR=0.174, target v2; NES=1.81, FDR=0.002). In agreement, high H2AX expressing tumors were associated with significant increase in MKI67 gene, which encodes cell proliferation marker Ki-67, as compared to H2AX low expressing tumor consistently in both TCGA (P<0.001) and METABRIC (P<0.001) (Figure 5C).
Figure 5.
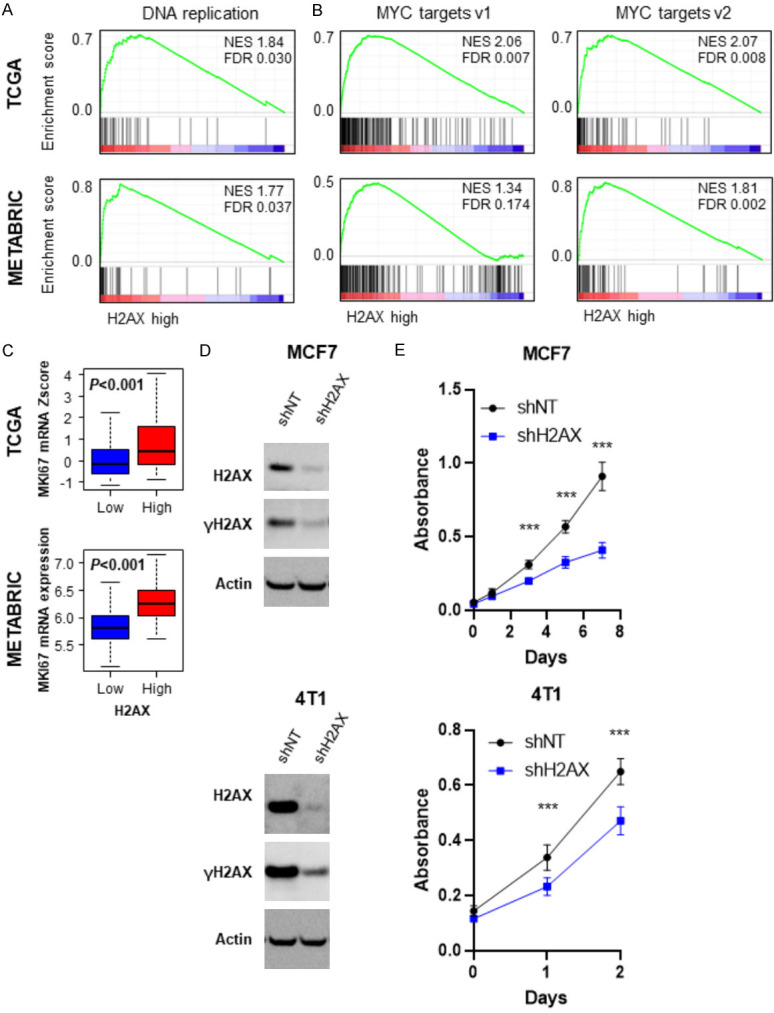
H2AX dependent proliferation rate in breast cancer. A. Gene set enrichment analysis (GSEA) of DNA replication pathway in comparison to high and low H2AX breast cancer in TCGA (upper) and METABRIC (lower) cohort. B. GSEA of MYC target genes in H2AX high and low breast cancers in TCGA (upper) and METABRIC (lower) cohort. C. MKI67 expression levels H2AX low and high breast cancer in TCGA (upper) and METABRIC (lower). H2AX high: n=55, H2AX low: n=1034 for TCGA, H2AX high: n=72, H2AX low: n=1351 for METABRIC. D. Immunoblot analysis of H2AX knockdown MCF7 and 4T1 cells. E. Cell proliferation rate of control and H2AX knockdown MCF7 and 4T1 cells (n=6, each). ***P<0.001.
Next, we generated H2AX knockdown cells of human breast cancer cell line, MCF7 and murine breast cancer cell line, 4T1 using different shH2AX. We found that knockdown of H2AX gene not only suppressed H2AX, but also γ-H2AX protein expression as well in both MCF7 and 4T1 cells (Figure 5D). H2AX knockdown suppressed cell proliferation as compared to the control shNT cells in both MCF7 and 4T1 cells (Figure 5E), implying that H2AX high expressing tumor has increased cell proliferation, which is not independent of γH2AX.
H2AX level is associated with metastasis in breast cancer
Since H2AX mRNA level was linked with cancer aggressiveness such as cell proliferation, we investigated whether H2AX mRNA expression is associated with breast cancer progression and metastasis. We compared H2AX mRNA expression between primary and metastatic cancer in both cell lines and patient samples. H2AX mRNA expression was significantly higher in the lung metastasis clone, LM2-4 as compared to its parental MDA-MB-231 cells in human breast cancer cell line (Figure 6A, P=0.042). Consistently, bone metastatic clone, 4T1.2, showed significantly higher H2AX expression as compared to its parental 4T1 cells in murine breast cancer cell line (Figure 6B, P<0.001). On the other hand, we were unable to obtain reproducible data on correlation of H2AX protein expression and metastatic clones (Figure S5). Similar findings were seen in the patient samples where H2AX mRNA expression was significantly higher in the metastatic tumors as compared to the primary tumors from same patients in GSE110590 cohort (Figure 6C, P<0.001). All of metastatic sites, including liver, lung and brain metastases showed higher H2AX expression as compared to the primary breast cancer (Figure 6C, P<0.001). Further, the proportion of patient with high H2AX expression showed higher in the advanced stage disease consistently in both TCGA and METABRIC cohorts (Figure 6D, 6E). We tested if H2AX knockdown impairs metastatic capability using mouse mastectomy model. Interestingly, H2AX knockdown reduced tumor growth in primary tumor before mastectomy (Figure S6A); however, there was no significant difference neither in metastatic burden which was quantified by the bioluminescence or mice survival (Figure S6B, S6C). These findings suggest that high expression of H2AX mRNA is associated with breast cancer progression and metastasis, however, the causality and mechanisms need to be further elucidated.
Figure 6.
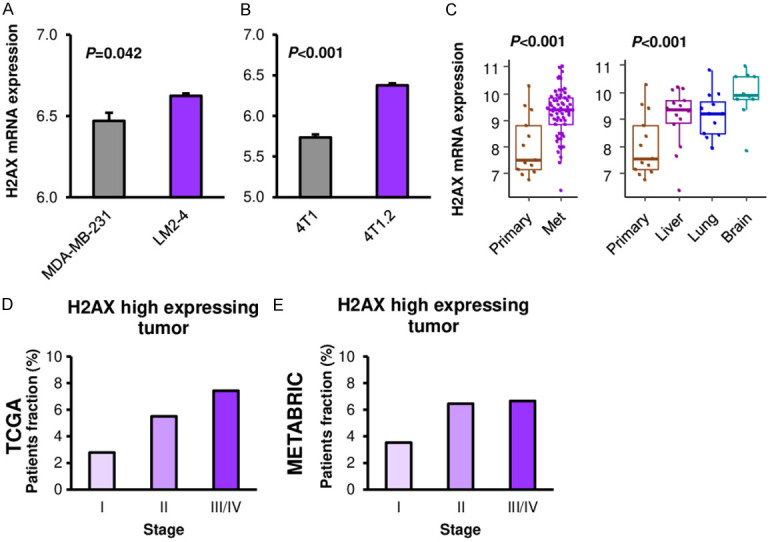
Association between H2AX and metastasis in breast cancer. A. H2AX expression comparison between parental (MDA-MB-231) and lung metastatic clone (LM2-4) in human breast cancer cell line (n=3, each). B. H2AX expression comparison between parental (4T1) and bone metastatic clone (4T1.2) in murine breast cancer cell line (n=3, each). C. H2AX expression comparison between primary (n=13) and metastatic breast cancers (liver: n=14, lung: n=12, brain: n=10) in breast cancer cohort GSE110590. D. H2AX high expressing tumor fraction by stage in TCGA breast cancer cohort. Stage I: n=180, stage II: n=618, stage III/IV: n=269. E. H2AX high expressing tumor fraction by stage in METABRIC breast cancer cohort. Stage I: n=368, stage II: n=604, stage III/IV: n=105.
Discussion
In the current study, we demonstrated that H2AX expression was associated with not only cancer recurrence, DFS, but also actual survival such as DSS and OS in breast cancer patients using two independent large patient cohorts. Similar to γ-H2AX, high H2AX tumor was associated with increased DNA repair. Although H2AX expression did not predict the chemotherapy response in pre-treatment tumors, it was significantly downregulated after effective chemotherapy and radiotherapy. We further found that high H2AX tumors were enriched for DNA replication and MYC targets gene sets, and showed increased MKI67 expression, which suggest enhanced cell proliferation. H2AX knockdown breast cancer cells demonstrated decreased cell proliferation as compared to the control cells, which further suggest that H2AX expression promotes cell proliferation. H2AX mRNA expression was higher in the metastatic clones of MDA-MB-231 and 4T1 cell lines as well as in the metastatic tumor compared to the primary breast cancer in the patient samples. Finally, high H2AX tumor proportion was higher in advanced AJCC cancer stage consistently in both TCGA and METABRIC cohorts, which implies that H2AX is associated with cancer progression and metastasis.
Phosphorylation of Ser-139 residue of H2AX generates γ-H2AX, which is a sensitive marker of DNA double-strand breaks and DNA repair. As we expected, H2AX high expressing tumors were enriched for DNA repair gene sets, which might be the result of increased γ-H2AX. The presence of double-strand break represents genomic instability; thus, it contributes to cancer progression [6]. Since cancer cells proliferate much faster than normal cells and results in damaged DNA accumulation, cancer cells need to repair the damaged DNA continuously, therefore, levels of H2AX mRNA or protein may be higher than normal cells. The level of H2AX may be critical for proliferation of cancer cells, but not for normal cells since normal cells do not increased damaged DNA burden. Indeed, higher γ-H2AX level predicts patient survival in several types of cancer [30-33], including breast cancer [12]. However, measurement of γ-H2AX levels require immunohistochemistry that is less objective, labor and cost intensive compared from H2AX mRNA expression analysis. Recently, we found that H2AX mRNA expression is associated with ovarian cancer patient prognosis [34]. To our knowledge, current study is the first to report that H2AX mRNA expression can predict patient survival in breast cancer.
Previously we found that DNA damage increases levels of ROS through increased H2AX [7], which is in agreement with other reports that show upregulation of H2AX is associated with ROS generation [8-10]. While the others reported that increased ROS reduces H2AX level [35], enhanced cell proliferation by activated MYC signaling induces DNA damage that correlated with increased levels of ROS [11]. In this study, we demonstrated that higher expression of H2AX was associated with enhanced MYC signaling, however, the association of H2AX with ROS was seen only in one cohort, implying that the relationship between H2AX and ROS may be context dependent in clinical setting.
We also investigated the association of H2AX mRNA expression level with DNA damaging chemotherapy as well as radiation sensitivity, because H2AX level may reflect genomic instability. We expected that higher H2AX expressing tumor shows higher sensitivity to chemotherapy and radiotherapy; however, H2AX mRNA expression level did not predict the response of anthracycline or taxane based chemotherapy or radiotherapy. On the other hand, H2AX expression was downregulated after effective chemotherapy or radiotherapy. H2AX high expressing tumor represents highly proliferating tumor; thus, we cannot help but speculate that chemotherapy selectively kill these highly proliferating H2AX high expressing cells among heterogenous cancer cells, resulting in low H2AX after chemotherapy or radiotherapy.
We demonstrated that H2AX mRNA expression level is associated with cancer progression and metastasis. This finding is supported by previous reports in which it was shown that H2AX promotes metastasis through HIF1α activation [12], and metastatic tumor is known to have genomic instability [36]. Cancer cells with higher genomic instability may possess higher metastatic potential, which eventually forms metastasis. However, to our surprise, H2AX mRNA expression did not correlate with protein expression in 2 metastatic cell lines examined. We speculate that discrepancies in mRNA and protein data may possibly be due to protein stability and post transcriptional modifications. Further, H2AX knockdown did not change metastatic capability in mice model. This warrants further investigation for the elucidation of the detailed mechanism of H2AX mRNA expression and breast cancer progression and metastasis.
This study has limitations. Most of analyses were conducted through bioinformatics, thus, the exact mechanisms of how H2AX is associated with worse prognosis are not fully elucidated. Further experimental approach is needed to investigate H2AX roles in breast cancer. In addition, while it is interesting to examine whether the impact of H2AX mRNA expression on breast cancer cell proliferation and metastasis is independent of γ-H2AX, we were unable to show this since we do not have access to a tool that inhibits the production of γ-H2AX without disturbing H2AX mRNA expression.
In conclusion, extremely high expression of H2AX is associated with increased DNA repair, MYC signaling, cell proliferation, metastatic capability, and poor prognosis in breast cancer.
Acknowledgements
This research was funded by US National Institutes of Health/National Cancer Institute grant R01CA160688, R01CA250412, R37CA248018, US Department of Defense BCRP grant W81XWH-19-1-0674, as well as the Edward K. Duch Foundation and Paul & Helen Ellis Charitable Trust to K.T., and US National Cancer Institute cancer center support grant P30-CA016056 to Roswell Park Comprehensive Cancer Center, source of Roswell Park Comprehensive Cancer Center. RNA sequencing was conducted at the Genomic Shared resource and processed by the Bioinformatics Shared Resource, shRNA transduction for MCF7 was conducted by Renae Holtz at the Gene Modulation Shared Resource of Roswell Park Comprehensive Cancer Center.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Surveillance, Epidemilolgy, and End Results Program. https://seer.cancer.gov/statfacts/html/breast.html.
- 4.Katsuta E, Rashid OM, Takabe K. Clinical relevance of tumor microenvironment: immune cells, vessels, and mouse models. Hum Cell. 2020;33:930–937. doi: 10.1007/s13577-020-00380-4. [DOI] [PubMed] [Google Scholar]
- 5.Zschenker O, Kulkarni A, Miller D, Reynolds GE, Granger-Locatelli M, Pottier G, Sabatier L, Murnane JP. Increased sensitivity of subtelomeric regions to DNA double-strand breaks in a human cancer cell line. DNA Repair (Amst) 2009;8:886–900. doi: 10.1016/j.dnarep.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang MA, So EY, Simons AL, Spitz DR, Ouchi T. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012;3:e249. doi: 10.1038/cddis.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajan I, Jayasree PR, Kumar PR. Zerumbone induces mitochondria-mediated apoptosis via increased calcium, generation of reactive oxygen species and upregulation of soluble histone H2AX in K562 chronic myelogenous leukemia cells. Tumour Biol. 2015;36:8479–8489. doi: 10.1007/s13277-015-3583-z. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T, Kurose A, Halicka HD, Huang X, Traganos F, Darzynkiewicz Z. Nitrogen oxide-releasing aspirin induces histone H2AX phosphorylation, ATM activation and apoptosis preferentially in S-phase cells: involvement of reactive oxygen species. Cell Cycle. 2006;5:1669–1674. doi: 10.4161/cc.5.15.3100. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, Tanaka T, Kurose A, Traganos F, Darzynkiewicz Z. Constitutive histone H2AX phosphorylation on Ser-139 in cells untreated by genotoxic agents is cell-cycle phase specific and attenuated by scavenging reactive oxygen species. Int J Oncol. 2006;29:495–501. [PubMed] [Google Scholar]
- 11.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 12.Rezaeian AH, Li CF, Wu CY, Zhang X, Delacerda J, You MJ, Han F, Cai Z, Jeong YS, Jin G, Phan L, Chou PC, Lee MH, Hung MC, Sarbassov D, Lin HK. A hypoxia-responsive TRAF6-ATM-H2AX signalling axis promotes HIF1α activation, tumorigenesis and metastasis. Nat Cell Biol. 2017;19:38–51. doi: 10.1038/ncb3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takabayashi H, Wakai T, Ajioka Y, Korita PV, Yamaguchi N. Alteration of the DNA damage response in colorectal tumor progression. Hum Pathol. 2013;44:1038–1046. doi: 10.1016/j.humpath.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vera-Ramirez L, Sanchez-Rovira P, Ramirez-Tortosa CL, Quiles JL, Ramirez-Tortosa M, Lorente JA. Transcriptional shift identifies a set of genes driving breast cancer chemoresistance. PLoS One. 2013;8:e53983. doi: 10.1371/journal.pone.0053983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, Martin M, Cotrina J, Gomez H, Hubbard R, Chacon JI, Ferrer-Lozano J, Dyer R, Buxton M, Gong Y, Wu Y, Ibrahim N, Andreopoulou E, Ueno NT, Hunt K, Yang W, Nazario A, DeMichele A, O’Shaughnessy J, Hortobagyi GN, Symmans WF. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snipstad K, Fenton CG, Kjaeve J, Cui G, Anderssen E, Paulssen RH. New specific molecular targets for radio-chemotherapy of rectal cancer. Mol Oncol. 2010;4:52–64. doi: 10.1016/j.molonc.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams S, Chakravarthy AB, Donach M, Spicer D, Lymberis S, Singh B, Bauer JA, Hochman T, Goldberg JD, Muggia F, Schneider RJ, Pietenpol JA, Formenti SC. Preoperative concurrent paclitaxel-radiation in locally advanced breast cancer: pathologic response correlates with five-year overall survival. Breast Cancer Res Treat. 2010;124:723–732. doi: 10.1007/s10549-010-1181-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel MB, He X, Hoadley KA, Hoyle A, Pearce JB, Garrett AL, Kumar S, Moylan VJ, Brady CM, Van Swearingen AE, Marron D, Gupta GP, Thorne LB, Kieran N, Livasy C, Mardis ER, Parker JS, Chen M, Anders CK, Carey LA, Perou CM. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest. 2018;128:1371–1383. doi: 10.1172/JCI96153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebos JM, Lee CR, Bogdanovic E, Alami J, Van Slyke P, Francia G, Xu P, Mutsaers AJ, Dumont DJ, Kerbel RS. Vascular endothelial growth factor-mediated decrease in plasma soluble vascular endothelial growth factor receptor-2 levels as a surrogate biomarker for tumor growth. Cancer Res. 2008;68:521–529. doi: 10.1158/0008-5472.CAN-07-3217. [DOI] [PubMed] [Google Scholar]
- 24.Munoz R, Man S, Shaked Y, Lee CR, Wong J, Francia G, Kerbel RS. Highly efficacious nontoxic preclinical treatment for advanced metastatic breast cancer using combination oral UFT-cyclophosphamide metronomic chemotherapy. Cancer Res. 2006;66:3386–3391. doi: 10.1158/0008-5472.CAN-05-4411. [DOI] [PubMed] [Google Scholar]
- 25.Bolin C, Sutherland C, Tawara K, Moselhy J, Jorcyk CL. Novel mouse mammary cell lines for in vivo bioluminescence imaging (BLI) of bone metastasis. Biol Proced Online. 2012;14:6. doi: 10.1186/1480-9222-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katsuta E, Rashid OM, Takabe K. Murine breast cancer mastectomy model that predicts patient outcomes for drug development. J Surg Res. 2017;219:310–318. doi: 10.1016/j.jss.2017.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katsuta E, Oshi M, Rashid OM, Takabe K. Generating a murine orthotopic metastatic breast cancer model and performing murine radical mastectomy. J Vis Exp. 2018 doi: 10.3791/57849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 29.Kawashima S, Kawaguchi N, Taniguchi K, Tashiro K, Komura K, Tanaka T, Inomata Y, Imai Y, Tanaka R, Yamamoto M, Inoue Y, Lee SW, Kawai M, Tanaka K, Okuda J, Uchiyama K. γ-H2AX as a potential indicator of radiosensitivity in colorectal cancer cells. Oncol Lett. 2020;20:2331–2337. doi: 10.3892/ol.2020.11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turinetto V, Pardini B, Allione A, Fiorito G, Viberti C, Guarrera S, Russo A, Anglesio S, Ruo Redda MG, Casetta G, Cucchiarale G, Destefanis P, Oderda M, Gontero P, Rolle L, Frea B, Vineis P, Sacerdote C, Giachino C, Matullo G. H2AX phosphorylation level in peripheral blood mononuclear cells as an event-free survival predictor for bladder cancer. Mol Carcinog. 2016;55:1833–1842. doi: 10.1002/mc.22431. [DOI] [PubMed] [Google Scholar]
- 31.Mei L, Hu Q, Peng J, Ruan J, Zou J, Huang Q, Liu S, Wang H. Phospho-histone H2AX is a diagnostic and prognostic marker for epithelial ovarian cancer. Int J Clin Exp Pathol. 2015;8:5597–5602. [PMC free article] [PubMed] [Google Scholar]
- 32.Matthaios D, Foukas PG, Kefala M, Hountis P, Trypsianis G, Panayiotides IG, Chatzaki E, Pantelidaki E, Bouros D, Karakitsos P, Kakolyris S. gamma-H2AX expression detected by immunohistochemistry correlates with prognosis in early operable non-small cell lung cancer. Onco Targets Ther. 2012;5:309–314. doi: 10.2147/OTT.S36995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakai T, Shirai Y, Sakata J, Korita PV, Matsuda Y, Takamura M, Ohashi R, Nagahashi M, Ajioka Y, Hatakeyama K. Alteration of p53-binding protein 1 expression as a risk factor for local recurrence in patients undergoing resection for extrahepatic cholangiocarcinoma. Int J Oncol. 2011;38:1227–1236. doi: 10.3892/ijo.2011.959. [DOI] [PubMed] [Google Scholar]
- 34.Saravi S, Katsuta E, Jeyaneethi J, Amin HA, Kaspar M, Takabe K, Pados G, Drenos F, Hall M, Karteris E. H2A histone family member X (H2AX) is upregulated in ovarian cancer and demonstrates utility as a prognostic biomarker in terms of overall survival. J Clin Med. 2020;9:2844. doi: 10.3390/jcm9092844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruosso T, Mieulet V, Cardon M, Bourachot B, Kieffer Y, Devun F, Dubois T, Dutreix M, Vincent-Salomon A, Miller KM, Mechta-Grigoriou F. Chronic oxidative stress promotes H2AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO Mol Med. 2016;8:527–549. doi: 10.15252/emmm.201505891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seton-Rogers S. Genomic instability: the sting of metastasis. Nat Rev Cancer. 2018;18:137. doi: 10.1038/nrc.2018.16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


