Abstract
Major depression is a complex disorder with no single, direct causal mechanism. Morbidity has been linked to genetic processes, developmental history, and unique environmental exposures. Epigenetic mechanisms, especially DNA methylation, are also likely important factors in the pathogenesis of major depressive disorder (MDD). A community-based twin sample has many advantages for epigenetic studies, given the shared genetic and developmental histories of same-sex twin pairs. This article describes the rationale and study design for the Mood and Methylation Study in which 133 twin pairs (101 monozygotic and 32 dizygotic), both discordant and concordant for lifetime history of MDD, were evaluated on a large number of variables related to MDD. The twins also provided blood samples for an epigenome-wide association study of differentially methylated regions (DMR) relevant to MDD. Although MDD is typically considered a disorder of the central nervous system, it is unfeasible to obtain a large sample of brain tissues. However, epigenetic variation is not limited to the affected tissue but can also be detected in peripheral blood leukocytes. Thus, this study focused on monocytes for the major analyses. Additional plans for the study include gene expression analysis from the same set of twins using RNA-seq and validation of significant DMRs in postmortem brain tissues from a separate sample. Moreover, sufficient samples have been collected to perform future ‘multi-omic’ analyses, including metabolome, microbiome, and transcriptome. Our long-term goal is to understand how epigenomic and other ‘omic’ factors can be manipulated for diagnostic, preventive, and therapeutic purposes for MDD and its related conditions.
Keywords: depression, methylation, epigenome, microbiome, transcriptome, metabolome, twins
Major depressive disorder (MDD) is a complex and sometimes devastating condition that affects millions of people across the globe every year. To date, no single, direct causal mechanism has been identified in MDD but MDD morbidity has been linked to genetic and epigenetic processes, developmental history, and unique environmental exposures. Epigenetic mechanisms, especially DNA methylation, are increasingly being recognized as factors in the pathogenesis of MDD, and several published studies from both small and large samples have shown depression-related differences in methylation of genes related to inflammation, G-protein coupled receptor protein signaling, and axon guidance among others (Byrne et al., 2013; Crawford et al., 2018; Davies et al., 2014; El-Sayed et al., 2012; Sabunciyan et al., 2012; Shimada et al., 2018; Story Jovanova et al., 2018; Uddin et al., 2011; Zhao et al., 2013).
DNA methylation involves the addition of a methyl group to the fifth carbon of a cytosine base. It occurs most frequently within CpG islands (i.e., genomic regions that contain a high frequency of CpG sites) in the promoter regions of many genes, although most CpG islands are unmethylated. Unlike the DNA sequence itself, which is stable during development, DNA methylation is highly dynamic and often induced by exposure to a range of factors such as nutrition, behavior, and chemicals (Dolinoy & Jirtle, 2008; Dolinoy et al., 2007). These factors can directly affect methylation, chromatin remodeling, and other epigenetic pathways, resulting in alterations of epigenome and subsequent gene expression, thereby contributing to disease etiology (Dolinoy & Jirtle, 2008; Dolinoy et al., 2007; Jiang et al., 2004; Tsankova et al., 2007). However, the biological pathways linking aberrant methylation to MDD remain largely unknown, either because the function of the methylated genes is not known or because the methylation results have not been compared to actual differences in gene expression or other mechanisms. This uncertainty hampers our ability to implement early diagnosis, prevention, and treatment for this debilitating disorder.
To address these challenges, we designed a twin study known as the Mood and Methylation Study (MMS). This study recruited monozygotic (MZ) and dizygotic (DZ) twin pairs discordant or concordant for a lifetime history of MDD, and collected detailed clinical phenotypes for MDD, MDD-related traits, and environmental factors for each twin participant. Biospecimens were also collected for the primary methylome and transcriptome analyses but also to promote future multi-omic studies. Discordant MZ twin pairs provide a natural experiment to study epigenetic differences in MDD, because MZ twins match exactly on genetic background, age, and sex, thus totally eliminating the effects of these important confounders (Fraga et al., 2005; Relton & Davey Smith, 2010). In addition, identical twins share developmental environment, providing further control for confounding by early life experience, which is known to have a long-lasting impact on the epigenetic plasticity of the human genome (Rutten & Mill, 2009). Therefore, a discordant co-twin control approach provides a powerful model for epigenomic research.
MDD is typically considered a disorder of the central nervous system, and it is known that DNA methylation is, to some extent, cell-type- and tissue-specific. But, whatever the degree of tissue specificity, it is unfeasible to obtain a large sample of brain tissues for epigenetic analyses. That said, there is increasing evidence that leukocytes may be a useful cell model to evaluate epigenetic changes, because epigenetic variation may not be limited to the affected tissue but can also be detected in peripheral blood leukocytes (Crawford et al., 2018; Story Jovanova et al., 2018). In fact, the study of peripheral blood may have some advantages over the use of brain tissue in that they are likely to accumulate fewer epigenetic changes induced by disease-related external factors, which could be confounding factors in postmortem studies (Pidsley & Mill, 2011). Importantly, blood samples are much easier to obtain and could be used for large-scale epidemiologic studies, tracking changes associated with depression as the disease develops (Story Jovanova et al., 2018). Although a great deal more research is needed, it appears that the patterns of methylation across CpG sites in brain and peripheral blood leukocytes are similar (Murphy et al., 2008; Yuferov et al., 2011) and correlations of DNA methylation profiles between different human tissues are reasonably high and can be evaluated using publicly available tools such as the Blood–Brain Epigenetic Concordance (BECon) application (Bromberg et al., 2010; Brown & Robinson, 2000; Edgar et al., 2017; Hannon et al., 2015). Because there are several cell types within leukocytes, we decided to isolate one type — monocytes — for this study.
Thus, we designed our study to first conduct an epigenome-wide association study (EWAS) using monocyte DNA extracted from whole blood in twin pairs discordant for lifetime history of MDD. In an effort to replicate significant differentially methylated regions (DMRs) in the central nervous system, we obtained clinically well-characterized postmortem brain tissues from the Stanley Medical Research Institute’s Depression Collection, with control samples from non-depressed brains. We also downloaded brain DNA methylation data (HumanMethylation450K BeadChip) through the publicly available Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/info/datasets.html) database GSE41826. Finally, we planned to determine the functional impact of DMRs via gene expression analyses using RNA-seq with postmortem brain gene expression data (RNA-seq) through GEO database GSE101521. With this combination of approaches, we anticipate discovering epigenetically dysregulated genomic regions for MDD, in addition to uncovering functional epigenetic alterations that may play a causative role in MDD pathogenesis.
Although results along these lines are still expected to provide novel epigenetic targets for diagnosis, prevention, and therapeutic intervention in MDD, given the advance of both theory and technology since initial submission and funding, the final methods for this study were structured to allow a study initially focused on epigenomics to become a ‘multi-omics’ study. That is, a sufficient volume of blood has been drawn, processed, and stored both to complete the proposed study and to allow for future metabolome and transcriptome studies. In addition, stool sample collection was added to the protocol to facilitate analysis of the gut microbiome and its association with both MDD and MDD-related DMRs. Because each of the omics has both genetic and environmental contributions, we also decided to recruit dizygotic twin pairs and pairs that are concordant for MDD (see Table 1 for a breakdown of the sample by zygosity and concordance). This combination of twin pairs allows for maximum flexibility in answering important questions about the heritability of individual phenotypes and the simultaneous effects of genes and environment on multiple phenotypes. Finally, the twins in this sample are community volunteers interested in participating in research and significant efforts are made by the Washington State Twin Registry to say in contact with the volunteers. It will therefore be possible to return a large percentage of them in the future for longitudinal analysis and new collection of MRI analyses to assess the connectome.
TABLE 1.
Study Recruitment Goals
| Zygosity | Sex | Lifetime history MDD | Enrollment goal | Final sample size |
|---|---|---|---|---|
| Dizygotic | Female | Concordant | 20 | 12 |
| Discordant | 20 | 12 | ||
| Male | Concordant | 10 | 3 | |
| Discordant | 10 | 5 | ||
| Monozygotic | Female | Concordant | 20 | 15 |
| Discordant | 60 | 54 | ||
| Male | Concordant | 10 | 7 | |
| Discordant | 30 | 25 |
Methods
Washington State Twin Registry Description and Involvement
Recruitment for the present study was completed via the community-based Washington State Twin Registry (WSTR; previously known as the University of Washington Twin Registry). Complete details of the construction and current components of the WSTR are available elsewhere (Afari et al., 2006; Strachan et al., 2013). Briefly, recruitment into the WSTR itself relies on initial identification of twins from the Washington State Department of Licensing (DOL), which asks all driver license and state ID applicants: ‘Are you a twin (Y/N)?’ Based on a data-sharing agreement between DOL and the University, the names and contact information of all twin respondents are then sent to the WSTR. Twins are contacted with information about the Registry and an enrollment survey. After both members of the pair complete the survey, the pair is enrolled in the Registry. The Registry contacts the twins multiple times each year with birthday cards, newsletters, and study invitations in order to keep contact information current. At the time recruitment began, the average age of enrolled twins was 37, and 41% of the twins reported being married. Approximately 4% of the Registry reported being Hispanic and 86% reported being White. All other racial categories were <4%. In terms of educational achievement, 25% completed high school or GED, an additional 34% had some college/vocational school, and 33% had a bachelor’s degree or higher.
Twin Sampling and Recruitment for the Mood and Methylation Study
All participants provided informed consent for the study procedures and all procedures have been reviewed and approved by the University of Washington Institutional Review Board. Enrollment into the study required at least one member of the pair to have a lifetime history of MDD as measured by expert structured interview. Non-depressed twins were required to have no lifetime history of a major depressive episode and to have Beck Depression Inventory (BDI-II) scores below 13 at prescreening (Beck et al., 1996). In addition, both members of the pair had to be willing to come to the University of Washington Medical Center for the in-person blood draw and data collection visit. Provision of stool samples, urine, saliva, and toenail clippings was optional. The primary exclusionary conditions for twin pairs were schizophrenia, bipolar disorder I and II, current substance use disorder, cancer, uncontrolled sleep apnea, and certain autoimmune, and uncontrolled endocrine disorders known to be associated with depressive symptoms. In order to avoid problems related to population stratification, only adult, same-sex, non-Hispanic, White twin pairs who were reared together, were enrolled.
Recruitment
In order to enroll participants from the WSTR into the study, we sent an introductory letter to potentially eligible WSTR members via postal mail and/or email. Participants in the Registry have already agreed to be approached for research projects by Twin Registry staff (all WSTR members were aged 18+ at the time this study began). We then followed up with those who expressed an interest with a prescreen survey (phone or online depending on participant preference) using approved scripts to screen for interest and eligibility. Once both members of the twin pair completed the prescreening without triggering any exclusionary criteria, each member was individually interviewed via telephone by a PhD-level psychologist using the relevant sections of the Structured Clinical Interview, Research Version, for the DSM-IV (SCID-RV), which was the latest version available at the time the study began (First et al., 1995). The SCID-RV is a semi-structured interview guide for making Axis I DSM-IV diagnoses. For the purposes of this study, participants completed the Mood Episodes, Psychotic and Associated Symptoms, Psychotic Disorders, Mood Disorders, Substance Use Disorders, and Anxiety Disorders modules. They did not complete the Somatoform Disorders, Eating Disorders, or Adjustment Disorders modules. Any questions or concerns about the depression phenotype or exclusionary psychiatric conditions were referred to the senior clinician for evaluation and final decision-making.
Figure 1 shows the enrollment funnel for the study and Table 1 shows the breakdown by sex and zygosity. At the prescreen stage, the primary reasons for exclusion were having an exclusionary medical condition (e.g. recent cancer, history of autoimmune disease), or that one or both members of a pair were not interested in participating. At the clinical interview stage, the primary reasons for exclusion were no history of MDD in either twin, current substance use disorder, and history of bipolar illness. In terms of overall numbers, we anticipated being able to recruit twin pairs in locations other than western Washington using a contract service to provide in-home consent documentation along with specimen and questionnaire data collection. However, with the need to isolate monocytes, the contract service proved prohibitively expensive and we were forced to adjust our recruitment targets down (Table 1).
FIGURE 1.
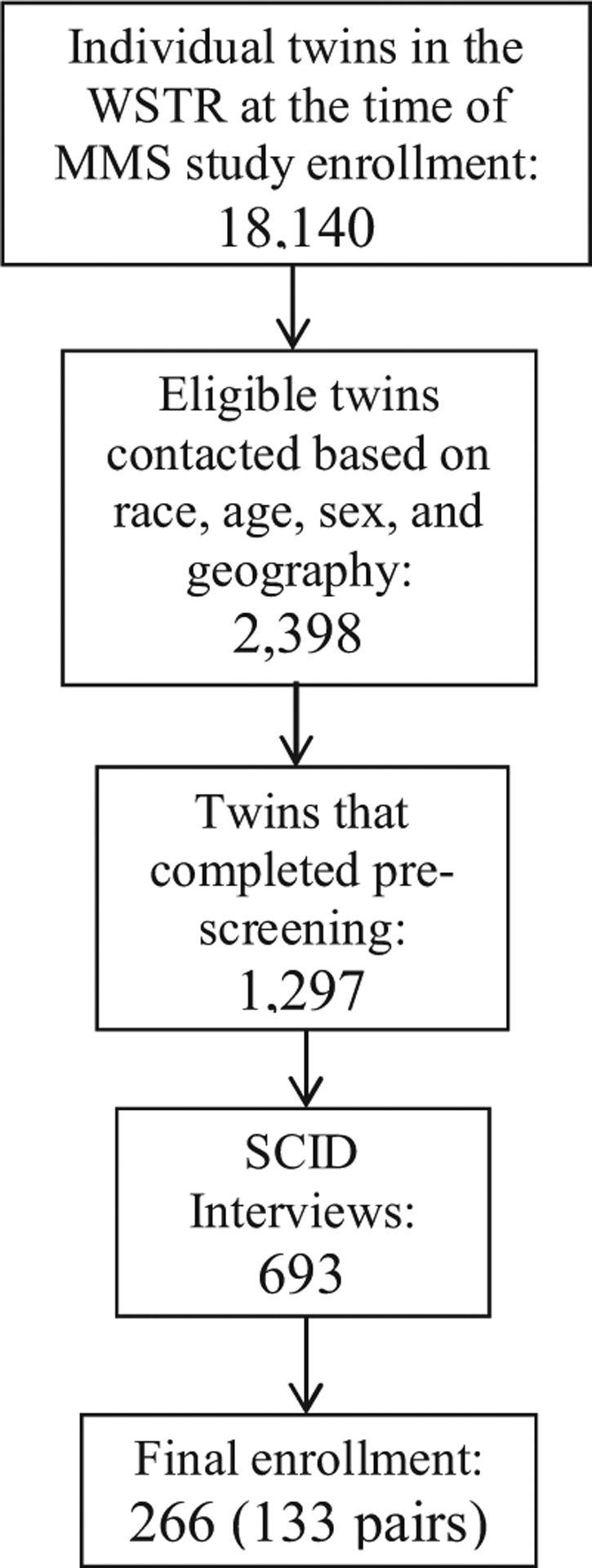
Enrollment funnel for the Twin MMS study.
Biospecimen and Phenotype Data Collection
Table 2 shows the interviews and questionnaires administered over the course of the study and Table 3 shows the outline of the biospecimen collection procedures.
TABLE 2.
Study Measures and Administration
| Prior to recruitment | ||
| ||
| Prescreening | ||
|
|
|
| Clinical interview | ||
| ||
| At-home collection | ||
|
|
|
| In-person study visit | ||
|
|
|
TABLE 3.
Biospecimen Collections
| Tissue type | Amount | Storage | Location |
|---|---|---|---|
| Monocyte DNA | 2–4 tubes each at 50–100 μl | −80°C | UW |
| Monocyte RNA | 6–8 tubes each at 15–20 μl | −80°C | UW |
| RNA PAXgene | 4 tubes at 10 ml | −80°C | UW |
| DNA PAXgene | 1 tube at 10 ml | −80°C | UW |
| Serum | 2 tubes each at .5 ml, 1 ml, and 2ml | −80°C | UW |
| Plasma | 2 tubes each at .5 ml and 1 ml 4 tubes each 2 ml and 5 ml |
−80°C | UW |
| EDTA whole blood | 1 tube at 4 or 6 ml | −80°C | NIMH |
| Stool | 2 tubes | −80°C | UW |
| Saliva | 3 tubes | −80°C | UW |
| Urine | 4 tubes at 5 ml 5 tubes at 2 ml |
−80°C | UW |
At-home procedures.
Prior to coming in for the in-person study visit, participants received a package that contained questionnaires along with stool, saliva, and toenail sample collection kits. The questionnaires included two measures of depression symptom severity, the BDI-II mentioned above and the Quick Inventory of Depressive Symptomatology (QIDS-SR16). The BDI-II is a self-administered 21-item scale that has acceptable sensitivity and specificity with regards to DSM criteria for MDD. The QIDS is also self-administered and was designed to assess the severity of depressive symptoms (Rush et al., 2003). The questionnaire packet also included measures of lifetime exposure to stressful events that may influence epigenetic variations leading to disease (Franklin et al., 2010; Waterland, 2009). Exposure to childhood traumatic stress was measured with the Early Trauma Inventory (Bremner et al., 2000). This self-administered instrument assesses not only the frequency and nature of traumatic events, but also the emotional impact of the stressor on the individual immediately after and across the life span, and the impact on social and occupational functioning. We also measured hostility using the Cook–Medley Hostility Inventory (Barefoot et al., 1989) and anger by means of the Spielberger’s Anger Expression Inventory (Spielberger, 1988). These factors are related to incidence of MDD (Keilp et al., 2006) and might also influence epigenetic alterations (Waterland, 2009). Finally, we measured social and emotional support by means of the Enhancing Recovery in Coronary Heart Disease (ENRICHD) Social Support Inventory, a five-item scale assessing both social and emotional support (ENRICHD Investigators, 2001). Table 4 shows initial demographic and clinical data broken down by zygosity.
TABLE 4.
Demographic and Clinical Data by Zygosity
| Monozygotic twins (n = 202) | Dizygotic twins (n = 64) | |
|---|---|---|
| Age at visit (yrs) | 38.22±15.45 | 43.87±16.41 |
| Gender (female) | 68.31% | 75.00% |
| Education (bachelor’s degree or higher) | 62.37% | 53.13% |
| Income (less than $20,000) | 8.91% | 3.13% |
| Lifetime history of depression | 60.89% | 73.43% |
| Current MDD | 6.93% | 10.94% |
| BDI-II score | 5.99±5.84 | 7.32±6.61 |
| QIDS score | 4.75±3.09 | 5.02±3.28 |
| Use of antidepressants | 40.11% | 62.26% |
| Use of antipsychotics | 2.20% | 7.54% |
| ACE score | 4.89±4.96 | 5.02±5.21 |
| ACE (≥4) | 51.50% | 44.52% |
Note: BDI-II = Beck Depression Inventory-II; QIDS = Quick Inventory of Depressive Symptomatology; ACE = Adverse Childhood Experiences.
The self-collection stool sample kits consisted of a collection bowl that sits on the participant’s toilet along with two tubes that have a scoop built onto the cap. The tubes have formalin fixative inside in order to stabilize the sample, along with small glass beads to aid in mixing the fixative into the sample. The participant used the cap scoop to collect a small amount of stool from the collection bowl and placed it inside the tubes. The tubes were then repeatedly inverted to distribute the fixative and stored in the participant’s freezer until they attended the in-person visit. The soluble stool collection bags were flushed down the toilet.
Saliva collection was also self-administered three times within the same day. The first was upon waking in the morning, the second was between 4:30 and 6:00 pm, and the third was prior to bedtime. The kits were absorbant cotton balls that were placed in the mouth until wet, then placed into sealed bags and brought to us at the in-person visit. For toenail collection, we included clippers and a pre-labeled plastic bag for participants to cut a sample from each nail on one foot, and place them in the bag to return to us at the in-person visit.
In-person procedures.
Participants came to the University of Washington Medical Center for anthropometric measurement along with blood and urine collection. The twins were permitted, but not required, to come to the visit together. Trained hospital staff completed the physical and physiological measures and drew up to 140 ml of blood into the various tubes. Participants also provided a urine sample, which was aliquoted and stored for future use. The blood samples were then processed immediately. From up to 110 ml of whole blood, we aliquoted plasma and serum. We then extracted DNA and RNA from monocytes isolated from the other peripheral blood mononuclear cells (PBMCs) with the Monocyte Isolation Kit II from Miltenyi Biotec. This isolation is achieved through negative selection. Non-monocytes are labeled with biotin-conjugated antibodies and Biotin Microbeads. The magnetically labeled cells stay in the column while the unlabeled monocytes pass through. Then the DNA and RNA were extracted with the AllPrep DNA/RNA/miRNA Universal Kit from Qiagen. This extraction started with the lysing of the monocyte cell pellet and binding of the DNA and RNA onto respective columns. The columns were then treated with specific buffers to remove unwanted proteins. The DNA/RNA were finally eluted from the column and the concentration determined using the NanoDrop Spectrophotometer.
DNA Methylation and Gene Expression Analysis
Using the collected peripheral blood monocytes, we are currently conducting an EWAS on MDD in 133 MZ and DZ pairs. DNA methylation is currently being assayed using the Infinium MethylationEPIC BeadChip. Compared to whole-genome sequencing, the BeadChip covers a small proportion of CpG sites. However, those sites are well distributed across the genome and the chip provide a cost-effective way to begin investigating methylation and disease. Gene expression—also using monocytes from the same sample of twins—will be measured by RNA-seq. Statistical analyses will be conducted to identify differentially methylated genes/regions (DMRs) associated with MDD. An initial power analysis indicated that our sample size has 80% power to detect a minimum of 9.7% methylation difference between cases and controls, assuming covariates accounting for 20% of phenotypic variability (Tsai & Bell, 2015). Functional importance of the identified DMR genes will be validated by linking to gene expression and further confirmed in postmortem brain tissue using available GTEx datasets. Sophisticated bioinformatics analyses will also be performed to investigate the potential functional role of the identified genes in MDD pathology. Also, given the collected specimens (e.g., plasma, stool samples), we plan to generate multi-omics datasets for all MMS twins, and conduct integrated omics analyses to identify novel biomarkers and biological pathways for MDD using a systems biology approach. DNA sample and the omics datasets will be publicly accessible through the NIMH biorepository or NIH dbGaP.
Future Directions
The methods described above are primarily dedicated to addressing the scientific aims of our original grant proposal. However, this sample of twins provides tremendous opportunity to evaluate MDD from a multi-omics perspective, and investigators interested in collaboration are encouraged to contact us. Our group has begun a pilot analysis of the gut microbiome from the MZ discordant pairs based on 16s RNA identification of gut microbiome diversity. Additional proposed research seeks to characterize MDD-associated changes in the plasma metabolome associated with microbial metabolism of neurotransmitters, neuroactive compounds, and neurometabolites and to measure differences in bacterial endotoxin exposure, measured as lipopolysaccharide binding protein, associated with leaky gut in MDD.
We anticipate that this twin sample will reveal promising directions for research on MDD treatment and prevention, but we also recognize that any significant results will require extensive replication and validation in samples different from the one described here. Greater racial diversity will be critical, along with additional focus on age-related effects and differences in early childhood exposures.
Acknowledgment
This research was funded by the National Institute of Mental Health (R01 MH097018: Zhao, Strachan).
Footnotes
Conflicts of Interest
None.
References
- Afari N, Noonan C, Goldberg J, Edwards K, Gadepalli K, Osterman B, … Buchwald D (2006). University of Washington twin registry: Construction and characteristics of a community-based twin registry. Twin Research and Human Genetics, 9, 1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barefoot JC, Dodge KA, Peterson BL, Dahlstrom WG, & Williams RBJ (1989). The Cook-Medley hostility scale: Item content and ability to predict survival. Psychosomatic Medicine, 51, 46–57. [DOI] [PubMed] [Google Scholar]
- Beck AT, Steer RA, & Brown GK (1996). BDI-II. Beck Depression Inventory: Second edition. San Antonio, TX: The Psychological Corporation. [Google Scholar]
- Bremner JD, Vermetten E, & Mazure CM (2000). Development and preliminary psychometric properties of an instrument for the measurement of childhood trauma: The early Trauma inventory. Depression and Anxiety, 12, 1–12. [DOI] [PubMed] [Google Scholar]
- Bromberg A, Levine J, Belmaker RH, & Agam G (2010). Hyperhomocysteinemia does not affect global DNA methylation and nicotinamide N-methyltransferase expression in mice. Journal of Psychopharmacology, 25, 976–981. [DOI] [PubMed] [Google Scholar]
- Brown CJ, & Robinson WP (2000). The causes and consequences of random and non-random X chromosome inactivation in humans. Clinical Genetics, 58, 353–363. [DOI] [PubMed] [Google Scholar]
- Byrne EM, Carrillo-Roa T, Henders AK, Bowdler L, McRae AF, Heath AC, … Wray NR (2013). Monozygotic twins affected with major depressive disorder have greater variance in methylation than their unaffected co-twin. Translational Psychiatry, 3, e269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford B, Craig Z, Mansell G, White I, Smith A, Spaull S, … Murphy TM (2018). DNA methylation and inflammation marker profiles associated with a history of depression. Human Molecular Genetics, 27, 2840–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Krause L, Bell JT, Gao F, Ward KJ, Wu H, … Wang J (2014). Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biology, 15, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, & Jirtle RL (2008). Environmental epigenomics in human health and disease. Environmental and Molecular Mutagenesis, 49, 4–8. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, & Jirtle RL (2007). Epigenetic gene regulation: Linking early developmental environment to adult disease. Reproductive Toxicology, 23, 297–307. [DOI] [PubMed] [Google Scholar]
- Edgar RD, Jones MJ, Meaney MJ, Turecki G, & Kobor MS (2017). BECon: A tool for interpreting DNA methylation findings from blood in the context of brain. Translational Psychiatry, 7, e1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed AM, Halossim MR, Galea S, & Koenen KC (2012). Epigenetic modifications associated with suicide and common mood and anxiety disorders: A systematic review of the literature. Biology of Mood & Anxiety Disorders, 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENRICHD Investigators. (2001). Enhancing Recovery in Coronary Heart Disease (ENRICHD): Baseline characteristics. American Journal of Cardiology, 88, 316–322. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Williams JBW, & Gibbon M (Eds.). (1995). Structured clinical interview for DSM IV-Pationet edition (SCID-P). Washington, DC: American Psychiatric Press. [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, … Esteller M (2005). Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America, 102, 10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, … Mansuy IM (2010). Epigenetic transmission of the impact of early stress across generations. Biological Psychiatry, 68, 408–415. [DOI] [PubMed] [Google Scholar]
- Hannon E, Lunnon K, Schalkwyk L, & Mill J (2015). Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics, 10, 1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Bressler J, & Beaudet AL (2004). Epigenetics and human disease. Annual Review of Genomics and Human Genetics, 5, 479–510. [DOI] [PubMed] [Google Scholar]
- Keilp JG, Gorlyn M, Oquendo MA, Brodsky B, Ellis SP, Stanley B, & Mann J (2006). Aggressiveness, not impulsiveness or hostility, distinguishes suicide attempters with major depression. Psychological Medicine, 36, 1779–1788. [DOI] [PubMed] [Google Scholar]
- Murphy BC, O’Reilly RL, & Singh SM (2008). DNA methylation and mRNA expression of SYN III, a candidate gene for schizophrenia. BMC Medical Genetics, 9, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, & Mill J (2011). Epigenetic studies of psychosis: Current findings, methodological approaches, and implications for postmortem research. Biological Psychiatry, 69, 146–156. [DOI] [PubMed] [Google Scholar]
- Relton CL, & Davey Smith G (2010). Epigenetic epidemiology of common complex disease: Prospects for prediction, prevention, and treatment. PLoS Medicine, 7, e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush AJ, Tirivedi MH, Ibrahim HM, Carmody TJ, Arnow B, Klein DN, … Keller MB (2003). The 16-item Quick Inventory of Depressive Symptomatology (QIDS) Clinician Rating (QIDS-C) and Self-Report (QIDS-SR): A psychometric evaluation in patients with chronic major depression. Biological Psychiatry, 54, 573–583. [DOI] [PubMed] [Google Scholar]
- Rutten BP, & Mill J (2009). Epigenetic mediation of environmental influences in major psychotic disorders. Schizophrenia Bulletin, 35, 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE, … Consortium G. (2012). Genome-wide DNA methylation scan in major depressive disorder. PLoS One, 7, e34451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Otowa T, Miyagawa T, Umekage T, Kawamura Y, Bundo M, … Sasaki T (2018). An epigenome-wide methylation study of healthy individuals with or without depressive symptoms. Journal of Human Genetics, 63, 319–326. [DOI] [PubMed] [Google Scholar]
- Spielberger CD (1988). Professional manual for the state-trait anger expression inventory. Tampa, FL: University of South Florida. [Google Scholar]
- Story Jovanova O, Nedeljkovic I, Spieler D, Walker RM, Liu C, Luciano M, … Amin N (2018). DNA methylation signatures of depressive symptoms in middle-aged and elderly persons: Meta-analysis of multiethnic epigenome-wide studies. JAMA Psychiatry, 75, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan E, Hunt C, Afari N, Duncan G, Noonan C, Schur E, … Buchwald D (2013). University of Washington twin registry: Poised for the next generation of twin research. Twin Research and Human Genetics, 16, 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PC, & Bell JT (2015). Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. International Journal of Epidemiology, 44, 1429–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, & Nestler EJ (2007). Epigenetic regulation in psychiatric disorders. Nature Reviews Neuroscience, 8, 355–367. [DOI] [PubMed] [Google Scholar]
- Uddin M, Koenen KC, Aiello AE, Wildman DE, de los Santos R, & Galea S (2011). Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychological Medicine, 41, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA (2009). Is epigenetics an important link between early life events and adult disease?. Hormone Research in Paediatrics, 71, 13–16. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Nielsen DA, Levran O, Randesi M, Hamon S, Ho A, … Kreek MJ (2011). Tissue-specific DNA methylation of the human prodynorphin gene in post-mortem brain tissues and PBMCs. Pharmacogenetics and Genomics, 21, 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Goldberg J, Bremner JD, & Vaccarino V (2013). Association between promoter methylation of serotonin transporter gene and depressive symptoms: A monozygotic twin study. Psychosomatic Medicine, 75, 523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]


