Abstract
Living organisms are continuously challenged by changes in their environment that can propagate to stresses at the cellular level, such as rapid changes in osmolarity or oxygen tension. To survive these sudden changes, cells have developed stress-responsive mechanisms that tune cellular processes. The response of Saccharomyces cerevisiae to osmostress includes a massive reprogramming of gene expression. Identifying the inherent features of stress-responsive genes is of significant interest for understanding the basic principles underlying the rewiring of gene expression upon stress. Here, we generated a comprehensive catalog of osmostress-responsive genes from 5 independent RNA-seq experiments. We explored 30 features of yeast genes and found that 25 (83%) were distinct in osmostress-responsive genes. We then identified 13 non-redundant minimal osmostress gene traits and used statistical modeling to rank the most stress-predictive features. Intriguingly, the most relevant features of osmostress-responsive genes are the number of transcription factors targeting them and gene conservation. Using data on HeLa samples, we showed that the same features that define yeast osmostress-responsive genes can predict osmostress-responsive genes in humans, but with changes in the rank-ordering of feature-importance. Our study provides a holistic understanding of the basic principles of the regulation of stress-responsive gene expression across eukaryotes.
INTRODUCTION
Sudden environmental changes, such as an increase in osmolarity, affect all organisms and compromise cell fitness and survival (1). Adaptive responses of Saccharomyces cerevisiae to a broad range of stresses have been widely studied due to its suitability as a model organism and its biotechnological applications. A well-characterized stress in S. cerevisiae is hyperosmotic stress -an increase in solute concentration of the media—which has been used as a paradigm of adaptive stress responses (2). Upon hyperosmotic stress, S. cerevisiae cells display a global adaptive response by regulating virtually all aspects of their physiology, including the accumulation of the osmoprotectant glycerol and other osmolytes, the regulation of cell cycle progression, and a massive reprogramming of gene expression. Central to this response is the highly conserved High-Osmolarity Glycerol (HOG) pathway, orchestrated by the Hog1/p38 Stress Activated Protein Kinase (SAPK) (2).
Upon osmostress, mRNA biogenesis is perturbed. The transcription, stability and translation of housekeeping genes decrease while Hog1 transiently accumulates in the nucleus to trigger the expression of osmostress-responsive genes (3). To induce osmostress-responsive genes, Hog1 stimulates transcription initiation directly associating with target genes and promoting the recruitment of the chromatin structure remodeling complex (RSC), histone modifiers, and the transcriptional machinery (e.g. RNA pol II and specific transcription factors (TFs)) (3). Furthermore, Hog1 promotes the transcriptional elongation of osmostress-responsive genes (4,5). The massive transcriptional response of yeast to stress, a phenomenon called the environmental stress response (ESR), promotes proper adaptation to several stresses. Essentially, the ESR comprises a set of genes commonly regulated by several independent stresses, such as oxidative, heat or osmotic stress (6). The ESR includes the upregulation of genes involved in energy-generating functions, detoxification, transport, and molecular chaperones, as well as the downregulation of genes related to growth processes, such as ribosome biogenesis and translation (7). Beyond the ESR, each type of stress triggers a specific transcriptional signature. For instance, osmostress induces the upregulation of genes related to glycerol biosynthesis (6,8). The genome-wide relevance of the ESR and the specific osmostress response were initially defined using DNA microarray assays (e.g. (6,8)). Since its first characterization, the hyperosmotic transcriptional landscape has been updated with tiling arrays (e.g. (9)) and RNA-seq (e.g., (5,10,11)).
The characterization of stress genes in terms of their biophysical and biochemical properties has remained elusive and limited to certain features through direct or indirect observations. For instance, ESR-upregulated genes are generally less conserved (12), have long mRNA half-lives (13) and long 5’UTRs (14). However, a systematic and comprehensive study of the features characterizing stress-responsive genes has not yet been performed. This is an important step towards understanding gene expression regulation and improving the tools used in synthetic biology and biotechnology. Moreover, it is unclear to what extent yeast and mammalian stress-responsive genes are conserved and, akin to yeast, there is a lack of a systematic characterization of stress-gene features. This has particular implications in biomedical research as stress responses are intrinsic to disease initiation or progression (15).
Here, we performed a transcriptome-wide comprehensive interrogation of the features characterizing osmostress-responsive genes. We identified a subset of 13 defining properties of osmostress genes and found that, although these genes are not conserved from yeast to humans, the minimal core features of yeast retain their predictive power for human genes.
MATERIALS AND METHODS
Yeast strains
Saccharomyces cerevisiae strain BY4741 (MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0) and its derivative YGM61 (HOG1::KanMX4) were used in this study.
Cell growth and stress
Wild-type and hog1 mutant (YGM61) strains were grown to early log phase in the absence (control) or presence of 0.4 M NaCl for 15 min. For each condition, a total of 15 ml of cells were harvested and flash-frozen in liquid nitrogen prior to RNA extraction. For each condition, three biological triplicates were done simultaneously.
RNA extraction and library prep
Total RNA extraction was done by hot phenol as reported previously (9). The quality of the extracted total RNA was assessed using a 2100 Bioanalyzer system (Agilent, 5067-1511) with an RNA integrity score (RIN > 8). Total RNA was used to generate whole gene, strand-specific libraries.
Experiment 1 (this study #1)
A total of 1 μg of RNA was used for library preparation using the TruSeq stranded mRNA v2 (Illumina) following manufacturer instructions. Quality control of generated libraries was performed with bioanalyzer and library quantification was done using the KAP kit (KapaBiosystems) prior to sequencing on an Illumina HiSeq2500 with 50 bp single-end reads.
Experiment 2 (this study #2)
A total of 5 μg of RNA were used as an input for the poly(A) selection module (Lexogen SKU: 039.100), following the manufacturer's instructions. Purified poly(A) RNA was used for library prep using the total CORALL RNA kit (Lexogen; SKU: 095.24), following the manufacturer's instructions. The number of cycles of library amplification was determined using a 1:10 dilution of the resulting libraries using the PCR Add-on for Illumina (Lexogen, SKU: 020.96). The resulting libraries were quantified by Qubit DNA High Sensitivity Assay (Thermo Fisher, Q33230) and assessed using the Bioanalyzer High Sensitivity DNA kit (Agilent, 5067-4626). All libraries were pooled at an equimolar ratio and sequenced with a single read (×50 cycles) using an Illumina HiSeq2500.
Read alignment and counting
S. cerevisiae
Sequencing reads were aligned to the S. cerevisiae genome (Ensembl, R64-1-1 v89) using HISAT2 (16) (v2.1.0) using the–rna-strandness parameter to account for strand-specific information, which differed between the libraries. The ‘F’ and ‘R’ labels were used to indicate single-end stranded and reversely stranded reads, respectively. For paired-end libraries, we used ‘RF’ ((5): ‘R’; (10): ‘RF’; (11): ‘R’; This study #1: ‘R’; This study #2: ‘F’). Reads were assigned to gene-level features (Ensembl, R64-1-1 v89 GTF) using featureCounts (17) (v1.5.1) in strand-specific mode (-s 1 for stranded and -s 2 for reversely stranded reads; (5): 2; (10): 2; (11): 2; this study #1: 2; this study #2: 1). Experiment 2 (this study #2) dataset reads contain unique molecule identifiers (UMIs) that were not used for the analysis for consistency with the rest datasets. For non-coding RNAs analyses, reads were assigned using the annotation from (18) and (19), containing Cryptic Unstable Transcripts (CUTs) and Stable Unnanotated Transcripts (SUTs).
Homo sapiens
Data from osmostressed (100 mM NaCl, 3 h) HeLa cervical cancer cells (20) were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE117699. Sequencing reads (reversely stranded) were aligned to the Homo sapiens genome (UCSC, hg38) using Tophat2 (21) (v2.1.0). Reads were assigned to gene-level features (GENCODE v24 GTF) using featureCounts (17) (v1.5.1) in strand-specific mode (-s 2). Processed raw counts from T47D breast cancer cells (22) upon hyperosmotic stress (110 mM NaCl, 3 h) were obtained from GEO under accession number GSE111904. Processed raw counts from HEK293 human embryonic kidney cells (23) upon hyperosmotic stress (80 mM KCl, 1 h) were obtained from GEO under accession number GSE152059. HeLa oxidative stress (60 μM H2O2, 8 h) and endoplasmic reticulum (ER; 0.5 μg/ml tunicamycin, 8 h) stress data (24) were downloaded from GEO under accession number GSE113171. HeLa hypoxia (1% oxygen, 16 h) data (25) were downloaded from https://osf.io/4a6tg/.
Definition of stress-responsive genes
S. cerevisiae
The catalog of osmostress-responsive genes in yeast was defined using differential expression analysis, performed with DESeq2 (26) (v1.26.0). Raw read counts from the five different experiments were combined and analyzed to assess the effect of the NaCl treatment while accounting for between experiment variability: ∼experiment + stress_treatment. Genes displaying an FDR ≤ 0.05 and |log2(fold change)| ≥ 1 were included in the catalog. As part of the quality control, batch effect removal for principal component analysis (PCA) was performed with the removeBatchEffect function from limma (27) (v3.42.2).
Homo sapiens
HeLa, T47D and HEK293 RNA-seq raw counts were analyzed individually with DESeq2 (26) (v1.26.0) with a single factor experimental design: ∼stress_treatment. Differentially expressed genes were defined as genes displaying an FDR ≤ 0.05 and |log2(fold change)| ≥ 1.
Definition of Hog1 dependency
Hog1-dependent genes were defined with yeast cells lacking Hog1 by using DESeq2 (v1.26.0) with the following design with interaction term: ∼hog1_genotype + stress_condition + hog1_genotype:stress_condition. We selected genes with FDRinteraction ≤ 0.05 and |log2(fold changeinteraction)| ≥ 0.25 that overlapped with stress-upregulated genes and stress-downregulated genes defined in the same experiment, enforcing opposite log2(fold change) directions (i.e., we considered genes less upregulated and less downregulated in HOG1 KO).
Functional enrichment analysis
Functional enrichment analysis of osmostress-responsive consensus genes was performed using g:Profiler (https://biit.cs.ut.ee/gprofiler/gost) (28) with default settings. Results for GO:Biological Processes were downloaded in Generic Enrichment Map format and visualized with the EnrichmentMap (29) app for Cytoscape (30).
Gene features characterization: S. cerevisiae
Features of genes were collected from previous publications and online sources or collected for the first time for this study. Full details of features already known can be found in the corresponding publications.
Features related to the 5’UTR and 3’UTR sequences were computed based on the annotation of (31), which was downloaded in GFF3 format from the SGD genome browser.
Broad conservation (32)
A measure of the number of species in which a gene has orthologs. The higher the number, the more conserved a gene is through evolution in the 86 species considered. Orthology relationships were derived from InParanoid (33) (v7).
Codon adaptation index (CAI) (32)
Reflects the degree of similarity in codon usage of a gene towards the codon usage of highly expressed genes.
Codon pair bias (this study)
A measure of the bias in the frequency of specific codon pairs (dicodons) given the frequency of their individual codons. To compute this measure, we implemented the formulas given by (34), (Supplementary Figure S1 in the original manuscript) in R. Briefly, we first computed a Codon Pair Score that indicates whether a given dicodon is over- or under-represented in the yeast coding genome. We then averaged the Codon Pair Score of each dicodon of a gene to generate the per-gene codon pair bias score.
Non-synonymous to synonymous substitution (dN/dS) (32)
A measure of gene conservation. Represents the ratio of non-synonymous over synonymous mutations for coding regions of S. cerevisiae as compared to sensu strictu yeast species (S. paradoxus, S. bayanus and S. mikatae).
GC content (this study)
Measured as the percentage of G and C occurrences in a given sequence. For simplicity of use, we applied the str_count() function from the seqinr package (35) (v4.2.4).
mRNA length (this study)
Computed using base R. Represents the number of nucleotides within the CDS of a gene (from start to end codon) using the Ensembl, R64-1-1 v89 annotation. 5’UTR and 3’UTR length were computed using the annotation of (31).
Effective number of codons (Nc) (32)
A measure of codon usage bias that quantifies the departure of codon usage of a gene from equal usage of synonymous codons. Low Nc values indicate strong codon bias and high values indicate low bias.
Genetic interaction degree (32)
Number of genetic connections of a gene derived from combinatorial perturbation genetics experiments.
Fitness defect (32)
A score that reflects the effect of the genetic perturbation of a gene on cell growth.
Yeast conservation (32)
Analog measure of broad conservation but restricted to a set of 23 Ascomycota fungi.
Co-expression degree (32)
Number of genes co-expressed with a given gene, obtained from a co-expression network. Only genes above the 95th percentile in co-expression were considered.
Distance-to-median (DM) (this study)
A mean-independent measure of RNA abundance variability in yeast single-cell RNA-seq data (36). We normalized single-cell RNA-seq data with median-based size factors using the computeSizeFactors() function from DESeq2 (26) (v1.26.0). We then computed distance-to-median using the DM() function from the scran package (37) (v1.14.6). This measure removes the relationship between CV2 and mean expression of an RNA by fitting a median-based trend to the log-transformed CV2 against the log-transformed mean.
Expression level (32)
A measure of RNA abundance in S. cerevisiae growing in basal conditions using high-density oligonucleotide arrays.
Expression variation (32)
A measure of the variance in the expression of a gene across different microarray experiments, including distinct growth conditions and replicates.
mRNA half-life (13)
A measure of the time needed to reduce the abundance of a given mRNA by half, which was determined using metabolic labeling with 4-thiouracil (4-TU).
Number of transcription factors (this study)
Total number of transcription factors associated with a given gene according to the YEASTRACT database (38) with documented evidence (DNA and expression evidence). TF motif discovery analysis of Hog1-dependent genes was performed using the HOMER tool (39) (v4.11.1). We looked for known TF motifs 400 bp upstream of Hog1-dependent genes using Hog1-independent genes as a background. TF motif enrichment analysis of osmostress-upregulated SUTs was performed using the AME tool from the MEME suite (40). The enrichment was performed considering 200 bp upstream of SUTs from the YEASTRACT database (38) using shuffled input sequences as control (default for AME).
PARS score (41)
Measures the degree of RNA secondary structure as captured by deep sequencing of the RNA treated with structure-specific enzymes. Here we displayed the mean PARS score per gene, where higher scores indicate higher structure.
Variance stabilized transformation (VST) residual variance (this study)
Another alternative measure of RNA abundance variability. It is the residual variance from a regularized negative binomial regression, computed on yeast single-cell data (42) using the vst() function from the sctransform package (43) (v0.3.1).
Protein disorder (32)
Proportion of unstructured residues of a protein, as computed by Disopred2 (44).
Prionic properties (45)
A measure of the prion-like character of a protein predicted by a hidden-Markov model algorithm trained on experimentally determined prion proteins. Briefly, Prion-Like AminoAcid (PLAAC) software (46) identifies prion sub-sequences inside a protein and generates a per protein log-likelihood ratio (LLR) score. For S. cerevisiae we used the already precomputed table for the whole proteome on the PLAAC webpage (http://plaac.wi.mit.edu/Scer-all-proteins-2014–05–17.xls).
Multifunctionality (32)
A measure of the number of functions carried out by a protein defined as the number of GO terms associated with it.
Number of domains (32)
Number of regions of a protein identified as domains by Pfam.
Number of unique domains (32)
Number of unique protein domains defined by Pfam after filtering out repeated domains within a protein.
Protein-protein interaction degree (PPI) (32)
Number of physical interactions of a given protein reported in BioGRID v.2.0.58 (47) restricted to the following terms: Affinity Capture-MS, Affinity Capture-RNA, Affinity Capture-Western, Biochemical Activity, Co-crystal Structure, Co-fractionation, Co-localization, Co-purification, Far Western, FRET, PCA, Protein-peptide, Protein-RNA and Reconstituted Complex and Two-hybrid.
Relative synonymous codon usage (RSCU) (48)
Measure of non-uniform codon usage of synonymous codons. RSCU values are the number of times a particular codon is observed, relative to the number of times that the codon would be observed for a uniform synonymous codon usage (i.e. all the codons for a given amino-acid have the same probability).
Interpro domains
Interpro domains associated to genes were downloaded through the BiomaRt's getBm function (49) (v2.45.9). Enrichment of a given Interpro domain in stress-upregulated or downregulated genes was assessed with an hypergeometic test.
Gene features characterization Homo sapiens
As humans have a more complex transcriptome annotation than yeast, with some genes having several annotated transcripts per gene, we decided to keep only the main transcript per gene by selecting ‘principal’ transcripts from the APPRIS annotation (50) accessed through the BiomaRt's getBm function (49) (v2.45.9). CDS, 5’UTR and 3’UTR sequences were obtained using the getSequence function (49) from BiomaRt() (v2.45.9). The set of features gathered for humans was restricted to those deemed important in the modeling approach in yeast.
Broad conservation (this study)
As in yeast, we computed the number of species in which a human gene has an ortholog, for a total number of 100 species. To this end, we used the ‘hom.Hs.inp.db’ Bioconductor package (33) (v7).
Codon pair bias (this study)
Computed as for yeast.
GC content (this study)
Computed as for yeast.
mRNA length (this study)
Computed using base R. Represents the number of nucleotides contained within 5’UTR, 3’UTR and start and end codons from GRCh38.p13 genome assembly annotation downloaded from Ensembl v101 (ensemble release).
Distance-to-median (this study).
Computed as for yeast using HeLa single-cell dataset (51). GEO database accession number: GSE129447.
mRNA half-life (52)
Data obtained from (52), using BRIC-seq. Briefly, this methodology labels RNAs with 5’-bromo-uridine and then measures the decrease in RNA levels with sequencing.
Number of transcription factors (this study)
Total number of unique transcription factors associated with each gene extracted from ENCODE. For its computation, a set of regulatory interactions for 181 TFs was downloaded from Harmonizome (53); (https://amp.pharm.mssm.edu/static/hdfs/harmonizome/data/encodetfppi/attribute_set_library_crisp.gmt.gz).
Protein disorder (this study)
Proportion of unstructured residues in a protein obtained from the Database of Disordered Proteins Predictions (D2P2) (54). The D2P2 database gathers the results of several protein structure prediction software packages. We accessed the whole D2P2 programmatically using the fromJSON() function from the rjson package (v0.2.20) and then parsed the json objects to obtain disordered residues. Residues were considered disordered when there is a consensus between > 75% of the predictors in the D2P2 database at a given position. We selected the longest protein isoform possible per gene and used human gene identifiers from Ensembl v63 for compatibility with D2P2.
PPI degree (this study)
As for yeast, a measure of the number of interactors of a given protein obtained from BioGRID (47) (v.3.5.181). We downloaded the whole BioGRID dataset (https://downloads.thebiogrid.org/File/BioGRID/Release-Archive/BIOGRID-3.5.181/BIOGRID-ALL-3.5.181.tab2.zip) and processed it with R to keep the same experimental evidence as for yeast and to consider only human interactions (taxonomy id: 9606).
Comparison of stress features
We assessed differences in the values of the features between osmostress-responsive genes (upregulated and downregulated) and unresponsive genes using pairwise Wilcoxon tests with the pairwise.wilcox.test R function. We computed FDR for multiple testing correction with p.adjust(method = ‘fdr’) R function. We then summarized the results in categories (‘Equal’, ‘Lower’ and ‘Higher’) based on the median value of the feature in each group and the result of the test (FDR ≤ 0.05). We assigned ‘Higher’ if FDR ≤ 0.05 and the value of the feature in the stress group was higher than the value in the unresponsive group. We assigned ‘Lower’ if FDR ≤ 0.05 and the value of the feature in the stress group was lower than the value in the unresponsive group. The remaining features were assigned the ‘Equal’ label.
Stress features filtering for modeling
Multinomial logistic regression requires comprehensive observations (i.e. each feature to be measured in each gene) and low levels of collinearity. For the 30 features in yeast, there were complete observations for only 1164 genes. Moreover, some of these observations were related or redundant (e.g. the complementary measures of RNA abundance variability or mRNA abundance and codon adaptation index). We performed a pairwise correlation analysis between the features and discarded those with a Pearson correlation >0.35, removing the feature in the pair with the lowest number of observations. We selected broad conservation instead of yeast conservation (same number of observations) since broad conservation comprises more species and is more informative. We chose DM over VST residual variance, since, in our data, this measure better removes the relationship of mRNA abundance variability with mean expression. We further removed fitness and PARS scores (5’UTR and 3’UTR) because they have the lowest number of observations (3466, 2679 and 2882, respectively) and significantly impact the number of complete observations for modeling. Finally, we removed co-expression degree, a feature expected to be strongly associated with the stress genes, that we used as positive control for the modeling. We used the resulting set of 13 unique features for modeling, with higher information content, for a total number of 3828 complete observations. Pairwise Pearson correlations were computed using the rcorr(type = ‘Pearson’) function from the Hmisc (v4.4.2) package.
Stress feature modeling
We used (Multinomial) logistic regression and random forest models for both S. cerevisiae and H. sapiens using features as predictors for classifying genes in three categories: upregulated, downregulated or unresponsive. For each model, we assessed the performance of the model and the relevance of the features. When performance was measured, we divided the data into 70% training and 30% testing. The performance of the model was measured by the Area Under the Curve (AUC) of the receiver operating characteristic (ROC) curve using prediction(), performance() and plot() functions from the ROCR package (55) (v1.0.11). The importance of the features was assessed differently for multinomial logistic regression and random forest models:
Multinomial logistic regression
Multinomial regression was performed with the multinom() function from the nnet package (56) (v7.3.14). Logistic regression was performed with the glm(family = binomial(link = ‘logit’)) function from the stats package. We used the following three approaches to assess the importance of the features:
AIC of the univariable model
We fitted a model for each feature and used the Akaike information criterion (AIC) to compare the features.
AIC of the multivariable model (leave-one-out approach)
We fitted a model with all the features, registered the AIC and then compared it with the AIC of the model after leaving a feature out.
Coefficient of the multinomial regression
We fitted a multivariable model with the scaled data and compared the coefficient of the multinomial regression for each feature. This coefficient indicates the increase or decrease in the log-likelihood of a gene being predicted as stress-responsive (upregulated or downregulated) over unresponsive with an increase of one (scaled) unit in the feature.
Random forest
Random forest modeling was performed using the randomForest(importance = T) function from the randomForest package (57) (v4.6.14). The importance of the features was measured as the mean decrease in the accuracy of the model after permuting the values of each feature.
TCGA data analysis
The Cancer Genome Atlas (TCGA) level 3 RNA-seq and copy number alteration (CNA) data were downloaded from Broad GDAC Firehose (https://gdac.broadinstitute.org/), release 2016-01-28. The RNA-seq data contain the scaled estimate expression for each type of sample and tumor type, which we rounded to an integer number for subsequent use with DESeq2. We kept only tumor-normal matched samples and only primary solid tumors (n = 1484) by using TCGA barcodes (01 and 11 for sample types). We then performed differential expression analysis with DESeq2 for each tumor with a paired design: ∼patient + condition. The CNA data describes homozygous and heterozygous loss, copy number neutral, and low and high gain states. CNA proportion was computed by dividing the number of samples with CN alterations (amplification and deletion) by the total number of samples considered. Fold changes and CNA proportion were graphically represented with the Heatmap() function from the ComplexHeatmap package (58) (v2.2.0).
RESULTS
OsmoAtlas: a comprehensive catalog of osmo-responsive transcriptome
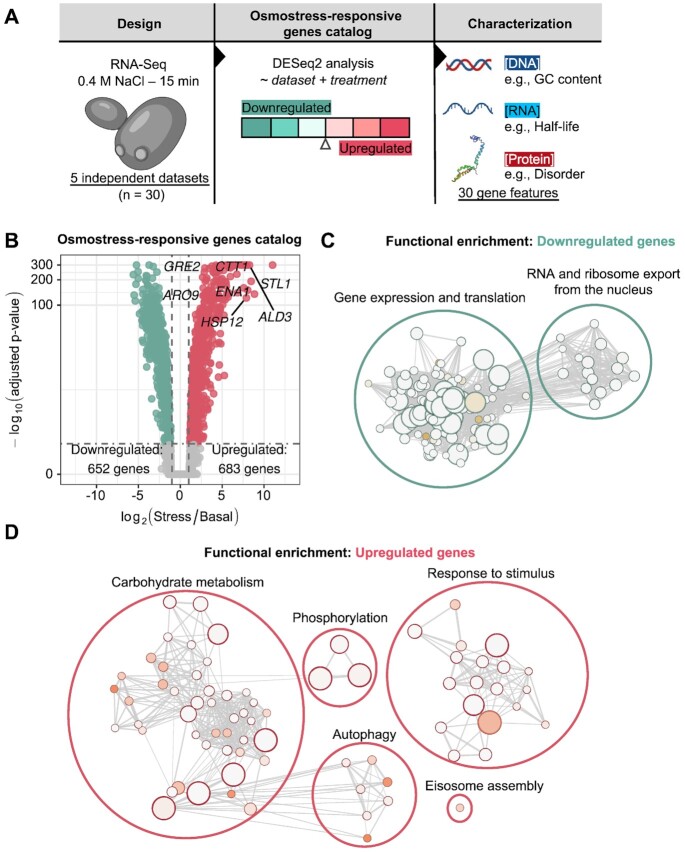
To study features that define osmostress-responsive genes in yeast, we first created a comprehensive catalog of transcripts whose expression is regulated in response to osmostress. We generated two independent RNA-seq datasets of cells subjected or not to osmostress (0.4 M NaCl, 15 min). Moreover, we collected data from three published RNA-seq datasets performed in triplicates with identical experimental conditions and genetic background (BY4741) (5,10,11). This dataset is a high replicate compendium, with a total of 30 samples (15 per condition) (Figure 1A). This experimental design captures the highest expression peak of the osmoadaptive response, which is turned on and off within 30 minutes, coinciding with activation/phosphorylation of Hog1 and its nuclear localization. We performed an integrative approach to correct for sample bias and found that samples in a principal component analysis (PCA) showed clear stress-dependent clustering, where an osmostress signature drove the variance in the data up to 96% when considering ORFs and 51% when considering ncRNAs (Supplementary Figure S1A and B). To generate a complete gene catalog, we included both protein coding (ORFs) annotated in the yeast genome (saccer3, SGD) together with non-coding (ncRNAs). These include stable unannotated transcripts (SUTs) and cryptic unstable transcripts (CUTs) from steady-state conditions (18) and upon osmotic stress (19). We defined as osmostress-responsive those transcripts whose expression shows at least a 2-fold differential expression upon NaCl treatment considering all the datasets together (see Materials and Methods).
Figure 1.
A high confidence, RNA-seq-based catalog of osmostress-responsive genes. (A) Experimental and computational outline of the generation and characterization of the catalog. (B) Volcano plot showing the number of genes that pass the criteria (|log2(fold change)| ≥ 1 and FDR ≤ 0.05) to be included in the catalog of consensus upregulated or downregulated genes. Positive controls are placed in the graph and, as expected, they appear as the most upregulated genes. Genes upregulated upon stress are shown in red and those downregulated in green. C-D) Functional enrichment regarding GO: biological Process of osmostress-responsive (C) downregulated and (D) upregulated genes compared to unresponsive genes. Nodes represent functional terms and their size reflects the size of the term. Nodes are connected if they share a significant number of genes. Significantly enriched terms are clustered by similarity and then manually annotated. Color-filled circles represent FDR values from functional enrichment analysis. Darker colors represent smaller FDR values. Panel (A) has figures created with BioRender.com.
The OsmoAtlas yielded a high-confidence osmostress-consensus list of upregulated (683 ORFs; 103 ncRNAs) and downregulated (652 ORFs, 63 ncRNAs) transcripts (Figure 1B and Supplementary Figure S1C). We uncovered that 8.7% of ncRNAs were differentially expressed upon stress (upregulated: 103, downregulated: 63). This ratio was similar to the one observed for the coding transcriptome (18.7% of osmo-regulated genes) (Supplementary Tables 1 and 2), indicating a highly dynamic and proportional regulation of the non-coding transcriptome upon stress. We also defined a control gene set (unresponsive) with genes with unperturbed expression during stress (5629 genes) (Figure 1B, Supplementary Table 1). The whole catalog correlates with previously published microarrays (Supplementary Figure S1D; Spearman's rho = 0.75) (6) or tiling arrays (Supplementary Figure S1E; Spearman's rho = 0.69) (9) performed in similar experimental conditions.
We further characterized the catalog of osmostress-responsive ORFs in terms of annotated cellular functions. Expected categories such as carbohydrate metabolism, response to stimulus and phosphorylation were enriched in upregulated genes, while gene expression and translation-related processes were enriched in downregulated genes (7) (Figures 1C and 1D). In addition, functions such as autophagy and eisosome assembly, which have been linked to osmostress (59,60), were also found enriched in upregulated genes (Figure 1D). These functions are not equally represented across previous transcriptome-wide studies, thus indicating the high-resolution and comprehensiveness of our gene catalog. This upgraded catalog of osmo-responsive genes serves to define common features of osmostress-responsive genes with high statistical power.
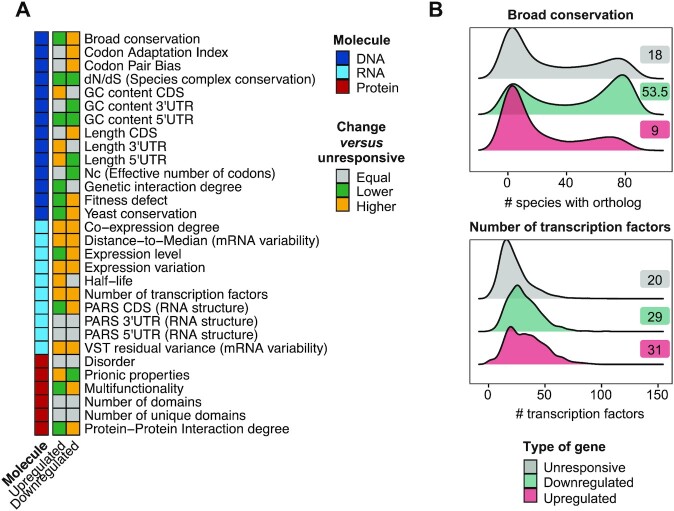
Stress-responsive genes display different features at all levels of gene expression
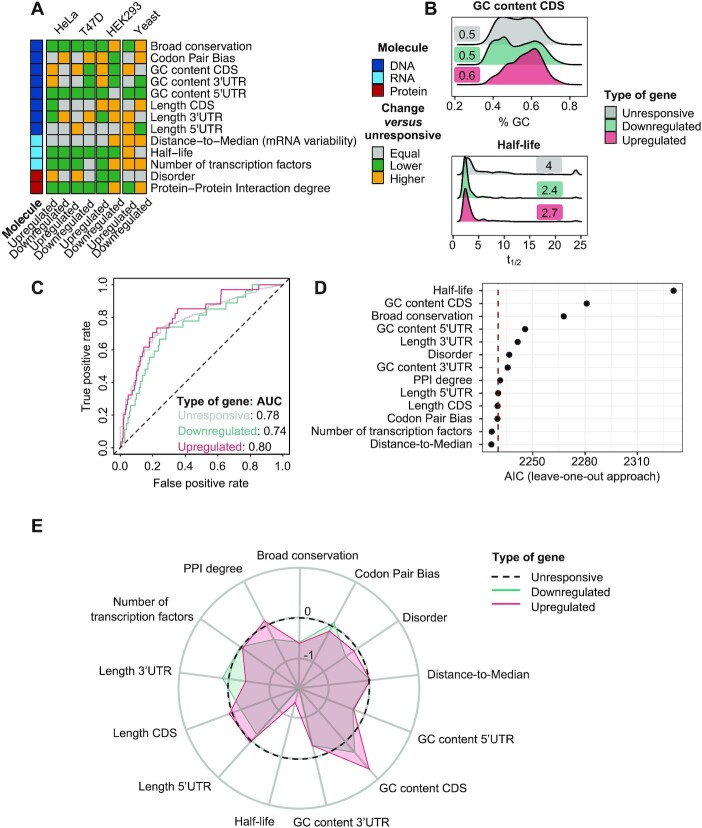
To identify the intrinsic features of osmostress-responsive genes, we compiled a comprehensive set of 30 gene features (Figure 2A), 9 of which were computed in this study and 21 obtained from public data sources (Supplementary Table 3). We classified these features on the basis of their association to DNA (n = 14), RNA (n = 10) or protein (n = 6). We then compared them between osmostress-responsive genes (up- and down-regulated) and unresponsive genes using a pairwise Wilcoxon test. We found that 25/30 features (83%) were significantly different (FDR ≤ 0.05; see Materials and Methods) for at least one group of osmostress-responsive genes (up- or down-regulated) and 16 of those (53%) were significantly different in both gene groups (up- and down-regulated) (Figure 2A and S2A; Supplementary Table 4). For example, broad conservation—a measurement of conservation across 100 species from yeast to humans- showed low values in upregulated genes and high values in downregulated genes, as compared to unresponsive genes (Figure 2B). Interestingly, the dN/dS ratio (ratio of non-synonymous over synonymous mutations) of osmostress-responsive genes within S. cerevisiae and 3 closely related species was generally lower than the dN/dS ratio of unresponsive genes thus indicating higher conservation of stress genes between proximate species (Figure 2A and S2A). Remarkably, overall, the number of unique transcription factors targeting a gene (TFs), co-expression degree and gene-expression variation of osmostress-responsive genes were high compared to unresponsive genes (Figure 2A, 2B and S2A). Thus, with this analysis, we recover known features of stress genes (12–14), which proves the robustness of our analysis, and identify new features of osmostress-responsive genes. These results suggest that osmostress-responsive genes have evolved distinct features.
Figure 2.
Stress-responsive genes display different features in all levels of regulation. (A) Heatmap summarizing the results for individual Wilcoxon tests assessing differences between osmostress-responsive and unresponsive genes for the indicated features. Gray indicates features that are unchanged, yellow features that are higher in osmostress-responsive genes, and green features that are higher in osmostress-responsive genes. Features are ordered first by molecular group and then alphabetically. The molecule of regulation to which the feature belongs is shown in the left column. Features related to DNA are indicated in dark blue, RNA in light blue and Protein in red. (B) Density plot shows the distribution of two selected features (broad conservation and number of TFs) for each group of genes. X-axis represents the value of a given feature and Y-axis the kernel density estimate. Labels show the median value of a given feature in each group of genes.
To provide a more detailed picture of osmoresponsive genes, we have assessed other features benefiting of a high-level of granularity. In this regard, we measured individual codon usage (48) in the top 500 most expressed genes (highly optimized for translation), and compared it with unresponsive, downregulated, and upregulated genes. As expected, highly expressed genes showed a specific codon usage. Interestingly, osmoresponsive genes, albeit being repressed under steady-state conditions, displayed a similar codon usage to unresponsive and downregulated genes (Supplementary Figure S2B). Furthermore, we explored the distribution of stress genes across chromosomes and found practically even distribution for all of them (Supplementary Figure S2C). We further characterized the landscape of RNA structure at the nucleotide level by analyzing PARS score distribution across 20 nt window, instead of the mean PARS score per gene. We observed a higher structure of downregulated CDS mRNA and a more disordered structure of upregulated CDS mRNAs. As previously reported, UTR regions displayed less secondary structure than coding regions, whereas the start codon displayed minimal PARS score, consistent with an increased accessibility. While an increase in structure at the coding regions was expected, downregulated genes were more structured than upregulated genes, for which the periodicity in structure observed in the first 200 nt was inverse (Supplementary Figure S2D). Last, we explored the diversity of protein domains within differentially expressed genes. By applying a hypergeometrical test using annotated InterPro domains (61) we found that, in addition to the expected motifs related to stress-upregulated genes (sugar/inositol transporter and heat shock protein, among others), all five yeast proteins that contain the UBX domain together with several arrestin-like domain appeared enriched in upregulated genes (Supplementary Figure S2E). Thus, it could be plausible that UBX or arrestin-like domains could contribute osmoadaptation with yet uncharacterized mechanisms.
Statistical modeling ranks the most distinctive features of stress genes
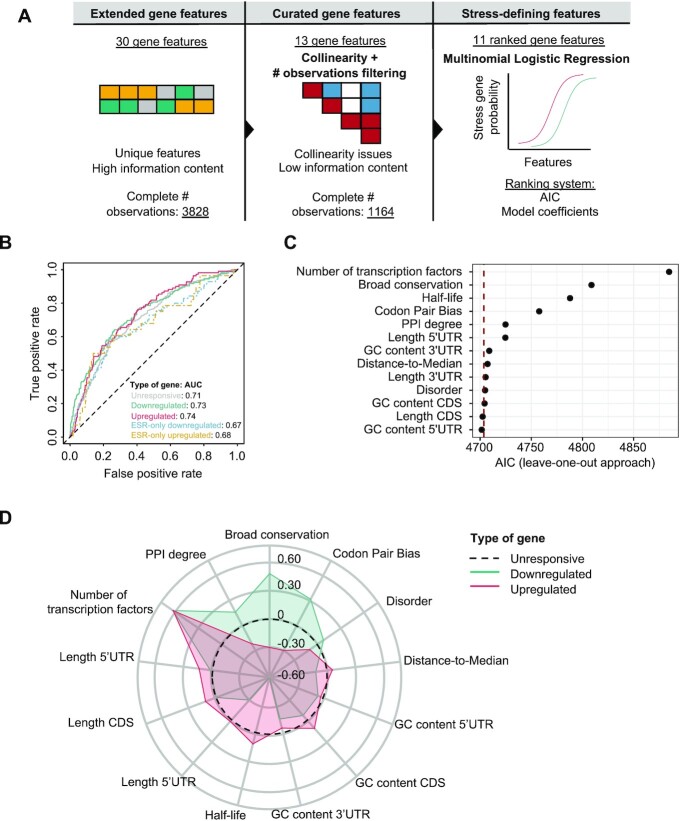
We used a data-driven approach to identify and rank the key features of osmostress-responsive genes. To this end, we trained a multinomial logistic regression model to classify genes into three categories: upregulated, downregulated or unresponsive, using gene features as predictor variables. We then evaluated the contribution of each feature to the model in terms of the AIC or by using coefficients of regression (Figure 3A).
Figure 3.
Identification of osmostress-specific gene features through statistical modeling. (A) Outline of the process of filtering and statistical modeling for prioritization of osmostress-responsive gene features. First, we gathered a set of n = 30 features with some degree of redundancy, collinearity and with some of them having a low number of observations. Briefly, the workflow starts with an unrestricted list of gene features, which is further prioritized to modeling by retaining the most relevant ones. These are then used for multinomial logistic regression to obtain a ranked list of significant gene properties. (B) ROC curve for the multinomial logistic regression model. The model is trained using 70% of the data and tested in the remaining 30%. AUC of the ROC curve is displayed in the legend. ESR-only genes are defined with data from (6). (C) Dot plot showing the AIC of the multinomial logistic regression after removing each variable from the model, one at a time. Features (Y-axis) are ordered on the basis of importance by the AIC leave-one-out approach. The dashed red line indicates the AIC of the model with all the features (full model). (D) Radial plot showing the coefficient of the multinomial logistic regression for each scaled feature. Red shows the comparison done using upregulated responsive genes and green using the downregulated genes.
Before fitting our models, we filtered predictor variables by balancing two aspects related to gene features: given the complexity of eukaryotic genomes, certain features are correlated between each other (e.g. codon adaptation index and mRNA structure), which may introduce bias due to collinearity issues and some of the features are not available or measured for all genes, yet the regression model needs complete observations (i.e. only 1164 genes display complete observations for all 30 features). Therefore, we prioritized modeling of only non-redundant features that were measured for the largest number of the genes (Figure 3A). We calculated the pairwise Pearson correlation among all features and retained the feature with the greatest number of observations when the correlation coefficient of the pair exceeded 0.35 (Supplementary Figure S3A) (see methods). This resulted in 13 unique ‘minimal features’ with high information content measured in a set of 3828 out of a total of 6692 genes (i.e. 57% of the yeast genes).
Next, we applied various multinomial logistic regression models to the ‘minimal feature set’. The multivariable model (MVM), which includes all the ‘minimal features’, showed an AUC of the ROC curve of 0.71 for unresponsive genes, 0.73 for downregulated genes and 0.74 for upregulated genes (Figure 3B), thus indicating predictive performance with just the ‘minimal features’. To assess the specificity of our model to osmoresponsive or ESR genes, we created a set of osmoresponsive-independent ESR genes by gathering ESR genes defined by (6) and removing the osmostress genes defined in the same study (sorbitol 1M, 15 min). We obtained AUCs of 0.67 and 0.68 for ESR genes not classified as osmostress-responsive, pointing to the potential generalization capabilities of the model (Figure 3B), hence suggesting that ESR genes share intrinsic properties to enable their multilayered regulation.
We subsequently assessed the individual contribution of these features and ranked them on the basis of their contribution to the quality of the MVM with a leave-one-out approach. Briefly, we removed one feature at a time from the model and measured the change in the AIC with respect to the full model. A positive change in the AIC denotes a decrease in the quality of the model after removal of a given feature, thus indicating that a specific feature is important for classification. Conversely, a negative change in the AIC denotes that a feature does not improve the classification and can therefore be removed from the model. We found that 11/13 selected features (all except CDS length and GC content of the 5’ UTR) were beneficial to the model's quality when comparing with the full model AIC. The five most important features were: the number of TFs targeting a gene, broad conservation, mRNA half-life, codon pair bias, and degree of protein-protein interaction (PPI) (Figure 3C).
As an alternative approach, we ranked the 13 features by their AIC in individual multinomial regressions (univariable model, UVM). Four out of the five top features remained similarly ranked (number of TFs, broad conservation, PPI degree and codon pair bias), but mRNA half-life dropped in the ranking, being replaced by distance-to-median, a measure of RNA abundance variability (Supplementary Figure S3B). To further validate our approach, we added co-expression degree to the MVM as a positive control. Co-expression degree is a measure of the correlation in expression, and it is expected to be high for coordinated transcriptional programs, such as the transcriptional response to osmostress. Accordingly, co-expression degree is one of the most important features for stress-responsive genes and was correctly prioritized as the first ranked feature when added to the model (Supplementary Figure S3C).
To verify that the relevance of each feature persisted when using a different modeling framework, we built a random forest model with the same set of genes (class) and features (predictors). After training, the random forest exhibited AUCs of 0.71, 0.76 and 0.76 for unresponsive, upregulated and downregulated genes, respectively (Supplementary Figure S3D). Reassuringly, four out of five of the features previously identified (half-life, number of TFs, broad conservation and codon pair bias) were again among the top 5 (Figure 3C versus Supplementary Figure S3E). Surprisingly, the random forest also prioritized CDS length as an important feature (fourth in the ranking), while the multinomial regression did not.
Finally, to quantitatively assess the direction and magnitude of change for each of these features, we compared the coefficients of the MVM for up- and down-regulated genes (Figure 3D). This analysis yielded known observations, such as upregulated genes being less conserved (12), having longer 5’ UTRs (14) and higher mRNA half-lives (13). Furthermore, it revealed new features for upregulated genes, such as a higher number of TFs and higher expression variability, while downregulated genes showed higher codon pair bias. Overall, we identified a variety of patterns, with certain features displaying similar quantitative changes in up- and down-regulated genes (e.g. number of TFs) and others with completely opposite patterns (e.g. broad conservation) (Figure 3D).
In summary, by using complementary approaches (univariable and multivariable multinomial logistic regression and random forest), our modeling analyses consistently identified the number of TFs, broad gene conservation and codon pair bias as the most distinctive features of osmostress-responsive genes. Specially, the number of TFs and broad gene conservation ranked in the very top in the three approaches (number of TFs: first in three models; broad conservation: second in MVM and UVM and third in random forest). Two of the three modeling approaches revealed degree of PPI, and mRNA half-life as top 5 key stress features. In addition, our analysis identified the direction and magnitude of change of known and novel osmostress-responsive gene features.
Distinctive features of Hog1-dependent genes
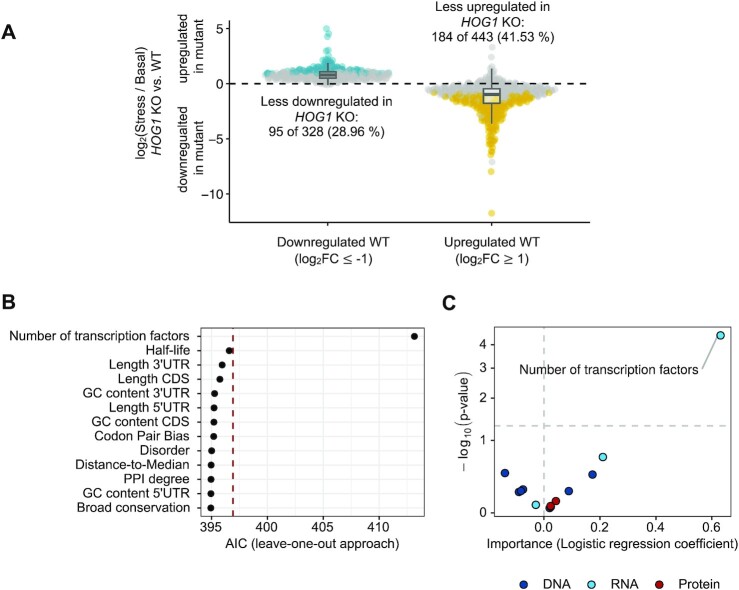
The Hog1 SAPK is crucial for transcriptional reprogramming during osmoadaptation, exerting a multilayered control of gene expression by directly associating to target genes to regulate all steps of the transcription cycle (3). Our experimental design (0.4 M NaCl, 15 min) was meant to capture the peak of expression and defined the contribution of Hog1 to the response. Thus, we aimed to provide further insights into the transcriptional response to osmostress by assessing the features that define Hog1-regulated genes. We generated a new dataset to determine Hog1 dependency upon hyperosmotic stress. Wild-type and hog1-deficient cells were subjected or not (basal) to hyperosmotic stress (0.4 M NaCl, 15 min), and mRNA abundance was determined by RNA-seq (n = 3 for each condition). As expected, a PCA revealed that the effect of hog1 deletion in basal conditions was negligible, whereas its impact upon osmostress was substantial (Supplementary Figure S4A). In basal conditions, only four genes changed their expression at least two-fold by hog1 deletion, as described before with DNA microarray assays (Supplementary Figure S4B) (62). We then first defined Hog1-dependent genes by comparing gene expression in response to stress in hog1-deficient cells with respect to wild-type cells. We considered a gene to be Hog1-regulated when the change in expression upon exposure to stress varied by 0.25 (with respect to log2(fold change)) in the presence of HOG1 (log2(fold changeinteraction) ≥ 0.25 and FDR ≤ 0.05; see Materials and Methods). Taking into account that a gene should change at least two-fold to be considered an osmostress-responsive gene (high stringency), these criteria yielded a total of 279 genes (184 upregulated versus 95 downregulated) Hog1-dependent genes (36% of all responsive genes identified in this experiment) (Figure 4A). We validated our defined gene sets by assessing the association of RNA Pol II and Hog1 by ChIP-seq using available data (19). As expected, the Z-score of RNA Pol II association increased upon stress to upregulated genes and dissociated from downregulated genes (Supplementary Figure S4C). Hog1-dependent genes displayed a stronger RNA Pol II association, which correlated with Hog1 recruitment (Supplementary Figure S4D). These results were in agreement with previous reports and validated the accuracy of the criteria used to classify gene sets (19).
Figure 4.
Features of Hog1-dependent genes. (A) Boxplots show the results of the differential expression analysis of the RNA-seq data (this study) from wild-type (WT) and hog1Δ cells in response to osmostress (0.4 M NaCl, 15 min). Horizontal lines in the boxplot indicate the median and dashed black lines a relative absolute log2(fold change) of 0 (|log2(fold changeinteraction)|). Yellow (less upregulated) and blue (less downregulated) indicate the genes that pass the threshold (FDR ≤ 0.05 and |log2(fold changeinteraction)| ≥ 0.25) to be considered as affected in the HOG1 KO upon stress and thus considered as Hog1-dependent genes. (B) Dot plot showing the AIC of the logistic regression model comparing upregulated Hog1-dependent genes against Hog1-independent genes after removing each variable from the model, one at a time. (C) Volcano plot showing the coefficient of the logistic regression comparing upregulated Hog1-dependent responsive genes against Hog1-independent responsive genes and the significance of each property. Features are colored based on their layer of regulation (dark blue: DNA; light blue: RNA; Protein: red) and those that pass the significance threshold (P-value ≤ 0.05) are labeled.
Next, we explored which features differentiate stress-upregulated Hog1-dependent and -independent genes. Like the previous analyses, we performed logistic regressions using an MVM with a leave-one-out approach (Figure 4B), as well as a UVM (Supplementary Figure S4E). Both models consistently found that the number of TFs was the most important feature to define Hog1-dependent genes. A random forest model corroborated these findings (Supplementary Figure S4F), with an increased number of TFs being the only relevant feature in the prediction of Hog1-dependency (Figure 4C).
Since the number of TFs appeared as the most important feature distinguishing upregulated Hog1-independent from upregulated Hog1-dependent genes, we sought to further explore it. To identify those regulatory elements that might be relevant for Hog1 dependency, we used motif discovery analysis. We identified 47 motifs enriched among the promoters of Hog1-dependent genes compared to promoters of Hog1-independent genes (Supplementary table 5). Among these motifs were motifs of TFs downstream of the Hog1 pathway, including Msn2/4 and Sko1 (63).Taken together, our results indicate that a higher number of TFs may also be a crucial feature for Hog1-dependent genes. Several TF motifs were found to be enriched among the promoters of Hog1-dependent genes, suggesting that the palette of TFs used by Hog1 to modulate gene expression is greater than previously anticipated (3,63).
Additionally, we defined the dependency of the osmosresponsive ncRNAs of the Hog1 SAPK. Despite the dynamic range of ncRNAs being smaller than for coding genes, the criteria used for defining Hog1 dependency was the same as the used for coding genes for coherence. Similar to coding genes, we observed a higher dependency on Hog1 of non-coding upregulated genes respect downregulated, detecting 25.86% of upregulated ncRNAs that required the presence of Hog1 for proper expression (Supplementary Figure S4G and H). To deepen the understanding of the TFs regulating Hog1-dependent ncRNAs, we characterized the enrichment of TFs motifs upstream of osmostress-upregulated SUTs (the ncRNAs with highest transcriptional changes upon stress) and found a rather limited set of transcription factors from which Sko1 scored as the first hit (Supplementary Table 6), suggesting its preferential use for osmoresponsive ncRNA expression.
Predictive features of yeast osmo-responsive genes are conserved in humans
To assess the conservation of yeast stress-dependent genes features in humans, we applied the same modeling approach as for yeast. First, we defined a set of osmostress-responsive genes from a previously published RNA-seq experiment in HeLa cervical cancer cells treated with hyperosmotic stress at a concentration and stress duration that balance transcriptional responses and cell survival (100 mM NaCl, 3 h) (20). HeLa cells are widely used to study hyperosmotic stress response (e.g. (20,64)). As for yeast, we considered up- and down-regulated genes whose expression varied by at least 2-fold upon stress (Supplementary Table 7). In total, we identified a total of 600 downregulated and 1286 upregulated genes. A functional enrichment analysis of these genes revealed similar processes than in yeast such as upregulated cellular responses to external stresses (chemical stress, stimulus, immune processes), signalling components and a downregulation of transcription. Futhermore, we found new activities specific to human upregulated genes such as terms related to cell movement, cell adhesion and cell communication and related with pathological processes such as amyloid fibril formation or cell population proliferation (Supplementary Table 8).
We then compiled features of human genes (see Materials and Methods) and performed a comparative analysis. We included all of the features that reached the final modeling step in yeast (i.e. the ‘minimal features’) (Supplementary table 9). We found that 11/13 (85%) features showed a significant difference in any direction when analyzing stress-responsive compared to unresponsive genes (Figure 5A, B and S5A; Supplementary Table 10). Additionaly, to assess the extensibility of our model to other cell types (cancerous and non-cancerous), we compared the behaviour of stress-features between HeLa, T47D (22)—NaCl, 3 h and HEK293 (23)—80 mM KCl, 1 h. We found a high degree of coincidences in stress-features (19/26 for T47D and 15/26 for HEK293) (Figure 5A) with a similar ranking-order of feature-importance (Figures S5E and F). Moreover, whether trends observed in yeast were retained in humans seemed to be partially dependent on the molecular identity of the features (DNA or RNA). Changes of DNA features tended to be more frequently similar between the two organisms than those of RNA features, albeit not statistically significant (P = 0.12, Fisher's exact test) given our low statistical power (coincident features: 7 out of 16 DNA features, 0 out of 6 RNA features; Figure 5A).
Figure 5.
Predictive features of yeast osmostress-responsive genes are conserved in humans. (A) Heatmap summarizing the results for individual Wilcoxon tests assessing differences between osmostress-responsive and unresponsive genes in terms of a variety of gene features. Organism/cell line of origin is shown at the top and upregulated and downregulated gene features in columns, as in Figure 2A. (B) Density plot shows the distribution of two selected features (GC content of the CDS and mRNA half-life) for each group of genes in humans. X-axis represents the value of a given feature and Y-axis the kernel density estimate. Labels show the median value of a given feature in each group of genes. (C) ROC curve assessing the performance of a multinomial logistic regression classifier of human osmostress-responsive genes using yeast predictive features for the classification. AUC for each category is displayed in the graph legend. For this plot analysis, we used 70% of the genes as a training set and 30% as a validation set. (D) Dot plot showing the AIC of the multinomial logistic regression after removing each variable from the model, one at a time. (E) Radial plot showing the coefficient of the multinomial logistic regression for each scaled feature. Red indicates the comparison done using upregulated responsive genes and green using the downregulated genes.
We used these features to train a multinomial logistic regression to predict osmostress-responsive genes. Similar to the yeast model, the AUCs in the human model were of 0.78, 0.80 and 0.74 for unresponsive, upregulated and downregulated genes, respectively (Figure 5C), thereby indicating predictive power in humans with the minimal set of yeast stress features. Again, a random forest classifier showed a similar performance for all gene groups (unresponsive: 0.81, upregulated: 0.79, downregulated: 0.80) (Supplementary Figure S5B). We then applied the previously described AIC-based approach for both an MVM using the leave-one-out approach (Figure 5D) and a UVM for the multinomial logistic regression, as well as a random forest (Supplementary Figure S5C and D). For the MVM, the top 5 ranked variables by AIC were (in order of relevance): mRNA half-life, GC content CDS, broad conservation, GC content 5’UTR and 3’UTR length. In addition, GC content of the 3’UTR, protein disorder, degree of PPI and 5’UTR length were relevant for predicting osmostress-responsive genes in human (Figure 5D). On the other hand, the number of TFs, CDS length, codon pair bias and distance to median were irrelevant to the model. When comparing these findings to the UVM and the random forest, two features remained consistent among the top 5 of each model (Supplementary Figure S5C and D). These variables were GC content of the CDS and mRNA half-life.
As for yeast, we used the coefficients of the MVM to quantitatively assess the direction and magnitude of change for each of the features in up- and down-regulated genes. In upregulated osmostress-responsive genes, GC content of the CDS increased whereas conservation and half-life appeared to decrease in osmostress-responsive genes compared to unresponsive ones (Figure 5E). Thus, features that are predictive of osmostress-responsive genes in yeast are also predictive of it in humans although with a different rank-ordering of feature-importance.
Stress-responsive genes are dysregulated in cancer
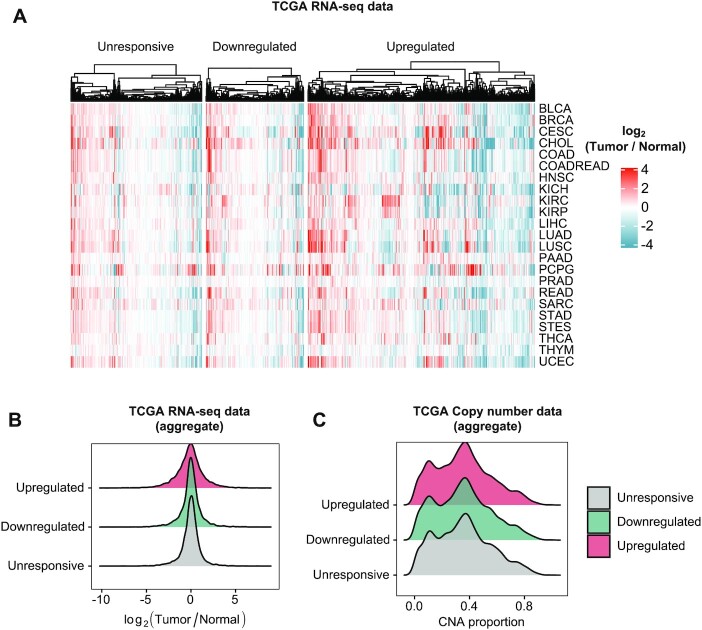
Given the broad physiological relevance of the stress response, we next considered its role in human pathologies. We focused on cancer, a clinically relevant scenario where cells are challenged by dramatic microenvironmental fluctuations that might cause chronic stress (65). To this end, we studied the RNA abundance patterns of osmostress-responsive genes that we defined in HeLa across matched tumor-normal pairs (n = 1484) from 23 cancer types to control for confounders such as age and sex (data from TCGA). Upregulated osmostress-responsive genes were almost ubiquitously dysregulated in human cancers (Figure 6A and B). To examine whether the observed dysregulation of osmostress-upregulated gene expression is a function of genetic changes in cancer, we analyzed copy number alteration (CNA) data from the TCGA. However, we did not observe any changes in copy number that could explain the expression patterns of the osmostress-responsive genes (Figure 6C and S6A). These results suggest that the dysregulation we observed in gene expression results from the adaptive processes that occur during tumorigenesis, rather than a function of changes in cancer driver gene selection. Since we defined osmostress-responsive genes using a tumoral cell line, the TCGA enrichment could be biased due to tumor-derived stresses (e.g. hypoxia, nutrient deprivation). To assess whether the observed dysregulation is cancerous cell independent and driven also by osmostress-specific genes (e.g. not only general stress response factors), we generated a set of osmostress specific (i.e. do not overlap with oxidative, hypoxia or ER stress upregulated genes) and non-HeLa specific (i.e. overlap with T47D or HEK293 upregulated genes) genes and observed that they displayed similar dysregulation levels (Supplementary Figure S6B and C).
Figure 6.
Stress-responsive genes are dysregulated in cancer. (A) Heatmap shows the log2(fold change) of the comparison between tumor and normal RNA-seq data of TCGA patients (only tumor-normal matched samples are included, n = 1484). Human stress-responsive genes from Figure 5 are shown as columns of the heatmap. TCGA tumor types included in the analysis are displayed as rows of the plot. Tumor type abbreviations can be found at https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations. Red and blue tones indicate positive and negative fold changes (genes up- and down-regulated in tumoral samples), respectively. (B) Density plot shows the distribution of log2(tumor/normal) expression values from TCGA RNA-seq data for each group of genes. All tumor types from panel (A) are aggregated for this plot. (C) Density plot shows the distribution of the copy number alteration (CNA) proportions from TCGA for each group of genes. All tumor types from panel (A) are aggregated for this plot.
DISCUSSION
In this study, we built a comprehensive characterization of the osmoresponsive transcriptome. To this end, we generated a catalog of osmostress-responsive genes in S. cerevisiae that include both the coding and non-coding transcriptome. Using this approach, we identified the specific features of these genes using computational modeling approaches. This characterization revealed a minimal set of features that can be used to predict osmostress-responsive genes in yeast and humans. In yeast, the most important features prioritized by all considered models are broad conservation and the number of TFs regulating a gene. Conversely, in humans, the consistently prioritized features are GC content of the CDS and mRNA half-life.
Certain features we identified as important for stress responsiveness have already been linked to stress in previous studies. For instance, (12) used 3 yeast species from the Saccharomycotina subphylum (400 million years of evolution) to show that non-conserved and duplicated genes (i.e. young genes) are more likely to be upregulated upon stress. In agreement, our analysis of broad conservation, which used 100 species from yeast to humans (1 billion years of evolution), reflects the same trend, namely that upregulated genes were less conserved than the rest of the genes and consistently ranks broad conservation among the top 5 features of stress-responsive genes. Alternatively, dN/dS analysis within S. cerevisiae and three closely related species (S. paradoxus, S. bayanis and S. mikatae) revealed the conservation of both, upregulated and downregulated osmostress-responsive genes. This could be because osmostress-responsive genes have coevolved under similar circumstances for closely related species than in more distant species, since environmental stresses are not a continuous selective pressure through evolution. Another example of a feature linked to osmostress-responsive genes is mRNA half-life. In this regard, (13) reported enrichment of carbohydrate catabolism genes among mRNAs with longer half-lives. This observation is in agreement with our findings of upregulated (carbohydrate catabolism) genes displaying higher mRNA half-lives. Furthermore, it is known that highly expressed genes, such as ribosomal proteins, are optimized for translation and thus display codon usage bias as well as some unique protein domains (66). Thus, it is unsurprising that we observed this optimization with codon usage bias measures such as codon adaptation index and Nc for downregulated stress genes, as these genes are enriched in ribosomal proteins. Thus, the accurate identification of several features known to be associated with stress genes demonstrates the validity of our approach for correctly recovering known features of osmostress-responsive genes.
In addition to known transcriptome-wide associations, we further identified features that were not previously associated with stress-responsive genes. Among these, we found a strong positive association between the number of TFs targeting a gene and the probability of this gene being osmostress-responsive. A higher number of interacting TFs may indicate higher reprogrammability, an essential trait for stress-responsive genes. This finding thus highlights the number of TFs as a prevalent gene expression regulator in yeast. To explore these findings, an interesting starting point is our observation that Hog1-upregulated genes have more (1.6 times) TFs than Hog1-independent ones. Due to the lack of features that can be associated and available for ncRNAs, we could not apply the model to identify ncRNA-features. Nonetheless, we could identify Sko1 as key regulator for ncRNA expression suggesting a major role for his TF, a well known Hog1-target, for global transcriptional reprogramming (67).
Hence, to further elucidate the role of the number of TFs in gene expression plasticity, a future research direction may be to search for the TFs most predictive of Hog1 dependence by modeling them individually or in combinations, possibly starting with the motifs revealed by our analysis. Newman and collaborators proposed that stress-upregulated proteins are noisier than proteins involved in protein biosynthesis (stress-downregulated) (68). Here, using yeast single-cell RNA-seq data (36), we showed that both up- and down-regulated stress genes present higher distance-to-median (DM) values, a mean independent measure of gene expression noise. This finding is consistent with an observation from the same study, in which the authors reported that genes with a higher dynamic range (i.e. those that are differentially expressed between different conditions) are more variable. In fact, our observation of osmostress-responsive genes being noisier suggests a link between gene expression responsiveness and gene expression noise in yeast, a finding recently reported in Drosophila (69). In addition, the number of TFs not only determines dynamic gene expression, as shown here, but also strongly determines gene expression noise (70). All together, these observations provide additional evidence of the role of TF networks as main determinants of gene expression variability across and within individuals.
Here, we addressed whether stress-responsive genes are conserved from yeast to humans and whether these genes are similar between these species. Since yeast and humans diverged ∼1 billion years ago (71) and were exposed to completely different environmental pressures, it is not surprising that we found poor conservation of osmostress-responsive genes from yeast to humans. In contrast, the ability to predict human osmostress-responsive genes using yeast-based features, despite the enormous evolutionary distance, suggests conservation of regulatory mechanisms of the osmostress response. For instance, broad conservation and mRNA half-life are among the top-ranked stress features in both species, although with opposite behaviours in the direction of change. Given the high degree of sequence and functional conservation of Hog1 (homolog of human p38) and downstream targets, it is plausible that DNA features are more likely to be conserved than RNA features for osmostress-responsive genes due to transcription being the first line of response to changes in proteome composition. While the results presented here support this notion, assessing such functional conservation and its extent is beyond the scope of this study.
The ranking of the features and their direction of change comparing up- versus downregulated stress-responsive genes are not completely conserved between yeast and humans. For instance, among the top 5 features in humans, GC content (especially GC content of the CDS) and mRNA half-life are the most predictive. The observed higher GC content of human upregulated-stress genes may be explained by recent discoveries. Previous studies indicate that GC-rich mRNAs are more efficiently translated and predominantly regulated at the mRNA decay level by the helicase DDX6 and the 5’-3’ exonuclease XRN1, whereas GC-poor genes are less efficiently translated, sequestered into P bodies and more controlled at the translational level (72). Moreover, GC content correlates with RNA structure, and it is known that, in addition to their roles in RNA decapping and degradation, enzymes such as the yeast homolog of DDX6 helicase, Dhh1 (73), and the exonuclease Xrn1 (74) regulate the translation of highly structured RNAs. During adaptation, upregulated stress genes need to be efficiently translated to ensure survival. Hence, a GC-rich CDS may be beneficial for selective translation of these genes, whose regulation may be mainly orchestrated by RNA decay. Further studies on the involvement of the individual contributing factors may shed light on the mechanisms regulating RNA translation and decay upon stress. We also observed an increase of GC in osmostress-responsive yeast genes, although this feature is not in the top ranking in yeast. This observation may indicate the conservation of RNA decay and translation control mechanisms between yeast and humans. Concordant to this hypothesis, in yeast, we observed highly-structured downregulated and less structured upregulated CDS (PARS score analysis). It could be plausible that a more structured RNA around the translation initiation site (TIS) for downregulated genes favored a tight translation control of these genes (41), whereas upregulated genes displayed a lower mRNA structure to facilitate translation initiation. However, research on this direction is needed to shed light on this possibility. Accordingly, key regulators such as DDX6 helicase (Dhh1 in yeast) are highly conserved. Moreover, we did not observe any relationship between gene expression variability and stress-responsive genes in humans. This finding is consistent with the reported lack of correlation between gene expression responsiveness and gene expression noise in mouse cells (75). Another interesting observation is that osmostress-downregulated genes displayed contrasting patterns in yeast and humans, with yeast downregulated genes showing a higher degree of PPI compared to unresponsive genes, whereas human genes show a lower PPI degree. Conversely, osmostress-upregulated genes in yeast and humans have a lower degree in PPI networks. Osmostress-upregulated genes are involved in stress-protective functions and could be expected to operate in exclusive functional modules with a few specific interactors. Alternatively, PPI networks are usually defined in basal conditions and thus PPI may be underrepresented for osmostress-upregulated genes. Further dedicated research may shed light on these aspects of the osmostress-response in yeast and humans.
In summary, we present a comprehensive set of yeast osmostress-responsive genes. Using this information, we defined the most distinctive features of osmostress-responsive genes, namely a higher number of TFs targeting their promoter region and their degree of conservation, in 100 species from yeast to humans. Remarkably, in humans, upregulated osmostress-responsive genes display a higher GC content of the CDS, which may implicate the involvement of the RNA decay machinery in their regulation. Strikingly, while comparison of yeast and human cells indicates that osmostress-responsive genes are not conserved, distinctive features defined in stress-dependent genes in yeast can be used to predict osmostress-responsive genes in humans, thus indicating that their regulatory mechanisms could be conserved. Our work provides a comprehensive resource of information that will be helpful for future studies of stress-responsive genes.
DATA AVAILABILITY
OsmoAtlas
We collected three published RNA-seq experiments and provided 2 additional experiments to generate an updated catalog of osmostress-responsive genes, including Hog1 dependency. We provide this catalog in an accessible and ready-to-use format to establish a reference for researchers in the field (Supplementary Table 1; OsmoAtlas GitHub page).
Gene features catalog
There is a lack of a unified resource or a compendium of gene-level features for yeast research. Here we computed, for the first time, 9 features and gathered the other 21 from different independent sources to build the largest catalog of yeast gene features. Similarly to the osmostress-responsive gene catalog, we provide this catalog of 30 gene features in a convenient format to reduce the time spent by researchers in collecting, preprocessing and homogenizing public data (Supplementary table 3; OsmoAtlas GitHub page).
Data availability
The computer code generated for this study is available in the GitHub repository (https://github.com/CellSignaling/OsmoAtlas_StressFeatures_2021).
ACCESSION NUMBERS
The raw sequencing data generated for this study is accessible under GEO (https://www.ncbi.nlm.nih.gov/geo/) with GSE171427 accession number.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the members of the P.C.B., F.P. and E.d.N. groups for critical feedback and insightful discussion. We thank E. Juárez-Escoto for proofreading the manuscript and contributing to the design of the figures. We thank the Biostatics / Bioinformatics Core Facility at IRB Barcelona for providing support on statistics and data analysis. The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author contributions: P.L., R.B., M.N.-R., F.P. and E.d.N. conceived the project. P.L., R.B. and C.H.L performed the computational analyses. M.N.R, G.M.-C. and C.S. designed and conducted the experiments. P.L., R.B., M.N.-R., P.C.B., F.P. and E.d.N. participated in the experimental design and data analysis. R.B., P.C.B., F.P. and E.d.N., supervised the work. P.L., R.B., M.N.-R., F.P. and E.d.N. wrote the manuscript with input from the other authors.
Contributor Information
Pablo Latorre, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain; Department of Experimental and Health Sciences, Universitat Pompeu Fabra (UPF), Barcelona, Spain.
René Böttcher, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain; Department of Experimental and Health Sciences, Universitat Pompeu Fabra (UPF), Barcelona, Spain.
Mariona Nadal-Ribelles, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain; Department of Experimental and Health Sciences, Universitat Pompeu Fabra (UPF), Barcelona, Spain.
Constance H Li, Departments of Human Genetics and Urology, Jonsson Comprehensive Cancer Center and Institute for Precision Health, University of California, Los Angeles, Los Angeles, CA 90095, USA; Department of Medical Biophysics, University of Toronto, Toronto, Canada.
Carme Solé, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain; Department of Experimental and Health Sciences, Universitat Pompeu Fabra (UPF), Barcelona, Spain.
Gerard Martínez-Cebrián, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain; Department of Experimental and Health Sciences, Universitat Pompeu Fabra (UPF), Barcelona, Spain.
Paul C Boutros, Departments of Human Genetics and Urology, Jonsson Comprehensive Cancer Center and Institute for Precision Health, University of California, Los Angeles, Los Angeles, CA 90095, USA; Department of Medical Biophysics, University of Toronto, Toronto, Canada.
Francesc Posas, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain; Department of Experimental and Health Sciences, Universitat Pompeu Fabra (UPF), Barcelona, Spain.
Eulàlia de Nadal, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain; Department of Experimental and Health Sciences, Universitat Pompeu Fabra (UPF), Barcelona, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
FI predoctoral fellowship and travel grant from Boehringer Ingelheim Fonds (to P.L.); Maria de Maeztu Postdoctoral Fellowship; La Caixa Junior Leader Fellow [LCF/BQ/PR20/11770001 to M.N.-R.]; Juan de la Cierva post-doctoral fellowship (to R.B.); FPI predoctoral fellowship (to G.M.-C.); National Institutes of Health [P30CA016042 to P.C.B.]; National Cancer Institute [1U01CA214194-01, 1U24CA248265-01 to P.C.B.]; Spanish Ministry of Economy and Competitiveness [BFU2017-85152-P to E.d.N., PGC2018-094136-B-I00 and FEDER to F.P.]; Catalan Government [2017 SGR 799 to E.N. and F.P.]; Unidad de Excelencia Maria de Maeztu [MDM-2014-0370 to the UPF]; F.P. and E.d.N. are recipients of an ICREA Acadèmia (Catalan Government); Spanish Ministry of Economy, Industry and Competitiveness (MINECO) through the Centres of Excellence Severo Ochoa award; CERCA Programme of the Catalan Government.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kultz D. Evolution of cellular stress response mechanisms. J. Exp. Zool. A Ecol. Integr. Physiol. 2020; 333:359–378. [DOI] [PubMed] [Google Scholar]
- 2. Saito H., Posas F.. Response to hyperosmotic stress. Genetics. 2012; 192:289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Nadal E., Posas F.. Osmostress-induced gene expression–a model to understand how stress-activated protein kinases (SAPKs) regulate transcription. FEBS J. 2015; 282:3275–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Proft M., Mas G., de Nadal E., Vendrell A., Noriega N., Struhl K., Posas F.. The stress-activated hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol. Cell. 2006; 23:241–250. [DOI] [PubMed] [Google Scholar]
- 5. Silva A., Cavero S., Begley V., Sole C., Bottcher R., Chavez S., Posas F., de N.E.. Regulation of transcription elongation in response to osmostress. PLoS. Genet. 2017; 13:e1007090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O.. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000; 11:4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gasch A.P. The environmental stress response: a common yeast response to diverse environmental stresses. Yeast Stress Responses. 2003; Springer; 11–70. [Google Scholar]
- 8. Posas F., Chambers J.R., Heyman J.A., Hoeffler J.P., de Nadal E., Arino J.. The transcriptional response of yeast to saline stress. J Biol. Chem. 2000; 275:17249–17255. [DOI] [PubMed] [Google Scholar]
- 9. Nadal-Ribelles M., Sole C., Xu Z., Steinmetz L.M., de Nadal E., Posas F.. Control of cdc28 CDK1 by a stress-induced lncRNA. Mol. Cell. 2014; 53:549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Studer R.A., Rodriguez-Mias R.A., Haas K.M., Hsu J.I., Vieitez C., Sole C., Swaney D.L., Stanford L.B., Liachko I., Bottcher R.et al.. Evolution of protein phosphorylation across 18 fungal species. Science. 2016; 354:229–232. [DOI] [PubMed] [Google Scholar]
- 11. Vieitez C., Martinez-Cebrian G., Sole C., Bottcher R., Potel C.M., Savitski M.M., Onnebo S., Fabregat M., Shilatifard A., Posas F.et al.. A genetic analysis reveals novel histone residues required for transcriptional reprogramming upon stress. Nucleic Acids Res. 2020; 48:3455–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doughty T.W., Domenzain I., Millan-Oropeza A., Montini N., de Groot P.A., Pereira R., Nielsen J., Henry C., Daran J.G., Siewers V.et al.. Stress-induced expression is enriched for evolutionarily young genes in diverse budding yeasts. Nat. Commun. 2020; 11:2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan L.Y., Mugler C.F., Heinrich S., Vallotton P., Weis K.. Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability. Elife. 2018; 7:e32536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin Z., Li W.H.. Evolution of 5' untranslated region length and gene expression reprogramming in yeasts. Mol. Biol. Evol. 2012; 29:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poljsak B., Milisav I.. Clinical implications of cellular stress responses. Bosn. J. Basic Med. Sci. 2012; 12:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 18. Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M.. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009; 457:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nadal-Ribelles M., Conde N., Flores O., Gonzalez-Vallinas J., Eyras E., Orozco M., de Nadal E., Posas F.. Hog1 bypasses stress-mediated down-regulation of transcription by RNA polymerase II redistribution and chromatin remodeling. Genome Biol. 2012; 13:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carbonell C., Ulsamer A., Vivori C., Papasaikas P., Bottcher R., Joaquin M., Minana B., Tejedor J.R., de N.E., Valcarcel J.et al.. Functional network analysis reveals the relevance of SKIIP in the regulation of alternative splicing by p38 SAPK. Cell Rep. 2019; 27:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amat R., Bottcher R., Le D.F., Vidal E., Quilez J., Cuartero Y., Beato M., de N.E., Posas F.. Rapid reversible changes in compartments and local chromatin organization revealed by hyperosmotic shock. Genome Res. 2019; 29:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosa-Mercado N.A., Zimmer J.T., Apostolidi M., Rinehart J., Simon M.D., Steitz J.A.. Hyperosmotic stress alters the RNA polymerase II interactome and induces readthrough transcription despite widespread transcriptional repression. Mol. Cell. 2021; 81:502–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rendleman J., Cheng Z., Maity S., Kastelic N., Munschauer M., Allgoewer K., Teo G., Zhang Y.B.M., Lei A., Parker B.et al.. New insights into the cellular temporal response to proteostatic stress. Elife. 2018; 7:e39054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frost J., Ciulli A., Rocha S.. RNA-seq analysis of PHD and VHL inhibitors reveals differences and similarities to the hypoxia response. Wellcome. Open. Res. 2019; 4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ritchie M.E., Phipson B., Wu D.I., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J.. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019; 47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Merico D., Isserlin R., Stueker O., Emili A., Bader G.D.. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010; 5:e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M.. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008; 320:1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koch E.N., Costanzo M., Bellay J., Deshpande R., Chatfield-Reed K., Chua G., D’Urso G., Andrews B.J., Boone C., Myers C.L.. Conserved rules govern genetic interaction degree across species. Genome Biol. 2012; 13:R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ostlund G., Schmitt T., Forslund K., Kastler T., Messina D.N., Roopra S., Frings O., Sonnhammer E.L.. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010; 38:D196–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coleman J.R., Papamichail D., Skiena S., Futcher B., Wimmer E., Mueller S.. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008; 320:1784–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Charif D., Lobry J.R.. SeqinR 1.0-2: a contributed package to the r project for statistical computing devoted to biological sequences retrieval and analysis. Structural Approaches to Sequence Evolution. 2007; Springer; 207–232. [Google Scholar]
- 36. Nadal-Ribelles M., Islam S., Wei W., Latorre P., Nguyen M., de Nadal E., Posas F., Steinmetz L.M.. Sensitive high-throughput single-cell RNA-seq reveals within-clonal transcript correlations in yeast populations. Nat. Microbiol. 2019; 4:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lun A.T., McCarthy D.J., Marioni J.C.. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with bioconductor. F1000Res. 2016; 5:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Monteiro P.T., Oliveira J., Pais P., Antunes M., Palma M., Cavalheiro M., Galocha M., Godinho C.P., Martins L.C., Bourbon N.et al.. YEASTRACT+: a portal for cross-species comparative genomics of transcription regulation in yeasts. Nucleic. Acids. Res. 2020; 48:D642–D649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-Regulatory elements required for macrophage and b cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bailey T.L., Johnson J., Grant C.E., Noble W.S.. The MEME suite. Nucleic Acids Res. 2015; 43:W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kertesz M., Wan Y., Mazor E., Rinn J.L., Nutter R.C., Chang H.Y., Segal E.. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010; 467:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nadal-Ribelles M., Islam S., Wei W., Latorre P.N.M., de Nadal E., Posas F., Steinmetz L.M.. Yeast Single-cell RNA-seq, cell by cell and step by step. Bio-protocol. 2019; 9:e3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hafemeister C., Satija R.. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019; 20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ward J.J., McGuffin L.J., Bryson K., Buxton B.F., Jones D.T.. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004; 20:2138–2139. [DOI] [PubMed] [Google Scholar]
- 45. Alberti S., Halfmann R., King O., Kapila A., Lindquist S.. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009; 137:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lancaster A.K., Nutter-Upham A., Lindquist S., King O.D.. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014; 30:2501–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oughtred R., Rust J., Chang C., Breitkreutz B., Stark C., Willems A., Boucher L., Leung G., Kolas N., Zhang F.. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021; 30:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sharp P.M., Tuohy T.M., Mosurski K.R.. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986; 14:5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Durinck S., Spellman P.T., Birney E., Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009; 4:1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rodriguez J.M., Maietta P., Ezkurdia I., Pietrelli A., Wesselink J.J., Lopez G., Valencia A., Tress M.L.. APPRIS: annotation of principal and alternative splice isoforms. Nucleic Acids Res. 2013; 41:D110–D117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu W., Zhang X., Guo Q.f., Yang J.w., Yang Y., Wei S.c., Su X.d.. HeLa-CCL2 cell heterogeneity studied by single-cell DNA and RNA sequencing. PLoS One. 2019; 14:e0225466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tani H., Mizutani R., Salam K.A., Tano K., Ijiri K., Wakamatsu A., Isogai T., Suzuki Y., Akimitsu N.. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012; 22:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rouillard A.D., Gundersen G.W., Fernandez N.F., Wang Z., Monteiro C.D., McDermott M.G., Ma’ayan A.. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. (Oxford). 2016; 2016:baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oates M.E., Romero P., Ishida T., Ghalwash M., Mizianty M.J., Xue B., Dosztanyi Z., Uversky V.N., Obradovic Z., Kurgan L.et al.. D(2)P(2): database of disordered protein predictions. Nucleic Acids Res. 2013; 41:D508–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sing T., Sander O., Beerenwinkel N., Lengauer T.. ROCR: visualizing classifier performance in R. Bioinformatics. 2005; 21:3940–3941. [DOI] [PubMed] [Google Scholar]
- 56. Venables W.N., Ripley B.D.. Exploratory multivariate analysis. Modern Applied Statistics with S. 2002; Springer; 301–330. [Google Scholar]
- 57. Liaw A., Wiener M.. Classification and regression by randomForest. R news. 2002; 2:18–22. [Google Scholar]
- 58. Gu Z., Eils R., Schlesner M.. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; 32:2847–2849. [DOI] [PubMed] [Google Scholar]
- 59. Prick T., Thumm M., Köhrer K., Häussinger D., Vom Dahl S.. In yeast, loss of hog1 leads to osmosensitivity of autophagy. Biochem. J. 2006; 394:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kabeche R., Howard L., Moseley J.B.. Eisosomes provide membrane reservoirs for rapid expansion of the yeast plasma membrane. J. Cell Sci. 2015; 128:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blum M., Chang H.Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A., Nuka G., Paysan-Lafosse T., Qureshi M., Raj S.et al.. The interpro protein families and domains database: 20 years on. Nucleic Acids Res. 2021; 49:D344–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. O’Rourke S.M., Herskowitz I.. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell. 2004; 15:532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Capaldi A.P., Kaplan T., Liu Y., Habib N., Regev A., Friedman N., O'Shea E.K. Structure and function of a transcriptional network activated by the MAPK hog1. Nat. Genet. 2008; 40:1300–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Peña-Oyarzun D., Troncoso R., Kretschmar C., Hernando C., Budini M., Morselli E., Lavandero S., Criollo A.. Hyperosmotic stress stimulates autophagy via polycystin-2. Oncotarget. 2017; 8:55984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bhandari V., Hoey C., Liu L.Y., Lalonde E., Ray J., Livingstone J., Lesurf R., Shiah Y.J., Vujcic T., Huang X.. Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet. 2019; 51:308–318. [DOI] [PubMed] [Google Scholar]
- 66. Velculescu V.E., Zhang L., Zhou W., Vogelstein J., Basrai M.A., Bassett D.E. Jr, Hieter P., Vogelstein B., Kinzler K.W. Characterization of the yeast transcriptome. Cell. 1997; 88:243–251. [DOI] [PubMed] [Google Scholar]
- 67. Proft M., Pascual-Ahuir A., de Nadal E., Arino J., Serrano R., Posas F.. Regulation of the sko1 transcriptional repressor by the hog1 MAP kinase in response to osmotic stress. EMBO J. 2001; 20:1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Newman J.R., Ghaemmaghami S., Ihmels J., Breslow D.K., Noble M., DeRisi J.L., Weissman J.S.. Single-cell proteomic analysis of s. cerevisiae reveals the architecture of biological noise. Nature. 2006; 441:840–846. [DOI] [PubMed] [Google Scholar]
- 69. Sigalova O.M., Shaeiri A., Forneris M., Furlong E.E., Zaugg J.B.. Predictive features of gene expression variation reveal mechanistic link with differential expression. Mol. Syst. Biol. 2020; 16:e9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Parab L., Pal S., Dhar R.. Transcription factor binding dynamics shape noise across biological processes. 2020; bioRxiv doi:28 July 2020, preprint: not peer reviewed 10.1101/2020.07.27.222596. [DOI]
- 71. Douzery E.J., Snell E.A., Bapteste E., Delsuc F.d., Philippe H.. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils?. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15386–15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Courel M., Clément Y., Bossevain C., Foretek D., Cruchez O.V., Yi Z., Bénard M., Benassy M.N., Kress M., Vindry C.. GC content shapes mRNA storage and decay in human cells. Elife. 2019; 8:e49708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jungfleisch J., Nedialkova D.D., Dotu I., Sloan K.E., Martinez-Bosch N., Brüning L., Raineri E., Navarro P., Bohnsack M.T., Leidel S.A.et al.. A novel translational control mechanism involving RNA structures within coding sequences. Genome Res. 2017; 27:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Blasco-Moreno B., de Campos-Mata L., Böttcher R., García-Martínez J., Jungfleisch J., Nedialkova D.D., Chattopadhyay S., Gas M.E., Oliva B., Pérez-Ortín J.E.. The exonuclease xrn1 activates transcription and translation of mRNAs encoding membrane proteins. Nat. Commun. 2019; 10:1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xiao L., Zhao Z., He F., Du Z.. Multivariable regulation of gene expression plasticity in metazoans. Open Biol. 2019; 9:190150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
OsmoAtlas
We collected three published RNA-seq experiments and provided 2 additional experiments to generate an updated catalog of osmostress-responsive genes, including Hog1 dependency. We provide this catalog in an accessible and ready-to-use format to establish a reference for researchers in the field (Supplementary Table 1; OsmoAtlas GitHub page).
Gene features catalog
There is a lack of a unified resource or a compendium of gene-level features for yeast research. Here we computed, for the first time, 9 features and gathered the other 21 from different independent sources to build the largest catalog of yeast gene features. Similarly to the osmostress-responsive gene catalog, we provide this catalog of 30 gene features in a convenient format to reduce the time spent by researchers in collecting, preprocessing and homogenizing public data (Supplementary table 3; OsmoAtlas GitHub page).
Data availability
The computer code generated for this study is available in the GitHub repository (https://github.com/CellSignaling/OsmoAtlas_StressFeatures_2021).
The computer code generated for this study is available in the GitHub repository (https://github.com/CellSignaling/OsmoAtlas_StressFeatures_2021).