Abstract
Here, we introduce carbocations (R3C+) as laser-initiated footprinting reagents for proteins. We screened seven candidates and selected trifluomethoxy benzyl bromide (TFBB) as an effective precursor for the electrophilic trifluomethoxy benzyl carbocation (TFB+) under laser (248 nm) irradiation on the fast photochemical oxidation of proteins (FPOP) platform. Initial results demonstrate that this electrophilic cation reagent affords residue coverage of nucleophilic amino acids including H, W, M and S. Further, the addition of TFB increases the hydrophobicity of the peptides so that separation of isomeric peptide products by reversed-phase LC is improved, suggesting opportunities for sub-residue footprinting. Comparison of apo- and holo-myoglobin footprints show that the TFB+ footprinting is sensitive to protein conformational change and solvent accessibility. Interestingly, because the TFB+ is amphiphilic, the reagent can potentially footprint membrane proteins as demonstrated for vitamin K epoxide reductase (VKOR) stabilized in a micelle. Not only does footprinting of the extra-membrane domain occur, but also some footprinting of the hydrophobic transmembrane domain is achieved owing to the interaction of TFB+ with the micelle. Carbocation precursors are stable and amenable for tailoring their properties and those of the incipient carbocation, enabling targeting their soluble or membrane-associated or embedded regions and distinguishing between the extra- and trans-membrane domains of membrane proteins.
Graphical Abstract
Protein conformational change and protein-small molecule interactions are critical for enabling biochemical and biomedical processes.1 Methods that can detect and map these processes are essential for understanding basic biology and in developing drugs that interact with proteins, especially membrane proteins.2 Mass spectrometry-based methods including hydrogen-deuterium exchange, cross-linking, footprinting of targeted amino acids,3,4 and fast footprinting with reactive species constitute four related approaches in structural proteomics that are sensitive and have relatively fast turnaround.5
Fast protein footprinting (achieving footprinting on the μs – ms time scale) is advantageous in structural proteomics because the reactive species it uses proteins so rapidly that conformational changes are not a complication.6 Fast photochemical oxidation of proteins (FPOP)6–8 and X-ray initiated free radical footprinting can follow protein fast conformational changes and identify protein/protein interfaces.9–14 A complementary strategy employing the FPOP platform is trifluoromethylation of proteins, affording also broad residue coverage.15 Other choices for fast protein footprinting are highly reactive carbene diradicals, which are also used in photoaffinity labeling.16 The commercially available photoleucine and 4,4-azipentanoic acid are example precursors for carbenes17,18 that enable footprinting of soluble proteins. Other carbene precursors are aryl diazirines,19 which afford transmembrane protein footprinting in a detergent micelle.
Recently, a free radical footprinter, perfluoroisopropyl iodide was developed that, together with tip sonication to assist partitioning into the membrane, provides 100% footprinting of Y and W for both the extramembrane domain and intramembrane domain of a membrane protein.20 Exploiting the advantage of tailoring the polarity of the precursor will enable new, highly reactive species and expand the repertoire of protein footprinters.
Carbocations are highly reactive intermediates with lifetimes as short as ~ns.21 Although their presence has been usually inferred over many years because their existence is often transitory in chemical reactions, they can be directly generated by laser flash photolysis of suitable neutral precursors, R-X (e.g., alkyl halides, acetates, aryl ethers) or charged precursors, R-X+ (e.g., ammonium R-NR3+ and phosphonium salts R-PR3+)22 They can be also directly observed in mass spectrometers where their lifetime is increased in vacuum where the concentration of nucleophiles is low.23 Carbocations are strong electrophiles that should react with the nucleophilic residues in proteins. There is only one reported footprinter based on a carbocation; that is, the Koshland’s reagent that labels W via an SN1 reaction.24 The quinoid resonance stabilizes the carbocation and leads to rapid labelling. The above development lays the foundation for carbocations as a new class of fast footprinters.
In this communication, we describe screening seven precursors that, upon laser irradiation, generate carbocations with a range of stability and reactivity and test their footprinting performance. From the seven candidates (see Figure S1 for detailed discussion), we selected 4-(trifluoromethoxy) benzyl bromide (TFBB) to generate the TFB+ carbocation (structure in Figure 1) in aqueous buffer solutions by using a pulsed laser suitable for fast protein footprinting. Specifically, we adopted the flow system in the FPOP platform to ensure ~ one laser shot to a solution “plug” passing through the transparent window of a silica capillary (Figure 1A). The production of the TFB+ intermediate can be efficiently accomplished by laser activation (248 nm). Its footprinting chemistry presumably occurs on a fast timescale because the lifetime of carbocations is short owing to competitive solvolysis (quenching of the carbocation by water, resulting in R-OH), ensuring capture of a protein’s native conformation. We also chose a benzyl bromide because the heterolysis of the C-Br is ready achieved.22
Figure 1.
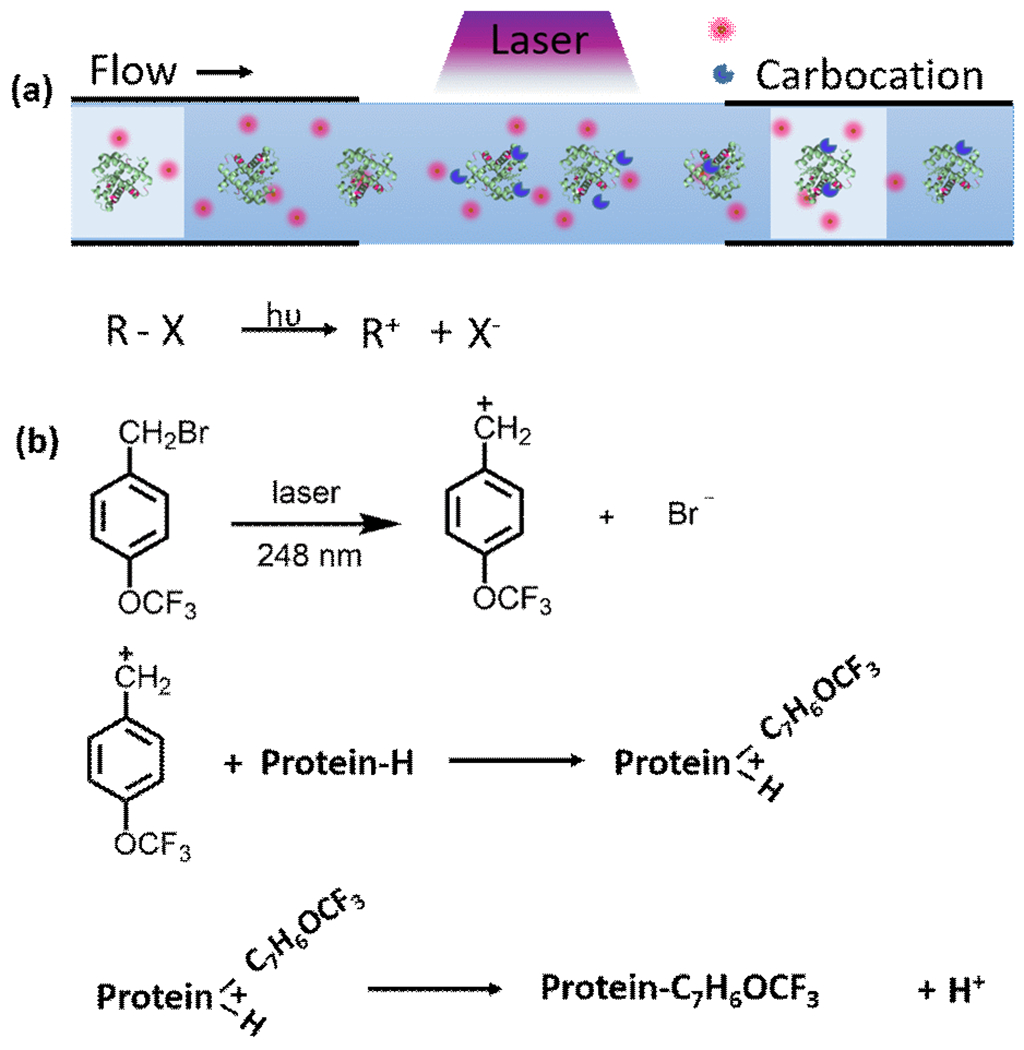
Approach for carbocation generation and footprinting. (a) Carbocation generation and footprinting in the FPOP flow system. (b) Proposed step-by-step mechanism of TFB+ footprinting. The TFB+ is generated by laser irradiation and then reacts by attack of the electron-rich sites in protein.
We showed that the TFB+ carbocation can footprint several nucleophilic residues in both a soluble protein and a membrane protein in a micelle. The generation and reactivity of a carbocation is determined by both the propensity of leaving group X and the stabilization of the carbocation R (Figure 1B). The spectroscopic properties of the precursor (UV absorbance and quantum yield to form ion pairs) must also play a role. As reflected by the UV-Vis absorbance spectrum (Figure S2), TFBB has high absorptivity at 248 nm. Although the trifluoromethoxy substituent is no more stabilizing of a positive charge (σ+ = 0.067) than a H substituent (σ+ = 0), the system presumably has an appropriate balance of properties to produce carbocations that modify proteins before they are “neutralized” by reacting with solvent.
TFBB can give a carbocation that specifically footprints proteins, affording a mass shift of 174.0365 (Figure S1 G). For the control (no laser irradiation), low TFB modification extents (< 10%, Figure S3) were found, presumably occurring by an SN2 reaction. Upon laser irradiation, the footprinting occurs as SN1 of increased efficiency (Figure 2) compared with no-laser applied group (Figure S3). Based on this, we propose a likely pathway for TFBB protein footprinting (Figure 1 b). That pathway involves a laser-assisted SN1 reaction to give the cation followed by its addition to nucleophilic sites of the protein. The laser-induced generation of the TFB+ carbocation is viewed as the trigger that accelerates protein footprinting.
Figure 2.
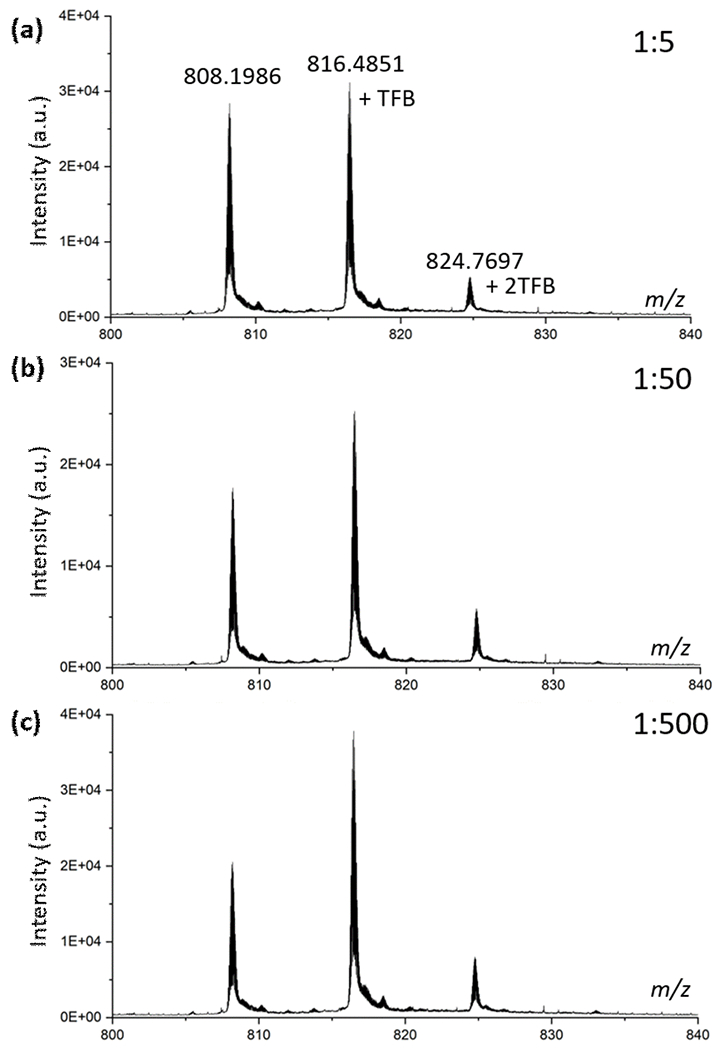
Mass spectra of TFBB footprinted myoglobin. (a)-(c) are results with different ratios of MB-to-TFBB.
Soluble Protein Footprinting.
Taking advantage of the unique mass shift of 174.0365, readily detectable by MS/MS, we located the TFB substitution on several residues in myoglobin (MB) including H, M, W and S (Figure S4 shows tryptic peptide coverage of MB, and Figures S5–18 show examples of EIC and MS/MS of unlabeled and labeled peptides). Of note, with a single TFB addition on H64 (Figure 3a), there is a significant shift to longer retention times in the reversed-phase HPLC peptide separation (Figure 3b), owing to the increased hydrophobicity introduced by addition of the carbocation. This sharply contrasts with the outcomes of footprinting by hydrophilic reagents (e.g., •OH).
Figure 3.
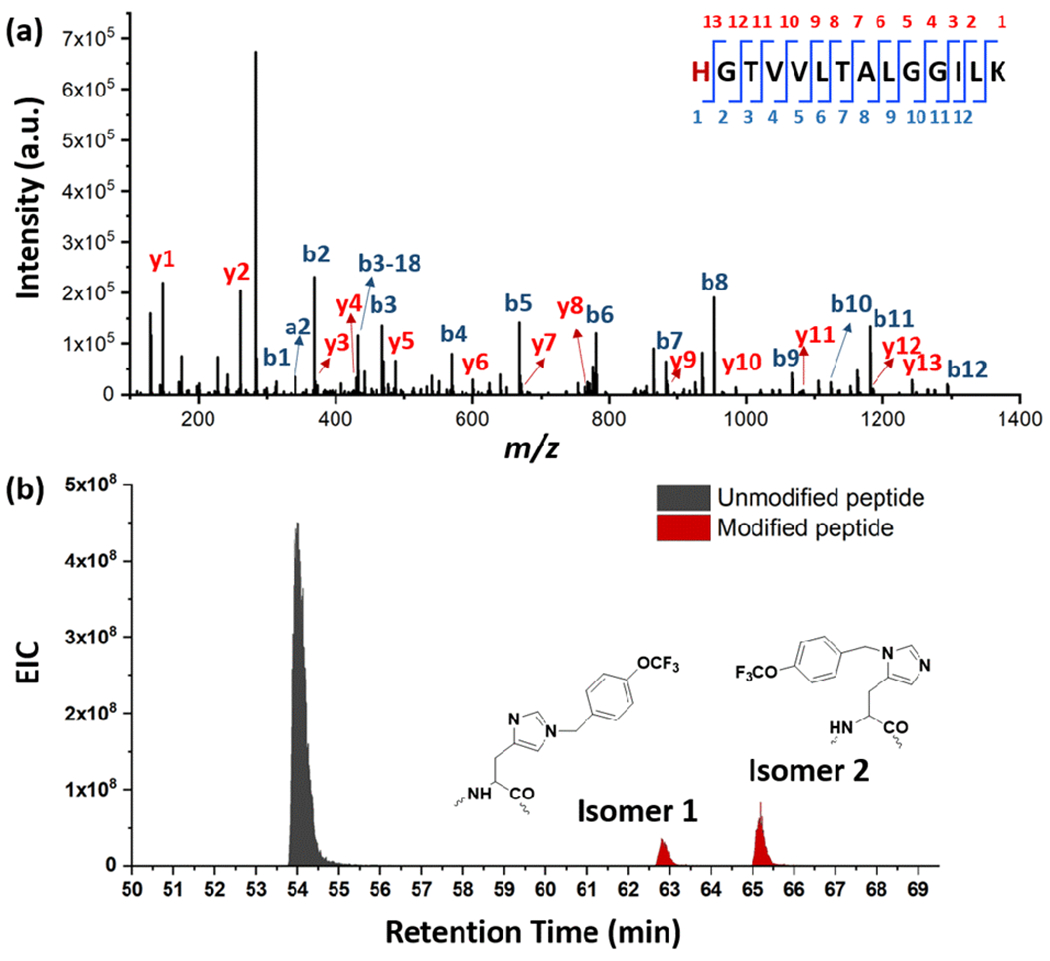
Footprinting of MB. (a) Product-ion (MS/MS) spectrum of the TFB-modified peptide 64-77 taken at a time determined from an extracted-ion chromatogram, EIC. The modification is on the N-terminal His, shown in red. (b) EIC of TFB-modified and unmodified peptide (64-77: HGTVVLTALGGILK) from MB.
The above feature of the TFBB reagent to increase hydrophobicity is general (Figure 3b) and adds considerably to the separability of isomeric peptides. For example, TFB-His64 gave two isomeric products owing to reactions presumably on the δN and εN of His64 (Figure S19 for mechanism); the two isomers are separated by HPLC without overlap and both are confirmed as His modification by MS/MS. This outcome shows the possibility of even sub-residue resolution in footprinting of proteins.
We then investigated whether the carbocation footprinting with TFB+ provides a meaningful differential measure of protein conformational change. We used as a test apo and holo-myoglobins (aMB/hMB) because they are simple proteins and structurally distinct. Furthermore, they have been commonly used in the development of other footprinting methods. MB usually undergoes broad modifications across the whole structure (Figure 4a). Comparisons of the footprinting yields for both apo and holo show that similar extents of TFB-modification occur for most regions of the MB as represented by the proteolytic peptides.
Figure 4.
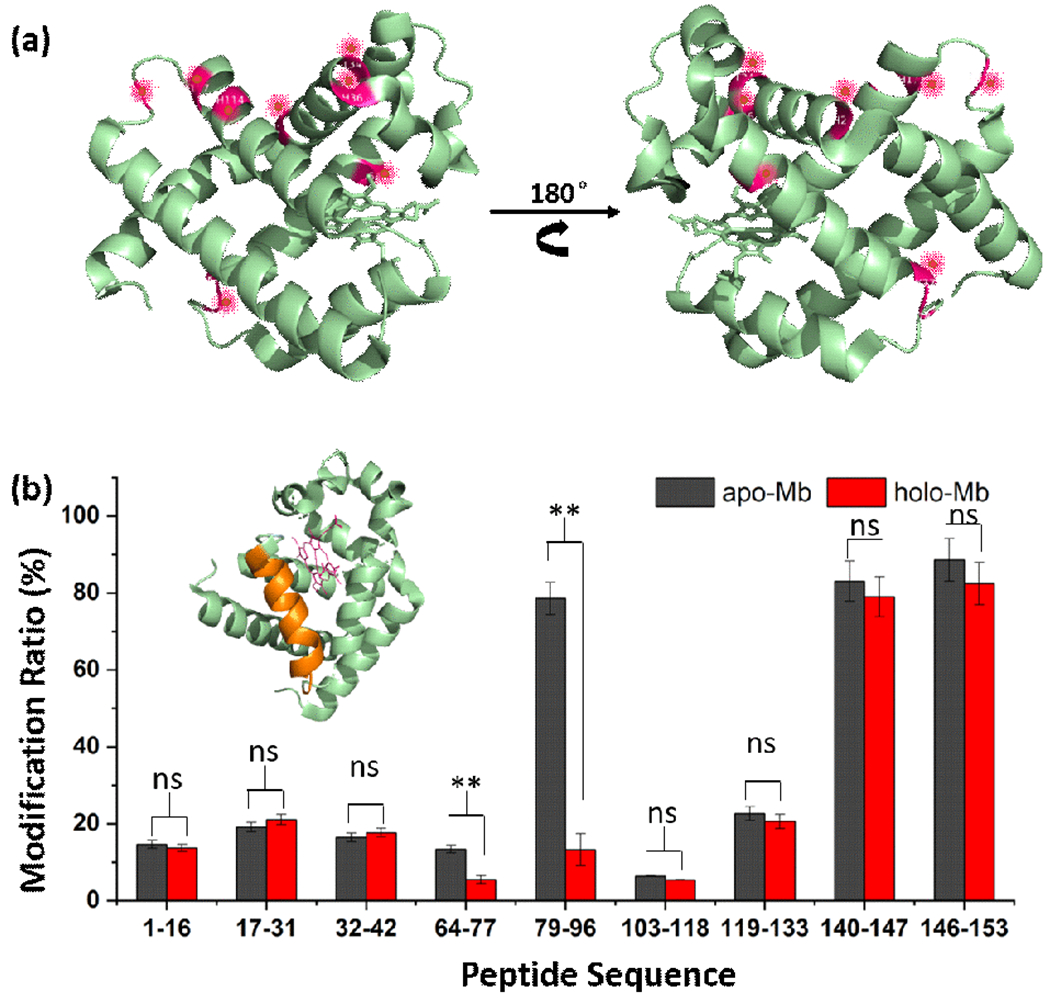
(a) Modification of holoMB (PDB ID: 1WLA). Red spots represent the modified residues. (b) Comparison of modification extents of constituent peptides from holo and apoMB. Significant differences in modification occur in the region represented by peptides 79-96 (orange color) where apoMB is not structured but holoMB contains a helix.
Nevertheless, there are significantly different modification extents for the region 79-96, which is disordered for the apoMB but is a helix for the holoMB (Figure 4b). The results are consistent with top-down HDX and NMR.25,26 MS/MS identified H93 to be the modified residue, which is involved in bonding the iron atom directly. The comparison of TFB-H93 from apo- and holoMB allows accurate determination of the binding site location. Furthermore, the modification of H64 between the two conformations of apo and holo is different. H64 is reported to bind to oxygen and may also play a role in the conformational change.
Membrane Protein Footprinting.
Membrane proteins are challenging for footprinting because their transmembrane regions are not solvent accessible. Thus, our ability to footprint membrane protein structure and interactions significantly lags that of their soluble counterparts. In a few studies, investigators have applied oxidative methodology or other reactive species such as trifluoromethyl radical and carbenes to integral membrane proteins.15,20,27,28 These studies report that significant labelling of amino acid side chains in the extramembrane or solvent-accessible regions of the proteins occurred. Recently, the use of aryldiazirine photoactivation to yield a carbene was reported to footprint the hydrophobic transmembrane region on an outer membrane porin protein F.19
Given the amphipathic nature of the TFB+ carbocation as governed by the hydrophobic CF3 group balanced by the positive charge, we postulated that the TFB+ may incorporate with detergent molecules and footprint the hydrophobic transmembrane domain in addition to the extra-cellular hydrophilic domain. We tested TFBB reagent on vitamin K epoxide reductase (VKOR) at pH 7.4 (PBS buffer with 0.03% DDM detergent to solubilize the protein). Most of the footprinting occurred on the extra-membrane region, either cytosolic or extra-cytosolic (Figure 5). TFB-modified peptides were also observed in the transmembrane region, even though the labeling efficiency was relatively low compared to the solvent-accessible domains. The MS/MS analysis revealed the locations of footprinting are mainly on M, S and W (for EIC and MS/MS, see Figure S20–29). This result indicates that TFB+ (or its precursor) interacts to some extent with the transmembrane domain, which is protected by the detergent. By increasing the hydrophobicity and reactivity of carbocation, higher modification ratio in transmembrane domain may be obtained in the future.
Figure 5.
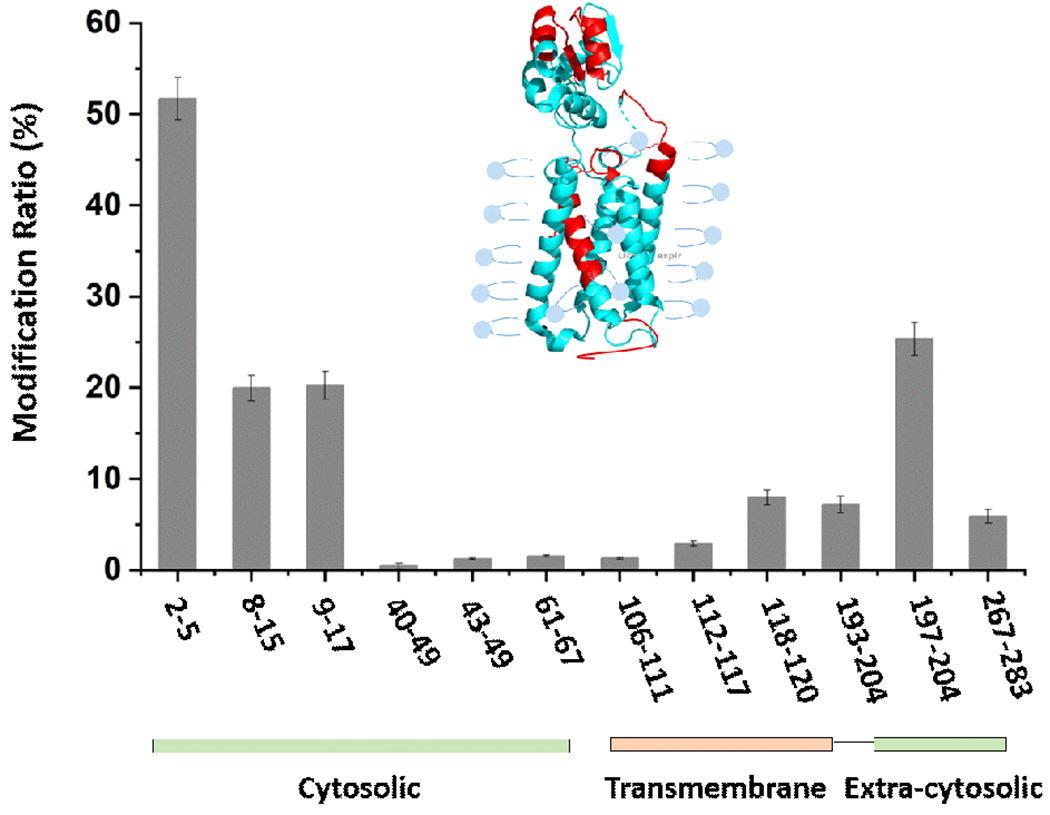
TFBB modification of a membrane protein VKOR in the micelle (PDB ID: 3KP9). The red color represents the modified peptides. The bar graph represents modification ratios on regional level as indicated by peptides designated on the x axis.
To provide evidence that the addition of TFBB in this footprinting condition is not detrimental to the native structure of VKOR, enzyme activity assays were performed (Figure S30). No significant change in enzyme activity was detected, indicating that the functional structure is not affected by TFBB.
In summary, we have shown that the TFB carbocation, derived from TFBB upon irradiation at 248 nm, is an efficient labeling reagent for footprinting both a soluble protein and a membrane protein solubilized in detergent. One pulse of UV laser irradiation for each plug of solution generates the carbocation. Because the modification chemistry is fast (the lifetime of the carbocation is short, and the laser frequency is controlled so that there is mainly one laser shot for every footprinting experiment), the protein conformation can be captured in its native state. Although we have no direct evidence that the laser produces only carbocations and no free radicals by homolysis, the nature of the product modifications on nucleophiles is consistent with predominant carbocation footprinting. Free radical processes would likely replace H with Br, as is observed in iodide radical footprinting,29 but this was not observed.
The increased hydrophobicity of the TFB-modified peptide increased the HPLC separation, demonstrating the future potential for sub-residue-resolution footprinting that could be complemented with ion mobility MS. More compelling, TFB carbocation and/or its precursor are amphiphilic, offering potential to footprint the transmembrane regions of integral membrane proteins. Currently, we are investigating methods to improve the footprinting efficiency for transmembrane domains by adjusting the functional group in the carbocation and looking more closely at mechanism.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health, Grant Nos. P41GM103422 and R24GM136766 to MLG, 1R01GM131008 to WL and MLG, and RO1DK099534 to WL. Authors are grateful to Protein Metrics for software.
Footnotes
Supporting Information
Experimental section; footprinting of seven carbocations on MB (Figure S1); UV-Vis absorbance spectrum (Figure S2); TFBB on MB without laser (Figure S3); sequence coverage of labeled MB (Figure S4); examples of EIC and MS/MS spectra from MB (Figure S5–S18); isomers of modified H (Figure S19); examples of EIC and MS/MS spectra from VKOR (Figure S20–S29); VKOR activity assay (Figure S30).
The Supporting Information is available free of charge on the ACS Publications website.
Conflict of Interest Disclosure:
MLG is an unpaid member of the scientific advisory boards of Protein Metrics and GenNext Technologies, two companies commercializing hardware and software for protein footprinting.
REFERENCES
- (1).Ha J-H; Loh SN Chemistry (Weinheim an der Bergstrasse, Germany) 2012, 18, 7984–7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Haffke M; Duckely M; Bergsdorf C; Jaakola V-P; Shrestha B Protein expression and purification 2020, 167, 105545. [DOI] [PubMed] [Google Scholar]
- (3).Zhao B; Zhuang J; Xu M; Liu T; Limpikirati P; Thayumanavan S; Vachet RW Analytical chemistry 2020, 92, 6637–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mendoza VL; Vachet RW Analytical chemistry 2008, 80, 2895–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Liu XR; Zhang MM; Gross ML Chemical reviews 2020, 120, 4355–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Liu XR; Rempel DL; Gross ML Nature protocols 2020, 15, 3942–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Rinas A; Mali VS; Espino JA; Jones LM Analytical chemistry 2016, 88, 10052–10058. [DOI] [PubMed] [Google Scholar]
- (8).Hambly DM; Gross ML Journal of the American Society for Mass Spectrometry 2005, 16, 2057–2063. [DOI] [PubMed] [Google Scholar]
- (9).Watson C; Sharp JS AAPS J 2012, 14, 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Minkoff BB; Blatz JM; Choudhury FA; Benjamin D; Shohet JL; Sussman MR Scientific reports 2017, 7, 12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Maleknia SD; Downard KM Chemical Society reviews 2014, 43, 3244–3258. [DOI] [PubMed] [Google Scholar]
- (12).Gupta S; Bavro VN; D’Mello R; Tucker SJ; Venien-Bryan C; Chance MR Structure 2010, 18, 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Xu G; Chance MR Chemical reviews 2007, 107, 3514–3543. [DOI] [PubMed] [Google Scholar]
- (14).Angel TE; Gupta S; Jastrzebska B; Palczewski K; Chance MR Proceedings of the National Academy of Sciences of the United States of America 2009, 106, 14367–14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ming Cheng; Bojie Zhang; Weidong Cui; Gross ML Angew. Chem. Int. Ed 2017, 56, 14007–14010. [Google Scholar]
- (16).Ge S-S; Chen B; Wu Y-Y; Long Q-S; Zhao Y-L; Wang P-Y; Yang S RSC Advances 2018, 8, 29428–29454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Jumper CC; Bomgarden R; Rogers J; Etienne C; Schriemer DC Analytical chemistry 2012, 84, 4411–4418. [DOI] [PubMed] [Google Scholar]
- (18).Zhang B; Rempel DL; Gross ML Journal of the American Society for Mass Spectrometry 2016, 27, 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Manzi L; Barrow AS; Hopper JTS; Kaminska R; Kleanthous C; Robinson CV; Moses JE; Oldham NJ Angewandte Chemie 2017, 56, 14873–14877. [DOI] [PubMed] [Google Scholar]
- (20).Cheng M; Guo C; Li W; Gross ML Angewandte Chemie International Edition 2021, 60, 8867–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Naredla RR; Klumpp DA Chemical reviews 2013, 113, 6905–6948. [DOI] [PubMed] [Google Scholar]
- (22).Ammer J; Mayr H Journal of Physical Organic Chemistry 2013, 26, 956–969. [Google Scholar]
- (23).Sun J; Liu H; Zhan L; Xiong C; Huang X; Xue J; Nie Z Analytical chemistry 2018, 90, 6397–6402. [DOI] [PubMed] [Google Scholar]
- (24).Koshland DE Journal of the American Chemical Society 1965, 87, 1126–1132. [DOI] [PubMed] [Google Scholar]
- (25).Pan J; Han J; Borchers CH; Konermann L Journal of the American Chemical Society 2009, 131, 12801–12808. [DOI] [PubMed] [Google Scholar]
- (26).Eliezer D; Wright PE Journal of molecular biology 1996, 263, 531–538. [DOI] [PubMed] [Google Scholar]
- (27).Lu Y; Zhang H; Niedzwiedzki DM; Jiang J; Blankenship RE; Gross ML Analytical chemistry 2016, 88, 8827–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Du Y; Duc NM; Rasmussen SGF; Hilger D; Kubiak X; Wang L; Bohon J; Kim HR; Wegrecki M; Asuru A; Jeong KM; Lee J; Chance MR; Lodowski DT; Kobilka BK; Chung KY Cell 2019, 177, 1232–1242 e1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen J; Cui W; Giblin D; Gross ML Journal of the American Society for Mass Spectrometry 2012, 23, 1306–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


