The asymmetric unit of the title compound comprises three 4-aminobenzoate ligands, two coordinated water molecules, a thulium metal ion and a water molecule of crystallization. The crystal structure features O—H⋯N, N—H⋯O, and O—H⋯O hydrogen-bonding interactions as well as C—H⋯π and off-set π–π stacking interactions. Hirshfeld surface analysis indicates that H⋯H contacts are the most significant contributors to the crystal packing.
Keywords: p-aminobenzoic acid, dinuclear thulium complex, single crystal, Hirshfeld analysis, crystal structure
Abstract
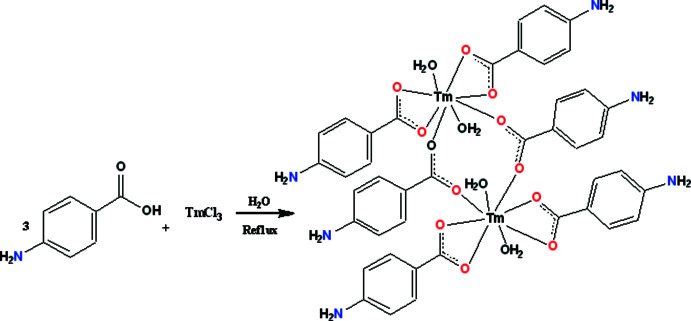
The asymmetric unit of the title compound, [Tm2(C7H6NO2)6(H2O)4]·2H2O, contains three 4-aminobenzoate (4ABA) ligands, two coordinated water molecules, a thulium metal ion, and a water molecule of crystallization. The overall structure of the complex (4ABA-Tm) is in the form of a dimer. In the dinuclear thulium complex, symmetry-relevant TmO8 coordination polyhedra are formed by the O atoms of two chelating 4-aminobenzoate ligands, the O atoms of two non-chelating 4-aminobenzoate ligands, and two water molecules. The Tm—O bond lengths range from 2.216 (3) to 2.471 (3) Å with the Tm⋯Tm separation in the dinuclear complex being 4.7863 (5) Å. The crystal structure features O—H⋯N, N—H⋯O, and O—H⋯O hydrogen-bonding interactions. Further stabilization of the crystal packing is due to C—H⋯π and off-set π–π stacking interactions. Hirshfeld surface analysis indicates that H⋯H contacts are the most significant contributors to the crystal packing (45.9%). In addition, a void analysis was performed to check the strength of the crystal packing.
Chemical context
The coordination chemistry of rare-earth metals has been widely studied, and the structures of a significant variety of complexes with diverse kinds of ligands have been reported (You et al., 2021 ▸). In particular, the lanthanide contraction along the series is of interest, and in a detailed analysis of this phenomenon using elements from the lanthanide series, p-aminobenzoic acid (HL) was found to be a very useful and biologically important ligand (Smith & Lynch, 2015 ▸). The carboxylate group of HL can be coordinated with the metals simultaneously in three different modes, namely chelating, bridging, and chelating-bridging (Ali et al., 2014 ▸). In the complexes of HL with alkali metals such as Na+ or K+, the ligand is not directly coordinated to the metal ion, but rather it is surrounded by coordinated water molecules (You et al., 2021 ▸). Both the carboxylic and amino groups of the ligand are coordinated to the metal in complexes with Ba2+, Ag+, Zn2+, Cd2+, and Ni2+ (Mamedov et al., 1982 ▸; Amirasłanov et al., 1982a
▸), while only the oxygen atoms of the carboxylic groups are coordinated to the metal ion in complexes of Sr2+, Mg2+, and Co2+ with this ligand (Amirasłanov et al., 1982b
▸; Sun et al., 2004 ▸). In comparison to the above coordination diversity, in the complexes of HL with rare-earth elements like Nd+3 and Sm+3 (Khiyalov et al., 1981 ▸; Mao & Lianq, 2016 ▸), only the nitrogen atom of the amino group is coordinated by the central metal atom, while in complexes of Lu+3 and Ho+3 with HL (Sun et al., 2004 ▸), the nitrogen atom of the amino group is not coordinated while the ligands are attached to the metal atom by the oxygen atoms of the carboxylate moiety. In this context, we report the synthesis, crystal structure, Hirshfeld surface, void, thermogravimetric and FT–IR analysis of the title compound, [Tm2(C7H6NO2)6(H2O)4]·2H2O, which is closely related to its Lu+3 and Ho+3 analogues (Sun et al., 2004 ▸).
Structural commentary
The asymmetric unit of the title compound 4ABA-Tm (Fig. 1 ▸) contains a centrosymmetric thulium dinuclear complex and one water molecule of crystallization. Each TmIII atom is octacoordinated by two chelating 4-aminobenzoate ligands, two bridging 4-aminobenzoate ligands and two coordinated water molecules. In the coordination sphere, bond lengths range from 2.216 (3) to 2.471 (3) Å, while bond angles range from 53.82 (10) to 161.28 (12)° (Table 1 ▸). The Tm⋯Tmi separation in the 4ABA-Tm complex is 4.7863 (5) Å (Table 1 ▸). The oxygen atoms O2 of the first 4-aminobenzoate chelate (ligand A, C1–C7/N1/O1/O2), O4 of the second 4-aminobenzoate chelate (ligand B, C8–C14/N2/O3/O4) and O5 of the 4-aminobenzoate non-chelate (ligand C, C15–C21/N3/O5/O6) show maximum deviations from their respective planes with values of 0.1748 (3) Å for O2, 0.3087 (3) Å for O4, and 0.1351 (3) Å for O5. Ligand B is twisted at a dihedral angle of 70.83 (7)° with respect to ligand A. The non-chelating ligand C is twisted at dihedral angles of 79.7 (9) and 72.7 (9)°, respectively, to the planes of ligands A and B. Intramolecular O—H⋯O hydrogen bonding (Table 2 ▸) involving OH from the non-coordinating water and the O atom (hydrogen-bond acceptor) of the chelating 4-aminobenzoate ligand stabilizes the molecular configuration.
Figure 1.
ORTEP view of 4ABA-Tm with ellipsoids drawn at a 30% probability level with H atoms shown as small circles of arbitrary radii.
Table 1. Selected geometric parameters (Å, °).
| Tm1—O5 | 2.216 (3) | Tm1—O3 | 2.374 (3) |
| Tm1—O6i | 2.223 (3) | Tm1—O8 | 2.382 (3) |
| Tm1—O7 | 2.293 (3) | Tm1—O4 | 2.457 (3) |
| Tm1—O2 | 2.329 (3) | Tm1—O1 | 2.471 (3) |
| O5—Tm1—O6i | 108.53 (13) | O3—Tm1—O8 | 124.48 (11) |
| O5—Tm1—O7 | 156.90 (13) | O5—Tm1—O4 | 80.34 (11) |
| O6i—Tm1—O7 | 84.40 (13) | O6i—Tm1—O4 | 142.65 (11) |
| O5—Tm1—O2 | 125.29 (12) | O7—Tm1—O4 | 78.06 (11) |
| O6i—Tm1—O2 | 81.97 (13) | O2—Tm1—O4 | 123.35 (10) |
| O7—Tm1—O2 | 74.42 (11) | O3—Tm1—O4 | 53.82 (10) |
| O5—Tm1—O3 | 80.05 (12) | O8—Tm1—O4 | 72.41 (10) |
| O6i—Tm1—O3 | 161.28 (12) | O5—Tm1—O1 | 75.80 (11) |
| O7—Tm1—O3 | 93.61 (13) | O6i—Tm1—O1 | 76.29 (11) |
| O2—Tm1—O3 | 79.58 (11) | O7—Tm1—O1 | 126.75 (11) |
| O5—Tm1—O8 | 78.30 (11) | O2—Tm1—O1 | 54.20 (10) |
| O6i—Tm1—O8 | 74.11 (12) | O3—Tm1—O1 | 90.29 (10) |
| O7—Tm1—O8 | 87.42 (11) | O8—Tm1—O1 | 131.47 (10) |
| O2—Tm1—O8 | 151.26 (11) | O4—Tm1—O1 | 139.92 (9) |
Symmetry code: (i)
 .
.
Table 2. Hydrogen-bond geometry (Å, °).
Cg3 is the centroid of the C16–C21 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O7—H7A⋯N3ii | 0.82 (1) | 1.95 (2) | 2.753 (6) | 169 (5) |
| O7—H7B⋯N2iii | 0.82 (1) | 2.18 (2) | 2.940 (5) | 154 (5) |
| O8—H8A⋯O1i | 0.82 (1) | 1.98 (1) | 2.791 (4) | 172 (4) |
| O8—H8B⋯O4iv | 0.82 (1) | 1.96 (1) | 2.780 (4) | 174 (4) |
| N1—H1B⋯O9v | 0.85 (1) | 2.06 (3) | 2.870 (8) | 158 (7) |
| N2—H2B⋯O9vi | 0.85 (1) | 2.24 (2) | 3.051 (7) | 161 (5) |
| N3—H3B⋯O5vii | 0.84 (1) | 2.46 (3) | 3.173 (6) | 144 (5) |
| N3—H3B⋯O8vii | 0.84 (1) | 2.56 (5) | 3.092 (5) | 122 (4) |
| O9—H9A⋯O3 | 0.83 (1) | 2.00 (1) | 2.828 (6) | 172 (7) |
| O9—H9B⋯O2viii | 0.84 (1) | 2.34 (6) | 2.849 (6) | 119 (5) |
| C11—H11⋯Cg3ix | 0.93 | 2.68 | 3.538 (5) | 155 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 ; (vi)
; (vi)
 ; (vii)
; (vii)
 ; (viii)
; (viii)
 ; (ix)
; (ix)
 .
.
Supramolecular features
The centrosymmetric dinuclear thulium complexes are linked through O—H⋯O, O—H⋯N and N—H⋯O hydrogen-bonding interactions. A C11 chain running along the b-axis direction is formed through O7—H7A⋯N3 H bonds while a loop is formed through the O7—H7A⋯N3 interactions. The water molecule of crystallization plays an important role in the stabilization of the crystal packing, acting as a hydrogen-bond donor and as well as a hydrogen-bond acceptor, connecting the centrosymmetric dinuclear thulium complex with each other. The hydrogen bonds lead to the formation of layers parallel to the bc plane (Fig. 2 ▸, Table 2 ▸). These layers are linked through C—H⋯π (Fig. 3 ▸), with H⋯π distance of 2.68 Å and off-set π–π stacking interactions (Fig. 4 ▸) with inter-centroid distances ranging from 3.661 (3) to 3.709 (3) Å, forming a three-dimensional network.
Figure 2.
Packing diagram of 4ABA-Tm. Selected H atoms are shown for clarity.
Figure 3.
Graphical representation of C—H⋯π interactions in 4ABA-Tm. Selected H atoms are shown while the water molecules are omitted for clarity.
Figure 4.
Graphical representation of off-set π–π interactions in 4ABA-Tm. H atoms and water molecules are not shown for simplicity.
Hirshfeld surface analysis
A Hirshfeld surface (HS) analysis was carried out using Crystal Explorer 21.5 (Spackman et al., 2021 ▸) in order to explore the non-covalent interactions in terms of the Hirshfeld surface and two-dimensional fingerprint plots. The HS of a molecule is the region in the crystal where the electron density relevant to the promolecule is greater than the electron density relevant to the procrystal (Spackman et al., 2009 ▸; Ashfaq et al., 2020 ▸). The Hirshfeld surface is constructed by employing colour coding to show the interatomic contacts that are shorter (red areas), equal to (white areas), or longer than (blue areas) the sum of the van der Waals radii (Ashfaq et al., 2021a ▸,b ▸). The red spots on the surface mapped over d norm (Fig. 5 ▸ a) indicate the involvement of atoms in hydrogen-bonding interactions. The HS mapped over shape-index (Fig. 5 ▸ b) is used to check for the presence of interactions such as C—H⋯π and π–π stacking (Ashfaq et al., 2021a ▸,b ▸). The existence of adjacent red and blue triangular regions around the aromatic rings conforms to the presence of π–π stacking interactions in the title compound.
Figure 5.
HS plotted over (a) d norm in the range −1.073 to 1.740 a.u. and (b) shape-index in the range −1 to 1 a.u.
Two-dimensional fingerprint plots provide unique information about the non-covalent interactions and the crystal packing in terms of the percentage contribution of the interatomic contacts (Spackman et al., 2002 ▸; Ashfaq et al., 2021a ▸,b ▸ ▸). Fig. 6 ▸ a shows the two-dimensional fingerprint plot for the overall interactions in 4ABA-Tm where d i and d e are the distances from the Hirshfeld surface to the nearest atom inside the Hirshfeld surface and outside it, respectively. The most important interatomic contact is H⋯H (Fig. 6 ▸ b) as it makes the highest contribution to the crystal packing (45.9%). Other major contributors are C⋯H (26.1%, Fig. 6 ▸ c) and O⋯H (15.5%, Fig. 6 ▸ d) interactions. The interatomic contacts that make comparatively smaller contributions in the crystal packing are shown in Fig. 6 ▸ e–l.
Figure 6.
Two-dimensional fingerprint plots of 4ABA-Tm for (a) all interactions and (b)–(l) individual interatomic contacts.
The response to applied stress or force mainly depends on the strength of the crystal packing in single crystals, which have a high mechanical strength as the molecules are strongly packed into them. To check whether the title compound is mechanically stable or not, a void analysis was performed. In order to calculate voids in the crystal packing, the electron densities of all of the atoms in the molecules present in the asymmetric unit are added up, the atoms being assumed to be spherically symmetric (Turner et al., 2011 ▸; Kargar et al., 2022 ▸). The volume of the void in the crystal packing of the title compound is 120.81 Å3 (Fig. 7 ▸), which infers that voids occupy 10.51% of the space and, hence, the molecules are strongly packed in the title compound.
Figure 7.
Graphical representation of 4ABA-Tm in (a) a view along the a axis and (b) a view along the b axis.
Infra-red spectroscopy
The structure of the newly synthesized complex was also investigated by FT–IR spectroscopy. It was found that the absorption bands of the –NH2 group appeared in the region of 3200 cm−1 while the absorption bands due to Tm—OH2 are visible in the region of 325 cm−1. The aromatic carbons show their absorption band at 1225 cm−1, while the Tm—O band is visible in the region of 650 cm−1. The absorption bands observed in the FT–IR spectrum of the free ligand in the regions of 1715 and 1435 cm−1 are caused by symmetric (νs) and asymmetric (νas) stretching of the carboxyl group, which are shifted to 1635 and 1436 cm−1, respectively, upon coordination with the TmIII metal ion. The difference between νs and νas is 199 cm−1, indicating that the carboxyl groups are coordinated to the central metal ion by chelate and bidentate-bridging coordination modes.
Thermogravimetric analysis
The title complex was further characterized by thermogravimetry. Thermolysis occurs in three stages. In the first stage, at a temperature of 20–200°C, intermolecular and coordinated water molecules are released, with a weight loss of 4.69%. The complex remains stable over the temperature 200–400°C. In the second stage, at a temperature of 400–600°C, the hydrocarbon residues are decomposed and simultaneously burned out. Thulium carbonate is formed in the last stage at a temperature between 600 and 800°C. The final product of decomposition above 800°C is metal oxide.
It is known that lanthanide carboxylates have good spectroscopic characteristics; they have enhanced thermal stability and are also resistant to moisture and oxygen in the air, which is of great importance in the production and operation of photoluminescent and electroluminescent devices based on them.
Database survey
A search of the Cambridge Structural Database (CSD, version 5.40; update February 2021; Groom et al., 2016) gave 206 hits, some of whose crystal structures are closely related to 4ABA-Tm. These include the yttrium (NADYEX), holmium (NADZAU), lutetium (NADZIC) and ytterbium (YENRAK01) complexes reported by Sun et al. (2004 ▸). Erbium (YUTNAE; Smith & Lynch, 2015 ▸) and terbium (NADXEW01; Ye et al., 2004 ▸) complexes were also found in the literature.
Synthesis and crystallization
The infra-red spectrum of 4ABA-Tm in the range 4000 to 250 cm−1 was recorded on an FT–IR Prestige 21 spectrophotometer after preparing the samples with KBr pellets. Thermal analysis was carried out using a NETSCHSTA-409 PC/PG derivatograph, TG, DTG and DTA curves were obtained in a static air atmosphere at a heating rate of 10°C min−1 from 20–800°C using platinum crucibles. Highly sintered Al2O3 was used as a reference. The elemental analysis for C, H, and N was performed using a Costech ECS 4010 CHNSO analyzer.
Preparation of the title complex
The reaction of aqueous solutions of TmCl3 and sodium p-aminobenzoate (1:3) yielded single crystals of tris-(p-aminobenzoato)thulium(III) dihydrate suitable for X-ray diffraction analysis. The mixture was refluxed for 30 minutes and then cooled to room temperature. After filtration, the filtrate was left for several days, covered with aluminum foil, until yellow prismatic crystals appeared. C42H48N6O18Tm2, M: 1262.72 g mol−1. Elemental analysis: calculated %: C:41.11; N: 6.25; Tm: 27.57: found %: C:41.24; N:6.72; Tm: 27.41.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. H atoms of all the water molecules and the amino groups of 4-aminobenzoate ligands were found by the careful inspection of residual electron-density peaks and positional parameters were refined using bond-length restraints (O—H = 0.82 Å, N—H = 0.85 Å) with U iso(H) = 1.5Ueq(O) or 1.2U eq(N). All other H atoms were refined at calculated positions using a riding-model approximation [C—H = 0.93 Å, U iso(H) = 1.2U eq(C)]. The highest positive and negative features in the final difference map are within 0.83 Å of the Tm atom.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C42H48N6O18Tm2 |
| M r | 1262.72 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 296 |
| a, b, c (Å) | 8.9659 (6), 10.9722 (7), 12.8027 (8) |
| α, β, γ (°) | 88.195 (3), 71.599 (3), 74.402 (3) |
| V (Å3) | 1149.10 (13) |
| Z | 1 |
| Radiation type | Mo Kα |
| μ (mm−1) | 3.92 |
| Crystal size (mm) | 0.32 × 0.18 × 0.16 |
| Data collection | |
| Diffractometer | Bruker Kappa APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.983, 0.986 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 13239, 4891, 4358 |
| R int | 0.037 |
| (sin θ/λ)max (Å−1) | 0.639 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.029, 0.064, 1.04 |
| No. of reflections | 4891 |
| No. of parameters | 343 |
| No. of restraints | 19 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 1.80, −1.07 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989022001116/zn2014sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022001116/zn2014Isup3.hkl
CCDC reference: 2118339
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors acknowledge support from the Department of Physics, University of Sargodha.
supplementary crystallographic information
Crystal data
| C42H48N6O18Tm2 | Z = 1 |
| Mr = 1262.72 | F(000) = 624 |
| Triclinic, P1 | Dx = 1.825 Mg m−3 |
| a = 8.9659 (6) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.9722 (7) Å | Cell parameters from 4358 reflections |
| c = 12.8027 (8) Å | θ = 2.5–27.0° |
| α = 88.195 (3)° | µ = 3.92 mm−1 |
| β = 71.599 (3)° | T = 296 K |
| γ = 74.402 (3)° | Prism, light yellow |
| V = 1149.10 (13) Å3 | 0.32 × 0.18 × 0.16 mm |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 4891 independent reflections |
| Radiation source: fine-focus sealed tube | 4358 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.037 |
| Detector resolution: 7.828 pixels mm-1 | θmax = 27.0°, θmin = 2.5° |
| ω scans | h = −11→9 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | k = −11→14 |
| Tmin = 0.983, Tmax = 0.986 | l = −16→16 |
| 13239 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.029 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.064 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0257P)2 + 0.6134P] where P = (Fo2 + 2Fc2)/3 |
| 4891 reflections | (Δ/σ)max = 0.001 |
| 343 parameters | Δρmax = 1.80 e Å−3 |
| 19 restraints | Δρmin = −1.07 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Tm1 | 0.63601 (2) | −0.01205 (2) | 0.13511 (2) | 0.02731 (7) | |
| O1 | 0.3377 (4) | 0.0164 (3) | 0.2017 (2) | 0.0380 (7) | |
| O2 | 0.4938 (4) | −0.1008 (3) | 0.2892 (2) | 0.0458 (8) | |
| O3 | 0.6097 (4) | 0.1341 (3) | 0.2770 (2) | 0.0468 (8) | |
| O4 | 0.8365 (3) | 0.1015 (2) | 0.1382 (2) | 0.0351 (6) | |
| O5 | 0.5346 (4) | 0.1682 (3) | 0.0675 (3) | 0.0518 (9) | |
| O6 | 0.3957 (5) | 0.1706 (3) | −0.0479 (3) | 0.0601 (10) | |
| O7 | 0.8305 (4) | −0.1673 (3) | 0.1803 (3) | 0.0469 (8) | |
| H7A | 0.9297 (15) | −0.183 (4) | 0.164 (4) | 0.056* | |
| H7B | 0.801 (5) | −0.221 (3) | 0.221 (3) | 0.056* | |
| O8 | 0.8252 (4) | −0.0313 (3) | −0.0465 (2) | 0.0372 (7) | |
| H8A | 0.783 (5) | −0.034 (4) | −0.094 (3) | 0.045* | |
| H8B | 0.9240 (14) | −0.053 (4) | −0.077 (3) | 0.045* | |
| N1 | −0.1604 (7) | −0.2330 (6) | 0.5608 (4) | 0.0795 (16) | |
| H1A | −0.163 (7) | −0.258 (6) | 0.625 (2) | 0.095* | |
| H1B | −0.248 (4) | −0.193 (6) | 0.549 (5) | 0.095* | |
| N2 | 0.8267 (6) | 0.6323 (4) | 0.3384 (3) | 0.0520 (11) | |
| H2A | 0.922 (3) | 0.629 (5) | 0.340 (4) | 0.062* | |
| H2B | 0.753 (4) | 0.677 (4) | 0.393 (3) | 0.062* | |
| N3 | 0.1667 (6) | 0.7572 (3) | 0.1054 (4) | 0.0595 (12) | |
| H3A | 0.184 (7) | 0.781 (5) | 0.162 (3) | 0.071* | |
| H3B | 0.209 (6) | 0.792 (5) | 0.049 (2) | 0.071* | |
| C1 | 0.3548 (6) | −0.0615 (4) | 0.2755 (3) | 0.0342 (9) | |
| C2 | 0.2186 (6) | −0.1081 (4) | 0.3440 (3) | 0.0350 (9) | |
| C3 | 0.0591 (6) | −0.0550 (4) | 0.3473 (3) | 0.0407 (10) | |
| H3 | 0.0364 | 0.0097 | 0.3013 | 0.049* | |
| C4 | −0.0683 (6) | −0.0961 (5) | 0.4179 (4) | 0.0502 (12) | |
| H4 | −0.1752 | −0.0594 | 0.4191 | 0.060* | |
| C5 | −0.0343 (7) | −0.1938 (5) | 0.4876 (4) | 0.0576 (15) | |
| C6 | 0.1255 (7) | −0.2498 (5) | 0.4815 (4) | 0.0558 (13) | |
| H6 | 0.1497 | −0.3172 | 0.5248 | 0.067* | |
| C7 | 0.2475 (6) | −0.2070 (4) | 0.4126 (4) | 0.0474 (11) | |
| H7 | 0.3544 | −0.2449 | 0.4110 | 0.057* | |
| C8 | 0.7365 (5) | 0.1669 (4) | 0.2249 (3) | 0.0330 (9) | |
| C9 | 0.7643 (5) | 0.2844 (4) | 0.2605 (3) | 0.0315 (9) | |
| C10 | 0.9155 (5) | 0.3092 (4) | 0.2208 (3) | 0.0349 (9) | |
| H10 | 1.0038 | 0.2487 | 0.1744 | 0.042* | |
| C11 | 0.9373 (6) | 0.4220 (4) | 0.2489 (3) | 0.0370 (10) | |
| H11 | 1.0401 | 0.4360 | 0.2225 | 0.044* | |
| C12 | 0.8074 (6) | 0.5145 (4) | 0.3160 (3) | 0.0379 (10) | |
| C13 | 0.6544 (6) | 0.4907 (4) | 0.3559 (3) | 0.0421 (11) | |
| H13 | 0.5658 | 0.5524 | 0.4006 | 0.051* | |
| C14 | 0.6336 (6) | 0.3768 (4) | 0.3299 (3) | 0.0379 (10) | |
| H14 | 0.5318 | 0.3614 | 0.3588 | 0.046* | |
| C15 | 0.4332 (5) | 0.2238 (3) | 0.0213 (4) | 0.0347 (9) | |
| C16 | 0.3553 (5) | 0.3619 (3) | 0.0476 (3) | 0.0260 (8) | |
| C17 | 0.3834 (5) | 0.4241 (4) | 0.1289 (3) | 0.0359 (10) | |
| H17 | 0.4461 | 0.3781 | 0.1700 | 0.043* | |
| C18 | 0.3196 (6) | 0.5536 (4) | 0.1497 (4) | 0.0425 (11) | |
| H18 | 0.3377 | 0.5938 | 0.2056 | 0.051* | |
| C19 | 0.2295 (5) | 0.6232 (3) | 0.0884 (4) | 0.0377 (10) | |
| C20 | 0.1950 (6) | 0.5613 (4) | 0.0106 (4) | 0.0431 (11) | |
| H20 | 0.1283 | 0.6070 | −0.0280 | 0.052* | |
| C21 | 0.2592 (5) | 0.4313 (4) | −0.0104 (3) | 0.0372 (10) | |
| H21 | 0.2370 | 0.3908 | −0.0642 | 0.045* | |
| O9 | 0.4466 (6) | 0.1607 (6) | 0.5070 (4) | 0.0969 (15) | |
| H9A | 0.499 (8) | 0.146 (6) | 0.4401 (15) | 0.116* | |
| H9B | 0.491 (9) | 0.094 (4) | 0.532 (5) | 0.116* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Tm1 | 0.03208 (11) | 0.02149 (9) | 0.02871 (10) | −0.00610 (7) | −0.01104 (7) | −0.00037 (6) |
| O1 | 0.0419 (18) | 0.0381 (15) | 0.0329 (15) | −0.0097 (13) | −0.0122 (13) | 0.0087 (12) |
| O2 | 0.0425 (19) | 0.0590 (19) | 0.0446 (18) | −0.0195 (16) | −0.0225 (15) | 0.0214 (15) |
| O3 | 0.054 (2) | 0.0528 (18) | 0.0336 (17) | −0.0295 (16) | −0.0005 (15) | −0.0096 (14) |
| O4 | 0.0367 (17) | 0.0360 (14) | 0.0327 (15) | −0.0106 (13) | −0.0096 (13) | −0.0083 (12) |
| O5 | 0.049 (2) | 0.0259 (14) | 0.071 (2) | −0.0004 (14) | −0.0142 (17) | 0.0154 (14) |
| O6 | 0.061 (2) | 0.0425 (18) | 0.074 (2) | −0.0184 (17) | −0.0109 (19) | −0.0271 (17) |
| O7 | 0.0337 (18) | 0.0453 (18) | 0.061 (2) | −0.0087 (15) | −0.0182 (17) | 0.0238 (15) |
| O8 | 0.0377 (17) | 0.0431 (16) | 0.0277 (15) | −0.0052 (14) | −0.0104 (13) | −0.0040 (12) |
| N1 | 0.096 (4) | 0.106 (5) | 0.052 (3) | −0.064 (4) | −0.014 (3) | 0.014 (3) |
| N2 | 0.077 (3) | 0.0324 (19) | 0.047 (2) | −0.025 (2) | −0.012 (2) | −0.0010 (17) |
| N3 | 0.047 (3) | 0.0211 (17) | 0.087 (3) | −0.0036 (17) | 0.006 (2) | −0.0008 (19) |
| C1 | 0.045 (3) | 0.0304 (19) | 0.028 (2) | −0.0130 (18) | −0.0115 (18) | 0.0008 (16) |
| C2 | 0.041 (3) | 0.035 (2) | 0.032 (2) | −0.0134 (19) | −0.0137 (18) | 0.0021 (17) |
| C3 | 0.046 (3) | 0.050 (3) | 0.032 (2) | −0.025 (2) | −0.011 (2) | 0.0033 (19) |
| C4 | 0.042 (3) | 0.073 (3) | 0.042 (3) | −0.027 (3) | −0.012 (2) | −0.005 (2) |
| C5 | 0.086 (4) | 0.069 (3) | 0.033 (3) | −0.055 (3) | −0.010 (3) | −0.001 (2) |
| C6 | 0.071 (4) | 0.058 (3) | 0.052 (3) | −0.032 (3) | −0.029 (3) | 0.019 (2) |
| C7 | 0.052 (3) | 0.047 (3) | 0.047 (3) | −0.018 (2) | −0.018 (2) | 0.010 (2) |
| C8 | 0.040 (2) | 0.037 (2) | 0.025 (2) | −0.0137 (19) | −0.0115 (18) | 0.0023 (16) |
| C9 | 0.041 (2) | 0.0311 (19) | 0.026 (2) | −0.0114 (18) | −0.0125 (17) | −0.0005 (15) |
| C10 | 0.041 (3) | 0.033 (2) | 0.029 (2) | −0.0115 (18) | −0.0075 (18) | −0.0031 (16) |
| C11 | 0.045 (3) | 0.040 (2) | 0.029 (2) | −0.020 (2) | −0.0084 (19) | 0.0022 (17) |
| C12 | 0.064 (3) | 0.0270 (19) | 0.027 (2) | −0.018 (2) | −0.016 (2) | 0.0047 (16) |
| C13 | 0.052 (3) | 0.030 (2) | 0.036 (2) | −0.006 (2) | −0.006 (2) | −0.0047 (17) |
| C14 | 0.043 (3) | 0.041 (2) | 0.028 (2) | −0.014 (2) | −0.0059 (18) | −0.0004 (17) |
| C15 | 0.031 (2) | 0.0228 (18) | 0.045 (2) | −0.0109 (17) | −0.0009 (18) | −0.0006 (17) |
| C16 | 0.029 (2) | 0.0219 (17) | 0.0278 (19) | −0.0073 (15) | −0.0100 (16) | −0.0011 (14) |
| C17 | 0.041 (3) | 0.031 (2) | 0.038 (2) | −0.0037 (18) | −0.0210 (19) | 0.0019 (17) |
| C18 | 0.044 (3) | 0.036 (2) | 0.050 (3) | −0.011 (2) | −0.017 (2) | −0.013 (2) |
| C19 | 0.035 (2) | 0.0201 (17) | 0.050 (3) | −0.0087 (17) | −0.0014 (19) | 0.0031 (17) |
| C20 | 0.043 (3) | 0.039 (2) | 0.043 (3) | −0.002 (2) | −0.016 (2) | 0.0157 (19) |
| C21 | 0.044 (3) | 0.040 (2) | 0.030 (2) | −0.0086 (19) | −0.0168 (19) | 0.0002 (17) |
| O9 | 0.067 (3) | 0.148 (5) | 0.066 (3) | −0.019 (3) | −0.016 (2) | 0.010 (3) |
Geometric parameters (Å, º)
| Tm1—O5 | 2.216 (3) | C3—C4 | 1.388 (6) |
| Tm1—O6i | 2.223 (3) | C3—H3 | 0.9300 |
| Tm1—O7 | 2.293 (3) | C4—C5 | 1.405 (7) |
| Tm1—O2 | 2.329 (3) | C4—H4 | 0.9300 |
| Tm1—O3 | 2.374 (3) | C5—C6 | 1.376 (6) |
| Tm1—O8 | 2.382 (3) | C6—C7 | 1.355 (7) |
| Tm1—O4 | 2.457 (3) | C6—H6 | 0.9300 |
| Tm1—O1 | 2.471 (3) | C7—H7 | 0.9300 |
| Tm1—C1 | 2.774 (4) | C8—C9 | 1.488 (5) |
| Tm1—C8 | 2.781 (4) | C9—C10 | 1.387 (6) |
| O1—C1 | 1.272 (5) | C9—C14 | 1.397 (5) |
| O2—C1 | 1.270 (5) | C10—C11 | 1.379 (5) |
| O3—C8 | 1.262 (5) | C10—H10 | 0.9300 |
| O4—C8 | 1.275 (5) | C11—C12 | 1.383 (6) |
| O5—C15 | 1.253 (5) | C11—H11 | 0.9300 |
| O6—C15 | 1.251 (5) | C12—C13 | 1.397 (6) |
| O6—Tm1i | 2.223 (3) | C13—C14 | 1.377 (6) |
| O7—H7A | 0.817 (10) | C13—H13 | 0.9300 |
| O7—H7B | 0.816 (10) | C14—H14 | 0.9300 |
| O8—H8A | 0.820 (10) | C15—C16 | 1.487 (5) |
| O8—H8B | 0.819 (10) | C16—C21 | 1.375 (5) |
| N1—C5 | 1.381 (7) | C16—C17 | 1.383 (5) |
| N1—H1A | 0.847 (10) | C17—C18 | 1.381 (5) |
| N1—H1B | 0.850 (10) | C17—H17 | 0.9300 |
| N2—C12 | 1.399 (5) | C18—C19 | 1.372 (6) |
| N2—H2A | 0.850 (10) | C18—H18 | 0.9300 |
| N2—H2B | 0.850 (10) | C19—C20 | 1.378 (6) |
| N3—C19 | 1.423 (5) | C20—C21 | 1.387 (6) |
| N3—H3A | 0.851 (10) | C20—H20 | 0.9300 |
| N3—H3B | 0.842 (10) | C21—H21 | 0.9300 |
| C1—C2 | 1.467 (6) | O9—H9A | 0.832 (10) |
| C2—C3 | 1.379 (6) | O9—H9B | 0.835 (10) |
| C2—C7 | 1.397 (6) | ||
| O5—Tm1—O6i | 108.53 (13) | O1—C1—C2 | 121.6 (4) |
| O5—Tm1—O7 | 156.90 (13) | O2—C1—Tm1 | 56.5 (2) |
| O6i—Tm1—O7 | 84.40 (13) | O1—C1—Tm1 | 62.9 (2) |
| O5—Tm1—O2 | 125.29 (12) | C2—C1—Tm1 | 171.2 (3) |
| O6i—Tm1—O2 | 81.97 (13) | C3—C2—C7 | 117.4 (4) |
| O7—Tm1—O2 | 74.42 (11) | C3—C2—C1 | 122.8 (4) |
| O5—Tm1—O3 | 80.05 (12) | C7—C2—C1 | 119.8 (4) |
| O6i—Tm1—O3 | 161.28 (12) | C2—C3—C4 | 121.4 (4) |
| O7—Tm1—O3 | 93.61 (13) | C2—C3—H3 | 119.3 |
| O2—Tm1—O3 | 79.58 (11) | C4—C3—H3 | 119.3 |
| O5—Tm1—O8 | 78.30 (11) | C3—C4—C5 | 119.4 (5) |
| O6i—Tm1—O8 | 74.11 (12) | C3—C4—H4 | 120.3 |
| O7—Tm1—O8 | 87.42 (11) | C5—C4—H4 | 120.3 |
| O2—Tm1—O8 | 151.26 (11) | C6—C5—N1 | 120.8 (5) |
| O3—Tm1—O8 | 124.48 (11) | C6—C5—C4 | 119.2 (5) |
| O5—Tm1—O4 | 80.34 (11) | N1—C5—C4 | 120.0 (6) |
| O6i—Tm1—O4 | 142.65 (11) | C7—C6—C5 | 120.2 (5) |
| O7—Tm1—O4 | 78.06 (11) | C7—C6—H6 | 119.9 |
| O2—Tm1—O4 | 123.35 (10) | C5—C6—H6 | 119.9 |
| O3—Tm1—O4 | 53.82 (10) | C6—C7—C2 | 122.3 (5) |
| O8—Tm1—O4 | 72.41 (10) | C6—C7—H7 | 118.8 |
| O5—Tm1—O1 | 75.80 (11) | C2—C7—H7 | 118.8 |
| O6i—Tm1—O1 | 76.29 (11) | O3—C8—O4 | 119.1 (4) |
| O7—Tm1—O1 | 126.75 (11) | O3—C8—C9 | 120.4 (4) |
| O2—Tm1—O1 | 54.20 (10) | O4—C8—C9 | 120.4 (4) |
| O3—Tm1—O1 | 90.29 (10) | O3—C8—Tm1 | 58.3 (2) |
| O8—Tm1—O1 | 131.47 (10) | O4—C8—Tm1 | 62.0 (2) |
| O4—Tm1—O1 | 139.92 (9) | C9—C8—Tm1 | 166.2 (3) |
| O5—Tm1—C1 | 101.68 (12) | C10—C9—C14 | 118.4 (4) |
| O6i—Tm1—C1 | 75.71 (12) | C10—C9—C8 | 121.6 (4) |
| O7—Tm1—C1 | 100.03 (12) | C14—C9—C8 | 119.8 (4) |
| O2—Tm1—C1 | 27.06 (11) | C11—C10—C9 | 121.1 (4) |
| O3—Tm1—C1 | 86.36 (11) | C11—C10—H10 | 119.4 |
| O8—Tm1—C1 | 148.00 (11) | C9—C10—H10 | 119.4 |
| O4—Tm1—C1 | 139.52 (10) | C10—C11—C12 | 120.6 (4) |
| O1—Tm1—C1 | 27.28 (10) | C10—C11—H11 | 119.7 |
| O5—Tm1—C8 | 75.79 (12) | C12—C11—H11 | 119.7 |
| O6i—Tm1—C8 | 169.50 (13) | C11—C12—C13 | 118.7 (4) |
| O7—Tm1—C8 | 88.48 (13) | C11—C12—N2 | 120.6 (4) |
| O2—Tm1—C8 | 103.56 (11) | C13—C12—N2 | 120.6 (4) |
| O3—Tm1—C8 | 26.88 (11) | C14—C13—C12 | 120.7 (4) |
| O8—Tm1—C8 | 97.92 (11) | C14—C13—H13 | 119.7 |
| O4—Tm1—C8 | 27.29 (11) | C12—C13—H13 | 119.7 |
| O1—Tm1—C8 | 114.19 (11) | C13—C14—C9 | 120.5 (4) |
| C1—Tm1—C8 | 113.24 (12) | C13—C14—H14 | 119.8 |
| C1—O1—Tm1 | 89.8 (3) | C9—C14—H14 | 119.8 |
| C1—O2—Tm1 | 96.4 (2) | O6—C15—O5 | 123.8 (4) |
| C8—O3—Tm1 | 94.8 (2) | O6—C15—C16 | 117.8 (4) |
| C8—O4—Tm1 | 90.7 (2) | O5—C15—C16 | 118.4 (4) |
| C15—O5—Tm1 | 145.2 (3) | C21—C16—C17 | 118.6 (3) |
| C15—O6—Tm1i | 157.5 (3) | C21—C16—C15 | 121.1 (3) |
| Tm1—O7—H7A | 134 (3) | C17—C16—C15 | 120.3 (3) |
| Tm1—O7—H7B | 118 (3) | C18—C17—C16 | 120.9 (4) |
| H7A—O7—H7B | 108 (4) | C18—C17—H17 | 119.6 |
| Tm1—O8—H8A | 113 (3) | C16—C17—H17 | 119.6 |
| Tm1—O8—H8B | 139 (3) | C19—C18—C17 | 120.3 (4) |
| H8A—O8—H8B | 106 (3) | C19—C18—H18 | 119.8 |
| C5—N1—H1A | 126 (4) | C17—C18—H18 | 119.8 |
| C5—N1—H1B | 107 (5) | C18—C19—C20 | 119.1 (4) |
| H1A—N1—H1B | 120 (4) | C18—C19—N3 | 121.7 (4) |
| C12—N2—H2A | 114 (4) | C20—C19—N3 | 119.2 (4) |
| C12—N2—H2B | 116 (3) | C19—C20—C21 | 120.5 (4) |
| H2A—N2—H2B | 111 (4) | C19—C20—H20 | 119.8 |
| C19—N3—H3A | 109 (4) | C21—C20—H20 | 119.8 |
| C19—N3—H3B | 110 (4) | C16—C21—C20 | 120.5 (4) |
| H3A—N3—H3B | 112 (4) | C16—C21—H21 | 119.7 |
| O2—C1—O1 | 119.0 (4) | C20—C21—H21 | 119.7 |
| O2—C1—C2 | 119.4 (4) | H9A—O9—H9B | 100 (4) |
| Tm1—O2—C1—O1 | −8.3 (4) | C14—C9—C10—C11 | 0.1 (6) |
| Tm1—O2—C1—C2 | 170.8 (3) | C8—C9—C10—C11 | 175.8 (4) |
| Tm1—O1—C1—O2 | 7.8 (4) | C9—C10—C11—C12 | −1.2 (6) |
| Tm1—O1—C1—C2 | −171.3 (3) | C10—C11—C12—C13 | 0.9 (6) |
| O2—C1—C2—C3 | 168.9 (4) | C10—C11—C12—N2 | −175.6 (4) |
| O1—C1—C2—C3 | −12.0 (6) | C11—C12—C13—C14 | 0.6 (6) |
| O2—C1—C2—C7 | −8.5 (6) | N2—C12—C13—C14 | 177.1 (4) |
| O1—C1—C2—C7 | 170.6 (4) | C12—C13—C14—C9 | −1.8 (6) |
| C7—C2—C3—C4 | 1.5 (6) | C10—C9—C14—C13 | 1.4 (6) |
| C1—C2—C3—C4 | −176.0 (4) | C8—C9—C14—C13 | −174.4 (4) |
| C2—C3—C4—C5 | 0.1 (7) | Tm1i—O6—C15—O5 | 23.7 (11) |
| C3—C4—C5—C6 | −2.4 (7) | Tm1i—O6—C15—C16 | −157.9 (6) |
| C3—C4—C5—N1 | 178.0 (5) | Tm1—O5—C15—O6 | −34.8 (8) |
| N1—C5—C6—C7 | −177.3 (5) | Tm1—O5—C15—C16 | 146.8 (4) |
| C4—C5—C6—C7 | 3.1 (8) | O6—C15—C16—C21 | −6.8 (6) |
| C5—C6—C7—C2 | −1.5 (8) | O5—C15—C16—C21 | 171.7 (4) |
| C3—C2—C7—C6 | −0.8 (7) | O6—C15—C16—C17 | 175.6 (4) |
| C1—C2—C7—C6 | 176.7 (4) | O5—C15—C16—C17 | −5.9 (6) |
| Tm1—O3—C8—O4 | −12.7 (4) | C21—C16—C17—C18 | −1.5 (7) |
| Tm1—O3—C8—C9 | 164.0 (3) | C15—C16—C17—C18 | 176.2 (4) |
| Tm1—O4—C8—O3 | 12.2 (4) | C16—C17—C18—C19 | −1.2 (7) |
| Tm1—O4—C8—C9 | −164.5 (3) | C17—C18—C19—C20 | 4.0 (7) |
| O3—C8—C9—C10 | 166.1 (4) | C17—C18—C19—N3 | −177.9 (4) |
| O4—C8—C9—C10 | −17.3 (6) | C18—C19—C20—C21 | −4.1 (7) |
| Tm1—C8—C9—C10 | −113.5 (12) | N3—C19—C20—C21 | 177.8 (4) |
| O3—C8—C9—C14 | −18.2 (6) | C17—C16—C21—C20 | 1.4 (6) |
| O4—C8—C9—C14 | 158.4 (4) | C15—C16—C21—C20 | −176.2 (4) |
| Tm1—C8—C9—C14 | 62.2 (13) | C19—C20—C21—C16 | 1.4 (7) |
Symmetry code: (i) −x+1, −y, −z.
Hydrogen-bond geometry (Å, º)
Cg3 is the centroid of the C16–C21 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7—H7A···N3ii | 0.82 (1) | 1.95 (2) | 2.753 (6) | 169 (5) |
| O7—H7B···N2iii | 0.82 (1) | 2.18 (2) | 2.940 (5) | 154 (5) |
| O8—H8A···O1i | 0.82 (1) | 1.98 (1) | 2.791 (4) | 172 (4) |
| O8—H8B···O4iv | 0.82 (1) | 1.96 (1) | 2.780 (4) | 174 (4) |
| N1—H1B···O9v | 0.85 (1) | 2.06 (3) | 2.870 (8) | 158 (7) |
| N2—H2B···O9vi | 0.85 (1) | 2.24 (2) | 3.051 (7) | 161 (5) |
| N3—H3B···O5vii | 0.84 (1) | 2.46 (3) | 3.173 (6) | 144 (5) |
| N3—H3B···O8vii | 0.84 (1) | 2.56 (5) | 3.092 (5) | 122 (4) |
| O9—H9A···O3 | 0.83 (1) | 2.00 (1) | 2.828 (6) | 172 (7) |
| O9—H9B···O2viii | 0.84 (1) | 2.34 (6) | 2.849 (6) | 119 (5) |
| C11—H11···Cg3ix | 0.93 | 2.68 | 3.538 (5) | 155 |
Symmetry codes: (i) −x+1, −y, −z; (ii) x+1, y−1, z; (iii) x, y−1, z; (iv) −x+2, −y, −z; (v) −x, −y, −z+1; (vi) −x+1, −y+1, −z+1; (vii) −x+1, −y+1, −z; (viii) −x+1, −y, −z+1; (ix) x+1, y, z.
Funding Statement
This work was funded by University of Sargodha.
References
- Ali, N., Tahir, M. N., Ali, S., Iqbal, M., Munawar, K. S. & Perveen, S. (2014). J. Coord. Chem. 67, 1290–1308.
- Amirasłanov, I. R., Musaev, F. N. & Mamedov, Kh. S. (1982a). Zh. Strukt. Khim. 23, 114.
- Amirasłanov, I. R., Musaev, F. N. & Mamedov, Kh. S. (1982b). Zh. Strukt. Khim. 23, 118.
- Ashfaq, M., Munawar, K. S., Bogdanov, G., Ali, A., Tahir, M. N., Ahmed, G., Ramalingam, A., Alam, M. M., Imran, M., Sambandam, S. & Munir, B. (2021a). J. Iran. Chem. Soc. pp. 1–9.
- Ashfaq, M., Tahir, M. N., Kuznetsov, A., Mirza, S. H., Khalid, M. & Ali, A. (2020). J. Mol. Struct. 1199, 127041.
- Ashfaq, M., Tahir, M. N., Muhammad, S., Munawar, K. S., Ali, A., Bogdanov, G. & Alarfaji, S. S. (2021b). ACS Omega, 6, 31211–31225. [DOI] [PMC free article] [PubMed]
- Bruker (2007). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Kargar, H., Fallah-Mehrjardi, M., Behjatmanesh-Ardakani, R., Munawar, K. S., Ashfaq, M. & Tahir, M. N. (2022). J. Mol. Struct. 1250, 131691.
- Khiyalov, M. S., Amiraslanov, I. R., Mamedov, K. S. & Movsumov, É. M. (1981). J. Struct. Chem. 22, 400–405.
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Mamedov, K. S., Movsumov, E. M., Amirasłanov, I. R. & Shkurpieło, A. I. (1982). Inst. Prikł. Fiziki, Kishiniev, p. 111.
- Mao, L. H. & Lianq, F. K. (2016). Chem. Res. Aplic. 18, 245–249.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Smith, G. & Lynch, D. E. (2015). Acta Cryst. E71, 1457–1461. [DOI] [PMC free article] [PubMed]
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spackman, M. A. & McKinnon, J. J. (2002). CrystEngComm, 4, 378–392.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst. 54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Sun, H. L., Ye, C. H., Wang, X. Y., Li, J. R., Gao, S. & Yu, K. B. (2004). J. Mol. Struct. 702, 77–83.
- Turner, M. J., McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2011). CrystEngComm, 13, 1804–1813.
- Ye, C. H., Sun, H. L., Wang, X. Y., Li, J. R., Nie, D. B., Fu, W. F. & Gao, S. (2004). J. Solid State Chem. 177, 3735–3742.
- You, F., Zhai, J., So, Y. M. & Shi, X. (2021). Inorg. Chem. 60, 1797–1805. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989022001116/zn2014sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022001116/zn2014Isup3.hkl
CCDC reference: 2118339
Additional supporting information: crystallographic information; 3D view; checkCIF report









