Abstract
Biophysical cues in the extracellular matrix (ECM) regulate cell behavior in a complex, non-linear, and interdependent manner. To quantify these important regulatory relationships and gain a comprehensive understanding of mechanotransduction, there is a need for high-throughput matrix platforms that enable parallel culture and analysis of cells in various matrix conditions. Here we describe a multi-well hyaluronic acid (HA) platform in which cells are cultured on combinatorial arrays of hydrogels spanning a range of elasticities and adhesivities. Our strategy utilizes orthogonal photopatterning of stiffness and adhesivity gradients, with the stiffness gradient implemented by a programmable light illumination system. The resulting platform allows individual treatment and analysis of each matrix environment while eliminating contributions of haptotaxis and durotaxis. In human mesenchymal stem cells (hMSCs), our platform recapitulates expected relationships between matrix stiffness, adhesivity and cell mechanosensing. We further applied the platform to show that as integrin ligand density falls, cell adhesion and migration depend more strongly on CD44-mediated interactions with the HA backbone. We anticipate that our system could bear great value for mechanistic discovery and screening where matrix mechanics and adhesivity are expected to influence phenotype.
Keywords: Mechanobiology, biomaterials, hyaluronic acid, combinatorial matrix arrays
Graphical Abstract

INTRODUCTION
Cells in tissue constantly sense and respond to biochemical and biophysical inputs from the microenvironment, including the extracellular matrix (ECM). Cell-ECM biophysical interactions profoundly impact many biological processes such as stem cell development,1 tumor progression,2,3 and immune responses.4 The relationships between individual matrix parameters and phenotype are often complex, non-linear, and interdependent. For example, matrix stiffness can drive cell behavior synergistically or antagonistically with adhesive ligand density, stress relaxation properties and surface wettability.5–7 Mesenchymal stem cell (MSC) phenotypes and transcriptomic profiles have also been reported to be non-linearly regulated by topography and stiffness.8–10 An important barrier to deconstructing this complex phase space has been the relative paucity of biomaterial platforms that allow for combinatorial or parallel deployment of many matrix conditions simultaneously.
Synthetic hydrogels are commonly used to mimic ECM due to their structural and mechanical similarities to native ECM and the versatility with which they can be chemically functionalized.11–14 Synthetic hydrogel platforms have been successfully applied to investigate cellular responses to various biophysical cues such as matrix mechanics,15–17 adhesivity,5,18,19 and topography.20,21 As interest has grown in understanding how matrix biophysical properties influence cell behavior, there has been much effort to engineer combinatorial hydrogel systems in which multiple ECM properties can be systematically and independently varied, analogous to experiments in which cultured cells are treated with systematic combinations of small molecules or growth factors to assess multifactorial interactions. For example, we and others have developed hydrogel platforms with dual gradients of stiffness and adhesive peptide density to investigate their coupling effects on adhesion, migration, and fate commitment.22–24 Other examples include hydrogels with orthogonal peptide gradients, stiffness and roughness gradients, and stiffness and wettability gradients.7,8,25,26 These high-throughput hydrogel platforms can be categorized into discrete hydrogel arrays24,27,28 and continuous gradient hydrogels.23,25,29 An important advantage of discrete arrays is the ability to isolate cells within a given set of matrix conditions, thereby eliminating cellular crosstalk across matrix conditions and permitting isolated extraction of cells and/or media from individual matrix conditions for downstream analysis.
Matrix arrays with hundreds or even thousands of gels on one device can be fabricated using dedicated robotic spotters.30,31 However, specialized instrumentation is needed to construct and read these arrays, and the extensive parallelization afforded by these platforms is not always required. It would also be advantageous to deploy multiplex hydrogel platforms in standard multi-well culture plate formats, for which a wealth of bio-analytical assays has been developed. While efforts have been made to engineer multi-well gel arrays, most notably by assembling gels directly in the wells of standard culture plates,32–34 most of these approaches have varied a single ECM parameter at a time (e.g. stiffness). As a result, there remains a strong need for ECM platforms that can be implemented in multi-well formats where multiple matrix parameters can be varied both independently and in parallel.
Here we describe a combinatorial, multi-well hydrogel array platform in which cells are cultured on hyaluronic acid (HA) gels with combinations of stiffness and adhesive RGD peptide density. HA is a common backbone of ECM-mimetic hydrogels in tissue engineering due to its biocompatibility and flexibility to chemical modifications.35,36 Methacrylated HA (HA-Me) hydrogels with tunable mechanical properties have been used by our lab and others to study cell- matrix interplays that are dependent on stiffness, adhesive ligand density, topography or other biophysical cues.37–40 Our approach innovates upon our previously described orthogonal photopatterning strategy, in which we created orthogonal, continuous gradients of stiffness and adhesive ligand density.22 Our new platform employs discrete ECM wells, made possible by replacing the previously documented gradient photomask with a custom-designed and programmable LED array.41,42 As a result, our platform allows individual treatment and extraction of cells from individual matrix environments while eliminating contributions of haptotaxis and durotaxis. We conducted proof-of-principle studies with human adipose-derived mesenchymal stem cells (hMSCs) to validate this platform and recapitulated the relationships between matrix stiffness and adhesivity on hMSC cell morphology. We further applied this platform to investigate the coupling effects of stiffness and adhesivity on human glioblastoma (GBM) cell spreading, adhesion, and migration in the presence or absence of the HA-binding receptor CD44.
EXPERIMENTAL SECTION
MATERIALS AND METHODS
Synthesis and Characterization of Methacrylated Hyaluronic Acid
Methacrylated HA (HA-Me) was synthesized as described previously.37 Briefly, HA powder (MW 66–90 kDa range; Lifecore Technologies) was dissolved at 1 wt% in deionized water, and a ten-fold molar excess of methacrylic anhydride (Sigma-Aldrich; relative to the HA disaccharide repeat unit) was added dropwise to the solution on ice. The pH was adjusted to 8–9 by 10 N NaOH and the reaction was maintained at 4 °C overnight. The next day, two-fold molar excess of methacrylic anhydride was added dropwise to the solution. The pH was adjusted to 8–9 by 10 N NaOH and the reaction was maintained at 4 °C for another day. Afterwards, methacrylated HA was precipitated from the aqueous solution by excess cold ethanol (Proof 200 ethanol, anhydrous, Koptec). The mixture was then centrifuged at 4,000 × g to recover the precipitate, which was redissolved in deionized water, frozen at −80 °C, and lyophilized. 1H-NMR spectra were collected at 400 MHz using a Bruker Avance AVB-400 instrument. The degree of methacrylation was then calculated as the ratio of the vinylic protons from the methacrylate group to the N-acetyl methyl protons from the HA backbone, normalized to the number of protons per group. A methacrylate to HA monomer ratio of ~70% was achieved (Figure S1).
Fabrication of HA Array
A lid with an array of polydimethylsiloxane (PDMS) (SYLGARDTM 184 silicone elastomer kit, Dow-Corning) posts was cast using acrylic molds. The acrylic mold was in turn custom-made by laser-cutting and stacking 1.5 mm-thick acrylic sheets (CLAREX Precision Thin Sheet, 1.5 mm, Astra Products). Specifically, a 1.5 mm-thick acrylic sheet was perforated with a H-series 20×12 Desktop CO2 Laser Engraver (Full Spectrum Laser) to generate an array of 3.3 mm-wide circular holes, with the distance between circles equal to that of the double-sided tape (Figure S2, Sheet A). A spacer acrylic sheet (Figure S2, Sheet B) with a rectangular cutout was placed on top of Sheet A, with another rectangular acrylic sheet (Figure S2, Sheet C) at the bottom of Sheet A. PDMS was poured into the mold, a glass slide (Fisherbrand, 12–550-A3) was placed on top, and the sandwich assembly was clamped together with binder clips. The PDMS was cured at 80°C for at least two hours before removing from the casting mold. The lid could be “recharged” for subsequent uses with 10 min plasma treatment (Harrick Plasma, PDC-32G) immediately before each use, for up to 10 reliable reuses.
A piece of double-sided tape (ARcare® 90106, Adhesives Research) was laser cut to generate an array of 3 mm-wide circular holes, with the distance between circles equal to the well- to-well distance of a standard 96-well plate. One side of the tape cover was removed, and the tape was affixed to a No. 1.5 cover glass (Ted Pella, Product #260423). Experiments performed in this manuscript utilized a 24 × 60 mm cover glass rectangle; however, glass may be cut to smaller dimensions using a pen-style glass cutter to accommodate the dimensions of customized gel arrays. A 1 mg/ml Poly-D-lysine (PDL, MW>300,000, Sigma-Aldrich) solution was used to coat the glass for 1 min followed by a quick rinse in double distilled H2O to facilitate HA hydrogel attachment.
HA-RGD solutions were made by mixing appropriate volumes of 10% wt HA-me, 4 mg/mL reconstituted RGD adhesive peptide (Ac-GCGYGRGDSPG-NH2, Anaspec) and PBS and then vortexing at room temperature overnight. Final HA-RGD precursor solutions had 5.71% wt HA- me but varied RGD conjugation concentrations. HA-RGD precursor solutions were mixed with 5% wt DTT (Thermo Fisher Scientific) and PBS to achieve a 0.15 thiol:HA repeat unit ratio and a final 5% wt HA-me. The hydrogel solution was immediately pipetted onto the PDL-treated glass surface (Figure 1A). The PDMS lid was then placed on top of the coverslip to form a sandwich assembly (Figure 1B). The assembly was allowed to sit at room temperature in a moisturized chamber overnight for hydrogel crosslinking. The following day, the assembly was soaked in 1X PBS for at least 15 minutes to detach the PDMS lid from the gel array. The double-sided tape cover was removed, and the exposed adhesive surface was used to adhere the gel array onto a bottomless 96-well black-wall culture plate (Greiner Bio-One International GmbH, #82051–528) and covered with a 96-well lid (Evergreen Scientific, 290–8019-01L) (Figure 1C–D). For cell differentiation assays and other long-term (>1 day) cell culture studies, sterile PBS was used at all times, plastic culture supplies were disinfected by UV radiation and the fabrication process occurred in a biosafety hood to minimize microbial contamination.
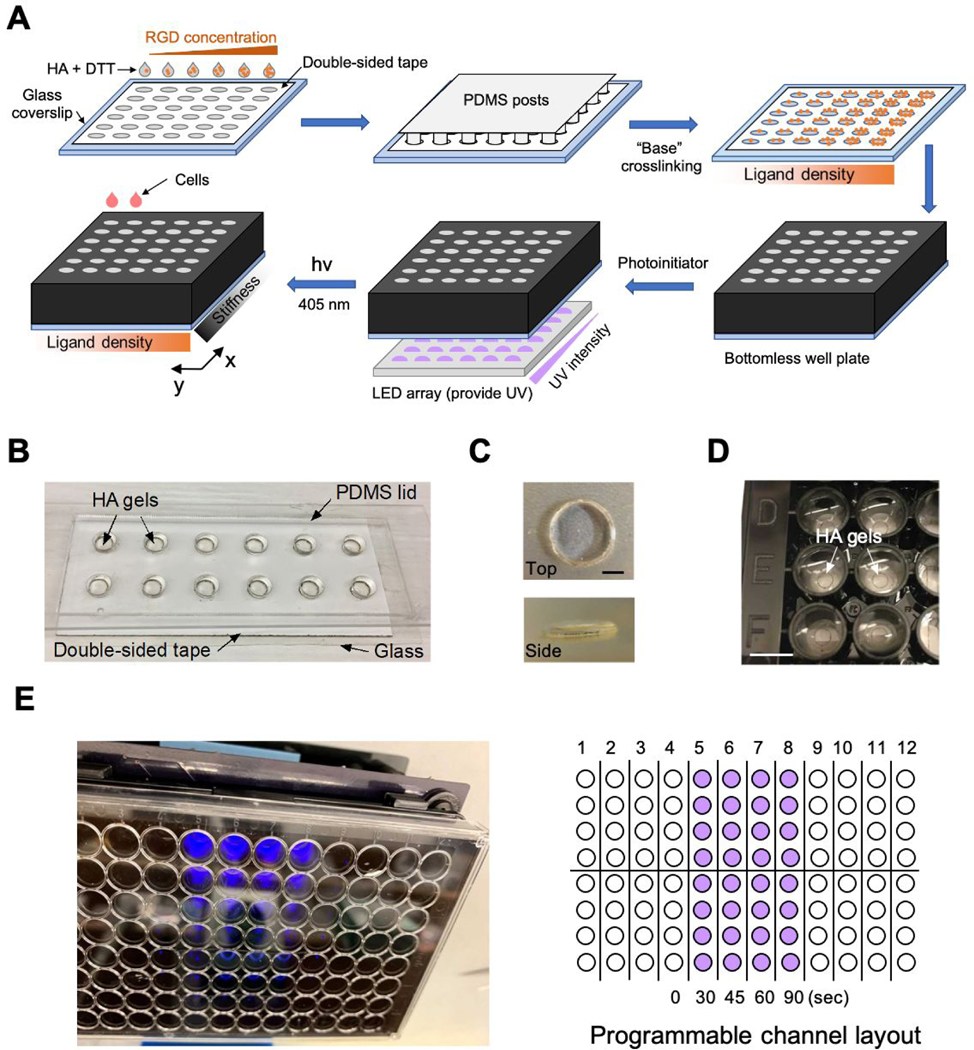
Figure 1.
Fabrication of the HA hydrogel array platform. (A) Workflow of the array fabrication. RGD density is controlled by mixing methacrylated HA precursor with varying peptide concentrations. Stiffness is determined by the accumulation of two crosslinking steps: first by DTT, and then by radical photocrosslinking. (B) The array in its “sandwich” assembly. (C) Top view (upper) and side view (lower) of one gel in the array. Each gel is ~3 mm wide and ~200 μm thick. Scale bar: 1 mm (D) Array assembled with a bottomless 96-well plate. Scale bar: 5 mm (E) The hydrogel array is photopatterned by a light activation at variable amplitudes (LAVA) device (left). The LAVA device can control 24 independent channels with programmable intensity and illumination time (right). Purple wells represent channels illuminated with UV light for various exposure times.
Stiffness Gradient Patterned by a Programmable Light Illumination System
A specialized illumination device for light activation at variable amplitudes (LAVA) was used for gel photostimulation (Figure 1E). The LAVA device was designed for 96-well plate illumination as described previously.41,42 Briefly, the illumination intensity of a surface-mount 405 nm LED (SMT405R, Marubeni) placed at the center of each well was controlled with pulse-width modulation. User-defined intensities for each well were programmed through a graphical user interface to allow 405 nm illumination in the [0 – 33.5] μW/mm2 intensity range. The LAVA device allowed independent control of 24 channels, so that clusters of four neighboring wells were controlled simultaneously in the 96-well format (Figure 1E).
Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP, Allevi) was dissolved in PBS to make a 0.5 wt % solution. 100 μL of freshly-made LAP solution was added to each well and the gels were soaked with LAP on a shaker for exactly 1 hr at room temperature. The array was then placed on the LAVA device and exposed to 405 nm UV light (intensity measured at the center of hydrogel =15 μW/mm2) for various amounts of time programmed by previously described software.41 Afterwards, the LAP solution was immediately removed and the array was washed with 1X PBS for 20 min on a shaker to remove excess LAP.
Atomic Force Microscope Characterization of Hydrogel Array
For AFM measurements, the hydrogel array was first attached to a removable plastic chamber that held the LAP solution during photocrosslinking on the LAVA device. Afterwards, the LAP solution was replaced with 1X PBS. AFM was performed with a Veeco Catalyst Bioscope (Bruker Corporation, Camarillo, California, USA). The gels were indented using a pyramidal-tipped probe (DNP-10, Bruker AFM Probes) with cantilever spring constants of 0.12–0.24 N/m, as measured by thermal calibration. Typically, 5 force curves per hydrogel were collected at different positions at least 100 μm apart from each other. Elastic moduli of the gels were calculated from force curves using a modified Hertz model, as described.43
Cell Culture
Human primary adipose-derived mesenchymal stem cells (hMSC) were purchased from ATCC (PCS-500–001), maintained and grown in accordance with the manufacturer’s recommendations. Cells were cultured in mesenchymal growth medium supplemented with corresponding growth supplements (ATCC) and used at passage number < 8. U-87 MG human glioblastoma (GBM) cells were obtained from the University of California, Berkeley Tissue Culture Facility, which sources its cultures directly from ATCC. U-87 MG cells were cultured in DMEM (Thermo Fisher Scientific), 1% (vol/vol) MEM non-essential amino acids (Thermo Fisher Scientific), and 1% (vol/vol) sodium pyruvate (Thermo Fisher Scientific). Cells were passaged every 5 days using 0.25% trypsin-EDTA (Thermo Fisher Scientific) and used at passage number < 30. Cells were screened on a monthly basis for mycoplasma and authenticated every six months by Short Tandem Repeat (STR) analysis at the University of California Cell Culture Facility. We used our previously developed CD44 KO and non-targeting U-87 MG cell lines, fabricated using CRISPR-based gene editing.44
hMSC Spreading Assay and Immunostaining
hMSCs were seeded on the hydrogels at a density of 5,000 – 7,500 cells/cm2 and allowed to attach and grow for 16 – 20 hrs. Cells were then fixed with 4% paraformaldehyde (PFA, Sigma-Aldrich) in 1X PBS for 15 min at room temperature, permeabilized with 0.5% (vol/vol) Triton X-100 (EMD Millipore, 9410) in PBS, and blocked using 5% (vol/vol) goat serum (Thermo Fisher Scientific) in PBS. YAP was visualized using YAP (D8H1X) XP® rabbit monoclonal antibody (Cell Signaling Technology). Actin filaments were visualized using AlexaFluor-546 labeled phalloidin (Thermo Fisher Scientific). Cell nuclei were visualized using 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI, Sigma-Aldrich). Fluorescence imaging was performed on a Nikon Eclipse Ti-E epifluorescence microscope. YAP nucleus/cytoplasm (nuc/cyto) intensity ratio was measured as the average intensity of YAP signal inside the nucleus divided by the average intensity of an area adjacent to the nucleus region, of similar size as the nucleus. Morphometric analysis of projected cell area was performed by thresholding the phalloidin fluorescence images to define the cell boundaries and applying automated particle shape analysis in ImageJ. Cells in clumps or near the outer edge of the gel were excluded from analysis.
hMSC Differentiation and Staining
hMSCs were seeded on the HA hydrogel array at a density of 20,000 cells/cm2 and allowed to grow in expansion medium for 1 day. Cells were then shifted to a mixed differentiation medium, consisting of a 1:1 mixture of osteogenic and adipogenic differentiation media (StemPro kits, Thermo Fisher Scientific). Cells were maintained for 7 days in these conditions with media change every 3–4 days.
Adipogenic differentiation of PFA-fixed hMSCs was assessed by lipid droplet staining of BODIPY™ 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene, Invitrogen), at a 2 μM working concentration. The cells were then washed extensively with PBS prior to imaging on a Nikon Eclipse Ti-E epifluorescence microscope. After fixation and permeabilization, osteogenic differentiation of cells was assessed by alkaline phosphatase activity using the SigmaFast BCIP/NBT assay (Sigma-Aldrich). A working solution of BCIP/NBT was prepared as per manufacturer’s instructions, added to the cells, incubated for 15 min at room temperature, and then washed extensively in 1X PBS. Alkaline phosphatase staining was identified as a dark purple color and imaged on an Olympus IX50 inverted fluorescence phase contrast microscope, with images captured by a Canon EOS Rebel T3 digital SLR camera.
U-87 Cell Spreading Assay
Cells were harvested and seeded onto HA gel arrays at a density of 6,000 cells/cm2 and incubated for at least 6 hrs. Cells were imaged using a 10X UPLFN objective lens and the MuviCyte Live-Cell Imaging System (PerkinElmer, Massachusetts, USA) in a 37 °C, CO2 controlled chamber. At least 20 cells were randomly selected for cell spreading quantification. Cells in multicellular clusters or near the outer edge of the gel were excluded from analysis. Cells with zero visible protrusions were labeled as “round” while cells with one or more protrusions were labeled as “spread.”
U-87 Centrifugal Adhesion Assay
Centrifugal adhesion assay was performed based on our previously reported protocol.38,44 Briefly, cells were harvested and seeded onto HA gels at a density of 6,000 cells/cm2 and incubated for at least 6 hrs. Prior to centrifugation, wells were filled with the addition of fresh medium, and cell culture plates were sealed with an adhesive plate sealer (Microseal ‘B’ Adhesive Seals, BioRad). The plate was then inverted and centrifuged for 5 minutes at 100 × g. Cells remaining on hydrogels were then fixed by paraformaldehyde (PFA) and stained with DAPI. Images were captured using the MuviCyte Live-Cell Imaging System. At least 3 unique microscopic fields were randomly imaged per well using a 10X objective lens. Automated thresholding analysis of the DAPI images was performed on ImageJ to determine a total count of the number of cells in each field of view.
U-87 Migration assay
Cells were seeded onto HA gels at a density of 6,000 cells/cm2 and incubated for at least 6 hrs. Then, migration assays were performed by imaging cells at 15-minute intervals for 8 hrs using the Muvicyte Live-Cell Imaging System with a 10× objective in a 37 °C, CO2 controlled chamber. The ImageJ Manual Tracking was used to track cell movements in each frame and calculate an average cell speed. Cells that were dividing, in multicellular clusters, or near the outer edge of the gel were excluded from analysis.
Real-time Polymerase Chain Reaction (RT-PCR)
Prior to cell seeding, a ring-shaped tape cover was laser cut from the double-sided tape and adhered to the bare tape region in each well of the gel array. Cells were then seeded onto the array and allowed to differentiate as described above. At the conclusion of the differentiation assay, the tape cover and all cells attached to the cover was removed by tweezers, while cells attached to the hydrogels remained in the wells. Cells were then lysed, and RNA was extracted following the manufacturer’s protocol (ReliaPrep™ RNA Miniprep systems, Promega Corporation). cDNA was synthesized using qScript™ cDNA SuperMix (Quantabio) following the manufacturer’s protocol. Real-time PCR reactions were prepared with a SYBR Green qPCR Master Mix (Bimake) and the PCR cycle was performed on a BioRad CFX Connect Real-Time System™.
Primer sequences used in RT-PCR of hMSC: ALP (ACC ACC ACG AGA GTG AAC CA; CGT TGT CTG AGT ACC AGT CCC, Elimbio), FABP4 (ACG AGA GGA TGA TAA ACT GGT GG; GCG AAC TTC AGT CCA GGT CAA C, Genewiz), GAPDH (GTC AAG GCT GAG AAC GGG AA; AAA TGA GCC CCA GCC TTC TC, Elimbio).
Quantification and statistical analysis
Statistical significance was calculated by the indicated tests using GraphPad/Prism, and statistical details can be found in figure legends. For figures in which multiple comparison analysis are displayed by letters, conditions with a common letter are not significantly different according to the stated statistical test at the 5% level of significance (p < 0.05).
RESULTS
HA Hydrogel Array Fabrication and Characterization
Our hydrogel array platform is assembled by casting HA hydrogel precursors in two-dimensional array patterns on a glass substrate that is later affixed to a bottomless multi-well plate. This fabrication method results in individually accessible wells, each containing a single gel. Key to this approach is the use of laser-cut double-sided tape, which serves both as a template for casting the HA gel precursor solution and as an adhesive to affix the glass to the bottomless multi-well plate. We began by using a laser cutter to perforate the double-sided tape to generate a two-dimensional array of circular holes corresponding to the desired dimension and distribution of gels. We then exposed one adhesive side of the patterned double-sided tape and attached it to an appropriately sized glass coverslip. Methacrylated HA hydrogel precursors were mixed with DTT and then pipetted into the shallow ‘wells’ formed by the union of the patterned double-sided tape and the glass surface (Figure 1A). A lid of PDMS posts was applied to flatten gel interfaces during initial crosslinking (Figure 1B). The resulting hydrogels are ~200 μm thick, approximately the same thickness as the double-sided tape, and 3 mm wide, approximately the same diameter as the circular holes patterned in the double-sided tape (Figure 1C). Following gelation and removal of the PDMS lid, the top adhesive surface of the tape was exposed and used to adhere the glass to a bottomless multi-well plate (Figure 1D). The union of the bottomless well plate, double sided tape, and glass formed a tight seal that prevents media leakage and exchange between neighboring wells (Figure S3). For this study, we designed the array in a 96-well plate format, but the fabrication technique can be readily adapted to other plate types (e.g. 6, 12, 24-well) by customizing the dimensions of the patterned double-sided tape and PDMS posts.
The array platform varies elasticity across wells in the x-direction and independently varies adhesive ligand density (adhesivity) along wells in the y-direction (Figure 1A). Prior to crosslinking, we conjugated various concentrations of RGD peptide (GCGYGRGDSPG) to methacrylated HA precursors (Figure S1) through thiol-ene reactions, generating RGD-modified HA (HA-RGD) precursors with varying RGD densities. The HA-RGD then underwent a two-step crosslinking reaction to generate the stiffness gradient. In the first step, HA-RGD was mixed with DTT at a thiol to HA unit ratio (T/H ratio) of 0.15 to create a relatively soft “base” gel. The second crosslinking step involved a tunable illumination process using a previously developed device featuring light activation at variable amplitudes (LAVA).41,42 On the LAVA device, individual hydrogel-containing wells were irradiated with a programmable 405 nm LED in the presence of LAP photoinitiator (Figure 1E). In our study, we illuminated the gel with a UV intensity of 15 μW/mm2 and adjusted the illumination time to achieve varying levels of crosslinking.
To characterize the stiffness gradient generated by the two-step crosslinking reaction we used atomic force microscopy (AFM). Young’s moduli were extracted by fitting the resulting force-indentation curves to a Hertz model, as described previously.43 The hydrogel stiffness was highly correlated with the illumination time, spanning a dynamic range of 12 to 91 kPa with 0 to 90 seconds of LED illumination (Figure 2). An extended illumination time can increase the modulus to above 200 kPa. Unless specified otherwise, the following studies utilized gel arrays within the 12–91 kPa range to mirror the modulus of most human tissues, which falls between 500 Pa and 100 kPa.45,46 As illumination time increased, we observed a broader distribution of moduli across gels of a given formulation, indicating increasing heterogeneity in crosslinking. We have previously observed that HA hydrogels in this stiffness range exhibit relatively modest stiffness- dependent variations in mesh size, indicating that swelling does not appreciably alter the effective concentration of HA monomers.37 There was no significant difference in modulus between HA- RGD gels and bare HA gels under the same illumination condition (Figure S4), suggesting that the HA backbone retained sufficient unreacted methacrylate groups after RGD conjugation for the two-step crosslinking process.
Figure 2.
Stiffness gradient on the HA hydrogel array after LAP-induced photocrosslinking of “base” 5% wt HA-me gels with 0.5 mM RGD. Intensity of 405 nm UV radiation was fixed at 15 μW/mm2 Gels were indented with a pyramid-tipped AFM probe and elastic moduli were calculated from at least 5 different locations on each individual gel to obtain an average elastic modulus per gel. For each condition, 5 individual gels were fabricated and measured in independent experiments (n=5). After 0, 30, 45, 60 and 90 seconds of radiation, the gel modulus (mean ± standard deviation (SD)) are 12±3 kPa, 21±1 kPa, 30±4 Pa, 48± 9 kPa and 91±22 kPa. All values were rounded to the nearest kPa. Statistical significance between neighboring conditions was evaluated using unpaired t-tests (**p<0.01). Boxes represent 25th and 75th percentiles and whiskers represent min and max.
Stiffness and Ligand Density Coupled to Modulate hMSC Mechanosensing and Morphology
To demonstrate that our hydrogel array platform can recapitulate the effects of ECM stiffness and adhesive ligand density on cell morphology and mechanotransduction, we cultured adipose-derived hMSCs on our platform and evaluated cell spreading and Yes-Associated Protein (YAP) localization. YAP is a transcriptional co-activator that localizes to the nucleus under mechanical tension, where it interacts with co-factors to regulate the expression of a variety of target genes.47,48 Specifically, YAP has been reported to preferentially localize to the nucleus of hMSCs cultured on stiff matrices but remain cytoplasmic for hMSCs on soft matrices. YAP nuclear localization has also been reported in cells cultured on matrices containing high ligand density, consistent with the notion that stiffness and ligand density can act synergistically to regulate cytoskeletal and focal adhesion assembly.49 hMSC morphologies are also highly sensitive to matrix stiffness and adhesive ligand density in vitro.18,50,51 We hypothesized that on our platform, increased stiffness and ligand density would enhance YAP nucleus localization and lead to increased cell area.
To evaluate hMSC sensitivity to stiffness and RGD density gradients, we made a 3×5 array with 3 RGD densities (0.1, 0.25, and 0.5 mM RGD in the HA-RGD precursors) and 5 elasticities (12, 21, 30, 48, and 91 kPa). hMSCs were seeded onto the array platform and allowed to attach for 16 – 20 hrs. The time window was chosen to ensure adequate cell attachment while minimizing any interference from cell division. After cell fixation and permeabilization, YAP localization was characterized by immunostaining. Cell shape and area were evaluated by phalloidin staining of the F-actin cytoskeleton. We excluded clustered cells from our analysis to minimize confounding effects of cell-cell interactions.
As expected, YAP localization was significantly affected by hydrogel stiffness and RGD density (Figure 3A–C). Stiff gels and high-RGD gels promoted YAP nuclear localization, shown as an overlap of YAP and DAPI staining (Figures 3A and S5). We quantified YAP nuclear/cytoplasmic (nuc/cyt) ratio for each individual cell by calculating the ratio of the average YAP intensity in the nucleus region and the average intensity in the cytoplasmic region of similar size that is adjacent to the nucleus. Some cells, especially those on soft gels with low RGD density, had rounded morphologies that made it challenging to discern the cytoplasm from the nucleus. These cells were thus excluded from the analysis. As stiffness increased from 12 to 91 kPa, YAP nuc/cyt ratio increased significantly on both low RGD and medium RGD gels (Figure 3B). On gels with high RGD density, while statistical differences between low and high stiffness still persisted, the gaps in their median values were narrowed. When stiffness was below 48 kPa, YAP was enriched in the nucleus as RGD density increased (Figure 3C). On 91 kPa gels, however, YAP localization was not sensitive to ligand density changes.
Figure 3.
YAP localized to hMSC nucleus as matrix stiffness and RGD density increased. (A) High stiffness/high RGD density promotes YAP nuclear localization. The nucleus is visualized with DAPI (blue). Arrows highlight cells with YAP nuc/cyt > 1.0. Scale bars: 50 μm. (B) Violin plot of YAP nuc/cyt ratios of individual cells on the array, grouped by RGD density. For each matrix condition, data was pooled from 3 independent experiments, as no systematic difference was observed among the triplicates. At least 40 cells were analyzed per matrix condition in each independent experiment, except on some low RGD or soft conditions where the overall number of hMSCs attached was under 40. (C) Violin plot of the same dataset as panel B, grouped by stiffness. a,b, c statistical families show p<0.05 from Kruskal-Wallis test for multiple comparison of non- normally distributed data. N equals to the total number of cells in the dataset. Horizontal lines represent Q1 - Q3.
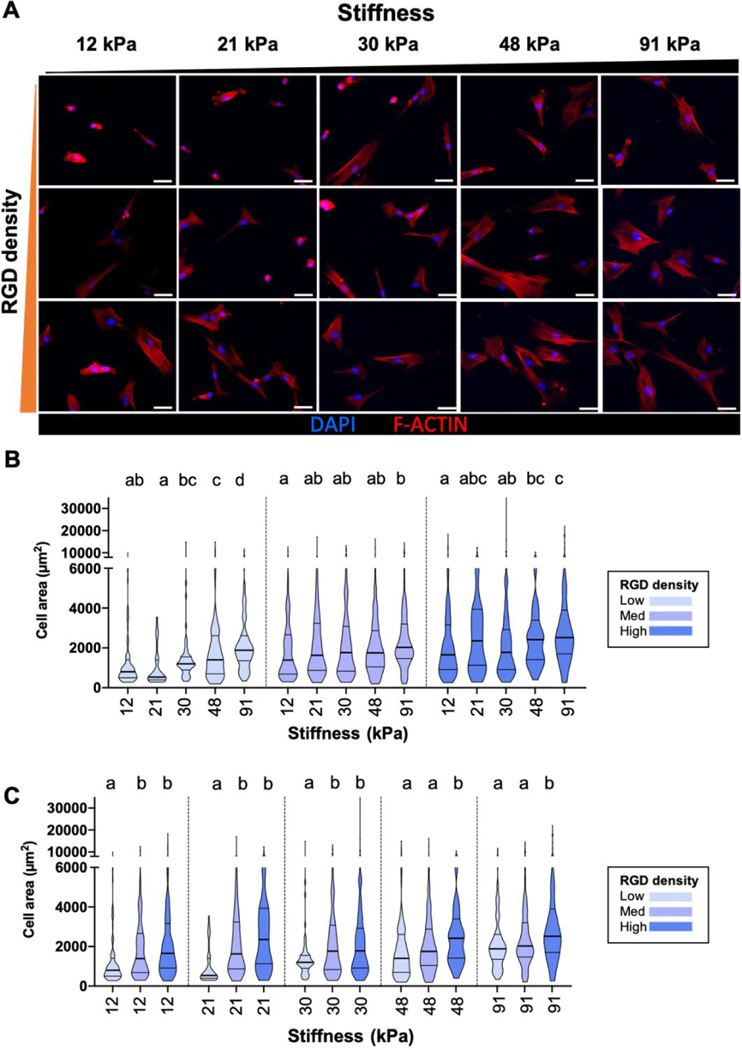
Further, hMSC spreading generally increased as stiffness and RGD density increased, along with the assembly of increasingly prominent stress fibers (Figure 4A). Stiffness dependent- spreading was seen on all three RGD densities (Figure 4B) and ligand density-dependent spreading was observed across all stiffness conditions (Figure 4C). Cell area is most sensitive to stiffness at lowest RGD density. As has been observed previously, ECM stiffness and ligand density synergistically facilitate hMSC spreading.5,22‘23 Thus, the array platform successfully recapitulates expected effects of ECM stiffness and adhesivity on YAP localization and cell spreading.
Figure 4.
hMSC spreading area increased as matrix stiffness and RGD density increased. (A) High stiffness/high RGD density promotes hMSC spreading. The F-actin network is visualized with phalloidin (red) and the nucleus is visualized with DAPI (blue). Scale bars: 50 μm. (B) Violin plot of cell area (μm2) of individual cells on the array, grouped by RGD density. For each matrix condition, data was pooled from 3 independent experiments, as we did not observe systematic difference between the triplicates. At least 40 cells were analyzed per matrix condition for each independent experiment, except on some low RGD or soft conditions where the overall number of hMSCs attached was under 40. (C) Violin plot of the same dataset as panel B, grouped by the same stiffness. a, b, c statistical families show p<0.05 from Kruskal-Wallis test for multiple comparison of non-normally distributed data. N equals to the total number of cells in the dataset. Horizontal lines represent Q1 - Q3.
Human Mesenchymal Stem Cell Differentiation on the Array
We next asked if the stiffness and RGD density gradients on the array platform could direct mechanosensitive hMSC lineage commitment. It has previously been observed that hMSCs have a strong tendency to differentiate into adipocytes on soft matrices, while stiff hydrogels largely promote osteogenesis.15,52–54 Using a gradient HA hydrogel with varying stiffness and fibronectin density, we also demonstrated that soft, low fibronectin density matrices biased hMSC toward adipogenesis, while stiff, high fibronectin density matrices supported osteogenesis.22 We therefore hypothesized that a similar trend would be observed on this multi-well array platform.
We first evaluated whether the array was permissive for hMSC adipogenesis and osteogenesis in the presence of the corresponding induction media. We exposed the cells to a 1:1 mixture of adipogenic and osteogenic differentiation media. After 7 days of differentiation, we analyzed adipogenesis and osteogenesis by quantifying lipid droplet area and alkaline phosphatase activity, respectively. On low and medium RGD gels, despite the initial attachment and spreading, the majority of hMSCs migrated to form cell clusters (Figure S6A), suggesting that cell-cell interactions may be replacing matrix effects. We did not observe cell migration-driven clustering on high RGD gels. The 1:1 mixed induction media induced both lineages simultaneously. Surprisingly, we did not see a strong stiffness-dependency of either adipogenic or osteogenic efficiency from 12 to 91 kPa on high RGD gels. Similar lipid droplet area and osteogenesis ALP activity were found on all stiffness conditions (Figure S6B–C). We also observed large variations of the adipogenesis across experimental replicates despite consistent gel fabrication (Figure S6B). Such batch-to-batch variation again pointed to the heterogeneity of hMSCs in their differentiation potency. We have also measured the expression levels of adipogenesis marker FABP4 and osteogenesis marker ALP across the stiffness range, yet we did not see any stiffness dependence of expression levels (data not shown).
Stiffness and Ligand Density Coupled to Modulate U-87 Mechanosensing and Morphology
We have previously shown that U-87 MG GBM cells adhere to HA via CD44 and that this interaction becomes an increasingly important mechanism of adhesion on matrices with low RGD density.44 Furthermore, we have shown that CD44/HA adhesion is intrinsically sensitive to matrix modulus.38 While both HA-CD44 binding and integrins serve as important elements of mechanical control, the interaction between these two signaling pathways and their influence on cell behavior remains poorly understood. Using our HA gel array, we systematically investigated the effects of stiffness and RGD density on cell phenotype and migration to further explore the interplay between HA matrix stiffness and RGD ligand concentration on U-87 cell behavior. We hypothesized that increased stiffness and RGD density would synergistically increase GBM cell spreading, adhesion and motility. We also hypothesized that cells lacking CD44 would display a decreased ability to attach, spread, and migrate on 2D HA hydrogels and that this phenotype that could be rescued on matrix with high stiffness and high RGD density.
To investigate the role of CD44 in GBM cell sensitivity to stiffness and RGD density gradients, we fabricated a 3×3 array with three different RGD densities (0.02, 0.15, and 0.5 mM RGD in the HA-RGD precursors) and 3 different elasticities (12, 30, and 91 kPa). We utilized our previously generated CD44 KO and non-targeting U-87 MG cells.44 We seeded U-87 cells onto the array platform and performed phase imaging to observe morphological changes of GBM cells on HA hydrogels of varying stiffness and RGD densities. Cell spreading was indirectly quantified by counting the number of round cells, which we defined as cells exhibiting a circular morphology and lacking visible protrusions when imaging with a 10X phase objective (Figure 5A). We observed that across gel conditions, CD44 KO cells generally had a higher percentage of round cells compared to non-targeting cells, supporting the hypothesis that CD44 supports cell spreading on HA. As we hypothesized, increasing stiffness and/or RGD density was associated with a decreasing percentage of round cells in both non-targeting and CD44 KO cells (Figures 5B and S7). Interestingly, on soft gels, CD44 KO cells displayed significantly more round cells than non-targeting cells on gels with low RGD density but had a similar fraction of round cells on gels with high RGD density (Figure 5C). There were no differences in the percentage of round cells across cell types on the stiffest gels, regardless of RGD density, suggesting that CD44-mediated signaling may influence cell spreading on soft matrices and conditions with low RGD density more than on stiff hydrogels or in conditions with high RGD density.
Figure 5.
U-87 spreading increases with increasing RGD density and stiffness. (A) Representative phase image of U-87 cells on HA hydrogels. Yellow circles outline cells labeled as round that have no visible protrusions using a 10X objective. Scale bar: 100 μm. (B) 3D column bar graph of cell spreading quantification for U-87 cells interacting with HA hydrogels of varying stiffness and RGD density. 6 fields of view were analyzed across three independent experiments (n = 6). (C) Cell spreading quantification for U-87 cells on HA hydrogels, a subset of the dataset presented in Figure 5B. a, b, and c represent statistical families with a significant difference of p<0.05 by ANOVA followed with Tukey-Kramer multiple comparisons test. Black lines represent mean and SD.
U-87 Adhesion on Soft HA Matrices is Enhanced by CD44-HA Binding
To more directly test the role of CD44 in facilitating cell adhesion on HA matrices of varying stiffness and RGD density, we measured cell adhesion on our gel array using a previously established centrifugal adhesion assay.38,44 We found that CD44 KO cells displayed reduced adhesion compared to non-targeting cells across all gel conditions, with 30 kPa high RGD and 91 kPa high RGD gels as the two exceptions (Figures 6A and S8). In contrast to the cell spreading trends, increasing RGD density did not rescue adhesion in the CD44 KO cells on soft gels (Figure 6B). Increasing RGD density on stiff gels, however, did rescue adhesion in cells lacking CD44, supporting our hypothesis that an abundance of integrin ligands on stiff HA hydrogels can compensate for the absence of CD44-based adhesions.
Figure 6.
Cell adhesion and migration are dependent on HA hydrogel stiffness, RGD density, and CD44 expression. (A) 3D column bar graph of centrifugal adhesion assay for U-87 cells interacting with HA hydrogels of varying stiffness and RGD ligand density (n = 6). (B) Centrifugal adhesion assay quantification for U-87 cells on HA hydrogels. n=6 total fields of view analyzed from three independent experiments. Black lines represent mean and SD. a, b, and c represent statistical families with a significant difference of p<0.05 by ANOVA followed with Tukey-Kramer multiple comparisons test. (C) 3D column bar graph showing random 2D migration speeds of U-87 cells on HA hydrogels with varying stiffness and RGD density (n=60). (D) Random 2D migration speeds of U-87 cells on HA hydrogels. n = 60 total cells analyzed from three independent experiments. a, b, and c represent statistical families with a significant difference of p<0.05 by Kruskal-Wallis test for multiple comparison of non-normally distributed data followed with Dunn’s multiple comparisons. Horizontal lines represent Q1 - Q3.
CD44-mediated Signaling is Important for U-87 Migration on Stiff HA Hydrogels
We next explored the relative roles of HA hydrogel stiffness and RGD density on 2D cell migration. We used time-lapse microscopy to track cell speed on our HA array platform 6 hours after seeding. Increasing stiffness and/or increasing RGD density increased cell migration speed across all cell lines (Figures 6C and S9). However, CD44 KO cells generally migrated more slowly than non-targeting cells for a given matrix modulus and RGD concentration. On the softest gels, increasing RGD density rescued migration of CD44 KO cells relative to non-targeting cells (Figure 6D). Interestingly, CD44 KO cells migrated more slowly than non-targeting cells on the stiffest hydrogels across all RGD densities tested. These findings suggest a role for CD44 in facilitating cell migration on stiff HA hydrogels, even when ample integrin ligand is present.
DISCUSSION
We have developed a multi-well 2D hydrogel array platform that allows parallel culture and analysis of cells on various combinations of matrix stiffness and adhesivity. We successfully incorporated orthogonal stiffness and ligand density gradients on the array by programmable UV photopatterning. Proof-of-principle studies of human MSCs validate that the platform can recapitulate expected stiffness and ligand density-sensitivity of YAP localization and spreading. We further applied the platform to reveal context-dependent contributions of CD44 on GBM cell spreading, adhesion, and migration.
A common approach to develop multi-well hydrogel array platforms has been to fabricate hydrogel precursors directly within wells and then crosslink them in situ.32–34 Hydrogels may also be crosslinked outside of the multi-well plates, cut into desired shapes, and later affixed to the multi-well plate.55,56 While these approaches have proven enormously valuable, it can be challenging to use these methods to vary multiple gel parameters in a single plate, since each gel must be individually fabricated. Our system provides an alternative approach that involves fabricating the gel array initially on a glass coverslip and then taping the substrate to a bottomless well plate. This aspect of our platform shares some conceptual connections with a previous PDMS screening platform in which various micro- and nano-topographies were presented in a standard multi-well format.57 An important innovation of our approach is the use of laser-cut double-sided tape to both pattern the array and anchor the multi-well chamber to the device floor. Importantly, the laser-cut pattern is fully customizable and can accommodate hydrogel arrays of arbitrary dimensions and shapes depending on the multi-well plate format used. The platform also highlights the application of a programmable LAVA device, which allows for more precise tuning of photopatterning conditions than photomasks, as UV intensity and exposure time can be adjusted in a continuous manner.41,42 Further, LEDs on the LAVA device are expected to produce extremely consistent intensity from one use to another, maximize adsorption efficiency and reduce batch-to-batch variation of stiffness patterning compared to traditional broad-spectrum UV light sources.
The array platform captured the synergic effect of ECM stiffness and RGD peptide density on promoting YAP nuclear localization and cell spreading. In addition, our observation that YAP localization is insensitive to stiffness (12–48 kPa) on low RGD gels echoes recent reports that YAP localization was insensitive to stiffness at very high or low fibronectin density,49 as both ligand density and stiffness induce YAP translocation to cytoskeletal tension and aVp3-integrin adhesion through similar mechanisms. For similar reasons, YAP localization is insensitive to ligand density on very stiff gels (91 kPa). Interestingly, YAP nuc/cyt ratio and cell area of hMSCs did not follow a normal distribution under all conditions but resembled long-tail distributions. Even on hydrogels with the highest stiffness and ligand density, there is a subpopulation of hMSCs that favors cytoplasmic localization of YAP or is not well spread. Also, a small percentage of cells on soft, low RGD density gels are still capable of spreading or have YAP enriched in the nucleus. The non-normal distribution may reflect intrinsic heterogeneities within the hMSC population, as has been observed elsewhere.58
This study also provides insight into the role of CD44 in U-87 cell spreading, adhesion and motility. While both CD44 KO and non-targeting U-87 cells appeared sensitive to matrix stiffness and RGD density on our HA gel array, CD44 KO cells were generally less spread and weakly adhered than non-targeting cells. As expected, the deficiency of CD44 KO cells was exacerbated on gels with low modulus and RGD density. However, increasing RGD density and/or stiffness generally restored cell spreading and adhesion to that of non-targeting cells. Interestingly, on soft matrices with high RGD density, CD44 KO cells displayed decreased adhesion strength to HA-hydrogels despite a classical “spread” 2D morphology. Previous studies have demonstrated that CD44 can complement and potentiate signaling from other surface receptors, including RGD- specific integrins, to drive cell spreading.59,60 In this study, CD44 KO cells displayed decreased adhesion on soft matrices regardless of RGD density, highlighting the importance of both CD44- and integrin-based adhesions in particular matrix conditions.
While cell migration speeds on our HA gel array generally increased with increasing substrate stiffness and RGD density, gel conditions facilitating the highest cell adhesion did not support the fastest cell migration. These results are consistent with the well-known biphasic dependence of cell migration on the strength of adhesion to the surrounding ECM.61,62 Furthermore, a previous study demonstrated a biphasic relationship between CD44 protein expression and cell migration rates, with intermediate CD44 expression leading to the fastest overall motility in cell culture and poorest survival in human GBM.63 On our platform, CD44 KO cells generally exhibited slower migration speeds than non-targeting cells across gel conditions. However, increasing RGD density restored migration speeds of CD44 KO cells to that of nontargeting cells on the softest gels, but not on the stiffest gels. Previous work from our laboratory has demonstrated that CD44 is sufficient to drive the formation of tension-bearing protrusions (microtentacles) that enable motility in the absence of integrin ligands.44 Using our HA array platform, we found that CD44 is required to effectively migrate on stiff substrates, further highlighting the importance of CD44-HA interactions in the generation of cellular tension. Thus, the importance of CD44 in cell behavior appears to be context-dependent; on soft HA matrices, CD44 is particularly important for cell adhesion and on stiff HA matrices it is important for cell migration.
One surprising result from our study is that while gels in our platform were permissive of hMSC osteogenic and adipogenic differentiation, we did not observe an appreciable matrix-dependent bias in lineage commitment. There are a number of potential explanations for this result. First, the soluble factors in the induction medium may have overridden the effects of matrix stiffness and thus induced significant adipogenesis on conditions as stiff as 91 kPa. We expect that a more comprehensive consideration of adhesive ligands and media formulation would produce a regime in which one sees stiffness-dependent differentiation. Second, despite seeding identical numbers of cells on each gel, we could not control cell density and cell clustering throughout the course of the experiment. For example, stiff gels routinely resulted in ~20% higher cell density on day 7 of differentiation, presumably due to greater proliferation and/or increased attachment on these gels. High cell density, which is expected to reduce average cell area and promote adipogenesis,64,65 may therefore have partially rescued adipogenesis on stiff gels. Third, the softest hydrogels (12 kPa) may still be too stiff to significantly suppress osteogenesis or stimulate adipogenesis. Previous reports on various hydrogel platforms suggested an optimal modulus range for adipogenesis from several hundred pascals to 5 kPa.22,29,54 The lowest stiffness (12 kPa) in our study was based on a DTT to HA monomer ratio of 0.15, and lower ratio of DTT crosslinker may cause gel swelling and compromise cell imaging qualities. However, the “base” gel stiffness may be further reduced by optimizing gel composition, HA degree of functionalization, or crosslinker types. Lastly, the broad distributions of hMSC phenotypes for a given matrix condition may reflect intrinsic heterogeneities within the hMSC population. The lack of an expected stiffness-dependence differentiation on the gel array platform could be a consequence of the heterogeneity that exists between MSC donors, tissue sources, culture methods, and even individual cells, as observed elsewhere.58
The fabrication process of the array platform has significant room for further optimization. For example, the RGD density gradient in this system was achieved by manually mixing HA and RGD in various ratios, which may become prohibitively labor-intensive for particularly parallelized arrays. This limitation could be addressed in the future by adapting dual photopatterning as described previously,22,23,25 where the ECM ligand is conjugated to the backbone by photoreaction. An additional area with room for improvement is greater compatibility with dynamic, high-resolution imaging modalities. While we successfully measured cell migration speeds using low-magnification phase-contrast live imaging, higher-magnification imaging with shorter-working distance objectives would likely require a thinner cover glass and/or thinner double-sided tape.
Despite these limitations, we envision that our platform could be adapted to a wide range of materials and applications beyond HA-based hydrogels since this platform could in principle accommodate any hydrogel formulation that can be affixed to glass, such as polyacrylamide- and poly(ethylene-glycol) (PEG)-based hydrogels. Moreover, combinatorial gradients of additional cell adhesion ligands and other ECM parameters (e.g. viscoelasticity, topography) could also potentially be incorporated to the platform through judicious choice of materials and photopatterning strategy. Changing the hydrogel composition, degree of functionalization, and crosslinking modalities may further extend the upper and lower limits of hydrogel stiffness.
Here we describe a multi-well hydrogel platform to investigate the complex and interdependent relationships of two biophysical cues, matrix stiffness and integrin-based adhesivity, on cell behavior. Our strategy utilizes a programmable light illumination system to fabricate gels with defined stiffness and adhesivity conditions. The platform allows individual treatment and analysis of matrix conditions while eliminating contributions of haptotaxis and durotaxis. The HA gel array recapitulates expected relationships between matrix stiffness, adhesivity and cell mechanosensing in hMSCs. Additionally, we further applied the platform to reveal context-dependent contributions of CD44 in GBM cell spreading, adhesion and migration. We anticipate that our system could bear great value for mechanistic discovery and drug screening where matrix mechanics and adhesivity are expected to influence phenotype.
CONCLUSIONS
We have developed a combinatorial HA-based hydrogel array in a standard multi-well plate format, with orthogonal gradients of matrix elasticity and adhesive ligand density. These gels support the osteogenic and adipogenic differentiation of adipose-derived hMSCs, with cell spreading and YAP localization strongly responding to both matrix stiffness and RGD peptide density. Our array platform also reveals a context-dependent role of CD44 in GBM cell mechanosensitive behavior. The deployment of ECM-mimetic hydrogels in a familiar, user- friendly, and parallelized format should facilitate mechanistic discovery and screening applications.
Supplementary Material
Acknowledgments
FUNDING SOURCES
This work was supported by awards from the National Institutes of Health (R01GM122375 to SK, R01NS074831 to SK and DVS, R01DK118940 to AS and SK, and R01CA227136 to SK) and the National Science Foundation (Graduate Research Fellowship to EAA).
Footnotes
SUPPORTING INFORMATION
Supporting Information. Me-HA reaction scheme and 1H-NMR spectrum, PDMS acrylic mold fabrication, dye leakage experiment, elastic modulus of RGD-modified and unmodified HA hydrogels, digitally magnified YAP localization images, hMSC adipogenesis and osteogenesis characterization, U-87 cell spreading quantification, U-87 centrifugal adhesion quantification, U-87 2D random migration quantification.
REFERENCES
- 1.Vining KH & Mooney DJ Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol 18, 728–742 (2017). 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei SC & Yang J. Forcing through Tumor Metastasis: The Interplay between Tissue Rigidity and Epithelial-Mesenchymal Transition. Trends Cell Biol. 26, 111–120 (2016). 10.1016/Zj.tcb.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J. & Kumar S. Biophysical Regulation of Cancer Stem/Initiating Cells: Implications for disease mechanisms and translation. Curr. Opin. Biomed. Eng 87–95 (2018). doi:10.1016/j.cobme.2017.02.006.Biophysical https://doi.org/10.1016/j.cobme.2017.02.006.Biophysical. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Upadhyaya A. Mechanosensing in the immune response. Semin. Cell Dev. Biol 71, 137–145 (2017). 10.1016/j.semcdb.2017.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engler A, Bacakova L, Newman C, Hategan A, Griffin M. & Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys. J 86, 617–628 (2004). 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong Z, Szczesny SE, Caliari SR, Charrier EE, Chaudhuri O, Cao X, Lin Y, Mauck RL, Janmey PA, Burdick JA & Shenoy VB Matching material and cellular timescales maximizes cell spreading on viscoelastic substrates. Proc. Natl. Acad. Sci 115, E2686 LP-E2695 (2018). 10.1073/pnas.1716620115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Q, Ge L, Guimaraes CF, Kuhn PT, Yang L. & van Rijn P. Development of a Novel Orthogonal Double Gradient for High-Throughput Screening of Mesenchymal Stem Cells-Materials Interaction. Adv. Mater. Interfaces 5, 4–11 (2018). 10.1002/admi.201800504. [DOI] [Google Scholar]
- 8.Hou Y, Yu L, Xie W, Camacho LC, Zhang M, Chu Z, Wei Q. & Haag R. Surface Roughness and Substrate Stiffness Synergize To Drive Cellular Mechanoresponse. Nano Lett. 20, 748–757 (2020). 10.1021/acs.nanolett.9b04761. [DOI] [PubMed] [Google Scholar]
- 9.Darnell M, Gu L. & Mooney D. RNA-seq reveals diverse effects of substrate stiffness on mesenchymal stem cells. Biomaterials 181, 182–188 (2018). 10.1016/j.biomaterials.2018.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Liu R, Gu Y. & Ding J. Nonmonotonic Self-Deformation of Cell Nuclei on Topological Surfaces with Micropillar Array. ACS Appl. Mater. Interfaces 9, 18521–18530 (2017). 10.1021/acsami.7b04027. [DOI] [PubMed] [Google Scholar]
- 11.Geckil H, Xu F, Zhang X, Moon S. & Demirci U. Engineering hydrogels as extracellular matrix mimics. Nanomedicine (Lond). 5, 469–484 (2010). 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J. & Marchant RE Design properties of hydrogel tissue-engineering scaffolds. Expert Rev. Med. Devices 8, 607–626 (2011). 10.1586/erd.11.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyburz KA & Anseth KS Synthetic mimics of the extracellular matrix: how simple is complex enough? Ann. Biomed. Eng 43, 489–500 (2015). 10.1007/s10439-015-1297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing H, Lee H, Luo L. & Kyriakides TR Extracellular matrix-derived biomaterials in engineering cell function. Biotechnol. Adv 42, 107421 (2020). 10.1016/j.biotechadv.2019.107421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J. & Mooney DJ Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat. Mater 9, 518–526 (2010). 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H-P, Lippens E, Duda GN & Mooney DJ Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater 15, 326–334 (2016). 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia T, Liu W. & Yang L. A review of gradient stiffness hydrogels used in tissue engineering and regenerative medicine. J. Biomed. Mater. Res. A 105, 1799–1812 (2017). 10.1002/jbm.a.36034. [DOI] [PubMed] [Google Scholar]
- 18.Rowlands AS, George PA & Cooper-White JJ Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am. J. Physiol. Cell Physiol 295, C1037–44 (2008). 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 19.Kilian KA & Mrksich M. Directing Stem Cell Fate by Controlling the Affinity and Density of Ligand-Receptor Interactions at the Biomaterials Interface. Angew. Chemie Int. Ed 51, 4891–4895 (2012). 10.1002/anie.201108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson MJ, Jiang Y, Yanez-Soto B, Liliensiek S, Murphy WL & Nealey PF Arrays of topographically and peptide-functionalized hydrogels for analysis of biomimetic extracellular matrix properties. J. Vac. Sci. Technol. B 30, 06F903 (2012). 10.1116/L4762842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Z, Hu K, Sun J, Zhang T, Zhang Q, Song L, Zhang X. & Gu N. Fabrication of Hydrogel with Cell Adhesive Micropatterns for Mimicking the Oriented Tumor- Associated Extracellular Matrix. ACS Appl. Mater. Interfaces 6, 10963–10968 (2014). 10.1021/am5023946. [DOI] [PubMed] [Google Scholar]
- 22.Rape AD, Zibinsky M, Murthy N. & Kumar S. A synthetic hydrogel for the high-throughput study of cell-ECM interactions. Nat. Commun 6, (2015). 10.1038/ncomms9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong X, Jiang J, Zhu D. & Yang F. Hydrogels with Dual Gradients of Mechanical and Biochemical Cues for Deciphering Cell-Niche Interactions. ACS Biomater. Sci. Eng 2, 845–852 (2016). 10.1021/acsbiomaterials.6b00074. [DOI] [PubMed] [Google Scholar]
- 24.Hansen TD, Koepsel JT, Le NN, Nguyen EH, Zorn S, Parlato M, Loveland SG, Schwartz MP & Murphy WL Biomaterial arrays with defined adhesion ligand densities and matrix stiffness identify distinct phenotypes for tumorigenic and nontumorigenic human mesenchymal cell types. Biomater. Sci 2, 745–756 (2014). 10.1039/C3BM60278H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vega SL, Kwon MY, Song KH, Wang C, Mauck RL, Han L. & Burdick JA Combinatorial hydrogels with biochemical gradients for screening 3D cellular microenvironments. Nat. Commun 9, 614 (2018). 10.1038/s41467-018-03021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Policastro GM, Li Q, Zheng J, Jacquet R, Landis WJ & Becker ML Concentration-Dependent hMSC Differentiation on Orthogonal Concentration Gradients of GRGDS and BMP-2 Peptides. Biomacromolecules 17, 1486–1495 (2016). 10.1021/acs.biomac.6b00088. [DOI] [PubMed] [Google Scholar]
- 27.Guermani E, Shaki H, Mohanty S, Mehrali M, Arpanaei A, Gaharwar AK & Dolatshahi-Pirouz A. Engineering complex tissue-like microgel arrays for evaluating stem cell differentiation. Sci. Rep 6, 30445 (2016). 10.1038/srep30445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson TC, Mathew S, Candiello JE, Goh SK, Kumta PN & Banerjee I. Development of an Alginate Array Platform to Decouple the Effect of Multiparametric Perturbations on Human Pluripotent Stem Cells During Pancreatic Differentiation. Biotechnol. J 13, 1–11 (2018). 10.1002/biot.201700099. [DOI] [PubMed] [Google Scholar]
- 29.Hadden WJ, Young JL, Holle AW, McFetridge ML, Kim DY, Wijesinghe P, Taylor-Weiner H, Wen JH, Lee AR, Bieback K, Vo B-N, Sampson DD, Kennedy BF, Spatz JP, Engler AJ & Choi YS Stem cell migration and mechanotransduction on linear stiffness gradient hydrogels. Proc. Natl. Acad. Sci. U. S. A 114, 5647–5652 (2017). 10.1073/pnas.1618239114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolatshahi-Pirouz A, Nikkhah M, Gaharwar AK, Hashmi B, Guermani E, Aliabadi H, Camci-Unal G, Ferrante T, Foss M, Ingber DE & Khademhosseini A. A combinatorial cell-laden gel microarray for inducing osteogenic differentiation of human mesenchymal stem cells. Sci. Rep 4, 3896 (2014). 10.1038/srep03896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A. & Lutolf MP 3D niche microarrays for systems-level analyses of cell fate. Nat. Commun 5, 4324 (2014). 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mih JD, Sharif AS, Liu F, Marinkovic A, Symer MM & Tschumperlin DJ A multiwell platform for studying stiffness-dependent cell biology. PLoS One 6, 1–10 (2011). 10.1371/journal.pone.0019929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Díaz-Bello B, Monroy-Romero AX, Pírez-Calixto D, Zamarrón-Hernández D, Serna-Marquez N, Vázquez-Victorio G. & Hautefeuille M. Method for the Direct Fabrication of Polyacrylamide Hydrogels with Controlled Stiffness in Polystyrene Multiwell Plates for Mechanobiology Assays. ACS Biomater. Sci. Eng 5, 4219–4227 (2019). 10.1021/acsbiomaterials.9b00988. [DOI] [PubMed] [Google Scholar]
- 34.Hribar KC & Buckley P. Apparatus for patterning hydrogels into multi-well plates. 1, (2018). [Google Scholar]
- 35.Highley CB, Prestwich GD & Burdick JA Recent advances in hyaluronic acid hydrogels for biomedical applications. Curr. Opin. Biotechnol 40, 35–40 (2016). 10.1016/jxopbio.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Wolf KJ & Kumar S. Hyaluronic Acid: Incorporating the Bio into the Material. ACS Biomater. Sci. Eng 5, 3753–3765 (2019). 10.1021/acsbiomaterials.8b01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ananthanarayanan B, Kim Y. & Kumar S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials 32, 7913–7923 (2011). 10.10167/j.biomaterials.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y. & Kumar S. CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility. Mol. Cancer Res 12, 1416–1429 (2014). 10.1158/1541-7786.MCR-13-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim IL, Khetan S, Baker BM, Chen CS & Burdick JA Fibrous hyaluronic acid hydrogels that direct MSC chondrogenesis through mechanical and adhesive cues. Biomaterials 34, 5571–5580 (2013). 10.1016/j.biomaterials.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cosgrove BD, Mui KL, Driscoll TP, Caliari SR, Mehta KD, Assoian RK, Burdick JA & Mauck RL N-cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nat. Mater 15, 1297–1306 (2016). 10.1038/nmat4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Repina NA, McClave T, Johnson HJ, Bao X, Kane RS & Schaffer DV Engineered Illumination Devices for Optogenetic Control of Cellular Signaling Dynamics. Cell Rep. 31, 107737 (2020). 10.1016/jxelrep.2020.107737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Repina NA, Johnson HJ, McClave T, Kane RS & Schaffer DV Protocol to Fabricate Engineered Illumination Devices for Optogenetic Control of Cellular Signaling Dynamics. STAR Protoc. 1, 100141 (2020). 10.1016/Zj.xpro.2020.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacKay JL & Kumar S. Measuring the Elastic Properties of Living Cells with Atomic Force Microscopy Indentation BT - Cell Imaging Techniques: Methods and Protocols. in (eds. Taatjes DJ & Roth J) 313–329 (Humana Press, 2013). doi:10.1007/978-1-62703-056-4_15 https://doi.org/10.1007/978-1-62703-056-4_15. [DOI] [PubMed] [Google Scholar]
- 44.Wolf KJ, Shukla P, Springer K, Lee S, Coombes JD, Choy CJ, Kenny SJ, Xu K. & Kumar S. A mode of cell adhesion and migration facilitated by CD44- dependent microtentacles. Proc. Natl. Acad. Sci. U. S. A 117, (2020). 10.1073/pnas.1914294117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engler AJ, Sen S, Sweeney HL & Discher DE Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 126, 677–689 (2006). 10.10167/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 46.Nemir S. & West JL Synthetic materials in the study of cell response to substrate rigidity. Ann. Biomed. Eng 38, 2–20 (2010). 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- 47.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N. & Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–184 (2011). 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 48.Das A, Fischer RS, Pan D. & Waterman CM YAP nuclear localization in the absence of cell-cell contact is mediated by a filamentous actin-dependent, Myosin Iland Phospho-YAP-independent pathway during extracellular matrix mechanosensing. J. Biol. Chem 291, 6096–6110 (2016). 10.1074/jbc.M115.708313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanton AE, Tong X, Lee S. & Yang F. Biochemical Ligand Density Regulates Yes- Associated Protein Translocation in Stem Cells through Cytoskeletal Tension and Integrins. ACS Appl. Mater. & interfaces 11, 8849–8857 (2019). 10.1021/acsami.8b21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park JS, Chu JS, Tsou AD, Diop R, Tang Z, Wang A. & Li S. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-p. Biomaterials 32, 3921–3930 (2011). 10.1016/j.biomaterials.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JP, Kassianidou E, MacDonald JI, Francis MB & Kumar S. N-terminal specific conjugation of extracellular matrix proteins to 2-pyridinecarboxaldehyde functionalized polyacrylamide hydrogels. Biomaterials 102, 268–276 (2016). 10.1016/j.biomaterials.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao W, Li X, Liu X, Zhang N. & Wen X. Effects of substrate stiffness on adipogenic and osteogenic differentiation of human mesenchymal stem cells. Mater. Sci. Eng. C 40, 316–323 (2014). 10.1016/j.msec.2014.03.048. [DOI] [PubMed] [Google Scholar]
- 53.Ye K, Wang X, Cao L, Li S, Li Z, Yu L. & Ding J. Matrix Stiffness and Nanoscale Spatial Organization of Cell-Adhesive Ligands Direct Stem Cell Fate. Nano Lett. 15, 4720–4729 (2015). 10.1021/acs.nanolett.5b01619. [DOI] [PubMed] [Google Scholar]
- 54.Lee J, Abdeen AA, Tang X, Saif TA & Kilian KA Matrix directed adipogenesis and neurogenesis of mesenchymal stem cells derived from adipose tissue and bone marrow. Acta Biomater. 42, 46–55 (2016). 10.1016/Zj.actbio.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Semler EJ, Lancin PA, Dasgupta A. & Moghe PV Engineering hepatocellular morphogenesis and function via ligand-presenting hydrogels with graded mechanical compliance. Biotechnol. Bioeng 89, 296–307 (2005). 10.1002/bit.20328. [DOI] [PubMed] [Google Scholar]
- 56.Syed S, Karadaghy A. & Zustiak S. Simple polyacrylamide-based multiwell stiffness assay for the study of stiffness-dependent cell responses. J. Vis. Exp 2015, 1–12 (2015). 10.3791/52643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu J, Gondarenko AA, Dang AP, Bashour KT, O’Connor RS, Lee S, Liapis A, Ghassemi S, Milone MC, Sheetz MP, Dustin ML, Kam LC & Hone JC High-Throughput Mechanobiology Screening Platform Using Micro- and Nanotopography. Nano Lett. 16, 2198–2204 (2016). 10.1021/acs.nanolett.5b04364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson A, Hodgson-Garms M, Frith JE & Genever P. Multiplicity of mesenchymal stromal cells: Finding the right route to therapy. Front. Immunol 10, 1–8 (2019). 10.3389/fimmu.2019.01112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chopra A, Murray ME, Byfield FJ, Mendez MG, Halleluyan R, Restle DJ, Raz-Ben Aroush D, Galie PA, Pogoda K, Bucki R, Marcinkiewicz C, Prestwich GD, Zarembinski TI, Chen CS, Puré E, Kresh JY & Janmey PA Augmentation of integrin-mediated mechanotransduction by hyaluronic acid. Biomaterials 35, 71–82 (2014). 10.1016/j.biomaterials.2013.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mandal K, Raz-Ben Aroush D, Graber ZT, Wu B, Park CY, Fredberg JJ, Guo W, Baumgart T. & Janmey PA Soft Hyaluronic Gels Promote Cell Spreading, Stress Fibers, Focal Adhesion, and Membrane Tension by Phosphoinositide Signaling, Not Traction Force. ACS Nano 13, 203–214 (2019). 10.1021/acsnano.8b05286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA & Horwitz AF Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness [published erratum appears in Nature 1997 Jul 10;388(6638):210]. Nature 385, 537–540 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Pathak A. & Kumar S. From molecular signal activation to locomotion: An integrated, multiscale analysis of cell motility on defined matrices. PLoS One 6, (2011). 10.1371/journal.pone.0018423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klank RL, Decker Grunke SA, Bangasser BL, Forster CL, Price MA, Odde TJ, SantaCruz KS, Rosenfeld SS, Canoll P, Turley EA, McCarthy JB, Ohlfest JR & Odde DJ Biphasic Dependence of Glioma Survival and Cell Migration on CD44 Expression Level. Cell Rep. 18, 23–31 (2017). 10.1016/jxelrep.2016.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McBeath R, Pirone DM, Nelson CM, Bhadriraju K. & Chen CS Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment. Dev. Cell 6, 483–495 (2004). 10.1016/S1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 65.Wiesner M, Berberich O, Hoefner C, Blunk T. & Bauer-Kreisel P. Gap junctional intercellular communication in adipose-derived stromal/stem cells is cell density-dependent and positively impacts adipogenic differentiation. J. Cell. Physiol 233, 3315–3329 (2018). 10.1002/jcp.26178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.