Abstract
Circulating tumor cell (CTC) clusters are present in cancer patients with severe metastasis, resulting in poor clinical outcomes. However, CTC clusters have not been studied as extensively as single CTCs, and the clinical utility of CTC clusters remains largely unknown. In this study, we aim sought to explore the feasibility of NanoVelcro Chips to simultaneously detect both single CTCs and CTC clusters with negligible perturbation to their intrinsic properties in neuroendocrine tumors (NETs). We discovered frequent CTC clusters in patients with advanced NETs and examined their potential roles, together with single NET CTCs, as novel biomarkers of patient response following peptide receptor radionuclide therapy (PRRT). We observed dynamic changes in both total NET CTCs and NET CTC cluster counts in NET patients undergoing PRRT which correlated with clinical outcome. These preliminary findings suggest that CTC clusters, along with single CTCs, offer a potential non-invasive option to monitor the treatment response in NET patients undergoing PRRT.
Keywords: Circulating tumor cells, Circulating tumor cell clusters, Nanostructured substrates, Neuroendocrine tumor, Peptide receptor radionuclide therapy
1. Introduction
As one of the major sources of liquid biopsy (Crowley et al., 2013), circulating tumor cells (CTCs) (Pantel et al., 2010; Speicher and Pantel, 2014) are cancer cells that escape from either primary or metastatic sites into blood circulation; thus, CTCs are often regarded as the cellular origin of new metastatic lesions. While detecting and enumerating (Cristofanilli et al., 2004; Riethdorf et al., 2007; Shaffer et al., 2007) single CTCs has demonstrated prognostic value in several types of solid tumors, it was recently reported that once clustered CTCs were observed in blood, cancer patients would almost certainly experience severe metastasis (Aceto et al., 2014) and poor prognostic outcomes (Hou et al., 2012). With this in mind, we envision that greater diagnostic, prognostic, and clinical utilities could be achieved by synergizing the potentials of both single CTCs and CTC clusters. However, existing technologies are mainly designed and optimized for separately detecting single CTCs (Shields et al., 2015) and CTC clusters (Sarioglu et al., 2015) because of the challenges in retaining the intrinsic properties (e.g., morphology and intactness) of single CTCs and CTC clusters simultaneously. Therefore, there is an unmet need to develop new technologies capable of detecting both single CTCs and CTC clusters while preserving their intrinsic properties.
Our group pioneered the concept of “NanoVelcro” Chips (Hsiao et al., 2014; Lin et al., 2014; Sekine et al., 2011; Wang et al., 2009; Zhang et al., 2012), where immunoaffinity agent-coated nanostructured substrates substantially improved “stickiness”, allowing for selective capture of CTCs and other types of circulating rare cells (e.g., circulating nucleated fetal cells (Hou et al., 2017; Afshar et al., 2021)) from blood. Conceptually, NanoVelcro Assays are analogous to Velcro™: the CTC surfaces covered with nanoscale cell-surface components (a.k.a., microvilli) and the substrate embedded with nanostructures can be regarded as the upper and lower strips of a Velcro fastener. When a CTC comes in contact with the substrate, the microvilli entangle with the nanostructures, dramatically increasing contact surface area to facilitate immunoaffinity agent-mediated CTC capture. By innovating previous designs over the past decade, we have created five generations of NanoVelcro Assays (Chen et al., 2016; Dong et al., 2020; Jan et al., 2018), each with different clinical utilities. Potential clinical utilities of the NanoVelcro CTC enumeration assay were demonstrated in disease staging and prognostic prediction for prostate cancer (Chen et al., 2015; Lu et al., 2013), pancreatic cancer (Court et al., 2015; Court et al., 2018a,b), and hepatocellular carcinoma (Court et al., 2018a,b). Up to this point, however, the feasibility of applying NanoVelcro Chips for enrichment and detection of CTC clusters remained unexplored.
Recognizing the aforementioned unmet need and the general applicability of our NanoVelcro Chips, we explored the applicability of NanoVelcro Chips for simultaneous enrichment and detection of both single CTCs and CTC clusters with negligible perturbation to their intrinsic properties, such as intactness and size distribution (Fig. 1). In this study, neuroendocrine tumors (NETs) were chosen as the disease model since we had previously observed the presence of both single CTCs and CTC clusters in many blood samples collected from advanced NET patients (Supplementary Fig. S1). In light of the reduction in EpCAM expression that is known to result from CTC cluster/sphere formation (Hyun et al., 2016), we first demonstrated that the performance of the NanoVelcro Assay is negligibly affected by the EpCAM expression levels of CTCs. Using artificial CTC cluster samples, we examined how the flow rate affected the capture efficiency of single cells and clusters of various sizes, as well as their spatial distribution within the three microchannels of a NanoVelcro Chip. We also verified the necessity of herringbone patterns in the PDMS chaotic mixer and embedded nanostructures in the anti-EpCAM-grafted SiNWS to achieve the expected capture performance. Using an optimal flow rate of 0.5 mL/h, we utilized NanoVelcro Chips for the enrichment and detection of both NET CTC clusters and single NET CTCs in a series of blood samples (n = 82, Table S1) collected from 21 patients with a variety of advanced NETs before and across four cycles of peptide receptor radionuclide therapy (PRRT). We observed dynamic changes in both total NET CTC and NET CTC cluster count in all NET patients undergoing PRRT. These results exhibited strong correlations with clinical responses in a subset of examined patients.
Fig. 1.
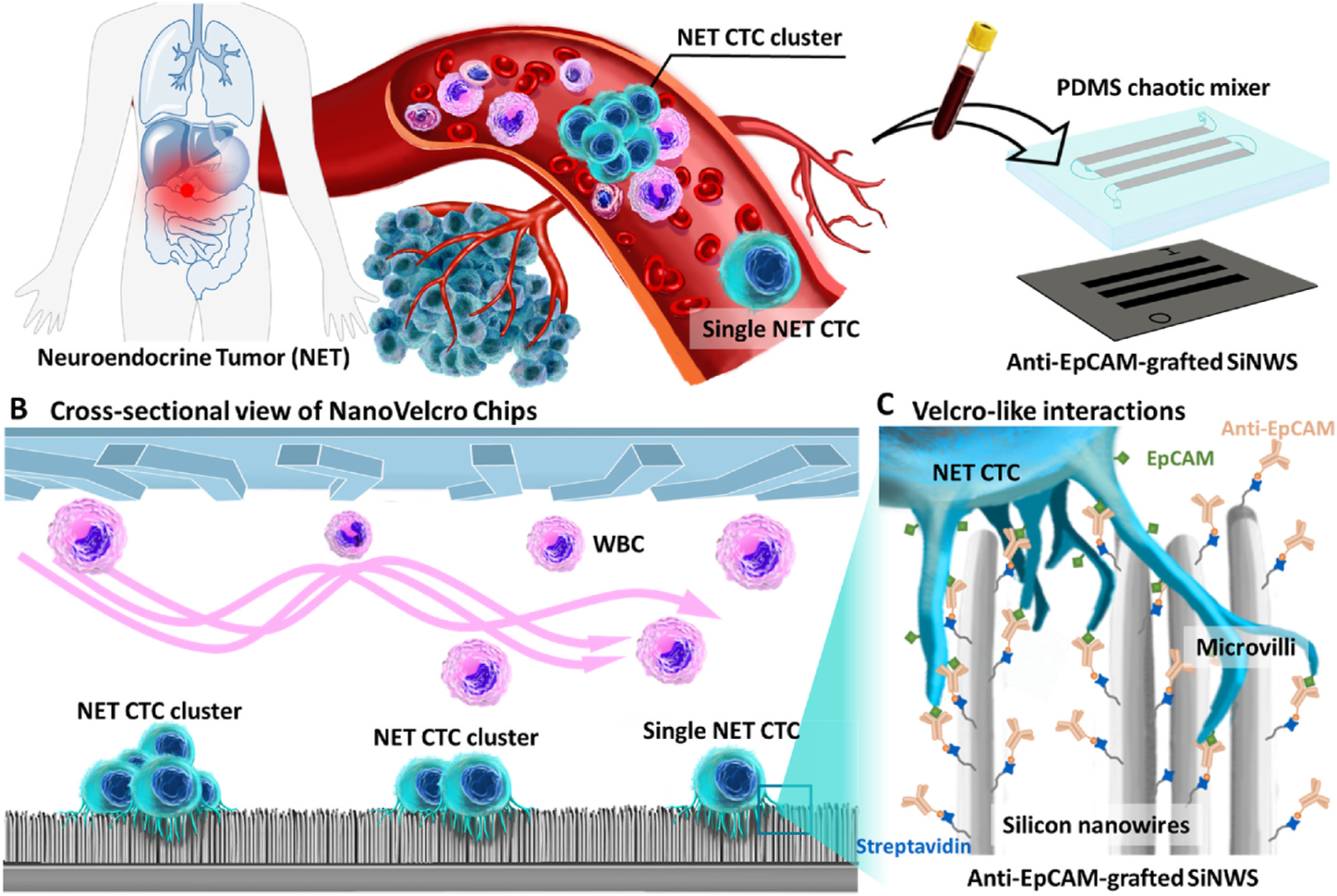
NanoVelcro Chips for enrichment and detection of both single CTCs and CTC clusters in NET patients’ blood. A) The majority of patients with advanced NETs have both CTCs and CTC clusters present in their peripheral blood. B) Cross-sectional view of NanoVelcro Chips. C) “Velcro-like” interactions between nanoscale cell-surface components (a.k.a., microvilli) and the anti-EpCAM-grafted SiNWS dramatically increase contact surface area, facilitating immunoaffinity-mediated capture of single NET CTCs and NET CTC clusters.
Neuroendocrine tumors are rare and arise from the diffuse neuroendocrine system. Clinically, they are often challenging to manage due to their late presentation with extensive metastasis and limited treatment options. PRRT is a promising molecular targeted radiotherapy recently approved for treating advanced gastroenteropancreatic NETs after failure of other therapies (Hicks et al., 2017; Kwekkeboom et al., 2005). Current imaging modalities and serum biomarkers used in NETs have limited effectiveness in monitoring disease status and treatment outcomes. NET CTCs offer a potential non-invasive solution for monitoring patients undergoing therapy, including PRRT. By combining the potentials of both single NET CTCs and NET CTC clusters, we hypothesized that such non-invasive information could complement existing imaging modalities and enable better monitoring of advanced NETs.
2. Experimental section
2.1. Fabrication of NanoVelcro Chips
NanoVelcro Chip is composed of two functional components: (i) a lithographically patterned SiNWS (grafted with anti-EpCAM capture agent) and (ii) a PDMS chaotic mixer. According to our previously published procedure (Wang et al., 2009, 2011), SiNWS were fabricated by combining photolithographic patterning and Ag nanoparticle-templated wet etching (Peng et al., 2002). Anti-EpCAM coating was also conducted following a previously established protocol (Wang et al., 2009), see Supplementary Information 1 and Supplementary Fig. S2.
2.2. Cell line culture and flow cytometry
Cancer cell lines, i.e., PC3 prostate cancer, A549 non-small cell lung cancer, Huh-7 hepatocellular carcinoma, SkBr-3 breast cancer, MCF-7 breast cancer and LNCAP prostate cancer, were obtained from American Type Culture Collection (ATCC, Manassas, VA). The pancreatic neuroendocrine tumor (PanNET) cell line BON-1 was a kind gift from Dr. Mark Hellmich (University of Texas Medical Branch, Galveston, USA). The cell lines were tested and found negative for Mycoplasma contamination. Sodium citrate (0.015 M sodium citrate with 0.135 M KCl) was used to detach the cultured cells for preparing single cell suspension (Nie et al., 2014). For each cell line, one million cells were fixed with 4% paraformaldehyde for 10 min, stained with EpCAM antibodies for 30 min, and then stained with FITC fluorophore secondary antibody for another 30 min at room temperature (RT). After washing twice with PBS, we collected 10,000 cells for EpCAM expression level detection using flow cytometry. Data were then analyzed using FlowJo and presented as histogram profiles.
2.3. Artificial CTC cluster sample preparation containing cell clusters and single cells
Sphere-forming BON-1 cell clusters were used to test the performance of NanoVelcro Chips in capturing cell clusters. The sphere-forming culture clusters were obtained using modified methods according to the previously published protocol (Cai et al., 2016; Wong et al., 2012). A single-cell suspension of 10,000 BON-1 cells was seeded in each well of a low adhesion 6-well plate. Cell sphere-formation was observed by inverted microscopy. When cell spheres expanded about 50-fold or so, the supernatant was collected and centrifuged at 1,000 rpm for 5 min, and the cell spheres were washed with phosphate buffer solution (PBS) twice. The single cells and cell clusters were pre-stained with Vybrant™ DiO green fluorescent dye. Then, the prepared BON-1 cell clusters/cells (ca. 500 cell per sample) were spiked into the PBMCs isolated from 2 mL of healthy donor whole blood to prepare artificial CTC cluster samples.
2.4. Optimization of NanoVelcro Chips for capturing CTC clusters
NanoVelcro Chips were initially developed and optimized for capturing single CTCs. We optimized the flow rate of NanoVelcro Chips for capturing CTC clusters using prepared artificial CTC cluster samples. The samples were injected into the NanoVelcro Chips at flow rates of 0.1, 0.2, 0.5, 1.0, and 2.0 mL h−1, controlled by syringe pumps (Legato 200, KD Scientific). The single cells and cell clusters captured in NanoVelcro Chips were fixed with 4.0% formaldehyde (PFA, Electron Microscopy Sciences) in PBS (200 μL). After nuclear staining with DAPI, all the stained chips were scanned and imaged under a fluorescence microscope (Nikon 90i). The single cells, cell clusters, and cells in each cluster captured on the chips were counted. The matched outflow cells collected in tubes were centrifuged, smeared in glass slides, and then stained with DAPI for enumeration.
2.5. Characterization of BON-1 cell clusters
For SEM characterization of BON-1 cell clusters, the SiNWS was separated from the chip after cell capture using artificial CTC cluster samples. The SiNWS with captured BON-1 cell clusters were fixed in 4% PFA for 1 h, followed by sequential dehydration through 30%, 50%, 75%, 85%, 95%, and 100% ethanol solutions for 10 min each. After overnight lyophilization, the samples were sputter-coated with gold at room temperature. The images were recorded with a ZEISS Supra 40VP SEM at an accelerating voltage of 10 keV.
2.6. Clinical samples collection and blood processing
All the participants in this study were enrolled between May 2019 -September 2020. NET patients who met the criteria of PRRT were enrolled in this study, and patients who had severe mental diseases were excluded. All patients and healthy donors provided written informed consent for this study according to the IRB protocol (IRB #10–001785) at UCLA. Peripheral blood samples were collected in acid citrate dextrose (ACD) vacutainer tubes (8 mL) and were processed within 4 h of withdrawal. The PBMCs were isolated according to our previously published protocol (Jan et al., 2019) from 2 mL of whole blood and were run through the NanoVelcro Chips using the optimized protocol.
2.7. Immunostaining and identification of NET CTC clusters and single NET CTCs
After CTC capture using NanoVelcro Chips on patients’ blood samples, the CTC samples were fixed by 4% PFA and then subjected to a 4-color ICC staining (Court et al., 2018a,b) with DAPI, anti-CK, anti-synaptophysin (anti-SYP) (Miettinen, 1987), and anti-CD45 for identification of NET CTC clusters and single NET CTCs. The substrates were then scanned with a fluorescence microscope (Nikon 90i). An automatic scan was carried out under 20 × magnification with DAPI, FITC, TRITC, and Cy5 channels corresponding to nuclear, CK, SYP, and CD45 staining, respectively. Immobilized cells were imaged using the Nikon Ni fluorescence microscope with NIS-Element imaging software. When analyzing the multi-channel ICC micrographs, WBCs were identified according to ICC criteria (DAPI+/CK-/SYP-/CD45+), and NET CTCs were identified according to ICC criteria (DAPI+/CK+/SYP+/CD45-) with intact nuclear morphology. NET CTC clusters were defined as aggregated NET CTCs with at least two NET CTCs attached to each other.
3. Results and discussion
3.1. The performance of NanoVelcro Chips is negligibly affected by the EpCAM expression levels of CTCs
Considering the reduced EpCAM expression observed in CTC clusters/spheres (Hyun et al., 2016), we first examined how the capture performance of NanoVelcro Chips would be negatively impacted by reduced CTC EpCAM expression levels (Fig. 2A). As shown in Fig. 2B, the six other tumor cell lines exhibited different levels of EpCAM expression, and BON-1 NET cells harvested from the sphere-forming conditions showed a small yet noticeable reduction in their EpCAM expression level in comparison to those cultured under regular conditions. Capture studies with these artificial CTC samples (Fig. 2C) showed that NanoVelcro Chips demonstrated a consistent CTC-capture efficiency (90.60 ± 1.00% - 98.35 ± 1.01%, n = 3), which was unaffected by the EpCAM expression levels of spiked NET or other tumor cells. This result contrasts with other existing immunoaffinity-mediated CTC capture systems (e.g., CellSearch®) (Wit et al., 2015), whose CTC capture performance is dependent on the EpCAM expression level of the target cells. We attribute this unique advantage to NanoVelcro Chips’ “Velcro-like” operating mechanism (Fig. 1C) – the entanglement between the microvilli structures on CTCs’ surfaces and the densely packed silicon nanowires on SiNWS results in significantly increased contact surface area. This increases the amount of binding interactions between EpCAM (on CTC surfaces) and anti-EpCAM (on SiNWS), even when the CTCs express low quantities of EpCAM. In contrast, there is much less contact surface area between CTC microvilli structures and flat substrates (used in most CTC capture systems), rendering capture performance dependent on CTC EpCAM expression (see the schematic illustration in Supplementary Fig. S3). To test the essential roles of immunoaffinity-mediated interactions in NanoVelcro Chips, we conducted a control study by introducing the artificial sample containing BON-1 NET cells into NanoVelcro Chips without anti-EpCAM coating. The control devices showed considerably reduced capture performance (Supplementary Fig. S4), which demonstrates the necessity of molecular recognition in NanoVelcro Chips to achieve specific “Velcro-like” interactions.
Fig. 2.
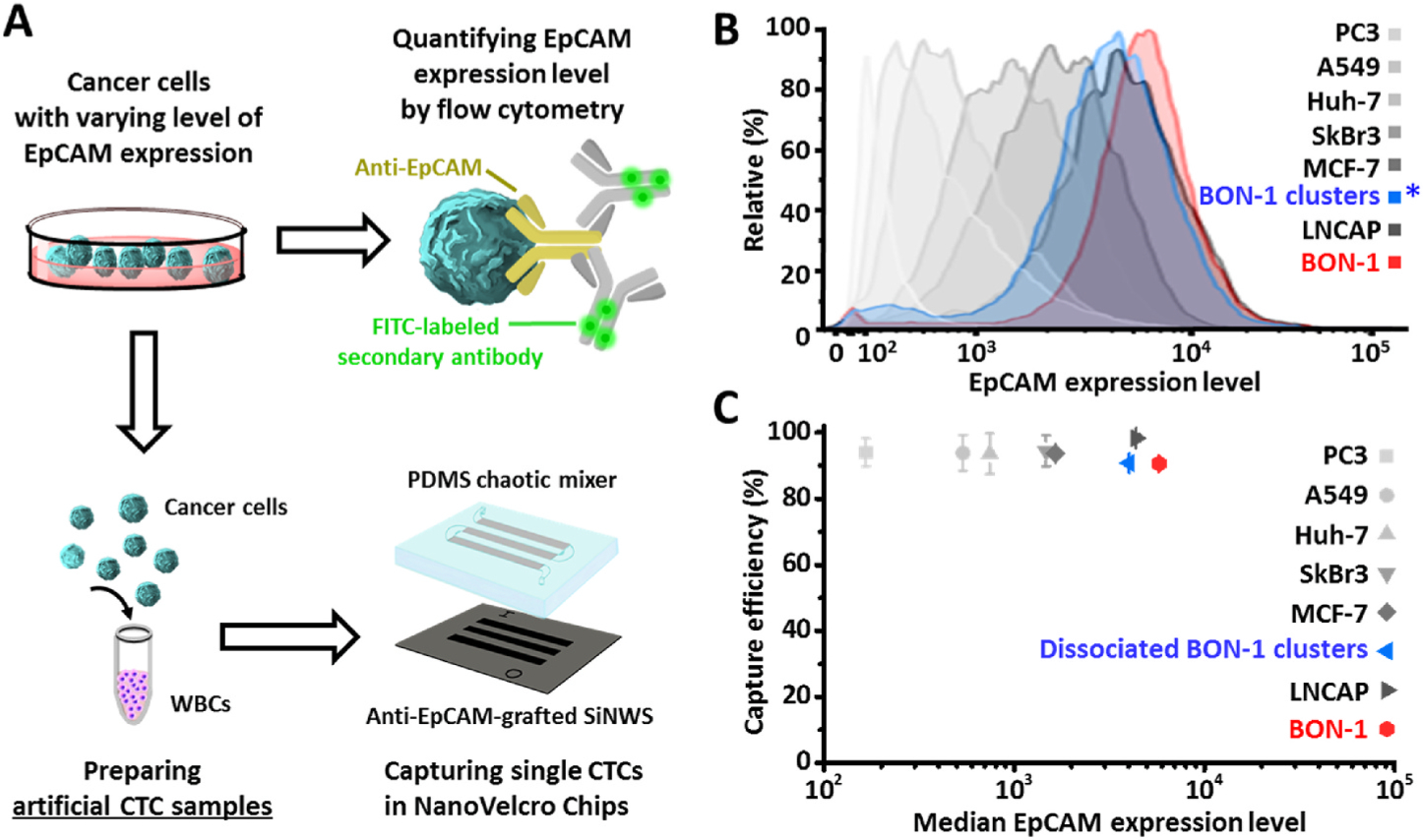
NanoVelcro Chips exhibit superb performance for capturing cancer cells with varying levels of EpCAM expression. A) Artificial CTC samples were prepared by mixing healthy donor’s PBMCs with NET or six other tumor cells. *Before spiking, aliquoted cell samples were first dissociated into single-cell suspensions, followed by FACS analysis and cell capture. B) FACS analysis confirmed their EpCAM expression levels. C) NanoVelcro Chips, which are composed of patterned SiNWS grafted with anti-EpCAM capture agent and a PDMS chaotic mixer, demonstrated a consistent CTC-capture efficiency, which was negligibly affected by the EpCAM expression levels of spiked NET or other tumor cells. Data are presented as the mean ± SD of three independent assays.
3.2. NanoVelcro Chips were optimized for simultaneous enrichment and detection of NET CTC clusters and single NET CTCs
We further explored the use of NanoVelcro Chips for simultaneous enrichment and detection of both NET CTC clusters and single NET CTCs with negligible perturbation to their intrinsic properties, including both intactness and size distributions. In a typical sphere-forming BON-1 cell sample used in artificial NET CTC cluster sample preparation, 70% of the cells were presented in clusters, which exhibited multidirectional aggregation morphologies containing 2 to 40 cells (Supplementary Fig. S5). A general workflow for the optimization of NanoVelcro Chips is depicted in Fig. 3A.
Fig. 3.
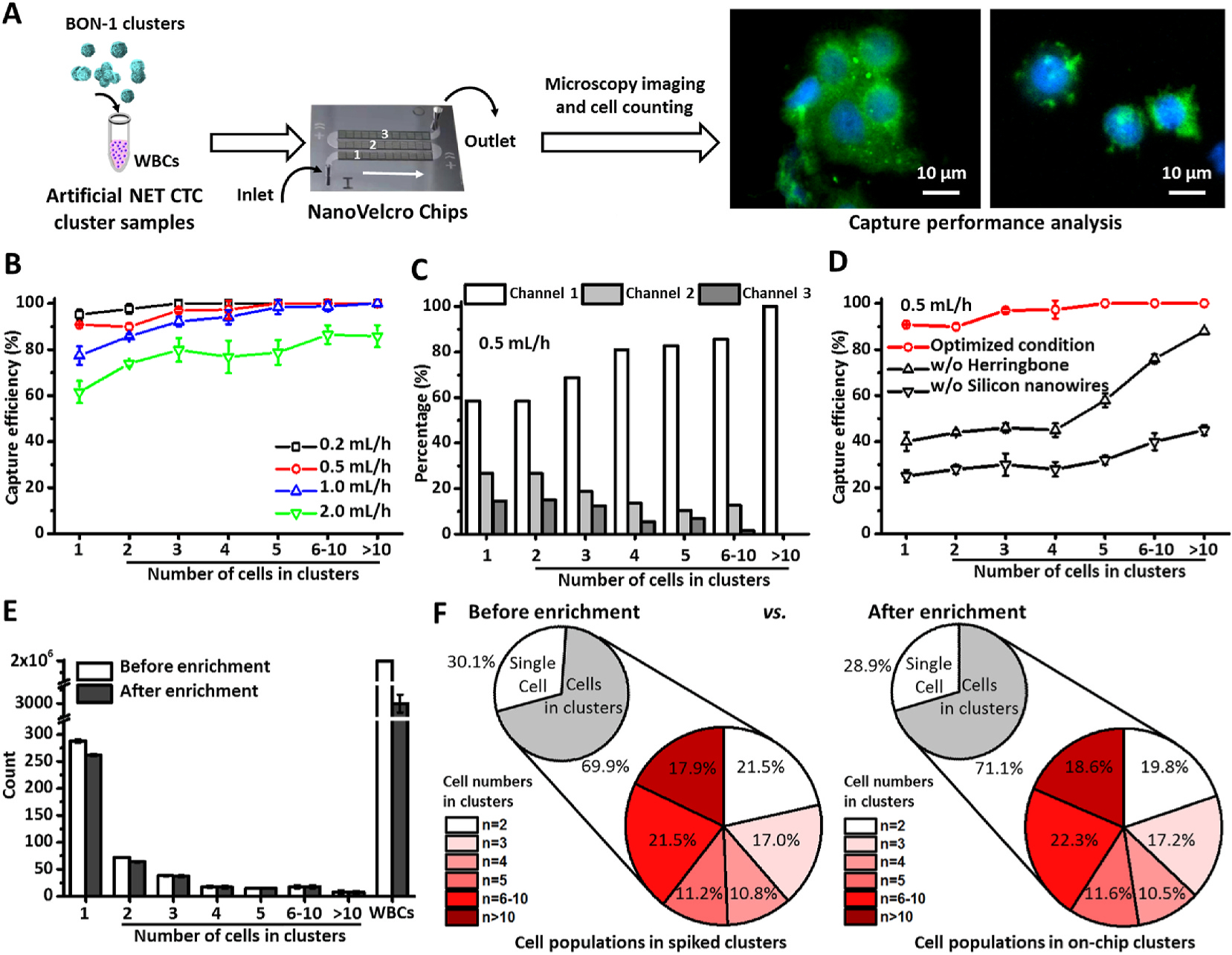
Optimization of NanoVelcro Chips for simultaneous enrichment and detection of single NET CTCs and NET CTC clusters using artificial CTC cluster samples. A) A general workflow developed for optimization of NanoVelcro Chips. B) Efficiencies for capturing single BON-1 cells and BON-1 clusters were studied at flow rates of 0.2, 0.5, 1.0, and 2.0 mL/h. Data are presented as the mean ± SD of three independent assays. C) A representative distribution of single BON-1 cells and BON-1 clusters in the three channels of NanoVelcro Chips under the optimal flow rate of 0.5 mL/h. D) Capture efficiencies observed for NanoVelcro Chips and the control devices. Data are presented as the mean ± SD of three independent assays. E) Side-by-side comparison of the distributions of single BON-1 cells and BON-1 clusters (of different sizes) characterized before and after enrichment in NanoVelcro Chips. Data are presented as the mean ± SD of three independent assays. F) Representative pie charts showing the distribution of single BON-1 cells and BON-1 clusters before (left) and after (right) enrichment.
We first examined (Fig. 3B) how the flow rate (0.2, 0.5, 1.0, and 2.0 mL/h) affected the capture efficiencies for single BON-1 cells and BON-1 clusters of different sizes. We observed that the majority of single BON-1 cells and BON-1 clusters were captured in the first microchannels of NanoVelcro Chips (Fig. 3C), indicating that the incorporation of the three microchannels in the current device design guarantees efficient capture of both single CTCs and clusters. Similar spatial distributions of single BON-1 cells and BON-1 clusters were also observed at different flow rates (Supplementary Figs. S6A–C). Overall, we found that 1) NanoVelcro Chips exhibited better capture performance for larger sized BON-1 clusters than smaller clusters and single BON-1 cells, and 2) higher flow rates led to reduced capture performance for both single cells and clusters. To minimize the assay run time while still maximizing assay performance, the flow rate of 0.5 mL/h was chosen. Under this optimal flow rate, we achieved capture efficiency of 100% for large CTC clusters (n≥5) and 89.90 ± 1.43% to 97.22 ± 3.93% for single cells or smaller CTC clusters (n<5). The capture efficiency of our NanoVelcro Chips for capturing CTC clusters is higher than most CTC cluster enrichment platforms that were previously reported (Table S1) (Au et al., 2017; Reduzzi et al., 2021; Sarioglu et al., 2015). We also tested the critical roles of the herringbone patterns in the PDMS chaotic mixer and the embedded silicon nanowires on the anti-EpCAM-grafted SiNWS. The data summarized in Fig. 3D indicated that the control devices lacking either the herringbone patterns or the embedded silicon nanowires demonstrated significantly reduced capture performance for single BON-1 cells and for BON-1 clusters. Finally, we studied how the enrichment process in NanoVelcro Chips could perturb the integrity of BON-1 clusters. Fig. 3E shows a side-by-side comparison of the size distributions of single BON-1 cells and BON-1 clusters characterized before and after enrichment in NanoVelcro Chips (n = 3). Representative pie charts shown in Fig. 3F offer a different view of the cell/cluster distribution shown in Fig. 3E. We noted that the distribution after enrichment almost entirely mirrored that of before enrichment, suggesting that NanoVelcro Chips are highly efficient without compromising the preservation of CTC clusters’ intrinsic properties.
3.3. Characterization of "Velcro-like" interactions at the interfaces between BON-1 clusters/cells and SiNWS by scanning electron microscopy (SEM)
To illustrate how the “Velcro-like” operating mechanism facilitates immunoaffinity-mediated capture of BON-1 clusters and single BON-1 cells onto SiNWS of NanoVelcro Chips, scanning electron microscopy (SEM) was employed to characterize the interfaces between BON-1 clusters/cells and SiNWS after they were captured on SiNWS. A comparison experiment was performed in parallel using control devices (flat substrates) without the embedded silicon nanowires. Fig. 4A–C shows representative SEM images of a large cluster (composed of more than 10 BON-1 cells), a two-cell cluster, and a single BON-1 cell captured on the SiNWS of NanoVelcro Chips. We observed tight interactions between the long microvilli of BON-1 cells and densely packed silicon nanowires on SiNWS, i.e., the characteristic “Velcro-like” interactions at the interfaces between BON-1 clusters/cells and SiNWS. In contrast, no “Velcro-like” interactions were found at the interfaces between BON-1 clusters/cells and flat substrates (Fig. 4D). These SEM images suggest that the “Velcro-like” operating mechanism – originally developed to facilitate immunoaffinity-mediated capture of single CTCs – can also be effectively adopted for immunoaffinity-mediated capture of CTC clusters.
Fig. 4.
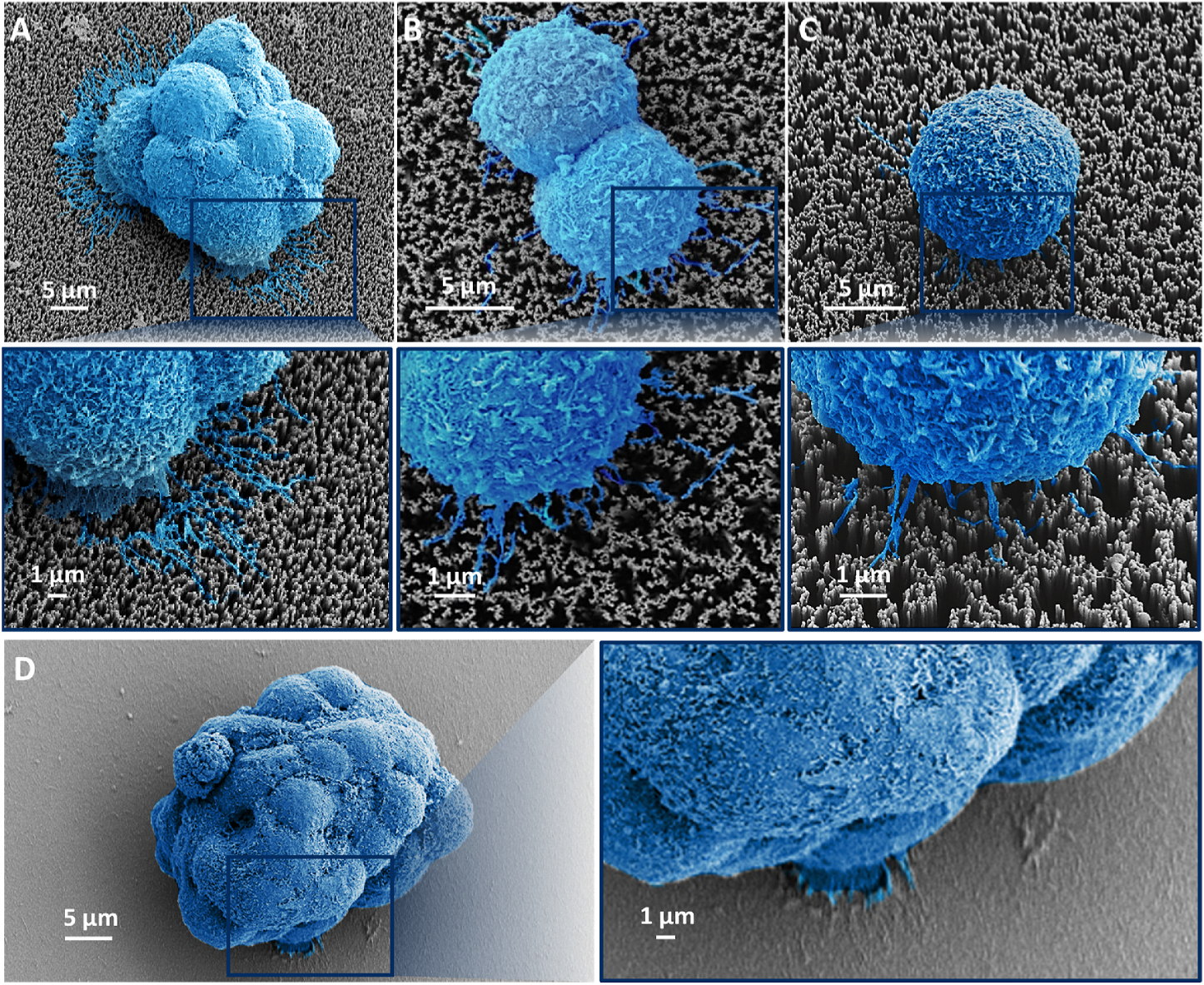
Representative SEM images of A) a large BON-1 cluster (composed of more than 10 cells), B) a two-cell cluster, C) a single BON-1 cell captured on SiNWS of NanoVelcro Chips, and D) a large cell cluster (>10 cells) captured on a flat substrate with the same surface chemistry (Fig. 3d).
3.4. Enumeration of CTC clusters and single CTCs in advanced NET patients
Under the optimal conditions identified above, we applied the usage of NanoVelcro Chips in the enumeration of both NET CTC clusters and single NET CTCs in the blood samples collected from advanced NET patients enrolled for PRRT. Eighty-two peripheral blood samples were collected from 21 advanced NET patients serially before initiating PRRT and then right after the 1st to 4th cycle of PRRT, respectively, as shown in Fig. 6A. In a control cohort, 16 random blood samples were collected from healthy donors. Representative fluorescent microscopy images of a single NET CTC, as well as NET CTC clusters including two CTCs, four CTCs, five CTCs and more than ten CTCs isolated in NanoVelcro Chips from patient samples are shown in Fig. 5. It is critical to note that unlike CTCs of other epithelial origins, which often exhibit larger sizes than WBCs, all of the single NET CTCs (median = 6.6 μm) and CTCs in NET clusters (median = 6.1 μm) captured by NanoVelcro Chips exhibit significantly smaller sizes than the surrounding WBCs (median = 9.7 μm) (Supplementary Fig. S7). The size difference between single NET CTCs (smaller than WBCs) and NET CTC clusters (aggregates are larger than WBCs) underscored the need to develop a technology (i.e., NanoVelcro Chip) that can capture both single NET CTCs and NET CTC clusters without any size bias.
Fig. 6.
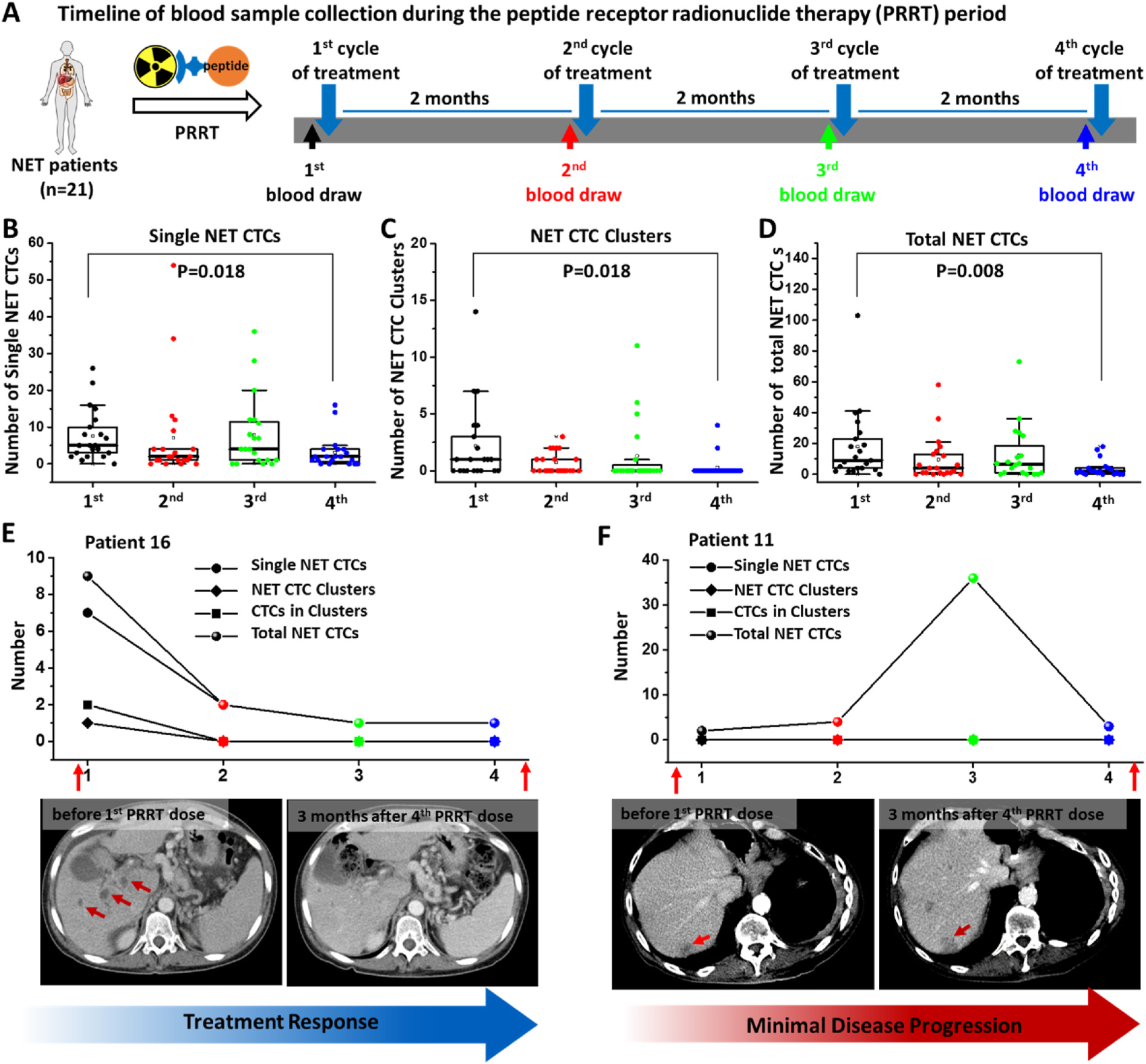
Dynamic changes of NET CTC clusters, single NET CTCs, and total CTCs in 21 advanced NET patients before and after their 1st to 4th cycle of PRRT. A) Timeline of blood draws in advanced NET patients taken before each of their four PRRT cycles. Box plots showing the dynamic changes of B) single NET CTCs, C) NET CTC clusters, and D) total NET CTCs per 2 mL blood sample from 21 advanced NET patients before and across 4 PRRT cycles. Significant differences between the 1st cycle and 4th cycle were evaluated using a paired t-test. The dynamic changes in the number of single NET CTCs, NET CTC clusters, CTCs in clusters, and total NET CTCs observed for representative NET patients who exhibited a remarkable treatment response (E, Patient 16) and disease progression (F, Patient 11).
Fig. 5.
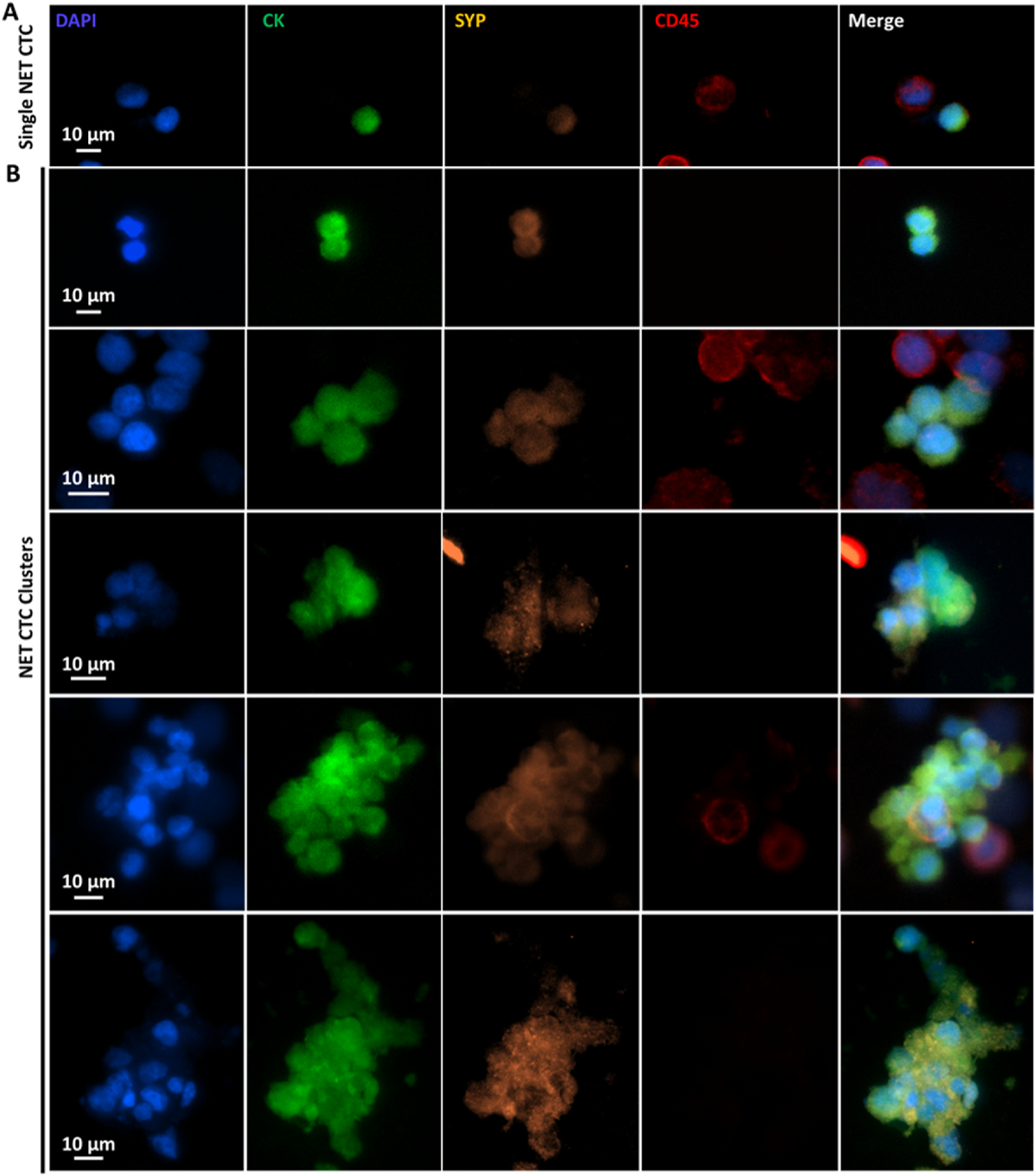
Characterization and enumeration of NET CTC clusters and single NET CTCs isolated from the blood samples of metastatic NET patients. Representative fluorescent micrographs of A) single NET CTCs, as well as B) NET CTC clusters including two CTCs, four CTCs, five CTCs, and more than ten CTCs found in advanced NET patients.
3.5. NET CTC clusters and single NET CTCs for monitoring treatment responses in advanced NET patients during PRRT
The enumeration results, i.e., the number of single NET CTCs, NET CTC clusters, and total NET CTCs (combining the numbers of NET CTCs in the clusters and single NET CTCs) are summarized in Fig. 6B–D. Overall, before PRRT therapy, NET CTC clusters were found in a majority of patients (15/21) and single NET CTCs were found in all patients (21/21). As depicted in the box plots (Fig. 6B–D), patients exhibited significant decreases in the number of single NET CTCs (P = 0.018), NET CTC clusters (P = 0.018), and total NET CTCs (P = 0.008) after 4 PRRT treatments, potentially reflecting a reduction in their systemic tumor burden as a result of PRRT. No NET CTC clusters or single NET CTCs were detected in the 16 healthy controls. No correlation was observed between number of NET CTC clusters or number of CTC in clusters before PRRT with TNM staging of the NET patients (Supplementary Fig. S8). The raw data of enumeration and detailed clinical information is provided in Supplementary Tables S2–S4. In Fig. 6E, we tracked the treatment of patient 16 before and after 4 PRRT treatments. The dynamic changes are plotted for single NET CTCs, NET CTC clusters, CTCs in clusters, and total NET CTCs detected in this patient’s blood. Computed tomography (CT) images of the patient showed that prior to PRRT, the patient exhibited multiple metastases in the liver. Following the initiation of PRRT, the patient’s CTC number reduced progressively over the course of PRRT, and a CT image 3 months after completing PRRT showed a significant reduction in hepatic tumor burden in conjunction with the reduction observed in single NET CTCs, NET CTC clusters, CTCs in clusters, and total NET CTCs detected in this patient’s blood. At the time of data cutoff, the patient’s treatment response with PRRT was durable. In contrast, a minimal increase in single NET CTCs was observed in another patient (patient 11) without CTC cluster after the 4th cycle of PRRT, correlating with disease progression noted on CT imaging 3 months after PRRT completion (Fig. 6F).
4. Conclusions
In this study, we exploited the use of NanoVelcro Chips to enrich and to detect both single NET CTCs and NET CTC clusters simultaneously with negligible perturbation of their intrinsic properties, such as intactness and size distributions. Here, we used NETs as a disease model, where we examined the potential role of NET CTCs and NET CTC clusters as novel biomarkers to aid in the evaluation of advanced NET patients’ treatment responses during the course of PRRT. We observed dynamic changes in the numbers of single NET CTCs, NET CTC clusters, and total NET CTCs in NET patients before and across PRRT, which strongly correlated with clinical treatment responses. This suggests that analysis of single NET CTCs and NET CTC clusters offers a potential non-invasive option to monitor treatment response in patients undergoing PRRT. Due to the relatively low incidence of NETs, our clinical validation study is limited by the relatively small single cohort and will require further validation in a larger group of patients with NETs.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (R21CA240887, R21 CA235340, U01 CA198900, U01 EB026421, R01 CA246304, R01 CA253651 and R01 CA255727) and a seed grant from the University of California Pancreatic Cancer Consortium. The pancreatic neuroendocrine tumor (PanNET) cell line BON-1 was provided by Dr. Mark Hellmich (University of Texas Medical Branch, Galveston, USA). Dr. N. Sun gratefully acknowledges financial support from Collaborative Innovation Center of Suzhou Nano Science and Technology. Dr. N. Sun, Y. Yang and H. Miao contributed equally.
Footnotes
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Following the guideline of UCLA Conflict of Interest Review Committee (CIRC), the authors would like to disclose: (i) The intellectual property used in this study has been licensed to CytoLumina Technologies Corp.; and (ii) Hsian-Rong Tseng has financial interests in CytoLumina Technologies Corp. given his role as the company’s founder.
CRediT authorship contribution statement
Na Sun: wrote and revised the manuscript with input, performed chip optimization using artificial samples, performed CTC isolation from clinical samples using optimized NanoVelcro Chips, performed SEM characterization, performed cell enumeration with assistance, analyzed data and made figures with assistance. Yingying Yang: performed chip optimization using artificial samples, performed CTC isolation from clinical samples using optimized NanoVelcro Chips, performed the collection of blood samples and clinical information with assistance, performed cell enumeration with assistance. Hui Miao: performed chip optimization using artificial samples, performed CTC isolation from clinical samples using optimized NanoVelcro Chips, performed cell enumeration with assistance. Peter Redublo: wrote and revised the manuscript with input, performed CTC isolation from clinical samples using optimized NanoVelcro Chips, performed the collection of blood samples and clinical information with assistance, performed cell enumeration with assistance. Hongtao Liu: performed chip optimization using artificial samples. Wenfei Liu: performed chip optimization using artificial samples. Yen-Wen Huang: performed chip optimization using artificial samples. Pai-Chi Teng: performed fluorescence-activated cell sorting analysis. Ceng Zhang: performed fluorescence-activated cell sorting analysis, performed cell enumeration with assistance. Ryan Y. Zhang: performed fluorescence-activated cell sorting analysis, performed cell enumeration with assistance. Matthew Smalley: wrote and revised the manuscript with input. Peng Yang: provided input and advice on the project. Shih-Jie Chou: performed cell culture, performed fluorescence-activated cell sorting analysis. Kevin Huai: performed chip optimization using artificial samples. Zhicheng Zhang: provided input and advice on the project. Yi-Te Lee: wrote and revised the manuscript with input, provided input and advice on the project. Jasmine J. Wang: wrote and revised the manuscript with input, provided input and advice on the project. Jing Wang: performed cell enumeration with assistance. Icy Y. Liang: wrote and revised the manuscript with input. Tiffany X. Zhang: analyzed data, and made figures with assistance. Dongyun Zhang: performed the collection of blood samples and clinical information with assistance. Li Liang: provided input and advice on the project. Paul S. Weiss: provided input and advice on the project. Edwin M. Posadas: performed the collection of blood samples and clinical information with assistance. Timothy Donahue: performed the collection of blood samples and clinical information with assistance. J. Randolph Hecht: performed the collection of blood samples and clinical information with assistance. Martin S. Allen-Auerbach: performed the collection of blood samples and clinical information with assistance. Emily K. Bergsland: performed the collection of blood samples and clinical information with assistance. Thomas A. Hope: performed cell enumeration with assistance. Renjun Pei: designed the study and oversaw project execution, wrote and revised the manuscript with input. Yazhen Zhu: designed the study and oversaw project execution, wrote and revised the manuscript with input. Hsian-Rong Tseng: designed the study and oversaw project execution, wrote and revised the manuscript with input, analyzed data and made figures with assistance. Anthony P. Heaney: designed the study and oversaw project execution, wrote and revised the manuscript with input, performed the collection of blood samples and clinical information with assistance.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113854.
References
- Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, Brannigan BW, Kapur R, Stott SL, Shioda T, Ramaswamy S, Ting DT, Lin CP, Toner M, Haber DA, Maheswaran S, 2014. Cell 158 (5), 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar Y, Dong J, Zhao P, et al. , 2021. Nat. Commun. 12, 4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au SH, Edd J, Stoddard AE, Wong KHK, Fachin F, Maheswaran S, et al. , 2017. Sci. Rep. 7, 2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Peng T, Wang J, Zhang J, Hu H, Tang D, Chu C, Yang T, Liu H, 2016. Cell. Physiol. Biochem. 39 (4), 1421–1432. [DOI] [PubMed] [Google Scholar]
- Chen JF, Ho H, Lichterman J, Lu YT, Zhang Y, Garcia MA, Chen SF, Liang AJ, Hodara E, Zhau HE, Hou S, Ahmed RS, Luthringer DJ, Huang J, Li KC, Chung LW, Ke Z, Tseng HR, Posadas EM, 2015. Cancer 121 (18), 3240–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Zhu Y, Lu YT, Hodara E, Hou S, Agopian VG, Tomlinson JS, Posadas EM, Tseng HR, 2016. Theranostics 6 (9), 1425–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court CM, Ankeny JS, Hou S, Tseng HR, Tomlinson JS, 2015. Expert Rev. Mol. Diagn. 15 (11), 1491–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court CM, Ankeny JS, Sho S, Winograd P, Hou S, Song M, Wainberg ZA, Girgis MD, Graeber TG, Agopian VG, Tseng HR, Tomlinson JS, 2018a. Ann. Surg Oncol. 25 (4), 1000–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court CM, Hou S, Winograd P, Segel NH, Li QW, Zhu Y, Sadeghi S, Finn RS, Ganapathy E, Song M, French SW, Naini BV, Sho S, Kaldas FM, Busuttil RW, Tomlinson JS, Tseng HR, Agopian VG, 2018b. Liver Transplant. 24 (7), 946–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF, 2004. N. Engl. J. Med. 351 (8), 781–791. [DOI] [PubMed] [Google Scholar]
- Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A, 2013. Nat. Rev. Clin. Oncol. 10 (8), 472–484. [DOI] [PubMed] [Google Scholar]
- Dong J, Chen JF, Smalley M, Zhao M, Ke Z, Zhu Y, Tseng HR, 2020. Adv. Mater. 32 (1), e1903663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks RJ, Kwekkeboom DJ, Krenning E, Bodei L, Grozinsky-Glasberg S, Arnold R, Borbath I, Cwikla J, Toumpanakis C, Kaltsas G, Davies P, Horsch D, Tiensuu Janson E, Ramage J, 2017. Antibes consensus conference. Neuroendocrinology 105 (3), 295–309. [DOI] [PubMed] [Google Scholar]
- Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, Priest LJ, Greystoke A, Zhou C, Morris K, Ward T, Blackhall FH, Dive C, 2012. J. Clin. Oncol. 30 (5), 525–532. [DOI] [PubMed] [Google Scholar]
- Hou S, Chen JF, Song M, Zhu Y, Jan YJ, Chen SH, Weng TH, Ling DA, Chen SF, Ro T, Liang AJ, Lee T, Jin H, Li M, Liu L, Hsiao YS, Chen P, Yu HH, Tsai MS, Pisarska MD, Chen A, Chen LC, Tseng HR, 2017. ACS Nano 11 (8), 8167–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao Y-S, Luo S-C, Hou S, Zhu B, Sekine J, Kuo C-W, Chueh D-Y, Yu H.-h., Tseng H-R, Chen P, 2014. Small 10, 3012–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun KA, Koo GB, Han H, Sohn J, Choi W, Kim SI, Jung HI, Kim YS, 2016. Oncotarget 7 (17), 24677–24687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YJ, Chen JF, Zhu Y, Lu YT, Chen SH, Chung H, Smalley M, Huang YW, Dong J, Chen LC, Yu HH, Tomlinson JS, Hou S, Agopian VG, Posadas EM, Tseng HR, 2018. Adv. Drug Deliv. Rev. 125, 78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YJ, Yoon J, Chen JF, Teng PC, Yao N, Cheng S, Lozano A, Chu GCY, Chung H, Lu YT, Chen PJ, Wang JJ, Lee YT, Kim M, Zhu Y, Knudsen BS, Feng FY, Garraway IP, Gao AC, Chung LWK, Freeman MR, You S, Tseng HR, Posadas EM, 2019. Theranostics 9 (10), 2812–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwekkeboom DJ, Mueller-Brand J, Paganelli G, Anthony LB, Pauwels S, Kvols LK, O’Dorisio TM, Valkema R, Bodei L, Chinol M, Maecke HR, Krenning EP, 2005. J. Nucl. Med. 46 (Suppl. 1), 62S–66S. [PubMed] [Google Scholar]
- Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S, Ke Z, Tseng HR, 2014. Acc. Chem. Res. 10, 2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YT, Zhao L, Shen Q, Garcia MA, Wu D, Hou S, Song M, Xu X, Ouyang WH, Ouyang WW, Lichterman J, Luo Z, Xuan X, Huang J, Chung LW, Rettig M, Tseng HR, Shao C, Posadas EM, 2013. Methods 64 (2), 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen M, 1987. Arch. Pathol. Lab Med. 111 (9), 813–818. [PubMed] [Google Scholar]
- Nie Y, Walsh P, Clarke DL, Rowley JA, Fellner T, 2014. PLoS One 9 (1), e88012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K, Alix-Panabieres C, 2010. Trends Mol. Med. 16 (9), 398–406. [DOI] [PubMed] [Google Scholar]
- Peng KQ, Yan YJ, Gao SP, Zhu J, 2002. Adv. Mater. 14 (16), 1164–1167. [Google Scholar]
- Reduzzi C, Di Cosimo S, Gerratana L, Motta R, Martinetti A, Vingiani A, D’Amico P, Zhang Y, Vismara M, Depretto C, et al. , 2021. Cancers 13, 2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K, 2007. Clin. Cancer Res. 13 (3), 920–928. [DOI] [PubMed] [Google Scholar]
- Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK, Miyamoto DT, Luo X, Bardia A, Wittner BS, Ramaswamy S, Shioda T, Ting DT, Stott SL, Kapur R, Maheswaran S, Haber DA, Toner M, 2015. Nat. Methods 12 (7), 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine J, Luo SC, Wang S, Zhu B, Tseng HR, Yu HH, 2011. Adv. Mater. 23 (41), 4788–4792. [DOI] [PubMed] [Google Scholar]
- Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, Anand A, Smith K, Maslak P, Doyle GV, Terstappen LW, Lilja H, Heller G, Fleisher M, Scher HI, 2007. Clin. Cancer Res. 13 (7), 2023–2029. [DOI] [PubMed] [Google Scholar]
- Shields C.W. t., Reyes CD, Lopez GP, 2015. Lab Chip 15 (5), 1230–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher MR, Pantel K, 2014. Nat. Biotechnol. 32 (5), 441–443. [DOI] [PubMed] [Google Scholar]
- Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR, 2011. Angew. Chem. Int. Ed. 50 (13), 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR, 2009. Angew. Chem. Int. Ed. 48 (47), 8970–8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wit S, Dalum G, Lenferink AT, Tibbe AG, Hiltermann TJ, Groen HJ, Rijn CJ, Terstappen LW, 2015. Sci. Rep. 5, 12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C, Vosburgh E, Levine AJ, Cong L, Xu EY, 2012. JoVE 66, e4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q, He R, Hong L, Liu W, Guo S, Liu K, Tseng HR, Xiong B, Zhao XZ, 2012. Adv. Mater. 24 (20), 2756–2760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


