Abstract
The determination of antimicrobial susceptibility of a clinical isolate, especially with increasing resistance, is often crucial for the optimal antimicrobial therapy of infected patients. Nucleic acid-based assays for the detection of resistance may offer advantages over phenotypic assays. Examples are the detection of the methicillin resistance-encoding mecA gene in staphylococci, rifampin resistance in Mycobacterium tuberculosis, and the spread of resistance determinants across the globe. However, molecular assays for the detection of resistance have a number of limitations. New resistance mechanisms may be missed, and in some cases the number of different genes makes generating an assay too costly to compete with phenotypic assays. In addition, proper quality control for molecular assays poses a problem for many laboratories, and this results in questionable results at best. The development of new molecular techniques, e.g., PCR using molecular beacons and DNA chips, expands the possibilities for monitoring resistance. Although molecular techniques for the detection of antimicrobial resistance clearly are winning a place in routine diagnostics, phenotypic assays are still the method of choice for most resistance determinations. In this review, we describe the applications of molecular techniques for the detection of antimicrobial resistance and the current state of the art.
The determination of antimicrobial susceptibility of a clinical isolate is often crucial for the optimal antimicrobial therapy of infected patients. This need is only increasing with increasing resistance and the emergence of multidrug-resistant microorganisms (88, 89, 91). Testing is required not only for therapy but also to monitor the spread of resistant organisms or resistance genes throughout the hospital and community. Standard procedures and breakpoints have been defined to predict therapeutic outcome both in time and at different geographic locations. In some cases the presence of a resistance gene is highly predictive for clinical outcome of antimicrobial therapy. For example, the presence of a β-lactamase in Neisseria gonorrhoeae correlates well with the outcome of penicillin treatment. However, the presence of a resistance gene does not necessarily lead to treatment failure (198), because the level of expression may be to low. For example, β-lactamase production among members of the Enterobacteriaceae is common, but the development of resistance is dependent on the mode and level of expression (180, 183).
Resistance can be caused by a variety of mechanisms: (i) the presence of an enzyme that inactivates the antimicrobial agent; (ii) the presence of an alternative enzyme for the enzyme that is inhibited by the antimicrobial agent; (iii) a mutation in the antimicrobial agent's target, which reduces the binding of the antimicrobial agent; (iv) posttranscriptional or posttranslational modification of the antimicrobial agent's target, which reduces binding of the antimicrobial agent; (v) reduced uptake of the antimicrobial agent; (vi) active efflux of the antimicrobial agent; and (vii) overproduction of the target of the antimicrobial agent. In addition, resistance may be caused by a previously unrecognized mechanism. On the other hand, a gene which is not expressed in vitro may be expressed in vivo.
Nucleic acid-based detection systems offer rapid and sensitive methods to detect the presence of resistance genes and play a critical role in the elucidation of resistance mechanisms. During the last decade, nucleic acid-based detection systems have expanded tremendously and are becoming more accessible for clinical microbiology laboratories. This accessibility is not limited to the detection and identification of microorganisms but is extended to the detection of properties of these microorganisms, such as virulence factors and antimicrobial resistance. The application of nucleic acid-based technology is particularly useful for slow-growing or nonculturable microorganisms and for the detection of point mutations or certain genotypes. Nucleic acid-based technology can be divided into hybridization systems and amplification systems, although most amplification technologies are also partly based on hybridization technology.
All these factors complicate the debate regarding the determination of phenotypic versus genotypic resistance. The objective of the present review is to discuss examples where molecular techniques were used to detect antimicrobial resistance as part of diagnostic microbiology. The most commonly used or new molecular methods will be described first, followed by the applications of these techniques to detect resistance.
MOLECULAR TECHNIQUES USED IN CLINICAL MICROBIOLOGY
Molecular techniques have been under development for the last 30 years, but progress throughout the last decade has been particularly rapid. In this section, we review the molecular techniques which have been used to detect antimicrobial resistance determinants as well as the techniques that hold great promise for the near future.
Hybridization is one of the oldest molecular techniques and is based on the fact that in nucleic acids a cytosine forms base pairs with a guanine and an adenine forms base pairs with either a thymidine (in DNA) or a uracil (in RNA). In hybridization, the DNA in a sample is rendered single stranded and allowed to combine with a single-stranded probe. Early hybridizations were performed with target DNA immobilized on a nitrocellulose membrane, but nowadays a variety of different solid supports, including magnetic beads, are used. Other variations include the binding of a capture probe to a solid support. After binding of the target, the probe can hybridize. Probes can be labeled with a variety of reporters, including radioactive isotopes, antigenic substrates, enzymes or chemiluminescent compounds. For an overview of hybridization see references (344) and (158). Despite the fact that hybridization is a relatively old technology, new developments lead to new applications. One important development is molecular beacons (discussed below).
PCR is well known and will be discussed only briefly. Two years after its first description by Mullis (202), and thus only 15 years ago, the first diagnostic application of PCR was published by Saiki et al. (281). The technique became broadly used after the introduction of a thermostable DNA polymerase from Thermus aquaticus (Taq DNA polymerase) (280) and the development of automated oligonucleotide synthesis and thermocyclers. PCR involves cycles of heating the sample for denaturing, annealing of the primers, and elongation of the primers by a thermostable DNA polymerase. In theory, each round of amplification gives a doubling of the number of DNA target molecules, but the process is seldom 100% efficient because of the presence of inhibitors, and in later rounds of amplification DNA polymerase may become limited.
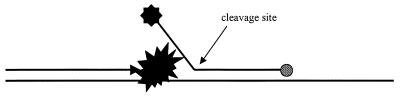
However, during the last few years, new developments in labeling technology have expanded the applicability of PCR. One such development was the use of 5′-fluorescence-labeled oligonucleotides that were blocked at their 3′ ends, thereby preventing elongation by DNA polymerase. Besides this special oligonucleotide, PCR has two traditional oligonucleotides which function as primers and are chosen in regions flanking the special oligonucleotide. The special oligonucleotide hybridizes with the target and is removed by the 5′→3′ exonuclease activity of Taq DNA polymerase during primer extension, resulting in enhanced fluorescence that can easily be detected (Fig. 1) (121).
FIG. 1.
Schematic representation of 5′→3′ exonuclease cleavage of a 5′-labeled (small black star) probe with a 3′-phosphate (grey circle) extension blocker. After cleavage of the label from the probe by DNA polymerase (large black star), the small labeled fragments generated can be separated from the larger probe.
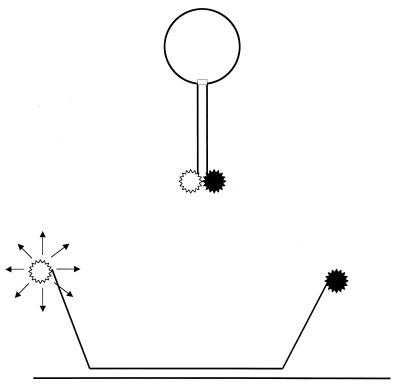
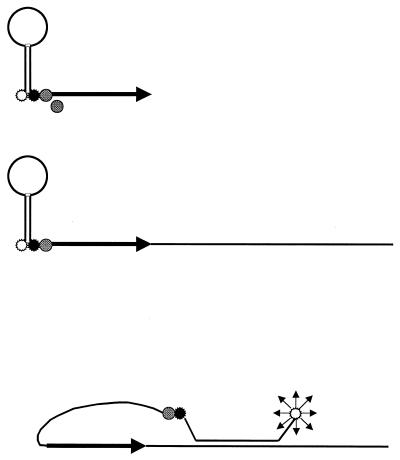
Another advance is the development of molecular beacons. Basically, molecular beacons are hairpin-shaped oligonucleotide probes with a fluorophore attached and a molecule that quenches this fluorescence when it is next to the fluorophore. On hybridization with the target, the fluorophore and quenching molecule are spatially separated and fluorescence is possible (Fig. 2) (360, 361). In principle, the use of fluorophores with different emission spectra makes it possible to discriminate multiple targets. The addition of molecular beacons to PCR amplifications makes possible real-time monitoring of amplification. Furthermore, it allows a relatively easy quantitative PCR (360, 361). This technology has now been commercially realized with the TaqMan (ABI/Perkin-Elmer Corp., Foster City, Calif.) and Lightcycler (Roche Molecular Biochemicals, Mannheim, Germany) systems. A variation on this theme is the Scorpions primer (Oswell Research Products, Southampton, United Kingdom) (393). In this PCR-based method, the primer, probe, and fluorescent label are integrated into one molecule and form part of a homogeneous (closed-tube) assay (Fig. 3).
FIG. 2.
Principle of molecular beacons. The stem of the hairpin is less stable than the hybridization of the specific probe (loop region) with its target (top). Hybridization leads to denaturation of the stem and the physical separation of the fluorophore (white star) and its quencher (black star), allowing fluorecence to occur (bottom).
FIG. 3.
The Scorpions primer is an extension of molecular beacons. To a molecular beacon, a blocker (grey circle) and PCR primer are added (top). The blocker prevents the copying of the molecular beacon part of the molecule. After one round of amplification (middle), the molecular beacon extension of the primer is able to hybridize with the newly synthesized DNA strand, allowing fluorescence (bottom).
In PCR-single-strand conformation polymorphism (PCR-SSCP), the PCR amplication product is denatured into two single-stranded molecules and subjected to nondenaturing polyacrylamide gel electrophoresis. Under nondenaturing conditions, the single-stranded DNA (ssDNA) molecule has a secondary structure that is determined by the nucleotide sequence, buffer conditions, and temperature. The mobility of the ssDNA molecule depends on both its size and secondary structure. ssDNAs at different positions in the gel indicate a difference in sequence. The technique was originally described for the detection of mutations in oncogenes and allelic variants in human genes (228, 229). PCR-SSCP is capable of detecting more than 90% of all single-nucleotide changes in a 200-nucleotide fragment (115).
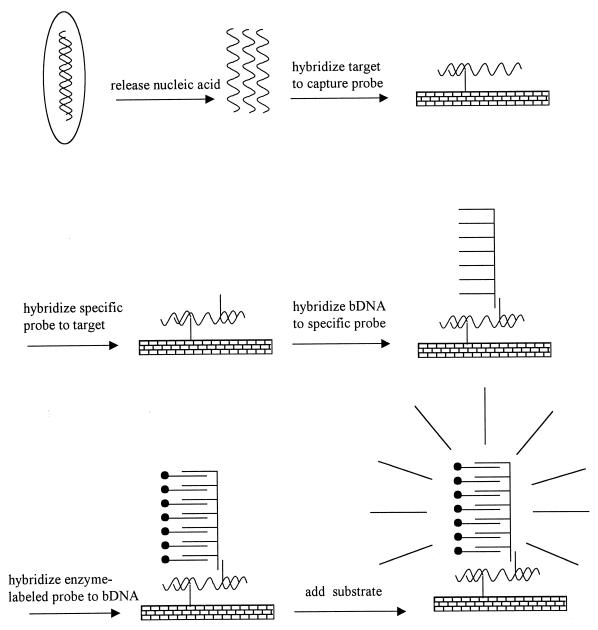
Branched DNA (bDNA) was developed by Chiron Corp. and uses multiple hybridization sites for enzyme-coupled probes (221, 285) (Fig. 4). Target-specific probes bound to a solid surface are allowed to capture target ssDNA. A second probe is allowed to hybridize with the target. This probe has a 5′ extension that does not hybridize with the target. This extension can hybridize with a bDNA probe. This probe has a bristle-like structure. At least 15 bristles are attached to each probe, and as many as three alkaline phosphatase reporter molecules can bind to each bristle. Using multiple target-specific probes for each target nucleic acid, up to 1,700 enzyme molecules can be bound to a single target molecule. A signal is generated by the addition of a chemiluminescent substrate. The sensitivity of the system is in the range of 103 to a 105 thousand target molecules.
FIG. 4.
Schematic presentation of bDNA amplification. For details, see the text. Reprinted from M. N. Widjojoatmodjo, Diagnosis of infections based on DNA amplification: obstacles and solutions, Academic thesis, University of Utrecht, Utrecht, The Netherlands, 1995, with permission of the author.
DNA sequencing is almost universally performed by dideoxy sequencing (287) and is a well-known technique. Technological developments over the past few years, largely driven by the genome-sequencing efforts, have led to advances in DNA sequencing. These developments, such as the ability to read longer sequences faster and cheaper, brought DNA sequencing within the capabilities of at least some diagnostic laboratories and is the method of choice for determining the resistance of human immunodeficiency virus to antiviral drugs (301).
DNA arrays and DNA chips are based on the principle of hybridization. DNA arrays and chips are devices which allow the mass screening of sequences. In contrast to conventional hybdrization assays, where target DNA is blotted onto a membrane, a large collection of probes is bound to a solid surface. The target DNA is generally tagged with a fluorescent label, and hybridization is detected by using an epifluorescence microscope. In DNA arrays cDNA fragment probes are usually used, whereas DNA chips employ oligonucleotides. Arrays are larger and generally use either a nylon membrane type of material or glass as the solid surface, whereas DNA chips use either glass or silicon. The fragment probes are applied to the solid surface after they are generated, whereas oligonucleotides are either applied after synthesis or synthesized in situ. Various schemes for applying probes onto solid surfaces have been reported (see, e.g., references 43, 217, 275, and 310). On DNA arrays, cDNAs or PCR products are attached to a solid surface and used for large-scale assessment of gene expression by measuring mRNA levels. On DNA chips, most often oligonucleotides are used which can be used not only for measurement of gene expression but also for sequencing (sequencing by hybridization) (43). The sequencing strategy can be explained as follows. A complete set of 65,536 octamer probes, each in a separate spot on a solid phase, is mixed with a 12-mer target oligonucleotide, AGCCTAGCTGAA. When only perfect hybridization is considered, five of the probes will bind the target oligonucleotide. Alignment of these overlapping octamer probes will reconstruct the complement of the 12-mer target (Fig. 5). In its first application (43), 256 tetranucleotides were generated in situ on a solid-phase support by photolithographic techniques. The DNA chip generated proved to be specific for the detection of complementary octanucleotides which were fluorescently labeled. The production of more than 400,000 different 20-mer oligonucleotides on a 1.6-cm2 glass slide should be feasible (101). For reviews, see references 94 and 105.
FIG. 5.
Principle of oligonucleotide array sequencing. Alignment of the overlapping probes reconstructs the complement of the original target (see the text for details).
Besides the molecular techniques described above, a number of other amplification techniques are used in the clinical microbiology laboratory, although not to detect antibiotic resistance determinants. These techniques include the DNA amplification techniques of strand displacement amplification (377, 378) and ligase chain reaction (16, 19, 25, 398, 405) and the RNA amplification techniques of Qβ replication (156) and self-sustained sequence replication or nucleic acid-based sequence amplification (55, 148). This latter method can be modified to amplify DNA (99).
The variety of molecular techniques used for diagnostic applications demonstrate that no universal technique exists which is optimal for detection of nucleic acids. The choice of a particular technique is also dependent on the information required or the targets under consideration, but some techniques are more favored than others. New techniques continue to be developed that involve a new approach to amplification, hybridization, formats, and labels (158).
ANTIBIOTIC RESISTANCE IN MYCOBACTERIUM TUBERCULOSIS
Introduction
In the wake of the human immunodeficiency virus epidemic and the breakdown of medical services in several Eastern European countries, the incidence of tuberculosis is rising rapidly. Of note, the treatment of tuberculosis is threatened by the emergence of multidrug-resistant strains of Mycobacterium tuberculosis. M. tuberculosis is usually treated with only a limited number of antimicrobial agents, the most important ones being rifampin, isoniazid, streptomycin, and ethambutol. Resistance to rifampin is conferred by mutations resulting in at least eight amino acid substitutions in the RpoB subunit of RNA polymerase (335). Isoniazid acts by inhibiting an oxygen-sensitive pathway in the mycolic acid biosynthesis of the cell wall. At least four genes have been described to be involved in resistance to isoniazid: the katG gene, which encodes a catalase; the inhA gene, which is the target for isoniazid; and the oxyR gene and neighboring aphC gene and their intergenic region (739). Streptomycin resistance has been associated with mutations in the rrs gene encoding 16S rRNA and the rspL gene encoding the S12 ribosomal protein (69, 196, 208). Ethambutol resistance is associated with an altered EmbB protein (2, 322), a protein involved in the synthesis of the cell wall component arabinogalactan.
Because the organism is slow growing, traditional diagnosis is time-consuming. Traditional phenotypic determination of resistance may take up to 10 weeks after referral of a sample to the laboratory, but both commercial and in-house amplification assays can greatly improve the detection time. Therefore, it is not surprising that within the past 8 years a multitude of different resistance assays based on molecular techniques were specifically developed for M. tuberculosis. However, many laboratories have had trouble with the technical rigor imposed by these assays (222). For review of mycobacterial resistance, see reference 128.
Rifampin Resistance
One of the first assays for the detection of rifampin resistance using PCR-SSCP was published by Telenti et al. (336). In a second paper the assay was more extensively evaluated both in a manual format with radioactively labeled amplification products and with 5′-fluorescein-labeled primers for detection on an automated DNA sequencer (337). Evaluation of the results showed that all 17 of the then known mutations in the rpoB gene leading to resistance could be detected. Equally important, the assays could be applied to minimally grown cultures in Bactec 12B medium with a growth index of ≤100 or on sputa with at least 10 organisms per field at a magnification of ×250. This clearly established the potential of PCR-SSCP as a powerful technique for the early detection of antimicrobial drug resistance in M. tuberculosis. The application of rifampin resistance detection by PCR-SSCP to cerebrospinal fluid specimens from patients with tuberculosis of the central nervous system also yielded excellent results (289).
PCR-SSCP requires careful control over electrophoresis conditions, which is difficult to achieve in many laboratories. This recognition led to a comparison of PCR-SSCP and dideoxy fingerprinting (84). Dideoxy fingerprinting is in fact an extension of SSCP. After PCR amplification of the gene fragment of interest, a second PCR is performed with a radioactively labeled primer. A dideoxynucleotide is added, which leads to chain termination similar to that obtained in dideoxy sequencing. The products are then analyzed in a similar manner to that in SSCP. Because more fragments are generated, differences between the susceptible and resistance types are more easily obtained in accordance with conventional susceptibility testing and PCR-SSCP. A drawback of this method is its use of a radioactive label. However, by using fluorescent labels, this assay can probably be adapted for use with an automated sequencer.
However, Kim et al. (150) observed that PCR-SSCP reported some isolates as resistant whereas their phenotype was susceptible, but in these isolates the part of the rpoB gene that was amplified contained a silent mutation and a deletion of two amino acids. Apparently, these mutations do not affect the susceptibility to rifampin. These authors therefore concluded that sequencing probably could rule out false-positive results.
Direct testing of a clinical specimen for resistance to rifampin by PCR without prior species determination is believed to be difficult because of the high levels of homology reported between different mycobacterial species, but Whelen et al. (392) devised a rpoB-based seminested amplification which was specific for M. tuberculosis. The assay correctly identified 21 of 24 culture-positive specimens, 13 of which were acid-fast smear negative in a panel of 51 clinical specimens. Three specimens were false-positive and tested negative after aerosol carryover was eliminated. This assay demonstrates that concurrent resistance determination and species identification are possible. However, it also clearly illustrates the dangers of (hemi)nested PCR assays and the need to carefully eliminate potential contamination of amplification reactions by unintended target DNA. SSCP analysis and automated sequencing of these isolates both showed that one isolate was resistant to rifampin and the others were susceptible, in accordance with phenotypic testing.
This group also analyzed the results of the heminested PCR using sputum samples and heteroduplex analysis (401). A total of 655 sputa were tested. The assay correctly detected 41 of 44 culture-positive sputa. In addition, 19 culture-negative sputa were identified as positive. Three assay-positive isolates belonged to either M. avium or M. tuberculosis. Thirty-five sputa which contained non-M. tuberculosis complex bacteria were negative in the assay as well as all other samples. The heteroduplex assay identified 39 of the 44 culture-positive isolates as rifampin susceptible, whereas Bactec radiometric susceptibility testing reported 38 susceptible isolates. The study showed the feasibility of a PCR-based assay directly with sputum samples for both identification and rifampin resistance determination. This is especially true for patients with either smear-positive untreated tuberculosis or suspected of having multidrug-resistant tuberculosis.
A modification of the heteroduplex method has also been described for the detection of rpoB-associated rifampin resistance (209). The region of interest was amplified by PCR with primers which contained the T7 or SP6 RNA polymerase promoter. The amplified fragments were then transcribed using either T7 or SP6 RNA polymerase. The resulting RNA was then hybridized to a reference RNA molecule (e.g., susceptible type sequence) and treated with RNase. In the case of a mismatch, the heteroduplex was cleaved whereas a homoduplex could not be cleaved. This assay reached 100% sensitivity and 96% specificity and took less then 24 h to perform after receipt of an isolate or a smear-positive sample.
Another modification of the heteroduplex assay is the double-gradient (DG) variant of the original denaturing gradient gel electrophoresis (DGGE). DG-DGGE uses both a temperature and a polyacrylamide gradient to optimize separation. DG-DGGE was able to detect all rifampin-resistant isolates among a set of 117 isolates (290). Direct testing on clinical samples showed that the rpoB-specific PCR was positive for 54 of the 84 IS6110 PCR-positive isolates (IS6110 is specific for M. tuberculosis). Of the rpoB PCR-positive isolates, 30 that were classified as susceptible by DG-DGGE were also susceptible in a conventional assay. The remaining 24 isolates were classified as rifampin resistant, but 1 isolate proved to be susceptible by conventional testing. The single discrepancy was due to the presence of at least two clones, one of which carried a mutation in the rpoB gene. Analysis of 48 cerebrospinal fluid specimens showed complete agreement between genotypic and phenotypic testing. The results obtained by direct testing of clinical specimens showed that the type of specimen can have an important effect on the outcome of a molecular assay. This underscores the need for controls and appropriate sample preparation methods.
Analysis of rpoB gene amplification products is possible not only by SSCP or heteroduplex analysis but also by a line probe assay (LiPA; Innogenetics) (63). In the nested PCR, the inner primers were biotinylated and nine different probes were immobilized on a nitrocellulose membrane in addition to a color control and an M. tuberculosis probe. The DNA was directly isolated from sputum and lymph node biopsy specimens. The results of this assay correctly matched classical resistance testing in 65 of 67 isolates. However, 2 isolates of a total of 23 phenotypically resistant isolates gave a susceptible hybridization pattern. DNA sequencing proved the absence of mutations in the sequence used for the assay, thus suggesting another mechanism of rifampin resistance, although this mechanism was not clarified. This underscores that although PCR assays may be valuable tools, they are not absolute in their outcome and unrecognized mechanisms of resistance may lead to therapeutic failure.
A second study (278) evaluated the ability of LiPA to detect mutations in the rpoB gene of 107 M. tuberculosis and 52 non-M. tuberculosis isolates in pure culture, as well as 61 and 203 unidentified clinical isolates that were rifampin resistant and susceptible, respectively. No discrepancies with sequencing were observed. All susceptible isolates were correctly identified. Only four resistant isolates yielded a resistant phenotype, but no mutation was observed in both sequencing and LiPA. It was not clear whether mutations elsewhere in the rpoB gene or a different mechanism of resistance were involved. In another evaluation of this assay, sputum and bronchoalveolar lavage fluid from only two patients were used. Within 48 h, the assay correctly identified mutations in the rpoB gene associated with resistance as confirmed by sequencing (104). A fourth evaluation of the assay with 30 M. tuberculosis isolates showed a 93.3% concordance between culture and LiPA, whereas sequencing was completely concordant with culture (192).
A South African study (149) into the detection of rifampin resistance compared the results from heteroduplex analysis with sequencing and LiPA. Sequencing revealed that some mutations in the rpoB gene apparently do not lead to resistance to rifampin, and a few isolates did not have mutations in the analyzed region. LiPA correlated with the sequence results. Heteroduplex hybridization results also agreed with DNA sequencing, although the presence or absence of some mutations was difficult to ascertain because of small differences in mobility when the mutant sequence was compared with the susceptible wild-type sequence. Despite these limitations, the authors concluded that the assay was a valuable tool for identifying and managing patients with multidrug-resistant M. tuberculosis.
The LiPA and the RNA-RNA heteroduplex assay (MisMatch Detect II; Ambion) for rifampin resistance were first compared with a phenotypic assay by using 16 M. tuberculosis isolates. The MisMatch assay missed one sample. A further evaluation was performed with 38 sputa and bronchoalveolar lavage fluid specimens and 21 isolates submitted by clinicians. The LiPA and MisMatch assay correlated with 36 and 38 of the primary samples, respectively, whereas all 21 isolates were classified correctly (383).
Another examination of the LiPA involved 75 clinical specimens from 70 patients suspected of having tuberculosis (193). A final diagnosis of tuberculosis was reached for 51 of these patients. In a nested PCR, only 31 specimens yielded a PCR product and 30 of these hybridized with the M. tuberculosis-specific probe of the LiPA. Nevertheless, culture yielded only 18 positive specimens. In the PCR-LiPA, only 13 of these culture-positive isolates yielded a positive result. For these same isolates, LiPA testing correlated with the resistance phenotype for 11 isolates. One isolate was phenotypically resistant but susceptible according to LiPA; however, sequencing did not reveal a mutation in rpoB. The other isolate was phenotypically susceptible but yielded a resistant genotype in LiPA, which was confirmed by sequencing.
Isoniazid Resistance
PCR-SSCP has also been used for the detection of katG-related resistance to isoniazid. A major problem, however, is that compared to rifampin resistance caused by mutations in the rpoB gene, the mutations causing isoniazid resistance are spread over a much longer sequence. This, combined with the fact that PCR-SSCP is most discriminatory when the amplified fragments are less than 400 bp, necessitates the analysis of multiple amplification fragments. In addition, all or part of the katG gene is lacking, meaning that appropriate controls are needed to distinguish the absence of amplification products from PCR inhibition. Initially, resistance was believed to be caused by complete deletion of the katG gene (6), but with the use of PCR and PCR-SSCP, it was shown that both deletion and mutation of the katG gene could lead to resistance (410). Heym et al. (118), who studied the relationship between mutations in the katG gene and isoniazid resistance, developed a discriminatory PCR-SSCP assay which involved 12 amplification products. Although this approach was successful, it is cumbersome to perform on a routine basis.
An extension of PCR-SSCP used Cleavage I, a structure-specific endonuclease (73). After PCR amplification, the DNA fragments were made single stranded and allowed to assume their secondary structures. After incubation with Cleavage I, the resulting fragments were analyzed by acrylamide gel electrophoresis. The technique allowed for a better discriminatory power than classical SSCP. Results with a 620-bp fragment of the katG gene showed the potential applicability of this technique to the detection of resistance in M. tuberculosis. This alternative technique may use larger fragments. This would alleviate the problem of the large number of PCR-SSCPs required for full coverage of the katG gene.
Multidrug Resistance
In a complex study, Nachamkin et al. (207) compared techniques to detect resistance caused by different resistance genes. For the katG gene, restriction fragment length polymorphism (RFLP) analysis of a 620-bp amplified PCR fragment was used. RFLP analysis detected the S315T mutation in 12 of 27 specimens, but other mutations were also responsible for isoniazid resistance. Rifampin resistance caused by mutations in the rpoB gene was analyzed by both heteroduplex analysis and sequencing. Heteroduplex analysis for the rpoB gene reached 76.2% sensitivity and 97.2% specificity compared to conventional testing, but sequencing reached 100% sensitivity and specificity. RFLP, despite its shortcomings, was also used to detect resistance to streptomycin caused by mutations in the rslP gene. Although the specificity reached 100%, the sensitivity was only 28.1%. Direct sequencing reached 99% specificity and 67.7% sensitivity. The authors concluded that the results only partly matched those of conventional testing. This is not entirely unexpected, because not all known mutations involved in resistance lead to a change in potential restriction sites. One mutation, although implicated in resistance, did not result in a resistant phenotype. The low sensitivity of both methods appeared to be related to other mechanisms of resistance to streptomycin, e.g., via the rrs gene. The RFLP results are in agreement with the results obtained by Temesgen et al. (338), who compared PCR-SSCP with RFLP for detection of the R463L mutation. The data obtained by these authors indicate that the appropriate molecular method must be chosen and that all resistance genes known to be involved in a particular resistance within a species should be investigated. Nevertheless, it cannot be excluded that unknown mechanisms play a role in resistance, and these mechanisms will be missed by these techniques.
The development of multidrug resistance in classical treatment options for M. tuberculosis has led to the consideration of alternative antimicrobial agents including fluoroquinolones. However, quinolone resistance was observed among clinical isolates and was easily induced with ciprofloxacin at 2 μg/ml (frequency, 1 in 107 to 108), although no induction of resistance was observed with ciprofloxacin at higher concentrations (333). Therefore, a PCR-SSCP was evaluated to distinguish fluoroquinolone-susceptible from fluoroquinolone-resistant isolates (333). The results demonstrated that discrimination between these isolates was possible.
Isolates often show multidrug resistance, and the number potential mutations involved means that the number of assays needed to cover them all can be quite large. Therefore, a number of PCR assays have been developed which do not directly determine the presence or absence of resistance-causing genes and mutations but either identify multiresistant strains by other properties or monitor the effect of chemotherapy. One such completely different approach to the determination of resistance in M. tuberculosis was taken by Cangelosi et al. (38). This group developed a reverse transcriptase PCR (RT-PCR) probe assay that was specific for M. tuberculosis precursor rRNA. Precursor rRNA carries terminal stems which are removed when mature rRNA subunits are formed. The number of these stems present in the bacterial cell is markedly affected by inhibition of RNA synthesis. Hybridization results showed that the assay was specific for M. tuberculosis, and resistance to rifampin and ciprofloxacin could correctly be predicted. As expected, no influence on the levels of precursor rRNA by isoniazid and ethambutol were seen. Instead of RT-PCR, quantitative PCR was also used to determine resistance to isoniazid. Bacteria were inoculated into medium containing different concentrations of isoniazid. After 1 to 3 weeks, more than a 1-log-unit difference in the amount of DNA was observed between isoniazid-resistant and susceptible isolates (1).
The problems with multiresistant M. tuberculosis isolates belonging to strain W, which was associated with several outbreaks in New York state, led to the development of a PCR assay to identify this strain (250). This assay used an internal control and primers to identify a specific fragment called NTF-1 and the orientation of this fragment, which were considered specific for this strain. The assay successfully identified all 48 W strain isolates among 193 isolates tested.
Molecular diagnostic techniques play an important role not only in detecting resistance but also in monitoring chemotherapy. This was demonstrated as early as 1994 in a small study (171). Sixteen patients with tuberculosis were monitored, who all became smear negative after 2 months of treatment. Treatment continued for 4 months. Although all the patients were smear negative after two months, four, two, and one patients were still positive in M. tuberculosis-specific PCR after 2, 3, and 6 months, respectively. These results demonstrated that in principle the identification of patients at high risk for a relapse is possible.
New Developments
Recent developments in molecular techniques such as molecular beacons and DNA chips are probably well suited for the detection of resistance in M. tuberculosis, and the first applications have been published and will be discussed below.
Piatek et al. (248) investigated the use of molecular beacons for the detection of mutations in rpoB. This group designed five fluorogenic probes which were used in five separate PCRs which were monitored in real time. In addition, a species-specific internal control and another complementary to a species-specific multicopy gene were used. The outcome correctly predicted the susceptibility of the 75 isolates tested. A potential drawback of this method is that new mutations might be missed. However, the big advantage of the technique is that the product formation can be analyzed visually without the need for additional equipment.
DNA chips are logical candidates for the analysis of mutations involved in resistance to antimicrobial agents used for tuberculosis treatment. Although the technique potentially holds great promise, it is still very expensive. Head et al. (116), in a limited study involving only nine rifampin-resistant isolates, investigated the application of DNA chip technology to the detection of rifampin resistance. The chip technology chosen sequenced the rpoB gene. Sequencing by chip technology identified two point mutations in all nine isolates. This group concluded that the technique is suitable for the detection of mutations.
Conclusion
Molecular techniques hold great promise for the detection of susceptibility and resistance to antimicrobial agents in M. tuberculosis. In fact, a number of useful assays already exist. However, the use of molecular techniques for the analysis of resistance is dependent on the prevalence of the resistance-causing mutation. With the current assays, this coverage appears incomplete and may vary between different geographical areas (see, e.g., reference 149). Nevertheless, molecular techniques may be especially helpful in the quick identification of multidrug-resistant isolates and the evaluation of culture-negative specimens. It should be kept in mind that isolates that are susceptible according to molecular assays may contain other mechanisms of resistance.
However, molecular techniques, owing to their extreme sensitivity, are prone to contamination, and their execution requires extra care as demonstrated (289).
RESISTANCE TO β-LACTAM ANTIBIOTICS
Mechanisms of Resistance
β-lactam antibiotics are among the most commonly used antimicrobial agents. They act on penicillin binding proteins (PBPs), which are involved in cell wall synthesis. Penicillin, a β-lactam antibiotic, was one of the first antibiotics. β-Lactam antibiotics are still the most widely used and diverse class of drugs used clinally, and new members are still being developed. It is therefore not surprising that resistance to many β-lactam compounds is commonplace and still evolving. Resistance is most often caused by the presence of β-lactamases, but mutations in PBPs resulting in reduced affinity for β-lactam antibiotics are also commonly observed. Resistance is less frequently caused by reduced uptake due to changes in the cell wall or active efflux. Several classification systems for β-lactamases have been published. One classification scheme is based on their nucleotide sequence, classes A through D (36). Class A, C, and D enzymes have a serine at their active site, while class B enzymes have four zinc atoms at their active site. Class A enzymes are highly active against benzylpenicillin. The extended-spectrum β-lactamases (ESBLs) also belong to this class. ESBLs also inactivate benzylpenicillins as well as some cephalosporins and/or monobactams. Class B β-lactamases are equally active against penicillins and cephalosporins, and at least some of these enzymes are able to inactivate carbapenems. Class C genes are usually inducible, but mutations can lead to overexpression. Class D is composed of the OXA-type enzymes, which are capable of hydrolyzing oxacillin. Genes encoding β-lactamases can located either on plasmids or the bacterial chromosome and are found among both gram-negative and gram-positive organisms. For purposes of discussion, we divide the β-lactamases into metallo-β-lactamases, ESBLs, and other β-lactamases. The β-lactamases in the last group are most common and were the first β-lactamases encountered in clinical practice. These β-lactamases will be referred to as common β-lactamases.
Methicillin-Resistant Staphylococci
Introduction of methicillin into medical practice in the early 1960s quickly resulted in the isolation of methicillin-resistant staphylococci. Methicillin and its analogues bind and inactivate the PBPs involved in cell wall synthesis. Low-level resistance is generally the result of β-lactamase overproduction, increased levels of intrinsic PBPs, or reduction of their binding affinity (17, 351). High-level resistance is always dependent on the expression of an alternative PBP (PBP2a) encoded by the mecA gene, which has low affinity for most β-lactam antibiotics (21, 113). The mecA gene is located on the chromosome. Expression of mecA is either constitutive or inducible by some β-lactam antibiotics, but not by methicillin or oxacillin, or heterogeneous, with only a few cells in a population expressing the gene (21).
Ten years ago a DNA fragment used as a probe for the detection of mecA in Staphylococcus aureus and coagulase-negative staphylococcus (CNS) isolates was described (11). The probe was tested with both a radioactive label and digoxigenin as a label. The results showed that the nonradioactive label performed as well as the radioactive label, bringing probes a step closer to routine use in a clinical laboratory. The probe assay was in complete agreement with the spread plate screening technique for assessing methicillin resistance in S. aureus isolates. Probe assay results for CNS did not correlate completely with the spread plate technique results, but differences with the broth microdilution and agar dilution methods were also observed. The interpretation of the result for CNS should take into account that the National Committee for Clinical Laboratory Standards (NCCLS) lowered the the breakpoint for oxacillin as determined by broth microdilution from 4 to 0.5 μg/ml (211). However, we were not able to reinterprete the results, because MICs for the isolates were not provided, but bigger differences between the techniques are probable. A year later, a digoxigenin-labeled fragment probe to mecA generated by PCR was described (178). Evaluation with isolates of S. aureus and CNS showed some discrepancies. Borderline-resistant S. aureus isolates (MIC, 8 μg/ml) were probe negative in some cases but produced β-lactamase. This β-lactamase may slowly hydrolyze oxacillin. The correlation for CNS was good, but when the new NCCLS breakpoint criteria were applied, some discrepancies resulted; isolates reported as resistant lacked the mecA gene.
In the past decade, several PCR assays have been described for the detection of methicillin-resistant S. aureus. Murakami et al. (204) evaluated their PCR against approximately 200 S. aureus isolates. Two isolates were mecA negative in the PCR and resistant to oxacillin but were not resistant to methicillin in a broth microdilution assay. Three isolates were mecA positive but susceptible to both methicillin and oxacillin. Evaluation against 100 CNS isolates showed a larger number of discrepancies, especially oxacillin-susceptible but mecA-positive isolates. However, these results would be different when the new NCCLS interpretative breakpoints for oxacillin and CNS were applied. In the same year Predari et al. compared the results of a mecA PCR with those obtained by dot blot hybridization and phenotypic testing for 74 CNS isolates (256). The PCR results correlated perfectly with phenotypic analysis when high inocula were used (108 CFU), an effect also observed by Hedin and Löfdahl (117). PCR performed better than hybridization did. This difference was believed to be the result of the loss of the mecA gene in a large proportion of the bacteria which belong to certain strains. These isolates also gave a weak amplification signal. Another study (349) investigated a total of 58 clinical isolates of S. aureus. Based on MICs of oxacillin and methicillin, 27 of these isolates were resistant and 1 isolate showed an oxacillin MIC of 32 μg/ml and a methicillin MIC of only 2 μg/ml. Thirty isolates were negative in the PCR, and 28 were positive for the mecA gene. These results were confirmed by a hybridization assay on HincII-restricted DNA probed with a mecA-specific 30-mer oligonucleotide. The PCR-positive isolates included not only the oxacillin-resistant but methicillin-susceptible isolate but also isolates with a methicillin MIC of 2 μg/ml and an oxacillin MIC of either 1 or 2 μg/ml. After preincubation of these isolates with ceftizoxime, the MICs of both methicillin and oxacillin increased for all but one isolate. This demonstrates that these isolates had an inducible phenotype. No oxacillin- or methicillin-resistant isolate was negative in either the PCR or hybridization assay.
An integrated PCR detection assay was published by Ubukata et al. (363). This group used a simple mecA PCR with one of the primers carrying a biotin group and the other primer carrying a dinitrophenol group. After PCR, the product was captured on a 96-well microtiter plate coated with streptavidin. After capture and washing, an anti-dinitrophenol antibody conjugated to alkaline phosphatase was added, followed by substrate. The sensitivity of the assay was >5 × 102 CFU for S. aureus and >5 × 103 CFU for CNS. A total of 97 S. aureus isolated and 64 CNS isolates were tested. One S. aureus isolate and 22 CNS isolates carried the mecA gene but were phenotypically susceptible using the older NCCLS breakpoints. The assay was also applied to the detection of mecA in staphylococcal isolates (n = 40) directly in blood culture bottles. Thirty-three isolates (18 S. aureus and 15 CNS) in the culture bottles were resistant to oxacillin, 31 of which were also positive in the PCR assay. Two CNS isolates were PCR negative. The authors conclude that the assay can be used with reasonable confidence in the clinical microbiology laboratory for the detection of MRSA in blood culture bottles. A possible cause of the difference may be the inhibition of the PCR by components of the blood or culture medium (119).
Comparison of mecA PCR with API ATB Staph (bioMérieux, Balme-les-Grottes, France), oxacillin disk test, agar dilution MIC test, and the BBL Crystal MRSA system (Becton Dickinson, Cockeysville, Md.) using the PCR as “gold standard” with 57 S. aureus isolates showed agreement with the first two methods, but the last two methods missed one isolate (379). However API ATB and the oxacillin disk test reported three isolates as methicillin resistant. The two other methods reported no methicillin-resistant isolates in mecA-negative isolates. The results were poor (negative predictive value of 68 and 82% for the BBL Crystal MRSA system and API ATB Staph, respectively) for 100 CNS isolates belonging to a number of different species. It was concluded that there was a good correlation with the oxacillin MIC for S. aureus in contrast to CNS, but it should be noted that this conclusion was based on the old breakpoints (211). No MIC data for individual isolates were provided, so no evaluation based on the current breakpoint could be made.
The PCR assays with only a single primer pair as described above are robust and simple to perform. However, these assays are vulnerable to inhibition. The addition of a second primer set designed to amplify a gene which is present in all isolates can solve this problem. The use of a species-specific primer also can confirm the identity of the isolate. However, the addition of extra primer pairs leads to increased complexity of the assay, which may be critical when testing is performed directly on clinical samples, although for optimized assays this risk is reduced. Brakstad et al. (32) developed a multiplex PCR in which both the mecA gene and the nuc gene, encoding the S. aureus-specific thermonuclease gene, were amplified. A total of 135 S. aureus isolates and 84 CNS isolates belonging to different species were evaluated. The amplification of nuc-specific sequences was in complete agreement with species determinations. The MICs of oxacillin for S. aureus agreed with the PCR results with the exception of one isolate, for which the MIC was 4 μg/ml. However, a number of discrepant results were reported for CNS using the older NCCLS breakpoints. Three isolates for which the MIC was 8 μg/ml were mecA negative, whereas two isolates for which the MICs were 2 and 1 μg/ml, respectively, were positive for mecA. However, when the new NCCLS breakpoint (211) for oxacillin resistance in CNS (≥0.5 μg/ml) is applied, seven isolates would be resistant but lacking the mecA gene. No false-positive results would be reported. This multiplex could be applied to samples from either blood culture bottles or whole blood. In the latter case, a sensitivity of 20 and 80 CFU per 3-μl sample was obtained for nuc and mecA products, respectively.
A variation using the S. aureus-specific coa (coagulase) gene was described recently (145). Complete agreement was observed between the results of the coa-specific PCR and conventional slide coagulase testing with S. aureus and CNS isolates. In contrast to most studies, there was also complete agreement between the mecA PCR and standard disk diffusion testing.
Salisbury et al. (283) developed a PCR for mecA which also included an internal control by amplifying 16S rRNA gene sequences. This study not only involved methicillin-resistant S. aureus (MRSA), methicillin-susceptible S. aureus (MSSA), methicillin-susceptible CNS (MSCNS), and methicillin-resistant CNS (MRCNS), but also involved high-β-lactamase producing S. aureus isolates. The results were in full agreement for MSSA, MRSA, and MRCNS, but one of the β-lactamase-overproducing isolates and three MSCNS isolates were also mecA positive. However, it should be noted that for CNS isolates the old breakpoint of 8 μg/ml for resistance was used. Because no MICs were listed in this study, no comparison with the new breakpoint was possible (211).
Using a multiplex PCR assay with the gyrA gene as an internal control, Zambardi et al. (407) evaluated 468 S. aureus isolates for methicillin resistance. Good results were obtained compared with MIC testing, the only exception being β-lactamase producers. When a part of the 16S rRNA gene was amplified as an internal control in conjunction with amplification of mecA, only four discrepant results were obtained when results for 228 S. aureus isolates were compared with those of agar dilution and disk susceptibility tests (93). Three of these discrepancies were due to β-lactamase hyperproducers. No discrepancies were observed for MSCNS isolates, but 11 mecA-positive isolates were phenotypically susceptible, again confirming findings with other studies of CNS isolates. This PCR was also compared with a hybridization assay (155). This assay was composed of a capture probe which was immobilized on magnetic beads and a acridinium ester-labeled probe for chemiluminescence detection. A bacterial lysate was mixed with the probes, and, after hybridization, excess material was easily removed after magnetization of the mixture. For S. aureus (n = 147), complete agreement between the methods was observed, while for CNS isolates (n = 253) only one mismatch was observed (one isolate was PCR negative). However, compared to disk diffusion, 14 isolates were CNS mecA negative but phenotypically resistant. Of these 14 isolates, 13 proved to be β-lactamase hyperproducers. Thirteen CNS isolates were mecA negative and phenotypically resistant. Only three of these isolates proved to be β-lactamase hyperproducers. In contrast to most other studies described, no phenotypically susceptible but mecA-positive isolates were observed.
A limited study by Kitagawa et al. (151) in Japan demonstrated the ability of PCR to directly detect MRSA in clinical specimen. This group developed a nested multiplex PCR for mecA and TSST-1 for use with 1-ml blood samples. The sensitivity of the PCR reached 103 CFU/ml of blood, and the result was obtained in 4 h, whereas culture took 48 h. A total of 35 patients with high fever or watery diarrhea who had undergone major gastrointestinal surgery and 6 healthy volunteers were examined. The PCR was positive for both products for isolates from 12 patients from whom MRSA was also cultured. None of the other patients or healthy volunteers were positive by culture or PCR. This result clearly showed the advantage of PCR over conventional culture, although the sample size was small.
Ünal et al. (365) developed a mecA PCR with the femA gene involved in peptiglycan synthesis of S. aureus as internal control. The results obtained by testing 79 clinical staphylococcal isolates showed that femA was S. aureus specific, and the results of PCR and phenotypic testing generally agreed. In a second study, results obtained using these primers for amplification and microdilution testing were compared. Among 1,450 S. aureus, 186 isolates were resistant to methicillin but 13 isolates for which the MIC was 4 μg/ml were mecA negative (366). A study in Turkey (153) using the same PCR method for the detection of mecA in both S. aureus and CNS was judged by the authors as usable for the clinical microbiology laboratory. In this study, hyperproducing β-lactamase strains demonstrating a methicillin-resistant phenotype but mecA-negative genotype were also reported.
Towner et al. (352) described a multiplex PCR based on the mecA and femB genes. The latter gene is involved in peptidoglycan synthesis and is believed to be absent in CNS. Primers for the former gene were labeled with either dinitrophenol or biotin, whereas the primers for the latter gene were labeled with either dinitrophenol or digoxigenin. Detection was performed using the Clearview (Unipath) immunoassay detection system involving a membrane with anti-biotin and anti-digoxigenin antibodies and blue latex beads coated with anti-dinitrophenol antibodies. A total of 480 S. aureus and CNS isolates were evaluated, and five discrepant results were obtained when femB PCR was compared with conventional identification. Of the 152 S. aureus isolates, 40 were considered MRSA by routine MIC testing whereas only 24 harbored the mecA gene. In addition, two isolates were mecA positive but methicillin- susceptible. A similar result was observed when the assay was used to detect MRSA in routine patient screening swabs.
bDNA signal amplification has also been used to detect the mecA gene (154). The results obtained with 416 clinical staphylocococcal isolates were 100% concordant with the results of a mecA PCR. The results of this assay was subsequently evaluated on BACTEC blood cultures with positive growth indices from 225 patients showing staphylococcus-like organisms on gram stain and compared to PCR (411); conventional identification showed 50 S. aureus and 175 CNS isolates. A total of 122 bottles (16 S. aureus and 106 CNS isolates) were positive in both mecA PCR and bDNA assays, and 102 bottles (including 33 with S. aureus) were negative in both assays. One S. aureus-containing blood culture was positive in the bDNA assay but PCR negative. This erroneous result remained after retesting and was confirmed by phenotypic testing. Despite this discrepancy, the bDNA test has the advantages that it does not require elaborate sample preparation, is not subject to inhibitors known to influence the PCR, and a result can be obtained in 6 h in a 96-well format.
In conclusion, the results obtained with various molecular techniques and isolates compared to phenotypic assays reflect the fact that mecA expression may be regulation dependent (21) but also that other mechanisms, may lead to low-level methicillin resistance (64). In addition, the population of MRSA may be composed of only a few clonal lineages (C. L. C. Wielders, M. Vriens, S. Brisse, E. D. J. Peters, A. T. A. Box, J. Verhoef, F. J. Sehmitz, and A. C. Fluit, submitted for publication), which may bias the results to some extent. Nevertheless, mecA PCR is the most robust and reliable way of detecting oxacillin-resistant staphylococci, and the results of this PCR assay are used as a “gold standard” to compare screening methods to identify MRSA (141, 342). Similarly, this is true for CNS, independent of whether the old or the new NCCLS breakpoint recommendations are followed. Alternative tests may provide a valuable addition, especially for quick initial screening.
Penicillin-Resistant Pneumococci
Resistance to penicillin in pneumococci is due to the presence of altered PBPs, especially PBP2, which have a reduced affinity for penicillin. β-Lactamases have never been detected in pneumococci (108). Streptococcus pneumoniae encodes six PBPs, termed PBP1a, PBP1b, PBP2a, PBP2b, PBP2x, and PBP3. In susceptible strains, these PBPs are highly conserved. The genes for low-affinity PBP1a, PBP2b, and PBP2x found in resistant isolates diverge significantly. These genes are called mosaic genes because part of the sequence is highly homologous to the counterpart in susceptible isolates whereas other parts are highly divergent (71, 162). The divergent regions appear to come from other related species, such as Streptococcus mitis (70, 71). In S. pneumoniae, PBP2b is the main target for penicillin but PBP2x is the target for cefotaxime (157).
Ubukata et al. (362) studied PBP2b-related pencillin resistance in 1,062 pneumococcal isolates. Three primer sets were developed: one set specific for the susceptible (wild-type) PBP2b and the other two sets specific for the forms of low-affinity PBP2b termed types A and B. In addition, a primer set was used for the lytA gene, which encodes autolysin and is S. pneumoniae specific. A total of 614 isolates from 621 phenotypically susceptible isolates yielded a product with the wild-type primers. Only 1.8% of the isolates for which the MIC was ≥0.125 μg/ml yielded a PCR product with the type A primers, and 70.3% yielded a product with the type B primers. The remaining isolates gave no PCR product. However, when only high-level resistance (MIC ≥1 μg/ml) was taken into account, 89.8% of 275 isolates showed a type B PCR product. Phenotypic analysis of the highly penicillin-resistant isolates, which yielded no PCR product (n = 123), showed the presence of a low-affinity PBP, indicating the presence of non-type A or B low-affinity PBP2bs. A comparable PCR was developed by a South African group (74), which used four different primers for the four known South African low-affinity PBP2b genes and a primer for the wild-type gene in combination with a primer chosen in a conserved region for all five types. PCR analysis of both cerebrospinal fluid and colonies obtained after culture of the cerebrospinal fluid specimens showed 100% concordance. These investigators also developed a seminested PCR for the detection of two resistance variants of the gene encoding PBP1a which are involved in high-level penicillin resistance. For 180 of 183 isolates, MIC data and PCR results were in agreement. Differences were caused by unknown mutations (75).
A somewhat different approach was taken by Jalal et al. (130). This group developed primers for the genes of the susceptible forms of PBP2b, PBP2x, and PBP1a. The latter gene is supposedly also present in resistant isolates and was used as an amplification control. A total of 230 pneumococcal isolates from different geographical locations were analyzed. PCR correctly identified 93% of 116 penicillin-susceptible isolates, 85% of the intermediate-resistant isolates, and all 49 highly penicillin-resistant isolates. Only two of the intermediate penicillin-resistant isolates were classified as susceptible by PCR analysis. A third approach using PCR followed by restriction enzyme digestion (PCR-RFLP) distinguished between susceptible and resistant genotypes of the gene for PBP2b (227). All susceptible isolates had identical RFLP patterns. This pattern was also observed in 14 of 30 intermediate-resistant isolates. A large number of different patterns were observed for both intermediate and high-level penicillin-resistant isolates.
The combined data from these PCR assays suggest that the presence of unknown mutations or mosaic genes for the PBPs can result in false-negative results for penicillin-resistant isolates. However, the formation of a PCR product using wild-type (susceptible) primers in a properly controlled PCR assay may be used as a screen for pencillin-resistant S. pneumoniae. The wild-type gene was not associated with high-level resistance.
Several groups used a variety of techniques including PCR in combination with restriction enzyme analysis (PCR-RFLP) to perform epidemiological typing studies on penicillin-resistant S. pneumoniae, but in none of the cases was PCR-RFLP sufficient (68, 98, 126, 188, 406). The results of these studies indicate widespread genetic exchange and clonal dissemination of strains.
Common β-Lactamases
Since the 1980s, a small but steady stream of studies has been published which applied molecular techniques to the detection of resistance caused by β-lactamases. Most of these studies investigated the spread of resistance determniants. For the purpose of discussion, we divided the β-lactamases into metallo-β-lactamases, ESBLs, and common β-lactamases.
One of the first probes described was used to investigate the relationship of TEM-1, which is the mostly frequenctly encountered β-lactamase among Escherichia coli isolates with other β-lactamases. The radiolabeled probe hybridized with TEM-2 and OXA-2 β-lactamase-encoding plasmids. No hybridization was observed with OXA-1-, OXA-3-, HMS-1-, SHV-1-, CARB-1 (PSE-4)-, and PSE-3-encoding plasmids (57), indicating that discrimination between genes encoding β-lactamases frequently encountered among gram-negative pathogens could be discriminated. This probe was also used in colony hybridization a assay of 328 isolates belonging to 11 gram-negative genera (140). TEM genes were detected in 53.6% of the isolates, and the results were 92.7% concordant with the results of isoelectric focusing. Sixteen isolates were positive in hybridization but yielded no result in isoelectric focusing. Although no explanation was offered for this discrepancy, the method was considered convenient for the rapid screening of isolates. Radiolabeled fragment probes were also used to detect the genes for TEM-1, SHV-1, OXA-1, OXA-2, PSE-1, PSE-2, and PSE-4 (127). Some of the probes cross-hybridized with related β-lactamase genes. Despite this drawback, the authors declared the probes a success. At least the probes are useful to indentify β-lactamase genes at the gene family level.
Ouellette and Roy (233) sequenced the OXA-1 β-lactamase gene, and from the data they developed a specific 15-mer oligonucleotide probe. The advantage of an oligonucleotide probe, if well designed, is that its hybridization is more specific than that of a fragment probe. In an extension of their work, a TEM family fragment probe was developed, together with two oligonucleotide probes which discriminated between the two (then known) members, TEM-1 and TEM-2, which differ by only one nucleotide (232). A danger noted by the authors is that when new TEM mutants arise, a negative hybridization may result due to the specificity of the oligonucleotides. No major difference between colony blotting and dot blotting of partially purified DNA was observed. The TEM-1- and OXA-1-specific oligonucleotides were used to assess the presence of these genes in 114 β-lactamase-producing isolates belonging to 16 species of gram-negative bacteria (231). The correlation of the hybridization with isoelectric focusing was 96 and 100% for the TEM-1 and OXA-1 probes, respectively. Five P. aeruginosa isolates with the same serotype originating from the same hospital reacted with the probe, but no silent TEM-1 gene could be detected. The authors suggested increasing the length of the probe to circumvent the problem. The study illustrated that with shorter oligonucleotides, the chance is increased that the same sequence in an unrelated gene will be present in a different organism. On the other hand, longer fragment probes often lack sufficient specificity and therefore also hybridize with (partly) related sequences. In the late 1980s and early 1990s, a number of fragment and oligonucleotide probes were found to detect specific β-lactamase genes including genes for TEM-1 (30, 40, 42), OXA-1 (30), SHV-1 (26), and AmpC (400).
The determination of the presence of TEM β-lactamases in bacteria directly in urine by filtering the urine over nitrocellulose followed by lysis of the bacteria, denaturation, and hybridization with a nonradioactive probe demonstrated problems with direct testing (40). One of the 81 samples tested was positive due to urine pigmentation. A false-negative result was obtained due to insufficient numbers of cells in the sample. One of the isolates did not express the β-lactamase. Whether the gene could be expressed in vivo is not clear, but the result illustrates the problem of phenotypic versus genotypic resistance determinations.
PCR products are generally detected by agarose gel electropheresis. A different approach to the detection of PCR products in a TEM-specific PCR was the use of an immunoassay. In this approach, a 16S rRNA-specific internal control primer and a TEM-specific primer are labeled with dinitrophenol, the second internal control primer is labeled with digoxigenin, and the second TEM-specific primer is labeled with biotin. A membrane is then coated with an anti-biotin detection line and an anti-digoxigenin control line. The PCR product is captured by the antibodies on the membrane and detected by blue latex beads coated with anti-dinitrophenol antibodies. Application of the assay to 477 E. coli isolates from urine samples showed 185 PCR immunoassay-positive isolates among 187 β-lactamase-producing isolates; after repeat testing, the two PCR immunoassay-negative isolates also became positive. All ampicillin-susceptible isolates except one were immunoassay negative. No explanation was found for this discrepancy (58). The major advantage of the assay is speed, with detection of the amplifcation products taking only minutes.
A very sensitive (104 to 105 CFU) and fast (4-h) chemiluminescent oligonucleotide hybridization assay for TEM-1 was designed for the detection of TEM-1 β-lactamase in N. gonorrhoea (286). The assay used 7 capture probes and 19 detection probes for the detection of TEM-1 and 10 capture probes and 26 labeling probes for the N. gonorrhoea-specific TEM-1 probe. No explanation for the large number of probes was given, and although the assay proved to be very specific, the large number of individual oligonucleotides required will make quality control a problem, especially with the current availability of PCR, which can more easily achieve similar levels of sensitivity. Nevertheless, the study showed the potential of signal amplification assays. In another study, a probe for TEM-1 was biotinylated and after hybridization streptavidin-coupled europium was added as fluorescent marker (59). The results demonstrated that 4 × 104 molecules could be detected.
In N. gonorrhoea, six related plasmids are responsible for β-lactamase production and only three of these, the Asia, Africa, and Toronto type plasmids, have been associated with epidemics. To differentiate among these plasmids, a PCR assay was developed that differentiated each plasmid type by its differently sized amplification products. The assay was 100% specific and sensitive and identified 16 isolates with the Asia plasmid, 41 isolates with the Africa plasmid, and 16 isolates with the Toronto plasmid, demonstrating the value of the assay for epidemiological typing of N. gonorrhoea β-lactamase plasmids (66). Whole-plasmid DNA probes were also used to investigate the prevalence of the ROB β-lactamase in Haemophilus influenzae (60). Before this investigation, only two isolates carrying the ROB β-lactamase were known. Hybridization of this probe and a TEM-1 probe with DNA from 161 clinical isolates from the United States showed that 13% of the isolates harbored the gene for the ROB β-lactamase and the others carried the TEM-1 gene. This investigation demonstrated that quick screening for the presence of newly discovered resistance genes can be performed easily with the use of probes. A PCR assay with primers specific for the TEM and ROB genes was developed to directly detect these genes in cerebrospinal fluid in combination with a 16S rRNA gene amplification (340). The 16S rRNA amplification product was identified by hybridization with an H. influenzae-specific probe. A few isolates, which were all ampicillin resistant, carried the TEM gene, but no ROB gene was detected. The authors noted that they had to use the native Taq DNA polymerase preparation, because the AmpliTaq preparations were contaminated with vector DNA containing TEM gene sequences. A PCR assay was also used to determine the presence of TEM and ROB-1 β-lactamase in 157 ampicillin-resistant H. influenzae isolates from Canada (305). The results for the ROB-1-specific PCR were confirmed by restriction enzyme analysis. Eleven isolates carried the ROB-1 gene, whereas the other isolates harbored a TEM gene. Although digestion with the restriction enzyme MboI allowed discrimination between TEM-1 and TEM-2, no TEM-2 β-lactamase genes were found.
Cephalosporinases are common β-lactamases among anaerobes. To obtain a better understanding of the distribution of cepA- and cfxA-encoded cephalosporinases among Bacteroides isolates, two specific oligonucleotides were designed (194). All 80 resistant isolates of Bacteroides fragilis carried the cepA gene, as well as 2 of 7 resistant Bacteroides distasonis isolates, 1 of 7 resistant Bacteroides vulgatus isolates, and 1 of 5 Bacteroides thetaiotaomicron isolates. Only one B. fragilis isolate harbored the cfxA gene compared to 20% of the B. fragilis group isolates. These results agreed with the isoelectric points for the corresponding β-lactamases.
Extended-Spectrum β-Lactamases
With few exceptions, ESBLs belong either to the TEM or SHV family of β-lactamase genes and are located on plasmids and ESBL are usually found in the Enterobacteriaceae. The β-lactamases belonging to one of the families differ only in a few amino acids, but these alterations influence the spectrum of acitivity of these enzymes. The vast majority of ESBLs are inhibited in vitro by clavulanic acid. The first ESBLs studied with probes belong to the TEM family. Point mutations in the TEM sequence which led to amino acid changes often broadened the spectrum of activity of these β-lactamases. Biotinylated oligonucleotide probes discriminated between TEM-1, TEM-3, and TEM-6 (348), and 12 radiolabeled primers were described that enabled the discrimination of TEM-1 to TEM-7 (185). The latter assay found 14 variants, including 7 new enzymes, among 265 clinical isolates. One of these enzymes had the same substrate profile and isoelectric point as TEM-2 but differed by a single amino acid change. The other enzymes (TEM-14 to TEM-19) were ESBLs which had novel combinations of known amino acid substitutions. The use of radioactively labeled fragment probes also led to the identification of novel ESBLs (13), although these these probes are less sensitive for the detection of mutations which are responsible for the extended substrate range. In some cases these mutations lead to the appearance or disappearance of restriction sites. Amplification of the relevant part of the gene by PCR followed by restriction enzyme analysis can thus indicate the presence or absence of some specific TEM or SHV derived ESBLs (12, 224).
PCR-SSCP has also been applied to the study of ESBLs. Five SHV-type ESBLs were analyzed by PCR-SSCP with satisfactory results, although only few isolates were tested (205). These results were later extended to include SHV-7 and the ability to detect more than one SHV-type gene in a single isolate (206). Another investigation applied PCR-SSCP to TEM-1, TEM-2, TEM-30, and TEM-32 to TEM-39 with good results (319). The results showed the presence of a novel TEM ESBL (TEM-58), which was confirmed by sequencing.
In Turkey, the distribution of the PER-1 ESBL was investigated (368). This ESBL is not related to either TEM or SHV ESBLs. A total of 72, 92, and 362 isolates of Acinetobacter, Klebsiella, and Pseudomonas, respectively, from eight university hospitals were studied by colony hybridization. The enzyme was not present in Klebsiella isolates, but 46% of the Acinetobacter isolates and 11% of the P. aeruginosa isolates carried the gene for this β-lactamase. Southern blot analysis with a PER-1 gene-specific probe showed that the gene is present on a number of different DNA fragments. The PER-1 β-lactamase currently appears to be restricted to Turkey, but the high prevalence and the presence in different clones suggest a high potential for spread.
Metallo-β-Lactamases
The blaIMP encoded metallo-β-lactamase efficiently hydrolyzes both carbapenems and cephalosporins. The enzyme was found in gram-negative rods in Japan (9, 306) and is located on an integron. Integrons are genetic elements which are associated with multidrug resistance (90) (see “Multidrug resistance” below). A total of 54 highly ceftazidime-resistant isolates were therefore tested with blaIMP-specific PCR; 22 isolates (9 P. aeruginosa, 9 Serratia marcescens, 2 Alcaligenes xylosoxidans, 1 Pseudomonas putida, and 1 Klebsiella pneumoniae) were identified which carried the gene. PCR assays for the integron integrase gene intI3 and the aac(6′)-Ib aminoglycoside resistance gene associated with the original isolates showed that these genes were well conserved. Rapid PCR detection of novel genes is therefore helpful in the early recognition of emerging resistance.
Conclusion
The study of β-lactamase resistance for diagnostic purposes is restricted mainly to epidemiology. The main reasons are the overwhelming number of different β-lactamases and their variants and the fact that most organisms treated with β-lactam antibiotics can be easily cultured. Furthermore the most important pathogenic species treated with β-lactam antibiotics are generally susceptible to at least one class of β-lactam antibiotics, despite the ubiquitous nature of β-lactamases (89). This, coupled with the cost of most molecular assays, puts these assays in an unfavorable position compared to phenotypic assays for detection of resistance. The introduction of direct sequencing and DNA chips may change this balance, especially for the detection of ESBLs.
RESISTANCE TO AMINOGLYCOSIDES
Mechanisms of Resistance
Aminoglycosides such as gentamicin, tobramycin, amikacin, and streptomycin are commonly used antimicrobial agents in the treatment of infections by both gram-negative and gram-positive organisms. Aminoglycosides bind to the ribosomes and thus interfere with protein synthesis. Resistance to these antimicrobial agents is widespread, with more than 50 aminoglycoside-modifying enzymes already described. Most of these genes are associated with gram-negative bacteria. Depending on their type of modification, these enzymes are classified as aminoglycoside acetyltransferases (AAC), aminoglycoside adenylyltransferases (also named aminoglycoside nucleotidyltransferases [ANT]), and aminoglycoside phosphotransferases (APH). A number with or without either a prime or double prime denotes the position of the modification on the substrate. A subclassification of these enzymes depends on the aminoglycoside substrates that are modified. The gene names follow this pattern, but a further subclassification is made when different genes encode enzymes that have the same substrate profile. Aminoglycosides modified at amino groups by AAC enzymes or at hydroxyl groups by ANT or APH enzymes lose their ribosome-binding ability and thus no longer inhibit protein synthesis (241). Besides aminoglycoside-modifying enzymes, efflux systems and rRNA mutations have been described (291, 309; for reviews, see references 257, 291, and 309).
Staphylococci
The main mechanism of aminoglycoside resistance in staphylococci is drug inactivation by cellular aminoglycoside-modifying enzymes. Several distinct gene loci encoding such modifying enzymes have been characterized in staphylococci.
Resistance to gentamicin and concomitant resistance to tobramycin and kanamycin in staphylococci is mediated by a bifunctional enzyme displaying AAC(6′) and APH(2′′) activity (195, 364). The aac(6′)-Ie+aph(2′′) gene encodes this bifunctional enzyme and is locateded on the composite transposon Tn4001. Tn4001-like elements are widely distributed in both S. aureus and CNS. Tn4001 has been found on pSK1 family plasmids, conjugative plasmids such as pSK41, occasionally on β-lactamase/heavy-metal resistance plasmids such as pSK23, and also in various chromosomal locations (241) (see also “Multidrug resistance” below).
Resistance to neomycin, kanamycin, tobramycin, and amikacin in staphylococci is mediated by an ANT(4′)-I enzyme encoded by ant(4′)-Ia. This gene is often carried on small plasmids, integrated into larger conjugative plasmids such as pSK41 and subsequently into the mec region of the chromosome of some S. aureus isolates, probably as a result of IS257-mediated recombination events (10, 37, 323). Additionally, a variety of other plasmids encoding ANT(4′)-I activity have been detected (241).
Resistance to neomycin and kanamycin by an APH(3′)-III enzyme has also been described for staphylococci. The aph(3′)-IIIa gene responsible for this phenotype is carried on the transposon Tn5405, known to be located on both the chromosome and plasmids (65).
Vanhoof et al. (373) developed PCRs for the detection of the most important aminoglycoside resistance genes aac(6′)-Ie+aph(2′′), aph(3′)-IIIa-3, and ant(4′)-Ia(373). These authors investigated the prevalence of these genes in 37 MRSA isolates collected from 1980 to 1985 and in 81 MRSA isolates from 1991 to 1992. All isolates were obtained from 10 Belgian hospitals. PCR results corresponded well to those obtained in the radiochemical phosphocellulose paper binding assay (373). The gene aac(6′)-Ie+aph(2′′) was the gene encountered most frequently. The prevalence of aph(3′)-IIIa-3 decreased significantly in the period from 1991 to 1992, while ant(4′)-Ia was found solely in isolates from this period.
Since PCR had been shown to be a reliable tool for the identification of aminoglycoside-modifying enzyme genes in staphylococci (373), this method was used by Schmitz et al. to detect the aac(6′)-Ie+aph(2′′), aph(3′)-IIIa and ant(4′)-Ia genes in 363 aminoglycoside-resistant staphylococci and, by inference, the enzymes they encode (293). The staphylococci were derived from 19 different hospitals. The data were used to assess their distribution among S. aureus and CNS isolates. Oligonucleotide primers for use in a multiplex PCR were selected using published DNA sequences from Vanhoof et al. (373). Among isolates of S. aureus, the most prevalent resistance gene was aac(6′)-Ie+aph(2′′), found in 76% of MRSA and 50% of MSSA isolates. The least common was aph(3′)-IIIa, occurring in 7% of MRSA and 13% of MSSA isolates. Similarly, 67% of MRCNS isolates carried aac(6′)-Ie+aph(2′′), while this gene was detected in only 32% of MSCNS isolates. As in S. aureus, the least common aminoglycoside resistance gene, aph(3′)-IIIa, was associated more with MSCNS (50% of isolates compared with 20% of MRCNS isolates); the reverse was true for the other two genes. aph(3′)-IIIa was considerably more prevalent among aminoglycoside-resistant isolates of CNS than among S. aureus isolates (25 and 8%, respectively) (293).
Recently, Hotta et al. used PCR to detect the aac(6′)-Ie+aph(2′′) gene in 23 arbekacin-resistant MRSA isolates. All arbekacin-resistant MRSA isolates possessed the gene, while 10 other MRSA strains lacking the gene were arbekacin sensitive (124).
Due to the limited number of aminoglycoside resistance-encoding genes in staphylococci, a PCR assay is a useful tool to detect this resistance even when no multiplex PCR is used.
Enterococci and Streptococci
The application of molecular assays to the detection of aminoglycoside resistance in enterococci and streptococci is limited to epidemiological studies. An early study using hybridization with an unnamed gentamicin resistance determinant from Enterococcus faecalis dates back to 1986 (164). A more recent study used probes to investigate approximately 500 enterococcal isolates for the presence of genes encoding aminoglycoside-modifying enzymes (234). The results were correlated with the enzymes expected to be present on the basis of phenotype. The agreement between phenotype and genotype was 87.6% for ANT(6) and 100% for APH(2′′)-AAC(6′), APH(3′), and ANT(4′). The discrepancy for the ANT(6) is most probably due to other mechanisms of resistance. Except for streptomycin resistance [ANT(6)], the correlation between hybridization results, resistance, and aminoglyoside-modifying enzyme are excellent.
A study (369) of the presence of high-level aminoglycoside resistance in 73 enterococci and 54 group A streptococci compared a high-load disk method, tube macrodilution, and PCR. The PCR was specific for the aadC [ANT(4′)], aac(6′)-Ie+aph(2′′) [AAC(6′)-APH(2′′)], and aphA3 [APH(3′)] genes. Only one streptococcal isolate displayed high-level resistance to streptomycin and kanamycin; PCR revealed the presence of the aphA3 gene. A total of 27 enterococcal isolates showed high-level aminoglycoside resistance. Seven of these isolates carried the aac(6′)-Ie+aph(2′′) gene alone, 15 isolates carried the apha3 gene, 3 isolates carried both, and 2 isolates were PCR negative. The two negative isolates were resistant only to streptomycin and probably had another mechanism of resistance. This study showed that with a limited number of PCR assays, a clear picture of the distribution of genes responsible for high-level aminoglycoside resistance in enterococci and streptococci in The Netherlands could be obtained. Kaufhold et al., who used PCR and nonradioactively labeled oligonucleotides, also demonstrated the presence of aac(6′)-Ie+aph(2′′) genes in both enterococcal and streptococcal species (144).
A small-scale hybridization investigation with seven highly gentamicin-resistant enterococcal isolates from widespread geographical areas in the United States revealed the presence of the aac(6′)-Ie+aph(2′′) gene in six of these isolates. In four cases the gene was located on distinct plasmids, and in two cases it was on the chromosome. These findings suggested that although the resistance determinants are related, no clonal spread of the bacteria had taken place (347). A probe against the same gene was used to study its dissemination among isolates from three continents (403). In contrast to the previous study, these authors concluded that the gene was present on highly related plasmids. Clearly the gene is widespread, but apparently it can be part of a number of different plasmids, one of which seems to be more prevalent. However, a study of gentamicin-resistant U.S. enterococcal isolates of animal origin did not show the presence of the aac(6′)-Ie+aph(2′′) gene (346). Apparently, the mechanism of resistance is different in these animals. It is not clear whether this gene has spread from human isolates to animal isolates or vice versa.
Gram-Negative Bacteria
One of the first applications of probes to monitor the spread of an aminoglycoside-modifying enzyme was described by Tenover et al. (341). This group identified an ANT(2′′) enzyme responsible for low-level resistance to gentamicin, tobramycin, and kanamycin. The gene was discovered on plasmids of 68 and 150 kb. To monitor the spread of the gene, a radioactively labeled probe was used for Southern hybridization. This method was highly specific and more sensitive than enzymatic methods for the detection of the ANT(2′′) gene in clinical isolates with complex aminoglycoside resistance phenotypes. A similar probe was constructed by Groot-Obbink et al. (107) The research by Tenover et al. was extended with an aacC1 [AAC(3)-I] specific probe (343). Of 219 gentamicin-resistant isolates, only 6 carried the gene. A similar study for the AAC(3)-V enzyme, which confers resistance to gentamicin was performed at Vanderbilt University Medical Center (18). The gene, originally present in an Serratia marcescens isolate, became incorporated in other species of Enterobacteriaceae. In 25 of 30 cases the gene was present on a conjugative plasmid. A probe specific for the aacC2 gene demonstrated that 86% of 86 multiple-drug-resistant isolates of Enterobacteriaceae implicated in a 18-month hospital epidemic in The Netherlands carried this gene (374). The aacC1 and aacC2 gene were also studied by a Spanish group with a radioactive fragment probe in a Southern blot format (7). A total of 315 gentamicin-resistant gram-negative bacteria were evaluated. Hybridization with the aacC1 probe was observed with 39 isolates, the aacC2 probe hybridized with 146 isolates, and 26 isolates hybridized with both probes. The aacC1 gene was most frequently encountered in P. aeruginosa isolates, whereas the other gene was more frequently seen in members of the Enterobacteriaceae and Acinetobacter spp. Another gene, aac(6′)-Ic, from S. marcescens was also studied using a nonradioactive dot-blot format (318); the results were confirmed by PCR. The gene was found only in S. marcescens isolates, not in 180 isolates belonging to 28 other species of gram-negative bacteria.
Tran Van Nieu et al. (355) made a more systematic search for the aacA4 gene and its product, which leads to amikacin resistance. A total of 70 resistant clinical isolates belonging to six genera of Enterobacteriaceae, Pseudomonas, Acinetobacter, and Achromobacter from four continents were analyzed by both probe and immunoblot analyses using a polyclonal rabbit anti-AAC(6′)-1b serum. Positive hybridization and immunological reactions were observed for 44% of the isolates. The enzyme was most frequent in the genera Klebsiella, Escherichia, and Enterobacter and was absent in Achromobacter and Providencia. The molecular mass of the enzyme varied between 24 and 26 kDa. This indicates that, although this probe may be useful as an epidemiological tool for amikacin resistance, sequence heterogeneity must be present.
PCR assays were also developed to detect genes for AAC(3)-I, AAC(3)-II, and AAC(3)-IV) (375). The specificity of the reaction was confirmed by restriction enzyme analysis in which the amplification product of each assay contained an unique restriction site. A PCR specific for 16S rRNA genes served as positive control. The assay confirmed that 63 isolates of Enterobacteriaceae previously suspected of carrying the aacC2 gene indeed possessed the gene. PCR was also used to identify the aadB gene encoding the ANT(2′′) enzyme (371). To ensure specificity, an oligonucleotide probe was used. Although the PCR technique could be used directly on colonies without DNA isolation, a drawback was its use of a radioactive probe for confirmation. A similar approach was successfully used to identify four different aacA genes (372).
A number of studies focused on aminoglycoside resistance among Acinetobacter spp. One of the first studies (251) investigated the presence of genes for AAC(6′)-I-type enzymes responsible for resistance to amikacin, tobramycin, and netilmicin but susceptibility to gentamicin. Both hybridization and PCR were used and led to the same results. Of the 51 isolates investigated, 19 carried the aac(6′)-Ib gene whereas the remaining isolates carried aac(6′)-Ih. The aac(6′)-Ig gene was present only in Acinetobacter haemolyticus, but all 10 isolates carried this gene. A set of degenerate primers allowed the detection all these genes by PCR. In addition, the related aac(6′)-Ij gene, also observed in Acinetobacter spp., was detected. Seward et al. (308) investigated the presence of aminoglycoside-modifying enzyme-encoding genes [aac(3)-Ia, aac(6′)-Ib, ant(2′′)-Ia, ant(3′′)-Ia, and aph (3′)-VIa] in Acinetobacter spp. by enzyme profiling, hybridization, and PCR. Often these genes proved to be present in various combinations. Remarkably, these authors describe the presence of completely different genes from those found in the study by Ploy et al. (251). A clear explanation cannot be given, but geographical differences in the sources of the isolates may play a role, thereby underscoring the fact that results obtained by one study cannot automatically be extrapolated to other situations. Genes encoding aminoglycoside-modifying enzymes were also used for epidemiological purposes (223) rather than to assess the presence or absence of resistance determinants. Three multiplex PCR assays covering a total of seven aminoglycoside resistance genes and the 16S rRNA genes were compared to pulsed field gel electrophoresis (PFGE). PCR analysis showed the presence of seven clusters instead of the eight PFGE clusters. Despite this single discrepancy, the authors concluded that the technique was useful as an epidemiological tool, especially because results could be obtained in less than 8 h compared to several days for for PFGE typing.
Conclusion
In conclusion, aminglycoside resistance determination by molecular techniques has received only limited attention. However, this is not entirely unexpected for a number of reasons. Most infections treated with aminoglycosides, especially with gentamicin, tobramycin, and amikacin, are caused by fast-growing organisms such as those belonging to the the family Enterobacteriaceae. Thus, the advantage of speed is less relevant for these species, particularly when the costs of traditional resistance determination and molecular techniques are compared. In addition, a multitude of different aminoglycoside resistance genes for gram-negative bacteria exist, making full coverage by probes or PCR a difficult exercise. DNA chips potentially may provide an answer because all genes can be represented on a single chip, but the cost, at least for the time being, will be prohibitive.
RESISTANCE TO FLUOROQUINOLONES
Mechanisms of Resistance
Fluoroquinolone antibiotics exert their antibacterial effects by inhibition of certain bacterial topoisomerase enzymes, namely, DNA gyrase (bacterial topoisomerase II) and topoisomerase IV. These essential bacterial enzymes alter the topology of double-stranded DNA (dsDNA) within the cell (72, 80, 122, 123, 257).
DNA gyrase and topoisomerase IV are heterotetrameric proteins composed of two subunits, designated A and B. The genes encoding the A and B subunits are referred to as gyrA and gyrB (DNA gyrase) or parC and parE (DNA topoisomerase IV [grlA and grlB in S. aureus]) (72, 80, 122, 123, 257). DNA gyrase is the only enzyme that can effect supercoiling of DNA. Inhibition of this activity by fluoroquinolones is associated with rapid killing of the bacterial cell. Topoisomerase IV also modifies the topology of dsDNA, but while DNA gyrase seems to be important for maintenance of supercoiling, topoisomerase IV is predominantly responsible for the separation of daughter DNA strands during cell division (72, 80, 122, 123, 257).
Mechanisms of bacterial resistance to fluoroquinolones fall into two principal categories, alterations in drug target enzymes and alterations that limit the permeation of drug to the target. The target enzymes are most commonly altered in domains near the enzyme active sites, and in some cases reduced drug binding affinity has been demonstrated (123).
In gram-negative organisms, DNA gyrase seems to be the primary target for all quinolones. In gram-positive organisms, topoisomerase IV or DNA gyrase is the primary target depending on the fluoroquinolone considered; i.e., the quinolone structure determines the mode of antibacterial action. Thus, the primary target seems to depend on the bacterial species as well as on the quinolone structure (72, 80, 122, 123, 257).
Alterations of target enzymes appear to be the most dominant factors in expression of resistance to quinolones. The small region from codons 67 to 106 of GyrA in E. coli, the most extensively studied organism, was designated the quinolone resistance-determining region (QRDR). The QRDR is near Tyr122, which is transiently covalently bound to DNA phosphate groups during the enzyme's DNA strand-passing reactions. In almost all instances, amino acid substitutions within the QRDR involve the replacement of a hydroxyl group with a bulky hydrophobic residue. This suggests that mutations in gyrA induce changes in the binding-site conformation and/or charge that may be important for quinolone-DNA gyrase interaction (72, 80, 122, 123, 257).
Although quinolones are thought to interact primarily with the A subunit of DNA gyrase, mutations have also been discovered in the B subunit which also confer quinolone resistance in some species such as E. coli. However, the frequency of gyrB mutations, compared to gyrA, has been shown to be relatively low in most species (72, 80, 122, 123, 257).
Topoisomerase IV alterations have now been characterized in several bacteria. The ParC subunit of topoisomerase IV is homologous to GyrA, and the ParE subunit is homologous to GyrB. Particularly highly conserved in the topoisomerase IV homologs are the QRDR domains of GyrA and GyrB, a conservation that predicts similarity of drug interaction and resistance mutations for the two enzymes. The largest body of information concerning the role of topoisomerase IV in quinolone resistance in gram-positive bacteria comes from studies of S. aureus and S. pneumoniae. Mutations encoding single amino acid changes in ParC (GrlA) have clustered in the amino terminus, with Ser80 and Glu84 alterations in S. aureus and Ser79 and Asp 83 alterations in S. pneumoniae (72, 80, 122, 123, 236, 237, 257, 294). Alterations in ParE appear to be uncommon in resistant laboratory and clinical strains, but parE has been evaluated less often than parC and gyrA in studies of resistant isolates.
Recent reviews (72, 122, 123) gave an overview of mutations in the GyrA and GyrB subunit of DNA gyrase and the ParC and ParE subunit of topoisomerse IV, which are associated with quinolone resistance. It should be pointed out that alterations in the QRDR of DNA gyrase and topoisomerase IV are normally associated with higher MICs than for to wild-type strains. Nevertheless, such alterations do not always increase the MICs, especially of the newer fluoroquinolones, such as gemifloxacin, gatifloxacin, and moxifloxacin, above currently used NCCLS breakpoints.
DNA gyrase and topoisomerase IV are both located in the cytoplasm of the bacterial cell. To reach their targets, fluoroquinolone antibiotics must traverse the cell envelope. Changes in the cell envelope of gram-negative bacteria, particularly in the outer membrane, have been associated with decreased uptake and increased resistance to fluoroquinolones. Decreased uptake has not been demonstrated to be a mechanism of resistance in gram-positive bacteria (72, 80, 122, 123, 257). So far, no genetic tests are available to detect a decreased quinolone uptake by the bacteria.
Increased efflux as a mechanism of fluoroquinolone resistance, mainly low-level resistance, has been found in fluoroquinolone-resistant gram-positive and gram-negative bacteria (177). Efflux pumps in gram-positive bacteria are multidrug transporters which are driven by the electrochemical proton gradient. Neyfakh et al. (215) determined that the efflux pump NorA of S. aureus belongs to the major facilitator superfamily and is very similar to BmrA of Bacillus subtilis. None of these efflux systems has been studied for diagnostic purposes until now.
Detection of Resistance
Various methods have been reported to detect point mutations in target genes, including sequence-specific oligonucleotide probe hybridization (82), sequencing of the target genes (105), RFLP (320, 321), radioisotopic or nonradioisotopic SSCP analysis (114, 230, 325, 350), mismatch amplification mutation assay PCR (398), and allele-specific PCR in combination with RFLP (100).
Fasching et al. developed probes to detect mutations in staphylococcal gyrA genes associated with resistance, but the probes have proved difficult to use, and not all resistant strains demonstrate differential binding of the probes (82).
Most of the studies have employed DNA sequencing of PCR-amplified DNA for the detection of alterations in the target enzymes. The primers, however, are mostly species specific. Thus, PCR sequencing tests for quinolone resistance are available only in laboratories that have DNA sequencing capability. Since many laboratories do not have the equipment, time, or expertise to sequence genes for the investigation of mutations relevant to fluoroquinolone resistance, other methods have been used for the specific detection of mutations in the genes or alterations in the enzymes.
Tokue et al. (350) studied 36 S. aureus isolates by nonradioisotopic SSCP (nRI-SSCP) for the presence of point mutations in the gyrA gene. Direct DNA-sequencing analysis of the PCR-amplified DNA fragments confirmed the results obtained by nRI-SSCP. These authors identified seven mutational types which were separated from the wild type in a single electrophoretic step within 3 h after PCR amplification. The authors concluded that the use of the nRI-SSCP method allows relatively rapid analysis of DNA from a large number of strains. Since the electrophoretic pattern is theoretically specific for each mutation, identification of a specific mutation is possible by comparison of the mobilities of the sample DNAs with that of control DNA carrying known mutations. Thus, nRI-SSCP analysis is a simple and useful method not only for the detection of point mutations associated with quinolone resistance but also for the investigation of epidemiologic markers.
In an extended study, Wang et al. (380) analyzed mutations in the grlA and gyrA genes of 344 clinical strains of S. aureus by combinations of nRI-SSCP, restriction fragment length analysis, and direct sequencing. Five types of single-point mutations and four types of double mutations were observed in the grlA genes, while four types of single-point mutations and four types of double mutations were found in the gyrA genes. Six of nine types of grlA mutations and all types of gyrA mutations were distinguishable from the wild type by nRI-SSCP analysis. Although the power of direct sequencing was greater for the detection of gyrA mutations, the authors concluded that SSCP analysis is a rapid, simple, and effective method for detection of point mutations in both the grlA gene and the gyrA gene of S. aureus strains.
Meanwhile, several investigators have applied SSCP analysis to the detection of mutations, mainly in gyrA, in several bacterial species (47, 203, 230, 325, 331, 350). In the most recent study, Takenouchi et al. (332) studied gyrA point mutations in 335 clinical P. aeruginosa isolates by nRI-SSCP analysis and direct sequencing. By SSCP analysis, 18 band patterns could be differentiated, with each pattern corresponding to a distinct mutation. The band patterns were reproducible and distinct from each other. Since SSCP analysis is simple and rapid, it might also be suitable for epidemiological surveillance of organisms involved in outbreaks or for epidemiological resistance studies.
A mismatch amplification mutation assay (MAMA) PCR protocol (44) was developed by Zirnstein et al. (412) that detects the most commonly encountered gyrA mutation in quinolone-resistant Campylobacter jejuni isolates. Since the Thr86-Ile alteration was the most commonly encountered alteration leading to fluoroquinolone resistance, the authors have developed a MAMA protocol for the detection of this special alteration. A conserved forward primer, MAMAgyrA1, and a reverse mutation detection primer, MAMAgyrA5, were used together in a PCR to generate a 256-bp PCR product that was a positive indication of the presence of the special alteration Thr86-Ile in C. jejuni gyrA. Primer GZgyrA4, a conserved reverse primer, was used in conjunction with primer MAMAgyrA1, to produce a positive PCR control product of 368 bp with any C. jejuni gyrA gene. Isolates with the wild-type amino acid 86 codon were not amplified with the reverse mutation primer MAMAgyrA5, whereas isolates with a mutated amino acid 86 codon generated a 256-bp PCR product with the MAMAgyrA5 reverse mutation primer and the MAMAgyrA1 forward conserved primer. Conserved primers GZgyrA4 and MAMAgyrA1 generated a 368-bp gyrA PCR product with DNA isolated from all isolates. When the MAMA protocol is used, isolates must be confirmed as C. jejuni, since false-positive PCR products can result when C. coli isolates are used. While the MAMA PCR assay described by Zirnstein et al. (412) is undoubtedly simpler than DNA sequencing for use in determining the presence of mutations relevant for fluoroquinolone resistance, it does have a disadvantage. Other alterations, besides the most commonly encountered Thr86-Ile alteration, cannot be detected in gyrA. However, it should be simple to develop additional MAMA PCR mutation detection primers. The MAMA PCR (44, 412) method is a simple and rapid alternative to SSCP and DNA sequencing for the detection of important alterations within the QRDRs.
Giraud et al. developed a rapid assay combining allele-specific PCR (100, 159, 213) and RFLP (AS-PCR-RFLP) for the screening of point mutations responsible for all amino acid changes encoded by the Salmonella enterica sarovor Typhimurium gyrA gene at codons 81, 83 and 87. These alterations were most frequently encountered in fluoroquinolone-resistant clinical and laboratory strains. The PCR amplification was performed with three primers. The forward primer, STGYRA1, and the reverse primer, STGYRA-HinfI/87, were expected to produce a 195-bp fragment with a HinfI restriction site at the codon corresponding to Ser83. As previously described for E. coli (235), the reverse primer STGYRA-HinfI/87, whose sequence is different by 1 base from the gene sequence, introduced an artificial HinfI cleavage site including the Asp87 codon according to the primer-specified restriction site modification method (109). A second allele-specific forward primer, AS-81, whose 3′-terminal nucleotide corresponds to the first nucleotide of codon 81, permitted the amplification of an 80-bp fragment only in the presence of this nucleotide, a fragment that also contained both the natural and the artificial HinfI cleavage sites. The AS-PCR-RFLP seems to be a simple and rapid alternative to SSCP and DNA sequencing for the detection of important alterations within the QRDRs. In contrast to the MAMA PCR method, several mutations can be detected. This method has the potential to be used as a quick screening method in laboratories where systematic sequencing of the target genes is not suitable.
Conclusion
For several reasons, the study of quinolone resistance for diagnostic purposes is restricted mainly to epidemiology and basic research. There are several reasons. Until now, only alterations in the target enzymes have been studied by using genetic tests. Changes in susceptibility to fluoroquinolones associated with decreased uptake or increased efflux have not been analyzed by genetic tests employed for diagnostic use in the laboratory. It has been pointed out that alterations in the QRDR of DNA gyrase and topoisomerase IV are normally associated with higher MICs compared to those for wild-type strains (137, 139, 292). Nevertheless, such alterations do not always increase the MICs, especially of the newer fluoroquinolones such as gemifloxacin, gatifloxacin, and moxifloxacin, above currently used NCCLS breakpoints. Thus, detection of alterations in the QRDRs does not always correlate with resistance to newer fluoroquinolones. Furthermore, due to the continuing development of newer quinolones with increasing potency, some bacteria with alterations in the QRDRs that are resistant to older fluoroquinolones may remain susceptible to the new, more potent agents by clinical-breakpoint criteria. Thus, in view of the newer quinolones, the screening for alterations in the QRDRs is only of limited interest.
RESISTANCE TO MLS ANTIBIOTICS
Mechanisms of Resistance
Macrolide, lincosamide, and streptogramin (MLS) antibiotics are chemically distinct inhibitors of bacterial protein synthesis. Intrinsic resistance to MLSB (including streptogramin B) antibiotics in gram-negative bacilli is due to low permeability of the outer membrane to these hydrophobic compounds. Three different mechanisms of acquired MLS resistance have been found in gram-positive bacteria (165, 168, 384–387).
The first mechanism of macrolide resistance described was due to posttranscriptional modifications of the 23S rRNA by the adenine-N6-methyltransferase. Target modification alters a site in 23S rRNA common to the binding of MLSB antibiotics. Modification of the ribosomal target confers cross-resistance to MLSB antibiotics (MLSB resistant phenotype) and remains the most frequent mechanism of resistance. In general, genes encoding these methylases have been designated erm (erythromycin ribosome methylation). For nomenclature, see the recent review in reference 272. Streptogramin A-type antibiotics are unaffected by erm genes, and synergy between the two components of streptogramin against MLS-resistant strains is maintained. Expression of MLSB resistance can be constitutive or inducible. The character of resistance is not related to the class of erm determinant but depends on the sequence of the regulatory region upstream from the structural gene for the methylase (22, 181, 182, 303, 388). Expression of MLS resistance in staphylococci may be constitutive or inducible. When expression is constitutive, the strains are resistant to all MLSB-type antibiotics. When expression is inducible, the strains are resistant to 14- and 15-membered macrolides only. The 16-membered macrolides, the commercially available lincosamides, and the streptogramin antibiotics remain active. MLS resistance in streptococci can also be expressed constitutively or inducibly. However, unlike the case in staphylococci, various macrolides or lincosamides may act as inducers to various degrees. Thus, in streptococci, whether inducible or constitutive, ribosomal methylation leads to cross-resistance among MLSB antibiotics. Good reviews of regulation of the erm genes can be found in references 165, 384, 385, and 387.
A number of different antibiotic resistance genes code for efflux proteins, which pump the antibiotic out of the cell or the cellular membrane, keeping intracellular concentrations low and ribosomes free from antibiotic. In early years, most macrolide resistance was mediated by the presence of erm genes. However, more recently, efflux mechanisms as well as other mechanisms have been found in increasing frequency in certain gram-positive populations (14, 52, 184, 272, 276, 312, 327, 329, 396, 397, 402). Three different efflux gene classes have been described for gram-positive cocci: mef, msr, and vga.
The mef (macrolide efflux) genes have been found in a variety of gram-positive genera (184). Many of these genes are associated with conjugative elements located in the chromosome and are readily transferred conjugally across species and genus barriers (184). Two mef genes have been characterized in detail, the mefA gene, which was found in Streptococcus pyogenes, and the mefE gene, which was found in S. pneumoniae. Streptococci carrying only the mef genes display the M phenotype, namely, resistance to macrolides but susceptibility to lincosamide and streptogramin B antibiotics (272).
The msr genes differ from the mef genes because they confer resistance to both macrolides and streptogramin B antibiotics (MS phenotype). The msrA and msrB genes were found in S. aureus (276, 277). In addition to the msrA efflux pumps, two efflux systems have been identified in staphylococci that confer resistance to streptogramin A antibiotics, vga and vgaB (272). A lincomycin-specific efflux pump encoded by lmrA has been found in Streptomyces lincolnensis 78–11 (409).
A variety of other mechanisms which usually confer resistance to only macrolides, lincosamides, or streptogramins (A or B) have been described (165, 168, 272, 384–387), with diagnostic tests being available for only some of the genes encoding these mechanisms. The genes encoding enzymes which hydrolyze streptogramin B (vgb and vgbB genes) or modify the antibiotic by adding an acetyl group (acetyltransferases) to streptogramin A (vat, vatB, vatC, satA, and satG genes) have been described and detected in different assays.
Unlike target modification, which causes resistance to structurally distinct antibiotics, enzymatic inactivation confers resistance mostly only to structurally related drugs. The enzymes EreA and EreB (ereA and ereB genes), which hydrolyze the lactone ring of the macrocyclic nucleus, and the phosphotransferases (type I [mphA] and type II), which inactivate macrolides by introducing a phosphate on the 2′-hydroxyl-group of the amino sugar, have been found in members of the family Enterobacteriaceae and in S. aureus.
Several lincomycin nucleotidyltransferases have also been identified, linA in Staphylococcus haemolyticus, linA′ in S. aureus, and linB in Enterococcus faecium (165, 168, 272, 384–387).
Staphylococci
Several authors have screened clinical isoaltes of erythromycin-resistant S. aureus and CNS for genes encoding resistance to macrolides, lincosamides and streptogramins (3–5, 15, 77, 131, 133, 147, 179, 212, 218, 297, 345, 390, 391). In a recent study (297), the prevalence of the macrolide resistance genes, ermA, ermB, ermC, msrA/msrB, ereA, and ereB was analyzed using PCR in 851 unrelated clinical erythromycin-resistant S. aureus isolates, comprising 358 MSSA and 493 MRSA isolates, which had been sent from 24 different European university hospitals. Oligonucleotide primers for use in the PCRs were selected from the DNA sequences published by Sutcliffe et al. (326). Primers specific for conserved regions of the 16S rRNA gene were used as additional internal controls (106). Genomic DNA was isolated and two multiplex PCRs (primer set for ermA, ermB, and ermC, together with msrA/msrB, as well as a primer set for ereA and ereB in a second separate PCR) were performed as described by Sutcliffe et al. (326). After confirmation of the presence of an erm gene, single PCRs were performed to verify the class of the erm gene, either ermA, ermB, or ermC. The most prevalent resistance gene in S. aureus was ermA (67%), followed by ermC (23%) and msrA/msrB (6%). Less common were ermB and ereB, each occurring in 0.6% of the erythromycin-resistant S. aureus isolates tested. The ereA gene was not detected in any of the isolates. The ermA gene was more common in MRSA isolates (88% in MRSA and 38% in MSSA isolates), while ermC was more common in MSSA isolates (5% in MRSA vs 47% in MSSA isolates). Within the S. aureus collection, ermA was predominant in strains expressing a constitutive MLSB phenotype while ermC was predominant in MSSA isolates with an inducible MLSB phenotype. One erm gene was detected in 84% of the S. aureus isolates tested, whereas the combination ermA and ermC was found in only 3% of the isolates. In general, these observations are in line with the findings of Lina et al. (179), who studied 144 MLSB-resistant S. aureus strains originating from French hospitals in 1995. Using PCR, they found that the ermA gene was more common in MRSA isolates (57.6%), mainly in strains with a constitutive MLSB expression, than in MSSA isolates (5.6%). However, ermC was more common in MSSA isolates (20.1%), mainly in strains with an inducible expression, than in MRSA isolates (4.9%) (179). Similar findings were also reported from Denmark (390, 391), where ermA and ermC genes were responsible for erythromycin resistance in 98% of the 428 S. aureus isolated studied. The ermA gene was solely responsible for erythromycin resistance until 1971, and then ermC became dominant between 1984 and 1988. In accordance with the observations from Denmark, Nicola et al. detected the ermA gene in 15 of 16 erythromycin-resistant S. aureus isolates originating from the United States and isolated between 1958 and 1969 (218). Thus, ermC has only recently become prevalent in the S. aureus population. The ereB gene, coding for a macrolide-inactivating enzyme, was found only in MRSA isolates expressing the constitutive MLSB phenotype (1%). Neither ereA nor a combination of ereB and another macrolide-resistant determinant was found (297). Macrolide resistance by efflux due to the msrA/msrB gene was found only in MSSA isolates (13%). This is somewhat in contrast to the results of Lina et al. (179), who detected the msrA/msrB gene in both MSSA and MRSA isolates. Moreover, they found the gene to be prevalent in only 2.1% of the 144 S. aureus strains tested. In line with their observations, however, no combination of msrA/msrB and another macrolide resistance determinant was found by Schmitz et al. (297). In CNS isolates, ermC was the most predominant gene in MRCNS and MSCNS, followed by ermA. These results from Lina et al. (179), who investigated 150 CNS isolates by PCR, confirmed those obtained by Eady et al. (77), who documented the predominance of ermC in clinical and commensal CNS isolates. Macrolide resistance due to efflux encoded by msrA was more prevalent in CNS (14.6%) than in S. aureus.
The incidence of staphylococci with lincomycin resistance but without resistance to macrolides and streptogramins is usually low. In a study by Lina et al. (179), only 1 of 144 S. aureus isolates and 7 of 150 CNS isolates had the linA/linA′ gene.
Concerning resistance to streptogramins, Lina et al. (179) detected 10 S. aureus strains and 3 CNS isolates displaying resistance to quinupristin-dalfopristin and pristinamycin. The resistance was always associated with resistance to type A streptogramins encoded by vat or vatB genes and occurred in combination with erm genes, resulting in MLSB and streptogramin A resistance. The vga gene conferring decreased susceptibility to type A streptogramins was present alone in three CNS isolates.
Allignet and el Sohl used PCR to analyze the presence of vga, vat, and vatB (encoding resistance to type A streptogramin), and vgb (encoding resistance to type B streptogramins) in 56 staphylococcal isolates displaying resistance to type A streptogramin (5). Since none of the genes could be detected in 48 isolates, it was concluded that there is at least one type A streptogramin resistance mechanism in staphylococci that has not yet been characterized. In a similar study by Allignet et al. (3), 12 clinical S. aureus isolates displaying resistance to type A streptogramin were screened by PCR for the presence of vat, vatB, vga, and satA. Since five isolates carried none of these genes, it was again concluded that there must be at least one additional type A streptogramin resistance mechanism in staphylococci.
Pneumococci
Erythromycin resistance in S. pneumoniae has increased considerably over the last few years, especially in North America (67, 138) and Europe (298, 299). Epidemiological surveys have shown that some erythromycin-resistant strains of pneumococci and group A streptococci are not co-resistant to lincosamides and streptogramin B antibiotics, the MLSB resistant phenotype typical of strains carrying an erm methylase (135, 311, 312). These clinical strains have been shown to display the M phenotype and possess the mefE gene in pneumococci and the mefA gene in group A streptococci (272).
Several authors have screened clinical S. pneumoniae strains for the presence of the macrolide efflux gene mefE and the bisomal methylase gene ermAM (ermB) (14, 163, 184, 189, 219, 311). While the efflux mechanism is the predominant form of macrolide resistance in the United States (61% of 114 erythromycin-resistant isolates tested [311]), macrolide resistance due to the presence of the ermB gene seems to be the predominant form in Europe and Japan (14, 163, 190, 219).
Besides the academic interest in the prevalence of these two genes, their presence in an erythromycin-resistant isolate may also have implications in terms of therapeutic choices. If an isolate carries a mefE gene, clindamycin can be considered, whereas the presence of an ermB gene would preclude consideration of a lincosamide. Furthermore, mutations in the 23S rRNA and alterations in ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. In a recent study, Tait-Kamradt et al. (330) analyzed the sequences of 23S rRNAs in the mutant and parental strains and found individual changes of C2611A, C2611G, A2058G, and A2059G (E. coli numbering) in four mutants. Mutations at these residues in domain V of 23S rRNA have been noted to confer erythromycin resistance in other species. Furthermore, two of the mutants which had no change in their 23S rRNA sequences had changes in a highly conserved strech of amino acids in ribosomal protein L4. One mutant contained a single amino acid change (G69C), while the other mutant had a 6-base insert, resulting in two amino acids (S and Q) being inserted between amino acids Q67 and K68 (330). The mutations in the 23S rRNA are located in residues which are important for binding of macrolides and maintaining the confirmation of the peptidyltransferase region. The presence of alterations in the L4 ribosomal protein is consistent with the interpretation that this protein is in contact with or near the peptidyltransferase region in domain V of 23S rRNA. Thus, this alteration may act indirectly to alter 23S rRNA confirmation. Although the frequency of these mutations and alterations is unknown, these additional resistance mechanisms should be considered when undertaking molecular resistance detection.
Streptococci
In streptococci, MLSB resistance is commonly mediated by genes belonging to the ermAM (ermB) class of genes. Recently, a novel erm gene, ermTR, was detected (307). In addition, macrolide efflux encoded by the mefA gene has been described (53). Several authors have screened group A, C, and G streptococci by PCR for the presence of these genes (62, 142, 143, 244, 252).
Kataja et al. (142) analyzed 45 erythromycin-resistant S. pyogenes isolates from Finland. All M phenotype isolates possessed the mefA gene, while all but one isolate displaying the MLSB phenotype had the erythromycin resistance methylase gene ermTR. One isolate with a constitutive macrolide resistance phenotype contained the ermB gene. Similar results were reported by De Azavedo et al. (62). These authors analyzed 67 erythromycin-resistant group A streptococci from Canada. Of these, 47 (70%) were susceptible to clindamycin and were found by PCR to possess the mefA gene. Of the other 20 strains, 18 and 2 showed inducible and constitutive resistance, respectively, to clindamycin. Nineteen of these strains were shown by PCR to possess the ermTR gene, and a single constitutively resistant strain harbored an ermB gene. Similar investigations were performed with strains from Spain by Perez-Trallero et al. (244) and Portillo et al. (252). These authors have analyzed 256 and 40 erythromycin-resistant S. pyogenes isolates, respectively; they found that 96.1 and 90% of the isolates, respectively, possessed the mefA gene.
In a study by Kataja et al. (143), 21 erythromycin-resistant group C and 32 group G streptococcal isolates from Finland were analyzed for the presence of resistance genes. While 95% of the group C streptococci possessed the M phenotype with detectable mef genes, 91 and 6% of the group G streptococci were inducible or constitutive MLSB resistant, respectively. The ermTR gene was found in all isolates with an inducible phenotype; of two isolates with a constitutive phenotype, one had an ermTR gene and the other had an ermB gene.
Enterococci
In a recent study by Schmitz et al. (297), 75 European erythromycin-resistant Enterococcus faecium isolates were screened by PCR for resistance genes. ermB was the most prevalent resistance gene found, followed by ermA (93 and 4%, respectively). The combination of ermA and ermB was detected in 2 of 75 isolates (3%). Jensen et al. (131) have recently analyzed 113 erythromycin-resistant enterococcal isolates of human and animal origin and found the ermB gene to be prevalent in 88% of the enterococci tested.
Bozdogan et al. (31) have described a new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in E. faecium. The linB gene was found in 15 lincosamide-resistant clinical isolates of E. faecium.
Jensen et al. (132) and Hammerum et al. (111) screened for the satA and vgb genes. Jensen et al. screened 51 streptogramin-resistant E. faecium strains from fecal samples of humans and animals. The satA gene was detected in 14 strains from humans and animals, and the vgb gene was found only in one human isolate. These genes have previously been found only in streptogramin-resistant isolates from hospitalized patients.
Werner and Witte (389) characterized a new enterococcal gene, satG, encoding a putative acetyltransferase conferring resistance to type A streptogramin compounds. Using PCR, the new satG gene could be detected in different enterococcal species from sewage, broiler samples, and poultry manure. In addition, 9 of 62 quinupristin-dalfopristin-resistant E. faecium isolates from German hospitals were positive for satG.
Helicobacter pylori
Helicobacter pylori infections can be effectively treated with macrolides in combination with another antibiotic such as omeprazol, amoxicillin, tetracycline, or metronidazole. The prevalence of macrolide-resistant strains seems to be increasing. The major cause of macrolide resistance in H. pylori is the lack of binding of the macrolides to the 23S rRNA components of the bacterial ribosome due to a modification of the target site by methylation or point mutations in the peptidyltransferase region of domain V of the 23S RNA. H. pylori contains two copies of the 23S ribosomal DNA (rDNA) gene, and at least five distinct point mutations have been reported (370) that are associated with macrolide resistance. Conventional methods such as disk diffusion, E-test, or microbroth dilution are time-consuming for determination of macrolide resistance of H. pylori; NCCLS now has recommendations for Helicobacter (211a). Several authors have described DNA-based diagnostic methods that offer a rapid alternative approach for macrolide susceptibility testing (28, 186, 187, 225, 249, 324, 370).
Recently, van Doorn et al. (370) evaluated a PCR-based reverse hybridization system (research prototype kit INNO-LiPA for H. pylori resistance) for the simultaneous detection of 23S rDNA point mutations. PCR products were analyzed by reverse hybridization in a reverse-hybridization LiPA. This assay is based on hybridization with a number of oligonucleotide probes that are immobilized as parallel lines on a nitrocellulose strip. Fifty-seven H. pylori strains were tested by PCR-LiPA, DNA sequencing, RFLP, and/or hybridization with oligonucleotide probes. The results were highly concordant, but PCR-LiPA appears to be more sensitive for the simultaneous detection of multiple mutants. This PCR method provides accurate information about different 23S rDNA mutants, even if they represent only a small portion of the bacterial population. Therefore, it could be particularly suitable for monitoring the development of resistance during antibiotic therapy.
Stone et al. (324) have developed an assay based on PCR followed by the oligonucleotide ligation assay (OLA) for the rapid detection of point mutations within the 23S rRNA gene of H. pylori strains. PCR-OLA discriminates sequence variations (transitions, transversions, and deletions) by constructing probes to identify the variation of interest. This is different from using restriction enzymes to identify point mutations, because restriction enzymes are limited to sequence specificity. The authors have used PCR-OLA to determine the prevalence of 23S rRNA gene mutations. Susceptible H. pylori isolates were wild type at positions 2143 and 2144, while 93% of the resistant isolates contained A-to-G mutations at either position and 7% of the isolates contained A-to-C mutations at position 2143. The MIC for 86% of the resistant isolates with an A2143 mutation was ≥64 μg of clarithromycin per ml, and that for 89% of the resistant isolates with an A2144 mutation was ≤32 μg/ml. The detection of resistance markers, which could potentially be performed directly on biopsy material, could help to direct the treatment regimen for the patient, i.e., to use or not to use macrolides in therapy.
In the study by Occhialini et al. (225), a PCR amplification with consecutive sequencing or RFLP was used. These authors used RFLP to detect the mutations in the 23S rRNA genes without sequencing by analyzing the occurrence of special restriction sites. These techniques have been used also by others. Marais et al. (187) and Pina et al. (249) have used a colorimetric hybridization in the liquid phase to detect the mutation at the molecular level after PCR amplification. All of the techniques applied for the detection of point mutations can be used with biopsy specimens or infected gastric juice.
Other Species
The use of macrolide monotherapy leads to resistance in Mycobacterium avium as a result of mutations in the 23S rRNA. Nash and Inderlied have described an adaptation of the commercially available RNA/RNA duplex mismatch assay (Ambion, Austin, Tex.) for the detection of mutation leading to macrolide resistance (210). This assay utilizes a PCR with primers containing a promoter site for RNA polymerase. The PCR products are transcribed, and the resulting RNA product is hydrized with probe DNA. Duplexes are then treated with RNase A; the duplex will be cut at the location of a mismatch caused by a mutation. The sensitivity and specificity of the assay for macrolide resistance-related mutations in the 23S rRNA of M. avium was 100%, and the assay could be performed within 24 h.
Furthermore, the distribution of MLS resistance genes has been studied in other bacterial species. Roberts et al. screened clinical and commensal erythromycin-resistant N. gonorrhoeae isolates for ermB, ermC, and ermF genes, which could be detected in parallel to tet(M) in most of the isolates (267). In clindamycin-resistant isolates of Clostridium difficile, the ermB gene was detected by using probes and primers (134). The isolates studied by Johnson et al. (134) were responsible for outbreaks of diarrhea in the United States. Roberts (264) studied the distribution of MLS resistance genes in anaerobic bacteria. ermF genes were detected in clinical isolates of Bacteroides spp. by using probes (86). Recently, Chung et al. screened for the presence of the ermF gene in human and animal bacteria (51). Finally, erythromycin resistance determinants were found in isolates of Treponema denticola (266) and Actinobacillus pleuropneumoniae (382) by using probes and PCR.
Conclusion
The study of MLS resistance genes for diagnostic purposes is restricted mainly to epidemiology and basic research. However, there are two exceptions, the detection of mef genes coding for macrolide efflux pumps in streptococci and the detection of point mutations in the 23S rRNA of H. pylori resulting in macrolide resistance.
The detection of the mef genes without detection of any other macrolide resistance determinant offers therapeutic options, since clindamycin can be considered as an alternative antibiotic in treatment. The detection of macrolide resistance in H. pylori by DNA-based techniques can very quickly direct the antibiotic treatment of the patient, while classical test methods are quite slow. Otherwise, the usefulness of genetic methods to screen for MLS resistance genes is Otherwise, the usefulness of genetic methods to screen for MLS resistance genes is limited. This is mainly due to the diversity of resistance mechanisms and resistance genes resulting in MLS resistance.
RESISTANCE TO GLYCOPEPTIDES
Mechanisms of Resistance
Vancomycin and teicoplanin are glycopeptide antibiotics of clinical interest. The antimicrobial activity is due to binding to d-alanyl–d-alanine side chains of peptidoglycan or its precursors, thereby preventing cross-linking of the peptidoglycan chain (261). Antimicrobial activity of glycopeptide antibiotics is largely limited to gram-positive microorganisms; the peptidoglycan in gram-negative bacteria inside the outer cell membrane cannot be reached by the glycopeptide molecule. Not all gram-positive organisms are susceptible to the glycopeptide antimicrobial agents. Natural (“wild-type”) resistance occurs in gram-positive cocci such as Leuconostoc spp. and in gram-positive rods such as Lactobaccillus spp. In the latter, the side chain of the peptidoglycan molecule consists of a d-alanyl–d-lactate chain, which has less affinity to glycopeptide antibiotics (24). Because many enzymes are involved in the synthesis and cross-linking of peptidoglycan, it was for a long time deemed unlikely that resistance in naturally susceptible species would develop based on changes in the peptidoglycan-side chain.
However, despite this expectation, outbreaks of vancomycin-resistant enterococci have been reported since 1988 (167, 367). Genetic analysis during the last decade has revealed many details of the mechanisms involved (166). Phenotypic VanA is the most common type of resistance and confers high-level resistance to vancomycin and teicoplanin. The presence of the vanA gene cluster results in the production of VanA, a novel d-Ala–d-Ala ligase, resulting in the rebuilding of the peptidoglycan side chain to express the d-alanyl–d-lactate type, which has less affinity to glycopeptides. Other proteins in this gene cluster that are necessary for resistance include VanH and VanX. Phenotypic VanB confers moderate levels of resistance to vancomycin and susceptibility to teicoplanin in vitro. Vancomycin induction of the vanB gene cluster results in glycopeptide resistance mediated by the same basic mechanism as for VanA. Phenotypic VanC resistance is intrinsic in Enterococcus gallinarum (vanC-1 gene), E. casseliflavus (vanC-2 gene), and E. flavescens (vanC-3 gene), leading to expression of d-alanyl–d-serine, a similar peptidoglycan precursor, as shown in the recently described VanE resistance in an E. faecalis strain (85). An additional type, VanD, has also been described (245). Recently, molecular homology has been described between the VanA gene cluster and genes from the glycopeptide-producing microorganisms Amycolatopsis orientalis and Streptomyces toyocaensis (190). This finding suggests that genes that were originally present to protect antibiotic-producing microorganisms against self-destruction jumped to other species, resulting in selective advantage under glycopeptide treatment pressure.
Diminished heterogeneous susceptibility of CNS to teicoplanin was reported soon after this agent became available for MIC determination (302). In fact, Sieradzki (315) suggested from experiments with pre-antibiotic era isolates that heterogeneous teicoplanin phenotypes are intrinsic to the species. Most strains with low-level teicoplanin resistance were identified as S. haemolyticus, but other CNS species have been involved as well (317). Although some high teicoplanin MIC results have been attributed to technical factors in susceptibility testing (such as choice of medium and inoculum) (83), some CNS strains did also have vancomycin MICs above current NCCLS breakpoint for susceptibility (302). In 1996, an S. aureus strain with a vancomycin MIC of 8 μg/m was isolated from a patient in Japan (120). Subsequently, several other S. aureus strains with diminished glycopeptide susceptibility were reported (125, 313, 317). Although experimental transfer of the enterococcal VanA system to S. aureus (220) on the skin of mice has been reported, this resistance mechanism has not been identified in staphylococcal strains isolated from patients. Analysis of these clinical strains revealed that most were phenotypically heterogeneous in their expression of diminished vancomycin susceptibility. A vancomycin-resistant mutant strain appeared to have an unusually thick peptidoglycan layer acting as a sink for vancomycin, thereby increasing the MIC above therapeutic levels (314). Analysis of staphylococci with diminished glycopeptide susceptibility has suggested several possible mechanisms (23, 112, 199, 201) to explain why some S. aureus strains are predisposed to display heterogeneous glycopeptide resistance given favorable circumstances (e.g., long-term glycopeptide treatment of infections of prosthetic devices that remain in situ [376]). However, no molecular assays are yet available for the detection of glycopeptide resistance in staphylococci.
Detection of Resistance
Although currently used phenotypic methods are able to detect high-level glycopeptide resistance, detection of low-level resistance and differentiation between different Van types are more difficult by phenotypic methods (48, 54). The first molecular assays to detect glycopeptide resistance in enterococci were described approximately 5 years ago. Dutka-Malen et al. (76) described primers for the detection of vanA, vanB, and vanC-1 and a primer set that detected vanC-2 and vanC-3 in a multiplex PCR. Also in that year a second set of primers specific for vanA, vanB, and vanC-1 was published by Miele et al. (197). Two years later a third set of primers for use in a multiplex PCR was published by Patel et al. (240). This last group used restriction enzyme analysis of the PCR product as confirmation of the gene type. A total 100 clinical isolates were tested, and 63 isolates yielded an amplification product. Ten isolates contained vanA, 30 contained vanB, 12 contained vanC-1, 6 contained vanC-2, and 1 contained both vanA and vanC-1. The MICs of vancomycin for the vanA and vanB genotypes were ≥64 μg/ml, the MICs for the vanC types were 4 to 8 μg/ml, and the MICs for isolates which were PCR negative were 2 to 4 μg/ml.
Satake et al. (288) tested the primers described by Dutka-Malen et al. (76) directly on material obtained by rectal or perirectal swabbing. DNA was extracted and analyzed in the multiplex PCR. However, a discrepancy was observed for vanA between the multiplex and single PCR. In multiplex PCR, 59 specimens were vanA positive, whereas in the single PCR, 77 specimens were positive. However, 87 specimens were vancomycin resistant by conventional testing. One isolate was vanA positive in the PCR but susceptible by conventional testing. No vanB-positive isolates were identified. Despite this difference, the authors concluded that PCR might be an alternative to culture for surveillance of vancomycin-resistant enterococci. This conclusion can be challenged. The performance of the multiplex PCR is poor, and an 88.5% sensitivity for the PCR with the vanA-specific primer set may be considered too low. Furthermore, the performance of the other other primer sets has not been tested. The same primers or slightly modified versions were used more successfully in other studies (247, 273, 279), although 100% sensitivity was not reached, most probably due to PCR inhibition. Specificity was very high. A few other studies reported PCR assays with 100% sensitivity (253, 260). The discrepancies between these studies may reflect differences in the number of isolates tested, the type of sample used, and the need to carefully control the composition of multiplex PCR incubations.
Besides PCR, cycling probe technology has also been used to detect the vanA and vanB genes. In this technique, a DNA-RNA-DNA hybrid probe is used. After hybridization the RNA part is susceptible to RNase H degradation and results in a cleaved probe that can be detected easily. The results obtained with this assay showed that 11 isolates of a total of 440 clinical isolates were were discrepant, but PCR confirmed the results obtained with the probe (200).
The use of molecular techniques to detect transposon Tn1546 that largely encodes VanA resistance for epidemiological purposes has been reviewed recently (79).
Conclusion
Susceptibility testing of glycopeptide-resistant enterococci by molecular analysis of the van gene cluster is feasible; in fact, some PCR assays proved to be superior to conventional susceptibility testing and PCR-analyzed strains have been used as standards in assessing the ability of different methods to identify glycopeptide-resistant enterococci (48). Coombs et al. (56) recently recommended the use of PCR to detect the presence of the different van gene clusters in vancomycin-resistant enterococci. The present labor-intensive method of amplification techniques prevented the widespread application glycopeptide resistance in routine diagnostics, but PCR-based schemes appear promising as efficient and reliable methods for the surveillance of vancomycin-resistant enterococci.
RESISTANCE TO TETRACYCLINES
Mechanisms of Resistance
Tetracyclines probably penetrate bacterial cells by passive diffusion. Tetracycline acts by binding to the 30S ribosomal subunit, resulting in the inhibition of protein synthesis (49, 50, 152, 300). A growing number of bacterial species have acquired resistance to the bacteriostatic activity of tetracycline. In 1989, a note describing the nomenclature for tetracycline resistance determinants which employed letters of the English alphabet was published by Levy et al. (176). To date, at least 24 tetracycline resistance (Tet) determinants and three oxytetracycline resistance (Otr) determinants, first found in oxytetracyline-producing Streptomyces, have been described and characterized, with new Tet determinants being identified continually (172–174, 263, 265, 334). A note with an overview of the nomenclature has recently been published (175).
Most of the resistance genes code for one of the two important mechanisms of tetracycline resistance, either efflux or ribosomal protection. These two widespread mechanisms of bacterial resistance to tetracycline do not destroy the compound. Efflux is mediated by energy-dependent efflux pumps; the other important mechanism involves an elongation factor G-like protein that confers ribosome protection. Oxidative destruction of tetracycline has been found in a few species. Nevertheless, the enzymatic inactivation of the antibiotic is not thought to be important in nature (39, 172–176, 263, 265, 300, 334). An overview of the distribution of the better-studied tetracycline resistance determinants among gram-negative and gram-positive genera has been given in recent reviews by Roberts (263, 265).
The efflux proteins exchange a proton for a tetracycline-cation complex and are antiporter systems. Efflux determinants from gram-negative bacteria (TetA to TetE, TetG, and TetH) have a common genetic organization that is different from that in gram-positive bacteria. They all contain a structural gene and a repressor gene that are expressed in opposite directions from overlapping operator regions. The gram-positive tetK and tetL genes encoding tetracycline efflux proteins are regulated by mRNA attenuation in a similar way to that described for gram-positive erm genes encoding rRNA methylase and cat genes encoding chloramphenical acetyltransferases (172–176, 263, 265, 300, 334). The efflux proteins confer resistance to tetracycline but not to minocycline.
Protection of the ribosome from the action of tetracycline as a mechanism of tetracycline resistance was discovered in streptococci. Tetracycline resistance can result from the production of a protein that interacts with the ribosome such that protein synthesis is unaffected by the presence of the antibiotic. The determinants TetM, TetO, TetB(P), TetQ, TetS, TetT, TetW, and OtrA confer resistance to tetracycline, doxycycline, and minocycline (172–176, 263, 265, 300, 334).
The tetX gene codes for an enzyme which inactivates tetracyclines. This enzyme is a novelty because it is the first and only enzyme described which inactivates tetracyline (175, 334). Neither the prevalence nor the clinical significance has been studied so far.
According to data from Roberts (263, 265), specific tetracycline resistance genes have been identified in at least 32 gram-negative and 22 gram-positive genera. Nineteen gram-negative genera have been identified which carry only one or more efflux genes. In addition, there are nine genera which carry a single ribosomal protection gene. One genus (Bacteroides) can carry one of two ribosomal protection genes (tetM or tetQ) and the tetX gene coding for an inactivating enzyme. The remaining three genera carry both efflux and ribosomal protection genes, often in combination.
Among the gram-positive genera, 10 have been identified which carry only ribosomal protection genes. Of these, seven carry the tetM gene, while isolates of the remaining three genera carry two of the tetM, tetO, or tetQ genes. The remaining ten genera carry both efflux and ribosomal protection genes, often in combination (263, 265).
Detection of Resistance
In 1989, Levy et al. proposed DNA-DNA hybridization as the standard for grouping related genes coding for tetracycline resistance (176). Since then, only a limited number of genes have been sequenced, with the exception of those belonging to the M class. The result is that DNA-DNA hybridization is still used as a standard for determining different classes of tetracycline resistance genes, rather than DNA sequencing of individual structural genes. As a consequence, the detection of the tet genes for diagnostic and epidemiological purposes was based on DNA-DNA hybridization until the early 1990s. Probes were either radiolabeled or nonradiolabeled (digoxigenin was often used as label) and contained either a plasmid containing a special tet gene fragment or the fragment alone (175, 263, 265, 334).
Several authors have used DNA-DNA hybridization for screening and epidemiological purposes in a diversity of genera (20, 34, 46, 172, 239, 255, 268–270, 272, 282, 334). The microorganisms tested originated either from clinical samples or from animals. Using probes containing Tet determinants, the presence and the location of special tet genes was screened in a diversity of microorganisms from human sources. More recently, isolates of staphylococci (27, 282), E. faecium (20), E. faecalis (46), Peptostreptococcus species (262), Listeria monocytogenes (255), bacteria isolated from the urogenital tract (268), Campylobacter (216), Ureaplasma urealyticum (34), Actinomyces viscosus, Eubacterium lentum, Mobiluncus curtisii and Mobiluncus mulieris (270), and Mycobacterium and Streptomyces species (239) were screened. Using probes, the authors analyzed the presence of different Tet determinants within the different genera. In general, there was a good correlation between the determination of MICs and the presence of tet genes. As an example, Trzcinski et al. (356) analyzed the expression of tetracycline resistance in 66 MRSA isolates. In that study, they used PCR to screen for the presence of four tetracycline resistance determinants, tetK, tetL, tetM, and tetO. Of the 66 tetracycline-resistant MRSA isolates, mainly from Eastern Europe, 24 displayed tetM only, 21 had tetK only, and 21 had both tetK and tetM. The authors also determined the MICs of tetracycline, doxycycline, and minocycline. All of the isolates carrying the tetM gene showed in vitro resistance to both tetracycline and minocycline. Furthermore, the MICs were higher for the isolates harboring both the tetK and tetM genes than for the isolates containing just one of the genes. Despite the results of their susceptibility tests, Trzcinski et al. suggested that all tetracycline-resistant S. aureus isolates also be considered doxycycline resistant and that all tetM-positive isolates be treated as resistant to all tetracyclines. Similar data were reported by Warsa et al. (381) and Bismuth et al. (27). These authors also detected the tetL gene in 5 of 183 tetracycline-resistant staphylococci tested (27).
In addition, the location of the tet genes, chromosomal or extrachromosomal, on transferable elements was determined. The majority of the Tet determinants are frequently associated with either conjugative or mobilizable elements, which may partially explain their wide distribution among bacterial species (176, 263, 265, 334). Tet determinants are often associated with other antibiotic resistance genes, especially the erm genes (272). The ermF gene is often linked with the tetQ gene, while the ermB gene is often linked with the tetM gene (272).
In addition, probes have been used to analyze the presence and distribution of tet genes in bacteria from nonhuman sources. More recently, authors have tested gram-negative fecal isolates from pigs (169), C. jejuni and C. coli from chickens and clinical samples (170), Campylobacter species isolated from swine and cattle (206), and staphylococci from the skin of pigs (304).
Tetracycline-resistant bacteria containing tet genes are found in pathogens, opportunistic pathogens, and members of the normal flora. Tetracycline-resistant bacteria can be isolated from humans, animals, food, and the environment (176, 263, 265, 334). The nonpathogens in each of the ecosystems may play an important role as reservoirs for the antibiotic resistance genes. Several authors used PCR to detect special tet genes in bacteria from clinical samples (29, 129, 160, 161, 226, 238, 266, 271, 359, 381, 394). PCR was evaluated with various genera and different clinical samples, e.g., vaginal swab specimen from women with vaginitis (270), semen specimens from men with prostatitis (271), and the oral microflora of patients with adult periodontitis (160, 169, 226). In addition, PCR was sed for the detection of tetracycline resistance genes in slow-growing or difficult-to-culture bacteria such as N. gonorrhoeae (129, 358), Mycoplasma hominis, U. urealyticum (29), and T. denticola (266).
Roberts et al. (271) used degenerate oligonucleotide primers in a PCR assay to detect the tetM and tetO genes. Each of 44 clinical isolates and 8 laboratory strains, representing 20 different species carrying either TetM or TetO determinants, gave appropriate PCR products with the two primer sets. PCR products hybridized with radiolabeled TetM or TetO probes. The PCR assay was then used to evaluate vaginal swab specimens from women with vaginitis and semen specimens from men with prostatitis. Seven of eight vaginal samples and five of eight semen samples exhibited PCR products that hybridized with the radiolabeled probes, suggesting the presence of tetM and/or tetO genes. The authors concluded that PCR-based detection of tetM and/or tetO genes holds promise for evaluation of urogenital specimens (271). One year later, Roberts and coworkers developed a PCR assay for the detection of the tetK and/or tetL genes (238). Forty-three isolates representing 11 genera carrying TetK and/or TetL determinants were evaluated by PCR. All isolates gave positive PCR products which hybridized with labeled TetK or TetL probes. Four vaginal samples, previously positive with the TetM/O PCR assay, were also positive with the TetK/L assay. This was the first report of the detection of TetK and TetL determinants in clinical samples (238).
Lacroix and Walker reported the development of PCR assays for the detection of tetM and tetQ genes in the microflora associated with adult periodontitis (160, 161). Subgingival plaque samples were collected from 68 patients with adult periodontitis, enumerated on Trypticase soy blood agar plates with or without tetracycline, and incubated. Each colony morphotype was examined for the presence of tetM and tetQ genes. Approximately 15% of the 210 isolates subcultured that had resistance to tetracycline and belonged to different species contained tetQ, and 60% contained tetM. All of the tetQ-containing isolates were gram-negative anaerobic bacilli and included all of the Prevotella and Bacteroides isolates (160, 161).
Since high-level resistance to tetracycline in N. gonorrhoeae has become a therapeutic problem in some parts of the world, tests are required to enable its rapid detection. Ison et al. (129) developed a PCR on whole bacterial cells to detect tetM genes. A total of 109 tetracycline-resistant N. gonorrhoeae isolates, with resistance confirmation by hybridization with the tetM probe, were tested. tetM was detected by PCR in all strains and exhibited some genetic variation that could be of use for epidemiological typing. Turner et al. (359) examined the epidemiology of the tetM gene by using PCR. A single-tube PCR was developed which distinguishes between the American and Dutch variants of the tetM gene. A PCR product was obtained from all N. gonorrhoeae isolates with high-level resistance to tetracycline. A total of 387 isolates produced a 778-bp product (American-type tetM) and 131 produced a 443-bp PCR product (Dutch-type tetM). The authors concluded that PCR product variations in amplifying the tetM gene could be used in epidemiological studies describing the spread of different tetracycline-resistant N. gonorrhoeae isolates.
Tetracycline resistance in M. hominis and U. urealyticum has been associated with the tetM genes and has been increasing in incidence. Blanchard et al. (29) developed a rapid method for detection of the tetM gene based on PCR with amplification of a 397-bp product and verification of specificity using the enzyme TaqI. Analysis of 49 M. hominis and 42 U. urealyticum tetracycline-resistant isolates indicated that PCR could be clinically useful for determination of tetracycline sensitivity.
A similar epidemiological study of S. pneumoniae was carried out by Widdowson et al. (394). These authors detected the tetO gene in five isolates of S. pneumoniae resistant to tetracycline but lacking tetM, the most frequently occurring tet gene in S. pneumoniae (395).
Conclusion
The study of tetracycline resistance genes is interesting for diagnostic purposes, especially in organisms which carry only one or two Tet determinants. Furthermore, it would be useful to differentiate quickly between efflux-mediated resistance and ribosomal protection, since efflux proteins confer resistance to tetracycline but not to minocycline. In addition, detection of different tet genes is useful for epidemiology and basic research. Otherwise, the usefulness of genetic methods to screen for tetracycline resistance genes is limited. This is mainly due to the diversity of genes resulting in tetracycline resistance in some genera.
RESISTANCE TO TRIMETHOPRIM
Trimethoprim is an analog of dihydrofolic acid, an essential component in the synthesis of amino acid and nucleotides that competitively inhibits the enzyme dihydrofolate reductase (DHFR). Resistance can be caused by a number of mechanisms including overproduction of the host DHFR, mutations in the structural gene for DHFR, and the acquisition of a gene (dfr) encoding a resistant DHFR enzyme. The latter mechanism is the most important in clinical isolates. At least 15 DHFR enzyme types are known based on their properties and sequence homology (291).
Only a few papers have been published which describe the use of molecular techniques for the detection of trimethoprim resistance. All studies used the techniques for epidemiological purposes. One of the first studies investigated the presence of type I and type II DHFR in diverse gram-negative isolates showing high levels of trimethoprim resistance (87). Nine isolates did not hybridize with either probe, 17 hybridized with the type I probe, 11 hybridized with the type II probe, and 5 hybridized with both probes. Type I DHFR genes were sometimes found on the chromosome, in contrast to type II DHFR genes. A biotinylated fragment probe showed that 68 of 83 resistance plasmids obtained from trimethoprim-resistant members of the Enterobacteriaceae from Nottingham, United Kingdom, and its hospital carried the DHFR type I gene (41). Two similar studies using biotinylated probes investigated the presence of DFHR type I, II, and V genes in Enterobacteriaceae from persons with urinary tract infections in Greece (357, 358). Only 1 of 64 E. coli isolates hybridized with the DHFR type V-specific probe, while 21 E. coli isolates and 12 of 65 other isolates belonging to the Enterobacteriaceae hybridized with the DHFR type I-specific probe. Hybridization with the DHFR type II-specific probe was observed for 51 isolates, 7 of which also hybridzed with the type I-specific probe. Biotinylated probes were also developed for DHFR type Ib and IV genes (258, 353).
Radioactively labeled fragment probes were also used to detect DHFR type I, II, III, and V DNA in 374 enterobacteria from persons with urinary tract infections in Taiwan (45). DHFR type I was found in 45.4% of the isolates, DHFR type V was found in 10.4% of the isolates, whereas no hybridization was observed with the probes specific for DHFR type II and III. The DHFR type was predominantly found in E. coli, and the DFHR type V was frequently seen in Enterobacter spp. A more extended study also using radioactively labeled probes was performed on trimethoprim-resistant isolates from India (328). Fragment probes specific for DHFR types Ia, IIa, III, IV, V, and VII were used. In addition, an oligonucleotide probe to distinguish dfr5 from dfr1b was used. DHFR type Ia was the most prevalent, but DHFR V was most often associated with transferable trimethoprim resistance. The genes for DHFR type Ib and type IV were also present. A study by Gibreel and Sköld using PCR showed that 90% of C. jejuni isolates, believed to be endogenously resistant to trimethoprim, contained either dfr9, dfr1, or both (95).
Comparable studies were also performed for CNS isolates. A probe developed from a conjugative S. aureus trimethoprim resistance plasmid hybridized with chromosomal DNA of 14 isolates, plasmid DNA from 9 isolates, and both plasmid and chromosomal DNA from 1 isolate, and no hybridization was observed with 5 isolates (92). Similar results were obtained in a comparable study in Australia (339). However, no specific resistance determinant was named. These results indicate that the trimethoprim resistance determinants in staphylococci are widespread and can be located both chromosomally and on plasmids.
In conclusion, molecular assays for the detection of trimethoprim resistance are not very well developed. The few assays described use fragment probes, which are not always able to discriminate between the sometimes closely related dfr genes. Nevertheless, appropriate molecular techniques are valuable to establish the mechanism responsible for trimethoprim resistance. The presence of an alternative DHFR, especially of type I, II, or V, is the most common mechanism of trimethoprim resistance for Enterobacteriaceae, whereas the determinant(s) responsible for the resistance were not identified, although these are widespread.
RESISTANCE TO CHLORAMPHENICOL
Chloramphenicol binds to the 50S ribosomal subunit and inhibits the peptidyltransferase step in protein synthesis. Resistance to chloramphenicol is generally due to inactivation of the antibiotic by a chloramphenicol acetyltransferase. The cat genes of gram-negative and gram-positive bacteria show little homology, and a variety of different enzymes have been described. The gene is most commonly found on plasmids. Sometimes decreased outer membrane permeability or active efflux is observed in gram-negative bacteria (291).
Detection of chloramphenicol resistance by molecular techniques has not been widely studied. Two early studies (242, 243) looked at cross-hybridization of cat determinants from staphylococcal plasmids and a group B Streptococcus plasmid with streptococal, enterococcal, and pneumonoccal isolates. Although cross-resistance was observed, no clear-cut conclusions about the presence of specific cat determinants could be made. The catP and catQ genes from Clostridium difficile and C. perfringens were studied using fragment probes (274). The results indicated that the genes were limited to the genus Clostridium. However, it should be noted that the study was extremely limited in terms of the number of species and isolates analyzed. A second study described the use of degenerate primers in a PCR assay to enable the amplification of an internal fragment of the cat genes in gram-positive cocci (354). The assay was able to detect three classes of cat gene present in staphylococci, catP, and a streptococcal cat gene. catQ was not detected. The six cat genes accounted for the chloramphenicol resistance in 12 streptococci tested, but only 3 cat genes accounted for the resistance in 10 enterococcal isolates.
In conclusion, detection of chloramphenicol resistance determinants has received little attention, because chloramphenicol is little used for the treatment of severe infections. Only a limited number of small-scale studies have used molecular techniques to investigate the distribution of cat genes.
RESISTANCE TO MUPIROCIN
The elimination of staphylococci, particularly MRSA, from the nares plays a crucial role in infection control protocols. Currently, one of the most effective topical agents for eradication of nasal carriage of MRSA is mupirocin (78). Mupirocin (pseudomonic acid A) is an analogue of isoleucine that competitively binds to isoleucyl-tRNA synthetase (IRS), inhibiting protein synthesis (81). Resistance to mupirocin is phenotypically divided into two groups, low-level (MICs, 4 to 256 μg/ml) and high-level (MICs, ≥512 μg/ml). Low- and high-level resistance has been detected in both S. aureus and CNS (96, 254, 295). Low-level resistance to mupirocin in most cases is probably due to mutations in the host IRS. Isolates resistant to high levels of mupirocin contain an additional biochemically distinct IRS enzyme that is less sensitive to inhibition by mupirocin (96). The mupA gene, which codes for the mupirocin-resistant IRS, is only 30% similar to the host IRS at the amino acid level. mupA is known to be carried on transferable plasmids that vary in size in both S. aureus and CNS isolates that are epidemiologically unrelated (81, 96, 254). Since the concentrations achieved locally are likely to be in the region of 20,000 μg/ml, it is unlikely that low-level resistance will have a major impact on staphylococcal clearance. However, for high-level mupirocin-resistant staphylococci, it is unlikely that mupirocin applied topically will eradicate the organism (81, 254). Thus, rapid detection of high-level mupirocin-resistant staphylococci is important.
In a first attempt, Rahman et al. constructed probes from a 4.05-kb EcoRI fragment of a mupirocin resistance plasmid (259). A 751-bp internal part of this fragment hybridized with DNA from all of 36 independent high-level mupirocin-resistant staphylococci tested. In most instances, the probes detected an EcoRI fragment of approximately 4 kb. The probes did not hybridize with DNA from low-level resistant strains or from strains sensitive to mupirocin (259). Woodford et al. extended these studies and determined the location of the mupA gene (404). The gene was carried on plasmids of variable size, some of which were transferable in vitro. DNA hybridization of genomic DNA from 85 isolates showed that mupA was located on EcoRI fragments of seven different sizes. The authors concluded that the assay applied could be used not only for the detection of high-level mupirocin resistance but also as a potential epidemiological tool with which to monitor the spread of high-level mupirocin resistance in S. aureus, especially MRSA (404).
Recently, Anthony et al. described a PCR assay for rapid detection of high-level mupirocin-resistant staphylococci (8). The authors designed primers to amplify a 456-bp region of the mupA gene. PCR correctly classified 103 of 107 isolates in accordance with the results of agar dilution. For two isolates, the phenotypic results may have been erroneous. In one originally PCR-negative isolate, a 456-bp product was detected when retested using a lowered annealing temperature. The authors concluded that PCR of the mupA gene is a useful, rapid method for detecting high-level mupirocin resistance in staphylococci (8). PCR seems to be a useful method for the detection of high-level mupirocin resistance in staphylococci, which excludes the use of this topical agent for eradication of MRSA.
MULTIDRUG RESISTANCE
Often bacterial isolates are multidrug resistant. However, the vast majority of studies have looked at the detection and diagnosis of resistance caused by one class of antimicrobial agents while others have looked at the detection of resistance to multiple antimicrobial agents.
Integrons are a special case of multidrug resistance. Integrons are genetic elements that contain the genetic determinants of a site-specific recombination system that recognizes and captures mobile gene cassettes. An integron contains an integrase and an adjacent recombination site. Gene cassettes can be integrated by the integrase at the recombination site, and multiple gene cassettes can be present in one integron (110). Four classes of integron have been described. Class 1 integrons are often associated with the sulfonamide resistance gene sulI and are the most common integrons. Class 2 integron are associated with Tn7. Only one class 3 integron has been described, and class 4 is limited to Vibrio cholerae. Integrons are found almost exclusively in gram-negative bacteria, with one known exception. At least 60 gene cassettes have been described for class 1 integrons. The majority of genes encode antibiotic disinfectant resistance, including resistance to aminoglycosides, penicillins, cephalosporins, trimethoprim, tetracycline, erythromycin, and chloramphenicol (for an overview, see reference 86). Only a few epidemiological studies using integron-specific PCRs have been described, but in general the gene cassettes present were not characterized due to the wide range of possibilities and combinations. Epidemiological studies of Enterobacteriaceae in France and The Netherlands (136, 284) showed that over half of the isolates tested carried an integron. These results can be more or less extended to the rest of western Europe (191). A German study (296) of 278 consecutive blood isolates belonging to 11 species of gram-negative bacteria showed that 13% of the isolates belonging to 6 six species carried an integron. A Chilean study showed that the majority of integrons found in Acinetobacter baumannii isolates belonged to class 2 (102). Although class 1 integrons themselves are not mobile, they are often associated with plasmids and Tn21, whereas class 2 is associated with Tn7.
Transposons are well known for their capacity to carry multiple antimicrobial resistance genes, but only a few molecular studies of the epidemiology of transposons have been published. The presence of Tn7 was often examined in studies using probes to detect trimethoprim resistance. The study by the group in Greece showed that Tn7 was present in 19 of 57 E. coli isolates from persons with urinary tract infections. Tn7 was most often associated with DHFR type I (357). A study of trimethoprim resistance in India also investigated Tn7 (328). All plasmids carrying DFHR type Ia also hybridized with the Tn7-specific probe, whereas plasmids harboring DHFR type Ib or V hybridized with a probe specific for Tn21-like transpsons. A Taiwanese study observed that 7.8% of 374 trimethoprin-resistant isolates also carried Tn7, but no link with the DFHR types studied was made (45). It is not known whether these isolates harbor integrons.
The best-studied multidrug resistance elements are those which are often harbored by MRSA (316). Australian MRSA isolates may harbor up to three different plasmids: small (1.6-kb) cryptic plasmids, 4.5-kb chloramphenicol resistance plasmids, and 20- to 42-kb plasmids which variously encode resistance to antiseptics and disinfectants, trimethoprim, penicillin, gentamicin, tobramycin, and kanamycin. The last three resistances are encoded on Tn4001. The same transposon, although with some slight differences, was also detected in isolates in North America.
Multidrug resistance can also often be caused by reduced expression of porins and changes in the cell, which cause reduced uptake or the expression of efflux pumps. These efflux pumps generally have a rather broad substrate specificity and are able to handle antimicrobial agents of several different classes (334). However, to our knowledge no diagnostic molecular assays have been described for these efllux pumps.
Multidrug resistance may also be a problem in microorganisms which are difficult to culture. Chlamydia pneumoniae is an important human pathogen, but determining its antibiotic susceptibility is problematic; this, in combination with the often nonspecific symptoms, makes appropriate antibiotic treatment difficult. The immunofluorescence test used for antimicrobial susceptibility testing is subjective. Khan et al. (146) therefore proposed a molecular assay based on viability. Because mRNA has limited stability, it is soon destroyed in dead organisms. For the assay, the mRNA encoding the DnaK protein was chosen as target for RT-PCR. The results of the amplification were analyzed using an internal probe. Doxycycline, tetracycline, erythromycin, and ciprofloxacin inhibited mRNA production. However, the MICs obtained were two- to fourfold higher than those obtained in the immunofluorescence assay. The MBCs of erythromycin, ciprofloxacin, and doxycycline were twofold higher in the RT-PCR than in the conventional assay. No difference was observed for tetracycline. The differences may be caused by the fact that antibiotic concentrations are sufficient to prevent the formation of inclusion bodies but not to stop replication completely. Another explanation is that the appearance of inclusion bodies is altered by antibiotics and they are no longer recognized. Trimethoprim and sulfamethoxazole had no effect on the RT-PCR, in agreement with the immunofluorescence assay. The assay is very useful for the detection of resistance in C. pneumoniae, but it should be kept in mind that only a limited number of antibiotics can be tested in this assay. A comparison of the phenotypic and genotypic testing in relation to clinical outcome would yield interesting data about the validity of the different MBCs obtained.
Although multidrug resistance is a major problem, the detection of genetic elements which carry multiple resistance genes for either diagnostic or epidemiological purposes is rather limited.
CONCLUDING REMARKS
Molecular analysis of bacterial resistance has yielded a wealth of information during the last decade. With the aid of molecular amplification techniques, great progress has been made in our knowledge of the distribution and spread of resistance markers among the species. However, the early expectation that molecular techniques would surpass phenotypic susceptibility testing in routine diagnostic susceptibility testing has not (yet) been realized. Challenges that remain include the variety of point mutations or genes leading to resistance and the labor-intensive nature of current amplification methods. DNA chip technology combined with automated amplification techniques has the potential to meet these challenges. However, the development of DNA chips containing a broad range of resistance markers that are usable for many different species remains a formidable challenge and requires a broader knowledge of resistance markers than is currently available. Whereas molecular assays for several resistance markers are robust and reliable (e.g., mecA in staphylococci and vanA in enterococci), assays for other resistance markers are often not known or lack field testing. In addition, to detect emerging new resistance mechanisms, phenotypic (MIC) testing remains necessary in the foreseeable future.
REFERENCES
- 1.Afghani B, Stutman H R. Quantitative-competitive polymerase chain reaction for rapid susceptibility testing of Mycobacterium tuberculosis to isoniazid. Biochem Mol Med. 1997;60:182–186. doi: 10.1006/bmme.1997.2573. [DOI] [PubMed] [Google Scholar]
- 2.Alcaide F, Pfyffer G E, Telenti A. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob Agents Chemother. 1997;41:2270–2273. doi: 10.1128/aac.41.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allignet J, Aubert S, Morvan A, el Sohl N. Distribution of genes encoding resistance to streptogramin A and related compounds among staphylococci resistant to these antibiotics. Antimicrob Agents Chemother. 1996;40:2523–2528. doi: 10.1128/aac.40.11.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allignet J, Liassine N, el Sohl N. Characterization of a staphylococcal plasmid related to pUB110 and carrying two novel genes, vatC and vgbB, encoding resistance to streptogramins A and B and similar antibiotics. Antimicrob Agents Chemother. 1998;42:1794–1798. doi: 10.1128/aac.42.7.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allignet J, el Solh N. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob Agents Chemother. 1995;39:2027–2036. doi: 10.1128/aac.39.9.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altamirano M, Marostenmaki J, Wong A, FitzGerald M, Black W A, Smith J A. Mutations in the catalse-peroxidase gene from isoniazid-resistant Mycobacterium tuberculosis isolates. J Infect Dis. 1994;169:1162–1165. doi: 10.1093/infdis/169.5.1162. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez M, Mendoza M C. Molecular epidemiology of two genes encoding 3-N-aminoglycoside acetyltransferases AAC(3)I and AAC(3)II among gram-negative bacteria from a a Spanish hospital. Eur J Epidemiol. 1993;9:650–657. doi: 10.1007/BF00211441. [DOI] [PubMed] [Google Scholar]
- 7a.Ambler R P. The structure of β-lactamases. Philos Trans R Soc London Ser B. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 8.Anthony R M, Connor A M, Power E G, French G L. Use of polymerase chain reaction for rapid detection of high-level mupirocin resistance in staphylococci. Eur J Clin Microbiol Infect Dis. 1999;18:30–34. doi: 10.1007/s100960050222. [DOI] [PubMed] [Google Scholar]
- 9.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Archer G L, Niemeyer D M. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 1994;2:343–347. doi: 10.1016/0966-842x(94)90608-4. [DOI] [PubMed] [Google Scholar]
- 11.Archer G L, Pennell E. Detection of methicillin resistance in staphylococci by using a DNA probe. Antimicrob Agents Chemother. 1990;34:1720–1724. doi: 10.1128/aac.34.9.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arlet G, Brami G, Décrè D, Flippo A, Gaillot O, Lagrange P H, Philippon A. Molecular characterization by PCR-restriction fragment length polymorphism of TEM β-lactamases. FEMS Microbiol Lett. 1995;134:203–208. doi: 10.1111/j.1574-6968.1995.tb07938.x. [DOI] [PubMed] [Google Scholar]
- 13.Arlet G, Philippon A. Construction by polymerase chain reaction and intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB) FEMS Microbiol Lett. 1991;82:19–26. doi: 10.1016/0378-1097(91)90414-6. [DOI] [PubMed] [Google Scholar]
- 14.Arpin C, Canron M H, Noury P, Quentin C. Emergence of mefA and mefE genes in beta-haemolytic streptococci and pneumococci in France. J Antimicrob Chemother. 1999;44:133–134. doi: 10.1093/jac/44.1.133. [DOI] [PubMed] [Google Scholar]
- 15.Arthur M, Molinas C, Mabilat C, Courvalin P. Detection of erythromycin resistance by the polymerase chain reaction using primers in conserved regions of erm rRNA methylase genes. Antimicrob Agents Chemother. 1990;34:2024–2026. doi: 10.1128/aac.34.10.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci USA. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barg N, Chambers H, Kernodle D. Borderline susceptibility to antistaphylococcal penicillins is not conferred exclusively by the hyperproduction of beta-lactamase. Antimicrob Agents Chemother. 1991;35:1975–1979. doi: 10.1128/aac.35.10.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barg N L. Construction of a probe for the aminoglycoside 3-V-acetyltransferase gene and detection of the gene among endemic clinical isolates. Antimicrob Agents Chemother. 1988;32:1834–1838. doi: 10.1128/aac.32.12.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barringer K, Orgel L, Wahl G, Gingeras T R. Blunt-end and single-stranded ligations by Escherichia coli ligase: influence on an in vitro amplication scheme. Gene. 1990;89:117–122. doi: 10.1016/0378-1119(90)90213-b. [DOI] [PubMed] [Google Scholar]
- 20.Bentorcha F, De Cespédès G, Horaud T. Tetracycline resistance heterogeneity in Enterococcus faecium. Antimicrob Agents Chemother. 1991;35:808–812. doi: 10.1128/aac.35.5.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger-Bächi B. Expression of resistance to methicillin. Trends Microbiol. 1994;2:389–393. doi: 10.1016/0966-842x(94)90617-3. [DOI] [PubMed] [Google Scholar]
- 22.Beyer D, Pepper K. The streptogramin antibiotics: update on their mechanisms of action. Exp Opin Investig Drugs. 1998;7:591–599. doi: 10.1517/13543784.7.4.591. [DOI] [PubMed] [Google Scholar]
- 23.Billot-Klein D, Gutmann L, Bryant D, Bell D, van-Heijenoort J, Grewal J, Shlaes D M. Peptidoglycan synthesis and structure in Staphylococcus haemolyticus expressing increasing levels of resistance to glycopeptide antibiotics. J Bacteriol. 1996;178:4696–4703. doi: 10.1128/jb.178.15.4696-4703.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Billot-Klein D, Gutman L, Sable S, Guittet E, van Heijenoort J. Modification of peptidoglycan precursors is a common feature of the low-level vancomycin-resistant VANB-type Enterococcus D366 and the naturally glycopeptide-resistant species Lactobacillus casei, Pediococcus pentosaceus, Leuconostoc mesenteroides, and Enterococcus gallinarum. J Bacteriol. 1994;176:2398–2405. doi: 10.1128/jb.176.8.2398-2405.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birkenmeyer L G, Mushahwar I K. DNA probe amplification methods. J Virol Methods. 1991;35:117–126. doi: 10.1016/0166-0934(91)90127-l. [DOI] [PubMed] [Google Scholar]
- 26.Bisessar U, James R. Molecular cloning of the Shv-1 β-lactamase gene and construction of an Shv-1 hybridization probe. J Gen Microbiol. 1988;134:835–840. doi: 10.1099/00221287-134-3-835. [DOI] [PubMed] [Google Scholar]
- 27.Bismuth R, Zilhao R, Sakamoto H, Guesdon J L, Courvalin P. Gene heterogeneity for tetracycline resistance in Staphylococcus spp. Antimicrob Agents Chemother. 1990;34:1611–1614. doi: 10.1128/aac.34.8.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjorkholm B, Befrits R, Jaup B, Engstrand L. Rapid PCR detection of Helicobacter pylori-associated virulence and resistance genes directly from gastric biopsy material. J Clin Microbiol. 1998;36:3689–3690. doi: 10.1128/jcm.36.12.3689-3690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchard A, Crabb D M, Dybvig K, Duffy L B, Cassel G H. Rapid detection of tetM in Mycoplasma hominis and Ureaplasma urealyticum by PCR: tetM confers resistance to tetracycline but not necessarily to doxycycline. FEMS Microbiol Lett. 1992;74:277–281. doi: 10.1016/0378-1097(92)90442-q. [DOI] [PubMed] [Google Scholar]
- 30.Boissinot M, Mercier J, Levesque R C. Development of natural and synthetic DNA probes for OXA-2 and TEM-1 β-lactamases. Antimicrob Agents Chemother. 1987;31:728–734. doi: 10.1128/aac.31.5.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bozdogan B, Berrezouga L, Kuo M S, Yurek D A, Farley K A, Stockman B J, Leclercq R. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob Agents Chemother. 1999;43:925–999. doi: 10.1128/aac.43.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brakstad O G, Maeland J A, Tveten Y. Multiplex polymerase chain reaction for detection of genes for Staphylococcus aureus thermonuclease and methicillin resistance and correlation with oxacillin resistance. APMIS. 1993;101:681–688. doi: 10.1111/j.1699-0463.1993.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 33.Brenwald N P, Gell M J, Wise R. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of S. pneumoniae. Antimicrob Agents Chemother. 1998;42:2032–2035. doi: 10.1128/aac.42.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunet B, Barbeyrac B, Renaudin H, Bebear C. Detection of tetracycline-resistant strains of Ureaplasma urealyticum by hybridization assays. Eur J Microbiol Infect Dis. 1989;8:636–638. doi: 10.1007/BF01968147. [DOI] [PubMed] [Google Scholar]
- 35.Broskey J, Coleman K, Gwynn M N, McCloskey L, Traini C, Voelker L, Warren R. Efflux and target mutations as quinolone resistance mechanisms in clinical isolates of Streptococcus pneumoniae. J Antimicrob Chemother. 2000;45(Suppl. 1):95–99. doi: 10.1093/jac/45.suppl_3.95. [DOI] [PubMed] [Google Scholar]
- 36.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structures. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byrne M E, Gillespie M T, Skurray R A. 4′,4′′-Adenylyltransferase activity on conjugative plasmids isolated from Staphylococcus aureus is encoded on an integrated copy of pUB110. Plasmid. 1991;24:70–75. doi: 10.1016/0147-619x(91)90008-k. [DOI] [PubMed] [Google Scholar]
- 38.Cangelosi G A, Brabant W H, Britschgi T A, Wallis C K. Antimicrob. Agents Chemother. 1996. Detection of rifampin- and ciprofloxacin-resistant Mycobacterium tuberculosis by using species-specific assays for precursor rRNA. Antimicrob Agents Chemother. 1996;40:1790–1795. doi: 10.1128/aac.40.8.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carballo M, Ng L K, Dillon J R. Detection of the tetM determinant in Neisseria gonorrhea using a non-radioactively labelled oligonucleotide probe. Mol Cell Probes. 1994;8:205–208. doi: 10.1006/mcpr.1994.1028. [DOI] [PubMed] [Google Scholar]
- 40.Carter G I, Towner K J, Pearson N J, Slack R C B. Use of a non-radioactive hybridization assay for direct detection of gram-negative bacteria carrying TEM β-lactamase genes in infected urine. J Med Microbiol. 1989;28:113–117. doi: 10.1099/00222615-28-2-113. [DOI] [PubMed] [Google Scholar]
- 41.Carter G I, Towner K J, Slack R C B. Rapid detection of a specific trimethoprim resistance gene using a biotinylated DNA probe. J Antimicrob Chemother. 1987;20:335–341. doi: 10.1093/jac/20.3.335. [DOI] [PubMed] [Google Scholar]
- 42.Carter G I, Towner K J, Slack R C B. Detection of TEM beta-lactamase genes by non-isotopic spot hybridisation. Eur J Clin Microbiol. 1987;6:406–409. doi: 10.1007/BF02013095. [DOI] [PubMed] [Google Scholar]
- 43.Caviani Pease A, Solas D, Sullivan E J, Cronin M T, Holmes C P, Fodor S P A. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cha R S, Zarbl H, Keohavong P, Thilly W G. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 45.Chang L-L, Chang S-F, Chow T-H, Wu W-J, Chang J-C. The distribution of the DHFR genes in trimethoprim-resistant urinary tract isolates from Taiwan. Epidemiol Infect. 1992;109:453–462. doi: 10.1017/s0950268800050445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charpentier E, Gerbaud G, Courvalin P. Presence of the Listeria resistance gene tet(S) in Enterococcus faecalis. Antimicrob Agents Chemother. 1994;38:2330–2335. doi: 10.1128/aac.38.10.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charvalos E, Peteinaki E, Spyridaki I, Manetas S, Tselentis Y. Detection of ciprofloxacin resistance mutations in Campylobacter jejuni gyrA by nonradioisotopic single-strand confirmation polymorphism and direct DNA sequencing. J Clin Lab Ann. 1996;10:129–133. doi: 10.1002/(SICI)1098-2825(1996)10:3<129::AID-JCLA3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y S, Marshall S A, Winokur P L, Coffman S L, Wilke W W, Murray P R, Spiegel C A, Pfaller M A, Doern G V, Jones R N. Use of molecular and reference susceptibility testing methods in a multicenter evaluation of MicroScan dried overnight gram-positive MIC panels for detection of vancomycin and high-level aminoglycoside resistances in enterococci. J Clin Microbiol. 1998;36:2996–3001. doi: 10.1128/jcm.36.10.2996-3001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chopra I. Tetracycline analogs whose primary target is not the bacterial ribosome. Antimicrob Agents Chemother. 1994;38:637–640. doi: 10.1128/aac.38.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chopra I, Hawky P M, Hinton M. Tetracyclines, molecular and clinical aspects. J Antimicrob Chemother. 1992;29:245–277. doi: 10.1093/jac/29.3.245. [DOI] [PubMed] [Google Scholar]
- 51.Chung W O, Werckenthin C, Schwarz S, Roberts M C. Host range of the ermF rRNA methylase gene in human and animal bacteria. J Antimicrob Chemother. 1999;43:5–14. doi: 10.1093/jac/43.1.5. [DOI] [PubMed] [Google Scholar]
- 52.Clancy J, Dib-Hajj F, Petitpas J W, Yuan W. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae. Antimicrob Agents Chemother. 1997;41:2719–2723. doi: 10.1128/aac.41.12.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 54.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 56.Coombs G W, Kay I D, Steven R A, Pearman J W, Bertolatti D, Grubb W B. Should genotypic testing be done on all phenotypically vancomycin-resistant enterococci detected in hospitals? J Clin Microbiol. 1999;37:1229–1230. doi: 10.1128/jcm.37.4.1229-1230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooksey R C, Clark N C, Thornsberry C. A gene probe for TEM type β-lactamases. Antimicrob Agents Chemother. 1985;28:154–156. doi: 10.1128/aac.28.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Curran R, Talbot D C S, Towner K J. A rapid immunoassay method for the direct detection of PCR products: application to detection of TEM β-lactamase genes. J Med Microbiol. 1996;45:76–78. doi: 10.1099/00222615-45-1-76. [DOI] [PubMed] [Google Scholar]
- 59.Dahlén P, Syvänen A C, Hurskainen P, Kwiatkowski M, Sund C, Ylikoski J, Söderlund H, Lövgren T. Sensitive detection of genes by sandwich hybridization and time-resolved fluorometry. Mol Cell Probes. 1987;1:159–168. doi: 10.1016/0890-8508(87)90024-7. [DOI] [PubMed] [Google Scholar]
- 60.Daum R S, Murphy-Corb M, Shapira E, Dipp S. Epidemiology of Rob β-lactamase among ampicillin-resistant Haemophilus influenzae isolates in the United States. J Infect Dis. 1988;157:450–455. doi: 10.1093/infdis/157.3.450. [DOI] [PubMed] [Google Scholar]
- 61.Davies T A, Kelly L M, Pankuch G A, Credito K L, Jacobs M R, Appelbaum P C. Antipneumococcal activities of gemifloxacin compared to those of nine other agents. Antimicrob Agents Chemother. 2000;44:304–310. doi: 10.1128/aac.44.2.304-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Azavedo J C, Yeung R H, Bast D J, Duncan C L, Borgia S B, Low D E. Prevalence and mechanisms of macrolide resistance in clinical isolates of group A streptococci from Ontario, Canada. Antimicrob Agents Chemother. 1999;43:2144–2147. doi: 10.1128/aac.43.9.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Beenhouwer H, Lhiang Z, Jannes G, Mijs W, Machtelinckx L, Rossau R, Traore H, Portaels F. Rapid detection of rifampicin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tubercle Lung Dis. 1995;76:425–430. doi: 10.1016/0962-8479(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 64.de Lencastre H, Figuereido A M S, Urban C, Rahal J, Tomasz A. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 1991;35:632–639. doi: 10.1128/aac.35.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Derbise A, Dyke K G, El Solh N. Characterization of a Staphylococcus aureus transposon Tn5407 carrying the aminoglycoside resistance genes, aphA-3 and aadE. Plasmid. 1996;35:174–188. doi: 10.1006/plas.1996.0020. [DOI] [PubMed] [Google Scholar]
- 66.Dillon J R, Li H, Yeung K-H, Aman T A. A PCR assay for discriminating Neisseria gonorrhoea β-lactamase-producing plasmids. Mol Cell Probes. 1999;13:89–92. doi: 10.1006/mcpr.1998.0216. [DOI] [PubMed] [Google Scholar]
- 67.Doern G, V, Pfaller M A, Kugler K, Freeman J, Jones R N. Prevalence of antimicrobial resistance among respiratory tract isolates of Streptococcus pneumoniae in North America: 1997 results from the SENTRY antimicrobial surveillance program. Clin Infect Dis. 1998;27:764–770. doi: 10.1086/514953. [DOI] [PubMed] [Google Scholar]
- 68.Doit C, Denamur E, Picard B, Geslin P, Elion J, Bingen E. Mechanisms of the spread of penicillin resistance in Streptococcus pneumoniae strains causing menigitis in children in France. J Infect Dis. 1996;174:520–528. doi: 10.1093/infdis/174.3.520. [DOI] [PubMed] [Google Scholar]
- 69.Douglas J, Steyn L M. A ribosomal gene mutation in streptomycin-resistant Mycobacterium tuberculosis isolates. J Infect Dis. 1993;167:1505–1506. doi: 10.1093/infdis/167.6.1505. [DOI] [PubMed] [Google Scholar]
- 70.Dowson C G, Coffey T J, Kell C, Whiley R A. Evolution of penicillin resistance in Streptococcus pneumoniae: the role of Streptococcus mitis in the formation of a low affinity PBP2b in Streptococcus pneumoniae. Mol Microbiol. 1993;9:635–643. doi: 10.1111/j.1365-2958.1993.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 71.Dowson C G, Hutchinson A, Brannigan J A, George R C, Hansman D, Liñares J, Tomasz A, Smith J M, Spratt B G. Horizontal transfer of pencillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drlica K, Zhao X L. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drobniewski F A, Wilson S M. The rapid diagnosis of isoniazid and rifampin resistance in Mycobacterium tuberculosis—a molecular story. J Med Microbiol. 1998;47:189–196. doi: 10.1099/00222615-47-3-189. [DOI] [PubMed] [Google Scholar]
- 74.Du Plessis M, Smith A M, Klugman K P. Rapid detection of penicillin-resistant Streptococcus pneumoniae in cerebrospinal fluid by seminested-PCR strategy. J Clin Microbiol. 1998;36:453–457. doi: 10.1128/jcm.36.2.453-457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du Plessis M, Smith A M, Klugman K P. Application of pbp1A PCR in identification of penicillin-resistant Streptococcus pneumoniae. J Clin Microbiol. 1999;37:628–632. doi: 10.1128/jcm.37.3.628-632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eady E A, Ross J I, Tipper J L, Walters C E, Cove J H, Noble W C. Distribution of genes encoding erythromycin ribosomal methylases and an erythromycin efflux pump in epidemiologically distinct groups of staphylococci. J Antimicrob Chemother. 1993;31:211–217. doi: 10.1093/jac/31.2.211. [DOI] [PubMed] [Google Scholar]
- 78.Eltringham I. Mupirocin resistance and methicillin-resistant Staphylococcus aureus (MRSA) J Hosp Infect. 1997;35:1–8. doi: 10.1016/s0195-6701(97)90162-6. [DOI] [PubMed] [Google Scholar]
- 79.Endtz H P, van den Braak N, Verbrugh H, van Belkum A. Vancomycin resistance: stautus quo and quo vadis. Eur J Clin Microbiol Infect Dis. 1999;18:683–690. doi: 10.1007/s100960050379. [DOI] [PubMed] [Google Scholar]
- 80.Everett M J, Piddock L J V. Mechanisms of resistance to fluoroquinolones. In: Kuhlmann J, Dahlhoff A, Zeiler H J, editors. Quinolone antibacterials 1998. Berlin, Germany: Springer-Verlag KG; 1998. pp. 259–297. [Google Scholar]
- 81.Farmer T H, Gilbart J, Elson S W. Biochemical basis of mupirocin resistance in strains of Staphylococcus aureus. J Antimicrob Chemother. 1992;30:587–596. doi: 10.1093/jac/30.5.587. [DOI] [PubMed] [Google Scholar]
- 82.Fasching C E, Tenover F C, Slama T G, Fisher L M, Sreedharan S, Oram M, Willard K, Sinn L M, Geerding D N, Peterson L R. GyrA mutations in ciprofloxacin resistant, methicillin resistant Staphylococcus aureus from Indiana, Minnesota and Tennessee. J Infect Dis. 1991;164:976–979. doi: 10.1093/infdis/164.5.976. [DOI] [PubMed] [Google Scholar]
- 83.Felmingham D, Solomonides K. The effect of medium and inoculum on the activity of vancomycin and teicoplanin against coagulase-negative staphylococci. J Antimicrob Chemother. 1987;33:2019–2022. doi: 10.1093/jac/20.4.609. [DOI] [PubMed] [Google Scholar]
- 84.Felmlee T A, Liu Q, Whelen C, Williams D, Sommer S S, Persing D H. Genotypic detection of Mycobacterium tuberculosis rifampin resistance: comparison of single-strand conformation polymorphism and dideoxy fingerprinting. J Clin Microbiol. 1995;33:1617–1623. doi: 10.1128/jcm.33.6.1617-1623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fines M, Perichon B, Reynolds P, Sahm D, Courvalain P. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. J Clin Microbiol. 1999;43:2161–2164. doi: 10.1128/aac.43.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fletcher H M, Macrina F L. Molecular survey of clindamycin and tetracycline resistance determinants in Bacteroides species. Antimicrob Agents Chemother. 1991;35:2415–2418. doi: 10.1128/aac.35.11.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fling M E, Walton L, Elwell L P. Monitoring of plasmid-encoded, trimethoprim-resistant dihydrofolate reductase genes: detection of a new resistant enzyme. Antimicrob Agents Chemother. 1982;22:882–888. doi: 10.1128/aac.22.5.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fluit A C, Jones M E, Schmitz F J, Acar J, Gupta R, Verhoef J for the Sentry Participants Group. Antimicrobial resistance among Urinary Tract Infection (UTI) isolates in Europe: results from the SENTRY Antimicrobial Surveillance Program 1997. Antonie Leeuwenhoek. 2000;77:147–152. doi: 10.1023/a:1002003123629. [DOI] [PubMed] [Google Scholar]
- 89.Fluit A C, Jones M E, Schmitz F-J, Acar J, Gupta R, Verhoef J for the SENTRY Participants Group. Bacteremia in European hospitals, incidence and antimicrobial susceptibility. Clin Infect Dis. 2000;30:454–460. doi: 10.1086/313710. [DOI] [PubMed] [Google Scholar]
- 90.Fluit A C, Schmitz F-J. Class 1 integrons: gene cassettes, and epidemiology. Eur J Clin Microbiol Infect Dis. 1999;18:761–770. doi: 10.1007/s100960050398. [DOI] [PubMed] [Google Scholar]
- 91.Fluit A C, Schmitz F-J, Jones M E, Acar J, Gupta R, Verhoef J for the SENTRY Participants Group. Antimicrobial resistance among community-acquired pneumoniae isolates in Europe: first results from the SENTRY antimicrobial surveillance program 1997. Int J Infect Dis. 1999;3:153–156. doi: 10.1016/s1201-9712(99)90037-1. [DOI] [PubMed] [Google Scholar]
- 92.Galetto D W, Johnston J L, Archer G L. Molecular epidemiology of trimethoprim resistance among coagulase-negative staphylococci. Antimicrob Agents Chemother. 1987;31:1683–1688. doi: 10.1128/aac.31.11.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Geha D J, Uhl J R, Gustaferro C A, Persing D H. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32:1768–1772. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gerhold D, Rushmore T, Caskey C T. DNA chips: promising toys have become powerful tools. Trends Biochem Sci. 1999;24:168–173. doi: 10.1016/s0968-0004(99)01382-1. [DOI] [PubMed] [Google Scholar]
- 95.Gibreel A, Sköld O. High-level resistance to trimethoprim in clinical isolates of Campylobacter jejuni by acquisition of foreign genes (dfr1 and dfr9) expressing drug-insensitive dihydrofolate reductases. Antimicrob Agents Chemother. 1998;42:3059–3064. doi: 10.1128/aac.42.12.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gilbart J, Perry C, Slocombe B. High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob Agents Chemother. 1993;37:32–38. doi: 10.1128/aac.37.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gill M J, Brenwald N P, Wise R. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in S. pneumoniae. Antimicrob Agents Chemother. 1999;43:187–1189. doi: 10.1128/aac.43.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gillespie S H, McHugh T D, Hughes J E, Dickens A, Kyi M S, Kelsey M. An outbreak of penicillin-resistant Streptococcus pneumoniae investigated by a polymerase chain reaction based genotyping method. J Clin Pathol. 1997;50:847–851. doi: 10.1136/jcp.50.10.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gingeras T R, Whitfield K M, Kwoh D Y. Unique features of the self-sustained sequence replication (3SR) reaction in in vitro amplification of nucleic acids. Ann Biol Clin. 1990;48:498–501. [PubMed] [Google Scholar]
- 100.Giraud E, Brisabois A, Martel J L, Chaslus-Dancla E. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob Agents Chemother. 1999;43:2131–2137. doi: 10.1128/aac.43.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goffeau A. Molecular fish on chips. Nature. 1997;385:202–203. doi: 10.1038/385202a0. [DOI] [PubMed] [Google Scholar]
- 102.Gonzalez G, Sossa K, Bello H, Dominguez M, Mella S, Zemelman R. Presence of integrons in isolates of different biotypes of Acinetobacter baumannii from Chilean hospitals. FEMS Microbiol Lett. 1998;161:125–128. doi: 10.1111/j.1574-6968.1998.tb12937.x. [DOI] [PubMed] [Google Scholar]
- 103.Goswitz J J, Willard K E, Fasching C E, Peterson L R. Detection of gyrA gene mutations associated with ciprofloxacin resistance in methicillin-resistant Staphylococcus aureus: analysis by polymerase chain reaction and automated direct DNA sequencing. Antimicrob Agents Chemother. 1992;36:1166–1169. doi: 10.1128/aac.36.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goyal M, Shaw R J, Banerjee D K, Coker R J, Robertson B D, Young D B. Rapid detection of multidrug-resistant tuberculosis. Eur Respir J. 1997;10:1120–1124. doi: 10.1183/09031936.97.10051120. [DOI] [PubMed] [Google Scholar]
- 105.Granjeaud S, Bertucci F, Jordan B R. Expression profiling: DNA arrays in many guises. Bioessays. 1999;21:781–790. doi: 10.1002/(SICI)1521-1878(199909)21:9<781::AID-BIES10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 106.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR-primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Groot Obbink D J, Ritchie L J, Cameron F H, Mattick J S, Ackerman V P. Construction of a gentamicin resistance gene probe for epidemiological studies. Antimicrob Agents Chemother. 1985;28:96–102. doi: 10.1128/aac.28.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hakenbeck R, Tarplay M, Tomasz A. Multiple changes of penicillin-biniding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980;17:364–371. doi: 10.1128/aac.17.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Haliassos A, Chomel J C, Tesson L, Baudis M, Kruh J, Kaplan J C, Kitzis A. Modifications of enzymatically amplified DNA for the detection of point mutations. Nucleic Acids Res. 1989;17:3606. doi: 10.1093/nar/17.9.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 111.Hammerum A M, Jensen L, Bogo L, Aarestrup F M. Detection of the satA gene and transferability of virginiamycin resistance in Enterococcus faecium from food animals. FEMS Microbiol Lett. 1998;168:145–151. doi: 10.1111/j.1574-6968.1998.tb13267.x. [DOI] [PubMed] [Google Scholar]
- 112.Hanaki H, Labischinski H, Inaba Y, Kondo N, Murakami H, Hiramatsu K. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J Antimicrob Chemother. 1998;42:315–320. doi: 10.1093/jac/42.3.315. [DOI] [PubMed] [Google Scholar]
- 113.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hayashi K. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 1991;1:34–38. doi: 10.1101/gr.1.1.34. [DOI] [PubMed] [Google Scholar]
- 115.Hayashi K. PCR-SSCP: a method for detection of mutations. Genet Anal Tech Appl. 1992;9:73–79. doi: 10.1016/1050-3862(92)90001-l. [DOI] [PubMed] [Google Scholar]
- 116.Head S R, Parikh K, Rogers Y-H, Bishai W, Goelet P, Boyce-Jacino M T. Solid-phase sequence scanning for drug resistance detection in tuberculosis. Mol Cell Probes. 1999;13:81–87. doi: 10.1006/mcpr.1998.0212. [DOI] [PubMed] [Google Scholar]
- 117.Hedin G, Löfdahl S. Detecting methicillin-resistant Staphylococcus epidermidis—disc diffusion, broth breakpoint or polymerase chain reaction? APMIS. 1993;101:311–318. doi: 10.1111/j.1699-0463.1993.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 118.Heym B, Alzari P M, Honoré N, Cole S T. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazed resistance in Mycobacterium tuberculosis. Mol Microbiol. 1995;15:235–245. doi: 10.1111/j.1365-2958.1995.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 119.Higuchi R. Simple and rapid sample preparation of samples for PCR. In: Ehrlich G A, editor. PCR technology: principles and applications for DNA amplification. New York, N.Y: Stockton Press; 1989. pp. 31–38. [Google Scholar]
- 120.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 121.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hooper D C. Bacterial topoisomerases, anti-topoisomerases and anti-topoisomerase resistance. Clin Infect Dis. 1998;27:54–63. doi: 10.1086/514923. [DOI] [PubMed] [Google Scholar]
- 123.Hooper D C. Mechanisms of fluoroquinolone resistance. Drug Resist Updates. 1999;2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 124.Hotta K, Ishikawa J, Ishii R, Saitoii F, Kira K, Arakawa Y, Ike Y. Necessity and usefulness of detection by PCR of mecA and aac(6′)-Ie+aph(2′′) genes for identification of arbekacin resistant MRSA. Jpn J Antibiot. 1999;52:525–532. [PubMed] [Google Scholar]
- 125.Howe R A, Bowker K E, Walsh T R, Feest T G, MacGowan A P. Vancomycin-resistant Staphylococcus aureus. Lancet. 1998;351:602. doi: 10.1016/S0140-6736(05)78597-4. [DOI] [PubMed] [Google Scholar]
- 126.Hsueh P-R, Teng L-J, Lee L-N, Yang P-C, Ho S-W, Luh K-T. Dissemination of high-level penicillin- extended-spectrum cephalosporin-, and erythromycin-resistant Streptococcus pneumoniae clones in Taiwan. J Clin Microbiol. 1999;37:221–224. doi: 10.1128/jcm.37.1.221-224.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huovinen S, Huovinen P, Jacoby G A. Detection of plasmid-mediated β-lactamases with DNA probes. Antimicrob Agents Chemother. 1988;32:175–179. doi: 10.1128/aac.32.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Interlied C B, Salfinger M. Antimycobacterial agents and susceptibility tests. In: Murray P R, Baron E J, Pfaller M A, Tenover F R, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 1308–1326. [Google Scholar]
- 129.Ison C A, Tekki N, Gill M J. Detection of the tetM determinant in Neisseria gonorrhoeae. Sex Transm Dis. 1993;20:329–333. [PubMed] [Google Scholar]
- 130.Jalal H, Organji S, Reynolds J, Bennett D, O'Mason E, Jr, Millar M R. Determination of penicillin susceptibility of Streptococcus pneumoniae using the polymerase chain reaction. Mol Pathol. 1997;50:45–50. doi: 10.1136/mp.50.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jensen L B, Frimondt-Moller N, Aarestrup F M. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol Lett. 1999;170:151–158. doi: 10.1111/j.1574-6968.1999.tb13368.x. [DOI] [PubMed] [Google Scholar]
- 132.Jensen L B, Hammerum A M, Aerestrup F M, van den Bogaard A E, Stobberingh E E. Occurrence of satA and vgb genes in streptogramin-resistant Enterococcus faecium isolates of animal and human origins in the Netherlands. Antimicrob Agents Chemother. 1998;42:3330–3331. doi: 10.1128/aac.42.12.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jenssen W D, Thakker-Varia S, Dubin D T, Weinstein M P. Prevalence of macrolides-lincosamides-streptogramin B resistance and erm gene classes among clinical strains of staphylococci and streptococci. Antimicrob Agents Chemother. 1987;31:883–888. doi: 10.1128/aac.31.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johnson S, Samore M H, Farrow K A, Killgore G E, Tenover F C, Lyras D, Rood J I, DeGirolami P, Baltch A L, Rafferty M E, Pear S M, Gerding D N. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N Engl J Med. 1999;25:1645–1651. doi: 10.1056/NEJM199911253412203. [DOI] [PubMed] [Google Scholar]
- 135.Johnston N J, de Azavedo J C, Kellner J D, Low D E. Prevalence and characterization of the mechanisms of macrolide, lincosamide and streptogramin resistance in isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2425–2426. doi: 10.1128/aac.42.9.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jones M E, Peters E, Weersink A, Fluit A, Verhoef J. Widespread occurrence of integrons causing multiple resistance in bacteria. Lancet. 1997;349:1742–1743. doi: 10.1016/S0140-6736(05)62954-6. [DOI] [PubMed] [Google Scholar]
- 137.Jones M E, Sahm D F, Martin N, Scheuring S, Heisig P, Thornsberry C, Köhrer K, Schmitz F J. The Prevalence of gyrA, gyrB, parC and parE mutations in clinical isolates S. pneumoniae with decreased susceptibilities to different fluoroquinolones and originating from worldwide surveillance studies during the 1997–1998 respiratory season. Antimicrob Agents Chemother. 2000;44:462–466. doi: 10.1128/aac.44.2.462-466.2000. , [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jones R N, Cormican M G, Wanger A. Clindamycin resistance among erythromycin-resistant Streptococcus pneumoniae. Diagn Microbiol Infect Dis. 1996;25:201–204. doi: 10.1016/s0732-8893(96)00100-9. [DOI] [PubMed] [Google Scholar]
- 139.Jorgensen J H, Weigel L M, Ferraro M J, Swenson J M, Tenover F C. Activities of newer fluoroquinolones against S. pneumoniae clinical isolates including those with mutations in the gyrA, parC and parE loci. Antimicrob Agents Chemother. 1999;43:329–334. doi: 10.1128/aac.43.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jouvenot M, Deschaseaux M L, Royez M, Mougin C, Cooksey R C, Michel-Briand Y, Adessi G L. Molecular hybridization versus isoelectric focusing to determine TEM-type β-lactamases in gram-negative bacteria. Antimicrob Agents Chemother. 1987;31:300–305. doi: 10.1128/aac.31.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kampf G, Weist K, Swidinsky S, Kegel M, Rüden H. Comparison of screening methods to identify methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1997;16:301–307. doi: 10.1007/BF01695635. [DOI] [PubMed] [Google Scholar]
- 142.Kataja J, Huovinen P, Skurnik M, Seppala H. Erythromycin resistance genes in group A streptococci in Finland. The Finnish study group for antimicrobial resistance. Antimicrob Agents Chemother. 1999;43:48–52. doi: 10.1128/aac.43.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kataja J, Seppala H, Skurnik M, Sarkkinen H, Huovinen P. Different erythromycin resistance mechanisms in group C and group G streptococci. Antimicrob Agents Chemother. 1998;42:1493–1494. doi: 10.1128/aac.42.6.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kaufhold A, Podbielski A, Horaud T, Ferrieri P. Identical genes confer high-level resistance to gentamicin upon Enterococcus faecalis, Enterococcus faecium, and Streptococcus agalactiae. Antimicrob Agents Chemother. 1992;36:1215–1218. doi: 10.1128/aac.36.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kearns A M, Seiders P R, Wheeler J, Freeman R, Steward M. Rapid detection of methicillin-resistant staphylococci by multiplex PCR. J Hosp Infect. 1999;43:33–37. doi: 10.1053/jhin.1999.0631. [DOI] [PubMed] [Google Scholar]
- 146.Khan M A, Potter C W, Sharrard R M. A reverse transcriptase-PCR based assay for in-vitro antibiotic susceptibility testing of Chlamydia pneumoniae. J Antimicrob Chemother. 1996;37:677–685. doi: 10.1093/jac/37.4.677. [DOI] [PubMed] [Google Scholar]
- 147.Khan S A, Nawaz M S, Khan A A, Cerniglia C E. Simultaneous detection of erythromycin-resistant methylase genes ermA and ermC from Staphylococcus spp. by multiplex PCR. Mol Cell Probes. 1999;13:381–387. doi: 10.1006/mcpr.1999.0265. [DOI] [PubMed] [Google Scholar]
- 148.Khow D Y, Davis G R, Whitfield K M, Chapelle H L, DiMichele L J, Gingeras T R. Transcription-based amplification system and detection of amplified human immunodeficiency virus type 1 with a bead-based sandwich hybridization format. Proc Natl Acad Sci USA. 1989;86:1173–1177. doi: 10.1073/pnas.86.4.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kiepiela P, Bishop K, Kormuth E, Roux L, York D F. Comparison of PCR-heteroduplex characterization by automated DNA sequencing and line probe assay for the detection of rifampicin resistance in Mycobacterium tuberculosis isolates from KwaZulu-Natal, South Africa. Microb Drug Resist. 1998;4:263–269. doi: 10.1089/mdr.1998.4.263. [DOI] [PubMed] [Google Scholar]
- 150.Kim B-J, Kim S-Y, Park B-H, Lyu M-A, Park I-K, Bai G-H, Kim S-J, Cha C-Y, Kook Y-H. Mutations in the rpoB gene of Mycobacterium tuberculosis that interfere with PCR-single-strand conformation polymorphism analysis for rifampin susceptibility testing. J Clin Microbiol. 1997;35:492–494. doi: 10.1128/jcm.35.2.492-494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kitagawa Y, Ueda M, Ando N, Endo M, Ishibiki K, Kobayashi Y, Arai T, Kitajima M. Rapid diagnosis of methicillin-resistant Staphylococcus aureus bacteremia by nested polymerase chain reaction. Ann Surg. 1996;224:665–671. doi: 10.1097/00000658-199611000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Klein N C, Cunha B A. Tetracyclines. Med Clin North Am. 1995;79:789–801. doi: 10.1016/s0025-7125(16)30039-6. [DOI] [PubMed] [Google Scholar]
- 153.Kocagöz S, Ünal S. The practical use of PCR for rapid detection of methicillin resistance among staphylococcal clinical isolates from Turkish hospitals. J Clin Microbiol. 1997;35:2188–2189. doi: 10.1128/jcm.35.8.2188-2189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kolbert C P, Arruda J, Varga-Delmore P, Zheng X, Lewis M, Kolberg J, Persing D H. Branched-DNA assay for the detection of mecA gene in oxacillin-resistant and oxacillin-sensitive staphylococci. J Clin Microbiol. 1999;36:2640–2644. doi: 10.1128/jcm.36.9.2640-2644.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kolbert C P, Connolly J E, Lee M J, Persing D H. Detection of the staphylococcal mecA gene by chemiluminescent DNA hybridization. J Clin Microbiol. 1995;33:2179–2182. doi: 10.1128/jcm.33.8.2179-2182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kramer F R, Lizardi P M. Replicatable RNA reporters. Nature. 1989;339:401–402. doi: 10.1038/339401a0. [DOI] [PubMed] [Google Scholar]
- 157.Krauss J, van der Linden M, Grebe T, Hakenbeck R. Penicillin-binding proteins 2x and 2b as primary PBP targets in Streptococcus pneumoniae. Microb Drug Resist. 1996;2:183–186. doi: 10.1089/mdr.1996.2.183. [DOI] [PubMed] [Google Scholar]
- 158.Kricka L J. Nucleic acid detection technologies—labels, strategies, and formats. Clin Chem. 1999;45:453–458. [PubMed] [Google Scholar]
- 159.Kwow S, Kellogg D E, McKinney N, Spasic D, Goda L, Levenson C, Sninsky J J. Effects of primer-template mismatches on the polymerase chain reaction: human deficiency virus type 1 model studies. Nucleic Acids Res. 1990;18:999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lacroix J M, Walker C B. Detection and incidence of the tetracycline resistance determinant tetM in the microflora associated with adult periodonitis. J Periodontol. 1995;66:102–108. doi: 10.1902/jop.1995.66.2.102. [DOI] [PubMed] [Google Scholar]
- 161.Lacroix J M, Walker C B. Detection and prevalence of the tetracycline-resistance determinant tetQ in the microbiota associated with adult periodonitis. Oral Microbiol Immunol. 1996;11:282–288. doi: 10.1111/j.1399-302x.1996.tb00182.x. [DOI] [PubMed] [Google Scholar]
- 162.Laible G, Spratt G B, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–20002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 163.Latini L, Ronchetti M P, Merolla R, Guglielmi F, Bajaksouzian S, Villa M P, Jacobs M R, Ronchetti R. Prevalence of mefE, erm and tet(M) genes in Streptococcus pneumoniae strains from Central Italy. Int J Antimicrob Agents. 1999;13:29–33. doi: 10.1016/s0924-8579(99)00097-7. [DOI] [PubMed] [Google Scholar]
- 164.LeBlanc D J, Inamine J M, Lee L N. Broad geographical distribution of homologus erythromycin, kanamycin, and streptomycin resistance determinants among group D streptococci of human and animal origin. Antimicrob Agents Chemother. 1986;29:549–555. doi: 10.1128/aac.29.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Leclerq R, Courvalin P. Resistance to glycopeptides in enterococci. Clin Infect Dis. 1997;24:545–556. doi: 10.1093/clind/24.4.545. [DOI] [PubMed] [Google Scholar]
- 167.Leclerq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 168.Leclercq R, Giannattasio R B, Jin H J, Weisblum B. Bacterial resistance to macrolide, lincosamide and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee C, Langlois B E, Dawson K A. Detection of tetracycline resistance determinants in pig isolates from three herds with different histories of antimicrobial agent exposure. Appl Environ Microbiol. 1993;59:1467–1472. doi: 10.1128/aem.59.5.1467-1472.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lee C Y, Tai C L, Lin S C, Chen Y T. Occurrence of plasmids and tetracycline resistance among Campylobacter jejuni and Campylobacter coli isolated from whole market chickens and clinical samples. International J Food Microbiol. 1994;24:161–170. doi: 10.1016/0168-1605(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 171.Levée G, Glaziou P, Gicquel B, Chanteau S. Follow-up of tuberculosis patients undergoing standard anti-tuberculosis chemotherapy by using polymerase chain reaction. Res Microbiol. 1994;145:5–8. doi: 10.1016/0923-2508(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 172.Levy S B. Tetracycline resistance determinants are widespread. ASM News. 1988;54:418–421. [Google Scholar]
- 173.Levy S B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Levy S B. The antibiotic paradox: how miracle drugs are destroying the miracle. New York, NY: Plenum Press; 1992. [Google Scholar]
- 175.Levy S B, McMurry L M, Barbosa, T. M. T M, Burdett V, Courvalin P, Hillen W, Roberts M C, Rood J I, Taylor D E. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Levy S B, McMurry L M, Burdett V, Courvalin P, Hillen W, Roberts M C, Taylor D E. Nomenclature for tetracycline resistance determinants. Antimicrob Agents Chmeother. 1989;33:1373–1374. doi: 10.1128/aac.33.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lewis K, Hooper D C, Ouelette M. Multidrug resistance pumps provide broad defense. ASM News. 1997;63:605–610. [Google Scholar]
- 178.Ligozzi M, Rossolini G M, Tonin E A, Fontana R. Nonradioactive DNA probe for detection of gene for methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1991;35:575–578. doi: 10.1128/aac.35.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lina G, Quaglia A, Reverdy M E, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–1066. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Livermore D M. Clinical significance of beta-lactamase induction and stable derepression in Gram-negative rods. Eur J Clin Microbiol. 1987;6:439–445. doi: 10.1007/BF02013107. [DOI] [PubMed] [Google Scholar]
- 181.Lodder G, Schwarz S, Gregory P, Dyke K. Tandem duplication in ermC translational attenuator of the macrolide-lincosamide-streptogramin B resistance plasmid pSES6 from Staphylococcus equorum. Antimicrob Agents Chemother. 1996;40:215–217. doi: 10.1128/aac.40.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lodder G, Werckenthin C, Schwarz S, Dyke K. Molecular analysis of naturally occurring ermC-encoding plasmids in staphylococci isolated from animals with and without previous contact with macrolide/lincosamide antibiotics. FEMS Immunol Med Microbiol. 1997;18:7–15. doi: 10.1111/j.1574-695X.1997.tb01022.x. [DOI] [PubMed] [Google Scholar]
- 183.Lui P Y F, Hall L C M, Livermore D M. Survey of the prevalence of beta-lactamases among 1000 Gram-negative bacilli isolated consecutively at the Royal London Hospital. J Antimicrob Chemother. 1992;30:429–447. doi: 10.1093/jac/30.4.429. [DOI] [PubMed] [Google Scholar]
- 184.Luna V A, Coates P, Eady E A, Cove J H, Nguyen T T, Roberts M C. A variety of gram-positive bacteria carry mobile mef genes. J Antimicrob Chemother. 1999;44:19–25. doi: 10.1093/jac/44.1.19. [DOI] [PubMed] [Google Scholar]
- 185.Mabilat C, Courvalin P. Development of “oligotyping” for characterization and molecular epidemiology of TEM β-lactamases in members of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1991;34:2210–2216. doi: 10.1128/aac.34.11.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Maeda S, Yoshida H, Ogura K, Kanai F, Shiratori Y, Omata M. Helicobacter pylori specific nested PCR assay for the detection of 23S rRNA mutation associated with clarithromycin resistance. Gut. 1998;43:317–321. doi: 10.1136/gut.43.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Marais A, Monteiro L, Occhialini A, Pina M, Lamouliatte H, Megraud F. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut. 1999;44:463–467. doi: 10.1136/gut.44.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Marchese A, Ramirez M, Schito G C, Tomasz A. Molecular epidemiology of penicillin-resistant Streptococcus pneumoniae isolates recovered in Italy from 1993 to 1996. J Clin Microbiol. 1998;36:2944–2949. doi: 10.1128/jcm.36.10.2944-2949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Marchese A, Tonoli E, Debbia E A, Schito G C. Macrolide resistance mechanisms and expression of phenotypes among Streptococcus pneumoniae circulating in Italy. J Antimicrob Chemother. 1999;44:461–464. doi: 10.1093/jac/44.4.461. [DOI] [PubMed] [Google Scholar]
- 190.Marshall C G, Lessard L A, Park I, Wright G D. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob Agents Chemother. 1998;42:2215–2220. doi: 10.1128/aac.42.9.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Martinez-Freijo P A, Fluit C, Schmitz F-J, Grek V S C, Verhoef J, Jones M E. Class I integrons in gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998;42:689–696. doi: 10.1093/jac/42.6.689. [DOI] [PubMed] [Google Scholar]
- 192.Marttila H J, Soini H, Vyshnevskiy B I, Otten T F, Vasilyef A V, Huovinen P, Viljanen M K. Rapid detection of rifampin-resistant Mycobacterium tuberculosis by sequencing and line probe assay. Scand J Infect Dis. 1998;30:129–132. doi: 10.1080/003655498750003492. [DOI] [PubMed] [Google Scholar]
- 193.Marttila H J, Soini H, Vyshnevskaya E, Vyshnevskiy B I, Otten T F, Vasilyef A V, Viljanen M K. Line probe assay in the rapid detection of rifampin-resistant Mycobacterium tuberculosis directly from clinical specimens. Scand J Infect Dis. 1999;31:269–273. doi: 10.1080/00365549950163563. [DOI] [PubMed] [Google Scholar]
- 194.Mastrantonio P, Cardines R, Spigaglia P. Oligonucleotide probes for the detection of cephalosporinases among Bacteroides strains. Antimicrob Agents Chemother. 1996;40:1014–1016. doi: 10.1128/aac.40.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Matsumura M, Karakura T, Imanaka T, Aiba S. Enzymatic and nucleotide sequence studies of a kanamycin inactivating enzyme encoded by a plasmid from thermophilic bacilli in comparison with that encoded by plasmid pUB110. J Bacteriol. 1984;160:413–420. doi: 10.1128/jb.160.1.413-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Meier A, Kirschner P, Bange F-C, Vogel U, Böttger E C. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob Agents Chemother. 1994;38:228–223. doi: 10.1128/aac.38.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Miele A, Bandera M, Goldstein B. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob Agents Chemother. 1995;39:1772–1778. doi: 10.1128/aac.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Milatovic D, Braveny I. Development of resistance during antibiotic therapy. Eur J Clin Microbiol. 1987;6:234–244. doi: 10.1007/BF02017607. [DOI] [PubMed] [Google Scholar]
- 199.Milewski W M, Boyle-Vavra S, Moreira B, Ebert C C, Daum R S. Overproduction of a 37-kilodalton cytoplasmic protein homologous to NAD+-linked d-lactate dehydrogenase associated with vancomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:166–172. doi: 10.1128/aac.40.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Modrusan Z, Marlowe C, Wheeler D, Pireyedi M, Bryan R N. Detection of vancomycin resistant genes vanA and vanB by cycling probe technology. Mol Cell Probes. 1999;13:223–331. doi: 10.1006/mcpr.1999.0242. [DOI] [PubMed] [Google Scholar]
- 201.Moreira B, Boyle-Vavra S, deJonge B L, Daum R S. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:1788–1793. doi: 10.1128/aac.41.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 203.Munakata N, Deguchi T, Kawamura T, Yasuda M, Kimura M, Okano Y, Hayashi K. Molecular charaterization of thirteen gyrA mutations conferring nalidixic acid resistance in Bacillus subtilis. Mol Gen Genet. 1994;244:97–103. doi: 10.1007/BF00280192. [DOI] [PubMed] [Google Scholar]
- 204.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.M'Zali F-H, Gascoyne-Binzi D M, Heritage J, Hawkey P M. Detection of mutations conferring extended-spectrum activity on SHV β-lactamases using polymerase chain reaction single strand conformational polymorphism (PCR-SSCP) J Antimicrob Chemother. 1996;37:797–802. doi: 10.1093/jac/37.4.797. [DOI] [PubMed] [Google Scholar]
- 206.M'Zali F-H, Heritage J, Gascoyne-Binzi D M, Snelling A M, Hawkey P M. PCR single strand conformational polymorphism can be used to detect the gene encoding SHV-7 extended-spectrum β-lactamase and to identify different SHV genes within the same strain. J Antimicrob Chemother. 1998;41:123–125. doi: 10.1093/jac/41.1.123. [DOI] [PubMed] [Google Scholar]
- 207.Nachamkin I, Kang C, Weinstein M P. Detection of resistance to isoniazid, rifampin, and streptomycin in clinical isolates of Mycobacterium tuberculosis by molecular methods. Clin Infect Dis. 1997;24:894–900. doi: 10.1093/clinids/24.5.894. [DOI] [PubMed] [Google Scholar]
- 208.Nair J, Rouse D A, Bai G-H, Morris S L. The rspL gene and streptomycin resistance in single and multiple drug-resistant strains of Mycobacterium tuberculosis. Mol Microbiol. 1993;10:521–517. doi: 10.1111/j.1365-2958.1993.tb00924.x. [DOI] [PubMed] [Google Scholar]
- 209.Nash K A, Gaytan A, Inderlied C B. Detection of rifampin resistance in Mycobacterium tuberculosis by use of a rapid, simple, and specific RNA/RNA mismatch assay. J Infect Dis. 1997;176:533–536. doi: 10.1086/517283. [DOI] [PubMed] [Google Scholar]
- 210.Nash K A, Inderlied C B. Rapid detection of mutations associated with macrolide resistance in Mycobacterium avium complex. Antimicrob Agents Chemother. 1988;40:1748–1750. doi: 10.1128/aac.40.7.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Supplement Tables. Wayne, Pa: NCCLS; 1999. [Google Scholar]
- 211a.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A5. 5th ed. Wayne, Pa: NCCLS; 2000. [Google Scholar]
- 212.Nawaz M S, Khan A A, Cerniglia C E. Detection of erythromycin resistant methylase gene by polymerase chain reaction. Mol Cell Probes. 1997;11:317–322. doi: 10.1006/mcpr.1997.0121. [DOI] [PubMed] [Google Scholar]
- 213.Newton C R, Graham A, Heptinstall L E, Powell S J, Summers C, Kalsheker N, Smith J C, Markham A F. Analysis of any point mutation in DNA. The amplification refractory mutation system. Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Neyfakh A A. The multidrug efflux transporter of Bacillus subtilis is a structural and functional homolog of the Staphylococcus NorA protein. Antimicrob Agents Chemother. 1992;36:484–485. doi: 10.1128/aac.36.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Neyfakh A A, Borsch C M, Kaatz G W. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob Agents Chemother. 1993;37:128–129. doi: 10.1128/aac.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ng L K, Stiles M E, Taylor D E. DNA probes for identification of tetracycline resistance genes in Campylobacter species isolated from swine and cattle. Antimicrob Agents Chemother. 1987;31:1669–1674. doi: 10.1128/aac.31.11.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Nguyen C, Rocha D, Granjeaud S, Baldit M, Bernard K, Naquet P, Jordan B R. Differential gene expression in the murine thymus assayed by quantitative hybridization of arrayed cDNA clones. Genomics. 1995;29:207–216. doi: 10.1006/geno.1995.1233. [DOI] [PubMed] [Google Scholar]
- 218.Nicola F G, McDougal L K, Biddle J W, Tenover F C. Characterization of erythromycin-resistant isolates of Staphylococcus aureus recovered in the United States from 1958 through 1969. Antimicrob Agents Chemother. 1998;42:3024–3027. doi: 10.1128/aac.42.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nishijima T, Saito Y, Aoki A, Toriya M, Toyonaga Y, Fujii R. Distribution of mefE and ermB genes in macrolide-resistant strains of Streptococcus pneumoniae and their variable susceptibility to various antibiotics. J Antimicrob Chemother. 1999;43:637–643. doi: 10.1093/jac/43.5.637. [DOI] [PubMed] [Google Scholar]
- 220.Noble W C, Virani Z, Cree R G A. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;93:195–98. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 221.Nolte F S. Branched DNA signal amplification for direct quantitation of nucleic acid sequences in clinical specimens. Adv Clin Chem. 1998;33:201–235. doi: 10.1016/s0065-2423(08)60209-7. [DOI] [PubMed] [Google Scholar]
- 222.Noordhoek G T, van Embden J D A, Kolk A H J. Reliability of nucleic acid amplification for detection of Mycobacterium tuberculosis: an international collaborative quality control study among 30 laboratories. J Clin Microbiol. 1996;34:2522–2525. doi: 10.1128/jcm.34.10.2522-2525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Noppe-Leclercq I, Wallet F, Haentjens S, Courcol R, Simonet M. PCR detection of aminoglycoside resistance genes: a rapid molecular typing method for Acinetobacter baumannii. Res Microbiol. 1999;150:317–322. doi: 10.1016/s0923-2508(99)80057-6. [DOI] [PubMed] [Google Scholar]
- 224.Nuesch-Inderbinen M T, Hachler H, Kayser F H. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the Etest. Eur J Clin Microbiol Infect Dis. 1996;15:398–402. doi: 10.1007/BF01690097. [DOI] [PubMed] [Google Scholar]
- 225.Occhialini A, Urdaci M, Doucet-Populaire F, Bebear C M, Lamouliatte H, Megraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Olsvik B, Olsen I, Tenover F C. Detection of tet(M) and tet(O) using the polymerase chain reaction in bacteria isolated from patients with periodontal disease. Oral Microbiol Immunol. 1995;10:87–92. doi: 10.1111/j.1399-302x.1995.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 227.O'Neil A M, Gillespie S H, Whiting G C. Detection of penicillin susceptibility in Streptococcus pneumoniae by pbp2b PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 1999;37:157–160. doi: 10.1128/jcm.37.1.157-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Orita M, Iwahana H, Kanazawa K, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by electrophoresis as single-strand conformation polymorphisms Proc. Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Orita M, Suzuki Y, Sekiya T, Hayashi K. Rapid and sensitive detection of point-mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989;5:874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 230.Oubdesselam S, Hooper D C, Tankovic J, Soussy C J. Detection of gyrA and gyrB mutations in quinolone-resistant clinical isolates of Escherichia coli by single-strand conformational polymorphism analysis and determination of levels of resistance conferred by two different single gyrA mutations. Antimicrob Agents Chemother. 1995;39:1667–1670. doi: 10.1128/aac.39.8.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ouellette M, Paul G C, Philippon A M, Roy P H. Oligonucleotide probes (TEM-1, OXA-1) versus isoelectric focusing in β-lactamase characterization of 114 resistant strains. Antimicrob Agents Chemother. 1988;32:397–399. doi: 10.1128/aac.32.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ouellette M, Rossi J J, Bazin R, Roy P H. Oligonucleotide probes for the detection of TEM-1 and TEM-2 β-lactamase genes and their transposons. Can J Microbiol. 1986;33:205–211. doi: 10.1139/m87-035. [DOI] [PubMed] [Google Scholar]
- 233.Ouellette M, Roy P H. Analysis by using DNA probes of the OXA-1 β-lactamase gene and its transposon. Antimicrob Agents Chemother. 1986;30:46–51. doi: 10.1128/aac.30.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ounissi H, Derlot E, Carlier C, Courvalin P. Gene homogeneity for aminoglycoside-modifying enzymes in gram-positive cocci. Antimicrob Agents Chemother. 1990;34:2164–2168. doi: 10.1128/aac.34.11.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ozeki S, Deguchi T, Yasuda M, Nakano M, Kawamura T, Nishino Y, Kawada Y. Development of a rapid assay for detecting gyrA mutations in Escherichia coli and determination of incidence of gyrA mutations in clinical strains isolated from patients with complicated urinary tract infections. J Clin Microbiol. 1997;35:2315–2319. doi: 10.1128/jcm.35.9.2315-2319.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Pan X S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Pan X S, Fisher L M. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Pang Y, Bosch T, Roberts M C. Single polymerase chain reaction for the detection of the tetracycline-resistant determinants tetK and tetL. Mol Cell Probes. 1994;8:417–422. doi: 10.1006/mcpr.1994.1059. [DOI] [PubMed] [Google Scholar]
- 239.Pang Y, Brown B A, Steingrube V A, Wallace R J, Jr, Roberts M C. Tetracycline resistance determinants in Mycobacterium and Streptomyces species. Antimicrob Agents Chemother. 1994;38:1408–1412. doi: 10.1128/aac.38.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Patel R, Uhl J R, Kohner P, Hopkins M K, Cockerill F R., III Multiplex PCR detection of vanA, vanB, vanC-1, and vanC2/3 genes in enterococci. J Clin Microbiol. 1997;35:703–707. doi: 10.1128/jcm.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Paulsen I T, Firth N, Skurray R A. Resistance to antimicrobial agents other than β-lactams. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. London, United Kingdom: Churchill Livingstone, Inc.; 1997. pp. 175–212. [Google Scholar]
- 242.Pepper K, le Bouguénec C, de Cespédès G, Horaud T. Dispersal of a plasmid-borne chloramphenicol resistance gene in streptococcal and enterococcal plasmids. Plasmid. 1986;16:195–203. doi: 10.1016/0147-619x(86)90057-0. [DOI] [PubMed] [Google Scholar]
- 243.Pepper K, de Cespédès G, Horaud T. Heterogeneity of chromosomal genes encoding chloramphenicol resistance in streptococci. Plasmid. 1988;19:71–74. doi: 10.1016/0147-619x(88)90065-0. [DOI] [PubMed] [Google Scholar]
- 244.Perez-Trallero E, Urbieta M, Montes M, Ayestaran I, Marimon J M. Eur. J. Clin. Microbiol. Infect. Dis. 17:25–31. 1998. Emergence of Streptococcus pyogenes strains resistant to erythromycin in Gipuzkoa, Spain. [DOI] [PubMed] [Google Scholar]
- 245.Perichon B, Reynolds P, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997;41:2016–2018. doi: 10.1128/aac.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Pestova E, Beyer R, Cianciotto N P, Noskin G A, Peterson L R. Contribution of topoisomerase IV and DNA gyrase mutations in Streptococcus pneumoniae to resistance to novel fluoroquinolones. Antimicrob Agents Chemother. 1999;43:2000–2004. doi: 10.1128/aac.43.8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Petrich A K, Luinstra K E, Groves D, Chernesky M A, Mahony J B. Direct detection of vanA and vanB genes in clinical specimens for rapid identification of vancomycin-resistant enterococci (VRE) using multiplex PCR. Mol Cell Probes. 1999;13:275–281. doi: 10.1006/mcpr.1999.0250. [DOI] [PubMed] [Google Scholar]
- 248.Piatek A S, Tyagi S, Pol A C, Telenti A, Miller L P, Russell Kramer F, Alland D. Molecular beacon sequence analysis for detection drug resistance in Mycobacterium tuberculosis. Nat Biotechnol. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 249.Pina M, Occhialini A, Monteiro L, Doermann H P, Megraud F. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol. 1998;36:3285–3290. doi: 10.1128/jcm.36.11.3285-3290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Plikaytis B B, Marden J L, Crawford J T, Woodley C L, Butler W R, Shinnick T M. Multiplex PCR assay specific for the multidrug-resistant strain W of Mycobacterium tuberculosis. J Clin Microbiol. 1994;32:1542–1546. doi: 10.1128/jcm.32.6.1542-1546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ploy M-C, Giamarellou H, Bourlioux P, Courvalin P, Lambert T. Detection of aac(6′)-I genes in amikacin-resistant Acinetobacter spp. by PCR. Antimicrob Agents Chemother. 1994;38:2925–2928. doi: 10.1128/aac.38.12.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Portillo A, Lantero M, Gastanares M J, Ruiz-Larrea F, Torres C. Macrolide resistence phenotypes and mechanisms of resistance in Streptococcus pyogenes in La Rioja, Spain. Int J Antimicrob Agents. 1999;13:137–140. doi: 10.1016/s0924-8579(99)00104-1. [DOI] [PubMed] [Google Scholar]
- 253.Poulsen R L, Pallesen L V, Frimodt-Moller N, Espersen F. Detection of clinical vancomycin-resistant enterococci in Denmark by multiplex PCR and sandwich hybridization. APMIS. 1999;107:404–412. doi: 10.1111/j.1699-0463.1999.tb01573.x. [DOI] [PubMed] [Google Scholar]
- 254.Poupard J A. Update on mupirocin resistance. J Chemother. 1995;7(Suppl.3):71–74. [PubMed] [Google Scholar]
- 255.Poyard-Salmeron C, Trieu-Cuot P, Carlier C, MacGowan A, McLauchlin J, Courvalin P. Genetic basis of tetracycline resistance in clinical isolates of Listeria monocytogenes. Antimicrob Agents Chemother. 1992;36:463–466. doi: 10.1128/aac.36.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Predari S C, Ligozzi M, Fontana R. Genotypic identification of methicillin-resistant coagulase-negative staphylococci by polymerase chain reaction. Antimicrob Agents Chemother. 1991;35:2568–2573. doi: 10.1128/aac.35.12.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Quintiliani R, Courvalin P. Mechanisms of resistance to antimicrobial agents. In: Murray P R, Baron E J, Pfaller M A, Tenover F R, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 1308–1326. [Google Scholar]
- 258.Qumsieh M J, Young H-K. Cloning of the type Ib trimethoprim-resistant dihydrofolate reductase gene and preparation of a specific biotynilated probe. J Antimicrob Chemother. 1991;27:707–712. doi: 10.1093/jac/27.6.707. [DOI] [PubMed] [Google Scholar]
- 259.Rahman M, Noble W C, Dyke K G. Probes for the study of mupirocin resistance in staphylococci. J Med Microbiol. 1993;39:446–449. doi: 10.1099/00222615-39-6-446. [DOI] [PubMed] [Google Scholar]
- 260.Reed R P, Sinickas V G, Lewis C, Byron K A. A comparison of polymerase chain reaction and phenotyping for rapid speciation of enterococci and detection of vancomycin resistance. Pathology. 1999;31:127–132. doi: 10.1080/003130299105313. [DOI] [PubMed] [Google Scholar]
- 261.Reynolds P E. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1989;8:943–950. doi: 10.1007/BF01967563. [DOI] [PubMed] [Google Scholar]
- 262.Roberts M C. Tetracycline resistance in Peptostreptococcus species. Antimicrob Agents Chemother. 1991;35:1682–1684. doi: 10.1128/aac.35.8.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Roberts M C. Epidemiology of tetracycline resistance determinants. Trends Microbiol. 1994;2:353–357. doi: 10.1016/0966-842x(94)90610-6. [DOI] [PubMed] [Google Scholar]
- 264.Roberts M C. Distribution of tetracycline and macrolides-lincosamides-streptogramin B (MLS) genes in anaerobic bacteria. Clin Infect Dis. 1995;20:367–369. doi: 10.1093/clinids/20.supplement_2.s367. [DOI] [PubMed] [Google Scholar]
- 265.Roberts M C. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 266.Roberts M C, Chung W O, Roe D E. Characterization of tetracycline and erythromycin resistance determinants in Treponema denticola. Antimicrob Agents Chemother. 1996;40:1690–1694. doi: 10.1128/aac.40.7.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Roberts M C, Chung W O, Roe D, Xia M, Marquez C, Borthagaray G, Whittington W L, Holmes K K. Erythromycin-resistant Neisseria gonorrhoeae and oral commensal Neisseria spp. carry known rRNA methylase genes. Antimicrob Agents Chemother. 1999;43:1367–1372. doi: 10.1128/aac.43.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Roberts M C, Hillier S L. Genetic basis of tetracycline resistance in urogenital bacteria. Antimicrob Agents Chemother. 1990;34:261–264. doi: 10.1128/aac.34.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Roberts M C, McFarland M V, Mullany P, Mulligan M E. Characterization of the genetic basis of antibiotic resistance in Clostridium difficile. J Antimicrob Chemother. 1994;33:419–429. doi: 10.1093/jac/33.3.419. [DOI] [PubMed] [Google Scholar]
- 270.Roberts M C, Moncla B J, Hillier S L. Characterization of unusual tetracycline-resistant gram-positive bacteria. Antimicrob Agents Chemother. 1991;35:2655–2657. doi: 10.1128/aac.35.12.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Roberts M C, Pang Y, Riley D E, Hillier S L, Berger R C, Krieger J N. Detection of tetM and tetO tetracycline resistance genes by polymerase chain reaction. Mol Cell Probes. 1993;5:387–93. doi: 10.1006/mcpr.1993.1057. [DOI] [PubMed] [Google Scholar]
- 272.Roberts M C, Sutcliffe J, Courvalin P, Jensen L B, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823–2830. doi: 10.1128/aac.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Roger M, Faucher M C, Forest P, St. Antoine P, Coutlee F. Evaluation of a vanA-specific PCR assay for detection of vancomycin-resistant Enterococcus faecium during a hospital outbreak. J Clin Microbiol. 1999;37:3348–3349. doi: 10.1128/jcm.37.10.3348-3349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Rood J I, Jefferson S, Bannam T L, Wilkie J M, Mullany P, Wren B W. Hybridization analysis of three chloramphenicol resistance determinants from Clostridium perfringens and Clostridium difficile. Antimicrob Agents Chemother. 1989;33:1569–1574. doi: 10.1128/aac.33.9.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Rose S D. Application of a novel microarraying system in genomics research and drug discovery. J Assoc Lab Autom. 1998;3:53–56. [Google Scholar]
- 276.Ross J I, Eady E A, Cove J H, Baumberg S. Identification of a chromosomally encoded ABC-transporter system with which the staphylococcal erythromycin exporter MsrA may interact. Gene. 1995;153:93–98. doi: 10.1016/0378-1119(94)00833-e. [DOI] [PubMed] [Google Scholar]
- 277.Ross J I, Eady E A, Cove J H, Cunliffe W J, Baumberg S. Inducible erythromycin-resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol. 1990;4:1207–1214. doi: 10.1111/j.1365-2958.1990.tb00696.x. [DOI] [PubMed] [Google Scholar]
- 278.Rossau R, Traore H, De Beenhouwer H, Mijs W, Jannes G, De Rijk P, Portaels F. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob Agents Chemother. 1997;41:2093–2098. doi: 10.1128/aac.41.10.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Sahm D F, Free L, Smith C, Eveland M, Mundy L M. Rapid characterization schemes for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 1997;35:2026–2030. doi: 10.1128/jcm.35.8.2026-2030.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Saiki R K, Gelfand D H, Stoffel S, scharf S J, Higuchi R, Horn G T, Mullis K B, Ehrlich H A. Primer-directed enzymatic amplification with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 281.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Ehrlich H A. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for the diagnosis of sickle-cell anemia. Science. 1988;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 282.Sakamoto H, Zilhao R, Bismuth R, Courvalin P, Guesdon J L. Non-radioactive gene probes for the detection of tetracycline and/or minocycline resistance in staphylococci. Mol Cell Probes. 1988;2:321–330. doi: 10.1016/0890-8508(88)90015-1. [DOI] [PubMed] [Google Scholar]
- 283.Salisbury S M, Sabatini L M, Spiegel C A. Identification of methicillin-resistant staphylococci by multiplex polymerase chain reaction assay. Am J Clin Pathol. 1997;107:368–373. doi: 10.1093/ajcp/107.3.368. [DOI] [PubMed] [Google Scholar]
- 284.Sallen B, Rojoharison A, Desvarenne S, Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of Enterobacteriaceae. Microb Drug Resist. 1995;1:195–202. doi: 10.1089/mdr.1995.1.195. [DOI] [PubMed] [Google Scholar]
- 285.Sanchez-Pescador R, Stempien M S, Urdea M S. Rapid chemiluminescent nucleic acid assays for detection of TEM-1 β-lactamase-mediated penicillin resistance in Neisseria gonorrhoea and other bacteria. J Clin Microbiol. 1988;26:1934–1938. doi: 10.1128/jcm.26.10.1934-1938.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Sanchez-Pescador R, Stempien M S, Urdea M S. Rapid chemiluminescent nucleic acid assays for detection of TEM-1 β-lactamase-mediated penicillin resistance in Neisseria gonorrhoeae and other bacteria. J Clin Microbiol. 1988;26:1934–1938. doi: 10.1128/jcm.26.10.1934-1938.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Satake S, Clark N, Rimland D, Nolte F S, Tenover F C. Detection of vancomycin-resistant enterococci in fecal samples by PCR. J Clin Microbiol. 1997;35:2325–2330. doi: 10.1128/jcm.35.9.2325-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Scarpellini P, Braglia S, Brambilla A M, Dalessandro M, Cichero P, Gori A, Lazzarin A. Detection of rifampin resistance by single-strand conformation polymorphism analysis of cerebrospinal fluid of patients with tuberculosis of the central nervous system. J Clin Microbiol. 1997;35:2802–2806. doi: 10.1128/jcm.35.11.2802-2806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Scarpellini P, Braglia S, Carrera P, Cedri M, Chicero P, Colombo A, Crucianelli R, Gori A, Ferrari M, Lazzarin A. Detection of rifampin resistance in Mycobacterium tuberculosis by double gradient-denaturing gradient gel electrophoresis. Antimicrob Agents Chemother. 1999;43:2550–2554. doi: 10.1128/aac.43.10.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Schmitz F-J, Fluit A C. Mechanisms of resistance. In: Armstrong D, Cohen S, editors. Infectious diseases. C. V. London, United Kingdom: Mosby, Ltd.; 1999. pp. 7.2.1–7.2.14. [Google Scholar]
- 292.Schmitz F-J, Fluit A C, Brisse S, Verhoef J, Köhrer K, Milatovic D. Molecular epidemiology of quinolone resistance and comparative in vitro activities of new quinolones against European Staphylococcus aureus isolates. FEMS Immunol Med Microbiol. 1999;24:281–287. doi: 10.1111/j.1574-695X.1999.tb01400.x. [DOI] [PubMed] [Google Scholar]
- 293.Schmitz F-J, Fluit A C, Gondolf M, Beyrau R, Lindenlauf E, Verhoef J, Heinz H-P, Jones M E. The prevalence of aminoglycoside-resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J Antimicrob Chemother. 1999;43:253–259. [PubMed] [Google Scholar]
- 294.Schmitz F-J, Jones M E, Hofmann B, Hansen B, Scheuring S, Lückefahr M, Fluit A C, Verhoef J, Hadding U, Heinz H-P, Köhrer K. Characterization of grlA, grlB, gyrA and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus in relation to minimal inhibitory concentrations of ciprofloxacin. Antimicrob Agents Chemother. 1998;42:1249–1252. doi: 10.1128/aac.42.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Schmitz F-J, Lindenlauf E, Hofmann B, Fluit A C, Verhoef J, Heinz H-P, Jones M E. The prevalence of low and high-level mupirocin-resistance in staphylococci from 19 European hospitals. J Antimicrob Chemother. 1998;42:489–495. doi: 10.1093/jac/42.4.489. [DOI] [PubMed] [Google Scholar]
- 296.Schmitz F-J, Martinez-Freijo P, Theis S, Fluit A C, Verhoef J, Heinz H-P, Jones M E. Prevalence of class 1 integrons and association with decreased antibiotic susceptibility in German gram-negative blood culture isolates. Clin Microbiol Infect. 1995;5:436–438. doi: 10.1111/j.1469-0691.1999.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 297.Schmitz F-J, Sadurski R, Kray A, Boss M, Geisel R, Köhrer K, Verhoef J, Fluit A C. Prevalence of macrolide resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J Antimicrob Chemother J Antimicrob Chemother. 2000;45:921–923. doi: 10.1093/jac/45.6.891. [DOI] [PubMed] [Google Scholar]
- 298.Schmitz F-J, Verhoef J, Fluit A C the SENTRY Participants Group. Comparative activity of 27 antimicrobial compounds against 698 Streptococcus pneumoniae isolates originating from 20 European university hospitals. Eur J Clin Microbiol Infect Dis. 1999;18:450–453. doi: 10.1007/s100960050318. [DOI] [PubMed] [Google Scholar]
- 299.Schmitz F-J, Verhoef J, Fluit A C the SENTRY Participants Group. Prevalence of resistance to MLS antibiotics in 20 European university hospitals participating in the European SENTRY surveillance program J. Antimicrob Chemother. 1999;43:783–792. doi: 10.1093/jac/43.6.783. [DOI] [PubMed] [Google Scholar]
- 300.Schnappinger D, Hillen W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996;165:359–69. doi: 10.1007/s002030050339. [DOI] [PubMed] [Google Scholar]
- 301.Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 1999;37:2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Schwalbe R S, Stapleton J T, Gilgan P H. Emerergence of vancomycin resistance in coagulase-negative staphylococci. N Engl J Med. 1987;316:927–931. doi: 10.1056/NEJM198704093161507. [DOI] [PubMed] [Google Scholar]
- 303.Schwarz S, Lange C, Werckenthin C. Molecular analysis of the macrolide-lincosamide resistance gene region of a novel plasmid from Staphylococcus hyicus. J Med Microbiol. 1998;47:63–70. doi: 10.1099/00222615-47-1-63. [DOI] [PubMed] [Google Scholar]
- 304.Schwarz S, Noble W C. Tetracycline resistance genes in staphylococci from the skin of pigs. J Appl Bacteriol. 1994;76:320–326. doi: 10.1111/j.1365-2672.1994.tb01635.x. [DOI] [PubMed] [Google Scholar]
- 305.Scriver S R, Walmsley S L, Kau C L, Hoban D J, Brunton J, McGeer A, Moore T C, Witwicki E, Low D E Canadian Haemophilus Study Group. Determination of antimicrobial susceptibilities of Canadian isolates of Haemophilus influenzae and characterization of their β-lactamases. Antimicrob Agents Chemother. 1994;38:1678–1680. doi: 10.1128/aac.38.7.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Senda K, Areca Y, Akimbo S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. PCR detection of metallo-β-lactamase genes (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J Clin Microbiol. 1996;34:2909–2913. doi: 10.1128/jcm.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Seppala H, Skrunik M, Soini H, Roberts M C, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–262. doi: 10.1128/aac.42.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Seward R J, Lambert T, Towner K J. Molecular epidemiology of aminoglycoside resistance in Acinetobacter spp. J Med Microbiol. 1998;47:455–462. doi: 10.1099/00222615-47-5-455. [DOI] [PubMed] [Google Scholar]
- 309.Shaw K J, Rather P N, Hare R S, Miller G H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Shena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 311.Shortridge V D, Doern G V, Brueggemann A B, Beyer J M, Flamm R K. Prevalence of macrolide resistance mechanisms in Streptococcus pneumoniae isolates from a multicenter antibiotic resistance surveillance study conducted in the united states in 1994–1995. Clin Infect Dis. 1999;29:1186–1189. doi: 10.1086/313452. [DOI] [PubMed] [Google Scholar]
- 312.Shortridge V D, Flamm R K, Ramer N, Beyer J, Tanaka S K. Novel mechanism of macrolide resistance in Streptococcus pneumoniae. Diagn Microbiol Infect Dis. 1996;26:73–78. doi: 10.1016/s0732-8893(96)00183-6. [DOI] [PubMed] [Google Scholar]
- 313.Sieradzki K, Roberts R B, Haber S W, Tomasz A. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection N. Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 314.Sieradzki K, Tomasz A. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J Bacteriol. 1997;179:2557–2566. doi: 10.1128/jb.179.8.2557-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Sieradzki K, Villari P, Tomasz A. Decreased susceptibilities to teicoplanin and vancomycin among coagulase-negative methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1998;42:100–107. doi: 10.1128/aac.42.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Skurray R A, Rouch D A, Lyon B R, Gillespie M T, Tennent J M, Byrne M E, Messerotti L J, May J W. Multiresistant Staphyloccus aureus: genetics and evolution of epidemic Australian strains. J Antimicrob Chemother. 1988;21(Suppl. C):19–38. doi: 10.1093/jac/21.suppl_c.19. [DOI] [PubMed] [Google Scholar]
- 317.Smith T L, Pearson M L, Wilcox K R, Cruz C, Lancaster M V, Robinson-Dunn B, Tenover F C, Zervos M J, Band J D, White E, Jarvis W R. Emergence of vancomycin resistance in Staphylococcus aureus. N Engl J Med. 1999;340:493–501. doi: 10.1056/NEJM199902183400701. [DOI] [PubMed] [Google Scholar]
- 318.Snelling A M, Hawkey P M, Heritage J, Downey P, Bennett P M, Holmes B. The use of a DNA probe and PCR to examine the distribution of the aac(6′)-Ic gene in Serratia marcescens and other Gram-negative bacteria. J Antimicrob Chemother. 1993;31:841–854. doi: 10.1093/jac/31.6.841. [DOI] [PubMed] [Google Scholar]
- 319.Speldooren V, Heym B, Labia R, Nicolas-Chanoine M-H. Discriminatory detection of inhibitor-resistant β-lactamases in Escherichia coli by single-strand conformation polymorphism-PCR. Antimicrob Agents Chemother. 1998;42:879–884. doi: 10.1128/aac.42.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Sreedharan S, Oram M, Jensen B, Peterson L R, Fisher L M. DNA gyrase gyrA mutations in ciprofloxacin resistant strains of Staphylococcus aureus: close similarity with quinolone resistance mutations in Escherichia coli. J Bacteriol. 1990;172:7260–7262. doi: 10.1128/jb.172.12.7260-7262.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Sreedharan S, Peterson L R, Fisher L M. Ciprofloxacin resistance in coagulase-positive and -negative staphylococci: role of mutations at serine 84 in DNA gyrase A protein of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1991;35:2151–2154. doi: 10.1128/aac.35.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Sreevatsan S, Stockbauer K E, Pan X, Kreiswirth B N, Moghazeh S L, Jacobs W R, Jr, Telenti A, Musser J. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob Agents Chemother. 1997;41:1677–1681. doi: 10.1128/aac.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Stewart P R, Dubin D T, Chikramane S G, Inglis B, Matthews P R, Poston S M. IS257 and small plasmid insertions in the mec region of th chromosome of Staphylococcus aureus. Plasmid. 1994;31:12–20. doi: 10.1006/plas.1994.1002. [DOI] [PubMed] [Google Scholar]
- 324.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamm R K, Grahem D Y, Ghoneim A T, Tanaka S K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Sugano K, Kyogoku A, Fukayama N, Ohkura H, Shimosato Y, Sekiya T, Hayashi K. Rapid and simple detection of c-Ki-ras2 gene codon 12 mutations by nonradioisotopic single-strand conformation polymorphism analysis. Lab Investig. 1993;68:362–366. [PubMed] [Google Scholar]
- 326.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Tait S, Aymes S G B. Trimethoprim resistant dihydrofolate reductases in normal faecal flora isolated in India. Epidemiol Infect. 1994;113:247–258. doi: 10.1017/s0950268800051670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. MefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Tait-Kamradt A, Davies T, Cronan M, Jacobs M R, Appelbaum P C, Sutcliffe J. Mutations in 23S RNA and alterations in ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob Agents Chemother. 2000;44:2118–2125. doi: 10.1128/aac.44.8.2118-2125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Takenouchi T, Ishij C, Sugawara M, Tokue Y, Ohya S. Incidence of various gyrA mutants in 451 Staphylococcus aureus strains isolated in Japan and their susceptibilities to 10 fluoroquinolones. Antimicrob Agents Chemother. 1995;39:1414–1418. doi: 10.1128/aac.39.7.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Takenouchi T, Sakagawa E, Sugawara M. Detection of gyrA mutations among 335 Pseudomonas aeruginosa strains isolated in Japan and their susceptibilities to fluoroquinolones. Antimicrob Agents Chemother. 1999;43:406–409. doi: 10.1128/aac.43.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Takiff H E, Salazar L, Guerrero C, Philipp W, Mun Huang W, Kreiswirth B, Cole S T, Jacobs W R, Jr, Telenti A. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Taylor D E, Chau A. Tetracycline resistance mediated by ribosomal protection. Antimicrob Agents Chemother. 1996;40:1–5. doi: 10.1128/aac.40.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Telenti A, Honoré N, Bernasconi C, March J, Ortega A, Heym B, Takiff H E, Cole S T. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J Clin Microbiol. 1997;35:719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Telenti A, Imboden A, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 337.Telenti A, Imboden P, Marchesi F, Schmidhein T, Bodmer T. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob Agents Chemother. 1993;37:2054–2058. doi: 10.1128/aac.37.10.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Temesgen Z, Satoh K, Uhl J R, Kline B C, Cockerill F R., III Use of polymerase chain reaction single-strand conformation polymorphism (PCR-SSCP) analysis to detect a point mutation in the catalase-peroxidase gene (katG) of Mycobacterium tuberculosis. Mol Cell Probes. 1997;11:59–63. doi: 10.1006/mcpr.1996.0077. [DOI] [PubMed] [Google Scholar]
- 339.Tennent J M, Young H-K, Lyon B R, Aymes S G B, Skurray R A. Trimethoprim resistance determinants encoding a dihydrofolate reductase in clinical isolates of Staphylococcus aureus and coagulase-negative staphylococci. J Med Microbiol. 1988;26:67–73. doi: 10.1099/00222615-26-1-67. [DOI] [PubMed] [Google Scholar]
- 340.Tenover F C, Bo Huang M, Rasheed J K, Persing D H. Development of PCR assays to detect ampicillin resistance genes in cerebrospinal fluid samples containing Haemophilus influenzae. J Clin Microbiol. 1994;32:2729–2737. doi: 10.1128/jcm.32.11.2729-2737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Tenover F C, Gootz T D, Gordon K P, Tompkins L S, Young S A, Plorde J J. Development of a DNA probe for the structural gene of the 2′′-O-adenyltransferase amino-glycoside-modifying enzyme. J Infect Dis. 1984;150:678–687. doi: 10.1093/infdis/150.5.678. [DOI] [PubMed] [Google Scholar]
- 342.Tenover F C, Jones R N, Swenson J M, Zimmer B, McAllister S, Jorgensen J H. Methods for improved detection of oxacillin resistance in coagulase-negative staphylococci: results of a multicenter study. J Clin Microbiol. 1999;37:4051–4058. doi: 10.1128/jcm.37.12.4051-4058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Tenover F C, Phillips K L, Gilbert T, Lockhart P, O'Hara P J, Plorde J J. Development of a DNA probe from the deoxyribonucleotide sequence of a 3-N-aminoglycoside acetyltranseferase [AAC(3)-I] resistance gene. Antimicrob Agents Chemother. 1989;33:551–559. doi: 10.1128/aac.33.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Tenover F C, Unger E R. Nucleic acid probes for the detection and identification of infectious agents. In: Pershing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 3–25. [Google Scholar]
- 345.Thakker-Varia S, Jensen W D, Moon-McDermott L, Weinstein M P, Dubin D T. Molecular epidemiology of macrolide-lincosamide-streptogramin B resistance in Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob Agents Chemother. 1987;31:735–743. doi: 10.1128/aac.31.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Thal L A, Chow J W, Mahayni R, Bonilla H, Perri M B, Donabedian S A, Silverman J, Taber S, Zervos M J. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrob Agents Chemother. 1995;39:2112–2115. doi: 10.1128/aac.39.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Thal L A, Chow J W, Patterson J E, Perri M B, Donabedian S, Clewell D B, Zervos M J. Molecular characterization of highly gentamicin-resistant Enterococcus faecalis isolates lacking high-level streptomycin resistance. Antimicrob Agents Chemother. 1993;37:134–137. doi: 10.1128/aac.37.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Tham T N, Mabilat C, Courvalin P, Guesdon J-L. Biotinylated oligonucleotide probes for the detection and the characterization of TEM-type extended broad spectrum β-lactamases in Enterobacteriaceae. FEMS Microbiol Lett. 1990;69:109–116. doi: 10.1016/0378-1097(90)90423-n. [DOI] [PubMed] [Google Scholar]
- 349.Tokue Y, Shoji S, Satoh K, Watanabe A, Motomiya M. Comparison of a polymerase chain reaction assay and a conventional microbiologic method for detection of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:6–9. doi: 10.1128/aac.36.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Tokue Y, Sugano K, Saito D, Noda T, Ohkura H, Shimosato Y, Sekiya T. Detection of novel mutations in the gyrA gene of Staphylococcus aureus by nonradioisotopic single-strand confirmation polymorphism analysis and direct DNA sequencing. Antimicrob Agents Chemother. 1994;38:428–431. doi: 10.1128/aac.38.3.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Tomasz A, Drugeon H B, de Lencastre H M, Jabes D, McDougall L, Bille J. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP2a gene and contain normal penicillin-binding protein with modified penicillin-binding capacity. Antimicrob Agents Chemother. 1989;33:1869–1874. doi: 10.1128/aac.33.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Towner K J, Talbot D C S, Curran R, Webster C A, Humphreys H. Development and evaluation of a PCR-based immunoassay for the rapid detection of methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1998;47:607–613. doi: 10.1099/00222615-47-7-607. [DOI] [PubMed] [Google Scholar]
- 353.Towner K J, Young H-K, Amyes S G B. Biotinylated DNA probes for trimethoprim-resistant dihydrofolate reductases types IV and V. J Antimicrob Chemother. 1988;22:285–291. doi: 10.1093/jac/22.3.285. [DOI] [PubMed] [Google Scholar]
- 354.Traced P, de Cespédès G, Bentorcha F, Delbos F, Gaspar E, Horaud T. Study of heterogeneity of chloramphenicol acetyltransferase (CAT) genes in streptococci and enterococci by polymerase chain reaction: characterization of a new CAT determinant. Antimicrob Agents Chemother. 1993;37:2593–2598. doi: 10.1128/aac.37.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Tran Van Nieu G, Bordon F, Collatz E. Incidence of an aminoglycoside 6-N-acetyltransferase, AAC(6′)-1b, in amikacin-resistant clinical isolates of gram-negative bacilli, as determined by DNA-DNA hybridization and immunoblotting. J Med Microbiol. 1992;36:83–88. doi: 10.1099/00222615-36-2-83. [DOI] [PubMed] [Google Scholar]
- 356.Trzcinski K, Cooper B S, Hryniewicz W, Dowson C G. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2000;45:763–70. doi: 10.1093/jac/45.6.763. [DOI] [PubMed] [Google Scholar]
- 357.Tsakris A, Johnson A P, Legakis N J, Tzouvelekis L S. Prevalence of the type I and type II DHFR genes in trimethoprim-resistant urinary isolates of Escherichia coli from Greece. J Antimicrob Chemother. 1993;31:665–671. doi: 10.1093/jac/31.5.665. [DOI] [PubMed] [Google Scholar]
- 358.Tsakris A, Vatopoulos A C, Johnson A P, Pitt T L, Legakis N J, Tzouvelekis L S. Prevalence of a plasmid-mediated type II dihydrofolate reductase gene among trimethoprim-resistant urinary pathogens in Greek hospitals. J Antimicrob Chemother. 1992;29:405–413. doi: 10.1093/jac/29.4.405. [DOI] [PubMed] [Google Scholar]
- 359.Turner A, Gough K E, Leeming J P. Molecular epidemiology of tetM genes in Neisseria gonorrhoeae. Sex Transm Dis. 1999;75:60–66. doi: 10.1136/sti.75.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Tyagi S, Bratu D P, Russell Kramer F. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 361.Tyagi S, Russell Kramer F. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 362.Ubukata K, Asahi Y, Yamane A, Konno M. Combinational detection of autolysin and penicillin-binding protein 2B gene of Streptococcus pneumoniae by PCR. J Clin Microbiol. 1996;34:592–596. doi: 10.1128/jcm.34.3.592-596.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Ubukata K, Nakagami S, Nitta A, Yamane A, Kawakami S, Sugiura M, Konno M. Rapid detection of the mecA gene in methicillin-resistant staphylococci by enzymatic detection of polymerase chain reaction products. J Clin Microbiol. 1992;30:1728–1733. doi: 10.1128/jcm.30.7.1728-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Ubukata K, Yamashita N, Gotoh A, Konno M. Purification and characterization of aminoglycoside-modifying enzymes from Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1984;25:754–759. doi: 10.1128/aac.25.6.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Ünal S, Hoskins J, Flokowitsch J E, Wu C Y, Preston D, Skatrud P L. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol. 1992;30:1685–1691. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Ünal S, Werner K, DeGirolami P, Barsanti E, Eliopoulos G. Comparison of tests for detection of methicillin-resistant Staphylococcus aureus in a clinical microbiology laboratory. Antimicrob Agents Chemother. 1994;38:345–347. doi: 10.1128/aac.38.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Uttley A H C, Collins C H, Naidoo J, George R C. Vancomycin-resistant enterococci. Lancet. 1988;i:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 368.Vahaboglu H, Öztürk R, Aygün G, Co_kunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik I, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamase among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.van Asselt G J, Vliegenthart J S, Petit P L C, van de Klundert J A M, Mouton R P. High-level aminoglycoside resistance among enterococci and group A streptococci. J Antimicrob Chemother. 1992;30:651–659. doi: 10.1093/jac/30.5.651. [DOI] [PubMed] [Google Scholar]
- 370.van Doorn L J, Debets-Ossenkopp Y J, Marais A, Sanna R, Megraud F, Kusters J G, Quint W G. Rapid detection, by PCR and reverse hybridization, of mutations in the Helicobacter pylori 23S rRNA gene, associated with macrolide resistance. Antimicrob Agents Chemother. 1999;43:1779–1782. doi: 10.1128/aac.43.7.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Vanhoof R, Content J, Van Bossuyt E, Dewit L, Hannecart-Pokorni E. Identification of the aadB gene coding for aminoglycoside-2′′-O-nucleotidyltransferase, ANT(2′′), by means of the polymerase chain reaction. J Antimicrob Chemother. 1992;29:365–374. doi: 10.1093/jac/29.4.365. [DOI] [PubMed] [Google Scholar]
- 372.Vanhoof R, Content J, Van Bossuyt E, Nulens E, Sonck P, Depuydt F, Hubrechts J M, Maes P, Hannecart-Pokorni E. Use of the polymerase chain reaction (PCR) for the detection of aacA genes encoding aminoglycoside-6′-N-acetyltransferases in reference strains and Gram-negative clinical isolates from two Belgian hospitals. J Antimicrob Chemother. 1993;32:23–35. doi: 10.1093/jac/32.1.23. [DOI] [PubMed] [Google Scholar]
- 373.Vanhoof R, Godard C, Content J, Nyssen H J, Hannecart-Pokorni E. Detection by polymerase chain reaction of genes encoding aminoglycoside-modifying enzymes in methicillin-resistant Staphylococcus aureus isolates of epidemic phage types. J Med Microbiol. 1994;41:282–290. doi: 10.1099/00222615-41-4-282. [DOI] [PubMed] [Google Scholar]
- 374.Vliegenthart J S, Ketelaar-van Gaalen P A G, van de Klundert J A M. Nucleotide sequence of the aacC2 gene, a gentamicin resistance determinant involved in a hospital epidemic of multiply resistant members of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1989;33:1153–1159. doi: 10.1128/aac.33.8.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Vliegenthart J S, Ketelaar-van Gaalen P A G, van de Klundert J A M. Identification of three genes coding for aminoglycoside-modifying enzymes by means of the polymerase chain reaction. J Antimicrob Chemother. 1990;25:759–765. doi: 10.1093/jac/25.5.759. [DOI] [PubMed] [Google Scholar]
- 376.Waldvogel F A. New resistance in Staphylococcus aureus. N Engl J Med. 1999;340:556–557. doi: 10.1056/NEJM199902183400709. [DOI] [PubMed] [Google Scholar]
- 377.Walker G T, Fraiser M S, Schram J L, Little M C, Nadeau J G, Malinowski D P. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Walker G T, Little M C, Nadeau J G, Shank D D. Isothermal in vitro amplification of DNA by restriction enzyme/DNA polymerase system. Proc Natl Acad Sci USA. 1992;89:392–396. doi: 10.1073/pnas.89.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Wallet F, Roussel-Delvallez M, Courcol R J. Choice of a routine method for detecting methicillin-resistance in staphylococci. J Antimicrob Chemother. 1996;37:901–909. doi: 10.1093/jac/37.5.901. [DOI] [PubMed] [Google Scholar]
- 380.Wang T, Tanaka M, Sato K. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob Agents Chemother. 1998;42:236–240. doi: 10.1128/aac.42.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Warsa U C, Nonoyama M, Ida T, Okamoto R, Okubo T, Shimauchi C, Kuga A, Inoue M. Detection of tetK and tetM in Staphylococcus aureus of Asian countries by the polymerase chain reaction. J Antibiot. 1996;49:1127–1132. doi: 10.7164/antibiotics.49.1127. [DOI] [PubMed] [Google Scholar]
- 382.Wasteson Y, Roe D E, Falk K, Roberts M C. Characterization of tetracycline and erythromycin resistance in Actinobacillus pleuropneumoniae. Vet Microbiol. 1996;48:41–50. doi: 10.1016/0378-1135(95)00130-1. [DOI] [PubMed] [Google Scholar]
- 383.Watterson S A, Wilson S M, Yates M D, Drobniewski F A. Comparison of three molecular assays for rapid detection of rifampin resistance in Mycobacterium tuberculosis. J Clin Microbiol. 1998;36:1969–1973. doi: 10.1128/jcm.36.7.1969-1973.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Weisblum B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother. 1995;39:797–805. doi: 10.1128/aac.39.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Weisblum B. Macrolide resistance. Drug Resist Update. 1998;1:29–41. doi: 10.1016/s1368-7646(98)80212-4. [DOI] [PubMed] [Google Scholar]
- 387.Weisblum B. Resistance to macrolide-lincosamide-streptogramin antibiotics. In: Fischetti V A, editor. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 1999. pp. 682–698. [Google Scholar]
- 388.Werckenthin C, Schwarz S, Westh H. Structural alterations in the translational attenuator of consecutively expressed ermC genes. Antimicrob Agents Chemother. 1999;43:1681–1685. doi: 10.1128/aac.43.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Werner G, Witte W. Characterization of a new enterococcal gene, satG, encoding a putative acetyltransferase conferring resistance to streptogramin A compounds. Antimicrob Agents Chemother. 1999;43:1813–1814. doi: 10.1128/aac.43.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Westh H, Hougaard D M, Vuust J, Rosdahl V T. Prevalence of erm gene classes in erythromycin-resistant Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob Agents Chemother. 1995;39:369–373. doi: 10.1128/aac.39.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Westh H, Hougaard D M, Vuust J, Rosdahl V T. erm genes in erythromycin-resistant Staphylococcus aureus and coagulase-negative staphylococci. APMIS. 1995;103:225–232. [PubMed] [Google Scholar]
- 392.Whelen A C, Felmlee T A, Hunt J M, Williams D L, Roberts G D, Stockman L, Persing D H. Direct genotypic detection of Mycobacterium tuberculosis rifampin resistance in clinical specimens by using single-tube heminested PCR. J Clin Microbiol. 1995;33:556–561. doi: 10.1128/jcm.33.3.556-561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Whitcombe D, Theaker J, Guy S P, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 394.Widdowson C A, Klugman K P. The molecular mechanisms of tetracycline resistance in the pneumococcus. Microb Drug Resist. 1998;4:79–84. doi: 10.1089/mdr.1998.4.79. [DOI] [PubMed] [Google Scholar]
- 395.Widdowson C A, Klugman K P, Hanslo D. Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2891–2893. doi: 10.1128/aac.40.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Widdowson C A, Klugman K P. Emergence of the M phenotype of erythromycin-resistant pneumococci in South Africa. Emerg Infect Dis. 1998;4:277–281. doi: 10.3201/eid0402.980216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Widdowson C A, Klugman K P. Molecular mechanisms of resistance to commonly used non-betalactam drugs in Streptococcus pneumoniae. Semin Respir Infect. 1999;14:255–268. [PubMed] [Google Scholar]
- 398.Wiedemann M, Wilson W J, Czajka J, Luo J, Barany F, Batt C A. Ligase chain reaction (LCR)—overview and applications. PCR Methods App. 1994;3:S51–S64. doi: 10.1101/gr.3.4.s51. [DOI] [PubMed] [Google Scholar]
- 399.Reference deleted.
- 400.Willard K E, Moddy J A, Peterson L R. A general ampC active site oligonucleotide probe for Gram-negative rods. Mol Cell Probes. 1991;5:97–102. doi: 10.1016/0890-8508(91)90002-2. [DOI] [PubMed] [Google Scholar]
- 401.Williams D L, Spring L, Gillis T P, Salfinger M, Persing D H. Evaluation of a polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin Infect Dis. 1998;26:446–450. doi: 10.1086/516313. [DOI] [PubMed] [Google Scholar]
- 402.Wondrack L, Massa M, Yang B V, Sutcliffe J. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob Agents Chemother. 1996;40:992–998. doi: 10.1128/aac.40.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Woodford N, Morrison D, Cookson B, George R C. Comparison of high-level gentamicin-resistant Enterococcus faecium isolates from different continents. Antimicrob Agents Chemother. 1993;37:681–684. doi: 10.1128/aac.37.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Woodford N, Watson A P, Patel S, Jevon M, Waghorn D J, Cookson B D. Heterogeneous location of the mupA high-level mupirocin resistance gene in Staphylococcus aureus. J Med Microbiol. 1998;47:829–835. doi: 10.1099/00222615-47-9-829. [DOI] [PubMed] [Google Scholar]
- 405.Wu D Y, Wallace R B. The ligase amplification reaction (LAR)-amplification of specific DNA sequences using sequential rounds of template-dependent ligation. Genomics. 1989;4:560–569. doi: 10.1016/0888-7543(89)90280-2. [DOI] [PubMed] [Google Scholar]
- 406.Yoshida R, Hirakata Y, Kaku M, Tomono K, Maesaki S, Yamada Y, Kamihira S, Jacobs M R, Appelbaum P C, Kohno S. Genetic analsysis of serotype 23F Streptococcus pneumoniae isolates from several countries by pencillin-binding protein fingerprinting and pulsed-field gel electrophoresis. Chemother. 1999;45:158–165. doi: 10.1159/000007178. [DOI] [PubMed] [Google Scholar]
- 407.Zambardi G, Reverdy M E, Bland S, Bes M, Feney J, Fleurette J. Laboratory diagnosis of oxacillin resistance in Staphylococcus aureus by a multiplex-polymerase chain reaction assay. Diagn Microbiol Infect Dis. 1994;19:25–31. doi: 10.1016/0732-8893(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 408.Zeller V, Janoir C, Kitzis M D, Gutmann L, Moreau N J. Active efflux as a mechnism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1973–1978. doi: 10.1128/aac.41.9.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Zhang H Z, Schmidt H, Piepersberg W. Molecular cloning and characterization of two lincomycin-resistance genes, lmrA and lmrB, from Streptomyces lincolnensis 78–11. Mol Microbiol. 1992;6:2147–2157. doi: 10.1111/j.1365-2958.1992.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 410.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tubeculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 411.Zheng X, Kolbert C P, Varga-Delmore P, Arruda J, Lewis M, Kolberg J, Cockerill F R, Persing D H. Direct mecA detection from blood culture bottles by branched-DNA signal amplification. J Clin Microbiol. 1999;37:4192–4193. doi: 10.1128/jcm.37.12.4192-4193.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Zirnstein G, Li Y, Swaminathan B, Angulo F. Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J Clin Microbiol. 1999;37:3276–3280. doi: 10.1128/jcm.37.10.3276-3280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]