Summary
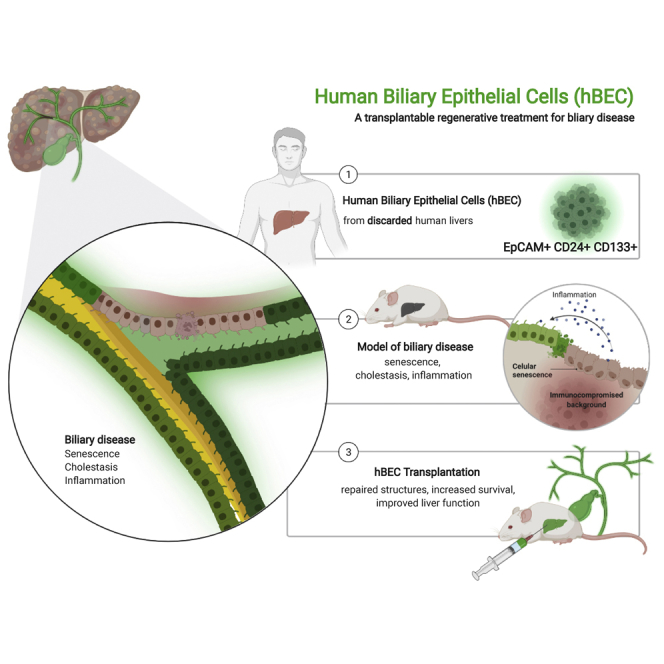
Biliary diseases can cause inflammation, fibrosis, bile duct destruction, and eventually liver failure. There are no curative treatments for biliary disease except for liver transplantation. New therapies are urgently required. We have therefore purified human biliary epithelial cells (hBECs) from human livers that were not used for liver transplantation. hBECs were tested as a cell therapy in a mouse model of biliary disease in which the conditional deletion of Mdm2 in cholangiocytes causes senescence, biliary strictures, and fibrosis. hBECs are expandable and phenotypically stable and help restore biliary structure and function, highlighting their regenerative capacity and a potential alternative to liver transplantation for biliary disease.
Keywords: liver, bile duct, biliary epithelial cells, cholangiocytes, progenitor cells, liver transplant, senescence, organoid, regeneration
Graphical abstract
Highlights
-
•
Human biliary epithelial cells (hBECs) can be isolated from discarded human livers
-
•
hBECs show regenerative properties when grafted into a biliary disease mouse model
-
•
Mice transplanted with hBECs regenerate bile ducts and show improved liver function
-
•
hBECs can be cultured in good manufacturing process conditions for clinical use
In this manuscript, Forbes and colleagues isolate and expand human biliary epithelial cells (hBECs) from discarded livers. Upon transplantation into a mouse model of biliary disease, hBECs regenerate and repair damaged bile ducts, offering a potential therapy for biliary disease.
Introduction
Biliary epithelial cells (BECs), termed cholangiocytes, have been shown in murine models to be capable of regenerating both themselves and hepatocytes when liver regeneration is profoundly impaired (Raven et al., 2017). This population of bipotent cells can be isolated, expanded, and transplanted to repopulate the damaged liver parenchyma, differentiating into mature hepatocytes in order to restore liver function when the endogenous regenerative mechanisms are exhausted (Raven et al., 2017; Lu et al., 2015; Huch et al., 2013, 2015; Sampaziotis et al., 2021; Kurial and Willenbring, 2021; Inada et al., 2020). Evidence of biliary regeneration from transplanted murine cells of cholangiocyte and hepatocyte origin has been demonstrated (Tarlow et al., 2014; Carpino et al., 2015; Schaub et al., 2018), highlighting the therapeutic potential of BECs in the management of liver conditions. However, the use of human BECs (hBECs) in clinical settings remains limited (Sampaziotis et al., 2017).
Although human cholangiocytes (containing a fraction of hBECs) can be isolated, expanded in vitro, and transplanted into the liver and the extrahepatic biliary tract (Huch et al., 2013; Dorrell et al., 2011; Li et al., 2017; Nevi et al., 2017), the mixed populations used showed no evidence of regenerative potential, resolution of biliary disease, or improvement in survival.
In this study, we show that hBECs, as a subset of cholangiocytes isolated from discarded human livers, defined by their expression of EpCAM, CD24, and CD133, are a highly expandable and phenotypically stable population. hBECs were transplanted into an immunodeficient model of biliary disease, based on the conditional deletion of Mdm2 in keratin 19 (K19)-positive cholangiocytes, which consistently reproduces traits of biliary deterioration and progressive liver injury (Ferreira-Gonzalez et al., 2018). hBECs engraft into the mouse liver resulting in resolution of biliary strictures, regression of hepatic fibrosis, and a reduction in overall mortality. These results demonstrate the potential of hBECs as a transplantable regenerative treatment for biliary disease.
Results
hBECs isolated from discarded human livers expand in vitro and maintain a stable phenotype
Primary hBECs were isolated from discarded human livers obtained from deceased organ donors. These livers were initially intended to be used for transplantation but were unable to be transplanted because of logistical reasons or deemed unsuitable because of excess steatosis or fibrosis following clinical assessment.
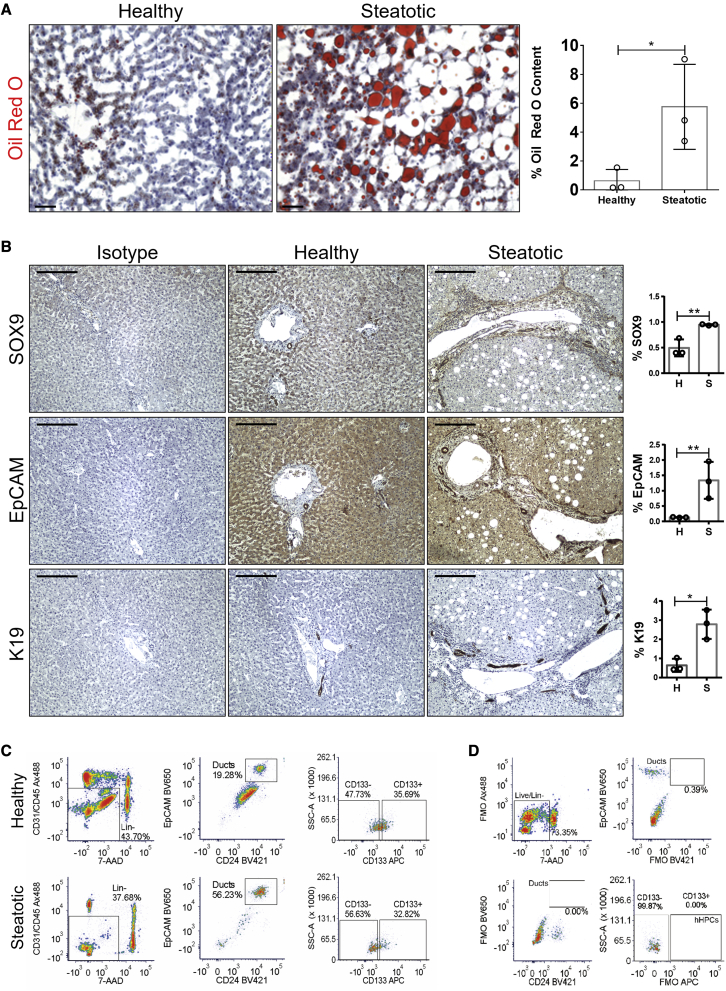
To determine the level of neutral triglycerides and lipids present, we performed oil red O staining (Figure 1A), which allowed us to classify, in concordance with the clinical assessment, the livers as histologically healthy or steatotic. First, we investigated whether these discarded livers could provide a source of hBECs. As ductular reactions (DRs) are thought to include putative stem cells (Lu et al., 2015; Boulter et al., 2012; Williams et al., 2014), we characterized this phenomenon.
Figure 1.
hBEC characterization in steatotic and healthy livers
(A) Left: oil red O staining in healthy and steatotic human rejected livers. Scale bars, 100 μm. Right: quantification of the percentage of oil red O. ∗ denotes p < 0.05 (mean ± SEM), Student’s t test between healthy (n = 3) and steatotic livers (n = 3).
(B) Isotype control, healthy, and steatotic human livers stained for SOX9, EpCAM, and K19. Scale bars, 250 μm. Far right: total pixel quantification expressed as percentage in healthy (H) and steatotic (S) human livers. Steatotic livers have significantly increased levels of SOX9, EpCAM, and K19 in comparison with healthy livers. ∗ denotes p < 0.05 (mean ± SEM), Student’s t test (n = 3).
(C) Representative gating strategy for isolation of EpCAM+/CD24+/CD133+ hBECs from whole healthy and steatotic liver digests. hBECs were defined as live, non-hematopoietic single cells that expressed EpCAM and CD24. This population was further subdivided into CD133− and CD133+ fractions.
(D) Fluorescence minus one (FMO) control staining for setting positive staining gates for the isolation of EpCAM+/CD24+/CD133+ hBECs.
Immunohistochemical staining and subsequent quantification of established markers of BECs, including K19, EpCAM, or SOX9 (Raven et al., 2017; Lu et al., 2015; Carpentier et al., 2011; Li et al., 2017; Rodrigo-Torres et al., 2014), confirmed that steatotic livers have significantly increased numbers of cholangiocytes and a more established DR than healthy controls (Figure 1B).
To identify and characterize hBECs with therapeutic potential, candidate hBEC populations were fluorescence-activated cell sorted (FACS) from liver tissue following mechanical and enzymatic digestion. We applied a sorting strategy that we have previously described for isolating murine BECs with bipotential capacity (Lu et al., 2015). Epithelial populations were enriched by selecting cells that were CD45−/CD31−, and then hBECs were identified by positive staining of EpCAM and CD24. We then sorted two candidate hBEC populations: CD45−/CD31−/EpCAM+/CD24+/CD133− (CD133−) and CD45−/CD31−/EpCAM+/CD24+/CD133+ (CD133+) (Figures 1C and 1D).
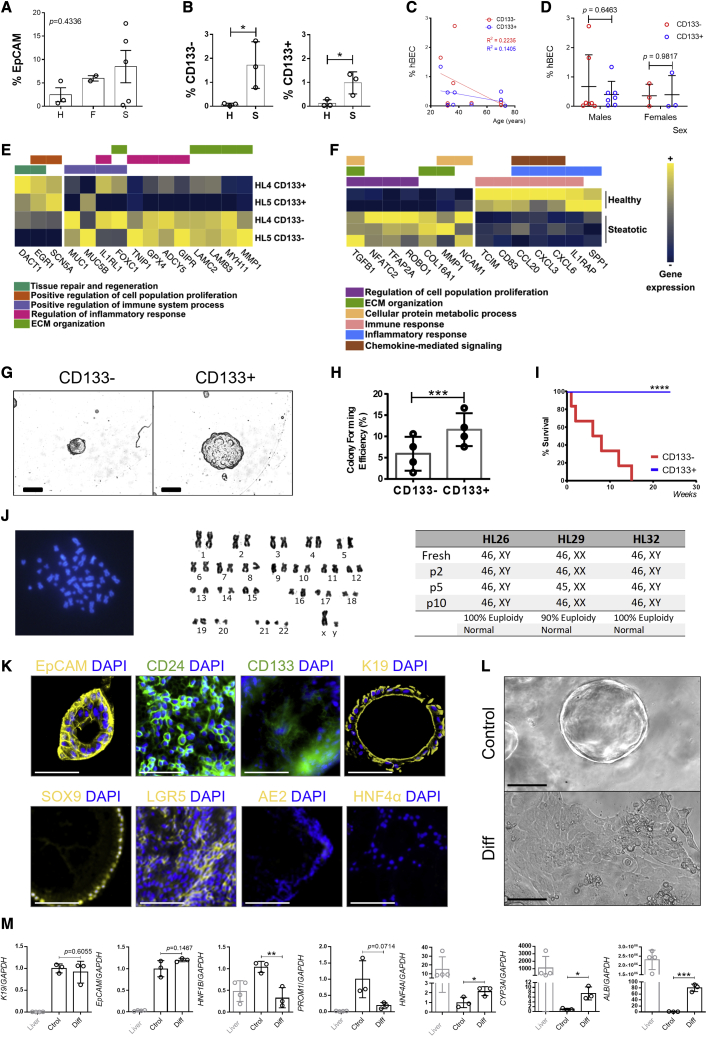
While the percentage of EpCAM+ cells did not change in healthy, steatotic, or fibrotic livers (Figure 2A), there was an increase of both CD133− and CD133+ hBECs in steatotic livers compared with healthy livers (Figure 2B), suggesting that this is a hBEC-related response. We did not find any correlations between donor age (Figure 2C), gender (Figure 2D), and the total number of CD133− and CD133+ candidate hBECs isolated.
Figure 2.
CD133- and CD133+ hBEC characterization in steatotic and healthy livers
(A) Total percentage of EpCAM+ cells in healthy (H), fibrotic (F), and steatotic (S) livers show no significant differences (p = 0.4336). One-way ANOVA.
(B) Total percentage of CD133− and CD133+ hBECs isolated from healthy (H) and steatotic (S) livers showing a significant increase in steatotic livers. ∗ denotes p < 0.05 (mean ± SEM), Student’s t test.
(C) Correlation of the percentage of isolated CD133− and CD133+ hBECs according to the age of the donor (years). The graph shows a non-significant trend to decrease (R2 = 0.2235 and R2 = 0.1405 for CD133− and CD133+ populations, respectively). Linear regression at 95% confidence intervals (n = 9 livers of diverse etiologies, 27 to 70 years old).
(D) Correlation of the percentage of isolated CD133− and CD133+ hBECs according to the sex of the donor shows no significant differences (mean ± SEM), Student’s t test (n = 7 males, 3 females).
(E) Heatmap of normalized expression values across genes significantly differentially expressed between CD133+ and CD133− hBECs in two healthy livers (HL4 and HL5). In yellow, relative upregulation; blue, relative downregulation.
(F) Heatmap of normalized expression values across genes significantly differentially expressed between CD133+ hBECs in healthy (n = 2) and steatotic livers (n = 3). In yellow, relative upregulation; blue, relative downregulation.
(G) Bright field image showing morphological differences of CD133− and CD133+ hBEC populations in a three-dimensional Matrigel culture. Scale bars, 100 μm.
(H) CD133+ population displays significantly increased colony-forming efficiency in comparison with the CD133− population. ∗∗∗ denotes p < 0.001 (mean ± SEM), Student’s t test (n = 4 biological replicates).
(I) Percentage of survival of CD133− and CD133+ hBECs over the course of time. ∗∗∗∗p < 0.0001 (mean ± SEM), Mantel-Cox test.
(J) Left: chromosomes of the CD133+ hBECs cultured for over 6 months. Right: karyotype of hBECs isolated from donor livers from passage 0 to passage 10 (n = 3).
(K) CD133+ cells expanded in Matrigel culture and immunostained for cholangiocyte, hepatocyte, and progenitor cell markers. CD133+ hBECs express K19, EpCAM, and SOX9 cholangiocyte markers, as well as the progenitor markers LGR5, CD24, and CD133, while lacking hepatocyte markers (HNF4α) and mature biliary markers (AE2). Scale bars, 100 μm.
(L) Brightfield images of CD133+ hBEC organoids cultured in 3D Matrigel spheres in standard expansion media (Control) and differentiation media (Diff). Scale bars, 100 μm.
(M) Expression of genes associated with a mature cholangiocyte phenotype (cytokeratin 19 K19, EPCAM, HNF1B), hepatocytes (HNF4A, CYP3A, ALB), and progenitor cells (PROM1) normalized to GAPDH. Data include total human liver for reference (liver, gray labeled), hBECs cultured in 3D Matrigel spheres in standard expansion media (Ctrol), and differentiation media (Diff). All results displayed as relative fold increase compared to controls. ∗ denotes p < 0.05, ∗∗p < 0.005 (mean ± SEM), Student’s t test. (n = 3–4 per group).
To characterize isolated hBECs, we performed whole-transcriptome analysis with RNA sequencing (RNA-seq) of healthy and steatotic livers. Differential expression analysis of healthy liver samples revealed markers of tissue repair and regeneration EGR1 and DACT1 to be significantly upregulated in CD133+ versus CD133− hBECs (p = 0.0405 and p = 0.0223, respectively). We observed upregulation of genes associated with proliferation (SCN5A, p = 0.0143), combined with the downregulation of genes associated with regulation of immune system process (such as MUC1, MUC5B), inflammatory response (IL1LR1, GPX4, ADCY5), and extracellular matrix organization (LAMB3, MMP1) (Figure 2E). Further gene set enrichment analysis (GSEA) showed significant depletion of inflammatory response, allograft rejection, Wnt β-catenin, and Notch signaling pathways (Figures S1A and S1B), highly suggestive of an enhanced regenerative potential in CD133+ hBECs.
We further explored the regenerative potential of CD133+ hBECs isolated from steatotic and healthy livers. Genes significantly upregulated in steatotic liver-derived CD133+ hBECs include those associated with cell proliferation, metabolism, and extracellular matrix organization. Genes significantly downregulated are associated with inflammatory responses and chemokine-mediated signaling (Figure 2F). GSEA revealed significant depletion of inflammatory response, reactive oxygen species pathway, and apoptosis (Figures S1C and S1D). This combination of proliferative and anti-inflammatory profiles suggests that the regenerative potential of hBECs increases under steatotic conditions.
We further analyzed the transcriptomic profile of CD133+ hBECs using publicly available cell transcriptomic data from Sampaziotis and colleagues (Sampaziotis et al., 2021). In comparison with EpCAM+ hBECs, expression of known progenitor cell markers (such as TUSC2, MDH2, or AGRN) is detectable in a greater proportion of EpCAM+CD133+ hBECs (Figures S2A and S2B), indicating that EpCAM+CD133+ hBECs have an enhanced progenitor-like profile. Similarly, a higher proportion of EpCAM+CD133+ hBECs show expression of genes associated with cell differentiation and positive regulation of cell proliferation (GO:0008283 and GO:0008284, respectively) (Figure S2C). Furthermore, a higher proportion of EpCAM+CD133+ hBECs show expression of genes associated with cholangiocyte proliferation (GO:1990705) (Figure S2D) and hepatocyte proliferation (GO:0072574) (Figure S2E), indicating an increased proliferative capacity and the bipotential ability of the EpCAM+CD133+ hBECs in comparison with EpCAM+ hBEC population.
To address the phenotype and stability of the cells in vitro, we performed transcriptomic analysis of hBECs freshly isolated from human donor livers and cultured during several passages. Expression of mature cholangiocyte markers is sustained over the course of time while maintaining a progenitor-like phenotype. hBECs do not acquire a hepatocyte phenotype in these conditions, although a small level of HNF4α expression at gene level is noted (Figure S2F). Similarly, over the course of passages, hBECs cultured in different conditions retain markers of stem cell and cholangiocyte proliferation, with minimal expression of hepatocyte proliferation markers (Figure S2G). Moreover, the percentage of CD133+ cells remains stable during culture (Figure S2H). These results suggest that hBEC phenotype is maintained during in vitro culture.
We then evaluated the profile of EpCAM+CD133+ compared to EpCAM+CD133− BECs using the publicly available single-cell transcriptomic data from Sampaziotis and colleagues (Sampaziotis et al., 2021). Our analysis of these data shows that the proportion of EpCAM+CD133− BECs with detectable expression of known proliferation-associated genes is lower than that of EpCAM+CD133+ BECs, suggesting that EpCAM+CD133− BECs have a decreased proliferative profile in comparison with EpCAM+CD133+ BECs (Figure S2I). Similarly, a lower proportion of EpCAM+CD133− BECs show expression of genes associated with cholangiocyte proliferation (GO:1990705) (Figure S2J) and hepatocyte proliferation (GO:0072574) (Figure S2K), further indicating an overall decreased proliferative capacity in comparison with EpCAM+CD133+ BECs.
We then investigated the colony-forming potential of both CD133− and CD133+ hBECs, plating both populations at clonal density in Matrigel (Figures 2G, S3A, and S3B). CD133+ hBECs had significantly increased colony-forming efficiency compared to CD133− hBECs (Figures 2H and S3C–S3E), forming colonies that underwent serial passages and survived beyond 15 weeks culture (Figure 2I) while retaining a normal diploid karyotype (Figure 2J).
Further characterization showed that CD133+ hBECs grown as organoids express cholangiocyte and progenitor markers such as K19, SOX9, EpCAM, STEM121, and LGR5 and lack hepatocyte markers such as albumin (ALB) and HNF4α (Figures 2K and S3F–S3I). hBEC organoids are actively proliferating (as assessed by KI67 and PCNA immunofluorescence, Figures S3F and S3G), retain membrane integrity (Figure S3J), and can be traced by the presence of human anti-mitochondrial staining (hAMA, Figure S3K).
CD133+ hBECs were also able to differentiate into a hepatocyte lineage (Lu et al., 2015), displaying distinctive morphological (Figure 2L) and transcriptional signatures (Figure 2M), indicating similar differentiating potential between human and murine BECs (Lu et al., 2015; Hay et al., 2008). We further characterized the plasticity of the CD133+ hBEC population by analyzing known markers of mature hepatocytes (Albumin, CYP2C9, TTR) and cholangiocytes (SOX9, aquaporin, HNF1B) in the EpCAM+CD133+ population in the transcriptomic data available from Sampaziotis and colleagues (Sampaziotis et al., 2021). In agreement with our data, the results indicate that a high proportion of CD133+ hBECs co-express mature markers of hepatocytes and cholangiocytes (78% of CD133+ hBECs co-express both ALB and SOX9, 78% co-express ALB and aquaporin and 59% co-express ALB and HNF1B, Figure S4A), potentially indicating a bipotential phenotype. Differentiated hBECs acquire markers of mature hepatocytes (CYP2D6, HNF4A) while losing markers associated with mature cholangiocytes, such as HNF1B (Figures S4B and S4C). Finally, differentiated hBECs increase ALB secretion (Figure S4D) in comparison with undifferentiated control hBECs, suggesting that hBECs acquire a functional hepatocyte-like phenotype.
hBEC transplantation mediates injury resolution and repair in an immunocompromised murine model of biliary disease
To assess the regenerative capacity of the CD133+ hBEC population in vivo, we transplanted the expanded hBECs into an immunocompromised murine model of biliary disease. The Krt19CreERMdm2fl/fl Rag2−/− Il2rg−/− model is based on the conditional deletion of Mdm2, a key negative regulator of p53, in cholangiocytes. After tamoxifen induction, the activated CreERT2-recombinase floxes out Mdm2, leading to the accumulation of p53 and p21 in K19+ cholangiocytes, establishing an irreversible cell-cycle arrest and senescence phenotype (Ferreira-Gonzalez et al., 2018). As previously shown, cellular senescence aggravates biliary injury established by the administration of 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) diet, mimicking the phenotypic traits of biliary diseases such as primary sclerosing cholangitis (PSC) and primary biliary cholangitis (PBC) (Ferreira-Gonzalez et al., 2018). The immunodeficient phenotype of the model (Rag2−/− Il2rg−/−) establishes a permissive niche for the transplantation of human cells (Kenney et al., 2016; Richmond and Su, 2008).
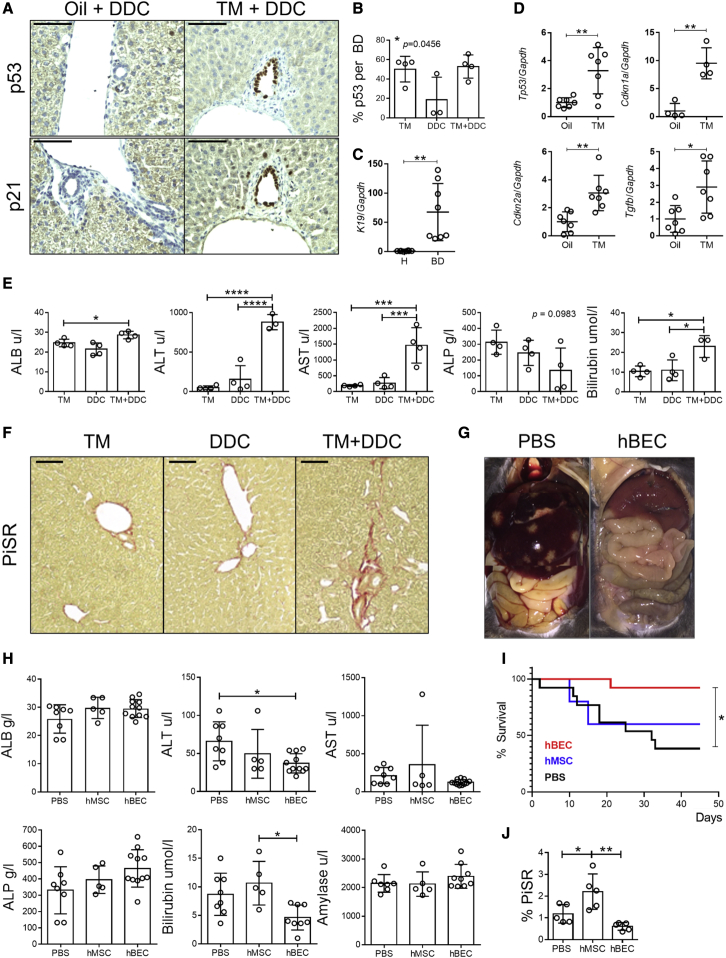
In our study, the Krt19CreERMdm2fl/fl Rag2−/− Il2rg−/− model, in combination with a short pulse of DDC diet, displayed markers of senescence in cholangiocytes, such as p53 and p21 (Figures 3A and 3B). Isolation of hepatocytes and bile ducts in our model (Figure 3C) showed significant upregulation of markers associated with cell-cycle arrest (Tp53 [p53], Cdkn1a [p21], Cdkn2a [p16]) and paracrine senescence (Tgfb) in the bile ducts (Figure 3D). Increased levels of serum transaminases (Figure 3E) alongside increased levels of fibrosis (assessed by PicroSirius Red staining, Figure 3F) suggest that the combination of the genetic deletion of Mdm2 and the administration of DDC diet provided a phenotype characteristic of advanced stages of biliary disease.
Figure 3.
CD133+ hBECs transplant in an immunodeficient model of biliary disease
(A) Immunostaining for p53 and p21 in control animals (Oil+DDC) versus induced animals (TM+DDC) (n = 3–4). Scale bars, 120 μm.
(B) Quantification of p53-positive cells per bile duct in tamoxifen (TM), DDC diet (DDC) and induced mice (administered with tamoxifen and DDC diet, TM+DDC). ∗ denotes p < 0.05 (mean ± SEM), one-way ANOVA (n = 3–4).
(C) Confirmation of cholangiocyte (K19) gene expression in isolated bile ducts (BDs) and absence in hepatocytes (H) normalized to Gapdh. ∗∗ denotes p < 0.005 (mean ± SEM), Student’s t test (n = 7).
(D) Gene expression analysis of isolated BDs in control (oil) versus induced (TM) mice shows a significant increase in senescent markers in the later population (Trp53, Cdkn1a, Cdkn2a, and Tgfb, normalized to Gapdh). ∗ denotes p < 0.05, ∗∗ denotes p < 0.005 (mean ± SEM), Student’s t test (n = 7).
(E) Transaminase analysis in TM, DDC, and induced mice (TM+DDC). ∗ denotes p < 0.05, ∗∗∗ denotes p < 0.001, ∗∗∗∗ denotes p < 0.001 (mean ± SEM), one-way ANOVA (n = 4).
(F) PicroSirius red staining (PiSR) increases in induced mice (TM+DDC), (n = 3). Scale bars, 250 μm.
(G) Representative postmortem image of peritoneum in PBS control and hBEC-transplanted Krt19CreERMdm2fl/flRag2−/−Il2rg−/− mice (n = 4).
(H) Transaminase analysis in PBS control (PBS), human adipose tissue-derived mesenchymal stem cells (hMSCs), and hBEC-transplanted mice (hBEC). ∗ denotes p < 0.05 (mean ± SEM), one-way ANOVA (n = 5–8).
(I) Survival analysis of mice receiving hBEC (n = 13), PBS (n = 13), or hMSC (n = 5). ∗ denotes p < 0.05 log-rank (Mantel-Cox) test.
(J) PiSR quantification in PBS control (PBS), hMSC-transplanted mice (hMSC), and hBEC-transplanted mice (hBEC) shows a significant reduction of fibrosis. ∗ denotes p < 0.05 (mean ± SEM), one-way ANOVA (n = 5).
As the Krt19CreERMdm2fl/fl Rag2−/− Il2rg−/− model harbors an immunodeficient background permissive for the transplant of human cells, we used it as a platform for the transplantation of hBECs. Following the induction of senescence and biliary injury, 1 × 106 CD133+ hBECs or a carrier control (PBS) was transplanted via intrasplenic injection. Gross postmortem examination of livers from hBEC-transplanted Krt19CreERMdm2fl/fl Rag2−/− Il2rg−/− mice revealed fewer intrahepatic lesions and a general reduction in hepato-splenomegaly in comparison with PBS controls (Figure 3G).
After hBEC transplantation in the Krt19CreERMdm2fl/fl Rag2−/− Il2rg−/− model, bilirubin levels decrease compared to PBS or a cellular control consisting of human adipose tissue-derived mesenchymal stem cells (hMSCs) (Figure 3H). Moreover, hBEC-transplanted mice show a significantly increased survival rate (Figure 3I) and decreased levels of fibrosis (Figure 3J) in comparison with the PBS control or transplanted hMSCs.
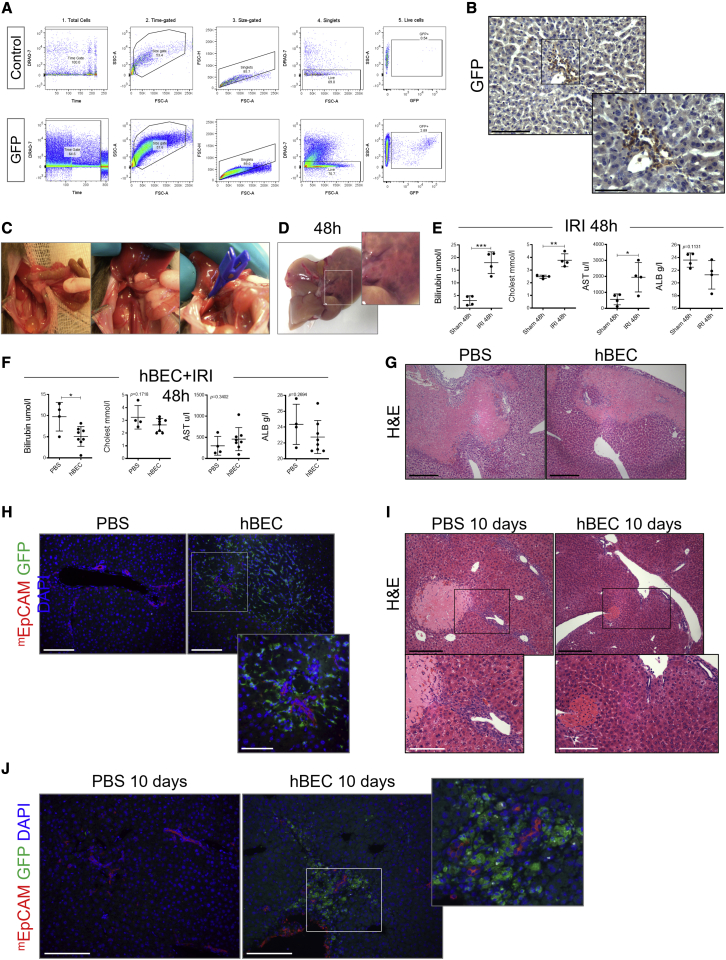
To test the regenerative efficacy of the cells in a second model, we also transplanted hBECs in an immunocompromised mouse model of ischemia reperfusion injury (IRI) (Abe et al., 2009; Rampes and Ma, 2019). For this set of experiments, we generated stable GFP-positive hBEC lines via GFP-lentiviral transduction (Figures 4A and 4B), which allowed us to trace hBECs after transplantation much more efficiently.
Figure 4.
CD133+ hBEC transplant in the IRI immunocompromised model
(A) FACS characterization of the GFP-positive hBECs in comparison with control hBECs.
(B) Immunohistochemistry of GFP hBECs transplanted in the Krt19CreERMdm2fl/flRag2−/−Il2rg−/− mice model of biliary disease. Scale bars, 120 μm.
(C) Model of ischemia reperfusion depicting the main stages of the surgery and clamping, which is maintained for 45 min.
(D) Liver injury in ischemic lobes in the late phase of injury after 48 h of reperfusion.
(E) Liver function biochemistry (bilirubin, cholesterol, aspartate aminotransferase [AST], and ALB) after 48 h of IRI in comparison with Sham controls. ∗ denotes p < 0.05, ∗∗ denotes p < 0.005, ∗∗∗ denotes p < 0.001 (mean ± SEM), Student’s t test, (n = 4).
(F) Liver function biochemistry after 48 h of IRI+ hBEC transplant in comparison with PBS controls. ∗ denotes p < 0.05 (mean ± SEM), Student’s t test, (n = 4 PBS and 8 hBECs).
(G) H&E staining in hBEC transplanted versus PBS controls. Scale bars, 250 μm.
(H) Immunohistochemistry for GFP-positive hBECs (green) and mouse-specific EpCAM (red) showing infiltration of hBECs in biliary regions. Scale bars, 250 μm; below, focused area (120 μm).
(I) H&E staining in hBEC transplanted versus PBS controls. Scale bars, 250 μm.
(J) Immunohistochemistry for GFP-positive hBECs (green) and mouse-specific EpCAM (red) showing infiltration of hBECs in biliary regions. Scale bars, 120 μm.
During IRI (Figure 4C), the ischemic insult causes functional changes that facilitate cellular injury. Reperfusion of the liver exacerbates the initial injury, which can be further divided into two phases—an early phase and a late phase (which last up to 48 h after reperfusion)—and is associated with ischemic-type biliary lesions (Figure 4D) and an increase in bilirubin and cholesterol (Figure 4E) following liver transplantation (Cursio and Gugenheim, 2012).
hBEC transplantation in IRI decreases the levels of bilirubin at 48 h post ischemia (Figure 4F) while decreasing histological damage (Figure 4G). GFP-positive hBECs engraft in the damaged areas in close proximity to the host’s cholangiocytes (Figure 4H). 10 days after IRI, when damage subdues and hepatic function is restored, hBEC-transplanted mice exhibit less necrosis and normal biliary structures in comparison with the control mice in which the ductular reaction still persists (Figure 4I). After 10 days, engrafted hBECs are still in close contact with the host biliary tract (Figure 4J). Altogether, these results suggest that hBECs ameliorate the phenotype of the models and showcase the potential regenerative response of hBECs in the context of biliary disease.
Transplanted hBECs engraft in close proximity to damaged biliary tracts and reduce biliary injury
Next, we investigated the biological mechanisms of hBEC transplantation and engraftment. Upon hBEC transplantation, the Krt19CreERMdm2fl/fl Rag2−/− Il2rg−/− biliary disease model shows restoration of the intrahepatic biliary anatomy and reduction of polymorphonuclear infiltrates and parenchymal damage (Figure 5A).
Figure 5.
Histological characterization in the hBEC-transplanted Krt19CreERMdm2fl/flRag2−/−Il2rg−/− biliary disease model
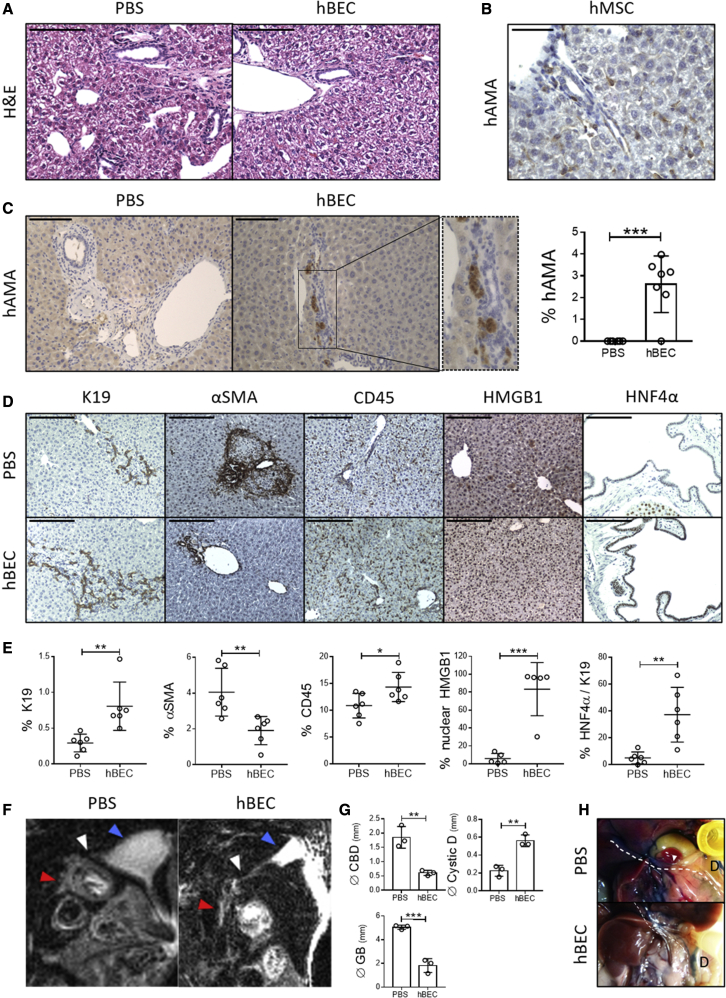
(A) H&E staining in the Krt19CreERMdm2fl/flRag2−/−Il2rg−/− biliary disease model reveals a pro-inflammatory profile in animals treated with PBS (PBS) and a normal parenchymal organization in the hBEC-transplanted mice (hBEC). Scale bars, 120 μm.
(B) Human anti-mitochondrial antibody immunostaining (hAMA) in tissue-derived human mesenchymal stem cell (hMSC)-transplanted Krt19CreERMdm2fl/flRag2−/−Il2rg−/− mice. Scale bars, 60 μm.
(C) hAMA immunostaining in PBS carrier (PBS) and hBEC-transplanted mice (hBEC). Far right: quantification of hBECs per total number of cells. ∗∗∗ denotes p < 0.001 (mean ± SEM), Student’s t test (n = 6 PBS and n = 7 hBECs). Scale bars, 120 μm.
(D) Immunostaining for K19 (ductular reaction), αSMA (activated hepatic stellate cells), CD45 (immune cells), HMGB1 (mediator of inflammation), and HNF4α (hepatocyte marker) in PBS carrier (PBS) and hBEC-transplanted mice (hBEC). Scale bars, 120 μm (n = 6).
(E) Quantification of the immunostainings in PBS carrier (PBS) and hBEC-transplanted mice (hBEC). Far right; quantification of HNF4α per K19+ cells. ∗ denotes p < 0.05, ∗∗ denotes p < 0.005, ∗∗∗ denotes p < 0.001 (mean ± SEM), Student’s t test (n = 6).
(F) MRCP. Sagittal section view on PBS control (PBS) and hBEC-transplanted mice (hBEC). Blue arrows indicate the position of the gallbladder, white arrows the cystic duct, and red arrows the common bile duct (CBD). Stricturing of the CBD is observed in the PBS group.
(G) Quantification of common bile duct (Ø CBD), cystic duct (Ø Cystic D), and gallbladder (Ø GB) diameters (in mm) in PBS control (PBS) and hBEC-transplanted mice (hBEC). ∗∗ denotes p < 0.005, ∗∗∗ denotes p < 0.001 (mean ± SEM), Student’s t test (n = 3).
(H) Methylene blue retrograde gallbladder injection in PBS control (PBS) and hBEC-transplanted mice (hBEC). Dashed line indicates CBD, D indicates duodenum, and the white arrow indicates the ampulla of vater (where CBD drains into the duodenum). Absence of methylene blue in the duodenum in the PBS group is indicative of obstruction in the biliary tract (n = 3).
Human anti-mitochondrial immunostaining (hAMA, specific for transplanted cells of human origin) reveals that transplanted control hMSCs engraft in the mouse parenchyma in an irregular stochastic pattern (Figure 5B), suggesting a diffuse infiltration. On the contrary, hBECs appear to engraft nearby the native mouse biliary tract (Figure 5C), suggesting that upon transplantation, hBECs undergo targeted migration. We observed no hBEC engraftment in other tissues, such as spleen or lung (Figure S5A).
Upon transplantation, hBECs appear to adopt a biliary-like morphology (Figure S5B) and can be found in close proximity to the native murine biliary tract (Figure S5C). hBECs also appear to proliferate after transplantation, as observed by the presence of proliferation markers such as KI67 (Figure S5D) and PCNA (Figure S5E).
Interestingly, upon transplantation in the IRI model (where the hepatic parenchyma is damaged as a consequence of the ischemic injury), we observed that a small number of GFP-positive hBECs can be found in distant areas from the biliary tract, expressing markers of mature hepatocytes (such as CYP2D6) (Figure S5F). In the GFP-positive hBECs that engrafted near the host biliary tracts, we did not observe expression of other hepatocyte markers such as HNF4α (Figure S5G).
We then characterized the histological response of the livers after hBEC transplantation and observed increased levels of K19+ cells, reduced levels of αSMA (indicative of a reduced stellate cell activation) and increased numbers of CD45+ cells compared to PBS controls (Figures 5D and 5E). We also observed a shift of high-mobility group box 1 (HMGB1) from the cytoplasm to the nucleus of hepatocytes (Figures 5D and 5E), alongside a significant reduction of pro-inflammatory cytokines in serum (Figure S6A) and liver (Figure S6B), suggesting the presence of anti-inflammatory mechanisms upon hBEC transplantation.
After hBEC transplantation, whole liver tissue gene expression indicated a significant increase in human ALB levels and a trend to increase for markers associated with cholangiocytes (EpCAM p = 0.0964, mean ± SEM) (Figure S6C). Murine mRNA analysis revealed significant increased levels of Notch3 and Hnf4a, alongside trends to increase of K19 (p = 0.1846, mean ± SEM) and Notch2 (p = 0.0569, mean ± SEM) (Figure S6D), suggesting that host regenerative response can be partially attributed to an hBEC-dependent ductular reaction response (see Figure 5D, K19 immunohistochemistry panel).
Interestingly, upon hBEC transplantation, K19+ cholangiocytes expressed HNF4α, suggesting the activation of bipotential pro-regenerative mechanisms. In particular, partial HNF4α expression in the common bile duct (CBD) suggests that part of the regenerative response seen in this model is due to expression changes in the extrahepatic areas of the biliary tract (Figures 5D and 5E).
To explore the involvement of the extrahepatic biliary system, magnetic resonance cholangiopancreatography (MRCP) was performed in PBS controls and hBEC-transplanted mice. The Krt19CreERMdm2fl/fl Rag2−/− Il2rg−/− biliary disease model presents markers of senescence such as p21 in large bile ducts, suggesting that extrahepatic areas of the model are also targeted during the induction of the senescent phenotype (Figure S6E). This model treated with PBS carrier presents cystic duct obliteration, stricturing, and dilation of the gallbladder and CBD (Figure 5F). Conversely, the CBD and gallbladder diameter were significantly decreased, while the cystic duct increases in hBEC-transplanted mice, suggesting that transplantation improves stricturing of extrahepatic areas of the biliary tract (Figures 5F and 5G). This result was further confirmed by retrograde injection of methylene blue in the gallbladder. In the PBS control group, the presence of strictures in the biliary tract prevented drainage into the duodenum, while hBEC-transplanted mice displayed a normal bile flow (Figure 5H). GFP-positive hBEC transplanted in our biliary disease model can also be found in the CBD (Figure S6F), suggesting that hBECs may partially rescue the damage in the model via repairing the stricturing or providing structural support.
hBECs can be isolated and expanded using GMP-compliant conditions
Alongside our current conventional hBEC isolation protocol, we have developed an isolation and culture process that is compliant with current good manufacturing process (GMP) regulation. This standard research procedure has been upscaled and adapted for use on larger sections of human liver, facilitating an increased yield of GMP-compliant hBECs (Figure S7).
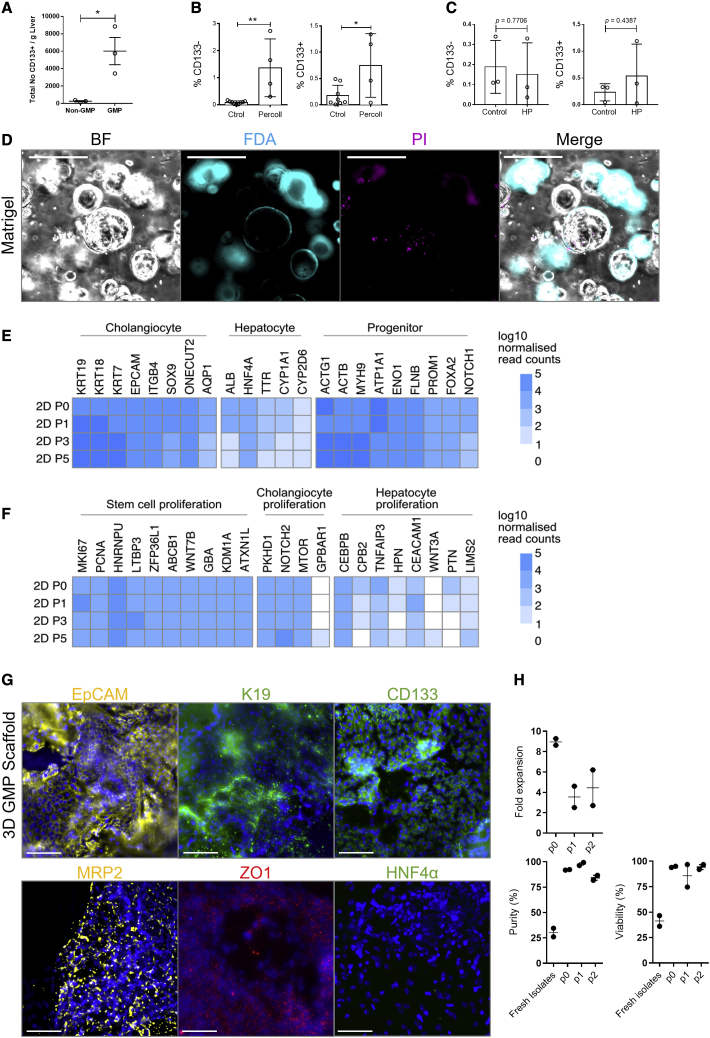
The process uses automation steps and GMP-compliant reagents for liver disaggregation, followed by clinical-grade magnetic bead sorting. Cells were isolated in GMP-compliant conditions, and the total number of CD133+ hBECs were quantified per gram of fresh human liver. The number of total CD133+ hBECs significantly increased in comparison with the standard research protocol from a mean of 260.33 hBECs (mean ± SEM) to 6,015.33 hBECs per gram of liver (p = 0.0213) (Figure 6A).
Figure 6.
hBECs can be isolated and expanded in vitro in GMP-compliant conditions
(A) Total number of CD133+ hBECs per gram of liver isolated in non-GMP- and GMP-compliant conditions. ∗ denotes p < 0.05 (mean ± SEM), Student’s t test (n = 3).
(B) Total percentage of CD133− and CD133+ hBECs isolated using the standard protocol (Ctrol) or a protocol including Percoll gradient isolation. ∗∗ denotes p < 0.005 (mean ± SEM), Student’s t test (n = 4).
(C) Total percentage of CD133− and CD133+ hBECs isolated using fresh livers (Control) or livers perfused under hypothermic conditions. Student’s t test (n = 3).
(D) CD133+ hBECs isolated in GMP-compliant conditions can be cultured in vitro as organoids in Matrigel spheres. From left, right: bright field depicting morphology of hBECs, FDA (cyan) showing live cells, propidium iodide (PI) (magenta) showing dead cells. Far right: merge. Scale bars, 50 μm.
(E) Heatmap of normalized expression values across genes associated with cholangiocytes, hepatocytes, or progenitor cell populations. These markers were analyzed in hBECs isolated from two donor livers maintained in 2D (GMP-compliant) in vitro culture conditions. P0 represents freshly isolated hBECs. Darker blue represents higher normalized gene expression.
(F) Heatmap of normalized expression values across genes associated with stem cell, cholangiocyte, and hepatocyte proliferation.
(G) hBECs cultured in 3D (GMP-compliant) scaffolds express markers of mature cholangiocytes (EpCAM, K19, CD133, MRP2, and ZO1) but not mature hepatocytes (HNF4α) (n = 5). Scale bars, 100 μm.
(H) Growth kinetics of isolated hBECs from donor livers were cultured on GMP-compliant conditions through a series of passages. Doubling time, purity, and viability of hBECs from isolation to passage 2 (p2) (N = 2).
Further sequential improvements of the isolation procedure (including incorporating Percoll gradients; Kegel et al., 2016), significantly increased the number of CD133+ and CD133− hBECs and reduced cell debris (Figure 6B). Alternative methods for the preservation of the liver, such as ex situ hypothermic oxygenated machine perfusion (Schlegel et al., 2019; Dutkowski et al., 2014), did not affect the total number of CD133− and CD133+ hBECs isolated (Figure 6C), suggesting that although this method preserves the overall function of the liver, it does not modify the preservation conditions necessary for the maintenance of hBECs.
We then confirmed the ability of GMP-compliant CD133+ hBECs to form colonies and expand in vitro. When plated in Matrigel, GMP-compliant CD133+ hBECs were able to expand and remained viable (Figure 6D). Our transcriptomic analysis suggests that the phenotype of hBECs is also maintained during culture in two-dimensional (2D) GMP-compatible conditions. Expression of mature cholangiocyte markers is sustained over the course of time while maintaining a progenitor-like phenotype. hBECs do not acquire a hepatocyte phenotype in these conditions, although a small level of HNF4α expression at gene level is noted (Figure 6E). Similarly, over the course of passages, hBECs cultured in different conditions retain markers of stem cell and cholangiocyte proliferation, with minimal expression of hepatocyte proliferation markers (Figure 6F). In three-dimensional (3D) GMP-compliant in vitro culture, hBECs retain markers associated with cholangiocyte expression (EpCAM, K19, CD133, MRP2, ZO1) (Figure 6G), suggesting that GMP-compliant hBECs retain their phenotype over the course of time, independent of in vitro culture conditions. We have also analyzed the stability of hBECs cultured in GMP-compliant 2D conditions through a series of passages. Doubling time was calculated at each passage after seeding onto a plate and indicated rapid expansion in the first culture phase, which reduced in subsequent passages. hBEC purity increased significantly after the first culture phase and, along with viability, remained consistently high in subsequent passages (Figure 6H).
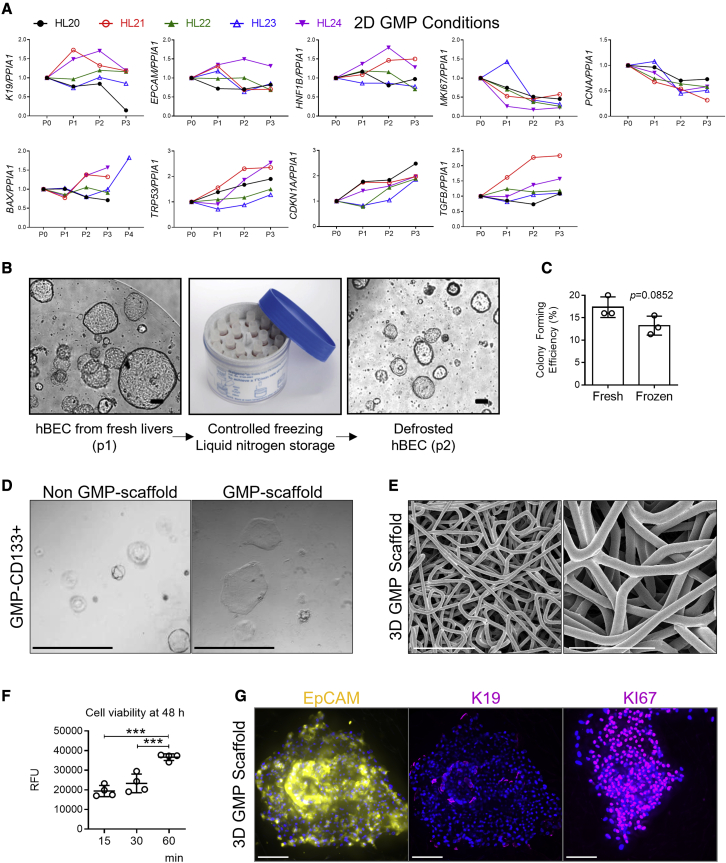
As a decrease in proliferation of hBECs was observed during culture in GMP-compatible conditions, we characterized the expression of known markers of proliferation (MKI67, PCNA) and cellular senescence (TRP53, CDKN1A). hBECs cultured in GMP-compliant conditions progressively decreased expression of markers of proliferation while increasing markers of senescence, suggesting an exhaustion of proliferative capacities. However, the hBEC phenotype (assessed by K19, EpCAM, and HNF1B expression) was maintained over the course of passages (Figure 7A).
Figure 7.
GMP-compatible conditions: Stability and freezing
(A) Gene expression of genes associated with mature cholangiocytes (K19, EpCAM, HNF1B), proliferation (MKI67, PCNA), induced apoptotic cell death (BAX), and cellular senescence (TRP53, CDKN1A, and TGFB) normalized to PPIA1 for hBECs isolated from 5 human donor livers (HL20 to HL24) over the course of three passages in 2D GMP-compliant scaffolds. Data normalized to freshly isolated hBECs.
(B) CD133+ cells isolated from fresh livers (here depicted at passage 1) can be stored long term in liquid nitrogen and later cultured again (here depicted at passage 2) as organoids in non-GMP-compliant conditions (n = 4 biological replicas). Scale bar, 100 μm.
(C) Colony-forming efficiency of fresh and frozen hBECs show no significant differences. Student’s t test (n = 3).
(D) CD133+ hBECs isolated in GMP-compliant conditions can be cultured as organoids in non-GMP conditions in Matrigel spheres (left). They can be cultured as well in two-dimensional GMP-compliant conditions (right). Both images represent passage 2 after isolation from fresh livers (n = 4 biological replicas). Scale bar, 60 μm.
(E) Polycaprolactone scaffolds electron microscopy. Scale bars, 100 μm (left), 50 μm (right).
(F) hBECs isolated in GMP-compliant condition viability after 48 h culture in polycaprolactone scaffolds. y axis indicates time allowed for hBEC seeding in the scaffolds. y axis indicates RFU (relative fluorescence units). ∗∗∗ denotes p < 0.001 (mean ± SEM), one-way ANOVA (n = 4).
(G) hBECs isolated in GMP-compliant conditions and cultured for 1 week in polycaprolactone scaffolds retain cholangiocellular markers (EpCAM and K19) and show signs of proliferation as seen by KI67 immunofluorescence (n = 3 biological replicas). Scale bars, 100 μm.
Sections of the discarded livers can be frozen and hBECs can be effectively isolated after long-term storage with similar colony-formation efficiency (Figures 7B and 7C). Alternatively, expanded hBECs can be stored in liquid nitrogen for an indefinite period and thawed to be used in culture again (Figure 7C), suggesting that hBECs can be maintained for long periods of time.
Finally, CD133+ hBECs can be cultured and expanded in vitro in 2D (Figure 7D) or 3D printed GMP-compliant scaffolds (Figure 7E) that maintained the cell viability (Figure 7F) while proliferating and expressing hBEC-related markers (Figure 7G). Therefore, GMP-compliant hBECs can be isolated in a highly efficient manner and expanded in vitro. These results suggest that both liver sections and hBEC cultures can be isolated, expanded, and stored in GMP conditions and can be used as a source of hBECs ready to apply in clinical settings when required.
Discussion
Biliary disease encompasses conditions that cause cholangiocyte pathology (Lazaridis et al., 2004; Nakanuma, 2012), including PBC (Carey et al., 2015), PSC (Dyson et al., 2018), biliary atresia (Hartley et al., 2009), Alagille syndrome (Saleh et al., 2016), and ischemic cholangiopathies (Nakanuma, 2012; Deltenre and Valla, 2006) arising after liver transplantation. Biliary disease is characterized by cholangiocyte loss, fibrosis, cholestasis, and ductopenia, eventually leading to liver failure.
Despite the worldwide increasing incidence of these pathologies and their substantial morbidity, mortality, and associated costs, current treatment options are limited. Current therapeutics only slow the progression of the disease and do not repair the already-damaged structures. Therefore, many patients eventually require a liver transplant as a life-saving procedure. The international shortfall between available donor organs and the number of patients requiring a transplant is significant, meaning patients often deteriorate and succumb while awaiting a transplantation, and current predictions indicate that this trend will continue (Ryckman et al., 2008). Therefore, alternatives to transplantation are required.
Regenerative cellular therapy with the potential to repair the injured biliary tree and relieve the symptoms offers an interesting option for the treatment of these patients either as a curative approach or as a bridge to organ transplantation. BECs, regardless of subpopulation and origin, have demonstrated great potential as in vitro systems for the culture and differentiation of liver cells (e.g., human umbilical vein-derived hepatic progenitor cells [Inada et al., 2020] and Lgr5+ primary adult liver stem cells [Huch et al., 2015]), among others. Furthermore, BECs have demonstrated therapeutic potential as a transplantable therapy in several models of liver disease, as recently exemplified (Sampaziotis et al., 2021).
Here, we show evidence of the regenerative potential of CD133+ hBECs, which, upon transplantation, are able to mediate repair and restore functionality of damaged biliary tracts, offering a potential alternative to alleviate the burden of biliary disease.
We have adapted our previous experience in the isolation of murine hepatic progenitor cells within the BEC compartment (Lu et al., 2015) to human livers. These donated livers were discarded because (1) the organ is too steatotic or otherwise damaged to be considered for transplantation or (2) the transplantation is not able to proceed for various technical reasons. This technique is therefore maximizing the utilization of this critical resource.
hBECs, characterized by EpCAM+ CD24+ CD133−/+ expression, can be isolated from all discarded livers but are particularly abundant in steatotic livers. These data are in accordance with human biopsy data that correlates the presence of biliary expansion (ductular reaction) with the progression of non-alcoholic fatty liver disease (Richardson et al., 2007).
Our RNA-seq analysis provides further insight into the phenotypic and functional differences between hBEC populations in healthy and steatotic conditions. Moreover, our data demonstrate that CD133+ hBECs have significantly higher colony-forming efficiency, as well as survival capacity, in comparison with CD133− hBECs, suggesting that despite heterogeneity within the biliary epithelial cell compartment (Strazzabosco and Fabris, 2008; Kanno et al., 2000), the CD133+ population offers a higher regenerative potential.
Isolated CD133+ hBECs of biliary origin are characterized by markers associated with liver progenitor cells showing expression of key regenerative genes such as DACT1, SOX9 (Yin, 2017; Kawaguchi, 2013), and FOXA (Li et al., 2009; McDaniel et al., 2017) as well as upregulation of biological pathways associated with stem cell pluripotency, proliferation, and repair. Furthermore, hBECs are characterized by the significant upregulation of TGFβ signaling pathway components, which have been previously shown to play a pivotal role in liver and biliary regeneration (Ferreira-Gonzalez et al., 2018).
We have tested the regenerative potential of hBECs in a murine model of biliary disease, characterized by the presence of cellular senescence, inflammation, fibrosis, and stricturing of the biliary tract. Upon transplantation, hBECs decreased mortality and levels of fibrosis while improving biochemical liver function as assessed by the levels of transaminases.
Furthermore, hBECs engrafted in the immediacies of the damaged biliary tracts and repaired biliary architecture, reducing the levels of activated hepatic stellate cells and HMGB1-mediated inflammation. The transplantation and engraftment of the human cells triggered a recipient ductular response with expansion of host ductular cells, suggesting a potential paracrine effect of the transplanted cells. This was associated with an increase in inflammatory cells adjacent to the host ductular response.
hBEC transplantation engrafted near the damaged biliary system, mediated resolution of extrahepatic biliary strictures, and improved the overall function of the liver, suggesting that the hBEC regenerative effects are not restricted to the intrahepatic biliary tract. Locating hBECs within the extrahepatic biliary system may help provide additional mechanistic insight into the regenerative potential of hBECs, whether that be structural restoration of a damaged biliary tract or intrahepatic pro-regenerative paracrine effects.
Our current clinical translation protocol demonstrates that GMP-compliant hBECs can be isolated in a clinically relevant procedure and can be further expanded in vitro. These results suggest that liver sections can be frozen for later processing and that hBECs can be isolated and expanded in GMP-compliant conditions and subsequently cryopreserved for later use. These cryopreserved cells retain function after thawing, providing strong evidence that hBECs could be manufactured and supplied for therapeutic use in clinical trials for relevant conditions, and offers a previously unappreciated cellular therapeutic approach for treatment of liver disease.
In summary, we have identified a subpopulation of human liver BECs that can be isolated from damaged organs and cultured long term without phenotypic drift under strict GMP-compatible conditions. Following transplantation, there is a significant improvement in animal survival, fibrosis, inflammation, and biochemical liver function through engraftment of the cells in the proximities of the biliary tracts. This engraftment mediates a pro-regenerative effect on the native cells that promotes repair and resolution of liver damage. This demonstration of a human cell therapy leading to the rescue of animals from biliary disease offers future clinical therapeutic opportunities for the use of these cells as a regenerative therapy for biliary disease.
Limitations of the study
hBECs represent a heterogeneous population, with the bipotential ability to become hepatocytes or cholangiocytes depending on the transplanted niche. Our model presented here focused on the hBEC biliary phenotype. However, further comprehensive studies of bipotential state are needed. Future studies will require the use of different murine models of hepatic and biliary damage to understand the behavior of hBECs in each scenario. If, upon transplantation in a model of hepatic damage, hBECs are able to engraft, repopulate, and repair the injury, this would suggest the use of hBECs as a potential cell therapy for types of liver disease other than biliary disease.
The results presented here are an experimental approach of a potential human therapy for biliary disease. Therefore, adapting this procedure to the clinical practice will need to define the optimal route of injection. Although hepatic artery, portal vein, and the biliary system are all possible routes for cell transplant, each one of them will have unique challenges to overcome in the context of an injured liver.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD31 Ax488 (for FACS isolation) | Biolegend | Cat#303109 (WM59); RRID:AB_493075 |
| CD45 Ax488 (for FACS isolation) | Biolegend | Cat#103121 (30-F11); RRID:AB_493532 |
| EpCAM BV650 (for FACS isolation) | Biolegend | Cat#324226 (9C4); RRID:AB_2562735 |
| CD24 BV421 (for FACS isolation) | Biolegend | Cat#311122 (ML5); RRID:AB_2561691 |
| CD133 APC (for FACS isolation) | Miltenyi Biotech | Cat#130-098-829 (AC133); RRID:AB_2660883 |
| αSMA | Sigma-Aldrich | Cat#A2547; RRID:AB_476701 |
| AE2 | Abcam | Cat#Ab140953 |
| ALB | Abcam | Cat#Ab2406; RRID:AB_303048 |
| CD45 | R&D | Cat#MAB114; RRID:AB_357485 |
| CK19 | Novocastra | Cat#NCL-CK19: RRID:AB_563799 |
| EpCAM | Abcam | Cat#Ab32392; RRID:AB_732181 |
| EpCAM (mouse specific) | Abcam | Cat#Ab221552 |
| CD133 | Abcam | Cat#Ab19898; RRID:AB_470302 |
| CD24 | Biolegend | Cat#101803; RRID:AB_312836 |
| GFP | Abcam | Cat#Ab13970; RRID:AB_300798 |
| HMGB1 | Abcam | Cat#Ab18256; RRID:AB_444360 |
| HNF4α | R&D | Cat#PP-H1415-00; RRID:AB_2263954 |
| Human Mitochondria | Merck | Cat#MAB1273; RRID:AB_94052 |
| Ki67 | Abcam | Cat#Ab16667; RRID:AB_302459 |
| LGR5 | Abcam | Cat#Ab75732; RRID:AB_1310281 |
| MRP2 | Thermo Fisher Scientific | Cat#MA532687; RRID:AB_2809964 |
| PCNA | Abcam | Cat#Ab29; RRID:AB_303394 |
| P53 | Abcam | Cat#Ab26; RRID:AB_303198 |
| P21 | Santa Cruz | Cat#SC-471; RRID:AB_632123 |
| STEM121 | Takara | Cat#Y40410; RRID:AB_2801314 |
| Sox9 | Merck | Cat#AB5535; RRID:AB_2239761 |
| ZO-1 | Santa Cruz | Cat#SC-33725; RRID:AB_628459 |
| Biotinylated Anti-Rabbit IgG (H+L) (secondary Ab) | Vector Lab. | Cat#BA-1000; RRID:AB_2313606 |
| Biotinylated Anti-Mouse IgG (H+L) (secondary Ab) | Vector Lab. | Cat#BA-9200; RRID:AB_2336171 |
| Biotinylated Anti-Rat IgG (H+L) (secondary Ab) | Vector Lab. | Cat#BA-9400; RRID:AB_2336202 |
| Anti-Mouse IgG(H+L)-555 (secondary Ab) | Thermo Fisher Scientific | Cat#A31570; RRID:AB_2536180 |
| Anti-Rabbit IgG(H+L)-488 (secondary Ab) | Thermo Fisher Scientific | Cat#A21206; RRID:AB_2535792 |
| Anti-Rabbit IgG(H+L)-555 (secondary Ab) | Thermo Fisher Scientific | Cat#A31572; RRID:AB_162543 |
| Anti-Rat IgG(H+L)-555 (secondary Ab) | Thermo Fisher Scientific | Cat#A21434; RRID:AB_141733 |
| Mouse IgG (Isotype Control) | Vector Lab. | Cat#I-2000; RRID:AB_2336354 |
| Rat IgG (Isotype Control) | Vector Lab. | Cat#I-4000; RRID:AB_2336356 |
| Rabbit IgG (Isotype Control) | Vector Lab. | Cat#I-1000 RID:AB_2336355 |
| Biological samples | ||
| Human livers unsuitable for donation preserved at 4°C in University of Wisconsin preservation fluid | NHS Organ Donation | Project No. 2015/0408 (LREC) 15/SS/0218 |
| Chemicals, peptides, and recombinant proteins | ||
| Belzer MPS UW Machine Perfusion Solution | Bridge-to-life | Cat#BTLBUW-1000 |
| Formaldehyde | VWR | Cat#9713.901 |
| Methanol | Fisher Scientific | Cat#M/3900/17 |
| Isopropanol | VWR | Cat#20842.323 |
| Chloroform | Sigma-Aldrich | Cat#C2432 |
| Glacial Acetic acid | Fisher Scientific | Cat#10365020 |
| KCl | GIBCO | Cat#529552 |
| NaCl | Sigma-Aldrich | Cat#S5886 |
| EDTA | Sigma-Aldrich | Cat#E6758 |
| EGTA | Sigma-Aldrich | Cat#E4378 |
| Trizma® base | Sigma-Aldrich | Cat#93362 |
| Triton X | Sigma-Aldrich | Cat# T8787 |
| Tween-20 | Sigma-Aldrich | Cat#P1379 |
| Phosphate Buffer Saline (PBS) | Sigma-Aldrich | Cat#D8537 |
| Hanks Balanced Salt Solution | GIBCO | Cat#14025-050 |
| Advanced DMEM/F-12 | GIBCO | Cat#12634010 |
| StemMACS MSC Culture medium | Miltenyi Biotec | Cat#130-091-680 |
| Liver Digest Media | GIBCO | Cat#17703034 |
| Liver Perfusion Media | GIBCO | Cat#17701038 |
| Penicillin /Streptomycin | GIBCO | Cat#15140-122 |
| Glutamine | GIBCO | Cat#25030-024 |
| Bovine Fetal Calf Serum | GIBCO | Cat# 10270106 |
| L-Glutamine | GIBCO | Cat# 25030081 |
| Versene 1:5000 (1x) | Thermo Fisher Scientific | Cat# 15040-033 |
| TryPLE Express | GIBCO | Cat#12604-013 |
| Dispase | STEMCELL technologies | Cat#07913 |
| Liberase MNP-S GMP grade | Roche | Cat#06297790001 |
| Percoll | Sigma-Aldrich | Cat#P1644 |
| CellBanker 2 | Amsbio | Cat#11891 |
| HEPES pH7 | GIBCO | Cat#15630-056 |
| A-83-01 | Sigma-Aldrich | Cat#SML0788 |
| B27 Supplement | GIBCO | Cat#12587010 |
| N2 Supplement | GIBCO | Cat#15630-056 |
| Forskolin | Tocris | Cat#1099 |
| Nicotinamide | Sigma-Aldrich | Cat#N0636 |
| Human EGF | Peprotech | Cat#AF-100-15-100 |
| Human FGF-10 | Peprotech | Cat#100-26-100 |
| Human HGF | Peprotech | Cat#100-39 |
| Human BMP-7 | Peprotech | Cat#120-03 |
| Human FGF-19 | Peprotech | Cat#100-32 |
| rhWnt-3a | R&D | Cat#5036-WN/CF |
| Gastrin | Tocris | Cat#3006 |
| N-Acetylcysteine (NAC) | Sigma-Aldrich | Cat#A9165 |
| Dexamethasone | Sigma-Aldrich | Cat#D2915 |
| Human R-Spondin 1 | Peprotech | Cat#120-38 |
| Y27632 | Sigma-Aldrich | Cat#Y0503 |
| Noggin | Peprotech | Cat#120-10c |
| DAPT | Sigma-Aldrich | Cat#D5942 |
| Corning Matrigel GFP Basement Membrane Matrix | Corning | Cat#354230 |
| Biolaminin 521 LN | BioLamina AB | Cat#LN521 |
| Polycaprolactone (avg. Mn. 80000) | Sigma-Aldrich | Cat#440744 |
| Human Platelet Lysate | STEMCELL technologies | Cat#06960 |
| Tamoxifen | Sigma-Aldrich | Cat#T5648 |
| Sunflower seed oil | Sigma-Aldrich | Cat#S5007 |
| Carprofen | Pfizer | Cat# |
| Ketamine | Pfizer | Cat# |
| Medetomidine | Orion Pharma | Cat# |
| Heparin Sodium Salt | Sigma-Aldrich | Cat# H3149 |
| Glutaraldehyde | Sigma-Aldrich | Cat#g6257 |
| Protein Block | Abcam | Cat#Ab64226 |
| Halt Protease Inhibitor | Thermo Fisher Scientific | Cat#78430 |
| BLOXALL block (peroxidase-alkaline phosphatase) | Vector Lab. | Cat#Sp-6000 |
| Avidin/Biotin block | Thermo Fisher Scientific | Cat#004303 |
| Vectastain ABC reagent R.T.U. | Vector Laboratories | Cat#PK-7100 |
| DAB+ Chromogen System | Dako | Cat#K3468 |
| Picric Acid | Sigma-Aldrich | Cat#P6744 |
| Fast Green | Sigma-Aldrich | Cat#F7258 |
| Direct Red | Sigma-Aldrich | Cat#365548 |
| Oil Red O | Sigma-Aldrich | Cat#O0625 |
| Fluorescein Diacetate (FDA) | Thermo Fisher Scientific | Cat#F7378 |
| Propidium Iodide | Sigma-Aldrich | Cat#P4170 |
| Methylene Blue | Sigma-Aldrich | Cat#M4159 |
| DAPI | Sigma-Aldrich | Cat#D9542 |
| Fluoromount-G, with DAPI | Thermo Fisher Scientific | Cat#00-4959-52 |
| DPX mountant media | Sigma-Aldrich | Cat#06522 |
| CliniMACS CD133 microbeads | Miltenyi Biotec | Cat#172-01 |
| Critical commercial assays | ||
| BCA protein assay | Thermo Fisher Scientific | Cat#23227 |
| MycoAlert Assay Control set | Lonza | Cat#LT07-518 |
| SYTOX-AAAdvanced Dead Cell Stain Kit | Thermo Fisher Scientific | Cat# S10349 |
| Cell Titer-Blue | Promega | Cat#G8080 |
| Human Albumin ELISA kit | Alpha Diagnostic | Cat#1190 |
| SMARTer Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian | Takara Bio | Cat#634411 |
| DNA HS kit | Agilent | Cat#23225 |
| Poly-A mRNA magnetic isolation module | NEB | Cat#E7490 |
| NEBNEXT Ultra II Directional RNA Library Prep Kit | NEB | Cat#E6440 |
| RNeasy Micro kit | QIAGEN | Cat#74004 |
| RNeasy Mini kit | QIAGEN | Cat#74106 |
| QuantiTect Reverse Transcription Kit | QIAGEN | Cat#205313 |
| QuantiFast SYBR Green PCR kit | QIAGEN | Cat#204056 |
| V-PLEX Proinflammatory Panel 1 mouse kit | Meso Scale Diagnostic | Cat#K15048D-1 |
| V-PLEX Cytokine Panel 1 Mouse Kit | Meso Scale Diagnostic | Cat#K15245D |
| AMPure XP beads | Beckman Coulter | Cat#A63880 |
| Deposited data | ||
| hBEC seq analysis | This paper | GEO: GSE155498 |
| Experimental models: Cell lines | ||
| Human Mesenchymal Stem Cells (hMSC) isolated from liposuction waste and visceral adipose tissue | Scottish National Blood Transfusion service | N/A |
| Experimental models: Organisms/strains | ||
| Mouse line: K19CreERTMDM2fl/fl Rag2−/−Il2rg−/− |
This paper | N/A |
| Oligonucleotides | ||
| Primers for mouse genotyping | This paper | Table S2 |
| QIAGEN primers for RT-qPCR (Human) | This paper | Table S6 |
| QIAGEN primers for RT-qPCR (Mouse) | This paper | Table S6 |
| Software and algorithms | ||
| FlowJo (v10.7.1) | Becton Dickinson | N/A |
| Columbus Image Analysis | Perkin Elmer | N/A |
| TIBCO Spotfire software | Tibco software Inc | N/A |
| FastQC (v.0.11.9) | Andrews, 2010 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc |
| MultiQC (v.1.3.dev0) | Ewels et al., 2016 | N/A |
| Cutadapt (v.1.16) | Martin, 2011 | N/A |
| STAR (v.2.7.1a) | Dobin et al., 2013 | N/A |
| Rsubread Bioconductor package (v.2.0.1) | Liao et al., 2019 | N/A |
| DESeq2 (v.1.26.0) | Love et al., 2014 | N/A |
| GSEA PreRanked (v.4.0.3) | Subramanian et al., 2005 | N/A |
| MSigDB Hallmark Gene Set | Liberzon et al., 2015 | |
| Gene Ontology project | Ashburner et al., 2000 | http://geneontology.org/ |
| AmiGO tool (v.2.5.13) | Carbon et al., 2009 | http://amigo.geneontology.org/amigo |
| ArrayExpress (E-MTAB-8495) | Sampaziotis et al., 2021 | N/A |
| OSCA Bioconductor workflow | Amezquita et al., 2020 | N/A |
| EmptyDrops | Lun et al., 2019 | N/A |
| Q-Imaging, Image Pro premier software | Media Cybernetics | QCAM version |
| Fiji ImageJ | GNU General Public License | https://imagej.net/software/fiji/ |
| Prism software version 5.0a | GraphPad | https://www.graphpad.com/support/prism-5-updates/ |
| Other | ||
| Tibbs Cannulas | DTR Medical | Cat#TAC20SO |
| Vicryl 6/0 | Ethicon | Cat#W9981 |
| GentleMACS C-tubes | Miltenyi Biotec | Cat#130-093-237 |
| Disposable sterile scalpels | Swann-Morton | Cat#0503 |
| Cell strainer 70 um | Fisher Scientific | Cat#22363548 |
| 12 well Suspension Culture plate | Greiner Bio-One | Cat# 665102 |
| Corning Cell Culture Flasks | Corning | Cat#CLS431081 |
| RM1 + 0.1% DDC (P) | Special Diets Services | Cat#824943 |
| Rat and Mouse No. 1 Maintenance (RM1) diet | Special Diets Services | Cat#801151 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stuart J Forbes (stuart.forbes@ed.ac.uk).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Experimental model and subject details
Clinical material
Human livers were initially accepted for transplantation and procured with the intent to transplant by a team of the UK National Retrieval Service. Following procurement, the liver grafts were deemed unsuitable by the consultant surgeon and declined by all UK liver transplant centers. The most common reasons for declining were logistical, excessive fat content, fibrosis or poor function. All grafts were initially preserved in University of Wisconsin preservation fluid at 4°C. Sample size and donor liver characteristics are stated in Table S1.
Livers were offered via NHS Organ Donation and Transplantation national research offer process. Ethical approval for the use of human livers was received from Lothian Research and Ethics Committee (LREC) reference number 15/SS/0218, Lothian Research and Development (Project No. 2015/0408), National Health Service Blood and Transplant (NHSBT) ethics committee and Research Innovation and Novel Technologies Advisory Group (RINTAG). Organ procurement and research utilization of the livers was undertaken in accordance with the United Kingdom’s Human Tissue Act (Scotland, 2006) and registered with NHS Lothian Tissue Governance.
Animal models
K19CreERTMdm2fl/fl mice were bred with Rag2−/− Il2rg−/− constitutive knockout (Taconic, 411-F, 4111-M) producing the K19CreERTMdm2fl/flRag2−/− Il2rg−/− murine line. The animals used in this study are C57BL/6 background, mix of males and females aged within 12–24 weeks at the start of the experiments. All animal genotyping was outsourced commercially to Transnetyx, Inc (TN, USA) using the primers stated.
Mice were housed in a specific pathogen-free environment in open cages (NKP, M3-sloping front) with Aspen chips as bedding at 21 °C. Mice were kept under standard conditions with a 14-h day cycle and access to food (irradiated RM3P) and water ad libitum. Power calculations are not routinely performed. However, animal numbers were chosen to reflect the expected magnitude of response taking into account the variability observed in previous experiments. Mice were randomly allocated to each experimental group and males/females were equally distributed. All animal experiments were carried out under procedural guidelines and severity protocols within the UK, with ethical permission from the Animal Welfare and Ethical Review Body (AWERB) and the Home Office (UK). Primers for mouse genotyping are stated in Table S2.
Method details
Human liver hypothermic perfusion
Upon arrival, livers were split and the left lateral section (Couinaud Segments II and III) was flushed with PBS and transferred to the lab for cell isolation as controls for the perfused segments. The extended right liver grafts (segments I, IV-VIII) were prepared for perfusion by flushing the portal vein with 1000 mL of cold (0-4°C) Belzer MPS UW Machine Perfusion Solution (Bridge-to-life, Ltd). When the caval effluent was clear the grafts were connected to the Liver Assist device (Organ Assist, NL) for two hours of ex-situ hypothermic machine perfusion, allowing dual oxygenated perfusion via the portal vein and the hepatic artery using two centrifugal pumps to provide a continuous venous flow and a pulsatile arterial flow at 60 bpm (500 mL/min of 100% oxygen on each of the two membrane oxygenators). The system is pressure controlled allowing autoregulation of the flow through the liver, limited to a mean of 25 mmHg for the hepatic artery and 5 mmHg for the portal vein. After two hours of machine perfusion the grafts were removed from the device and used for subsequent analysis.
Human liver processing and cryopreservation
Human livers and perfused grafts were dissected, with samples of common bile duct, gallbladder and lobes. Samples were taken for histological preservation in 4% formaldehyde, methacarn (60% Methanol, 30% Chloroform, 10% Glacial Acetic Acid) or snap frozen on dry ice.
Liver lobes were diced into 0.5 cm3 pieces, mixed with cryopreservation solution (CellBanker 2, Amsbio) and stored in cryovials at −80°C for 72 h, then transferred to liquid nitrogen for long term storage.
hBEC isolation
Cryopreserved sections or fresh samples of tissue were defrosted at 37°C in a water bath before mechanical digestion (minced to 1 mm3 pieces), followed by enzymatic digestion for 30 min at 37°C in a solution of Collagenase (2 μg/mL, GIBCO)-Dispase (2 μg/mL, GIBCO)-Bovine Fetal Calf Serum (2% FCS, GIBCO) in Advanced DMEM/F-12 (GIBCO) supplemented with 1% Penicillin/Streptomycin (Pen/Strep, GIBCO) and 1% L-Glutamine (GIBCO). When the intrahepatic bile ducts could be observed under the microscope, the sample was centrifuged at 100xg for 5 min, and incubated in Versene (Thermo Fisher Scientific) at 37°C for 1 h. Dissociated cells were filtered through a 40 μm cell strainer and washed in Advanced DMEM/F-12. Cells were resuspended in Advanced DMEM/F-12 and underlayed with an equal volume of 20% and 50% (v/v) Percoll (Sigma). Following centrifugation at 1800xg for 30 min at 4°C, the hBEC rich fraction at the interface of the 20% and 50% Percoll layers was collected, washed and re-suspended in Phosphate Buffer Saline (PBS, GIBCO) with 2% FCS and 100 mM EDTA (Sigma-Aldrich) for antibody staining.
Cells were incubated with primary antibodies. Samples were stained with SYTOX-AADvanced Dead Cell Stain Kit (Thermo Fisher Scientific) and antibody-defined populations sorted with a BD FACSAria Fusion (BD Biosciences).
hBEC antibodies for FACS isolation are stated in Table S3.
To determine the CD133-positive phenotype of the cells over the course of time in culture, cells were stained with anti-human-CD133-phycoerythrin (PE), anti-human-EpCAM-BrilliantViolet650 (BV), anti-human-CD24-BV421, anti-human-CD31-AlexaFluor488 (AF488) and anti-human-CD45-AF488 antibodies with DRAQ7 used to exclude dead cells from analyses. All appropriate single stains and fluorescence minus one (FMO) controls were also performed and cells were acquired on a BD LSR Fortessa flow cytometer. Analysis was performed using FlowJo software (V10.7.1) with cell populations gated using dotplots. Cells were initially gated to exclude debris based on SSC-A (side scatter-area) versus FSC-A (forward scatter-area), single cells identified by FSC-H (forward scatter-height) versus FSC-A and the live cell population classified by gating for the DRAQ7 negative population. The AF488 negative population was gated for to exclude CD31+ and CD45+ cells.
hBEC isolation at large scale from fresh liver
One cut surface (lobes VII and VIII) was cut from fresh human livers and large veins and arteries cannulated with Tibbs cannulas (DTR Medical) and sutured in place using 6/0 Vicryl (Ethicon). Cannulated liver was placed on a perfusion chamber (Argyll innovations, Edmonton) and perfused with Liver warm Digest Media (LDC, GIBCO) for 2 h at 50ml/min using a peristaltic pump. LDC was maintained at 37°C with a blood warmer Protherm II (Biegler). Tissue was disaggregated with forceps and transferred to gentleMACS C-tubes (Miltenyi Biotec) for subsequent procedures.
GMP-conformant isolation of hBEC
Disposable sterile scalpels (Swann-Morton) were used to mince the human livers into 2-3 mm3 chunks. Using a set 1.5 h program (37C_h_TDK_1cus) on the GentleMACS Octo Dissociator (Miltenyi Biotec), liver chunks were further digested by transferring 2-3 g of liver into gentleMACS C-tubes (Miltenyi Biotec) in 15 mL of Hanks’ Balanced Salt Solution (Sigma-Aldrich) with 1.2 Wünsch units Liberase MNP-S GMP grade (Roche) at 37°C. After digestions, samples were diluted 6 times with cold PBS + 5 mM EDTA and passed through a 70 μm cell strainer (Corning). Samples were spun at 300xg for 10 min, supernatant aspirated and remaining cells labeled with clinical grade CliniMACS CD133 microbeads (Miltenyi Biotec). CD133-expressing cells were positively selected by passing cells through a LS column on a MidiMACS magnetic field (Miltenyi Biotec). Retained cells were considered CD133-positive hBEC. Efficiency of isolation was determined using flow cytometry.
hBEC culture
Sorted hBEC were grown as organoids by plating onto suspension culture plates (Greiner Bio-One) in hemispheres of growth factor reduced Matrigel (Corning) and grown in hBEC Expansion Medium. For the first three days of culture medium was supplemented with 100 ng/mL Noggin and 100 ng/mL Wnt, as previously described (Huch et al., 2013, 2015). Medium was changed every three days and cells passaged by mechanical disruption once confluent every 7 to 10 days. Cells were routinely tested for mycoplasma contamination using MycoAlert Assay Control set (Lonza).
For GMP-compliant culture cells were cultured in 2D in 6 well plates covered in 1 mg/mL Laminin 1 (Biolamina).
For long-term storage, organoids were washed twice with Advanced DMEM/F-12 (GIBCO) to remove Matrigel, mixed with cryopreservation solution (CellBanker 2, Amsbio) and stored in cryovials at −80°C for 72 h, then transferred to liquid nitrogen for long term storage. hBEC expansion medium reagents and concentrations are stated in Table S4.
hMSC culture
hMSC isolated from liposuction waste and visceral adipose tissue were provided by the Scottish National Blood Transfusion service as a frozen cell stock. Cells were defrosted and plated in Cell Culture Flasks (Corning) with MSC culture medium (Stem MACS, Miltenyi Biotec) supplemented with 5% Human Platelet Lysate (Stem Cell Technologies), 2 units/mL Heparin (Sigma-Aldrich) and 1% Pen/Strep (GIBCO). Cells were routinely tested for mycoplasma contamination using MycoAlert Assay Control set (Lonza).
hBEC culture in three-dimensional polycaprolactone scaffolds
Electrospun polycaprolactone (PCL, Avg. Mn: 80000, Sigma) scaffolds were produced using the IME EC-DIG electrospinning apparatus according to a previously described method (Burton et al., 2017). Scaffolds of 12 mm were then washed three times in 70% ethanol and freeze-dried overnight and then exposed to oxygen plasma using a plasma coater (Harrick Plasma). Scaffolds were placed into the plasma coating vacuum chamber and coated for 30 s at 500 mTorr and at medium RF power. Scaffolds were placed into PBS solution in 12 well plates.
hBEC in Matrigel (Corning) were collected from culture and washed three times with Advanced DMEM/F-12 supplemented with 1%Pen/Strep and 1% Glut, to eliminate Matrigel and then centrifuged at 350xg for 5 min. Pellets were seeded into the scaffolds and after 30 min covered with hBEC Expansion Medium. Cell viability was assessed using the CellTiter-Blue (Promega) as per manufacturer’s instructions. Briefly, the scaffolds were removed from the culture well and placed into 300 mL of a 1:5 mixture of CellTiter-Blue reagent and complete media respectively. The scaffolds were then incubated at 37°C and 5% CO2 in the reagent mixture for 2 h. After incubation, 100 mL of the supernatant reagent mixture was placed into a black 96-well microplate. Fluorescence at ex. 525nm em. 580-640nm was measured using a Modulus II Microplate reader and results normalized to a negative control.
PCL scaffolds were sputter coated for 1 min using a gold-palladium filament with an EmScope SC500A sputter coating device. Scaffolds were imaged using a Hitachi S4700 scanning electron microscope with an accelerating voltage of 15 kV.
hBEC single-cell colony forming assay
384 well plates were prefilled with 20 μl of Matrigel/well using the VIAFLO 96/384 semi-automated pipette (Integra BioSciences). Single hBEC were then individually sorted into each well using BD FACSAria Fusion cell sorter (BD Biosciences) and incubated in hBEC Expansion Medium. Plates were imaged on day 1 and every 7 days thereafter using the Operetta CLS High-Content Analysis System (Perkin Elmer). Data analysis was performed using Columbus Image Analysis (Perkin Elmer) and TIBCO Spotfire software (Tibco software Inc).
hBEC differentiation
hBEC were grown as organoids in hemispheres of growth factor reduced Matrigel (Corning) in hBEC expansion media for 5 days, washed with Advanced DMEM/F-12 and cultured in hBEC differentiation medium. Differentiation medium was changed every three days for 15 days. hBEC Differentiation Medium reagents and concentrations are stated in Table S5.
Enzyme-linked Immunosorbent assay (ELISA) detection of albumin
Conditioned medium from cultured cells was stored at −80C for further analysis. Stored medium was thawed at room temperature and tested for albumin concentration using Human Albumin ELISA kit (Alpha Diagnostic), following manufacturer’s protocol. Absorbance at 450 nm was read using a spectrophotometer (SPECTROstar, Omega).
RNA sequencing
For RNA-seq library preparation of the isolated hBECs presented in Figures 2E and 2F, 1ng of each total RNA sample was fragmented to a size appropriate for sequencing based on the level of degradation assessed using an Agilent Bioanalyser (Agilent), and cDNA was generated using the SMARTer Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian (Takara Bio). AMPure XP beads (Beckman Coulter) were then used to purify the cDNA library. Depletion of ribosomal cDNA was achieved using ZapR and R-probes. The library fragments originating from rRNA (18S and 28S) and mitochondrial rRNA (m12S and m16S) were cut by ZapR in the presence of R-probes (mammalian-specific). R-probes were hybridized to ribosomal RNA and mitochondrial rRNA sequences derived from the human mitochondrial genome and are therefore strictly human specific. Uncleaved fragments were then enriched by 15 cycles of PCR before a final purification using AMPure XP beads (Beckman Coulter). Completed libraries were quantified using the Qubit 2.0 Fluorometer and the Qubit dsDNA HS assay and assessed for quality and size distribution of library fragments using the Agilent Bioanalyser and the DNA HS kit (Agilent). Libraries were multiplexed in two equimolar pools and sequenced on two flow cells on an Illumina NextSeq 550 sequencer (Illumina) at the Edinburgh Clinical Research Facility (ECRF), Western General Hospital, Edinburgh, UK.
For library preparation presented in Figures 6E, 6F, S2F, and S2G, 500ng of each total RNA sample was used after quality and integrity were assessed using the Agilent Bioanalyser Instrument. Poly-A containing mRNA molecules were purified from total RNA using Poly-A mRNA magnetic isolation module (NEB) following manufacturer’s protocol. cDNA was then generated from mRNA fragments using the NEBNEXT Ultra II Directional RNA Library Prep Kit (NEB) and purified using AMPure XP beads. cDNA fragments were enriched by 11 cycles of PCR with unique dual indexes to allow multiplexed sequencing before a final purification using AMPure XP beads. Completed libraries were quantified as above. Libraries were combined in an equimolar pool of 9 and sequenced on a single P2 flow cell on an Illumina NextSeq 2000 sequencer at the ECRF.
RNA-seq analysis
Read quality of sequenced FASTQ files was assessed using FastQC (v.0.11.9) (Andrews, 2010) and MultiQC (v.1.3.dev0) (Ewels et al., 2016). For the isolated hBEC RNA-seq data, adaptor and poly-G sequences were trimmed using Cutadapt (v.1.16) (Martin, 2011) and low-quality bases (Phred < 20) were also trimmed; further quality assessment confirmed adaptor contamination of < 0.1% per sample. Sequence reads were aligned to the human reference genome (GRCh38.99) with STAR (v.2.7.1a) (Dobin et al., 2013). Data was imported into R and reads counted using the featureCounts function from the Rsubread Bioconductor package (v.2.0.1) (Liao et al., 2019); genes with no symbol were excluded from downstream analysis. For the isolated hBEC RNA-seq data, differential expression between appropriate groups was computed using DESeq2 (v.1.26.0) (Love et al., 2014). Gene Set Enrichment Analysis was computed by applying GSEA PreRanked (v.4.0.3) (Subramanian et al., 2005) with the MSigDB Hallmark Gene Set (Liberzon et al., 2015) to differential expression results. Gene sets with fewer than 15 genes or more than 500 genes were excluded from analysis; gene sets with FDR less than the default GSEA FDR threshold of 0.25 were judged to be significant. Additional gene functional annotations were extracted from the Gene Ontology project (Ashburner et al., 2000; The Gene Ontology Consortium, 2019) via the AmiGO tool (v.2.5.13) (Carbon et al., 2009). For heatmap visualization of RNA-seq data, read counts were normalized with respect to library size using the regularized log (rlog) transform (Love et al., 2014).
The RNA-seq data associated with this study has been deposited in NCBI Gene Expression Omnibus (accession number GEO: GSE155498).
Single-cell RNA-seq analysis
Publicly available scRNA-seq data deriving from primary tissue from intrahepatic bile ducts was downloaded from ArrayExpress (E-MTAB-8495) (Sampaziotis et al., 2021). Demultiplexing of scRNA-seq data, alignment and barcode counting were performed using 10X Genomics Cell Ranger (v5.0.0) with reference dataset GRCh38/2020-A. Unfiltered UMI count matrices from Cell Ranger were used as input for downstream analysis following the OSCA Bioconductor workflow (Amezquita et al., 2020). EmptyDrops (Lun et al., 2019) was applied to remove cells predicted to contain only ambient RNA, at the default FDR of 0.1%. Cholangiocytes were then isolated following the method of Sampaziotis, et al. (Sampaziotis et al., 2021), where cells with > 3 counts for at least one of the human biliary markers EpCAM, KRT7 or KRT19 were retained for downstream analysis; samples with fewer than 50 retained cells were removed from analysis. Cells were classified as EpCAM+, EpCAM+CD133+, or EpCAM+CD133- based on detectable expression of these genes.
Induction of the mouse model
To induce BEC-senescence, recombination of loxP sites was induced with three doses of 180mg/kg of tamoxifen (Sigma-Aldrich) in sunflower seed oil (Sigma-Aldrich) by oral gavage on alternate days. Control mice received the equivalent volume of sunflower oil.
To induce biliary injury, mice were given 0.1% 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) mixed with Rat & Mouse No Maintenance (RM1) diet (Special Diet Services) as previously described (Dorrell et al., 2011). DDC diet was administered for 2 days after last tamoxifen injection. Although the literature describes prolonged periods of DDC feeding in different mouse models, the K19CreERTMdm2fl/flRag2−/− Il2rg−/− line only tolerated 1 week of DDC diet due to its C57BL/6J background. This decision takes into consideration restrictions of a maximum total weight loss of 20%.
For the IRI model, briefly: laparotomy was performed under isoflurane anesthesia in fasted (16-18 h) mice. Using two moistened cotton swabs, intestines were carefully externalized and liver lobes lifted and separated. Using an atraumatic clip (Fine Science Tools), portal vein, hepatic artery and bile duct were clamped above the branching to the right lateral lobe for 45 min (Abe et al., 2009). Following ischemia, the clamp was carefully removed and 500 μl of warm sterile saline were administered to the peritoneal cavity to replenish any fluid loss during surgery (Abe et al., 2009). Mice were sutured and kept for 48 h or 10 days after surgery.
hBEC and hMSC transplant
For hBEC transplantation, media was aspirated from the cultures and Matrigel spheres washed three times in Advanced DMEM/F-12. Matrigel was manually disrupted using a pipette, placed on ice to allow the Matrigel to dissolve and washed to eliminate any remaining Matrigel. Clusters of cells were centrifuged at 350xg for 5 min and incubated with Versene (Thermo Fisher Scientific) at 37°C for 45 min until they were dissociated into a single cell suspension. Cells were washed in PBS and 1 × 106 hBEC resuspended for transplantation in 100 μl of sterile PBS. hMSC were dissociated using TrypLE Express (GIBCO) as per manufacturer’s instructions and 1 × 106 cells resuspended for transplantation in 100 μl of sterile PBS.
Cells were injected intrasplenically after laparotomy as previously described (Lu et al., 2015) after senescence and DDC injury, or after Ischemia-Reperfusion injury. Briefly, a small subcostal incision was performed on the left flank under isoflurane anesthesia, the spleen was identified and delivered through the incision. Cells were injected with a BD MicroFine insulin syringe (BD) and the injection site covered with haemostatic dressing (Surgicel, Ethicon). The spleen was returned to the abdomen and the skin closed using 6/0 Vicryl suture (Ethicon). The transplantation control group received 100 μL PBS only. Mice recovered in a heated cage at 30°C with mash diet and received post-operative analgesia at 12 and 24 h after recovery (Carprofen, Pfizer).
Sample collection
Mice were killed according to UK Home Office regulations and blood collected by cardiac puncture. Livers were perfused in situ with PBS through the portal vein and harvested. Organs were harvested and either directly frozen at −80°C or fixed in 10% formalin (in PBS) for 12 h prior to embedding in paraffin blocks.
Serum analysis
Blood was collected via cardiac puncture after confirming mouse death and centrifuged at 6000xg 10 min at 4°C. Serum analysis used commercial kits according to the manufacturer’s instructions: serum albumin (Alb), aspartate aminotransferase (AST) and alkaline phosphatase(ALP) (Randox laboratories), Alanine transaminase (ALT), total bilirubin and Amylase (Alpha Laboratories). All kits were adapted for use on a Cobas FARA centrifugal analyzer (Roche Diagnostics).
Isolation of murine intra-hepatic bile ducts
Mice were injected with 35 μl of each Ketamine (Pfizer) and Medetomidine (Orion Pharma) and received a midline laparotomy. The portal vein was cannulated and perfused with 50 mL of Liver Perfusion Media (GIBCO) followed by 50 mL of Liver Digest Media (GIBCO) at 100 mL/h at 37°C. The liver was then dissected and placed in Liver Digest Media at 37°C until the parenchymal tissue manually separated from the biliary tree. Bile ducts were manually collected and washed in PBS twice.
RT-qPCR
Cells or tissue were homogenized and lysed using the RNeasy Micro kit (QIAGEN) or RNeasy Mini kit (QIAGEN) respectively as per manufacturer’s instructions. RNA quantity and quality were assessed using a spectrophotometer (NanoDrop ND-1000, Marshall Scientific). cDNA was prepared using QuantiTect Reverse Transcription Kit (QIAGEN) following manufacturer’s instructions. Real time-qPCR was performed using QuantiFast SYBR Green PCR kit (QIAGEN) on a LightCycler 480 II (Roche) with commercial primers from QIAGEN’s QuantiTect. Each gene expression was assessed with its own standard curve and normalized using GAPDH as housekeeping gene. Samples were run in triplicate. Primers for RT-qPCR (both mouse and Human) are stated in Table S6.
Protein isolation
75 mg of frozen mouse liver was mixed with 0.5 mL of ice cold tissue lysis buffer: 150 mM NaCl (Sigma-Aldrich), 20 mM Tris (Sigma-Aldrich), 1 mM EDTA (Sigma-Aldrich), 1 mM EGTA (Sigma-Aldrich), 1% Triton X (Sigma-Aldrich) and 2x Protease Inhibitor cocktail (Sigma-Aldrich) in PBS adjusted to pH 7.5. Samples were homogenized using a Tissue-Tearor (BioSpec products), rotated at 4°C for 20 min and centrifuged at 20000xg for 10 min. Supernatant was collected and BCA protein assay (Thermo Fisher Scientific) performed to calculate protein concentration.
Cytokine analysis
Meso Scale Discovery (MSD) multi-spot electrochemiluminescence assay system was used to quantify cytokine levels in blood serum or liver protein. V-PLEX Proinflammatory Panel 1 Mouse Kit (K15048D-1, Meso Scale Diagnostics) and V-PLEX Cytokine Panel 1 Mouse Kit (K15245D, Meso Scale Diagnostics) were used according to manufacturer’s instructions.
Histology, immunohistochemistry. and immunofluorescence
Tissues were fixed O/N with formalin, washed and transferred to tissue cassettes and paraffin blocks using standard methods. Paraffin sections (4 μm) were cut and stained using the antibodies listed. hBEC grown as organoids were fixed in 4% Formaldehyde and 1% Glutaraldehyde (Sigma-Aldrich), washed with PBS and permeabilized with 0.1% Tween-20 (Sigma-Aldrich) for 10 min. Cells were then sequentially incubated with Protein Block (Spring Bioscience), primary antibody, Alexa Fluor fluorescent-secondary antibody (Life Tech) and DAPI (Sigma-Aldrich). For tissue immunohistochemistry, sodium citrate pH 6.0 or Tris-EDTA pH 9.0 were used for antigen retrieval. Sections were blocked for endogenous peroxidase and alkaline phosphatase activity (BLOXALL, Vector), endogenous Avidin/Biotin (LifeTech) and nonspecific protein binding (Protein Block, Spring Bioscience). Primary antibodies, followed by species-specific secondary biotinylated antibodies (Vector Laboratories), VECTASTAIN ABC reagent, R.T.U. (Vector Laboratories) and DAB chromogen (Dako) were sequentially applied. Cells on polycaprolactone scaffolds were washed twice with PBS and stained on site. For immunofluorescence, primary antibodies were detected using fluorescent-conjugated secondary antibodies (Alexa Fluor, LifeTech). Sections were mounted with DAPI-containing media (Southern Biotech) or counterstained with DAPI. Isotype controls were used at the same concentration as the corresponding primary antibody. Antibodies, concentrations and method for antigen retrieval are stated in Table S7.
Hematoxylin and eosin staining
Sections were stained routinely and mounted in fluorescence-free DPX mountant media (Sigma-Aldrich).
PicroSirius red staining
Sections were stained using Picric Acid (P6744), Fast Green (F7258) and Direct Red (365548, all from Sigma-Aldrich).
Oil red O staining
Frozen liver human sections were allowed to equilibrate at room temperature. Oil red O (Sigma-Aldrich) stock solution (0.3% w/v in isopropanol) was prepared, diluted (3:2, stock:water) and sections stained for 10 min. Sections were counterstained in hematoxylin and mounted using aqueous mounting medium.
Karyotype
hBEC growing as organoids were washed with fresh media and Colcemid (GIBCO) was added for 2.5 h. hBEC were detached as stated before, washed with PBS, and incubated in 75 mM KCl (GIBCO) 15 min at RT. hBEC were resuspended in fixative (3:1 methanol:acetic acid), spread onto glass slides and mounted with DAPI-Fluoromount-G (Southern Biotech). Images were acquired using a Nikon Eclipse e600 and Retiga 2000R camera (Q-Imaging, Image Pro premier software) and quantified using Fiji ImageJ.
Fluorescein Diacetate (FDA)/Propidium Iodide (PI) staining
hBEC in polycaprolactone scaffolds were incubated 5 min at 37°C with 5 mg/mL FDA (Thermo Fisher) and 2 mg/mL PI (Sigma-Aldrich) prepared in PBS, washed and imaged using a Nikon Eclipse Ti Inverted microscope (Nikon).
Microscopy and cell counting
Images were acquired using a Nikon Eclipse e600 and Retiga 2000R camera (Q-Imaging, ImagePro premier software) or Perkin Elmer Operetta High content imaging system. Cells on polycaprolactone scaffolds were imaged using a Nikon Eclipse Ti Inverted microscope (Nikon). Oil Red O images were acquired in the AxioScan Z.1 (Zeiss) and analyzed in Fiji ImageJ using a macro instruction.
Histological sections were assigned a randomized blinded code prior to quantification, and the randomization decoded at the time of the final data analysis. For single-cell quantification in tissue, images were acquired at x20 magnification, and analyzed using Fiji ImageJ or Columbus Image Data Storage and Analysis System (Perkin Elmer) software. Cells were identified based on DAPI/Hoechst stained-nuclei, morphology and specific staining for each population. Illumination correction and background normalization was performed using the sliding parabola module. For each experiment identical thresholds were used in all images for assigning nuclei to a specific population. For pixel analysis, ImagePro premier software was used to select regions of positivity and automatically analyzed using a macro-instruction. Results are expressed as the mean percentage of positive pixels per field. PicroSirius Red analysis used the AxioScan Z.1 (Zeiss) to acquire tiled images at x20 magnification. Images were then analyzed using a standard color threshold in Fiji ImageJ. Scale bars as per individual figures.
Murine magnetic resonance cholangio-pancreatography (MRCP)
MRCP was performed using a 7 Tesla horizontal bore NMR spectrometer (Agilent Technologies) equipped with a high-performance gradient insert (60 mm inner diameter), maximum gradient strength 1000 mT/m. A 33 mm diameter birdcage volume coil (RAPID Biomedical GmbH) was used for radio frequency transmission and signal reception.
Following hBEC or control transplants mice were anaesthetized with 1.8% Isoflurane in oxygen/air (50/50, 1 l/min) and placed in a cradle (RAPID Biomedical GmbH). Rectal temperature (37°C) and respiration rate were monitored throughout the experiments. After verifying correct position and instrument optimization, mice were killed by increasing the isoflurane concentration to 5% and a three-dimensional fast spin echo scan was acquired with the following parameters: repetition time 300 ms, echo time 60 ms, field of view 39x26x26 mm, acquisition matrix 192x128x128 (resolution: 0.203x0.203x0.203 mm), echo train length 10 and 4 signal averages. The total scan time was 33 min.
Images were analyzed using Fiji ImageJ. Common bile duct and fall bladder images were processed as maximum intensity projections and diameters of maximum bile duct, maximum cystic duct and maximum gall bladder were measured.
Ink retrograde injection
Mouse duodenum was clamped and gall bladder cannulated with a catheter. 20 μl of Methylene Blue (Sigma-Aldrich) were slowly injected in a retrograde fashion. Images were acquired using a SMZ800 stereoscope (Nikon).
Quantification and statistical analysis
A minimum of four mice were included per experimental group. Power calculations are not routinely performed; however, animal numbers were chosen to reflect the expected magnitude of response considering the variability observed in previous experiments. Mice were randomly allocated to each experimental group and males/females equally distributed.
For cell culture experiments, a minimum of four independent biological replicates were performed.
Prism software version 5.0a (GraphPad Software, Inc) was used for all statistical analysis. Normal distribution of data was determined using D’Agostino and Pearson Omnibus normality test with Welch’s correction if variances differed (f test). For parametric data, data significance was analyzed using a two-tailed unpaired Student’s t test. Non-parametric data was analyzed using Mann-Whitney test. In cases where more than two groups were being compared, then a one-way ANOVA (with Bonferroni correction) was used. Survival curves were calculated using Log-Rank comparison (Mantel-Cox test) and Gehan-Breslow-Wilcoxon test.
Statistical significance was assumed at p < 0.05. Data is presented as mean ± standard error of the mean (SEM). N refers to biological replicates.
Routinely qPCR experiments were performed in technical triplicates of multiple biological replicates. For representative images 3-4 liver lobes were examined histologically in at least 3 biological replicates; further details referring to the specific numbers of biological replicates for each experiment can be found in the figure legend.
Acknowledgments
This work was supported by the UK Medical Research Council. S.F.-G., T.Y.M., and K.T. were funded by Developmental Pathway Funding MRC, MR/P016839/1. J.M.H. was funded by the Edinburgh Clinical Academic Track Cancer Research UK PhD fellowship and the Edinburgh and Lothians Health Foundation Liver Transplant Fund. This work has made use of the resources provided by the Edinburgh Compute and Data Facility (ECDF).
Author contributions
Conceptualization and design, J.M.H., S.F.-G., W.Y.-L., and S.J.F.; data generation, J.M.H., S.F.-G., T.Y.M., A.M.K., H.E., K.T., M.T.M., D.R.-T., B.J.D., V.L.G., C.A.-H., W.Y.-L., J.P.T., M.A.J., E.O., P.J.S.L., L.C., R.E.A., T.S.R.B., A.R.F., J.D.M.C., G.C.O., D.C.H., and A.C.; data analysis and interpretation, J.M.H., S.F.-G., B.J.D., K.T., J.P.T., A.M.K., W.Y.-L., E.O., M.A.J., D.R.-T., P.J.S.L., L.C., A.R.F., J.D.M.C., D.C.H., G.C.O., and S.J.F.; manuscript preparation, S.F.-.G.; writing – review & editing, S.F.-G., D.R.-T., A.M.K., and S.J.F.; funding acquisition, S.J.F.
Declaration of interests
S.J.F. and J.D.M.C. are founders and scientific advisors of Resolution Therapeutics Ltd (not related to this study). D.C.H. is a founder, director, and shareholder at Stemnovate Limited and Stimuliver ApS (not related to this study).
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper self-identifies as living with a disability.
Published: March 3, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stem.2022.02.006.
Supplemental information
Data and code availability
-
•
The hBEC RNA-seq data have been deposited at NCBI Gene Expression Omnibus and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abe Y., Hines I.N., Zibari G., Pavlick K., Gray L., Kitagawa Y., Grisham M.B. Mouse model of liver ischemia and reperfusion injury: method for studying reactive oxygen and nitrogen metabolites in vivo. Free Radic. Biol. Med. 2009;46:1–7. doi: 10.1016/j.freeradbiomed.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amezquita R.A., Lun A.T.L., Becht E., Carey V.J., Carpp L.N., Geistlinger L., Marini F., Rue-Albrecht K., Risso D., Soneson C., et al. Orchestrating single-cell analysis with Bioconductor. Nat. Methods. 2020;17:137–145. doi: 10.1038/s41592-019-0654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high-throughput sequence data.https://www.bioinformatics.babraham.ac.uk/projects/fastqc [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter L., Govaere O., Bird T.G., Radulescu S., Ramachandran P., Pellicoro A., Ridgway R.A., Seo S.S., Spee B., Van Rooijen N., et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat. Med. 2012;18:572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton T.P., Corcoran A., Callanan A. The effect of electrospun polycaprolactone scaffold morphology on human kidney epithelial cells. Biomed. Mater. 2017;13:015006. doi: 10.1088/1748-605X/aa8dde. [DOI] [PubMed] [Google Scholar]
- Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S., AmiGO Hub. Web Presence Working Group AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey E.J., Ali A.H., Lindor K.D. Primary biliary cirrhosis. Lancet. 2015;386:1565–1575. doi: 10.1016/S0140-6736(15)00154-3. [DOI] [PubMed] [Google Scholar]
- Carpentier R., Suñer R.E., van Hul N., Kopp J.L., Beaudry J.B., Cordi S., Antoniou A., Raynaud P., Lepreux S., Jacquemin P., et al. Embryonic ductal plate cells give rise to cholangiocytes, periportal hepatocytes, and adult liver progenitor cells. Gastroenterology. 2011;141:1432–1438, 1438.e1–1438.e4. doi: 10.1053/j.gastro.2011.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpino G., Cardinale V., Renzi A., Hov J.R., Berloco P.B., Rossi M., Karlsen T.H., Alvaro D., Gaudio E. Activation of biliary tree stem cells within peribiliary glands in primary sclerosing cholangitis. J. Hepatol. 2015;63:1220–1228. doi: 10.1016/j.jhep.2015.06.018. [DOI] [PubMed] [Google Scholar]
- Cursio R., Gugenheim J. Ischemia-Reperfusion Injury and ischemic-type biliary lesions following liver transplantation. J. Transplant. 2012;2012:164329. doi: 10.1155/2012/164329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltenre P., Valla D.C. Ischemic cholangiopathy. J. Hepatol. 2006;44:806–817. doi: 10.1016/j.jhep.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C., Erker L., Schug J., Kopp J.L., Canaday P.S., Fox A.J., Smirnova O., Duncan A.W., Finegold M.J., Sander M., et al. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 2011;25:1193–1203. doi: 10.1101/gad.2029411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkowski P., Schlegel A., de Oliveira M., Müllhaupt B., Neff F., Clavien P.A. HOPE for human liver grafts obtained from donors after cardiac death. J. Hepatol. 2014;60:765–772. doi: 10.1016/j.jhep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Dyson J.K., Beuers U., Jones D.E.J., Lohse A.W., Hudson M. Primary sclerosing cholangitis. Lancet. 2018;391:2547–2559. doi: 10.1016/S0140-6736(18)30300-3. [DOI] [PubMed] [Google Scholar]
- Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Gonzalez S., Lu W.Y., Raven A., Dwyer B., Man T.Y., O’Duibhir E., Lewis P.J.S., Campana L., Kendall T.J., Bird T.G., et al. Paracrine cellular senescence exacerbates biliary injury and impairs regeneration. Nat. Commun. 2018;9:1020. doi: 10.1038/s41467-018-03299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J.L., Davenport M., Kelly D.A. Biliary atresia. Lancet. 2009;374:1704–1713. doi: 10.1016/S0140-6736(09)60946-6. [DOI] [PubMed] [Google Scholar]
- Hay D.C., Fletcher J., Payne C., Terrace J.D., Gallagher R.C.J., Snoeys J., Black J.R., Wojtacha D., Samuel K., Hannoun Z., et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc. Natl. Acad. Sci. USA. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S.W., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J., et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M.M.A., Ellis E., van Wenum M., Fuchs S.A., de Ligt J., et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada H., Udono M., Matsuda-Ito K., Horisawa K., Ohkawa Y., Miura S., Goya T., Yamamoto J., Nagasaki M., Ueno K., et al. Direct reprogramming of human umbilical vein- and peripheral blood-derived endothelial cells into hepatic progenitor cells. Nat. Commun. 2020;11:5292. doi: 10.1038/s41467-020-19041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno N., LeSage G., Glaser S., Alvaro D., Alpini G. Functional heterogeneity of the intrahepatic biliary epithelium. Hepatology. 2000;31:555–561. doi: 10.1002/hep.510310302. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y. Sox9 and programming of liver and pancreatic progenitors. J. Clin. Invest. 2013;123:1881–1886. doi: 10.1172/JCI66022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel V., Deharde D., Pfeiffer E., Zeilinger K., Seehofer D., Damm G. Protocol for isolation of primary human hepatocytes and corresponding major populations of non-parenchymal liver cells. J. Vis. Exp. 2016;e53069:e53069. doi: 10.3791/53069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney L.L., Shultz L.D., Greiner D.L., Brehm M.A. Humanized Mouse Models for Transplant Immunology. Am. J. Transplant. 2016;16:389–397. doi: 10.1111/ajt.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurial S.N.T., Willenbring H. Emerging cell therapy for biliary diseases. Science. 2021;371:786–787. doi: 10.1126/science.abg3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis K.N., Strazzabosco M., Larusso N.F. The cholangiopathies: disorders of biliary epithelia. Gastroenterology. 2004;127:1565–1577. doi: 10.1053/j.gastro.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Li Z., White P., Tuteja G., Rubins N., Sackett S., Kaestner K.H. Foxa1 and Foxa2 regulate bile duct development in mice. J. Clin. Invest. 2009;119:1537–1545. doi: 10.1172/JCI38201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dorrell C., Canaday P.S., Pelz C., Haft A., Finegold M., Grompe M. Adult mouse liver contains two distinct populations of cholangiocytes. Stem Cell Reports. 2017;9:478–489. doi: 10.1016/j.stemcr.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.Y., Bird T.G., Boulter L., Tsuchiya A., Cole A.M., Hay T., Guest R.V., Wojtacha D., Man T.Y., Mackinnon A., et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat. Cell Biol. 2015;17:971–983. doi: 10.1038/ncb3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T., Marioni J.C., participants in the 1st Human Cell Atlas Jamboree EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20:63. doi: 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- McDaniel K., Meng F., Wu N., Sato K., Venter J., Bernuzzi F., Invernizzi P., Zhou T., Kyritsi K., Wan Y., et al. Forkhead box A2 regulates biliary heterogeneity and senescence during cholestatic liver injury in mice. Hepatology. 2017;65:544–559. doi: 10.1002/hep.28831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanuma Y. Tutorial review for understanding of cholangiopathy. Int. J. Hepatol. 2012;2012:547840. doi: 10.1155/2012/547840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevi L., Cardinale V., Carpino G., Costantini D., Di Matteo S., Cantafora A., Melandro F., Brunelli R., Bastianelli C., Aliberti C., et al. Cryopreservation protocol for human biliary tree stem/progenitors, hepatic and pancreatic precursors. Sci. Rep. 2017;7:6080. doi: 10.1038/s41598-017-05858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampes S., Ma D. Hepatic ischemia-reperfusion injury in liver transplant setting: mechanisms and protective strategies. J. Biomed. Res. 2019;33:221–234. doi: 10.7555/JBR.32.20180087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven A., Lu W.Y., Man T.Y., Ferreira-Gonzalez S., O’Duibhir E., Dwyer B.J., Thomson J.P., Meehan R.R., Bogorad R., Koteliansky V., et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017;547:350–354. doi: 10.1038/nature23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M.M., Jonsson J.R., Powell E.E., Brunt E.M., Neuschwander-Tetri B.A., Bhathal P.S., Dixon J.B., Weltman M.D., Tilg H., Moschen A.R., et al. Progressive fibrosis in nonalcoholic steatohepatitis: association with altered regeneration and a ductular reaction. Gastroenterology. 2007;133:80–90. doi: 10.1053/j.gastro.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Richmond A., Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Dis. Model. Mech. 2008;1:78–82. doi: 10.1242/dmm.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Torres D., Affò S., Coll M., Morales-Ibanez O., Millán C., Blaya D., Alvarez-Guaita A., Rentero C., Lozano J.J., Maestro M.A., et al. The biliary epithelium gives rise to liver progenitor cells. Hepatology. 2014;60:1367–1377. doi: 10.1002/hep.27078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckman F.C., Bucuvalas J.C., Nathan J., Alonso M., Tiao G., Balistreri W.F. Outcomes following liver transplantation. Semin. Pediatr. Surg. 2008;17:123–130. doi: 10.1053/j.sempedsurg.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Saleh M., Kamath B.M., Chitayat D. Alagille syndrome: clinical perspectives. Appl. Clin. Genet. 2016;9:75–82. doi: 10.2147/TACG.S86420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaziotis F., Justin A.W., Tysoe O.C., Sawiak S., Godfrey E.M., Upponi S.S., Gieseck R.L., 3rd, de Brito M.C., Berntsen N.L., Gómez-Vázquez M.J., et al. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat. Med. 2017;23:954–963. doi: 10.1038/nm.4360. [DOI] [PubMed] [Google Scholar]
- Sampaziotis F., Muraro D., Tysoe O.C., Sawiak S., Beach T.E., Godfrey E.M., Upponi S.S., Brevini T., Wesley B.T., Garcia-Bernardo J., et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science. 2021;371:839–846. doi: 10.1126/science.aaz6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub J.R., Huppert K.A., Kurial S.N.T., Hsu B.Y., Cast A.E., Donnelly B., Karns R.A., Chen F., Rezvani M., Luu H.Y., et al. De novo formation of the biliary system by TGFβ-mediated hepatocyte transdifferentiation. Nature. 2018;557:247–251. doi: 10.1038/s41586-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A., Muller X., Kalisvaart M., Muellhaupt B., Perera M.T.P.R., Isaac J.R., Clavien P.A., Muiesan P., Dutkowski P. Outcomes of DCD liver transplantation using organs treated by hypothermic oxygenated perfusion before implantation. J. Hepatol. 2019;70:50–57. doi: 10.1016/j.jhep.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Strazzabosco M., Fabris L. Functional anatomy of normal bile ducts. Anat. Rec. (Hoboken) 2008;291:653–660. doi: 10.1002/ar.20664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow B.D., Finegold M.J., Grompe M. Clonal tracing of Sox9+ liver progenitors in mouse oval cell injury. Hepatology. 2014;60:278–289. doi: 10.1002/hep.27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.J., Clouston A.D., Forbes S.J. Links between hepatic fibrosis, ductular reaction, and progenitor cell expansion. Gastroenterology. 2014;146:349–356. doi: 10.1053/j.gastro.2013.11.034. [DOI] [PubMed] [Google Scholar]
- Yin C. Molecular mechanisms of Sox transcription factors during the development of liver, bile duct, and pancreas. Semin. Cell Dev. Biol. 2017;63:68–78. doi: 10.1016/j.semcdb.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The hBEC RNA-seq data have been deposited at NCBI Gene Expression Omnibus and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.