Abstract
Extreme loss of skeletal muscle overwhelms the natural regenerative capability of the body, results in permanent disability and substantial economic burden. Current surgical techniques result in poor healing, secondary injury to the autograft donor site, and incomplete recuperation of muscle function. Most current tissue engineering and regenerative strategies fail to create an adequate mechanical and biological environment that enables cell infiltration, proliferation, and myogenic differentiation. In this study, we present a nanoengineered skeletal muscle scaffold based on functionalized gelatin methacrylate (GelMA) hydrogel, optimized for muscle progenitors’ proliferation and differentiation. The scaffold was capable of controlling the release of insulin-like growth factor 1 (IGF-1), an important myogenic growth factor, by utilizing the electrostatic interactions with Laponite® nanoclays (NCs). Physiologically relevant levels of IGF-1 were maintained during a controlled release over two weeks. The NC was able to retain 50% of the released IGF-1 within the hydrogel niche, significantly improving cellular proliferation and differentiation compared to control hydrogels. IGF-1 supplemented medium controls required 44% more IGF-1 than the comparable NC hydrogel composites. The nanofunctionalized scaffold is a viable option for the treatment of extreme muscle injuries and offers scalable benefits for translational interventions and the growing field of clean meat production.
Keywords: Myogenesis, muscle tissue engineering, nanoclay, nanofunctionalized GelMA, controlled release, insulin-like growth factor
1. Introduction
Composing over 40% of the mass of the human body, skeletal muscle is responsible for the dynamic motion of the human body, supports internal organs, and maintains homeostasis.1, 2 Despite the regenerative capacity of skeletal muscle following minor injuries, extreme loss of muscle from trauma, surgical resection, or disease results in chronic and incomplete healing.3–5 Volumetric muscle loss (VML) describes an injury where more than 20% of the muscle mass is lost overwhelming the natural regenerative capacity of skeletal muscle.6 VML injuries are devoid of the local satellite cell population, lack the necessary extracellular matrix to enable cellular proliferation, and are in a chronically inflamed, pro-fibrotic state that results in functional disability.7 VML injuries contribute to the over $400 billion annual estimated economic burden of trauma in the United States.8
The current treatment options for VML include physical therapy, bracing, and surgical intervention.9, 10 Physical therapy has limited clinical functional recovery and is made impossible for some patients due to the severe nature of their injury.11 Orthoses and advanced bracing have provided improved function and enabled increased participation in rehabilitation programs.10, 11 However, custom prosthetics and bracing require custom fabrication and expensive hardware.10 Surgical intervention includes tissue debridement which can be followed by muscle autologous transplantation, transposition, or amputation.9 Unfortunately, muscle transfer is limited by donor site weakness, morbidity, and incomplete functional restoration.6, 12 The use of autografts also constrains the size and shape of reconstructed tissue.6 Further, skeletal muscle reconstruction is plagued by the technical experience required from the surgical team.13 Microsurgical techniques have improved patient outcomes, but autografts still suffer from incomplete innervation and vascularization of the transplant.6 These factors lead to a poor functional restoration of the patient, remaining significantly below pre-injured levels.6
Because of the complications associated with surgical correction and the incomplete functional restoration of autografts, regenerative medicine techniques have been explored as potential treatments for skeletal muscle injuries.14, 15 The proinflammatory and profibrotic wound environment can be modified through the delivery of soluble mediators such as growth factors, cells, and scaffolding materials.16–18
The addition of several myogenic growth factors has been shown to improve muscle regeneration.19 One of the most notable of these growth factors is insulin-like growth factor-1 (IGF-1). IGF-1 is a circulating growth hormone that is also synthesized in skeletal muscle during normal physiological conditions and upregulated after regenerating skeletal muscle injuries.3 IGF-1 protects cells from apoptosis, increases myoblast proliferation, increases myogenic differentiation, and increases hypertrophy.20–22 Despite the important role of IGF-1 in muscle maintenance and regeneration, the downregulation of its level post VML injury reduces the regenerative capacity of the damaged tissue.3 IGF-1 levels are downregulated in VML models (0.078 ng/mL) compared to cryoinjury models (4.129 ng/mL) where muscle regeneration can occur.3 Exogenous IGF-1 therapies have clinically failed due to bolus injections that do not retain adequate concentrations at the regeneration site.20, 23 Due to its effectiveness as a growth factor, a systemically delivered high dosage of IGF-1 poses a carcinogenic risk.23 To overcome the limitations of bolus injections, tissue engineers have sought to control the local delivery of IGF-1 in several skeletal muscle injury studies. Further, most in vitro mono cell cultures lack M1 macrophages that release IGF-1 as part of modulating skeletal muscle immune response.22
The delivery of myogenic and immunomodulatory cells has demonstrated improved benefits in injured muscle, but cell delivery is complicated by the difficulty of myogenic cell harvesting and in vitro cell expansion techniques that are needed to produce the necessary quantities of myogenic cells.19, 24 Further, the delivery of cells is limited in efficacy due to high stresses experienced during injection, low cellular residency at the injury site, and extremely low cellular engraftment.19 While non-myogenic cell sources are an alternative, appropriately controlling their differentiation remains challenging.19 Pairing IGF-1 delivery with myoblasts would enable facile control over cellular proliferation with fewer initial cells and replicate in vivo myogenic differentiation without the need for co-culture with immune cells.
Scaffolding materials can provide mechanical support to the wound, act as a delivery mechanism for growth factors and cells, and enable cellular migration. Because of this ability to improve IGF-1 release, Borselli et al. attempted to control the delivery of IGF-1 and VEGF through alginate microgels in ischemic and myotoxic injuries.25, 26 While the IGF-1 containing scaffolds resulted in improved histological and functional benefits, an undesired burst release of 70% of the encapsulated IGF-1 was reported after 1 day of incubation, and the total release of all the encapsulated IGF-1 was registered after 3 days.25, 26 This timescale does not match the 2–6 weeks healing period needed for skeletal muscle injuries.4 Further, the amount of IGF-1 used in these studies is several orders of magnitude above physiological levels and is not retained at the injury site. To address regeneration after extreme muscle loss, Tomblyn et al. and Passiperi et al. created keratin hydrogels loaded with IGF-1 and investigated their efficacy in a VML injury model.27, 28 While IGF-1 delivered in a VML injury did improve isometric torque over the levels in the non-treated group, the scaffold contained high concentrations of IGF-1 of 100 μg/mL, which is over 25,000 times larger than the normal concentration in regenerating skeletal muscle.3, 28 Most importantly, the release of IGF-1 from keratin scaffolds was purely dependent on the erosion of the scaffold (up to 70% in one week) and was incapable of sustainably releasing IGF-1 without losing the structural and mechanical features of the scaffold. As a result, inadequate myoblast migration into the scaffold, loss of scaffold after 12 weeks, and excessive fibrous scar tissue formation were reported.28
Beyond the undesirable burst release reported by Borselli et al., scaffolding materials like sodium alginate are not considered optimal because of their need for modification to incorporate integrin-binding sites and often inhibit cellular ingrowth.29 On the other hand, keratin degrades relatively rapidly, and consequently, its release of IGF-1 is dependent on erosion.27 Gelatin methacrylate (GelMA) is a chemically modified form of gelatin that enables cell proliferation due to naturally occurring integrin binding sites.30 It is photopolymerizable and easily tunable by the degree of functionalization or concentration.13, 31 GelMA has been demonstrated to support injured skeletal muscle, act as a protective carrier for skeletal muscle progenitors, and enable skeletal muscle differentiation.13, 32, 33 Recently, GelMA has been shown to adhere directly to a skeletal muscle injury creating a continuous interface with the injury and avoiding the complications of fixation of soft hydrogels.13, 32–34 However, GelMA derived from type A gelatin (isoelectric point ≈9) is not adequate to bind to electropositive growth factors like IGF-1 due to its positive charge at a pH of 7.4.35
To utilize the biological and mechanical characteristics of GelMA hydrogels for skeletal muscle tissue engineering and to address its limitations in binding IGF-1, herein, we developed a nanoengineered biocomposite scaffold that exploits the electrostatic binding capabilities of Laponite® nanoclays (NCs) to sustain the local release of IGF-1 at therapeutically optimized concentrations for improved myogenic proliferation and differentiation (Scheme 1). The large negative charge of Laponite® NC discs at physiologic pH is suitable for binding and controlling the release of positive proteins like IGF-1 and being incorporated into a positively charged polymeric network like GelMA.36 The GelMA concentration was first tuned to facilitate cell infiltration and proliferation. Next, the effects of varying sustained IGF-1 concentrations around and slightly above noncritical muscle regeneration levels were evaluated in terms of myoblast proliferation and differentiation. Then, the binding parameters of Laponite® NC discs were augmented to sustain the release of IGF-1 for maintaining local retention of the myogenic growth factor within hydrogels. The nanofunctionalized hydrogels were then studied for proliferative and differentiative capabilities using C2C12 myoblasts.
Scheme 1.
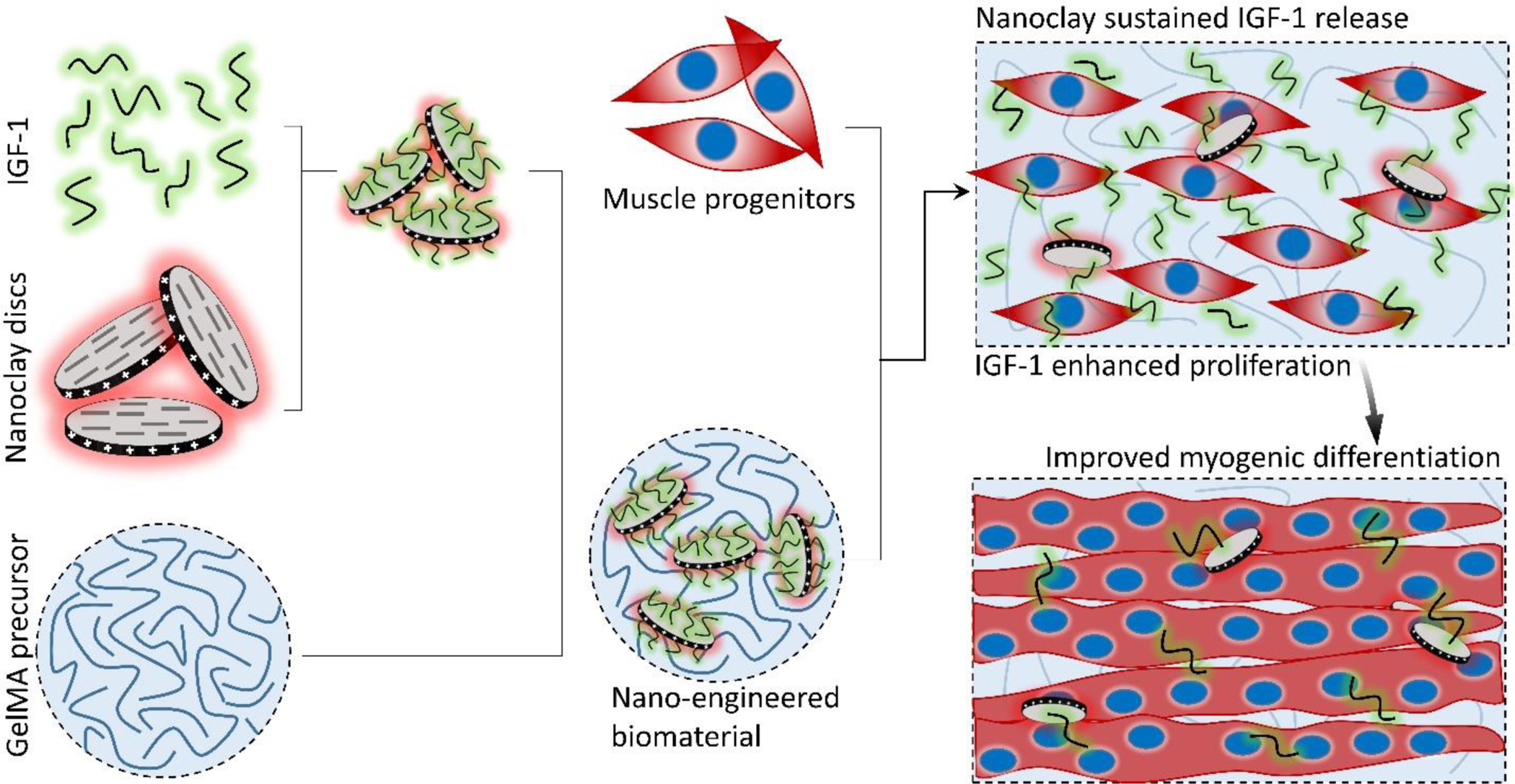
The proposed skeletal muscle tissue engineering scaffolds were composed of insulin-like growth factor-1 (IGF-1) bound to Laponite® nanoclay (NC) discs encapsulated in an optimized GelMA precursor. The IGF-1 bound NC discs encapsulated in the precursor GelMA were mixed with muscle progenitors (C2C12 myoblasts) and photocrosslinked. The scaffolds were cultured in proliferation medium until day 7 and then switched to differentiation medium until day 21.
2. Experimental
2.1. Materials
Gelatin from porcine skin 300 Bloom, Type A; methacrylic anhydride; and lithium phenyl-2,4,6-trimethyl-benzoyl phosphinate (LAP) photoinitiator were purchased from Sigma-Aldrich. Gibco™ Dulbecco’s Phosphate-Buffered Saline (DPBS), no calcium, no magnesium was purchased from ThermoFisher Scientific. Laponite® nanoclay (NC) was obtained from BYK Additives & Instruments.
2.2. GelMA synthesis
GelMA was synthesized using an established protocol.37 Gelatin from porcine skin, Type A, 300 Bloom, was dissolved in DPBS at 50°C using a magnetic stir bar at 200 rpm. A medium degree of functionalization was obtained by adding methacrylic anhydride dropwise at 1.25% (v/v) to the gelatin solution. The reaction occurred for 1 h at 50°C under constant stirring. Termination of the reaction was accomplished through a 5X dilution of 40°C DPBS. Excess salt and unreacted methacrylic anhydride were removed via dialysis against double deionized (DDI) water at 40°C for 1 week using a 12–14 kDa cutoff dialysis membrane (Thermo Fisher Scientific). The DDI water was changed twice daily. The solution was vacuum filtered using a 0.22 μm filter (Sigma-Aldrich) and frozen in 50 mL centrifuge tubes (Thermo Fisher Scientific) at −20 °C. The frozen GelMA was then lyophilized using a FreeZone® benchtop freeze dryer (Labconco®, Kansas City, MO) for 1 week. Lyophilized GelMA was stored at −20°C until use.
2.3. Myoblast cell culture, encapsulation, and differentiation
Immortalized mouse myoblast C2C12 (CRL-1772™, ATCC) were expanded in T75 flasks and cultured in proliferation medium: Dubulco’s Modified Eagle Medium (DMEM), high glucose, no glutamine, no calcium (Gibco); 10% (v/v) heat-inactivated fetal bovine serum (Gibco); and 1% (v/v) penicillin-streptomycin (Gibco). At 80% confluency, cells were suspended using a Trypsin-EDTA solution (Gibco). At passage 4, cells were used for encapsulation experiments by resuspending isolated cells in hydrogel prepolymer solutions. The GelMA prepolymer hydrogels contained ~0.07% (w/v) of lithium phenyl-2,4,6-trimethyl-benzoyl phosphinate (LAP) photoinitiator. This concentration of photoinitiator was used for all subsequent hydrogel experiments. Differentiation was induced via serum deprivation by changing the culture medium to DMEM with 2% (v/v) heat-inactivated horse serum (Gibco) and 1% penicillin-streptomycin.
2.4. Metabolic Assays
PrestoBlue™ (Invitrogen) viability assays were performed by first making a 10% reagent solution in a proliferation medium. The samples’ culture medium was replaced with the reagent solution and incubated for 1 hour at 37°C. After incubation, 100 μL of the supernatant medium was collected, placed in a 96 well plate, and analyzed for fluorescence intensity at 540 nm excitation and 600 nm emission using a Cytation 5 plate reader (BioTek). Sample signals were subtracted from reagent solution control signals.
2.5. Actin, Nuclear, and Immunofluorescent Microscopy
At each selected time point, cultured samples were washed in DPBS and fixed using a 4% paraformaldehyde solution in DPBS (Thermo Scientific). Samples were permeabilized in a 0.3% (v/v) Triton X solution in DPBS for 15 min at room temperature. After intermediate DPBS washes, the samples were blocked using a 1% bovine serum albumin and 0.1% TritonX solution in DPBS for 1 h at room temperature. Samples were stained for actin using AlexaFluor® 488 phalloidin (Invitrogen) at a ratio of 1:40 in the blocking solution and incubated for 1 h at room temperature. The concentrations of primary and secondary antibodies were optimized using control samples for increased signal-to-noise ratios. The lack of false-positive and non-specific staining was observed by testing primary and secondary antibodies in nondifferentiated samples and by adding only the secondary antibodies to differentiated samples (Figure S9a). Mouse anti-myosin heavy chain, sarcomere primary antibody (MF20, DSHB) at 3 μg/mL and rabbit anti-myogenin primary antibody (ab124800, Abcam) at 1 μg/mL in the blocking solution were incubated with the samples at 4°C overnight on a rocker plate. After intermediate washing, the following day, anti-mouse Alexa Fluor® 633 and anti-rabbit Alexa Fluor® Plus 555 secondary antibodies at 10 μg/mL in blocking solution were incubated at room temperature for one hour on a shaker plate. Finally, the nuclei were stained using DAPI (4′,6-diamidino-2-phenylindole) (Abcam) dye at a ratio of 1:1000 in the blocking solution for 5 min at room temperature. Samples were imaged using an inverted fluorescent Axio Observer 7 microscope.
2.6. In vitro GelMA concentration optimization
C2C12 myoblasts were suspended at 2M cells/mL in prepolymer GelMA hydrogels reconstituted in DMEM at concentrations of 3%−8% (w/v). 125 μL of cell-laden prepolymer hydrogel samples were pipetted and evenly distributed into the bottom of 48 well tissue culture plates (Thermo Fisher). The hydrogel samples were crosslinked from a distance of 17 mm using 365 nm light at 120 lumens from a penlight (JowBeam). Cells were cultured for 6 days in a proliferation medium. On days 1 and 3, PrestoBlue™ (Invitrogen) viability assays were performed as described (n = 4). Samples were stained for actin and nuclei as described. Cells were imaged within the hydrogel and at the gels surface to assess cell infiltration.
2.7. IGF-1 concentration optimization
Similarly, GelMA prepolymer solutions with 2 M C2C12 cells/mL were distributed and crosslinked in the bottom of 48 well tissue culture plates. The proliferation medium was supplemented with none (0 ng/mL), 1 ng/mL, 10 ng/mL, and 100 ng/mL of recombinant mouse IGF-1 (R&D Systems). On day 7, the medium was changed to a differentiation medium supplemented by the same amount of IGF-1. Cell proliferation was measured using PrestoBlue™ viability assay on days 1, 3, and 5. Cells were fixed, stained, and imaged for actin and nuclei on days 1, 3, 7, 14, and 21. The day 14 and 21 samples were also stained and imaged for myosin heavy chain as described. Myogenic differentiation was quantified by a blinded experimenter taking 3 coded random 10X images from each well (the average counting as a single representative replicate), processing using ImageJ (NIH, Bethesda, MD), decoding each sample, and statistically analyzing to compare each experimental group. Myosin heavy chain staining images were analyzed for tube number and tube width at day 21 of culture (n =3). Due to the nature of the micrographs being from a single focal plane, myotubes were only counted and measured for those in the focal plane.
2.8. Laponite® preparation and characterization
Laponite® NC was exfoliated by dissolution in varying DPBS solutions at 0.1% (w/v). For almost all experiments, NCs were allowed to exfoliate for 24 h before use.
2.8.1. Scanning electron microscopy (SEM):
Dry and aqueous NC SEM samples were placed or pipetted, respectively, onto carbon tape and baked at 50°C to remove moisture. Samples were observed using a Hitachi TM1000 SEM.
2.8.2. Zeta potential measurements:
The zeta potential of NC samples at 0.05% (w/v) before (0 h) and after (24 h) exfoliation in DDI water or DPBS solutions were analyzed using a dynamic light scattering analyzer (Zetasizer Nano ZS90, Malvern Instruments Ltd, UK) (n = 3). To minimize the effect of settling, the samples were mixed immediately before analysis. The zeta potential of NC samples after exfoliation in DDI water and DPBS before and after binding with protein for 1 h at 3000 rpm using a vortex mixer (Thermo Fisher Scientific) (n = 3) were analyzed.
2.8.3. Size analysis:
The size of exfoliated NCs in varying DPBS concentrations was measured using a dual light scattering analyzer (90 Plus Particle Size Analyzer, Brookhaven Instrument Corporation).
2.9. Protein binding studies
IGF-1 was bound to exfoliated NCs at 0.05% (w/v) in varying salt concentrations (0–100% DPBS) and at varying NC to protein ratios (0.25–50) for 1 h at 3000 rpm using a vortex mixer at 4°C (n = 4). Samples were run through Amicon ultra-0.5 100 kDa centrifugal filter units to separate proteins bound and not bound to NC. Collected supernatants were analyzed for unbound IGF-1 concentration using mouse IGF-1 DuoSet ELISA (R&D Systems). The percentage of bound protein was calculated as the difference between the mass of the measured protein unbound in the supernatant and the initial mass of protein added to each sample normalized to the initial mass of protein added to each sample:
Final NC bound groups were performed in 10% DPBS solution at 0.05% (w/v) NC with 3,333.3 ng/mL and 10,000 ng/mL IGF-1 for the 1X and 3X groups, respectively.
2.10. Protein release studies
IGF-1 bound to NC samples were diluted 10 times in either DPBS or GelMA prepolymer solutions to bring protein concentrations to 333.33 ng/mL for the NC+ and 1X groups. The 3X group was diluted to 1,000 ng/mL in GelMA prepolymer. Diluted prepolymer and free samples were pipetted at a volume of 200 μL (initial vehicle volume) into cell culture inserts and crosslinked as previously described (n = 4). The cell culture inserts were placed into 48 well tissue culture plates and filled with 0.5 mL of DPBS solution, sealed, and incubated at 37°C. The entire DPBS solution in the well plates was removed at various time points over 15 days and replaced with new DPBS. Released protein was measured using an IGF-1 DuoSet ELISA as previously described. The cumulative release was calculated by adding the total protein released from previous time points. The percentage of release was calculated by normalizing to the initial protein mass added to each sample. The mass of protein release was normalized to the initial liquid volume (composed of GelMA prepolymer or DPBS for the nonencapsulated groups) of 200 μL, resulting in the units of ng/mL.
2.11. 1H NMR characterization
The degree of methacrylation and crosslinking degree of the GelMA prepolymer, GelMA hydrogel, and GelMA/NC hydrogel was assessed using a Bruker Avance III 300 MHz NMR spectrometer. Before analysis, the lyophilized samples were dissolved in a mixture of solvents (D2O, DMSO-d6), and then all the appeared signals in the spectrums were normalized to the phenylalanine signal at δ = 7.00–7.5 ppm, using TopSpin 4.0.6 software. The ratio representing the summation of the methacrylate peaks (0.26 + 0.27) at δ = 5.3–5.7 ppm to the phenylalanine peak (1.00) yields a value of 53% signifying the degree of methacrylation for GelMA prepolymer.
As for the crosslinking degree, H1 NMR results revealed the methacrylate groups at 5.3–5.7 ppm were incorporated in the crosslinked network of samples. As seen in Figure S8, the signals related to methacrylate groups (black arrows) decreased for both GelMA and GelMA/NC hydrogels compared to GelMA prepolymer, representing a decrease in the number of free methacrylate groups after crosslinking. Accordingly, the crosslinking degree for hydrogels was calculated based on the following equation38:
2.12. Characterization of bulk nanofunctionalized hydrogels
2.12.1. Compression testing:
Discs of GelMA hydrogels with and without NC at 0.05% (w/v) were crosslinked into PDMS cylindrical molds with a diameter of 10 mm and a height of 4.5 mm. A positively charged glass slide was placed on top of the prepolymer before crosslinking to cap the mold and act as a transportation base and to enable easy mold removal (n = 4). The hydrogel cylinders were compressed using an Electroforce 3220 (TA Instruments, New Castle, DE) mechanical tester at a rate of 0.0166 mm/s. The compression modulus of each sample was determined from the slope of the first 10% of the stress-strain curve.
2.12.2. Swelling:
The swelling characteristics of GelMA discs with and without 0.05% (w/v) NCs were assessed by first crosslinking prepolymer solutions using a cylindrical PDMS mold with a diameter of 10 mm and height of 4.5 mm onto an 80 μm cell strainer (n = 4). The cell strainers with cylindrical hydrogel were submerged in DPBS in 6 well plates. The mass of only the hydrogels was calculated by subtracting the basket alone from the mass of the baskets after hydrogel-molded crosslinking. The change in mass was calculated by similarly subtracting the mass of the hydrogel at several time points over 1 week from the initial basket weight of each sample. Samples were blotted dry at each timepoint to remove excess DPBS.
2.12.3. Degradation:
Similar to the swelling assay, the degradation of GelMA discs with and without 0.05% (w/v) NCs was evaluated by molding the same dimensioned hydrogel discs onto cell strainers (n = 4). The samples were incubated in a 2 U/mL collagenase solution to simulate expedited degradation conditions as reported elsewhere.39 The mass change was measured and calculated as in the swelling assay.
2.12.4. Scanning electron microscopy (SEM):
Molded and crosslinked GelMA hydrogel samples with and without 0.05% NCs were flash-frozen through submersion in liquid nitrogen and immediately lyophilized for 48 h. The lyophilized samples were broken to reveal inner structural morphology and mounted on carbon tape. The samples were sputter-coated using gold-palladium for 30 s. Images were captured using a Hitachi TM1000 SEM.
2.13. In vitro nanoclay (NC) GelMA myogenic characterization
Various combinations of cell-laden GelMA with and without NCs and with and without IGF1 were cultured over 21 days: GelMA (GelMA-), GelMA loaded with NCs (GelMA/NC-), GelMA encapsulating NC with bound IGF-1 at 333.3 ng/mL with respect to the crosslinked gel (GelMA/NC+ (1X)), GelMA encapsulating NC with three times the mass of bound IGF-1 at 1,000 ng/mL with respect to the crosslinked gel (GelMA/NC+ (3X)), GelMA incorporating IGF-1 at 333.3 ng/mL (GelMA+ (1X)), and GelMA with IGF-1 supplemented at 10 ng/mL in the media (GelMA Media+). First, NCs were bound to IGF1 as previously described; NC devoid groups underwent the same mixing steps but the NC volumes were replaced with DPBS. The IGF-1 bound NCs were then mixed and diluted 10-fold into a 4% (w/v) GelMA prepolymer solution with 0.06875% LAP in proliferation medium. Detached C2C12 myoblasts were dispersed into the prepolymer solutions at 2 M cells/mL. Volumes of 125 μL of the prepolymer solutions were pipetted, evenly distributed, and crosslinked into the bottom of 48 well tissue culture plates as described above. Samples were cultured with proliferation medium with medium changes every 2 days. On day 7, the medium was changed to a differentiation medium. On days 1 and 3, PrestoBlue™ assays were performed as described (n = 4). Samples were fixed, stained, and imaged for actin and nuclei, myogenin, and myosin on days 3, 7, 14, and 21. Images were captured on an Axio Observer 7 microscope. Myogenic differentiation was quantified by a blinded experimenter taking 3 coded random 10X images from each well (the average counting as a single representative replicate), processing using ImageJ (NIH, Bethesda, MD), decoding each sample, and statistically analyzing to compare each experimental group. Myosin heavy chain staining images were analyzed for tube number and tube length at day 21 of culture (n =3). Due to the nature of the micrographs being from a single focal plane, myotubes were only counted and measured from end to end for those in the focal plane.
2.14. Statistical analyses
Data are presented as mean ± standard deviation. Statistical analysis was performed using GraphPad Prism 9.0 software (San Diego, CA). Column statistical analysis was performed using two-tailed Student’s t-tests. Paired values were accounted for and incorporated into analyses. Grouped statistical analysis was performed using analyses of variance (ANOVA) testing. ANOVA results that proved significant, p < 0.05, were assessed for pairwise comparison using Tukey’s multiple comparison post-hoc testing or more relevant tests. Significance is displayed as p < 0.05 with *, p < 0.01 with **, p < 0.001 with ***, and p < 0.0001 with ****.
3. Results
3.1. Optimization of GelMA hydrogel concentration
To first develop the nano-functionalized GelMA scaffold, we optimized the GelMA concentration for cell permissibility. A medium degree of methacrylation GelMA hydrogel of varying concentration (3–8 % w/v) was used to encapsulate C2C12 myoblasts and studied for their capacity for proliferation and migration (Figure S1). To assess the proliferation of the myoblasts, we exposed the samples to a PrestoBlue™ viability assay at day 1 and day 3 of cell culture (Figure S1a). There was no significant difference among the tested samples on day 1, but on day 3, higher metabolic activity was observed for the lower GelMA concentration samples. The highest metabolic activity was observed in the 3% (w/v) samples followed by 4% and 5% (w/v). After 6 days of culture, the samples were stained for actin (green) and nuclei (blue) to visualize cellular morphology (Figure S1b). Images were captured on the gel surface and inside of the gel. The 3% sample did not survive cell culture conditions past day 3 and was deemed unsuitable for further experimentation. The 4% (w/v) samples showed similar cell morphology on the surface and inside of the gel. The myoblasts had a normal stretched morphology with large aspect ratios with networks penetrating the entire depth of the gel. At concentrations of 5% (w/v) and above, the cells were unable to spread and form networks within the gels. In the samples with a higher concentration of GelMA, the myoblasts proliferated only on the surfaces to form dense networks on the hydrogel interface with the cell culture medium. The cells on the hydrogel surface of the higher concentration GelMA samples displayed a flatter cellular morphology. The individual cells that remained in the gels had round architectures with indistinguishable actin and nuclear regions. Therefore, a concentration of 4% (w/v) GelMA was selected as the best option that can balance cell proliferation and permissibility and was used for the remainder of the experiments.
3.2. IGF-1 concentration optimization for GelMA encapsulated myoblasts
We secondly optimized the IGF-1 concentration in the scaffold environment by exposing C2C12s encapsulated in 4% (w/v) GelMA hydrogels to varying concentrations of IGF-1 supplemented in the cell culture medium. The concentrations ranged from 1–100 ng/mL to study the effect of IGF-1 at concentrations near and above normal physiological levels in regenerating skeletal muscle in noncritical injuries (Figure 1).3 The levels of IGF-1tested had no significant effects on cell viability at day 1 and day 3 of cell culture compared to 2D controls or IGF-1 free GelMA (Figure S2). Serum-deprived differentiation was initiated on day 7 of culture and continued for 14 days until day 21 of culture. The proliferation ability of the groups was again assessed using a PrestoBlue™ viability kit (Figure 1a). On day 1, the level of proliferation in the 100 ng/mL IGF-1 group was detected to be significantly higher than all other groups. This trend continued onto day 3 and day 5. On day 5, the 100 ng/mL group matched the 2D control cells signal while a statistically significant difference was also identified between the 10 ng/mL group and 1 ng/mL group. Samples were stained for actin (green) and nuclei (blue) at days 1, 3, 7, 14, and 21 (Figure 1b). At the early time points, networks of myotubes were observed in the 10 and 100 ng/mL samples. On day 7, local areas of cell alignment were observed in the 10 and 100 ng/mL samples. One and two weeks after the induction of differentiation, the samples were stained for myosin heavy chain (MHC) (purple), an indicator of functional myotube formation (Figure 2a). MHC myotubes were evident in all samples after 7 days of differentiation. No qualitative trend was noticed between groups at day 7 of differentiation. On day 14 of differentiation, more MHC myotubes were visible. The groups with higher concentrations of IGF-1 (10 and 100 ng/mL) resulted in a higher average number of myotubes compared to GelMA only controls that did not contain IGF-1 (Figure 2b). The 100 ng/mL IGF-1 samples also resulted in significantly thicker myotubes than the GelMA only controls (Figure 2c).
Figure 1.
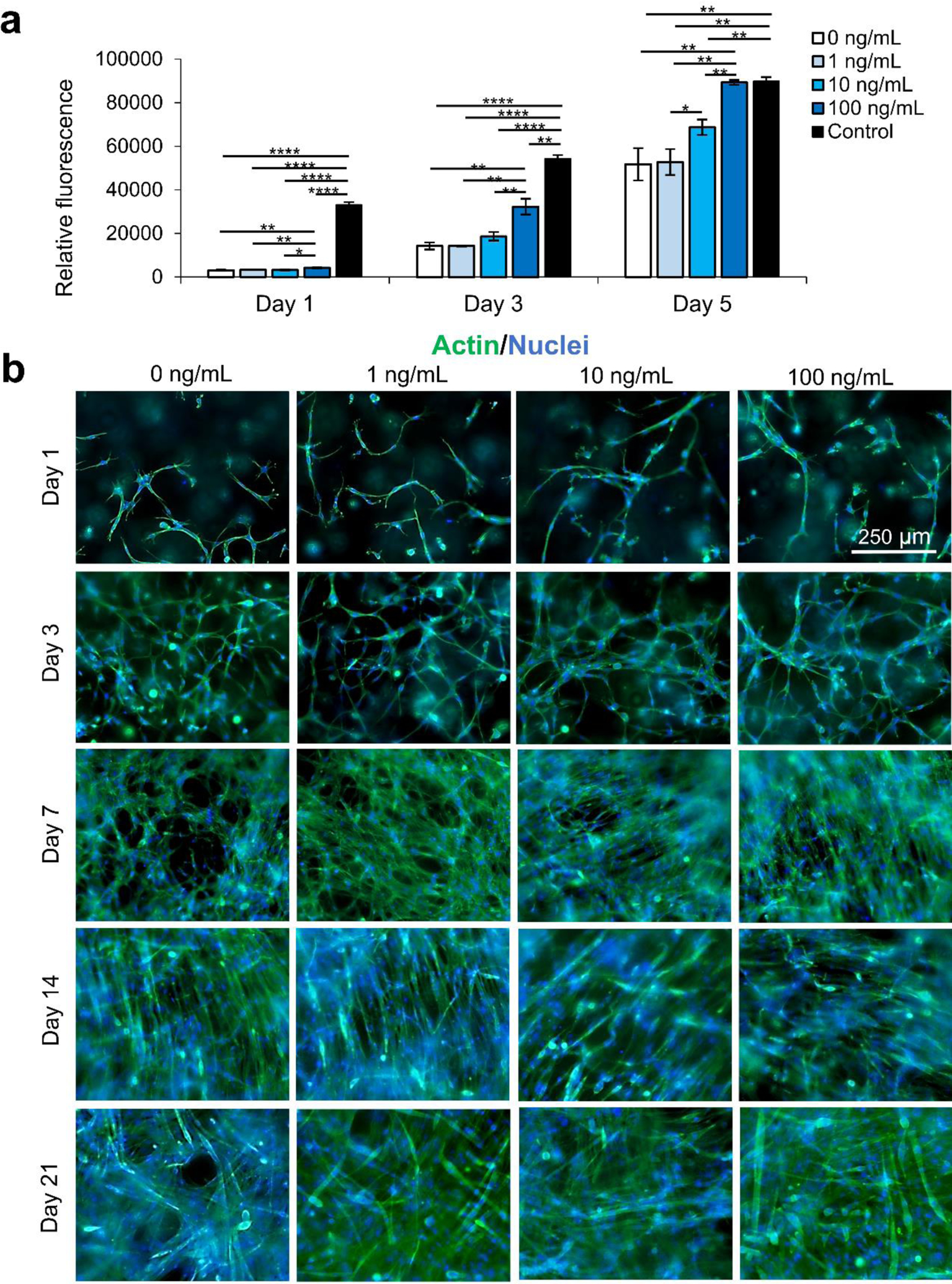
IGF-1 concentration optimization for improving proliferation of the C2C12 cells encapsulated in 4% GelMA hydrogel. a) PrestoBlue™ viability assay assessed the proliferation ability of myoblasts exposed to 0–100 ng/mL of IGF-1. IGF-1 appeared to have an increasing dose-dependent relationship with a higher cell proliferation rate. b) The morphology of C2C12s exposed to varying levels of IGF-1 in GelMA hydrogels: actin (green) nuclei (blue) staining. Scale bars are 250 μm for all micrographs.
Figure 2.
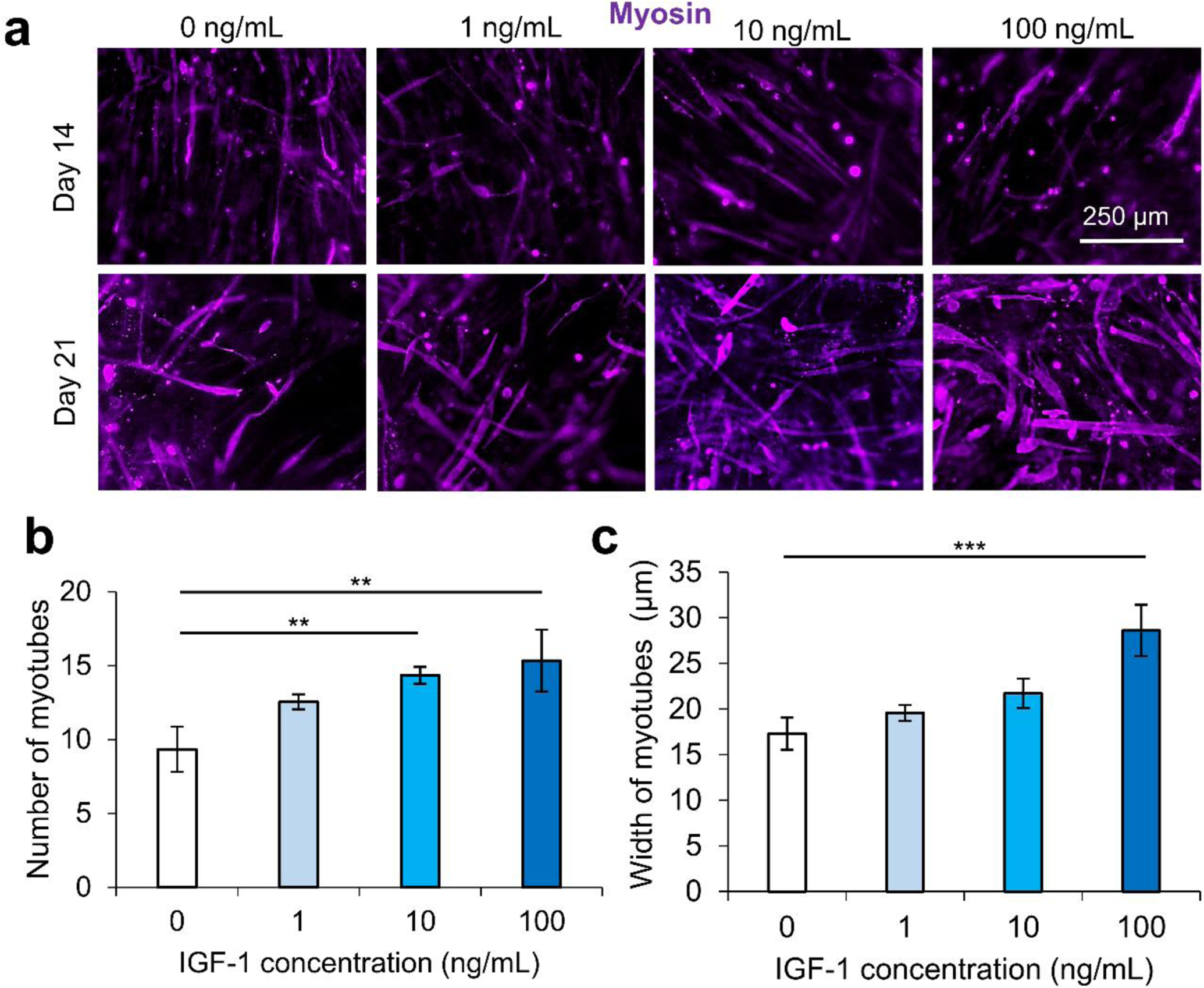
IGF-1 concentration optimization effect on the differentiation of C2C12 cells encapsulated in 4% GelMA hydrogel. a) Myosin heavy chain (purple) staining at day 14 and day 21, demonstrating the differentiation of myoblasts within GelMA hydrogels exposed to different IGF-1 levels. b) Quantification of the number of myotubes from varying concentrations of IGF-1. Higher levels of IGF-1 led to higher numbers of multinucleated myotubes. c) The quantification of myotube thickness. The highest concentration of IGF-1 (100 ng/mL) resulted in thicker, more mature myotubes compared to controls without IGF-1. Scale bars are 250 μm for all micrographs.
3.3. Nanoclay (NC) electrostatic property analysis
Laponite® nanoclay (NC) discs have been implemented in this study as protein delivery vehicles. NC was integrated into the GelMA hydrogel to enable sustained delivery of the IGF-1. To do so, we optimized different binding parameters of Laponite® NC and IGF-1 for protein binding and release (Figure 3). The NC comes as a dry powder composed of stacked discs bound together with sodium ions (Figure 3a). The sodium ions are released when the NC is placed into solution (Figure 3b,c). Figure 3 Exfoliation is partially prevented in high ionic solutions due to flocculation and enabled in low ionic solutions (Figure S3). Therefore, we tittered the concentration of Dulbecco’s Phosphate-Buffered Saline (DPBS), referred to as 100% at its 1X concentration of 150 mM. After dispersion in the solvent for 24 h, samples were dried on conductive tapes and imaged under scanning electron microscopy (SEM, Figure S3). In pure DPBS, the nanoclay discs remain as large chunks with a rough surface (Figure 3b). As the salt concentration is reduced to 50%, the nanoclay begins to exfoliate, forming micrometer-sized clusters (Figure S3). In a 10% DPBS solution, the exfoliation is enabled, resulting in a smooth phase with dispersed particles (Figure 3c and Figure S3 green call out). Secondary and tertiary structures of the NC were observed in the 10% DPBS solution (Figure S3 red call out).
Figure 3.
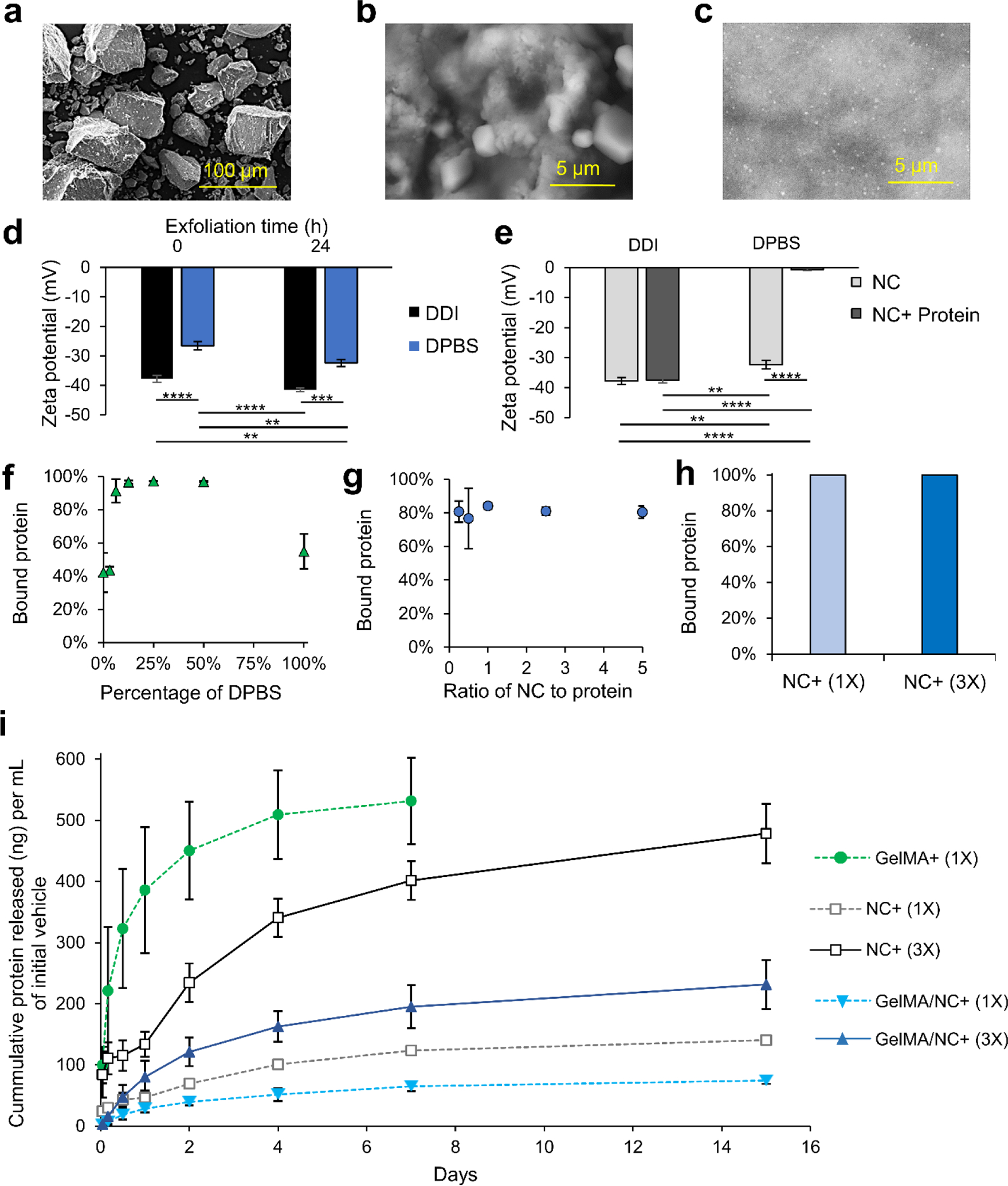
The interaction of Laponite® nanoclay (NC) with proteins and optimization for enhanced binding and sustained release. Scanning electron microscopy (SEM) of a) NC powder (scale bar: 100 μm), b) partial NC exfoliation in 100% DPBS (scale bar: 5 μm), and c) full NC exfoliation in 10% DPBS (scale bar: 5 μm). d) Increased zeta potential was observed for NC in both double-deionized (DDI) water and DPBS over a 24 h period demonstrating continual exfoliation. e) The zeta potential of NC in DDI water did not significantly change, while the zeta potential of NC in DPBS became approached zero after mixing with protein. f) Protein binding to NC solutions was highly dependent on salt solution. Near 100% binding was observed in 10–50% DPBS, while lower percentages of protein binding were detected at 0% and 100% DPBS concentrations. g) NC protein binding was not affected by increasing or decreasing the ratio of NC to protein. h) The application of 0.05% (w/v) of NC in a 10% DPBS solution yielded almost complete protein binding regardless of the protein concentration (1X and 3X correspond with 333 ng/mL and 1,000 ng/mL final concentration of protein encapsulated in GelMA, respectively). i) The mass of IGF-1 released normalized to the initial vehicle (DPBS or gel) volume over 15 days from GelMA alone (GelMA+ (1X)), protein-bound NC alone and not encapsulated (NC+ (1X) and (3X)), and protein-bound NC encapsulated in GelMA (GelMA /NC+ (1X) and GelMA/NC+ (3X)). The hydrogel only (GelMA+ (1X) group resulted in a quick release of almost all of the initially bound protein over 7 days. All NC groups slowed the release and continuously released protein over 15 days. The NC groups that were not encapsulated had a quicker release than the encapsulated groups of the same initial protein concentration. The hydrogel-encapsulated groups, GelMA /NC+ (1X) and GelMA/NC+ (3X), had similar release profiles regardless of the initial concentration of bound protein and retained a significant portion of the IGF-1 within the scaffolds. The 3X groups released almost 3 times the protein per volume of initial gel to the 1X groups.
The size of the NC complexes in varying ionic concentrations was quantified using dual light scattering (Figure S4). In 100% DPBS, the NC are unable to exfoliate and the measured unsettled particles were around 2 μm in size with a narrow distribution (Figure S4i). As the salt concentration is reduced to 50%, the size of the particle gradually decreases to about 1.6 μm with a similar narrow distribution (Figure S4i). Through the further reduction of the salt to 10%, three distinct distributions appear at ranges of 2–10 nm, 100–420 nm, and 550–4200 nm, representing primary single NCs, secondary structures, and tertiary structures, respectively (Figure S4ii–iii). The three distinct peaks of the 10% solution are similar to the 0% pure double-deionized (DDI) water with minor shifts in size and differences in amplitude (Figure S4ii–iii). The addition of 10% salt led to more individual suspended NC discs (2–10 nm) than the 0% solvent (Figure S4iii).
To better decipher the interaction of positively charged growth factors with NC discs, the electrostatic properties of the NC discs in different conditions were evaluated using zeta potential measurements (Figure 3d,e). First, the effect of time was studied by measuring the differences in zeta potential immediately after making the NC solution, and after 24 h incubation at room temperature (Figure 3d). Both the 100% DPBS and double deionized (DDI) water solutions experienced decreases in their zeta potentials, becoming more negative, indicating further exfoliation. The unexfoliated NC in 100% DPBS solution resulted in a drop from −26.6 mV to −32.4 mV. This was less negative before and after incubation than the NC in DDI solution which had a zeta potential of −37.8 mV to −41.5 mV. These results indicated more NC exfoliation in the DDI solution compared to the 100% DPBS solution, which correlated with the SEM observations. Exfoliation has been shown to be completed about 24 h after incubation at room temperature.40 For this reason, samples used for binding were exfoliated for at least 24 h. The solutions were then evaluated to assess the protein binding capability to NC (Figure 3 e–j). Measuring the zeta potential before and after the addition of protein demonstrated an increase in zeta potential following the protein addition. However, the zeta potential of NCs exfoliated in DDI experienced a less extreme increase in zeta potential after the addition of protein compared to NCs exfoliated in 100% DPBS, most likely due to the formation of an impeding double layer from excessive sodium ions (Figure 3e). The zeta potential of the DDI NCs insignificantly decreased from −37.8 mV to −37.5 mV, while the zeta potential of the 100% DPBS NCs significantly decreased from −26.6 to −4.2 mV. The move toward neutral in the zeta potential indicates the coverage of the negative flat surface charge of the NCs due to protein binding.
3.4. Nanoclay (NC) protein binding and release studies
To optimize the protein binding ability, we mixed NCs exfoliated in varying concentrations of DPBS with protein and assessed unbound protein via enzyme-linked immunosorbent assays (ELISAs; Figure 3f). Similar to the zeta potential measurements, the DDI (0% DPBS) solution did not bind protein more effectively. The results further demonstrated that increasing the concentration of salt to 10–50% increased the protein binding to near 100%. These results correlate with the increased exfoliation of Laponite® with small concentrations of ions in the solvent. Therefore, a 10% DPBS was selected as the solution for binding proteins to NC. We next assessed the effect of NC to protein ratio in the 10% DPBS solution (Figure 3g and Figure S5). From a mass ratio of NC to the protein of 1:4 to 4:1, no significant difference in binding ability was observed (Figure 3g). In a larger spectrum, no increase in binding was observed in NC to protein ranges up to 50:1 (Figure S5). No significant difference in binding ability was observed (Figure S5). With this information, 0.05% (w/v) NCs in a 10% DPBS solution was selected as the protein binding solution. Protein was bound at 10,000 ng/mL and 3,333 ng/mL and then diluted to 1,000 ng/mL and 333.3 ng/mL in GelMA, respectively referred to as 1X and 3X from here forward. The 3X and 1X protein concentrations were determined by preliminary release data to ensure that the protein release would sustain protein levels within the range of the initial IGF-1 concentration optimization experiments. The ELISA results confirmed almost all protein was bound to the NC in these conditions (Figure 3h).
The release kinetics of the IGF-1 bound to NCs and encapsulated in GelMA were then studied. Five different groups were considered in the release kinetics studies: IGF-1 in GelMA (GelMA+ (1X), 1X protein-bound NC in DPBS (NC+ (1X)), 3X protein-bound NC in DPBS (NC+ (3X)), 1X protein-bound NC encapsulated in GelMA (GelMA/NC+ (1X)), and 3X protein-bound NC encapsulated in GelMA (GelMA/NC+ (3X)) (Table 1). The final concentration of IGF-1 was 333.33 ng/mL and 1,000 ng/mL, respectively for 1X and 3X groups. The same volume of the samples was added to cell culture inserts, crosslinked, and interfaced with DPBS solution to allow protein release for 15 days (Figure 3i and Figure S6). The GelMA+ (1X) group released almost all of the initial protein encapsulated after 7 days. All of the groups utilizing NC experienced a slowed but continuous growth factor release over the 15 days. The unencapsulated NC+ (1X) and (3X) protein-bound groups more quickly released the loosely bound IGF-1 relative to the encapsulated groups (Figure 3i). Within each subset of NC alone or NC encapsulated in GelMA, the percentage of IGF-1 released over time from the 1X and 3X protein levels was similar (Figure S6). However, the three times loaded IGF-1 mass in the NC 3X groups resulted in almost exactly three times the mass release of protein per unit volume of initial gel compared to the 1X groups (Figure 3i). The 50% difference in the released percentages and masses of IGF-1 between the unencapsulated NC+ (1X and 3X) and encapsulated GelMA/NC+ (1X and 3X) groups indicates the ability of the NC to retain the eluted IGF-1 in the hydrogel network (Figure 3i and Figure S6).
Table 1:
Experimental groups used in protein release kinetics
| Group Name | Composition | IGF-1 concentration (ng/mL) |
|---|---|---|
| GelMA+ (1X) | IGF-1 freely mixed and encapsulated into GelMA hydrogel | 333.33 |
| NC+ (1X) | IGF-1 bound to Laponite® NC in DPBS | 333.33 |
| NC+ (3X) | IGF-1 bound to Laponite® NC in DPBS | 1000 |
| GelMA/NC+ (1X) | IGF-1 bound to Laponite® NC encapsulated in GelMA hydrogel | 333.33 |
| GelMA/NC+ (3X) | IGF-1 bound to Laponite® NC encapsulated in GelMA hydrogel | 1000 |
3.5. Composite hydrogel material properties
To verify that the release of IGF-1 within the scaffold was not due to erosion of the scaffold as seen in keratin VML scaffolds, we studied the swelling characteristics of GelMA hydrogel while submerged in DPBS, with and without the presence of NC (the same condition as the in vitro IGF-1 protein release).27 Both GelMA and GelMA/NC groups experienced swelling over 1.5 days, increasing their masses by almost 60% (Figure 4a). Hydration of GelMA prepolymer solutions has been observed by several studies.13 No significant degradation in DPBS was observed in either group, and no significant difference was observed in the hydrogel with the addition of NCs. The lack of degradation in DPBS over 7 days indicated that the controlled release of IGF-1 was due to the diffusion-dependent release from the NC. To study the degradation of the scaffolds under extreme conditions, a 2 U/mL collagenase solution was used to investigate mass reduction differences between GelMA with and without the addition of NCs (Figure 4b). Both GelMA and GelMA/NCs experienced a bit of swelling after an hour, but we observed steady degradation after the initial swelling. No significant differences were observed in the degradation rate with and without NCs, and the samples were completely dissolved after 27 h. The degradation of the GelMA is consistent with the supporting literature.39 Therefore, under normal physiological conditions present in VML injuries, containing lower levels of collagenase, the GelMA/NC composite scaffolds are expected to maintain structural support while enabling cell permissibility. Scaffolds composed of similar concentrations of GelMA have demonstrated stability after implantation into VML injury models.13, 32
Figure 4.
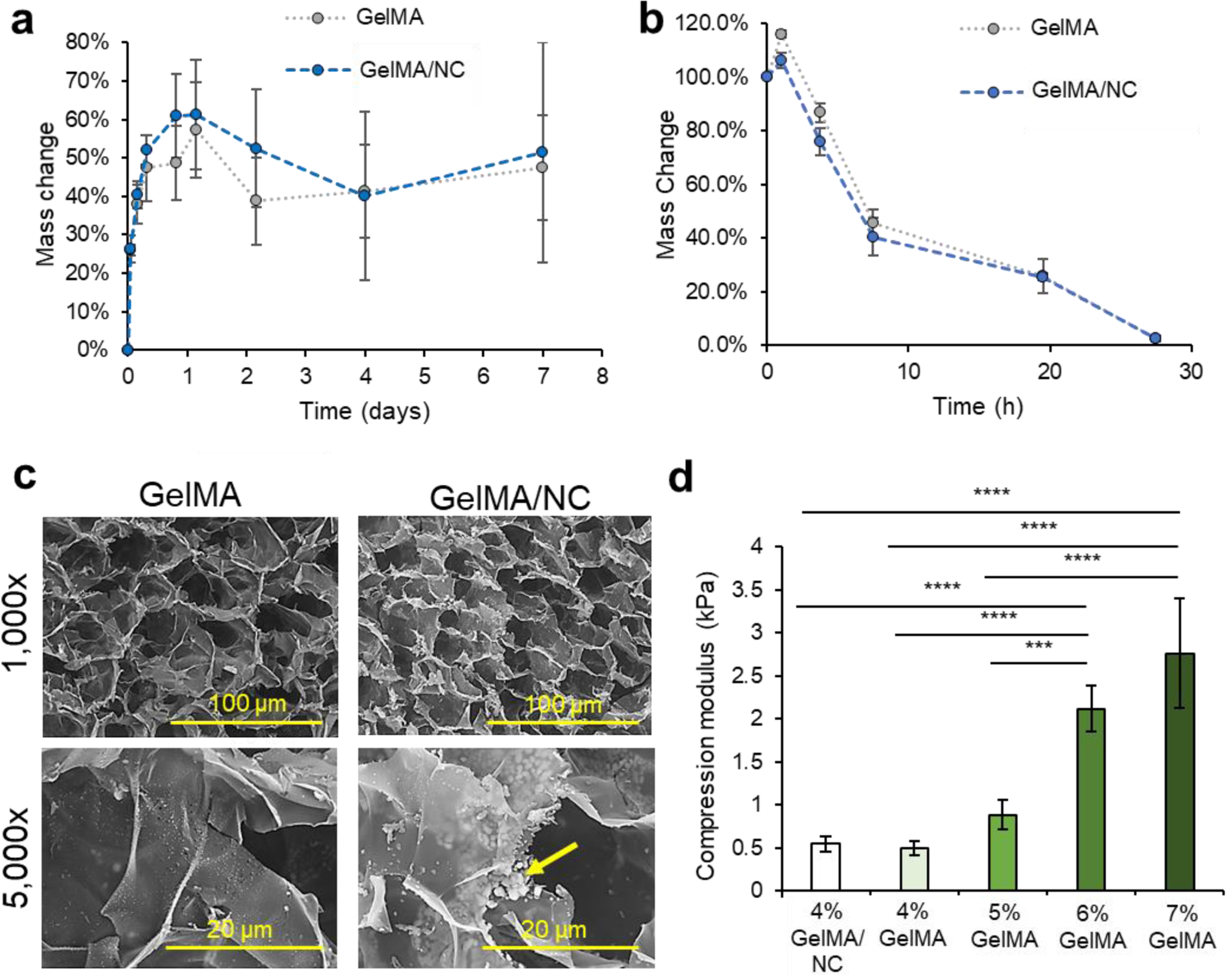
Material properties of GelMA/NC hydrogel composite. a) A swelling assay demonstrated similar absorption characteristics of GelMA and GelMA/NC discs in DPBS. Hydrogel mass was increased for 1 day and then was retained for up to 7 days. b) An accelerated degradation assay in a 2 U/mL collagenase solution yielded similar degradation profiles of GelMA and GelMA/NC hydrogels. Total degradation in both conditions was observed after 27 h. c) SEM micrographs of GelMA and GelMA/NC demonstrated similar porous morphologies at 1,000X and 5,000X (scale bars of 100 μm and 20 μm, respectively). Pore sizes were of the same order of magnitude. The GelMA/NC group did display powdered clusters of NC (yellow arrow) in between GelMA layers as demonstrated at 5,000x. d) During muscle contraction, implanted scaffolds undergo compressive strengths, so to evaluate the ability of the scaffold to withstand compressive loading, GelMA, and GelMA/NC discs were evaluated for compressive modulus. The addition of NC in the 4% GelMA hydrogel did not statistically increase the stiffness of the hydrogel. The 4% GelMA hydrogels were significantly less stiff than the 5%, 6%, and 7% samples.
To further assess any changes with the addition of NC to GelMA, cross-sections of hydrogel samples were observed using SEM (Figure 4c). Both GelMA and GelMA NC have similar pore sizes on the order of 10–40 um at 1,000X magnification. No microscale morphology changes were observed which is consistent with 1H NMR analysis of the 53% degree of functionalized GelMA (Figure S7). 1H NMR analysis revealed similar levels of crosslinking degree of GelMA and GelMA/NC hydrogels: 61% and 70%, respectively. Despite the lack of major structural change, at 5,000x, clusters of apparent NCs were visibly located between the thin layers of the GelMA hydrogel. These clusters were not visible on the GelMA-only samples. The dispersed clusters of NCs match the observed distribution of NCs within the hydrated GelMA samples (Figure S8). However, the NC distribution shown in Figure S8 may not account for smaller, submicrometer NC complex visualization due to weak signal and post encapsulation binding with ethidium homodimer-1.
The compressive modulus of the GelMA NC hydrogels was assessed and compared to NC free GelMA at varying concentrations from 4–7% (w/v) (Figure 4d). We compressed hydrogel discs of 10 mm in diameter, and their compressive modulus was calculated from the slope of the stress-strain plot over the first 10% of strain. A reduction of the compressive modulus was observed with a decrease in GelMA mass concentration (w/v). While a 7% GelMA has more rigidity to support normal loading, the lower compressive modulus of 4% GelMA hydrogels (and lower concentrations) is what enables cellular ingrowth. A statistically significant difference was not observed between the 4% GelMA samples with (0.543 ± 0.084 kPa) and without (0.497 ± 0.081 kPa) NCs.
3.6. Bulk nanoengineered GelMA/NC composite proliferation analysis
The optimized nanoengineered hydrogel scaffold was subsequently assessed for its cell viability and proliferative ability (Figure S9 and Figure 5). Various GelMA hydrogel groups were used to encapsulate C2C12 myoblasts: GelMA (GelMA-), GelMA loaded with NC (GelMA/NC-), GelMA encapsulating NC with bound IGF-1 at 333.3 ng/mL with respect to the crosslinked gel (GelMA/NC+ (1X)), GelMA encapsulating NC with three times the mass of bound IGF-1 at 1,000 ng/mL with respect to the crosslinked gel (GelMA/NC+ (3X)), GelMA with IGF-1 at 333.3 ng/mL (GelMA+ (1X)), and GelMA with IGF-1 supplemented at 10 ng/mL in the media (GelMA/Media+) (Table 2). Crosslinked samples were cultured in a proliferation medium until day 7. Almost all of the cells in the encapsulated groups were alive and did not have different viability than control groups (Figure S9). A PrestoBlue™ metabolic assay was used to assess the proliferation of myoblasts within the hydrogels at days 1 and 3 (Figure 5a). The only statistically significant groups on day 1 of culture were the GelMA/NC+ (1X) and GelMA/NC+ (3X) groups, indicating an increase in metabolic activity in the NC control release gels. While no statistically significant increase was observed at day 1 between the GelMA/NC+ (1X) and GelMA/NC+ (3X) groups, the GelMA/NC+ (3X) group had a larger relative fluorescence of 16,511.75 ± 993.99 compared to the GelMA/NC+ (1X) group at a fluorescence of 14,598.25 ± 1,337.00. Similar trends were observed at day 3 with the GelMA/NC+ (3X) group having the largest signal at 54,596.25 ± 1,443.89. The GelMA/NC+ (3X) group was statistically significant from all groups at day 3. The GelMA/NC+ (1X) group then had the next highest signal at 46,860.50 ± 948.46 followed closely by the GelMA/NC- group at 44,996.50 ± 2,574.47. The increase in proliferative ability with the NC vehicle encapsulated in the GelMA is supported by the demonstrated ability of NC to manipulated myogenic behavior.41
Figure 5.
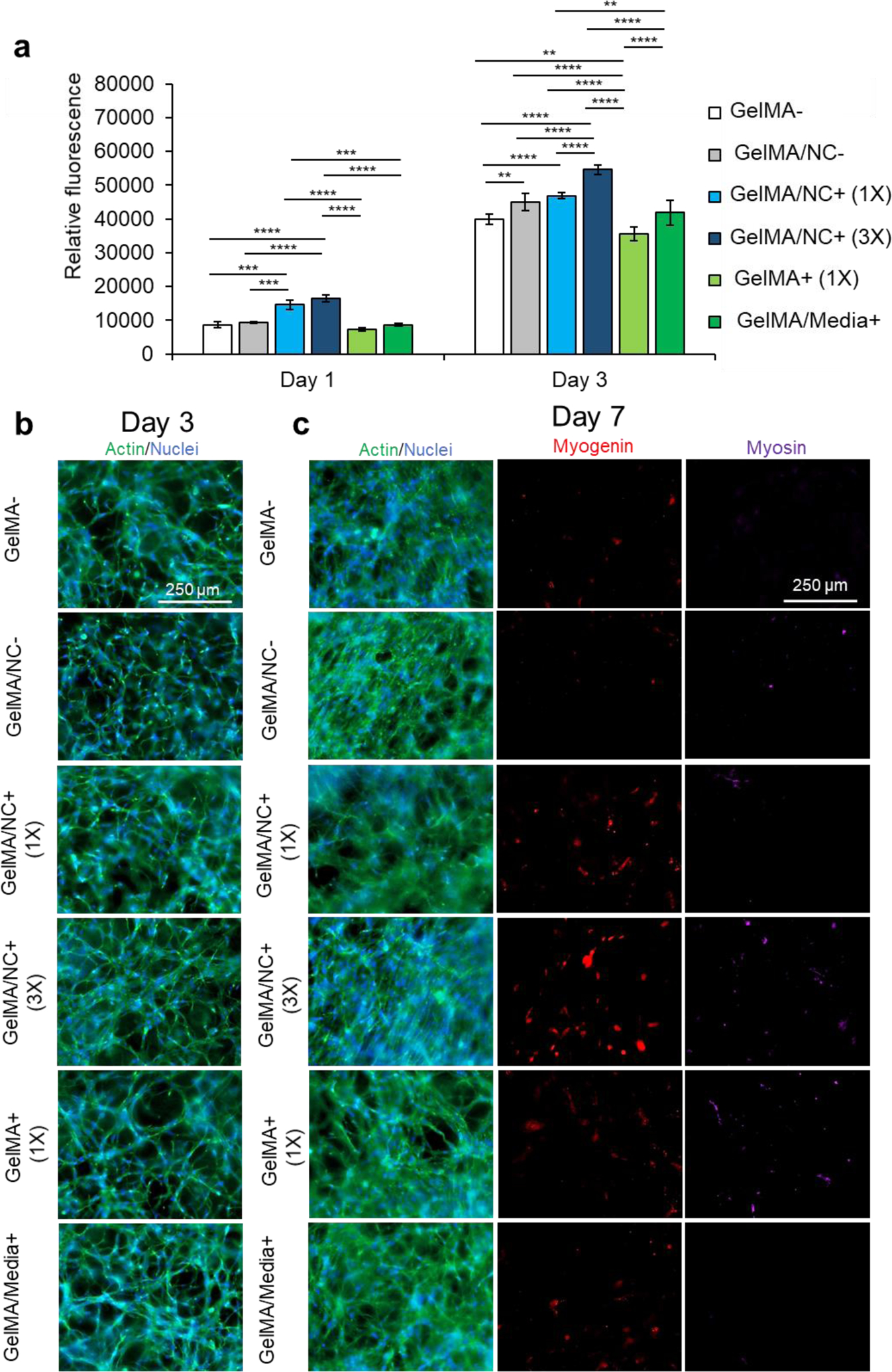
Proliferation studies within NC and control hydrogel conditions. (a) A PrestoBlue™ viability assay was used to determine the proliferative ability of C2C12 myoblasts encapsulated in varying GelMA-based hydrogel conditions: pristine GelMA (GelMA-), GelMA loaded with NC (GelMA/NC-), GelMA encapsulating 1X concentration of IGF-1 bound to NC (GelMA/NC+ (1X)), GelMA encapsulating 3X concentration of IGF-1 bound to NC (GelMA/NC+ (3X)), GelMA encapsulating IGF-1 (GelMA+ (1X)), and GelMA with IGF-1 added to the culture media (GelMA/Media+). On day 1, the GelMA/NC+ (1X) and GelMA/NC+ (3X) groups demonstrated the highest metabolic activity. On day 3 of culture, the GelMA/NC+ (3X) group had the most metabolic activity and was significantly different from all other groups. The lowest group was the GelMA+ (1X) group followed by the GelMA- control group. Exogenous IGF-1 added to the media in the GelMA/Media+ group did not appear to significantly increase the proliferation ability of the myoblasts relative to the GelMA- control. The GelMA/NC- control group demonstrated that the NC vehicle alone resulted in an increase in metabolic activity in association with previous studies. No significant difference was observed between the GelMA/NC- and the GelMA/1X groups on day 3. b) Actin (green) and nuclear (blue) staining of the hydrogel groups at 3 days of culture demonstrates the proliferative ability to vary IGF-1 delivery mechanisms. c) Actin (green) and nuclear (blue), myogenin (red), and myosin (purple) immunofluorescent images of myoblasts at day 7 of culture. Even without serum-deprived initiation of differentiation, the GelMA/NC+ (3X) condition initiated C2C12 differentiation displayed by the myogenin positive cells. Without induced differentiation, no apparent functional myotubes formed as evidenced by the negative myosin staining. Scale bars of 250 μm for all micrographs.
Table 2:
Experimental groups used in the in vitro analysis of bulk nanoengineered scaffolds
| Group Name | Composition | IGF-1 concentration (ng/mL) |
|---|---|---|
| GelMA- | GelMA hydrogel alone | 0 |
| GelMA/NC- | NC encapsulated in GelMA hydrogel | 0 |
| GelMA/NC+ (1X) | IGF-1 bound to Laponite® NC encapsulated in GelMA hydrogel | 333.33 |
| GelMA/NC+ (3X) | IGF-1 bound to Laponite® NC encapsulated in GelMA hydrogel | 1000 |
| GelMA+ (1X) | IGF-1 freely mixed and encapsulated into GelMA hydrogel | 333.33 |
| GelMA/Media+ | GelMA hydrogel alone with IGF-1 supplemented in the culture medium | 10 in the culture medium |
The hydrogel samples were imaged for actin (green) and nuclei (blue) at day 3 (Figure 5b) and actin and nuclei (green and blue), myogenin (red), and MHC (purple) at day 7 (Figure 5c). The proliferation of cells appeared the highest in the 3X sample (Figure 5b). Some groups like the GelMA/Media+ appear to have an increase in cell number, but a portion of the increase in cell number is due to migration to the IGF-1 rich media interface at the surface of the hydrogel. Similar local areas of cellular alignment are observed to some extent in all groups with the most alignment observed in GelMA/NC- and GelMA/NC+ (3X). Staining against myogenin, an intermediate myogenic differentiation marker, demonstrated that the GelMA/NC+ (3X) group was capable of initiating differentiation without serum deprivation (Figure 5c). This is due to the self-differentiation through cell communication as a result of high proliferation in this group and is similar to extremely dense 2D control samples before induction of differentiation (Figure S11). However, the self-induced differentiation in group 3X was early and did not include more myosin-positive cells than the other groups.
3.7. Bulk nanoengineered GelMA/NC composite differentiation analysis
Subsequently, all the groups underwent serum-deprived differentiation on day 7 of culturing and were cultured for a total of 21 days (Figure 6). The samples were stained for actin (green) and nuclei (blue), myogenin (red), and myosin heavy chain (purple) on day 14, one week after differentiation (Figure 6a). Intermediate differentiation noted that the GelMA/NC+ (3X) and GelMA/NC- group still had the most myogenin-positive cells at day 14. However, no clear trends are visible between the other groups. On day 21 of culture, actin and nuclei were stained as well as myosin (Figure 6b). Several single MHC-positive cells that were not yet fused to form multinucleated myotubes were visible in all groups; single MHC-positive cells have been shown to be a sign of incomplete differentiation.42 To investigate the possibility of false positive staining of cultures, 2D controls were used to demonstrate that non-differentiated day 3 cells were not stained with the used antibodies (Figure S10b). Further, day 21 differentiated controls also demonstrated that single cell MHC-positive cells were found, albeit at a lower concentration, in highly confluent and differentiated samples (Figure S10c(i)). The least myosin-positive fibers were found in the GelMA- control with an average of 10 myosin heavy chain positive tubes (Figure 6c). Only the GelMA/NC+ (1X) group (20 average tubes) and GelMA/NC+ (3X) group (21 average tubes) were statistically significant from the GelMA- group (p = 0.0257 and p = 0.0083, respectively). Despite the lack of a significant difference in average tube number, there was a difference in average tube length (Figure 6d). The longest fibers were found in the GelMA/NC+ (3X) group (622 ± 25 μm) and then the GelMA/NC+ (1X) group (487 ± 21 μm). The shortest fibers (220 ± 5 μm) were found in the GelMA- group. By day 21 of cell culture, most of the GelMA/NC+ (1X) and GelMA/NC+ (3X) samples had portions of each scaffold that were contracted from the internal stresses generated from the differentiated myotubes. This phenomenon was not observed in any other groups.
Figure 6.
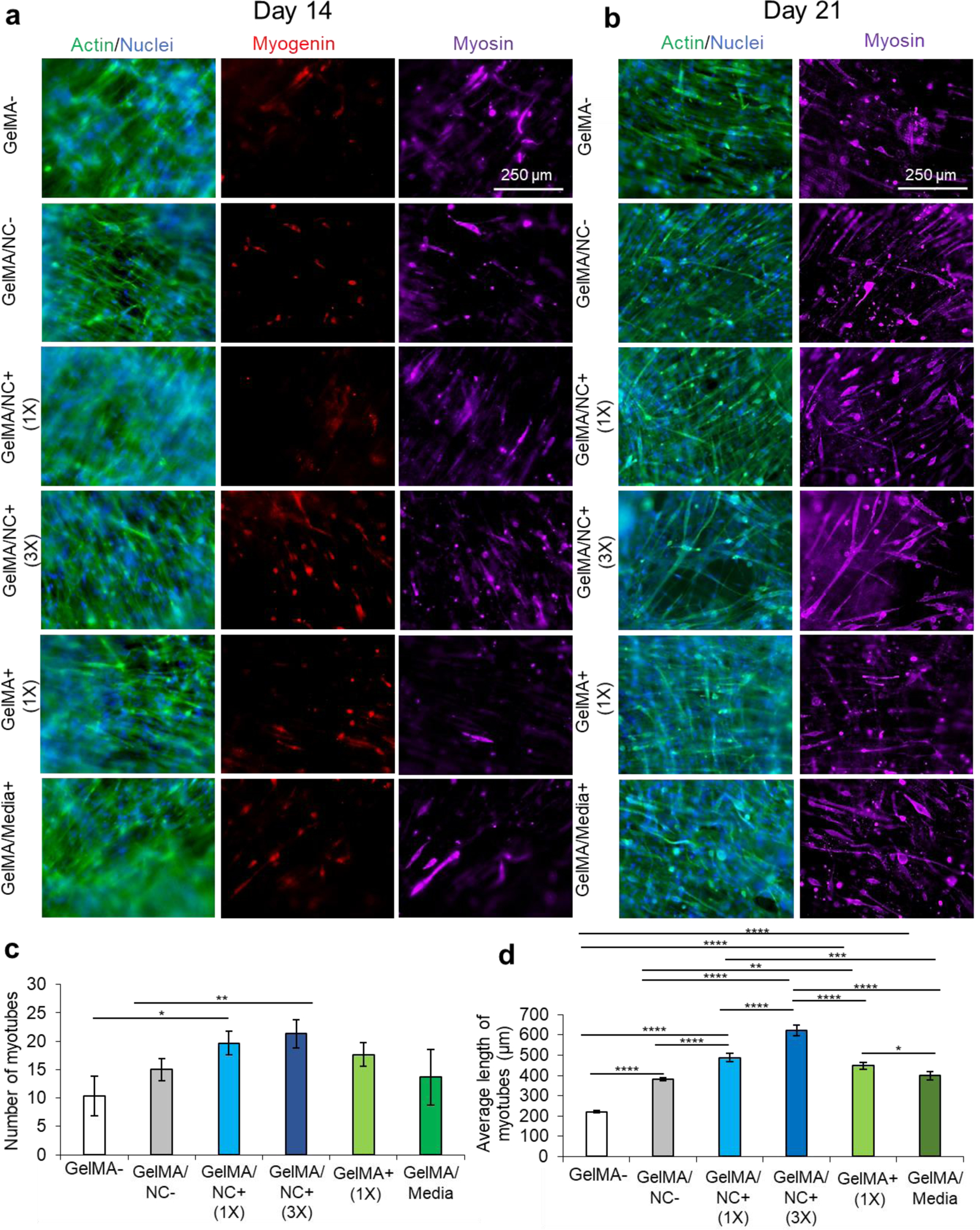
Immunofluorescent imaging of C2C12 differentiation at 14 and 21 days of culture. a) Immunofluorescent staining of actin (green) and nuclei (blue), myogenin (red), and myosin heavy chain (MHC) (purple) at 14 days of culture and 7 days after serum-starved induced differentiation. On day 14, the GelMA/NC+ (3X) condition has the most myosin-positive myotubes. b) Immunofluorescent staining of actin (green) and nuclei (blue); and myosin (purple) at 21 days of culture and 14 days after serum-starved induced differentiation. The longest myosin positive myotubes formed in the GelMA/NC+ (3X) samples. c) Quantified number of MHC myotubes per micrograph. The GelMA/NC+ (1X) and GelMA/NC+ (3X) had significantly more myotubes per micrograph. d) Quantified average length of MHC myotubes per micrograph. All groups had longer myotubes than the GelMA- control. The longest myotubes were found in the GelMA/NC+ (1X) and the GelMA/NC+ (3X) groups. Scale bars of 250 μm for all micrographs.
4. Discussion
GelMA hydrogels are capable of supporting the compressive loads of skeletal muscle and offer cell-binding sites.32, 37 However, until recently, the factors affecting the cell permissibility of these and other 3D hydrogels have not been widely explored.43–45 While it has been demonstrated that stiffer 2D substrates with elastic moduli around 11 kPa offer the best myogenic differentiation, the 3D cell culture environment is more complex.46, 47 To combat this, recent attempts have been made at retaining the mechanical rigidity of GelMA hydrogels and similar hydrogels while changing their microscale architecture to include pores and other migratory pathways using techniques such as bead sintering.43 However, these techniques require higher concentrations of the base hydrogel to enable scaffold fidelity, which prevents cell infiltration into the bulk gel and motivates cells to migrate around into the microporous topography. This can altogether be avoided by reducing the stiffness of GelMA hydrogel to resemble the soft and cell permissive environments of acute soft tissue injuries.44 It has been observed that higher concentrations of GelMA hydrogels do not allow cells to migrate and proliferate throughout the scaffold resulting in lower cellular interaction and differentiation.45 Further, higher hydrogel concentrations reduce diffusion of growth factors which motivates migration to the hydrogel surface and higher cellular proliferation on the 2D hydrogel interface with the culture medium.44, 47 To reduce the stiffness of GelMA, the concentration and the degree of functionalization can be reduced. A medium degree of functionalization has been shown to effectively crosslink and demonstrate higher stiffness while reducing degradation compared to the same weight percentages of GelMA with lower degrees of functionalization. Our hydrogel optimization results reaffirm that lower weight concentrations of GelkMA hydrogel improve myogenic proliferation and permissibility throughout the scaffold (Figure S1).
IGF-1 is a key regulator of myoblast proliferation, myofiber hypertrophy, and immune modulation of the skeletal muscle wound in vivo.20–22 However, the effect of IGF-1 concentration tittered over a physiology range on the proliferation and differentiation of skeletal muscle in a 3D scaffold has not been deeply investigated by previous researchers. Bolus IGF-1 delivery of free IGF-1 has failed to adequately maintain needed IGF-1 levels for increased proliferation, differentiation, and functional recovery.20, 23 Additionally, IGF-1 is often encapsulated in scaffolds at extremely high doses.25–28 We sought to explore the optimization of IGF-1 concentrations within our cell permissive GelMA hydrogel in a range up to 2 orders of magnitude above native IGF-1 concentrations found in healthy, noncritical skeletal muscle regeneration.3 Our varied range of IGF-1 from 0 to 100 ng/mL in the culture media of our selected 4% (w/v) GelMA hydrogel demonstrated a dose-dependent proliferative nature of myoblasts due to IGF-1 concentration (Figure 1a). As expected, higher levels of IGF-1 were able to increase the number and thickness of MHC myotubes (Figure 2). IGF-1 increased differentiation by increasing the density of the cellular network during the proliferative phase and post-differentiative hypertrophy.
Building upon the studied effect of IGF-1 within physiological conditions, the challenge was to sustain and improve the local retention of IGF-1 within a hydrogel network without media supplementation, combating the limitations of previous clinical trials and tissue engineered constructs and generating a translatable scaffold. To do this, Laponite® NC was used to electrostatically bind and release IGF-1. Laponite® NCs are biocompatible, degrade into nontoxic ionic products, and have been reported to increase myogenic differentiation.36, 41 Laponite® NCs require exfoliation from their manufactured stacked state and this is most effectively modulated through control of ionic concentration in the dissolution solvent (Figure 3). Through exfoliation in solvents, the interstitial sodium ions are released and allow the negatively charged faces and positively charged edges to interact with each other, polymeric networks, or proteins (Figure 3b,c). Ionic levels around 20 mM have shown improved exfoliation by increasing the removal of sodium through the presence of cations.48, 49 These structures often form a house of cards structure depending on solvent condition but still facilitate protein binding and release.36 The ability of the NCs to electrostatically bind with protein was improved by using small concentrations of added salt (Figure 3f). A concentration of 10% (v/v) DPBS which approximates the ideal ionic concentrations for exfoliation while preventing flocculation (<20 mM) was used for the remainder of the studies.48, 49 The exfoliated NCs proved to be a suitable protein carrier by binding protein at high NC to protein ratios (Figure 3a–h). The NC prevented a burst release as seen with GelMA alone (GelMA+ (1X)) group and with other carriers used with IGF-1 (Figure 3i).25 Further, the NC enabled control over the mass of protein released by adjusting the initial mass of protein bound. Also, when encapsulated in GelMA hydrogel, the NC retained IGF-1 within the scaffold.
To ensure that the release mechanisms of the NC hydrogels were independent of degradation, a swelling assay was performed on bulk hydrogel samples under the same conditions applied during the release experiments (Figure 4a). Unlike keratin scaffolds, the GelMA and GelMA/NC samples did not degrade to release the IGF-1.27 However, GelMA and GelMA/NC hydrogels are subjected to degradation under collagenase exposure within 27 h (Figure 4b). The ability for degradation using natural enzymes is the key to maintaining the ability of the skeletal muscle to remodel the implanted scaffold at the appropriate rate for tissue replacement and regeneration. At the morphological level, we did not observe any changes to the normal porous morphology of the hydrogels with the addition of NCs (Figure 4c). We did not observe a change in the compressive modulus of 4% GelMA hydrogel with the addition of NC, yet the concentration-dependent stiffness of GelMA hydrogels correlates with the inability of GelMA hydrogels higher than 4% to retain encapsulated cells and to promote cell proliferation within the hydrogel (Figure 4d).
The NCs demonstrated their ability to maintain the bioactivity of the loaded IGF-1 on the proliferation of myoblasts (Figure 5). NCs within a hydrogel without IGF-1 also increased proliferation over hydrogel only controls further highlighting the role of NC-based materials in skeletal muscle applications and the importance of nanocarrier selection for tissue application (Figure 5a).36, 41 IGF-1 continued to demonstrate a dose-dependent effect on proliferation which should be balanced with off-target effects. Notably, the NC mediated-controlled release of IGF-1 resulted in higher proliferation than IGF-1 loaded into the GelMA hydrogel without NC vehicles (GelMA+ (1X)) and GelMA cultured in the presence of IGF-1 supplemented medium (GelMA/Media+). The retention of the IGF-1 in the hydrogel network enables the retention of myogenic cells to proliferate within the matrix (Figure 5b). Additionally, the GelMA/NC+ (3X) group displayed the ability of this scaffold to initiate differentiation without the need for serum deprivation (Figure 5c).
After the initiation of differentiation, the higher levels of proliferation from the engineered NC groups resulted in higher and better differentiation as indicated by myogenin and myosin staining (Figure 6). All the control gels loaded with IGF-1 and/or NC groups had a higher number of myotubes than the GelMA- (not loaded with NCs or IGF-1) condition, indicating the effect of both Laponite® NCs and IGF-1 on skeletal muscle differentiation (Figure 6b,c). Not only were there more MHC myotubes in the GelMA/NC+ (1X) and GelMA/NC+ (3X) groups, they also resulted in an average longer more mature MHC myotubes than other groups (Figure 6b and 5d). Most importantly, the controlled release of IGF-1 loaded into NCs better improved cell proliferation and subsequent differentiation than IGF-1 encapsulated into GelMA hydrogel. In the same manner, the presence of NCs not loaded with IGF-1 showed a better cell response than the medium supplemented IGF-1 condition.
The improved differentiation of the NC nanoengineered scaffolds has implications for tailored growth factor delivery and translation of myogenic scaffolds for a variety of applications. Most notably, the 0.5 mL of 10 ng/mL IGF-1 supplemented medium that was changed every two days (as was done in this study) requires 44% more IGF-1 in the GelMA/Media+ condition compared to the IGF-1 encapsulated in the GelMA+ (1X) condition. The reduction in the use of myogenic growth factors has further potential benefits in the rapidly growing clean meat industry. Further, medium supplemented scaffolds are only relevant to in vitro cultured constructs and are not beneficial after implantation into a patient. The retentive ability of the NC mediated-controlled release of IGF-1 enables the reduction of total utilized IGF-1 relative to previous studies, limiting system exposure, and improving myogenic proliferation and differentiation. The lower mass of IGF-1 in NC bound GelMA offers scalable monetary savings when used in complex printing applications that require higher volumes of culture medium that are required for clinical applications.9
5. Conclusions
We developed a skeletal muscle scaffold that was optimized to enable cellular ingrowth, proliferation, and differentiation relevant for critically sized defects and extreme mass loss such as VML injury. For this purpose, GelMA-based hydrogel was selected because of its native cell-binding sites, versatility in terms of surgical application, and controlled degradation. The 4% (w/v) optimized GelMA concentration enabled higher cell infiltration and retention compared to higher concentrations. Physiological relevant concentrations of IGF-1, due to its role in skeletal muscle regeneration, were optimized and loaded into the hydrogel to evaluate the most effective myogenesis. While the proliferative effects of IGF-1 were verified to be dose-dependent, lower concentrations than the values reported in previous findings demonstrated effective myogenesis. Laponite® NC was used to electrostatically bind to IGF-1 and control its delivery for retention within the hydrogel structure for improving myogenic expansion and differentiation while negating the negative effects of burst release or reliance on scaffold degradation. We controlled the exfoliation by modifying ionic concentration to tune the binding to IGF-1. In vitro release studies demonstrated a continual release of IGF-1 from encapsulated NC over 15 days and resulted in almost half of the released IGF-1 being retained within the hydrogel. We further demonstrated that varying levels of IGF-1 could be bound to a fixed concentration of NC for tuning the quantity of protein release. The use of NCs did not affect the morphological, mechanical, swelling, or degradation properties of the GelMA hydrogels, an improvement over the erosion-dependent release of the most common scaffolds used to deliver IGF-1 for the treatment of VML injuries. The slow release of IGF-1 from the NC encapsulated hydrogels also resulted in higher cell proliferation compared to the other tested conditions. The engineered composite hydrogel offers an effective, tunable scaffold that enables the delivery of myogenic growth factors. It is suitable for several microfabrication and delivery techniques for the treatment of severe skeletal muscle loss.
Supplementary Material
Acknowledgments
The financial support from the National Institutes of Health (AR077132, AR073822) and the University of Connecticut are gratefully acknowledged. C.R. would like to acknowledge the financial support from the Polish Ministry of Science and Higher Education through the scholarship for outstanding young scientists and from the Foundation for Polish Science (FNP).
References
- 1.Rivas DA and Fielding RA, in Encyclopedia of Human Nutrition (Third Edition), ed. Caballero B, Academic Press, Waltham, 2013, DOI: 10.1016/B978-0-12-375083-9.00188-4, pp. 193–199. [DOI] [Google Scholar]
- 2.Hall J, Guyton and Hall Textbook of Medical Physiology, Elsevier, 13th edn., 2015. [Google Scholar]
- 3.Nuutila K, Sakthivel D, Kruse C, Tran P, Giatsidis G and Sinha I, Wound Repair and Regeneration, 2017, 25, 408–413. [DOI] [PubMed] [Google Scholar]
- 4.Järvinen TAH, Järvinen TLN, Kääriäinen M, Kalimo H and Järvinen M, The American Journal of Sports Medicine, 2005, 33, 745–764. [DOI] [PubMed] [Google Scholar]
- 5.Scully D, Naseem KM and Matsakas A, Acta Physiologica, 2018, 223, e13071. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Saul D, Böker KO, Ernst J, Lehman W and Schilling AF, Biomed Res Int, 2018, 2018, 1984879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grasman JM, Zayas MJ, Page RL and Pins GD, Acta Biomater, 2015, 25, 2–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corona BT, Henderson BEP, Ward CL and Greising SM, Physiological Reports, 2017, 5, e13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samandari M, Quint J, Rodríguez-delaRosa A, Sinha I, Pourquié O and Tamayol A, Advanced Materials, 2021, n/a, 2105883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grogan BF, Hsu JR and C. Skeletal Trauma Research, JAAOS - Journal of the American Academy of Orthopaedic Surgeons, 2011, 19. [Google Scholar]
- 11.Greising SM, Dearth CL and Corona BT, Cells Tissues Organs, 2016, 202, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman JW and Browe DP, in Bio-Instructive Scaffolds for Musculoskeletal Tissue Engineering and Regenerative Medicine, eds. Brown JL, Kumbar SG and Banik BL, Academic Press, 2017, DOI: 10.1016/B978-0-12-803394-4.00008-2, pp. 187–199. [DOI] [Google Scholar]
- 13.Russell CS, Mostafavi A, Quint JP, Panayi AC, Baldino K, Williams TJ, Daubendiek JG, Hugo Sánchez V, Bonick Z, Trujillo-Miranda M, Shin SR, Pourquie O, Salehi S, Sinha I and Tamayol A, ACS Applied Bio Materials, 2020, 3, 1568–1579. [DOI] [PubMed] [Google Scholar]
- 14.Gao G and Cui X, Biotechnology Letters, 2016, 38, 203–211. [DOI] [PubMed] [Google Scholar]
- 15.Ostrovidov S, Salehi S, Costantini M, Suthiwanich K, Ebrahimi M, Sadeghian RB, Fujie T, Shi X, Cannata S, Gargioli C, Tamayol A, Dokmeci MR, Orive G, Swieszkowski W and Khademhosseini A, Small, 2019, 15, 1805530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fallahi A, Yazdi IK, Serex L, Lesha E, Faramarzi N, Tarlan F, Avci H, Costa-Almeida R, Sharifi F, Rinoldi C, Gomes ME, Shin SR, Khademhosseini A, Akbari M and Tamayol A, ACS Biomaterials Science & Engineering, 2020, 6, 1112–1123. [DOI] [PubMed] [Google Scholar]
- 17.Sicari BM, Rubin JP, Dearth CL, Wolf MT, Ambrosio F, Boninger M, Turner NJ, Weber DJ, Simpson TW, Wyse A, Brown EH, Dziki JL, Fisher LE, Brown S and Badylak SF, Sci Transl Med, 2014, 6, 234ra258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costantini M, Testa S, Rinoldi C, Celikkin N, Idaszek J, Colosi C, Barbetta A, Gargioli C and Święszkowski W, in Biofabrication and 3D Tissue Modeling, The Royal Society of Chemistry, 2019, DOI: 10.1039/9781788012683-00184, pp. 184–215. [DOI] [Google Scholar]
- 19.Qazi TH, Duda GN, Ort MJ, Perka C, Geissler S and Winkler T, J Cachexia Sarcopenia Muscle, 2019, 10, 501–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad SS, Ahmad K, Lee EJ, Lee Y-H and Choi I, Cells, 2020, 9, 1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tonkin J, Temmerman L, Sampson RD, Gallego-Colon E, Barberi L, Bilbao D, Schneider MD, Musarò A and Rosenthal N, Molecular Therapy, 2015, 23, 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tidball JG, Nature Reviews Immunology, 2017, 17, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philippou A and Barton ER, Growth Horm IGF Res, 2014, 24, 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.del Carmen Ortuño-Costela M, García-López M, Cerrada V and Gallardo ME, Journal of Cellular and Molecular Medicine, 2019, 23, 3784–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borselli C, Storrie H, Benesch-Lee F, Shvartsman D, Cezar C, Lichtman JW, Vandenburgh HH and Mooney DJ, Proceedings of the National Academy of Sciences, 2010, 107, 3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borselli C, Cezar CA, Shvartsman D, Vandenburgh HH and Mooney DJ, Biomaterials, 2011, 32, 8905–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomblyn S, Pettit Kneller EL, Walker SJ, Ellenburg MD, Kowalczewski CJ, Van Dyke M, Burnett L and Saul JM, J Biomed Mater Res B Appl Biomater, 2016, 104, 864–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passipieri JA, Baker HB, Siriwardane M, Ellenburg MD, Vadhavkar M, Saul JM, Tomblyn S, Burnett L and Christ GJ, Tissue Eng Part A, 2017, 23, 556–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samandari M, Alipanah F, Majidzadeh-A K, Alvarez MM, Trujillo-de Santiago G and Tamayol A, Applied Physics Reviews, 2021, 8, 021404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mostafavi A, Quint J, Russell C and Tamayol A, in Biomaterials for Organ and Tissue Regeneration, eds. Vrana NE, Knopf-Marques H and Barthes J, Woodhead Publishing, 2020, DOI: 10.1016/B978-0-08-102906-0.00023-4, pp. 499–528. [DOI] [Google Scholar]
- 31.Nuutila K, Samandari M, Endo Y, Zhang Y, Quint J, Schmidt TA, Tamayol A and Sinha I, Bioactive Materials, 2022, 8, 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quint JP, Mostafavi A, Endo Y, Panayi A, Russell CS, Nourmahnad A, Wiseman C, Abbasi L, Samandari M, Sheikhi A, Nuutila K, Sinha I and Tamayol A, Advanced Healthcare Materials, 2021, n/a, 2002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jodat YA, Zhang T, Al Tanoury Z, Kamperman T, Shi K, Huang Y, Panayi A, Endo Y, Wang X, Quint JP, Arnaout A, Kiaee K, Hassan S, Lee J, Flores Huidobro Martinez A, Ochoa SL, Lee KJ, Calabrese M, Carlucci A, Tamayol A, Sinha I, Pourquié O and Shin WR, Nature Portfolio, 2021, DOI: 10.21203/rs.3.rs-146091/v1. [DOI] [Google Scholar]
- 34.Samandari JPQM, and Tamayol A, in Musculoskeletal Tissue Engineering Elsevier, 1st edn., 2021. [Google Scholar]
- 35.Wang H, Hansen MB, Löwik DWPM, van Hest JCM, Li Y, Jansen JA and Leeuwenburgh SCG, Advanced Materials, 2011, 23, H119–H124. [DOI] [PubMed] [Google Scholar]
- 36.Gaharwar AK, Cross LM, Peak CW, Gold K, Carrow JK, Brokesh A and Singh KA, Advanced Materials, 2019, 31, 1900332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N and Khademhosseini A, Biomaterials, 2015, 73, 254–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Assmann A, Vegh A, Ghasemi-Rad M, Bagherifard S, Cheng G, Sani ES, Ruiz-Esparza GU, Noshadi I, Lassaletta AD, Gangadharan S, Tamayol A, Khademhosseini A and Annabi N, Biomaterials, 2017, 140, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Lang Q, Yildirimer L, Lin ZY, Cui W, Annabi N, Ng KW, Dokmeci MR, Ghaemmaghami AM and Khademhosseini A, Advanced healthcare materials, 2016, 5, 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karpovich A VM, Sapronova N, Sukharev V, Ivanov V, Orient J Chem, 2016, 32(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poussard S, Decossas M, Le Bihan O, Mornet S, Naudin G and Lambert O, Int J Nanomedicine, 2015, 10, 1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki A, Pelikan Richard C and Iwata J, Molecular and Cellular Biology, 2015, 35, 1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seymour AJ, Shin S and Heilshorn SC, Advanced Healthcare Materials, 2021, n/a, 2100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaudhuri O, Cooper-White J, Janmey PA, Mooney DJ and Shenoy VB, Nature, 2020, 584, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deshmukh DV, Pasquero N, Rathore G, Zvick J, Bar-Nur O, Dual J and Tibbitt MW, Bioeng Transl Med, 2020, 5, e10181–e10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engler AJ, Sen S, Sweeney HL and Discher DE, Cell, 2006, 126, 677–689. [DOI] [PubMed] [Google Scholar]
- 47.Yamada KM and Sixt M, Nature Reviews Molecular Cell Biology, 2019, 20, 738–752. [DOI] [PubMed] [Google Scholar]
- 48.Sheikhi A, Afewerki S, Oklu R, Gaharwar AK and Khademhosseini A, Biomater Sci, 2018, 6, 2073–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afghah F, Altunbek M, Dikyol C and Koc B, Scientific Reports, 2020, 10, 5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


