Abstract
Background:
Gastrointestinal symptoms are common in Coronavirus Disease 2019 (COVID-19), related to infection of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) of intestinal cells through the angiotensin converting enzyme 2 (ACE2) receptor in the brush border. Also, patients are treated with multiple antibiotics. Therefore, an increase in gut dysbiosis and in the prevalence of Clostridium difficile infection (CDI) is expected in patients with COVID-19.
Methods:
A PubMed search was conducted using the terms “gut microbiota,” “gut mycobiota,” “dysbiosis” AND “COVID-19”; “Clostridium difficile,” “Clostridioides difficile” AND “COVID-19”; “probiotics,” “bacteriotherapy AND COVID-19.” Only case series, observational and experimental studies were included.
Results:
A total of 384 papers were retrieved and 21 fulfilled selection criteria. Later, a new paper was identified, thus 22 papers were reviewed. Main findings: (1) gut bacterial dysbiosis has been found in fecal samples of COVID-19 patients, with enrichment of opportunistic organisms and decrease of beneficial commensals such as Faecalibacterium prausnitizii. Dysbiosis is related to inflammatory markers and illness severity. (2) There is evidence for abnormal gut barrier and bacterial translocation with a negative impact in the lungs. (3) Fungal dysbiosis correlating with pulmonary mycobiota, has also been found. (4) There is controversy in the CDI rates among COVID-19 patients versus controls and pandemic versus prepandemic era. (5) There is no available evidence yet to support bacteriotherapy in COVID-19. (6) Fecal microbiota transplantation (FMT) has been proposed for COVID-19, although there is no evidence to support it. Also, FMT can be safely used during the pandemic for CDI if strict screening protocols for donors and fecal product are implemented.
Conclusions:
In COVID-19 there is bacterial and fungal dysbiosis that correlates with systemic and pulmonary inflammation, and illness severity. Further investigations are warranted to determine the efficacy of bacteriotherapy and FMT for modulating gut dysbiosis in COVID-19.
Key Words: coronavirus disease 2019, gut bacterial dysbiosis, gut fungal dysbiosis, gut mycobiota, gut-lung axis, gut-barrier disruption, bacteriotherapy, fecal microbiota transplantation
BACKGROUND
The Coronavirus Disease 2019 (COVID-19) produced by the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), was recognized by the CDC as producing gastrointestinal (GI) symptoms following reports initially from China and afterwards from other parts of the world, about these clinical manifestations.1 The COVID-19 GI symptoms ranged from nausea, vomiting, heartburn/reflux, abdominal pain, bloating, diarrhea, and anorexia.2,3 GI symptoms could be present before the typical manifestations of COVID-19 of fever, headache, and dyspnea; appeared during the illness; and in some cases even be the only symptoms (ie, atypical manifestations), retarding the diagnosis of COVID-19 by 2 days.4 In addition, the presence of SARS-CoV-2 viral RNA was also reported in feces, even persisting for longer times after clearance of the RNA from the upper respiratory tract,5,6 although, not necessarily correlating with GI symptoms.5 Besides the above findings, there have been reports of inflammatory processes in the digestive tract such as the presence of aphthous lesions and even ulcerations, together with GI bleeding,7 thickening of the gut wall resembling that of inflammatory bowel disease and even elevation of fecal calprotectin levels.8,9 These changes may be related to the angiotensin converting enzyme 2 (ACE2) required by the SARS-CoV-2 to infect the human cells; and the highest expression of this receptor in the human body is in the brush border of the intestinal cells.10 In addition, there is a well-known close relationship between the integrity and immune regulation of the intestinal mucosal barrier and a healthy balance in the gut microbiome.11,12 Disrupting this balance can contribute to systemic inflammation.12 Considering the above, it is plausible to presume that gut dysbiosis is present in patients with COVID-19. Also, in COVID-19, the so called “gut-lung axis” is thought to be an underlying mechanism for which several bacterial metabolites and bacterial fragments may impact the immune response in the lungs.13 Thus, it has been postulated that manipulation of the microbial patterns through the use of probiotics and dietary fibers consumption may contribute to reduce inflammation and strengthen the immune system response in COVID-19 infection.14 Despite all of the above, the relationship between gut microbes and COVID-19 patients is not well understood. In addition, considering that hospitalized patients with COVID-19 are treated with multiple antibiotics, one may expect an increased prevalence of Clostridioides difficile infection (CDI). Therefore, we sought to review the literature for reports of gut dysbiosis and other related factors in patients with COVID-19, as well as the frequency of CDI, and evidence for gut microbiota modulation.
METHODS
A literature search was conducted to determine the presence of original studies analyzing the presence of gut bacterial microbiota and/or fungal (mycobiota) dysbiosis and related factors in patients with COVID-19, and the presence of CDI in those patients. In addition, to review reports on the presence of CDI in those patients. Thus, on May 14, 2021, a search was conducted on PubMed using the terms “gut microbiota,” or “gut mycobiota,” or “dysbiosis” AND “COVID-19,” as well as “Clostridium difficile,” or “Clostridioides difficile” AND “COVID-19,” or “probiotics AND COVID-19,” or “bacteriotherapy AND COVID-19.” Only case series, observational and experimental studies, were included. Case reports, review papers, editorials or hypothesis generating papers, were excluded. The abstracts of the retrieved papers were initially reviewed by 4 authors (L.L-G., M.E.C-B., W.H-C., M.S.), and a meeting was done to conciliate the final papers that were to be included in this review.
RESULTS
The gut microbiota/mycobiota search retrieved 216 papers of which 10 papers fulfilled the selection criteria, while the C. difficile search retrieved 40 papers of which 9 were selected, and in bacteriotherapy and probiotics, 128 papers were retrieved, of which only 1 paper was selected. In addition, when finalizing the drafting of this review, a new paper on bacteriotherapy focused on GI symptoms was published and we considered that it was an important study to include as well as one paper on microbiota that had not been identified with our search engine. The gut microbiota search identified 2 papers on fecal microbiota transplantation (FMT) and 1 that analyzed the fecal microbiota and afterwards studied the effect of FMT. Thus, a total of 22 papers were reviewed. Based on the topics of the selected papers, we classified them into the following subjects: (1) gut bacterial microbiota in COVID-19; (2) bacterial translocation/gut barrier dysfunction in COVID-19; (3) gut mycobiota in COVID-19; (4) C. difficile and COVID-19; (5) Oral bacteriotherapy in patients with COVID-19; (6) FMT for COVID-19 and C. difficile during the pandemic.
Gut Bacterial Microbiota in COVID-19
Seven studies analyzed the gut microbiota in patients with COVID-1915–21 (Table 1). The first study by Zuo and colleagues, conducted metagenomic sequencing of fecal samples of 15 patients with COVID-19 in Hong Kong to identify changes in the microbiota during hospitalization, its association with disease severity and with the presence of fecal SARS-CoV-2 RNA. The COVID-19 patients showed significant abnormalities in the microbiota characterized by enrichment of opportunistic organisms and decrease in beneficial commensals. The abundance of microorganisms such as Coprobacillus, Clostridium ramosum, and Clostridium hathewayi (normal inhabitants of the gut microbiome sometimes classified as opportunistic organisms for causing bacteremia) correlated with severity of COVID-19; while the abundance of Faecalibacterium prausnitzii (an important commensal-butyrate producing organism necessary for maintaining the gut barrier and immunity with anti-inflammatory properties), inversely correlated with disease severity. During hospitalization, Bacteroides dorei, Bacteroides thetaiotaomicron, Bacteroides massiliensis, and Bacteroides ovatus were also diminished. The importance of the latter ones is that they have been related to the regulation of the expression of the ACE2 receptor in the murine gut.15 In addition, Zuo et al15 showed that Bacteroides spp. were inversely correlated with the levels of SARS-CoV-2 in fecal samples; suggesting a protective effect of Bacteroides spp. against the SARS-CoV-2 infection of the gut cells. Another study from Ganzhou City, China, by 16S sequencing, reported that gut microbiome composition of discharged COVID-19 patients differed from that of the general population at the phylum level. These differences were characterized by a lower proportion of Firmicutes (41.0%) and Actinobacteria (4.0%), and a higher proportion of Bacteroidetes (42.9%) and Proteobacteria (9.2%).16
TABLE 1.
Studies Reporting Bacterial and Fungal Gut Dysbiosis in Patients With COVID-19
| References | Region, Country | Subjects/Patients (n) | Analyzed Sample | Microbiota Findings | Details of COVID-19 Microbiota |
|---|---|---|---|---|---|
| Bacterial dysbiosis | |||||
| Zuo et al15 | Hong Kong | COVID-19 (15) | Feces | Enrichment of opportunistic bacteria Decrease of commensal symbiotics | Coprobacillus, Clostridium ramosum, Clostridium hathewayi: Correlated with severity Faecalibacterium prausnitzii: Inversely correlated with severity Decrease of Bacteroides spp.: inversely correlated with SARS-CoV-2 in feces |
| Liu et al16 | Ganzhou City, China | COVID-19 after hospital discharge (11) | Feces | Microbiome composition differed from that of the general population | Higher proportion of Bacteroidetes and Proteobacteria Lower proportion of Firmicutes and Actinobacteria |
| Gu et al17 | Zhejiang, China | COVID-19 (30) A-H1N1 (24) HC (30) | Feces | COVID-19 showed increase in relative abundance of opportunistic pathogens and decrease in commensal symbionts A-H1N1 lower diversity and microbial composition vs. COVID-19 | Increase in Streptococcus, Rothia, Veillonella, Actinomyces Fusicatenibacter, Romboutsia, Intestinibacter, Actinomyces, Erysipelatoclostridium, distinguished COVID-19 vs. HC Streptococcus, Rothia, Veillonella, Actinomyces, correlated with CRP, D-dimer |
| Mazzarelli et al18 | Rome, Italy | COVID-19+ pneumon. hospitalized in ICU (9) COVID-19+ pneumon. in IDW (6) Hospitalized controls (8) | Rectal swabs | ICU patients displayed lower richness w/o difference in diversity Microbiota differences in COVID-19 with different disease severity compared with controls | IDW had increase in Proteobacteria ICU had decrease of Fusobacteria, Spirochetes vs. Controls ICU vs. IDW, increase in Staphylococcaceae, Microbacteriaceae, Micrococcaceae, Pseudonocardiaceae, Erysipelotrichales, others: and decrease in Carnobacteriaceae, Pectobacteriaceae, Moritellaceae, Selenomonadaceae, Micromonosporaceae, Coriobacteriaceae |
| Tang et al19 | Wuhan, China | COVID-19 general (20) COVID-19 severe (19) COVID-19 critically ill (18) | Feces | Decrease in abundance of beneficial butyrate bacteria | Independently of severity, decrease in Faecalibacterium prausnitzii, Clostridium butyricum, Clostridium leptum, Eubacterium rectale Critically ill, increase in Enterococcus/Enterobacteriaceae ratio |
| Yeoh et al20 | Hong Kong | COVID-19 basal (100) After resolution (30/100) Non-COVID-19 controls (78) | Feces collected for 30 d | Decrease in immunomo-dulatory commensals Microbiome composition correlated with inflammatory markers | Decrease in Faecalibacterium prausnitzii, Eubacterium rectale, Bifidobacteria |
| Chen et al22 | Zhejiang, China | COVID-19 (30) in acute (illness to viral clearance), convalescence (viral clearance to 2 weeks after hospital discharge), postconvalescence (6 mo after) phases non-COVID-19 controls (30) | Feces | Lower microbiota richness in acute phase in COVID-19 vs. non-COVID-19 controls Increase in richness from acute to convalescence period Reduction in postconvalescence richness was associated with inflammatory markers, reduction in pulmonary function, higher ICU admissions | α-Diversity by microbiota richness (Chao 1 index), was reduced in COVID-19 PCoA of Bray-Curtis distance analysis demonstrated that overall microbial composition of patients with COVID-19 deviated from the non-COVID-19 controls (analysis of similarities, R=0.201, P=0.001) |
| Lv et al21 | Zhejiang, China | COVID-19 in hospitalization, after discharged (56) HC (47) | Feces | COVID-19 patients enriched with metabolites that should be absorbed, cannot be synthesized or are harmful Decrease in microbe-related compounds | Sucrose (should be absorbed), D-pinitol (cannot be synthesized), correlated with either Actinomyces, Sphingomonas, Rothia, Streptococcus parasanguinisthe; 2-palmitoyl-glycerol (should be metabolized) negatively correlated with Aspergillus sp. Oxalic acid negatively with Aspergillus |
| Fungal dysbiosis | |||||
| Zuo et al23 | Hong Kong | COVID-19 (30) Community acquired pneumon. (9) Healthy controls (30), all 2-3/times per week | Feces | COVID-19 hospitalized patients had more heterogenous mycobiome than controls Increase mycobiome diversity, opportunistic fungal pathogens | COVID-19 enrichment by 20% in Candida albicans but absent in healthy controls, Candida auris, Aspergillus flavus Aspergillus flavus and niger were detected even after clearance of nasopharingeal SARS-CoV-2 Aspergillus spp. present in respiratory secretions linked to fecal presence |
| Lv et al24 | Hangzhou, China | COVID-19 (67) A-H1N1 (35) HC (48) | Feces | Similar Diversity in COVID-19 vs. HC; higher in both groups than in H1N1 No difference according to COVID-19 severity Total amount of fungi either in COVID-19 or H1N1 was higher than in HC In COVID-19 and A-H1N1 some altered gut fungi were associated with altered gut bacteria (mutualism, commensalism competition?) | COVID-19 depletion in phylum, Ascomycota (Aspergillaceae, such as Penicillium citrinum, Penicillium polonicum, Aspergillus spp.) Basidiomycota (Malassezia yamatoensis, Rhodotorula mucilaginosa, Moesziomyces aphidis, Trechispora sp., Wallemia sebi) Mucoromycota (Mucor racemosus) Ascomycota and its members were negatively correlated with Lachnospiraceae and its genera Agathobacter, Dorea, Roseburia; Ruminococcaceae and its genera Butyricicoccus, Faecalibacterium; Eggerthella, Veillonella Mucoromycota positively correlated with Peptostreptococcaceae, Bifidobacterium, Fusicatenibacter, Intestinibacter, and Aspergillus with Agathobacter In COVID-19 patients, Aspergillus niger was associated with diarrhea; Penicillium citrinum inversely correlated CRP; Rhodotorula mucilaginosa negatively correlated with circulating ACE |
ACE indicates angiotensin converting enzyme; COVID-19, Coronavirus Disease 2019; CRP, C reactive protein; HC, healthy controls; ICU, intensive care unit; IDW, infectious disease wards; PCoA, principal coordinate analysis; Pneumon, pneumonia.
In a cross-sectional study from Zhejiang, China, among 30 patients with COVID-19, 24 patients with influenza A-H1N1 and 30 controls, Gu and colleagues analyzed the differences in the fecal microbiota by 16S ribosomal RNA gene V3-V4 region sequencing. Compared with healthy controls (HC), the COVID-19 patients presented an increase in the relative abundance of opportunistic bacteria such as Streptococcus, Rothia, Veillonella, and Actinomyces and a decrease in beneficial commensal symbionts. Five final biomarkers (Fusicatenibacter, Romboutsia, Intestinibacter, Actinomyces, Erysipelatoclostridium) were distinguished between the COVID-19 group and HC, with ROC-plot area under the curve (AUC) value of 0.89 [95% confidence interval (CI): 0.8-0.97]. In contrast, the influenza A-H1N1 patients showed lower diversity and different overall microbial composition compared with the COVID-19 patients.17 In addition, 7 final biomarkers (Streptococcus, Fusicatenibacter, Collinsella, Dorea, Agathobacter, Eubacterium hallii group, Ruminococcus torques group) distinguish the 2 cohorts, with an AUC of 0.94 (95% CI: 0.87-1.00). Furthermore, Fusicatenibacter, Roseburia, and Ruminococcaceae UCG−013, were depleted in COVID-19 compared with HC and were negatively correlated with C reactive protein (CRP), procalcitonin, or D-dimer levels. In contrast, CRP and D-dimer levels positively correlated with COVID-19-enriched bacteria (Streptococcus, Rothia, Veillonella, and Actinomyces). The authors suggested that the gut microbiota has the potential as a diagnostic biomarker, can correlate with systemic inflammation, and a treatment target for COVID-19.17
Mazzarelli and colleagues reported about the gut microbiota composition in COVID-19 pneumonia patients that were hospitalized in the intensive care unit (COVID-19 ICU) or in the infectious disease wards (COVID-19 IDW), in Rome, Italy. Using 16S rRNA gene sequencing of rectal swabs, the COVID-19 ICU patients were found to have a decrease in the microbial richness (Chao1 index) compared with that of COVID-19 IDW and on non-COVID-19 hospitalized controls. In contrast, there were no differences in species numbers and evenness of species abundance (Shannon index). At the phylum level, the COVID-19 ICU demonstrated an increase of Proteobacteria compared with controls. Also, a decrease of Fusobacteria and Spirochetes was found, with the latter decreased in COVID-19 ICU patients versus controls. These findings suggest a difference in rectal microbiota according to COVID-19 severity, which in the future may serve as biomarkers for patients stratification.18 Tang and colleagues, in Wuhan China, analyzed fresh stool specimens by quantitative polymerase chain reaction (q-PCR) in 20 patients with general, 19 with severe, and 18 with critical illness, COVID-19. They also reported that abundance of beneficial butyrate bacteria such as F. prausnitzii, Clostridium butyricum, Clostridium leptum, and Eubacterium rectale, were significantly decreased in patients with COVID-19 independently of their severity. In addition, the abundance of conditional pathogenic bacteria such as Enterobacteriaceae, decreased as the concentration of Enterococcus increased with severity. Accordingly, the Enterococcus/Enterobacteriaceae ratio significantly increased in critical patients, suggesting that this ratio can be used to predict death in critically ill patients.19
In a study conducted in 2-hospital cohorts in Hong Kong, by Yeoh and colleagues, blood, and serial stool samples from patients with COVID-19 were collected for 30 days after the resolution of the viral infection and were compared with samples of non-COVID-19 controls, for the same period. The gut microbiome composition was characterized by shotgun sequencing of total DNA extracted from stools. They found that commensals with a known immunomodulatory role such as F. prausnitzii, Eubacterium rectale, and Bifidobacteria, were still underrepresented in patients 30 days after disease resolution. Furthermore, this microbiome composition was concordant with the elevation of plasma inflammatory cytokines and blood markers, including CRP, lactate dehydrogenase, aspartate aminotransferase and gamma-glutamyl transferase. These markers are considered to be involved in the modulation of host inflammatory responses in COVID-19, again showing that composition of intestinal microbiota might determine the disease severity.20
Chen and colleagues, in Zhejiang, China, conducted a longitudinal study by 16S rDNA sequencing to monitor the gut microbiota abnormalities in patients with COVID-19 in 3 time points: the acute phase (from the beginning of the infectious disease until the viral clearance), during convalescence (from the viral clearance until 2 wk after the hospital discharge) and postconvalescence (6 mo after hospital discharge). In this study, the gut microbiota richness determined by the Chao 1 index, was lower during the acute phase compared with that of noninfected controls. In addition, there was a nonsignificant increase in the Chao 1 index from the acute phase to the convalescence and postconvalescence time points. Further, in the postconvalescence, patients were divided in 2 groups based on a low (≤259, n=15) or high (>259, n=15) Chao 1 index. Patients with a reduction in the postconvalescence richness were found to have higher levels of CRP, higher frequency of ICU admissions, and use of high flow oxygen therapy, as well as a reduction in pulmonary function. Also, the microbiota richness was not restored after 6 months of recovery. These findings also support the concept that gut microbiota composition during COVID-19 is related to the pathogenesis of the acute pulmonary damage.22 Several mechanisms were postulated to explain this relationship, including bacterial translocation from the gut to the lungs, and an immune modulatory effect mediated by the gut microbiota metabolites. Therefore, the authors suggested that manipulating the gut microbiota could be an important strategy in the treatment of patients with COVID-19 to accelerate their recovery.22
To investigate the relationship of the intestinal infection of SARS-CoV-2 with digestion and absorption, Lv and colleagues, also in Zhejiang, China, used gas chromatography-mass spectrometry, to analyze the fecal metabolome of 56 patients with SARS-CoV-2 infection during hospitalization and after discharge. They reported that COVID-19 patient feces were enriched with important nutrients that should be metabolized or absorbed, such as sucrose and 2-palmitoyl-glycerol; diet-related components that cannot be synthesized by humans, such as 1,5-anhydroglucitol and D-pinitol; and harmful metabolites, such as oxalate. In contrast, some purine metabolites, low-water-soluble long-chain fatty acids, compounds rarely occurring in nature such as D-allose and D-arabinose, and microbe-related compounds such as 2,4-di-tert-butylphenol, were depleted in the feces of COVID-19 patients. In addition, some of these molecules correlated with gut microbes. For example, sucrose and D-pinitol correlated with at least one among Actinomyces, Sphingomonas, Rothia, and Streptococcus parasanguinisthe, while 2-palmitoyl-glycerol negatively correlated with Aspergillus sp., and oxalic acid negatively with Aspergillus. Although the alterations in the fecal metabolome among COVID-19 patients reported in this study may reflect malnutrition and intestinal inflammation, the relationship with gut microbiota abnormalities needs to be further examined.21
Bacterial Translocation/Gut Barrier Dysfunction in COVID-19
Prasad and colleagues studied the gut barrier dysfunction by analyzing the plasma microbiome in COVID-19 patients admitted to Birmingham Hospital in Alabama. Plasma samples were studied by rRNA 16S sequencing for circulating microbiome, metatranscriptome, and intestinal permeability markers, in 30 COVID-19 patients and 16 HC. The COVID-19 patients were divided in 2 groups based on plasma samples availability: the first group included 14 patients who were analyzed for the circulating microbiome, and the second group of 16 patients for permeability markers. The first group presented a microbial disturbance in the bloodstream with higher rates of Proteobacteria, Firmicutes and Actinobacteria. Also, there was a predominance of gram-negative bacteria (Acinetobacter, Nitrospirillum, Cupriavidus, Pseudomonas, Aquabacterium, Burkholderia, Caballeronia, Parabhurkholderia, Brevibacterium, and Sphingomonas) over gram-positive ones (Staphylococcus and Lactobacillus). The level of fatty acid-binding protein-2 (FABP2), a marker of intestinal barrier damage; and peptidoglycan and lipopolysaccharides, markers of gut microbial peptide translocation into the systemic circulation, were significantly increased in COVID-19 patients compared with HC.25 These findings suggest that an abnormal gut barrier may represent a source of bacteremia which could contribute to worsen COVID-19 disease outcomes,25 and support the theory of the so called “gut-lung axis.”
Although the studies described in sections 1 and 2 are heterogenous in their studied samples and methodological analysis, some data consistently emerges across them. For example, studies using 16S sequencing found higher proportions of phylum Proteobacteria and Bacteroidetes among patients with COVID-19.16,18,25 Another consistent finding among these patients is a decrease in the beneficial F. prausnitzii, both by metagenomic sequencing and q-PCR.19,20,23 Finally, microbial richness was also diminished among COVID-19 patients.18,22
Gut Mycobiota in COVID-19
Two studies of fungal microbiome (mycobiota) were identified23,24 (Table 1). The first study by Zuo and colleagues in Hong Kong, investigated the fecal mycobiome by shotgun metagenomics. They studied 30 patients with COVID-19, 9 patients with community acquired pneumonia, and 30 HC. Stool samples were collected 2 to 3 times per week from the moment when they were hospitalized until discharged and clearance of SARS-CoV-2 from nasopharyngeal samples. COVID-19 patients had an increase in opportunistic fungal pathogens such as Candida albicans (significantly enriched in 20% of COVID-19 ill patients, but absent in HC), Candida auris, and Aspergillus flavus. The latter one, and Aspergillus niger, were detected even after clearance of SARS-CoV-2 and the resolution of respiratory symptoms. It seems that hospitalized patients have more heterogeneous gut mycobiome than healthy people, a finding that might represent a higher risk for severe pneumonia.23 Furthermore, Aspergillus species were reported in respiratory tract secretions linked to its fecal presence in a subset of patients who presented cough during hospitalization. For its part, Candida albicans colonization is supposed to aggravate inflammation in the gut and nongut tissues.23
The second study by Lv and colleagues in Hangzhou, China, investigated 67 patients with COVID-19, 35 H1N1 infected patients and 48 HC, using internal transcribed spacer (ITS) 3-ITS4 sequencing, to analyze the association between the gut mycobiota and clinical features. Their results showed that both the fungal α-diversity and the relative abundance of the most altered taxa in the fecal gut mycobiome of COVID-19 patients and H1N1-infected patients, were significantly lower than those in HC.24 The COVID-19 and H1N1-infected patients demonstrated depletion of Aspergillus and Penicillium. However, in COVID-19 patients, intestinal mycobiota profiles were similar in those with mild and severe symptoms. Also, in COVID-19 patients, Aspergillus niger was significantly associated with diarrhea, while Penicillium citrinum had a negative correlation with CRP. Also, Rhodotorula mucilaginosa was highly negatively correlated with blood angiotensin-converting enzyme levels. Finally, there was no significant difference in the number of gut fungi between COVID-19 patients at admission and discharge. These findings confirm that fungal gut dysbiosis takes place both in COVID-19 and H1N1 infected patients. Also, in COVID-19, this fungal dysbiosis does not improve after hospitalization.24
The explanation for fungal dysbiosis in COVID-19 is not completely elucidated. However, the authors discussed the evidence reporting that bacterial dysbiosis,26 tissue damage and the presence of an inflammatory environment can cause fungal overgrowth in the gut.27 In contrast, species and overgrowth of fungi such as Candida or Malassezia, have been shown to be related to Crohn’s disease as well as to exacerbate experimental colitis in mouse models.28 Thus, considering the presence of gut microbial dysbiosis reported in COVID-19 as well as the tissue inflammation, further studies are warranted to determine if fungal dysbiosis is a cause or a consequence of COVID-19.
C. difficile and COVID-19
Eight retrospective observational studies reported on CDI in COVID-19, focusing on the rates of coinfection and risk factors, as well as on the CDI rates before and after the pandemic (Table 2).29–36 For example, Sandhu and colleagues in Detroit, Michigan, reported an increase in the CDI rate in their institution during March-April 2020 compared with the January-February 2020 period. They also described nine patients, mean age 75, with coinfection of SARS-CoV-2 and C. difficile confirmed by PCR. CDI was established at different times: 2 patients had diarrhea and were diagnosed with COVID-19 coinfection at admission, while 7 patients were diagnosed during treatment for COVID-19. The median time from COVID-19 to CDI diagnosis in the 7 patients was 6 days. This group was severely ill and presented multiple underlying conditions including high blood pressure and diabetes. Three of them received antibiotics in the month before admission, and 8 during hospitalization. The most used antibiotics were cefepime, ceftriaxone, meropenem and azithromycin, however, 1 patient not receiving any antibiotic, was already colonized with C. difficile. Four patients died during hospitalization. The authors highlighted the importance of antibiotic stewardship, especially in high-risk patients such as the elderly. They also remarked that symptoms of CDI can interfere with a timely diagnosis of COVID-19, or vice-versa, as both conditions may present similar manifestations. Accordingly, appropriate testing for the 2 diseases is warranted.29 Lewandowski and colleagues conducted a retrospective, single-center study in Warsaw, Poland, of 441 patients with SARS-CoV-2 infection and 2961 patients from the prepandemic era. There was a significant increase of CDI during the COVID-19 pandemic (10.9%) compared with the period before (2.6%). Identified risk factors were comorbidities such as cardiovascular, chronic kidney, and nervous system diseases, as well as onset of abdominal symptoms during hospitalization, length of hospital stay, age, and treatment with antibiotics, with the only exception of azithromycin, which showed no effect.30
TABLE 2.
Studies Reporting C. difficile Coinfection in Patients with COVID-19 and Associated Risk Factors
| References | Region, Country | Subjects/Patients (n) | Pre-COVID-19 CDI Rates | COVID-19 Pandemic CDI Rates | Findings and Identified Risk Factors |
|---|---|---|---|---|---|
| Sandhu et al29 | Michigan, USA | COVID-19 and CDI (9) | 3.32/10,000 patient days | 3.6/10,000 patient days | Increased CDI rates in the pandemic CDI appeared after 6 d of COVID-19 diagnosis and patients were severely ill Symptoms such as diarrhea common to both CDI and COVID-19, can interfere with a timely diagnosis of each one |
| Lewandowski et al30 | Warsaw, Poland | COVID-19 (441) and prepandemic controls (2961) | 10.9% | 2.6% | Risk factors for Co-infection: Comorbidities (cardiovascular, chronic kidney disease, nervous system); Onset of abdominal symptoms during hospitalization; Length of hospitalization stay; Older age; Antibiotics |
| Luo Y et al37 | New York, USA | COVID-19 pandemic (NR) and prepandemic controls (NR) | — | — | No difference in HO-CDI SIR before and during the pandemic Nonsignificant fewer tests for CDI during the pandemic Trend toward higher percentage of positive tests |
| Laszkowska et al31 | New York, USA | Among hospitalized patients (4973) tested for GI Infections (311): COVID-19 positive (204) and COVID-19 negative patients (107) | 5.1% | 8.2% | CDI in COVID-19 vs. controls, P=0.33 Any GI infection in COVID-19 (10%) vs. controls (22%), P<0.01 CDI did not appear to be a significant contributor to diarrhea |
| Sehgal et al32 | Minnesota, USA | COVID-19 and CDI patients (21) | NR | NR | The majority presented underlying conditions and previous antibiotic exposure No clear relationship between CDI and COVID-19: CDI diagnosed at admission for COVID-19: 19%; CDI after COVID-19: 57%; COVID-19 within 4 wk after CDI: 23.9% |
| Granata et al33 | Italy | COVID-19 and CDI (38), and COVID-19 controls (114) | NR | NR | HO-CDI: 84.2% vs. community onset: 15.8% Risk factors for CDI coinfection: Cardiovascular disease; Immunosupression; Previous transplantation; Prior hospitalization; Antibiotics; PPIs; Steroids in the previous 2 mo; Antibiotics during hospitalization for COVID-19 |
| Allegretti et al38 | Massachusetts, United States | COVID-19 CDI tests (97) and all inpatients CDI tests in 2019 (2984) | 5.3% | 5.2% | Number of antibiotics prescribed were not significantly different between CDI and non-CDI patients All CDI patients were exposed to at least 2 antibiotics prior to the infection No significant differences in laboratory values, demographics, comorbidities or symptoms |
| Ponce-Alonso et al34 | Madrid, Spain | COVID-19 (2337) | 34 cases Incidence density: 2.68/10,000 patient days | 12 cases Incidence density: 8.54/10,000 patient days | Incidence density pre vs. pandemic: P=0.000257 C. difficile testing was reduced by 9.8% during the COVID-19 pandemic vs. control period Authors hypothesized that a 70% reduction in CDI was due to the reduction in patient mobility pre (587.61/1000 vs. COVID-19 period (300.86/1000 patient days), P<0.0001, and increased hygiene measures |
| Bentivegna et al35 | Rome, Italy | Discharged patients pre (1467) and during COVID-19 pandemic (150) | Incidence: 0.033 | Incidence: 0.047 | HO-CDI incidence in 2020 vs. 2017: OR=2.98, P=0.002; vs. 2018: OR=2.27, P=0.023; vs. 2019: OR=2.07, P=0.047 COVID-19 wards in 2020 showed higher HO-CDI incidence vs. COVID-19 free wards (NS) |
| Ochoa-Hein et al36 | Mexico City, Mexico | CDI cases before (56), and during the COVID-19 pandemic (2) | Incidence: 9.3/1000 Monthly range: 1.9-20.6 | Incidence:1.4/1000 Monthly range: 0-5.2 | An increase of hand hygiene mean adherence pre: 66.1% vs. COVID-19 pandemic: 94.7% Use of antibiotics in HO-CDI: 90.7% vs. patients without HO-CDI after conversion COVID-19 pandemic: 92.5%, NS Probably the better adherence to hygiene measures had an impact on the overall HCDA-CDI rates |
| Hazel et al39 | United Kingdom | HCFA-CDI cases in 2018 (14), 2019 (27) and during the COVID-19 pandemic (9) | 2.24 per 10,000 BDU (2018) and 4.24 (2019) | 2.15 per 10,000 BDU (2020) | Hospital admissions and HCFA-CDI rates were significantly lower during the COVID-19 pandemic Hand hygiene scores were higher in 2020 |
BDU indicates bed days used; CDI, C. difficile infection; HC, healthy controls; HCDA-CDI, health care facility associated CDI; HO-CDI SIR, hospital onset CDI standardized infection ratio; NR, not reported; NS, not significant; OR, odds ratio; PPIs, proton pump inhibitors.
Another retrospective cohort by Luo and colleagues, in New York, compared patients from the prepandemic era (February-June 2019) with a COVID-19 pandemic (February-June 2020) group, finding that fewer tests for C. difficile were given during the pandemic. The testing decrease was thought to be related to symptoms like diarrhea, being attributed to COVID-19. In contrast to the previous studies, there were no differences in hospital onset CDI rates despite a trend toward increased high-risk factors such as antibiotic exposures.37 Laszkowska and colleagues studied 4973 hospitalized patients at 2 centers in New York City, from which 311 were tested for GI infections (204 COVID-19 positive and 107 COVID-19 negative). The COVID-19 patients were less likely to test positive for any GI infection (10% vs. 22%), and there were no differences according to illness severity.31 Similar to the Luo et al study,37 there were no differences in the CDI rates.31 This study also compared the trend of GI PCR and C. difficile testing before and after the beginning of the pandemic (February to April 2020), showing a dramatic decrease of almost 50% of testing for general enteric infections, while testing for C. difficile maintained its prepandemic levels. The authors suggested that CDI should be considered as a differential diagnosis for diarrhea, regardless of the COVID-19 pandemic, especially in patients with a longer hospital stay as well as in COVID-19 patients.31
Sehgal and colleagues, reported a retrospective study in Rochester, Minnesota, of 21 patients, mean age 71, with COVID-19 and CDI that were confirmed by PCR. All but 2 patients presented underlying comorbidities such as hypertension, dyslipidemia, type 2 diabetes mellitus, neoplasia, and hypothyroidism, among others. Four patients with CDI were diagnosed with COVID-19 at the time of admission, 12 during hospitalization, and 5 were within 4 weeks after the CDI. Seventy-six percent of the patients had been exposed to antibiotics before the CDI, and 38% were on immunosuppression. In addition, 33% had other co-infections besides C. difficile, including Campylobacter jejuni, urinary tract infection with Enterobacter, enteropathogenic E. coli, and opportunistic infections such as Cytomegalovirus viremia or Pseudomona aeruginosa and Cryptococcus neoformans in the respiratory tract. Eventually all resolved the CDI with a standard vancomycin treatment-cycle except 6 patients, who needed a longer therapy. Of importance, the patients in this series had not been treated with antibiotics for COVID-19, hence, CDI appears to have developed due to prior exposure.32
Granata and colleagues in Italy, prospectively compared COVID-19 coinfected with C. difficile versus COVID-19 controls without C. difficile, and followed them up to 30 days from their discharge. Of 40,415 patients admitted in the 8 participant hospitals, 38 COVID-19 and CDI patients with an age range of 53 to 97, were identified. Of them, 32 were hospital-onset and 6 community-onset CDI cases. Elevated inflammatory markers and abnormal blood chemistry results were common, although lower mean albumin values were the only difference in COVID-19 with CDI, compared with controls. In 84% of the patients, the onset of CDI occurred after the COVID-19 diagnosis. Risk factors for C. difficile included comorbid cardiovascular diseases, immunosuppression or previous transplantations, prior hospitalization, antibiotic use, proton pump inhibitors (PPIs), and steroids in the previous 2 months, as well as antibiotic medications for COVID-19 treatment during hospitalization.33 In contrast, a retrospective cohort study was made to evaluate the prevalence, clinical characteristics, and outcomes of CDI among 390 hospitalized patients with COVID-19 across 9 hospitals in Massachusetts. They collected stool samples for a glutamate dehydrogenase and enzyme-linked immunosorbent assay (ELISA) and immunoassay (EIA) testing for CDI and collected patient demographic characteristics and used medications. Mortality and other outcomes were compared between SARS-CoV-2 infected patients with and without CDI. They found that the number of prescribed antibiotics were not different between CDI and non-CDI patients, but all CDI patients were exposed to at least 2 antibiotics before CDI diagnosis. Also, they highlight that none of CDI patients debuted with diarrhea. No significant differences were found in laboratory values (inflammation markers), demographics, comorbidities or presenting symptoms (diarrhea, nausea, vomiting, abdominal pain, fever, dyspnea, and sore throat) (Fig. 1).38
FIGURE 1.
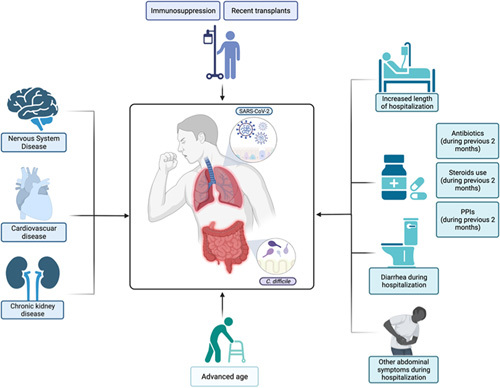
The figure depicts the factors that have been associated with Clostridium difficile coinfection in patients with SARS-CoV-2. PPIs indicates proton pump inhibitors. Created with Biorender.com.
Four studies analyzed the effects of the COVID-19 pandemic preventive measures on the incidence of the health care facility-associated CDI (HCFA-CDI) (Table 2). Ponce-Alonso et al in Madrid, Spain, retrospectively compared the incidence density of HCFA-CDI (March 11 to May 11, 2020) with the same period during the previous year and assessed the antibiotic use and patient mobility between wards. They reported that the rate of C. difficile testing in hospitalized patients was reduced by 9.8% in the COVID-19 period (1.6 per 1000 hospital stays) compared with the control period (5.1 per 1000 hospital stays). Twelve HCFA-CDI cases were identified, almost 3 times less than the previous year which included 34 cases. Patient mobility also suffered a significant drastic reduction during the COVID-19 period, as well as the number of surgical interventions. In contrast, use of antibiotics was slightly higher during the pandemic with third generation cephalosporins and macrolides being the most used.34 The authors hypothesized that in the context of no antibiotic reduction, the almost 70% reduction in C. difficile could be the result of the extraordinary reinforcement of all infection control measures, including patient isolation, universal personal protection equipment, limited patient mobility, and constant cleaning, and the increased adherence to these measures by health care workers.34 A study by Bentivegna and colleagues in Rome, Italy, compared data of discharged patients between March and June 2020 with available data from the previous 3 years, considering the implemented strategies that were adopted because of the COVID-19 pandemic. They found that the HCFA-CDI incidence was significantly lower during the index period compared with 2017 and 2019, even though there were higher incidence-rates in the COVID-19 compared with the non-COVID-19 wards. Once again, the data suggests that SARS-CoV-2 infection may be a possible risk factor for CDI, however, implementing strict hygiene protocols may have had a major role in its reduction.35 Similarly, Hazel and colleagues also compared the HCFA-CDI available data from March to May, 2020, with the same periods in 2018 and 2019. Fifty HCFA-CID patients were identified: 13 in 2018, 27 in 2019 and 9 in 2020 (3 with COVID-19 coinfection). When compared with the previous 2 years, the HCFA-CDI rates, as well as the hospital admissions for CDI, were significantly lower, while hand hygiene scores were higher during the COVID-19 pandemic.39 Ochoa-Hein and colleagues in Mexico, analyzed the HCFA-CDI rates from January to February 2020 and from April to July 2020. In the first period, 56 cases (9.3 cases per 10,000) of HCFA-CDI were identified, and only 2 cases (1.4 cases per 10,000) were identified in the second period, demonstrating a great reduction of the overall infection rate. This study measured the adherence to hand hygiene before and after the COVID-19 hospital conversion, finding an increase from 66.1% to 94.7%. Also, before the COVID-19 conversion, 46.3% of the HCFA-CDI cases were women with a median age of 47.5 years, that reported hospitalization in the previous month and had recent abdominal surgery (29.6%) or an active solid organ neoplasia (25.9%), used antibiotics (90.7%) or PPIs (73.5%). Conversely, the 2 HCFA-CDI cases after the COVID-19 hospital conversion were women with COVID-19 who had previously used antibiotics; 1 with generalized lupus erythematosus and 1 with diabetes mellitus. Once more, the authors highlighted that a better adherence to hygiene measures was a crucial factor in the reported data.36
Oral Bacteriotherapy in Patients With COVID-19
One paper on bacteriotherapy for modulating gut microbiota in hospitalized patients with COVID-19 was identified. It is a retrospective observational cohort of 200 COVID-19 adults with severe pneumonia, by Ceccarelli and colleagues in the Lazio region of Italy. Of them, 112 received the best available therapy (BAT) (ie, low–molecular-weight heparin plus one or more of hydroxychloroquine, azithromycin, antivirals, and tocilizumab), and 88 were treated with BAT plus an oral bacteriotherapy supplement (Sivomixx a multi-strain product containing 5 strains of Lactobacilli, 2 strains of Bifidobacteria, and 1 strain of Streptococcus thermophilus). Mortality rates were significantly lower in the BAT and bacteriotherapy versus the BAT group (11% vs. 30%). By multivariate analysis, age older than 65, CRP >41.8 mg/L, platelets <150,000 mmc, and cardiovascular events, were associated with the increased risk of mortality, while oral bacteriotherapy was an independent factor for a reduced mortality risk.40
Although not retrieved by our search engine, when finalizing the drafting of this paper, a new paper on bacteriotherapy on COVID-19 was published and it was important to include it in this review.41 It is a retrospective study of 70 patients with COVID-19 hospitalized between March 9th and April 4th, 2020 in Rome, Italy. Forty-two patients received hydroxychloroquine, antibiotics, and tocilizumab, alone or in combination, and a second group of 28 subjects received the same therapy combined with Sivomixx. The groups were similar in clinical parameters and symptoms, however, the bacteriotherapy induced remission of diarrhea in all the patients within 7 days, the majority within 72 hours, compared with less than half of the patients without bacteriotherapy, as well as other systemic symptoms. Also, the other symptoms including fever, asthenia, headache, myalgia, and dyspnea, presented similar trends. In addition, the estimated risk for developing respiratory failure was eight-fold lower in the group with bacteriotherapy, and there was a trend for more patients transferred to the ICU for mechanical ventilation or for a lethal outcome in the group without bacteriotherapy (Table 3).41
TABLE 3.
Oral Bacteriotherapy and Fecal Microbiota Transplantation (FMT) in Patients With COVID-19
| References | Region, Country | Subjects/Patients (n) | Intervention (n) | Aims | Main Findings |
|---|---|---|---|---|---|
| Oral bacteriotherapy for COVID-19 | |||||
| Ceccarelli et al40 | Lazio, Italy | COVID-19 (200) with severe pneumonia | BAT (112), BAT+Sivommixx (88) | Retrospective analysis of mortality rates | Lower mortality rates with combined therapy vs. BAT alone (11% vs. 30%) Increased mortality factors: age >65, CRP> 41.8 mg/L, platelets <150.000/mmc Oral bacteriotherapy was an independent factor for lower mortality |
| d’Ettorre et al41 | Rome, Italy | COVID-19 (70) | Hydroxicloroquine, and/or antibiotics, and/or Tocilizumab (42) Same + Sivommixx (28) | Oral bacteriotherapy as complementary therapeutic strategy to avoid the progression of COVID-19 | Bacteriotherapy induced remission of diarrhea fever, asthenia, headache, myalgia, and dyspnea in all patients vs. half of the not supplemented group |
| FMT for CDI during the COVID-19 pandemic | |||||
| Liu et al16 | Ganzhou City, China | COVID-19 1 mo after being discharged from the hospital (11); GI symptoms: constipation, diarrhea, abdominal pain, gastralgia, acid reflux, gastrectasia (5/11) | FMT by 10 oral capsules/day×4 consecutive days | Investigate the potential benefit over GI symptoms | GI symptoms improved after FMT Altered peripheral lymphocytes: Decreased naive B cells (P=0.012); Increased memory B cells (P=0.001); Increased nonswitched B cells (P=0.012) FMT partially restored gut dysbiosis by increasing the relative abundance of phylum Actinobacteria (15.0%), reduced Proteobacteria (2.8%); increased genera Bifidobacterium and Faecalibacterium |
| Ianiro et al42 | Rome, Italy | Recurrent or refractory CDI (21) | FMT for CDI during the COVID-19 pandemic | To report outcomes of a FMT service that adapted its operational workflow to prevent SARS-CoV-2 transmission | No recurrence of CDI after FMT in 18 that were followed for 8 wk It was possible to maintain standard volumes, efficacy and safety of FMT for CDI during the COVID-19 pandemic |
| Olesen et al43 | Cambridge Massachusetts, USA | Abstract model of FMT donors, simulating their donation schedule, SARS-CoV-2 infection incidence, and COVID-19 disease course | Estimate the utility of different testing strategies (PCR with nasopharyngeal swabs, stool-based PCR, donor serology tests, or a combination of those assays | Mathematical model to determine the effectiveness of the testing strategies | The risk that a released donation is virus-positive varied approx. proportionally with the incidence of infection: a 10-fold increase in incidence led to 10-fold increased risk The more stringent testing strategies (symptoms checks, nasopharyngeal swabs, serology tests, testing every stool) the lower the probability of releasing a virus-positive stool for donation |
BAT indicates best available therapy (low–molecular-weight heparin plus one or more of hydroxychloroquine, azithromycin, antivirals, and tocilizumab); CDI, clostridium difficile infection; FMT, fecal microbiota transplantation; GI, gastrointestinal; Sivomixx, an oral a multistrain product containing 5 strains of Lactobacilli, 2 strains of Bifidobacteria, and 1 strain of Streptococcus thermophiles.
FMT for COVID-19 and C. difficile and During the Pandemic
FMT is now accepted in the treatment of severe or recurrent CDI, as a therapy to modulate the gut microbiota.44 Therefore, it is plausible to consider the use of FMT in patients with COVID-19 to restore the gut microbiota and improve GI symptoms, and even for the treatment of CDI. Notwithstanding, FMT is currently limited due to concerns of potential transmission of SARS-CoV-2.45 Three studies on FMT were identified in the search for gut microbiota and bacteriotherapy in COVID-19 (Table 3).16,42,43
A single-center pilot-study in Ganzhou City, China by Liu and colleagues, previously cited in the Gut microbiota in COVID-19 section, recruited patients after being released from the hospital to participate in this protocol of 10 FMT oral capsules per day, during 4 continuous days. Five reported GI symptom improvement after FMT. Furthermore, FMT restored dysbiosis by increasing the relative abundance of Actinobacteria by 15.0% and decreasing Proteobacteria by 2.8%, at the phylum level; and significantly increasing Bifidobacterium, Faecalibacterium, and Collinsella, at the genera level.16
In relation to FMT for C. difficile, Ianiro and colleagues reported a single-center prospective observational cohort study from Rome, Italy, conducted from March to July 2020, in 26 patients with recurrent or refractory CDI. After a first Telemedicine videoconsultation, 3 patients decided to be treated with an alternative option rather than FMT and 2 declined the procedure.42 Hence, only 21 patients were treated with FMT. The product used in 13 cases had already been available in the stool bank from before the COVID-19 pandemic, 2 other donors were previously known in the stool bank and were recalled for stool donation, and 2 new donors were enrolled for collecting stool product. The latter 4 donors were negative for SARS-CoV-2 by nasopharyngeal swab, and none had a COVID-19 diagnosis during the study period. In total, there were 26 FMT sessions in these patients; an initial session in all of them and sequential infusions in three patients because of a lack of response to the first FMT. Eighteen patients completed an 8-week follow-up after FMT, with no recurrence of C. difficile after the procedure. The remaining patients were reported as “ongoing” in the follow-up. There were no serious adverse events or new COVID-19 infection reports. The researchers mentioned that upon following the general safety practices and the adaption of the working protocols for FMT, they managed to maintain the volumes and outcomes of the pre-COVID-19 era to guarantee a high level of safety. The implemented changes were mainly based on virtual consultation, as opposed to in-person visits, and a meticulous screening of the donors to avoid a possible transmission of the virus by feces. The screening included evaluating symptomatology, potential exposure to COVID-19, serological examination, and follow-up for COVID-19, as well as a 30-day quarantine of donated feces with product aliquots frozen at −80°C. After concluding the quarantine period, the corresponding donors were assessed again for symptoms or exposure to COVID-19, as well as PCR testing of both stored and recent fecal samples.42 Direct stool testing for SARS-CoV-2 is mentioned as a more direct and safer way to prevent FMT-related transmission. One limitation of the latter measure is that currently, PCR testing for SARS-CoV-2 in stools is not available.
Finally, Olesen and colleagues proposed a mathematical model to simulate the utility of different testing strategies for FMT donors in the COVID-19 pandemic. The authors calculated the effect of PCR with nasopharyngeal swabs, stool-based PCR tests, donor serology tests or a combination of them. The analysis revealed that the risk of a donation being SARS-CoV-2-positive is proportional to the incidence of infection in the general population, and more stringent strategies are related with lower risk of SARS-CoV-2 infected fecal donations for FMT. However, the more sensitive the strategies were, the less specific. The proportion of an evaluation based only on donor symptoms had the highest amount of contaminated donations, while the combination of PCR swab and stools serological testing, had the fewest ones. They suggested that the most appropriate strategy should be selected by a balance between stringency and resources considerations.43
CONCLUSIONS
GI manifestations are very common among patients with COVID-19 and there is several evidence supporting an inflammatory process in the intestinal epithelium, thus it is plausible to expect the presence of bacterial and fungal abnormalities in the gut. Further, the use of antibiotics in the treatment of these patients can have a negative impact on the gut microbiota and increase the risk for CDI. Therefore, bacteriotherapy and FMT are potential treatments for COVID-19. Accordingly, we have conducted a literature review of these factors, that can be summarized as follows: (1) by analyzing fecal samples, several studies have confirmed the presence of gut bacterial dysbiosis in patients with COVID-19, characterized by enrichment of opportunistic organisms and decrease in beneficial commensals such as F. prausnitizii. In addition, enrichment of opportunistic organisms or depletion of beneficial bacteria was positively or negatively associated with inflammatory markers such as CRP; dysbiosis was also correlated with the severity of the illness. (2) There is evidence for gut-barrier disfunction with bacterial translocation in patients with COVID-19. However, more studies are warranted to determine if the dysbiosis is the original factor disrupting the epithelial barrier or if the gut inflammation with subsequent gut hyperpermeability drives the gut dysbiosis. (3) There is also gut fungal dysbiosis, again with enrichment of opportunistic pathogens and depletion of fungal commensals, with some evidence suggesting a correlation between the gut and pulmonary mycobiota. The mechanism for fungal dysbiosis in COVID-19 deserves to be further investigated as well, and although could be related to gut inflammation, mutualism, or competition with bacterial dysbiosis can also explain it. In Figure 2 we have proposed a model of gut bacterial and fungal dysbiosis in patients with COVID-19, the relationship with intestinal wall inflammation, gut barrier dysfunction and the “gut-lung axis,” as well as potential treatments. (4) Although some studies reported an increase in the prevalence rate of CDI among patients with COVID-19 compared with patients hospitalized for other diseases, and between the pandemic and the pre-pandemic era, the data is controversial. Some data suggest that implementation of hospital preventive measures against COVID-19, may have positively impacted in decreasing the CDI rates, despite the risk of antibiotics in these patients. In addition, it is important to highlight that in patients with COVID-19 and diarrhea, a differential diagnosis is required to establish if these symptoms are clinical manifestations of COVID-19, CDI, or from any other overlapped GI infection. (5) Considering all the above, it is plausible to consider the use of bacteriotherapy by means of probiotics, to modulate dysbiosis and decrease the systemic inflammatory response in COVID-19. However, there is no available evidence yet to support this measure. (6) Finally, in terms of FMT, there is no evidence to support its use in COVID-19. As for treatment of C. difficile, implementing strict screening methods of donors, and testing for SARS-CoV-2 in donors and donated fecal product, can safely protect recipients of COVID-19 infection through FMT.
FIGURE 2.
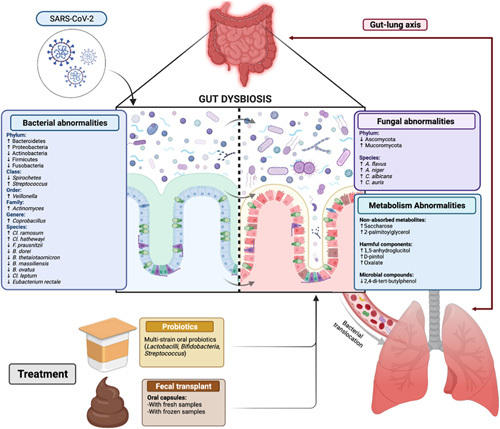
The figure summarizes the gut bacterial and fungal abnormalities that have been reported in patients with SARS-CoV-2 infection, according to the taxonomic classification. Metabolic abnormalities such as nonabsorbed metabolites, increased harmful components, possibly related to dysbiosis, are also shown. The relationship with gut inflammation and epithelial permeability is also depicted. On the left there is a normal epithelium depicted in blue with a controlled microbiota, while on the right side there is inflammation depicted in red with an increase in epithelial permeability and bacterial and fungal dysbiosis. The gut barrier disruption is associated with bacterial and fungal translocation to the lungs through the so called “gut-lung axis.” Proposed treatment approaches include bacteriotherapy/probiotics or fecal microbiota transplantation (FMT), that warrant further investigations. Created with Biorender.com.
Finally, we might have had some limitations in finding other papers as we did not include other search terms such as SARS-CoV-2. However, we wanted to focus on the presence of bacterial or fungal dysbiosis and coinfection with C. difficile in patients with the clinical illness (COVID-19). In addition, the studies reported in this review were conducted in different countries of the world, with different diet and environmental factors which may have influenced the basal microbiota and mycobiota independently of the presence of COVID-19. Furthermore, the results may vary with the evolving viral strains of SARS-CoV-2 and the changing predominant strain during different waves of the pandemic, which warrant future investigations.
Footnotes
The authors declare that they have nothing to disclose.
Contributor Information
Laura Linares-García, Email: llinaresg2008@hotmail.com.
María E. Cárdenas-Barragán, Email: elena_csb472@hotmail.com.
Winston Hernández-Ceballos, Email: winherceb001@gmail.com.
Carlos S. Pérez-Solano, Email: sebaselite-.300@hotmail.com.
Alizon S. Morales-Guzmán, Email: sujey_alizon@hotmail.com.
Danielle S. Miller, Email: daniellesm100@gmail.com.
Max Schmulson, Email: maxjulio@prodigy.net.mx;mschmulson@gmail.com.
REFERENCES
- 1.Centers for Disease Control and Prevention CDC. Symptoms of Coronavirus. Coronavirus Disease 2019 (COVID-19). Volume 2020. 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html. Accessed February 21, 2021.
- 2.Tian Y, Rong L, Nian W, et al. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment Pharmacol Ther. 2020;51:843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmulson M, Davalos MF, Berumen J. Beware: Gastrointestinal symptoms can be a manifestation of COVID-19. Rev Gastroenterol Mex. 2020;85:282–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan L, Mu M, Gang Ren H, et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol. 2020;115:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung KS, Hung IF, Chan PP, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the hong kong cohort and systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin L, Jiang X, Zhang Z, et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho A, Alqusairi R, Adams A, et al. SARS-CoV-2 gastrointestinal infection causing hemorrhagic colitis. Am J Gastroenterol. 2020;115:942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Effenberger M, Grabherr F, Mayr L, et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du M, Cai G, Chen F, et al. Multiomics evaluation of gastrointestinal and other clinical characteristics of COVID-19. Gastroenterology. 2020;158:2298–2301.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allam-Ndoul B, Castonguay-Paradis S, Veilleux A. Gut microbiota and intestinal trans-epithelial permeability. Int J Mol Sci. 2020;21:6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakaroun RM, Massier L, Kovacs P. Gut microbiome, intestinal permeability, and tissue bacteria in metabolic disease: perpetrators or bystanders? Nutrients. 2020;12:1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahlawat S, Asha, Sharma KK. Immunological co-ordination between gut and lungs in SARS-CoV-2 infection. Virus Res. 2020;286:198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conte L, Toraldo DM. Targeting the gut-lung microbiota axis by means of a high-fibre diet and probiotics may have anti-inflammatory effects in COVID-19 infection. Ther Adv Respir Dis. 2020;14:1753466620937170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Ye S, Zhu X, et al. Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged COVID-19 patients. J Med Case Rep. 2021;15:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu S, Chen Y, Wu Z, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis. 2020;71:2669–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzarelli A, Giancola ML, Farina A, et al. 16S rRNA gene sequencing of rectal swab in patients affected by COVID-19. PLoS One. 2021;16:e0247041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang L, Gu S, Gong Y, et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering (Beijing). 2020;6:1178–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeoh YK, Zuo T, Lui GC, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lv L, Jiang H, Chen Y, et al. The faecal metabolome in COVID-19 patients is altered and associated with clinical features and gut microbes. Anal Chim Acta. 2021;1152:338267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Gu S, Chen Y, et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2022;71:222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuo T, Zhan H, Zhang F, et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159:1302–1310.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv L, Gu S, Jiang H, et al. Gut mycobiota alterations in patients with COVID-19 and H1N1 infections and their associations with clinical features. Commun Biol. 2021;4:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasad R, Patton MJ, Floyd JL, et al. Plasma microbiome in COVID-19 subjects: an indicator of gut barrier defects and dysbiosis. bioRxiv [Preprint]. 2021:2021.04.06.438634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuo T, Wong SH, Cheung CP, et al. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat Commun. 2018;9:3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liguori G, Lamas B, Richard ML, et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J Crohns Colitis. 2016;10:296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Limon JJ, Tang J, Li D, et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe. 2019;25:377–388.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandhu A, Tillotson G, Polistico J, et al. Clostridioides difficile in COVID-19 patients, Detroit, Michigan, USA, March-April 2020. Emerg Infect Dis. 2020;26:2272–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewandowski K, Rosołowski M, Kaniewska M, et al. Clostridioides difficile infection in coronavirus disease 2019 (COVID-19): an underestimated problem? Pol Arch Intern Med. 2021;131:121–127. [DOI] [PubMed] [Google Scholar]
- 31.Laszkowska M, Kim J, Faye AS, et al. Prevalence of Clostridioides difficile and other gastrointestinal pathogens in patients with COVID-19. Dig Dis Sci. 2021;66:4398–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sehgal K, Fadel HJ, Tande AJ, et al. Outcomes in patients with SARS-CoV-2 and Clostridioides difficile coinfection. Infect Drug Resist. 2021;14:1645–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Granata G, Bartoloni A, Codeluppi M, et al. The burden of Clostridioides difficile infection during the COVID-19 pandemic: a retrospective case-control study in Italian Hospitals (CloVid). J Clin Med. 2020;9:3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponce-Alonso M, Sáez de la Fuente J, Rincón-Carlavilla A, et al. Impact of the coronavirus disease 2019 (COVID-19) pandemic on nosocomial Clostridioides difficile infection. Infect Control Hosp Epidemiol. 2021;42:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bentivegna E, Alessio G, Spuntarelli V, et al. Impact of COVID-19 prevention measures on risk of health care-associated Clostridium difficile infection. Am J Infect Control. 2021;49:640–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochoa-Hein E, Rajme-López S, Rodríguez-Aldama JC, et al. Substantial reduction of healthcare facility-onset Clostridioides difficile infection (HO-CDI) rates after conversion of a hospital for exclusive treatment of COVID-19 patients. Am J Infect Control. 2021;49:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo Y, Grinspan LT, Fu Y, et al. Hospital-onset Clostridioides difficile infections during the COVID-19 pandemic. Infect Control Hosp Epidemiol. 2021;42:1165–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allegretti JR, Nije C, McClure E, et al. Prevalence and impact of Clostridioides difficile infection among hospitalized patients with coranavirus disease 2019. JGH Open. 2021;5:622–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hazel K, Skally M, Glynn E, et al. The other “C”: Hospital-acquired Clostridioides difficile infection during the coronavirus disease 2019 (COVID-19) pandemic. Infect Control Hosp Epidemiol. 2021:1–2. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceccarelli G, Borrazzo C, Pinacchio C, et al. Oral bacteriotherapy in patients with COVID-19: a retrospective cohort study. Front Nutr. 2020;7:613928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.d’Ettorre G, Ceccarelli G, Marazzato M, et al. Challenges in the management of SARS-CoV2 infection: the role of oral bacteriotherapy as complementary therapeutic strategy to avoid the progression of COVID-19. Front Med (Lausanne). 2020;7:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ianiro G, Bibbò S, Masucci L, et al. Maintaining standard volumes, efficacy and safety, of fecal microbiota transplantation for C. difficile infection during the COVID-19 pandemic: a prospective cohort study. Dig Liver Dis. 2020;52:1390–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olesen SW, Zaman A, Osman M, et al. Modeling donor screening strategies to reduce the risk of severe acute respiratory syndrome coronavirus 2 transmission via fecal microbiota transplantation. Open Forum Infect Dis. 2020;7:ofaa499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López-Vidal Y, Soto-Hernández JL, Orduña P, et al. Allogenic fecal microbiota transplantation in Clostridioides difficile infection: a case series in Mexico. Biomed J Sci Tech Res. 2021;36:28840–28845. [Google Scholar]
- 45.Ianiro G, Mullish BH, Kelly CR, et al. Screening of faecal microbiota transplant donors during the COVID-19 outbreak: suggestions for urgent updates from an international expert panel. Lancet Gastroenterol Hepatol. 2020;5:430–432. [DOI] [PMC free article] [PubMed] [Google Scholar]


