Abstract
Background:
Maternal exposure to environmental chemicals during pregnancy can influence various maternal and offspring health parameters. Modification of maternal metabolism by environmental exposure may be an important pathway for these impacts. However, there is limited evidence regarding exposure to a wide array of chemicals and the metabolome during pregnancy.
Objectives:
We investigated the relationship between the urinary exposome and metabolome during pregnancy.
Methods:
Urine samples were collected in the first and third trimesters from 1,024 pregnant women recruited in prenatal clinics in Jiangsu Province, China. The exposome was analyzed using the first trimester sample with ultra-high performance liquid chromatography–high resolution accurate mass spectrometry (UHPLC-HRMS) and inductively coupled plasma mass spectrometry. The metabolome was analyzed using the third trimester sample with UHPLC-HRMS. We evaluated associations between each of 106 exposures in the first trimester with 139 metabolites in the third trimester.
Results:
We identified 1,245 significant associations (, Bonferroni correction) between chemical exposures and maternal metabolism during pregnancy. Among elements, the largest number of the significant metabolic associations were observed for magnesium, and among organic compounds, for 4--octylphenol. We used exposome–metabolome associations to explore mechanisms underlying published associations between prenatal chemical exposures and offspring health outcomes. This integration of the literature with our results suggests that reported associations between 10 analytes and birth weight, gestational age, fat deposition, neurobehavioral development, immunological disorders, and hypertension may be partially mediated by metabolites associated with these exposures.
Discussion:
This high-dimensional analysis of the urinary exposome and metabolome identified many associations between chemical exposures and maternal metabolism during pregnancy. Integration of these associations with the literature on health outcomes of exposure suggests that environmental modulation of the maternal metabolome may play a role in the association between prenatal exposure on pregnancy and child health outcomes. https://doi.org/10.1289/EHP9745
Introduction
Pregnancy is a critical period for maternal and offspring health. The developing fetus and the pregnant mother are both especially susceptible to exposure to environmental chemicals (Barr et al. 2007). Many chemicals can pass through the placenta into the fetus (Barr et al. 2007). Exposures during pregnancy may lead to adverse outcomes in both mother (Wang et al. 2020) and fetus (Buckley et al. 2016; Chiu et al. 2018; Laine et al. 2015; Wang et al. 2019) that manifest both early and much later in life.
Most studies of the impact of prenatal exposure have examined a small number of specific chemicals (Tsai et al. 2021). There is growing interest in examining a larger range of exposures at once. This has led to the concept of measuring the chemical exposome to take a comprehensive and agnostic approach toward assessment of chemical exposures (Vermeulen et al. 2020). Application of the exposome concept to pregnancy would enable the assessment of the burden of exposure to a larger range of chemicals, which is important because vulnerability to environmental exposures is enhanced in the prenatal period (Vrijheid et al. 2014).
Environmental epidemiological studies have identified many associations between exposure to individual chemicals during pregnancy and maternal (Wang et al. 2020) and offspring health outcomes (Buckley et al. 2016; Chiu et al. 2018; Laine et al. 2015; Wang et al. 2019), but the underlying mechanisms remain largely unknown. Metabolomics systematically profiles metabolites, which are the endogenous small molecule substrates, intermediates, and products of cell metabolism in a biological sample (Nicholson and Lindon 2008). This comprehensive approach can identify metabolic signatures of chemical exposures that could play a role in disease etiology (Ramirez et al. 2013). Notably, metabolomics has been recognized as an omics technology that may be most closely relevant to disease phenotypes (Guijas et al. 2018). For example, metabolomic platforms assess clinically applicable indicators, such as steroid hormones and cholesterol (Kliesch 2014). Moreover, the small molecules interrogated by metabolomics approaches include nutrients and their metabolites that are involved in disease pathogenesis in humans and animals (Wang et al. 2011a, 2011b). Dramatic metabolic changes take place in pregnancy, including increased protein synthesis from amino acids to enable fetal growth and steroid hormone synthesis to support maintenance of the pregnancy (Duggleby and Jackson 2002; Noyola-Martínez et al. 2019). Therefore, metabolomics is of increasing interest in understanding both the normal physiology and pathology of pregnancy (Souza et al. 2019). In genetic studies, metabolomics has been used to suggest mechanisms that may underlie associations with health outcomes in humans (Suhre et al. 2011).
Application of exposomic and metabolomic technologies together in pregnancy could identify metabolomic signatures of a broad range of exposures that could shed light on potential health effects of these exposure, as well as their underlying mechanisms. However, to our knowledge, there are no published studies dedicated to this design. Exposures early in pregnancy that impact the metabolome across pregnancy may be especially injurious to the fetus (Dencker and Eriksson 1998). Taking advantage of longitudinal data to examine associations between exposures in the first trimester and the metabolome in the third trimester has the potential to explore causal associations. We conducted a chemical exposome- and metabolome-wide association study in a cohort of pregnant women. We comprehensively examined various chemical exposures in the first trimester and maternal metabolites in the third trimester during pregnancy to identify environmentally determined urinary metabotypes. Similar to an approach used to examine the role of metabolomics in genetic associations with health outcomes (Suhre et al. 2011), we integrated associations identified in the literature on health effects of the implicated analytes in offspring to exploring potential mechanisms underlying diseases and biological process related to chemical exposure during pregnancy.
Methods
Study Participants and Sample Collection
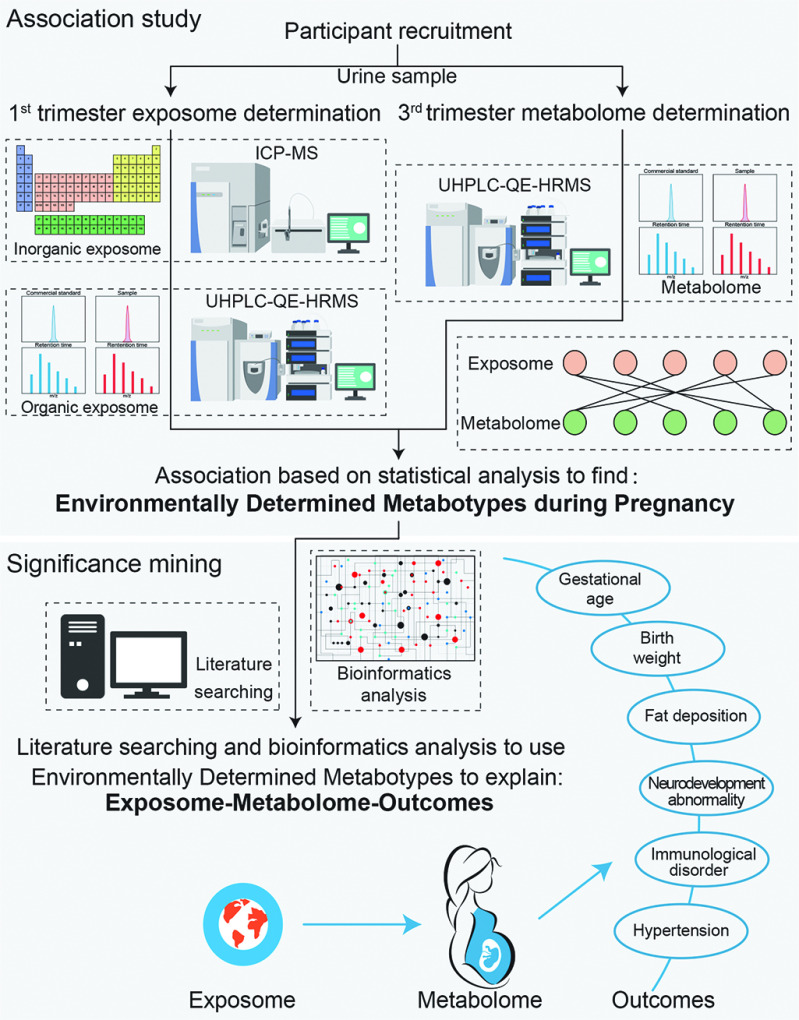
From April 2013 to July 2016, 1,532 pregnant women were recruited in the first trimester of pregnancy from the prenatal clinics during their first visits for the physical examinations during pregnancy in Jiangsu Province, China. Women eligible for enrollment were of age and reported neither assisted reproduction nor human immunodeficiency virus infection. Midstream urine samples were collected from participants in their first and third trimesters of pregnancy and stored at . The urine samples were collected in the morning at hours. A total of 1,453 women provided a urine sample in the first trimester, and 1,084 of these women also provided a urine sample in the third trimester. After excluding the 60 women with insufficient sample volume, 1,024 women with complete demographic information and sufficient urine were available for organic exposome analysis in the first trimester and metabolome analysis in the third trimester (Figure S1). The study design is shown in Figure 1. Written informed consent was obtained from each participating woman, and this study was approved by the institutional review board of Nanjing Medical University.
Figure 1.
Study Design. Note: ICP-MS, inductively coupled plasma mass spectrometry; UHPLC-QE-HRMS, ultra-high performance liquid chromatography–Q Exactive hybrid quadrupole-orbitrap high resolution mass spectrometry.
Maternal Interviews
Questionnaires administered to each participant in person in the first, second, and third trimesters elicited information on age, ethnicity, parity, height, weight before pregnancy, smoking, alcohol consumption, and education level. The options on the questionnaire were to select Han, who are the majority in our study area, or to write in other ethnicities. Only one ethnicity could be selected. Questions on fixed characteristics, such as age, ethnicity, parity, height, weight before pregnancy, and education level were asked at baseline. The questions on changeable characteristics, such as smoking and alcohol consumption status, were asked at every visit. Maternal body mass index (BMI) was calculated as self-reported weight before pregnancy (in kilograms) divided by height (in meters squared). Information about age, parity, height, and weight before pregnancy was confirmed by medical records. The medical records were reviewed for all women. If the information differed, we contacted the women for clarification.
Organic Exposome Analysis
The exposome includes both organic and inorganic chemicals (i.e., elements). The exposome refers to chemicals with no known metabolic function in the human body or that are not natural components of human body (pollutants and nonnutritive plant chemicals) (Misra 2020). Analysis of the organic exposome was conducted with ultra-high performance liquid chromatography Ultimate 3000 system (Dionex, Germering, Germany)–Q Exactive hybrid quadrupole-orbitrap high resolution mass spectrometry (UHPLC-QE-HRMS) (Thermo Fisher Scientific), which covered major kinds of bioactive environmental chemicals, including pesticides, antimicrobial agents, phthalates, phenols, phytoestrogens, parabens, food metabolites, smoking metabolites, fire retardants, personal care products, and perfluorochemicals (CDC 2015, 2019). The sample preparation, instrumental analysis, chemical identification and relative levels detection and normalization were performed as previously described (Hu et al. 2016). Briefly, the urine sample was mixed with methanol and a set of stable isotope-labeling internal standards, such as creatinine-d3, nicotinic-d4 acid, -(4-hydroxyphenyl)acetamide-2,2,2-d3, and -benzoyl-d5-glycine. After centrifugation, the supernatant was dried in Centrivap and reconstituted before instrumental analysis. The instrument was set at a 70,000 resolution with full-scan acquisition ranging from m/z 70 to 1,050. A multistep gradient was used with mobile phase A of 0.1% formic acid in ultra-pure water and mobile phase B of acetonitrile acidified with 0.1% formic acid; the gradient operated at a flow rate of over a run time of 15 min. All samples were analyzed in random fashion to avoid bias from the injection order. The chemical identification was based on the accurate mass and the retention time compared with the commercial standards using the author-constructed library with TraceFinder (version 3.1; Thermo Fisher Scientific). The relative levels of each chemical were normalized by a ratio relative to stable isotope-labeling internal standard peak areas (Chen et al. 2014). Quality control and blank samples were analyzed together with study samples. A total of 1,024 urine samples in the first trimester were analyzed (Figure S1).
Inorganic Exposome Analysis
The elementomics quantitative analysis was performed with an iCAP Qc inductively coupled plasma mass spectrometry (ICP-MS) instrument (Thermo Scientific) according to our previous report (Silver et al. 2018). Briefly, of urine and nitric acid were mixed for 3 h, and the samples were mixed with water to the volume of . After centrifugation, the supernatant was subjected to ICP-MS analysis. The limit of detection was defined as three times of the standard deviation (SD) of the obtained concentrations in the blank with 10 replicates. Quality control and blank samples were analyzed with study samples. Because of the amount of sample available, a total of 963 urine samples in the first trimester were analyzed for the inorganic exposome profiling.
Metabolomics Analysis
The metabolome refers to organic chemicals with known metabolic function in the human body or that are natural components of the human body (Rinschen et al. 2019). Analysis of the metabolome in urine with UHPLC-QE-HRMS was performed as previously described (Hu et al. 2016). The sample preparation, instrumental analysis, chemical identification, and relative levels detection and normalization were conducted according to the standard procedure of organic chemical profiling described above in the “Organic Exposome Analysis” section. Quality control and blank samples were analyzed in parallel with study samples. A total of 1,024 urine samples in the third trimester were analyzed (Figure S1).
Statistical Analysis
Urinary creatinine (CR) concentrations obtained by UHPLC-QE-HRMS analysis were used for correcting the variations of concentrations caused by fluctuations in urine concentration by dividing the exposomic and metabolomic analyte by CR; the adjustment of urinary chemical concentration by CR is useful not only for environmental chemical exposure data but also for metabolome data (Blydt-Hansen et al. 2014). The chemicals with a detection rate of were included in the statistical analysis. The 106 exposome chemicals retained for statistical analysis included 59 organic chemicals and 47 elements. A total of 139 metabolites were retained for statistical analysis. The inclusion and classification of organic exposome were based on data from PubChem (https://pubchem.ncbi.nlm.nih.gov/), Human Metabolome Database (HMDB; https://hmdb.ca/) and Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/). The classification of elements was based on the reference Maret and Copsey (2012). The inclusion and classification of metabolome were based on data from HMDB and KEGG. When metabolites belonged to more than one classification, the classification was selected based on the relevance and the simplification of classification for data interpretation. The comparison of elements in our study and those in a nationally representative U.S. sample of nonsmoking adult females reported in the U.S. National Exposure Report was based on whether the urinary geometric mean of each element in our study was within the 95% confidence interval of the corresponding urinary geometric mean in the U.S. National Exposure Report (CDC 2019). The exposome data of ICP-MS and UPLC-QE were next categorized before analysis (Ernst et al. 2019). When the detection rate of a chemical exceeded 66.7%, tertiles were created. If the detection rate of a chemical was , the undetectable samples were assigned to the lowest category, and the remaining samples were divided into two equal categories. The dichotomization of metabolite level was according to the following procedure: If the detection rate of a metabolite was , two equal categories were made; if the detection rate of a metabolite was , the undetectable samples were assigned to the low category, and the remaining samples were assigned to high category.
We used polytomous logistic regression to explore the associations between exposome and metabolome with the adjustment for maternal age, BMI before pregnancy, parity, and education (Biesheuvel et al. 2008). Because nearly all women were of Han ethnicity and very few () reported either smoking or drinking, these variables were not included in models. Maternal age and BMI were included as continuous variables; parity and education were included as binary variables. The selection of covariates was based on the biological considerations and according to previous reports (Shapiro et al. 2015). Multiple comparison was accounted for by the Bonferroni correction for 14,734 tests (, ). Environmentally determined urinary metabotypes were defined as the urinary metabolites in the third trimester significantly associated with the exposome in the first trimester ( from the polytomous logistic regression models). These environmentally determined urinary metabotypes are the metabolites in urine that might be altered by early exposure to environmental factors. To test the robustness of the these environmentally determined urinary metabotypes, two kinds of sensitivity analyses were next conducted. First, we addressed potential selection bias due to missing data and attrition by applying inverse probability weights (IPWs) to all models (Narduzzi et al. 2014). We considered the entire population (all the 1,532 pregnant women) and calculated the probability of nonmissing information using a logistic regression model, where the response was the nonmissingness and the covariates were its possible predictors, including maternal age, BMI, parity, and education (Tables 1 and S1). The weight of each subject was given by the inverse of the predicted probability. The analysis was performed only on the nonmissing observations using the weighted polytomous logistic regression model. Second, we excluded smokers and drinkers from the population and analyzed these environmentally determined urinary metabotypes with the same polytomous logistic regression settings. The statistical analysis was carried out in R stats package (version 3.6.0; R Development Core Team). Polytomous model used nnet package (version 7.3.12; R Development Core Team), and IPW used ipw package (version 1.0.11; R Development Core Team). Partial least squares-discriminant analysis (PLS-DA) was conducted using the package Mixomics (version 6.10.9; R Development Core Team). All the exposome data were included as the and each metabolite was included as , and the default setting of the package was used. The variable importance in the projection (VIP) in PLS-DA was calculated to find significant associations with a VIP . PLS-DA was used for identifying the relationships between all chemicals in the exposome and each metabolite in one model, and the output VIP indicated that this chemical in the exposome had above-average influence on this metabolite and that their association was potentially important from the exposome perspective. The metabolomic pathways were visualized with iPath (https://pathways.embl.de/) and the MetScape (version 3.1.3; National Center for Integrative Biomedical Informatics) plugin for Cytoscape software (version: 3.6.1; Cytoscape Consortium). The change rate was calculated by the number of significant metabolites divided by the number of metabolites profiled in this major metabolic pathway for the major metabolic pathway enrichment.
Table 1.
Demographic information of pregnant women in Jiangsu Province from April 2013 to July 2016 ().
| Maternal characteristic | or (%) |
|---|---|
| Maternal age (y) | |
| Ethnicity | |
| Han | 999 (97.6) |
| Other | 25 (2.4) |
| Maternal height (cm) | |
| Weight before pregnancy (kg) | |
| BMI before pregnancy () | |
| Parity | |
| 0 | 814 (79.5) |
| 210 (20.5) | |
| Education (y) | |
| 446 (43.6) | |
| 578 (56.4) | |
| Smokinga | |
| Yes | 23 (2.2) |
| No | 1,001 (97.8) |
| Alcohol consumptiona | |
| Yes | 40 (3.9) |
| No | 984 (96.1) |
Note: These data were complete for all participants. BMI, body mass index.
Reported “yes” at least one time in the first, second, or third trimester.
Literature Search for Interpreting Health Implications of Identified Associations
We took a similar approach to evaluate the health implications of identified associations between exposures and metabolites as was taken in a study that integrated genome-wide association study (GWAS) findings with metabolomics to provide insights into metabolic mechanisms for GWAS associations with common diseases (Suhre et al. 2011). We searched for studies from the databases of PubMed and Google Scholar published before May 2020. Search terms included pregnancy, prenatal and perinatal, a list of the standard names of chemicals and metabolites with significant associations in our study, and various maternal and offspring health parameters, such as gestational age, birth weight, cardiovascular disease, metabolic disorder, neurodevelopment abnormality, neurogenesis, immunological disorder, reproduction abnormality, and cancer. A manual search based on the database results was also conducted. This literature search was intended to provide a current understanding about whether and how the exposure or the presence of a metabolite during pregnancy was associated with the abovementioned health outcomes. Inclusion of literature in our study was based on the following criteria: a) the publication reported a statistically significant association between an exogenous chemical or a metabolite during pregnancy and the health outcome, b) the associations and their directions in the included publication were reliable and supported by other independent biological evidence to avoid including controversial associations that might be contrary to consensus and contain publication bias through careful consideration of true and nontrue relationships, and c) the directions of associations between maternal exposure to exogenous chemicals and outcomes from literature, between maternal metabolites and outcomes from literature, and between maternal exposure to exogenous chemicals and metabolites identified in our study were consistent for explaining the exposome–metabolome-outcome pathway. The explanation of the logic is shown in Figure S2. Three reviewers were involved in study selection, data extraction, and validity assessment, which helped minimize the potential for bias and error. Disagreements were resolved by discussion, and all three reviewers came to consensus.
Results
Demographic Information
The 1,024 pregnant women ranged in age from 19 to 43 y. They were generally lean (mean BMI before , ). Nearly four-fifths (79.5%) of the mothers were primiparous, and more than half (56.4%) had received an education of above high school level (Table 1).
Prevalence of Exposure to Exposome Analytes
The coverage of urinary exposome and metabolome can be found in Figure 2, Table S2, and Excel Table S1. The exposome detection results are summarized in Figure 3 and Table S2. A total of 21 classes of organic chemicals and elements were identified in the first trimester urine. Among organic chemicals, median detectable rates were above or near 50% for organophosphate pesticides, amide pesticides, phthalates, parabens, nicotine and its breakdown product, the perfluorochemical perfluorobutane sulfonic acid, the personal care product glycolic acid, and the plasticizer and fire retardant triphenylphosphate, and various phytoestrogens (Figure 3A). Among the elements, the macro elements, such as calcium, showed the highest concentrations, whereas a large number of unclassified elements showed the lowest concentrations (Figure 3B). Only 1/106 () exposome variables had a detection rate of (actual value of 8.5%, chloroneb). The detection rate was generally higher for the metabolome analytes than for the exposome with only 1/139 (0.7%) metabolome variables with a detection rate of (actual value of 7.9%, ursodeoxycholic acid). When compared with the urinary concentrations of elements in nationally representative U.S. nonsmoking adult female populations reported in the U.S. National Exposure Report (CDC 2019), the elements manganese, arsenic (As), lead, strontium, cesium, barium (Ba), uranium (U), and thallium were higher in our study; the elements cobalt, molybdenum (Mo), cadmium, and tin were similar; and antimony was lower (CDC 2019) (Table S2).
Figure 2.
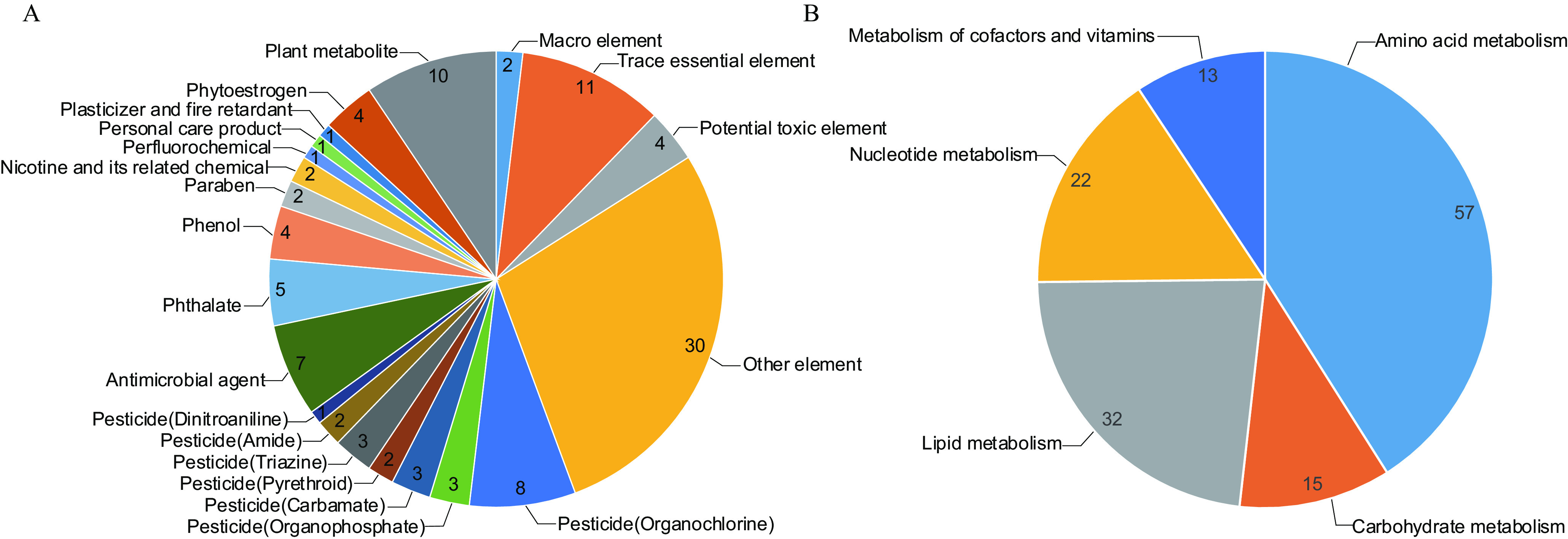
The coverage of the exposome and the metabolome. (A) The distribution of exposome based on chemical/element classification. (B) The distribution of metabolome based on metabolism classification. Among organic chemicals detected by the ultra-high performance liquid chromatography–Q Exactive hybrid quadrupole-orbitrap high resolution mass spectrometry platform, we defined the exposome as those exposures with no known metabolic function in the human body or which are not natural components of human body (pollutants and nonnutritive plant chemicals, organic exposome). The exposome also includes elements (inorganic exposome). The definition of the metabolome is organic chemicals with known metabolic function in the human body or which are natural components of the human body. The inclusion and classification of organic exposome were based on data from PubChem (https://pubchem.ncbi.nlm.nih.gov/), HMDB (https://hmdb.ca/), and KEGG (https://www.genome.jp/kegg/). The classification of elements was based on the reference Maret and Copsey (2012). The inclusion and classification of metabolome were based on data from HMDB and KEGG. The data underlying this figure can be found in Excel Table S1. Note: HMDB, Human Metabolome Database; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 3.
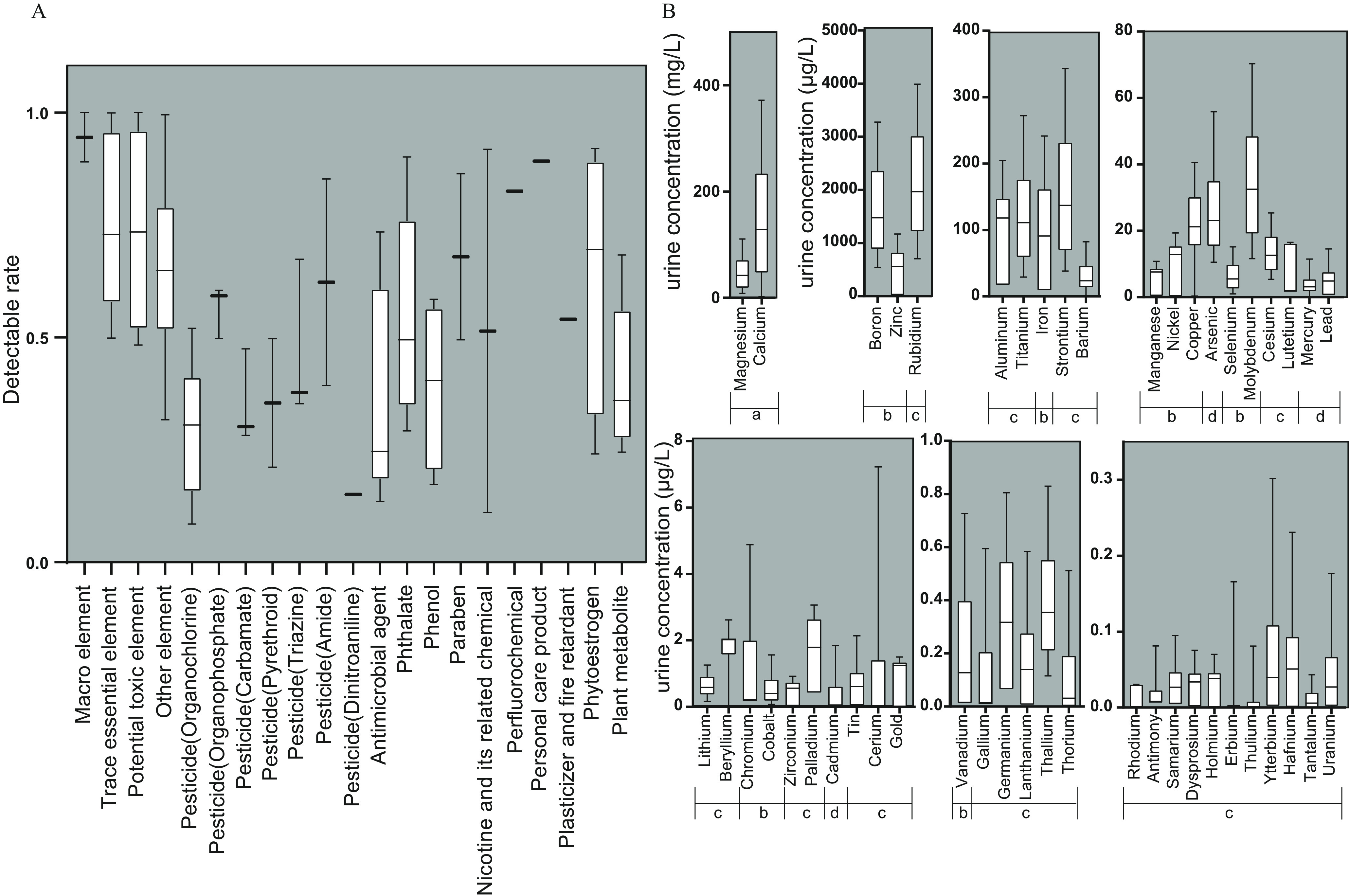
The analytical results of the exposome in urine in the first trimester of pregnant women in Jiangsu Province from April 2013 to July 2016. (A) The detectable rates of exposome based on chemical classification (). (B) The element concentrations. The data are presented as interquartile range box plots (). The top and bottom of each box represents the 75th and 25th percentile, respectively. Also shown are the median (—) separating the inner quartiles, the “whiskers” from the 90th value to the 10th value. The was imputed with the value of LOD/2 for calculation. The presentation of the element data was ordered by the concentration level and atomic number. The data underlying this figure can be found in Table S2. Note: a, macro element; b, trace essential element; c, other element; d, potential toxic element; LOD, limit of detection.
Results of the Exposome and Metabolome-Wide Association Analysis: Environmentally Determined Urinary Metabotypes
As shown in Figures 4 and S3 and Excel Table S2, among 14,734 associations tests, after Bonferroni correction, there were 1,245 significant associations between the exposome in the first trimester and the urinary metabolome in the third trimester (Excel Table S2). All of the 1,245 significant associations remained significant in the sensitivity analysis, using IPW for addressing potential selection bias (Bonferroni correction for 1,245 tests cutoff, ). A total of 1,244 of the 1,245 significant associations remained significant in a sensitivity analysis by excluding 57 smokers and drinkers (Bonferroni correction cutoff for 1,245 tests, ), but the association between Ba and capric acid did not reach the Bonferroni threshold () (Excel Table S2). By using the exposure mixtures analysis method, we found 1,235/1,245 significant associations (99.2%) remained significant with in the PLS-DA (Excel Table S2). Given the robustness of the results, all the 1,245 significant associations were considered as environmentally determined urinary metabotypes and put into the following analysis.
Figure 4.

Heatmap of Bonferroni-14,734–corrected -values regarding selected associations between the inorganic or organic exposome and the metabolome in urine from pregnant women in Jiangsu Province from April 2013 to July 2016. (A) The associations between macro elements and the metabolome. (B) The associations between organophosphate pesticides and the metabolome. When the association was in the positive direction, the blue scale was used for visualizing . When the association was in the negative direction, the red scale was used for visualizing . The association was adjusted by maternal age, BMI before pregnancy, parity, and education using polytomous logistic regression with the Bonferroni correction. * indicates Bonferroni-corrected . The sample size for the association analysis between the organic exposome and the metabolome was 1,024; the sample size for the association analysis between inorganic exposome and metabolome was 963. The data underlying this figure can be found in Excel Table S2, and the heatmap for all associations can be found in Figure S3. Note: AMP, adenosine monophosphate; BMI, body mass index; cAMP, cyclic adenosine monophosphate; CoA, coenzyme A; LysoPC, lysophosphatidlycholine; Mg, Magnesium; Ca, Calcium; IMP, 2-isopropyl-6-methylpyrimidin-4-ol; TCP, 3,5,6-trichloro-2-pyridinol; PHO, phosphamidon; TCA, the citric acid cycle.
We summarized the environmentally determined urinary metabotypes based on classes of organic chemicals and elements (Figure 5). Among the 106 exposome chemicals, 56 exposome chemicals (24 elements and 32 organic chemicals) were significantly associated with at least one metabolite in the metabolome. The classes are described in Figure 2 and Excel Table S1. The macro elements, atrazine and its metabolite deisopropyl-atrazine, and organophosphate pesticides showed relatively consistent associations with the urinary metabolome in classes of exposures (see a subset of results in Figure 4 and all results in Figure S3). There were metabolic associations in almost all classes of exposures. Generally, among all the elements, the macro element, magnesium (Mg) had the largest number of significant metabolic associations (38), followed by titanium (Ti) with the second largest number (34). Among the trace elements, Mo had the largest number of metabolic associations (33). Among toxic elements, As had the largest number of metabolic associations (29). Among the organic compounds, the largest number (73) of metabolic associations was seen for 4--octylphenol. Among the pesticides, silafluofen (a pyrethroid) had the largest number of metabolic associations (69).
Figure 5.
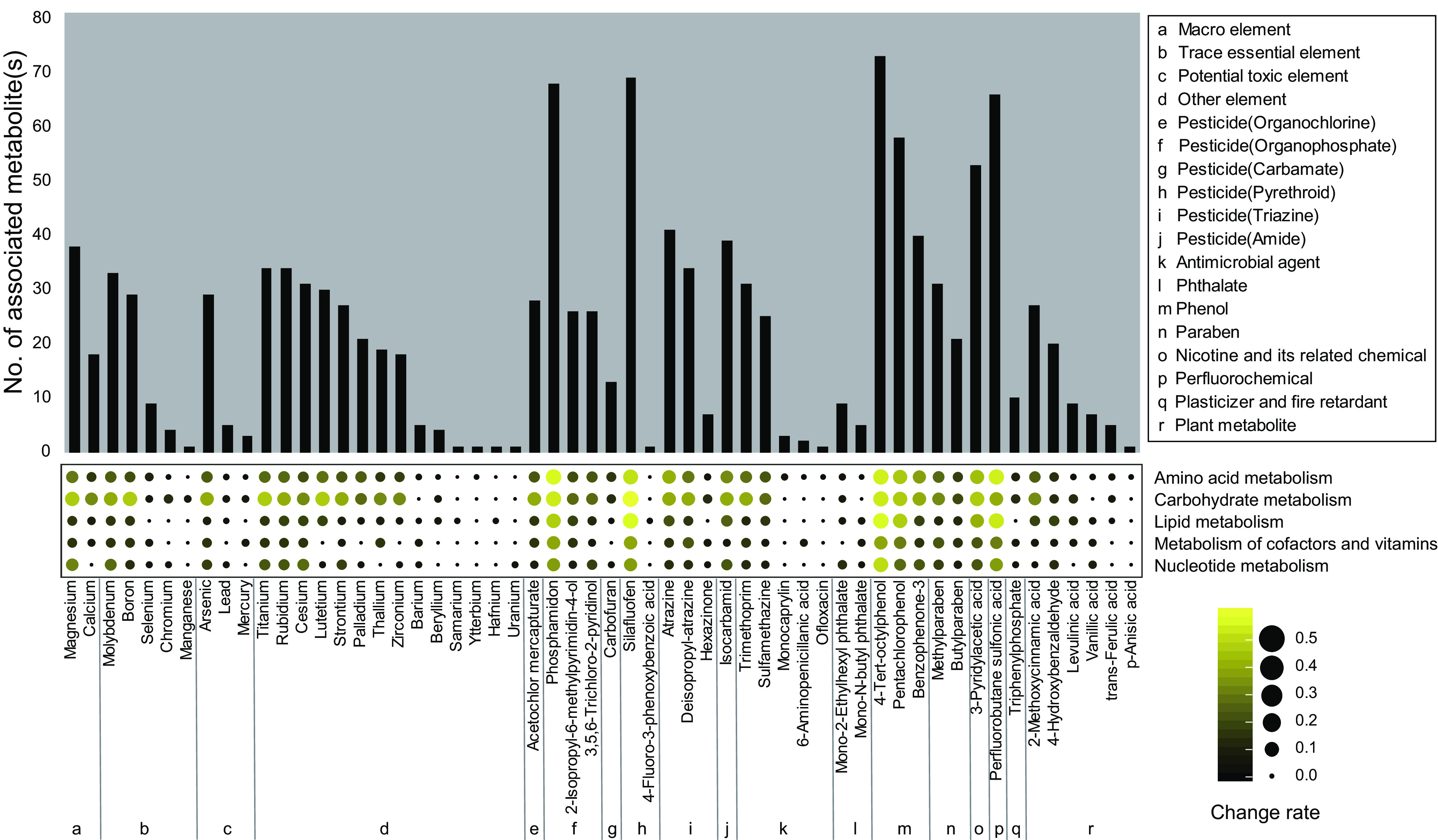
The summary of environmentally determined urinary metabotypes according to the exposome and their change rates in metabolic major pathways of pregnant women in Jiangsu Province from April 2013 to July 2016. Change rate is equal to the number of significant metabolites divided by the number of metabolites profiled in this metabolic major pathway. The sample size for the environmentally determined urinary metabotypes according to the organic exposome was 1,024; the sample size for the environmentally determined urinary metabotypes according to the inorganic exposome was 963. The data underlying this figure can be found in Figure S3 and Excel Table S2.
Based on the major metabolic pathway classification, the change rates were generally highest for carbohydrate metabolism among inorganic and organic exposome-determined urinary metabotypes (Figure 5). We next summarized the environmentally determined urinary metabotypes based on the metabolic network (Figures 6 and S4). Among the 139 metabolites, 96 metabolites were significantly associated with at least one exposome chemical. The enrichment was based on the comparison between the proportion of the number of significant associated exposome chemicals in four classifications—a) macro and trace essential element, b) potential toxic and other element, c) organic pollutant, and d) plant metabolite and phytoestrogen) for each metabolite and the proportion of the original number of detected exposome chemicals in these same four classifications (for original proportion, see Excel Table S1). Generally, organic pollutants were disproportionally associated with the urinary metabolome. Specifically, within carbohydrate metabolism, the pathway of the citric acid cycle was sensitive to the exposome. The most significant metabolite was the central one in this pathway: citric acid, which was also the metabolite with the largest number (37) of associations with exposome. Within lipid metabolism, the top six most sensitive metabolites included the long-chain fatty acids capric acid (number: 31) and dodecanoic acid (number: 24), as well as carnitine (number: 14), a major metabolite involved in fatty acid degradation. The other most sensitive metabolites were unsaturated fatty acids and their related metabolites docosahexaenoic acid (number: 21) and prostaglandin E2 (number: 16), which are bioactive metabolites involved in various events during pregnancy. In addition, a number of metabolites in amino acid metabolism (41) and nucleotide metabolism (16) were significantly associated with the exposome.
Figure 6.
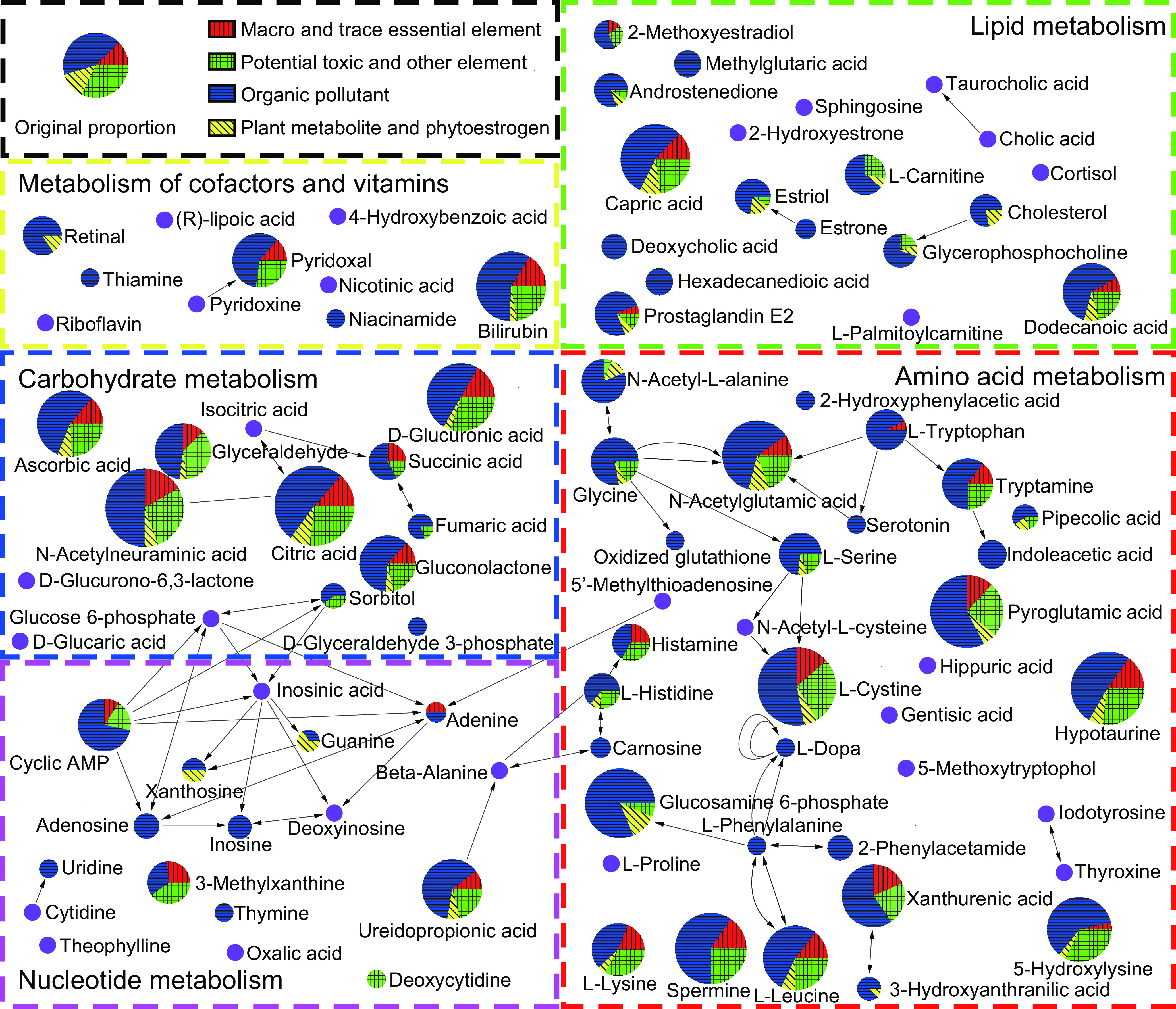
Connection of environmentally determined urinary metabotypes of pregnant women in Jiangsu Province from April 2013 to July 2016 based on the KEGG metabolic pathways. The figure was built by MetScape (version 3.1.3; National Center for Integrative Biomedical Informatics) plugin for Cytoscape software (version: 3.6.1; National Center for Integrative Biomedical Informatics). The sample size for the environmentally determined urinary metabotypes according to the organic exposome was 1,024; the sample size for the environmentally determined urinary metabotypes according to the inorganic exposome was 963. The pie chart named “Original proportion” shows the original constituent ratios of numbers of profiled chemicals in the exposome classified into macro and trace essential element, potential toxic and other element, organic pollutant, and plant metabolite and phytoestrogen. The pie charts in the pathway were built based on constituent ratios of numbers of chemicals in the exposome that were significantly associated with this metabolite classified into macro and trace essential element, potential toxic and other element, organic pollutant, and plant metabolite and phytoestrogen, and the size of the pie charts reflects the number of chemicals in the exposome that were significantly associated with this metabolite. Other profiled metabolites without significant association with any exposome chemical in our study were colored purple in the pathway map. The data underlying this figure can be found in Figure S3 and Excel Table S2. Note: cAMP, cyclic adenosine monophosphate; KEGG, Kyoto Encyclopedia of Genes and Genomes; PGE2, prostaglandin E2.
Environmentally Determined Urinary Metabotypes Revealed the Potential Mechanisms Underlying Exposome-Induced Maternal and Offspring Outcomes
Based on our literature search covering various maternal and offspring outcomes, gestational age, birth weight, fat deposition, neurodevelopment abnormality, immunological disorder, and hypertension were identified as the outcomes where the directions of significant associations from the literature for maternal exposure to exogenous chemicals and outcomes and for maternal metabolite and outcome were consistent with directions of association identified in our study for maternal exposure to exogenous chemicals and metabolite. Details about how the consistency was determined are in Figure S2. These can potentially be used for explaining the metabolic mechanisms underlying the outcomes caused by prenatal exposure to exogenous chemicals.
Among all the outcomes searched, we identified the largest number of potential mechanisms for birth weight (Figure 7; Excel Table S3). This synthesis of the literature with our findings suggests that the effects of 10 analytes on birth weight, gestational age, fat deposition, neurobehavioral development, immunological disorders, and hypertension might be mediated by metabolites that we found to be associated with them and which in the literature were related to these outcomes with a direction consistent with our associations (Figure 7; Excel Table S3).
Figure 7.
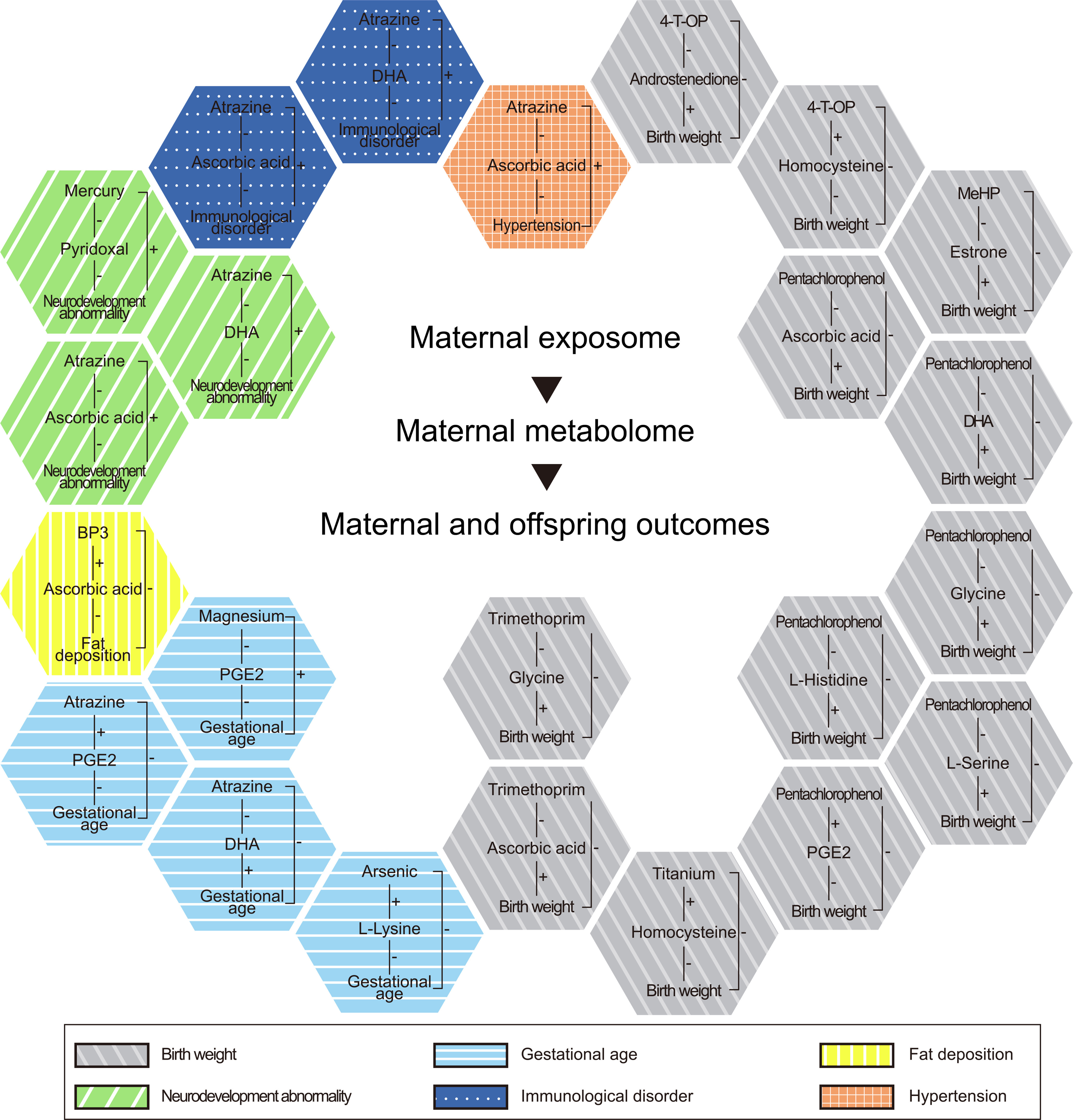
Associations between the exposome and maternal and offspring outcomes that might be mediated by environmentally determined urinary metabotypes. The graph was established by using the literature search strategy described in the “Methods” section. The maternal and offspring outcomes that had consistent associations with the exposome and the metabolome during pregnancy are shown in different colors and textures. The directions of associations between exposome/metabolome and outcomes are indicated as “” and “–”. The detailed results underlying this graph can be found in Excel Table S3. Note: BP3, benzophenone-3; DHA, docosahexaenoic acid; 4-T-OP, 4-tert-octylphenol; MeHP, mono(2-ethylhexyl) phthalate; PGE2, prostaglandin E2.
Discussion
By using exposome-wide analysis (Figure 2), our study provides extensive data regarding organic and inorganic exposure burden in a pregnant population in China (Figure 3). The finding regarding the urinary element concentration comparison between our study and U.S. National Exposure Report was also supported by the comparison with the available data of urinary concentrations reported in The human urine metabolome database (https://urinemetabolome.ca/metabolites) (Table S2), except for U, which has not been included in the database. The heightened exposure to heavy metals in our study population are consistent with the industrialization and resulting heavy metal pollution in China (Shifaw 2018). This supports the importance of studying the health effects of heavy metals exposure in pregnant women in China and other locations undergoing similar trajectories of industrialization. Based on our exposome profiling, 59 organic chemicals, including 45 pollutants, were detectable in our study of pregnant women in China, raising concern about the potential health issues caused by these exposures during pregnancy. Pesticides (i.e., organophosphates and amides), phthalates, parabens, nicotine and its related chemical perfluorobutane sulfonic acid, glycolic acid, triphenylphosphate, and phytoestrogens showed median detection rates above or near 50%, consistent with previous reports of elevated exposure to some of these chemicals among pregnant women in China (Gao et al. 2017; Tang et al. 2015).
Metabolomics is recognized as most closely relevant to disease phenotypes in the omics family (Guijas et al. 2018). Therefore, metabolic disruption caused by exogenous chemical exposure in pregnancy has been studied in relation to pregnancy outcomes in humans (Yang et al. 2020; Zhao et al. 2020). In our study, we identified 1,245 significant associations between chemical exposure in the first trimester and metabolism in the third trimester, providing potential insights into the association between the exposome in early pregnancy and later metabolic perturbations (Figure S3; Excel Table S2). Many of the chemicals we identified as related to metabolic changes, to our knowledge, have not been studied in relation to health effects in the setting of pregnancy. Therefore, given the current and deepening understanding of biological significance of metabolites in pregnant women, the associations we identified may be useful for prediction of risk from these exposures during pregnancy (Figures 5 and 6). Among all the exposures, 4--octylphenol showed the largest number of metabolic associations (Figure S3). This aligns with previous reports that 4--octylphenol is one of the most potent endocrine-disrupting chemicals in vitro (Bonefeld-Jørgensen et al. 2007; Routledge and Sumpter 1997) . Among all the elements, Ti had the second largest number of significant metabolic associations. Ti alloy is widely used in sporting goods, daily necessities, medical supplies, and building materials (Markowska-Szczupak et al. 2020). Titanium dioxide () is a white pigment used in coatings, plastics, inks, paper, chemical fibers, ceramics, rubbers, cosmetics, spices, sunscreens, soaps, toothpastes, and food additives. There is rising concern over exposure to nanoparticles (NPs) during critical windows, such as pregnancy (Wu et al. 2020). Notably, the steroid hormone metabolites, including cholesterol, estriol, androstenedione, and estrone, were disproportionally related to the exposome. They were predominantly associated with pesticides, such as atrazine, phenols and phthalates, which are major endocrine-disrupting chemicals (Roy et al. 2009). Figure S4 depicts the metabolic connection of the major findings of our study, which highlight current findings in our study in the whole metabolism network, providing suggested avenues for future study on the effects of maternal exposome on metabolome during pregnancy.
Metabolic changes during pregnancy can be used to explain the mechanisms underlying the relationships between chemical exposure and maternal and offspring health, as recently described in humans (Yang et al. 2020). We integrated the metabolic associations with exposures identified in our data with published reports of associations of exposures and health outcomes in pregnancy to explore mechanisms, generating a large volume of information on possible mechanisms underlying the relationships between chemicals and various outcomes (Figure 7). With respect to birth weight, for 4--octylphenol, which had the largest number of metabolic associations in our study (Figure 5), exposure during pregnancy has been reported to be related to decreased birth weight in humans (Lv et al. 2016). Exposure in the first trimester was negatively related to levels of androstenedione in the third trimester in our study, and androstenedione/dehydroepiandrosterone during pregnancy has been reported to be positively related to birth weight in male offspring in humans (Mitsui et al. 2018), suggesting prenatal 4--octylphenol exposure might decrease birth weight through decreasing androstenedione in the third trimester. Androstenedione belongs to the steroid hormone biosynthesis pathway, and 4--octylphenol has an potent endocrine-disrupting effect in vitro (Bonefeld-Jørgensen et al. 2007; Routledge and Sumpter 1997) that could explain the observed association between 4--octylphenol and androstenedione in our study. Exposure to 4--octylphenol in the first trimester was positively related to homocysteine in the third trimester in our study. Homocysteine during pregnancy has been reported to be negatively related to birth weight in humans (Liu et al. 2020), suggesting prenatal 4--octylphenol exposure might decrease birth weight through increasing homocysteine in later pregnancy. Similarly, with respect to gestational age, the relationships of three exogenous chemicals, including As, atrazine, and Mg, on gestational age might be mediated by their associated metabolites, including l-Lysine, docosahexaenoic acid, and prostaglandin E2 (Excel Table S3). With respect to fat deposition, benzophenone-3 during pregnancy has been reported to be negatively related to fat deposition during childhood in humans (Buckley et al. 2016). This could be potentially explained by its positive association in our study with ascorbic acid, which has been reported to have a negative association with fat deposition in offspring in humans (Horan et al. 2016). With respect to neurodevelopment, exposure to atrazine (Rastegar-Moghaddam et al. 2019) and mercury (Hg) (Wang et al. 2019) during pregnancy has been reported to be related to deficits in neonatal neurobehavioral development in a mouse model and humans, respectively. In our study, these chemicals were negatively associated with ascorbic acid, docosahexaenoic acid, and pyridoxal. In humans and rodent models, ascorbic acid (Nam et al. 2019; Sirasanagandla et al. 2014), docosahexaenoic acid (Hibbeln et al. 2007; Jayashankar et al. 2012), and pyridoxal (Virk et al. 2018) have been reported to exert positive effects on neurobehavioral development. Thus, it is possible that some of the deleterious effects of atrazine and Hg could be explained by the toxicant-induced metabolic alternations. With respect to immunological disorders, in mouse models, atrazine exposure during pregnancy increases immunopotentiation in offspring (Rowe et al. 2006). In humans, inverse (i.e., protective) relationships have been reported between ascorbic acid (West et al. 2012) and docosahexaenoic acid (Olsen et al. 2008) in pregnancy and infant allergic outcomes. In our study, atrazine was negatively related to ascorbic acid and docosahexaenoic acid, suggesting possible mediating mechanisms for the reported adverse effects of this exposure in the experimental study. With respect to hypertension, in a rat model, exposure to atrazine during pregnancy elevated blood pressure in childhood and adulthood (Rogers et al. 2014). In another rat model, ascorbic acid during pregnancy protected against hypertension in offspring (Wang et al. 2016). In our study, atrazine was negatively related to ascorbic acid, suggesting a potential role for this micronutrient in the association between prenatal atrazine exposure and offspring hypertension. In reviewing the literature, we added the criterion that the associations and their directions in the included publication should be biologically reasonable and supported by other evidence. We believe that we did not have a biased selection of literature because what was included was generally consistent with the majority of studies. Meanwhile, we included papers reporting metabolite levels in blood, whereas our measurement was of levels in urine. However, the association between outcome and metabolite in the cited article might reflect the association between outcome and the exposure to these metabolites in the body, which could be reflected by blood as well as urine because urine production involves filtration of the blood. The positive correlation of concentrations of glycine between urine and blood was supported by previous reports (Oshima et al. 2019). As more associations between prenatal exposure and pregnancy and offspring outcomes are reported, the metabolic associations identified in our study will be useful for exploring potential mechanisms of these findings.
Our goal for this study was to perform exploratory analyses to establish a knowledge base that could be useful to others. Therefore, having the individual contaminant and metabolite association might be more robust and interpretable for our study design. The type of exploratory study we conducted was modeled on a previous paper that used this approach to study the metabolome in relation to metabolomic outcomes and genetic variation (Suhre et al. 2011). However, the development and use of exposure mixtures analysis methods, such as dimension reduction analysis, are of increasing interest. Therefore, we further used PLS-DA to identify the key associations by analyzing all of the exposome information together. We found the almost all significant associations in the individual analysis remained so in the PLS-DA, suggesting that the environmentally determined urinary metabotypes identified in this study were robust from both individual and mixed perspectives. The study of the associations between exposome and metabolome with a full use of exposure mixture analysis and data reduction approaches is an important and interesting topic for future work.
Our study had some limitations. We analyzed only on a spot urine sample rather than repeated samples for exposure measurement, which can lead to misclassification. Because of the difficulty of implementation, there was no requirement of fasting for urine collection in our study, which also might increase the uncertainty of measurement and cause the misclassification of chemical exposure. However, if this misclassification is nondifferential, as we would expect, it would generally lead to attenuation of observed associations (Mendiola et al. 2010). Although food intake could cause some metabolic changes, the urine collection time and food intake time were very close in our study, which should decrease the effect of nonfasting condition on the urine metabolome. Nonfasting samples are also commonly used samples in metabolomics-based epidemiological studies in women during early pregnancy (Voerman et al. 2020). Another limitation is that we were not able to quantify the concentration of organic chemicals in our study. The profiling of chemical exposure in pregnant women without quantitative analysis is still new technology (Wang et al. 2018). Development of exposome technologies that can measure the concentration of organic chemicals should be a priority in the future. Ideally, we would have individual data on all of the health outcomes considered in the literature integration. These data were not available in our study. Therefore, given the fact that metabolome abnormalities are often in the upstream of diseases (Lee-Sarwar et al. 2020), we used the approach in previous large-scale integration of metabolomic data with genomic data to identify potential metabolic mechanisms for health effects of genetic variants in humans (Suhre et al. 2011), which also did not include individual data on the health outcomes considered. This strategy of integrating information from the literature information provided comprehensive insights into metabolome mediated various outcomes affected by genetic factors (Suhre et al. 2011). The first trimester of pregnancy is a vulnerable period of development when chemical exposures may be especially important and have long-lasting effects (Hu et al. 2006). Meanwhile, in the third trimester, a number of maternal (Salonen Ros et al. 2001) and fetal abnormalities (Drukker et al. 2021) related to pregnancy become apparent. A previous report also found that pregnancy outcomes included in the “Literature Search for Interpreting Health Implications of Identified Associations” section were closely related to metabolome changes in the third trimester (Heazell et al. 2012). Therefore, we studied the exposome in the first trimester and correlated it to the metabolome in the third trimester; this design also greatly increased the likelihood that the exposure predated the metabolic outcomes. However, if perturbations of metabolism resulting from exposure are short term, our study design would not capture these. That is one of the limitations of our study. Our study had a number of important strengths, including the large sample size, longitudinal cohort study design, and comprehensive measurement of the urinary exposome and metabolome, as well as their integration.
In conclusion, by conducting an exposome- and metabolome-wide association study during pregnancy, we provide a comprehensive assessment of exposure to elements and organic chemicals exposure in a large sample of pregnant women. This study identified many associations between chemical exposures and maternal metabolism during pregnancy from an omics-wide perspective with large sample size and interpreted potential health implications of identified associations. This integration advances our knowledge of the environmental basis of metabolic variation in pregnant women.
Supplementary Material
Acknowledgments
We thank L. Li, Y. Li, and M. Shi of the National Institutes of Health/National Institute of Environmental Health Sciences (NIH/NIEHS) Biostatistics and Computational Biology Branch and J. Ward and J.-L. Li of the NIEHS Integrative Bioinformatics Support Group for their expert advice. S.J.L. is supported by the Intramural Research Program of the NIH/NIEHS (ZO1 ES49019). M.C., Y.G., R.H., J.D., J.Z., T.C., X.W., and Y.X. are supported by the National Natural Science Foundation of China (81872650 and 81630085) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- Barr DB, Bishop A, Needham LL. 2007. Concentrations of xenobiotic chemicals in the maternal-fetal unit. Reprod Toxicol 23(3):260–266, PMID: , 10.1016/j.reprotox.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Biesheuvel CJ, Vergouwe Y, Steyerberg EW, Grobbee DE, Moons KGM. 2008. Polytomous logistic regression analysis could be applied more often in diagnostic research. J Clin Epidemiol 61(2):125–134, PMID: , 10.1016/j.jclinepi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Blydt-Hansen TD, Sharma A, Gibson IW, Mandal R, Wishart DS. 2014. Urinary metabolomics for noninvasive detection of borderline and acute T cell-mediated rejection in children after kidney transplantation. Am J Transplant 14(10):2339–2349, PMID: , 10.1111/ajt.12837. [DOI] [PubMed] [Google Scholar]
- Bonefeld-Jørgensen EC, Long M, Hofmeister MV, Vinggaard AM. 2007. Endocrine-disrupting potential of bisphenol A, bisphenol A dimethacrylate, 4-n-nonylphenol, and 4-n-octylphenol in vitro: new data and a brief review. Environ Health Perspect 115(suppl 1):69–76, PMID: , 10.1289/ehp.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley JP, Herring AH, Wolff MS, Calafat AM, Engel SM. 2016. Prenatal exposure to environmental phenols and childhood fat mass in the Mount Sinai Children’s Environmental Health Study. Environ Int 91:350–356, PMID: , 10.1016/j.envint.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (Centers for Disease Control and Prevention). 2015. The Fourth National Report on Human Exposure to Environmental Chemicals. February 2015. Atlanta, GA: CDC. [Google Scholar]
- CDC. 2019. The Fourth National Report on Human Exposure to Environmental Chemicals, Updated Tables. January 2019. Atlanta, GA: CDC. [Google Scholar]
- Chen M, Rao RSP, Zhang Y, Zhong CX, Thelen JJ. 2014. A modified data normalization method for GC-MS-based metabolomics to minimize batch variation. Springerplus 3:439, PMID: , 10.1186/2193-1801-3-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Bellavia A, James-Todd T, Correia KF, Valeri L, Messerlian C, et al. . 2018. Evaluating effects of prenatal exposure to phthalate mixtures on birth weight: a comparison of three statistical approaches. Environ Int 113:231–239, PMID: , 10.1016/j.envint.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dencker L, Eriksson P. 1998. Susceptibility in utero and upon neonatal exposure. Food Addit Contam 15(suppl):37–43, PMID: , 10.1080/02652039809374613. [DOI] [PubMed] [Google Scholar]
- Drukker L, Bradburn E, Rodriguez GB, Roberts NW, Impey L, Papageorghiou AT. 2021. How often do we identify fetal abnormalities during routine third-trimester ultrasound? A systematic review and meta-analysis. BJOG 128(2):259–269, PMID: , 10.1111/1471-0528.16468. [DOI] [PubMed] [Google Scholar]
- Duggleby SL, Jackson AA. 2002. Protein, amino acid and nitrogen metabolism during pregnancy: how might the mother meet the needs of her fetus? Curr Opin Clin Nutr Metab Care 5(5):503–509, PMID: , 10.1097/00075197-200209000-00008. [DOI] [PubMed] [Google Scholar]
- Ernst A, Brix N, Lauridsen LLB, Olsen J, Parner ET, Liew Z, et al. . 2019. Exposure to perfluoroalkyl substances during fetal life and pubertal development in boys and girls from the Danish National Birth Cohort. Environ Health Perspect 127(1):17004, PMID: , 10.1289/EHP3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Zhang YW, Huang K, Yan SQ, Mao LJ, Ge X, et al. . 2017. Urinary concentrations of phthalate metabolites in early pregnancy associated with clinical pregnancy loss in Chinese women. Sci Rep 7(1):6800, PMID: , 10.1038/s41598-017-06450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijas C, Montenegro-Burke JR, Warth B, Spilker ME, Siuzdak G. 2018. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat Biotechnol 36(4):316–320, PMID: , 10.1038/nbt.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazell AE, Bernatavicius G, Warrander L, Brown MC, Dunn WB. 2012. A metabolomic approach identifies differences in maternal serum in third trimester pregnancies that end in poor perinatal outcome. Reprod Sci 19(8):863–875, PMID: , 10.1177/1933719112438446. [DOI] [PubMed] [Google Scholar]
- Hibbeln JR, Davis JM, Steer C, Emmett P, Rogers I, Williams C, et al. . 2007. Maternal seafood consumption in pregnancy and neurodevelopmental outcomes in childhood (ALSPAC study): an observational cohort study. Lancet 369(9561):578–585, PMID: , 10.1016/S0140-6736(07)60277-3. [DOI] [PubMed] [Google Scholar]
- Horan MK, McGowan CA, Gibney ER, Byrne J, Donnelly JM, McAuliffe FM. 2016. Maternal nutrition and glycaemic index during pregnancy impacts on offspring adiposity at 6 months of age—analysis from the ROLO randomised controlled trial. Nutrients 8(1):7, PMID: , 10.3390/nu8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Téllez-Rojo MM, Bellinger D, Smith D, Ettinger AS, Lamadrid-Figueroa H, et al. . 2006. Fetal lead exposure at each stage of pregnancy as a predictor of infant mental development. Environ Health Perspect 114(11):1730–1735, PMID: , 10.1289/ehp.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Chen M, Wu W, Lu J, Zhao D, Pan F, et al. . 2016. Gene-gene and gene-environment interactions on risk of male infertility: focus on the metabolites. Environ Int 91:188–195, PMID: , 10.1016/j.envint.2016.02.025. [DOI] [PubMed] [Google Scholar]
- Jayashankar S, Glover CN, Folven KI, Brattelid T, Hogstrand C, Lundebye AK. 2012. Cerebral gene expression and neurobehavioural responses in mice pups exposed to methylmercury and docosahexaenoic acid through the maternal diet. Environ Toxicol Pharmacol 33(1):26–38, PMID: , 10.1016/j.etap.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Kliesch S. 2014. Diagnosis of male infertility: diagnostic work-up of the infertile man. Eur Urol Suppl 13(4):73–82, 10.1016/j.eursup.2014.08.002. [DOI] [Google Scholar]
- Laine JE, Bailey KA, Rubio-Andrade M, Olshan AF, Smeester L, Drobná Z, et al. . 2015. Maternal arsenic exposure, arsenic methylation efficiency, and birth outcomes in the Biomarkers of Exposure to ARsenic (BEAR) pregnancy cohort in Mexico. Environ Health Perspect 123(2):186–192, PMID: , 10.1289/ehp.1307476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Sarwar KA, Lasky-Su J, Kelly RS, Litonjua AA, Weiss ST. 2020. Metabolome–microbiome crosstalk and human disease. Metabolites 10(5):181, PMID: , 10.3390/metabo10050181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Luo D, Wang Q, Ma Y, Ping L, Wu T, et al. . 2020. Serum homocysteine and folate concentrations in early pregnancy and subsequent events of adverse pregnancy outcome: the Sichuan Homocysteine study. BMC Pregnancy Childbirth 20(1):176, PMID: , 10.1186/s12884-020-02860-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Wu C, Lu D, Qi X, Xu H, Guo J, et al. . 2016. Birth outcome measures and prenatal exposure to 4-tert-octylphenol. Environ Pollut 212:65–70, PMID: , 10.1016/j.envpol.2016.01.048. [DOI] [PubMed] [Google Scholar]
- Maret W, Copsey M. 2012. Metallomics: whence and whither. Metallomics 4(10):1017–1019, PMID: , 10.1039/c2mt90041f. [DOI] [PubMed] [Google Scholar]
- Markowska-Szczupak A, Endo-Kimura M, Paszkiewicz O, Kowalska E. 2020. Are titania photocatalysts and titanium implants safe? Review on the toxicity of titanium compounds. Nanomaterials (Basel) 10(10):2065, PMID: , 10.3390/nano10102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola J, Jørgensen N, Andersson AM, Calafat AM, Ye X, Redmon JB, et al. . 2010. Are environmental levels of bisphenol A associated with reproductive function in fertile men? Environ Health Perspect 118(9):1286–1291, PMID: , 10.1289/ehp.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra BB. 2020. The chemical exposome of human aging. Front Genet 11:574936, PMID: , 10.3389/fgene.2020.574936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui T, Araki A, Goudarzi H, Miyashita C, Ito S, Sasaki S, et al. . 2018. Relationship between adrenal steroid hormones in cord blood and birth weight: the SAPPORO Cohort, Hokkaido Study on Environment and Children’s Health. Am J Hum Biol 30(4):e23127, PMID: , 10.1002/ajhb.23127. [DOI] [PubMed] [Google Scholar]
- Nam SM, Seo JS, Nahm SS, Chang BJ. 2019. Effects of ascorbic acid on osteopontin expression and axonal myelination in the developing cerebellum of lead-exposed rat pups. Int J Environ Res Public Health 16(6):983, PMID: , 10.3390/ijerph16060983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narduzzi S, Golini MN, Porta D, Stafoggia M, Forastiere F. 2014. Inverse probability weighting (IPW) for evaluating and “correcting” selection bias. [In Italian]. Epidemiol Prev 38(5):335–341, PMID: . [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC. 2008. Systems biology: metabonomics. Nature 455(7216):1054–1056, PMID: , 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- Noyola-Martínez N, Halhali A, Barrera D. 2019. Steroid hormones and pregnancy. Gynecol Endocrinol 35(5):376–384, PMID: , 10.1080/09513590.2018.1564742. [DOI] [PubMed] [Google Scholar]
- Olsen SF, Østerdal ML, Salvig JD, Mortensen LM, Rytter D, Secher NJ, et al. . 2008. Fish oil intake compared with olive oil intake in late pregnancy and asthma in the offspring: 16 y of registry-based follow-up from a randomized controlled trial. Am J Clin Nutr 88(1):167–175, PMID: , 10.1093/ajcn/88.1.167. [DOI] [PubMed] [Google Scholar]
- Oshima S, Shiiya S, Nakamura Y. 2019. Combined supplementation with glycine and tryptophan reduces purine-induced serum uric acid elevation by accelerating urinary uric acid excretion: a randomized, single-blind, placebo-controlled, crossover study. Nutrients 11(11):2562, PMID: , 10.3390/nu11112562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez T, Daneshian M, Kamp H, Bois FY, Clench MR, Coen M, et al. . 2013. Metabolomics in toxicology and preclinical research. ALTEX 30(2):209–225, PMID: , 10.14573/altex.2013.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar-Moghaddam SH, Mohammadipour A, Hosseini M, Bargi R, Ebrahimzadeh-Bideskan A. 2019. Maternal exposure to atrazine induces the hippocampal cell apoptosis in mice offspring and impairs their learning and spatial memory. Toxin Rev 38(4):298–306, 10.1080/15569543.2018.1466804. [DOI] [Google Scholar]
- Rinschen MM, Ivanisevic J, Giera M, Siuzdak G. 2019. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol 20(6):353–367, PMID: , 10.1038/s41580-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JM, Ellis-Hutchings RG, Grey BE, Zucker RM, Norwood J Jr, Grace CE, et al. . 2014. Elevated blood pressure in offspring of rats exposed to diverse chemicals during pregnancy. Toxicol Sci 137(2):436–446, PMID: , 10.1093/toxsci/kft248. [DOI] [PubMed] [Google Scholar]
- Routledge EJ, Sumpter JP. 1997. Structural features of alkylphenolic chemicals associated with estrogenic activity. J Biol Chem 272(6):3280–3288, PMID: , 10.1074/jbc.272.6.3280. [DOI] [PubMed] [Google Scholar]
- Rowe AM, Brundage KM, Schafer R, Barnett JB. 2006. Immunomodulatory effects of maternal atrazine exposure on male Balb/c mice. Toxicol Appl Pharmacol 214(1):69–77, PMID: , 10.1016/j.taap.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy JR, Chakraborty S, Chakraborty TR. 2009. Estrogen-like endocrine disrupting chemicals affecting puberty in humans—a review. Med Sci Monit 15(6):RA137–145, PMID: . [PubMed] [Google Scholar]
- Salonen Ros H, Lichtenstein P, Bellocco R, Petersson G, Cnattingius S. 2001. Increased risks of circulatory diseases in late pregnancy and puerperium. Epidemiology 12(4):456–460, PMID: , 10.1097/00001648-200107000-00016. [DOI] [PubMed] [Google Scholar]
- Shapiro GD, Dodds L, Arbuckle TE, Ashley-Martin J, Fraser W, Fisher M, et al. . 2015. Exposure to phthalates, bisphenol A and metals in pregnancy and the association with impaired glucose tolerance and gestational diabetes mellitus: the MIREC study. Environ Int 83:63–71, PMID: , 10.1016/j.envint.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Shifaw E. 2018. Review of heavy metals pollution in China in agricultural and urban soils. J Health Pollut 8(18):180607, PMID: , 10.5696/2156-9614-8.18.180607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver MK, Arain AL, Shao J, Chen M, Xia Y, Lozoff B, et al. . 2018. Distribution and predictors of 20 toxic and essential metals in the umbilical cord blood of Chinese newborns. Chemosphere 210:1167–1175, PMID: , 10.1016/j.chemosphere.2018.07.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirasanagandla SR, Rooben RK, Rajkumar Narayanan SN, Jetti R. 2014. Ascorbic acid ameliorates nicotine exposure induced impaired spatial memory performances in rats. West Indian Med J 63(4):318–324, PMID: , 10.7727/wimj.2013.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza RT, Mayrink J, Leite DF, Costa ML, Calderon IM, Rocha Filho EA, et al. . 2019. Metabolomics applied to maternal and perinatal health: a review of new frontiers with a translation potential. Clinics (Sao Paulo) 74:e894, PMID: , 10.6061/clinics/2019/e894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wägele B, et al. . 2011. Human metabolic individuality in biomedical and pharmaceutical research. Nature 477(7362):54–60, PMID: , 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Chen M, Zhou K, Chen D, Yu J, Hu W, et al. . 2015. Prenatal lignan exposures, pregnancy urine estrogen profiles and birth outcomes. Environ Pollut 205:261–268, PMID: , 10.1016/j.envpol.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Tsai TL, Wang SL, Hsieh CJ, Wen HJ, Kuo CC, Liu HJ, et al. . 2021. Association between prenatal exposure to metals and atopic dermatitis among children aged 4 years in Taiwan. JAMA Netw Open 4(10):e2131327, PMID: , 10.1001/jamanetworkopen.2021.31327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen R, Schymanski EL, Barabási AL, Miller GW. 2020. The exposome and health: where chemistry meets biology. Science 367(6476):392–396, PMID: , 10.1126/science.aay3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virk J, Liew Z, Olsen J, Nohr EA, Catov JM, Ritz B. 2018. Pre-conceptual and prenatal supplementary folic acid and multivitamin intake, behavioral problems, and hyperkinetic disorders: a study based on the Danish National Birth Cohort (DNBC). Nutr Neurosci 21(5):352–360, PMID: , 10.1080/1028415X.2017.1290932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voerman E, Jaddoe VWV, Uhl O, Shokry E, Horak J, Felix JF, et al. . 2020. A population-based resource for intergenerational metabolomics analyses in pregnant women and their children: the Generation R Study. Metabolomics 16(4):43, PMID: , 10.1007/s11306-020-01667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijheid M, Slama R, Robinson O, Chatzi L, Coen M, van den Hazel P, et al. . 2014. The Human Early-Life Exposome (HELIX): project rationale and design. Environ Health Perspect 122(6):535–544, PMID: , 10.1289/ehp.1307204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Gerona RR, Schwartz JM, Lin T, Sirota M, Morello-Frosch R, et al. . 2018. A suspect screening method for characterizing multiple chemical exposures among a demographically diverse population of pregnant women in San Francisco. Environ Health Perspect 126(7):077009, PMID: , 10.1289/EHP2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wu W, Li H, Cao L, Wu M, Liu J, et al. . 2019. Relation of prenatal low-level mercury exposure with early child neurobehavioral development and exploration of the effects of sex and DHA on it. Environ Int 126:14–23, PMID: , 10.1016/j.envint.2019.02.012. [DOI] [PubMed] [Google Scholar]
- Wang J, Yin N, Deng Y, Wei Y, Huang Y, Pu X, et al. . 2016. Ascorbic acid protects against hypertension through downregulation of ACE1 gene expression mediated by histone deacetylation in prenatal inflammation-induced offspring. Sci Rep 6:39469, PMID: , 10.1038/srep39469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. . 2011a. Metabolite profiles and the risk of developing diabetes. Nat Med 17(4):448–453, PMID: , 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gao D, Zhang G, Zhang X, Li Q, Gao Q, et al. . 2020. Exposure to multiple metals in early pregnancy and gestational diabetes mellitus: a prospective cohort study. Environ Int 135:105370, PMID: , 10.1016/j.envint.2019.105370. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. . 2011b. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472(7341):57–63, PMID: , 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CE, Dunstan J, McCarthy S, Metcalfe J, D’Vaz N, Meldrum S, et al. . 2012. Associations between maternal antioxidant intakes in pregnancy and infant allergic outcomes. Nutrients 4(11):1747–1758, PMID: , 10.3390/nu4111747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Chen L, Chen F, Zou H, Wang Z. 2020. A key moment for TiO2: prenatal exposure to TiO2 nanoparticles may inhibit the development of offspring. Ecotoxicol Environ Saf 202:110911, PMID: , 10.1016/j.ecoenv.2020.110911. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang M, Lu T, Chen S, Sun X, Guan Y, et al. . 2020. Metabolomics study and meta-analysis on the association between maternal pesticide exposome and birth outcomes. Environ Res 182:109087, PMID: , 10.1016/j.envres.2019.109087. [DOI] [PubMed] [Google Scholar]
- Zhao H, Zheng Y, Zhu L, Xiang L, Zhou Y, Li J, et al. . 2020. Paraben exposure related to purine metabolism and other pathways revealed by mass spectrometry-based metabolomics. Environ Sci Technol 54(6):3447–3454, PMID: , 10.1021/acs.est.9b07634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.