Abstract
Huntington disease (HD) is a neurodegenerative disease caused by a trinucleotide repeat expansion in the HTT gene encoding an elongated polyglutamine tract in the huntingtin (HTT) protein. Expanded mutant HTT (mHTT) is toxic and leads to regional atrophy and neuronal cell loss in the brain, which occurs earliest in the striatum. Therapeutic lowering of mHTT in the central nervous system (CNS) has shown promise in preclinical studies, with multiple approaches currently in clinical development for HD. Quantitation of mHTT in the cerebrospinal fluid (CSF) is being used as a clinical pharmacodynamic biomarker of target engagement in the CNS. We have previously shown that the CNS is a major source of mHTT in the CSF. However, little is known about the specific brain regions and cell types that contribute to CSF mHTT. Therefore, a better understanding of the origins of CSF mHTT and whether therapies targeting mHTT in the striatum would be expected to be associated with significant lowering of mHTT in the CSF is needed. Here, we use complementary pharmacological and genetic-based approaches to either restrict expression of mHTT to the striatum or selectively deplete mHTT in the striatum to evaluate the contribution of this brain region to mHTT in the CSF. We show that viral expression of a mHTT fragment restricted to the striatum leads to detectable mHTT in the CSF. We demonstrate that targeted lowering of mHTT selectively in the striatum using an antisense oligonucleotide leads to a significant reduction of mHTT in the CSF of HD mice. Furthermore, using a transgenic mouse model of HD that expresses full length human mHTT and wild type HTT, we show that genetic inactivation of mHTT selectively in the striatum results in a significant reduction of mHTT in the CSF. Taken together, our data supports the conclusion that the striatum contributes sufficiently to the pool of mHTT in the CSF that therapeutic levels of mHTT lowering in the striatum can be detected by this measure in HD mice. This suggests that CSF mHTT may represent a pharmacodynamic biomarker for therapies that lower mHTT in the striatum.
Keywords: Huntington disease, biomarker, neurodegeneration, huntingtin, cerebrospinal fluid, antisense oligonucleotide
Introduction
Huntington disease (HD) is a fatal neurodegenerative disease that affects an estimated 13.7 per 100,000 individuals in the general population (Fisher and Hayden, 2014). HD is caused by a cytosine-adenine-guanine trinucleotide expansion in the HTT gene which codes for an elongated polyglutamine (polyQ) tract in the huntingtin (HTT) protein (1993). Expanded mutant HTT (mHTT) is ubiquitously expressed throughout the body where it disrupts multiple cellular processes and causes widespread cellular dysfunction (Zuccato et al., 2010). Over time, mHTT-induced toxicity in the central nervous system (CNS) leads to robust regional atrophy and neuronal cell loss in the cortex and basal ganglia, which occurs earliest and is most severe in the caudate nucleus and putamen (collectively the striatum) (Vonsattel et al., 1985).
Lowering levels of mHTT in the central nervous system (CNS) improves HD-like behavioural phenotypes and prevents regional forebrain atrophy in rodent models of HD (Caron et al., 2020b; Harper et al., 2005; Kordasiewicz et al., 2012; Southwell et al., 2018). These preclinical studies have provided compelling evidence to support the advancement of multiple HTT lowering modalities, including: antisense oligonucleotide (ASO; NCT03761849, NCT03225833, NCT03225846) and RNA interference (RNAi) -based therapeutics (NCT04120493) into clinical trials for HD. However, the identification of sensitive and robust pharmacodynamic (PD) biomarkers to accurately assess the magnitude and region of HTT target engagement in the brain are critical for the success of these trials.
We and others have previously developed analogous ultrasensitive immuno-assays to detect mHTT in the cerebrospinal fluid (CSF), and showed that mHTT in the CSF increases with disease progression (Rodrigues et al., 2020; Southwell et al., 2015; Wild et al., 2015). Moreover, we demonstrated that lowering mHTT in the brains of HD mice is reflected by a correlative reduction of mHTT in the CSF (Southwell et al., 2015). This work contributed to the use of CSF mHTT as an exploratory biomarker of target engagement in the first-in-human phase I/IIa clinical trial testing a HTT-targeted ASO (RG6042, RO7234292, tominersen) in patients with early HD (Tabrizi et al., 2019). Notably, results from this trial showed a dose-dependent reduction of mHTT in the CSF in ASO treated individuals, suggesting target engagement in the CNS (Tabrizi et al., 2019). However, lowering of mHTT in the CSF has not been shown to be associated with clinical efficacy outcomes. Preliminary data from the recently halted phase III trial evaluating the efficacy of tominersen in patients with early HD (NCT03761849) showed that a reduction of mHTT in the CSF alone may not predict clinical benefit.
ASO and RNAi-based approaches to lower HTT in clinical trials differ in their routes of administration and are expected to result in different regional patterns of mHTT lowering in the CNS of humans. ASOs targeting HTT to either lower total HTT or selectively reduce mHTT are being delivered by intrathecal infusion in clinical trials for HD. In non-human primates, intrathecal infusion of HTT-targeted ASOs leads to a gradient of ASO distribution in the brain, with brain regions in close proximity to CSF flow being exposed to higher concentrations of ASO, and thus higher HTT target engagement, compared to deeper brain structures more distant from the CSF, where target engagement is minimal (Kordasiewicz et al., 2012; Southwell et al., 2018). These findings suggest that the ASO-mediated reduction of mHTT observed in the CSF of HD patients may reflect more target engagement in the superficial regions of the CNS, such as the spinal cord and cortex, compared to deeper brain structures, such as the basal ganglia.
Alternatively, an adeno-associated virus (AAV) serotype 5 encoding an engineered microRNA targeting the HTT transcript (AAV5-miHTT; AMT-130) is being delivered by stereotaxic infusion into the caudate and putamen in a clinical trial for HD (NCT04120493). Infusion of AAV5-miHTT into the caudate and putamen of a transgenic minipig model of HD resulted in a significant reduction of mHTT throughout the brain with the highest magnitude of mHTT lowering observed at the site of injection, the striatum (Valles et al., 2021). This broad mHTT lowering in the CNS was accompanied by a significant reduction of mHTT in the CSF up to 12 months after administration of AAV5-miHTT (Valles et al., 2021). However, the relative contribution of the striatum to CSF mHTT remains unknown.
Therefore, elucidating whether the striatum contributes to mHTT in the CSF may be critical for the accurate interpretation of clinical target engagement data. If mHTT from the striatum contributes to mHTT in the CSF, then changes seen in clinical trials that target the striatum may reflect therapeutic activity in this region of the brain. Here, we investigate the contribution of the striatum to the pool of mHTT in the CSF. We demonstrate that viral expression of an amino-terminal fragment of mHTT with transduction restricted to the striatum leads to detectable mHTT in the CSF. We also utilize complementary pharmacological and genetic approaches to deplete mHTT selectively in the striatum to investigate the contribution of this brain region to the mHTT in the CSF. We show that mHTT lowering restricted to the striatum using a HTT-targeted ASO leads to a significant reduction of CSF mHTT in young but not older mice, suggesting an interaction with HD-like regional forebrain atrophy. Additionally, we show that genetic inactivation of the mHTT transgene specifically in the striatum using the regulator of G protein signaling 9 (Rgs9)-Cre line crossed to Hu97/18 HD mice leads to progressive reduction of mHTT in the CSF, suggesting that therapeutic levels of HTT lowering in the striatum will likely be detectable in the CSF. These findings will be helpful for the interpretation of clinical mHTT lowering data, and suggest that CSF mHTT will be a valuable clinical biomarker for therapeutic approaches that target the striatum.
Materials and Methods
Animals:
Experiments were performed using FVB/NJ, BACHD (Gray et al., 2008), Hu97/18 (Southwell et al., 2013) and Rgs9-Cre (Dang et al., 2006) mice. Approximately equal numbers of male and female mice were used for all experiments. Animals were maintained under a 12 hour (hr) light:12 hr dark cycle in a clean barrier facility and given ad libitum access to food and water. Experiments were performed with the approval of the Animal Care Committee at the University of British Columbia (A16–0130, A20–0107) and Institute Animal Care and Use Committee at the University of Central Florida (17–32, 2019–07, 2020–50).
Lentivirus:
A lentiviral genome encoding mHTT exon 1 with 103 Qs fused to green fluorescent protein (mHTTex1-GFP) (Southwell et al., 2009) was packaged into a high titer lentivirus by Applied Biological Materials, Inc (Vancouver, BC).
Intrastriatal (IS) lentivirus injections:
Mice were anesthetized with isoflurane and secured into a stereotaxic frame (Stoelting). The scalp was shaved, sterilized with betaine and 70% ethanol and an incision made along the mid-line. The skull was dried to enhance visibility of sutures and landmarks. A dental drill was used to make a bilateral burr holes at 0.8 mm anterior and 1.8 mm lateral to Bregma. A Hamilton syringe with a 30 gauge (G) needle was loaded with either 1 μL of sterile PBS and 4 μL of virus diluted in sterile PBS to a titer of 1E108 IU/mL for convection enhanced delivery (CED) or 2 μL of virus diluted in sterile PBS to a titer of 1E108 IU/mL for non-CED restricted to the striatum. The needle was lowered 3.5 mm ventrally through the burr hole into the striatum and the virus injected at 0.5 μL/minute (min) for CED or 0.2 μL/min for non-CED using an UltraMicroPump with Micro4 controller. The needle was left in place for 5 min and then withdrawn slowly. This process was repeated for the other hemisphere.
Antisense oligonucleotide (ASO) targeting human HTT:
For all ASO studies, a gapmer with a phosphorothioate-modified backbone containing locked nucleic acid (LNA) wing modifications complementary to the human HTT transcript was used (+T*+A*+A*+T*+A*C*G*T*A*A*G*T*G*T*+C*+A*+C*+A*+A; + = LNA modifications and * = phosphorothioate modifications), as previously described (Southwell et al., 2015). The ASO was commercially synthesized (Qiagen) at a 10 mg scale and purified by HPLC. Lyophilized ASO was resuspended in Dulbecco’s phosphate-buffered saline (PBS; Gibco, catalog # 14190–144) and incubated at 37°C for 2 hr. ASO concentrations were calculated based on the equation: Concentration (M) = OD260 x dilution factor/ ε260 where ε260 = extinction coefficient of oligo at 260 nm. For ASO treatments, the molecule was diluted to the indicated concentration in sterile PBS in a total volume of either 10 μL for intracerebroventricular (ICV) injections or 0.5 μL for IS injections and administered as described below.
ICV ASO injections:
ICV injections of ASO were performed as previously described (Southwell et al., 2014).
IS ASO injections:
Mice were anesthetized and prepared for IS injections as described above. A Hamilton syringe with a 30G needle loaded with 0.5 μL of HTT ASO diluted to the appropriate dose in sterile PBS was lowered to a depth of 3.5 mm ventrally through the burr hole into the striatum and injected at a rate of 0.1 μL/min using an UltraMicroPump with Micro4 controller. The needle was left in place for 5 min and then withdrawn slowly. For bilateral intrastriatal injections, this process was repeated for the other hemisphere.
CSF collection:
Mouse CSF was collected as a terminal procedure through the cisterna magna into a 50 μl Hamilton syringe with 12° bevel needle using an adapted stereotaxic frame and UltraMicroPump with Micro4 controller as previously described (Caron et al., 2020a; Southwell et al., 2015).
Tissue collection and processing:
Mice allotted for terminal molecular and biochemical analysis were anesthetized using 2,2,2-Tribromoethanol (Avertin, Sigma Aldrich, catalog # T48402) and CSF was collected as described above. Blood was then collected via cardiac puncture for plasma isolation. Brains were removed and placed on ice for ~1 min to increase tissue rigidity. Brains were then microdissected into different regions (striatum, cortex, hippocampus, cerebellum), frozen in liquid N2 and stored at −80°C until use.
Mice allotted for histology were perfused transcardially with PBS containing 10 IU/mL heparin followed by 4% paraformaldehyde (PFA) in PBS. Brains were removed and post-fixed in 4% PFA in PBS for 24 hr at 4°C. The following day, brains were transferred into 30% sucrose containing 0.01% NaN3. Once equilibrated, brains were frozen on dry ice, mounted in Tissue-TEK O.C.T. embedding compound (Sakura, catalog # 4583), and cut via cryostat (Leica CM3050S) into a series of either 25 μm (immunohistochemistry) or 10 μm (fluorescence in situ hybridization) coronal sections free-floating in PBS with 0.01% NaN3.
HTT immunoblotting:
Quantitative immunoblotting to resolve full length HTT alleles was performed as previously described (Caron et al., 2021). Briefly, 50 μg of total protein was resolved on 10% bis:acrylamide (1:200) gels, transferred to 0.45 μm nitrocellulose and probed with mouse 1CU-4C8 anti-HTT (Fisher Scientific, catalog # MAB2166) and rabbit anti-calnexin (Sigma, catalog # C4731) as a loading control. Immunoblotting for mHTT was performed by resolving 50 μg of total protein on 3–8% Tris-Acetate precast gels (NuPAGE, Invitrogen, catalog # EA0375). Protein was transferred to 0.45 μm nitrocellulose and probed with mouse 1C2 anti-expanded polyQ antibody (Millipore Sigma, catalog # MAB1574) and rabbit anti-β tubulin (Abcam, catalog # ab6046) as a loading control. Immunoblotting for HTTex1-GFP was adapted from (Southwell et al., 2008). Briefly, 50 μg of total protein was resolved on 4–12% Bis-Tris precast gels (NuPage, Invitrogen, catalog #NP0336) using MOPS running buffer (NuPage, Invitrogen, catalog #NP0001). Protein was transferred to 0.45 μm nitrocellulose and probed with rabbit anti-GFP (Invitrogen, catalog # A-11122) and mouse anti-β tubulin (Abcam, catalog # ab131205) loading control. Primary antibodies were detected with IR dye 800CW goat antimouse (Rockland, catalog # 610–131-007) and AlexaFluor 680 goat anti-rabbit (Molecular Probes, catalog # A21076)- labelled secondary antibodies. All blots were scanned using the LiCor Odyssey Infrared Imaging system. Densitometry on band intensities was performed using the LiCor Image Studio Lite software. GFP band integrated optical density (IOD) values were normalized to the loading control IOD values and then to the mean striatal GFP values for each mode of administration. HTT band IOD values were normalized to the loading control IOD values and then to the same allele for PBS treated mice (HTT ASO experiments) or Cre negative mice (genetic inactivation experiments) on the same membrane.
Immunohistochemistry (IHC):
IHC for HTTex1-GFP distribution was performed using rabbit anti-GFP primary antibody (Invitrogen, catalog #A-11122) and goat anti-rabbit AlexaFluor-488 (Invitrogen, catalog #A-11008) secondary antibody as previously described (Caron et al., 2020a).
Fluorescence in situ hybrization (FiSH):
For ASO distribution following intrastriatal administration, a probe complementary to the ASO sequence containing internal LNA as well as 5’ and 3’ fluorescein amidite modifications was synthesized by Qiagen. Lyophilized probes were resuspended in DEPC-treated water (Invitrogen, catalog # AM9920) to a concentration of 25μM. A circle was drawn on glass slides using a delimiting pen, and glass slides were washed 2 × 3 min with PBS in a coplin jar with constant agitation. Slides were then incubated in acetylation buffer (0.06N HCl, 1.35% (v/v) triethanolamine, 0.25% (v/v) acetic anhydride in DEPC-treated water) for 5 min and washed for 2 × 3 min in PBS. Hybridization buffer (50% (v/v) formamide, 5x sodium-saline citrate buffer (SSC; Sigma, catalog # S6639), 0.5 μg/μL yeast tRNA (Invitrogen, catalog # AM7119) and 1x Denhardt’s solution (Sigma, catalog # D2532) in DEPC-treated water, pH 6.5) was then added to sections, covered with Parafilm and incubated at 42°C for 30 min. Parafilm and hybridization buffer were then removed and fresh hybridization buffer containing 2.5 pmol of FiSH probe was added to each slide, covered with Parafilm, and incubated at 42°C for 1 hr. Hybridization buffer was then removed and slides were washed 3 × 10 min in 1x SSC buffer at 50°C, and 1 × 5 min in 2x SSC buffer at room temperature. Sections were then mounted using ProLong Gold Antifade mountant with DAPI (Thermo Fisher, catalog # P36931).
Fluorescence microscopy:
Imaging of HTTex1-GFP distribution was performed using a 5x objective using a Zeiss Axio Observer A1 fluorescence microscope, Zeiss AxioCam ICc 3 camera and AxioVision 4.8.1 software. Imaging of ASO distribution in the brain was performed using a 2.5x objective (Zeiss) using a Zeiss Axio Vert.A1 microscope, Zeiss AxioCam ICm1 CCD camera and Zeiss Zen 2.3 software. Individual images were stitched together using the automated Photomerge tool in Adobe Photoshop CC 2018.
Immunoprecipitation – flow cytometry (IP-FCM):
For measurement of mHTT or mHTTex1GFP in CSF and plasma, HDB4E10/MW1 or GFP IP-FCM, respectively, was performed as previously described (Caron et al., 2020a; Southwell et al., 2015).
Experimental design and statistical analyses:
All statistical analyses were performed using GraphPad Prism v9 (GraphPad). All experiments involving animals included approximately the same number of male and female mice. The number of animals (biological replicates; N) used for each experiment is reported in the figure legends. All bar graphs are presented as scatter dot plots to demonstrate data complexity and variability. Group data from averages of multiple animals are represented as means ± standard error of the mean. Alpha values of <0.05 were considered significant for all analyses. Exact p values for each statistical test are presented in the text and corresponding figure legends.
Groupwise comparisons were assessed using either the student’s t-test (2 groups) or analysis of variance (ANOVA; more than 2 groups). Post hoc analyses for ANOVA tests were performed using the Tukey test (one-way ANOVA or two-way ANOVA) or the Sidak test (two-way ANOVA) to correct for multiple comparisons. Correlation comparisons were assessed using simple linear regression analysis.
Results
Lentiviral delivery of mHTTex1-GFP restricted to the striatum results in detectable fusion protein in the CSF
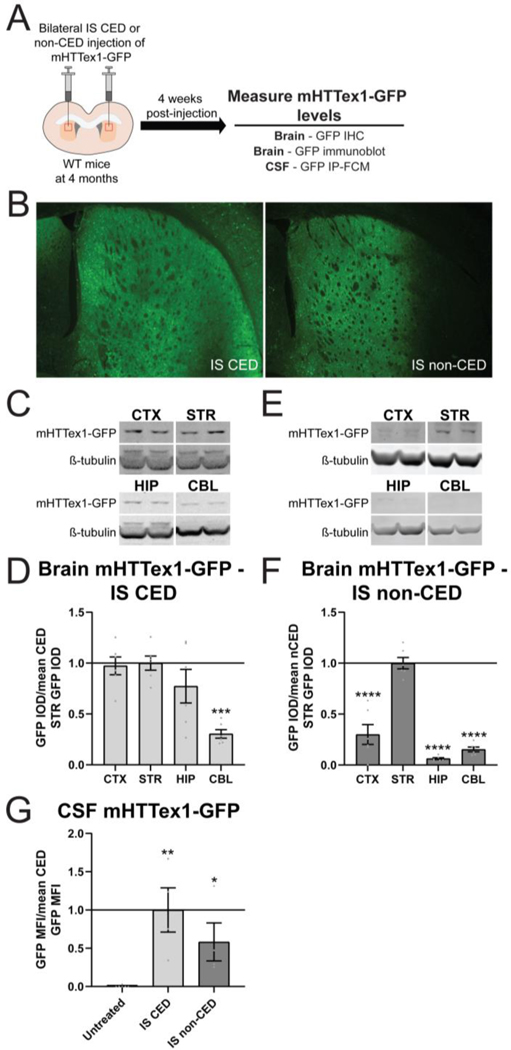
We have previously shown that convection enhanced delivery (CED) of a lentivirus encoding mHTTex1-GFP into the striatum of wild type (WT) mice leads to broad transduction of cells throughout the striatum, as well as in the thalamus, hippocampus and deeper layers of cortex (Caron et al., 2020a). CED resulted in a significant detectable mHTTex1-GFP in the CSF, peaking during the process of neurodegeneration at 3–4 weeks post-injection and declining by 6 weeks post-injection (Caron et al., 2020a). To evaluate the contribution of mHTT derived from the striatum to the pool of mHTT in the CSF, we compared broad forebrain transduction by intrastriatal CED of lentiviral mHTTex1-GFP to restricted striatal transduction achieved by administering half the titer of virus into the striatum by non-CED (Figure 1A). Four weeks after injection, brains were collected for immunohistochemistry (IHC) and immunoblot to evaluate mHTTex1-GFP distribution, and CSF was collected to measure fusion protein levels by GFP IP-FCM (Figure 1A). GFP immunoreactivity confirmed broad forebrain transduction following CED and restricted striatal transgene expression following non-CED (Figure 1B). Quantitation of GFP signal in the brain following CED of mHTTex1-GFP showed similar fusion protein levels in the cortex, striatum and hippocampus, with significantly less measured in the cerebellum (Figure 1C and D. One-way ANOVA: p=0.0003. Tukey’s multiple comparison test: ***p=0.0005 compared to striatum). In contrast, non-CED of mHTTex1-GFP resulted in significantly higher fusion protein levels in the striatum compared to other brain regions (Figure 1E and F. One-way ANOVA: p<0.0001. Tukey’s multiple comparison test: ****p<0.0001 compared to striatum). In the CSF, we measured mHTTex1-GFP signal following broad delivery as well as delivery restricted to the striatum (Figure 1G. One-way ANOVA: p= 0.0078. Tukey’s multiple comparison test: compared to untreated **p=0.0026 IS CED, *p=0.0463 IS non-CED), demonstrating a striatal contribution to CSF mHTT in the context of robust neurodegeneration.
Figure 1. Lentiviral delivery of mHTTex1-GFP restricted to the striatum results in detectable fusion protein in the CSF.
A) Graphical experimental overview where 4 month old wild type (WT) mice received bilateral intrastriatal (IS) injections with mHTTex1-GFP lentivirus via either convection enhanced delivery (CED) or non-CED. Four weeks later, brains were collected for immunohistochemistry (IHC) and immunoblot to evaluate mHTTex1-GFP distribution and CSF was collected to measure fusion protein levels by GFP IP-FCM. B) IHC on brains injected with mHTTex1-GFP lentivirus shows broad forebrain expression following CED or restricted striatal expression following non-CED. C) Representative immunoblots (probed with GFP antibody, A-11122) of the cortex (CTX), striatum (STR), hippocampus (HIP) and cerebellum (CBL) from 5 month old WT mice following bilateral IS CED injections of mHTTex1-GFP. D) Relative quantification of mHTTex1-GFP levels in the brain 4 weeks after IS CED injection of mHTTex1-GFP shows broad distribution of the fusion protein (N=6. One-way ANOVA: p=0.0003. Tukey’s multiple comparison test: ***p=0.0005 compared to STR). E) Representative immunoblots of the CTX, STR, HIP and CBL from 5 month old WT mice following bilateral IS non-CED injections of mHTTex1-GFP. F) Relative quantification of mHTTex1-GFP levels in the brain 4 weeks after IS non-CED injection of mHTTex1-GFP shows distribution of fusion protein is predominantly restricted to the STR (N=6. One-way ANOVA: p<0.0001. Tukey’s multiple comparison test: ****p<0.0001 compared to STR). G) Relative quantification of mHTTex1-GFP CSF normalized to mean CED GFP MFI shows that in comparison to untreated mice, the fusion protein is detected in the CSF following both broad CED and striatum restricted non-CED (N=6 for untreated, N=4 for IS CED and IS non-CED. One-way ANOVA p= 0.0078. Tukey’s multiple comparison test: IS CED **p=0.0026, IS non-CED *p=0.0463 compared to untreated).
Selective mHTT lowering in the striatum using a HTT-targeted ASO
We have previously shown that a unilateral ICV injection with ascending doses of an ASO targeting the human HTT transcript results in a dose-dependent reduction of HTT in the brain of HD mice at 4 weeks post-injection (Southwell et al., 2015). Here, we delivered ascending doses (15–150 μg) of the same HTT-targeted ASO (HTT ASO) via a single, unilateral ICV injection in 2 month old BACHD mice, which harbour a full-length, human mutant HTT transgene (Figure 2A). Four weeks after treatment, we quantified the magnitude of mHTT lowering in different brain regions, including the cortex, striatum, hippocampus and cerebellum by quantitative immunoblotting (Caron et al., 2021). We observed a potent reduction of mHTT in the cortex, striatum, hippocampus and cerebellum (Figure 2B) and measured a significant dose-dependent reduction of mHTT in all brain regions assessed (Figure 2C. Two-way ANOVA: treatment p<0.0001, brain region p<0.0001, interaction p=0.0110. Tukey’s multiple comparison test: ****p<0.0001 HTT ASO compared to PBS). Consistent with the HTT ASO targeting a sequence unique to the human HTT transcript, no lowering of mouse Htt was observed with the HTT ASO at the highest dose tested, 150 μg (Supplementary figure 1A. Two-way ANOVA: treatment p=0.2061, brain region p=0.9749, interaction p=0.9749). As such, only mHTT levels are reported for subsequent experiments with HTT ASO treatment in BACHD animals.
Figure 2. Selective mHTT lowering in the striatum using a HTT-targeted ASO.
A) Graphical experimental overview where 2 month old BACHD animals received a unilateral ICV injection of either PBS or ascending doses HTT ASO (15–150 μg). Four weeks after ICV injections, treated animals were harvested and mHTT levels in different brain regions were quantified by immunoblot. B) Representative immunoblots (probed with total HTT antibody, MAB2166) of the cortex (CTX), striatum (STR), hippocampus (HIP) and cerebellum (CBL) from 3 month old BACHD animals following a unilateral intracerebroventricular (ICV) injection with either PBS or 150 μg HTT ASO. C) Relative quantification of mHTT levels in the brain following ICV injections with ascending doses of HTT ASO (15–150 μg) at 2 months shows dose-dependent lowering of mHTT in all brain regions 4 weeks post-treatment at 3 months of age (N=21 for PBS, N=17 for 15 μg HTT ASO, N=4 for 50 and 150 μg HTT ASO. Two-way ANOVA: treatment p<0.0001, brain region p<0.0001, interaction p=0.0110. Tukey’s multiple comparison test: HTT ASO ****p<0.0001 compared to PBS). D) Graphical experimental overview where 2 month old BACHD animals received bilateral IS injections of either PBS or ascending doses HTT ASO (0.15–15 μg). Four weeks after IS injections, treated animals were harvested and mHTT levels in different brain regions were quantified. E) Representative immunoblots of the CTX, STR, HIP and CBL following bilateral IS injections with either PBS or 0.5 μg HTT ASO. F) Relative quantification of mHTT in different brain regions following bilateral IS injections with ascending doses of a HTT ASO at 2 months of age shows potent lowering of mHTT that is restricted to the striatum with 0.5 μg HTT ASO at 4 weeks post-treatment (N=26 for PBS, N=8 for 0.15 μg HTT ASO, N=18 for 0.5 μg HTT ASO, N=21 for 5 μg HTT ASO, N=12 for 15 μg HTT ASO Two-way ANOVA: treatment p<0.0001, brain region p<0.0001, interaction p<0.0001. Tukey’s multiple comparison test: 0.5 μg HTT ASO CTX p=0.6082, STR ****p<0.0001, HIP p=0.9997, CBL p>0.9999 compared to PBS). G) Fluorescence in situ hybridization with a probe complementary to the HTT ASO sequence 72 hr after IS administration with 0.5 μg HTT ASO shows staining restricted to the STR. A dashed white line was added to highlight the boundaries of the striatum. IOD = integrated optical density.
Guided by the ICV dosing, we next wanted to investigate whether we could restrict mHTT lowering to the striatum following treatment with the HTT ASO. To test this, we performed bilateral IS infusions with either PBS or HTT ASO at doses ranging from 0.15–15 μg and quantified mHTT levels in different brain regions by immunoblotting at 4 weeks post-injection (Figure 2D). Treatment with 0.5 μg HTT ASO resulted in significant mHTT reduction in the striatum, without dramatically lowering mHTT levels in other brain regions (Figure 2E and F. Two-way ANOVA: treatment p<0.0001, brain region p<0.0001, interaction p<0.0001. Tukey’s multiple comparison test: cortex p=0.6082, striatum ****p<0.0001, hippocampus p=0.9997, cerebellum p>0.9999 compared to PBS). In contrast, treatment with 5 and 15 μg of HTT ASO induced significant mHTT lowering in the cortex and hippocampus, as well as the striatum (Figure 2F. Tukey’s multiple comparison test: 5 or 15 μg HTT ASO ****p<0.0001 compared to PBS), whereas 0.15 μg HTT ASO failed to significantly lower mHTT in any brain region (Figure 2F. Tukey’s multiple comparison test: cortex p=0.7554, striatum p=0.8524, hippocampus p=0.9951, cerebellum p=0.0513 compared to PBS). No reduction of mHTT in the cerebellum was measured following IS delivery of HTT ASO at any dose (Figure 2F). We therefore used 0.5 μg HTT ASO for all subsequent IS ASO studies to restrict mHTT lowering to the striatum. We further validated the restricted distribution of the HTT ASO following IS administration by FiSH using a probe complementary to the HTT ASO sequence (Figure 2G).
Localized mHTT lowering restricted to the striatum leads to a reduction of CSF mHTT
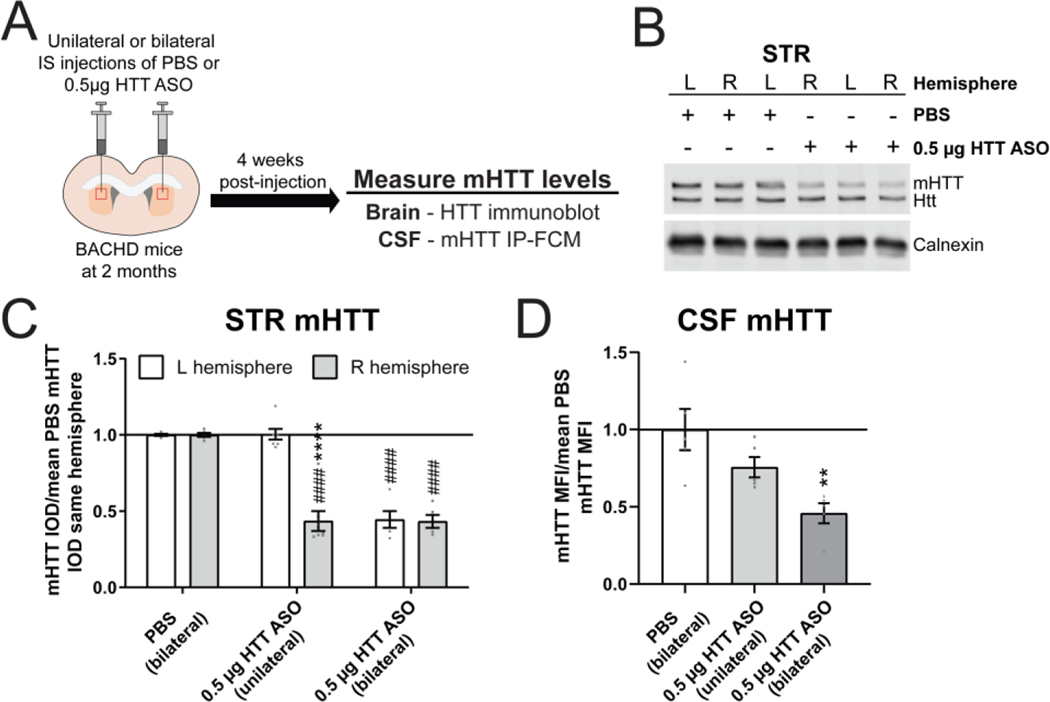
To evaluate the striatal contribution of mHTT to the mHTT in the CSF, we compared unilateral (right hemisphere only) and bilateral IS administration of either PBS or 0.5 μg HTT ASO at 2 months of age and quantified brain and CSF mHTT levels 4 weeks post-injection, at 3 months of age (Figure 3A). We measured a significant reduction of mHTT restricted to the right striatal hemisphere following unilateral IS HTT ASO treatment, whereas no mHTT reduction was observed in the un-injected left striatal hemisphere (Figure 3B and C. Two-way ANOVA: treatment p<0.0001, hemisphere p<0.0001 and interaction p<0.0001. Sidak’s multiple comparison test: unilateral HTT ASO left compared to right hemisphere ****p<0.0001, PBS right hemisphere compared to unilateral HTT ASO right hemisphere ####p<0.0001). As expected, significant mHTT lowering was observed in both hemispheres following bilateral IS HTT ASO treatment (Figure 3C. Sidak’s multiple comparison test: PBS right hemisphere compared to bilateral HTT ASO right hemisphere ####p<0.0001, PBS left hemisphere compared bilateral HTT ASO left hemisphere ####p<0.0001). The magnitude to mHTT lowering in the right striatal hemisphere following either unilateral or bilateral IS HTT ASO treatment was nearly identical (Figure 3C. Sidak’s multiple comparison test: unilateral vs bilateral HTT ASO right hemisphere p=0.9985) and consistent with the mHTT lowering observed in the dose-response study (Figure 2E). Quantitation of mHTT MFI in the CSF by IP-FCM showed a significant reduction of mHTT levels with bilateral but not with unilateral mHTT lowering in the striatum (Figure 3D. One-way ANOVA: p=0.0053. Tukey’s multiple comparison test: bilateral IS HTT ASO **p=0.0040, unilateral IS HTT ASO p=0.2011 compared to PBS). Notably, mHTT CSF levels in animals that received a unilateral IS HTT ASO treatment were reduced approximately half as much as levels in mice that received bilateral IS HTT ASO treatment (Figure 3D. 24% vs 54% reduction of CSF mHTT. Tukey’s multiple comparison test: unilateral compared to bilateral IS HTT ASO p=0.1016). These data demonstrate an additive reduction of CSF mHTT following bilateral compared to unilateral IS injection, and suggests a proportionate contribution of both striatal hemispheres to mHTT detected in the CSF.
Figure 3. Localized mHTT lowering restricted to the striatum leads to a reduction of CSF mHTT.
A) Graphical experimental overview where 2 month old BACHD animals received either unilateral or bilateral IS injections with either PBS or 0.5 μg HTT ASO. Four weeks after IS injections, treated animals were harvested and mHTT levels in the STR and CSF were quantified. B) Representative immunoblot and C) relative quantification of mHTT levels in both STR hemispheres following unilateral (right hemisphere only) or bilateral IS injections with either PBS or 0.5 μg HTT ASO (N=5 for PBS, N=7 for unilateral 0.5 μg HTT ASO, N=5 for bilateral 0.5 μg HTT ASO. Two-way ANOVA: treatment p<0.0001, hemisphere p<0.0001 and interaction p<0.0001. Sidak’s multiple comparison test: unilateral HTT ASO L vs R hemisphere ****p<0.0001, bilateral 0.5 μg HTT ASO L vs R hemisphere p=0.9966, PBS R hemisphere vs unilateral 0.5 μg HTT ASO R hemisphere ####p<0.0001, PBS L hemisphere vs unilateral 0.5 μg HTT ASO L hemisphere p=0.9976, PBS R hemisphere vs bilateral 0.5 μg HTT ASO R hemisphere ####p<0.0001, PBS L hemisphere vs bilateral HTT 0.5 μg ASO L hemisphere ####p<0.0001). D) Relative quantification of mHTT levels in the CSF normalized to the mean of PBS mHTT median fluorescence intensity (MFI) shows an additive reduction of CSF mHTT with bilateral compared to unilateral IS injection of 0.5 μg HTT ASO, highlighting the proportionate contribution of both STR hemispheres to CSF mHTT (N=5 for each treatment condition. One-way ANOVA: p=0.0053. Tukey’s multiple comparison test: compared to PBS treated **p=0.0040 bilateral, p=0.2011 unilateral 0.5μg HTT ASO). IS = intrastriatal, L = left, R = right, CTX = cortex, STR = striatum, HIP = hippocampus, CBL = cerebellum, CSF = cerebrospinal fluid, IOD = integrated optical density.
CSF mHTT levels are dynamic following mHTT lowering restricted to the striatum
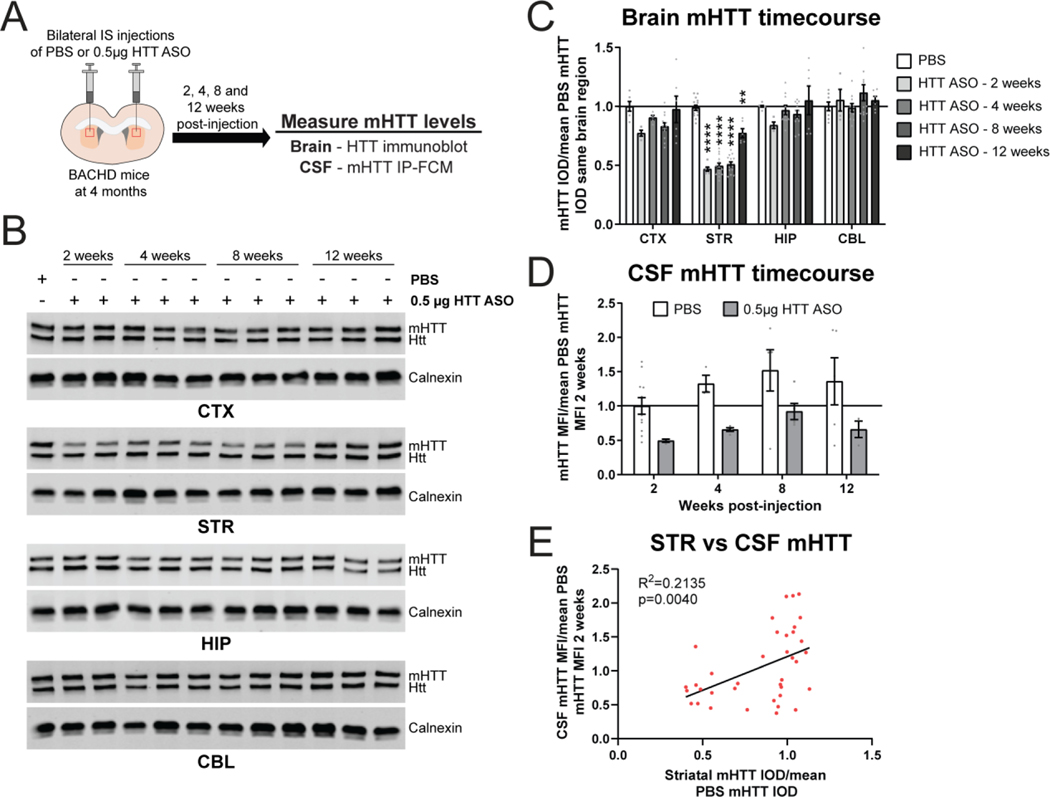
We next evaluated the kinetics of CSF mHTT changes in response to mHTT lowering restricted to the striatum at different timepoints after treatment. To test this, 4 month old BACHD mice received bilateral IS injections of either PBS or 0.5 μg of HTT ASO, and we collected brains and CSF at 2, 4, 8 and 12 weeks post-injection for quantitation of mHTT (Figure 4A). Treatment with 0.5 μg of HTT ASO resulted in a significant reduction of mHTT in the striatum at all timepoints compared to PBS treatment (Figure 4B and C. Two-way ANOVA: treatment p<0.0001, timepoint p<0.0001, interaction p<0.0001. Tukey’s multiple comparison test: 0.5 μg HTT ASO compared to PBS **p=0.0094, ****p<0.0001). Levels of mHTT in other brain regions were not significantly reduced at any timepoint. We observed a significant effect of HTT ASO-mediated striatal mHTT lowering on CSF mHTT, but this did not reach posthoc significance at any one timepoint (Figure 4D. Two-way ANOVA: treatment p=0.0004, timepoint p=0.1714, interaction p=0.9723. Sidak’s multiple comparison test: 0.5 μg HTT ASO compared to PBS at 2 weeks p=0.3285, 4 weeks p=0.2252, 8 weeks p=0.1623 and 12 weeks p=0.1561). We also observed a trend towards an increase in CSF mHTT over time following both PBS and HTT ASO treatment, but these did not reach significance (Figure 4D. Sidak’s multiple comparison test: PBS 2 compared to 4 weeks p=0.6901, 2 compared to 8 weeks p=0.1637, 2 compared to 12 weeks p=0.4615; HTT ASO 2 compared to 4 weeks p=0.9645, 2 compared to 8 weeks p=0.5752, 2 compared to 12 weeks p=0.9691; Test for linear trend: PBS p=0.1171, HTT ASO p=0.0855). Comparing mHTT levels in the striatum and CSF at all timepoints revealed a modest but significant positive correlation between these variables (Figure 4E. Linear regression: R2=0.2135, p=0.0040). These data show that CSF mHTT levels are dynamic in response to mHTT lowering in the striatum and suggest a correlation between mHTT levels in both compartments.
Figure 4. CSF mHTT levels are dynamic following mHTT lowering restricted to the striatum.
A) Graphical experimental overview where 4 month old BACHD animals received bilateral IS injections of either PBS or 0.5μg of HTT ASO. Animals were collected at 2, 4, 8 and 12 weeks post-injection and mHTT levels in the brain and CSF were quantified. B) Representative western blots and C) relative quantification of mHTT in the CTX, STR, HIP and CBL at different timepoints following bilateral IS injections with either PBS or 0.5μg HTT ASO (N=6–14 for PBS, N=4–6 for 0.5 μg HTT ASO - 2 weeks, N=5–14 for 0.5 μg HTT ASO - 4 weeks, N=12 for 0.5 μg HTT ASO - 8 weeks, N=6 for 0.5 μg HTT ASO - 12 weeks. Two-way ANOVA: treatment p<0.0001, timepoint p<0.0001, interaction p<0.0001. Tukey’s multiple comparison test: compared to PBS **p=0.0094, ****p<0.0001). D) Relative quantification of mHTT levels in the CSF normalized to the mean of 2 week PBS mHTT MFI shows that CSF mHTT levels are dynamic following selective reduction of mHTT in the STR (N=3–11 for PBS, N=3–5 for 0.5 μg HTT ASO per timepoint. Two-way ANOVA: treatment p=0.0004, timepoint p=0.1714, interaction p=0.9723. Sidak’s multiple comparison test: 2 weeks p=0.3285, 4 weeks p=0.2252, 8 weeks p=0.1623 and 12 weeks p=0.1561 0.5μg HTT ASO compared to PBS; Test for linear trend: PBS p=0.1171, HTT ASO p=0.0855). E) Comparison of STR mHTT and CSF mHTT levels in matched samples at all timepoints following bilateral IS injections with either PBS or 0.5μg HTT ASO shows a significant correlation between these variables (Linear regression: R2=0.2135, p=0.0040). IS = intrastriatal, CTX = cortex, STR = striatum, HIP = hippocampus, CBL = cerebellum, CSF = cerebrospinal fluid, IOD = integrated optical density, MFI = median fluorescence intensity.
Selective mHTT lowering in the striatum at multiple ages correlates with a reduction of CSF mHTT
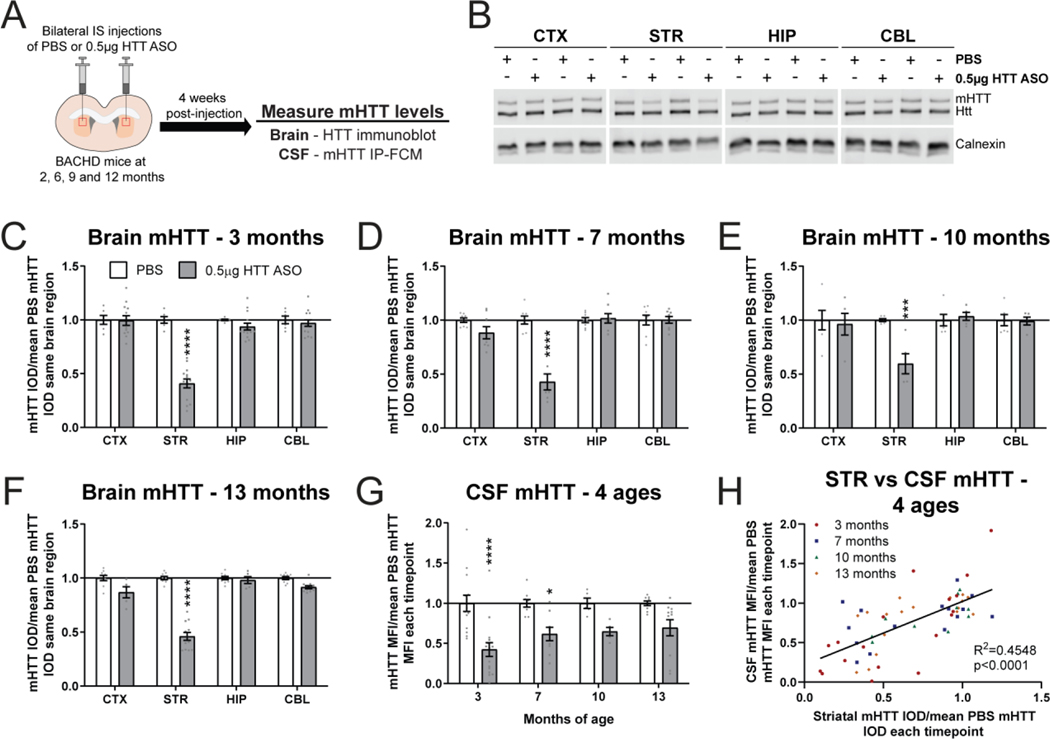
To evaluate the effect of age on the contribution of striatal mHTT to CSF mHTT, we performed bilateral IS injections with either PBS vehicle or 0.5 μg HTT ASO in BACHD animals at either 2, 6, 9 or 12 months of age and quantified brain and CSF mHTT levels 4 weeks post-injection, at 3, 7, 10 and 13 months of age, respectively (Figure 5A). The oldest age was selected since BACHD mice show robust striatal atrophy and degeneration of neurons in the striatum at 12 months of age (Gray et al., 2008; Pouladi et al., 2012). Consistent with IS HTT ASO treatment at younger ages, we observed potent mHTT lowering in the striatum at 13 months of age following HTT ASO treatment (Figure 5B). Quantitation of mHTT in different brain regions following treatment showed a significant reduction of mHTT in the striatum at 3 months (Figure 5C. Two-way ANOVA: treatment p<0.0001, brain region p<0.0001, interaction p<0.0001. Sidak’s multiple comparison test: ****p<0.0001 compared to PBS), 7 months (Figure 5D. Two-way ANOVA: treatment p=0.0005, brain region p<0.0001, interaction p<0.0001. Sidak’s multiple comparison test: ****p<0.0001 compared to PBS) 10 months (Figure 5E. Two-way ANOVA: treatment p=0.0313, brain region p=0.0067, interaction p=0.0067. Sidak’s multiple comparison test: ***p=0.0004 compared to PBS), and 13 months of age (Figure 5F. Two-way ANOVA: treatment p<0.0001, brain region p<0.0001, interaction p<0.0001. Sidak’s multiple comparison test: ****p<0.0001 compared to PBS). The magnitude of mHTT lowering in the striatum following IS HTT ASO treatment was not significantly different at any age assessed (41–59% mHTT reduction. Supplementary figure 2A. Two-way ANOVA: age p=0.3183, treatment p<0.0001, interaction p=0.3183). In the CSF, we measured a significant reduction of mHTT with HTT ASO treatment which reached posthoc significance at 3 and 7 months of age but not at 10 or 13 months of age, although a trend towards reduction was observed at the 2 older ages (Figure 5G. Two-way ANOVA: treatment p<0.0001, age p=0.3541, interaction p=0.3541. Sidak’s multiple comparison test: 3 months ****p<0.0001, 7 months *p=0.0207, 10 months p=0.1908 and 13 months p=0.0726 compared to PBS). Despite a similar magnitude of mHTT lowering in the striatum at all ages, there was a significant linear trend towards an increase in CSF mHTT levels in HTT ASO treated animals from 3 and 13 months of age (Supplementary figure 2B. Sidak’s multiple comparison test: HTT ASO 3 vs 7 months p=0.3580, 3 vs 10 months p=0.4139, 3 vs 13 months p=0.0750; Test for linear trend: HTT ASO p=0.0269). Comparing relative mHTT levels in the striatum to matched mHTT CSF following bilateral IS injections with either PBS or 0.5 μg HTT ASO revealed a significant positive correlation at all ages (Figure 5H. Linear regression R2=0.4548, p<0.0001). Together these data demonstrate that mHTT originating from the striatum contributes to the pool of CSF mHTT in BACHD mice at different ages but that this contribution may be influenced by the onset of neuropathological abnormalities at older ages.
Figure 5. Selective mHTT lowering in the striatum at multiple ages correlates with a reduction of CSF mHTT.
A) Graphical experimental overview where 2, 6, 9 and 12 month BACHD animals received bilateral IS injections of either PBS or 0.5μg HTT ASO. Four weeks after IS injections, treated animals were harvested and mHTT levels in the brain and CSF were quantified at 3, 7, 10 and 13 months, respectively. B) Representative immunoblot of HTT in the CTX, STR, HIP and CBL of 13 month old BACHD mice following bilateral IS injections with either PBS or 0.5μg HTT ASO. Relative quantification of mHTT in different brain regions of C) 3 month (N=6 for PBS, N=14 for 0.5 μg HTT ASO. Two-way ANOVA: treatment p<0.0001, brain region p<0.0001, interaction p<0.0001. Sidak’s multiple comparison test: ****p<0.0001), D) 7 month (N=8 for PBS, N=8 for 0.5 μg HTT ASO. Two-way ANOVA: treatment p<0.0001, brain region p<0.0001, interaction p<0.0001. Sidak’s multiple comparison test: ****p<0.0001), E) 10 month (N=6 for PBS, N=5 for 0.5 μg HTT ASO. Two-way ANOVA: treatment p=0.0313, brain region p=0.0067, interaction p=0.0067. Sidak’s multiple comparison test: ***p=0.0004), and F) 13 month old treated BACHD animals (N=9 for PBS, N=6–12 for 0.5 μg HTT ASO. Two-way ANOVA: treatment p<0.0001, brain region p<0.0001, interaction p<0.0001. Sidak’s multiple comparison test: ****p<0.0001) shows consistent mHTT lowering restricted to the striatum. G) Relative quantification of mHTT in the CSF normalized to the mean of PBS mHTT MFI at each timepoint shows a significant reduction of CSF mHTT following bilateral IS with 0.5μg HTT ASO at 3 and 7 months but not at 10 and 13 months of age (N=5–14 for PBS, N=5–16 for 0.5 μg HTT ASO per age. Two-way ANOVA: treatment p<0.0001, age p=0.3541, interaction p=0.3541. Sidak’s multiple comparison test: 3 months ****p<0.0001, 7 months *p=0.0207, 10 months p=0.1908 and 13 months p=0.0726 0.5μg HTT ASO compared to PBS; Test for linear trend: HTT ASO p=0.0269). H) Comparison of STR mHTT and CSF mHTT levels at all ages following bilateral IS injections with either PBS or 0.5μg HTT ASO (Linear regression: R2=0.4548, p<0.0001). IS = intrastriatal, CTX = cortex, STR = striatum, HIP = hippocampus, CBL = cerebellum, CSF = cerebrospinal fluid, IOD = integrated optical density, MFI = median fluorescence intensity.
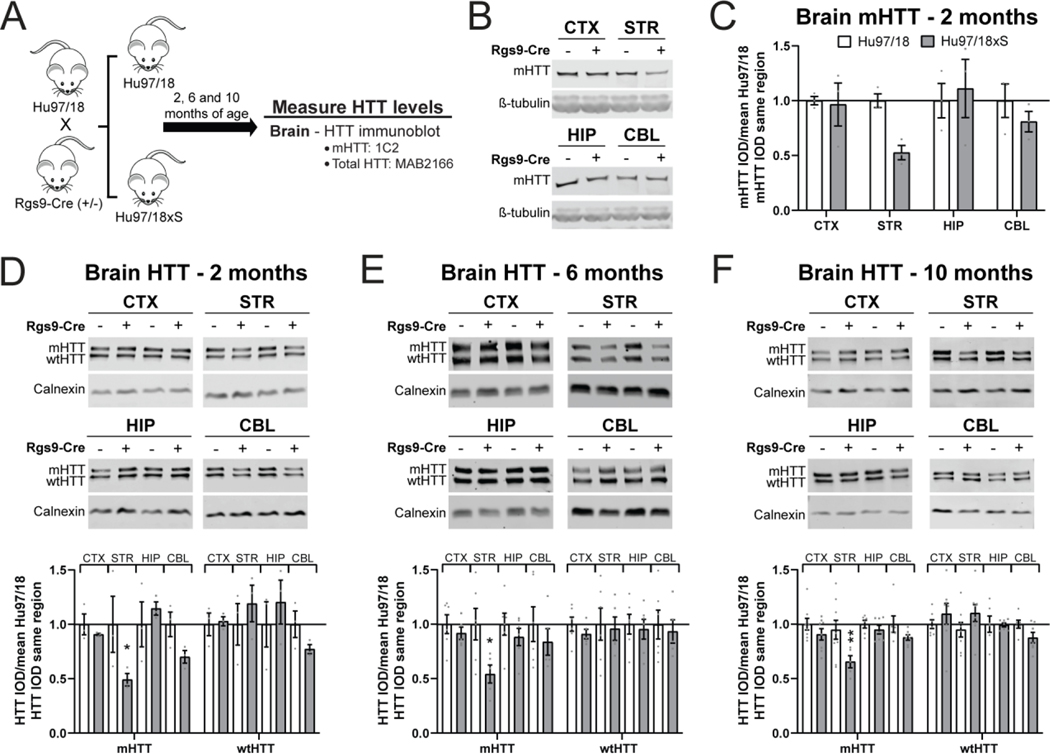
Rgs9-Cre selectively inactivates mHTT in the striatum of Hu97/18 mice
We next evaluated whether constitutive genetic inactivation of mHTT restricted to the striatum would alter levels of mHTT in the CSF. To achieve this, we utilized the Hu97/18 mouse model of HD, which expresses human, full-length mHTT (BACHD) and wild type HTT (wtHTT; YAC18) transgenes at approximately endogenous levels on the mouse Htt knockout background (Southwell et al., 2013). The BACHD mHTT transgene carries loxP sites flanking exon 1 where mHTT can be inactivated in Cre-expressing cell lineages (Gray et al., 2008; Wang et al., 2014). Here, we have crossed the Hu97/18 mouse model of HD to the Rgs9-Cre line, which expresses Cre recombinase under the control of the Rgs9 promoter and is predominantly restricted to medium spiny neurons (MSNs) of the striatum (Dang et al., 2006; Wang et al., 2014) (Figure 6A). This cross yielded littermate mice with genotypes Hu97/18;Rgs9-Cre (−/−), which are equivalent to the parent Hu97/18 line, or Hu97/18;Rgs9-Cre (+/−), which have mHTT selectively inactivated in the striatum, hereafter called Hu97/18xS.
Figure 6. Rgs9-cre selectively inactivates mHTT in the striatum of Hu97/18 mice.
A) Graphical experimental overview where the Hu97/18 line was crossed to the Rgs9-Cre line to produce Hu97/18;Rgs9-Cre (−/−) (Hu97/18) and Hu97/18;Rgs9-Cre (+/−) (Hu97/18xS) littermates. Animals were collected at 2, 6 and 10 months of age for measurement of HTT in the brain. B) Representative immunoblots probed with the 1C2 antibody, raised against expanded polyglutamine, and C) relative quantification of mHTT levels in the CTX, STR, HIP and CBL of Hu97/18 and Hu97/18xS littermates at 2 months of age (N=3 per genotype. Two-way ANOVA: genotype p= 0.1798, brain region p= 0.2721, interaction p= 0.2721. Sidak’s multiple comparison test: striatum p= 0.1442). D-F) Representative immunoblots probed with total HTT antibody MAB2166 (top) and relative quantification (bottom) of HTT in the CTX, STR, HIP and CBL of Hu97/18 and Hu97/18xS littermates at D) 2 months (N=3 per genotype. Two-way ANOVA: genotype p= 0.0381, brain region p= 0.1269, interaction p= 0.0628. Sidak’s multiple comparison test: *p= 0.0374), E) 6 months (N=6 per genotype. Two-way ANOVA: genotype p= 0.0136, brain region p= 0.3418, interaction p= 0.3418. Sidak’s multiple comparison test: *p= 0.0280), and F) 10 months of age (N=8 per genotype. Two-way ANOVA: genotype p= 0.0022, brain region p= 0.0322, interaction p= 0.2331. Sidak’s multiple comparison test: **p= 0.0070). CTX = cortex, STR = striatum, HIP = hippocampus, CBL = cerebellum, IOD = integrated optical density.
We first evaluated the regional selectivity of mHTT inactivation in this line by immunoblotting lysates from the cortex, striatum, hippocampus and cerebellum from 2 month old animals probed with the 1C2 antibody, specific for expanded polyQ (Figure 6B). This antibody was used for assessment of the previously characterized BACHD;Rgs9-Cre cross (Wang et al., 2014). We observed selective reduction of mHTT protein in the striatum, though the genotype effect failed to reach significance, potentially due to the small number of animals assessed (Figure 6C. Two-way ANOVA: genotype p= 0.1798, brain region p= 0.2721, interaction p= 0.2721. Sidak’s multiple comparison test: striatum p= 0.1442). We next assessed both alleles of HTT by allelic separation immunoblotting using an antibody against total HTT, MAB2166, and observed dramatic reduction of mHTT in the striatum, but not in the cortex, hippocampus or cerebellum at 2 months (Figure 6D. Two-way ANOVA: genotype p= 0.0381, brain region p= 0.1269, interaction p= 0.0628. Sidak’s multiple comparison test: striatum *p= 0.0374), 6 months (Figure 6E. Two-way ANOVA: genotype p= 0.0136, brain region p= 0.3418, interaction p= 0.3418. Sidak’s multiple comparison test: striatum *p= 0.0280) and 10 months of age (Figure 6F. Two-way ANOVA: genotype p= 0.0022, brain region p= 0.0322, interaction p= 0.2331. Sidak’s multiple comparison test: striatum **p= 0.0070). At 2 months of age, a trend toward reduced mHTT was observed in the cerebellum, though this failed to reach post hoc significance (Sidak’s multiple comparison test: cerebellum p= 0.4341). This trend was seen in the wtHTT allele as well, which lacks loxP sites in the transgene, suggesting that it is not a direct result of Cre-mediated inactivation of the mHTT transgene. There was no effect of genotype on wtHTT protein at any age. Together these data demonstrate selective striatal inactivation of mHTT in Hu97/18xS mice at multiple ages. The magnitude and regional specificity of mHTT inactivation in the striatum is consistent with the previously reported BACHD;Rgs9-Cre mouse line (Wang et al., 2014).
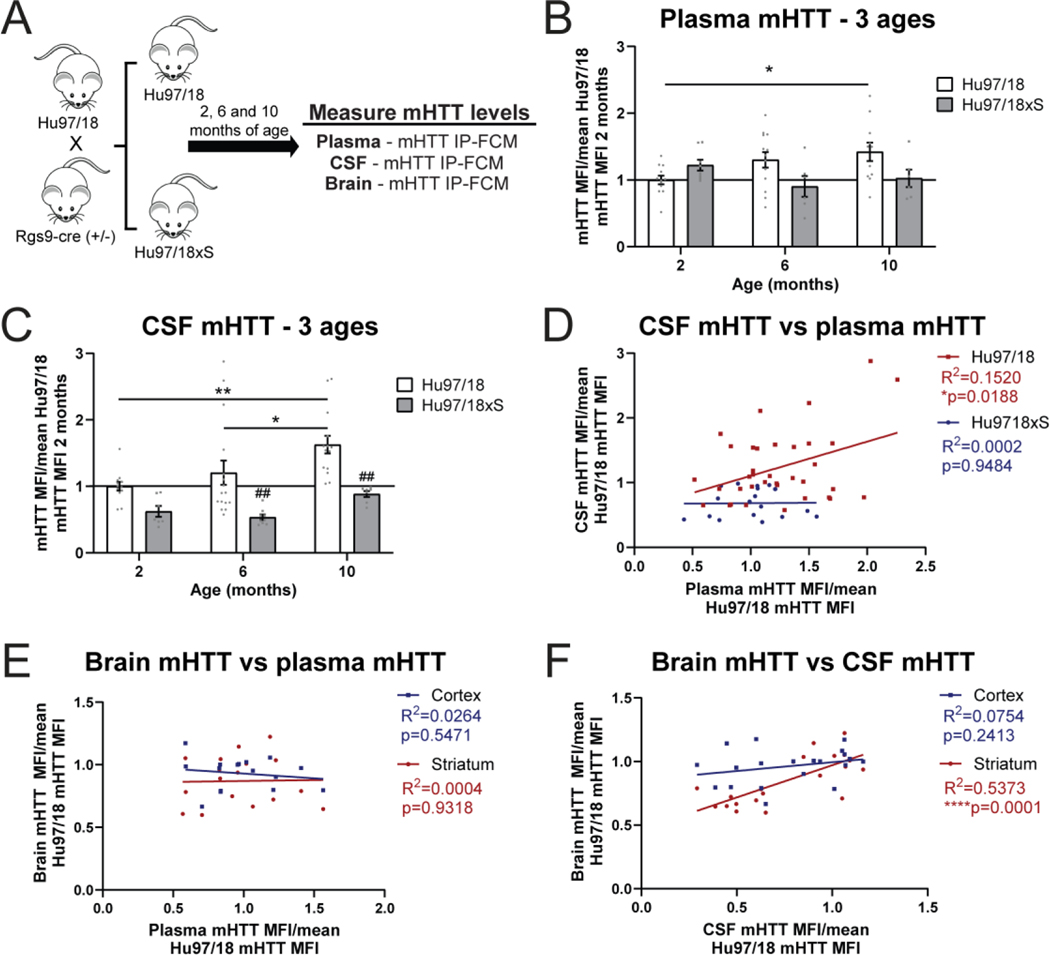
Selective genetic inactivation of mHTT in the striatum results in a correlative reduction of CSF mHTT but not plasma mHTT
We next measured mHTT levels in the CSF and plasma of Hu97/18 and Hu97/18xS mice at 2, 6 and 10 months of age by IP-FCM (Figure 7A). We observed an age-dependent increase in plasma mHTT protein in Hu97/18 mice that reached post hoc significance between 2 and 10 months of age (Figure 7B. Two-way ANOVA: genotype p= 0.0613, age p= 0.5640, interaction p= 0.0152. Sidak’s multiple comparison test: Hu97/18 2 months vs 10 months *p= 0.0175; Test for linear trend Hu97/18 *p= 0.0112, Hu97/18xS p=0.2068). Additionally, while there was a trend toward a genotype effect, this did not reach post hoc significance at any one age (Sidak’s multiple comparison test: 2 months p= 0.4430, 6 months p= 0.0800, 10 months p= 0.0945). Taken together these data, particularly the observed significant interaction between genotype and age on plasma mHTT levels, suggest that there is likely a striatal contribution to circulating mHTT protein in the blood, but it may not be proportionally large enough for the assessment of CNS mHTT changes primarily in the basal ganglia.
Figure 7. Selective genetic inactivation of mHTT in the striatum results in a correlative reduction of CSF mHTT but not plasma mHTT.
A) Graphical experimental overview where Hu97/18 and Hu97/18xS littermates were collected at 2, 6, and 10 months of age for quantification of mHTT in the CSF, plasma and brain by IP-FCM. B) Natural history of plasma mHTT with all values normalized to the mean mHTT MFI for 2 month old Hu97/18 mice (N=11–13 for Hu97/18, N=6–8 for Hu97/18xS per age. Two-way ANOVA: genotype p= 0.0613, age p= 0.5640, interaction p= 0.0152. Sidak’s multiple comparison test: Hu97/18 2 months vs 10 months *p= 0.0175; Hu97/18 vs Hu97/18xS at 2 months p= 0.4430, 6 months p= 0.0800, 10 months p= 0.0945; Test for linear trend: Hu97/18 *p= 0.0112, Hu97/18xS p=0.2068). C) Natural history of CSF mHTT with all values normalized to the mean mHTT MFI for 2 month old Hu97/18 mice (N=12–16 for Hu97/18, N=7–9 for Hu97/18xS per age. Two-way ANOVA: genotype p <0.0001, age p= 0.0078, interaction p=0.4469. Sidak’s multiple comparison test: Hu97/18 2 months vs 6 months: p= 0.4744, 2 months vs 10 months **p= 0.0026, 6 months vs 10 months *p= 0.0370; Hu97/18xS 2 months vs 6 months p= 0.9245, 2 months vs 10 months p= 0.5338, 6 months vs 10 months p= 0.2905; Hu97/18 vs Hu97/18xS at 2 months p= 0.2357, 6 months ##p= 0.0025, 10 months ##p= 0.0025; Test for linear trend: Hu97/18 **p=0.0057, Hu97/18xS **p=0.0049). D) Comparison of CSF and plasma mHTT levels shows a significant relationship in Hu97/18, but not Hu97/18xS mice (Linear regression: Hu97/18 R2= 0.1520, *p= 0.0188, Hu97/18xS R2= 0.0002, p= 0.9484). E) Comparison of plasma and brain mHTT IP-FCM MFI signal from Hu97/18 and Hu97/18xS mice at all ages does not show a relationship between plasma mHTT and STR or CTX mHTT (Linear regression: striatum R2= 0.0004, p= 0.9318; cortex R2= 0.0264, p= 0.5417). F) Comparison of CSF and brain mHTT mHTT IP-FCM MFI signal from Hu97/18 and Hu97/18xS mice at all ages demonstrates a significant correlation between CSF mHTT and striatal, but not cortical mHTT (Linear regression: STR R2= 0.5373, ****p= 0.0001; CTX R2= 0.07541, p= 0.2413). CTX = cortex, STR = striatum, CSF = cerebrospinal fluid, MFI = median fluorescence intensity.
We observed a significant age-dependent increase in CSF mHTT protein in Hu97/18 mice (Figure 7C. Two-way ANOVA: genotype p <0.0001, age p= 0.0078, interaction p= 0.4469. Sidak’s multiple comparison test: Hu97/18 2 months vs 6 months: p= 0.4744, 2 months vs 10 months **p= 0.0026, 6 months vs 10 months *p= 0.0370; Test for linear trend: Hu97/18 **p=0.0057), which is consistent with the previously reported disease stage/burden-dependent increase in CSF mHTT observed in HD gene positive individuals (Byrne et al., 2018; Rodrigues et al., 2020; Southwell et al., 2015; Wild et al., 2015). We did not observe age-dependent increases in CSF mHTT in Hu97/18xS mice (Sidak’s multiple comparison test: Hu97/18xS 2 months vs 6 months p= 0.9245, 2 months vs 10 months p= 0.5338, 6 months vs 10 months p= 0.2905). However, a linear trend across all 3 ages similar to that seen for Hu97/18 mice was observed (Test for linear trend: Hu97/18xS **p=0.0049), suggesting that the overall mHTT reduction by striatal inactivation may confound age-dependent assessments of CSF mHTT in the Hu97/18xS genotype. Supporting this is the finding that while CSF and plasma mHTT protein levels are correlated in Hu97/18 mice (Figure 7D. Linear regression: R2= 0.1520, *p= 0.0188), this relationship is not apparent in Hu97/18xS mice (Linear regression: R2= 0.0002, p= 0.9484).
We measured a significant reduction of mHTT in the CSF of Hu97/18xS compared to Hu97/18 mice at 6 and 10 months of age, however this failed to reach post hoc significance at 2 months of age (Figure 7C, Sidak’s multiple comparison test: 2 months p= 0.2357, 6 months ##p= 0.0025, 10 months ##p= 0.0025), a time point known to precede forebrain atrophy in the Hu97/18 line (Southwell et al., 2017). This suggests an interaction between neurodegeneration and the striatal contribution to CSF mHTT. Comparison of brain and plasma mHTT IP-FCM signal from Hu97/18 and Hu97/18xS mice at all ages did not reveal a significant relationship between plasma mHTT and either striatal or cortical mHTT (Figure 7E. Linear regression: striatum R2= 0.0004, p= 0.9318; cortex R2= 0.0264, p= 0.5417). Conversely, a comparison of brain and the CSF mHTT IP-FCM signal from Hu97/18 and Hu97/18xS mice at all ages showed a significant correlation between CSF mHTT and striatal, but not cortical mHTT (Figure 7F. Linear regression: striatum R2= 0.5373, ****p= 0.0001; cortex R2= 0.07541, p= 0.2413) demonstrating a clear relationship between the amount of mHTT protein in the striatum and in the CSF. These data further validate that a sufficiently measurable portion of mHTT in the CSF originates from the striatum and that CSF mHTT changes may reflect striatal mHTT changes.
Discussion
This study provides the first evidence of the relative contribution of mHTT originating from a specific brain region to the pool of CSF mHTT. We have previously demonstrated that the CNS is the major source of CSF mHTT (Southwell et al., 2015) and that active mechanisms, including cellular secretion and glymphatic clearance, as well as neurodegeneration contribute to mHTT in the CSF (Caron et al., 2020a). HD is characterized by progressive neurodegeneration in the cortex and basal ganglia, which is most severe in the striatum, making this brain region an important target for HTT lowering therapies. We therefore sought to investigate whether mHTT originating in cells of the striatum contribute to the pool of mHTT in CSF to 1) enhance our understanding of the CNS sources of CSF mHTT, 2) determine if CSF mHTT represents a useful PD biomarker of target engagement in the striatum and 3) to improve our interpretation of target engagement data for HTT lowering clinical trials in HD that specifically target the striatum.
Lentiviruses infect both dividing and non-dividing cells and allow for the transduction of discreet brain regions (Naldini et al., 1996). Using an optimized IS non-CED method, we were able to predominantly restrict transduction of the mHTTex1-GFP lentivirus to the striatum. Despite injecting half the viral titer compared to CED, we were able to detect the mHTTex1-GFP fusion in the CSF 4 weeks post-injection, a time point we have previously shown to have peak transgene levels detectable in CSF and ongoing neurodegeneration (Caron et al., 2020a). This demonstrates that during acute neuron death, mHTT that is produced in striatal cells is released to the CSF. While this is compelling data, this model system undergoes more robust and rapid neuron death than is observed in HD. Additionally, this model system relies upon exogenous delivery of a small, amino-terminal fragment of mHTT, so we next moved into more physiologically relevant model systems expressing a full-length, human mHTT transgene.
To accomplish this, we utilized complementary pharmacological and genetic approaches to selectively deplete mHTT levels in the striatum of HD mice. We optimized IS delivery of a HTT ASO to reproducibly restrict mHTT lowering to the striatum. We showed that unilateral or bilateral lowering of mHTT in the striatum resulted in an incremental reduction of CSF mHTT, which suggested a proportional contribution of both striatal hemispheres to CSF mHTT. In animals that received bilateral IS HTT ASO, we observed that 55–60% reduction of mHTT in both striatal hemispheres resulted in a measurable and significant reduction of mHTT in the CSF at 3 months of age. Since this age precedes any reported striatal atrophy in the BACHD model (Gray et al., 2008), the observed reduction of CSF mHTT following HTT ASO-mediated mHTT lowering in the striatum likely represents mHTT target engagement. As such, the contribution of mHTT from the striatum to CSF mHTT at this age may predominantly reflect an intracellular source driven by active clearance mechanisms, including cellular secretion and glymphatic clearance (Caron et al., 2020a), rather than an extracellular source resulting from neurodegenerative processes.
We next performed a time course to evaluate the kinetics of CSF mHTT levels in response to mHTT lowering in the striatum up to 12 weeks after treatment. We showed a significant effect of HTT ASO treatment on CSF mHTT although it did not reach post hoc significance at any one timepoint. We did however observe a modest but significant linear correlation between mHTT levels in the striatum and the CSF at all timepoints with the largest reduction of mHTT in both compartments being measured at 2 weeks post-injection. We also observed a trend towards an increase of CSF mHTT in both PBS and HTT ASO treated animals up to 12 weeks post-injection but this did not reach significance. Longitudinal CSF mHTT levels in HD mutation carriers increase over time with disease progression (Rodrigues et al., 2020). However, the time course experiment was performed over a relatively short period of time (12 weeks) and was done in animals treated at 4 months, prior to the onset of overt neuropathological abnormalities in the model (Gray et al., 2008). Therefore, it is perhaps not surprising that we did not detect a significant increase in CSF mHTT during this relatively short window.
Lowering mHTT in the striatum at multiple ages resulted in a consistent magnitude and regional-restriction of mHTT lowering in the striatum at all ages. This treatment resulted in a significant reduction of mHTT in the CSF when assessed at 3 and 7 months of age and a trend towards a reduction was observed at 10 and 13 months of age, but this did not reach post hoc significance. Despite the similar magnitude of mHTT lowering in the striatum at all ages, there was a significant linear trend towards an increase in CSF mHTT levels in HTT ASO treated animals from 3 and 13 months of age. At 12 month of age, BACHD mice show robust regional atrophy and degeneration of neurons in the striatum (Gray et al., 2008; Pouladi et al., 2012). One possible explanation for the blunted reduction of CSF mHTT effect after HTT ASO treatment at 12 months of age, is that lowering of mHTT in the striatum initiated at this timepoint may improve neuronal health and/or help preserve neurons which could result in less mHTT released from degenerating cells. It is also possible that ASO-mediated striatal mHTT lowering initiated at a more advanced stage when neurodegeneration is extensive may be less efficacious. A potential limitation of this experiment is that CSF from each age group was analyzed independently, preventing us from comparing cross-sectional mHTT CSF levels over the natural history in these animals. Despite this caveat, we can conclude that the mHTT originating from the striatum can be detected in the CSF.
As a secondary approach, we leveraged loxP sites flanking exon1 of the expanded mutant HTT transgene in Hu97/18 mice and crossed these animals to Rgs9-Cre mice to selectively inactivate mHTT expression in the striatum. The Rgs9-cre line has previously been crossed to the BACHD line to selectively deplete mHTT in the striatum (Wang et al., 2014). We validated these findings and further characterized the selectivity of the inactivation of mHTT in the striatum. At all ages, the magnitude of striatal mHTT reduction was similar whereas mHTT levels in the cortex and hippocampus were unchanged. While it did not reach significance at any age, a trend toward reduced cerebellar mHTT was observed, most strongly at 2 months of age. However, this effect was also observed for the wtHTT allele, which lacks the loxP sites, suggesting that it is not a result of transgene recombination in the cerebellum, but may rather be related to changes in gene expression that result from having a healthier striatum. Regardless of this potential cerebellar effect, the selective inactivation of the mHTT transgene in the striatum of mice expressing the Cre recombinase driven by the Rgs9 promoter was validated.
In the CSF, we observed a significant reduction of mHTT in the Hu97/18xS mice at 6 and 10 months of age, however this failed to reach post hoc significance at 2 months of age, a time point known to precede forebrain atrophy in the Hu97/18 line (Southwell et al., 2017). It is interesting to note that we observed a progressive genotypic effect that got stronger with age whereas as we saw the opposite trend in experiments with the IS HTT ASO. In contrast to the transient reduction of mHTT in the IS HTT ASO experiments, the Hu97/18xS cross results in a constitutive inactivation of mHTT in the striatum. Therefore, we may expect reduced striatal atrophy in Hu97/18xS animals at 10 months of age. This is supported by data from the original BACHD/Rgs9-Cre cross study that showed a 36% increase (not statistically significant) in striatal volume compared to BACHD mice at 12 months of age when mHTT was constitutively inactivated selectively in the striatum (Wang et al., 2014). Therefore, since neurodegeneration increases mHTT in CSF, it is perhaps not surprising that less mHTT would be detected in the CSF at 10 months in the cross. This data suggests an interaction between neurodegeneration and the striatal contribution to CSF mHTT.
We also observed a significant age-dependent increase in CSF mHTT in Hu97/18 mice. This is the first report of natural history of mHTT in CSF of mice, and the recapitulation of what has been seen in humans provides validation for the use of mouse models for the study of mHTT and other CSF biomarkers. In Hu97/18xS mice, the age-dependent increase in CSF mHTT was observed, but appeared delayed with the trend only apparent at 10 months of age and absent at 6 months of age. This may reflect the timing of HD-like neurodegeneration, which occurs earliest in the striatum. Thus, in animals with greatly reduced striatal mHTT, the disease-dependent increase in CSF mHTT may be delayed. Conversely, it may just be more difficult to distinguish differences in protein level in these animals with lower amounts of CSF mHTT. However, the observed correlation between plasma and CSF mHTT in Hu97/18 mice that is lost in Hu97/18xS mice where striatal neurodegeneration is expected to be greatly reduced indicates a neurodegenerative mechanism for these changes. This is further supported by the observed correlation between CSF mHTT and striatal mHTT. The R2 value of 0.5373 suggests that as much as half of the variation in CSF mHTT is driven by the level of striatal mHTT. Considering that the striatum accounts for approximately 6–7% of the brain mass of mice, this suggests a potential regional bias toward striatal contribution, which may be driven by neurodegenerative processes.
Despite CSF mHTT being a promising biomarker of CNS target engagement, there is to date no evidence to suggest that lowering of mHTT in the CSF is associated with clinical efficacy outcomes in HD. Data from the recently halted phase III clinical trial evaluating the efficacy of tominersen (NCT03761849) suggests that a reduction of CSF mHTT alone does not predict clinical benefit. One possible explanation is that there was inadequate target engagement in the striatum and that the reduction of CSF mHTT observed in the trial may have been due to mHTT lowering in other brain regions, though it is also possible that a toxic effect of the drug prevented clinical benefit.
Here, we have conclusively shown that reduction of mHTT in the striatum is associated with decreases in mHTT in the CSF. However, a decrease in mHTT in the CSF may occur even if the striatum is not specifically targeted with mHTT lowering approaches. In other words, a decrease in mHTT in the CSF cannot on its own be taken as evidence that mHTT is being lowered in the striatum. However, if the striatum is targeted specifically, our data support that a decrease in mHTT in the CSF is a proxy measure for striatal mHTT lowering.
Conclusions
While further studies to investigate the contribution of other brain regions will be required to determine the magnitude of the striatal component of CSF mHTT, our findings do demonstrate a substantial striatal contribution to CSF mHTT. Though the striatum makes up a smaller percentage of total brain mass in humans than in mice, there is also more striatal neurodegeneration in the human HD brain than is observed in FL human mHTT transgenic HD models, which more closely recapitulate early disease phenotypes. Thus, the striatum is still expected to be a significant contributor to CSF mHTT, at least in prodromal and early manifest individuals which are expected to have ongoing striatal neurodegeneration. Taken together, our data demonstrates that a measurable portion of mHTT in the CSF originates from cells of the striatum. These findings support the use of CSF mHTT as a PD biomarker for assessing target engagement of targeted therapeutics designed to lower mHTT in the striatum.
Supplementary Material
A) Relative quantification of mouse Htt levels in the brain following ICV injections with either PBS or 150μg HTT ASO at 2 months of age does not lower Htt in any brain region at 4 weeks post-treatment (N=4 per treatment condition. Two-way ANOVA: treatment p=0.2061, brain region p=0.9749, interaction p=0.9749). ICV = intracerebroventricular, CTX = cortex, STR = striatum, HIP = hippocampus, CBL = cerebellum, IOD = integrated optical density.
A) Relative quantification of mHTT levels in the STR following IS injection with either PBS or 150μg HTT ASO at 2, 6, 9 and 12 months of leads to a similar magnitude of mHTT lowering 4 weeks post-treatment, at 3, 7, 10 and 13 months of age (N=6–14 for 3 months, N=8 for 7 months, N= 5–6 for 10 months, N=9–12 for 13 months per treatment condition. Two-way ANOVA: age p=0.3183, treatment p<0.0001, interaction p=0.3183). B) Relative quantification of mHTT in the CSF normalized to the mean of PBS mHTT MFI at each timepoint shows a strong trend towards an increase in CSF mHTT levels in HTT ASO treated animals between 3 and 13 months of age (N=14–16 for 3 months, N=9 for 7 months, N= 5 for 10 months, N=9–11 for 13 months per treatment condition. Two-way ANOVA: treatment p<0.0001, age p=0.3541, interaction p=0.3541. Sidak’s multiple comparison test: HTT ASO 3 vs 7 months p=0.3580, 3 vs 10 months p=0.4139, 3 vs 13 months p=0.0750). IS = intrastriatal, STR = striatum, CSF = cerebrospinal fluid, IOD = integrated optical density, MFI = median fluorescence intensity.
Highlights.
Striatal viral expression of mutant huntingtin (mHTT) is detectable in the CSF
Targeted striatal lowering of mHTT leads to a reduction of CSF mHTT in HD mice
Inactivation of mHTT in the striatum results in a reduction of CSF mHTT in HD mice
CSF mHTT is a pharmacodynamic biomarker for therapies lowering mHTT in the striatum
Acknowledgments
The authors would like to thank Yuqing Li and X. William Yang for providing Rgs9-Cre mice, Ali Khoshnan for the lentiviral genome, Paul H. Patterson and the Developmental Studies Hybridoma Bank at the University of Iowa for the MW1 antibody, and Mark Wang, Qingwen Xia, Morgan Rook, Renee Ouellette, and Mazzen Eldeeb for technical assistance and support.
Funding
Support for N.S.C. provided by a Canadian Institutes of Health Research (CIHR) Postdoctoral Fellowship and the Huntington’s Disease Society of America (HDSA) Berman/Topper HD Career Development Fellowship. Project operational support for M.R.H. provided by a CIHR Foundation grant (FDN 154278). M.R.H. is a Canada Research Chair in Human Genetics and Molecular Medicine. Project operational support for A.L.S. provided by the HDSA and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (R01NS116099). The funding agencies listed were not involved in the design of the study; collection, analysis, and interpretation of the data; or writing of the manuscript.
Abbreviations
- AAV
adeno-associated virus
- ASO
antisense oligonucleotide
- CED
convection-enhanced delivery
- CSF
cerebrospinal fluid
- CNS
central nervous system
- FiSH
fluorescence in situ hybridization
- GFP
green fluorescent protein
- HTT
huntingtin
- HD
Huntington disease
- ICV
intracerebroventricular
- IHC
immunohistochemistry
- IP-FCM
immunoprecipitation and flow cytometry
- IS
intrastriatal
- LNA
locked nucleid acid
- MFI
median fluorescence intensity
- mHTT
mutant huntingtin
- MSN
medium spiny neuron
- PBS
phosphate-buffered saline
- PD
pharmacodynamic
- PFA
paraformaldehyde
- polyQ
polyglutamine
- Rgs9
Regulator of G protein signaling 9
- RNAi
RNA interference
- WT
wild-type
Footnotes
Competing interests
MRH currently serves on the public boards of Ionis Pharmaceuticals, Xenon Pharmaceuticals, Aurinia Pharmaceuticals and 89bio. The other authors declare no competing interests.
Ethics approval and consent to participate
No human participants, tissues or data were used in this study. Experiments using animals were performed with the approval of the Animal Care Committee at the University of British Columbia (A16–0130, A20–0107) and the Institute Animal Care and Use Committee at the University of Central Florida (17–32, 2019–07, 2020–50).
Availability of data and material
The datasets supporting the conclusions of this article are included within the article and its additional files.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 72, 971–83. [DOI] [PubMed] [Google Scholar]
- Byrne LM, et al. , 2018. Evaluation of mutant huntingtin and neurofilament proteins as potential markers in Huntington’s disease. Sci Transl Med. 10. [DOI] [PubMed] [Google Scholar]
- Caron NS, et al. , 2021. Reliable Resolution of Full-Length Huntingtin Alleles by Quantitative Immunoblotting. J Huntingtons Dis. [DOI] [PubMed] [Google Scholar]
- Caron NS, et al. , 2020a. Mutant huntingtin is cleared from the brain via active mechanisms in Huntington disease. J Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron NS, et al. , 2020b. Potent and sustained huntingtin lowering via AAV5 encoding miRNA preserves striatal volume and cognitive function in a humanized mouse model of Huntington disease. Nucleic Acids Res. 48, 36–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang MT, et al. , 2006. Disrupted motor learning and long-term synaptic plasticity in mice lacking NMDAR1 in the striatum. Proc Natl Acad Sci U S A. 103, 15254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher ER, Hayden MR, 2014. Multisource ascertainment of Huntington disease in Canada: prevalence and population at risk. Mov Disord. 29, 105–14. [DOI] [PubMed] [Google Scholar]
- Gray M, et al. , 2008. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 28, 6182–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, et al. , 2005. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A. 102, 5820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordasiewicz HB, et al. , 2012. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 74, 1031–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L, et al. , 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A. 93, 11382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouladi MA, et al. , 2012. Marked differences in neurochemistry and aggregates despite similar behavioural and neuropathological features of Huntington disease in the full-length BACHD and YAC128 mice. Hum Mol Genet. 21, 2219–32. [DOI] [PubMed] [Google Scholar]
- Rodrigues FB, et al. , 2020. Mutant huntingtin and neurofilament light have distinct longitudinal dynamics in Huntington’s disease. Sci Transl Med. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell AL, et al. , 2008. Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J Neurosci. 28, 9013–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell AL, et al. , 2009. Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington’s disease. J Neurosci. 29, 13589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell AL, et al. , 2018. Huntingtin suppression restores cognitive function in a mouse model of Huntington’s disease. Sci Transl Med. 10. [DOI] [PubMed] [Google Scholar]
- Southwell AL, et al. , 2014. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol Ther. 22, 2093–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell AL, et al. , 2017. A novel humanized mouse model of Huntington disease for preclinical development of therapeutics targeting mutant huntingtin alleles. Hum Mol Genet. 26, 1115–1132. [DOI] [PubMed] [Google Scholar]
- Southwell AL, et al. , 2015. Ultrasensitive measurement of huntingtin protein in cerebrospinal fluid demonstrates increase with Huntington disease stage and decrease following brain huntingtin suppression. Sci Rep. 5, 12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell AL, et al. , 2013. A fully humanized transgenic mouse model of Huntington disease. Hum Mol Genet. 22, 18–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi SJ, et al. , 2019. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N Engl J Med. 380, 2307–2316. [DOI] [PubMed] [Google Scholar]
- Valles A, et al. , 2021. Widespread and sustained target engagement in Huntington’s disease minipigs upon intrastriatal microRNA-based gene therapy. Sci Transl Med. 13. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, et al. , 1985. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 44, 559–77. [DOI] [PubMed] [Google Scholar]
- Wang N, et al. , 2014. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington’s disease. Nat Med. 20, 536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild EJ, et al. , 2015. Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington’s disease patients. J Clin Invest. 125, 1979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, et al. , 2010. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 90, 905–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Relative quantification of mouse Htt levels in the brain following ICV injections with either PBS or 150μg HTT ASO at 2 months of age does not lower Htt in any brain region at 4 weeks post-treatment (N=4 per treatment condition. Two-way ANOVA: treatment p=0.2061, brain region p=0.9749, interaction p=0.9749). ICV = intracerebroventricular, CTX = cortex, STR = striatum, HIP = hippocampus, CBL = cerebellum, IOD = integrated optical density.
A) Relative quantification of mHTT levels in the STR following IS injection with either PBS or 150μg HTT ASO at 2, 6, 9 and 12 months of leads to a similar magnitude of mHTT lowering 4 weeks post-treatment, at 3, 7, 10 and 13 months of age (N=6–14 for 3 months, N=8 for 7 months, N= 5–6 for 10 months, N=9–12 for 13 months per treatment condition. Two-way ANOVA: age p=0.3183, treatment p<0.0001, interaction p=0.3183). B) Relative quantification of mHTT in the CSF normalized to the mean of PBS mHTT MFI at each timepoint shows a strong trend towards an increase in CSF mHTT levels in HTT ASO treated animals between 3 and 13 months of age (N=14–16 for 3 months, N=9 for 7 months, N= 5 for 10 months, N=9–11 for 13 months per treatment condition. Two-way ANOVA: treatment p<0.0001, age p=0.3541, interaction p=0.3541. Sidak’s multiple comparison test: HTT ASO 3 vs 7 months p=0.3580, 3 vs 10 months p=0.4139, 3 vs 13 months p=0.0750). IS = intrastriatal, STR = striatum, CSF = cerebrospinal fluid, IOD = integrated optical density, MFI = median fluorescence intensity.