Abstract
The palatine tonsils (hereinafter referred to as “tonsils”) serve as a reservoir for viral infections and play roles in the immune system's first line of defense. The aims of this study were to establish tonsil epithelial cell–derived organoids and examine their feasibility as an ex vivo model for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. The tonsil organoids successfully recapitulated the key characteristics of the tonsil epithelium, including cellular composition, histologic properties, and biomarker distribution. Notably, the basal layer cells of the organoids express molecules essential for SARS-CoV-2 entry, such as angiotensin-converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2) and furin, being susceptible to the viral infection. Changes in the gene expression profile in tonsil organoids revealed that 395 genes associated with oncostatin M signaling and lipid metabolism were highly upregulated within 72 h after SARS-CoV-2 infection. Notably, remdesivir suppressed the viral RNA copy number in organoid culture supernatants and intracellular viral protein levels in a dose-dependent manner. Here, we suggest that tonsil epithelial organoids could provide a preclinical and translational research platform for investigating SARS-CoV-2 infectivity and transmissibility or for evaluating antiviral candidates.
Keywords: Tonsil epithelial organoid, Tonsil tissue, SARS-CoV-2, Antiviral, Transcriptome
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, was first identified on December 2019 in Wuhan, China [1,2]. The emerging virus spread rapidly around the world, and the World Health Organization (WHO) declared the outbreak as a pandemic in March 2020. SARS-CoV-2 belongs to genus Betacoronavirus, which also includes the highly pathogenic Middle East respiratory syndrome (MERS)- and SARS-CoVs [3]. Infection with SARS-CoV-2, which is mainly transmitted through the upper respiratory tract, causes diverse disease symptoms ranging from mild sickness (including fever, cough, sore throat, and loss of olfaction or taste) to severe illness (including pneumonia and even death) [1,4,5]. Currently, mRNA- and adenoviral vector–based vaccines have been approved for prevention of SARS-CoV-2 infection, and antivirals such as remdesivir (polymerase inhibitor) and PF-07321332 (protease inhibitor) have been developed for treatment of infected patients [[6], [7], [8], [9], [10]]. However, viral variants with higher transmissibility or pathogenicity have been isolated with increasing frequency worldwide. Regarding the emerging and re-emerging rate of mutant CoVs, a suitable ex vivo SARS-CoV-2 infection model system is indispensable for development of broad-spectrum antivirals as well as universal vaccines or for understanding SARS-CoV-2 pathogenesis in a personalized manner [[11], [12], [13], [14]].
Several reports have described the recapitulation of human organoids for SARS-CoV-2 infection and demonstrated their feasibility for analyses of antiviral activity or tissue susceptibility. For instance, human pluripotent stem cell (hPSC)-derived organoids have proven valuable for investigation of SARS-CoV-2 tropism [15,16]. In addition, SARS-CoV-2 infection has been evaluated in human intestinal, liver ductal, lung, and brain organoids [[17], [18], [19], [20], [21]]. However, most of these approaches use synchronized stem cells or human tissues obtained through conventional open surgeries, imposing limitations on clinical sample availability and research in personalized medicine. To address these issues, we attempted to establish an alternative organoid system for which i) human tissues could be reliably accessible with reduced physical burdens, ii) organoids can be maintained for a considerable period of time, and iii) SARS-CoV-2 can robustly replicate through multiple rounds of infection. Among easily accessible tissues, we selected the tonsils for establishment of human organoids and infection with SARS-CoV-2, as they have the potential to satisfy the unmet needs described above. From a clinical point of view, tonsils are also appropriate source material due to their susceptibility to diverse respiratory viruses including influenza virus, respiratory syncytial virus, and even coronavirus [[22], [23], [24]].
The tonsils are two oval-shaped immunologic organs in the back of the oral cavity [24] (Fig. S1A). Histologically, they are composed of mucosa-lining epithelium and underlying parenchymal B or T cell lymphocytes [24]. Multilayered, stratified tonsil epithelium are covered with differentiated epithelial cells at the superficial layer, which are generated from renewable stem-like epithelial cells in the bottom layer [25]. Unfortunately, a tonsil organoid model has not yet been established due to the difficulty of culturing tonsillar epithelial cells that can be stably maintained over long-term serial passage [25]. In addition, small laboratory animals, such as mouse and hamster, are not available for optimization of organoid construction because they lack orthologous organs [26]. However, establishment of an organoid model from tonsil tissues was expected to be achievable by applying advanced technologies for the long-term 3D culture of epithelial stem cells from human-derived specimens [27,28]. This approach might be valuable in efforts to understand cellular responses to viral infection and antiviral activity, as organoids are composed of diverse differentiated or proliferating cell types and can reconstitute the physiological conditions of human organs [[29], [30], [31], [32], [33], [34], [35]]. In this study, we sought to establish tonsil epithelial cell–derived organoids with morphological and molecular biological features comparable to those of epithelium of human tonsil tissues, and to examine their feasibility as an ex vivo model for SARS-CoV-2 infection.
2. Results
2.1. Establishment of human tonsil epithelial organoids
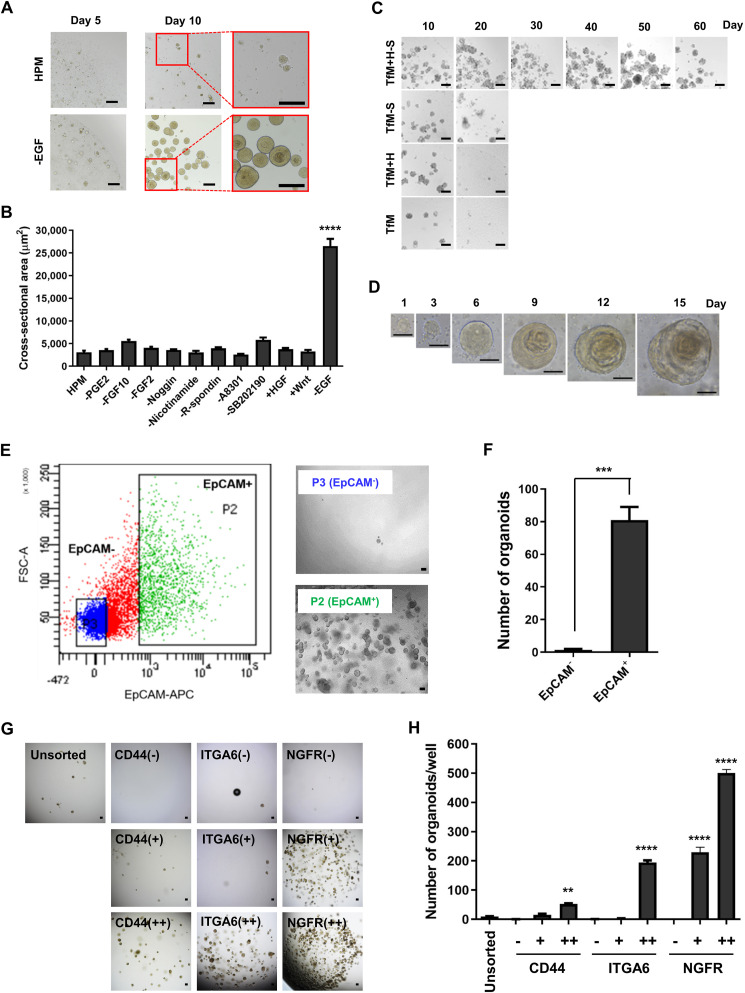
To generate organoids from human tonsil epithelium, enzymatically dissociated cells from tonsil tissues were embedded in Matrigel. According to previous reports, they were cultured either in human colon organoid medium (HCM) or human prostate organoid medium (HPM) supplemented with growth factors including Noggin, fibroblast growth factor 2 (FGF2), FGF10, and R-spondin-1 (RSPO1), which have been identified to play pivotal roles in epithelial stem cell differentiation and proliferation (Fig. S1A and Table S1) [36,37]. HPM yielded 2-fold more 3D organoids within 10 days than HCM (Fig. S1B, C). To achieve more efficient organoid formation, the composition of the medium was optimized by seeding dissociated tonsil epithelial cells in various conditioned HPM that contained or omitted individual components associated with cell proliferation. The largest organoids appeared with the highest organoid-forming efficiency when cultured in HPM in the absence of epidermal growth factor (EGF), which was subsequently defined as tonsil organoid formation medium (TfM) (Fig. 1 A and B, and Fig. S1D). Microscopic and histological analyses revealed that organoids cultured in TfM developed stratified squamous epithelial structures resembling those of tonsil tissues between days 10 and 15 (Fig. S2A and B). However, TfM limited subculture to two passages, and the cells were not available for additional expansion (Fig. S2C). To resolve this issue, we further modified TfM by removing SB202190, a selective p38 MAPK inhibitor, but by adding hepatocyte growth factor (HGF). These changes did not affect organoid size at passage 1, but well-shaped organoids were successfully expanded beyond five passages for 60 days. This optimized medium was defined as tonsil organoid expansion medium (TeM) (Fig. 1C and D; Fig. S2D and E). Moreover, it did not affect reproducibility in the organoid formation after thawing of their cryopreservation (Fig. S3).
Fig. 1.
Generation and characterization of human tonsil epithelial organoids. Media compositions were optimized for long-term expansion of the tonsil organoids, and biomarkers essential for organoid formation were detected by cell-sorting analysis. (A) Comparison of formation of tonsil organoids cultured in human prostate media (HPM) with or without EGF (-EGF). Bright-field microscopic images were obtained on days 5 and 10. Areas in red rectangles are enlarged on right. (B) Size of organoids cultured in HPM containing various combinations of media components. Values are expressed as means ± SEM of three independent experiments. ****, p < 0.0001 (Two-way ANOVA with Dunnett's multiple comparison test). (C) Microscopic images of human tonsil organoids cultured in TfM (tonsil organoid formation medium), TfM with HGF (TfM + H), TfM without SB202190 (TfM-S), or TfM with HGF but without SB202190 (TfM + H-S) for 60 days. (D) Time course of tonsil organoids growth in TeM (tonsil organoid expansion medium, TfM + H-S) for 15 days. (E) Flow cytometry analysis showing EpCAM+ (P2 fraction) or EpCAM− cell (P3 fraction) populations in tonsil tissues. Each fraction was cultured in TeM, and their images were observed on day 15. (F) Organoids cultured with P3 (EpCAM−) and P2 (EpCAM+) cells as shown in (E) were counted. Data from three independent experiments were analyzed by unpaired t-test and ordinary one-way ANOVA. ***, p < 0.001. (G) Comparison of organoid formation efficiency between biomarker-high (++), -low(+), -negative(−) expressing cells on the basis of CD44, ITGA6 and NGFR levels. (H) Number of formed organoids, presented as means ± SEM of the three different donors. Data were analyzed by ordinary one-way ANOVA followed by Dunnett's multiple comparison test. n.s., not significant. **, p < 0.01; ****, p < 0.0001. In the panels (A), (C), (D), (E), and (G), scale bars represent 100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
It has been reported that epithelial cell adhesion molecule (EpCAM), a representative marker for epithelial cells, is expressed in the basal layer of both epithelium of several tissues, including tonsil, and diverse epithelial organoids (Fig. S4A) [[38], [39], [40], [41], [42]]. To investigate whether the tonsil organoids originated from EpCAM+ cells, we compared organoid formation ability of different cell populations from tonsil tissues on the basis of EpCAM expression. Flow cytometric analysis revealed that in contrast to EpCAM− cells, EpCAM+ cells facilitated robust organoid formation (Fig. 1E and F). This result suggested that tonsil organoids cultured in TeM are generated from EpCAM+ cells, verifying their epithelium-like property.
Among CD44, integrin alpha 6 (ITGA6) and nerve growth factor receptor (NGFR), that are abundantly expressed in stem cells and involved in other organoid formation, we analyzed their effects on tonsil epithelial organoid formation [[43], [44], [45], [46]]. Each subpopulation was isolated from E-cadherin+ cells by flow cytometric cell sorting and then cultured in TeM (Fig. S5). The number of resulting organoids showed that it was highest in NGFR++ cells, followed by NGFR+ cells, ITGA6++ cells, CD44++ cells in that order. On the other hand, few organoids were obtained in the absence of any of these factors or in the lower level of CD44 (CD44+) or ITGA (ITGA+) (Fig. 1G and H). Cell sorting analysis indicated that expression of the three molecules, CD44, ITGA6 and NGFR, are required for differentiation and maturation of tonsil epithelial cells into organoids.
2.2. Histological and immunohistological characterization of human tonsil epithelial organoids
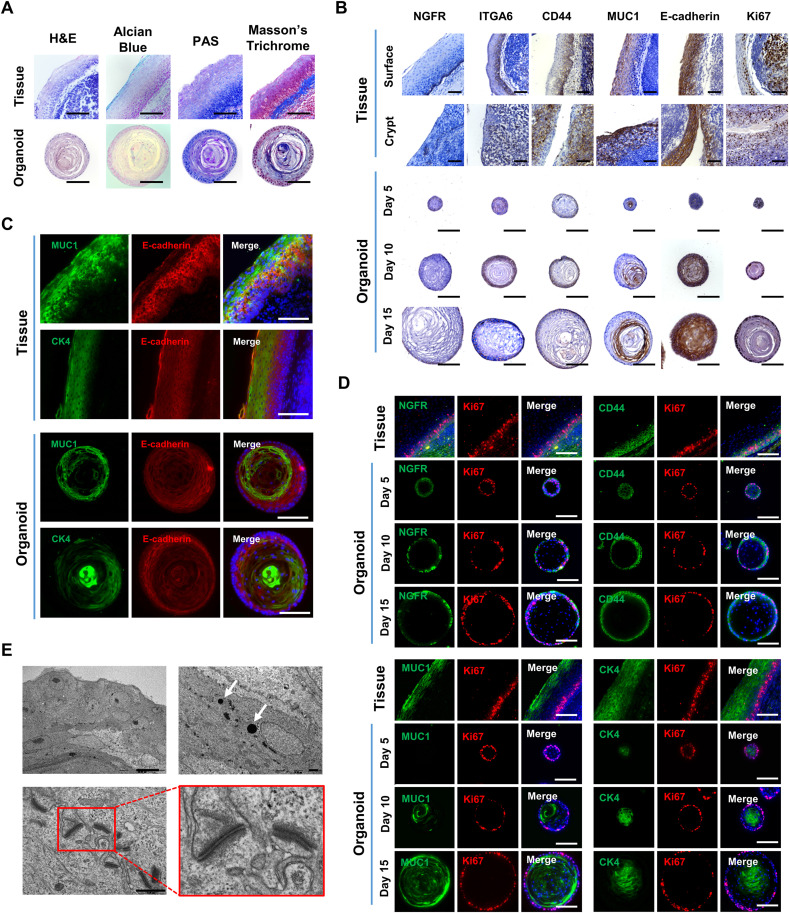
To determine whether the tonsil organoids could recapitulate the human original tonsil tissues as depicted in Supplementary Figure S4A, we explored their structural features and biomarker distribution. Staining of histological sections with hematoxylin and eosin (H&E), Alcian blue, periodic acid–Schiff (PAS), and Masson's trichrome revealed circular stratified squamous organoids with a biochemical composition similar to that of tonsil tissues (Fig. 2 A). In detail, we compared histochemical composition of multilayered cellular components in the tonsil organoids during their maturation with those of tissue epithelium either from tonsil surface or from crypt. The results revealed that basal layer cells of tonsil surface or crypt epithelium expressing NGFR, ITGA6, or CD44 are localized on the outer side of the organoids between days 10 and 15, whereas suprabasal or superficial layer cells expressing MUC1 were mainly detected in the interior of tonsil organoids (Fig. 2B). In addition, E-cadherin, a ubiquitous epithelial cell marker, was evenly distributed, whereas Ki67, a proliferation marker, was predominantly expressed at the spherical boundary within the organoids, indicating active cell proliferation in that zone. These histological data suggested that tonsil organoid maturation was successfully achieved from a single cell of tonsil tissue, yielding a highly ordered stratified epithelium within 15 days in TeM.
Fig. 2.
Histological, immunohistological and ultrastructural analyses of tonsil epithelial organoids. (A) Histology of human tonsil epithelium tissues and organoids on day 15. Each sample was stained with H&E for morphological analysis, Alcian blue to visualize the mucosal layer and mucous-producing goblet cells, PAS to visualize mucosubstances, and Masson's trichrome to visualize connective matrix. (B) Immunohistological analyses of surface and crypt areas of human tonsil epithelium tissues along with tonsil organoids on days 5, 10 and 15. Each sample was immuno-stained with antibodies specific for epithelium markers, including NGFR, ITGA6, CD44, MUC1, and E-cadherin, as well as a cell proliferation marker, Ki67. (C) Immunofluorescence to compare distribution of suprabasal/superficial layer markers, MUC1 and CK4 (green), between the tonsil tissues and the organoids on day 15; the epithelial marker E-cadherin (red), was used as a control. Merged images with Hoechst 33342 staining (blue) are shown in the right-hand panels. (D) Immunofluorescence images showing a time course of organoid development on days 5, 10, and 15. Representative markers (green) for basal layer cells (NGFR and CD44; upper panels) and suprabasal/superficial layer cells (MUC1 and CK4; lower panels) were labeled with the corresponding antibodies; Ki67 (red) was used as a control for active proliferation. Merged images with Hoechst 33342 staining (blue) are shown in the right-hand panels. (E) TEM images illustrating ultrastructural morphology of human tonsil epithelial organoids. Keratohyaline granules are indicated by white arrows (right upper panel). The image of desmosome–tonofilament complexes in the red rectangle in the left lower panel is enlarged on right. In all panels except for (E), scale bar is 100 μm. In (E), scale bars are 10 μm (upper left) and 1 μm (upper right and lower left). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The systematic organization of different cell types was further examined by immunofluorescence microscopy. For immunohistological experiments, organoids as well as tonsil tissues were co-stained with antibodies against specific markers representing individual tissue layers. First, we sought to determine the distribution of epithelial cells expressing MUC1, CK4, NGFR, CK5 and CD44 by using E-cadherin as a marker for the overall area of the epithelium. As expected, both MUC1- and CK4-expressing cells were detected on the inner sides of organoids, which correspond to the suprabasal/superfical layers of the tissues (Fig. 2C), whereas NGFR-, CK5-, and CD44-expressing cells were detected on the outer ring of organoids, which corresponds to the basal layer of the tissue (Fig. S4B). We further examined the time course of tonsil organoid maturation, where Ki67 was used as a cell proliferation marker. Consistent with the immunohistochemistry data before (Fig. 2C, Fig. S4B), basal layer markers of tonsil tissue, including NGFR and CD44, as well as the suprabasal/superficial layer markers, including MUC1 and CK4, accumulated at the outer side and the inner side, respectively, completing maturation between days 10 and 15 (Fig. 2D). Importantly, colocalization of NGFR or CD44 with Ki67 in the organoids revealed that the essential factors required for tonsil organoid formation (Fig. 1G and H) are continuously expressed at the proliferating cells throughout the differentiation and maturation period. In addition, a tonsil crypt-specific marker, cytokeratin 7 (CK7), was expressed at the inner layer cells of the mature organoids, whereas proliferating cell nuclear antigen (PCNA) was expressed in the outer border cells (Fig. S4C). Given this finding, it is assumed that the organoids might be differentiated along the crypt–surface axis by placing crypt-derived cells at the less proliferative inner space [47]. In line with the biomarker-based histological analyses, electron micrographs of the organoids at day 15 revealed typical ultrastructural features of stratified squamous epithelium, with characteristic elements such as keratohyalin granules and desmosome-tonofilament complexes [48] (Fig. 2E). Collectively, the histological, immunohistological and ultramicroscopic images strongly suggested that the outer and inner domains of tonsil organoids recapitulate the basal and suprabasal/superficial layers of the primary tonsil epithelium, respectively.
2.3. Gene expression profiles in human tonsil epithelial organoids and their response to a pathogen-derived immune stimulant
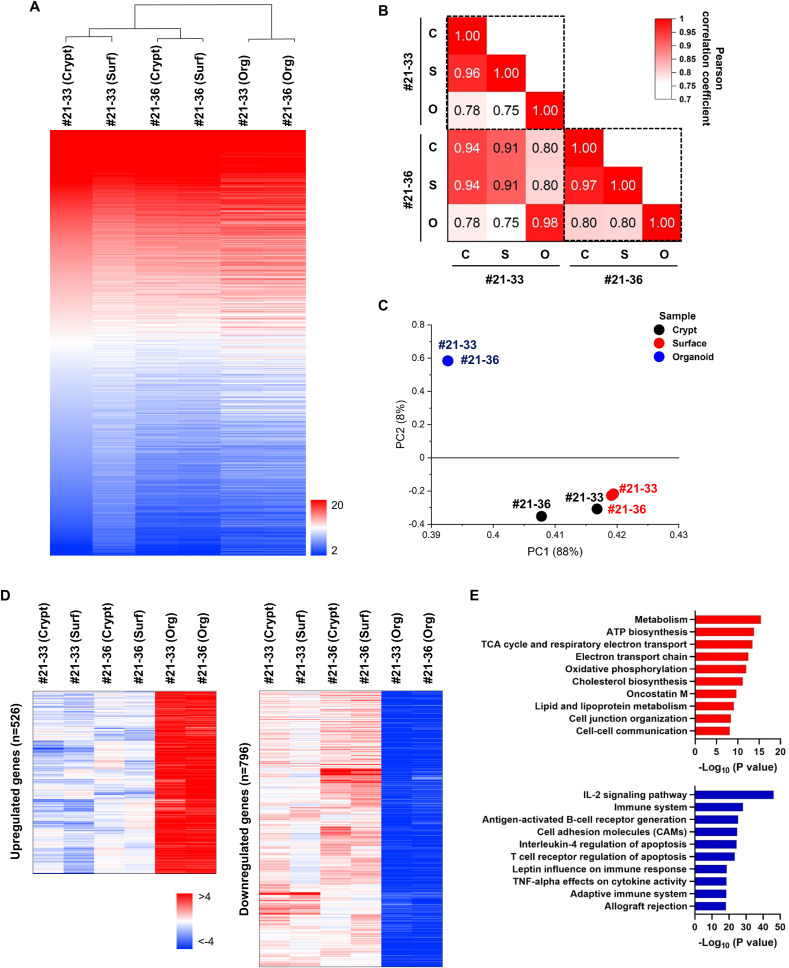
We investigated how similar the mRNA transcriptome of tonsil epithelial organoids was to that of tonsil tissues by comparing whole-genome expression levels of tonsil organoids from two donors (nos. 21–33 and 21–36) with their paired tissues, i.e., tonsil crypt and surface samples separately (Fig. S6). Heatmap analysis and hierarchical clustering of the high-throughput data from 21,448 genes showed high correlation coefficients between crypt and surface samples from a same donor (r = 0.96 for donor 21–33; r = 0.97 for donor 21–36) (Fig. 3 A and B). Even between the two donors, this gene expression profile was well correlated, showing r values above 0.91. It was interesting that the tonsil organoids showed donor-independent homogeneity (r = 0.98) but with relatively reduced correlation coefficient values, ranging from 0.75 to 0.80, to the tissue samples. The results suggested that gene expression profile became further synchronized when tonsil epithelial cells are matured into organoids.
Fig. 3.
Gene expression profile analysis in tonsil epithelial organoids. (A) Heatmap showing the gene expression pattern of the tonsil organoids (Org) cultured for 15 days and their matched tonsil crypt and surface (Surf) tissues from two donors 21–33 and 21–36. Gene expression levels (ranging from 2 to 20) were pseudo-temporally ordered and the samples were clustered based on the similarity of the gene expression pattern. (B) Pairwise Pearsons' correlation coefficients between samples (r). (C) Principal component analysis (PCA) plot of microarray samples. PCA scores on PC1 (88%) and PC2 (8%) are pointed with black, red and blue dots, accounting for crypt, surface and organoid samples, respectively, from the two donors. (D) Heatmaps showing differentially expressed genes (DEGs) in tonsil organoids compared to tonsil tissues (with > 4-fold changes). Upregulated genes are colored in red (left panel), while downregulated genes are in blue (right panel). (E) Gene ontology (GO) enrichment analysis of differentially expressed genes. The bar graphs indicate the top 10 upregulated (red) and downregulated (blue) GO biological processes in the tonsil epithelial organoids. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Although overall gene expression of tonsil organoids was overlapped with that of tissues, we wondered which genes are differently expressed in organoids. Principal component analysis (PCA) plot showed that mRNA expression of the majority of genes (PC1, 88%) was similar between organoids and tissues (Fig. 3C). However, in a second major population of genes (PC2, 8%), organoids had a distinctive cluster from crypt and surface tissue samples. Specifically, 526 genes were upregulated but 796 genes were downregulated in organoids when compared to tissues (Fig. 3D). Gene ontology and pathway analysis revealed that metabolic reprogramming occurred in the organoids by stimulating ATP synthesis, tricarboxylic acid cycle (TCA) and cholesterol synthesis and in parallel, cell-cell communication was increased possibly for 3D morphogenesis (Fig. 3E, upper) [49]. In contrast, immune system was suppressed by downregulating IL-2 signaling pathway, antigen-activated B-cell receptor generation and cell adhesion molecules (Fig. 3E, lower). This inhibition might be related to biased differentiation of tonsil organoids to epithelial cells rather than to immune cells in the optimized media, TeM.
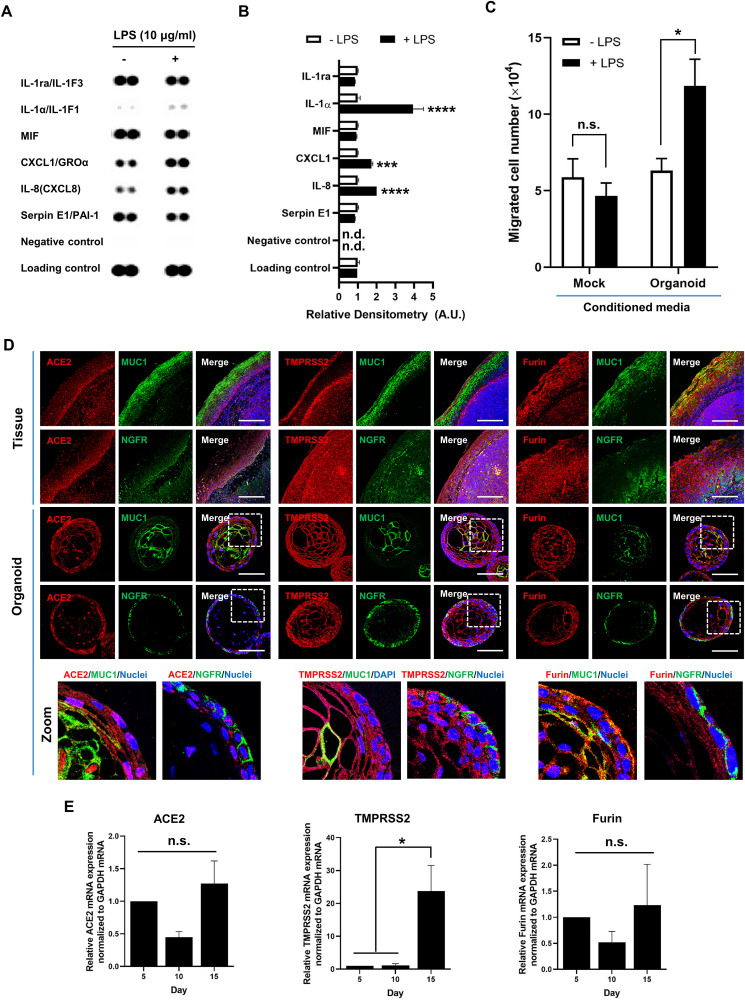
As a preliminary functional study, we examined how the organoids’ innate immunity responds to a pathogen-derived molecule. It has been elucidated that microbial challenge stimulates tonsil epithelium to secrete C-X-C motif chemokines, including CXC ligand 1 (CXCL1) [50]. After treatment of organoids with lipopolysaccharide (LPS), a cell wall component from Gram-negative bacteria that binds to Toll-like receptor 4 (TLR4), we quantified several secretory cytokines. Culture supernatants of mock- or LPS-treated organoids were compared using membrane-based cytokine array (Fig. S7). Quantification of band intensity of the immunoblots revealed that interleukin-1 alpha (IL-1α), CXCL1 and IL-8 were significantly enhanced in the culture supernatants at 24 h after LPS treatment (Fig. 4 A and B). We then tested whether secretion of the chemokines or cytokines from tonsil organoids could affect cell migration. Counting of migrated cell number revealed that the migration index of HL-60 cells increased by approximately 2.6-fold when they were exposed to conditioned medium harvested from LPS-treated organoids compared to LPS-untreated organoids (Fig. 4C). As expected, mock-cultured medium without organoids had no effect on cell migration ability, irrespective of LPS treatment. The results are consistent with those of tissue samples from patients with tonsillitis, in which surface and crypt epithelium contributes to inflammatory processes as a defense mechanism for restricting microbial attack through secretion of chemokines as well as through facilitating neutrophil migration [50]. Our data confirms that the tonsil epithelial organoids are able to deploy their innate immune machinery in the presence of a TLR4-stimulating pathogen-associated molecular pattern (PAMP), LPS, in a similar way as tonsil tissues.
Fig. 4.
Innate immune response and SARS-CoV-2 (co)receptor expression in tonsil epithelial organoids. (A) Human cytokine array for secreted proteins from the tonsil organoids before (−) and after (+) LPS treatment at a concentration of 10 μg/mL for 24 h. (B) Changes in cytokine secretion levels were determined by densitometric analysis of the blots shown in (A). Values relative to the control levels (- LPS) are expressed as means ± SEM of three independent experiments. Statistical analysis was performed by two-way ANOVA with Sidak's pairwise multiple comparison test. ***, p < 0.001; ****, p < 0.0001. n.d., not detected. A.U., arbitrary units. (C) Effect of LPS-stimulated culture supernatants on cell migration. HL-60 cells were treated with mock- or tonsil organoid culture medium before (- LPS) and after LPS stimulation (+LPS). Numbers of migrated cells are presented as means ± SEM of the three independent experiments. Two-way ANOVA with Sidak's multiple comparison test was used for statistical analysis. n.s., not significant. *, p < 0.05. (D) Immunofluorescence staining of ACE2 (left), TMPRSS2 (middle) and furin (right) (red) expressed in tonsil organoids on day 15 as well as in tonsil tissues. NGFR and MUC1 (green) are stained as epithelial markers for the basal and suprabasal/superficial layers, respectively. Merged images with Hoechst 33342 staining (blue) are presented on the right side of each panel. Zoom images of white rectangles are displayed at the bottom. Scale bars are 100 μm. (E) Quantitative RT-PCR to detect ACE2 (left), TMPRSS2 (middle) and Furin (right) mRNA transcripts on days 5, 10 and 15 in tonsil organoids originating from three different donors. Statistical analysis was performed by ordinary one-way ANOVA with Tukey's multiple comparison test. n.s., not significant. *, p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Angiotensin-converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2) and furin have been identified to be essential for SARS-CoV-2 viral entry to the target cells [[51], [52], [53]]. Specifically, ACE2 as a receptor binds to the viral spike protein, while TMPRSS2 and furin are involved in spike protein maturation for membrane fusion. Before investigating susceptibility of tonsil organoids to SARS-CoV-2 infection, it was necessary to determine whether these receptor or co-receptor proteins are expressed in the organoids. Immunofluorescence analysis, in which tonsil tissues were used as a control, exhibited that ACE2 is localized on the outer side, consisting of NGFR-positive basal layer cells, but less on the inner side, which consisted of MUC1-expressing suprabasal/superficial layer cellular components (Fig. 4D). In contrast, TMPRSS2 and furin showed relatively homogenous expression throughout the whole area of organoid sections. Time-course mRNA expression data from three different donors revealed that TMPRSS2 mRNA was upregulated between days 10 and 15; by contrast, ACE2 or Furin mRNA was comparably expressed for 15 days (Fig. 4E). Accordingly, it can be expected that tonsil organoids could be susceptible to SARS-CoV-2 infection.
2.4. SARS-CoV-2 infection of human tonsil epithelial organoids
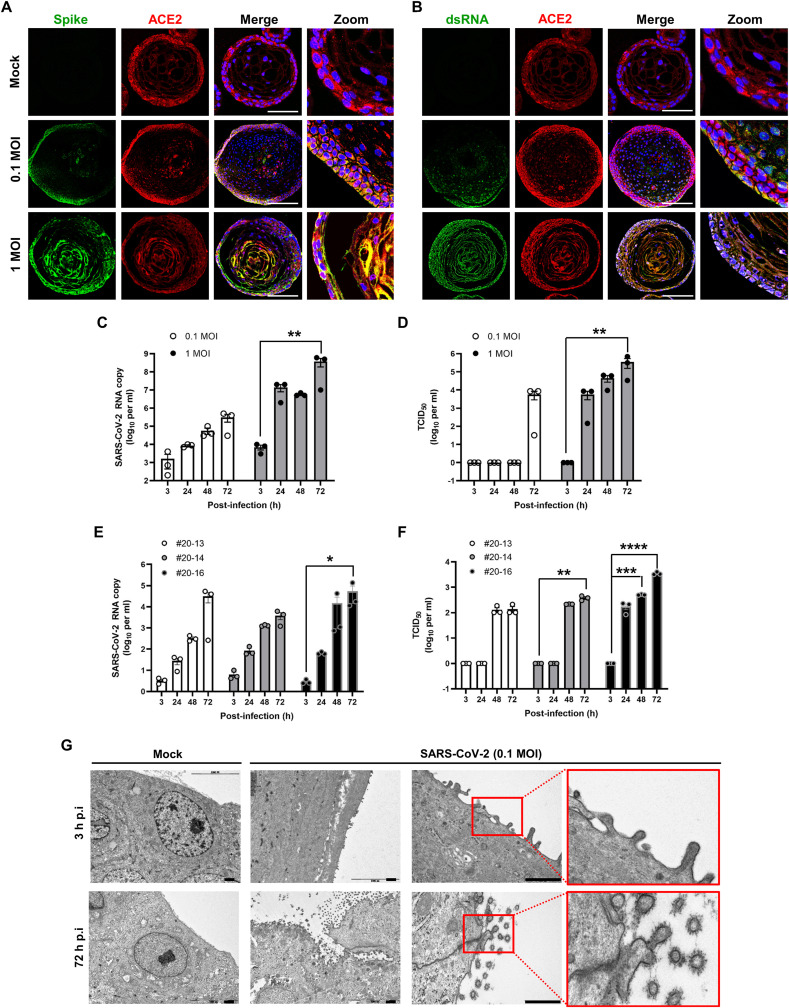
To determine whether the tonsil epithelial organoids are susceptible to SARS-CoV-2 infection, the organoids after 7 days were incubated with SARS-CoV-2 at an MOI of 0.1 or 1, in which mock infections were performed as a negative control to exclude the possibility of donor-derived viral infection. Viral protein expression and RNA replication were monitored at day 2 post-infection by probing with anti-spike protein and anti-dsRNA antibodies, respectively, by co-staining the SARS-CoV-2 receptor, ACE2. As the viral infection visualized in human lung epithelial Calu-3 cells (Fig. S8), fluorescence microscopy images from the tonsil organoids revealed viral spike protein (Fig. 5 A) and dsRNA (Fig. 5B) in the ACE2-expressing layers at both viral titers. Interestingly, ACE2 distribution was expanded from the basal layer into the suprabasal/superficial layer as SARS-CoV-2 robustly replicated.
Fig. 5.
Infection and amplification of SARS-CoV-2 in human tonsil epithelial organoids. (A, B) Immunofluorescence staining for viral spike protein (A) and dsRNA (B) (green) and a cellular receptor, ACE2 (red), in tonsil epithelium organoids on day 3 after SARS-CoV-2 infection at MOI of 0 (Mock), 0.1, or 1. Scale bar is 100 μm. Merged images with nuclei (blue) and their zooms are presented in the right panels. (C) Quantitative RT-PCR for detecting viral RNA in the culture supernatants of tonsil organoids infected with SARS-CoV-2 at an MOI of 0.1 or 1 at various time points after infection. (D) Determination of the amount of infectious viral particle in culture supernatants of SARS-CoV-2–infected tonsil organoids from the same donor of (C). Viral titers were measured by infection of fresh Vero CCL-81 cells with organoid culture medium for 2 days and by staining with anti-spike protein antibody, as described in Materials and Methods. (E, F) Both viral RNA copies (E) and infectious SARS-CoV-2 titers (F) in culture supernatants were determined independently from three additional tonsil organoids (donors 20-13, 20-14 and 2016). Two-way ANOVA with Dunnett's multiple comparison test was used for statistical analysis. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. (G) TEM analysis showing accumulation of SARS-CoV-2 particles on the apical surface of tonsil organoids on day 3 (lower) but not on 3 h (upper) after infection at an MOI of 0.1. Specific in the red rectangle are enlarged at right to show budding-out of viral particles on 72 h post-infection. Mock-infected samples are used as a control for showing pathogen-free samples. Scale bars are 1 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As a next step, we assessed release of amplified viral particles from infected cells by quantification of viral RNA copies and determination of 50% tissue culture infective dose (TCID50). Quantitative RT-PCR revealed a time-dependent increase in viral RNA levels, which was greater at 1 MOI than 0.1 MOI (Fig. 5C). Consistent with this result, an increase in infectious viral titer was observed 72 h after the lower-titer challenge but at 24, 48, and 72 h after the higher-titer challenge (Fig. 5D). This viral amplification was further verified in an independent experiment using three additional tonsil organoids established from different donors (Fig. 5E and F). Even given the variation in susceptibility among donors, efficiency of SARS-CoV-2 replication increased continuously during 72 h post-infection. To visualize the generation of progeny viral particles at the final stage of the virus life cycle, we performed transmission electron microscopy (TEM) on SARS-CoV-2–infected tonsil epithelial organoids at 3 and 72 h post-infection at an MOI of 0.1. We observed a high density of amplified viral particles with the typical crown-like appearance on the external surface of the apical membrane in the SARS-CoV-2–infected organoids at 72 h, but not in mock-infected or 3 h post-infection samples (Fig. 5G). Viral diameter was distributed between 99.7 and 179.6 nm. Taken together, these results suggested that SARS-CoV-2 can infect the tonsil epithelial organoids, from which progeny viruses are released through viral protein expression and RNA-dependent RNA replication.
2.5. Activation of oncostatin M signaling and lipid metabolic remodeling but suppression of innate immune signaling by SARS-CoV-2 infection in the tonsil epithelial organoids
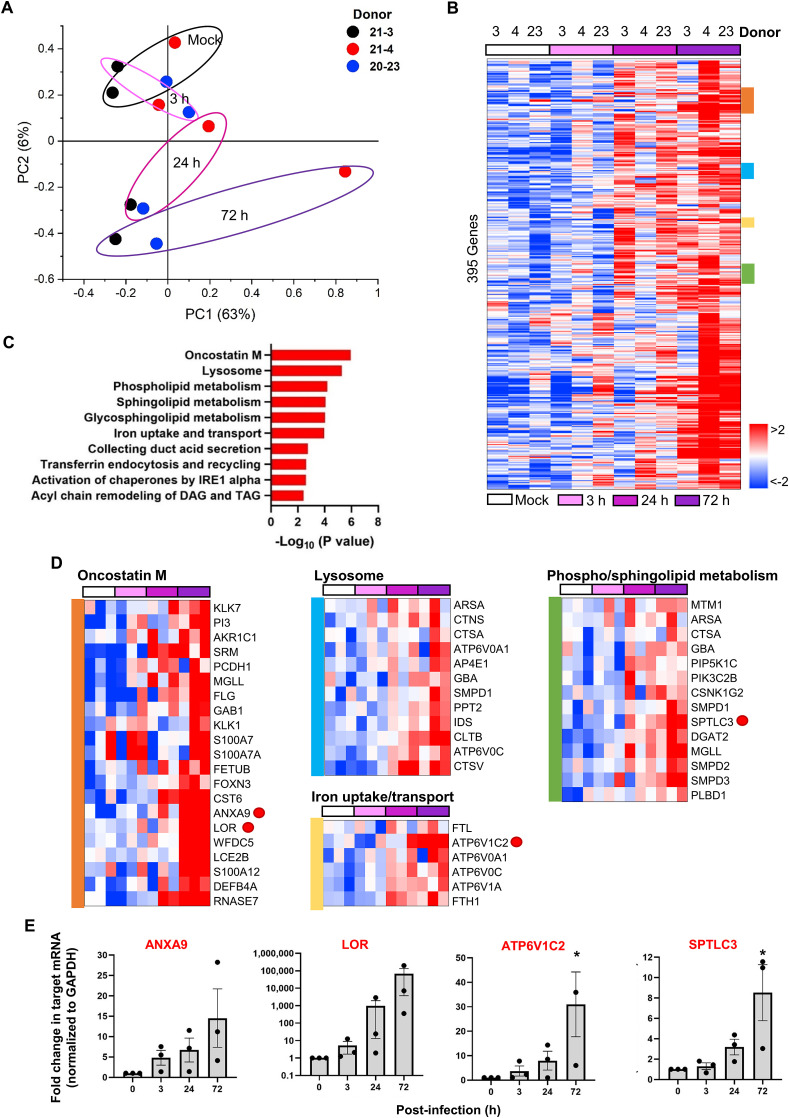
To perform Gene Ontology (GO) analysis affected by SARS-CoV-2 infection, the virus was highly purified by ultracentrifugation, that removes secretory molecules, such as pro-inflammatory cytokines, existing in the viral stock [54]. We compared mRNA expression levels of 21,449 genes of the organoids from three donors (21-3, 21-4 and 20–23) at 3, 24, and 72 h after infection. Their infection with SARS-CoV-2 was quality-controlled by qRT-PCR using the culture supernatant (Fig. S9). PCA plot showed that gene expression profile was similar in all donors before infection (Mock), but it became discriminated in a donor- and also time-dependent manner, with a greatest deviation in the donor 21-4 at 72 h post-infection (Fig. 6 A). Among the analyzed genes, 395 genes were upregulated, which are involved in oncostatin M signaling, lysosomal pathway, iron transport and lipid metabolism (Fig. 6B–D). Their upregulation at the transcriptional level was confirmed by qRT-PCR of ANXA9, LOR, ATP6V1C2 and SPTLC3 mRNA, representatively (Fig. 6E). It is worthy of note that oncostatin M signaling pathway is highly activated in all virus-infected organoids. Given that this signaling was also stimulated in the tonsil organoids compared to their epithelial tissues (Fig. 3E), it seems that SARS-CoV-2 infection intensively coordinates cellular differentiation or proliferation, subsequently supporting its robust amplification there [55,56]. It was found that mRNA transcripts responsible for lipidomic remodeling are systematically upregulated, potentially being resulted from increased utilization of membrane fusion, lipid rafts, lipid droplets, endosomes and exosomes during the virus life cycle [57].
Fig. 6.
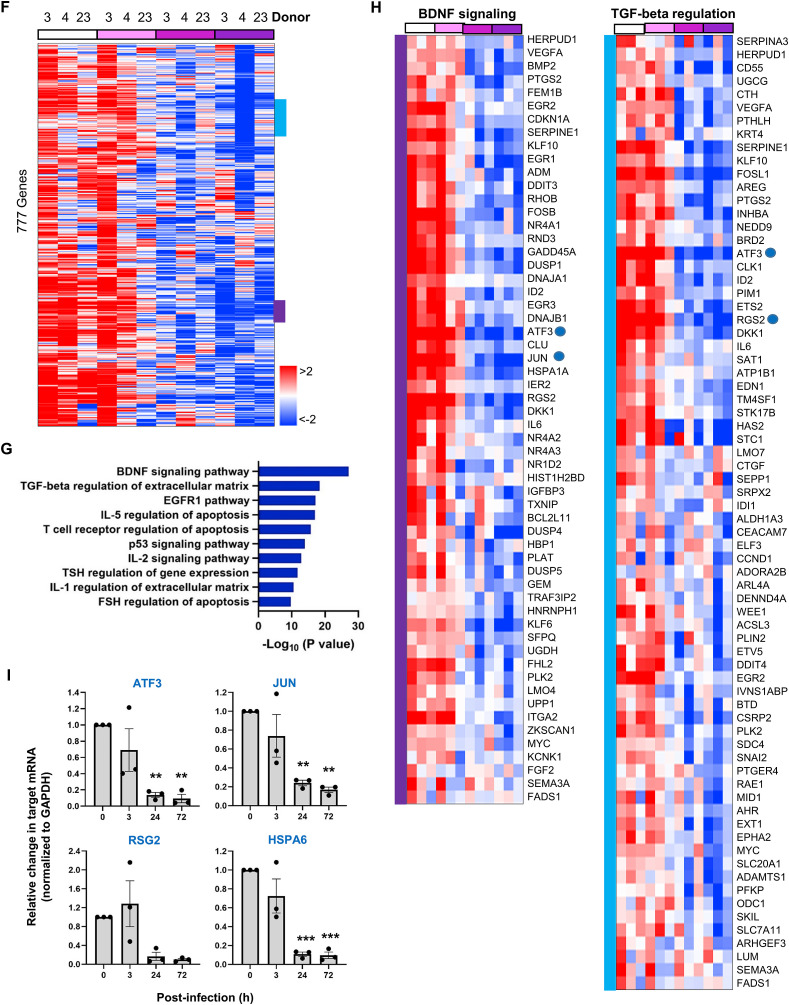
Gene expression changes in human tonsil epithelial organoids in response to SARS-CoV-2 infection. (A) PCA plot of time-course microarray samples from three different donors, 21-3 (black), 21-4 (red) and 20–23 (blue). PCA scores on PC1 (63%) and PC2 (6%) are grouped on the basis of time course, 0 (Mock), 3, 24 and 72 h post-infection. (B) Heatmap showing 395 genes upregulated after SARS-CoV-2 infection in tonsil epithelial organoids from three different donors, 21-3 (simply 3), 21-4 (4), and 20–23 (23). Red and blue represent upregulation and downregulation, respectively (with > 2-fold changes). (C) Top 10 enriched GO categories of differentially upregulated genes in SARS-CoV-2-infected samples. (D) Representative upregulated gene clusters colored in (B) are enlarged with their gene lists. (E) Quantitative RT-PCR of upregulated genes, ANXA9, LOR, ATP6V1C2 and SPTLC3, which are randomly selected among DEGs, marked with red spots in (D). Values are presented as means ± SEM of three independent experiments from the three donors. (F) Heatmap showing 777 genes downregulated after SARS-CoV-2 infection in tonsil epithelial organoids from three different donors, 21-3 (3), 21-4 (4), and 20–23 (23). (G) Top 10 enriched GO categories of differentially downregulated gene in SARS-CoV-2-infected samples. (H) Representative downregulated gene clusters colored in (F) are enlarged with their gene lists. (I) Quantitative RT-PCR of downregulated genes, ATF3, JUN, RSG2 and HSPA6, which are randomly selected among DEGs, marked with blue spots in (D). HSPA6 is downregulated in the IL-5 regulation pathway, thus not marked with a dot. Values are presented as means ± SEM of three independent experiments from the three donors. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In contrast, 777 genes were markedly decreased at 72 h post-infection in the infected organoids (Fig. 6F). They are responsible for brain-derived neurotrophic factor (BDNF) signaling, cell growth or innate immune responses (Fig. 6G and H). Quantitative RT-PCR analysis of ATF3, JUN, RSG2 and HSPA6 mRNA showed their time-dependent decreases, ensuring reliability of the GO analysis (Fig. 6G and I). In accordance with a recent publication that reported a correlation between low serum BDNF level and severity of SARS-CoV-2 infection in patients [58], its signaling pathway was significantly suppressed in our organoid system. Overall, the transcriptome analysis suggested that SARS-CoV-2 infects the tonsil epithelial organoids and robustly self-amplifies there by remodeling cellular lipid metabolism but by suppressing host defense mechanism such innate immunity or growth factor signaling. This immunocompromised or immunosuppressed property of tonsil epithelial organoids acquired by viral infection might be related to its role as a reservoir for diverse viral infection or latency [59].
2.6. Evaluation of antiviral activity of remdesivir against SARS-CoV-2 infection in the tonsil epithelial organoids
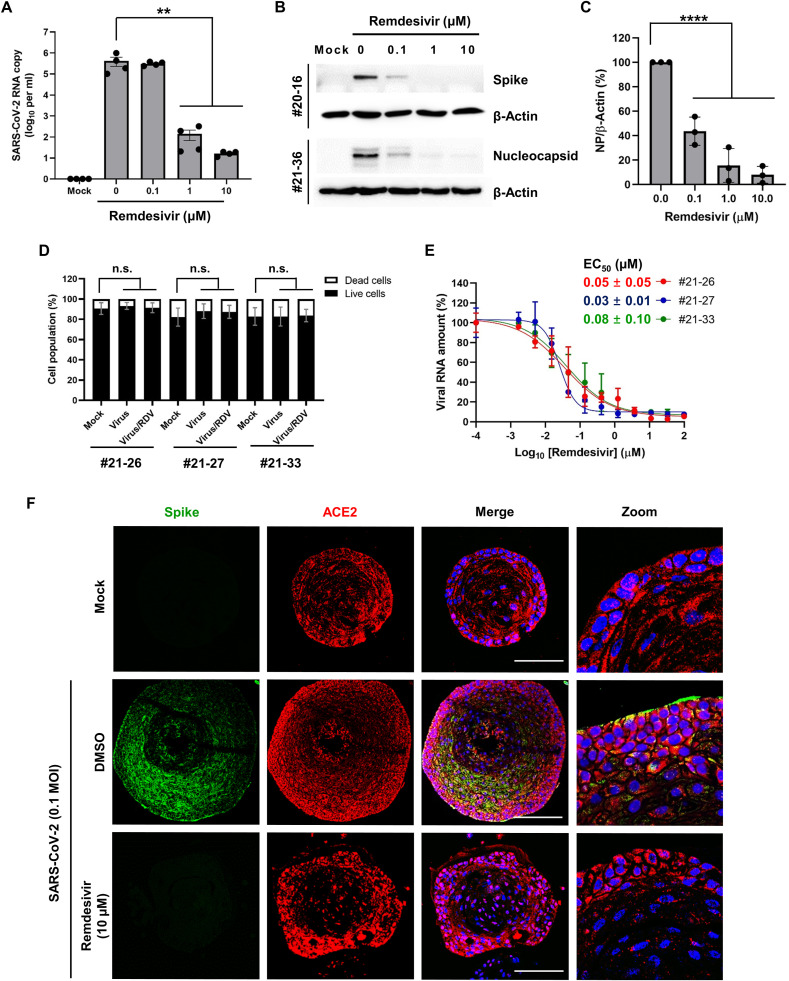
To examine whether SARS-CoV-2 infection could be inhibited by an antiviral compound in the organoids, we treated virus-infected samples with increasing concentrations of remdesivir, a prodrug nucleoside analog that inhibits viral RNA replication. On day 2 after treatment, a dose-dependent reduction in viral RNA level as well as viral spike and nucleocapsid protein levels was observed in dose-dependent manner (Fig. 7 A–C). To test its reliability, tonsil epithelial organoids from three different donors (21–26, 21–27 and 21–33) were additionally prepared. Cell viability test by microscopically counting live and dead cells revealed that neither SARS-CoV-2 at an MOI of 0.1 nor remdesivir at a maximum dose of 100 μM did affect cell viability at day 2 post-infection (Fig. 7D). Dose response analysis by measuring viral RNA in the culture supernatants exhibited antiviral activity of remdesivir in all SARS-CoV-2-infected organoids with half-maximal effective concentration (EC50) values ranging from 0.03 to 0.08 μM (Fig. 7E). These values have more improved antiviral activity when compared to those in our cell culture system, where it had EC50 values of 35.4 μM in Vero 81 cells and 0.1–1 μM in Calu-3 cells [60]. This difference might be caused by variations in antiviral assay methods or viral growth kinetics in the cell types used. Confocal microscopy decisively visualized reduction of viral protein expression by remdesivir in the SARS-CoV-2-infected tonsil organoid (Fig. 7E). Our data emphasize that the tonsil epithelial organoids are competent for evaluation of the antiviral efficacy of small molecules before they are translated to clinical trials.
Fig. 7.
Evaluation of antiviral activity of remdesivir in SARS-CoV-2-infected tonsil organoids. (A) Quantitative RT-PCR showing inhibition of viral RNA titers on day 2 after treatment of SARS-CoV-2–infected organoids with increasing concentrations of remdesivir (0.1, 1, or 10 μM). Mock: non-infected control. RNA copies are presented as means ± SEM of four independent experiments using tonsil organoids from a single donor (no. 20-16). Statistical analysis was performed by ordinary one-way ANOVA with Dunnett's multiple comparison test. **, p < 0.01. (B) Western blot analysis showing reduction of viral spike and nucleocapsid proteins in two different organoids samples (donors 20-16 and 21–36, respectively) in the presence of remdesivir. Beta-actin was used as a loading control. (C) Reduction of nucleocapsid protein by remdesivir was quantified from Western blot analysis with three different organoid samples (donors 21-16, 21–34 and 21–36). Statistical analysis was performed by two-way ANOVA with Dunnett's multiple comparison test. ****, p < 0.0001. (D) Cell viability test. Organoids from three different donors (21–26, 21–27 and 22–33) were mock infected or infected with SARS-CoV-2 alone (MOI, 0.1) or in the presence of 100 μM remdesivir. On day 2, organoids from three different wells were dissociated with trypsin-EDTA and individually stained with trypan blue. Live and dead cells were counted by using an automated microscopic cell counter. Statistical analysis was performed by two-way ANOVA with Sidak's multiple comparison test. n.s., statistically not significant. (E) Organoid samples from three different donors (21–26, 21–27 and 22–33) were infected with SARS-CoV-2 (MOI, 0.1) and treated with increasing concentrations of remdesivir. On day 2 post-infection, RNA samples were harvested from culture supernatants for qRT-PCR against the viral nucleocapsid gene. EC50 values are calculated in three independent experiments and expressed as mean ± SD. (F) Confocal miscopy showing reduction of viral spike protein by remdesivir. Tonsil epithelial organoids (donor 21–36) were mock-infected (upper) or infected with SARS-CoV-2 at an MOI of 0.1. The infected organoids were treated with DMSO (middle) or remdesivir (10 μM; lower) for 2 days. Viral spike protein (blue) and its binding receptor ACE2 (red) were stained with their specific antibodies. Nuclei (blue) were counter-stained with Hoechst 33342. Merged images and their zooms are displayed on the right side. Scale bar is 100 μm.(For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. Discussion
The oropharynx, located behind the oral cavity and beneath the nasopharynx, functions in digestion and respiration. It consists of four parts: the tonsils, the base of the tongue, the soft palate, and the posterior oropharyngeal wall. The tonsils (also called the palatine tonsils or the oropharyngeal tonsils) contain large populations of immune and epithelial cells. This cellular diversity could account for the tonsils’ multifunctional roles in front-line defense against bacteria or viruses invading the nasal or oral cavity, as well as in their function as reservoirs for infection by pathogens including Epstein-Barr virus, adenoviruses, influenza A and B viruses, herpes simplex virus, rhinovirus, enterovirus, and even human papillomavirus [[61], [62], [63]]. The two major proteins required for SARS-CoV-2 entry, ACE2 and TMPRSS2, have been reported to be expressed in the squamous epithelium lining oropharyngeal tonsillar tissue [64]. This motivated us to determine whether tonsil tissue is susceptible to the SARS-CoV-2 infection. However, the lack of a culture system for tonsillar epithelial cells has deterred in-depth cytopathologic or virological studies of the pandemic pathogen. In the present study, we were able to establish a reliable culture system for human tonsil epithelial organoids by optimizing culture medium composition, thereby recapitulating the dominant features observed in in vivo tonsil epithelium, such as stratified squamous epithelial layers and appropriate distributions of tonsillar biomarkers (Fig. 1, Fig. 2). The major population of genes (88%) of the organoids follow a transcriptome profile of tonsil epithelial tissues, both crypt and surface (Fig. 3). The tonsil epithelial organoids expressed the major components, such as ACE2, TMPRSS2 and furin, which are essential for viral attachment and fusion during the entry step, and were highly susceptible to SARS-CoV-2 infection (Fig. 4, Fig. 5). Consequently, the virus was vigorously amplified through multiple rounds of infection, resulting in abundant secretion of progeny viral particles. Our findings suggest that the tonsil epithelium could be one among the major target organs for primary infection of SARS-CoV-2, and that its organoids are a desirable ex vivo model for investigation of viral susceptibility or transmissibility.
‘Cytokine storm’ is an initial cause of acute respiratory distress syndrome (ARDS), multi-organ failure, and severe pneumonia, which result in considerably mortality among COVID-19 patients [65]. This hyperactive inflammatory response is primarily associated with abrupt release of pro-inflammatory or inflammatory mediators, including IL-6, TNF-α, and CXCL8 (also named IL-8) [1]. However, SARS-CoV-2 infection does not trigger ‘cytokine storm’ in children (a mean age of 7.3 year) with COVID-19, not affecting the levels of plasma pro-inflammatory cytokines [66]. From these contradictory findings, our cDNA microarray analysis stood for the latter by showing that SARS-CoV-2 infection suppresses NF-κB-related pro-inflammatory cytokine pathways in the tonsil epithelial organoid at 72 h post-infection (Fig. 6F–I). Thus, we assume that among the upper respiratory tract tissues, the tonsil epithelium might be an optimized place for SARS-CoV-2 infection, through which they could play a pivotal role as the antigen presenting cells in the circumstances with abundant immune cells. Recently, preparation of immune organoids from tonsils and their remodeling adaptive immunity have been established by L. E. Wager et al. [67]. As a further study combining this technology, we are going to explore how the adaptive immunity is educated and lipid metabolic reprogramming changes when tonsil immune cell–derived organoids are co-cultured with SARS-CoV-2-infected autologous epithelial organoids. This approach could contribute to elucidating immunological tripartite crosstalk between tonsil epithelial cells, immune cells, and SARS-CoV-2, providing a comprehensive understanding not only of the host defense mechanism from the standpoint of both innate and adaptive immunity, but also of viral immune evasion strategies counteracting host immunity.
We evaluated the therapeutic efficacy of remdesivir in the tonsil organoid model, demonstrating its value as a drug candidate screening platform for development of novel SARS-CoV-2 therapies. This organoid system has advantages as an alternative to animal models, particularly when they target host factors involved in the viral life cycle or immune regulation. It is nontrivial because the efficacy of those antivirals could be underestimated (or impossible to assess) in animals due to low genetic homology or null function in model organisms. This is the first study to report the generation of human tonsil epithelial organoids and their suitability for infection by a respiratory virus, especially SARS-CoV-2. From the clinical perspective, tonsils are a readily accessible intraoral organ from which specimens are frequently underutilized after surgery; by contrast, obtaining samples from the kidney, liver, bronchus, or gut requires invasive and aggressive procedures. Thus, tonsil epithelial organoids can be suggested as an attractive ex vivo model for fundamental or applied research on infections by SARS-CoV-2 and potentially other pathogenic microbes.
4. Materials and methods
4.1. Human specimens
This study was approved by the institutional review board of Konkuk University Hospital (IRB no. KUH1110073) and carried out with the written consent of all donors. Total 37 donors consisting of 18 females and 19 males had chronic tonsillitis with an average age of 26.3 years between 13 and 64. Whole tonsils were collected in saline after tonsillectomy and decontaminated by immersion for 1 h at 4 °C in Earle's balanced salt solution (EBSS) with sodium bicarbonate (Sigma-Aldrich, St. Louis, MO, USA) with 1 × Antibiotic–Antimycotic (Thermo Fisher Scientific, Waltham, MA, USA), 100 units/mL of penicillin, 100 μg/mL of streptomycin, and 250 ng/mL amphotericin B (Gibco, Grand Island, NY, USA).
4.2. Formation of organoids from human tonsil tissues
The tonsil samples were chopped and washed with Dulbecco's phosphate-buffered saline (D-PBS; Welgene, Daegu, Korea) and then enzymatically digested with 1 mg/mL collagenase II (Gibco) in advanced DMEM/F12 (Gibco) for 2 h at 37 °C. After digestion, isolated cells were embedded in Matrigel (Corning Inc., Corning, NY, USA) in a 48-well plate (SPL Inc., Seongnam, Korea) and incubated at 37 °C for 10 min to polymerize the matrices. Tonsil organoids were cultured in advanced DMEM/F12 supplemented with Antibiotic–Antimycotic, Glutamax (Thermo Fisher Scientific), B27 (Invitrogen, Carlsbad, CA, USA), 10% conditioned media from Cultrex HA-R-spondin1-Fc 293T cells (R&D Systems, Minneapolis, MN, USA), and the following growth factors: 50 ng/mL recombinant murine HGF (Peprotech, Rocky Hill, NJ, USA), 100 ng/mL noggin (ProSpec, St. Paul, MN, USA), 20 nM A83-01 (Sigma), 50 ng/mL human FGF10 (ATGen, Seongnam, Korea), 20 ng/mL human bFGF (Peprotech), 10 μM prostaglandin E2 (BioGems, Westlake Village, CA, USA), and 10 mM nicotinamide (Sigma). For passage, organoids were dissociated by incubation in 0.25% trypsin-EDTA (Invitrogen, Waltham, Massachusetts, USA) every 7–10 days depending on the number and size of organoids. For the first 2 days at every passage, 10 μM Y-27632 (Tocris Biosciences, Bristol, UK) was added to the culture medium.
4.3. Clonal organoid formation assay from cells isolated by fluorescence-activated cell sorting (FACS)
To determine the origin of cells forming the organoids, cells dissociated from tissues as mentioned above were stained with anti-E-cadherin (cat no. AF748; R&D Systems, Minnesota, United States) or anti-NGFR (cat no. sc-13577; Santa Cruz Biotechnology, Dallas, TX, USA) and their secondary antibody, Alexa Fluor 488–labeled donkey anti-goat IgG (cat no. A-11055; Invitrogen) or goat anti-mouse IgG (cat no. A-32723; Invitrogen). For direct labeling, FITC-conjugated anti-CD44 (cat no. 555478; BD Biosciences, Heidelberg, Germany), PE-conjugated anti-ITGA6 (cat no. 12-0495-83; BioLegend, San Diego, CA, USA), PE/Cyanine7-conjugated anti-NGFR Antibody (cat no. 345110; BioLegend) and APC-conjugated anti-EpCAM (cat no. 130-113-260; Miltenyi Biotec, Bergisch Gladbach, Germany) antibodies were used (Table S2). They were individually sorted using an S3e Cell Sorter (Bio-Rad, Hercules, CA, USA) and cultured in Matrigel with TeM for 15 days.
4.4. SARS-CoV-2 amplification and purification
SARS-CoV-2 (BetaCoV/Korea/KCDC03/2020) provided by the Korea Disease Control and Prevention Agency (Cheongju, Korea) was amplified in Vero E6 cells (American Type Culture Collection, Rockville, MD, USA) through three passages in DMEM (HyClone, South Logan, UT, USA). After centrifugation of the supernatants at 3000 g for 10 min, the viral stock was stored at −80 °C before use.
For host transcriptome analysis, SARS-CoV-2 was purified through ultracentrifugation according to a previous report with some modifications [68]. Briefly, the virus stock was loaded onto 20% sucrose dissolved in TNE buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl and 1 mM EDTA) and centrifuged at 175,000 g for 3 h at 4 °C using Optima XPN-100 Ultracentrifuge with SW 32 Ti rotor (Beckman Coulter, Brea, CA, USA). The viral pellet was resuspended in PBS (1/2 of the starting volume). Viral titer was determined by plaque assay before use. All experiments with infectious SARS-CoV-2 were performed within a Biosafety Level 3 (BL3) facility in KRICT.
4.5. Infection of tonsil organoids with SARS-CoV-2
Tonsil epithelial organoids containing 6 × 104 cells at day 7 of passage 2 were suspended at 37 °C for 1 h in 5 μL TeM for infection with the same volume of SARS-CoV-2 at an MOI of 0.1 or 1. Unadsorbed virus was removed by washing twice with 1 mL of advanced DMEM/F12 with centrifugation at 1000 g for 10 s. Each sample of mock-infected or SARS-CoV-2-infected organoids was embedded in 20 μL Matrigel on a 48-well plate and cultured in 300 μL TeM supplemented with 10 μM Y-27632. The supernatants were harvested at 3, 24, 48, and 72 h post-infection for viral RNA quantification or infectious viral titration. For cDNA microarray analysis, cell lysates were harvested for RNA extraction.
4.6. Histology, immunohistochemistry, and immunofluorescence
Tissues and organoids were fixed in 4% paraformaldehyde (PFA; Bio Solution, Suwon, Korea) and embedded in paraffin. Paraffin sections (5 μm thick) were deparaffinized in xylene and hydrated in a graded series of ethanol. They were then stained using H&E, Alcian blue (Agilent Technologies, Santa Clara, CA, USA), PAS staining kit (Abcam, Cambridge, MA, USA), or Masson's trichrome staining kit (Dako, Carpinteria, CA, USA). For immunohistochemistry, antigen retrieval was performed by incubating in sodium citrate buffer (10 mM sodium citrate with 0.05% Tween 20, pH 6.0) at 95 °C for 30 min. Endogenous peroxidase was blocked by incubating in 3% H2O2 in methanol for 10 min. After blocking with 1% BSA in PBS, sections were incubated at 4 °C overnight with primary antibody and then incubated at room temperature for 30 min with a biotinylated secondary antibody (anti-mouse IgG or anti-rabbit IgG) using the Vectastain ABC kit (Vector Laboratories, Burlingame, CA, USA). Sections were stained according to the avidin–biotin complex method using the Vectastain DAB kit (Vector Laboratories).
For immunofluorescence analysis, fixed samples were cryoprotected by immersion in PBS containing 30% sucrose and 0.1% sodium azide at 4 °C, and then embedded in optimal cutting temperature (OCT) compound (Sakura-Finetek, Torrance, CA, USA). Rapidly frozen sections (4 μm thick) were pre-blocked with 5% normal horse serum (Vector Laboratories) in PBS at room temperature for 2 h, followed by incubation at 4 °C overnight with primary antibodies specific for cellular biomarkers, viral spike protein, or dsRNA. They were then incubated with fluorescently-labeled secondary antibodies at room temperature for additional 2 h. Nuclei were counterstained with Hoechst 33342 (1 μg/mL; Sigma). Acquisition of confocal images was performed on an LSM 880 confocal microscope (Carl Zeiss, Jena, Germany). Information about antibodies used in immunofluorescence analysis is summarized in Table S2.
4.7. Transmission electron microscopy (TEM)
To observe ultrastructure of tonsil organoids or release of progeny SARS-CoV-2 particles from infected organoids, TEM analysis was performed according to a previous report with some modifications [69]. Briefly, tonsil organoids were fixed in Karnovsky's fixative solution (2% glutaraldehyde, 2% paraformaldehyde, 0.5% CaCl2 in 0.1 M phosphate buffer, pH 7.4) and embedded in the epoxy resin after post-fixation with 1% OsO4 dissolved in 0.1 M phosphate buffer. Ultrathin sections were processed using an ultramicrotome (EM UC7; Leica, Wetzlar, Germany) and placed on a copper grid stained with 6% uranyl and lead citrate. The samples were observed using a JEM-1011 transmission electron microscope (JEOL, Tokyo, Japan).
4.8. Microarray analysis
To analyze the genetic similarity between tonsil tissues, crypt or surface, and organoids or whole-genome expression changes resulting from SARS-CoV-2 infection, total RNA was extracted from tissue and organoids using the MagListo 5 M Cell Total RNA Extraction Kit (Bioneer, Daejeon, Korea) or Trizol reagent (Invitrogen). mRNA was reverse-transcribed into cDNA using GeneChip Whole Transcript (WT) cDNA Synthesis and Amplification kit (Affymetrix, Santa Clara, CA, USA). After fragmentation of cDNA, it was biotin-labeled using the GeneChip WT Terminal labeling kit (Affymetrix) and hybridized to the Affymetrix Gene Chip Array (Affymetrix) at 45 °C for 16 h. Sample quality and gene expression profiling were certified at Macrogen (Seoul, Korea). To assess global gene expression profiles, we performed clustering analysis based on the centered correlation coefficient between transcriptomes of different samples. Gene set enrichment was analyzed using the Ingenuity Pathway Analysis software (IPA; Qiagen, Hilden, Germany).
To evaluate gene expression changes observed in microarray analysis, target mRNA expression was quantified by qRT-PCR. Total RNA was reverse-transcribed with random hexameric oligonucleotide primers using SuperScript III First-Strand Synthesis System (Invitrogen) and cDNA was amplified with target-specific primers and SYBR Green Master Mix the CFX96 (Bio-Rad) using Touch real-time PCR instrument (Bio-Rad). Their expression was normalized to GAPDH gene. All primer sequences used in this study are listed in Table S3.
4.9. Cytokine array and chemotaxis assay
The cytokine profile assay was performed using the Proteome Profiler Human Cytokine Array kit (R&D Systems, Minneapolis, MN, USA). Culture supernatants from LPS-treated (Sigma) or -untreated tonsil organoids were collected and incubated with precoated Proteome Profiler array membrane at 4 °C overnight. The membrane was then washed and incubated with streptavidin-horseradish peroxidase (HRP) buffer for 30 min. After treatment with Chemi Reagent Mix (provided in the kit), dot densities were imaged on an LAS4000 (GE Healthcare, Chicago, IL, USA) and quantified using the ImageJ software (https://imagej.nih.gov/ij/). To perform the chemotaxis assay, HL-60 cells (Korean Cell Line Bank, Seoul, Korea) were differentiated into granulocytes by culture in the presence of 1.3% DMSO (Duchefa Biochemie, Haarlem, The Netherlands) for 7 days, as described previously [70]. Cell monolayers grown on Transwell inserts (Corning) at a density of 5 × 105 cells/well in RPMI 1640 medium (Gibco) containing 0.5% BSA were exposed to supernatants that were collected from LPS-treated or -untreated organoids. After 1 h, migrated HL-60 cells were counted using a LUNA Automated Cell Counter (Logos Biosystems, Annandale, VA, USA).
4.10. Viral RNA quantification and TCID50 determination
Viral RNA was purified from the culture supernatant at the indicated time points using Viral RNA Purification Kit (Qiagen). One-step RT-PCR targeting the viral nucleocapsid (N) gene was performed using a diagnostic kit (PCL Inc., Seoul, Korea) and the CFX96 Touch real-time PCR instrument (Bio-Rad). In all experimental sets, RNAs purified from serial dilutions of a virus stock with quantified plaque titers were used as a standard to determine the absolute viral RNA copy number in the samples. In parallel, Vero CCL-81 cells were seeded at a density of 2 × 104 cells per well in 96-well plates and then treated with serially-diluted culture supernatants from SARS-CoV-2-infected organoids at an MOI of 0.1 or 1. On day 2 post-infection, the cells were fixed and permeabilized with chilled acetone:methanol (1:3) solution at room temperature for 10 min. Viral spike (S) protein was probed using anti-S antibody (Genetex, Irvine, CA, USA) and Alexa Fluor 488–conjugated goat anti-mouse IgG antibody (Invitrogen) and cellular nuclei were stained with 4’,6-diamidino-2-phenylindole (Invitrogen) as described previously report [71]. Fluorescence images were captured and analyzed on an Operetta High-content Screening System (PerkinElmer, Waltham, MA, USA) and quantified using the built-in Harmony software. Compared with the control infection (100%), dilution folds resulting in 50% infection were calculated to determine the 50% tissue culture infectious doses (TCID50).
4.11. Antiviral activity of remdesivir against SARS-CoV-2 in tonsil organoids
Antiviral assays were performed according to our previous report with some modifications [60]. Briefly, 7-day–cultured tonsil organoids were infected with SARS-CoV-2 at an MOI of 0.1 and embedded in 20 μL of fresh TeM plus Matrigel (6 × 104 cells per well in 48-well plates). Remdesivir (purity 99.84%; MedChemExpress, Monmouth Junction, NJ, USA) was 10-fold serially diluted in TeM from 10 to 0.1 μM and used to treat SARS-CoV-2-infected organoids. For determining EC50 values, which are chemical concentrations to reduce viral RNA titers by 50%, three-fold serial dilutions of remdisivir (from 100 μM to 1.7 nM) were treated to the SARS-CoV-2-infected organoids from three different donors in triplicate. On day 2, the culture supernatants were harvested to measure the amount of viral RNA by quantitative RT-PCR. In parallel, cell lysates were collected from four wells using RIPA buffer (LPS solution, Daejeon, Korea), and viral protein expression levels were compared by Western blot analysis. The immuno-transferred membrane was probed with anti-spike primary antibody (GeneTex, Irvine, CA, USA) or anti-nucleocapsid antibody (Sino Biological, Beijing, China) followed by HRP–conjugated anti-mouse goat IgG as the secondary antibody (Invitrogen).
4.12. Statistical analysis
Statistical significance of differences was analyzed using GraphPad Prism software package, version 8.4.3 (GraphPad, San Diego, CA, USA). Unpaired two-tailed t-tests were used for comparisons of two groups. In general, two-way ANOVA with Dunnett's or Sidak's multiple comparison tests were used to compare multiple groups, but if needed, one-way ANOVA was performed followed by Dunnett's or Tukey's multiples comparison tests. p < 0.05 was considered significant. Unless otherwise specified, all experiments were performed in triplicate and their results are presented as means ± SEM from three independent experiments.
Author contributions
M.K. Y.C.L. and J.Y. designed and supervised the study. H.K.K., H.K., D.H.B., W.H.C. and J.-Y.P. cultured organoids. H.K.K, H.K., D.H.B.. and W.H.C. conducted the histological staining. K.H.K., H.W.H., B.L., and G.C. analyzed the genomics data. M.K.L, Y.J. and J.S.S performed SARS-CoV-2-related experiments in a biosafety level 3 facility in KRICT. Y.C.L. provided the human tissues and clinical specimens. H.K.K., S.I.H., M.K., Y.C.L. and J.Y. wrote the manuscript.
Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (HR16C0002, HI18C2458 to J.Y.), by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science & ICT (MSIT), Republic of Korea (2018R1D1A1A02050030 to J.Y., 2016R1A5A2012284, 2018R1D1A1A02086084 and 2021R1A2C2004382 to Y.C.L.), by the 3D-TissueChip Based Drug Discovery Platform Program through the Korea Evaluation Institute of Industrial Technology funded by the Ministry of Commerce, Industry and Energy (20009773 to J.Y.), by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (NRF-2020M3A9I2108564 to M.K.) and by an intramural research funding from KRICT (SI2232-20 to M.K.). The SARS-CoV-2 resource (NCCP No., 43326) was provided by the National Culture Collection for Pathogens, Republic of Korea. The authors declare no competing financial interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2022.121460.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. https://www.ncbi.nlm.nih.gov/pubmed/31986264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui D.S., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., McHugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. https://www.ncbi.nlm.nih.gov/pubmed/31953166E.I Azhar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu X., Yu C., Qu J., Zhang L., Jiang S., Huang D., Chen B., Zhang Z., Guan W., Ling Z., Jiang R., Hu T., Ding Y., Lin L., Gan Q., Luo L., Tang X., Liu J. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur. J. Nucl. Med. Mol. Imag. 2020;47(5):1275–1280. doi: 10.1007/s00259-020-04735-9. https://www.ncbi.nlm.nih.gov/pubmed/32107577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delgado-Roche L., Mesta F. Oxidative stress as key player in severe acute respiratory syndrome coronavirus (SARS-CoV) infection. Arch. Med. Res. 2020;51(5):384–387. doi: 10.1016/j.arcmed.2020.04.019. https://www.ncbi.nlm.nih.gov/pubmed/32402576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver S.E., Gargano J.W., Marin M., Wallace M., Curran K.G., Chamberland M., McClung N., Campos-Outcalt D., Morgan R.L., Mbaeyi S., Romero J.R., Talbot H.K., Lee G.M., Bell B.P., Dooling K. The advisory committee on immunization practices' interim recommendation for use of pfizer-BioNTech COVID-19 vaccine - United States, december 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69(50):1922–1924. doi: 10.15585/mmwr.mm6950e2. https://www.ncbi.nlm.nih.gov/pubmed/33332292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliver S.E., Gargano J.W., Marin M., Wallace M., Curran K.G., Chamberland M., McClung N., Campos-Outcalt D., Morgan R.L., Mbaeyi S., Romero J.R., Talbot H.K., Lee G.M., Bell B.P., Dooling K. The advisory committee on immunization practices' interim recommendation for use of moderna COVID-19 vaccine - United States, december 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;69(5152):1653–1656. doi: 10.15585/mmwr.mm695152e1. https://www.ncbi.nlm.nih.gov/pubmed/33382675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver S.E., Gargano J.W., Scobie H., Wallace M., Hadler S.C., Leung J., Blain A.E., McClung N., Campos-Outcalt D., Morgan R.L., Mbaeyi S., MacNeil J., Romero J.R., Talbot H.K., Lee G.M., Bell B.P., Dooling K. The advisory committee on immunization practices' interim recommendation for use of janssen COVID-19 vaccine - United States, february 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70(9):329–332. doi: 10.15585/mmwr.mm7009e4. https://www.ncbi.nlm.nih.gov/pubmed/33661860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. https://www.ncbi.nlm.nih.gov/pubmed/34726479 [DOI] [PubMed] [Google Scholar]
- 10.Rubin D., Chan-Tack K., Farley J., Sherwat A. FDA approval of remdesivir - a step in the right direction. N. Engl. J. Med. 2020;383(27):2598–2600. doi: 10.1056/NEJMp2032369. https://www.ncbi.nlm.nih.gov/pubmed/33264539 [DOI] [PubMed] [Google Scholar]
- 11.Muik A., Wallisch A.K., Sanger B., Swanson K.A., Muhl J., Chen W., Cai H., Maurus D., Sarkar R., Tureci O., Dormitzer P.R., Sahin U. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371(6534):1152–1153. doi: 10.1126/science.abg6105. https://www.ncbi.nlm.nih.gov/pubmed/33514629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidd M., Richter A., Best A., Cumley N., Mirza J., Percival B., Mayhew M., Megram O., Ashford F., White T., Moles-Garcia E., Crawford L., Bosworth A., Atabani S.F., Plant T., McNally A. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab082. https://www.ncbi.nlm.nih.gov/pubmed/33580259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giandhari J., Pillay S., Wilkinson E., Tegally H., Sinayskiy I., Schuld M., Lourenco J., Chimukangara B., Lessells R., Moosa Y., Gazy I., Fish M., Singh L., Sedwell Khanyile K., Fonseca V., Giovanetti M., Carlos Junior Alcantara L., Petruccione F., de Oliveira T. Early transmission of SARS-CoV-2 in South Africa: an epidemiological and phylogenetic report. Int. J. Infect. Dis. 2021;103:234–241. doi: 10.1016/j.ijid.2020.11.128. https://www.ncbi.nlm.nih.gov/pubmed/33189939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontanet A., Autran B., Lina B., Kieny M.P., Karim S.S.A., Sridhar D. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397(10278):952–954. doi: 10.1016/S0140-6736(21)00370-6. https://www.ncbi.nlm.nih.gov/pubmed/33581803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F., Zhang T., Kim T.W., Harschnitz O., Redmond D., Houghton S., Liu C., Naji A., Ciceri G., Guttikonda S., Bram Y., Nguyen D.T., Cioffi M., Chandar V., Hoagland D.A., Huang Y., Xiang J., Wang H., Lyden D., Borczuk A., Chen H.J., Studer L., Pan F.C., Ho D.D., tenOever B.R., Evans T., Schwartz R.E., Chen S. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27(1):125–136. doi: 10.1016/j.stem.2020.06.015. https://www.ncbi.nlm.nih.gov/pubmed/32579880 e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Y., Yang L., Duan X., Duan F., Nilsson-Payant B.E., Yaron T.M., Wang P., Tang X., Zhang T., Zhao Z., Bram Y., Redmond D., Houghton S., Nguyen D., Xu D., Wang X., Uhl S., Huang Y., Johnson J.L., Xiang J., Wang H., Pan F.C., Cantley L.C., tenOever B.R., Ho D.D., Evans T., Schwartz R.E., Chen H.J., Chen S. Identification of candidate COVID-19 therapeutics using hPSC-derived lung organoids. bioRxiv. 2020 doi: 10.1101/2020.05.05.079095. https://www.ncbi.nlm.nih.gov/pubmed/32511403 [DOI] [Google Scholar]
- 17.Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., Bram Y., Richardson C., Zhu J., Zhao Z., Redmond D., Houghton S., Nguyen D.T., Xu D., Wang X., Jessurun J., Borczuk A., Huang Y., Johnson J.L., Liu Y., Xiang J., Wang H., Cantley L.C., tenOever B.R., Ho D.D., Pan F.C., Evans T., Chen H.J., Schwartz R.E., Chen S. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589(7841):270–275. doi: 10.1038/s41586-020-2901-9. https://www.ncbi.nlm.nih.gov/pubmed/33116299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H., Cai J.P., Yip C.C., Chan I.H., Wong K.K., Tsang O.T., Chan K.H., Chan J.F., To K.K., Chen H., Yuen K.Y. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26(7):1077–1083. doi: 10.1038/s41591-020-0912-6. https://www.ncbi.nlm.nih.gov/pubmed/32405028 [DOI] [PubMed] [Google Scholar]
- 19.Salahudeen A.A., Choi S.S., Rustagi A., Zhu J., van Unen V., de la O.S., Flynn R.A., Margalef-Catala M., Santos A.J.M., Ju J., Batish A., Usui T., Zheng G.X.Y., Edwards C.E., Wagar L.E., Luca V., Anchang B., Nagendran M., Nguyen K., Hart D.J., Terry J.M., Belgrader P., Ziraldo S.B., Mikkelsen T.S., Harbury P.B., Glenn J.S., Garcia K.C., Davis M.M., Baric R.S., Sabatti C., Amieva M.R., Blish C.A., Desai T.J., Kuo C.J. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 2020;588(7839):670–675. doi: 10.1038/s41586-020-3014-1. https://www.ncbi.nlm.nih.gov/pubmed/33238290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao B., Ni C., Gao R., Wang Y., Yang L., Wei J., Lv T., Liang J., Zhang Q., Xu W., Xie Y., Wang X., Yuan Z., Liang J., Zhang R., Lin X. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11(10):771–775. doi: 10.1007/s13238-020-00718-6. https://www.ncbi.nlm.nih.gov/pubmed/32303993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B.Z., Chu H., Han S., Shuai H., Deng J., Hu Y.F., Gong H.R., Lee A.C., Zou Z., Yau T., Wu W., Hung I.F., Chan J.F., Yuen K.Y., Huang J.D. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30(10):928–931. doi: 10.1038/s41422-020-0390-x. https://www.ncbi.nlm.nih.gov/pubmed/32753756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faden H., Callanan V., Pizzuto M., Nagy M., Wilby M., Lamson D., Wrotniak B., Juretschko S., St George K. The ubiquity of asymptomatic respiratory viral infections in the tonsils and adenoids of children and their impact on airway obstruction. Int. J. Pediatr. Otorhinolaryngol. 2016;90:128–132. doi: 10.1016/j.ijporl.2016.09.006. https://www.ncbi.nlm.nih.gov/pubmed/27729119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nave H., Gebert A., Pabst R. Morphology and immunology of the human palatine tonsil. Anat. Embryol. 2001;204(5):367–373. doi: 10.1007/s004290100210. https://www.ncbi.nlm.nih.gov/pubmed/11789984 [DOI] [PubMed] [Google Scholar]
- 24.Perry M., Whyte A. Immunology of the tonsils. Immunol. Today. 1998;19(9):414–421. doi: 10.1016/s0167-5699(98)01307-3. https://www.ncbi.nlm.nih.gov/pubmed/9745205 [DOI] [PubMed] [Google Scholar]
- 25.Kang S.Y.C., Kannan N., Zhang L., Martinez V., Rosin M.P., Eaves C.J. Characterization of epithelial progenitors in normal human palatine tonsils and their HPV16 E6/E7-induced perturbation. Stem Cell Rep. 2015;5(6):1210–1225. doi: 10.1016/j.stemcr.2015.09.020. https://www.ncbi.nlm.nih.gov/pubmed/26527383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casteleyn C., Breugelmans S., Simoens P., Van den Broeck W. The tonsils revisited: review of the anatomical localization and histological characteristics of the tonsils of domestic and laboratory animals. Clin. Dev. Immunol. 2011;2011:472460. doi: 10.1155/2011/472460. https://www.ncbi.nlm.nih.gov/pubmed/21869895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J., Haft A., Vries R.G., Grompe M., Clevers H. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494(7436):247–250. doi: 10.1038/nature11826. https://www.ncbi.nlm.nih.gov/pubmed/23354049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. https://www.ncbi.nlm.nih.gov/pubmed/19329995 [DOI] [PubMed] [Google Scholar]
- 29.Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23(5):393–410. doi: 10.1016/j.molmed.2017.02.007. https://www.ncbi.nlm.nih.gov/pubmed/28341301 [DOI] [PubMed] [Google Scholar]
- 30.Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. https://www.ncbi.nlm.nih.gov/pubmed/21889923 [DOI] [PubMed] [Google Scholar]
- 31.Clevers H. Modeling development and disease with organoids. Cell. 2016;165(7):1586–1597. doi: 10.1016/j.cell.2016.05.082. https://www.ncbi.nlm.nih.gov/pubmed/27315476 [DOI] [PubMed] [Google Scholar]
- 32.Ren W., Lewandowski B.C., Watson J., Aihara E., Iwatsuki K., Bachmanov A.A., Margolskee R.F., Jiang P. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl. Acad. Sci. U. S. A. 2014;111(46):16401–16406. doi: 10.1073/pnas.1409064111. https://www.ncbi.nlm.nih.gov/pubmed/25368147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M., Liu Y., Chen Y.G. Generation of 3D human gastrointestinal organoids: principle and applications. Cell Regen. 2020;9(1):6. doi: 10.1186/s13619-020-00040-w. https://www.ncbi.nlm.nih.gov/pubmed/32588198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartfeld S., Bayram T., van de Wetering M., Huch M., Begthel H., Kujala P., Vries R., Peters P.J., Clevers H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148(1):126–136. doi: 10.1053/j.gastro.2014.09.042. https://www.ncbi.nlm.nih.gov/pubmed/25307862 e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M.M., Ellis E., van Wenum M., Fuchs S.A., de Ligt J., van de Wetering M., Sasaki N., Boers S.J., Kemperman H., de Jonge J., Ijzermans J.N., Nieuwenhuis E.E., Hoekstra R., Strom S., Vries R.R., van der Laan L.J., Cuppen E., Clevers H. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160(1–2):299–312. doi: 10.1016/j.cell.2014.11.050. https://www.ncbi.nlm.nih.gov/pubmed/25533785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drost J., van Jaarsveld R.H., Ponsioen B., Zimberlin C., van Boxtel R., Buijs A., Sachs N., Overmeer R.M., Offerhaus G.J., Begthel H., Korving J., van de Wetering M., Schwank G., Logtenberg M., Cuppen E., Snippert H.J., Medema J.P., Kops G.J., Clevers H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521(7550):43–47. doi: 10.1038/nature14415. https://www.ncbi.nlm.nih.gov/pubmed/25924068 [DOI] [PubMed] [Google Scholar]
- 37.Karthaus W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C.M., Gao D., Begthel H., Sachs N., Vries R.G.J., Cuppen E., Chen Y., Sawyers C.L., Clevers H.C. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–175. doi: 10.1016/j.cell.2014.08.017. https://www.ncbi.nlm.nih.gov/pubmed/25201529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buzhor E., Harari-Steinberg O., Omer D., Metsuyanim S., Jacob-Hirsch J., Noiman T., Dotan Z., Goldstein R.S., Dekel B. Kidney spheroids recapitulate tubular organoids leading to enhanced tubulogenic potency of human kidney-derived cells. Tissue Eng. Part A. 2011;17(17–18):2305–2319. doi: 10.1089/ten.TEA.2010.0595. https://www.ncbi.nlm.nih.gov/pubmed/21542667 [DOI] [PubMed] [Google Scholar]
- 39.Itoh T. Stem/progenitor cells in liver regeneration. Hepatology. 2016;64(2):663–668. doi: 10.1002/hep.28661. https://www.ncbi.nlm.nih.gov/pubmed/27227904 [DOI] [PubMed] [Google Scholar]
- 40.Jung P., Sato T., Merlos-Suarez A., Barriga F.M., Iglesias M., Rossell D., Auer H., Gallardo M., Blasco M.A., Sancho E., Clevers H., Batlle E. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 2011;17(10):1225–1227. doi: 10.1038/nm.2470. https://www.ncbi.nlm.nih.gov/pubmed/21892181 [DOI] [PubMed] [Google Scholar]
- 41.Kessler M., Hoffmann K., Brinkmann V., Thieck O., Jackisch S., Toelle B., Berger H., Mollenkopf H.J., Mangler M., Sehouli J., Fotopoulou C., Meyer T.F. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 2015;6:8989. doi: 10.1038/ncomms9989. https://www.ncbi.nlm.nih.gov/pubmed/26643275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trzpis M., McLaughlin P.M., de Leij L.M., Harmsen M.C. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am. J. Pathol. 2007;171(2):386–395. doi: 10.2353/ajpath.2007.070152. https://www.ncbi.nlm.nih.gov/pubmed/17600130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zha K., Yang Y., Tian G., Sun Z., Yang Z., Li X., Sui X., Liu S., Zhao J., Guo Q. Nerve growth factor (NGF) and NGF receptors in mesenchymal stem/stromal cells: impact on potential therapies. Stem Cell. Trans. Med. 2021;10(7):1008–1020. doi: 10.1002/sctm.20-0290. https://www.ncbi.nlm.nih.gov/pubmed/33586908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo C., Liu H., Zhang B.H., Cadaneanu R.M., Mayle A.M., Garraway I.P. Epcam, CD44, and CD49f distinguish sphere-forming human prostate basal cells from a subpopulation with predominant tubule initiation capability. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0034219. https://www.ncbi.nlm.nih.gov/pubmed/22514625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos C.P., Lapi E., Martinez de Villarreal J., Alvaro-Espinosa L., Fernandez-Barral A., Barbachano A., Dominguez O., Laughney A.M., Megias D., Munoz A., Real F.X. Urothelial organoids originating from Cd49f(high) mouse stem cells display Notch-dependent differentiation capacity. Nat. Commun. 2019;10(1):4407. doi: 10.1038/s41467-019-12307-1. https://www.ncbi.nlm.nih.gov/pubmed/31562298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krebsbach P.H., Villa-Diaz L.G. The role of integrin alpha6 (CD49f) in stem cells: more than a conserved biomarker. Stem Cell. Dev. 2017;26(15):1090–1099. doi: 10.1089/scd.2016.0319. https://www.ncbi.nlm.nih.gov/pubmed/28494695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woods R.S.R., Keegan H., White C., Tewari P., Toner M., Kennedy S., O'Regan E.M., Martin C.M., Timon C.V.I., O'Leary J.J. Cytokeratin 7 in oropharyngeal squamous cell carcinoma: a junctional biomarker for human papillomavirus-related tumors. Cancer Epidemiol. Biomarkers Prev. 2017;26(5):702–710. doi: 10.1158/1055-9965.EPI-16-0619. https://www.ncbi.nlm.nih.gov/pubmed/28082347 [DOI] [PubMed] [Google Scholar]
- 48.Howie A.J. Scanning and transmission electron microscopy on the epithelium of human palatine tonsils. J. Pathol. 1980;130(2):91–98. doi: 10.1002/path.1711300205. https://www.ncbi.nlm.nih.gov/pubmed/7365575 [DOI] [PubMed] [Google Scholar]
- 49.Ellison D., Mugler A., Brennan M.D., Lee S.H., Huebner R.J., Shamir E.R., Woo L.A., Kim J., Amar P., Nemenman I., Ewald A.J., Levchenko A. Cell-cell communication enhances the capacity of cell ensembles to sense shallow gradients during morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 2016;113(6):E679–E688. doi: 10.1073/pnas.1516503113. https://www.ncbi.nlm.nih.gov/pubmed/26792522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sachse F., Ahlers F., Stoll W., Rudack C. Neutrophil chemokines in epithelial inflammatory processes of human tonsils. Clin. Exp. Immunol. 2005;140(2):293–300. doi: 10.1111/j.1365-2249.2005.02773.x. https://www.ncbi.nlm.nih.gov/pubmed/15807854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. https://www.ncbi.nlm.nih.gov/pubmed/32142651 e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell. Mol. Life Sci. 2004;61(21):2738–2743. doi: 10.1007/s00018-004-4242-5. https://www.ncbi.nlm.nih.gov/pubmed/15549175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Sign. Trans. Target Ther. 2020;5(1):92. doi: 10.1038/s41392-020-0184-0. https://www.ncbi.nlm.nih.gov/pubmed/32532959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neufeldt C.J., Cerikan B., Cortese M., Frankish J., Lee J.Y., Plociennikowska A., Heigwer F., Prasad V., Joecks S., Burkart S.S., Zander D.Y., Subramanian B., Gimi R., Padmanabhan S., Iyer R., Gendarme M., El Debs B., Halama N., Merle U., Boutros M., Binder M., Bartenschlager R. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-kappaB. Commun. Biol. 2022;5(1):45. doi: 10.1038/s42003-021-02983-5. https://www.ncbi.nlm.nih.gov/pubmed/35022513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patra T., Meyer K., Geerling L., Isbell T.S., Hoft D.F., Brien J., Pinto A.K., Ray R.B., Ray R. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog. 2020;16(12) doi: 10.1371/journal.ppat.1009128. https://www.ncbi.nlm.nih.gov/pubmed/33284859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernard C., Merval R., Lebret M., Delerive P., Dusanter-Fourt I., Lehoux S., Creminon C., Staels B., Maclouf J., Tedgui A. Oncostatin M induces interleukin-6 and cyclooxygenase-2 expression in human vascular smooth muscle cells : synergy with interleukin-1beta. Circ. Res. 1999;85(12):1124–1131. doi: 10.1161/01.res.85.12.1124. https://www.ncbi.nlm.nih.gov/pubmed/10590238 [DOI] [PubMed] [Google Scholar]
- 57.Casari I., Manfredi M., Metharom P., Falasca M. Dissecting lipid metabolism alterations in SARS-CoV-2. Prog. Lipid Res. 2021;82:101092. doi: 10.1016/j.plipres.2021.101092. https://www.ncbi.nlm.nih.gov/pubmed/33571544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Azoulay D., Shehadeh M., Chepa S., Shaoul E., Baroum M., Horowitz N.A., Kaykov E. Recovery from SARS-CoV-2 infection is associated with serum BDNF restoration. J. Infect. 2020;81(3):e79–e81. doi: 10.1016/j.jinf.2020.06.038. https://www.ncbi.nlm.nih.gov/pubmed/32569603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang L., Xu D., Huang Q., Yang G., Zhang M., Bi J., Shan J., Li E., He S. Characterization of tonsil microbiota and their effect on adenovirus reactivation in tonsillectomy samples. Microbiol. Spectr. 2021;9(2) doi: 10.1128/Spectrum.01246-21. https://www.ncbi.nlm.nih.gov/pubmed/34668748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jang Y., Shin J.S., Lee M.K., Jung E., An T., Kim U.I., Kim K., Kim M. Comparison of antiviral activity of gemcitabine with 2'-fluoro-2'-deoxycytidine and combination therapy with remdesivir against SARS-CoV-2. Int. J. Mol. Sci. 2021;22(4) doi: 10.3390/ijms22041581. https://www.ncbi.nlm.nih.gov/pubmed/33557278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brook I. The clinical microbiology of Waldeyer's ring. Otolaryngol. Clin. N. Am. 1987;20(2):259–272. https://www.ncbi.nlm.nih.gov/pubmed/3299209 [PubMed] [Google Scholar]
- 62.Chatterjee S., Do Kang S., Alam S., Salzberg A.C., Milici J., van der Burg S.H., Freeman W., Meyers C. Tissue-specific gene expression during productive human papillomavirus 16 infection of cervical, Foreskin, and tonsil epithelium. J. Virol. 2019;93(17):e00915–e00919. doi: 10.1128/JVI.00915-19. https://www.ncbi.nlm.nih.gov/pubmed/31189705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mellin H., Dahlgren L., Munck-Wikland E., Lindholm J., Rabbani H., Kalantari M., Dalianis T. Human papillomavirus type 16 is episomal and a high viral load may be correlated to better prognosis in tonsillar cancer. Int. J. Cancer. 2002;102(2):152–158. doi: 10.1002/ijc.10669. https://www.ncbi.nlm.nih.gov/pubmed/12385011 [DOI] [PubMed] [Google Scholar]
- 64.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist S.R., Gralinski L.E., Schafer A., Dang H., Gilmore R., Nakano S., Sun L., Fulcher M.L., Livraghi-Butrico A., Nicely N.I., Cameron M., Cameron C., Kelvin D.J., de Silva A., Margolis D.M., Markmann A., Bartelt L., Zumwalt R., Martinez F.J., Salvatore S.P., Borczuk A., Tata P.R., Sontake V., Kimple A., Jaspers I., O'Neal W.K., Randell S.H., Boucher R.C., Baric R.S. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182(2):429–446. doi: 10.1016/j.cell.2020.05.042. https://www.ncbi.nlm.nih.gov/pubmed/32526206 e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the 'Cytokine Storm' in COVID-19. J. Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. https://www.ncbi.nlm.nih.gov/pubmed/32283152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qian G., Zhang Y., Xu Y., Hu W., Hall I.P., Yue J., Lu H., Ruan L., Ye M., Mei J. Reduced inflammatory responses to SARS-CoV-2 infection in children presenting to hospital with COVID-19 in China. EClinicalMedicine. 2021;34:100831. doi: 10.1016/j.eclinm.2021.100831. https://www.ncbi.nlm.nih.gov/pubmed/33880437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wagar L.E., Salahudeen A., Constantz C.M., Wendel B.S., Lyons M.M., Mallajosyula V., Jatt L.P., Adamska J.Z., Blum L.K., Gupta N., Jackson K.J.L., Yang F., Roltgen K., Roskin K.M., Blaine K.M., Meister K.D., Ahmad I.N., Cortese M., Dora E.G., Tucker S.N., Sperling A.I., Jain A., Davies D.H., Felgner P.L., Hammer G.B., Kim P.S., Robinson W.H., Boyd S.D., Kuo C.J., Davis M.M. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 2021;27(1):125–135. doi: 10.1038/s41591-020-01145-0. https://www.ncbi.nlm.nih.gov/pubmed/33432170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J., Fang S., Xiao H., Chen B., Tam J.P., Liu D.X. Interaction of the coronavirus infectious bronchitis virus membrane protein with beta-actin and its implication in virion assembly and budding. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0004908. https://www.ncbi.nlm.nih.gov/pubmed/19287488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. https://www.ncbi.nlm.nih.gov/pubmed/32358202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Millius A., Weiner O.D. Chemotaxis in neutrophil-like HL-60 cells. Methods Mol. Biol. 2009;571:167–177. doi: 10.1007/978-1-60761-198-1_11. https://www.ncbi.nlm.nih.gov/pubmed/19763966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jang Y., Shin H., Lee M.K., Kwon O.S., Shin J.S., Kim Y.I., Kim C.W., Lee H.R., Kim M. Antiviral activity of lambda-carrageenan against influenza viruses and severe acute respiratory syndrome coronavirus 2. Sci. Rep. 2021;11(1):821. doi: 10.1038/s41598-020-80896-9. https://www.ncbi.nlm.nih.gov/pubmed/33436985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.