Abstract
Human natural killer (NK) and innate lymphoid cells (ILCs) include diverse specialized phenotypic and functional subsets that reflect their roles as innate immune effector cells present in tissue and circulation. In recent years, significant advances have been made in better defining their tissue resident phenotypes, developmental pathways, and phenotypic plasticity. Here we offer a brief review of new insights into human NK cell diversity specifically defined by next generation sequencing and single-cell transcriptomic studies and integrate these into our current models of human NK cell developmental trajectories and mature subsets. These studies highlight both a deeper understanding of innate lymphoid cell differentiation and homeostasis and underscore critical questions that remain outstanding in the field.
Keywords: natural killer cell, hematopoiesis, scRNA-Seq
Introduction
Human natural killer (NK) cells can be isolated from multiple tissues including peripheral blood (PB), liver, spleen, bone marrow, lung, lymph node, and uterus [1-3]. While it is becoming increasingly evident that many of these sites can host both tissue resident and transitory cells, the primary sites of NK cell development are most frequently defined as bone marrow and secondary lymphoid tissue (SLT) [4-8]. Bone marrow is the site of adult hematopoietic precursors that give rise to common lymphoid precursors from which innate lymphoid cells (ILCs) and NK cells are generated. The isolation of an early NK cell precursor and NK cell developmental intermediates from secondary lymphoid tissue suggests that an early precursor exits from the bone marrow and seeds SLT [6,8]. In addition to the earliest CD34dim CD45RA+ integrin β7bright precursor that is found in PB and SLT [8], more restricted ILC precursors are also found at low frequency in circulation [9-11]. While the identification and classification of innate lymphoid cell subsets have greatly increased our understanding of NK cell ontology [12,13], many outstanding questions remain about the trajectory and sites of human NK cell development.
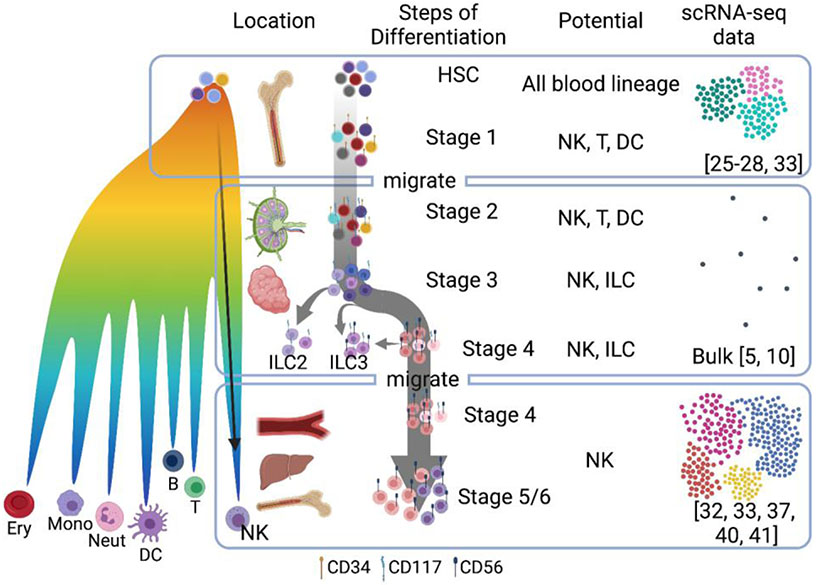
Human NK cell development is frequently described as a linear model beginning with the earliest precursor (stage 1) and culminating in terminally mature CD56dim CD16+ (stage 5) or CD56dim CD16+ CD57+ (stage 6) cells [14]. Briefly, stage 1 cells are defined as CD34+ CD117− CD94− precursors, found in bone marrow and SLT, that are not responsive to IL-15 [15]. Stage 1 NK cells gain CD117 expression and become IL-15 responsive stage 2 cells, yet retain multilineage potential and can give rise to T cells or DC cells in addition to NK cells when cultured ex vivo [6]. Stage 3 cells are defined by loss of CD34 expression and commitment to the NK/ILC lineage, with CD56 expression marking divergence of conventional NK cells and ILC3 precursors and CD56 negative cells representing the emerging ILC2 population [10]. At stage 4, NK cells become CD56bright and start expressing NK cell markers and cytokines, and further commit to the conventional NK cell lineage as stage 4B NK cells can be defined by acquisition of NKp80 [16]. About 10% of NK cells in PB are stage 4B CD56bright cells, whereas terminally differentiated stages 5 and 6 NK cells are the predominant subsets found in PB where they comprise ~90% of NK cells [16-18]. Stages 4 to 6 NK cells are also present in tissue, such as lung, gut, and tonsil, where they express NK cell maturation markers similar to corresponding NK cell subsets in PB. However, despite phenotypic similarities between PB and tissue-resident NK cells, unique transcriptional signatures reveal the cellular environment as a source of NK cell heterogeneity within the same apparent developmental stage [5,19,20].
With the identification of ILC subsets and further dissection of the stages of human NK cell development, previous linear models of NK cell development have given rise to branching models that represent both lineage plasticity and dominant lineage trajectories. Single-cell transcriptomics studies have particularly provided important insight into the relationship between NK cell subsets, particularly in bone marrow, which is thought to be the primary site of human NK cell development, and PB, a common source of circulating mature NK cells. Similar studies have used single-cell approaches to describe the heterogeneity of ILC1, ILC2, ILC3, NK cells, and lymphoid tissue inducer (LTi) ILCs in different organs [2,13,20]. Since ILC subtypes and NK cells share many cell surface markers and exhibit lineage plasticity, it is important to delineate ILC and NK cell subsets when considering single cell datasets. Most notably, NK cells are the only cytotoxic ILC subset and can be distinguished by expression of molecules relating to effector function. In addition, the development of ILC subsets depends on distinct transcription factors; ILC1, ILC2, ILC3 require GATA3, whereas NK cell development depends on EOMES [21]. Recent reviews dissect such ILC diversity and their developmental landscape in relation to conventional NK cell subsets in detail [12,21,22]. Here, we will keep our focus on understanding the heterogeneity of conventional NK cells and their precursors.
Redefining hematopoiesis with single-cell sequencing
Hematopoiesis is classically described as a tree-like model where HSCs unidirectionally give rise to more lineage-restricted progenitors leading to terminal differentiation [23]. This model is commonly supported by experiments in which specific subpopulations of blood cells are isolated with antibodies and differentiated to show their limited lineage potential. Recent scRNA-seq studies have revealed a more continuous transcriptional differentiation landscape that lacks classically defined precursor stages (Fig. 1) [24-28], (reviewed in [29]). The differences between more discrete surface marker expression patterns and transcriptome-wide measures that define heterogeneous intermediates suggest that cells undergo much more subtle and complex transcriptional calculation that precedes a surface marker phenotype. Single-cell sequencing technologies enable us to study such lineage priming at high resolution in addition to defining cellular heterogeneity and trajectories of development that may not be inferred by lineage potential experiments.
Figure 1. scRNA-seq reveals the developmental landscape of NK cells.
Left: a revised model of hematopoiesis with continuous cell states. Right: our current knowledge of NK cell differentiation steps including references for human datasets discussed in the text.
With this in mind, we can consider what such experiments tell us about the developmental trajectory of human NK cells. Several studies have performed scRNA-seq to interrogate hematopoiesis using bone marrow cells from a single time point. Due to the relatively fast turnover and abundance of precursors in the bone marrow, most steps from progenitor to mature blood cells can be captured without sequential sample collection and aligned along pseudotime [27,28]. However, as shown in the bone marrow human cell atlas data [27], T cells and NK cells appear disconnected from the bone marrow landscape that identifies developmental lineages from the HSCs. Such discontinuity signifies that due to the spatial separation of the developing T cells in the thymus and NK cells presumably in SLT or other tissues, intermediate populations are extremely rare in the bone marrow, which presents a unique challenge in mapping their developmental landscape. Whether this discontinuity supports extramedullary sites of NK cell development as the dominant sites in adults, or is a technical feature of not sequencing enough cells to define intermediate populations, is unclear.
Capturing NK cell developmental subsets
Murine NK cells are thought to predominantly develop in the bone marrow (reviewed in [30]). The trajectory of murine NK cell development has been captured by isolating CD3e− CD122+cells, thought to contain both immature and mature NK cells, and performing scRNA-seq [31]. This approach identified five NK developmental subsets: one immature, one mature, and three transitional NK cell groups, and further defined mTORC1 and mTORC2 signaling as key determinants of NK cell maturation dictating movement between immature and transitional stages of development [31].
As human NK cells have been shown to develop in other sites besides bone marrow, fully understanding human NK cell development requires sequencing of multiple organs. Crinier et al. [32] performed scRNA-seq on NK cells from the spleen to enable comparison between mouse and human splenic NK cells. The earliest (CD56neg) NK cell precursors would not have been included in this study, as CD56+ cells from the human spleen were sequenced. Unbiased analysis demonstrated that the cells isolated and analyzed were primarily mature NK cell subsets analogous to CD56bright and CD56dim subsets [32]. However, as most NK cells detected by flow cytometry in spleen are stages 4 and 5 (CD56bright and CD56dim) [4], it is not surprising that earlier stages of NK cell development were not identified. Similarly, a comparison of ILC subsets from human tonsil included CD45+Lin−CD56+CD127−NKG2A+CD16− NK cells, which enabled the single-cell transcriptomic analysis of NK cells, but not early NK cell precursors [13]. Bulk RNA-seq data of early NK precursors from tonsil and blood have outlined developmental trajectories using phenotypically sorted subsets (Figure 1) [10,32]. However, scRNA-seq in this space is still required to populate this landscape further and delineate the developmental process.
While the trajectory of mature human NK cell development has not yet been captured in high resolution, a recent study has defined embryonic human NK cell trajectories within the fetal liver and bone marrow without discontinuity [33]. Using scRNA-seq, the authors show a smooth trajectory from hematopoietic stem cells and multipotent progenitors to NK cells and other hematopoietic cell types. Interestingly, while NK cells were identified, neither T cells nor ILCs were found in fetal bone marrow, validating the observation that T cell development is occurring in the thymus and further suggesting that fetal ILC development may similarly be occurring at other tissue sites. The differentiation trajectory analysis defined highly proliferative lymphoid-myeloid progenitors (LMPs) expressing genes that include IGLL1, HMGB2, and CD79B that were positioned upstream of mature NK cells and B cells, monocytes, and plasmacytoid dendritic cells. This study demonstrates how data from multiple donors and organs can be integrated to generate a comprehensive view of NK cell development. Collecting enough NK cells and progenitors from SLT without enrichment or FACS sorting presents a significant challenge due to their low frequency [34]. For example, a scRNA-seq experiment from mouse spleen with minimal selection defined 230 NK cells from 9,552 sequenced cells [35]. Therefore, strategic gating with developmental markers and pooling samples from multiple donors, as was done to interrogate fetal bone marrow and liver, may be necessary to overcome this difficulty but would likely provide a deeper understanding of the trajectory of human NK cell development in adult tissue.
Mapping diversity of mature NK cell subsets with novel transcriptional signatures
The diversity of NK cells at the protein level has been demonstrated at a single-cell level with high-dimensional flow cytometry and cytometry by time-of-flight (CyTOF) [3,36]. Single-cell transcriptomics provides a way of further dissecting the cells with transcriptome-wide measures and gaining a more comprehensive view of the identified subpopulations. Single-cell transcriptomics as applied to NK cell biology has been most frequently used to better understand the identity and origins of mature NK cell subsets. Crinier et al. generated scRNA-seq datasets from spleen, blood, and bone marrow NK cells to better define and understand the structure and heterogeneities of NK cell subsets [32,37]. In both humans and mice, NK cells most prominently clustered according to their tissue of origin, consistent with other studies using bulk RNA-Seq showing that phenotypically equivalent subsets from tissue and blood have unique transcriptional profiles [5,32]. Unbiased analysis revealed two clusters of human NK cells in the blood, hNK_Bl1, and hNK_Bl2, corresponding to known subsets of more cytotoxic CD56dim and cytokine producing CD56bright populations, respectively [5,32]. Spleen and bone marrow NK cells also showed cell clusters that aligned with CD56dim and CD56bright cells, but more importantly, showed additional subpopulations of relatively smaller sizes. The characteristics and functionality of the bone marrow subsets have been debated, but the clustering of subgroups is robust [37-39]. Gene ontology of the differentially expressed genes indicates that the differences between these NK cell subtypes are likely to be biologically meaningful.
The massively parallel and unbiased nature of scRNA-seq also enables discovering new transcriptional signatures within subpopulations, which can provide a robust way to identify CD56+ NK cells from other datasets with mixed cell populations. Crinier et al [32] used correlation with NKp46 expression to define a consensus NK cell gene signature with 13 genes. The NK core gene list from humans contains activating receptors (CD160,KLRC3 [NKG2E], CD244 [2B4], KLRF1 [NKp80]), the inhibitory receptor KLRC1 [NKG2A], antimicrobial proteins (GNLY), cytolytic proteins (PRF1), inflammatory cytokines (XCL2), and transcription factors, recapitulating the functional characteristics of NK cells. The consensus list for mouse NK cells has genes from similar categories; however, only two of the genes are shared between humans and mice (IL18RAP, PRF1). Such differences emphasize how human and mouse immune systems have divergently evolved to adapt to their environment while conserving the biological identity of NK cells.
Expression patterns of well-characterized genes are often used to define NK cell subpopulations. For example, IL7R, SELL, GZMK, and KLRC1 are commonly used to identify CD56bright NK cells [40,41]. In addition to known marker genes, clustering analysis reports new transcriptional signatures for each subpopulation. In particular, Yang et al. [41] identified CD44 and XCL1 as novel markers for CD56bright NK cells, and Smith et al. [40] discovered a shared signature between CD56bright and terminally differentiated NK cells; CD69, DUSP1, FOS, and JUN. Single-cell sequencing also provides an opportunity to identify smaller subpopulations that were impossible to detect in bulk data processing. Two studies have sequenced a greater number of blood NK cells after enriching them from PBMCs (3061 cells, [41]; 8462 cells, [40]) and further dissected this population into as many as seven clusters. In addition to CD56bright cells, both studies dissect CD56dim populations into active, mature, and terminally differentiated populations. Smith et al., with more than 8000 cells, describe additional minor populations, including type1 INF responding cells, cytokine-induced memory like (CIML) cells, and a novel NK cell population with decreased expression of ribosomal genes [40]. Smith et al. sequenced the greatest number of cells and included a cytokine-induced activation condition that increased the dimensionality of the data. With an in silico down-sampling experiment, they demonstrate that processing a lower number of cells results in a smaller number of clusters where the minor cell populations get absorbed into larger populations. This result highlights the importance of depth of the sequencing and breadth of the sample collection in discovering rare novel subsets with confidence.
Moving beyond the transcriptome
Recent studies have explored epigenetic mechanisms that shape NK cell heterogeneity by pairing sorted bulk RNA-seq with ChIP-seq or ATAC-seq [42,43]. These studies demonstrate that NK subpopulations are transcriptionally and epigenetically distinct and further identify the transcription factor Bcl11b, or an axis of transcription factors (TCF1-LEF1-MYC axis), that shape NK cell subpopulations [42,43]. Epigenetic surveys at the single-cell level are becoming available to show another aspect of such observations. Ranzoni et al. [33] performed scRNA-seq and scATAC-seq on fetal human liver and bone marrow cells. The authors discovered further heterogeneity within transcriptionally homogenous HSCs and multipotent progenitors. Specifically, ATAC-seq data revealed that HSCs/MPPs could have different activity levels of transcription factors that prime cells at promoters before lineage commitment can be transcriptionally detected. This study demonstrates how single-cell multi-omics studies can reveal a new level of heterogeneity within hierarchical epigenetic mechanisms that precedes transcriptional readout and adds further complexity to our re-designed model of hematopoiesis. Further, these multi-omics studies highlight how bulk sequencing, with its depth, can be used to detect and reveal the subtle differences between known subpopulations, and single-cell sequencing, with its breadth, can complement to reveal further heterogeneity within a cell population.
Conclusions
Hematopoiesis and immune cell development are processes of making cells of ever-increasing heterogeneity. With scRNA-seq, we have started to perceive the variety of cell types, developmental intermediates, and other aspects that were not previously described. As other single-cell sequencing technologies mature, we are gaining further insight into the transcriptomic and epigenetic complexity of developing NK cells. We now must move beyond observations to gain a better understanding of the functional significance of this complexity. In addition, as human NK cells undergo trafficking during development, multiple organs must be analyzed to map the developmental landscape. Finally, the spatial distribution of NK cells and their precursors in tissue, and their local cell-cell interactions, must be investigated to fully understand the complexity and road map of developing NK cells.
Acknowledgments
The authors wish to thank Drs. Bethany Mundy-Bosse and Aharon Freud for their critical reading of the manuscript and apologize to all authors whose work could not be included due to space constraints. This work was supported in part by NIH-NIAID R01137275 to EMM.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brownlie D, Scharenberg M, Mold JE, Hård J, Kekäläinen E, Buggert M, Nguyen S, Wilson JN, Al-Ameri M, Ljunggren H-G, et al. : Expansions of adaptive-like NK cells with a tissue-resident phenotype in human lung and blood. Proc National Acad Sci 2021, 118:e2016580118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marquardt N, Kekäläinen E, Chen P, Kvedaraite E, Wilson JN, Ivarsson MA, Mjösberg J, Berglin L, Säfholm J, Manson ML, et al. : Human lung natural killer cells are predominantly comprised of highly differentiated hypofunctional CD69–CD56dim cells. J Allergy Clin Immun 2017, 139:1321–1330.e4. [DOI] [PubMed] [Google Scholar]
- 3.Dogra P, Rancan C, Ma W, Toth M, Senda T, Carpenter DJ, Kubota M, Matsumoto R, Thapa P, Szabo PA, et al. : Tissue Determinants of Human NK Cell Development, Function, and Residence. Cell 2020, 180:749–763.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eissens DN, Spanholtz J, van der Meer A, van Cranenbroek B, Dolstra H, Kwekkeboom J, Preijers FWMB, Joosten I: Defining Early Human NK Cell Developmental Stages in Primary and Secondary Lymphoid Tissues. Plos One 2012, 7:e30930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegewisch-Solloa E, Seo S, Mundy-Bosse BL, Mishra A, Waldman EH, Maurrasse S, Grunstein E, Connors TJ, Freud AG, Mace EM: Differential Integrin Adhesome Expression Defines Human NK Cell Residency and Developmental Stage. J Immunol 2021, doi: 10.4049/jimmunol.2100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, Caligiuri MA: Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Medicine 2006, 203:1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scoville SD, Mundy-Bosse BL, Zhang MH, Chen L, Zhang X, Keller KA, Hughes T, Chen L, Cheng S, Bergin SM, et al. : A Progenitor Cell Expressing Transcription Factor RORγt Generates All Human Innate Lymphoid Cell Subsets. Immunity 2016, 44:1140–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, Hughes TL, Marburger TB, Sung J, Baiocchi RA, et al. : A Human CD34(+) Subset Resides in Lymph Nodes and Differentiates into CD56brightNatural Killer Cells. Immunity 2005, 22:295–304. [DOI] [PubMed] [Google Scholar]
- 9.Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, Serafini N, Puel A, Bustamante J, Surace L, et al. : Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell 2017, 168:1086–1100.e10. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Youssef Y, Robinson C, Ernst GF, Carson MY, Young KA, Scoville SD, Zhang X, Harris R, Sekhri P, et al. : CD56 Expression Marks Human Group 2 Innate Lymphoid Cell Divergence from a Shared NK Cell and Group 3 Innate Lymphoid Cell Developmental Pathway. Immunity 2018, 49:464–476.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bal SM, Golebski K, Spits H: Plasticity of innate lymphoid cell subsets. Nat Rev Immunol 2020, 20:552–565. [DOI] [PubMed] [Google Scholar]
- 12.Vivier E, Artis D, Colonna M, Diefenbach A, Santo JPD, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. : Innate Lymphoid Cells: 10 Years On. Cell 2018, 174:1054–1066. [DOI] [PubMed] [Google Scholar]
- 13.Björklund ÅK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, Sandberg R, Mjösberg J: The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol 2016, 17:451 460. [DOI] [PubMed] [Google Scholar]
- 14.Scoville SD, Freud AG, Caligiuri MA: Modeling Human Natural Killer Cell Development in the Era of Innate Lymphoid Cells. Front Immunol 2017, 8:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freud AG, Caligiuri MA: Human natural killer cell development. Immunol Rev 2006, 214:56–72. [DOI] [PubMed] [Google Scholar]
- 16.Freud AG, Keller KA, Scoville SD, Mundy-Bosse BL, Cheng S, Youssef Y, Hughes T, Zhang X, Mo X, Porcu P, et al. : NKp80 Defines a Critical Step during Human Natural Killer Cell Development. Cell Reports 2016, 16:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan A, Hong D-L, Atzberger A, Kollnberger S, Filer AD, Buckley CD, McMichael A, Enver T, Bowness P: CD56bright Human NK Cells Differentiate into CD56dim Cells: Role of Contact with Peripheral Fibroblasts. J Immunol 2007, 179:89–94. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, Freud AG, Caligiuri MA: Location and cellular stages of natural killer cell development. Trends Immunol 2013, 34:573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquardt N, Kekäläinen E, Chen P, Lourda M, Wilson JN, Scharenberg M, Bergman P, Al-Ameri M, Hård J, Mold JE, et al. : Unique transcriptional and protein-expression signature in human lung tissue-resident NK cells. Nat Commun 2019, 10:3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazzurana L, Czarnewski P, Jonsson V, Wigge L, Ringnér M, Williams TC, Ravindran A, Björklund ÅK, Säfholm J, Nilsson G, et al. : Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length single-cell RNA-sequencing. Cell Res 2021, 31:554–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stokic-Trtica V, Diefenbach A, Klose CSN: NK Cell Development in Times of Innate Lymphoid Cell Diversity. Front Immunol 2020, 11:813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diefenbach A, Colonna M, Romagnani C: The ILC World Revisited. Immunity 2017, 46:327–332. [DOI] [PubMed] [Google Scholar]
- 23.Weissman IL: Stem Cells Units of Development, Units of Regeneration, and Units in Evolution. Cell 2000, 100:157–168. [DOI] [PubMed] [Google Scholar]
- 24.Rodda LB, Lu E, Bennett ML, Sokol CL, Wang X, Luther SA, Barres BA, Luster AD, Ye CJ, Cyster JG: Single-Cell RNA Sequencing of Lymph Node Stromal Cells Reveals Niche-Associated Heterogeneity. Immunity 2018, 48:1014–1028.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, Hirche C, Lutz C, Buss EC, Nowak D, et al. : Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol 2017, 19:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, Winter D, Lara-Astiaso D, Gury M, Weiner A, et al. : Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 2015, 163:1663–1677. [DOI] [PubMed] [Google Scholar]
- 27.Hay SB, Ferchen K, Chetal K, Grimes HL, Salomonis N: The Human Cell Atlas bone marrow single-cell interactive web portal. Exp Hematol 2018, 68:51 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellin D, Loperfido M, Baricordi C, Wolock SL, Montepeloso A, Weinberg OK, Biffi A, Klein AM, Biasco L: A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun 2019, 10:2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watcham S, Kucinski I, Gottgens B: New insights into hematopoietic differentiation landscapes from single-cell RNA sequencing. Blood 2019, 133:1415 1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abel AM, Yang C, Thakar MS, Malarkannan S: Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front Immunol 2018, 9:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang C, Siebert JR, Burns R, Zheng Y, Mei A, Bonacci B, Wang D, Urrutia RA, Riese MJ, Rao S, et al. : Single-cell transcriptome reveals the novel role of T-bet in suppressing the immature NK gene signature. Elite 2020, 9:e51339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **32. Crinier A, Milpied P, Escalière B, Piperoglou C, Galluso J, Balsamo A, Spinelli L, Cervera-Marzal I, Ebbo M, Girard-Madoux M, et al. : High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity 2018, 49:971 986.e5. scRNA-seq of blood and spleen NK cells from both mouse and human identifies subpopulations and their transcriptional signatures and defines a consensus NK cell gene signature
- **33. Ranzoni AM, Tangherloni A, Berest I, Riva SG, Myers B, Strzelecka PM, Xu J, Panada E, Mohorianu I, Zaugg JB, et al. : Integrative Single-Cell RNA-Seq and ATAC-Seq Analysis of Human Developmental Hematopoiesis. Cell Stem Cell 2021, 28:472–487.e7. scRNA-seq and scATAC-seq of immunophenotypic blood cells from fetal liver and bone marrow shows epigenetic variations that precede transcriptional changes in development.
- 34.Melsen JE, Lugthart G, Lankester AC, Schilham MW: Human Circulating and Tissue-Resident CD56bright Natural Killer Cell Populations. Front Immunol 2016, 7:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, Darmanis S, Batson J, Botvinnik O, Chen MB, et al. : Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018, 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, et al. : Genetic and Environmental Determinants of Human NK Cell Diversity Revealed by Mass Cytometry. Sci Transl Med 2013, 5:208ra145–208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **37. Crinier A, Dumas P-Y, Escalière B, Piperoglou C, Gil L, Villacreces A, Vély F, Ivanovic Z, Milpied P, Narni-Mancinelli É, et al. : Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia. Cell Mol Immunol 2021, 18:1290–1304. scRNA-seq of human bone marrow NK cells reveals new mature NK cell subsets and their transcriptional signatures.
- 38.Crinier A, Escalière B, Narni-Mancinelli E, Vivier E: Reply to ‘Comment to: Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia.’ Cell Mol Immunol 2021, 18:1350–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melsen JE, Lugthart G, Dam MM van O, Schilham MW: Comment to: Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia. Cell Mol Immunol 2021, 18:1348–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40. Smith SL, Kennedy PR, Stacey KB, Worboys JD, Yarwood A, Seo S, Solloa EH, Mistretta B, Chatterjee SS, Gunaratne P, et al. : Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood Adv 2020, 4:1388–1406. scRNA-seq of human PB NK cells with stimulation reveals novel NK cell subpopulations and their transcriptional signatures.
- **41. Yang C, Siebert JR, Burns R, Gerbec ZJ, Bonacci B, Rymaszewski A, Rau M, Riese MJ, Rao S, Carlson K-S, et al. : Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun 2019, 10:3931. scRNA-seq of human PB and bone marrow NK cells reveals novel NK cell subpopulations and describes how their distributions differ in an individual with a pathogenic GATA2 variant.
- 42.Collins PL, Celia M, Porter SI, Li S, Gurewitz GL, Hong HS, Johnson RP, Oltz EM, Colonna M: Gene Regulatory Programs Conferring Phenotypic Identities to Human NK Cells. Cell 2019, 176:348–360.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes TD, Pandey RV, Helm EY, Schlums H, Han H, Campbell TM, Drashansky TT, Chiang S, Wu C-Y, Tao C, et al. : The transcription factor Bcl11b promotes both canonical and adaptive NK cell differentiation. Sci Immunol 2021, 6:eabc9801. [DOI] [PMC free article] [PubMed] [Google Scholar]