Abstract
The sequential activation of Nucleotide-binding oligomerization domain, Leucine rich Repeat and Pyrin domain containing protein 1 (Nlrp1) inflammasome, Caspase-1 (Casp1), and Caspase-6 (Casp6) is implicated in primary human neuron cultures and Alzheimer Disease (AD) neurodegeneration. To validate the Nlrp1-Casp1-Casp6 pathway in vivo, the APPSwedish/Indiana J20 AD transgenic mouse model was generated on either a Nlrp1, Casp1 or Casp6 null genetic background and mice were studied at 4–5 months of age. Episodic memory deficits assessed with novel object recognition were normalized by genetic ablation of Nlrp1, Casp1, or Casp6 in J20 mice. Spatial learning deficits, assessed with the Barnes Maze, were normalized in genetically ablated J20, whereas memory recall was normalized in J20/Casp1−/− and J20/Casp6−/−, and improved in J20/Nlrp1−/− mice. Hippocampal CA1 dendritic spine density of the mushroom subtype was reduced in J20, and normalized in genetically ablated J20 mice. Reduced J20 hippocampal dentate gyrus and CA3 synaptophysin levels were normalized in genetically ablated J20. Increased Iba1+-microglia in the hippocampus and cortex of J20 brains were normalized by Casp1 and Casp6 ablation and reduced by Nlrp1 ablation. Increased pro-inflammatory cytokines, TNF-α and CXCL1, in the J20 hippocampus were normalized by Nlrp1 or Casp1 genetic ablation. CXCL1 was also normalized by Casp6 genetic ablation. IFN-γ was increased and total amyloid β peptide was decreased in genetically ablated Nlrp1, Casp1 or Casp6 J20 hippocampi. We conclude that Nlrp1, Casp1, or Casp6 are implicated in AD-related cognitive impairment, inflammation, and amyloidogenesis. These results indicate that Nlrp1, Casp1, and Casp6 represent rational therapeutic targets against cognitive impairment and inflammation in AD.
Subject terms: Cognitive ageing, Neurological disorders, Inflammasome
Introduction
Alternative approaches are required for effective prevention and treatment of Alzheimer disease (AD). Current treatments, while helpful, have limited effects. Therefore, therapeutics against well-recognized AD pathologies are in development [1]. Our premise is that the underlying molecular mechanism of neuronal dysfunction in early AD-related cognitive decline and impairment would be an ideal therapeutic target that would obviate multifactorial causes, which initiate cognitive loss and subsequent dementia in aging individuals [2].
A degenerative pathway linking intraneuronal Nucleotide-binding oligomerization domain, Leucine rich Repeat and Pyrin domain containing protein 1 (Nlrp1) inflammasome and Caspase-1 (Casp1)-mediated neuroinflammation to Caspase-6 (Casp6)-mediated axonal degeneration has been identified in AD brains and human CNS primary neuron cultures [3, 4]. Active Casp6 is abundant in sporadic and familial AD brain neuritic plaques, neurofibrillary tangles, and neuropil threads [5, 6]. Its appearance in the entorhinal cortex and hippocampal Cornu Ammonis 1 (CA1) is associated with decreased episodic and semantic memory performance in non-cognitively impaired aged individuals [7, 8]. Conditional expression of human self-activated Casp6 in mouse hippocampal CA1 pyramidal neurons induces reversible age-dependent cognitive impairment associated with inflammation and neurodegeneration in the absence of plaques and tangles [9, 10]. In human CNS primary neuron cultures and cell lines, active Casp6 cleaves amyloid precursor protein (APP), Tau, and several AD-related synaptic and cytoskeletal neuronal proteins [6, 11–15], and caspases indirectly increase Aβ levels [16–18]. By itself, active Casp6 does not induce cell death [19, 20], but causes axonal degeneration in primary human cortical [21, 22] and NGF-deprived mouse sensory [23, 24] neurons. Active Casp6 microinjection induces synaptic transmission loss and degeneration of CA1 neurons [25], and active Casp6-expressing CA1 neurons cannot initiate long-term potentiation [10]. Together, this suggests that Casp6 activity is associated with neuronal dysfunction and mild cognitive impairment (MCI) preceding AD.
Casp6 is activated by Casp1 in human CNS primary neuron cultures [3]. Casp1 is implicated in cognitive decline and AD. Active Casp1 triggers the maturation of pro-inflammatory interleukin-1-β (IL-1β) [26]. Active Casp1 [27] and IL-1 [28] are increased in AD. IL-1 is associated with gliosis, memory impairment, pro-inflammatory cytokine production, and leukocyte infiltration [29]. CASP1 genetic variants are associated with lower IL-1β levels, better executive function performance, and a trend towards better memory function [30], and predict accelerated progression from MCI to AD [31]. The Casp1 inhibitor, VX-765, delays [32] or reverses [33] cognitive impairment, when administered pre-symptomatically or at symptom onset, respectively, in J20 APPSw/Ind AD mice. Furthermore, Casp1 ablation prevents memory impairment in J20 and APP/PS1 mice [27, 33].
Casp1 is activated by a functional Nlrp1 inflammasome in human neurons [4]. Nlrp1 immunopositive neurons are increased 25-30-fold in AD brains and Nlrp1 co-localizes with active Casp6. Non-synonymous NLRP1 SNPs are associated with AD [34]. Genetic ablation of Nlrp1 and Casp1 in mice prevents LPS-induced Casp6 activation [4].
Together, these studies suggest an association between the Nlrp1/Casp1/Casp6 neurodegenerative pathway and cognitive decline in aging and AD. However, the in vivo implication of these three genes has not been investigated in genetically identical AD mice at an early age to avoid indirect amyloid- or age-related secondary effects that would complicate data interpretation and conclusions. Here, we investigated the early role of Nlrp1, Casp1, and Casp6 in J20 cognitive deficits, inflammation, and Aβ production and deposition, by placing the J20 on a homozygous or heterozygous null background for each of the three genes.
Results
To assess the role of Nlrp1, Casp1, and Casp6 in AD, Nlrp1−/−, Casp1−/−, and Casp6−/− mice were bred to generate J20/ or WT/Nlrp1−/− or +/−, J20/ or WT/Casp1−/− or +/−, and J20/ or WT/Casp6−/− or +/− mice. PCR confirmed the deletion of one or two copies of Nlrp1, Casp1, or Casp6 from J20 and WT mice (Supplementary Fig. S1).
J20/ or WT/Nlrp1−/− or +/−, J20/ or WT/Casp1−/− or +/−, and J20/ or WT/Casp6−/− or +/− mice are competent to undergo memory assessments
Open field analyses assessed the competence of each cohort to undergo memory testing. Four- to five-month-old J20 mice did not show significant differences in distance traveled, quadrant entries, % time moving, or thigmotaxis compared to WT mice (Fig. 1A and Supplementary Fig. S2A–C). J20/Nlrp1−/− or +/− and J20/Casp1−/− mice showed slightly higher distance traveled and quadrant entries than WT mice, although their % time moving and in the periphery was normal. J20/Casp6−/− or +/− and WT/Nlrp1−/− or +/−, /Casp1−/− or +/−, and /Casp6−/− or +/− mice performed normally in all four measures. These results indicate that all fourteen mouse groups are competent to undergo memory testing.
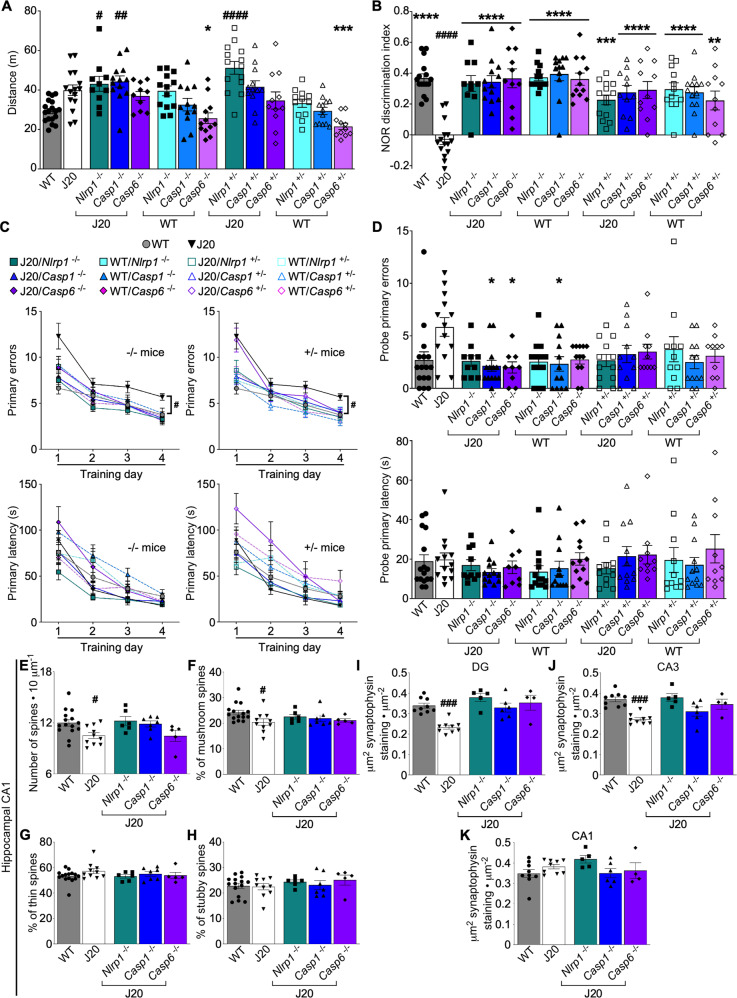
Fig. 1. Nlrp1, Casp1, or Casp6 genetic ablation prevents cognitive deficits in J20 mice.
A Distance traveled in open field task [F = 8.921, p < 0.0001, n = 18 WT, 15 J20, 10 J20/Nlrp1−/−, 13 J20/Casp1−/−, 10 J20/Casp6−/−, 13 WT/Nlrp1−/−, 12 WT/Casp1−/−, 12 WT/Casp6−/−, 14 J20/Nlrp1+/−, 12 J20/Casp1+/−, 11 J20/Casp6+/−, 12 WT/Nlrp1+/−, 13 WT/Casp1+/−, 11 WT/Casp6+/−]. B NOR DI [F = 8.081, p < 0.0001, n = 16 WT, 15 J20, 10 J20/Nlrp1−/−, 13 J20/Casp1−/−, 11 J20/Casp6−/−, 13 WT/Nlrp1−/−, 12 WT/Casp1−/−, 12 WT/Casp6−/−, 14 J20/Nlrp1+/−, 11 J20/Casp1+/−, 11 J20/Casp6+/−, 12 WT/Nlrp1+/−, 13 WT/Casp1+/−, 11 WT/Casp6+/−]. C Barnes maze learning acquisition showing primary errors (top) and primary latency (bottom) to reach target of +/− (right) and −/− (left) mice [Primary errors performance: Ftraining day = 142.5, p < 0.0001, Fgenotype = 4.455, p < 0.0001, 2-way ANOVA, n = 16 WT, 13 J20, 10 J20/Nlrp1−/−, 13 J20/Casp1−/−, 9 J20/Casp6−/−, 13 WT/Nlrp1−/−, 12 WT/Casp1−/−, 12 WT/Casp6−/−, 12 J20/Nlrp1+/−, 11 J20/Casp1+/−, 10 J20/Casp6+/−, 11 WT/Nlrp1+/−, 12 WT/Casp1+/−, 10 WT/Casp6+/−]. D Barnes maze probe showing primary errors (top) and primary latency (bottom) to blocked target [F = 1.915, p = 0.0324, n as in C]. E–H Golgi–Cox quantification (n = 15 WT, 10 J20, 6 J20/Nlrp1−/−, 7 J20/Casp1−/−, 5 J20/Casp6−/−) E Dendritic spine density, and the % of F mushroom, G thin, and H stubby dendritic spines, in hippocampal CA1 SR. Synaptophysin quantification (n = 9 WT, 9 J20, 5 J20/Nlrp1−/−, 6 J20/Casp1−/−, 4 J20/Casp6−/−) in hippocampal I DG [F = 12.10, p < 0.0001], J CA3 stratum lucidum (SLu) [F = 9.731, p < 0.0001], and K CA1 SR. Bars represent mean ± SEM of all mice per group; symbols denote performances of individual mice. For A, B, D–K, one-way ANOVA, Bonferroni’s post-hoc compared to WT (#) or J20 (*). # or *p < 0.05, ## or **p < 0.01, ### or ***p < 0.001, #### or ****p < 0.0001.
Nlrp1, Casp1, or Casp6 genetic ablation prevents episodic and spatial memory deficits in J20 mice
NOR discrimination indices (DI) were lower in J20 compared to WT mice confirming impaired episodic memory in 4- to 5-month-old J20 mice (Fig. 1B, Supplementary Table S1), as observed previously [33, 35]. J20/Nlrp1−/− or +/−, J20/Casp1−/− or +/−, and J20/Casp6−/− or +/− mice DI were normalized to WT levels. Episodic memory was unaltered in genetically ablated WT mice. These results confirm the importance of Nlrp1, Casp1, and Casp6 in APPSw/Ind-mediated episodic memory deficits and indicate that reducing gene expression by half is sufficient to eliminate impairment.
Spatial learning and memory retention were assessed in 4- to 5-month-old J20 mice with the Barnes maze. During the four-day training period to assess learning, J20, but not J20/Nlrp1−/− or +/−, J20/Casp1−/− or +/−, and J20/Casp6−/− or +/−, committed more errors to reach the target than WT mice by the end of the training period (Fig. 1C, Supplementary Table S1). Latencies to reach the target did not differ between any of the genotypes (Fig. 1C). During the probe test measuring spatial memory retention, J20 mice trended towards committing more errors (p = 0.065), but due to large variability in responses, this was not statistically significant (Fig. 1D). This lack of significant spatial memory deficits in J20 is likely due to their young age since J20 of 5-7 months clearly show spatial memory deficits [33, 35]. Nevertheless, J20/Casp1−/− and J20/Casp6−/− mice committed less errors than J20 mice. None of the genotypes altered probe latency. J20 showed a less pronounced preference to the target area than WT mice; their percentage of pokes to each hole was scattered among adjacent areas, creating a smaller peak at the target (Supplementary Fig. S3). This was normalized in J20/Nlrp1−/− or +/−, J20/Casp1−/− or +/−, and J20/Casp6−/− or +/− mice. All WT mice groups behaved normally. These results indicate that Casp1 or Casp6 ablations normalize spatial memory in J20 mice. J20/Nlrp1−/− or +/−, J20/Casp1+/−, and J20/Casp6+/− mice also improve learning and memory retention, although they do not quite reach normal levels.
Genetic ablation of Nlrp1, Casp1 or Casp6 normalizes J20 hippocampal CA1 dendritic spines, and CA3 and DG synaptophysin levels
Dendritic spine density and morphology were assessed with Golgi–Cox staining in the stratum radiatum (SR) of the hippocampal CA1 region (Supplementary Fig. S4A). Dendritic spine density (Fig. 1E) and the % of mushroom (Fig. 1F), but not thin or stubby (Fig. 1G, H), spine subtypes were significantly reduced in J20 compared to WT. Spine density and morphology were normalized in J20/Nlrp1−/−, J20/Casp1−/−, and J20/Casp6−/− mice compared to WT. Cell density in the CA1 pyramidal cell layer (PCL) was the same for all genotypes (Supplementary Fig. S4B), indicating that dendritic spine alterations in J20 mice were not related to neuronal loss.
Pre-synaptic synaptophysin immunostaining density was also measured in the hippocampus (Supplementary Fig. S4C). Synaptophysin levels decreased in J20 relative to WT in dentate gyrus (DG) (Fig. 1I) and CA3 (Fig. 1J), but not in CA1 (Fig. 1K). The genetic ablation of Nlrp1, Casp1, and Casp6 in the DG and CA3, restored J20 synaptophysin levels to WT levels. These results indicate that Nlrp1, Casp1, and Casp6 are involved in J20-mediated synaptic disruptions in dendritic spines and synaptophysin levels.
Nlrp1, Casp1, or Casp6 genetic ablation curtails ionized calcium binding adaptor molecule 1 (Iba1)+-microglial density in J20 mice
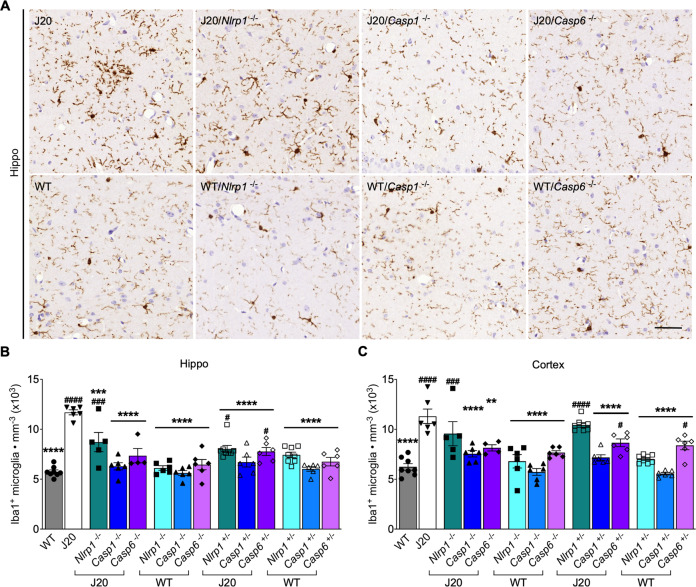
The number of Iba1+-microglia, a marker for inflammation, was quantitatively measured in the hippocampus (Fig. 2A) and cortex (Supplementary Fig. S5). Iba1+-microglia were twice as abundant in J20 than WT hippocampus and cortex (Fig. 2B, C and Supplementary Table S2). J20/Casp1−/− or +/− and J20/Casp6−/− hippocampal and cortical microglia decreased compared to J20 and normalized to WT levels, whereas J20/Casp6+/− Iba1+-microglia numbers were lower than J20 but still higher than the WT levels. J20/Nlrp1−/− or +/− hippocampal microglia were lower than J20 but higher than WT mice, while cortical Iba1+-microglia numbers remained similar to those of J20 mice. All genetically ablated WT groups had similar hippocampal and cortical Iba1+-microglia numbers, except for increased cortical microglia in WT/Casp6+/−. These results indicate a clear involvement of Nlrp1, Casp1 and Casp6 on APPSw/Ind-mediated increased hippocampal Iba1+-microglia, although J20/Nlrp1−/− or +/− and J20/Casp6+/− Iba1+-microglia levels were not normalized. In contrast, while Casp1 and Casp6 are involved in increased cortical microglia, Nlrp1 is not. These results suggest that another inflammasome may be more involved in the cortex.
Fig. 2. Nlrp1, Casp1, or Casp6 genetic ablation reduces activated microglia, but not astrocytes, in J20 mice.
A Representative micrographs of Iba1+ microglia in the hippocampal CA1. Scale bar = 50 μm. B Iba1+microglia quantitation from the PCL to the SLM in the hippocampal CA1 [F = 15.10, p < 0.0001, one-way ANOVA, n = 8 WT, 6 J20, 5 J20/Nlrp1−/−, 6 J20/Casp1−/−, 4 J20/Casp6−/−, 6 WT/Nlrp1−/−, 6 WT/Casp1−/−, 6 WT/Casp6−/−, 8 J20/Nlrp1+/−, 6 J20/Casp1+/−, 6 J20/Casp6+/−, 8 WT/Nlrp1+/−, 6 WT/Casp1+/−, 6 WT/Casp6+/−]. C Iba1+microglial quantitation in the cortical retrosplenial and S1 area [F = 15.35, p < 0.0001, one-way ANOVA, n as in B]. Bars represent mean ± SEM of all mice per group; symbols denote individual results. For B and C, Bonferroni’s post-hoc compared to WT (#) or J20 (*). #p < 0.05, ** p < 0.01, ### or *** p < 0.001, #### or **** p < 0.0001.
Genetic ablations do not alter glial fibrillary acidic protein immunopositive (GFAP+) astrocytes in J20 mice
GFAP+ staining density (Supplementary Fig. S6A) was approximately 6-fold higher in J20 hippocampus than cortex but did not differ significantly between WT and J20 mice (Supplementary Fig. S6B, C and Supplementary Table S2). Complete or partial Nlrp1, Casp1, or Casp6 ablation did not alter GFAP+ staining in J20 or WT mice. These results indicate that Nlrp1, Casp1, and Casp6 are not involved in J20-mediated astrogliosis.
Nlrp1, Casp1, or Casp6 genetic ablation normalizes C-X-C motif chemokine ligand 1 (CXCL1) and increases interferon gamma (IFN- γ) levels whereas Nlrp1 or Casp1 genetic ablation normalizes tumor necrosis factor alpha (TNF-α) in J20 hippocampi
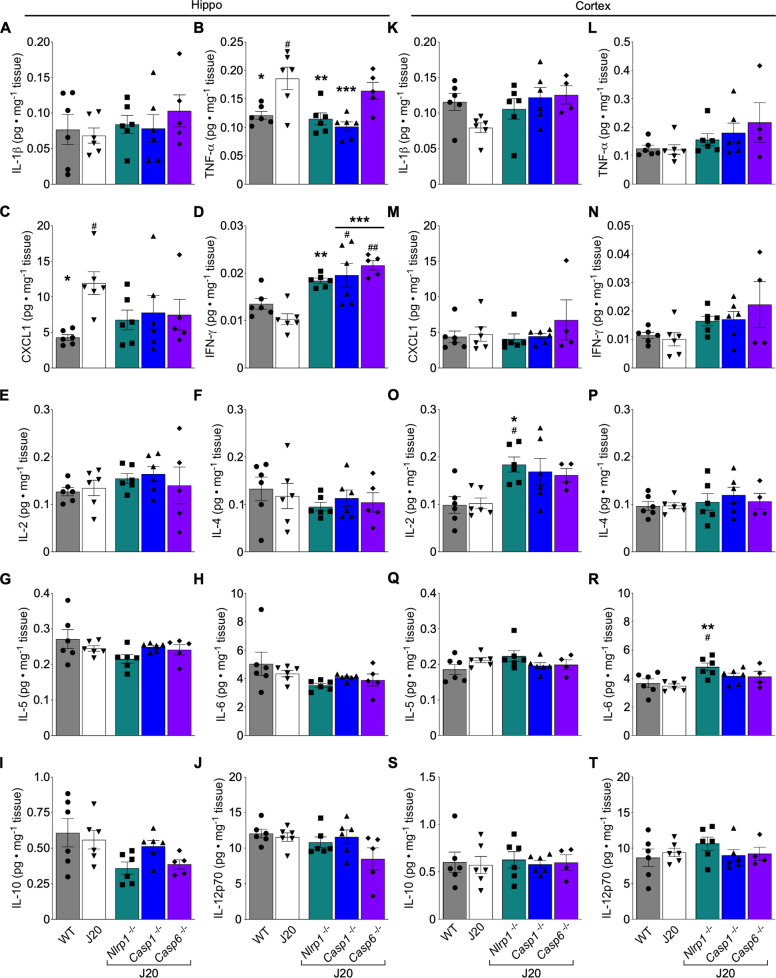
Cytokine production was quantitated in WT, J20, J20/Nlrp1−/−, J20/Casp1−/−, and J20/Casp6−/−. In the hippocampus (Fig. 3A–J), total IL-1β did not differ between any group (Fig. 3A). TNF-α (Fig. 3B) and CXCL1 (Fig. 3C) increased in J20 compared to WT mice. Complete ablation of Nlrp1 and Casp1, but not Casp6, normalized TNF-α (Fig. 3B), whereas Nlrp1, Casp1, and Casp6 ablation normalized CXCL1 levels (Fig. 3C). IFN-γ increased in all genetically ablated J20 hippocampi, but did not differ between WT and J20 mice (Fig. 3D). No changes were detected in hippocampal IL-2, IL-4, IL-5, IL6, IL-10, or IL12p70 (Fig. 3E–J). In the cortex (Fig. 3K–T), no differences in any of the cytokines were found between any of the groups except for increased IL-2 (Fig. 3O) and IL-6 (Fig. 3R) in J20/Nlrp1−/− compared to WT and J20. These results indicate that TNF-α and CXCL1 pro-inflammatory cytokines may be implicated in APPSw/Ind-mediated cognitive impairment and microglial activation. In addition, increased IFN-γ may contribute to the beneficial effect of the genetic ablation of Nlrp1, Casp1, and Casp6 in APPSw/Ind-mediated cognitive decline and microglial activation.
Fig. 3. Nlrp1, Casp1, or Casp6 genetic ablation alters pro-inflammatory cytokine levels in J20 hippocampi.
A-J Hippocampal protein levels (n = 6 WT, 6 J20, 6 J20/Nlrp1−/−, 6 J20/Casp1−/−, 5 J20/Casp6−/−) for A IL-1β, B TNF-α [F = 7.881, p = 0.0003] C CXCL1 (KC/GRO) [F = 2.699, p = 0.054], D IFN-γ [F = 10.26, p = 0.00005], E IL-2, F IL-4, G IL-5, H IL-6, I IL-10 [F = 2.975, p = 0.039], and J IL-12p70. Cortical protein levels (n = 6 WT, 6 J20, 6 J20/Nlrp1−/−, 6 J20/Casp1−/−, 4 J20/Casp6−/−) for K IL-1β, L TNF-α, M CXCL1 (KC/GRO), N IFN-γ, O IL-2 [F = 4.657, p = 0.0067], P IL-4, Q IL-5, R IL-6 [F = 4.359, p = 0.009], S IL-10, and T IL-12p70. Bars represent mean ± SEM of all mice per group; symbols denote individual results. One-way ANOVA, Bonferroni’s post-hoc compared to WT (#) or J20 (*) for A-T. # or *p < 0.05, ## or **p < 0.01, ***p < 0.001.
Nlrp1, Casp1, or Casp6 genetic ablation does not alter APP levels in J20 mice
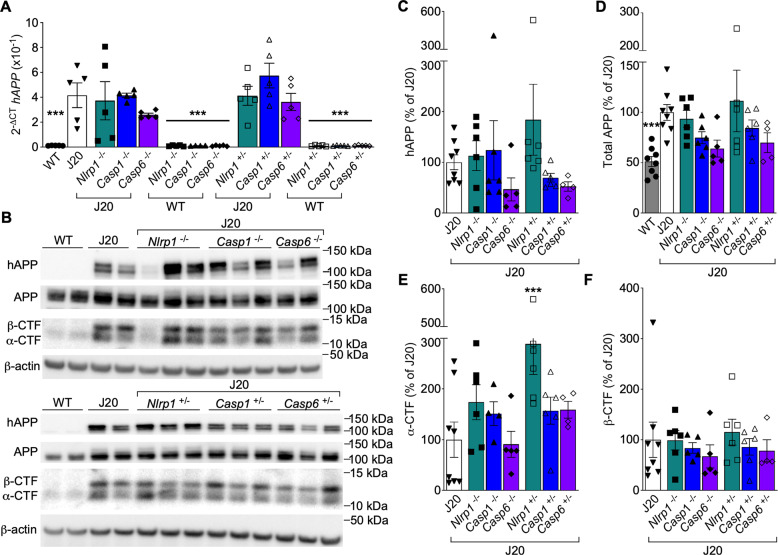
Human APP mRNA (Fig. 4A) and protein (Fig. 4B–D) levels increased in J20 relative to WT, but remained unaltered in J20/Nlrp1, /Casp1, or /Casp6 −/− or +/− brains. APP α C-terminal fragments (CTF) (Fig. 4B, E) and βCTF (Fig. 4B, F) did not differ between any of the J20 genotypes, except for increased αCTF in J20/Nlrp1+/− (Fig. 4E). Together, these results exclude the possibility that altered APP expression or processing by α- or β-secretase account for the beneficial effects of Nlrp1, Casp1, or Casp6 genetic ablation on J20 cognitive impairment and microglial activation.
Fig. 4. Nlrp1, Casp1, or Casp6 genetic ablation does not alter APP levels in J20 cortex.
A Human APP (hAPP) mRNA cortical levels measured with qRT-PCR [F = 11.45, p = <0.0001, n = 5 WT, 5 J20, 5 J20/Nlrp1−/−, 5 J20/Casp1−/−, 5 J20/Casp6−/−, 5 WT/Nlrp1−/−, 4 WT/Casp1−/−, 5 WT/Casp6−/−, 5 J20/Nlrp1+/−, 5 J20/Casp1+/−, 5 J20/Casp6+/−, 5 WT/Nlrp1+/−, 5 WT/Casp1+/−, 5 WT/Casp6+/−]. B Representative western blot images of hAPP, mouse and hAPP, αCTF, and βCTF in the cortex. Western blot quantification of C hAPP (n = 8 J20, 6 J20/Nlrp1−/−, 6 J20/Casp1−/−, 5 J20/Casp6−/−, 6 J20/Nlrp1+/−, 6 J20/Casp1+/−, 4 J20/Casp6+/−), D mouse APP and hAPP [F = 2.732, p = 0.02, n as in C plus 8 WT], E αCTF [F = 3.194, p = 0.014, n = 8 J20, 6 J20/Nlrp1−/−, 5 J20/Casp1−/−, 5 J20/Casp6−/−, 6 J20/Nlrp1+/−, 6 J20/Casp1+/−, 4 J20/Casp6+/−) and F βCTF (n as in E). Bars represent mean ± SEM of all mice per group; symbols denote individual results. One-way ANOVA, Dunnett’s post-hoc compared to J20 for A, C–F. ***p < 0.001.
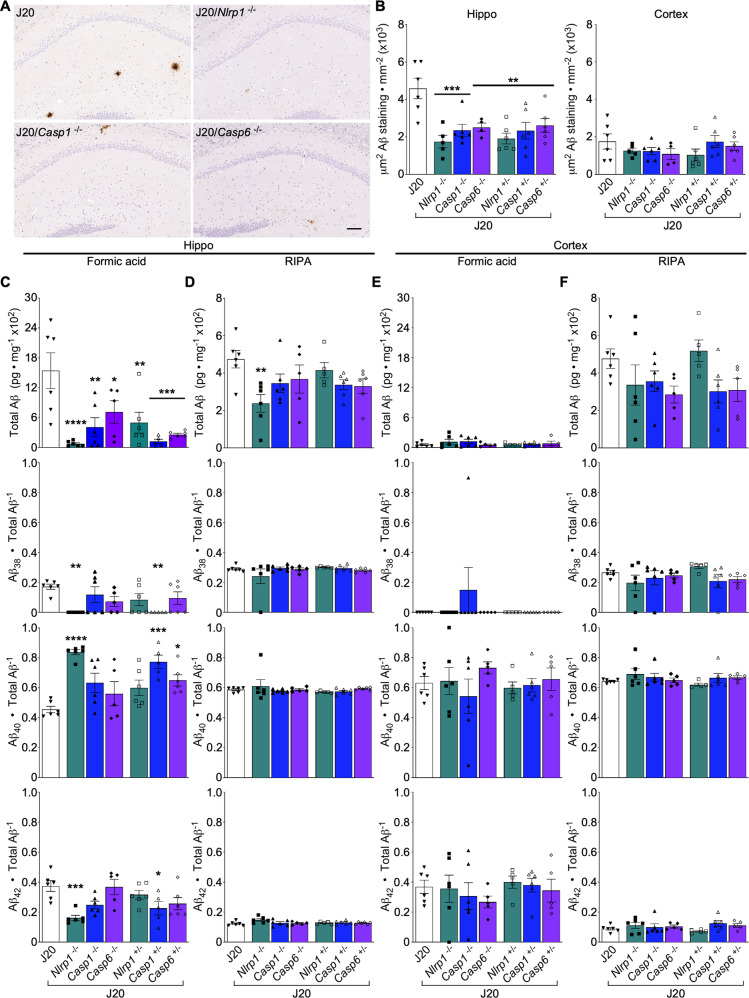
Nlrp1, Casp1, or Casp6 genetic ablation reduces hippocampal Aβ levels by 50% in J20 mice
Aβ deposits in 4-5-month-old J20, and partially or completely Nlrp1-, Casp1-, and Casp6-ablated J20 brains, were diffuse and mainly concentrated in the hippocampal stratum lacunosum moleculare (SLM) and DG (Fig. 5A, Supplementary Fig. S7A). Rare Aβ deposits were present in the cortical retrosplenial and primary and secondary motor areas (Supplementary Fig. S7B). Aβ deposits were approximately twice as abundant in J20 hippocampus than in cortex, and 50% lower in the hippocampus, but equivalent in the cortex, of partially or completely Nlrp1-, Casp1-, or Casp6-ablated J20 compared to J20 mice (Fig. 5B and Supplementary Table S3). Consistently, formic acid-soluble total Aβ, Aβ38, Aβ40, and Aβ42 levels decreased by more than 50% in the hippocampus after Nlrp1, Casp1, or Casp6 ablation in J20 mice (Fig. 5C, Supplementary Fig. S7C and Supplementary Table S3). Significant changes in the ratios of formic acid-soluble Aβ38, Aβ40, and Aβ42 subspecies relative to total Aβ were also observed in J20/Nlrp1−/− and J20/Casp1+/− mice: Aβ40/total Aβ increased at the expense of reduced Aβ38 and Aβ42 ratios (Fig. 5C). Hippocampal RIPA-soluble total Aβ, Aβ38, Aβ40, and Aβ42 levels decreased ~50% in J20/Nlrp1-/- mice (Fig. 5D, Supplementary Fig. S7D) but were unaltered in all other genetically ablated J20 mice.
Fig. 5. Nlrp1, Casp1, or Casp6 genetic ablation prevents amyloid deposition in J20 mice.
A Representative Aβ-stained micrographs of the hippocampus. Scale bar = 100 μm. B Aβ+ immunostaining density from the PCL to the SLM in the hippocampal CA1 (left) and cortical retrosplenial and S1 area (right) [Aβ staining density in the hippocampus: F = 6.005, p = 0.0003, n = 6 J20, 5 J20/Nlrp1−/−, 6 J20/Casp1−/−, 4 J20/Casp6−/−, 6 J20/Nlrp1+/−, 6 J20/Casp1+/−, 6 J20/Casp6+/−]. C–F Aβ protein ratios in the hippocampus and cortex (n = 6 J20, 6 J20/Nlrp1−/−, 6 J20/Casp1−/−, 5 J20/Casp6−/−, 6 J20/Nlrp1+/−, 6 J20/Casp1+/−, 6 J20/Casp6+/−) C Hippocampal formic acid-soluble total Aβ [F = 6.730, p = 0.0001], Aβ38/total Aβ [F = 3.254, p = 0.013], Aβ40/total Aβ [F = 7.189, p = 0.000058], and Aβ42/total Aβ [F = 5.026, p = 0.00093]. D Hippocampal RIPA-soluble total Aβ [F = 2.521, p = 0.04], Aβ38/total Aβ, Aβ40/total Aβ, and Aβ42/total Aβ. E Cortical formic acid-soluble total Aβ, Aβ38/total Aβ, Aβ40/total Aβ, and Aβ42/total Aβ. F Cortical RIPA-soluble total Aβ, Aβ38/total Aβ, Aβ40/total Aβ, and Aβ42/total Aβ. Bars represent mean ± SEM of all mice per group; symbols denote individual results. One-way ANOVA, Dunnett’s post-hoc compared to J20 for B-F. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Formic acid-soluble total Aβ levels were much lower in cortex than hippocampus (Fig. 5E). The complete or partial ablation of Nlrp1, Casp1, and Casp6 did not alter formic acid (Fig. 5E, Supplementary Fig. S7E) or RIPA-soluble (Fig. 5F, Supplementary Fig. S7F) Aβ38, Aβ40, and Aβ42 levels or their ratios to total Aβ in J20 cortex. These results indicate that Nlrp1, Casp1, and Casp6 contribute to the aggregated Aβ deposits in J20 hippocampi.
Iba1+-microglia predict episodic memory whereas IFNγ and CXCL1 predict spatial memory in J20
Multivariable linear regressions were performed to determine which hippocampal variable best predicted episodic and spatial memory deficits in J20 mice (Table 1). Analyses were performed on either data acquired from paraformaldehyde fixed or frozen brains. Measures of synaptophysin, Iba1+-microglia, and Aβ immunostaining from paraformaldehyde fixed hippocampus showed that only Iba1+-microglial cell density associated with NOR discrimination index scores (Table 1). None of the hippocampal biochemical markers in frozen brains, which included spine density, pro-inflammatory TNFα, IFNγ, and CXCL1 cytokines, and total formic acid-soluble Aβ protein levels, associated with NOR scores. Neither synaptophysin, Iba1+-microglia density, or Aβ levels obtained from paraformaldehyde-fixed tissues associated with Barnes maze probe primary errors (Table 1). From frozen brain data, only IFNγ and CXCL1 levels associated with Barnes maze spatial memory. Together, these results indicate that Iba1+-microglia predict episodic, whereas IFNγ and CXCL1 predict spatial, memory performance in J20.
Table 1.
Iba1+-microglia predict episodic memory whereas IFNγ and CXCL1 predict spatial memory in J20.
| Predicting NOR episodic memory deficits | |||||
|---|---|---|---|---|---|
| Marker (independent variable) | Coefficient | Standard error | t value | p value | |
| Histology markers | Intercept | 0.71 | 0.30 | 2.35 | 0.037 |
| Hippocampus | Synaptophysin+ staining | 0.13 | 0.27 | 0.47 | 0.65 |
| Iba1+ microglia | −0.50 | 0.21 | −2.38 | 0.035 | |
| Aβ+ staining | −0.059 | 0.036 | −1.62 | 0.13 | |
| Biochemistry markers | Intercept | 0.68 | 0.39 | 1.73 | 0.10 |
| Hippocampus | Spine Density | −0.049 | 0.031 | −1.60 | 0.13 |
| TNFα | −0.85 | 0.99 | −0.86 | 0.40 | |
| IFNγ | 13.56 | 8.27 | 1.64 | 0.12 | |
| CXCL1 | 0.0082 | 0.0093 | 0.89 | 0.39 | |
| Total Aβ | −0.12 | 0.075 | −1.62 | 0.13 | |
| Predicting Barnes maze spatial memory deficits | |||||
|---|---|---|---|---|---|
| Marker (independent variable) | Coefficient | Standard error | t value | p value | |
| Histology markers | Intercept | 1.75 | 1.53 | 1.15 | 0.27 |
| Hippocampus | Synaptophysin+ staining | −1.02 | 1.33 | −0.77 | 0.45 |
| Iba1+ microglia | 0.53 | 0.54 | 0.97 | 0.35 | |
| Aβ+ staining | −0.15 | 0.18 | −0.84 | 0.42 | |
| Biochemistry markers | Intercept | 4.019 | 1.16 | 3.45 | 0.0039 |
| Hippocampus | Spine Density | −0.012 | 0.092 | −0.13 | 0.90 |
| TNFα | 2.038 | 3.00 | 0.68 | 0.51 | |
| IFNγ | −95.17 | 25.64 | −3.71 | 0.0023 | |
| CXCL1 | −0.091 | 0.028 | −3.23 | 0.0060 | |
| Total Aβ | 0.18 | 0.23 | 0.79 | 0.44 | |
Linear regression model results determining which hippocampal variable associates with NOR episodic memory (top) and Barnes maze spatial memory (bottom) deficits. Analyses were performed on WT, J20, J20/Nlrp1−/−, J20/Casp1−/−, and J20/Casp6−/− mice with complete data only. Markers separated into two separate models based on paraformaldehyde fixed or frozen brain tissue processing. Model one combines immunohistochemical markers synaptophysin staining, Iba1+ microglia, and Aβ staining in the hippocampus (n = 16). Model two combines Golgi–cox-stained spine density, pro-inflammatory TNFα, IFNγ, and CXCL1 (cytokines that previously showed significant changes in J20 mice), and total formic acid-soluble Aβ protein levels (n = 21). The large values for Iba1, Aβ staining, and total Aβ were re-scaled to bring coefficients into an acceptable range but does not affect t or p values.
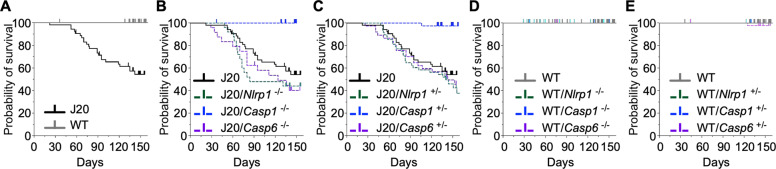
Casp1, but not Nlrp1 or Casp6, genetic ablation protects J20 mice from premature mortality
Only 40–50% of J20 mice survived at 5 months of age, despite being kept in a pathogen-free facility (Fig. 6A). J20 survival rate was unaltered by genetic ablation of Nlrp1 or Casp6 but improved to near 100% with genetic ablation of Casp1 (Fig. 6B, C). Survival was not affected in WT mice by any genetic ablations (Fig. 6D, E). These results indicate that Casp1, but not Nlrp1 and Casp6, is involved in J20 increased mortality rate.
Fig. 6. Casp1, but not Nlrp1 or Casp6, genetic ablation improves survival in J20 mice.
Kaplan–Meier survival curves and pairwise comparisons between A WT and J20 mice (p < 0.0001), B J20 and J20/Nlrp1−/− (p = 0.1807), J20/Casp1−/− (p = 0.0030), and J20/Casp6−/− (p = 0.2280) mice, C J20 and J20/Nlrp1+/− (p = 0.3548), J20/Casp1+/− (p < 0.0001), and J20/Casp6+/− (p = 0.6374) mice, D WT and WT/Nlrp1−/−, WT/Casp1−/−, and WT/Casp6−/− mice (no significant differences), and E WT and WT/Nlrp1+/−, WT/Casp1+/−, and WT/Casp6+/− mice (no significant differences).
Discussion
This study provides unrivaled evidence to validate the importance of Nlrp1, Casp1, and Casp6 in episodic and spatial memory in AD. The use of the J20 mouse model generated with only one mutant gene, the study of heterozygous or homozygous Nlrp1, Casp1, or Casp6 null mice on genetically identical J20 or WT backgrounds at an age where there isn’t significant accumulation of amyloid deposits, and the submission of all mouse groups to identical experimental conditions crystallize the role of Nlrp1, Casp1, and Casp6 in AD-related cognitive deficits. In support of our findings, five-month-old APP/PS1 AD mice submitted to Nlrp1 or Casp1 siRNA knock down [36], 8-month-old J20/Casp1−/− or +/− [33], 16-month-old APP/PS1/Casp1−/− [27], and 7-month-old 5xFAD/Casp6−/− mice [37] have all shown improved spatial memory. However, differences in their (1) transgenes (APPK670/M671L/PS1∆E9 in APP/PS1, APPK670/M671L/V717F in J20, and APPK670/M671L /I716V/V717I/PS1M146L, L286V in 5xFAD), (2) promoters directing expression in different cell types (mouse prion protein in APP/PS1, PDGFB in J20, and Thy1 in 5xFAD), (3) age tested (5 months for siRNA and 16 months for KO in APP/PS1, 7 months in 5xFAD and 8 months in J20) relative to the age of appearance of Aβ (8–10 months, 2 months, and 5–7 months, respectively), and (4) the use of only the Morris water maze to test memory, restrict the direct comparison of Nlrp1, Casp1, and Casp6 in AD-related pathophysiology.
These studies in J20 AD mice strongly support the suspected role of Nlrp1, Casp1, and Casp6 in human age-dependent and AD cognitive deficits. Four NLRP1 variants have been associated with AD [34]. While no association has been observed between CASP1 and AD [38], a rare CASP6 variant, encoding Casp6N73T with reduced Casp6 activity and less neurofunctional and neurodegeneration abilities than WT Casp6, has been associated with preserved hippocampal volume in a stable mild cognitively impaired individual [39]. NLRP1, CASP1, and CASP6 mRNAs are increased in AD brains [4, 27, 39–41]. Nlrp1, active Casp1, and active Casp6 proteins are increased in early and late stages of AD [4–8, 27]. Active Casp6 levels in the entorhinal cortex and CA1, two areas affected early in AD, correlate negatively with episodic and semantic memory performance in aged individuals [5, 8, 9]. The data is also consistent with the induction of Casp6-mediated neurodegeneration in human primary neurons over-expressing APPWT, APPSw, and APPLondon, three conditions associated with familial AD [22].
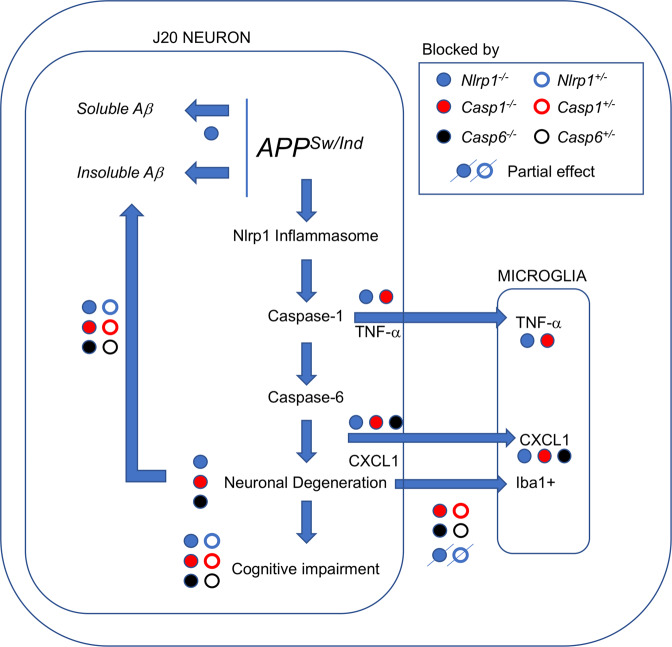
These data indicate that Nlrp1, Casp1, and Casp6 involvement in cognition occur through the same pathway. Sequential activation of Nlrp1, Casp1, and Casp6 were previously identified in serum-deprived human neurons [3, 4, 17], and in LPS-injected WT/Nlrp1−/− and WT/Casp1−/− mouse brains [4]. In this study, genetic ablation of all three genes almost identically corrected J20-mediated episodic and spatial memory deficits, CA1 dendritic spine density loss, and CA3 and DG synaptophysin immunoreactivity reduction. Because APPSw/Ind is expressed under a neuron-specific promoter [42, 43], and Casp6 is absent or very low in microglia [4], our results indicate that the Nlrp1-Casp1-Casp6 neurodegenerative pathway leads to J20 mouse brain neuron degeneration (Fig. 7). Consequently, genetic ablation of all three genes relieves J20 mouse cognitive impairment. The 50% reduction of insoluble Aβ by partial and complete Nlrp1, Casp1, or Casp6 ablation is consistent with increased Aβ production by stressed neurons [4, 17, 22, 44]. Aβ induces the Nlrp3 inflammasome in 16 month-old APP/PS1 mouse microglia [27]. Here, Aβ is unlikely activating microglia because genetic ablation of all three genes decrease Aβ levels to the same extent but only Casp1 and Casp6, not Nlrp1, genetic ablations completely normalize Iba1+-microglia levels. Furthermore, 4–5-month-old J20 mice have not yet accumulated significant levels of Aβ. Instead of microglial activation by Aβ, we propose that 4–5-month-old J20 Iba1+-microglia are the result of neuronal degeneration caused by the Nlrp1-Casp1-Casp6 pathway because neurodegeneration is always associated with glial activation and both Casp1 and Casp6 genetic ablations completely normalize Iba1+-microglia. The partial effect of Nlrp1 ablation on Iba1+-microglia levels suggest that neuronal APPSw/Ind expression can activate another inflammasome that leads to Casp1 and Casp6-mediated neurodegeneration. At this time, it’s unclear if increased TNF-α and CXCL1 levels are of neuronal or microglial origin [45, 46]. CXCL1 levels are normalized by Nlrp1, Casp1, or Casp6 genetic ablation suggesting a neuronal origin for CXCL1.
Fig. 7. Schematic diagram summarizing the results of Nlrp1, Casp1, and Casp6 genetic ablation on J20-mediated neurodegeneration and cognitive impairment.
Not all partial knock out animals were tested in all experiments. The absence of data for heterozygous null mice means these were not included in the experiment, not that the results were negative.
Multivariable linear regression analyses were performed in an attempt to determine which measures were associated with cognitive function in J20 mice. Hippocampal Iba1+-microglia was the only measure significantly associated with episodic memory performance, whereas IFNγ and CXCL1 were associated with spatial memory performance. These results indicate specific pathways for each memory type. Whether these associations are causative or ancillary to cognition remains to be confirmed but their role in cognition is supported by other studies. Iba1+-microglia correlated negatively with episodic memory in J20 mice [32] and decreased with cognitive function recovery in VX-765-treated J20 [33]. Microglia depletion or anti-inflammatory drug treatment decreased Iba1+-microglia and restored normal NOR performance and spine density in a Down Syndrome mouse model [47]. Similarly, here, depletion of Nlrp1, Casp1, and Casp6 restored cognition, spine density, synaptophysin levels and Iba1+-microglia levels. Hippocampal IFN-γ was elevated with Nlrp1, Casp1, and Casp6 genetic ablations in J20 indicating a possible contribution to alleviated cognitive deficits. Consistently, IFN-γ improves cognition in 5xFAD and APP/PS1 mice [48, 49]. Increased hippocampal CXCL1 in J20 was normalized in all genetically ablated J20. CXCL1 is a potent chemoattractant for neutrophils [50]. Interestingly, neutrophils stall blood flow in the small capillaries of APP/PS1, 5xFAD, and CRND8 AD mouse models, resulting in impaired memory [51]. Memory deficits are rapidly reversed by anti-Ly6G treatment which clears neutrophils from blood vessels.
Both partial and complete ablation of Casp1, but not Nlrp1 or Casp6, completely protected J20 mice against premature mortality. Here, approximately 40% of J20 mice died by 5 months of age despite being raised in a pathogen-free facility. The cause of death was not confirmed since mice always died at night on a normal light cycle when in their active phase. This suggests that previously reported non-convulsive epilepsy, which manifests itself in active mice, may be responsible for J20 premature mortality [52]. Elucidating the exact role of Casp1 in J20 premature mortality will require in depth studies but the results indicate another important function of Casp1 in AD pathophysiology. Lastly, these results indicate that the cause of premature mortality is not responsible for J20 mice cognitive deficits since J20/Nlrp1−/− or +/− and J20/Casp6−/− or +/− do not prevent early mortality but restore cognition.
Nlrp1, Casp1, or Casp6 inhibitors represent feasible treatments against cognitive impairment in MCI and AD. Inhibition of Casp1 and Casp6 appear compatible with normal human life. Rare naturally occurring CASP1 or CASP6 variants with very low Casp1 activity [53] or with reduced or no Casp6 catalytic activity [54], respectively, are not associated with human pathologies. Casp6 expression is hardly detectable in normal adult human brains [55].
In conclusion, these results highlight the importance of Nlrp1, Casp1, and Casp6 in AD cognitive deficits. Since each of these proteins are altered in AD brains, Nlrp1, Casp1, and Casp6 represent three rational therapeutic targets to prevent or treat cognitive impairment in AD.
Materials and methods
Experimental design and breeding
All animal procedures followed the Canadian Council on Animal Care guidelines and approved by the Comité de Déontologie de l’Expérimentation sur les Animaux (CDEA, Université de Montréal) and McGill University Animal Care committee. The J20 APPSw/Ind transgenic mouse model [56], which express the Swedish (670/671KM→NL) and Indiana (717V→F) human APP mutations, was chosen for this study because J20 mice display episodic memory deficits by 3–4 months, spatial memory and learning deficits by 6–7 months [35], increased total hippocampal Aβ42 levels by 2 months of age, Aβ plaque formation by 7 months [56], and synaptophysin loss by 2–6 months [56], thus allowing rapid assessment of the effect of Nlrp1, Casp1, and Casp6 on these features. Microglial inflammation and neuronal loss precede plaque deposition [57]. Thus, the J20 mouse represents an excellent mouse model to assess the implication of various genes in several aspects of AD pathophysiology. J20 mice (B6.Cg-Zbtb20Tg(PDGF-APPSwInd/20lms)/2Mmjax, stock No 006293, Jackson Laboratories, ME, USA) [56], were obtained on a C57BL/6J background. The following mice were crossed with J20 or WT mice to generate J20 and WT heterozygous breeders for each gene: Nlrp1b- (B6.129S6-Nlrp1btm1Bhk/J, stock No. 021301, Jackson Laboratories) [58], Casp1- (B6N/129S2-Casp1tm1Flv/J, stock No. 016621, Jackson Laboratories) [59], and Casp6- (B6.129S6-Casp6tm1Flv/J, stock No. 006236, Jackson Laboratories) [60]. Sperm of J20 heterozygous males for each gene was cryopreserved and used for IVF with WT/Nlrp1+/−, /Casp1+/−, and /Casp6+/− heterozygous females to generate WT/ and J20/Nlrp1−/− or +/−, /Casp1−/− or +/− and /Casp6−/− or +/−, in addition to J20 and WT control littermates. Mice were aged to 4 months at the Institut de Recherche en Immunologie et en Cancérologie (IRIC) at the Université de Montréal. Both males and females were used for these studies. Mice were group-housed (2–4 mice per cage) in standard macrolon cages under a 12-h light/dark cycle and controlled environmental conditions. Food and water were available ad libitum
Nlrp1, Casp1, and Casp6 cohorts were developed separately approximately one month apart. Casp1 and Casp6 cohorts were each developed in a single breeding experiment; Nlrp1 mice required two breeding experiments to obtain an adequate sample size. Mice were transferred from the IRIC to the home animal facility (where the study was conducted) two weeks prior to the start of the experiment. At the home facility, mice were housed under a 12-h reversed light-dark cycle with controlled environmental conditions and ad libitum food and water. Each cohort was separated into two groups: group one was behaviorally assessed at 4 months and subsequently processed at 4.5 months of age, while group two was behaviorally assessed immediately after at 4.5 months and processed at 5 months of age. WT and J20 experimental control mice were taken from each of the three different cohorts and merged as a single group for analysis.
PCR genotyping
Genotypes were determined by biopsy and PCR at 3–4 weeks of age. DNA was extracted from mice tail or ear punch tissue samples. All animals were genotyped for the presence of the transgene expressing mutant human APP, APPSwInd, and reaction functionality was demonstrated by simultaneous amplification of an internal control using the following primers: 5’-GGTGAGTTTGTAAGTGATGCC-3’, 5’-TCTTCTTCTTCCACCTCAGC-3’ yield the 360 bp hAPP and 5’-CAAATGTTGCTTGTCTGGTG-3’, 5’-GTCAGTCGAGTGCACAGTTT-3’ give the 200 bp internal control. To determine the Nlrp1, Casp1, Casp6 genotypes of animals generated for the J20/Nlrp1, J20/Casp1 and J20/Casp6 cohorts respectively, the following primers were used: 5’-TGCCTGCTCTTTACTGAAGG-3’, 5’-GCCCACAGCTCCC ATAAAT-3’ and 5’-TCCACAATTCTGTCCATTTCC-3’ to generate the 490 bp mutant and 792 bp WT Nlrp1 amplicons, 5’-TGCTAAAGCGCATGCTCCAGACTG-3’, 5’-GAGACATATAAGGGAGAA GGG-3’ and 5’-ATGGCACACCACAGATATCGG-3’ to obtain the 300 bp mutant and 500 bp WT Casp1 amplicons and 5’-GCCTTCTTGACGAGTTCTTCTGAGG-3’, 5’-CTCCACGGCCTAATGCAGTTCC TGG-3’ and 5’-CTGAGGGGCGGAGCACCTTTGCTG-3’ to produce the 340 bp mutant and 620 bp WT Casp6 amplicons. All primer sequences were obtained from Jackson Laboratories. Taq DNA polymerase (New England Biolabs, Whitby, ON, Canada) was used to amplify all DNA targets and products were visualized on 2% agarose gels stained with RedSafe (Froggabio, North York, ON, Canada).
Behavioral analysis
Each mouse underwent Open field testing followed by NOR 24 h later. The Barnes maze was assessed 24–48 h after completion of the NOR test. All behavioral testing were measured using an HVS2100 automated tracking system (HVS Image, Hampton, UK). Behavioral scoring was blinded to mouse genotype and treatment. Detailed behavioral methods have been previously described [33].
Open field
Mice were placed in an open field chamber, composed of a 40 cm × 40 cm plexiglass box with no ceiling and white floor, and allowed to explore for 5 min.
NOR
Twenty-four h after exposure to the open field chamber, mice were again put into the chamber and pre-exposed to two identical objects for 5 min. Two h after pre-exposure, mice were placed back into the chamber and exposed to one familiar object and one novel object for an additional 5 min. The discrimination index (DI) [(number of touches novel object – number of touches familiar object)/total touches] was assessed for each mouse.
Barnes maze
The Barnes maze platform was composed of a 91-cm diameter round platform, elevated 90 cm from the floor, and consisted of 20 holes (each 5 cm in diameter). All holes were blocked with the exception of one target hole that led to an escape box located under the platform. Visual cues, bright light, and white noise were used to motivate the mice to quickly find the escape box. Testing was done in 3 stages: Adaption phase (day 0) – mice were allowed to explore the platform for 60 s. Any mouse that did not find the escape box was guided and allowed to remain there for 90 s. Learning acquisition (days 1–4) – mice were trained to locate the target and enter the escape box within 180 s. Four trials per day, ~15 min apart, were performed for four consecutive days. Probe (day 5) - a single, 90 s trial was performed to measure primary latency and number of errors to reach the blocked target.
Immunohistochemistry and quantifications
Mice were anesthetized with isoflurane and perfused with ice-cold 4% paraformaldehyde in 0.1 M PBS (Sigma-Aldrich, Oakville, ON, Canada). Brains were collected for immunohistochemistry, post-fixed overnight in 10% neutral-buffered formalin (ThermoFisher Scientific, ON, Canada) and transferred to 70% ethanol, paraffin embedded, and sectioned at 4 μm.
Brain tissue slides underwent heat-induced antigen retrieval with Tris-EDTA buffer (pH 9.0) and immunostaining was performed using the Dako Autostainer Plus slide processor (Dako, ON, Canada) and EnVision Flex system (Agilent, Technologies, Mississauga, ON, Canada). Tissue slides were blocked with Dual Endogenous Enzyme Block and Serum-Free Protein Block, and immunostained with the following antibodies: 1:8000 mouse anti-synaptophysin (Sigma-Aldrich), 1:2000 rabbit anti-Iba1 (Wako, VA, USA), 1:8000 rabbit anti-GFAP (Dako), and 1:2000 rabbit anti-Aβ1-40 (F25276 [44]). Slides were incubated with an HRP-conjugated anti-mouse or anti-rabbit secondary antibody, and immunoreactivity was revealed with diaminobenzidine and counterstained with hematoxylin (Dako). Sections were digitally scanned with MIRAX SCAN (Zeiss, ON, Canada) and used for analyses. Representative images in this manuscript have only been cropped and did no undergo any post-processing.
Iba1+ microglia was quantitated using a modified version of an area-specific counting frame [61]. The number of Iba1+cells per mm3 from the PCL to the SLM in the anterior CA1 hippocampus, and in the retrosplenial and S1 area of the cortex, was estimated across a minimum of four sections, spaced 60 μm apart. Aβ1-40 and GFAP immunostaining density were measured in the SR and SLM of the CA1, and in the retrosplenial and S1 area of the cortex. Synaptophysin density was measured in the SR of the CA1, the stratum lucidum of the CA3, and in the molecular layer of the DG. Cell density quantification in the CA1 PCL was performed by counting the number of hematoxylin positive cell soma in brain sections stained for synaptophysin. Measurements were made across three to five sections using ImageJ software (NIH, MD, USA). Quantification was blinded to the genotype.
Golgi–Cox staining and quantification
Dendritic spines were stained with the FD Rapid GolgiStain™ kit from FD NeuroTechnologies (Columbia, MD, USA), according to the manufacturer’s instructions. Mice were isoflurane anaesthetized and sacrificed by cervical dislocation, the brains dissected out and rapidly rinsed with dH2O before incubation in impregnation solution for 2 weeks in the dark at room temperature. Tissues were then immerged in solution C for 3 days. Brains were sectioned at 100 μm, mounted on gelatin-coated slides (#PO101, FD NeuroTechnologies), air-dried for 2 days, stained according to the manufacturer’s protocol, and coverslipped with Permount (Thermo Fisher Scientific).
Brightfield microscopy images of dorsal CA1 pyramidal dendrites located in the SR were acquired at 400× magnification using MIRAX SCAN with an 40× objective. A minimum of 15 neurons, 3 dendrites per neuron with a length ranging 10–45 μm, was analyzed per mouse, totaling a minimum 1000 μm of dendrite length analyzed per mouse. Dendritic segments analyzed were located at least 25 µm from the soma and did not overlap with other cells. Dendritic spines were categorized into 3 groups based on their morphology: mushroom spines with a large head and a narrow neck, thin spines with a smaller head and a narrow neck, and stubby spines devoid of neck [62]. Results were expressed as total number of spines per 10 μm of an uninterrupted section and percentage of each morphological dendritic spine subtype. Tissue processing and quantifications were blinded to mouse genotype.
Protein extraction
Mice were isoflurane anesthetized and sacrificed by cervical dislocation. The hippocampus and cortex sample were dissected out and immediately frozen on dry ice. Brain proteins were extracted by homogenization in 5× volume/weight radioimmunoprecipitation assay (RIPA) buffer (50 mM Trist-HCl, pH 7.4, 1% NP-40, 150 mM NaCl, 0.25% Na-deoxycholate, 1 mM EDTA) with protease inhibitors (300 µM AEBSF, 2 µM Leupeptin, 2.4 µM Pepstatin A, 0.8 µM TLCK, 1 mM PMSF, 1 mM NaF, 1 mM Na3VO4) on ice with a tissue homogenizer (OMNI International, Kennesaw, GA, USA). Samples were centrifuged (20,000 × g, 4 °C) for 20 min, the supernatant recovered and protein concentration was quantified using the BCA method (ThermoFisher Scientific). Protein samples for western blot were prepared in LDS4x buffer (Invitrogen), with 50 mM DTT and protease inhibitors, at a protein concentration of 2 μg/μl and boiled for 5 min. RIPA-insoluble pellets for ELISA were further extracted in 70% formic acid in dH2O. The formic acid was evaporated in a speed vacuum and resulting pellet was solubilized in 200 mM Tris-HCl, pH 7.5.
Western blot and quantification
Polyacrylamide gel electrophoresis (PAGE) was used to separate 20 μg protein samples on 4–12% NuPAGE Bis-Tris gels (Invitrogen, Carlsbad, CA, USA) prior to protein transfer onto PVDF membranes (Bio-Rad, Mississauga, ON, Canada). Membranes were blocked in 5% non-fat dry milk in Tris-buffered saline with 0.1% Tween-20 and probed with the following primary antibodies in PBS blocking buffer (ThermoFisher): 1:500 mouse anti-human Aβ1-16 (6E10) (Biolegend, San Diego, CA, USA), 1:2000 rabbit anti-APP C-terminus (Sigma-Aldrich), and 1:5000 mouse anti-β-actin (Sigma-Aldrich). Immunoreactivity was detected with 1:5000 HRP-linked anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) or anti-rabbit (Agilent Technologies) secondary antibodies followed by enhanced chemiluminescence (ECL) (GE Healthcare Life Sciences, Mississauga, ON, Canada). Immunoreactive bands were visualized using the ImageQuant LAS 4000 imaging system (Fujifilm USA, Valhalla, NY, USA). Densitometric analyses were performed with Image J (NIH) and protein levels were normalized to β-actin.
ELISA
ELISA were performed on RIPA and formic acid samples described above using Meso Scale Discovery multiplex kits (Rockville, MD, USA). Mouse cytokines were measured using the mouse pro-inflammatory multiplex panel I, which measured IL-1β, TNF-α, CXCLI, IFN-γ, IL-2, IL-4, IL-5, IL6, IL-10, and IL12p70. Aβ was measured using the multiplex Aβ 6E10 panel, which analyzed Aβ38, Aβ40, and Aβ42. Samples and standards were run in duplicate and prepared according to the manufacturer’s protocols.
Statistical and regression analyses
Statistical analyses were performed using regular ANOVA with Bonferroni’s post-hoc showing significant differences compared to WT and J20 or Dunnett’s post-hoc compared to J20. To determine associations between behavioral performance and immunohistological markers, separate multivariable linear regressions were performed to determine the effect of hippocampal synaptophysin (sum of CA1, CA3 and DG staining), Iba1+ microglia, and Aβ+ staining density on NOR discrimination index scores and Barnes maze probe errors. Similarly, separate multivariable linear regressions were performed to determine the effect of the following biochemical markers on NOR discrimination index scores and Barnes maze probe errors: Golgi–Cox-stained spine density, pro-inflammatory cytokines TNFα, IFNγ, and CXCL1, and formic acid-soluble total Aβ levels. Interaction effects between the independent variables were also assessed for each regression model. Multicollinearity between independent variables was assessed by checking the variance inflation factor for each independent variable in the model. Furthermore, models assumptions on the errors (randomness, homogeneity of variance and normality of the distribution) as well as the presence of outliers were evaluated with a graphical analysis of residuals. All statistical tests were two-sided and performed at the p < 0.05 level of significance.
Survival probabilities were plotted on Kaplan–Meier curves and different genotypes were compared using the Mantel–Cox log-rank test. A Bonferroni corrected significance threshold of p < 0.05 ÷ k, where k is the number of group comparisons, was used to account for multiple comparisons.
Statistical calculations were performed using GraphPad Prism 8.0 software (GraphPad Software, CA, USA). Regression analyses were performed using SAS version 9.4 (SAS Institute Inc., Cary, NC, USA). All mice are indicated as individual points and expressed as mean ± SEM in all behavioral, immunohistological, and biochemical figures. Sample sizes were based on previous behavioral pilot studies that showed the minimum number of mice needed to produce a significant deficit in the J20 experimental line. Sample sizes and detailed statistical values are described in the figure legends.
Supplementary information
Acknowledgements
We acknowledge Dr. José Correa (McGill University) for his help and expertise in our statistical analyses. We thank Sébastien Harton and Dr. Julie Gervais at the IRIC at the Université de Montréal for help with breeding protocols.
Author contributions
Conceptualization, ALB; Methodology, ALB, AN, MLF and JF; Investigation, JF, AN and MLF; Formal Analysis, JF and AN; Writing – Original Draft, ALB, JF and MLF; Writing – Review and Editing, ALB, AN, MLF and JF; Visualization, ALB, AN, and JF; Project Administration, ALB and JF; Funding Acquisition, ALB; Supervision, ALB.
Funding
This work was supported by funds from the Canadian Institutes for Health Research 2011MOP-243413-BCA-CGAG-45097 and 201610PJT-377052-PJT-CFAF-45097, Leaders Opportunity Fund, Canadian Foundation for Innovation and Jewish General Hospital Foundation to ALB.
Data availability
The published article includes all data of individual mice tested as shown in the appropriate figures. Digital scans of full immunohistological staining and western blot membranes have been saved electronically and can be made available upon request and provision of a depository with sufficient memory to accept the files. Any additional information is available upon request.
Competing interests
The authors declare no competing interests.
Ethical
All procedures involving animals were in accordance with the Canadian Council on Animal Care guidelines and approved by the animal care committees at Université de Montréal (protocol #20-045) and McGill University (protocol #2014-7558).
Footnotes
Edited by L. Greene
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00881-1.
References
- 1.Cummings J, Lee G, Ritter A, Sabbagh M, Zhong K. Alzheimer’s disease drug development pipeline: 2020. Alzheimers Dement. 2020;6:e12050. doi: 10.1002/trc2.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gong CX, Liu F, Iqbal K. Multifactorial hypothesis and multi-targets for Alzheimer’s disease. J Alzheimers Dis. 2018;64:S107–17. doi: 10.3233/JAD-179921. [DOI] [PubMed] [Google Scholar]
- 3.Guo H, Petrin D, Zhang Y, Bergeron C, Goodyer CG, LeBlanc AC. Caspase-1 activation of caspase-6 in human apoptotic neurons. Cell Death Differ. 2006;13:285–92. doi: 10.1038/sj.cdd.4401753. [DOI] [PubMed] [Google Scholar]
- 4.Kaushal V, Dye R, Pakavathkumar P, Foveau B, Flores J, Hyman B, et al. Neuronal NLRP1 inflammasome activation of Caspase-1 coordinately regulates inflammatory interleukin-1-beta production and axonal degeneration-associated Caspase-6 activation. Cell Death Differ. 2015;22:1676–86. doi: 10.1038/cdd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albrecht S, Bogdanovic N, Ghetti B, Winblad B, LeBlanc AC. Caspase-6 activation in familial Alzheimer disease brains carrying amyloid precursor protein or presenilin I or presenilin II mutations. J Neuropathol Exp Neurol. 2009;68:1282–93. doi: 10.1097/NEN.0b013e3181c1da10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC. Active Caspase-6 and Caspase-6 cleaved Tau in neuropil threads, neuritic plaques and neurofibrillary tangles of Alzheimer’s Disease. Am J Pathol. 2004;165:523–31. doi: 10.1016/S0002-9440(10)63317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albrecht S, Bourdeau M, Bennett D, Mufson EJ, Bhattacharjee M, LeBlanc AC. Activation of caspase-6 in aging and mild cognitive impairment. Am J Pathol. 2007;170:1200–9. doi: 10.2353/ajpath.2007.060974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramcharitar J, Afonso VM, Albrecht S, Bennett DA, LeBlanc AC. Caspase-6 activity predicts lower episodic memory ability in aged individuals. Neurobiol Aging. 2013;34:1815–24. doi: 10.1016/j.neurobiolaging.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeBlanc AC, Ramcharitar J, Afonso V, Hamel E, Bennett DA, Pakavathkumar P, et al. Caspase-6 activity in the CA1 region of the hippocampus induces age-dependent memory impairment. Cell Death Differ. 2014;21:696–706. doi: 10.1038/cdd.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou L, Flores J, Noël A, Beauchet O, Sjostrom PJ, LeBlanc AC. Methylene blue inhibits Caspase-6 activity, and reverses Caspase-6-induced cognitive impairment and neuroinflammation in aged mice. Acta Neuropathol Commun. 2019;7:210. doi: 10.1186/s40478-019-0856-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamblin TC, Chen F, Zambrano A, Abraha A, Lagalwar S, Guillozet AL, et al. Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer’s disease. Proc Natl Acad Sci USA. 2003;100:10032–7. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horowitz PM, Patterson KR, Guillozet-Bongaarts AL, Reynolds MR, Carroll CA, Weintraub ST, et al. Early N-terminal changes and caspase-6 cleavage of tau in Alzheimer’s disease. J Neurosci. 2004;24:7895–902. doi: 10.1523/JNEUROSCI.1988-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klaiman G, Petzke TL, Hammond J, LeBlanc AC. Targets of caspase-6 activity in human neurons and Alzheimer disease. Mol Cell Proteom. 2008;7:1541–55. doi: 10.1074/mcp.M800007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokolowski JD, Gamage KK, Heffron DS, LeBlanc AC, Deppmann CD, Mandell JW. Caspase-mediated cleavage of actin and tubulin is a common feature and sensitive marker of axonal degeneration in neural development and injury. Acta Neuropathol Commun. 2014;2:16. doi: 10.1186/2051-5960-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halawani D, Tessier S, Anzellotti D, Bennett DA, Latterich M, LeBlanc AC. Identification of Caspase-6-mediated processing of the valosin containing protein (p97) in Alzheimer’s disease: a novel link to dysfunction in ubiquitin proteasome system-mediated protein degradation. J Neurosci. 2010;30:6132–42. doi: 10.1523/JNEUROSCI.5874-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gervais F, Xu D, Robertson G, Vaillancourt J, Zhu Y, Huang J, et al. Involvement of caspases in proteolytic cleavage of Alzheimer’s ß-amyloid precursor protein and amyloidogenic ß-peptide formation. Cell. 1999;97:395–406. doi: 10.1016/s0092-8674(00)80748-5. [DOI] [PubMed] [Google Scholar]
- 17.LeBlanc A, Liu H, Goodyer C, Bergeron C, Hammond J. Caspase-6 role in apoptosis of human neurons, amyloidogenesis, and Alzheimer’s disease. J Biol Chem. 1999;274:23426–36. doi: 10.1074/jbc.274.33.23426. [DOI] [PubMed] [Google Scholar]
- 18.Tesco G, Koh YH, Kang EL, Cameron AN, Das S, Sena-Esteves M, et al. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–37. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142:637–46. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klaiman G, Champagne N, LeBlanc AC. Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochim Biophys Acta. 2009;1793:592–601. doi: 10.1016/j.bbamcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Goodyer C, LeBlanc A. Selective and protracted apoptosis in human primary neurons microinjected with active caspase-3, -6, -7, and -8. J Neurosci. 2000;20:8384–9. doi: 10.1523/JNEUROSCI.20-22-08384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sivananthan SN, Lee AW, Goodyer CG, LeBlanc AC. Familial amyloid precursor protein mutants cause caspase-6-dependent but amyloid beta-peptide-independent neuronal degeneration in primary human neuron cultures. Cell Death Dis. 2010;1:e100. doi: 10.1038/cddis.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cusack CL, Swahari V, Henley HW, Ramsey JM, Deshmukh M. Distinct pathways mediate axon degeneration during apoptosis and axon-specific pruning. Nat Commun. 2013;4:1876. doi: 10.1038/ncomms2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–9. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Noël A, Zhou L, Foveau B, Sjostrom PJ, LeBlanc AC. Differential susceptibility of striatal, hippocampal and cortical neurons to Caspase-6. Cell Death Differ. 2018;25:1319–35. doi: 10.1038/s41418-017-0043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–50. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 27.Heneka MT, Kummer MP, Stutz A, Delekate A, Schwartz S, Vieira-Saecker A, et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493:674–8. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffin WS, Stanley LC, Ling C, White L, MacLeod V, Perrot LJ, et al. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer’s disease. Proc Natl Acad Sci USA. 1989;86:7611–5. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaftel SS, Griffin WS, O’Banion MK. The role of interleukin-1 in neuroinflammation and Alzheimer disease: an evolving perspective. J Neuroinflammation. 2008;5:7. doi: 10.1186/1742-2094-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trompet S, de Craen AJ, Slagboom P, Shepherd J, Blauw GJ, Murphy MB, et al. Genetic variation in the interleukin-1 beta-converting enzyme associates with cognitive function. The PROSPER study. Brain. 2008;131:1069–77. doi: 10.1093/brain/awn023. [DOI] [PubMed] [Google Scholar]
- 31.Pozueta A, Vazquez-Higuera JL, Sanchez-Juan P, Rodriguez-Rodriguez E, Sanchez-Quintana C, Mateo I, et al. Genetic variation in caspase-1 as predictor of accelerated progression from mild cognitive impairment to Alzheimer’s disease. J Neurol. 2011;258:1538–9. doi: 10.1007/s00415-011-5935-y. [DOI] [PubMed] [Google Scholar]
- 32.Flores J, Noël A, Foveau B, Beauchet O, LeBlanc AC. Pre-symptomatic Caspase-1 inhibitor delays cognitive decline in a mouse model of Alzheimer disease and aging. Nat Commun. 2020;11:4571. doi: 10.1038/s41467-020-18405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flores J, Noël A, Foveau B, Lynham J, Lecrux C, LeBlanc AC. Caspase-1 inhibition alleviates cognitive impairment and neuropathology in an Alzheimer’s disease mouse model. Nat Commun. 2018;9:3916. doi: 10.1038/s41467-018-06449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pontillo A, Catamo E, Arosio B, Mari D, Crovella S. NALP1/NLRP1 genetic variants are associated with Alzheimer disease. Alzheimer Dis Assoc Disord. 2012;26:277–81. doi: 10.1097/WAD.0b013e318231a8ac. [DOI] [PubMed] [Google Scholar]
- 35.Harris JA, Devidze N, Halabisky B, Lo I, Thwin MT, Yu GQ, et al. Many neuronal and behavioral impairments in transgenic mouse models of Alzheimer’s disease are independent of caspase cleavage of the amyloid precursor protein. J Neurosci. 2010;30:372–81. doi: 10.1523/JNEUROSCI.5341-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan MS, Tan L, Jiang T, Zhu XC, Wang HF, Jia CD, et al. Amyloid-beta induces NLRP1-dependent neuronal pyroptosis in models of Alzheimer’s disease. Cell Death Dis. 2014;5:e1382. doi: 10.1038/cddis.2014.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angel A, Volkman R, Royal TG, Offen D. Caspase-6 knockout in the 5xFAD model of Alzheimer’s disease reveals favorable outcome on memory and neurological hallmarks. Int J Mol Sci. 2020;21:1144. [DOI] [PMC free article] [PubMed]
- 38.Vazquez-Higuera JL, Rodriguez-Rodriguez E, Sanchez-Juan P, Mateo I, Pozueta A, Martinez-Garcia A, et al. Caspase-1 genetic variation is not associated with Alzheimer’s disease risk. BMC Med Genet. 2010;11:32. doi: 10.1186/1471-2350-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou L, Nho K, Haddad MG, Cherepacha N, Tubeleviciute-Aydin A, Tsai AP, et al. Rare CASP6N73T variant associated with hippocampal volume exhibits decreased proteolytic activity, synaptic transmission defect, and neurodegeneration. Sci Rep. 2021;11:12695. doi: 10.1038/s41598-021-91367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pompl PN, Yemul S, Xiang Z, Ho L, Haroutunian V, Purohit D, et al. Caspase gene expression in the brain as a function of the clinical progression of Alzheimer disease. Arch Neurol. 2003;60:369–76. doi: 10.1001/archneur.60.3.369. [DOI] [PubMed] [Google Scholar]
- 41.Saresella M, La Rosa F, Piancone F, Zoppis M, Marventano I, Calabrese E, et al. The NLRP3 and NLRP1 inflammasomes are activated in Alzheimer’s disease. Mol Neurodegener. 2016;11:23. doi: 10.1186/s13024-016-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu BH, Wang X, Ma YX, Wang S. CMV enhancer/human PDGF-beta promoter for neuron-specific transgene expression. Gene Ther. 2004;11:52–60. doi: 10.1038/sj.gt.3302126. [DOI] [PubMed] [Google Scholar]
- 43.Sasahara M, Fries JW, Raines EW, Gown AM, Westrum LE, Frosch MP, et al. PDGF B-chain in neurons of the central nervous system, posterior pituitary, and in a transgenic model. Cell. 1991;64:217–27. doi: 10.1016/0092-8674(91)90223-l. [DOI] [PubMed] [Google Scholar]
- 44.LeBlanc A. Increased production of 4 kDa amyloid beta peptide in serum deprived human primary neuron cultures: possible involvement of apoptosis. J Neurosci. 1995;15:7837–46. doi: 10.1523/JNEUROSCI.15-12-07837.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michael BD, Bricio-Moreno L, Sorensen EW, Miyabe Y, Lian J, Solomon T, et al. Astrocyte- and neuron-derived CXCL1 drives neutrophil transmigration and blood-brain barrier permeability in viral encephalitis. Cell Rep. 2020;32:108150. doi: 10.1016/j.celrep.2020.108150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Probert L. TNF and its receptors in the CNS: the essential, the desirable and the deleterious effects. Neuroscience. 2015;302:2–22. doi: 10.1016/j.neuroscience.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 47.Pinto B, Morelli G, Rastogi M, Savardi A, Fumagalli A, Petretto A, et al. Rescuing over-activated microglia restores cognitive performance in juvenile animals of the Dp(16) mouse model of down syndrome. Neuron. 2020;108:887–904 e12. doi: 10.1016/j.neuron.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He Z, Yang Y, Xing Z, Zuo Z, Wang R, Gu H, et al. Intraperitoneal injection of IFN-gamma restores microglial autophagy, promotes amyloid-beta clearance and improves cognition in APP/PS1 mice. Cell Death Dis. 2020;11:440. doi: 10.1038/s41419-020-2644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baruch K, Deczkowska A, Rosenzweig N, Tsitsou-Kampeli A, Sharif AM, Matcovitch-Natan O, et al. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer’s disease. Nat Med. 2016;22:135–7. doi: 10.1038/nm.4022. [DOI] [PubMed] [Google Scholar]
- 50.Boro M, Balaji KN. CXCL1 and CXCL2 regulate NLRP3 inflammasome activation via G-protein-coupled receptor CXCR2. J Immunol. 2017;199:1660–71. doi: 10.4049/jimmunol.1700129. [DOI] [PubMed] [Google Scholar]
- 51.Hernandez JCC, Bracko O, Kersbergen CJ, Muse V, Haft-Javaherian M, Berg M, et al. Neutrophil adhesion in brain capillaries reduces cortical blood flow and impairs memory function in Alzheimer’s disease mouse models. Nat Neurosci. 2019;22:413–20. doi: 10.1038/s41593-018-0329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson ECB, Ho K, Yu GQ, Das M, Sanchez PE, Djukic B, et al. Behavioral and neural network abnormalities in human APP transgenic mice resemble those of App knock-in mice and are modulated by familial Alzheimer’s disease mutations but not by inhibition of BACE1. Mol Neurodegener. 2020;15:53. doi: 10.1186/s13024-020-00393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luksch H, Romanowski MJ, Chara O, Tungler V, Caffarena ER, Heymann MC, et al. Naturally occurring genetic variants of human caspase-1 differ considerably in structure and the ability to activate interleukin-1beta. Hum Mutat. 2013;34:122–31. doi: 10.1002/humu.22169. [DOI] [PubMed] [Google Scholar]
- 54.Tubeleviciute-Aydin A, Zhou L, Sharma G, Soni IV, Savinov SN, Hardy JA, et al. Rare human Caspase-6-R65W and Caspase-6-G66R variants identify a novel regulatory region of Caspase-6 activity. Sci Rep. 2018;8:4428. doi: 10.1038/s41598-018-22283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Godefroy N, Foveau B, Albrecht S, Goodyer CG, LeBlanc AC. Expression and activation of caspase-6 in human fetal and adult tissues. PLoS One. 2013;8:e79313. doi: 10.1371/journal.pone.0079313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, et al. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–8. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright AL, Zinn R, Hohensinn B, Konen LM, Beynon SB, Tan RP, et al. Neuroinflammation and neuronal loss precede Abeta plaque deposition in the hAPP-J20 mouse model of Alzheimer’s disease. PLoS One. 2013;8:e59586. doi: 10.1371/journal.pone.0059586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovarova M, Hesker PR, Jania L, Nguyen M, Snouwaert JN, Xiang Z, et al. NLRP1-dependent pyroptosis leads to acute lung injury and morbidity in mice. J Immunol. 2012;189:2006–16. doi: 10.4049/jimmunol.1201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuida K, Lippke JA, Ku G, Harding MW, Livingston DJ, Su MS, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–3. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 60.Zandy AJ, Lakhani S, Zheng T, Flavell RA, Bassnett S. Role of the executioner caspases during lens development. J Biol Chem. 2005;280:30263–72. doi: 10.1074/jbc.M504007200. [DOI] [PubMed] [Google Scholar]
- 61.Gundersen HJ, Jensen EB. The efficiency of systematic sampling in stereology and its prediction. J Microsc. 1987;147:229–63. doi: 10.1111/j.1365-2818.1987.tb02837.x. [DOI] [PubMed] [Google Scholar]
- 62.Peters A, Kaiserman-Abramof IR. The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines. Am J Anat. 1970;127:321–55. doi: 10.1002/aja.1001270402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data of individual mice tested as shown in the appropriate figures. Digital scans of full immunohistological staining and western blot membranes have been saved electronically and can be made available upon request and provision of a depository with sufficient memory to accept the files. Any additional information is available upon request.