Abstract
Mutations in susceptibility alleles correlate with gut-inflammatory diseases, such as Crohn’s disease; however, this does not often impact the disease progression indicating the existence of compensatory genes. We show that a reduction in Foxo3a expression in IL-10-deficient mice results in a spontaneous and aggressive Crohn’s- like disease with 100% penetrance, which is rescued by deletion of myeloid cells, T cells and inhibition of mTORC1. In Foxo3a−/− IL-10−/− mice, there is poor cell death of myeloid cells in the gut, leading to increased accumulation of myeloid and T cells in the gut. Myeloid cells express high levels of inflammatory cytokines, and regulatory T cells are dysfunctional despite increased abundance. Foxo3a signaling represses the transcription of glutaminase (GLS/GLS2) to prevent over-consumption of glutamine by activated T cells and its conversion to glutamate that contributes to the TCA cycle and mTORC1 activation. Finally, we show that Foxo3a restricts the abundance of colitogenic microbiota in IL-10-deficient mice. Thus, by suppressing glutaminolysis in activated T cells Foxo3a mediates a critical checkpoint that prevents the development of fulminant gut inflammatory disease.
Subject terms: Cell death and immune response, Chronic inflammation
Introduction
Immune cells mediate surveillance against pathogens in different anatomical compartments. Since the gut is one of the most prominent mucosal environments harboring a massive microbiome, understanding the mechanisms that regulate immune responses in the gut is important. Immune responses are initiated by toll-like receptor (TLR) engagement which results in the activation of transcription factors (e.g., NF-κB) and the expression of pro- (e.g., TNFα) and anti- (e.g., IL-10) inflammatory cytokines [1]. IL-10 inhibits the transcription of pro-inflammatory cytokines, and this results in the inhibition of tissue toxicity, wound healing and tissue repair [2].
Forkhead box O (FOXO) transcription factors promote the expression of genes that regulate cell survival, cell division and energy utilization [3]. In mammals, there are four individual Foxo genes (Foxo1, Foxo3, Foxo4 and Foxo6), which have unique and/or redundant functions [3]. In the immune system, Foxo1 plays a dominant role in T cell response and the development of regulatory T cells [4–6] whereas Foxo3a has a redundant role [3, 7–9]. Foxo3a was originally identified as a tumor suppressor [10], but it also appears to regulate inflammation [11–13]. Reports on the molecular mechanisms through which Foxo3a modulates immune responses have been inconsistent [12, 14]. The acquired immune response in Foxo3a-deficient mice has been shown to be modulated modestly with no impact on the viral or bacterial burden [7, 8]. In contrast to Foxo3a, IL-10 is an anti-inflammatory cytokine that is expressed by myeloid cells and T cells and inhibits inflammatory responses [15]. Deficiency of IL-10 has been shown to result in spontaneous gut inflammatory disease [16] that is caused by poor functioning of regulatory T cells [17]. Modulation of cellular metabolism has been shown to play an important role in inflammatory responses [18]. Most of the insights in Foxo3a signaling have been derived from endothelial-, cancer- or neuronal cells, where Foxo3a has been shown to regulate metabolism [19]. Interestingly, IL-10 has recently been shown to regulate metabolism by inhibiting mTORC1 signaling [20].
Foxo3a signaling has been shown to negatively regulate the expression of IL-10 [11, 21]. Since the impairment in IL-10 signaling has been shown to promote susceptibility to Crohn’s disease [22, 23], and some strains of IL-10-deficient mice develop gut inflammation [16, 24] we generated mice with a double-deficiency of IL-10 and Foxo3a. We report that Foxo3a signaling restrains the metabolism in myeloid cells and T cells, and this maintains the function of regulatory T cells and prevents the accumulation of colitogenic microbiota and development of gut inflammation in Il-10−/− hosts.
Results
Loss of Foxo3a promotes the development of colitis in IL-10-deficient mice
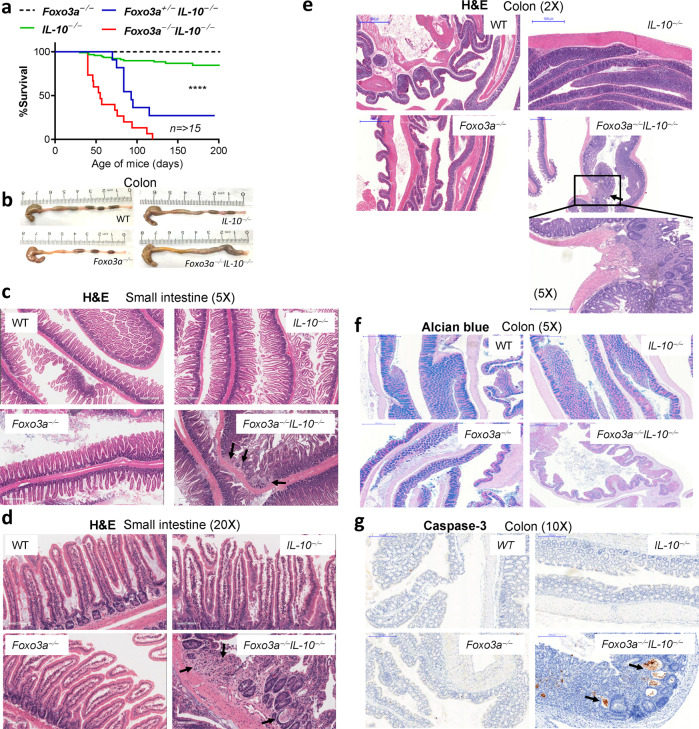
Foxo3a−/− mice express increased levels of IL-10 relative to WT mice [11]. We crossbred Foxo3a+/− Il-10+/− mice to generate Foxo3a-IL-10 double-deficient mice (Fig. S1a) and observed a typical Mendelian distribution of frequencies with ~6.5% of off-springs being Foxo3a−/− Il-10−/− (Fig. S1b). While the weights of the kidney, liver and small intestines were comparable across the four groups of mice, the weight of the colon was increased in the Foxo3a−/− Il-10−/− mice (Fig. S1c). We observed that littermates (males and females) that were deficient in both Foxo3a and IL-10 started losing weight spontaneously as of day 36 after birth, and by day 100 all mice reached a humane endpoint (>20% drop in weight, diarrhea, rectal bleeding, hunching, poor movement, and piloerection) (Fig. 1a and S1d). A heterozygosity of Foxo3a (Foxo3a+/−) was sufficient to trigger this phenotype in Il-10−/− mice albeit at a slightly delayed kinetics and penetrance. Minority of Il-10−/− and no Foxo3a−/− littermates developed this phenotype (Fig. 1a). The colons of Foxo3a−/−Il-10−/− mice appeared to be heavier and inflamed (Fig. 1b) in comparison to the other groups of mice. H&E staining of small intestines revealed focal erosion, loss of epithelial integrity and patchy inflammation (Fig. 1c, d). Patchy villous shortening (Fig. S1e) in association with reactive and regenerative epithelial changes were observed in the small bowel of double-knockout mice. This progressive disease in Foxo3a−/−Il-10−/− mice was characterized by the discontinuous pattern of inflammation throughout the length of the colon and demonstrated a mixed inflammatory infiltrate expanding into the mucosa and submucosa (Fig. 1e). Alcian blue staining of the small intestines and the colons demonstrated a reduction in the amount of mucin produced by the goblet cells in Foxo3a−/−Il-10−/− mice (Fig. 1f and S1f). In addition, cleaved caspase-3, indicative of cell death, was detected only in the crypts of Foxo3a−/−Il-10−/− mice (Fig. 1g and S1g, h). Masson Trichrome staining of tissue sections revealed increased deposition of collagen into the submucosa of Foxo3a−/−Il-10−/− mice, an indication of increased fibrosis (Fig. S1i). The type of inflammation observed in the Foxo3a−/−Il-10−/− mice is reminiscent of Crohn’s disease [22, 23]. Il-10−/− mice in the 129SvEv genetic background were reported to develop colitis spontaneously [16]. This, however, did not occur in mice on the C57BL/6 J and C57BL/10 genetic background [16, 24]. This suggests that there are additional mechanisms that control the development of gut inflammation in IL-10-deficient hosts.
Fig. 1. Foxo3a prevents the development of colitis in Il-10-deficient mice.
a Survival of mice monitored over a period of 200 days. b Representative images of the colons of the four groups of mice at 6 weeks after birth. c, d Representative images (H&E) of the small intestines of the four groups of mice. Arrows indicate areas of damage. e Representative images (H&E) of the colons of the four groups of mice. f Representative images of Alcian blue staining of the colons of the four groups of mice. g Representative images of the colons stained with anti-Caspase 3 antibody by IHC in the four groups of mice. Arrows indicate apoptotic crypts. Data are representative of a minimum of three mice per group. Data in (b-g) is from mice at 6 weeks after birth. The Mantel–Cox test was used for survival analysis.
Foxo3a prevents the disruption in the homeostasis of myeloid- and T- cells in IL-10-deficient hosts
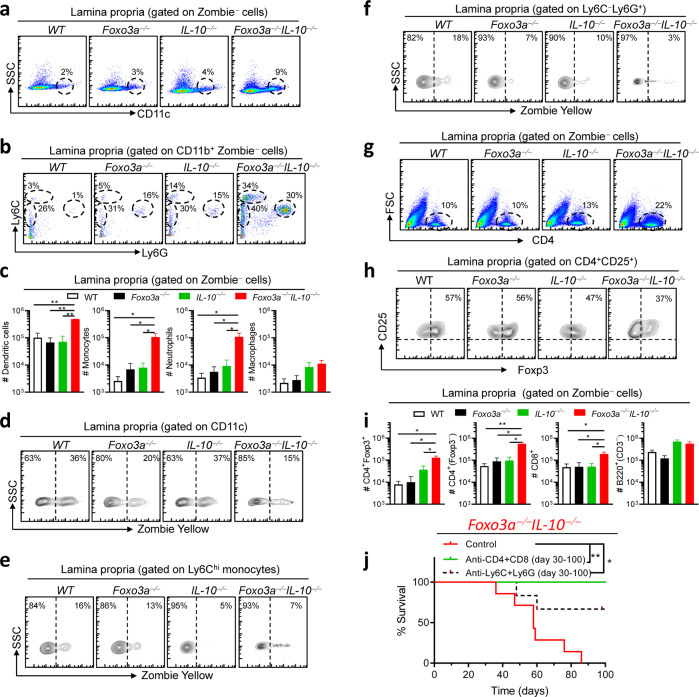
Given the aggressive gut phenotype that we observed in the double-deficient mice, we evaluated whether this correlated with increased numbers of various immune cell subsets in the colonic lamina propria. We observed that the numbers of infiltrating innate immune cells, including DCs (CD11c+), monocytes (Ly6Chi and Ly6Cint) and neutrophils (Ly6Ghi) were increased in Foxo3a−/−Il-10−/− mice in comparison to the other groups (Fig. 2a–c). Viability stain (Zombie) indicated poor cell death of the above-mentioned cell subsets in the lamina propria of Foxo3a−/−Il-10−/− mice (Fig. 2d–f), which explains the increased cell numbers in these mice. There were increased numbers of CD4+ and CD8+ T cells in the lamina propria of Foxo3a−/−Il-10−/− mice in comparison to the other groups (Fig. 2g, i). The overall number of T-regs (CD4+Foxp3+) was significantly higher in Foxo3a−/−Il-10−/− mice (Fig. 2i) although the proportion of T-regs was slightly reduced (Fig. 2h).
Fig. 2. Foxo3a controls the immune cell numbers in the colonic lamina propria.
a, b Representative FACS plots showing the gating strategy for myeloid cells in the colon lamina propria of the four groups of mice. c Total numbers of myeloid subsets in the colon lamina propria of the four groups of mice. d–f Representative FACS plot showing the viability of various myeloid cell subsets in the colon lamina propria in the four groups of mice. g, h Representative FACS plots showing the expression of CD4 and Foxp3 in the colonic lamina propria of the four groups of mice. i Total numbers of lymphoid subsets in the colonic lamina propria of the four groups of mice. j Survival of Foxo3a−/−IL10−/− mice depleted of myeloid cells or T cells using appropriate antibodies (mentioned in the methods section) given twice a week (200 μg/injection) from day 30 till day 100 of birth. The Mantel–Cox test was used for survival analysis. All graphs depict mean ± SEM. Statistics were done using One-way ANOVA (*P < 0.05, ***P < 0.001, and ****P < 0.0001).
In the spleens of Foxo3a−/−Il-10−/− mice, we observed that there was an expansion of the white pulp and increased extramedullary hematopoiesis (revealed by elevation in the numbers of megakaryocytes) (Fig. S2 a). Unlike in the lamina propria, the proportion of T cells was not modulated in the spleens of Foxo3a−/−Il-10−/− mice (Fig. S2b). However, the proportion of activated T cells (CD62Llow) among CD4+ and CD8+ T cells was highly upregulated in the spleens of Foxo3a−/−Il-10−/− mice (Fig. S2c, d). The proportion of T-regs was reduced in the spleens of Foxo3a−/− and Foxo3a−/−Il-10−/− mice (Fig. S2e), although this may be related to their migration to the gut. In the absence of Foxo3a, the MFI of Foxp3 was reduced in T-regs (Fig. S2f). In the spleen, we also observed a remarkable expansion of the myeloid cell compartment (CD11b+), especially macrophages (F4/80hiLy6Clow), neutrophils and monocytes (Fig. S2g–i). Biweekly depletion of myeloid cells (with anti-Ly-6C/G antibodies) or T cells (with anti-CD4 and anti-CD8 antibodies) resulted in potent rescue of the phenotype in Foxo3a−/−Il-10−/− mice (Fig. 2j), suggesting that both the myeloid and the T cell compartment perpetuate the disease in Foxo3a−/−Il-10−/− mice.
Foxo3a prevents the development of a hyperinflammatory response in the gut of IL-10-deficient mice
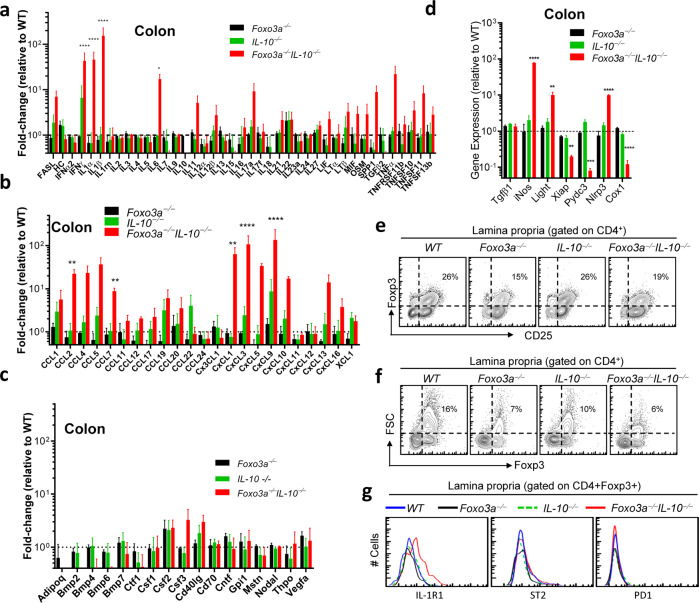
We performed a transcriptome analysis of cytokines and chemokines in the gut of the four groups of mice. The mRNA levels of critical pro-inflammatory cytokines such as IL-6, IFNγ, IL-1α and IL-1β and members of the TNFα-family, CCL- and CXCL- chemokine family were elevated in the colons of naive Foxo3a−/−Il-10−/− mice (Fig. 3a–c). Interestingly, the elevation in the transcription of these inflammatory cytokines/chemokines was not observed in the spleens (Fig. S3a–c). We also measured the mRNA expression (by qRT-PCR) of various pro-inflammatory genes and observed that the expression of iNOS, Light and Nlrp3, was significantly enhanced in the colons of Foxo3a−/−Il-10−/− mice (Fig. 3d). Interestingly, the expression of the anti-inflammatory genes such as Xiap, Pydc3 and Cox1, was reduced. While the proportion of Foxp3+ T-regs in the lamina propria of Foxo3a−/−Il-10−/− mice was slightly decreased (Figs. 3e, S3d), the proportion of activated non-T-reg CD4+ T cells (CD25+) was increased (Fig. 3e). The proportion of FSChi (forward scatter) T-regs was reduced in the lamina propria of Foxo3a−/−Il-10−/− mice (Fig. 3f and S3e). Finally, the T-regs in the lamina propria of Foxo3a−/−Il-10−/− mice expressed increased levels of IL-1R1 (Fig. 3g), indicative of exposure to inflammatory cytokines and instability of their inhibitory function [25]. Interestingly, splenic T-regs of Foxo3a−/−Il-10−/− mice did not display upregulation of IL-1R1 (Fig. S3f) which makes sense, since we did not observe any upregulation of inflammatory cytokines in the spleen (Fig. S3a–c).
Fig. 3. Foxo3a prevents the development of an inflammatory response in the colons of Il-10-deficient mice and promotes Foxp3 expression in regulatory T-cells.
a–c Expression of inflammatory cytokines and chemokines in the colons of the four groups of naïve mice measured by q-RT PCR (RT2 profiler array). Values are fold-change relative to WT colons. d Expression of pro- and anti-inflammatory mediators in the colons of the four groups of naïve mice measured by qRT-PCR. Values are fold-change relative to WT colons. Data are representative of four biological replicates per group, each being an average of 2 experimental replicates. e, f Representative FACS plots showing Foxp3 and CD25 expression in gated CD4+ T-cells in the lamina propria. g Representative histograms showing the expression for IL-1R1, PD-1 and ST2 in lamina propria CD4+ Foxp3+ T-cells. Data are representative of three biological replicates per group. e, g All graphs depict mean ± SEM. Statistics were done using One-way ANOVA (*P < 0.05, ***P < 0.001, and ****P < 0.0001).
Foxo3a restricts cytokine expression by myeloid- and T-cells of IL-10-deficient mice
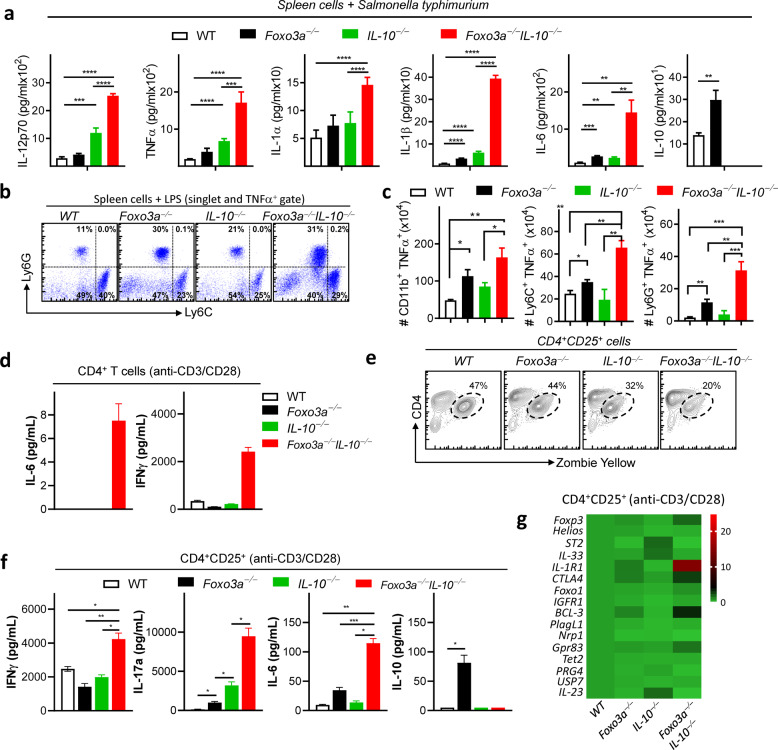
Infection of spleen cells with Salmonella typhimurium revealed that deficiency in both Foxo3a and Il-10 results in exacerbation of inflammatory cytokines (Fig. 4a). Measurement of intracellular TNFα expression by Flow Cytometry (Fig. S4a) indicated that neutrophils, monocytes, and macrophages produced TNFα (Fig. 4b, c). Purified naïve splenic CD4+ T-cells, deficient in both Foxo3a and IL-10, produced high levels of IL-6 and IFNγ at 24 h post stimulation in vitro in comparison to CD4+ T-cells from the other groups (Fig. 4d). To evaluate the suppressive function of purified CD25+CD4+ T-cells, we measured cytokine production at 72 h post stimulation. We observed that the CD25+CD4+ T-cells from Foxo3a−/−Il-10−/− mice underwent reduced cell death upon activation (Fig. 4e, S4b), which correlated with increased production of IFNγ, IL17, and IL-6 (Fig. 4f). Analysis of mRNA expression (by qRT-PCR) of various proteins involved in T-reg development and function revealed significant increase in the expression of Il-1r1 and bcl-3 in Foxo3a−/−Il-10−/− cells (Figs. 4g and S4c). Expression of BCL-3 and IL-1R1 on T-regs has been reported to impair their inhibitory function [25, 26]. The mRNA expression of Foxp3 and Foxo1, which have been shown to impact T-reg function, was not reduced in Foxo3a−/−Il-10−/− cells. These results indicate that the T-regs from Foxo3a−/−Il-10−/− mice are poor at restricting cytokine production by activated T cells.
Fig. 4. Foxo3a is required to suppress overt cytokine expression in the absence of IL-10.
a Cytokine levels measured in the supernatants of spleen cells (107cells/300 μl) infected with Salmonella Typhimurium (ST) for 6 h. b Representative FACS plots showing intracellular TNFα-producing myeloid cells subsets (gated on TNFa+) following LPS treatment of spleen cells of the four groups of naïve mice. c Total number of TNFα-producing myeloid cell subsets following LPS treatment of spleen cells of the four groups of mice. d Cytokine levels measured in the supernatants (24 h) of anti-CD3/CD28 activated CD4+ T-cells. e Representative FACS plot showing CD4 vs. Zombie yellow staining in purified CD25+ CD4+ T-cells activated with anti-CD3/CD28 for 72 h. f Cytokine levels measured in the supernatants (at 72 h) of purified CD25+ CD4+ T cells activated with anti- CD3/CD28 in the presence of rIL-2/rTGFβ. g Expression of various genes by qRT-PCR at 72 h post-activation with anti-CD3/CD28 and rIL-2/rTGFβ. Values are fold-change relative to WT cells. A minimum of 3 biological replicates per group have been used. All graphs depict mean ± SEM. Statistics were done using One-way ANOVA (*P < 0.05, ***P < 0.001 and ****P < 0.0001).
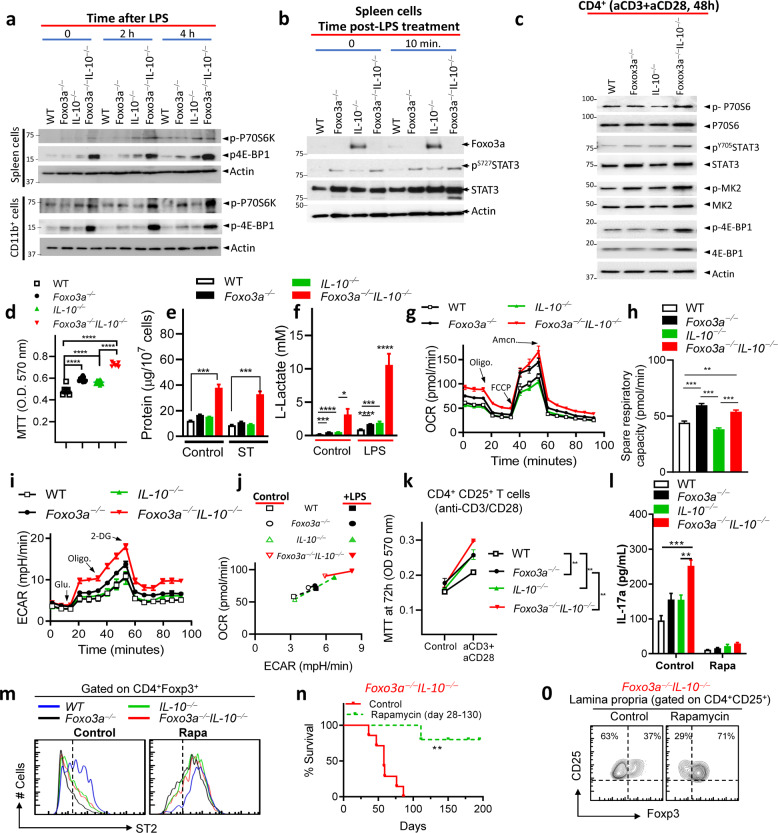
Foxo3a regulates mTORC1 signaling in myeloid cells and T cells
We evaluated the impact of Foxo3a on mTORC1 signaling since this complex plays a key role in metabolism [27]. Phosphorylation of the mTORC1 targets, ribosomal protein S6 kinase (S6K1) and eukaryotic initiation factor 4E-binding protein (4E-BP1), was higher in Foxo3a−/−Il-10−/− spleen cells, CD11b+ cells (Fig. 5a) and CD4+ T cells (Fig. 5c) in comparison to other groups of cells, suggesting that Foxo3a and IL-10 restrain mTORC1 signaling. Interestingly, we observed that the basal expression of Foxo3a was barely detectable in WT spleen cells, possibly due to suppression by IL-10, as Il-10−/− spleen cells showed increased expression of Foxo3a (Fig. 5b). We also observed that the expression of IL-10 was substantially increased in Foxo3a−/− cells (Fig. 4a, f). It has been previously reported that Foxo3a represses IL-10 expression by BCG-infected macrophages via direct binding to IL-10 promoter [21]. These results suggest that Foxo3a and IL-10 regulate each other negatively. We also observed that Foxo3a inhibited the phosphorylation of STAT3 in spleen cells and CD4+ T cells (Fig. 5b, c), perhaps due to Foxo3a-mediated inhibition of IL-10 and IL-6. Interestingly, colon biopsies of IBD-patients were shown to have elevated STAT3 signaling [28].
Fig. 5. Foxo3a controls mTOR activation to restrict gut inflammation in Il-10-deficient hosts.
a, b Western blots of spleen cells and purified splenic CD11b+ cells at various time intervals post-treatment with LPS (100 ng/ml). c Western blot of purified CD4+ T-cells at 48 h post stimulation with anti-CD3/CD28. d MTT uptake was evaluated in control (non-stimulated) spleen cells of various genotypes at 5 h post-culture in media. e Protein amount (μg) measured by BCA per 107 spleen cells before and after infection with ST in the four groups of mice. f L-lactate levels (mM) measured in control and LPS-treated spleen cells of mice at 6 h post-treatment. g, i, j Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured in the spleen cells using a Seahorse XF 96 flux analyzer. h Spare respiratory capacity of cells (pmol/min) was evaluated in the four groups of cells by evaluating the difference between basal ATP production and the maximal activity of the mitochondria. k MTT uptake was evaluated in anti-CD3/CD28-activated purified CD25+CD4+ T-cells at72h. l IL-17a secretion was measured in the supernatants of anti-CD3/CD28-activated CD25+CD4+ T-cells at 48 h post stimulation with and without treatment with Rapamycin (100 nΜ). m Representative FACS plot in gated CD4+Foxp3+ cells showing ST2 expression in cells described in panel l without and with Rapamycin treatment. n Survival of control-fed and rapamycin-fed Foxo3a−/− IL-10−/− mice (a minimum of 5 mice per group). o Representative FACS plot showing CD25 vs. Foxp3 expression in colonic lamina propria-purified CD4+ CD25+ cells in control-fed and rapamycin-fed Foxo3a−/− IL-10−/− mice. The Mantel–Cox test was used for survival analysis. All graphs depict mean ± SEM. Statistics were done using One-way ANOVA (*P < 0.05, ***P < 0.001 and ****P < 0.0001).
We have previously performed microarray analysis of WT and Foxo3a−/− macrophages and reported that the MAPK pathway is significantly elevated in Foxo3a−/− macrophages [11]. MK2 is the downstream kinase that is activated by P38MAPK and has been previously shown to promote cytokine expression by preventing mRNA degradation of various cytokines [29]. Based on these earlier findings, we looked at MK2 and found that its activation is enhanced in CD4+ T cells and spleen cells in the absence of Foxo3a (Fig. 5c and S5a).
We observed that the Foxo3a−/−Il-10−/− spleen cells displayed increased metabolism as evaluated by MTT uptake (Fig. 5d), the total protein amount per cell (Fig. 5e) and the global transcriptional activity (Fig. S5b). Increased MTT uptake by Foxo3a−/−Il-10−/− cells is indicative of elevated mitochondrial activity. We recently reported that Foxo3a regulates several genes in the glycolysis pathway [11], although there was no detectable impact on the production of L-lactate, the end product of glycolysis [11]. Moreover, it has been recently reported that IL-10 inhibits metabolism in macrophages [20]. Since we recently reported that Foxo3a inhibits IL-10 expression [11], we therefore considered the possibility that the lack of any detectable impact on L-lactate production in Foxo3a−/− cells may be due to compensatory increase in IL-10 expression in cells. Hence, we re-evaluated the levels of L-lactate, this time in all four genotypes. In control spleen cells, L-lactate was detectable only in Foxo3a−/− Il-10−/− mice, and it was increased substantially following LPS stimulation in contrast to the other experimental groups where the increase in L-lactate was not as potent (Fig. 5f).
We then measured the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in spleen cells using the Seahorse analyzer. The functional profile of mitochondria was evaluated by determining real-time changes in OCR during sequential treatment of cells with Oligomycin (Inhibitor of ATP synthase), Trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP) (H+ ionophore) and Antimycin/Rotenone (inhibitors of complexes I and III in the electron transport chain). The basal respiration rate (absence of inhibitors) was the highest in Foxo3a−/−Il-10−/− spleen cells, and it was enhanced in response to FCCP or LPS (Fig. 5g, j and S5e). After the addition of the Oligomycin to prevent electron transport through complex I-IV, OCR decreased in cells of the four groups; however, the residual OCR, which is representative of respiration due to proton-leakage, was still the highest in Foxo3a−/−Il-10−/− cells (Figs. 5g and S5e). ATP-linked respiration, which represents the difference between basal and leak respiration after injection of Oligomycin, was highest in Foxo3a−/− and Foxo3a−/−Il-10−/− spleen cells (Fig. 5g and S5 e). This is reflective of an increased ATP demand in the absence of Foxo3a. Foxo3a−/−Il-10−/− spleen cells displayed maximal respiration following addition of FCCP, which depolarizes the mitochondrial membrane potential and allows electron transport chain to function at the maximal rate. Spare respiratory capacity was significantly impacted by the deficiency of Foxo3a (Fig. 5h). ECAR was measured before and after the addition of glucose, Oligomycin and 2-Deoxyglucose. Our results show that Foxo3a−/−Il-10−/− cells undergo maximal ECAR in the absence or presence of LPS (Fig. 5 i, j and S5c, d).
The metabolic activity, as measured by MTT was the highest in Foxo3a−/−Il-10−/− CD4+CD25+ T cells (Fig. 5k), which correlated with increased secretion of IL-17 (Fig. 5l). Treatment of with Rapamycin (inhibitor of mTORC1) reduced IL-17a in all groups (Fig. 5l). We also measured the expression of ST2, which is known to maintain the stable regulatory function of T-regs [25], and observed that ST2 expression on T-regs was highly reduced in Foxo3a as well as IL-10- deficient cells, and this was restored by Rapamycin treatment (Fig. 5m). Finally, treatment of mice with Rapamycin resulted in a substantial rescue in the survival of Foxo3a−/−Il-10−/− mice (Fig. 5n) and an increase in T-regs (Fig. 5o).
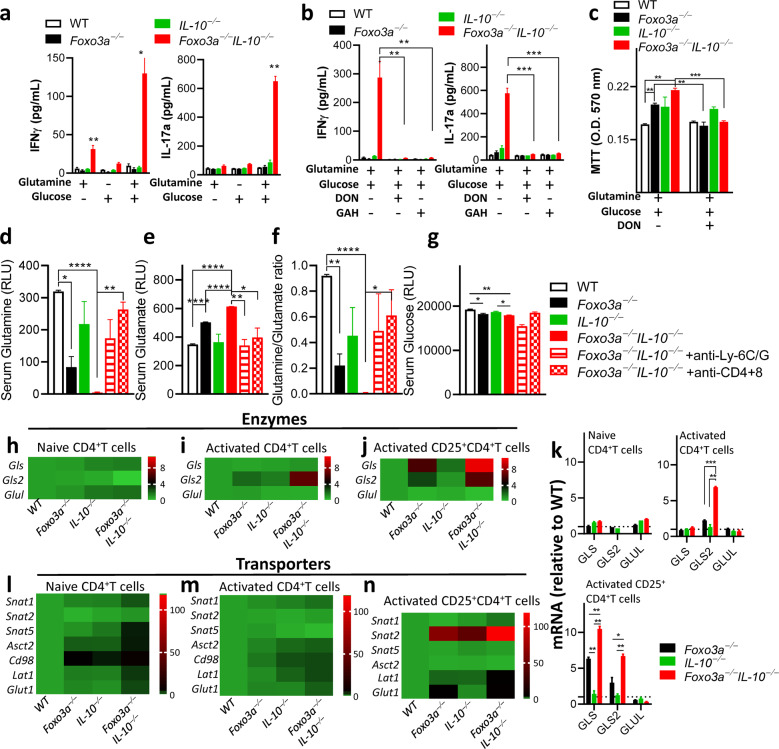
Foxo3a represses glutaminolysis in activated T cells
Besides glucose oxidation, T-cell activation induces a large increase in glutamine import by actively coordinating the up-regulation in the expression of glutamine transporters and activities of enzymes required to allow the use of glutamine as a Krebs cycle substrate in T cells [30]. Moreover, glutamine metabolism has been shown to differentially regulate the development of T cells into Th17 or Th1 subsets [31]. We measured cytokine production by CD4+ T cells upon supplementing the glucose/glutamine free media with either 2 mM glutamine, 10 mM glucose or both. Dialyzed FBS was used for cell culture. Addition of glutamine was sufficient to induce cytokine production by Foxo3a−/−Il-10−/− CD4+ T cells, and this was increased further by the addition of glucose (Fig. 6a and S6a). Expression of cytokines was suppressed by treatment of cells with the inhibitors of glutamine metabolism, 6-Diazo-5-oxo-L-norleucine (DON) or L-Glutamic acid γ-monohydroxamate (GAH) (Fig. 6b). Inhibiting glutamine metabolism also normalized mitochondrial activity in Foxo3a−/− and Foxo3a−/−Il-10−/− cells to WT levels and there was no impact in Il-10−/− CD4+ T cells (Fig. 6c).
Fig. 6. Foxo3a favors glutamine synthesis by repressing the expression of glutaminase and glutamine transporters.
a, b IFNγ and IL-17a levels were measured (at 72 h) in the supernatants of anti-CD3/CD28 activated CD4+ T-cells supplemented with either 10 mM glucose or 2 mM glutamine or both in the absence or presence of the glutamine antagonist DON (50 μM) and inhibitor of glutamine transport GAH (1 mM). c MTT uptake was evaluated in activated CD4+ T-cells in glucose/glutamine free media supplemented with either 10 mM glucose or 2 mM glutamine or both in the absence or presence of the inhibitors DON (50 μM) and GAH (1 mM). d–g Glutamine, glutamate, glutamine/glutamate ratio, and glucose levels were measured (as relative light units) in the sera of mice. h–j, l–n Heatmaps showing the expressions of major metabolic transporters and glutamine metabolism-associated enzymes (relative to wildtype cells) in non-activated and activated CD4+ and activated CD25+ CD4+ T-cells normalized to actin. Values are fold-change relative to WT cells. k Expressions of glutamine metabolism-associated enzymes by q-RT PCR normalized to actin. Values are fold-change relative to WT cells. All graphs depict mean ± SEM. Statistics were done using One-way ANOVA (*P < 0.05, ***P < 0.001 and ****P < 0.0001).
We evaluated the levels of glutamine and glutamate in the serum of 12-week-old mice to determine the impact of Foxo3a, IL-10 and cell depletions on glutamine metabolism globally. The levels of glutamine were significantly reduced in the serum of Foxo3a−/− mice in comparison to IL-10−/− mice (Fig. 6d). Interestingly, glutamine levels in the serum of Foxo3a−/−Il-10−/− mice were abolished, and these were significantly restored in mice that were depleted of myeloid cells and T cells (Fig. 6d). This suggests that T cells and myeloid cells consume glutamine to the extent that there is a depletion in glutamine levels systemically. Since glutaminolysis involves the conversion of glutamine to glutamate by the enzyme glutaminase (GLS/GLS2), we measured serum levels of glutamate and observed an inverse correlation with glutamine levels. Foxo3a−/− Il-10−/− mice had the highest levels of serum glutamate which were reduced by depletion of myeloid and T cells (Fig. 6e). These results suggest that glutamine is increasingly converted to glutamate in Foxo3a−/− Il-10−/− mice (Fig. 6d–f), which would enter the TCA cycle and promote mitochondrial activity. In addition to glutamine levels, the levels of serum glucose were modestly but significantly reduced in Foxo3a−/− Il-10−/− mice (Fig. 6g). Overall, the impact of Foxo3a on glutamine levels was profound, and the double deficiency of Foxo3a and IL-10 leads to massive decline in glutamine pools bordering on glutaminemia, and this was driven by overconsumption by myeloid and T cells.
We performed qRT-PCR to quantitate the expression of enzymes involved in the conversion of glutamine to glutamate and vice versa. We observed that the expression of Gls/Gls2 which convert glutamine to glutamate was markedly enhanced in activated Foxo3a−/−Il-10−/− cells with Foxo3a playing a dominant role (Fig. 6h–k). On the other hand, the expression of Glul (glutamine synthetase) which converts glutamate to glutamine was not modulated by Foxo3a or IL-10 (Fig. 6h–k). Based on this analysis, the equilibrium in Foxo3a−/−Il-10−/− cells would shift towards more glutamine consumption by T cells and increased entry into the TCA cycle, leading to increased mitochondrial activity.
We also measured the expression of key transporters involved in glutamine metabolism. We observed that both Foxo3a and IL-10 appear to modulate the transcription of major glutamine importers (SNAT 1,2, 5 and ASCT2) and exporters (LAT1/CD98 heterocomplex) in CD4+ T-cells (Fig. 6l–n). Naïve CD4+ T-cells isolated from the spleens of Foxo3a−/−Il-10−/− mice showed higher transcript levels for most of the transporters, and this possibly reflects enhanced glutamine metabolism in these cells in vivo (Figs. 6l and S6b). At 48 h post-activation of CD4+ T cells the expression of some of the transporters was still elevated in Foxo3a−/−Il-10−/− cells in comparison to other groups (Figs. 6m and S6c). Interestingly, in purified CD25+ T cells, the expression of SNAT2 was significantly elevated in Foxo3a−/− cells at 72 h post activation and this was elevated even further in Foxo3a−/−Il-10−/− cells (Figs. 6n and S6d).
Overall, these results indicate that Foxo3a represses glutaminolysis in T cells, which reaches a toxic state by an additional deficiency of IL-10.
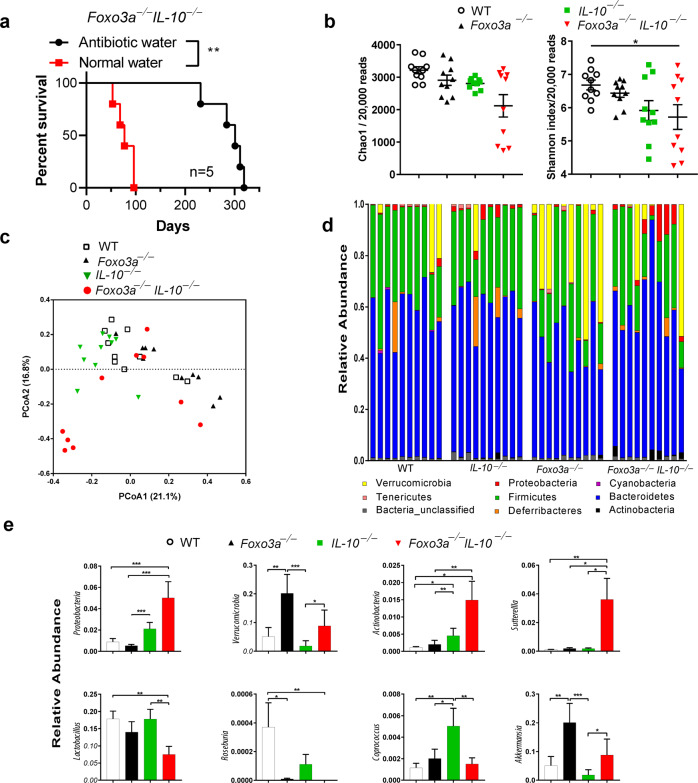
Foxo3a-deficiency promotes the abundance of colitogenic microbiota in IL-10−/− mice
Treatment of Foxo3a−/−IL-10−/− mice with antibiotics significantly increased their survival compared to untreated mice (Fig. 7a). Interestingly, treatment with antibiotics was not a permanent cure for these mice. These results indicate that Foxo3a might influence the gut microbiota of mice. Microbiota diversity was unchanged between WT, IL-10−/−, and Foxo3a−/− mice. However, the microbiota of Foxo3a−/−IL-10−/− mice exhibited a slightly reduced diversity compared to WT mice (Fig. 7b; P < 0.05). Beta diversity analysis showed that the microbiota of different samples was partially clustered based on the mice phenotype. Further analysis revealed that the tested mice phenotypes contributed to 21.039% of the microbiota variability among samples (R2 = 0.21039; P = 0.001; Fig. 7c). We also characterized the composition of gut microbiota in WT, IL-10−/−, Foxo3a−/− and Foxo3a−/−IL-10−/−mice. In general, 8 bacteria phyla were identified with 76–90% of reads assigned to Firmicutes and Bacteroidetes (Fig. 7d). The microbiota of Foxo3a−/− mice exhibited a significantly higher abundance of Verrucomicrobia (mean ± SEM = 20.16 ± 6.6%) as compared to that of WT (5.12 ± 3.15%) or IL-10−/− mice (1.83 ± 1.8%) (Fig. 7e). Proteobacteria were enriched in Foxo3a−/−IL-10−/− mice (5 ± 1.5%) as compared to WT (0.8 ± 0.3%) and Foxo3a−/− (0.5 ± 0.1%) and in IL-10−/− mice (2.1 ± 0.6%) compared to Foxo3a−/− mice. Foxo3a−/−IL-10−/− and IL-10−/− mice showed a higher relative abundance of Actinobacteria compared to WT and Foxo3a−/− mice (Fig. 7e). At the genus level, Sutterella was enriched in the microbiota of Foxo3a−/−IL-10−/− as compared to the other three phenotypes. On the other hand, Roseburia and Lactobacillus were depleted in Foxo3a−/−IL-10−/− microbiota in comparison with WT mice (Fig. 7e). The microbiota of IL-10−/− mice exhibited a higher abundance of Coprococcus compared to other phenotypes. Finally, the genus Akkermansia showed a higher relative abundance in the microbiota of Foxo3a−/− mice as compared to WT and IL-10−/− mice (Fig. 7e). A complete list of identified differential taxa are presented in (Fig. S7a–d). Overall, these results indicate the abundance of colitogenic microbiota in Foxo3a−/−Il-10−/− mice.
Fig. 7. FoxO3a regulates the abundance of gut microbiota.
a Survival of FoxO3a−/−IL-10−/−mice with and without antibiotic treatment. FoxO3a−/−IL-10−/− mice (n = 5; age 5–8 weeks of age) received a 2 month-course of antibiotic treatment consisting of Ciprofloxacin (0.225 mg/mL) and Metronidazole (0.45 mg/mL) in their drinking water. Then, they were maintained on antibiotic-free water for the remainder of the study. As a control group, 5 mice received antibiotic-free drinking water. The numbers of spontaneous deaths were recorded over the course of time. Data was analyzed using Mantel–Cox test. (*P < 0.05, ***P < 0.001, and ****P < 0.0001). b Diversity of intestinal microbiota in WT, IL-10−/−, FoxO3a−/−, and FoxO3a−/−IL-10−/−mice (n = 10 each). Chao1 estimates and Shannon indices were calculated from rarefied 20,000 reads per sample. Lines represent mean ± SEM. Data was analyzed using Kruskal-Wallis test and Two-stage Benjamini, Krieger, and Yekutieli FDR procedure (*P < 0.05). c Principal Coordinate Analyses (PCoA) based on Bray-Curtis distances. PCoA was conducted using QIIME 1.9.0 on 20,000 randomly selected reads per sample. The samples were colored by mice phenotype. PCoA1and PCoA2 represent the top two coordinates that captured the highest variability between samples and the percentage shown indicates the fraction of variation represented by each coordinate. Adonis analysis was used to test for the statistical significance of sample grouping by the mice phenotypes (R2 = 0.21039; P = 0.001). d Relative abundance of the identified phyla in each sample. Samples are grouped by mice phenotype (n = 10/group). e Relative abundance of differentially abundant taxa among the microbiota of WT, IL-10−/−, FoxO3a−/−, and FoxO3a−/− IL-10−/−mice. Lines represent mean. Data was analyzed using Kruskal-Wallis test and Two-stage Benjamini, Krieger, and Yekutieli FDR procedure (*P < 0.05, **P < 0.01, and ***P < 0.001).
Discussion
The enormity of the gut microbiome necessitates extensive regulation of the gut-immune interphase to prevent the development of an inflammatory response that can potentially damage the gut and systemic tissues. Crohn’s disease (CD) is a typical inflammatory disease of the gut, which affects any part of the GI tract with a predilection for the distal ileum and causes a patchy pattern of inflammation extending through the thickness of the bowel wall. Early onset CD (<6 years old) has been reported in children with mutations in Il-10/Il-l0R [22, 23] and Xiap [32]. In congruence with this, Il-10−/− mice were reported to develop colitis spontaneously [16]. This, however, was dependent on the genetic background of the mouse [16, 24] with the 129SvEv strain being susceptible and C57BL/6 J and C57BL/10 strains being resistant. We report here that Foxo3a signaling acts as a key molecular checkpoint to prevent the development of fulminant gut inflammatory disease in Il-10-deficient hosts.
Genome-wide association studies (GWAS) identified numerous susceptibility loci (e.g., NOD2, IL23A, ATG16L1, IL-10) in IBD [33]. However, attempts to correlate genetic susceptibility to disease outcome have not been successful. In a recent GWAS, in mild versus progressive CD patients, four different genetic loci were identified (XACT, MHC, Foxo3, IGFBP1-3) that correlate with disease progression [34]. Our results showing the critical regulatory role of Foxo3a in preventing the development of disease in Il-10−/− mice corroborate the human GWAS. We show that a heterozygosity of Foxo3a is enough to trigger the disease in Il-10−/− mice suggesting that the level of Foxo3a must be very low in wildtype mice so that a mere reduction of one copy is enough to trigger the disease.
Modulation of various susceptibility genes (e.g., NOD2, MyD88, TLR4, NLRP6, NLRP12 and CYLD) does not lead to spontaneous IBD in mice, unless the gut barrier is compromised by administration of dextran sodium sulfate (DSS) [35–37]. Il-10 is one of the few genes whose inactivation causes spontaneous Crohn’s disease-like phenotype, but this occurs only in the 129 Sv strain of mice and at much slower kinetics [16, 24]. The Foxo3a−/−IL-10−/− mice that we have generated develop an early onset, aggressive form of Crohn’s disease spontaneously, and these mice could potentially be used for development of therapeutics as they develop the disease earlier with 100% penetrance.
We have shown that colitis in Foxo3a−/−Il-10−/− mice is immune-related, such that depletion of either the innate or adaptive immune cells ameliorated their survival. We observed increased activation of p38 and STAT3, correlating with increased production of inflammatory cytokines. Transient activation of STAT3 is seen in healthy immune cells, but constitutive activation is associated with immune- activation in IBD patients [38] and development of Th17 phenotype [39]. Since both myeloid cell and T cell depletion ameliorate the disease, it is likely that the myeloid cells are responsible for compromising the function of regulatory T cells leading to increased T-cell activation. It is likely that the disease outcome is the result of modulation in the communication between these three cell types. However, it is also conceivable that there is a crosstalk between the immune cells, gut epithelial cells and the colitogenic microbiota which collectively results in fulminant Crohn’s like disease in the Foxo3a−/−IL-10−/− mice, and neutralization of one cell type or modulation of gut microbiota can offset this crosstalk in favor of host survival.
Initial studies on Foxo3a signaling in the immune system showed that Foxo3a restrains NFκB signaling in T cells; however, the mechanism was not clear, since the promoter region of NFκB did not contain a Foxo3a binding site [12]. On one hand, Foxo3a was shown to induce the activation-induced cell death of T cells in a cell intrinsic manner [8, 9, 40]. On the other hand, Foxo3a was also shown to restrict the expansion of T cell response during a viral infection through a cell extrinsic mechanism by lowering the expression of IL-6 by dendritic cells [7]. There is considerable redundancy in the role of various Foxo transcription factors in the immune system. Foxo1 appears to be the dominant member with Foxo3a playing a supportive role [4–6]. Although Foxo3a modulates the T cell response in some experimental models, Foxo3a has been reported to either attenuate or promote NF-κB signaling in different cell-types [12, 14, 41]. Thus, the consensus mechanism through which Foxo3a modulates immune responses has been inconsistent [12, 14] and the impact on the response and the host outcome is minimal [7, 8].
Lack or dysfunction in regulatory T-cells in mice and humans contributes to the development of autoimmune or inflammatory disease including colitis [42–44]. As with conventional naïve T cells, Foxo1 was shown to promote the development of regulatory T cells with Foxo3a playing a secondary role [3, 7–9]. Deletion of Foxo1 resulted in a significant loss of regulatory T cells, and additional deletion of Foxo3a resulted in the complete elimination of the regulatory T cell pool [5]. Regulatory T-cells are characterized by their expression for the fork-head transcription factor Foxp3, whose sustained expression is essential for their development and suppressive function [45]. Moreover, IL-10 production and signaling endow Tregs with the ability to suppress pathogenic Th17 cells in a STAT3-dependent manner [17]. Although we noticed a marked decrease in the expression of Foxp3 expression in the regulatory T cells of Foxo3a knockout mice, we did not notice any loss in the numbers of regulatory T cells or a decline in their ability to suppress T cell activation. We observed that their ability to suppress T cell activation was compromised only by concomitant deletion of IL-10. It was previously demonstrated that ex vivo expanded Tregs, high on forward scatter, display potent suppressive activities [46]. We have shown that Foxp3+ cells in Foxo3a−/− and Foxo3a−/− IL-10−/− colons are FSChigh, but only in the Foxo3a−/− IL-10−/− colons, regulatory T cells display upregulation of IL-1R1, suggesting an inflammatory Th17 polarizing milieu, which has been previously reported to destabilize regulatory T cells towards Th17 phenotype [25]. Remarkably, the absolute cell numbers of Tregs were increased in the colons of Foxo3a−/− IL-10−/− mice compared to the other groups suggestive of dysfunction through diminished suppressive capacity. The CD25+ T cells in all the experimental groups expressed similar levels of Foxo1 indicating that there wasn’t any compensatory modulation of Foxo1 expression in regulatory T cells in Foxo3a or IL-10 knockouts.
Th17 cells have been shown to favor glutaminolysis through up-regulating glutaminase, the first enzyme in the glutaminolysis pathway [31, 47]. Glutaminase catalyzes the conversion of glutamine to glutamate and exists as two isozymes: kidney-type GLS and liver-type GLS2, which have been shown to play opposing roles in cancer [48, 49]. We show that Foxo3a inhibits glutaminase expression in activated T-cells leading to suppression of glutaminolysis and consequent accumulation of glutamine. The expression of GLS and GLS2 was significantly upregulated in Foxo3a−/− upon activation, and there was no impact on the expression of glutamine synthetase (GLUL/GS) which converts glutamate to glutamine. This suggests that the increase in the levels of glutamate that we observed in Foxo3a−/− mice cannot be due to poor conversion to glutamine. GLS2 expression seems to be inducible as it was only detected after 48 h of activation. Glutamine consumption is markedly increased in Foxo3a−/−IL-10−/− deficient mice, such that their systemic glutamine levels are profoundly depleted. Clearly, the activated immune cells constitute a major source of glutamine utilization, since depleting them rescues glutamine levels in mice.
Metabolism has been shown to play a key role in promoting inflammatory responses [18]. Most of the insights in Foxo3a signaling have been derived from endothelial-, cancer- or neuronal- cells, where Foxo3a has been shown to regulate metabolism [19]. Our results indicate that mTOR signaling is exacerbated in Foxo3a−/−Il-10−/− cells as revealed by increased phosphorylation of the mTORC1 target ribosomal protein S6 kinase (S6K1). The phosphorylation (inhibition) of the translational repressor 4E-BP1 [50] was increased in Foxo3a−/− and Il-10−/− cells, and this was strongly increased in cells with double deficiency of both genes. IL-10 has recently been shown to inhibit mTORC1 signaling in a STAT3-DDIT4 dependent manner in macrophages [20]. Amino acids, such as glutamine, can be potent activators of mTOR signaling, and this is critical for T-cell activation [51]. In fact, in addition to requiring glucose metabolism as the primary metabolic adaptation, T-cells are particularly sensitive to glutamine. Following activation of T cells, glutaminolysis is up-regulated which allows glutamate to enter the TCA cycle and promote the proliferation of activated cells [31]. Glutamine metabolism has been shown to activate mTORC1 [52]. While glutamine is adequately synthesized to meet the metabolic needs, it can become conditionally essential during catabolic states. Systemic glutamine levels decline as glutamine utilization exceeds endogenous glutamine production. Rapamycin-treated Foxo3a-IL-10- deficient mice demonstrated significant improvement in survival and enhanced expansion of regulatory T-cells. In addition, Rapamycin seemed to strengthen ST2 expression in regulatory T cells. ST2, also known as IL-1RL1, is the receptor for IL-33 that activates a Th2 response in case of tissue damage [53]. In GATA3+ T regs, ST2/IL-33 signaling is shown to promote the expression of Foxp3 and the proper function of regulatory T cells [54–56]. Therefore, an impairment in both Foxo3a and IL-10 signaling results in aberrant immune metabolic responses leading to comprehensive mTORC1 activation potentially triggering an exaggerated inflammatory response in the gut.
Alterations in the gut microbiota play a pivotal role in driving the pathogenesis of numerous chronic inflammatory diseases including IBD [57, 58]. Loss of commensal homeostasis due to genetic deficiencies correlates with exacerbated colitis [36, 37]. We observed that the deficiency of Foxo3a resulted in an abundance of the phylum Verrucomicrobia, and more specifically its genus Akkermensia, which are anaerobic Gram-negative mucin-degrading bacteria that contribute to intestinal health by degrading mucin and producing short chain fatty acids supplied to mucin-producing goblet cells [59]. Akkermensia, induce the production of IL-10 and promote the intestinal barrier integrity [60]. This may suggest that in Foxo3a−/− mice Akkermensia may contribute to increased production of IL-10 and protection against colitis. In contrast, IL-10−/− mice did not display any major changes in the gut microbiome except that there was a slight increase in the abundance of the colitogenic Proteobacteria.
The microbiome of IBD patients is associated with decreased levels of resident Firmicutes and/or Bacteroides and an overabundance of Proteobacteria [61, 62]. At the phylum level, we observed a higher abundance of Actinobacteria and Proteobacteria, with increased Enterobacteriaceae, in Foxo3a−/−IL-10−/− mice. Proteobacteria are colitogenic strains that are significantly abundant in aggressive Crohn’s disease patients [63]. At the genus level, we observed that Sutterella was significantly enriched in Foxo3a−/−IL-10−/− mice. Sutterella has been shown to be increased in IBD patients [64, 65]. On the other hand, we observed that the abundance of Lactobacilli, which suppress production of pro-inflammatory cytokines [66], was particularly reduced in Foxo3a−/−IL-10−/− mice. Furthermore, we observed that Roseburia, a Gram-positive aerobic species of the Firmicutes phylum, was depleted in Foxo3a−/−IL-10−/− mice. Roseburia produce the short chain fatty acid butyrate, which is a key energy source for colonocytes that also signals through the G-protein coupled receptors to regulate immune response. Thus, depletion of butyrate negatively influences intestinal inflammation [67, 68].
In summary, our results indicate that Foxo3a restricts the abundance of colitogenic microbiota in IL-10-deficient hosts and inhibits glutaminolysis and mTORC1 activation thereby regulating the activation of myeloid cells and preventing them from impairing the function of regulatory T cells. This prevents the overt activation of Th17 cells and the development of gut inflammatory disease.
Methods
Mice
Foxo3a−/− mice were generated as previously described [8]. Il-10−/− and wildtype C57BL/6 J mice were obtained from the Jackson Laboratory (Bar Harbor, Maine, USA). Foxo3a and IL-10 double deficient mice (Foxo3a−/−Il-10−/−) were generated from mice heterozygous for Foxo3a and Il-10 (Foxo3a+/− Il-10+/− breeding pairs) or from mice deficient in Il-10 and heterozygous for Foxo3a (Foxo3a+/− Il-10−/− breeding pairs). The development of spontaneous intestinal inflammation was monitored in males and females and both sexes were equally distributed in the various groups of mice. Age-matched background mice were used for baseline comparison. Mice (minimum n = 15 per group) were monitored for signs of disease and euthanized when reaching humane endpoint characterized by the following criteria: significant weight loss, rectal bleeding, diarrhea, lethargy, hunched posture and piloerection.
Isolation of lamina propria leukocytes
Lamina propria leukocytes were isolated using a modified version of a previously described protocol [69]. Colons were placed in a petri dish containing ice cold PBS and cut open longitudinally into 0.5 cm pieces. Colon fragments were transferred to a Falcon tube and incubated in a pre-digestion HBSS solution containing 5 mM EDTA and 1 mM DTT at 37 °C for 20 min. at 250 rpm. After 20 min., the supernatant was poured off through a 70μm filter into a new Falcon tube. The colonic fragments were incubated two more times in the pre-digestion solution for a total of 3 incubations. Collected supernatants were discarded. Remaining colonic fragments were further minced and placed into a digestion solution containing 0.5 mg/ml collagenase D (Roche) and 0.5 mg/mL DNase I (Roche) at 37 °C for 20 min. at 250 rpm. Supernatants were discarded. Digestion was repeated another two times for a total of 3 rounds. Remaining tissue was disaggregated using an 18 G needle and syringe and added to the collected digestion supernatants. Complete media was added to top off collected supernatant and cells were washed with PBS and resuspended in 40% (v/v) isotonic Percoll (GE Life Sciences) in complete media. 40% Percoll cell suspension was layered onto an 80% Percoll layer and cells were centrifuged at 1000 x g for 20 mins with no brake. Lamina propria immune cells were obtained from the interphase between the two Percoll layers, and cells were washed and re-suspended in complete media prior to antibody staining.
Measurement of cytokines
The following cytokines were measured in supernatants by ELISA according to the manufacturer’s instructions: IL-1β (Mouse R&D Duo Set #DY401), IL-1α (Mouse R&D Duo Set #DY400), TNFα (BD #555268), IL-10 (BD #555252), IL-6 (BD 555240), IL-12p70 (BD #555256), IL-17a (Biolegend #432501), and IFNg (Biolegend #430801). For intracellular cytokine staining, cells were stimulated with LPS (100 ng/ml) and treated with Brefeldin A (1:1000, Biolegend #420601). After staining for surface markers, cells were fixed and permeabilized using the BD Cytofix/Cytoperm buffer system followed by staining with anti-TNF-PE-Cy7.
Flow cytometry staining
The 1–3 × 106 cells were transferred to 5 ml tubes (Fisher, catalog # 14-961-10) and washed with PBS twice. Zombie Yellow™ (BioLegend, San Diego, CA) was used to assess cell viability. Cells were incubated with Fc block (anti-CD16/32; BD, catalog #553142) for 10 min at 4 °C to prevent non-specific binding. Fluorochrome-tagged antibodies against various cell surface receptors were added in PBS-BSA followed by incubation for 30 min at 4 °C. After washing, samples were fixed with 1% paraformaldehyde before acquisition on flow cytometer (BD LSR Fortessa). Then data were analyzed using Kaluza software (Beckman Coulter, version 1.2) or FlowJo V10 (FlowJo).
For intracellular staining cells were first stained for surface markers. Then, they were fixed and permeabilized using Foxp3 Transcriptions Factor Fix/Perm Buffer Set (Biolegend #421403) followed by staining with anti-FoxP3-PE or APC.
The following fluorochrome -tagged antibodies were used against different surface and intracellular markers: anti-CD8-APC-eFluor780 (eBioscience #47-0081-82) or eF450 (eBioscience # 48-0081-82), anti-CD62L- PerCp Cy5.5 (eBioscience #25-0621-82), anti-CD11c-eF450 (eBioscience #48-0114-80), anti-B220-PerCP Cy5.5 (Biolegend #103235), anti-CD3ε – PE (Biolegend #100307) or eF-450 (eBioscience #48-0032-82), anti-CD4-APC (eBioscience 17-0042-82) or FITC (Biolegend #100405), anti-CD11b-APC (eBioscience #17-0112-82) or eF450 (eBioscience # 48-0112-82) anti-DX5-APC (eBioscience # 17-5971-81), anti-Ly6G-FITC (eBioscience #11-5931-82), anti-Ly6C-Pe (eBioscience 12-5932-82), anti-F4/80 - APC ef780 (eBioscience 47-4801-80), anti-IL1R1- BV421 (BD Biosciences #564387), anti-ST2- Per Cp Cy5.5 (eBioscience #46-9335-80) anti-PD-1-Pe (BD Pharmigen #561788), anti-CD25-PeCy7 (Biolegend #102015) or BV421 (Biolegend #102033), anti-Foxp3-APC (eBioscience #17-5773-80) or Pe (Biolegend #126403) and anti-TNF-PeCy7 (eBioscience #25-7321-80). Gating strategy employed: dendritic cells (CD11c+), neutrophils (CD11b + Ly6Ghi Ly6C-/int), monocytes (CD11b + Ly6G-/int Ly6Chi), macrophages (CD11b + Ly6G-/int Ly6Clo/int F4/80+), T-cells (CD11b- CD3+), B-cells (CD11b- B220+) and NK-cells (CD11b + DX5+).
Histopathology and immunohistochemistry
Various organs were harvested and fixed in neutral buffered 10% formalin, embedded in paraffin, and processed for routine and special histopathological examination. Vertical sections (5 μm thick) were stained with haematoxylin and eosin, Alcian Blue pH = 2.5, and Masson’s trichrome. Images were acquired on a Zeiss Axio Mirax Midi scanner. Paraformaldehyde-fixed, paraffin-embedded colon sections were de-paraffinized in xylene and rehydrated through a 100-70% ethanol gradient. Heat-induced epitope retrieval was done at 110 °C for 12 min with citrate buffer pH 6.0 and 3% hydrogen peroxidase was used to block endogenous peroxidases. Sections were blocked with Background Sniper blocking reagent (BioCare Medical, CA, USA) and then incubated with the Cleaved Caspase-3 antibody (cell signaling, #9661) at 1:500 dilution for 1 h at room temperature. The MACH4™ + DAB detection system was used for chromogenic staining according to the manufacturer’s instructions (BioCare Medical, CA, USA). Blinded samples were sent for evaluation of pathology at the Ottawa Hospital.
In vitro spleen cell culture
Spleens were homogenized using frosted-end glass slides (Fisher Scientific #12- 550-343). Red blood cells were lysed using RBC lysis buffer (Sigma-Aldrich #R7757) according to the manufacturer’s instructions. Spleen cells were washed and re-suspended in RPMI-1640 (Thermo Fisher Scientific #31800089) supplemented with 8% fetal bovine serum (FBS) (Gibco #12483020) and 55 μM β-mercaptoethanol (Thermo Fisher Scientific #21985-023) (with or without antibiotic). The media is referred to as R8. 107 cells were seeded per well in a 24-well plate. To ensure that the cells are properly adhered to the wells and to avoid stimulating them when stressed, the cells were incubated at 37 °C for 2 h. Next, cells were either treated with LPS (100 ng/ml) or infected with Salmonella typhimurium. Lower multiplicity of infection (10 MOI) was used for measurement of cytokines and immunoblotting. For the infection protocol, cells were centrifuged at 2500 rpm for 7 min to enhance bacterial uptake. Following incubation of 30 min at 37 °C, the media with the bacteria was aspirated out, and media containing 50 μg/ml Gentamicin (Thermo Fisher Scientific #15750-060) was added for 2 h to eliminate extracellular bacteria. Finally, the media was replaced with media containing a lower concentration of gentamicin (10 μg/mL). The infection was then allowed to proceed for desired timepoints.
Purification of CD4+ T-cells
CD4+ and CD4+CD25+ T cells were purified from the spleens of WT, Foxo3a−/−, IL-10−/− and Foxo3a−/− IL-10−/− mice using EasySep™ Mouse CD4+CD25+ Regulatory T Cell Isolation Kit II (STEMCELL # 18783). 100,000 cells were seeded in a 96 well plate and were stimulated with plate-bound 1 μg/ml anti-CD3ε (Βiolegend #100340) and 1 μg/ml anti-CD28 (Biolegend # 102115). For stimulation of regulatory T-cells, CD4+CD25+ T-cells were additionally stimulated with the addition of recombinant IL-2 and TGFβ (both at a concentration of 2.5 ng/mL). RNA and protein extracts were collected at different timepoints (24, 48 and 72H). Cells were either seeded in regular R8 or glutamine/glucose free RPMI (Biological Industries # 01-101-1A) supplemented with 8% dialyzed FBS (Thermofisher #A3382001), 55 μM 2-ME, and either glutamine or glucose or both and further treated with the following inhibitors: 6-Diazo-5-oxo-L-norleucine or DON (Sigma # D2141) and L-Glutamic acid γ-monohydroxamate or GAH (Sigma # G2253).
Glutamine and glucose measurements
Glutamine, glutamate, and glucose levels were measured using the Glutamine/Glutamate-Glo™ Assay and Glucose-Glo™ Assay kits from Promega (J8021 and J6021 respectively) according to the instructions of the manufacturer.
In vivo survival experiments
For the neutrophil/monocyte depletion experiment, InvivoPLUS grade Anti-Ly6C (Clone 1A8) and anti-Ly6C/G (clone: RB6-8C5) were obtained from BioXCell and injected i.p. (200 micrograms/mouse given twice per week from day 30 till 100). For CD4+/CD8+ T cell depletion, InvivoPLUS grade Anti-CD4 (Clone GK1.5) and anti-CD8 (clone: 2.43) were obtained from BioXCell and injected i.p. (200 micrograms/mouse given twice per week from day 30 till 100). For suppression of mTORC1 activation, Rapamycin was obtained from LC laboratories and injected i.p. at a dose of 5 mg/kg every 3 days from day 28 till day 130.
Purification of CD11b+ cells
CD11b+ cells were purified from the spleens of WT, Foxo3a−/−, Il-10−/−, and Foxo3a−/−Il-10−/−, mice as per manufacturer’s instructions using EasySep™ Mouse CD11b Positive Selection Kit II (STEMCELL #18970). 400,000 cells were seeded per well in a 24 well plate followed by treatment with LPS (100 ng/ml). Lysates were collected at various time intervals for Western blotting.
Measurement of inflammatory genes by qPCR
The 30 mg sections of colon and/or spleen were harvested from mice and immediately frozen in RNAlater stabilization reagent (Qiagen, Hilden, Germany) then homogenized for RNA extraction. Total tissue RNA was extracted using the RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions. The 5 μg high-quality total RNA was then reverse transcribed using the First Strand Synthesis Kit (Qiagen) and loaded onto an inflammatory cytokine and chemokine RT2 profiler array (Qiagen). Real-time PCR was done using RT² SYBR Green qPCR Mastermix (Qiagen) and the reactions were run on a Biorad CFX 384 Real time PCR System. All data was normalized to an average of five housekeeping genes (Gusb, Hprt, Hsp90ab1, Gapdh, and Actb). For measurement of the expression of specific transcripts by qRT-PCR, data was normalized to Actb. Wherever possible, exon–exon spanning primers were used to avoid amplification of genomic DNA. The primer sequences were generated using the NCBI primer tool. Primers used are mentioned in Supplementary Table.
Western blotting
Cells were lysed in 1% SDS lysis buffer containing 1% β-mercaptoethanol and boiled immediately after for 5 min. Lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis at 134 V for 75 min and then transferred to methanol-activated PVDF membrane (Biorad #1620177). The transfer was run at 100 V for one hour on ice. After transferring, the membranes were blocked in 5% bovine serum albumin in Tris Buffered Saline Solution (TBS-0.5 M Tris, 1.5 NaCl pH 7.6) with 0.5% Tween-20 for 1 h at room temperature on a rocker. The membranes were probed with primary antibodies, diluted in the same blocking buffer, overnight on a rocker at 4 °C. After incubation, the membranes were washed 5 times (5 min each) with TBS + 0.5% Tween-20 followed by incubation with a conjugated secondary antibody, diluted in the same blocking buffer, for 1 h at room temperature on a rocker. Finally, the membranes were washed 5 times and visualized using either Super Signal West Pico Plus (Thermo Scientific #34578) or the Super Signal Femto Plus (Thermo Scientific #34096). Antibodies used were as follows: phospho-p65 (Cell Signaling #3033), p65 (Cell Signaling #8242), phospho-p38MAPK (Cell Signaling #4511), p38MAPK (Cell Signaling #8690), p-Stat3 (Ser727) (Cell Signaling #9134), p-Stat3 (Tyr705) (Cell Signaling #9131 S), Stat3 (Cell Signaling #4904), p-4E-BP1 (Cell Signaling #2855), 4E-BP1 (Cell Signaling #9644), P62 (SQSTM1) (Cell Signaling #5114), p-P70S6K (Cell Signaling # 9234), P70S6K (Cell Signaling # 2708), Foxo3a (Cell Signaling # 12829), p-MK2 (Cell Signaling #30075), MK2 (Cell Signaling # 3042) and β-actin (Santa Cruz Biotechnology #sc-81178).
Bicinchoninic acid assay (BCA)
Protein concentration in whole cell lysates was measured using the BCA kit (Fisher # 23225) following the manufacturer’s instructions.
Glycolysis cell-based assay
L-lactate was measured in supernatants using the Glycolysis Cell-Based Assay kit from Cayman Chemical (Michigan, USA), according to manufacturer’s instructions.
MTT assay
Mitochondrial function was measured by adding MTT (3-[4,5-dimethylthiazol-2-yl]−2,5-diphenyltetrazolium bromide) to cells. The MTT reagent was diluted with R8 media at a final concentration 0.5 mg/ml and incubated at 37 °C. After 2 h, 5 mM HCl isopropyl alcohol were added to solubilize MTT crystals and absorbance was measured at a wavelength of 570 nm with a reference wavelength of 650 nm on a Molecular Devices FilterMax plate reader.
Mitochondrial bioenergetics assay
Wells of a 96-well Seahorse tissue culture plate (Seahorse Bioscience # 101085-004) were coated with cell adhesive Cell-Tak (Corning #354240) following the manufacturer’s instructions to promote adherence of cells. Bone-marrow derived macrophages were seeded into the wells at a density of 1.5 × 105 cells/well and incubated overnight at 37 °C. Next day, cells were treated with LPS (100 ng/ml) for 24 h. Splenocytes were seeded into the wells at a density of 8 × 105 cells/well and were incubated for 2 h at 37 °C followed by treatment with LPS (100 ng/mL) for ~5 h. The R8 medium was then removed, and cells were washed twice with warm Seahorse assay medium XF RPMI Medium pH 7.4 (Agilent Seahorse #103576-100). The medium was supplemented differently based on the assay performed: for both glycolysis and Mito stress tests, the media contained 1 mM sodium pyruvate and 2 mM Glutamine. Only media for Mito stress test was supplemented with 10 mM Glucose. Cells were then allowed to equilibrate at 37 °C for 1 h without CO2. Next, oxygen consumption rates (OCR, in pmol/min) or extracellular acidification rate (ECAR, in mph/min) were recorded on a Seahorse Bioscience XF96 Extracellular Flux Analyzer. Basal OCR and ECAR were recorded, as well as their fluctuations in response to sequential addition of various inhibitors in the following order: For Mito-stress test: Oligomycin (2 μM), FCCP (1 μM), and Rotenone/Antimycin A (1 μM); for glycolysis stress test: Glucose (10 mM), Oligomycin (2 μM) and 2-deoxyglucose (30 mM).
DNA extraction, V4-16S rRNA library construction and sequencing
Metagenomic DNA was extracted using MoBio PowerMag Soil DNA Isolation kit (Qiagen) as per its standard protocol and a KingFisher robot. The V4-16S rRNA sequencing was completed by Microbiome Insights, University of British Columbia, Vancouver, BC, Canada. The protocol details are described in the Supporting Materials and Methods.
Microbiota sequencing data analysis
The V4 region of 16 S rRNA gene was amplified using dual-barcoded primers (515 F 5ʹ-GTGCCAGCMGCCGCGGTAA-3ʹ, and 806 R 5ʹ-GGACTACHVGGGTWTCTAAT-3ʹ) and the amplicon library for sequencing was constructed as described before [70]. The amplicon libraries were pooled and paired-end sequenced with Illumina MiSeq platform using 300 bp MiSeq Reagent Kit v3. To control for contaminants, a template-free control and extraction kit reagents control were co-sequenced with the specimens. The demultiplexed raw reads are available at NCBI Sequence Read Archive under PRJNA623633.
Sequences were quality filtered and clustered into operational taxonomic units (OTUs) based on 97%-similarity using mothur software package (v. 1.39.5) [71] and the taxonomy was assigned to the generated OTUs using Greengenes (v. 13_8) as the reference database. Contaminant OTUs were identified and removed if their mean abundance in negative and extraction reagents controls is ≥25% of their mean abundance in the samples. Singleton and doubleton OTUs and OTUs that occur in less than 10% of the samples were removed. The remaining OTUs were rarefied into equal number of reads per sample (20,000) using QIIME 1.9.0 [72]. Alpha diversity was estimated with Chao1 estimate and Shannon index. Beta diversity among samples were calculated using Bray-Curtis distance and visualized using Principal Coordinate Analysis (PCoA). The contribution of different mice groups to the variability of gut microbiota community was assessed from the Bray-Curtis distance matrix using adonis [73] using 999 permutations. To identify differential taxa among mice groups, linear discriminant effect size analysis was conducted on the relative abundance of different taxa levels using LEfSe [74]. The taxa with log10 LDA score ≥2 and P < 0.05 were considered significant. When required Kruskal-Wallis test was applied for statistical analysis, and P values were corrected using Two-stage Benjamini, Krieger, and Yekutieli false discovery rate (FDR) procedure.
Data analysis and statistics
Data was analyzed in GraphPad Prism (GraphPad Software, California, USA). Survival curves were analyzed using Mantel–Cox test; all other data were analyzed by one-way ANOVA. All graphs represent mean ± s.e.m (*P < 0.05, ***P < 0.001, and ****P < 0.0001).
Supplementary information
Author contributions
Conceptualization, SS, JJ, and SH.; Methodology, SH, JJ, M-EH, TA, AB, RR, and SS; Investigation, SH, JJ, NN, AA, AB, and SP; Validation, NN, SP, WM; Writing—Original Draft, SH and SS; Writing—Review & Editing, SH, SS, WM, and TA.; Funding Acquisition, SS; Resources, SS and M-EH, Supervision, SS.
Funding
The work was funded by a grant from the Canadian Institutes of Health Research (#420007 to Dr. Subash Sad). Stephanie Hajjar was supported by Alexander Graham Bell-Canada Graduate Scholarship from NSERC.
Data availability
All data is available in the main text or the supplementary materials. The demultiplexed raw reads are available at NCBI Sequence Read Archive under PRJNA623633.
Competing interests
The authors declare no competing interests.
Ethical approval
All animal procedures were performed on approval by the University of Ottawa Animal Care and Veterinary Committee and maintained in accordance with the Canadian Council on Animal Care (CCAC) guidelines.
Footnotes
Edited by Y Shi
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00876-y.
References
- 1.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–26. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A. IL-10 inhibits cytokine production by activated macrophages. JImmunol. 1991;147:3815–22. [PubMed] [Google Scholar]
- 3.Hedrick SM, Hess Michelini R, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nat Rev Immunol. 2012;12:649–61. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim MV, Ouyang W, Liao W, Zhang MQ, Li MO. The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity. 2013;39:286–97. doi: 10.1016/j.immuni.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch’en IL, Stockmann C, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delpoux A, Marcel N, Hess Michelini R, Katayama CD, Allison KA, Glass CK, et al. FOXO1 constrains activation and regulates senescence in CD8 T cells. Cell Rep. 2021;34:108674. doi: 10.1016/j.celrep.2020.108674. [DOI] [PubMed] [Google Scholar]
- 7.Dejean AS, Beisner DR, Ch’en IL, Kerdiles YM, Babour A, Arden KC, et al. Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat Immunol. 2009;10:504–13. doi: 10.1038/ni.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzelepis F, Joseph J, Haddad EK, Maclean S, Dudani R, Agenes F, et al. Intrinsic role of FoxO3a in the development of CD8+ T cell memory. J Immunol. 2013;190:1066–75. doi: 10.4049/jimmunol.1200639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riou C, Yassine-Diab B, Van Grevenynghe J, Somogyi R, Greller LD, Gagnon D, et al. Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. JExpMed. 2007;204:79–91. doi: 10.1084/jem.20061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–25. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 11.Joseph J, Ametepe ES, Haribabu N, Agbayani G, Krishnan L, Blais A, et al. Inhibition of ROS and upregulation of inflammatory cytokines by FoxO3a promotes survival against Salmonella typhimurium. Nat Commun. 2016;7:12748. doi: 10.1038/ncomms12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin L, Hron JD, Peng SL. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–13. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 13.Lee JC, Espeli M, Anderson CA, Linterman MA, Pocock JM, Williams NJ, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155:57–69. doi: 10.1016/j.cell.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Zhang H, Chen Y, Fan L, Fang J. Forkhead transcription factor FOXO3a protein activates nuclear factor kappaB through B-cell lymphoma/leukemia 10 (BCL10) protein and promotes tumor cell survival in serum deprivation. J Biol Chem. 2012;287:17737–45. doi: 10.1074/jbc.M111.291708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore KW, O’Garra A, de-Waal MR, Vieira P, Mosmann TR. Interleukin-10. AnnuRevImmunol. 1993;11:165–90. doi: 10.1146/annurev.iy.11.040193.001121. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–78. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–65. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–36. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- 20.Ip WKE, Hoshi N, Shouval DS, Snapper S, Medzhitov R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. 2017;356:513–9. doi: 10.1126/science.aal3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouzeyen R, Haoues M, Barbouche MR, Singh R, Essafi M. FOXO3 Transcription Factor Regulates IL-10 Expression in Mycobacteria-Infected Macrophages, Tuning Their Polarization and the Subsequent Adaptive Immune Response. Front Immunol. 2019;10:2922. doi: 10.3389/fimmu.2019.02922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl J Med. 2009;361:2033–45. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirkov MU, Verstockt B, Cleynen I. Genetics of inflammatory bowel disease: beyond NOD2. Lancet Gastroenterol Hepatol. 2017;2:224–34. doi: 10.1016/S2468-1253(16)30111-X. [DOI] [PubMed] [Google Scholar]
- 24.Buchler G, Wos-Oxley ML, Smoczek A, Zschemisch NH, Neumann D, Pieper DH, et al. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota-host interactions. Inflamm Bowel Dis. 2012;18:943–54. doi: 10.1002/ibd.21895. [DOI] [PubMed] [Google Scholar]
- 25.Alvarez F, Istomine R, Shourian M, Pavey N, Al-Aubodah TA, Qureshi S, et al. The alarmins IL-1 and IL-33 differentially regulate the functional specialisation of Foxp3(+) regulatory T cells during mucosal inflammation. Mucosal Immunol. 2019;12:746–60. doi: 10.1038/s41385-019-0153-5. [DOI] [PubMed] [Google Scholar]
- 26.Reissig S, Tang Y, Nikolaev A, Gerlach K, Wolf C, Davari K, et al. Elevated levels of Bcl-3 inhibits Treg development and function resulting in spontaneous colitis. Nat Commun. 2017;8:15069. doi: 10.1038/ncomms15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J, Ji J, Yan XH. Cross-talk between AMPK and mTOR in regulating energy balance. Crit Rev Food Sci Nutr. 2012;52:373–81. doi: 10.1080/10408398.2010.500245. [DOI] [PubMed] [Google Scholar]
- 28.Degagne E, Pandurangan A, Bandhuvula P, Kumar A, Eltanawy A, Zhang M, et al. Sphingosine-1-phosphate lyase downregulation promotes colon carcinogenesis through STAT3-activated microRNAs. J Clin Investig. 2014;124:5368–84. doi: 10.1172/JCI74188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neininger A, Kontoyiannis D, Kotlyarov A, Winzen R, Eckert R, Volk HD, et al. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J Biol Chem. 2002;277:3065–8. doi: 10.1074/jbc.C100685200. [DOI] [PubMed] [Google Scholar]
- 30.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185:1037–44. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell. 2018;175:1780–95. doi: 10.1016/j.cell.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–4. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 33.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JC, Biasci D, Roberts R, Gearry RB, Mansfield JC, Ahmad T, et al. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat Genet. 2017;49:262–8. doi: 10.1038/ng.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karrasch T, Jobin C. NF-kappaB and the intestine: friend or foe? Inflamm Bowel Dis. 2008;14:114–24. doi: 10.1002/ibd.20243. [DOI] [PubMed] [Google Scholar]
- 36.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Wilson JE, Koenigsknecht MJ, Chou WC, Montgomery SA, Truax AD, et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol. 2017;18:541–51. doi: 10.1038/ni.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carey R, Jurickova I, Ballard E, Bonkowski E, Han X, Xu H, et al. Activation of an IL-6:STAT3-dependent transcriptome in pediatric-onset inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:446–57. doi: 10.1002/ibd.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–91. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan JA, Kim EH, Plisch EH, Peng SL, Suresh M. FOXO3 regulates CD8 T cell memory by T cell-intrinsic mechanisms. PLoSPathog. 2012;8:e1002533. doi: 10.1371/journal.ppat.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson MG, Larson M, Vidrine A, Barrios K, Navarro F, Meyers K, et al. FOXO3-NF-kappaB RelA Protein Complexes Reduce Proinflammatory Cell Signaling and Function. J Immunol. 2015;195:5637–47. doi: 10.4049/jimmunol.1501758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyara M, Amoura Z, Parizot C, Badoual C, Dorgham K, Trad S, et al. Global natural regulatory T cell depletion in active systemic lupus erythematosus. J Immunol. 2005;175:8392–8400. doi: 10.4049/jimmunol.175.12.8392. [DOI] [PubMed] [Google Scholar]
- 43.Pop SM, Wong CP, Culton DA, Clarke SH, Tisch R. Single cell analysis shows decreasing FoxP3 and TGFbeta1 coexpressing CD4+CD25+ regulatory T cells during autoimmune diabetes. J Exp Med. 2005;201:1333–46. doi: 10.1084/jem.20042398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mottet C, Uhlig HH, Powrie F. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J Immunol. 2003;170:3939–43. doi: 10.4049/jimmunol.170.8.3939. [DOI] [PubMed] [Google Scholar]
- 45.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–5. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 46.Zhou X, Wang J, Shi W, Brand DD, Liu Z, Fan H, et al. Isolation of purified and live Foxp3+ regulatory T cells using FACS sorting on scatter plot. J Mol Cell Biol. 2010;2:164–9. doi: 10.1093/jmcb/mjq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kono M, Yoshida N, Maeda K, Tsokos GC. Transcriptional factor ICER promotes glutaminolysis and the generation of Th17 cells. Proc Natl Acad Sci USA. 2018;115:2478–83. doi: 10.1073/pnas.1714717115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang L, Mou J, Shao B, Wei Y, Liang H, Takano N, et al. Glutaminase 1 expression in colorectal cancer cells is induced by hypoxia and required for tumor growth, invasion, and metastatic colonization. Cell death Dis. 2019;10:40. doi: 10.1038/s41419-018-1291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Liu J, Zhao Y, Yue X, Zhu Y, Wang X, et al. Glutaminase 2 is a novel negative regulator of small GTPase Rac1 and mediates p53 function in suppressing metastasis. Elife. 2016;5:e10727. doi: 10.7554/eLife.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr., et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5’-cap function. Nature. 1994;371:762–7. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 51.Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jewell JL, Kim YC, Russell RC, Yu FX, Park HW, Plouffe SW, et al. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015;347:194–8. doi: 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faustino LD, Griffith JW, Rahimi RA, Nepal K, Hamilos DL, Cho JL, et al. Interleukin-33 activates regulatory T cells to suppress innate gammadelta T cell responses in the lung. Nat Immunol. 2020;21:1371–83. doi: 10.1038/s41590-020-0785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiering C, Krausgruber T, Chomka A, Frohlich A, Adelmann K, Wohlfert EA, et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513:564–8. doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matta BM, Lott JM, Mathews LR, Liu Q, Rosborough BR, Blazar BR, et al. IL-33 is an unconventional Alarmin that stimulates IL-2 secretion by dendritic cells to selectively expand IL-33R/ST2+ regulatory T cells. J Immunol. 2014;193:4010–20. doi: 10.4049/jimmunol.1400481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turnquist HR, Zhao Z, Rosborough BR, Liu Q, Castellaneta A, Isse K, et al. IL-33 expands suppressive CD11b+ Gr-1(int) and regulatory T cells, including ST2L+ Foxp3+ cells, and mediates regulatory T cell-dependent promotion of cardiac allograft survival. J Immunol. 2011;187:4598–610. doi: 10.4049/jimmunol.1100519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mottawea W, Chiang CK, Muhlbauer M, Starr AE, Butcher J, Abujamel T, et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn’s disease. Nat Commun. 2016;7:13419. doi: 10.1038/ncomms13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin J, Noh JR, Chang DH, Kim YH, Kim MH, Lee ES, et al. Elucidation of Akkermansia muciniphila probiotic traits driven by Mucin depletion. Front Microbiol. 2019;10:1137. doi: 10.3389/fmicb.2019.01137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ottman N, Reunanen J, Meijerink M, Pietila TE, Kainulainen V, Klievink J, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One. 2017;12:e0173004. doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 62.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vester-Andersen MK, Mirsepasi-Lauridsen HC, Prosberg MV, Mortensen CO, Trager C, Skovsen K, et al. Increased abundance of proteobacteria in aggressive Crohn’s disease seven years after diagnosis. Sci Rep. 2019;9:13473. doi: 10.1038/s41598-019-49833-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S, et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep. 2017;7:9523. doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lavelle A, Lennon G, O’Sullivan O, Docherty N, Balfe A, Maguire A, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. 2015;64:1553–61. doi: 10.1136/gutjnl-2014-307873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L, Zou Y, Peng J, Lu F, Yin Y, Li F, et al. Lactobacillus acidophilus suppresses colitis-associated activation of the IL-23/Th17 axis. J Immunol Res. 2015;2015:909514. doi: 10.1155/2015/909514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parada Venegas D, De la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weigmann B, Tubbe I, Seidel D, Nicolaev A, Becker C, Neurath MF. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc. 2007;2:2307–11. doi: 10.1038/nprot.2007.315. [DOI] [PubMed] [Google Scholar]
- 70.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecology Package. R package 2016.
- 74.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the main text or the supplementary materials. The demultiplexed raw reads are available at NCBI Sequence Read Archive under PRJNA623633.