Abstract
Long noncoding RNAs (lncRNAs) are dysregulated in different cancer types, and thus have emerged as important regulators of the initiation and progression of human cancers. However, the biological functions and the underlying mechanisms responsible for their functions in gastric cancer (GC) remain poorly understood. Here, by lncRNA microarray, we identified 1414 differentially expressed lncRNAs, among which THAP7-AS1 was significantly upregulated in GC tissues compared with non-tumorous gastric tissues. High expression of THAP7-AS1 was correlated with positive lymph node metastasis and poorer prognosis. SP1, a transcription factor, could bind directly to the THAP7-AS1 promoter region and activate its transcription. Moreover, the m6A modification of THAP7-AS1 by METTL3 enhanced its expression depending on the “reader” protein IGF2BP1-dependent pathway. THAP7-AS1 promoted GC cell progression. Mechanistically, THAP7-AS1 interacted with the 1-50 Amino Acid Region (nuclear localization signal) of CUL4B through its 1-442 nt Sequence, and it promoted interaction between nuclear localization signal (NLS) and importin α1, and improved the CUL4B protein entry into the nucleus, repressing miR-22-3p and miR-320a expression by CUL4B-catalyzed H2AK119ub1 and the EZH2-mediated H3K27me3, subsequently activating PI3K/AKT signaling pathway to promote GC progression. Moreover, LV-sh-THAP7-AS1 treatment could suppress GC growth, invasion and metastasis, indicating that THAP7-AS1 may act as a promising molecular target for GC therapies. Taken together, our results show that THAP7-AS1, transcriptionally activated by SP1 and then modified by METTL3-mediated m6A, exerts oncogenic functions, by promoting interaction between NLS and importin α1 and then improving the CUL4B protein entry into the nucleus to repress the transcription of miR-22-3p and miR-320a.
Subject terms: Oncogenes, Epigenetics
Introduction
Gastric cancer (GC) is the fourth most common human malignant tumor worldwide [1]. Despite the fact that great advances have been made in the comprehensive treatment of GC, it remains the third leading cause of cancer-associated death worldwide, due to tumor invasion and metastasis [2]. Thus, characterizing the molecular mechanisms underlying GC occurrence and progression is essential for the effective treatment of this disease.
Long noncoding RNAs (lncRNAs) are commonly defined as RNAs longer than 200 nucleotides (nt) that do not encode proteins [3]. Regulation of lncRNA transcription is approximately as evolutionarily conserved as that of mRNA transcription in humans, further suggesting the importance of lncRNA in the genetic programming of complex mechanisms [4]. They are involved in numerous important biological processes, including the biological behavior of cancer cells [5]. Zheng et al. reported that the lncRNA HOX transcript antisense RNA (HOTAIR) promotes tumor invasion via inhibition of miR-206 expression [6]. A macrophage-specific lncRNA MAARS regulates apoptosis and atherosclerosis by tethering ELAVL1 in the nucleus [7]. Hence, functional lncRNAs may serve as potential therapeutic targets for cancer.
At present, the biological functions and underlying mechanisms responsible for the functions of lncRNAs in GC remain poorly understood, and few novel lncRNAs have been functionally characterized in this disease.
Materials and methods
Clinical specimens, cell culture, and transfection
A total of 72 fresh GC tissues were obtained from the Qilu Hospital of Shandong University (Shandong Province, China) and Shandong Provincial Hospital (Shandong, China) between 2012 and 2014. This study was approved by the Institute’s Research Ethics Committee of Shandong University and conducted in accordance with the ethical guidelines of the World Medical Association Declaration of Helsinki. Informed consent was written by all patients prior to this study. Cell authentication was confirmed by short tandem repeat profiling. The human GC cell lines MKN-45 and BGC823, and the embryonic kidney cell line 293T were obtained from the Chinese Academy of Sciences (Shanghai, China). Mycoplasma PCR testing of these cells was performed every month. Cells were cultured as previously described [8].
RNA extraction and quantitative PCR
Total RNA was extracted from fresh GC tissue and cell lines using Trizol reagent (Invitrogen, Carlsbad, CA, USA). RNA from the cytoplasmic and nuclear fractions of GC cells was extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, MA, USA), and were reverse transcribed into cDNA using a reverse transcriptase cDNA synthesis kit (Toyobo, Osaka, Japan). Quantitative PCR (qPCR) was performed to detect RNA expression levels using the SYBR Green PCR Kit (Roche, Basel, Switzerland).
RNA-seq gene expression profiling
Differential gene expression analysis of mRNA was performed between si-THAP7-AS1 or si-NC cancer cells. The mRNA expression profile was examined using Solexa pipeline v1.8 (Off‐Line Base Caller software, v1.8, Illumina, Foster City, CA, USA). The data were deposited to the NCBI’s GEO Repository and accessible to readers through Gene Expression Omnibus (GEO) series accession number GSE150539.
Cell proliferation assays
The EdU, MTS, and colony formation assays were used to observe cell proliferation, as previously described [9].
Cell migration and invasion assays
Cell migration and invasion assays were performed to evaluate the migration and invasion abilities of GC cells, as previously reported [10].
Transfection of THAP7-AS1 Smart Silencer
LncRNA Smart Silencer, a mixture of six siRNAs, synthesized from RiboBio (Guangzhou, China), was used to knock down the expression of THAP7-AS1. The target sequences of siRNAs are shown in Supplementary Table 1. The negative control (NC) Smart Silencer does not contain domains homologous to humans, mice, and rats. THAP7-AS1 Smart Silencer transfection was performed with Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
In vivo tumor formation assay
Nu/nu athymic nude mice (male, 3–4-weeks old) were purchased from Weitong lihua Biotechnology (Beijing, China) and were randomly divided into two groups. A total of 3 × 105 MKN-45 cells, stably transfected with THAP7 antisense RNA 1 (THAP7-AS1, a novel lncRNA) vector or empty vector, were inoculated subcutaneously into the axillary fossa or lateral tail vein of the mice (n = 6 each). Tumor growth was examined every 4 days, and tumor volumes were calculated using the following equation: V (mm3) = A × B2/2 (A, longitudinal diameter; and B, latitudinal diameter). Seven weeks after injection, the mice were sacrificed, and the tumors were excised and snap-frozen for subsequent RNA and protein extraction. Tumor and lung metastatic loci were confirmed histologically. Animal experiments were approved by the Committee for Animal Protection of Shandong University.
Fluorescence in situ hybridization and Immunofluorescence analysis
THAP7-AS1 FISH probe was synthesized by RiboBio Technology Co. Ltd. (Guangzhou, China). A fluorescence in situ hybridization (FISH) kit was used to detect the subcellular localization of THAP7-AS1 using a confocal microscope (Zeiss, Oberkochen, Germany) in accordance with the manufacturer’s protocol (RiboBio). The protein expression of cullin 4B (CUL4B) in MKN-45 and BGC-823 cells were determined by immunofluorescence with anti-CUL4B antibody (1:100; Abcam, Cambridge, MA, USA), as previously described [11].
Western blotting
The primary antibodies used for western blotting were as follows: mouse anti-CUL4B (SAB1406670; Merck), mouse anti-ribosomal protein LP0 (sc-293260; Santa Cruz), rabbit anti-Lamin B1 (AB0054; Abways), mouse anti-phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α-isoform (PIK3CA; sc-293172; Santa Cruz), mouse ant-PIK3CD (sc-55589; Santa Cruz), mouse ant-GFP (sc-9996; Santa Cruz), mouse ant-His (sc-8036; Santa Cruz), Rabbit anti-KPNA2(importin α1) (10819-1-AP; Proteintech), mouse anti-AKT3 (sc-134254; Santa Cruz), rabbit Phospho-PI3 Kinase (# 17366S; CST), rabbit Phospho-AKT (ab192623; Abcam), rabbit c-myc (10828-1-AP; Proteintech), rabbit METTL3 (15073-1-AP; Proteintech), rabbit Phospho-mTOR (CY5996; Abways), rabbit Palladin (10853-1-AP; Proteintech) and rabbit anti-AKT (ab18785; Abcam). Cytosolic and nuclear fractions were prepared from GC cells using Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China).
Construction of vectors and luciferase reporter assays
The full-length cDNA of human THAP7-AS1 was synthesized by Invitrogen and cloned into the pCDNA3.1 expression vector (Beauchamp Biology Co. Ltd., Shanghai, China). Using the University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/), the 2000 base pair (bp) sequence upstream of the transcription start site (TSS) was extracted. The potential THAP7-AS1 promoter regions (−2000/0, −1012/0, −509/0, −257/0, −210/0, −155/0, and −101/0) were PCR-amplified with the primer sequences shown in Supplementary Table 2 and were then inserted into the NheI/HindIII sites upstream of the firefly luciferase in the pGL3-Basic vector (Promega, Madison, WI, USA). All of the constructs were named based on the location of the promoter fragments relative to the TSS. To identify putative transcription factor (TF)-binding sites, the promoter sequence of THAP7-AS1 was analyzed using the ALGGEN program (http://alggen.lsi.upc.edu/). The full-length cDNA sequence of several TFs including SP1, ETS-domain containing protein (ELK1), CCAAT/enhancer-binding protein alpha (CEBPA), Krueppel-like factor 5 (KLF5), and early growth response protein 1 (EGR1) were PCR-amplified with the cDNA from MKN-45 cells and cloned into the pcDNA3.1 vector. The three single-site mutations (SP1–1(−416 to −407 bp) mut, SP1–2(−392 to −383 bp) mut and SP1–3(−295 to −287 bp) mut) plasmids, the two-site mutation (SP1–1,2 mut, SP1–1,3 mut, SP1–2,3 mut) and the three-site mutation (SP1–1-3 mut) plasmids were obtained by GENERAL BIO (Anhui, China). pEGFP-C1-CUL4B, pEGFP-C1-CUL4B 1–50, and pEGFP-C1-CUL4BΔ37-40 (NLS-delete) was kindly provided by professor Gong [12]. The importin α1 plasmid were purchased from Vigene Biosciences. The pmirGLO-PIK3CD, AKT3, and PIK3CA plasmids were generated in our previous study. All vectors were confirmed by direct sequencing. Luciferase activities were evaluated using the Dual-Luciferase Assay Kit (Promega) in accordance with the manufacturer’s instructions.
Co-immunoprecipitation (Co-IP)
Co-IP was conducted as following, briefly, the treated cells were collected and lysed in IP lysis buffer. One milligram of total protein were used for Co-IP and incubated for 1 h with anti-GFP or anti-His antibodies, and normal mouse IgG as a negative IP control, respectively. The mixtures were incubated overnight with protein A/G agarose beads, and then beads were washed at least 10 times, and treated and boiled for 5 min with 1× SDS sample buffer.
M6A MeRIP qRT–PCR of THAP7-AS1
The Magna m6A MeRIP Kit (Millipore) was used to determine the quantification of m6A-modified THAP7-AS1 levels according to the manufacturer’s instructions. Total RNA was isolated from MKN-45 and BGC-823 cells by Trizol extraction. Then, RNA was fragmented to 94 °C for 5 min, and immunoprecipitated with 10 μg anti-m6A antibody in 1 ml buffer containing RNase inhibitor. Protein A/G magnetic beads were washed, added to the mixture and incubated for 2 h at 4 °C with rotation. m6A RNA was eluted twice with 20 mM N6-methyladenosine 5ʹ-monophosphate sodium salt at 4 °C for 1 h and purified by RNA purification kit. The m6A enrichment was analyzed via RT-qPCR analysis.
RNA pull-down assays and mass spectrometry
The biotin-labeled THAP7-AS1 and antisense strands RNA were transcribed using T7 RNA polymerase in vitro (Ambio Life), followed by purification using the RNeasy Plus Mini Kit (QIAGEN) and treatment with RNase-free DNase I (QIAGEN). Biotinylated RNAs were incubated with protein extracted from MKN-45 cell and mixed with pre-cleared with streptavidin beads. Beads were collected by centrifugation and washed using RNA wash buffer. RNA-protein complexes were eluted, denaturation, and processed by SDS/PAGE followed by silver staining. Differential band specifically pulled down between biotinylated THAP7-AS1 and antisense strands RNA were excised from the gel, digested, and identified by mass spectrometry. To identify which specific region of THAP7-AS1 interacts with CUL4B, we truncated THAP7-AS1(Δ1 represents the region between 1–442 nucleotides (nt) of THAP7-AS1, which is the truncated 5′; Δ2 represents the region between 443–830 nt of THAP7-AS1; Δ3 represents the region between 831–1301 nt of THAP7-AS1, which is the truncated 3′).
RNA immunoprecipitation assay
The EZMagna RNA immunoprecipitation (RIP) Kit (17-701; Millipore) was used to investigate whether THAP7-AS1 could interact potential binding proteins CUL4B and importin α1. MKN-45 and BGC-823 cells were lysed in complete RIP lysis buffer contains Rnasin Inhibitor and the cell extract was incubated with magnetic beads conjugated with anti-CUL4B (SAB1406670; Millipore) antibodies, anti-importin α1(10819-1-AP; Proteintech) or control IgG (Millipore) for overnight at 4 °C. Next, the beads were washed using RIP buffer and incubated with Proteinase K for 30 min at 55 °C to remove the proteins. Finally, RT-qPCR assays were performed to detect the expression of THAP7-AS1 in purified RNA.
RNA polymerase II Inhibitor ActD chase assay
The si-IGF2BP1 was transiently transfected into gastric cancer cells using Lipofectamine 2000 (Invitrogen). After 48 h, Actinomycin D (ActD, 5 mg/mL) was added to the culture medium, followed by incubation for 15 min, 30 min, 1 h, 2 h, or 3 h. THAP7-AS1 stability in the ActD treatment group was analyzed by qPCR.
Chromatin immunoprecipitation assays
The chromatin immunopreciptation (ChIP) assay was performed in MKN-45 and BGC-823 cells, as previously described [13]. Briefly, cell lysates were sonicated, pre-cleared, and immunoprecipitated with SP1 (SAB1412220; Millipore), CUL4B (SAB1406670; Millipore), enhancer of zeste homologue 2 (EZH2; sc-137255; Santa Cruz), histone methylation occurring on the amino-terminal tail of the core histone H3 (H3K27me3; #9733; CST), monoubiquitinated histone H2A on lysine 119 (H2AK119ub1; #8240; CST), histone deacetylase 1 (HDAC1; sc-81598; Santa Cruz), HDAC2 (#57156; CST) antibodies or IgG (Millipore) as the control. Enrichment of the DNA template was analyzed by qPCR using primers specific for each target gene promoter (Supplementary Tables 2–4).
Immunohistochemistry
Immunohistochemical staining of SP1 in human GC tissues was performed. Positive SP1 immunostaining showed brown signals in the nuclei. SP1 staining intensity was graded as: 1 (weak), 2 (moderate), or 3 (strong). The staining score was calculated as follows: score (maximum of 300) = sum of 1 × percentage of weak, 2 × percentage of moderate, and 3 × percentage of strongly stained cells.
ShRNA-THAP7-AS1 lentiviral vector treatment in vivo
MKN-45 cells suspensions of 3 × 105 cells (0.2 ml) were submucosally injected into the left lateral of axilla region in 12 BALB/c mice (4-weeks old) (Weitong lihua, China). When the tumors were about 5.0–10.0 mm in diameter, 12 mice were randomly divided into two groups. Then, the same volume (20 μL) of LV-sh-THAP7-AS1 and LV-sh-NC were injected into several points intratumorally in two different groups, every 5 days for four injections. Tumor volumes were measured. Three weeks later the mice were sacrificed and tumors were removed, weighed, and subjected to IHC analysis of Ki67. Animal experiments were approved by the Committee for Animal Protection and Utilisation of Shandong University.
Statistical analysis
The difference between the two groups was assessed by the Student’s t-test. The chi-square test was used to analyze the association between THAP7-AS1 and the pathologic features of patients with GC. Correlations between SP1 and THAP7-AS1 were analyzed by Spearman rank correlation. A P value <0.05 was considered statistically significant. All data were statistically analyzed using GraphPad Prism 5 software (San Diego, CA, USA).
Results
THAP7-AS1 is upregulated in GC samples with lymph node metastasis
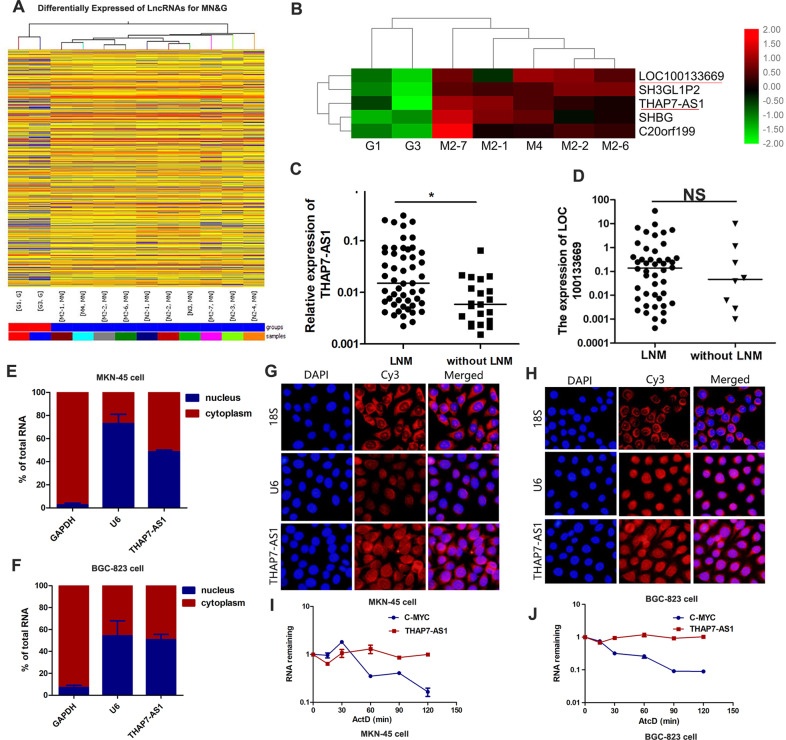
To identify the global lncRNA expression profiles in GC, a lncRNA microarray was used to analyze the differential expression of lncRNAs between 10 GC cases and 2 non-tumorous gastric tissues. In the GC cases, 425 lncRNAs were significantly downregulated, and 989 lncRNAs were significantly upregulated (fold change ≥2, P < 0.05, raw data >50; Fig. 1A, B). LncRNA expression profile data are available in the GEO database (GSE72307). Two lncRNAs including LOC100133669 and THAP7-AS1 were identified as candidates based on the following properties: (1) fold change ≥5; (2) P < 0.05; (3) the expression of lncRNAs was markedly upregulated in GC cases compared with non-tumorous gastric tissues. Interestingly, in 72 primary GC cases, THAP7-AS1 (Fig. 1C) rather than LOC100133669 (Fig. 1D) was significantly upregulated by 2.6-fold in GC cases with lymph node metastasis (LNM) compared to those without LNM.
Fig. 1. THAP7-AS1 is upregulated in GC samples with lymph node metastasis.
A, B Hierarchical clustering of differentially lncRNAs expression in 10 GC cases and 2 non-tumorous gastric tissues. Each row represents different group and each column represents the expression level of an individual lncRNA. Red represents upregulated genes, and green represents down-regulated genes. C THAP7-AS1 expression was detected in human GC tissues with lymph node metastasis (LNM) and without LNM. D LOC100133669 expression was detected in human GC tissues with lymph node metastasis (LNM) and without LNM. E–H Nuclear/cytoplasm fractionation (E–F) and RNA FISH assay (G–H, ×200) were performed to observe the cellular location of THAP7-AS1 in MKN-45 cells (E, G) and BGC-823 cells (F, H). I–J RNA Polymerase II Inhibitor Actinomycin D (ActD) Chase assay was used to observe the RNA stability of THAP7-AS1 in MKN-45 cells (I) and BGC-823 cells (J), with c-Myc as a positive control for short RNA half-life in GC cells. *P < 0.05, **P < 0.01, ***P < 0.001.
The Coding-Potential Assessment Tool (http://lilab.research.bcm.edu/cpat/index.php) and PyhloCSF (a comparative genomics method) indicated that THAP7-AS1 has very weak protein-coding potential (Supplementary Fig. 1A–B). In addition, the RNA pull-down assay indicated no interaction between THAP7-AS1 and ribosomal protein LP0 (Fig. 4B and Supplementary Fig. 2C). These results strongly indicate that THAP7-AS1 does not encode a protein. Nuclear/cytoplasm fractionation analysis demonstrated that THAP7-AS1 was located in both the cytoplasm and nucleus (Fig. 1E, F), which was confirmed by the RNA FISH assay (Fig. 1G, H). The RNA polymerase II inhibitor Actinomycin D (ActD) chase assay showed that THAP7-AS1 was stably expressed (Fig. 1I–J).
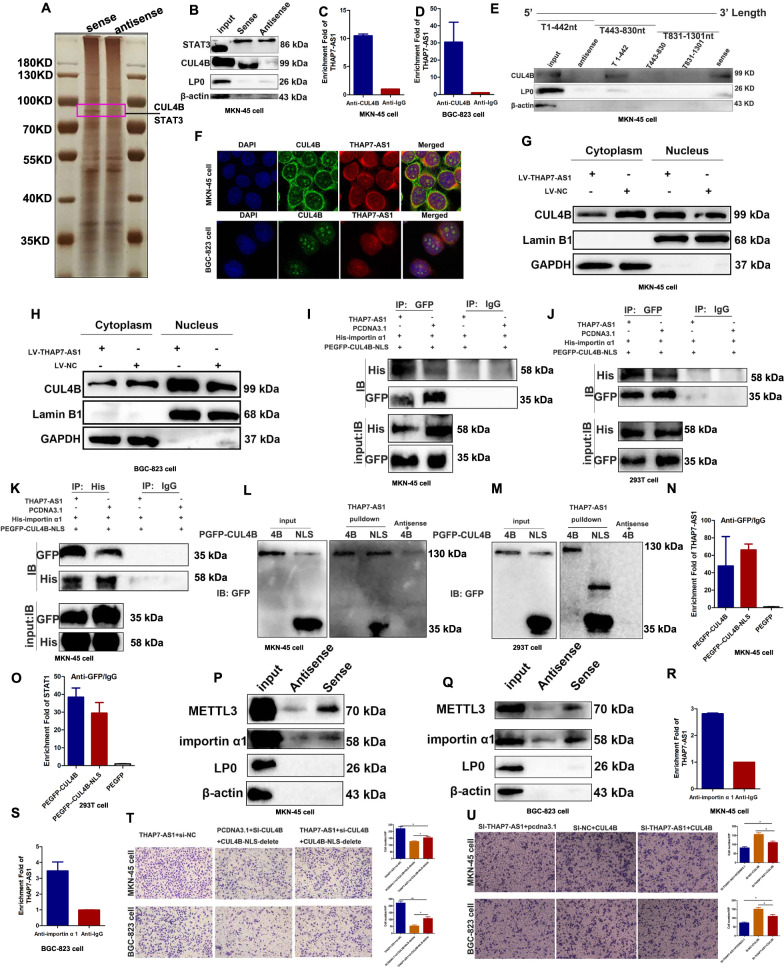
Fig. 4. THAP7-AS1 binds the nuclear localization signal region of CUL4B and mediates the entry of CUL4B into the nucleus.
A Silver staining of the THAP7-AS1–protein complex by incubation of biotinylated THAP7-AS1 with protein extracts from MKN-45 cells. The arrow shows the differential band that was present when the protein extracts were incubated with THAP7-AS1 when compared with antisense RNA. B Western blot analysis of the two proteins from the proteomics screen after pulldown shows the specific interaction of CUL4B but not STAT3 with THAP7-AS1 in MKN-45 cells. C–D RNA immunoprecipitation with an anti-CUL4B antibody was used to assess whether CUL4B binding to THAP7-AS1 in MKN-45 cells (C) and BGC-823 cells (D); IgG was used as the control. E Deletion mapping of the CUL4B-binding domain in THAP7-AS1. Up, diagrams of full-length THAP7-AS1 and the deletion fragments. Bottom, western blot analysis for CUL4B in protein samples pulled down by the different THAP7-AS1 constructs. F Confocal micrographs of THAP7-AS1 and CUL4B. DAPI, 4′,6-diamidino-2-phenylindole (×630). G–H The cytoplasmic and nuclear protein fractions in lentivirus THAP7-AS1 transfected MKN-45 (G) and BGC-823 (H) cells showed that CUL4B expression was decreased in cytoplasm and increased in the nucleus. I–J Co-immunoprecipitation assays of NLS region of CUL4B and importin α1 in MKN-45 (I) and 293T (J) cells. Both cells were transfected with plasmid for pcDNA3.1-THAP7-AS1 or pcDNA3.1, pEGFP-C1-CUL4B 1-50aa and pENTER-his-importin α1. Cell lysates were immunoprecipitated with IgG control antibody or GFP antibody and then immunoblotted with His antibody. K Co-immunoprecipitation assays of NLS region of CUL4B and importin α1 in MKN-45 (K) cells. Cells were transfected with plasmid for pcDNA3.1-THAP7-AS1 or pcDNA3.1, pEGFP-C1-CUL4B 1-50aa and pENTER-his-importin α1. Cell lysates were immunoprecipitated with IgG control antibody or His antibody and then immunoblotted with GFP antibody. L–M Western blot of truncated CUL4B proteins retrieved by in vitro-transcribed THAP7-AS1 in MKN-45 (L) and 293T (M) cells. N–O RIP assay was used to investigate whether NLS region of CUL4B could interact THAP7-AS1 in MKN-45 (N) and 293T (O) cells. Both cells were respectively transfected with plasmid for pEGFP-C1-CUL4B 1-50aa (NLS), pEGFP-C1-CUL4B and pEGFP-C1. Cell lysates were immunoprecipitated with IgG control antibody or GFP antibody. P–Q Western blot analysis of the METTL3 and importin α1 after pulldown shows the specific interaction of METTL3 and importin α1 with THAP7-AS1 in MKN-45 cells (P) and BGC-823 cells (Q). R–S. RNA immunoprecipitation with an anti-KPNA2 (importin α1) antibody was used to assess whether importin α1 binding to THAP7-AS1 in MKN-45 cells (R) and BGC-823 cells (S); IgG was used as the control. T–U Migration abilities of both cells transiently transfected with indicated RNA and plasmid were observed in both GC cells (×100). *P < 0.05, **P < 0.01, ***P < 0.001.
SP1 activates THAP7-AS1 transcription in GC cells
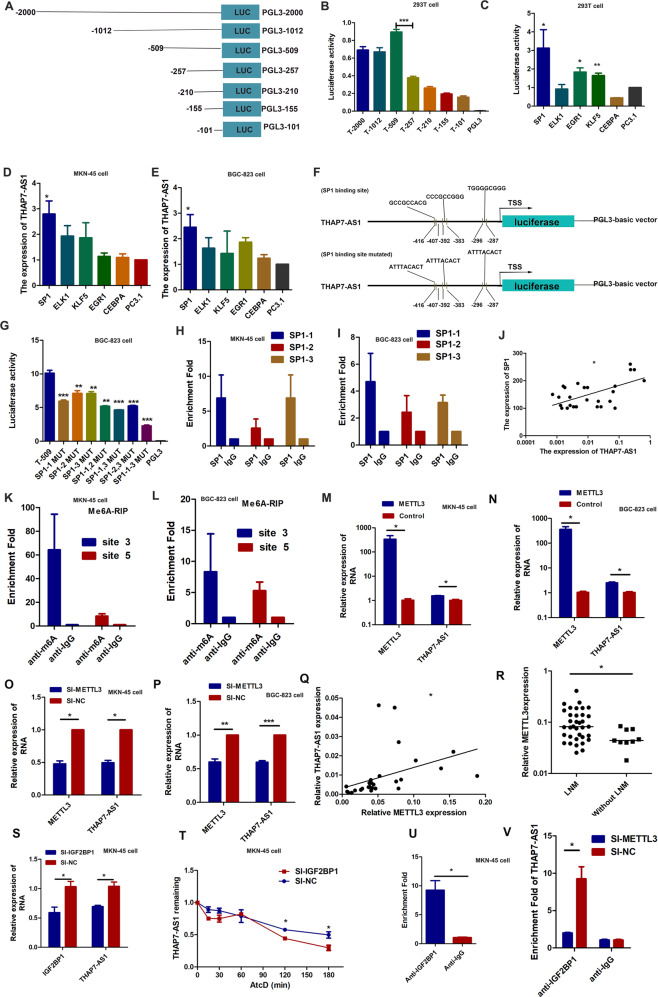
To explore the potential regulators involved in THAP7-AS1 upregulation, a detailed promoter analysis was performed. As shown in Fig. 2A, B, the pGL3-509/0 fragments displayed the highest promoter activity among the seven fragments. The promoter activity of the pGL3-257/0 region significantly decreased to 44% of that of the pGL3-509/0 region, indicating that the −509/−257 region was required for the full transcription of THAP7-AS1. To identify the TFs (Transcription Factor) binding to the promoter region of THAP7-AS1 in GC cells, the region from −509 to −257 was predicted by the online software ALGGEN program to obtain putative TFs and their binding sites. Five binding regions with high scores were found that contained three SP1-binding sites, four ELK1-binding sites, one CEBPA-binding site, six KLF5-binding sites, and two EGR1-binding sites. To determine their roles in regulating THAP7-AS1 transcription, these five vectors were, respectively, overexpressed in 293 T cells transiently transfected with the pGL3-509/0 construct. The promoter activities of pGL3-509/0 was markedly enhanced in the SP1, KLF5, and EGR1 overexpression group (Fig. 2C). However, only SP1 significantly promoted the expression of THAP7-AS1 (Fig. 2D, E). To observe the contribution of the three putative SP1-binding sites to the regulation of the THAP7-AS1 promoter, we introduced three single-site mutations (SP1–1(−416 to −407 bp) mut, SP1–2(−392 to −383 bp) mut, and SP1–3(−295 to −287 bp) mut), the two-site mutation (SP1–1,2 mut, SP1–1,3 mut, and SP1–2,3 mut) and the three-site mutation (SP1–1-3 mut) (Fig. 2F). The results showed that single-site mutations (SP1–1 mut, SP1–2 mut, and SP1–3 mut) plasmids reduced by 29% to 42% respectively in promoter activity compared with the wild-type control. The two-site mutation plasmids (SP1–1,2 mut, SP1–1,3 mut, SP1–2,3 mut) reduced by 54% to 60% respectively and the three-site mutation (SP1–1-3 mut) plasmids decreased by 70% in BGC-823 cells, MKN-45 cells, and 293T cells (Fig. 2G and Supplementary Fig. 1C–D). These data suggest that the three SP1-binding site act independently. Next, we designed three primers that encompassed the SP1-binding sites (Fig. 2F) and performed ChIP assays to determine if SP1 could bind to these three sites. As shown in Fig. 2H, I, SP1 could directly bind to the THAP7-AS1 promoter. The correlation of SP1 and THAP7-AS1 transcription was further elucidated in GC tissue samples, and the results showed that THAP7-AS1 expression was positively correlated with SP1 levels in GC (Fig. 2J). Taken together, these results showed that SP1 binds to the promoter of THAP7-AS1 and subsequently activates its transcription.
Fig. 2. THAP7-AS1, transcriptionally activated by SP1 and post- transcriptionally stabilized by METTL3-mediated m6A modification.
A Schematic diagram of the THAP7-AS1 promoter fragments spanning from −2,000/ −1,002/ −509/ −257/ −210/ −155/−101 to 0. This promoter fragments were cloned into the upstream of the firefly luciferase reporter gene in the pGL3-basic vector. B Transcriptional activity analysis of the potential THAP7-AS1 promoter fragments in 293T cells. C Luciferase activity assay demonstrated that SP1, EGR1 and KLF5 observably increased promoter activities of pGL3 − 509/0. D–E RT-qPCR assay showed SP1 enhanced the expression level of THAP7-AS1 in MKN-45 cells (D) and BGC-823 cells (E). (F). Three primers that covered the SP1 binding sites at the regions −416 to −407 bp (SP1-1), −392 to −383 bp (SP1-2), −295 to −287 bp (SP1-3) were designed (up). Schematic diagram of the luciferase reporter construct containing the human THAP7-AS1 promoter and the mutant construct (SP1-Mut-1/2/3) containing the basal promoter in which the presumed THAP7-AS1 binding site was mutated (bottom). G Luciferase activity of the THAP7-AS1 promoter was reduced when the three single site (SP1–1, SP1–2 and SP1–3), the two-site (SP1–1,2, SP1–1,3, SP1-2,3) and the three-site mutation (SP1–1-3) were mutated in BGC-823 cells. H–I ChIP-qPCR analysis showed higher fold enrichment of promoter amplicons of SP1 in anti-SP1 antibody group than that of IgG group in MKN-45 cells (H) and BGC-823 cells (I), indicating that SP1 could directly bind to THAP7-AS1 promoter. SP1-1, SP1-2 and SP1-3 respectively represent primer that covered the SP1 binding sites. J A significant positive correlation was found between the protein levels of SP1 and THAP7-AS1 in GC tissues. K–L Enrichment of m6A-modified THAP7-AS1 site 3 and 5 in MKN-45 (K) and BGC-823 cells (L). M–N Transcript levels of METTL3 and THAP7-AS1 in METTL3 and PcDNA3.1 MKN-45 cells (M) and BGC-823 cells (N). O–P Transcript levels of METTL3 and THAP7-AS1 in si-METTL3 and si-NC MKN-45 cells (O) and BGC-823 cells (P). Q A significant positive correlation was found between the METTL3 and THAP7-AS1 in GC tissues. R METTL3 expression was detected in human GC tissues with LNM and without LNM. S Transcript levels of IGF2BP1 and THAP7-AS1 in si-IGF2BP1 and si-NC MKN-45 cells. T MKN-45 cells transfected with SI-IGF2BP1 and control cells treated with Actinomycin D (ActD 5 μg/ml) for the indicated periods of time. THAP7-AS1 levels were analyzed by RT-qPCR. U RNA immunoprecipitation with an anti-IGF2BP1 antibody was used to assess whether IGF2BP1 binding to THAP7-AS1 in MKN-45 cells; IgG was used as the control. V RIP-qPCR using anti-IGF2BP1 antibody showed the affinity of THAP7-AS1 RNA to IGF2BP1 in si-METTL3 cells. *P < 0.05, **P < 0.01, ***P < 0.001.
METTL3-mediated m6A modification enhanced THAP7-AS1 expression via IGF2BP1-dependent stabilization of RNA
N6-methyladenosine (m6A) modification was reported to the most prevalent internal RNA modification and involved in the regulation of RNA degradation [14], stability [15], and splicing [16]. To evaluate the importance of m6A in THAP7-AS1 post-transcriptional regulation, sequence-based RNA adenosine methylation site predictor (SRAMP) was used to predict m6A modification sites in THAP7-AS1 sequences [17], which demonstrating seven putative m6A sites (Supplementary Fig. 1E). Next, we performed a methylated RNA immunoprecipitation (MeRIP) assay to shown that there existed m6A modification in THAP7-AS1 (Fig. 2K, L). Moreover, overexpression or knockdown of METTL3, an important component of the m6A methylase complex [18], rather than METTL14 (Supplementary Fig. 1F–G), dramatically increased or reduced the level of THAP7-AS1 in GC cells (Fig. 2M–P). The correlation of METTL3 and THAP7-AS1 was further determined in GC tissue samples, and the results showed that METTL3 expression was positively correlated with THAP7-AS1 levels (Fig. 2Q). Moreover, METTL3 was significantly upregulated in GC cases with LNM compared to those without LNM (Fig. 2R). The results indicated that METTL3 was a major m6A methyltransferase for THAP7-AS1. Furthermore, RNA-pulldown assay indicated that THAP7-AS1 could bind to METTL3 (Fig. 4P, Q). Next, Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs; including IGF2BP1/2/3) were chosen to investigate the potential role in METTL3-dependent upregulation of THAP7-AS1, as IGF2BPs are well-known RNA binding proteins to stabilize their target RNAs [19]. Our results showed that knockdown IGF2BP1, rather than IGF2BP2-3, led to decreased THAP7-AS1 expression (Fig. 2S and Supplementary Fig. 1H–I). To distinguish between a transcriptional and a post-transcriptional mechanism, we performed an RNA polymerase II Inhibitor ActD chase assay to assess the effects of IGF2BP1 on THAP7-AS1 stability, which showed poor THAP7-AS1 stability after IGF2BP1 depletion (Fig. 2T). In addition, RIP assay discovered that enrichment of THAP7-AS1 in anti-IGF2BP1 immunoprecipitates compared with the IgG control (Fig. 2U). Moreover, we found a notably reduced affinity of IGF2BP1 to THAP7-AS1 in METTL3-silenced MKN-45 cells (Fig. 2V). Collectively, our findings revealed that METTL3-mediated m6A modification enhanced THAP7-AS1 expression via IGF2BP1-dependent stabilization of RNA.
THAP7-AS1 promotes GC cell growth, migration, and invasion in vitro and in vivo
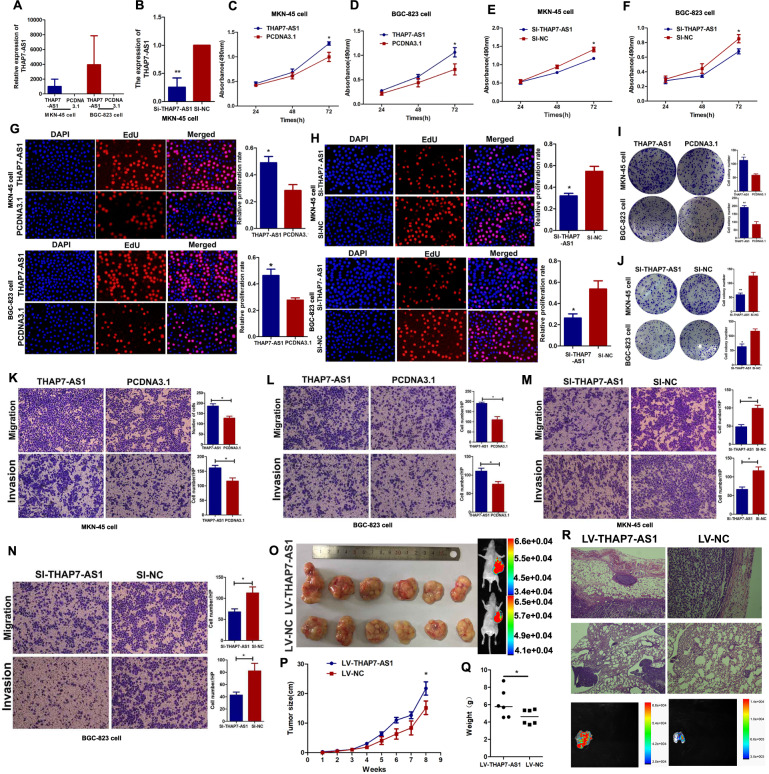
To investigate the biological functions of THAP7-AS1 in GC cells, THAP7-AS1 was overexpressed by transfection with a THAP7-AS1 overexpression plasmid (Fig. 3A) and was knocked down by transfection with THAP7-AS1 Smart Silencer (Fig. 3B and Supplementary Fig. 1J). The MTS, EdU, and colony formation assays showed that THAP7-AS1 overexpression enhanced MKN-45 and BGC-823 cell growth capabilities (Fig. 3C–D, G, I), while silencing of THAP7-AS1 inhibited the proliferation of both GC cell lines (Fig. 3E–F, H, J). In addition, THAP7-AS1 overexpression markedly enhanced the migration and invasion capabilities of MKN-45 and BGC-823 cells (Fig. 3K, L). By contrast, silencing of THAP7-AS1 abolished this effect (Fig. 3M, N). The growth-promoting effects of THAP7-AS1 were demonstrated by in vivo tumor growth assays. The tumor growth of THAP7-AS1-overexpressing xenografts was markedly promoted as demonstrated by the increased mean volume (Fig. 3O, P) and weight (Fig. 3Q) compared with the control group. Next, we found that the LV-THAP7-AS1 group showed local tumor invasion, with islands of cancer cells invading the adipose tissue. However, the tumor nodules in the control group were non-invasive or well encapsulated (Fig. 3R). Moreover, the number of metastatic foci in the lung of the LV-THAP7-AS1 group was larger than those of the control group (Supplementary Fig. 2A–B). Taken together, these findings suggest that THAP7-AS1 plays an important role in initiating the malignant phenotype of GC cells.
Fig. 3. THAP7-AS1 promote GC cells growth, invasion, and metastasis abilities in vitro and in vivo.
A The overexpression efficiency of THAP7-AS1 in both GC cells. B The stably knockdown efficiency of THAP7-AS1 in MKN-45 cells. C–F The MKN-45 (C, E) and BGC-823 (D, F) cell growth rates were evaluated with MTS proliferation assays. G–H MKN-45 (G) and BGC-823 (H) cells growth was determined by the EdU assay (×100). I–J Colony formation assays were performed to observe the cell colony formation ability of THAP7-AS1 and Si- THAP7-AS1–transfected cells. K–N Migration and invasion ability of the MKN-45 cells (K, M) and BGC-823 cells (L, N) by Transwell assays (×100). O The THAP7-AS1 stably expressing group or the negative control were used for tumorigenesis assay. Seven weeks post implantation, the tumors in Nu/Nu mice was observed by an in vivo imaging system. P–Q After subcutaneous injection of MKN-45 cells containing a stable overexpression of THAP7-AS1 or the negative control, tumor growth curves are shown (P). The tumor weights (Q) were measured when the mice were sacrificed and the tumors were excise. R Compared with the LV-NC group, more stroma, muscle invasion or lung metastasis was observed in the LV-THAP7-AS1 group. Representative stromal invasion was shown in H&E staining (top, ×100). Representative lung metastasis locus was shown in H&E staining (middle, ×100) and IVIS system imaging (bottom). *P < 0.05, **P < 0.01, ***P < 0.001.
THAP7-AS1 binds the nuclear localization signal region of CUL4B and mediates the entry of CUL4B into the nucleus
Recent studies have indicated that lncRNAs are involved in molecular regulatory pathways through their interactions with proteins [20, 21]. We speculated that THAP7-AS1 might interact with certain cellular proteins to regulate the malignant phenotype of GC cells. The RNA pull-down assay and MS analysis were performed to identify the THAP7-AS1–protein complex (Fig. 4A), with two potential binding proteins including CUL4B and STAT3 (signal transducer and activator of transcription 3) were selected for further binding validation based on the following criteria: (1) molecular weight of ~100 kD; (2) peptide score > 100; (3) reportedly involves in tumor progression. Our results showed that only CUL4B bound specifically to THAP7-AS1 in both GC cell lines (Fig. 4B–D and Supplementary Fig. 2C). We constructed a series of THAP7-AS1 truncation mutants to map its binding region to CUL4B and found that nt 1–442 at the 5′ end of THAP7-AS1 were required for its interaction with CUL4B (Fig. 4E). Furthermore, RNA FISH technology combined with immunofluorescence analysis demonstrated the co-localization of THAP7-AS1 and CUL4B protein in both GC cell lines (Fig. 4F). Next, we found that THAP7-AS1 did not regulate CUL4B expression (Fig. 5A, E and Supplementary Fig. 3D), suggesting that THAP7-AS1 was not involved in the post-transcriptional regulation of CUL4B. Interestingly, CUL4B expression was decreased in cytoplasm and increased in the nucleus after lentivirus THAP7-AS1 transfection (Fig. 4G, H), indicating that THAP7-AS1 promoted CUL4B protein entry into the nucleus. CUL4B protein domain mapping studies revealed that importin α1 binds the 1-50 amino acid (aa) region of CUL4B, which is reported to be the nuclear localization signal (NLS), and mediates the nuclear transport of CUL4B [12]. Our results indicated that THAP7-AS1 improved the interaction capabilities between NLS and importin α1 (Fig. 4I–K). And THAP7-AS1 binds directly to NLS region of CUL4B protein (Fig. 4L, M). In addition, RIP assay determined that NLS region of CUL4B could interact THAP7-AS1 in MKN-45 (Fig. 4N) and 293T (Fig. 4O) cells. Furthermore, RNA pulldown and RIP assay indicated that THAP7-AS1 could directly bind to importin α1 (Fig. 4P–S). These results suggested that THAP7-AS1 interacts with NLS region of CUL4B and importin α1, and promotes the interaction between CUL4B and importin α1. Next, a rescue experiment was performed to determine if THAP7-AS1 exerts biological functions through CUL4B nuclear localization. Interestingly, CUL4B knockdown rescued the migration abilities enhanced by THAP7-AS1. And we put back a CUL4B-NLS deletion after CUL4B knockdown, the reduced migration induced by CUL4B knockout could not be reversed (Fig. 4T), indicating that THAP7-AS1 promote GC progression in a way dependent on mediating CUL4B nuclear localization. Similarly, CUL4B restoration rescued the migration abilities reduced by THAP7-AS1 silencing (Fig. 4U), indicating that THAP7-AS1 promoted the malignant phenotype of GC cells by mediating the entry of CUL4B into the nucleus.
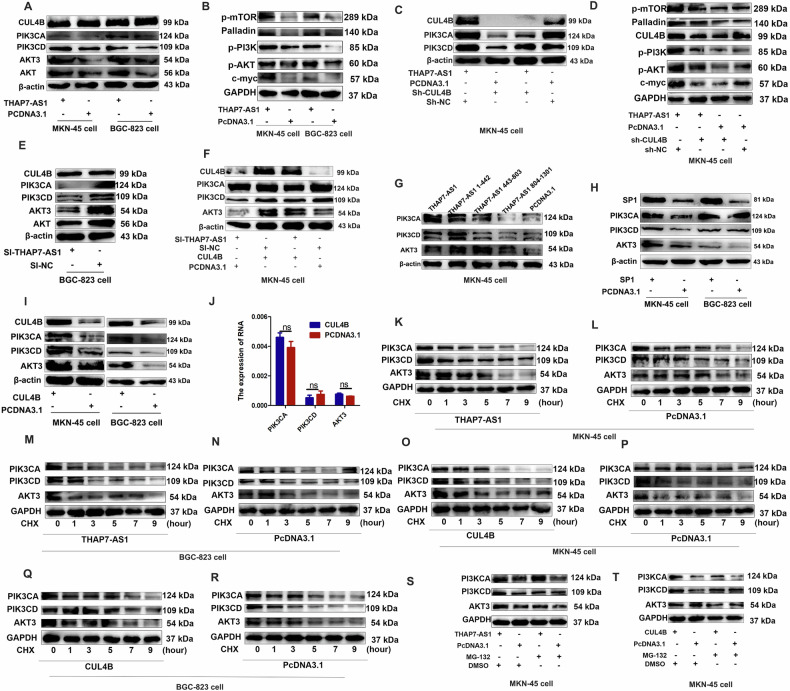
Fig. 5. THAP7-AS1/CUL4B complex initiates PI3K/AKT signaling.
A–B Western blot analysis of the different signaling modules downstream of THAP7-AS1 in both GC cells expressing THAP7-AS1 or control cells. C–D Western blot analysis of the PI3K/AKT signaling downstream of THAP7-AS1 in GC cells expressing THAP7-AS1 or control cells with or without CUL4B knockdown. E Western blot analysis of the different signaling modules downstream of THAP7-AS1 in GC cells expressing si-THAP7-AS1 or control cells. F Western blot analysis of the PI3K/AKT signaling module downstream of THAP7-AS1 in MKN-45 cells expressing si-THAP7-AS1 or control cells with or without transient transfection with PCDNA3.1-CUL4B. G Western blot analysis of the THAP7-AS1 fragments mediated downstream PI3K/AKT signaling of THAP7-AS1/ CUL4B in MKN-45 cells. H Western blot analysis of the different signaling downstream of THAP7-AS1 in both GC cells expressing SP1 or control cells. I Western blot analysis of the different signaling downstream of THAP7-AS1 in both GC cells expressing CUL4B or control cells. J The mRNA expression of PIK3CD, PIK3CA and AKT3 was detected by RT-qPCR assays. K–R MKN-45 cells (K–L and O–P) and BGC-823 cells (M–N and Q–R) transfected with THAP7-AS1(K, M) or CUL4B (O, Q) overexpression vector and control cells were treated with cycloheximide (CHX; 5 mg/mL) or vehicle for the indicated periods. PIK3CA, PIK3CD and AKT3 protein levels were analyzed by Western blotting. S–T MKN-45 cells transfected with THAP7-AS1 (S) or CUL4B (T) overexpression vector and control cells were treated with MG132 (5 mmol/L) or vehicle for 24 h. Cell lysates were analyzed by Western blotting.
THAP7-AS1/CUL4B complex initiates miR-22-3p/miR-320a-suppressed PI3K/AKT signaling
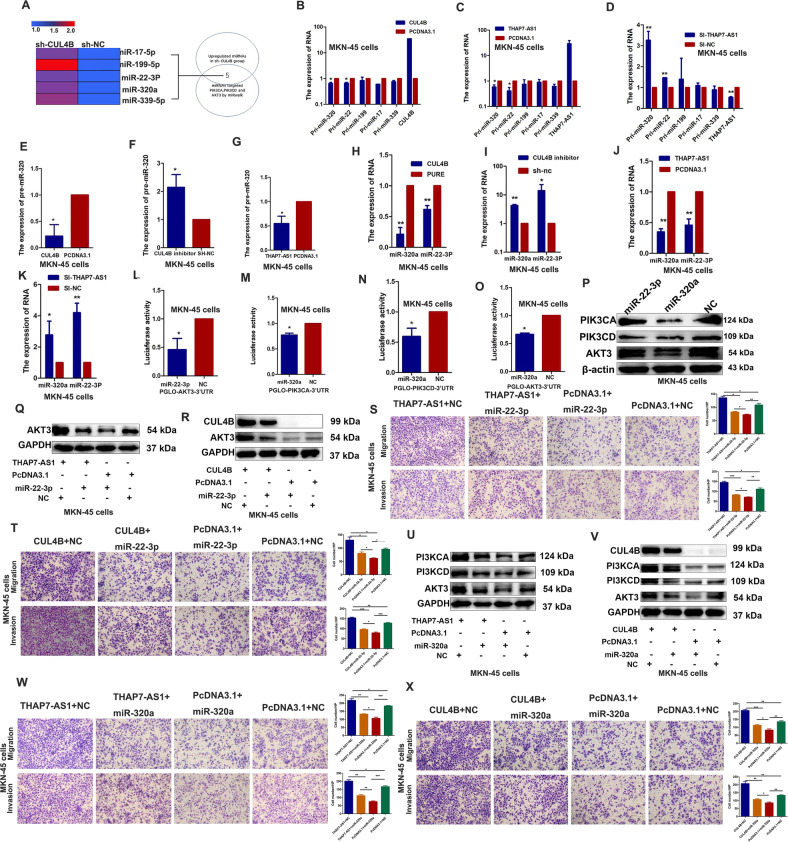
To identify the potential genes involved in THAP7-AS1/CUL4B complex-mediated biological functions, RNA sequence assay was performed between si-THAP7-AS1 cancer cells and control group. Compared with the control group, 6502 genes were upregulated and 5794 genes were downregulated in si-THAP7-AS1 cells (Supplementary Fig. 2D). GSEA (Gene Set Enrichment Analysis) indicated that ERBB2 signaling pathway was significantly enriched in THAP7-AS1 downregulated cancer cells (Supplementary Fig. 2E). THAP7-AS1 overexpression upregulated the protein expression of PIK3CA, PIK3CD, and AKT3 in the GC cell lines (Fig. 5A) and mouse xenograft tissues (Supplementary Fig. 3A). In addition, the expressions of p-PI3K, p-AKT, and the key target genes of the PI3K/AKT pathway including p-mTOR, palladin, and c-myc were increased in THAP7-AS1 overexpressed MKN-45 and BGC-823 cells (Fig. 5B), indicating that ERBB2/PI3K/AKT signaling was involved in THAP7-AS1/CUL4B-induced GC progression. And CUL4B knockdown reversed the enhanced expression of PIK3CA, PIK3CD, AKT3, p-PI3K, p-AKT, p-mTOR, palladin, and c-myc induced by THAP7-AS1 (Fig. 5C, D and Supplementary Fig. 3B–C). In addition, THAP7-AS1 silencing led to a decrease in the protein expression of PIK3CA, PIK3CD, and AKT3 (Fig. 5E and Supplementary Fig. 3D), whereas CUL4B overexpression reversed the reduced expression of PIK3CA, PIK3CD, and AKT3 induced by THAP7-AS1 knockdown (Fig. 5F and Supplementary Fig. 3E). Moreover, THAP7-AS1 and Δ1 (nt 1–442) but not Δ2 (nt 443–803) and Δ3 (nt 804–1301) upregulated the expression of PIK3CA, PIK3CD, and AKT3 (Fig. 5G and Supplementary Fig. 3F), suggesting that nt 1–442 were critical for the THAP7-AS1/CUL4B complex-mediated activation of PI3K/AKT signaling. SP1 overexpression also enhanced the expression of PIK3CA, PIK3CD, and AKT3 (Fig. 5H) due to SP1-mediated upregulation of THAP7-AS1 transcription. In addition, CUL4B upregulation also promoted the protein expression of PIK3CA, PIK3CD, and AKT3 (Fig. 5I). However, there was no correlation between CUL4B and PIK3CA, PIK3CD, or AKT3 at the mRNA level (Fig. 5J). These results indicated that the THAP7-AS1/CUL4B complex upregulated PIK3CA, PIK3CD, and AKT3 protein expression at the post-transcriptional level. CUL4B is an important component in cullin 4B-RING ubiquitin ligase (E3) complex, which regulates target protein’s degradation. In order to explore whether CUL4B nuclear localization could lead to decreased PI3K/AKT components degradation, the protein synthesis inhibitor cycloheximide (CHX) and proteasome inhibitor MG132 was respectively used to evaluate the effects of THAP7-AS1 and CUL4B on PIK3CA, PIK3CD, and AKT3 protein stability. The results indicated that THAP7-AS1 and CUL4B had no obvious effect on the half-life of PIK3CA, PIK3CD, and AKT3 in CHX or MG132 treatment group compared with that of control group (Fig. 5K–T and Supplementary Fig. 3G–H). These results suggest that CUL4B nuclear localization do not lead to decreased PI3K/AKT components degradation in a cullin 4B-RING ubiquitin ligase (E3) dependent way. Mounting evidence has shown that microRNAs (miRNAs) are negative post-transcriptional regulators in various human cancers [22]. Thus, we hypothesized that miRNAs might be involved in the upregulation of PIK3CA, PIK3CD, and AKT3 by CUL4B. Using the GSE database (GSE 90503), we integrated CUL4B-regulated miRNA gene expression data and miRNA bioinformatics analysis (Fig. 6A). Five miRNAs including miR-17, miR-19, miR-22-3p, miR-320a, and miR-339 were identified as candidates based on the fact: (1) upregulated in sh-CUL4B-MKN45 cells; (2) predicted to bind to the 3′ untranslated region of target PIK3CA, PIK3CD, and AKT3. Pri-miR-22-3p and pri-miR-320a expression was significantly decreased in CUL4B and THAP7-AS1-overexpressing cells (Fig. 6B, C and Supplementary Fig. 3I–J) and increased in THAP7-AS1 knockdown cells (Fig. 6D and Supplementary Fig. 3K). Furthermore, the expression of pre-miR-320a/miR-22-3p and mature miR-320a/miR-22-3p was consistent with that of pri-miR-320a/miR-22-3p in THAP7-AS1 and CUL4B overexpressing or knockdown GC cells (Fig. 6E–K and Supplementary Fig. 3L–R). Thus, miR-22-3p and miR-320a were selected for further study. MiR-22-3p suppressed the luciferase activity of pmirGLO-AKT3 (Fig. 6L and Supplementary Fig. 3S), and miR-320a inhibited the luciferase activity of pmirGLO-PIK3CA (Fig. 6M and Supplementary Fig. 3T), pmirGLO-PIK3CD (Fig. 6N and Supplementary Fig. 3U), and pmirGLO-AKT3 (Fig. 6O and Supplementary Fig. 3V). Subsequently, we demonstrated that miR-22-3p attenuated AKT3 protein expression, indicating that AKT3 was a direct target gene of miR-22-3p. In addition, miR-320a significantly decreased PIK3CA, PIK3CD, and AKT3 protein levels in both GC cell lines (Fig. 6P and Supplementary Fig. 3W), suggesting that PIK3CA, PIK3CD, and AKT3 were direct target genes of miR-320a. In order to investigate the effects of two miRNAs on PI3K/AKT expression and GC development, we conducted a series of reversal experiments. Our results showed that miR-22-3p reversed the increased AKT3 expression and GC migration or invasion abilities induced by THAP7-AS1 and CUL4B (Fig. 6Q–T and Supplementary Fig. 4A–D). Similarly, miR-320 reversed the enhanced expression of PIK3CA, PIK3CD, and AKT3 and GC migration or invasion abilities induced by THAP7-AS1 and CUL4B (Fig. 6U–X and Supplementary Fig. 4E–H). Thus, these results indicated that THAP7-AS1/CUL4B complex promoted PI3K/AKT components expression and GC development at least partly due to inhibition of miR-22-3p and miR-320a expression.
Fig. 6. THAP7-AS1/CUL4B complex initiates miR-22-3p/miR-320a -suppressed PI3K/AKT signaling.
A A Venn diagram depicting five miRNAs that were upregulated by CUL4B knockdown in MKN45 cells (GSE 90503), predicted to target PIK3CA, PIK3CD and AKT3 proteins by prediction algorithms. B–D Expression of primary miR-17, miR-19, miR-22-3p, miR-320a, and miR-339 expression was assessed by RT–qPCR in MKN-45 cells respectively transfected with CUL4B (B), THAP7-AS1 (C), and SI-THAP7-AS1 (D). E–G Expression of pre-miR-320a expression was assessed by RT–qPCR in MKN-45 respectively transfected with CUL4B (E), sh-CUL4B (F), and THAP7-AS1 (G). H–K Expression of mature miR-320a and miR-22-3p expression was assessed by RT–qPCR in MKN45 respectively transfected with CUL4B (H), sh-CUL4B (I), THAP7-AS1 (J), and si-THAP7-AS1 (K). L The results indicated a decrease in luciferase activity in MKN-45 cells transfected with PGLO-AKT3-3′UTR and miR-22-3p. M–O Luciferase activity showed a decrease in MKN-45 cells transfected with PGLO-PIK3CA-3′UTR (M), PGLO-PIK3CD-3′UTR (N), PGLO-AKT3-3’UTR (O) and miR-320a. P Western blot analyses were performed to confirm the PIK3CA, PIK3CD and AKT3 expression in MKN-45 cells transfected with miR-22-3p or miR-320a mimics. Q–R Western blotting analysis of the AKT3 in MKN-45 cells overexpressing THAP7-AS1 (Q) or CUL4B (R) with or without transient transfection with miR-22-3p mimics. S–T Migration and invasion abilities of MKN-45 cells transiently transfected with indicated RNA and plasmid were observed. (×100). U–V Western blotting analysis of the PIK3CA, PIK3CD and AKT3 in MKN-45 cells overexpressing THAP7-AS1 (U) or CUL4B (V) with or without transient transfection with miR-320a mimics. W–X Migration and invasion abilities of MKN-45 cells transiently transfected with indicated RNA and plasmid were observed. (×100). *P < 0.05, **P < 0.01, ***P < 0.001.
THAP7-AS1/CUL4B complex transcriptionally represses the expression of miR-22-3p/miR-320a by CUL4B-catalyzed H2AK119ub1 and EZH2-mediated H3K27me3
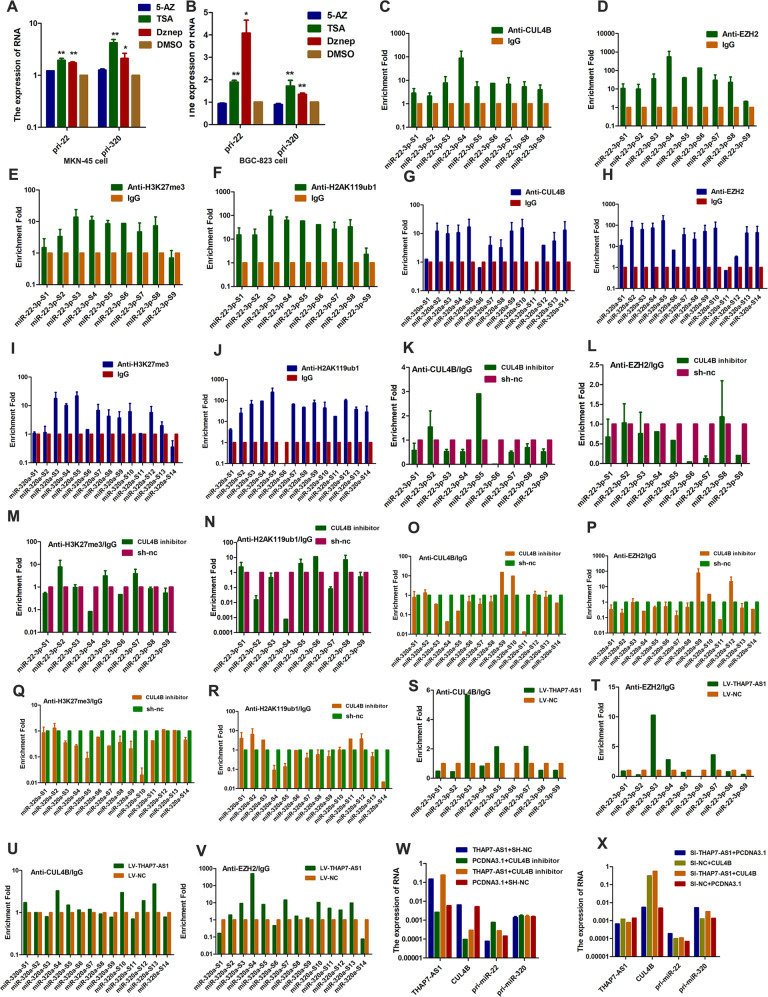
Recently, CUL4B was found to regulate the epigenetic inactivation of tumor suppressors by monoubiquitinating H2AK119 [23, 24]. EZH2, a catalytic subunit of PRC2, is critical for the formation of a stable and enzymatically active methyltransferase complex [25]. In order to detect whether the expression of miR-22-3p and miR-320a were regulated by above mentioned epigenetic modification, we treated GC cells with 5-Azacytidine (5-AZ), trichostatin A (TSA) and 3-deazaneplanocin A (Dznep). The expression of pri-miR-22-3p and pri-miR-320a was enhanced when cells were treated with TSA and Dznep (Fig. 7A, B), suggesting that the promoters of miR-22-3p and miR-320a were regulated by histone methylation and histone acetylation. To determine whether THAP7-AS1 regulates miR-22-3p and miR-320a expression levels by binding with CUL4B, we used ENCODE Histone Modification Tracks embedded in UCSC Genome Browser and found histone methylation enrichment peaks surrounding the miR-22-3p and miR-320a promoter region (Supplementary Fig. 5A). ChIP assays with antibodies against CUL4B, EZH2, H2AK119ub1, H3K27me3, HDAC1, and HADC2 or control IgG showed that CUL4B (Fig. 7C, G), EZH2 (Fig. 7D, H), H3K27me3 (Fig. 7E, I), and H2AK119ub1 (Fig. 7F, J), rather than HDAC1 (Supplementary Fig. 5B, D) and HDAC2 (Supplementary Fig. 5C, E), efficiently immunoprecipitated the promoter region of miR-22-3p and miR-320a, indicating that the promoter regions of miR-22-3p and miR-320a could be regulated by CUL4B-catalyzed monoubiquitinated H2AK119 and PRC2-mediated H3K27me3. Knockdown of CUL4B in cells led to a significant reduction of CUL4B (Fig. 7K, O), EZH2 (Fig. 7L, P), H3K27me3 (Fig. 7M, Q), and H2AK119ub1 (Fig. 7N, R) binding to the promoters of miR-22-3p and miR-320a, suggesting that CUL4B regulated the transcriptional repression of miR-22-3p and miR-320a by recruiting PRC2. In addition, THAP7-AS1 overexpression resulted in a significant increase of CUL4B (Fig. 7S, U) and EZH2 (Fig. 7T, V) binding to the promoters of miR-22-3p and miR-320a, indicating that THAP7-AS1 was involved in CUL4B-mediated PRC2 recruitment. Next, we found that CUL4B knockdown reversed the decrease in pri-miR-22-3p/pri-miR-320a caused by THAP7- AS1 overexpression and vice versa (Fig. 7W, X and Supplementary Fig. 5F). Taken together, our results showed that the THAP7-AS1/CUL4B complex repressed the expression of miR-22-3p/miR-320a at the transcriptional level by CUL4B-catalyzed H2AK119ub1 and EZH2-mediated H3K27me3.
Fig. 7. THAP7-AS1-CUL4B complex transcriptionally represses the expression of miR-22-3p/miR-320a by CUL4B-catalyzed H2AK119ub1 and EZH2-mediated H3K27me3.
A–B MKN-45 (A) and BGC-823 (B) cells were treated with 5-Azacytidine (5-AZ), Trichostatin A (TSA) and deazaneplanocinA (Dznep). C–F We used primers that were located ~1 kb upstream and downstream of the TSS of miR-22-3p (sites S1–S9) of CUL4B-binding sites. ChIP assays with an anti-CUL4B (C), anti-EZH2 (D), anti-H3K27me3 (E) and anti-H2AK119ub1 (F) or negative control (anti-IgG) antibodies showed these four proteins could bind to the miR-22-3p promoter in MKN45 cells. G–J We used primers that were located ~1 kb upstream and downstream of the TSS of miR-320a (sites S1–S14) of CUL4B-binding sites. ChIP assays with an anti-CUL4B (G), anti-EZH2 (H), anti-H3K27me3 (I), and anti-H2AK119ub1 (J) or negative control (anti-IgG) antibodies demonstrated these four proteins could bind to the miR-320a promoter in MKN45 cells. K–N ChIP assays of miR-22-3p promoter primers in CUL4B knockdown MKN-45 cell using indicated antibodies. O–R ChIP assays of miR-320 promoter primers in CUL4B knockdown MKN-45 cell using indicated antibodies. S–V ChIP assays of miR-22-3p (S–T) or miR-320 (U–V) promoter primers in LV-THAP7-AS1 MKN-45 cells using indicated antibodies. W–X RT-qPCR assay was performed to detect primary miR-22-3p and miR-320a in MKN-45 cells transiently transfected with indicated RNA and plasmid. *P < 0.05, **P < 0.01, ***P < 0.001.
THAP7-AS1 may serve as a prognostic and therapeutic biomarker for patients with GC
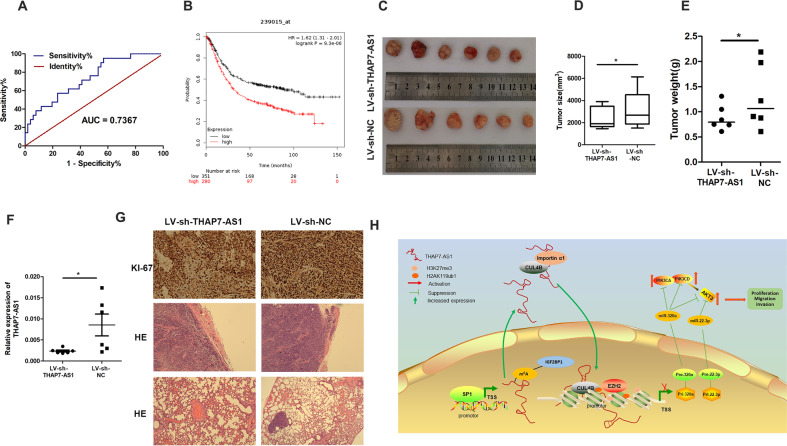
We further explore the correlation between THAP7-AS1 expression and clinical-pathological parameters in GC patients. Higher THAP7-AS1 expression was positively associated with a higher risk of LNM (P = 0.0149) and higher ACJJ stage (P = 0.0362) (Table 1). However, there was no association between THAP7-AS1 expression and tumor size, gender, differentiation, and tumor stage. A receiver operating characteristic (ROC) curve showed that the area under the curve value for THAP7-AS1, which was constructed to distinguish GC cases with and without LNM, reached up to 0.7367 (Fig. 8A, P = 0.0016). These data suggest that THAP7-AS1 is associated with GC metastasis. Using GSE62254 data set, we found that GC patients with high expression of THAP7-AS displayed a poor overall survival (Fig. 8B, n = 283, P = 0.00022).
Table 1.
Association of THAP7-AS1 expression with clinicopathological parameters.
| Variable | n | THAP7-AS1 expression | p value | |
|---|---|---|---|---|
| lower high | ||||
| Age (years) | p = 0.7274 | |||
| <62 | 38 | 20 | 18 | |
| ≥62 | 33 | 16 | 17 | |
| Missing | 1 | 0 | 1 | |
| Gender | p = 0.7432 | |||
| Male | 11 | 6 | 5 | |
| Female | 61 | 30 | 31 | |
| Tumor size (cm) | p = 0.6631 | |||
| <5 | 22 | 12 | 10 | |
| ≥ 5 | 43 | 21 | 22 | |
| Missing | 7 | 3 | 4 | |
| ACJJ stage | p = 0.0362 | |||
| I + II | 22 | 15 | 7 | |
| III + IV | 44 | 17 | 27 | |
| Missing | 6 | 4 | 2 | |
| T classification | p = 0.5490 | |||
| T1 | 3 | 2 | 1 | |
| T2 | 5 | 1 | 4 | |
| T3 | 9 | 4 | 5 | |
| T4 | 50 | 25 | 25 | |
| Missing | 5 | 4 | 1 | |
| Lymph node metastasis | p = 0.0149 | |||
| N0 | 21 | 15 | 6 | |
| N1 | 5 | 2 | 3 | |
| N2 | 18 | 11 | 7 | |
| N3 | 26 | 7 | 19 | |
| Missing | 2 | 1 | 1 | |
| Distant metastasis (M) | p = 0.4372 | |||
| Negative (M0) | 46 | 25 | 21 | |
| Positive (M1) | 22 | 9 | 13 | |
| Missing | 4 | 2 | 2 | |
| Differentiation | p = 0.5294 | |||
| Well | 1 | 1 | 0 | |
| Moderate | 35 | 18 | 17 | |
| Poor | 35 | 16 | 19 | |
| Missing | 2 | 1 | 1 | |
Fig. 8. THAP7-AS1 may serve as a prognostic and therapeutic biomarker for patients with GC.
A Receiver operating characteristic (ROC) curve was performed to evaluate whether THAP7-AS1 expression could be used to distinguish the patients with LNM. B The correlation between THAP7-AS1 expression and overall survival in GC patients was assessed by Kaplan–Meier survival analysis (GSE62254, n = 283, P = 0.00022). C After four times injection of LV-sh-THAP7-AS1 and LV-sh-NC, photographs of representative tumors are shown. D–E Compared with the LV-sh-NC group, the tumor size (D) and weight (E) were significantly lower in the LV-sh-THAP7-AS1-treated mice. F RT-qPCR assay showed that THAP7-AS1 expression was decreased in LV-sh-THAP7-AS1 group. G IHC of tumor tissues showed a lower positivity rate in Ki67 in LV-sh-THAP7-AS1 group than that of LV-sh-NC group(top, ×200). H&E staining was performed to observe local invasion (middle, ×100) and lung metastasis (bottom, ×100) in LV-sh-THAP7-AS1 and LV-sh-NC group. H Proposed functional action of THAP7-AS1/CUL4B complex in modulating GC progression. SP1 activated THAP7-AS1 transcription by directly binding to its promoter. METTL3-mediated m6A modification enhanced THAP7-AS1 expression via m6A-IGF2BP1-dependent pathway. nt 1–442 of THAP7-AS1 interact with NLS region of CUL4B and importin α1, and improve their combination between CUL4B and importin α1, and enhance the CUL4B protein entry into the nucleus. The THAP7-AS1/CUL4B complex activates PI3K/AKT signaling through transcriptionally repressing miR-22-3p and miR-320a, which promote GC cell progression.
To investigate whether THAP7-AS1 could play an inhibitory effect on preestablished tumor growth and metastasis, LV-sh-THAP7-AS1 and LV-sh-NC were injected intratumorally into several sites per xenograft tumor. Compared with the LV-sh-NC group, the tumor size and weight were significantly decreased in the LV-sh-THAP7-AS1-treated mice (Fig. 8C–F). And a lower positivity rate in Ki67 was also observed (Fig. 8G top). Furthermore, we demonstrated that the tumor nodules in the LV-sh-THAP7-AS1 group were well encapsulated. However, the LV-sh-NC group showed multi-site local invasion (Fig. 8G middle). Moreover, metastasis loci in the lungs were found in the LV-sh-NC control group rather than the LV-sh-THAP7-AS1 group (Fig. 8G bottom). These data suggested that LV-sh-THAP7-AS1 could suppress GC growth, invasion or metastasis, therefore acting as a potential prognostic and therapeutic target for GC therapy.
Discussion
Mounting evidence has demonstrated that lncRNAs participate in regulating pathologic and physiologic processes [26]. Here, a global lncRNA gene microarray was performed to select differentially expressed lncRNAs between GC tissues and normal gastric mucosa. THAP7-AS1 was found to be significantly upregulated in GC with LNM compared with those without LNM. And high THAP7-AS1 expression was correlated with higher LNM and ACJJ clinical stage. The ROC curve suggested that THAP7-AS1 may act as a biomarker for predicting LNM in patients with GC. Next, the regulators responsible for the abnormal expression of THAP7-AS1 were studied. We found that the SP1 transcription factor activated THAP7-AS1 expression by binding to the promoter region. Interestingly, SP1 promoted the expression of PIK3CA, PIK3CD, and AKT3 due to SP1-mediated THAP7-AS1 upregulation. M6A modification plays an important role in post-transcriptional regulation of RNAs [14]. Thus, we further explored the effect of m6A modification on THAP7-AS1 expression. Here we show that METTL3-mediated m6A modification enhanced THAP7-AS1 expression via m6A-IGF2BP1-dependent pathway.
Next, we demonstrated that THAP7-AS1 promoted GC growth and progression in vitro and in vivo, suggesting that THAP7-AS1 plays an oncogenic role in GC. LncRNAs may participate in chromatin remodeling complexes involved in gene silencing [27]. At present, the underlying molecular mechanisms of THAP7-AS1 in GC progression remain unknown. By RNA pull-down, MS, and RIP assay, we demonstrated that THAP7-AS1 directly binds to CUL4B, which was confirmed by FISH. CUL4B, a scaffold protein in the cullin 4B-RING E3 ligase complex [28], is upregulated in various human tumors and functions as an oncogene [23, 29, 30]. CUL4B can accumulate in the nucleus due to a functional NLS and its interaction with nuclear import receptor proteins [12]. We found that nt 1–442 at the 5′ end of THAP7-AS1 is required for its interaction with CUL4B. And THAP7-AS1 could interact with NLS region of CUL4B and importin α1. Furthermore, THAP7-AS1 improved the binding abilities between NLS and importin α1, and then enhanced the CUL4B protein entry into the nucleus, and its nuclear localization was required for biological function.
Next, RNA sequence assay showed that ERBB2 signaling pathway was significantly enriched in THAP7-AS1 downregulated cancer cells. We found that THAP7-AS1 and CUL4B could activate PI3K/AKT signaling, in which PIK3CA, PIK3CD, and AKT3 was involved. We confirmed that CUL4B nuclear localization did not lead to decreased PI3K/AKT components degradation. Next, we focused on a miRNA-based mechanism, as our data indicated that PIK3CA, PIK3CD, and AKT3 expression is induced by CUL4B via post-transcriptional regulation. We confirmed that AKT3 is a direct target gene of miR-22-3p and that PIK3CA, PIK3CD, and AKT3 are direct target genes of miR-320a. In addition, we found that CUL4B-catalyzed H2AK119ub1 and PRC2-mediated H3K27me3 are involved in the regulation of miR-22-3p and miR-320a expression. Increasing evidence has demonstrated that miR-22-3p and miR-320a act as tumor suppressors to inhibit cancer progression in various cancers [31–36]. In order to investigate the effects of two miRNAs on PI3K/Akt components expression and GC development, we conducted a series of reversal experiment. Our results indicated that miR-22-3p reversed the increased AKT3 expression and GC cells migration or invasion abilities induced by THAP7-AS1 and CUL4B. Similarly, miR-320 reversed the enhanced expression of PIK3CA, PIK3CD and AKT3 and GC migration or invasion abilities induced by THAP7-AS1 and CUL4B. Taken together, these data showed that THAP7-AS1 promotes CUL4B-induced miR-22-3p and miR-320a transcription repression, thereby upregulating PIK3CA, PIK3CD, and AKT3 expression to promote GC progression.
In summary, differential expression profiles of lncRNA were generated in human GC tissues. And THAP7-AS1 was identified as a novel lncRNA that promotes GC progression, which was transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification. Moreover, we demonstrated that THAP7-AS1 improved the combination between CUL4B and importin α1, and enhance the CUL4B protein entry into the nucleus. THAP7-AS1/CUL4B complex activates PI3K/AKT signaling through transcriptionally repressing miR-22-3p and miR-320a by CUL4B-catalyzed H2AK119ub1 and EZH2-mediated H3K27me3 (Fig. 8H). These results highlight the important functional interaction among lncRNAs, miRNAs, and proteins in GC and suggest that THAP7-AS1 may serve as a promising molecular target for GC therapies.
Supplementary information
Acknowledgements
We thank all the participants for their contributions to the study.
Author contributions
P.G. performed the conception and design of this manuscript. H.-T.L., Y.-X.Z., S.L., G.-H.Z., X.-Y.G., and W.-J.Z. collected clinical tumor samples. H.-T.L. and P.G. performed data analysis and interpretation. H.-T.L. and P.G. performed the manuscript writing. R.-R.M. provides experiment guidance. All authors were involved in writing the paper and had final approval of the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81672842, 82072665 and 31671427), the Taishan Scholars Program of Shandong Province (Grant No. ts201511096), Major projects of transformation of new and old kinetic energy in Shandong Province, Natural Science Foundation of Shandong Province (Grant No. ZR2020QH223) and Postdoctoral Innovation Project of Shandong Province (No.202001006).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. Microarray data were deposited in NCBI Gene Expression Omnibus (GEO, GSE150539).
Ethics statement
Clinical tissue samples were obtained from Qilu Hospital and Shandong Provincial Hospital. This study was approved by the Medical Ethics Committee of the Shandong University and performed in accordance with the Declaration of Helsinki. All participants have informed consent and their privacy has been fully protected. All of the animal experiments were conducted according to the Guidelines for Animal Health and Use (Ministry of Science and Technology, China, 2006). Animal experiments were approved by the Committee for Animal Protection of Shandong University.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: Mistakenly copied the PIK3CD strip of Figure 5A by combining the pictures.
Edited by V. D'Angiolella
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/9/2024
A Correction to this paper has been published: 10.1038/s41418-024-01335-0
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00879-9.
References
- 1.Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang S, et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17:69. doi: 10.1186/s12943-018-0820-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Q, Xiang S, Ma J, Hui P, Wang T, Meng W, et al. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol Oncol. 2018;12:799–813. doi: 10.1002/1878-0261.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng P, Yin Z, Wu Y, Xu Y, Luo Y, Zhang TC. LncRNA HOTAIR promotes cell migration and invasion by regulating MKL1 via inhibition miR206 expression in HeLa cells. Cell Commun Signal. 2018;16:5. doi: 10.1186/s12964-018-0216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simion V, Zhou H, Haemmig S, Pierce JB, Mendes S, Tesmenitsky Y, et al. A macrophage-specific lncRNA regulates apoptosis and atherosclerosis by tethering HuR in the nucleus. Nat Commun. 2020;11:6135. doi: 10.1038/s41467-020-19664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu HT, Liu S, Liu L, Ma RR, Gao P. EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell cycle progression in gastric cancer. Cancer Res. 2018;78:5877–5890. doi: 10.1158/0008-5472.CAN-18-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D, et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67:171–87. doi: 10.1002/hep.29405. [DOI] [PubMed] [Google Scholar]
- 10.Xing AY, Wang YW, Su ZX, Shi DB, Wang B, Gao P. Catenin-delta1, negatively regulated by miR-145, promotes tumour aggressiveness in gastric cancer. J Pathol. 2015;236:53–64. doi: 10.1002/path.4495. [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Cui M, Yang F, Li N, Jiang B, Yu Z, et al. A cullin 4B-RING E3 ligase complex fine-tunes pancreatic delta cell paracrine interactions. J Clin Investig. 2017;127:2631–46. doi: 10.1172/JCI91348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou Y, Mi J, Cui J, Lu D, Zhang X, Guo C, et al. Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J Biol Chem. 2009;284:33320–32. doi: 10.1074/jbc.M109.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arun G, Akhade VS, Donakonda S, Rao MR. mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Mol Cell Biol. 2012;32:3140–52. doi: 10.1128/MCB.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res. 2017;45:11356–70. doi: 10.1093/nar/gkx778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–83. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci USA. 2014;111:1002–7. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu H, Yang Y, Ji Q, Zhao W, Jiang B, Liu R, et al. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell. 2012;22:781–95. doi: 10.1016/j.ccr.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Liu R, Qiu R, Zheng Y, Huang W, Hu H, et al. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene. 2015;34:104–18. doi: 10.1038/onc.2013.522. [DOI] [PubMed] [Google Scholar]
- 25.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–55. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 26.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009;34:562–70. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian Y, Yuan J, Hu H, Yang Q, Li J, Zhang S, et al. The CUL4B/AKT/beta-catenin axis restricts the accumulation of myeloid-derived suppressor cells to prohibit the establishment of a tumor-permissive microenvironment. Cancer Res. 2015;75:5070–83. doi: 10.1158/0008-5472.CAN-15-0898. [DOI] [PubMed] [Google Scholar]
- 30.He YM, Xiao YS, Wei L, Zhang JQ, Peng CH. CUL4B promotes metastasis and proliferation in pancreatic cancer cells by inducing epithelial-mesenchymal transition via the Wnt/beta-catenin signaling pathway. J Cell Biochem. 2018;119:5308–23. doi: 10.1002/jcb.26643. [DOI] [PubMed] [Google Scholar]
- 31.Zou Q, Tang Q, Pan Y, Wang X, Dong X, Liang Z, et al. MicroRNA-22 inhibits cell growth and metastasis in breast cancer via targeting of SIRT1. Exp Ther Med. 2017;14:1009–16. doi: 10.3892/etm.2017.4590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Zhang K, Li XY, Wang ZM, Han ZF, Zhao YH. MiR-22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur Rev Med Pharm Sci. 2017;21:3598–604. [PubMed] [Google Scholar]
- 33.Feng X, Luo Q, Wang H, Zhang H, Chen F. MicroRNA-22 suppresses cell proliferation, migration and invasion in oral squamous cell carcinoma by targeting NLRP3. J Cell Physiol. 2018;233:6705–13. doi: 10.1002/jcp.26331. [DOI] [PubMed] [Google Scholar]
- 34.Guo MM, Hu LH, Wang YQ, Chen P, Huang JG, Lu N, et al. miR-22 is down-regulated in gastric cancer, and its overexpression inhibits cell migration and invasion via targeting transcription factor Sp1. Med Oncol. 2013;30:542. doi: 10.1007/s12032-013-0542-7. [DOI] [PubMed] [Google Scholar]
- 35.Lu C, Liao Z, Cai M, Zhang G. MicroRNA-320a downregulation mediates human liver cancer cell proliferation through the Wnt/beta-catenin signaling pathway. Oncol Lett. 2017;13:573–8. doi: 10.3892/ol.2016.5479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Lv Q, Hu JX, Li YJ, Xie N, Song DD, Zhao W, et al. MiR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biol Ther. 2017;18:142–51. doi: 10.1080/15384047.2017.1281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. Microarray data were deposited in NCBI Gene Expression Omnibus (GEO, GSE150539).