Abstract
Three vancomycin-dependent clinical isolates of Enterococcus faecalis of the VanB type were studied by determining (i) the sequence of the ddl gene encoding the host d-Ala:d-Ala ligase and the vanSB-vanRB genes specifying the two-component regulatory system that activates transcription of the vanB operon, (ii) the level of expression of resistance genes by using dd-dipeptidase activity as a reporter, and (iii) the proportions of the peptidoglycan precursors synthesized. Each strain had a mutation in ddl leading to an amino acid substitution (D295 to V; T316 to I) or deletion (DAK251-253 to E) at invariant positions in d-Ala:d-Ala, d-Ala:d-Lac, and d-Ala:d-Ser ligases. These mutations resulted in impaired host d-Ala:d-Ala ligases since only precursors terminating in d-Ala-d-Lac were synthesized under vancomycin-inducing conditions. Two types of vancomycin-independent revertants of one isolate were obtained in vitro after growth in the absence of vancomycin: (i) vancomycin-resistant, teicoplanin-susceptible mutants had a 6-bp insertion in the host ddl gene, causing the E251-to-EYK change that restored d-Ala:d-Ala ligase activity, (ii) constitutive vancomycin-resistant, teicoplanin-resistant mutants had substitutions (S232 to F or E247 to K) in the vicinity of the autophosphorylation site of the VanSB sensor and produced exclusively precursors ending in d-Ala-d-Lac. Vancomycin- and teicoplanin-dependent mutants obtained by growth in the presence of teicoplanin had an 18-bp deletion in VanSB, affecting residues 402 to 407 and overlapping the G2 ATP binding domain. The rapid emergence of vancomycin-independent revertants in vitro suggests that interruption of vancomycin therapy may not be sufficient to cure patients infected with vancomycin-dependent enterococci.
Resistance to glycopeptides in enterococci has become an increasingly worrisome problem in clinical practice. Resistance is observed most commonly in Enterococcus faecium, which is often resistant to other classes of antibiotics such as β-lactams and aminoglycosides, and to a lesser extent in Enterococcus faecalis (19). In these species, resistance to glycopeptides is mediated by mobile elements which encode the production of enzymes under the control of a two-component regulatory system. These enzymes result in the synthesis of late peptidoglycan precursors ending in d-alanyl-d-lactate (d-Ala-d-Lac) instead of d-alanyl-d-alanine (d-Ala-d-Ala) and in the elimination of precursors ending in d-Ala-d-Ala (for a review, see reference 4). Activity of glycopeptides is due to formation of high-affinity complexes between the antibiotics and the d-Ala-d-Ala residues of peptidoglycan precursors as they emerge through the membrane, which blocks the remaining steps of peptidoglycan synthesis. Therefore, the resistance mechanism consists of an alternative pathway of cell wall synthesis insensitive to glycopeptides.
VanA and VanB are the most common resistance phenotypes. In both, resistance results from the acquisition of a gene cluster encoding seven proteins. The functions of six of these proteins have been studied extensively (for a review, see reference 4). The sensor (VanS or VanSB) and the regulator (VanR or VanRB) constitute a two-component regulatory system responsible for recognition of the presence of glycopeptides in the culture medium and transcriptional activation of the resistance genes. Two proteins are essential for the production of peptidoglycan precursors ending in d-Ala-d-Lac, namely, a dehydrogenase (VanH or VanHB) which synthesizes d-Lac from pyruvate and a ligase (VanA or VanB) which ligates d-Lac to d-Ala. Two proteins (a dd-dipeptidase [VanX or VanXB] and a dd-carboxypeptidase [VanY or VanYB]) are responsible for the catabolism of d-Ala-d-Ala termini, by cleaving the d-Ala-d-Ala dipeptide or by removing the ultimate d-Ala from the pentapeptide precursor. VanA-type strains are characterized by a high level of resistance to vancomycin and teicoplanin because both antibiotics are inducers, whereas VanB-type strains remain susceptible to teicoplanin because this glycopeptide is not an inducer of the vanB operon.
More recently, enterococci that require the presence of vancomycin in the culture medium for effective growth have been isolated from patients treated for long periods with this antibiotic. E. faecalis strains with this phenotype have been isolated from urine or blood samples (14, 15, 28), and E. faecium strains have been isolated from feces or blood samples (7, 16, 31). Similar mutants have also been isolated from animals treated with vancomycin (5) or obtained in vitro by growing bacteria in the presence of the antibiotic (6, 26). The mechanism responsible for vancomycin dependence has been studied in two in vitro mutants. In each case, a unique mutation in the ddl gene encoding the host d-Ala:d-Ala ligase was found. The mutations were responsible for inactivation of the enzyme following synthesis of a truncated protein (26) or replacement of an amino acid essential for substrate binding (6). The bacteria grew only in the presence of vancomycin, since this antibiotic was required for induction of the resistance proteins and, therefore, for synthesis of peptidoglycan from d-Ala-d-Lac- instead of d-Ala-d-Ala-containing precursors.
In the present study, we report the mechanism of vancomycin dependence in clinical strains of E. faecalis with the vanB genotype isolated from three patients treated with vancomycin for prolonged periods. We also studied in vitro mutants of one of these strains which either reverted to a nondependent highly glycopeptide-resistant phenotype or became dependent on both vancomycin and teicoplanin for growth.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The isolates used in this work are described in Table 1 and Fig. 1. E. faecalis BM4386 is an isolate from the bloodstream of a patient at Hahnemann University (Philadelphia, Pa.) previously treated with vancomycin for a Staphylococcus aureus infection. BM4387 and TJ310 (15) are urinary tract isolates from patients at Thomas Jefferson University Hospital (Philadelphia, Pa.) who had received vancomycin. TJ282, a non-vancomycin-dependent strain (15), and TJ310 were isolated from one patient, but TJ282 was isolated several days earlier than TJ310 was.
TABLE 1.
Properties of the Enterococcus strains studied
| Strain (genotype) | Phenotypea | MIC (μg/ml)
|
Origin and/or Reference | ||
|---|---|---|---|---|---|
| Vm | Te | Te in the presence of Vm (10 μg/ml) | |||
| E. faecalis V583 (vanB) | Vmr Tes | 32 | 1 | 0.25b | 23 |
| E. faecium BM4147 (vanA) | Vmr Ter | 512 | 512 | 512 | 17 |
| E. faecalis BM4386 (vanB) | VmD | 1,024 | <1c | 1,024 | Clinical isolate |
| E. faecalis BM4387 (vanB) | VmD | 512 | <1c | 16 | Clinical isolate |
| E. faecalis TJ310 (vanB) | VmD | 512 | <1c | 64 | Clinical isolate (15) |
| E. faecalis TJ282 (vanB) | Vmr Tes | 16 | 1 | <0.25b | Clinical isolate (15) |
| E. faecalis BM4388 (vanB) | Vmr Tes | 512 | 1 | 8 | In vitro mutant |
| E. faecalis BM4389 (vanB) | Vmr Ter | 1,024 | 256 | 256 | In vitro mutant |
| E. faecalis BM4390 (vanB) | Vmr Ter | 1,024 | 256 | 256 | In vitro mutant |
| E. faecalis BM4391 (vanB) | VmD TeD | 256 | 512 | 512 | In vitro mutant |
D, dependent; r, resistant; s, susceptible; Te, teicoplanin; Vm, vancomycin.
Determined in the presence of 4 μg of Vm instead of 10 μg/ml, because of the low level of resistance to Vm in this strain.
Determined in the presence of 50 mM d-Ala-d-Ala.
FIG. 1.
Properties of the E. faecalis isolates. The scheme indicates the antibiotics used for growth (G) and selection (S) of the in vitro mutants, as well as the phenotype (D, dependent; r, low-level resistant; R, high-level resistant; S, susceptible; Te, teicoplanin; Vm, vancomycin). The substitutions in Ddl or VanSB are indicated below each strain.
Three types of mutants with distinct phenotypes were obtained by growing TJ310 under various conditions (Fig. 1 and Table 1). A teicoplanin-dependent derivative (BM4391) was selected by incubating TJ310 overnight in broth containing 10 μg of vancomycin (Eli Lilly, Saint-Cloud, France) per ml and 1 μg of teicoplanin (Marion Merrell-Dow, Levallois-Perret, France) per ml; the culture was then plated on agar containing 10 μg of teicoplanin per ml and screened for the inability to grow on unsupplemented medium. Nondependent revertants were selected for growth of TJ310 for 24 h in broth in the absence of antibiotics; the culture was then plated on unsupplemented medium and screened for growth on agar containing 10 μg of teicoplanin per ml. Two types of revertants were selected for the ability to grow on both unsupplemented and teicoplanin-containing medium (BM4389 and BM4390) or only on medium devoid of antibiotic (BM4388). Two to four mutants of each phenotype isolated from independent experiments were selected for further analysis.
The vancomycin-dependent clinical strains were grown in brain heart infusion (BHI; Difco Laboratories, Detroit, Mich.) supplemented with 10 μg of vancomycin per ml, and the teicoplanin-dependent in vitro mutant (BM4391) was grown in the same medium supplemented with 10 μg of teicoplanin per ml. The revertant mutants (BM4388, BM4389, and BM4390) and the nondependent clinical strain TJ282 were grown in unsupplemented BHI or in BHI containing 4 to 10 μg of vancomycin per ml for induction of the resistance genes.
Phenotypic and genotypic characterization of the strains.
The MICs of vancomycin and teicoplanin were determined after 48 h of incubation by the method of Steers et al. (27) with 105 CFU per spot on BHI or on BHI supplemented with 50 mM d-Ala-d-Ala (to determine the MICs of teicoplanin for the vancomycin-dependent strains) or with 4 or 10 μg of vancomycin per ml (to induce the expression of resistance genes). Susceptibility to antibiotics other than glycopeptides was determined by disk agar diffusion on BHI or BHI supplemented with 10 μg of vancomycin per ml (for vancomycin-dependent strains) or 10 μg of teicoplanin per ml (for teicoplanin-dependent BM4391). Enterococci were identified at the species level by a PCR assay (8) with DNA from E. faecalis V583 (vanB) and E. faecium BM4147 (vanA) as a positive control. The glycopeptide resistance genotype was determined by a PCR assay with the primers for vanB (12) and vanA (8) genes and confirmed by Southern hybridization of EcoRI-digested total DNA (21) with a vanB probe (11) labeled with [α-32P]dCTP (Radiochemical Center, Amersham, United Kingdom) by nick translation using E. faecalis V583 (vanB) as a positive control. All strains were found to be E. faecalis of the vanB genotype and did not contain the vanA gene (Table 1).
Zero-integrated field gel electrophoresis.
Genomic DNA embedded in agarose plugs (18) was digested overnight at 37°C with 100 U of NotI endonuclease. The restriction fragments were separated with a software-assisted zero-integrated field gel electrophoresis apparatus according to the manufacturer’s recommendations (Auto Base; Q-Life System Inc.). This apparatus was used with ROM card no. 3 that optimizes DNA fragment separation in the 8- to 500-kb range.
Amplification, cloning, and sequencing of the ddl, vanSB, and vanRB genes.
Purified total DNA was used as a template for amplification by PCR, using Pfu polymerase and the oligodeoxyribonucleotide primers for the ddl gene of E. faecalis (11) and the vanSB and vanRB genes (6). Amplification was carried out in a 100-μl volume containing 50 pmol of each oligonucleotide primer, 50 nmol of each 2′ deoxynucleoside 5′ triphosphate, reaction buffer, 100 ng of DNA, and 10 U of DNA Pfu polymerase. A total of 30 cycles of PCR were done; 1 cycle consisted of 1 min at 94°C for denaturation, 1 min at 54°C for annealing, and 2 min 30 s at 72°C for polymerization. The PCR products were purified from agarose gels (Sephaglas kit; Pharmacia, Uppsala, Sweden), cloned into pCR-Blunt (Invitrogen, Leek, The Netherlands), and sequenced by the dideoxy-chain termination method (24), using T7 DNA polymerase (Sequenase kit; U.S. Biochemical Corp., Cleveland, Ohio) and [α-35S]dATP (Amersham Radiochemical Center).
Extraction and analysis of peptidoglycan precursors.
Extraction and analysis of the peptidoglycan precursors were performed by the procedures of Reynolds and coworkers (22). In brief, enterococci were grown in BHI medium supplemented with yeast extract and glycopeptides (if induction of resistance was required). Ramoplanin was added to inhibit peptidoglycan synthesis, and incubation was continued for one half of the mean generation time to cause peptidoglycan precursor accumulation. Bacteria were then harvested, and cytoplasmic precursors were extracted and analyzed by high-pressure liquid chromatography.
dd-Dipeptidase activity.
VanXB activity was measured in cytoplasmic extracts of bacteria grown to an A600 of 0.7 in the absence (noninducing conditions) or presence (inducing conditions) of glycopeptide. The amount of d-Ala released from d-Ala-d-Ala was determined by using d-amino acid oxidase coupled to peroxidase for indicator reaction (2). One unit of activity was defined as the number of nanomoles of product formed at 37°C per minute per milligram of protein contained in the extract (as measured by the method of Bradford).
RESULTS AND DISCUSSION
Clinical isolates. (i) Characterization of the strains.
We have studied three isolates of vancomycin-dependent (VmD) E. faecalis of the vanB genotype. A vancomycin-resistant (Vmr) VanB-type E. faecalis TJ282 isolated from the same patient as TJ310 but isolated a few days earlier was included in the investigation. The three VmD E. faecalis strains isolated from different patients at various hospitals and at different times were considered different strains since they had distinct restriction fragment patterns following pulsed-field gel electrophoresis (Fig. 2) and differed in their susceptibility to antibiotics other than glycopeptides. TJ310 was resistant to gentamicin; BM4386 was resistant to gentamicin, streptomycin, and erythromycin; and BM4387 was also resistant to chloramphenicol. All were highly resistant to vancomycin and remained susceptible to teicoplanin when their growth was triggered by d-Ala-d-Ala (Table 1). However, they displayed variable levels of resistance to teicoplanin in the presence of 10 μg of vancomycin per ml to induce expression of the resistance genes.
FIG. 2.
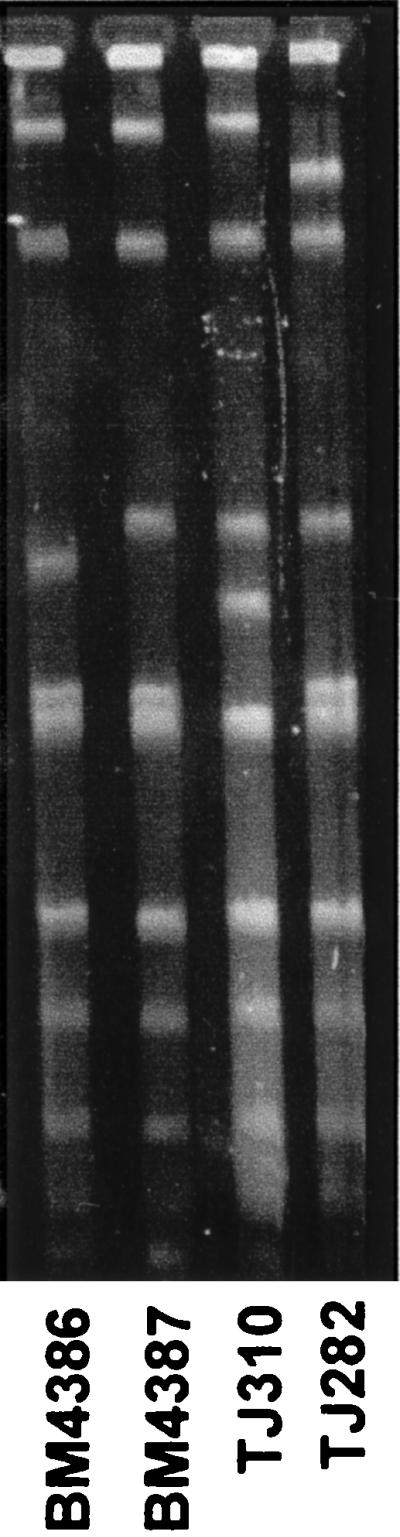
Pulsed-field gel electrophoresis of total DNA from clinical isolates digested by NotI.
In contrast to TJ310, TJ282 was not dependent on vancomycin for growth but was resistant to low levels of this antibiotic (Table 1). Like TJ310, TJ282 had the vanB gene and was resistant to gentamicin. However, it was also resistant to streptomycin and erythromycin and differed from TJ310 by two bands in its restriction fragment pattern (Fig. 2). A previously published pattern of TJ282 and TJ310 using SmaI digests also demonstrated two band differences between these isolates (15). Although TJ282 is probably clonally related to TJ310, it may not be its direct progenitor in vivo.
(ii) Sequence of the ddl gene.
It was recently shown that vancomycin dependence of mutants obtained in vitro was associated with mutations in the host ddl gene leading to synthesis of an inactive d-Ala:d-Ala ligase (6, 26). We therefore sequenced the ddl genes of the three VmD clinical isolates and Vmr TJ282. The ddl sequence of the latter was identical to that of E. faecalis V583 (11). In contrast, a mutation in the ddl gene of the VmD strains was found (Fig. 3). The ddl genes of BM4386 and BM4387 had a single point mutation at codon 295 (GAT to GTT) and 316 (ACT to ATT), leading to Asp-to-Val and Thr-to-Ile substitutions, respectively. The ddl gene of TJ310 had suffered a deletion of 6 bp (GAT GCA AAA to GAA) at codons 251 to 253, leading to the substitution of Asp-Ala-Lys by Glu.
FIG. 3.
Schematic representation of the d-Ala:d-Ala ligase of E. faecalis. The positions of the amino acids implicated in the binding of d-Ala1, d-Ala2, and ATP are indicated by black, dotted, and hatched bars, respectively. Substitutions relative to the deduced DDl sequence of TJ282 (and the corresponding mutations in parentheses) are indicated in bold italic characters.
(iii) Analysis of peptidoglycan precursors.
To ascertain whether the mutations had impaired d-Ala:d-Ala ligase activity, we determined the nature and the relative amounts of the peptidoglycan precursors synthesized by the hosts (Fig. 4, left panel). This was assessed by inhibiting peptidoglycan synthesis with the antibiotic ramoplanin for a short period when bacteria were in the exponential phase of growth and investigating the accumulation of the precursors that could no longer be used. In the absence of induction by vancomycin, only TJ282 was able to grow and synthesized quasiexclusively UDPMurNac pentapeptide. Induction by 4 μg of vancomycin per ml resulted in the activity of both glycopeptide-susceptible and -resistant pathways of peptidoglycan synthesis, as indicated by the presence of pentapeptide (34%), pentadepsipeptide (50%), and tetrapeptide (16%) in the precursor pool. As expected, VmD strains synthesized exclusively pentadepsipeptide (when these strains were grown in the presence of 10 μg of vancomycin per ml, confirming their exclusive reliance on the resistance pathway of peptidoglycan synthesis for growth).
FIG. 4.
Relative proportions of pentapeptide (cross-hatched bars), tetrapeptide (black bars), tripeptide (white bars), and pentadepsipeptide (hatched bars) synthesized by clinical isolates (left panel) and in vitro mutants obtained from TJ310 (right panel). Precursors were determined from noninduced bacteria (left bar of the pair of bars for each strain) or from bacteria induced (right bar of each pair of bars) and by growth in the presence of 10 μg of vancomycin per ml (BM4386, BM4387, TJ310, BM4389, and BM4390), 4 μg of vancomycin per ml (TJ282 and BM4388), or 10 μg of teicoplanin per ml (BM4391). NA, not applicable.
(iv) dd-Dipeptidase assays.
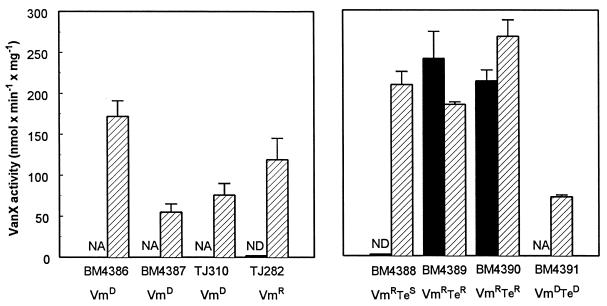
The expression levels of the resistance proteins were determined by measuring the VanXB dd-dipeptidase activity (2). This enzyme can serve as a reporter since the vanYB, vanW, vanHB, vanB, and vanXB genes are transcribed from a single promoter (10). As illustrated in Fig. 5 (left panel), the activity was negligible in uninduced TJ282. Although VmD strains produced exclusively resistant precursors, VanXB was synthesized at relatively low levels in BM4387 and TJ310, even though the resistance operon was switched on. VanXB and VanYB activities are not required in these strains because d-Ala-d-Ala and hence pentapeptide are not synthesized and therefore do not have to be removed. However, induction of these VmD strains resulted in expression of the resistance proteins at levels that correlated well with their teicoplanin resistance levels (Table 1), as already reported (2). This was not the case for Vmr TJ282, which was resistant to low levels of vancomycin but remained susceptible to teicoplanin, even after induction by vancomycin. The latter observation could be due to a relatively low level of expression of the resistance proteins, resulting in a pentadepsipeptide/pentapeptide ratio insufficient to increase the teicoplanin MIC (2).
FIG. 5.
VanXB dd-dipeptidase activity in clinical isolates (left panel) and in vitro mutants of TJ310 (right panel). The enzymatic activity was measured in extracts prepared from noninduced strains (black bars) or from strains induced (hatched bars) by growth in the presence of 10 μg of vancomycin per ml, except for BM4391 which was induced with 10 μg of teicoplanin per ml. Data are the means ± standard deviations from three independent experiments. NA, not applicable; ND, not detectable.
Taken together, these data confirm that vancomycin dependence results from impairment of the host d-Ala:d-Ala ligase activity. Peptidoglycan synthesis in these cells is reliant on production of the resistance proteins, which is dependent on the presence of vancomycin in the culture medium. In a previous study of a VmD mutant, it was reported that a nonsense mutation gave rise to a truncated protein, resulting in loss of d-Ala:d-Ala ligase activity (26). In the current investigation, the three clinical VmD isolates had a base pair substitution or deletion in the ligase gene. Such mutations can result in loss of substrate binding or catalytic activity if the substituted amino acid is at a critical functional position. This is illustrated by an in vitro VmD mutant of E. faecalis in which a Ser 319-to-Ile substitution (6) suppressed a hydrogen bond essential for the interaction of the enzyme with d-Ala-2 (13, 25). The three-dimensional structure of Escherichia coli Ddl cocrystallized with a S,R-methylphosphinate inhibitor and ATP has been determined (13), and this enzyme has been analyzed by mutagenesis experiments (25). The Asp 295 residue mutated in BM4386 corresponds to an invariant position in the ligases (9). It is believed to form a hydrogen bond via a water molecule with d-Ala-2 and to participate therefore in substrate recognition and/or catalysis, since substitution of the corresponding amino acid (Asp 257) in E. coli ligase totally abolished enzyme activity (25). The mutation at position 316 in BM4387 does not affect an amino acid previously reported to be directly implicated in enzyme activity. However, this Thr residue appears to be highly conserved in the d-Ala:d-Ala, d-Ala:d-Lac, and d-Ala:d-Ser ligases from various bacterial species (9), suggesting that it could also play an important role in either enzyme activity or conformation. Thr 316 is close to four other conserved residues (Glu 311, Asn 313, Ser 319, and Met 320), and Ser 281, which in E. coli corresponds to the penultimate amino acid, has a key role in the interaction with d-Ala-2 (25). By analogy with the observations made on the corresponding Lys 215 mutant in the E. coli ligase (13, 25), loss of the highly conserved Lys at position 253 of TJ310 ligase may have impaired the enzyme activity by suppressing a residue involved in the transfer of the phosphate group of ATP. The concomitant lack of amino acids at positions 251 and 252 in this strain affects nonconserved residues (9), and therefore, the potential impact of this deletion cannot be established.
In vitro mutants.
We obtained revertants of VmD strain TJ310 by growing TJ310 in the absence of vancomycin (Table 1 and Fig. 1) as described previously (7, 16). The mutants were classified according to their susceptibility (Vmr Tes; strain BM4388) or resistance (Vmr Ter BM4389 and BM4390) to teicoplanin. Vancomycin and teicoplanin-dependent mutant (VmD TeD) BM4391 was selected by growing TJ310 in the presence of 10 μg of vancomycin per ml and 1 μg of teicoplanin per ml. All the mutants displayed the same resistance phenotype to antibiotics other than glycopeptides as did the parental strain TJ310.
(i) Vancomycin-resistant, teicoplanin-susceptible revertants.
Loss of vancomycin dependence by Vmr Tes BM4388 prompted us to examine the sequence of the ddl gene. Compared with the ddl sequence of VmD parental strain TJ310, the sequence of the mutant had an insertion of 6 bp (TAC AAA) corresponding to positions 252 to 253 (Fig. 3). Interestingly, this mutation resulted in a triplet of amino acids at positions 251 to 253, of which two are different from those in wild-type E. faecalis ligase (Glu-Tyr instead of Asp-Ala), but Lys 253, thought to be essential for activity, was present in both. Most surprisingly, the same mutation was found in the four independent revertants sequenced, suggesting that only a few combinations of amino acids can restore ligase activity. As expected, in the absence of vancomycin, strain BM4388 synthesized peptidoglycan precursors qualitatively similar to those produced by TJ282 (Fig. 4, right panel). Following preincubation with 4 μg of vancomycin per ml, the proportions of pentapeptide and tetrapeptide, respectively, were 34 and 16% in Vmr TJ282 but only 12 and 5% in Vmr BM4388, and pentadepsipeptide represented 83% of the total precursors in the latter strain. This increase in the percentage of resistant precursors generated by VanHB and VanB correlated with higher VanXB activity (Fig. 5, right panel) and accounts for high-level vancomycin resistance of this strain. However, despite increased production of the resistance proteins, higher than in VmD BM4386, strain BM4388 was resistant to low levels of teicoplanin only under inducing conditions. This is due to the fact that almost complete elimination of the pentapeptide is required to achieve resistance to this antibiotic (2). No mutations were found in the vanRB and vanSB genes encoding the two-component regulatory system in this mutant (Fig. 6).
FIG. 6.
Schematic representation of the VanSB sensor. The putative membrane-associated sensor domain (white) containing transmembrane segments (black) and the cytosolic kinase domain (cross-hatched) are depicted. H, N, G1, F, and G2 refer to conserved amino acid motifs present in histidine kinases. Amino acids in the sequence underlined for TJ310 correspond to the G2 domain. The substitutions relative to VanSB of TJ310 (and the corresponding mutations in parentheses) are indicated in bold italic characters.
(ii) Vancomycin and teicoplanin-resistant revertants.
Vmr Ter BM4389 and BM4390 revertants had the same mutation in the ddl gene as their VmD parent TJ310 did (Fig. 3). They synthesized only pentadepsipeptide precursors, even in the absence of induction (Fig. 4, right panel), indicating that peptidoglycan synthesis used the resistance pathway exclusively. Moreover, VanXB enzymatic activity in the revertants was as high as that of induced BM4388, even under noninducing conditions (Fig. 5, right panel). The lack of pentapeptide precursors explains, as in VmD BM4386, the high level of teicoplanin resistance. Taken together, these data demonstrate that the resistance genes must be expressed constitutively. We therefore determined the sequences of the vanSB and vanRB genes which encode the two-component regulatory-system controlling expression of the resistance genes (1, 3, 10). In this system, the VanSB sensor is composed of a transmembrane sensor domain and a cytosolic kinase domain, which undergoes autophosphorylation on a conserved histidine residue in response to the presence of vancomycin in the culture medium (Fig. 6). The phosphate group is then transferred to a conserved residue of the VanRB regulator which, in its phosphorylated form, acts as a transcriptional activator of the resistance genes. Whereas the sensor domain of VanSB shows no significant homology with other sensor molecules of other two-component systems, including VanS of the vanA gene cluster, five blocks of the kinase domain (Fig. 6) appear highly conserved (10, 20). The H block is responsible for both autophosphorylation and kinase and phosphatase activities (corresponding to the fixation of phosphate on the sensor, its transfer on the regulator, or its removal from the regulator). G1 and G2 correspond to ATP-binding blocks, and the functions of the N and F blocks have not been characterized. In the absence of vancomycin, VanSB stimulates dephosphorylation of phospho-VanRB, which down-regulates transcription of the resistance genes. No mutations were found in the vanRB genes of BM4389 and BM4390. Sequencing of vanSB revealed point mutations GAG to AAG at codon 247 of BM4389 and TCT to TTT at codon 232 of BM4390, leading to Glu-to-Lys and Ser-to-Phe substitutions in the vicinity of the conserved H domain (Fig. 6). These mutations are distinct from those found in constitutive mutants of VmD strains which produced truncated inactive sensors (6). By analogy with other sensor molecules (e.g., EnvZ [29]), it could be inferred that the Ser 232-to-Phe substitution in BM4390 impairs dephosphorylation of VanRB by VanSB, leading to constitutive transcription of the resistance genes. Similarly, an in vitro constitutive mutant of a Vmr E. faecalis had a Thr-to-Lys substitution at position 237, which was also thought to be associated with a defect in phosphatase but not in kinase activity (6). No mutation corresponding to the Glu 247-to-Lys substitution in BM4389 has been described in other two-component regulatory systems. However, the same amino acid is present at the corresponding positions in PhoR and EnvZ, two other sensor molecules (10), and the localization close to the H domain may be critical for conformation and/or substrate recognition.
(iii) Vancomycin- and teicoplanin-dependent mutants.
VmD TeD mutant BM4391 had the same mutation in its ddl gene as the VmD progenitor TJ310 did (Fig. 3). This strain also synthesized only pentadepsipeptide precursors and had a low level of VanXB activity (right panels of Fig. 4 and 5). The only difference observed in comparison with parental TJ310 was an alteration of induction specificity, since BM4391 could grow in the presence of either vancomycin or teicoplanin. No mutation was found in the vanRB gene of BM4391, but a mutation was identified in its vanSB gene. This consisted of a 18-bp deletion (AGC AGA AAA AGC GGG CGA) affecting amino acids 402 to 407 and leading to loss of part of the conserved G2 domain. Since certain transmitters (AgrORF2, NarQ, and NarX) possess a single nucleotide binding domain, loss of the G2 block should not necessarily impair ATP binding. However, it has been proposed that the G2 block could also play a role in the modulation of enzyme conformation (20). If ATP were unable to bind at this site in BM4391, it might impair the stabilizing role of ATP on the molecule conformation without affecting enzyme activity (20). It is conceivable that this change could result in recognition of teicoplanin, since a single amino acid substitution in the sensor domain is sufficient to cause a similar change in induction specificity (6).
Emergence of VmD strains in clinical settings currently appears to be more of a curiosity than problem of clinical concern (28, 30). The inability of these bacteria to survive in the absence of vancomycin makes them, a priori, easy to eliminate, simply by discontinuing antibiotic therapy. However, VmD enterococci are rarely isolated in routine laboratory practice because of their particular nutritional requirements. Growth is obtained only on medium supplemented by d-Ala-d-Ala, vancomycin, or other inducers of vancomycin resistance gene expression (e.g., bacitracin and moenomycin). Moreover, VmD mutants have also been isolated from patients not receiving vancomycin. Therefore, they constitute a reservoir of vancomycin resistance genes which could be transferred to other bacteria. More importantly, VmD strains can easily revert, at least in vitro, to a nondependent, highly resistant phenotype, and may therefore require particular attention. Screening for VmD isolates in infected patients is therefore advisable.
ACKNOWLEDGMENTS
F.V.B. is collaborateur scientifique of the Belgian Fonds National de la Recherche Scientifique. We thank B. Périchon for constant technical advice to F.V.B. and M. Baptista for helpful discussions.
This work was supported in part by a Bristol-Myers Squibb Unrestricted Biomedical Research Grant in Infectious Diseases.
REFERENCES
- 1.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol. 1997;179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Depardieu F, Reynolds P E, Courvalin P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol. 1996;21:33–44. doi: 10.1046/j.1365-2958.1996.00617.x. [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur M, Reynolds P E, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 5.Aslangul E, Baptista M, Fantin B, Depardieu F, Arthur M, Courvalin P, Carbon C. Selection of glycopeptide-resistant mutants of VanB-type Enterococcus faecalis BM4281 in vitro and in experimental endocarditis. J Infect Dis. 1997;175:598–605. doi: 10.1093/infdis/175.3.598. [DOI] [PubMed] [Google Scholar]
- 6.Baptista M, Depardieu F, Reynolds P E, Courvalin P, Arthur M. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol Microbiol. 1997;25:93–105. doi: 10.1046/j.1365-2958.1997.4401812.x. [DOI] [PubMed] [Google Scholar]
- 7.Dever L L, Smith S M, Handwerger S, Eng R H K. Vancomycin-dependent Enterococcus faecium isolated from stool following oral vancomycin therapy. J Clin Microbiol. 1995;33:2770–2773. doi: 10.1128/jcm.33.10.2770-2773.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evers S, Casadewall B, Charles M, Dutka-Malen S, Galimand M, Courvalin P. Evolution of structure and substrate specificity in D-alanine-D-alanine ligases and related enzymes. J Mol Evol. 1996;42:706–712. doi: 10.1007/BF02338803. [DOI] [PubMed] [Google Scholar]
- 10.Evers S, Courvalin P. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. 1996;178:1302–1309. doi: 10.1128/jb.178.5.1302-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evers S, Reynolds P E, Courvalin P. Sequence of the vanB and ddl genes encoding D-alanine:D-lactate and D-alanine:D-alanine ligases in vancomycin-resistant Enterococcus faecalis V583. Gene. 1994;140:97–102. doi: 10.1016/0378-1119(94)90737-4. [DOI] [PubMed] [Google Scholar]
- 12.Evers S, Sahm D F, Courvalin P. The vanB gene of vancomycin-resistant Enterococcus faecalis V583 is structurally related to genes encoding D-Ala:D-Ala ligases and glycopeptide-resistance proteins VanA and VanC. Gene. 1993;124:143–144. doi: 10.1016/0378-1119(93)90779-3. [DOI] [PubMed] [Google Scholar]
- 13.Fan C, Moews P C, Walsh C T, Knox J R. Vancomycin-resistance: structure of D-alanine:D-alanine ligase at 2.3 Å resolution. Science. 1994;266:439–443. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- 14.Farrag N, Eltringham I, Liddy H. Vancomycin-dependent Enterococcus faecalis. Lancet. 1996;348:1581–1582. doi: 10.1016/S0140-6736(96)24049-8. [DOI] [PubMed] [Google Scholar]
- 15.Fraimow H S, Jundkind D L, Lander D W, Delso D R, Dean J L. Urinary tract infection with an Enterococcus faecalis isolate that requires vancomycin for growth. Ann Intern Med. 1994;121:22–26. doi: 10.7326/0003-4819-121-1-199407010-00004. [DOI] [PubMed] [Google Scholar]
- 16.Green M, Shlaes J H, Barbadora K, Shlaes D M. Bacteremia due to vancomycin-dependent Enterococcus faecium. Clin Infect Dis. 1995;20:712–714. doi: 10.1093/clinids/20.3.712. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 18.Miranda A G, Singh K V, Murray B E. DNA fingerprinting of Enterococcus faecium by pulsed-field gel electrophoresis may be a useful epidemiologic tool. J Clin Microbiol. 1991;29:2752–2757. doi: 10.1128/jcm.29.12.2752-2757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray B E. Diversity among multidrug-resistant enterococci. Emerg Infect Dis. 1998;4:37–47. doi: 10.3201/eid0401.980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 21.Quintiliani R, Jr, Evers S, Courvalin P. The vanB gene confers various levels of self-transferable resistance to vancomycin in enterococci. J Infect Dis. 1993;167:1220–1223. doi: 10.1093/infdis/167.5.1220. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds P E, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. Glycopeptide resistance mediated by transposon Tn1546 requires production of VanX for hydrolysis of D-alanyl-D-alanine. Mol Microbiol. 1994;13:1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 23.Sahm D F, Kissinger J, Gilmore M S, Murray P R, Mulder R, Solliday J, Clarke B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson A R. Sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y, Walsh C T. Active site mapping of Escherichia colid-Ala-d-Ala ligase by structure-based mutagenesis. Biochemistry. 1995;34:2768–2776. doi: 10.1021/bi00009a005. [DOI] [PubMed] [Google Scholar]
- 26.Sifaoui F, Gutmann L. Vancomycin dependence in a VanA-producing Enterococcus avium strain with a nonsense mutation in the natural d-Ala-d-Ala ligase gene. Antimicrob Agents Chemother. 1997;41:1409. doi: 10.1128/aac.41.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steers E, Foltz E L, Graves B S, Riden J. An inocula replicating apparatus for routine testing of bacterial susceptibility to antibiotics. Antibiot Chemother. 1959;9:307–311. [PubMed] [Google Scholar]
- 28.Stewart B, Hall L, Duke B, Ball D. Vancomycin-dependent enterococci: curious phenomenon or serious threat? J Antimicrob Chemother. 1997;40:734–735. doi: 10.1093/jac/40.5.734. [DOI] [PubMed] [Google Scholar]
- 29.Tokishita S, Kojima A, Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the transmembrane regions of the membrane-located protein kinase, EnvZ. J Biochem. 1992;111:707–713. doi: 10.1093/oxfordjournals.jbchem.a123823. [DOI] [PubMed] [Google Scholar]
- 30.Wilks M. Vancomycin-dependent enterococcus. Lancet. 1997;349:429–430. doi: 10.1016/S0140-6736(05)65052-0. [DOI] [PubMed] [Google Scholar]
- 31.Woodford N, Johnson A P, Morrison D, Hastings J G M, Elliot T S J, Worthington A, Tolley J L. Vancomycin-dependent enterococci in the United Kingdom. J Antimicrob Chemother. 1994;33:1066. doi: 10.1093/jac/33.5.1066. [DOI] [PubMed] [Google Scholar]