Abstract
Objective
Metaplasia arises from differentiated cell types in response to injury and is considered a precursor in many cancers. Heterogeneous cell lineages are present in the reparative metaplastic mucosa with response to injury, including foveolar cells, proliferating cells and spasmolytic polypeptide-expressing metaplasia (SPEM) cells, a key metaplastic cell population. Zymogen-secreting chief cells are long-lived cells in the stomach mucosa and have been considered the origin of SPEM cells; however, a conflicting paradigm has proposed isthmal progenitor cells as an origin for SPEM.
Design
Gastric intrinsic factor (GIF) is a stomach tissue-specific gene and exhibits protein expression unique to mature mouse chief cells. We generated a novel chief cell-specific driver mouse allele, GIF-rtTA. GIF-GFP reporter mice were used to validate specificity of GIF-rtTA driver in chief cells. GIF-Cre-RnTnG mice were used to perform lineage tracing during homoeostasis and acute metaplasia development. L635 treatment was used to induce acute mucosal injury and coimmunofluorescence staining was performed for various gastric lineage markers.
Results
We demonstrated that mature chief cells, rather than isthmal progenitor cells, serve as the predominant origin of SPEM cells during the metaplastic process after acute mucosal injury. Furthermore, we observed long-term label-retaining chief cells at 1 year after the GFP labelling in chief cells. However, only a very small subset of the long-term label-retaining chief cells displayed the reprogramming ability in homoeostasis. In contrast, we identified chief cell-originating SPEM cells as contributing to lineages within foveolar cell hyperplasia in response to the acute mucosal injury.
Conclusion
Our study provides pivotal evidence for cell plasticity and lineage contributions from differentiated gastric chief cells during acute metaplasia development.
Keywords: gastric metaplasia, gastric epithelial cell function, mucosal injury
Significance of this study.
What is already known on this subject?
Metaplasia arises from differentiated cell types in response to injury.
Mature chief cells have been considered the origin of spasmolytic polypeptide-expressing metaplasia (SPEM) cells, a key metaplastic cell population.
A conflicting paradigm has proposed isthmal progenitor cells as an origin for SPEM.
What are the new findings?
Mature chief cells, rather than isthmal progenitor cells, serve as the predominant origin of SPEM cells.
SPEM cells display lineage conversion capability to produce foveolar cells in response to acute injury.
How might it impact on clinical practice in the foreseeable future?
Our studies using a novel chief cell-specific driver mouse model clarify a number of conflicting questions relevant to the origin of metaplastic cells, which are precursors of gastric cancer.
Introduction
Metaplasia in the stomach, the appearance of abnormal mucous cell lineages, is commonly induced by environmental factors such as chronic infection and hormonal stimulation as well as responses to injury.1–4 Metaplastic cells are also considered precursors to many cancers.2 5–8 Intestinal-type gastric cancer is the most common type of gastric cancer and usually develops within fields of metaplasia. Metaplastic glands in the stomach corpus are composed of heterogeneous cell populations including foveolar (surface) cells, proliferating cells and spasmolytic polypeptide (TFF2) expressing metaplasia (SPEM) cells.9–11 SPEM cells are located at the base of the metaplastic glands after injury and are defined by coexpression of multiple markers, such as GSII-lectin, Tff2, Muc6, CD44v9 and AQP5.3 6 12 The SPEM cells appear quickly in response to acute injury after acid-secreting parietal cell loss in the stomach corpus mucosa. It has been suggested that the SPEM cells may not only contribute to the epithelial regeneration in response to injury, but can also enter a pro-carcinogenic programme leading to dysplasia.3 5 13 14
Chief cells are a differentiated cell lineage, which produce zymogen granules in the stomach mucosa, and a subpopulation of the chief cells also functions as ‘reserve’ stem cells in the stomach corpus mucosa.15–17 Over the past several decades, many studies have reported that zymogen-secreting mature chief cells present in the stomach corpus are the origin of SPEM cells through a transdifferentiation process, suggesting that the fully differentiated gastric chief cells can be reprogrammed into other cell types.6 15 18–20 However, a conflicting paradigm of SPEM cell origin has recently been suggested, proposing that gastric isthmal progenitor cells serve as the origin of SPEM cells.21–23 These conflicting results require more definitive analyses using a more representative chief cell-specific driver mouse model to resolve disparate interpretations between the conflicting paradigms of SPEM cell origin. In this study, we have generated a novel chief cell-specific driver mouse allele with a doxycycline (Dox) (tetracycline)-inducible rtTA system, inserted into the gastric intrinsic factor (GIF) locus (GIF-rtTA). Using the GIF-rtTA mouse allele, we defined the role of chief cells during metaplasia development as the predominant cellular origin of SPEM cells and described a new role for SPEM cells as a precursor to foveolar cell lineages within metaplastic glands.
Results
SPEM cells arise predominantly from transdifferentiation of chief cells and not from the expansion of proliferating isthmal progenitor cells
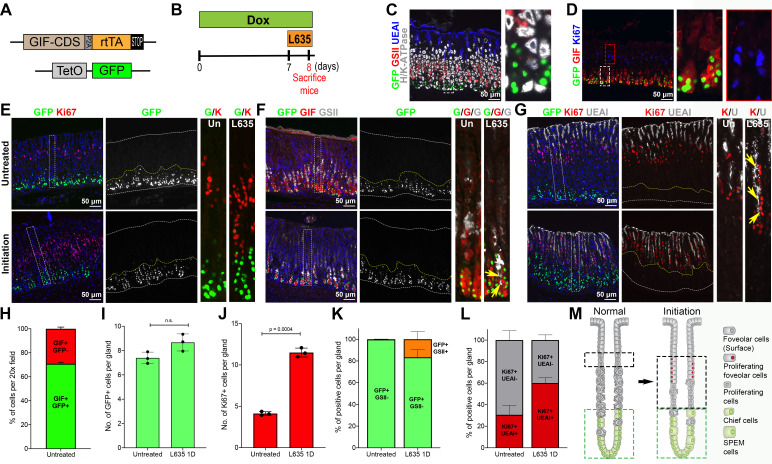
The GIF-rtTA transgenic mouse allele was generated by insertion of a DNA fragment of P2A-rtTA sequences at the end of the coding region of the GIF locus in C57BL/6 mice via a CRISPR/Cas9 approach (online supplemental figure S1A). To functionally validate the authenticity and efficiency of chief cell-specific targeting, we first crossed the GIF-rtTA allele mouse with the TetO-nucleusGFP (GIF-GFP) allele mouse (figure 1A). The GFP expression was assessed in the stomach mucosa of both the corpus and antrum of GIF-GFP reporter allele mice at 1 week after Dox administration (figure 1B and online supplemental figure S1C). GFP-expressing cells were observed in 89% of corpus glands (online supplemental figure S1E). The GFP-expressing cells were present only at the base of the glands where the mature chief cells are located (online supplemental figure S1C) and were only copositive with a chief cell marker, GIF, in the corpus (figure 1D). Further, the GFP-expressing cells were not copositive for any other cell lineage markers such as surface cell marker (UEAI), parietal cell marker (H/K-ATPase), mucous neck cell marker (GSII) (figure 1C) or proliferating cell/isthmal progenitor marker (Ki67) (figure 1D). Seventy-one per cent of GIF+ chief cells were copositive for GFP and those GIF +GFP+ cells were located at the very base of glands indicating that fully mature chief cells express GFP (figure 1H). It should be noted that we also observed GFP+ deep antral gland cells in 34% of antral glands (online supplemental figure S1C, E). However, the GFP was not observed in other organs such as liver, pancreas, intestine and lung, confirming the stomach tissue-specific expression of rtTA in the GIF-rtTA driver mouse allele (online supplemental figure S1B).
Figure 1.
Observation of GFP-positive chief cells during metaplasia initiation. (A) Diagram of the GIF-GFP allele used in this study. (B) Scheme of L635 treatment study in GIF-GFP mice. To first express GFP in chief cells, mice were administered doxycycline (Dox) water for 1 week before and throughout the L635 treatment. The GIF-GFP mice were treated with 1 dose of L635 following 1 week of Dox treatment to initiate the metaplastic process in the stomach mucosa. (C, D) Sections of the stomach tissues from GIF-GFP mice treated with Dox for 1 week were immunostained with antibodies against (C) GFP (green), GSII (mucus neck cells, red), UEAI (surface cells, blue) and H/K-ATPase (parietal cells, white) or (D) GFP (green), GIF (chief cells, red), Ki67 (proliferative cells/isthmal progenitor cells, blue). N=3 mice per group. (E–G) Sections of the stomach tissues from GIF-GFP mice treated without (untreated) or with 1 dose of L635 (initiation) at 1 week after the Dox treatment were immunostained with antibodies against (E) GFP (green) and Ki67 (red), (F) GFP (green), GIF (red) and GSII (white) or (G) GFP (green), Ki67 (red) and UEAI (white). Nuclei were counterstained with Hoechst (blue). Yellow arrows indicate cells copositive for GIF, GSII and GFP in panel F and cells copositive for UEAI and Ki67 in panel G. Dotted boxes depict regions enlarged. White dotted lines depict top and bottom of glands and yellow dotted lines depict the GFP+ cell zone. n=3 mice per group. (H) Quantitation of GIF and GFP copositive cells in GIF-GFP untreated mouse stomachs. The graph displays the percentage of GIF+/GFP+ (green) or GIF+/GFP− (red) cells per ×20 field. (I) Quantitation of GFP+ cells in GIF-GFP untreated or L635 1D mouse stomachs. The graph displays the number of GFP+ cells per gland. (J) Quantitation of K67+ cells in GIF-GFP untreated or L635 1D mouse stomachs. The graph displays the number of Ki67+ cells per gland. (K) Quantitation of GFP/GSII copositive cells in GIF-GFP untreated or L635 1D mouse stomachs. The graph displays the percentage of GFP+/GSII- (green) or GFP+/GSII+ cells (orange) per gland. (L) Quantitation of Ki67/UEAI copositive cells in GIF-GFP untreated or L635 1D mouse stomachs. The graph displays the percentage of Ki67+/UEAI+ (red) or of Ki67+/UEAI− cells (grey) per gland. Statistical significances were determined by unpaired Welch’s test (N=3 per group). (M) Schematic diagram of metaplasia initiation. Ggreen dotted lines depict the GFP+ chief cell zones and black dotted lines depict the proliferating cell zones in glands at the normal or metaplasia initiating stage. GIF, gastric intrinsic factor; n.s, not significant.
gutjnl-2021-325310supp001.pdf (886.9KB, pdf)
gutjnl-2021-325310supp005.pdf (63.8KB, pdf)
To examine whether SPEM cells arise from either chief cells or isthmal progenitor cells, we used the GIF-GFP reporter mouse allele and an acute atrophic injury model by administering the parietal cell toxic drug L635 to the GIF-GFP mice.24 25 We first administered Dox to GIF-GFP mice for 1 week to label chief cells with GFP and the GIF-GFP mice were then treated with 1 dose of L635 by oral gavage to induce parietal cell loss and initiate the metaplastic process (figure 1B). The GFP-positive cells distributed in the basal zone of transdifferentiating chief cells distinct from proliferating isthmal progenitor cells (figure 1E, yellow dotted lines). Although Ki67 +isthmal progenitor cells rapidly expanded on parietal cell loss after L635 treatment for one dose (figure 1E), similar numbers of GFP+ cells were still present at the base of L635-treated glands compared with the numbers in untreated glands (figure 1E–G, I). Notably, 17% of the GFP+ cells were copositive for GSII indicating that those copositive chief cells had already entered the transdifferentiation process (figure 1F, K). Although the number of Ki67+ cells increased 2.7-fold in the stomach mucosa of mice treated with 1 dose of L635 for (figure 1J), none of the Ki67-positive cells were copositive for GFP. Additionally, 60% of the Ki67-positive cells were proliferating ulex europaeus agglutinin I (UEAI)-lectin positive surface cells, which is an indication that the contribution of proliferating cells is supporting the expansion of the foveolar (surface cell) cell zone, rather than SPEM cells (figure 1G, yellow arrows, and 1 L). Thus, these results demonstrated that SPEM cell transdifferentiation in response to the injury arises predominantly from GFP-expressing chief cells, rather than from the expansion of proliferating isthmal progenitor cells (figure 1M).
Chief cells are long-lived gastric epithelial cells in the homoeostatic condition
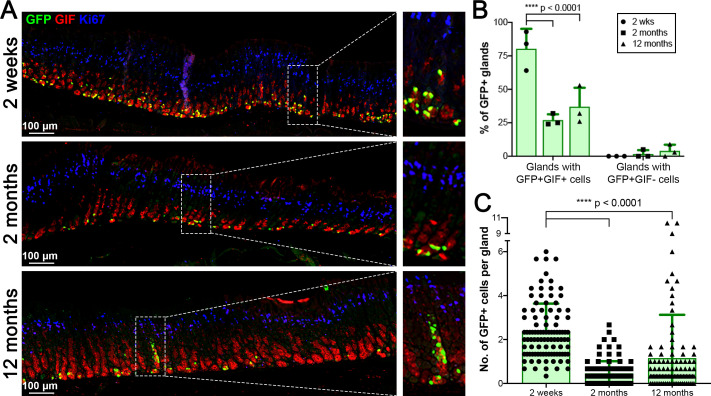
Chief cells are considered a long-lived cell population in the stomach mucosa and several previous reports have also suggested that a small subset of the chief cells functions as ‘reserve’ stem cells.16 17 26–28 We, therefore, performed a lineage tracing study from chief cells using a lineage tracing model, the GIF-Cre-RnTnG mouse allele, to examine the longevity of chief cells in homoeostasis and determine whether GFP marks all types of chief cells and if the GFP+ cells are also capable of self-renewal (figure 2). We first administered Dox to GIF-Cre-RnTnG mice for 1 week to label chief cell nuclei with GFP and the mice were sacrificed at 2 weeks and 2 to 12 months after Dox treatment. At 2 weeks after Dox treatment, we observed chief cells labelled with GFP. The GFP-labelled chief cells at 2 and 12 months were still present and copositive for GIF (figure 2A), indicating the presence of long-term label-retaining chief cells up to 1 year after the Dox treatment. While GFP-labelled cells were observed in about 80% of glands at 2 weeks, GFP-labelled cells were present in about 27% of glands at 2 months and the numbers of GFP-labelled cells were decreased (figure 2B). Only about 1.6 GFP-labelled cells per gland were present and those cells were restricted to the very base of glands (figure 2C). This distribution was similar to GFP-labelled cells observed at a year after the Dox treatment. Additionally, we observed that GIF-negative GFP-labelled cells were first present in about 1.7% of corpus glands at 2 months and scattered labelled cells were present in the neck region of corpus glands at 6 months (online supplemental figure S2A, B, arrows). The GIF-negative GFP-labelled cells were widely distributed in about 4% of corpus glands at 12 months after the Dox treatment (figure 2B and online supplemental figure S2A, arrows). Those GFP-labelled cells showed reprogramming into cells that were copositive for Ki67 (proliferating cells), UEAI (surface cells), GSII (mucus neck cells) and H/K-ATPase (parietal cells) (online supplemental figure S2C, D, arrows). These results confirm the long life of mature chief cells in the mucosal homoeostasis, but only a very small subset of mature chief cells functions as ‘reserve’ stem cells and is able to differentiate into various types of gastric cell lineages.
Figure 2.
Lineage tracing of GIF-expressing chief cells in normal stomachs. (A) Immunostaining for GFP (green), GIF (red) and Ki-67 (blue) at 2 weeks, 2 months and 12 months following Dox treatment in GIF-Cre-RnTnG mouse stomachs. GFP+GIF+ cells represent long-lived chief cells and GFP+GIF cells indicate a lineage derivation from mature chief cells (****p<0.0001). The dotted boxes indicate the enlarged region. N=3 mice per group. (B) Quantitation of GFP+ glands. (C) Quantitation of GFP+ cells per gland. One hundred glands from proximal corpus were counted to perform the quantitative analyses (n=3, ****p<0.0001). GIF, gastric intrinsic factor.
gutjnl-2021-325310supp002.pdf (769.6KB, pdf)
Chief cells directly give rise to SPEM cells during drug-induced metaplasia development
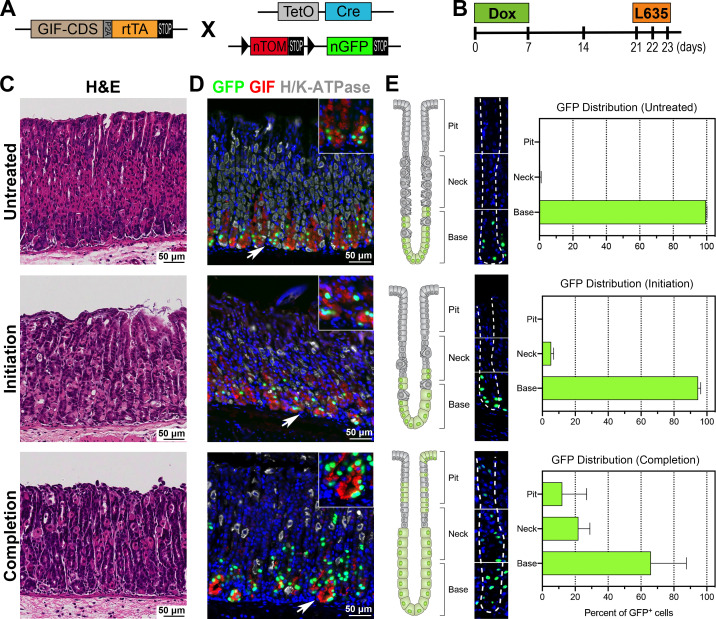
We next examined whether the chief cells directly give rise to SPEM cells using a lineage tracing model: the GIF-Cre-RnTnG mouse allele (figure 3A). Lineage tracing of chief cells with GFP was analysed 2 weeks after the 1 week Dox treatment to ensure the full maturation of the GFP-labelled chief cells.19 26 GFP-labelled cells were present in 71% of corpus glands and 20% of antral glands (online supplemental figure S1D, F). The GIF-Cre-RnTnG mice were then treated with L635 for either one or three doses to track the fate of the GFP-labelled chief cell lineage derivation during the metaplastic process (figure 3B, C). Lineage tracing of GFP-labelled chief cells in untreated mice revealed that nearly 100% of the GFP-labelled cells were located at the gland base. Additionally, all GFP-labelled cells were copositive for GIF (figure 3D), but not copositive for Ki67, confirming that no GIF transcription activity occurs in the isthmal progenitor cells (online supplemental figure S3A). The majority of GFP-labelled cells after 1 dose of L635 treatment, the initiation step of the metaplastic process, were still observed at the base of glands and were copositive for GIF (figure 3D, E). Also, the GFP-labelled cell zone remained distinct from the expanded isthmal progenitor cell zone (online supplemental figure S3A).
Figure 3.
Lineage tracing of GFP-labelled chief cells in metaplasia development. (A) Diagram of the GIF-Cre-RnTnG allele used in this study. (B) Scheme of L635 treatment study in GIF-Cre-RnTnG mice. To lineage label chief cells with GFP, mice were administered with Dox for 1 week on, 2 weeks off before the L635 treatment to fully mature the GFP-labelled chief cells. The GIF-Cre-RnTnG mice were treated without (untreated, N=4) or with one or three doses of L635 (initiation or completion, N=3). (C) H&E-stained stomach sections from GIF-Cre-RnTnG mice, untreated, L635-treated for one or three doses. (D) Sections of the stomach tissues from untreated, or L635 treated GIF-Cre-RnTnG mice were immunostained with antibodies against GFP (green), GIF (red) and H/K-ATPase (white). Nuclei were counterstained with Hoechst (blue). White arrows denote magnified insets. (E) Analysis of the distribution of GFP-labelled cells in different regions of glands in untreated or L635-treated GIF-Cre-RnTnG mouse stomachs. The corpus glands are illustrated on the left to show GFP-labelled cell position in different regions of glands (top, neck and base). A corpus gland stained for GFP (green) and Hoechst (blue) is at the centre. White dotted lines delineate a gland. Bar graphs display the percentage of GFP-labelled cells in different regions of gland. GIF, gastric intrinsic factor.
gutjnl-2021-325310supp003.pdf (427.1KB, pdf)
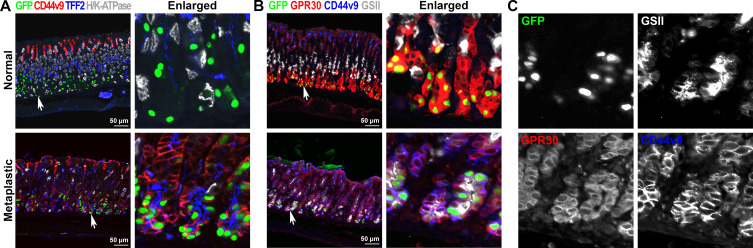
The GFP-labelled cells were visualised after three doses of L635 treatment, which represents the completion step of the metaplastic process, and extended from the base into the pit region of the metaplastic glands (figure 3D,E and figure 5A yellow dotted lines). The GFP-labelled cells located at the base of the glands were copositive for various SPEM markers such as GSII, CD44v9, TFF2 and GIF (figure 4A and online supplemental figure S3B). In addition, we also examined the expression of GPR30 protein, a new chief cell marker,29 after three doses of L635 treatment. In untreated normal corpus glands, the GPR30 protein was strongly expressed in chief cells and copositive for GFP (figure 4B). While it is known that the GPR30 transcriptional activity was quickly downregulated when metaplasia is developed in the stomach mucosa,29 the GPR30-expressing cells remained in the metaplastic glands (figure 4B, C) and were still copositive for GFP as well as two SPEM cell markers, GSII and CD44v9. This result supports the concept that the chief cells did not die after induction of parietal cell loss, but rather transdifferentiated into SPEM cells in response to the acute mucosal injury. Interestingly, GFP-labelled cells copositive for Ki67 were observed at the completion step (figure 5A) and 48% of the GFP-labelled cells were copositive for both GSII and Ki67, indicative of proliferating SPEM cells (online supplemental figure S3B, C). However, none of the GFP-labelled cells located close to the expanded proliferative cell zone were copositive for Ki67 at the initiation step (online supplemental figure S3A, white arrows). These data indicate that chief cells initiate the metaplastic process by transdifferentiating into SPEM cells, and then complete the metaplastic process by re-entering cell division. These steps reflect that the chief cells reprogram to complete the metaplastic conversion in response to injury30 31 and offer direct evidence for chief cells as the predominant origin of SPEM cells.16 19 25 32–34
Figure 4.
Immunofluorescence staining for SPEM markers in metaplastic glands. (A) Sections of the stomach tissues from GIF-Cre-RnTnG mice treated without (untreated) or with L635 for three doses (completion) were immunostained with antibodies against GFP (green), CD44v9 (red), TFF2 (blue) and H/K-ATPase (white). White arrows indicate enlarged area in panel. (B) Sections of the stomach tissues from GIF-Cre-RnTnG mice treated without (untreated) or with L635 for three doses (completion) were immunostained with antibodies against GFP (green), GPR30 (red), CD44v9 (blue) and GSII (white). White arrows indicate enlarged area in panel (C). GIF, gastric intrinsic factor; SPEM, spasmolytic polypeptide-expressing metaplasia.
Figure 5.
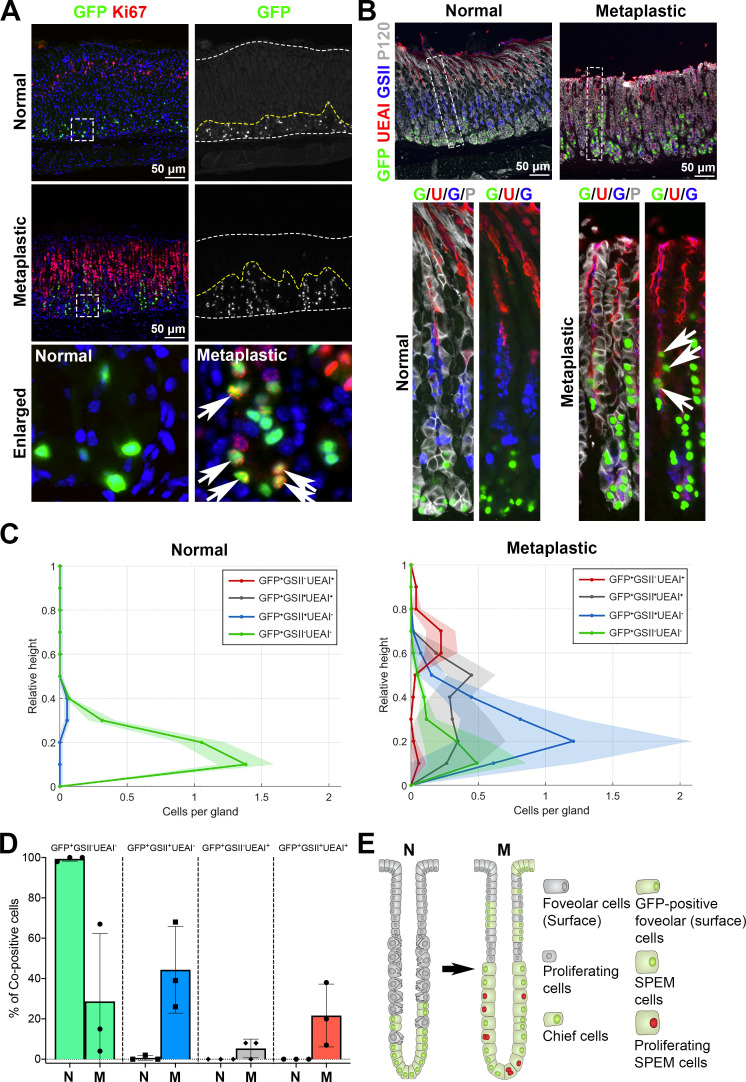
Lineage contribution of GFP-labelled chief cells in metaplastic glands. (A) Sections of the stomach tissues from untreated (normal) or L635-treated (three doses, metaplastic) GIF-Cre-RnTnG mice were immunostained with antibodies against GFP (green) and Ki67 (red). Nuclei were counterstained with Hoechst (blue). Dotted boxes depict enlarged regions. white dotted lines delineate mucosa regions and yellow dotted lines define the GFP+ cell zones. White arrows indicate GFP and Ki67 copositive cells. N=3 mice per group. (B) Sections of the stomach tissues from untreated or L635-treated (three doses, metaplastic) GIF-Cre-RnTnG mice were immunostained with antibodies against GFP (green), UEAI (red), GSII (blue) and p120 (membrane marker, white). White dotted boxes depict enlarged regions. White arrows indicate GFP and UEAI copositive cells. N=3 mice per group. (C) Analysis of relative distribution of GFP+ cells in untreated (normal) and L635 treated (three doses, metaplastic) mouse stomach mucosa. The distribution histograms display each GFP+ cells coimmunostained for UEAI and GSII in the corpus gland shown in B. GFP+GSII-UEAI+ cells (red) indicate GFP-labelled surface cells. GFP+GSII+UEAI+ cells (grey) indicate transitioning cells from SPEM cells to surface cells. The y-axis represents the corpus gland divided into 10% increments (1=top and 0=base). The x-axis represents the number of cells in each region per gland. more than 50 glands were analysed in each group. (D) Bar graphs display the percentage of GFP+ cells coimmunostained for UEAI and GSII in the corpus gland shown in B. N=3 mice per group. (E) Schematic diagram of lineage contribution of GFP-labelled chief cells in metaplasia development. GIF, gastric intrinsic factor; SPEM, spasmolytic polypeptide-expressing metaplasia.
Chief cell plasticity contributes to the foveolar cell hyperplasia in metaplastic glands
The lineage tracing of mature chief cells with GFP also revealed a dynamic contribution of SPEM cells during metaplasia development. At the completion step, GFP-labelled cells, which migrated into the pit area in metaplastic glands, were copositive for UEAI, a surface cell marker (figure 5B, white arrows). In normal glands, the GFP-labelled cells displayed a regional restriction to the base of glands and a homogeneous cell type. More than 99% of cells were negative for both UEAI and GSII (figure 5B, C, Green) and only a few GFP-labelled cells were copositive for GSII and were located in the upper area of the GFP+ cell zone, likely representing transitioning chief cells that may not be fully mature (figure 5C, blue and figure 5D). On the other hand, the GFP-labelled cells in metaplastic glands displayed regional redistribution to the top of glands and a variety of mucous cell types, copositive for GSII and/or UEAI (figure 5C, D, online supplemental figure S4A). 44% of GFP-labelled cells were copositive only for GSII (GFP +GSII+UEAI−, blue), indicating SPEM cells, and were located only in the base and neck areas. Interestingly, 22% of GFP-labelled cells located in the neck and pit area were copositive for UEAI (GFP +GSII-UEAI+, red) and Muc5ac (online supplemental figure S4B), indicating surface cells which were derived from GFP-labelled cells. Additionally, 5% of GFP-labelled cells were copositive for both GSII and UEAI (GFP +GSII+UEAI+, grey). This population was widely distributed throughout the gland and suggests that a minor SPEM cell population may undergo a cell type conversion to surface mucous cells. It is important to note that 29% of GFP-labelled cells were negative for both GSII and UEAI (GFP +GSII-UEAI−, green) and those cells were only present at the base of glands, indicating a subset of chief cells which did not undergo transdifferentiation.35 These results clearly reveal the dynamic activity and plasticity of SPEM cells derived from mature chief cells, and a gradual expansion of those SPEM cells from the bottom of glands towards the surface area after the completion of the chief cell transdifferentiation process (figure 5E). The SPEM cells which display a heterogeneous contribution in metaplastic glands, demonstrate features similar to the ulceration-associated cell lineage (UACL), a metaplastic lineage which arises from the bases of intestinal crypts identified in the human gastrointestinal track in response to severe damage, especially in the setting of Crohn’s disease.36 37 Our results indicate that that SPEM cells, similar to the pattern observed for UACL, can give rise to a different cell lineage, surface cells, and contribute to the cellular heterogeneity of metaplastic glands. Consequently, our lineage tracing studies suggest a new paradigm of a lineage contribution of SPEM cells to the foveolar hyperplasia. Since it is not yet clear whether the non-proliferative or proliferating SPEM cells are responsible for the SPEM cell conversion into surface cells, further studies to define the independent roles of the subsets of SPEM cells are required.
gutjnl-2021-325310supp004.pdf (485.3KB, pdf)
SPEM cells emerge from the chief cell transdifferentiation in response to ulceration
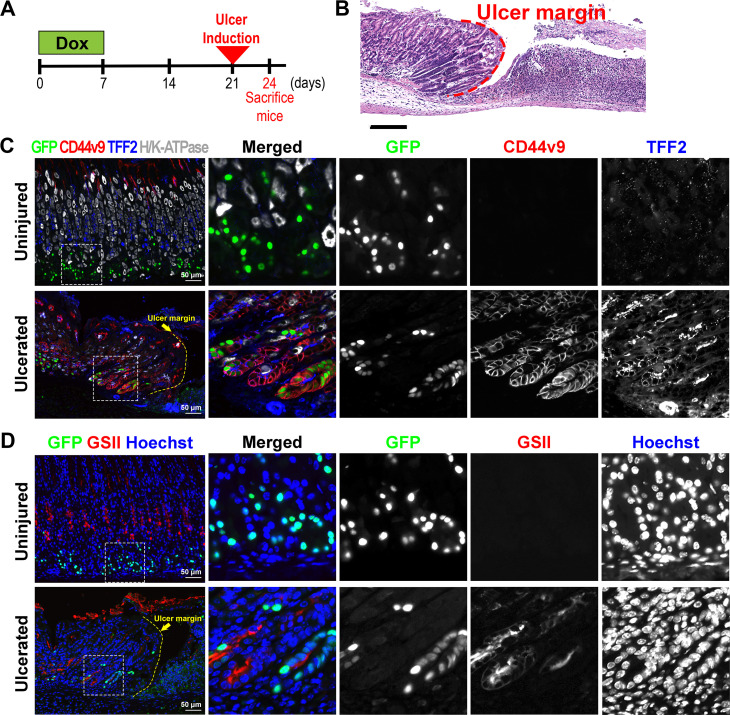
To address whether the chief cell transdifferentiation is a common feature of the emergence of metaplastic cells in response to acute injury in the stomach mucosa, we examined the GFP-labelled cells after acetic acid-induced ulceration in the stomach corpus. Ulcer healing in this acute mucosal injury model is also associated with a metaplastic reaction.13 We first administered Dox to the GIF-Cre-RnTnG mice for 1 week to label chief cells with GFP and induced acute mucosal ulceration 2 weeks after the Dox treatment, through application of acetic acid to the serosal surface of the mouse stomachs (figure 6A). H&E staining revealed that the stomach mucosa had ulceration with a distinct ulcer margin at the edge of a denuded mucosal area in the GIF-Cre-RnTnG mouse stomachs 3 days after acetic acid-ulcer injury (figure 6B, red dotted line). In uninjured mucosa, GFP-labelled chief cells were not copositive for any SPEM markers, such as CD44v9, TFF2 and GSII, as well as a parietal cell marker, H/K-ATPase, as expected (figure 6C, D). However, in the ulcerated mucosa, the GFP-labelled cells were still present at the bases of glands closely located adjacent to the ulcer margin and they were copositive for SPEM markers, including CD44v9, TFF2 and GSII (figure 6C, D), confirming that the SPEM cells, in response to the ulceration, were also derived from the chief cell transdifferentiation. Therefore, our results suggest that the chief cells are the predominant origin of SPEM cells emerging during the metaplastic process in response to acute injury.
Figure 6.
Lineage contribution of GFP-labelled chief cells in acetic acid-induced ulceration. (A) Scheme of acetic acid-induced ulceration in GIF-Cre-RnTnG mice. To first lineage label chief cells with GFP, mice received Dox for 1 week, followed by 2 weeks off before the gastric ulceration was induced. The stomach tissues were collected 3 days after the ulceration. (B) H&E-stained stomach sections from GIF-Cre-RnTnG mice at 3 days after acetic acid-induced injury. N=3 mice. Dotted line indicates ulcer margin. (C, D) Sections of the stomach tissues from uninjured or ulcerated lesions in the GIF-Cre-RnTnG mice were immunostained with (C) antibodies against GFP (green), CD44v9 (red), TFF2 (blue) and H/K-ATPase (white) and (D) antibodies against GFP (green) and GSII (red). Nuclei were counterstained with Hoechst. White dotted boxes depict enlarged regions and yellow dotted line indicates ulcer margin. GIF, gastric intrinsic factor.
Discussion
Although GIF is one of the most abundant and representative chief cell-specific markers in the mouse corpus, there has been no driver mouse model using the GIF locus in the field, which allows for cell type-specific lineage mapping. For this reason, the current conflicting paradigms of SPEM cell origin in the stomach have not been resolved. The GIF-rtTA mouse model which we generated is a true chief cell-specific driver allele in the stomach corpus and both the GIF-GFP reporter mouse and the GIF-Cre-RnTnG lineage tracing mouse exhibit a distinct chief cell population at the base of glands in normal stomach mucosa. In particular, the lineage tracing results revealed the longevity of chief cells without further reprogramming in homoeostasis up to 1 year after the GFP labelling. Only about 4% of the long-term label-retaining chief cells showed self-renewal capacity and differentiated into other types of gastric cell lineages, such as parietal cells and surface cells.
We have previously shown that the isthmal progenitor cells are distinct from the process of chief cell transdifferentiation and only contribute to the foveolar hyperplasia in response to Kras activation.38 Our results using both the GIF-GFP reporter and the GIF-Cre-RnTnG lineage tracing mouse alleles demonstrated that a conserved response to acute injury, where chief cells undergo transdifferentiation into SPEM cells. The lineage tracing also showed that the GFP-labelled cells migrate upwards to the foveolar/surface cell zone after the acute injury suggesting that chief cells also contribute to foveolar lineages in metaplastic glands as part of the phenotype of pyloric metaplasia.3 Therefore, our present study clearly confirms that chief cell transdifferentiation results in development of metaplasia through the expansion of the SPEM cell and surface cell compartments. However, since the GFP was prelabelled in chief cells before the acute injury, it is not clear whether the foveolar cells positive for GFP in the metaplastic glands originated from the SPEM cell differentiation or direct reprogramming of chief cells. It is also possible that chief cells may acquire the stem cell ability during the transdifferentiation process since there is a subpopulation in chief cells functioning as ‘reserve’ stem cells.
It has previously been reported that the mucosal recovery after gastric ulceration is associated with SPEM development, and SPEM cells may be responsible for wound healing after gastric injury.13 In the GIF-Cre-RnTnG mouse stomachs, about 30% of chief cells labelled with GFP in a gland did not transdifferentiate in response to the mucosal injury. However, we were not able to address whether the GFP-labelled SPEM cells also contributed to the ulcer recovery, because the GFP+ cells between the GFP +SPEM cell progeny and the GFP+ chief cells that did not undergo transdifferentiation were not distinguished in the recovered mucosa. A new SPEM cell marker, AQP5, has been identified which specifically labels SPEM cells in metaplastic glands in both mouse and human stomach corpus,12 and we believe the AQP5-CreERT driver mouse allele would be useful for further examination of the SPEM cell plasticity during the recovery phase after acetic acid-induced ulceration in the stomach corpus. In addition, mucus neck cells are progenitor cells of chief cells and not proliferative in homoeostatic glands. Since mucus neck cell hyperplasia is also observed in the setting of metaplasia development,39 we cannot rule out the possibility of neck cell contribution to the phenotype of SPEM cells in glands with pyloric metaplasia. Our current mouse model system is a chief cell-specific model and there are no mucus neck cell-specific driver mouse alleles available at present. Therefore, further studies are required to address whether the mucus neck cells also can acquire stem cell-like properties and contribute to SPEM cell production.
Collectively, our results conclude that the SPEM cells originate primarily from mature chief cells in response to injury even in the absence of any genetic alterations.32 Also, the chief cell-originated SPEM cells can orchestrate cellular reconstitution of normal gastric glands to metaplastic glands containing heterogeneous cell types after the completion of the reprogramming process into progenitor-like cells, which display not only proliferative activity, but also lineage conversion capability to produce surface cells. Furthermore, additional animal models beyond the acute mucosal injury, such as chronic injury models through Helicobacter felis or Helicobacter pylori infection7 40 or oncogenic gene activation6 would further clarify the roles of chief cell plasticity during metaplasia development and progression. In conclusion, cellular plasticity of mature chief cells as a differentiated gastric epithelial cell type is responsible for the diverse lineage distribution during metaplasia development following severe mucosal injury.
Methods
Experimental animal models
The care, maintenance and treatment of mice used in this study followed protocols approved by the Institutional Animal Care and Use Committee of Vanderbilt University Medical Center and each experimental group contained 2–4 mice as noted in the figure legends. Littermates from both sexes were randomly assigned to experimental groups. Mice were maintained in housing of five animals per cage with a 12-hour light/dark cycle and water and food at libitum.
Generation of the GIF-rtTA mouse allele
We designed a strategy for rtTA knock-in CRISPR/Cas9 system, targeting the GIF locus in mouse genome (online supplemental figure S1A). Two single guide (sg) RNAs were designed to target the GIF 3′-UTR in close proximity to the coding sequences (CDS). A single strand DNA (ssDNA, 976 bp) was designed as a donor to target the GIF 3′-UTR near the CDS. The Donor ssDNA contains a promoterless 2a-rtTA sequence flanked by two homologous arms at each end and was inserted at the end of the coding region of the GIF locus. The Donor ssDNA and synthetic guide RNA (sgDNA) were generated by Applied StemCell Company and they performed the pronuclear injections into mouse embryos and transferred the embryos into recipient mice. A microinjection cocktail was prepared by mixing of annealed crRNA:tracrRNA (20 ng/µL), Cas9 protein (20 ng/µL), and donor ssDNA (10 ng/µL). Microinjection was performed at the pronuclear stage and a total of six transgenic founders (F0 founders) were identified in two rounds of microinjection. Genomic DNAs from the tail tips of derived pups were extracted for use as a PCR template to screen for the transgene within 6 weeks after the injection. Genotyping was performed to identify the F1 positive mice that are the heterozygous founders. The primer sequences used for genotyping are listed in online supplemental table 1. Two F1 male mice were produced through breeding potential F0 founders to wild type C57BL/6 J mice. The two F1 male mice were then transferred to The Vanderbilt University to establish the GIF-rtTA founder line. The GIF-rtTA mouse allele created by Applied Stemcell was maintained as homozygous.
Transgenic mouse alleles
The generation of Gif-rtTA-TetO-H2BGFP (GIF-GFP) and Gif-rtTA-TetO-Cre-Gt(ROSA)26Sortm(CAG-tdTomato*,-EGFP*)Ees/J (GIF-Cre-RnTnG) mice is described as follows. The Gif-rtTA mice were crossed with TetO-H2BGFP sourced from the Jackson Laboratory (Bar Harbor, ME) for a successful breeding of a dual positive Gif-rtTA-TetO-H2BGFP (GIF-GFP) transgenic mouse allele. To establish the triple cross GIF-Cre-RnTnG mouse allele, the Gif-rtTA mice were crossed against TetO-Cre mice then, dual positive mice were crossed with Gt(ROSA)26Sortm(CAG-tdTomato*, −EGFP*)Ees/J to complete the allele of Gif-rtTA-TetO-Cre-Gt(ROSA)26Sortm(CAG-tdTomato*, −EGFP*)Ees/J (GIF-Cre-RnTnG). The GIF-GFP and GIF-Cre-RnTnG lines were maintained on a mixed background of C57BL/6 and CD1.
Genotyping
Ear punches were collected from mice at 3 weeks of age on weaning. Samples were lysed using a 5% mixture of Viagen Direct PCR and Viagen Proteinase K. Samples were lysed in solution overnight at 55°C and then incubated at 85°C for 1 hour the following day and iced for 15 min. Protocols for each individual genotyping used a master mix of 8 uL of distilled water, 10 uL GoTaq 2 x premix, 1 uL of 10 mM primer mixture per sample and 1 uL of template gDNA. Gif-rtTA 5’ and 3’ sequences use Platinum Hot Start II and Gif-rtTA WT primers use Promega GoTaq G2 Green MasterMix. Conditions for GIF-rtTA PCR are as follows: 1 initialisation cycle at 94°C for 2 min, 35 cycles at 94°C for 30 s, 58°C for 30 s, 72°C for 90 s and a final elongation at 72°C for 5 min. TetO-H2B-GFP uses Promega GoTaq G2 Green MasterMix. Conditions for TetO-H2B-GFP PCR are: 1 initialisation cycle at 94°C for 3 min, 32 cycles at 94°C for 30 s, 58°C for 30 s, 72°C for 40 s and a final elongation at 72°C for 5 min. Primer sequences for TetO-Cre use Promega GoTaq G2 Green MasterMix. Conditions for TetO-Cre PCR follow: 1initalisation cycle at 94°C for 2 min, 10 cycles at 94°C for 20 s, 65°C for 15 s, 68°C for 10 s, 28 cycles at 94°C for 15 s, 60°C for 15 s, 72°C for 30 s and a final elongation at 72°C for 2 min. RosanTnG primers use DreamTaq GreenPCR MasterMix 2 x. Conditions for RosanTNG PCR are as follows: 1 initialisation cycle at 94°C for 5 min, 35 cycles at 94°C for 30 s, 61°C for 60 s, 72°C for 1 min; 30 s and a final elongation at 72°C for 7 min. The primer sequences used for genotyping are listed in online supplemental table 1.
Drug treatment
At 6 weeks of age, GIF-GFP mice were treated with Dox water at a concentration of 0.2 mg/mL over the course of 1 week. Sequentially, L635 (synthesised by the Chemical Synthesis Core of the Vanderbilt Institute of Chemical Biology, Nashville, Tennessee, USA) was administered once a day for one or three doses by oral gavage (350 mg/kg) to mice maintained on Dox water treatment. The GIF-Cre-RnTnG mice were treated with Doxwater at a concentration of 0.2 mg/mL for 1 week. Mice were left for 2 weeks for complete GFP-labelled chief cell maturation and then treated with L635 once a day for one or three doses by oral gavage (350 mg/kg). Stomachs were resected from wildtype, GIF-GFP, GIF-Cre-RnTnG and washed in PBS. Samples were then fixed in 4% paraformaldehyde solution in PBS overnight at 4°C. The following day, the samples were trimmed and processed according to a standard histological protocol for paraffin embedding. We grouped the GIF-GFP or GIF-Cre-RnTnG mice from various litters for different groups for untreated and treated with L635 one dose or three doses for a single experiment and all the L635 treatment experiments were repeated two to four times to collect three to four stomach tissues per group from individual mice.
Ulcer induction
Gastric ulcers were induced in mice approximately 9 weeks of age by acetic acid under isoflurane anaesthesia based on a previously published protocol.13 Prior to induction, each mouse was dosed with ketoprofen analgesic (5–10 mg/kg) with follow-up doses each day until the completion of the experiment. A midline abdominal surgical incision was performed and the stomach was exposed for ulceration. Microcapillary tubes (0.8 mm in diameter) were filled with acetic acid (99%) and placed in direct contact with the exterior surface of the proximal stomach corpus region and were held in place for 25 s. Sterile gauze was used to wipe the stomach clean from any residual acid and the muscle and skin incisions were sutured in two separate layers. The mice were then maintained with standard food and water and checked daily until completion of the experiment.
Immunostaining
Paraffin tissues sections of 5 µm were deparaffinised in a series of histoclear washes and rehydrated through a progression of ethanol (100%, 95%, 75%). Antigen retrieval was performed using Dako target retrieval solution PH 6 in a pressure cooker for 15 min and cooled for 1 hour. Sections were incubated in Dako serum-free protein block solution at room temperature for 1.5 hours and primary antibodies were added to Dako antibody diluent with background reducing components and then applied to sections overnight at 4°C. The following day, slides were washed in PBS for 5 min three times, then incubated for 1 hour in secondary antibodies that were added in dako antibody diluent at room temperature for immunofluorescence staining. Nuclei counterstaining incubated for 5 min at room temperature using Hoechst at (0.2 ug/mL) in PBS and sections were washed three times in PBS, mounted and cover slipped.
Immunohistochemistry staining
ImmPRESS polymer detection reagent and ImmPACT DAB substrate kits from Vector Laboratories were used for primary antibody detection. The primary and secondary antibodies used for immunostaining are listed in online supplemental table 1. Fluorescence images were attained using a Zeiss Axio Imager M2, equipped with a SPOT Explorer camera using SPOT basic software at ×20 magnification. Preparation and overlay of fluorescence images were executed in Adobe Photoshop. Stomach tissues after the immunohistochemistry were scanned on an Ariol SL-50 slide scanner (Leica) at ×20 magnification.
Quantification and statistical analysis
Statistics
To quantify the GFP+ glands in GIF-GFP or GIF-Cre-RnTnG mouse stomachs, we counted 100 GFP+ glands in both the corpus and antrum zones in each mouse stomach tissue section. The average values of GFP +glands in the corpus or antrum from each mouse section were compared by Welch’s test (p<0.05 for significance). For the long term-lineage tracing study, we counted 100 glands from the proximal corpus in each mouse stomach tissue section after immunofluorescence staining for GFP, GIF and Ki67. Only GFP+GIF+ cells were considered GFP-labelled chief cells and GFP+GIF cells were considered cell lineages derived from mature chief cells. The average values of GFP+ glands or GFP+ cells per gland from each mouse section were compared by Welch’s test (p<0.05 for significance). For quantitation of GFP+ chief cells, three representative images of stained dual positive GFP and GIF stomach tissue corpus regions were taken from a paraffin section of each mouse stomach at ×20 magnification. The average values of GFP+GIF+ cells or GFP-GIF+ cells in the corpus from each mouse section were then compared by Welch’s test (p<0.05 for significance).
Distribution analysis
Experimental groups contained three to four mice. At least three representative images (>50 glands) of proximal stomach corpus were taken from each mouse at ×20 objective for analysis. Both the top and base of glands were identified and denoted, then glands were split into thirds (pit, neck, and base). Cell numbers in each region were determined manually using ImageJ. The results were reported as percent of cells in each region. For mucous cell type distribution, top of glands and base of glands were identified and gland height was normalised from 0 to 1 (1=top and 0=base of gland). A custom-built software was developed using MATLAB to analyse the quantitative spatial localisation of GFP positive cells. Immunolabeled cells for each marker were manually identified. The software divided the gastric gland into 10% increments. For each labelled cell, the height location within the gland was determined by calculating distance of the labelled nuclei to an increment within the denoted top and base of the gland. The results were normalised against the total number of glands and reported as cells per gland. Both the top and base of glands were identified and denoted, and glands were split into thirds (pit, neck and base). Cell numbers in each region were determined manually using ImageJ. The results were reported as per cent of cells in each region.
Acknowledgments
We thank the Vanderbilt Section of Surgical Sciences for support to generate the GIF-rtTA mouse allele and Dr. James Goldenring for providing L635.
Footnotes
Contributors: BC performed experiments, analysed the data and wrote the manuscript. ARM analysed the data and revised the manuscript. JAW analysed the data. ACE designed and performed the experiments, revised the manuscript. EC conducted and performed the experiments, analysed the data and wrote the manuscript.
Funding: These studies were supported by grants from the Department of Defense CA160399 and CA190172, NIH R37 CA244970 and pilot funding from VICC GI SPORE P50CA236733 (to EC). ARM was supported by NIH F31 DK117592. ACE was supported by NIH F32 DK111101 and KO1 DK121869. JAW was supported by NIH K25 CA204599.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information.
Ethics statements
Patient consent for publication
Not required.
References
- 1. Slack JMW. Metaplasia and transdifferentiation: from pure biology to the clinic. Nat Rev Mol Cell Biol 2007;8:369–78. 10.1038/nrm2146 [DOI] [PubMed] [Google Scholar]
- 2. Giroux V, Rustgi AK. Metaplasia: tissue injury adaptation and a precursor to the dysplasia-cancer sequence. Nat Rev Cancer 2017;17:594–604. 10.1038/nrc.2017.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldenring JR. Pyloric metaplasia, pseudopyloric metaplasia, ulcer-associated cell lineage and spasmolytic polypeptide-expressing metaplasia: reparative lineages in the gastrointestinal mucosa. J Pathol 2018;245:132–7. 10.1002/path.5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meyer AR, Goldenring JR. Injury, repair, inflammation and metaplasia in the stomach. J Physiol 2018;596:3861–7. 10.1113/JP275512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sáenz JB, Mills JC. Acid and the basis for cellular plasticity and reprogramming in gastric repair and cancer. Nat Rev Gastroenterol Hepatol 2018;15:257–73. 10.1038/nrgastro.2018.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi E, Hendley AM, Bailey JM, et al. Expression of activated Ras in gastric chief cells of mice leads to the full spectrum of metaplastic lineage transitions. Gastroenterology 2016;150:918–30. 10.1053/j.gastro.2015.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nomura S, Baxter T, Yamaguchi H, et al. Spasmolytic polypeptide expressing metaplasia to preneoplasia in H. felis-infected mice. Gastroenterology 2004;127:582–94. 10.1053/j.gastro.2004.05.029 [DOI] [PubMed] [Google Scholar]
- 8. Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol 2017;14:296–304. 10.1038/nrgastro.2017.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 2012;13:607–15. 10.1016/S1470-2045(12)70137-7 [DOI] [PubMed] [Google Scholar]
- 10. Chang ET, Gomez SL, Fish K, et al. Gastric cancer incidence among Hispanics in California: patterns by time, nativity, and neighborhood characteristics. Cancer Epidemiol Biomarkers Prev 2012;21:709–19. 10.1158/1055-9965.EPI-11-1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Siegel R, Naishadham D, Jemal A. Cancer statistics for Hispanics/Latinos, 2012. CA Cancer J Clin 2012;62:283–98. 10.3322/caac.21153 [DOI] [PubMed] [Google Scholar]
- 12. Lee SH, Jang B, Min J, et al. Upregulation of AQP5 defines spasmolytic polypeptide-expressing metaplasia (SPEM) and progression to incomplete intestinal metaplasia. Cellular and Molecular Gastroenterology and Hepatology. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engevik AC, Feng R, Choi E, et al. The development of spasmolytic Polypeptide/TFF2-Expressing metaplasia (SPEM) during gastric repair is absent in the aged stomach. Cell Mol Gastroenterol Hepatol 2016;2:605–24. 10.1016/j.jcmgh.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmidt PH, Lee JR, Joshi V, et al. Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab Invest 1999;79:639–46. [PMC free article] [PubMed] [Google Scholar]
- 15. Goldenring JR, Nam KT, Mills JC. The origin of pre-neoplastic metaplasia in the stomach: chief cells emerge from the mist. Exp Cell Res 2011;317:2759–64. 10.1016/j.yexcr.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leushacke M, Tan SH, Wong A, et al. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat Cell Biol 2017;19:774–86. 10.1038/ncb3541 [DOI] [PubMed] [Google Scholar]
- 17. Stange DE, Koo B-K, Huch M, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 2013;155:357–68. 10.1016/j.cell.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mills JC, Goldenring JR. Metaplasia in the stomach arises from gastric chief cells. Cell Mol Gastroenterol Hepatol 2017;4:85–8. 10.1016/j.jcmgh.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nam KT, Lee H-J, Sousa JF, et al. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology 2010;139:2028–37. 10.1053/j.gastro.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nozaki K, Ogawa M, Williams JA, et al. A molecular signature of gastric metaplasia arising in response to acute parietal cell loss. Gastroenterology 2008;134:511–22. 10.1053/j.gastro.2007.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hayakawa Y, Ariyama H, Stancikova J, et al. Mist1 expressing gastric stem cells maintain the normal and neoplastic gastric epithelium and are supported by a perivascular stem cell niche. Cancer Cell 2015;28:800–14. 10.1016/j.ccell.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayakawa Y, Fox JG, Wang TC. Isthmus stem cells are the origins of metaplasia in the gastric corpus. Cell Mol Gastroenterol Hepatol 2017;4:89–94. 10.1016/j.jcmgh.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kinoshita H, Hayakawa Y, Niu Z, et al. Mature gastric chief cells are not required for the development of metaplasia. Am J Physiol Gastrointest Liver Physiol 2018;314:G583–96. 10.1152/ajpgi.00351.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shimizu T, Sohn Y, Choi E, et al. Decrease in miR-148a expression during initiation of chief cell transdifferentiation. Cell Mol Gastroenterol Hepatol 2020;9:61–78. 10.1016/j.jcmgh.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meyer AR, Engevik AC, Willet SG, et al. Cystine/Glutamate antiporter (xCT) is required for chief cell plasticity after gastric injury. Cell Mol Gastroenterol Hepatol 2019;8:379–405. 10.1016/j.jcmgh.2019.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weis VG, Petersen CP, Weis JA, et al. Maturity and age influence chief cell ability to transdifferentiate into metaplasia. Am J Physiol Gastrointest Liver Physiol 2017;312:G67–76. 10.1152/ajpgi.00326.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karam SM, Leblond CP. Dynamics of epithelial cells in the corpus of the mouse stomach. III. Inward migration of neck cells followed by progressive transformation into zymogenic cells. Anat Rec 1993;236:297–313. 10.1002/ar.1092360204 [DOI] [PubMed] [Google Scholar]
- 28. Quante M, Marrache F, Goldenring JR, et al. TFF2 mRNA transcript expression marks a gland progenitor cell of the gastric oxyntic mucosa. Gastroenterology 2010;139:2018–27. 10.1053/j.gastro.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hata M, Kinoshita H, Hayakawa Y, et al. GPR30-Expressing gastric chief cells do not Dedifferentiate but are eliminated via PDK-Dependent cell competition during development of metaplasia. Gastroenterology 2020;158:1650–66. 10.1053/j.gastro.2020.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Willet SG, Lewis MA, Miao Z-F, et al. Regenerative proliferation of differentiated cells by mTORC1-dependent paligenosis. Embo J 2018;37:e98311. 10.15252/embj.201798311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burclaff J, Mills JC. Plasticity of differentiated cells in wound repair and tumorigenesis, part I: stomach and pancreas. Dis Model Mech 2018;11:dmm033373. 10.1242/dmm.033373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Radyk MD, Burclaff J, Willet SG, et al. Metaplastic cells in the stomach arise, independently of stem cells, via dedifferentiation or transdifferentiation of chief cells. Gastroenterology 2018;154:839–43. 10.1053/j.gastro.2017.11.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burclaff J, Osaki LH, Liu D, et al. Targeted apoptosis of parietal cells is insufficient to induce metaplasia in stomach. Gastroenterology 2017;152:762–6. 10.1053/j.gastro.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burclaff J, Willet SG, Sáenz JB, et al. Proliferation and differentiation of gastric mucous neck and chief cells during homeostasis and injury-induced metaplasia. Gastroenterology 2020;158:598–609. 10.1053/j.gastro.2019.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nam KT, O'Neal RL, Coffey RJ, et al. Spasmolytic polypeptide-expressing metaplasia (SPEM) in the gastric oxyntic mucosa does not arise from Lgr5-expressing cells. Gut 2012;61:1678–85. 10.1136/gutjnl-2011-301193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanby AM, Poulsom R, Singh S, et al. Hyperplastic polyps: a cell lineage which both synthesizes and secretes trefoil-peptides and has phenotypic similarity with the ulcer-associated cell lineage. Am J Pathol 1993;142:663–8. [PMC free article] [PubMed] [Google Scholar]
- 37. Hanby AM, Wright NA. The ulcer-associated cell lineage: the gastrointestinal repair kit? J Pathol 1993;171:3–4. 10.1002/path.1711710103 [DOI] [PubMed] [Google Scholar]
- 38. Choi E, Means AL, Coffey RJ, et al. Active Kras Expression in Gastric Isthmal Progenitor Cells Induces Foveolar Hyperplasia but Not Metaplasia. Cell Mol Gastroenterol Hepatol 2019;7:251–3. 10.1016/j.jcmgh.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petersen CP, Mills JC, Goldenring JR. Murine models of gastric corpus preneoplasia. Cell Mol Gastroenterol Hepatol 2017;3:11–26. 10.1016/j.jcmgh.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Noto JM, Peek RM. The gastric microbiome, its interaction with Helicobacter pylori, and its potential role in the progression to stomach cancer. PLoS Pathog 2017;13:e1006573. 10.1371/journal.ppat.1006573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2021-325310supp001.pdf (886.9KB, pdf)
gutjnl-2021-325310supp005.pdf (63.8KB, pdf)
gutjnl-2021-325310supp002.pdf (769.6KB, pdf)
gutjnl-2021-325310supp003.pdf (427.1KB, pdf)
gutjnl-2021-325310supp004.pdf (485.3KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information.