Abstract
Dickeya zeae, a plant soft-rot pathogen, possesses a type III secretion system (T3SS) as one of the major virulence factors, infecting a wide variety of monocotyledonous and dicotyledonous plants and causing serious losses to the production of economic crops. In order to alleviate the problem of pesticide resistance during bacterial disease treatment, compounds targeting at T3SS have been screened using a hrpA-gfp bioreporter. After screening by Multifunctional Microplate Reader and determining by flow cytometer, five compounds including salicylic acid (SA), p-hydroxybenzoic acid (PHBA), cinnamyl alcohol (CA), p-coumaric acid (PCA), and hydrocinnamic acid (HA) significantly inhibiting hrpA promoter activity without affecting bacterial growth have been screened out. All the five compounds reduced hypersensitive response (HR) on non-host tobacco leaves and downregulated the expression of T3SS, especially the master regulator encoding gene hrpL. Inhibition efficacy of the five compounds against soft rot were also evaluated and results confirmed that the above compounds significantly lessened the soft-rot symptoms caused by Dickeya dadantii 3937 on potato, Dickeya fangzhongdai CL3 on taro, Dickeya oryzae EC1 on rice, and D. zeae MS2 on banana seedlings. Findings in this study provide potential biocontrol agents for prevention of soft-rot disease caused by Dickeya spp.
Keywords: Dickeya, type III secretion system, HrpL, inhibitor, biocontrol
Introduction
Dickeya species are necrotrophic, Gram-negative plant pathogens that cause severe disease in a wide range of plant hosts. In particular, the diseases caused by different species in Dickeya genus on rice, banana, potato, and taro have become a major threat to agricultural production in recent years (Hussain et al., 2008; Hu et al., 2018; Li et al., 2020; Dong et al., 2021; Huang et al., 2021). There are currently 13 species in this genus, including Dickeya chrysanthemi, Dickeya dadantii, Dickeya dianthicola, Dickeya paradisiaca, Dickeya zeae, Dickeya solani, Dickeya fangzhongdi, Dickeya aquatic, Dickeya poaceaephila, Dickeya lacustris, Dickeya undicola, Dickeya oryzae, and Dickeya parazeae (Hu et al., 2018; Hugouvieux-Cotte-Pattat et al., 2019; Oulghazi et al., 2019; Wang et al., 2020; Hugouvieux-Cotte-Pattat and Van Gijsegem, 2021), and among them, D. zeae, formerly known as Erwinia chrysanthemi pv. zeae, can infect a wide variety of monocotyledons and dicotyledons (Hussain et al., 2008), causing severe soft rot in crops and ornamental plants worldwide. For the prevention and control of this pathogen and other pathogenic bacteria, the most widely used measure in fields is mainly through agriculturally antibiotic treatment. Antibiotics achieve the purpose of prevention and control by inhibiting the growth of or directly killing the pathogen. Several chemical compounds including copper, anthium dioxide, formaldehyde, chlorine dioxide, 8-hydroxyquinoline, cetalkonium chloride, benzalkonium chloride, gluytaraldehyde, and others were used to control Erwinias soft rot disease (Lund and Lyon, 1975; Letal, 1977; Wyatt and Lund, 1981). Previous studies have shown that kasugamycin, Virginiamycin, or fungicides (acetic acid, boric acid, and bleach) can reduce the incidence of soft rot (Czajkowski et al., 2011). In addition, benziothiazolinoe 3% wettable powder (WP), tetramycin 0.3% aqueous solution (AS), and bismerthiazol 20% WP had better control effect on potato soft rot (Liang et al., 2020). However, a long-term usage of antibiotics has led to the increasing drug resistance of strains, and the WHO has listed antibiotic resistance as one of the three most important public health threats of the 21st century (Rasko and Sperandio, 2010; WHO, 2014). In this content, it is very important to find new alternative solutions for bacterial disease control.
The pathogenicity of D. zeae mainly depends on different virulence factors, including extracellular polysaccharides (EPS), plant cell wall degrading enzymes (PCWDEs), phytotoxins, flagellin, and secretion systems (Zhou et al., 2015; Hu et al., 2018; Feng et al., 2019). Type III secretion system (T3SS) is highly conserved in Gram-negative bacteria, including animal pathogens such as Salmonella, Pseudomonas aeruginosa, Escherichia coli, Chlamydia yersinia, Shigella fowleri, etc. (Hueck, 1998; Keyser et al., 2008; Lu et al., 2018), and plant pathogens such as Erwinia, Pseudomonas syringae, Pectobacterium, Xanthomonas, Ralstonia, and Dickeya, etc. (Hueck, 1998; Rojas et al., 2004; Pique et al., 2015). In Dickeya spp., T3SS is encoded by dsp/hrp/hrc gene clusters and plays an extremely important role in pathogenicity (Yang et al., 2002; Yap et al., 2005; Zhou et al., 2015). hrp gene expression was inhibited in rich medium, but was induced in nutrient deficient medium as well as in plants (Wei et al., 1992). T3SS is mainly used to directly inject Type III secreted effectors (T3SEs) into host cells by forming a syringe-like Type III secreting device, leading to disease resistance of host plants and hypersensitive response (HR) of non-host plants (Boucher et al., 1987; Yang et al., 2002; Buttner, 2012). In Dickeya bacteria, the expression of T3SS is regulated by a master regulator, HrpL, whose expression and activity are controlled under several regulatory cascades (Zeng et al., 2012). On one hand, hrpL upregulates many hrp genes that encode the T3SS structural and functional proteins, such as hrpA, hrpN, and dspE. T3SS pilus, whose formation relies on a large number of HrpA subunits, is required for effector proteins translocating into plant cells and located at the downstream of the T3SS regulatory pathway (Jin and He, 2001; Zeng et al., 2010; Li et al., 2015). In soft rot Erwinia amylovora, the expression of hrpA is most influenced by HrpL among the HrpL regulon (McNally et al., 2012). On the other hand, the expression of hrpL is regulated by the HrpX/HrpY-HrpS-HrpL pathway at the transcriptional level and the GacS-GacA-RsmB-RsmA pathway at the post-transcriptional level (Yang et al., 2008; Zeng et al., 2012; Yuan et al., 2020). In addition to the above regulatory pathways, It is reported that the amount of functional rsmB transcripts in D. dadantii have been reduced by polynucleotide phosphorylase (PNPase), resulting in decreased hrpL mRNA stability (Zeng et al., 2010). Additionally, SlyA, a SlyA/MarR family regulator, regulates hrp genes of the HrpL regulon in parallel with HrpL in D. dadantii, which positively regulates the expression of hrpA and hrpN, and negatively regulates the expression of hrpL by downregulating hrpS and upregulating rsmA (Zou et al., 2012). Therefore, T3SS is critical in pathogen-host interaction.
Usage of T3SS inhibitors that do not affect pathogenic bacterial growth but effectively reduce their virulence has become one of the promising alternatives of antibiotic treatment for bacterial disease control (Marshall and Finlay, 2014; Charro and Mota, 2015). For instance, some small molecules that specifically inhibit T3SS synthesis or function have been identified as T3SS inhibitors (Yuan et al., 2020). In the exploration of T3SS inhibitors drugs targeting T3SS in animal pathogens Yersinia pseudotuberculosis, E. coli, Salmonella enterica, Chlamydia, and P. aeruginosa have been found, which include salicylidene acylhydrazides (Kauppi et al., 2003; Tree et al., 2009; Layton et al., 2010; Ur-Rehman et al., 2012; Anantharajah et al., 2017; Uusitalo et al., 2017), phenoxyacetamides (Aiello et al., 2010; Zhang et al., 2013; Bowlin et al., 2014), N-Hydroxybenzimidazoles (Kim et al., 2009; Marsden et al., 2016), caminosides (Linington et al., 2002, 2006), and naringenin (Vikram et al., 2011). The mechanism of the action of these inhibitors mainly includes inhibition of T3SS gene transcription, toxic protein secretion, and T3SS device assembly. Some plant-derived and chemically synthesized compounds have been identified as T3SS inhibitors in plant pathogens. In D. dadantii 3937, the expression of T3SS was induced by plant-derived compounds o-coumaric acid and t-cinnamic acid through the rsmB-RsmA pathway (Yang et al., 2008). Previous studies have shown that the plant phenolic compounds and derivatives p-coumaric acid (PCA), cinnamyl alcohol (CA),and trans-4-hydroxycinnamohydroxamic acid play a role in the inhibition of T3SS expression of D. dadantii 3937 (Li et al., 2009, 2015; Joshi et al., 2021). Plant phenolic compounds and derivatives such as o-coumaric acid, trans-2-phenylcyclopropane-1-carboxylic-acid, trans-2-methylcinnamic acid, and trans-2-methoxycinnamic acid as well as synthetic compound ethyl 2-nitro-3-arylacrylates, have been reported to affect the expression of X. oryzae T3SS genes (Fan et al., 2017; Jiang et al., 2019). Khokhani et al. (2013) found T3SS inhibitors (4-methoxy-cinnamic acid and benzoic acid) and T3SS inducers [trans-2-(4-hydroxyphenyl)-ethenylsulfonate] change the expression of E. amylovora T3SS through HrpS-HrpL pathway, and salicylic acid (SA) inhibits hrpA promoter activity. More recently, salicylidene acylhydrazide derivatives, which were described as inhibitors of T3SS in zoonoses, have been verified for their effects on management of plant diseases caused by Ralstonia solanacearum and P. syringae pv. tomato (Puigvert et al., 2019). In addition, plant-derived compounds eugenol, SA, chlorogenic acid, resveratrol, CA, and fumaric acid were reported as inducers of T3SS in R. solanacearum (Joshi et al., 2021), p-hydroxybenozic acid (PHBA) and vanillic acid have been identified as inhibitors of T3SS transcription in P. syringae pv. tomato (Kang et al., 2020).
In this study, we selected some plant natural compounds for screening of Dickeya T3SS inhibitors using a hrpA-gfp bioreporter by Multifunctional Microplate Reader, followed by flow cytometer. The compounds significantly repressed the hrpA promoter activity without affecting bacterial growth, were used for in-depth evaluation on their potential on prevention of bacterial soft rot of important crops.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains, plasmids, and primers used in this study are listed in Supplementary Table S1. Dickyea bacterial strains including D. zeae MS2 isolated from banana (Hu et al., 2018; Feng et al., 2019), D. oryzae EC1 isolated from rice (Zhou et al., 2011; Cheng et al., 2013; Lv et al., 2019), D. dadantii 3937 isolated from Saintpaulia ionantha (Kotoujansky et al., 1982; Boucher et al., 1987; Glasner et al., 2011), and D. fangzhongdai CL3 isolated from taro, were grown in LB or LS5 medium at 28°C. LS5 is a nutrient poor medium (Liao et al., 2014), which induces hrp gene expression. Escherichia coli and the derived strains were grown in LB medium at 37°C.
To construct the reporter strain of hrpA promoter, a fragment containing the promoter region of hrpA, harboring a putative hrp-box (GGAACCATCTCTTGCTATCTCCTACTTA), was amplified using the primer pair PhrpA-F-HindIII (5′-ggaattggggatcggaagcttCTGGCCCGGCAACATCCGT-3′) and PhrpA-R-BamHI (5′-gagctcggtacccggggatccGCAACTTCATGCTATCCATAG-3′; Supplementary Figure S1) with MS2 genomic DNA as the template. The fragment was purified using NucleoSpin® Gel and PCR Clean-up kit (MACHEREY-NAGEL GmbH & Co. KG, Düren Neumann Neander, Germany) and ligated with the HindIII/BamHI-digested pPROBE-NT plasmid (Miller et al., 2000) using ClonExpress® MultiS kit (Vazyme Biotech Co., Nanjing, China). The resultant plasmid was verified by DNA sequencing and transformed into the strain MS2 by triparental conjugation (Zhou et al., 2016), which was designated as MS2 (pPhrpA-gfp).
To generate gacA deletion mutants, upstream and downstream fragments of the deleted genes were respectively amplified using primers gacA-1 & gacA-2, and gacA-3 & gacA-4 listed in Supplementary Table S1, and purified with NucleoSpin Gel and PCR Clean-up kit. The fragments were fused with the BamHI and SpeI digested suicide plasmid pKNG101 using ClonExpress® MultiS kit (Vazyme Biotech Co., Nanjing, China). The construct was transformed into E. coli CC118λ competent cells and introduced into strain MS2 by triparental conjugation using the method described previously (Zhou et al., 2016). The resultant gacA deletion mutant was confirmed by PCR using the primer pair gacA-F & gacA-R.
Antibiotics were added at the following final concentrations when required: kanamycin (Km), 50 μg/ml; and streptomycin (Sm), 50 μg/ml.
Sources of the Screened Compounds
The compounds used in this work are listed in Table 1. All compounds were purchased from Guangzhou Dingguo Biotechnology Co. LTD, and dissolved in dimethyl sulfoxide (DMSO) to corresponding concentrations.
Table 1.
Screening for inhibitors of Dickeya zeae MS2 type III secretion system (T3SS) by Multifunctional Microplate Reader.
| ID | Compound (0.2 mM) | Chemical structures | Average MFI ± SDa | DMSO%b |
|---|---|---|---|---|
| Phytohormone | ||||
| IAA | Indole-3-acetic acid |
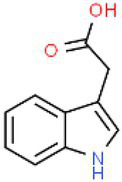
|
11,608.67 ± 137.42 | 96.58 |
| MeJA | Methyl Jasmonate |
|
12,568.33 ± 248.82 | 104.56 |
| SA | Salicylic acid |
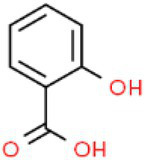
|
4,962.33 ± 77.69 | 41.28 |
| GA3 | Gibberellin A3 |
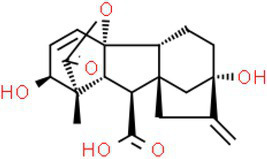
|
11,248.33 ± 178.20 | 93.58 |
| t-Z | Trans-zeatin |
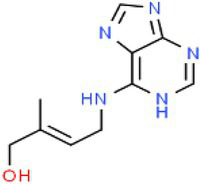
|
11,491.33 ± 436.43 | 95.60 |
| Other compounds | ||||
| HA | Hydrocinnamic acid |
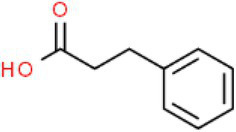
|
8,073.00 ± 87.43 | 67.16 |
| PHBA | p-Hydroxybenzoic acid |
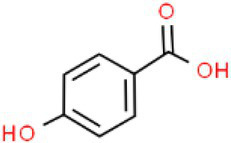
|
8,868.00 ± 62.07 | 73.77 |
| OCA | o-Coumaric acid |
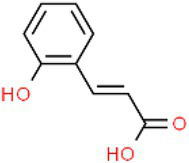
|
11,441.33 ± 170.00 | 95.18 |
| PCA | p-Coumaric acid |
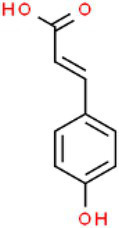
|
4,783.00 ± 168.59 | 39.79 |
| 2-CCA | 2-Chlorocinnamic acid |
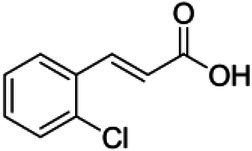
|
11,108.33 ± 85.23 | 92.41 |
| CA | Cinnamic alcohol |
|
6,198.00 ± 355.33 | 51.56 |
| AS | Acetosyringone |
|
17,213.00 ± 217.44 | 143.20 |
Average MFI ± SD of dimethyl sulfoxide (DMSO) was 12,020.33 ± 226.40.
Dickeya zeae MS2 cells carrying the green fluorescent protein (GFP) reporter pPhrpA were grown in LS5 supplemented with 0.2 mM compounds for 14 h. GFP mean fluorescence intensity (MFI) of bacterial cells was determined by Multifunctional Microplate Reader. Three replicates were used in each compound and three independent experiments were performed.
DMSO% represents the relative promoter activity of hrpA, calculated according to the formula: DMSO% = 100 × MFIcompound/MFIDMSO.
Detection of the Mean Fluorescence Intensity of hrpA Promoter Activity
MS2(pPhrpA-gfp) were cultured in LB medium supplemented with Km to OD600 of 2.0 by shaking at 200 rpm at 28°C, then transferred into LS5 medium added with kanamycin and 0.2 mM each tested compound listed in Table 1 at a ratio of 1:10, 100 μl of which was added into each well of polystyrene 96 well-tissue culture plate (Guangzhou Jet Bio-Filtration Co., Ltd., China). Three replicates were set for each culture. The plate was then placed in an incubator at 28°C and shaking-cultured in the dark at 200 rpm for 14 h. The fluorescence intensity and optical density (OD) of each well was detected by a Multifunctional Microplate Reader (Microplate reader, BioTek, United States). The excitation and emission wavelength of fluorescence intensity at 485 and 528 nm, respectively. The experiment was repeated three independent times. Equal volume of DMSO and LS5 medium was respectively used as the solvent control and blank control. The fluorescence intensity of LS5 medium supplemented with the compounds at the concentration of 0.2 mM was compared with that of LS5 medium and LS5 + DMSO to test whether individual compounds might have their own fluorescence.
To verify the screened inhibitor candidates, the hrpA promoter activity of the reporter strain was evaluated under corresponding concentrations by flow cytometer. Briefly, MS2(pPhrpA-gfp) was cultured in LB medium to OD600 of 2.0 at 28°C, and then transferred into LS5 medium containing the corresponding concentration of each compounds at a ratio of 1:100 and shaking-cultured in the dark at 200 rpm, 28°C for 14 h. Equal volume of DMSO was used as the solvent control. Three replicates were conducted for each compound treatment. Bacterial cells were resuspended in 1 × phosphate-buffered saline (PBS) to OD600 of 0.3. The fluorescence intensity of each treatment was detected by FACS-Caliber flow cytometer (CytoFLEX, Brea, CA, United States).
Measurement of Dickeya zeae MS2 Growth Treated With the Five Screened Compounds
To determine whether the screened compounds with effects on hrpA promoter activity have impacts on the growth of D. zeae MS2, we measured the optical densities of MS2 treated with different concentrations of the five compounds in nutrient-limited LS5 medium (used to measure the induced T3SS expression) using Bioscreen automatic growth curve analyzer (Bioscreen, Finland). Briefly, MS2 single colony was cultured in LB medium to OD600 of 2.0 at 28°C by shaking at 200 rpm, then transferred into LS5 medium added with the different concentrations of the tested compounds or DMSO at a ratio of 1:10, and 200 μl of which was dispended into each well of a 96 well-tissue culture plate. Growth of the bacterium was measured every 2 h at 600 nm for 44 h. At the same time, colony forming units (CFU) were counted by gradient dilution of the bacterial suspension at 14 h to analyze the effects of five compounds on bacterial growth. On each LB agar plate, 100 μl bacterial diluent was evenly spread and cultured at 28°C for 24 h. Plates with colony numbers between 30 and 300 were selected to count the CFU. The wide-type MS2 was used as the positive control. Three replicates were used in each compound and three independent experiments were performed.
Non-host Tobacco Plant Hypersensitive Response Assay
MS2 cells were grown in LB medium and shaking cultured at 200 rpm at 28°C to OD600 of 2.0, and then transferred into 10 ml of LS5 medium at a ratio of 1:100, shaking cultured till OD600 of 0.6. The bacterial cells were collected and suspended in LS5 medium added with optimal concentrations of the tested compounds or DMSO for incubation in the dark at 28°C for 4 h before leaf infiltration. Nicotiana tobacum K326 plants were used for HR assays (Hu et al., 2022), where 100 μl (4.8 × 106 CFU) of bacterial suspensions were press-infiltrated to tobacco leaves using 1.0 ml needleless syringes. HRs were photographed at 24 h. The area of lesions was measured using ImageJ software. Three replicates were used in each compound and three independent experiments were performed. Equal volume of MS2 + DMSO and LS5 + DMSO was used as the positive and negative control, respectively.
RNA Extraction and qRT-PCR Analysis
MS2 cells were cultured in LB medium to OD600 of 1.5 at 28°C by shaking at 200 rpm, and then transferred into LS5 medium containing optimal concentration of each compound at a ratio of 1:100 for shaking culture in the dark at 200 rpm at 28°C to OD600 of 0.8. Total MS2 RNA was isolated using SV total RNA isolated system kit (Promega, Madison, WI, United States), further purified using RNA clean kit (Qiagen, Hilden, Germany), and treated with DNaseI to degrade any possible DNA contamination, and then 1 μg of RNA was reverse transcribed using HiScriptII Q RT SuperMix Kit (Vazyme Biotech Co., Nanjing, China). The cDNA levels of different samples were quantified by real-time PCR (RT-PCR) using a SYBR Green Master Mix (Vazyme Biotech Co., Nanjing, China). For quantitative PCR, the cDNA template was diluted 40 times and 1 μl of cDNA was added to 20 μl of reaction system. Primer efficiency (between 90% and 107%) of each gene was determined using DNA standards at different concentrations. To calculate the relative expression level of target genes, the expression level of a housekeeping gene atpD was used as the internal control. The relative levels of gene expression were determined using the 2-ΔΔCT method (Livak and Schmittgen, 2001). All gene expression under compound treatment or no solvent (MS2) was compared with that of MS2 + DMSO using Student’s t-test analysis. Three technical replicates were used each time. The procedure for analysis on the expression of rpoN, hrpL, and gacA in wild-type MS2 and ΔgacA mutant was the same as above except that the strains were grown in LB medium. The expression of gacA in ΔgacA was used as a negative control. The primers of quantitative real-time PCR (qRT-PCR) are listed in Supplementary Table S1.
Pathogenicity Tests on Host Plants
To evaluate the efficacy of the five screened inhibitors on restraining the virulence or infection of Dickeya, different Dickeya bacteria including D. zeae MS2, D. oryzae EC1, D. dadantii 3937, and D. fangzhongdai CL3, were grown overnight in LB medium with shaking at 200 rpm at 28°C and then diluted into 10 ml of fresh LS5 at a ratio of 1:100 and grown to OD600 of 1.0. The bacterial cells were collected and suspended in LS5 medium supplemented with optimal concentrations of the tested compounds or DMSO. The cell suspensions were incubated in the dark at 28°C for 4 h before plant inoculation. Equal volume of MS2 + DMSO and LS5 + DMSO was used as the positive and negative control, respectively.
For inoculation on dicotyledonous potato and monocotyledonous taro, tubers were sliced in 5 mm-thickness and dried for about 20 min at room temperature. An aliquot of 2 μl (9.6 × 104 CFU) of D. dadantii 3937 bacterial cells were then inoculated on the center of potato slices, and 5 μl (2.4 × 105 CFU) of CL3 bacterial cells were inoculated on the center of taro slices. After inoculation, tissue slices were placed in a growth chamber with conditions of 28 ± 2°C and 75 ± 15% relative humidity for about 24 h. Image J software was used to measure the area of lesions.
To test the effect of the five compounds on controlling rice foot rot disease, eight rice seedlings were planted in each pot and grown for 3 weeks. Firstly, a 1.0 ml sterile needle syringe was used to stab the roots of the seedlings, 175 ml (8.4 × 109 CFU) of the EC1 and inhibitor mixture (incubated for 4 h) was poured into the pot. All pots were placed in the growth chamber with conditions of 28°C and 95% relative humidity with 12 h alternating light–dark cycles for 3 days. Three replicates were used in each compound and three independent experiments were performed. Equal volume of LS5 liquid medium was used as the blank control.
For inoculation on banana (Musa ABB), 3~4 leaf seedlings were selected and acclimated for 2 weeks prior at 25°C with 12 h alternating light–dark cycles before inoculation. MS2 bacterial suspensions in 500 μl (2.4 × 107 CFU) were injected into the center of banana pseudostem using a 1.0 ml needleless syringe. Plants were placed in the growth chamber with conditions of 28°C and 95% relative humidity with 12 h alternating light–dark cycles for 7 days. The severity of disease in rice and banana was assessed using the virulence scoring method described in our previous study (Feng et al., 2019; Hu et al., 2022).
Statistical Analysis
GraphPad Prism 8.4.3 software was used for statistical analysis. The results were analyzed by Student’s t-test.
Results
Identification of Small Molecule Compounds That Inhibit or Induce the hrpA Promoter Activity of MS2
As we all know, new drugs targeting T3SS that can block its function without affecting the growth and survival of bacteria have been found in many bacteria, which undoubtedly becomes a new strategy for the prevention and treatment of bacterial diseases. To screen T3SS inhibitors or inducers of D. zeae, a promoter-gfp fusion plasmid was firstly constructed, which includes a promoter of hrpA of D. zeae banana strain MS2 (Hu et al., 2018; Feng et al., 2019), a green fluorescent protein (GFP) encoding gene and a kanamycin resistance gene. Secondly, the resultant plasmid pPhrpA-gfp was transformed into the MS2 parental cells and grown in T3SS-inducing LS5 medium supplemented with each tested compound at a concentration of 0.2 mM for 14 h. Next, the mean fluorescence intensity (MFI) representing the promoter activity of hrpA was detected by microplate reader, and the compounds that impact the promoter activity of hrpA were preliminarily screened out. Among the compounds screened, five compounds at a concentration of 0.2 mM showed obvious inhibitory effect on hrpA promoter activity, which are SA, p-hydroxybenzoic acid (PHBA), CA, PCA, and hydrocinnamic acid (HA), respectively (Table 1). These five compounds are not self-fluorescent at the concentration of 0.2 mM (Supplementary Figure S2).
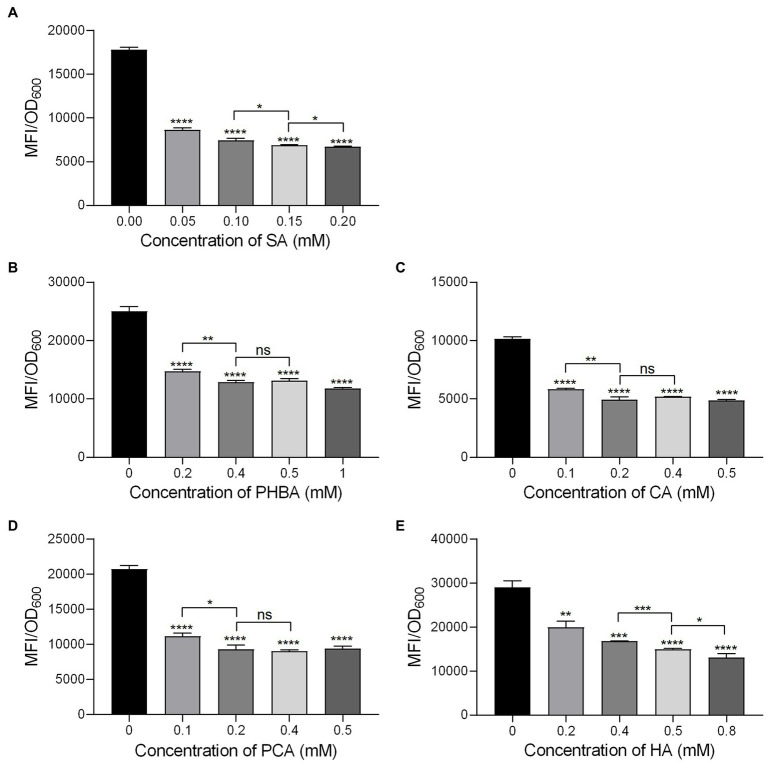
In order to determine the optimal concentration of the compounds to inhibit the hrpA promoter activity, the MFI of MS2(pPhrpA-gfp) treated with different concentrations of the five compounds was detected by microplate reader. The results showed that with the increase of concentration, the MFI value of MS2(pPhrpA-gfp) under the treatment of the five compounds decreased to different degrees (Figure 1). For compounds SA and HA, concentrations of 0.2 and 0.8 mM, respectively achieved strongest inhibitory activity of hrpA promoter in their corresponding assay (Figures 1A,E), but could affect the growth of MS2(pPhrpA-gfp) under these two concentrations (Supplementary Figure S3). About 0.2 mM PCA slightly promoted bacterial growth (Supplementary Figure S3). Therefore, the optimal concentrations of the five compounds acting on the hrpA promoter activity at 14 h of bacterial growth were determined as 0.15 mM SA, 0.4 mM PHBA, 0.2 mM CA, 0.2 mM PCA, and 0.5 mM HA (Figure 1; Supplementary Figure S3), which were used for the following study. Then the five compounds at respective optimal concentration were measured for their alterations in hrpA promoter activity through the Multifunctional Microplate Reader and FACS-Caliber flow cytometer, the results showed that compared with the DMSO control, the MFI decreased to less than 46.93 and 36.96%, respectively, and the inhibitory rate of hrpA promoter activity was more than 53.07 and 63.04%, respectively (Table 2), indicating that all the five compounds could significantly inhibit the hrpA promoter activity.
Figure 1.
The hrpA promoter activity of MS2 in LS5 medium supplemented with different concentrations of salicylic acid (SA; A), p-hydroxybenzoic acid (PHBA; B), cinnamyl alcohol (CA; C), p-coumaric acid (PCA; D), and hydrocinnamic acid (HA; E) at 14 h of growth. MS2(pPhrpA-gfp) cultures (OD600 of 2.0) were transferred into the LS5 medium supplemented with different concentrations of tested compounds or DMSO at a ratio of 1:10, and then dispended into a 96 well-tissue culture plate (100 μl per well) for cultivation in the dark for 14 h. MFI was measured by a Multifunctional Microplate Reader. DMSO was used as the positive control. Three replicates were used in each compound and three independent experiments were performed with similar results. For statistical analysis, GraphPad Prism 8.4.3 software was used to perform Student’s t-test. Asterisks indicate statistically significant differences (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
Table 2.
The hrpA promoter activity of MS2 in LS5 medium supplemented with optimal concentrations of compounds at 14 h of growth and the corresponding inhibition rates of the compounds toward hrpA promoter activity.
| Multifunctional Microplate Reader | Flow cytometer | |||||
|---|---|---|---|---|---|---|
| Compound | MFI ± SDa | DMSO%b | Inhibition rate% (100%-DMSO%) | MFI ± SDa | DMSO%b | Inhibition rate% (100%-DMSO%) |
| DMSO | 16,605.33 ± 75.61 | / | / | 11,520.60 ± 349.49 | / | / |
| 0.15 mM SA | 6,660.67 ± 35.16 | 40.11 | 59.89 | 2,201.97 ± 177.32 | 19.11 | 80.89 |
| 0.4 mM PHBA | 7,792.67 ± 108.04 | 46.93 | 53.07 | 4,258.57 ± 251.78 | 36.96 | 63.04 |
| 0.2 mM CA | 7,082.00 ± 55.46 | 42.65 | 57.35 | 4,005.77 ± 100.93 | 34.77 | 65.23 |
| 0.2 mM PCA | 6,569.00 ± 84.55 | 39.56 | 60.44 | 2,622.47 ± 154.13 | 22.76 | 77.24 |
| 0.5 mM HA | 7,549.67 ± 59.53 | 45.47 | 54.53 | 2,988.77 ± 118.56 | 25.94 | 74.06 |
Dickeya zeae MS2 cells carrying the GFP reporter pPhrpA were grown in LS5 supplemented with optimal concentration of each compound for 14 h. GFP MFI of bacterial cells was determined by Microplate Reader and Flow Cytometer. Three replicates were used in each compound and three independent experiments were performed.
DMSO% represents the relative promoter activity of hrpA, calculated according to the formula: %DMSO = 100 × MFIcompound/MFIDMSO.
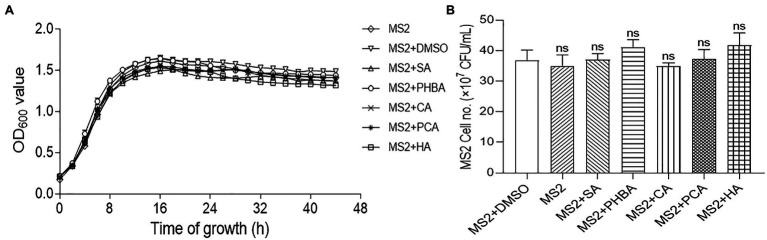
Growth of MS2 Is Not Affected by the Five Tested Compounds at Respective Optimal Concentrations
To exclude the possibility that the decrease in MFI was due to the influence of the compounds on bacterial growth, we tested the effects of the compounds on the growth of D. zeae MS2. Firstly, the growth rate of MS2 in LS5 medium supplemented with each of the compounds at optimal concentration or DMSO was measured by Bioscreen automatic growth curve analyzer. Results showed that compared with the untreated control, addition of either DMSO or compounds SA, PHBA, CA, PCA, or HA at their respective optimal concentration had no obvious impact on MS2 growth rate at different time points (Figure 2A). Secondly, the live bacterial cells of MS2 were also counted at 14 h of bacterial growth. Similar result was obtained that the five compounds had no significant effect on the CFU of MS2 compared with the medium or DMSO control (Figure 2B), indicating that the five compounds at respective optimal concentration do not affect the normal growth and survival of D. zeae MS2.
Figure 2.
Effects of SA (0.15 mM), PHBA (0.4 mM), CA (0.2 mM), PCA (0.2 mM), and HA (0.5 mM) on the growth of MS2 in LS5. (A) The growth curves of MS2 in LS5 supplemented with DMSO or tested compounds, respectively. MS2 culture (OD600 of 2.0) was transferred into LS5 medium supplemented with candidate concentrations of tested compounds at a ratio of 1:10, and then dispended into a 96 well-tissue culture plate (200 μl per well). Optical density (OD600) of bacterial cultures was recorded every 2 h during a 44-h period using a Bioscreen automatic growth curve analyzer. Each point indicates the average value of three replicates with error bars representing the SD. (B) Colony forming units (CFU) of MS2 at 14 h of growth in LS5 supplemented with DMSO or tested compounds, respectively. Bacterial suspensions were diluted in gradient, 100 μl of which was spread on LB agar plate and cultured at 28°C for 24 h. Plates with colony numbers between 30 and 300 were selected to count the CFU. MS2 + DMSO was used as the positive control. The data were the mean of three replicates and subjected to Student’s t-test analysis by Graphpad Prism 8.4.3 (ns, no statistical significance).
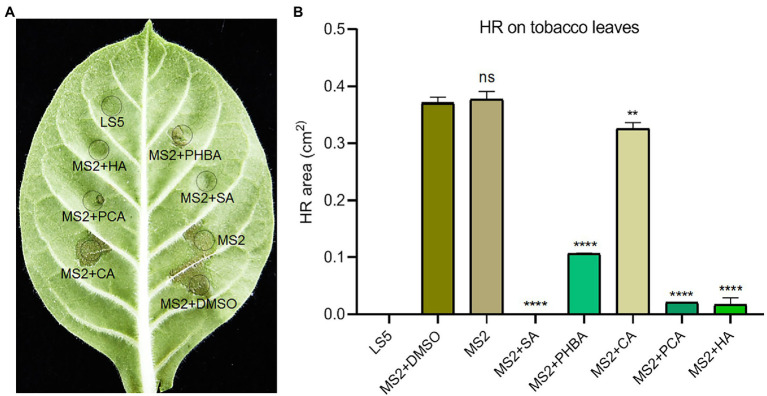
The Five Compounds Suppress HR Caused by MS2 on Tobacco
MS2 cells with functional T3SS can induce HR on non-host tobacco leaves. To test the influence of the five compounds on the elicitation of HR, we added the above five compounds to the MS2 suspensions at their respective optimal concentration for incubation for 2 h and then infiltrated them into tobacco leaves using sterile 1.0 ml needleless syringes. The MS2 or MS2 suspended in DMSO triggered visible HR symptoms on tobacco leaves after 24 h post inoculation (hpi), when addition of the five compounds significantly suppressed the HR reaction on tobacco leaves (Figure 3). Specifically, 0.15 mM SA completely inhibited the HR of MS2 on tobacco, followed by 0.5 mM HA and 0.2 mM PCA with up to 94 and 95% inhibition effect on HR, respectively (Figure 3). Among the five compounds, 0.2 mM CA got the weakest inhibition effect on HR response induced by MS2 (Figure 3).
Figure 3.
Effects of SA (0.15 mM), PHBA (0.4 mM), CA (0.2 mM), PCA (0.2 mM), and HA (0.5 mM) on the hypersensitive response (HR) induced by MS2 on tobacco leaves. (A) HR symptoms on Nicotiana tabacum K326 leaves. Cell suspensions of MS2 at OD600 of 0.6 were incubated in the dark with optimal concentrations of the tested compounds or DMSO for 4 h, then 100 μl of bacterial suspensions were press-infiltrated to tobacco leaves. HRs were photographed at 24 h. (B) The area of HR lesions. ImageJ software was used to measure the area of lesions. MS2 + DMSO was used as the positive control. The data were the mean of three replicates and subjected to Student’s t-test analysis by Graphpad Prism 8.4.3 (ns, no statistical significance, **p < 0.01, ****p < 0.0001).
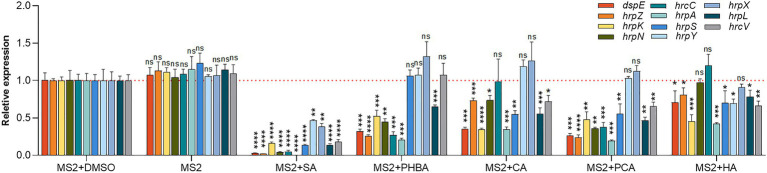
Inhibitory Effect of the Five Compounds on the Transcription of T3SS-Related Genes
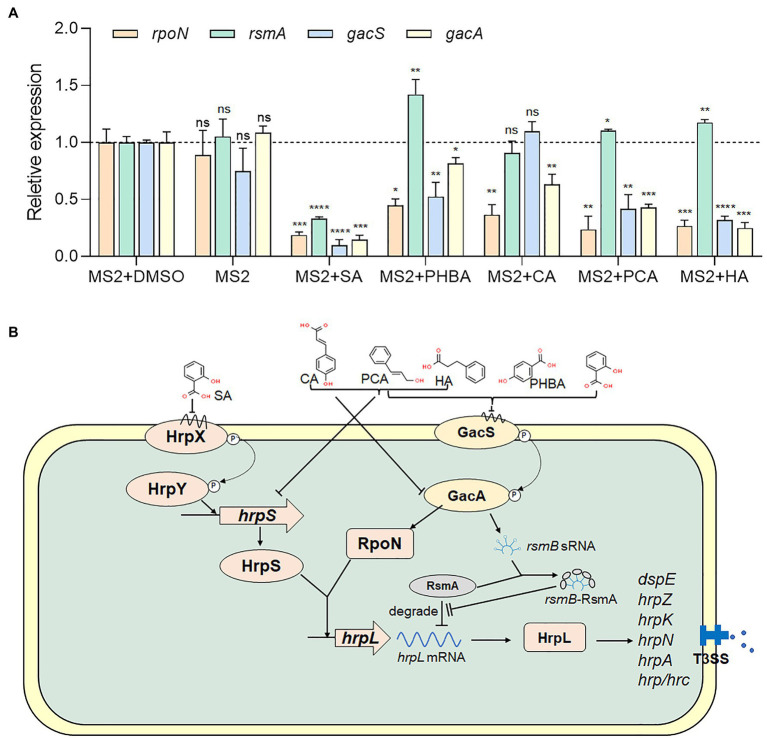
To understand whether the five screened compounds have inhibitory effects on the expression of T3SS-related genes of MS2, qRT-PCR was performed to measure relative transcriptional levels of some representative genes in the dsp/hrp/hrc gene clusters. The qRT-PCR results demonstrated that, compared with the MS2 + DMSO control, the expression levels of many T3SS tested genes were significantly altered when MS2 was incubated with the optimal dosage of each of the five molecules (Figure 4). In the presence of 0.15 mM SA, the mRNA expression levels of all the tested T3SS-related genes were significantly decreased, and in the presence of 0.4 mM PHBA, 0.2 mM CA, 0.2 mM PCA, or 0.5 mM HA, the transcription level of partial T3SS genes was significantly lower than that of the solvent control (Figure 4). It is worth noting that the mRNA level of hrpL was reduced by more than 35% in the presence of four inhibitors (SA, PHBA, CA, and PCA), in which, 0.15 mM SA reduced the mRNA level of hrpL by approximately 90%. HA in 0.5 mM slightly affected the expression of hrpL, and dramatically reduced the mRNA expression level of hrpA (Figure 4), suggesting that HA may directly actions on the hrpA. Our results indicated that SA, PHBA, CA, and PCA suppress T3SS gene expression probably through the T3SS master regulator HrpL. Current investigation about the regulation of T3SS reveals that the expression of HrpL is regulated by several regulatory pathways including the GacS/GacA-RsmB-RsmA pathway and the σ54-containing RNA polymerase holoenzyme RpoN, apart from the HrpX/HrpY-HrpS-RpoN pathway (Chatterjee et al., 2002; Yap et al., 2005; Yuan et al., 2020). qRT-PCR analysis indicated that the expression of rpoN and gacA is regulated by all of these five compounds in consistent pattern, while the expression of rsmA is only downregulated by 0.15 mM SA (Figure 5A). To determine whether there is any interaction between RpoN and GacA, qRT-PCR analysis was performed to measure the rpoN and gacA expression in the ΔgacA mutant (ΔrpoN was not obtained in this study). Results showed that mutation of gacA resulted in decreased expressions of the rpoN by 1.96-fold, which indicates that GacA positively regulates rpoN (Supplementary Figure S4). Similarly, we examined the hrpL expression and found that the transcript level of hrpL was downregulated by 4.2-fold in the ΔgacA mutant (Supplementary Figure S4). Thus, the mechanism mode for the T3SS-inhibiting compounds was drawn as Figure 5B.
Figure 4.
Effects of SA (0.15 mM), PHBA (0.4 mM), CA (0.2 mM), PCA (0.2 mM), and HA (0.5 mM) on the expression of T3SS-related genes in MS2. MS2 cells were cultured in LS5 medium added with optimal concentrations of the tested compounds or DMSO. RNA was collected at OD600 of 0.8. The cDNA levels of different samples were quantified by real-time PCR (RT-PCR) using a SYBR Green Master Mix. A housekeeping gene atpD was used as an endogenous control for data analysis. All gene expression under compound treatment or no solvent (MS2) was compared with that of MS2 + DMSO using Student’s t-test analysis (Graphpad Prism 8.4.3). Three independent tests were performed with similar results (ns, no statistical significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
Figure 5.
Regulation of SA, PHBA, CA, PCA, and HA on T3SS. (A) Effects of SA (0.15 mM), PHBA (0.4 mM), CA (0.2 mM), PCA (0.2 mM), and HA (0.5 mM) on the expression of rpoN, rsmA, gacS, and gacA. RNA was collected at a bacterial concentration of OD600 of 0.8. The cDNA levels of different samples were quantified by RT-PCR using a SYBR Green Master Mix. A housekeeping gene atpD was used as an endogenous control for data analysis. All gene expression under compound treatment or no solvent (MS2) was compared with that of MS2 + DMSO using Student’s t-test analysis (Graphpad Prism 8.4.3). Three independent tests were performed with similar results (ns, no statistical significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001). (B) Regulatory network controlling the Dickeya zeae T3SS. In Dickeya bacteria, the expression of T3SS is regulated by a master regulator, HrpL (Hu et al., 2022). On one hand, hrpL upregulates many hrp genes that encode the T3SS structural and functional proteins, such as hrpA, hrpN, and dspE (Hu et al., 2022). On the other hand, the expression of hrpL is regulated by the HrpX/HrpY-HrpS-HrpL pathway at the transcriptional level and the GacS-GacA-RsmB-RsmA pathway at the post-transcriptional level (Chatterjee et al., 2002; Yap et al., 2005; Yuan et al., 2020). ⊥, negative control; →, positive control.
The Five T3SS Inhibitors Have Good Performance on Alleviating Crop Soft Rot Caused by Dickeya
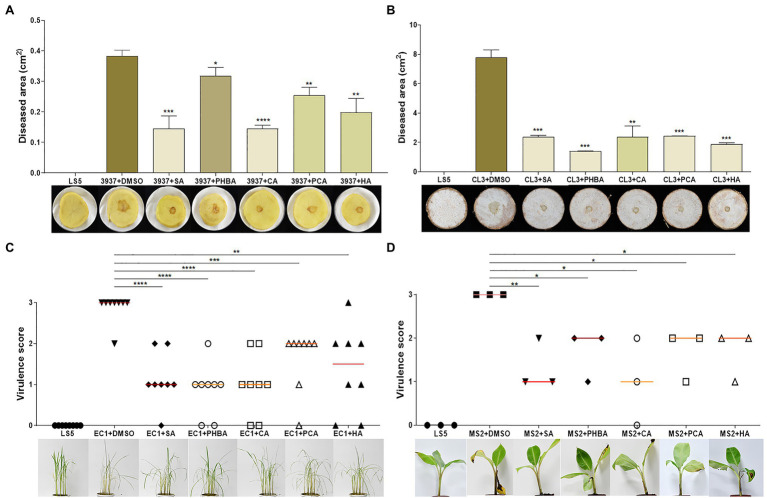
To test whether the above five T3SS inhibitors could affect the ability of the Dickeya to induce disease symptoms in soft rot diseases, we incubated the five compounds with several Dickeya pathogens isolated from different sources for 4 h and then inoculated them into dicotyledonous potato slices, and monocotyledonous banana and rice seedlings as well as taro slices. After 24 hpi, the potato and taro slices inoculated with Dickeya bacteria supplemented with the five T3SS inhibitors at their optimal concentration significantly reduced soft rot disease symptoms compared with those inoculated with controls of Dickeya + DMSO (Figures 6A,B).
Figure 6.
Inhibition of SA (0.15 mM), PHBA (0.4 mM), CA (0.2 mM), PCA (0.2 mM), and HA (0.5 mM) against crop soft rot disease on potato (A), taro (B), rice (C), and banana (D). Cell suspensions of Dickeya dadantii 3937, D. fangzhongdai CL3, D. oryzae EC1, and D. zeae MS2 at OD600 of 1.0 were incubated in the dark with optimal concentrations of the tested compounds or DMSO for 4 h. About 2 μl (9.6 × 104 CFU) of D. dadantii 3937 and 5 μl (2.4 × 105 CFU) of CL3 bacterial cells were respectively applied on the center of potato and taro slices, 500 μl (2.4 × 107 CFU) of MS2 bacterial cells were injected into the pseudostems of banana seedlings, and 175 ml (4.8 × 109 CFU) of EC1 bacterial cells were irrigated into pots with rice seedlings after needle punctures on stem bases. Three potato slices, taro slices, and banana seedlings, and eight rice seedlings were inoculated each time, and three independent tests were performed with similar results. MS2 + DMSO was used as the positive control. The data were subjected to Student’s t-test analysis by Graphpad Prism 8.4.3 (ns, no statistical significance, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
We further investigated the effect of D. oryzae EC1 on rice seedlings after treatment with five compounds. On EC1 + DMSO-infected rice, apparent disease symptoms were observed after 3 days, including the base of the stem appeared brown, the middle stem and leaves turned yellow and the leaves showed signs of water loss and wilting (Figure 6C). However, these symptoms were significantly reduced by treatment of the bacterial germs with five inhibitors (Figure 6C). Additionally, bacterial suspensions treated with compounds were injected into the center of banana pseudostem using a 1.0 ml syringe. After a week of incubation, the results showed that compared with the control groups, the soft rot symptoms caused by MS2 on banana seedlings were alleviated and weakened (Figure 6D).
Discussion
Dickeya is one of the top 10 important bacterial phytopathogens in the world, and the bacterial soft rot caused by Dickeya spp. often results in serious economic losses to crop yields, especially on rice, potato, banana, and taro (Mansfield et al., 2012; van der Wolf et al., 2014; Zhang et al., 2014; Li et al., 2020; Huang et al., 2021). Plant molecules play an important role in host–microbe interaction. An increasing number of studies have proven that plant natural compounds such as cinnamaldehyde, cinnamic, coumaric acid, SA, carvacrol, syringic, and catechol via many virulence pathways like quorum sensing (QS), T3SS, motility, biofilm formation, and exoenzyme activity directly affect pathogenicity of Pectobacterium and/or Dickeya (Li et al., 2009; Joshi et al., 2015, 2016, 2020, 2021; Jiang et al., 2021). T3SS is one of the key virulence factors in many Gram-negative bacteria including Dickeya. Since it is well conserved, T3SS could serve as a good candidate target for the development of novel antibacterial agents (Baron, 2010; Kline et al., 2012). In this work, five phytohormone and seven chemical compounds were evaluated for their suppression of T3SS gene expression in D. zeae MS2. Among these, five plant natural products SA, PHBA, CA, PCA, and HA suppressed T3SS gene expression, and we also demonstrated that these compounds are able to suppress the HR of MS2 on non-host tobacco leaves and disease symptoms on host crops. It would be important for agricultural production to have a deeper understanding of how they affect the function of the T3SS in Dickeya.
As a master regulator of T3SS, HrpL is transcriptionally regulated by the HrpX/HrpY-HrpS-RpoN pathway and post-transcriptionally regulated by the GacS/GacA-RsmB-RsmA pathway (Yuan et al., 2020). In which, RpoN, a σ54-containing RNA polymerase holoenzyme, interacts with the σ54 enhancer-binding protein HrpS and initiates the transcription of hrpL (Chatterjee et al., 2002; Yap et al., 2005; Figure 5B).
We further analyzed which specific pathway(s) is affected by the five screened compounds in D. zeae MS2. Results indicated that all the five compounds SA (0.15 mM), PHBA (0.4 mM), CA (0.2 mM), PCA (0.2 mM), and HA (0.5 mM) significantly inhibited the expression of the master regulator HrpL, and SA had the most obvious inhibitory effect on the expression of all of the tested T3SS genes (Figure 4). Besides transcriptional inhibition through the HrpX/HrpY-HrpS-RpoN pathway, it inactivated HrpL through the GacS/GacA-RsmB-RsmA pathway (Figure 5). As a signal molecule in plants, SA is required for the induction of systemic acquired resistance as well as the activation of defense responses against biotrophic and hemi-biotrophic pathogens (Bari and Jones, 2009). A number of studies have found that SA can change the motility, biofilm formation, exoenzyme activity, and the pathogenicity of bacteria (Joshi et al., 2015; Cattò et al., 2017; Ahmed et al., 2019). Furthermore, SA has also been reported to reduce the expression of vir genes and reset virulence via SghR/SghA pathway in Agrobacterium tumefaciens (Yuan et al., 2007; Anand et al., 2008; Wang et al., 2019). It has also been shown to be a QS inhibitor for multiple bacterial species, including P. aeruginosa, Pectobacterium, and A. tumefaciens (Chang et al., 2014; Subramoni et al., 2014; Joshi et al., 2016, 2020; Ahmed et al., 2019). Previous studies have found that SA affects the hrpA promoter activity of several bacterial pathogens, including E. amylovora and P. aeruginosa (Yamazaki et al., 2012; Khokhani et al., 2013). This study identified the specific pathways of SA affecting hrpA expression and thus the virulence of Dickeya spp. on crops.
Naturally occurring PHBA is produced mainly by plants. Early studies have shown that PHBA has antibacterial and antioxidant activities against a variety of Gram-positive and Gram-negative bacteria (Rice-Evans et al., 1996; Cho et al., 1998). Recently, it was found that PHBA significantly downregulated the transcription of some genes in hrp/hrc gene cluster of P. syringae pv. tomato DC3000, and alleviated the disease symptoms of tomato leaves (Kang et al., 2020). In this study, PHBA did not affect the growth of D. zeae MS2 or alter the hrpX, hrpY, and hrpS mRNA levels, but obviously reduced the mRNA levels of rpoN, gacS, gacA, and other downstream regulatory genes of hrpL (Figures 4, 5A), suggesting that PHBA may directly act on gacS to inhibit the transcription of hrpL.
Cinnamyl alcohol and PCA, like PHBA, did not play a role through the two-component signal transduction system HrpX/HrpY, but inhibited the expression of hrpS, rpoN, and gacS (CA did not affect gacS), indicating CA and PCA inhibit hrpL transcription through both hrpS and gacA. PCA has been shown to inhibit the transcription of D. dadantii T3SS through HrpX/HrpY-HrpS-HrpL regulatory pathway (Li et al., 2009), rather than through the global regulator GacS/GacA, but our data revealed significantly lower gacS and gacA mRNA levels in cells grown in LS5 medium supplemented with PCA in comparison with DMSO control (Figure 5A). The contradiction may be attributed to the different signal transduction pathways of different strains. These results suggest that PCA inhibits hrpL through both the HrpS-RpoN (but not HrpX/HrpY) and the GacS/GacA pathways, and consequently lowers the expression of HrpL regulon genes. CA has also been shown to affect the hrpA promoter activity of D. dadantii and E. amylovora, but whether they alter other T3SS gene expression has not been confirmed (Khokhani et al., 2013; Li et al., 2015).
Hydrocinnamic acid has been previously discovered to affect the hrpA promoter activity of E. amylovora (Khokhani et al., 2013). In P. aeruginosa, HA inhibited QS and its regulatory genes (Sharma et al., 2019). In this study, HA showed a similar regulatory pattern to PCA except the unobvious effect on the expression of hrpN and hrcC, and a weak effect on hrpY (Figure 4).
In P. syringae pv. tomato DC3000, gacA mutation significantly attenuated the transcription of rpoN, but regulatory mechanism was unknown (Chatterjee et al., 2003). Another study found that the expression of the gacA global regulatory gene was significantly increased during the entire growth cycle in a rpoN mutant of P. aeruginosa PAO1 (Heurlier et al., 2003). This indicates that gacA and rpoN are correlated in some bacteria. Our study revealed the coordinate regulation patterns of gacA and rpoN (Figure 5A), suggesting the correlation of them in regulating T3SS in D. zeae MS2. In the ΔgacA mutant, rpoN expression decreased by 1.96-fold, suggesting positive regulation of GacA on the transcription of rpoN (Supplementary Figure S4), similar to that in Erwinia spp., Pantoea stewartii, and P. syringae (Chatterjee et al., 2003; Tang et al., 2006). Whether RpoN regulates gacA needs further study.
Type III secretion system is typically characterized by direct injection of T3SEs into host cells, leading to disease resistance of host plants and HR reaction. In our previous study, 50 putative T3SEs were predicted in MS2 genome based on a combination of four state-of-the-art bioinformatic tools (i.e., Bastion3, BEAN2, DeepT3, and pEffect), and 13 of which was demonstrated to be positively regulated by HrpL (Hu et al., 2022). Among them, only six include hrp box (GGAACC/T-N15/16-C/T/GCACNNA) in their promoter regions, such as DspE, HrpN, HrpW, HrpZ, HrpK, and HrpJ (Hu et al., 2022). In Dickeya bacteria, only DspE, HrpN, and HrpW have been characterized to be secreted via T3SS (Glasner et al., 2011). Although, we did not test the secretion of these T3SEs, it is probably affected under the treatment of the five inhibitors, since the expression of dspE, hrpZ, hrpK, and hrpN, except hrpN by HA treatment, was significantly reduced (Figure 4). Furthermore, from our recent study, T3SEs also function as virulence factors promoting the development of tissue maceration on host plants (Hu et al., 2022). HR is a programmed cell death reaction. After syringe infiltration in the tobacco, we found that MS2 strain treated with the five compounds showed weaker HR compared to the control (Figure 3). Specifically, the inhibition rate of SA, PCA, and HA on HR at least 94% (Figure 3), consistent with the result that most T3SS gene expression is inhibited (Figure 4). PHBA and CA also suppressed the HR remarkably. Finally, virulence assays in planta were performed to evaluate the inhibitory effects of five inhibitors in suppressing soft rot symptoms caused by different Dickeya pathogens on host plants. According to our results, the five compounds showed particularly obvious inhibitory effects on virulence in different species of Dickeya bacteria (D. dadantii 3,937, D. fangzhongdai CL3, D. oryzae EC1, and D. zeae MS2; Figure 6). It has been previously reported that cinnamic acid and SA affect the QS machinery of Pectobacterium, and completely suppress disease symptoms (Joshi et al., 2016). In our study, five plant-derived compounds failed to completely suppressed soft rot symptoms. This might be because other virulence factors or regulatory factors were still functional.
It is an important way to develop biosafety control agents from plant or microbial origin that inhibit plant pathogenic bacteria T3SS. In our study, we have shown the inhibitory effects of five plant natural products SA, PHBA, CA, PCA, and HA on the T3SS of MS2 both in vitro and in planta. They significantly inhibited the hrpA promoter activity and reduced the expression level of T3SS genes without affecting the growth and survival of D. zeae MS2. Furthermore, these compounds have been proven to be effective in attenuating the soft rot symptoms caused by different species of Dickeya bacteria on host crops. We also elucidated their possible regulatory pathways. The regulatory pathways of all of the compounds identified that were active against D. zeae have never been identified previously. This study also reports the universal functions of plant natural products SA, PHBA, CA, PCA, and HA on reducing the virulence of soft rot Dickeya bacteria. These results indicate that they play an important role in host–microbe interaction and have the potential to be used as natural, safe, and effective plant-derived biocontrol agents to cure plant diseases caused by Dickeya pathogens.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
JZ conceived and designed the experiments. MH constructed the reporter plasmid MS2(pPhrpA-GFP). AH and XT screened the compounds and performed RNA extraction, cDNA synthesis, and qRT-PCR analysis. AH, MH, SC, and YX performed the pathogenicity tests and analyzed the results. AH, JZ, and MH wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work was financially supported by grants from the Key-Area Research and Development Program of Guangdong Province (2020B0202090001 and 2018B020205003), the Guangzhou Basic Research Program (202102080613), the National Natural Science Foundation of China (31972230), the Natural Science Foundation of Guangdong Province, China (2020A1515011534), the Science and Technology Planning Project of Shaoguan City (200805094530618), and the Cultivation of Guangdong College Students’ Scientific and Technological Innovation (pdjh2019b0082).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank all the students in the research group and the Integrate Microbiology Research Center, South China Agricultural University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.839025/full#supplementary-material
References
- Ahmed S., Rudden M., Smyth T. J., Dooley J. S. G., Marchant R., Banat I. M. (2019). Natural quorum sensing inhibitors effectively downregulate gene expression of Pseudomonas aeruginosa virulence factors. Appl. Microbiol. Biotechnol. 103, 3521–3535. doi: 10.1007/s00253-019-09618-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello D., Williams J. D., Majgier-Baranowska H., Patel I., Peet N. P., Huang J., et al. (2010). Discovery and characterization of inhibitors of Pseudomonas aeruginosa type III secretion. Antimicrob. Agents Chemother. 54, 1988–1999. doi: 10.1128/AAC.01598-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A., Uppalapati S. R., Ryu C. M., Allen S. N., Kang L., Tang Y., et al. (2008). Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 146, 703–715. doi: 10.1104/pp.107.111302, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharajah A., Buyck J. M., Sundin C., Tulkens P. M., Mingeot-Leclercq M. P., Van Bambeke F. (2017). Salicylidene acylhydrazides and kydroxyquinolines act as inhibitors of type three secretion systems in Pseudomonas aeruginosa by distinct mechanisms. Antimicrob. Agents Chemother. 61, e02566–e02616. doi: 10.1128/AAC.02566-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Jones J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0, PMID: [DOI] [PubMed] [Google Scholar]
- Baron C. (2010). Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 13, 100–105. doi: 10.1016/j.mib.2009.12.003, PMID: [DOI] [PubMed] [Google Scholar]
- Boucher C. A., Van Gijsegem F., Barberis P. A., Arlat M., Zischek C. (1987). Pseudomonas solanacearum genes controlling both pathogenicity on tomato and hypersensitivity on tobacco are clustered. J. Bacteriol. 169, 5626–5632. doi: 10.1128/jb.169.12.5626-5632.1987, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowlin N. O., Williams J. D., Knoten C. A., Torhan M. C., Tashjian T. F., Li B., et al. (2014). Mutations in the Pseudomonas aeruginosa needle protein gene pscF confer resistance to phenoxyacetamide inhibitors of the type III secretion system. Antimicrob. Agents Chemother. 58, 2211–2220. doi: 10.1128/AAC.02795-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner D. (2012). Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. doi: 10.1128/MMBR.05017-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattò C., Grazioso G., Dell'Orto S., Gelain A., Villa S., Marzano V., et al. (2017). The response of Escherichia coli biofilm to salicylic acid. Biofouling 33, 235–251. doi: 10.1080/08927014.2017.1286649, PMID: [DOI] [PubMed] [Google Scholar]
- Chang C. Y., Krishnan T., Wang H., Chen Y., Yin W. F., Chong Y. M., et al. (2014). Non-antibiotic quorum sensing inhibitors acting against N-acyl homoserine lactone synthase as druggable target. Sci. Rep. 4:7245. doi: 10.1038/srep07245, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charro N., Mota L. J. (2015). Approaches targeting the type III secretion system to treat or prevent bacterial infections. Expert Opin. Drug Discovery 10, 373–387. doi: 10.1517/17460441.2015.1019860, PMID: [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Cui Y., Chatterjee A. K. (2002). Regulation of Erwinia carotovora hrpL(Ecc) (sigma-L(Ecc)), which encodes an extracytoplasmic function subfamily of sigma factor required for expression of the HRP regulon. Mol. Plant-Microbe Interact. 15, 971–980. doi: 10.1094/MPMI.2002.15.9.971, PMID: [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Cui Y., Yang H., Collmer A., Alfano J. R., Chatterjee A. K. (2003). GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant-Microbe Interact. 16, 1106–1117. doi: 10.1094/MPMI.2003.16.12.1106, PMID: [DOI] [PubMed] [Google Scholar]
- Cheng Y. Y., Liu X. L., An S. W., Chang C. Q., Zou Y. Q., Huang L. H., et al. (2013). A nonribosomal peptide synthase containing a stand-alone condensation domain is essential for phytotoxin zeamine biosynthesis. Mol. Plant-Microbe Interact. 26, 1294–1301. doi: 10.1094/MPMI-04-13-0098-R, PMID: [DOI] [PubMed] [Google Scholar]
- Cho J. Y., Moon J. H., Seong K. Y., Park K. H. (1998). Antimicrobial activity of 4-hydroxybenzoic acid and trans 4-hydroxycinnamic acid isolated and identified from rice hull. Biosci. Biotechnol. Biochem. 62, 2273–2276. doi: 10.1271/bbb.62.2273, PMID: [DOI] [PubMed] [Google Scholar]
- Czajkowski R., Pérombelon M. C. M., van Veen J. A., van der Wolf J. M. (2011). Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: a review. Plant Pathol. 60, 999–1013. doi: 10.1111/j.1365-3059.2011.02470.x [DOI] [Google Scholar]
- Dong X. F., Fang L., Ye Z. Y., Zhu G. Q., Lai Q. Y., Liu S. R. (2021). Screening of biocontrol bacteria against soft rot disease of Colocasia esculenta (L.) schott and its field application. PLoS One 16:e0254070. doi: 10.1371/journal.pone.0254070, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S. S., Tian F., Li J. Y., Hutchins W., Chen H. M., Yang F. H., et al. (2017). Identification of phenolic compounds that suppress the virulence of Xanthomonas oryzae on rice via the type III secretion system. Mol. Plant Pathol. 18, 555–568. doi: 10.1111/mpp.12415, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L. W., Schaefer A. L., Hu M., Chen R. Y., Greenberg E. P., Zhou J. N. (2019). Virulence factor identification in the banana pathogen Dickeya zeae MS2. Appl. Environ. Microbiol. 85, e01611–e01619. doi: 10.1128/AEM.01611-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasner J. D., Yang C. H., Reverchon S., Hugouvieux-Cotte-Pattat N., Condemine G., Bohin J. P., et al. (2011). Genome sequence of the plant-pathogenic bacterium Dickeya dadantii 3937. J. Bacteriol. 193, 2076–2077. doi: 10.1128/JB.01513-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurlier K., Dénervaud V., Pessi G., Reimmann C., Haas D. (2003). Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185, 2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Li J. L., Chen R. T., Li W. J., Feng L. W., Shi L., et al. (2018). Dickeya zeae strains isolated from rice, banana and clivia rot plants show great virulence differentials. BMC Microbiol. 18:136. doi: 10.1186/s12866-018-1300-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Xue Y., Li C., Lv M., Zhang L. H., Parsek M. R., et al. (2022). Genomic and functional dissections of Dickeya zeae shed light on the role of type III secretion system and cell-wall degrading enzymes to host range and virulence. Microbiol. Spectr. 2:e0159021. doi: 10.1128/spectrum.01590-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. F., Chen Z. Q., Hu M., Xue Y., Liao L. S., Zhang L. H. (2021). First report of bacterial soft rot disease on taro caused by Dickeya fangzhongdai in China. Plant Dis. doi: 10.1094/PDIS-10-20-2225-PDN [Epub ahead of print] [DOI] [PubMed]
- Hueck C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. doi: 10.1128/MMBR.62.2.379-433.1998, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Jacot-des-Combes C., Briolay J. (2019). Dickeya lacustris sp. nov., a water-living pectinolytic bacterium isolated from lakes in France. Int. J. Syst. Evol. Microbiol. 69, 721–726. doi: 10.1099/ijsem.0.003208, PMID: [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Van Gijsegem F. (2021). Diversity within the Dickeya zeae complex, identification of Dickeya zeae and Dickeya oryzae members, proposal of the novel species Dickeya parazeae sp. nov. Int. J. Syst. Evol. Microbiol. 71:11. doi: 10.1099/ijsem.0.005059, PMID: [DOI] [PubMed] [Google Scholar]
- Hussain M. B., Zhang H. B., Xu J. L., Liu Q. G., Jiang Z. D., Zhang L. H. (2008). The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J. Bacteriol. 190, 1045–1053. doi: 10.1128/JB.01472-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Li H., Ahmed W., Xiang X. W., Song G. P., Cui Z. N. (2019). Discovery of ethyl 2-nitro-3-arylacrylates molecules as T3SS inhibitor reducing the virulence of plant pathogenic bacteria Xanthomonas. Front. Microbiol. 10:1874. doi: 10.3389/fmicb.2019.01874, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Zhang J., Yang Q., Sun D., Pu X., Shen H., et al. (2021). Antimicrobial activity of natural plant compound carvacrol against soft rot disease agent Dickeya zeae. Curr. Microbiol. 78, 3453–3463. doi: 10.1007/s00284-021-02609-3, PMID: [DOI] [PubMed] [Google Scholar]
- Jin Q., He S. Y. (2001). Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science 294, 2556–2558. doi: 10.1126/science.1066397, PMID: [DOI] [PubMed] [Google Scholar]
- Joshi J. R., Burdman S., Lipsky A., Yariv S., Yedidia I. (2016). Plant phenolic acids affect the virulence of Pectobacterium aroidearum and P. carotovorum ssp. brasiliense via quorum sensing regulation. Mol. Plant Pathol. 17, 487–500. doi: 10.1111/mpp.12295, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi J. R., Burdman S., Lipsky A., Yedidia I. (2015). Effects of plant antimicrobial phenolic compounds on virulence of the genus Pectobacterium. Res. Microbiol. 166, 535–545. doi: 10.1016/j.resmic.2015.04.004, PMID: [DOI] [PubMed] [Google Scholar]
- Joshi J. R., Khazanov N., Charkowski A., Faigenboim A., Senderowitz H., Yedidia I. (2021). Interkingdom signaling interference: the effect of plant-derived small molecules on quorum wensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 59, 153–190. doi: 10.1146/annurev-phyto-020620-095740, PMID: [DOI] [PubMed] [Google Scholar]
- Joshi J. R., Khazanov N., Khadka N., Charkowski A. O., Burdman S., Carmi N., et al. (2020). Direct binding of salicylic acid to Pectobacterium N-acyl-Hhomoserine lactone synthase. ACS Chem. Biol. 15, 1883–1891. doi: 10.1021/acschembio.0c00185, PMID: [DOI] [PubMed] [Google Scholar]
- Kang J. E., Jeon B. J., Park M. Y., Kim B. S. (2020). Inhibitory activity of sedum middendorffianum-derived 4-hydroxybenzoic acid and vanillic acid on the type III secretion system of Pseudomonas syringae pv. tomato DC3000. Plant Pathol. 36, 608–617. doi: 10.5423/PPJ.OA.08.2020.0162, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi A. M., Nordfelth R., Uvell H., Wolf-Watz H., Elofsson M. (2003). Targeting bacterial virulence. Chem. Biol. 10, 241–249. doi: 10.1016/S1074-5521(03)00046-2, PMID: [DOI] [PubMed] [Google Scholar]
- Keyser P., Elofsson M., Rosell S., Wolf-Watz H. (2008). Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against gram-negative bacteria. J. Intern. Med. 264, 17–29. doi: 10.1111/j.1365-2796.2008.01941.x, PMID: [DOI] [PubMed] [Google Scholar]
- Khokhani D., Zhang C., Li Y., Wang Q., Zeng Q., Yamazaki A., et al. (2013). Discovery of plant phenolic compounds that act as type III secretion system inhibitors or inducers of the fire blight pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 79, 5424–5436. doi: 10.1128/AEM.00845-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O. K., Garrity-Ryan L. K., Bartlett V. J., Grier M. C., Verma A. K., Medjanis G., et al. (2009). N-hydroxybenzimidazole inhibitors of the transcription factor LcrF in Yersinia: novel antivirulence agents. J. Med. Chem. 52, 5626–5634. doi: 10.1021/jm9006577, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline T., Felise H. B., Sanowar S., Miller S. I. (2012). The type III secretion system as a source of novel antibacterial drug targets. Curr. Drug Targets 13, 338–351. doi: 10.2174/138945012799424642, PMID: [DOI] [PubMed] [Google Scholar]
- Kotoujansky A., Lemattre M., Boistard P. (1982). Utilization of a thermosensitive episome bearing transposon TN10 to isolate Hfr donor strains of Erwinia carotovora subsp. chrysanthemi. J. Bacteriol. 150, 122–131. doi: 10.1128/jb.150.1.122-131.1982, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton A. N., Hudson D. L., Thompson A., Hinton J. C., Stevens J. M., Galyov E. E., et al. (2010). Salicylidene acylhydrazide-mediated inhibition of type III secretion system-1 in Salmonella enterica serovar Typhimurium is associated with iron restriction and can be reversed by free iron. FEMS Microbiol. Lett. 302, 114–122. doi: 10.1111/j.1574-6968.2009.01847.x, PMID: [DOI] [PubMed] [Google Scholar]
- Letal J. R. (1977). Efficacy of disinfestants against potato ring rot and blackleg bacteria. Am. J. Potato Res. 54, 405–409. [Google Scholar]
- Li J. L., Hu M., Xue Y., Chen X., Lu G. T., Zhang L. H., et al. (2020). Screening, identification and efficacy evaluation of antagonistic bacteria for biocontrol of soft rot disease caused by Dickeya zeae. Microorganisms 8:697. doi: 10.3390/microorganisms8050697, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hutchins W., Wu X. G., Liang C. R., Zhang C. F., Yuan X. C., et al. (2015). Derivative of plant phenolic compound inhibits the type III secretion system of Dickeya dadantii via HrpX/HrpY two-component signal transduction and Rsm systems. Mol. Plant Pathol. 16, 150–163. doi: 10.1111/mpp.12168, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Peng Q., Selimi D., Wang Q., Charkowski A. O., Chen X., et al. (2009). The plant phenolic compound p-coumaric acid represses gene expression in the Dickeya dadantii type III secretion system. Appl. Environ. Microbiol. 75, 1223–1228. doi: 10.1128/AEM.02015-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Xu J., Wang X. L., Zhang T., Xu J. S., Zhang H., et al. (2020). Control effect of 11 fungicides on potato soft rot. Plant Prot. 46, 309–315. doi: 10.16688/j.zwbh.2019278 [DOI] [Google Scholar]
- Liao L. S., Cheng Y. Y., Liu S. Y., Zhou J. N., An S. W., Lv M. F., et al. (2014). Production of novel antibiotics zeamines through optimizing Dickeya zeae fermentation conditions. PLoS One 9:e116047. doi: 10.1371/journal.pone.0116047, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linington R. G., Robertson M., Gauthier A., Finlay B. B., MacMillan J. B., Molinski T. F., et al. (2006). Caminosides B-D, antimicrobial glycolipids isolated from the marine sponge Caminus sphaeroconia. J. Nat. Prod. 69, 173–177. doi: 10.1021/np050192h, PMID: [DOI] [PubMed] [Google Scholar]
- Linington R. G., Robertson M., Gauthier A., Finlay B. B., van Soest R., Andersen R. J. (2002). Caminoside A, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Org. Lett. 4, 4089–4092. doi: 10.1021/ol0268337, PMID: [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262, PMID: [DOI] [PubMed] [Google Scholar]
- Lu C. H., Li J. F., Guo Z. X., Li T. H., Shen Y. M. (2018). Research progress of type III secretory system and its inhibitors. Chin. J. Antibiot. 43, 387–393. [Google Scholar]
- Lund B. M., Lyon G. D. (1975). Detection of inhibitors of Erwinia carotovora and E. herbicola on thin-layer chromatograms. J. Chromatogr. 110, 193–196. doi: 10.1016/s0021-9673(00)91229-9, PMID: [DOI] [PubMed] [Google Scholar]
- Lv M. F., Hu M., Li P., Jiang Z. D., Zhang L. H., Zhou J. N. (2019). A two-component regulatory system VfmIH modulates multiple virulence traits in Dickeya zeae. Mol. Microbiol. 111, 1493–1509. doi: 10.1111/mmi.14233, PMID: [DOI] [PubMed] [Google Scholar]
- Mansfield J., Genin S., Magori S., Citovsky V., Sriariyanum M., Ronald P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden A. E., King J. M., Spies M. A., Kim O. K., Yahr T. L. (2016). Inhibition of Pseudomonas aeruginosa ExsA DNA-binding activity by N-hydroxybenzimidazoles. Antimicrob. Agents Chemother. 60, 766–776. doi: 10.1128/AAC.02242-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N. C., Finlay B. B. (2014). Targeting the type III secretion system to treat bacterial infections. Expert Opin. Ther. Targets 18, 137–152. doi: 10.1517/14728222.2014.855199, PMID: [DOI] [PubMed] [Google Scholar]
- McNally R. R., Toth I. K., Cock P. J., Pritchard L., Hedley P. E., Morris J. A., et al. (2012). Genetic characterization of the HrpL regulon of the fire blight pathogen Erwinia amylovora reveals novel virulence factors. Mol. Plant Pathol. 13, 160–173. doi: 10.1111/j.1364-3703.2011.00738.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. G., Leveau J. H., Lindow S. E. (2000). Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13, 1243–1250. doi: 10.1094/MPMI.2000.13.11.1243, PMID: [DOI] [PubMed] [Google Scholar]
- Oulghazi S., Pedron J., Cigna J., Lau Y. Y., Moumni M., Van Gijsegem F., et al. (2019). Dickeya undicola sp. nov., a novel species for pectinolytic isolates from surface waters in Europe and Asia. Int. J. Syst. Evol. Microbiol. 69, 2440–2444. doi: 10.1099/ijsem.0.003497, PMID: [DOI] [PubMed] [Google Scholar]
- Pique N., Minana-Galbis D., Merino S., Tomas J. M. (2015). Virulence factors of Erwinia amylovora: a review. Int. J. Mol. Sci. 16, 12836–12854. doi: 10.3390/ijms160612836, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigvert M., Sole M., Lopez-Garcia B., Coll N. S., Beattie K. D., Davis R. A., et al. (2019). Type III secretion inhibitors for the management of bacterial plant diseases. Mol. Plant Pathol. 20, 20–32. doi: 10.1111/mpp.12736, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Sperandio V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117–128. doi: 10.1038/nrd3013, PMID: [DOI] [PubMed] [Google Scholar]
- Rice-Evans C. A., Miller N. J., Paganga G. (1996). Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 20, 933–956. doi: 10.1016/0891-5849(95)02227-9, PMID: [DOI] [PubMed] [Google Scholar]
- Rojas C. M., Ham J. H., Schechter L. M., Kim J. F., Beer S. V., Collmer A. (2004). The Erwinia chrysanthemi EC16 hrp/hrc gene cluster encodes an active Hrp type III secretion system that is flanked by virulence genes functionally unrelated to the Hrp system. Mol. Plant-Microbe Interact. 17, 644–653. doi: 10.1094/MPMI.2004.17.6.644, PMID: [DOI] [PubMed] [Google Scholar]
- Sharma S., Gopu V., Sivasankar C., Shetty P. H. (2019). Hydrocinnamic acid produced by Enterobacter xiangfangensis impairs AHL-based quorum sensing and biofilm formation in Pseudomonas aeruginosa. RSC Adv. 9, 28678–28687. doi: 10.1039/C9RA05725K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni S., Nathoo N., Klimov E., Yuan Z. C. (2014). Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front. Plant Sci. 5:322. doi: 10.3389/fpls.2014.00322, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Xiao Y., Zhou J. (2006). Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant-Microbe Inter. 19, 1159–1166. doi: 10.1094/MPMI-19-1159, PMID: [DOI] [PubMed] [Google Scholar]
- Tree J. J., Wang D., McInally C., Mahajan A., Layton A., Houghton I., et al. (2009). Characterization of the effects of salicylidene acylhydrazide compounds on type III secretion in Escherichia coli O157:H7. Infect. Immun. 77, 4209–4220. doi: 10.1128/IAI.00562-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ur-Rehman T., Slepenkin A., Chu H., Blomgren A., Dahlgren M. K., Zetterstrom C. E., et al. (2012). Pre-clinical pharmacokinetics and anti-chlamydial activity of salicylidene acylhydrazide inhibitors of bacterial type III secretion. J. Antibiot. 65, 397–404. doi: 10.1038/ja.2012.43, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusitalo P., Hägglund U., Rhöös E., Norberg H. S., Elofsson M., Sundin C. (2017). The salicylidene acylhydrazide INP0341 attenuates Pseudomonas aeruginosa virulence in vitro and in vivo. J. Antibiot. 70, 937–943. doi: 10.1038/ja.2017.64, PMID: [DOI] [PubMed] [Google Scholar]
- van der Wolf J. M., Nijhuis E. H., Kowalewska M. J., Saddler G. S., Parkinson N., Elphinstone J. G., et al. (2014). Dickeya solani sp. nov., a pectinolytic plant-pathogenic bacterium isolated from potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 64, 768–774. doi: 10.1099/ijs.0.052944-0, PMID: [DOI] [PubMed] [Google Scholar]
- Vikram A., Jesudhasan P. R., Jayaprakasha G. K., Pillai S. D., Jayaraman A., Patil B. S. (2011). Citrus flavonoid represses Salmonella pathogenicity island 1 and motility in S. typhimurium LT2. Int. J. Food Microbiol. 145, 28–36. doi: 10.1016/j.ijfoodmicro.2010.11.013, PMID: [DOI] [PubMed] [Google Scholar]
- Wang X., He S. W., Guo H. B., Han J. G., Thin K. K., Gao J. S., et al. (2020). Dickeya oryzae sp. nov., isolated from the roots of rice. Int. J. Syst. Evol. Microbiol. 70, 4171–4178. doi: 10.1099/ijsem.0.004265, PMID: [DOI] [PubMed] [Google Scholar]
- Wang C., Ye F. Z., Chang C. Q., Liu X. L., Wang J. H., Wang J. P., et al. (2019). Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals. Proc. Natl. Acad. Sci. U. S. A. 116, 22331–22340. doi: 10.1073/pnas.1903695116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z. M., Sneath B. J., Beer S. V. (1992). Expression of Erwinia amylovora hrp genes in response to environmental stimuli. J. Bacteriol. 174, 1875–1882. doi: 10.1128/jb.174.6.1875-1882.1992, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2014). Antimicrobial Resistance: Global Report On Surveillance. WHO, Geneva Switzerland, Available at: https://www.who.int/publications/i/item/9789241564748 (Accessed February 09, 2022).
- Wyatt G. M., Lund B. M. (1981). The effect of antibacterial products on bacterial soft rot of potatoes. Potato Res. 24, 315–329. [Google Scholar]
- Yamazaki A., Li J., Zeng Q., Khokhani D., Hutchins W. C., Yost A. C., et al. (2012). Derivatives of plant phenolic compound affect the type III secretion system of Pseudomonas aeruginosa via a GacS-GacA two-component signal transduction system. Antimicrob. Agents Chemother. 56, 36–43. doi: 10.1128/AAC.00732-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Gavilanes-Ruiz M., Okinaka Y., Vedel R., Berthuy I., Boccara M., et al. (2002). hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Mol. Plant-Microbe Interact. 15, 472–480. doi: 10.1094/MPMI.2002.15.5.472, PMID: [DOI] [PubMed] [Google Scholar]
- Yang S., Peng Q., San Francisco M., Wang Y., Zeng Q., Yang C. H. (2008). Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS One 3:e2973. doi: 10.1371/annotation/91170966-226f-4678-999e-22f2c4a6bb8d, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap M. N., Yang C. H., Barak J. D., Jahn C. E., Charkowski A. O. (2005). The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J. Bacteriol. 187, 639–648. doi: 10.1128/JB.187.2.639-648.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z. C., Edlind M. P., Liu P., Saenkham P., Banta L. M., Wise A. A., et al. (2007). The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc. Natl. Acad. Sci. U. S. A. 104, 11790–11795. doi: 10.1073/pnas.0704866104, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X. C., Yu M. D., Yang C. H. (2020). Innovation and application of the type III secretion system inhibitors in plant pathogenic bacteria. Microorganisms 8:1956. doi: 10.3390/microorganisms8121956, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q., Ibekwe A. M., Biddle E., Yang C. H. (2010). Regulatory mechanisms of exoribonuclease PNPase and regulatory small RNA on T3SS of Dickeya dadantii. Mol. Plant-Microbe Interact. 23, 1345–1355. doi: 10.1094/MPMI-03-10-0063, PMID: [DOI] [PubMed] [Google Scholar]
- Zeng Q., Laiosa M. D., Steeber D. A., Biddle E. M., Peng Q., Yang C. H. (2012). Cell individuality: the bistable gene expression of the type III secretion system in Dickeya dadantii 3937. Mol. Plant-Microbe Interact. 25, 37–47. doi: 10.1094/MPMI-05-11-0105, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang J. X., Shen H. F., Pu X. M., Lin B. R., Hu J. (2014). Identification of Dickeya zeae as a causal agent of bacterial soft rot in banana in China. Plant Dis. 98, 436–442. doi: 10.1094/PDIS-07-13-0711-RE, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang C., Wu X., Li Y., Liang C., Che Y., Gu L., et al. (2013). Synthesis and bioactivity of novel inhibitors for type III secretion system of Pseudomonas aeruginosa PAO1. Chinese J. Organ. Chem. 33:1309. doi: 10.6023/cjoc201302021 [DOI] [Google Scholar]
- Zhou J. N., Jiang Z. D., Zhang L. H. (2015). Research progress on pathogenic mechanism of bacterial soft rot pathogen Dickeya. Acta Phytopathol. Sin. 45, 337–349. doi: 10.13926/j.cnki.apps.2015.04.001 [DOI] [Google Scholar]
- Zhou J. N., Zhang H. B., Lv M. F., Chen Y. F., Liao L. S., Cheng Y. Y., et al. (2016). SlyA regulates phytotoxin production and virulence in Dickeya zeae EC1. Mol. Plant Pathol. 17, 1398–1408. doi: 10.1111/mpp.12376, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. N., Zhang H. B., Wu J. E., Liu Q. G., Xi P. G., Lee J., et al. (2011). A novel multidomain polyketide synthase is essential for zeamine production and the virulence of Dickeya zeae. Mol. Plant-Microbe Interact. 24, 1156–1164. doi: 10.1094/MPMI-04-11-0087, PMID: [DOI] [PubMed] [Google Scholar]
- Zou L. F., Zeng Q., Lin H. P., Gyaneshwar P., Chen G. Y., Yang C. H. (2012). SlyA regulates type III secretion system (T3SS) genes in parallel with the T3SS master regulator HrpL in Dickeya dadantii 3937. Appl. Environ. Microbiol. 78, 2888–2895. doi: 10.1128/AEM.07021-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.