Abstract
Background
Dynamic whole-body (D-WB) FDG PET/CT is a recently developed technique that allows direct reconstruction of multiparametric images of metabolic rate of FDG uptake (MRFDG) and “free” FDG (DVFDG). Multiparametric images have a markedly different appearance than the conventional SUV images obtained by static PET imaging, and normal values of MRFDG and DVFDG in frequently used reference tissues and organs are lacking. The aim of this study was therefore to: (1) provide an overview of normal MRFDG and DVFDG values and range of variation in organs and tissues; (2) analyse organ time-activity curves (TACs); (3) validate the accuracy of directly reconstructed MRFDG tissue values versus manually calculated Ki (and MRFDG) values; and (4) explore correlations between demographics, blood glucose levels and MRFDG values. D-WB data from 126 prospectively recruited patients (100 without diabetes and 26 with diabetes) were retrospectively analysed. Participants were scanned using a 70-min multiparametric PET acquisition protocol on a Siemens Biograph Vision 600 PET/CT scanner. 13 regions (bone, brain grey and white matter, colon, heart, kidney, liver, lung, skeletal muscle of the back and thigh, pancreas, spleen, and stomach) as well as representative pathological findings were manually delineated, and values of static PET (SUV), D-WB PET (Ki, MRFDG and DVFDG) and individual TACs were extracted. Multiparametric values were compared with manual TAC-based calculations of Ki and MRFDG, and correlations with blood glucose, age, weight, BMI, and injected tracer dose were explored.
Results
Tissue and organ MRFDG values showed little variation, comparable to corresponding SUV variation. All regional TACs were in line with previously published FDG kinetics, and the multiparametric metrics correlated well with manual TAC-based calculations (r2 = 0.97, p < 0.0001). No correlations were observed between glucose levels and MRFDG in tissues known not to be substrate driven, while tissues with substrate driven glucose uptake had significantly correlated glucose levels and MRFDG values.
Conclusion
The multiparametric D-WB PET scan protocol provides normal MRFDG values with little inter-subject variation and in agreement with manual TAC-based calculations and literature values. The technique therefore facilitates both accurate clinical reports and simpler acquisition of quantitative estimates of whole-body tissue glucose metabolism.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13550-022-00884-0.
Keywords: Dynamic whole-body PET, Multiparametric imaging, Normal values, Patlak
Introduction
Positron emission tomography (PET) using 18F-fluorodeoxyglucose (FDG) is a key part of diagnostics and follow-up of both malignant and non-malignant diseases. Conventional static FDG PET routine consists of a single whole-body (WB) pass and reconstruction of a standard uptake value (SUV) image representing the tracer distribution 60 min after tracer injection [1]. This snapshot of accumulated tracer is not without problems. SUV images are highly dependent on the timing of the scan with any delay incurring a variation of measured activity. Factors such as scanner calibration, non-perfect injections and even the subject’s body composition or blood glucose levels also influence the resulting images [2, 3]. Furthermore, static FDG PET images capture not only FDG-6-P retained in glucose consuming tissue but also a substantial background of unbound FDG in tissue and circulation, complicating the evaluation of vascularized organs.
Advances in PET scanner technology and software have introduced new possibilities [4, 5]. Dynamic whole-body (D-WB) PET/CT is a recently developed technique involving multiple WB passes and extraction of an arterial image-derived input function (IDIF), which provide the dynamic PET data needed for direct reconstruction of WB multiparametric images based on the linear Patlak analysis [6, 7]. Multiparametric imaging complements the standard SUV image with two new parametric images: One representing the metabolic rate of FDG into the tissue (MRFDG) and another representing the distribution volume of free FDG in the tissue (DVFDG).
Until now, there is sparse data regarding clinical applications of D-WB PET [8, 9]. A recent study published by our group demonstrated the feasibility of the technique in a clinical setting and also showed some promising results from the resulting MRFDG and DVFDG images including superior lesion target-to-background ratios as well as fewer false positive findings [8]. As multiparametric imaging becomes available in more PET facilities and in all PET/CT systems, nuclear medicine clinicians will need to familiarize themselves with MRFDG and DVFDG images. In particular, organ and tissue thresholds for physiological MRFDG and DVFDG values should be established to ease detection of pathology.
We therefore performed a review of our clinically acquired D-WB FDG PET scans to provide estimates of expected normal values and population-based variation of SUV, MRFDG and DVFDG in selected tissues and organs. In addition, we extracted organ and tissue time-activity curves (TACs), validated the multiparametric MRFDG and DVFDG values obtained by direct reconstruction against post-reconstruction manual TAC-based calculations and explored correlations between age, diabetes status, body composition and the multiparametric values.
Materials and methods
Patient population
This was a retrospective analysis of prospectively collected data. Included subjects were recruited from the entire cohort of patients referred for FDG PET/CT as part of their clinical diagnostic work-up or treatment response evaluation. Inclusion into the main D-WB protocol and study was solely based on whether patients were deemed fit to lie still for 70 min while in the PET/CT scanner. The study was approved by the local ethics committee in Region Midtjylland (1-10-72-188-19).
D-WB data from 126 individuals (100 patients without diabetes (Non-DM) and 26 patients with diabetes (DM)) were analysed. The study population’s indications for PET referral, sex and age are shown in Table 1.
Table 1.
PET study indication and population demographics
| Scan indication | Number of patients | Age distribution* | |||||
|---|---|---|---|---|---|---|---|
| Total | Male | Female | Total | Male | Female | ||
| Cancer of unknown primary origin | Non-DM | 6 | 1 | 5 | 54.5 [35–65] | 53 | 54.8 [35–65] |
| DM | 2 | 1 | 1 | 67 [59–75] | 75 | 59 | |
| Gastro-intestinal cancer | Non-DM | 3 | 2 | 1 | 57.3 [45–68] | 63.5 [59–68] | 45 |
| DM | 1 | 0 | 1 | 57 | – | 57 | |
| Head and neck cancer | Non-DM | 15 | 7 | 8 | 58.6 [23–81] | 54.7 [23–81] | 62 [49–72] |
| DM | 6 | 2 | 4 | 66.6 [55–77] | 73.5 [70–77] | 63.3 [55–70] | |
| Infection and inflammation | Non-DM | 22 | 13 | 9 | 50.4 [22–76] | 49.1 [22–71] | 52.2 [24–76] |
| DM | 5 | 3 | 2 | 60.6 [51–74] | 59.3 [52–64] | 62.5 [51–74] | |
| Lung cancer | Non-DM | 38 | 18 | 20 | 64.8 [49–81] | 66 [49–78] | 63.7 [51–81] |
| DM | 7 | 6 | 1 | 73.4 [69–77] | 74.2 [72–77] | 69 | |
| Lymphoma | Non-DM | 16 | 9 | 7 | 55.5 [18–85] | 53.6 [18–85] | 58 [37–72] |
| DM | 3 | 3 | 0 | 51.7 [28–64] | 51.7 [28–64] | – | |
| Uro-genital cancer | Non-DM | 0 | 0 | 0 | – | – | – |
| DM | 2 | 1 | 1 | 69 [63–75] | 75 | 63 | |
| Total | Non-DM | 100 | 50 | 50 | 58.4 [18–85] | 57.4 [18–85] | 59.3 [24–81] |
| DM | 26 | 16 | 10 | 65.4 [28–77] | 67.2 [28–77] | 62.6 [51–74] | |
*Values presented as mean [range]
Data acquisition and image reconstruction
Participants were scanned using a fully automated multiparametric PET/CT acquisition protocol (FlowMotion® Multiparametric PET, Siemens Healthineers, Knoxville, USA) on a Siemens Biograph Vision 600 PET/CT scanner (Siemens Healthineers, Knoxville, USA) with 26.2 cm axial field of view. In short, a 70-min multiparametric PET acquisition protocol was started at the time of a standardized injection of FDG (4 MBq/kg) using an Intego PET Infusion System (MEDRAD, Inc., Warrendale, PA, USA). The PET protocol consisted of 1) a 6-min dynamic scan with the bed fixed at the chest region including organs as heart, liver and most importantly aorta, and 2) a 64-min dynamic WB PET scan consisting of 16 continuous bed motion passes: 7 × 2-min WB passes followed by 9 × 5-min WB passes.
The automated multiparametric scan protocol automatically identified the aorta on the low-dose WB CT scan [10] and placed a VOI (1.6 mm3 cylinder) to extract the IDIF from the full dynamic PET series of the chest region. Such IDIF is robust and can be used to replace an arterial input function (AIF) for quantitative Patlak modelling [11].
Multiparametric images (MRFDG and DVFDG) were reconstructed using data from 40 to 70 min post injection (p.i.) and the IDIF. A standard-of-care static SUV image was reconstructed using data from 60 to 70 min p.i. (see example in Fig. 1). A more detailed description of the D-WB PET acquisition technique and image reconstruction parameters can be found in our previous publication [8].
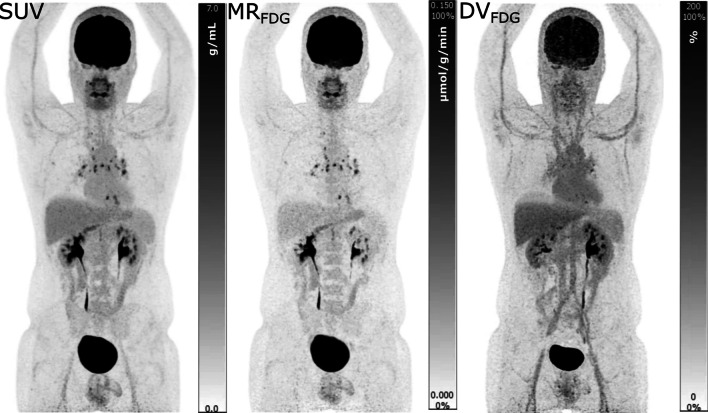
Fig. 1.
Example of SUV and Patlak images analysed in our study. In this case, a 34-year-old man with mediastinal sarcoidosis. Left column: static SUV images; Middle column: MRFDG images; right column: DVFDG images. The actual FDG uptake in the inflammatory sarcoid lymph nodes is evident on the MRFDG image, whereas the free FDG in the circulation is clearly visible on the DVFDG image
Interpretation of multiparametric WB FDG images
The Patlak model is currently the only model available for direct reconstruction of WB parametric images, but the use of the same irreversible model in the entire image does lead to a challenging image interpretation.
Assuming irreversible kinetics, Ki represent the steady-state rate of tracer uptake in the trapped compartment representing FDG-6-phosphate. For 18FDG, Ki can be multiplied by the subject’s blood glucose to obtain the metabolic rate of 18FDG, MRFDG = Ki × blood glucose, which can be divided with the lumped constant to obtain the metabolic rate of glucose, MRGlu = MRFDG/LC. The images and tables have not been corrected for LC, i.e. we used LC = 1, as it varies by tissue type (see Additional file 1: table S3 for a list of published values) and is not known for all tissues. Therefore, as of today, it is not really possible to generate a meaningful full WB parametric image of MRGlu, and for that reason we do not present these values. However, for tissues with well-established LC values, such as brain [12, 13], MRFDG can easily recalculated into MRGlu, e.g. for comparison with single-organ studies where LC have been used.
The apparent distribution volume, DVFDG, is approximately equal to the sum of the blood volume and distribution volume of the reversible components, which both varies between tissues and cannot be separated by the Patlak model [14]. A separate estimate of the blood volume would require full kinetic modelling. Thus, DVFDG reflects the combined distribution volume of free 18FDG in blood and tissue.
Clear interpretations of MRFDG and DVFDG values are based on the assumption of irreversible 18FDG kinetics. However, this assumption is not always met with notable exceptions as the liver and kidney. Thus, in multi-parametric WB images of MRFDG and DVFDG there are tissues with reversible FDG kinetics that obscure the physiologic interpretation of the voxel values. As an example, the liver is known to de-phosphorylate 18FDG-6P, and this reversible FDG kinetics led to lower MRFDG values (less irreversible trapping) and higher DVFDG (the reversible compartments include FDG-6P).
Image analysis and VOI delineation
We analysed 13 volumes of interests (VOIs) corresponding to different organ and tissue locations: bone, brain (white and grey matter), colon, heart, kidney, liver, lung, muscle (paravertebral and thigh), pancreas, spleen and stomach. Due to the differing morphologies of the selected organs and tissues, different approaches were used to outline representative VOIs, and care was taken to ensure that the outlined VOIs corresponded to areas not affected by pathology or visual artefacts. See Additional file 1: table S1 for the methodology used in each region.
Furthermore, we also analysed an area representative of pathology in these patients. In oncological patients, the VOI was placed on the primary tumour, while for patients referred for infectious or inflammatory disease, a VOI was placed in the area of greatest FDG uptake. See Additional file 1: table S1 for the methodology used. Excluded from this analysis were patients who had previously undergone surgery or who had been treated with chemo-/radiotherapy. Also excluded were patients where no signs of active disease were detected on the PET scan. The final analysis of pathological findings was therefore performed on 55 patients.
Multiparametric images were visually inspected using Hermes Gold Client v.2.5.0 (Hermes Medical Solutions AB, Stockholm, Sweden). VOI delineation of the multiparametric images was performed by AHD using PMOD® 4.0 (PMOD Technologies Ltd, Zürich, Switzerland). Semiquantitative values of SUVmax and SUVmean were obtained from the conventional PET reconstructions, whereas MRFDG and DVFDG values were extracted from the multiparametric images.
Time-activity curves
TACs were obtained from the 6-min dynamic scan of the chest region and the 16 D-WB passes. Analyses of brain regions were limited to patients scanned from the top of the skull. Therefore, the analysis of the grey and white matter was performed in a total of 75 patients (14 DM and 61 non-DM). The VOIs used for the manual TAC-based calculation in PMOD were the same that had been delineated and used to extract parameter values from multiparametric images.
Comparison of multiparametric and manual TAC-based MRFDG and DVFDG values
Kinetic parameter estimates derived from post-reconstruction TAC-based analyses can be biased compared to those from direct reconstruction of parametric images [15]. Thus, we compared kinetic parameter estimates using the two methods. From a subpopulation of 10 randomly selected patients, the D-WB series and IDIF were used to manually calculate MRFDG and DVFDG values. These TAC-based parameters were estimated by linear Patlak analysis in PMOD® 4.0, using the PKIN module. Measured blood sugar levels were used to calculate MRFDG. TAC-based MRFDG and DVFDG values were subsequently compared with values extracted from multiparametric images obtained using direct reconstruction using the FlowMotion® Multiparametric PET software from Siemens Healthineers.
Statistical analysis
The statistical analysis was performed using Stata 16 or GraphPad Prism 9.2.0. Statistical tests were used for group comparisons based on whether data were normally distributed and paired/unpaired. Pearson’s correlation analysis was performed for the relation between MRFDG and SUV values and blood glucose, patient age, weight, tracer dose and BMI. p values of < 0.05 were considered significant. Continuous group data are presented as mean ± SD or median (range) as appropriate. Time-series are presented as mean ± SEM.
Results
Organ and tissue quantitative and semi-quantitative FDG uptake
Normal values of SUVmean, SUVmax, MRFDG mean, Ki,mean, and DVFDG,mean (Table 2) were obtained for the bone, brain grey and white matter, colon, heart, kidney, liver, lung, skeletal muscle, pancreas, spleen and stomach. The same values were also obtained for the subpopulations of male and female patients (patients without diabetes Additional file 1: table S2A, patients with diabetes Additional file 1: table S2B).
Table 2.
Normal values of the population without diabetes (N = 100) and the population with diabetes (N = 26)
| Volume of interest | SUVmean* (g/mL) | SUVmax* (g/mL) | 100 × MRFDG* (µmol/g/min) | 100 × Ki * (mL/mL/min) | DVFDG* (%) | |
|---|---|---|---|---|---|---|
| Bone | Non-DM | 1.90 [1.12–4.87] | 2.72 [1.59–6.12] | 3.69 [1.08–9.09] | 0.67 [0.16–1.57] | 30.97 [3.50–51] |
| DM | 1.85 [1.12–2.61] | 2.79 [1.66–4.24] | 4.28 [0.64–90] | 0.63 [0.09–1.03] | 23.51 [6.24–60.12] | |
| Brain GM† | Non-DM | 8.08 [4.85–13.56] | 15.20 [8.66–46.50] | 17.46 [11.68–27.61] | 3.28 [2.02–5.40] | 81.18 [13.58–140.70] |
| DM | 6.69 [4.65–9.43] | 11.60 [8.59–19.28] | 16.80 [12.79–24.28] | 2.23 [1.62–3.92] | 68.79 [41.6–95.26] | |
| Brain WM† | Non-DM | 3.39 [2.09–5.29] | 5.86 [3.59–10.30] | 6.03 [4.02–9.53] | 1.09 [0.72–1.80] | 46.07 [15.29–87.4] |
| DM | 2.94 [2.17–3.68] | 4.88 [3.63–6.68] | 6.44 [4.20–8.27] | 0.86 [0.55–1.33] | 39.76 [29.12–67.61] | |
| Colon | Non-DM | 1.26 [0.52–6.36] | 2.38 [0.95–12.2] | 3.17 [1.24–21.23] | 0.57 [0.23–3.60] | 47.70 [17.45–174.4] |
| DM | 1.84 [0.87–5.72] | 3.17 [1.49–10.48] | 6.07 [0.68–20.55] | 0.74 [0.10–2.19] | 56.46 [35.80–163.7] | |
| Heart | Non-DM | 4.48 [0.72–14.50] | 9.53 [1.58–33.6] | 11.03 [0.35–42.6] | 1.82 [0.08–7.22] | 54.17 [16.84–191.70] |
| DM | 2.67 [0.99–13.24] | 7.01 [2.06–30.97] | 5.90 [0.14–43.46] | 0.86 [0.02–6.30] | 48.50 [28.53–123.80] | |
| Kidney | Non-DM | 2.03 [0.67–3.03] | 3.55 [1.32–4.77] | 3.81 [0.08–7.95] | 0.69 [0.01–1.45] | 95.54 [32.51–141] |
| DM | 2.31 [1.91–3.18] | 4.02 [3.11–6.05] | 4.49 [0.53–8.38] | 0.67 [0.08–1.10] | 99.98 [75–137.70] | |
| Liver | Non-DM | 2.26 [1.31–3.11] | 3.80 [2.51–5.69] | 2.02 [0.74–4.35] | 0.36 [0.12–0.73] | 83.80 [43.66–126] |
| DM | 2.19 [1.77–2.81] | 3.84 [2.89–5.17] | 2.76 [0.15–5.35] | 0.35 [0.03–0.54] | 80.43 [62.71–105.90] | |
| Lung | Non-DM | 0.40 [0.07–0.83] | 0.83 [0.34–1.46] | 0.35 [0.03–1.74] | 0.06 [0.01–0.30] | 15.18 [2.13–28.15] |
| DM | 0.41 [0.15–0.62] | 0.84 [0.5–1.19] | 0.51 [0.05–0.97] | 0.07 [0.01–0.11] | 15.01 [6.11–21.34] | |
| Muscle back | Non-DM | 0.65 [0.38–1.01] | 1.10 [0.76–1.99] | 0.89 [0.40–1.64] | 0.16 [0.08–0.30] | 15.19 [6.02–33.72] |
| DM | 0.69 [0.41–1.17] | 1.23 [0.73–2.16] | 1.16 [0.08–2.68] | 0.14 [0.01–0.38] | 15.89 [7.67–29.84] | |
| Muscle thigh | Non-DM | 0.55 [0.36–1.41] | 1.19 [0.72–3.11] | 0.98 [0.14–2.28] | 0.17 [0.03–0.39] | 13.41 [6.26–25.80] |
| DM | 0.54 [0.38–2.02] | 1.26 [0.81–4.39] | 1.06 [0.10–5.11] | 0.14 [0.02–0.82] | 15.33 [11.16–30.23] | |
| Pancreas | Non-DM | 1.68 [1.15–2.44] | 2.71 [1.64–4.29] | 3.40 [1.16–6.67] | 0.62 [0.26–1.15] | 68.72 [42.57–123.80] |
| DM | 1.89 [1.29–2.43] | 2.83 [2.04–3.43] | 4.62 [0.81–6.09] | 0.62 [0.12–0.80] | 70.05 [42.48–92.01] | |
| Spleen | Non-DM | 1.96 [1.41–5.93] | 2.83 [1.98–8.46] | 2.45 [1.18–15.30] | 0.44 [0.20–2.78] | 58.30 [10.07–111.60] |
| DM | 1.91 [1.55–2.20] | 2.90 [2.19–3.69] | 2.55 [0.37–5.41] | 0.35 [0.05–0.62] | 54.30 [41.57–87.58] | |
| Stomach | Non-DM | 2.26 [1.33–5.25] | 4.19 [2.43–9.72] | 5.88 [2.84–19.13] | 0.99 [0.49–3.24] | 79.63 [45.22–178.40] |
| DM | 2.20 [1.47–2.82] | 4.07 [2.79–5.38] | 6.87 [0.94–11.14] | 0.94 [0.14–1.41] | 78.38 [40.73–102.80] | |
*Values are median [min–max]; † Brain VOIs: Non-DM N = 61; DM N = 14. Note that MRFDG and Ki values are multiplied by 100
Lumped constant = 1. See Additional file 1: table S3 for information on the use of a tissue adjusted lumped constant for the calculation of MRFDG
SUVmax is included for comparison, as it is the most used PET metric in clinical practice
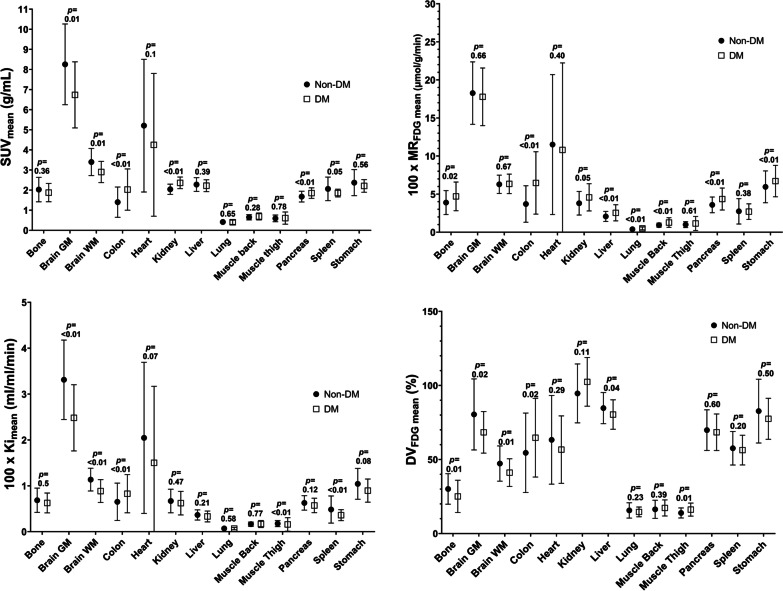
SUVmean and MRFDG,mean distribution plots are presented in Fig. 2. There was a remarkable similarity between the plots underlining the robustness of the quantitative values derived from the multiparametric images. Unsurprisingly, the organs that displayed the greatest MRFDG and by inference glucose metabolism were the heart and the grey matter of the brain, whereas the lowest measured glucose metabolism was observed in the lungs, an area of low tissue fraction.
Fig. 2.
Distribution plots of SUVmean, MRFDG,mean, Ki,mean and DVFDG,mean for the DM and Non-DM populations in 13 different types of tissue and organs. Plotted are the mean and standard deviation values. As seen, population variation was remarkably similar for SUV and MRFDG values. Calculated p-values between patients with DM and without DM are displayed above each organ
SUV, MRFDG and DVFDG values for different pathologies are displayed in Table 3. MRFDG values were significantly greater in most pathological lesions than in background tissue. However, no difference was observed between malignant and non-malignant pathological lesions.
Table 3.
Values from pathological findings (N = 55)
| Volume of Interest | N | SUVmean* (g/mL) | SUVmax* (g/mL) | 100 × MRFDG* (µmol/g/min) | 100 × Ki * (mL/mL/min) | DVFDG* (%) |
|---|---|---|---|---|---|---|
| Gastrointestinal cancer | 4 | 8.83 [6.58–14.70] | 15.52 [12.06–25.40] | 23.53 [14.72–31.49] | 4.31 [2.78–7.32] | 244.70 [125–360.40] |
| Head and neck cancer | 8 | 9.57 [5.19–22.00] | 17.02 [9.69–39.57] | 26.86 [11.47–68.31] | 4.14 [2.12–11.02] | 236.90 [88.61–620.30] |
| Infection and Inflammation | 13 | 4.60 [2.54–9.33] | 9.14 [4.67–17.88] | 14.85 [3.59–38.44] | 2.46 [0.84–6.41] | 126.70 [50.98–230] |
| Lung adenocarcinoma | 14 | 9.88 [1.38–24.3] | 15.07 [2.47–35.52] | 28.53 [4.05–77.55] | 5.06 [0.53–14.36] | 278.00 [43.38–762] |
| Lung squamous cell carcinoma | 9 | 14.30 [7.33–28.60] | 21.59 [2.45–44.48] | 31.27 [16.84–55.55] | 5.49 [3.01–10.48] | 272.90 [114–592] |
| Lymphoma | 7 | 7.40 [4.64–17.80] | 13.49 [8.05–34.50] | 23.13 [9.77–68.51] | 4.03 [1.74–12.46] | 248.80 [102.10–938] |
*Values are median [min–max]
Note that MRFDG and Ki values are multiplied by 100. Lumped constant = 1
SUVmax is included for comparison, as it is the most used PET metric in clinical practice
We next investigated the impact of diabetes on SUV, MRFDG and DVFDG values. As expected, patients with diabetes had significantly higher average blood glucose than patients with no history of diabetes (7.6 ± 1.5 mmol/L vs. 5.7 ± 0.7 mmol/L, p < 0.0001), whereas the two groups did not differ on age, sex and BMI distributions.
SUVmean was significantly lower in patients with diabetes in both brain areas and the heart and was greater in the colon, kidney, pancreas and spleen. By contrast, MRFDG in the brain and heart was not impacted by diabetes status, whereas patients with diabetes had significantly greater MRFDG in the bone, colon, kidney, liver, lungs, paravertebral muscle, pancreas, and stomach.
Time-activity curves
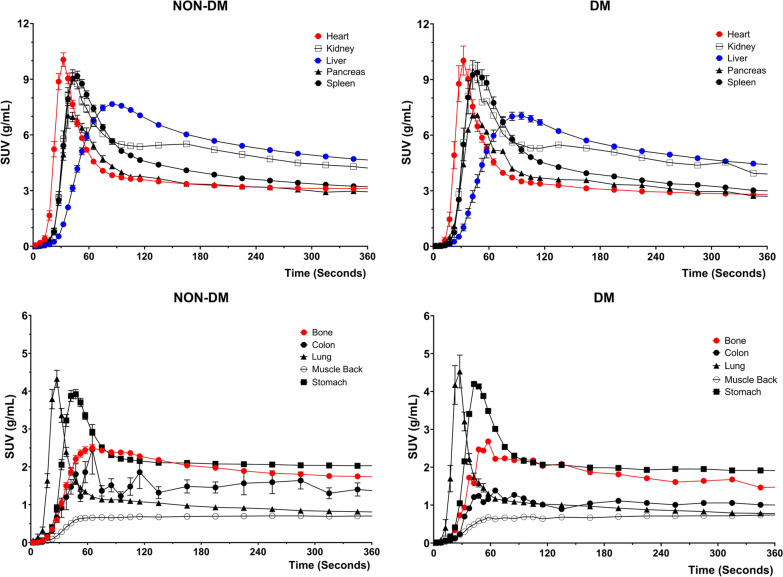
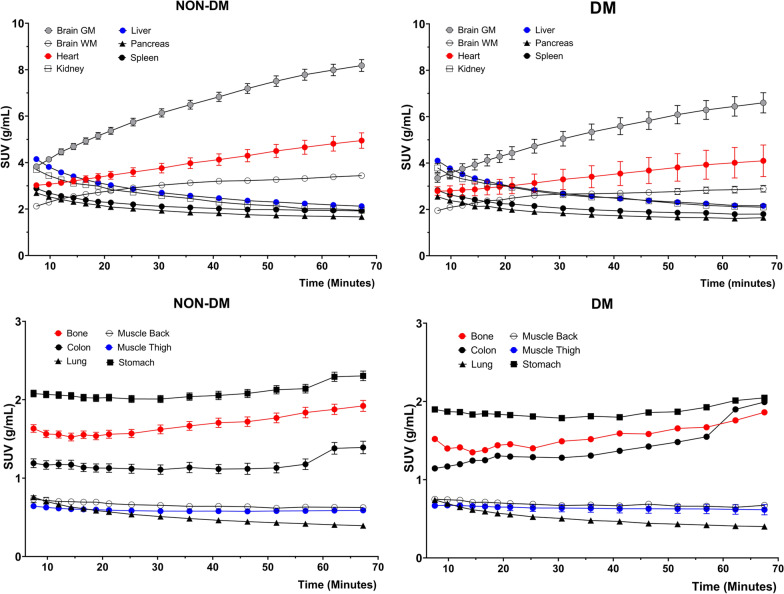
The D-WB dynamic series contain complete TACs of most presented organs and tissues as presented in Figs. 3 and 4. The TACs confirm previous findings of FDG kinetics, such as the largely increasing uptake by the brain and heart, and decreasing uptake by the kidneys, but also interestingly solidly increasing FDG activity over time in the bone marrow. The characteristics of the TAC shapes are reflected in the SUV, MRFDG, and DVFDG values in Fig. 2. Tissues with increasing TACs generally have greater MRFDG values than tissue with decreasing TACs.
Fig. 3.
Time-activity curves for the first 6 min of dynamic scanning over the chest area. Represented values are mean and SEM of all 126 patients. The panels on the left side correspond to the patients without diabetes; the panels on the right side to the patients with diabetes
Fig. 4.
Time-activity curves for the remaining 6–70 min of dynamic WB scanning. Represented values are mean and SEM of all 126 patients. The panels on the left side correspond to the patients without diabetes; the panels on the right side to the patients with diabetes
Comparison of multiparametric and TAC-based MRFDG values
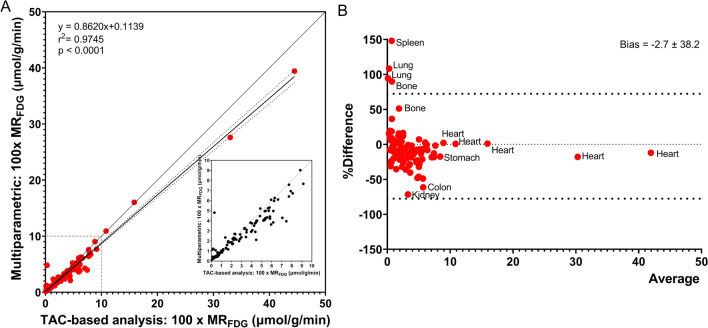
Figure 5 shows the results of the comparison study between the values extracted from the multiparametric images from direct reconstruction versus the manual post-reconstruction TAC-based calculations of the Patlak model performed in PMOD’s PKIN module. The analysis showed an excellent correlation (r2 = 0.97, p < 0.0001), with a tendency towards higher manual TAC-based MRFDG values (bias 3% in the Bland–Altman analysis).
Fig. 5.
A Correlation between multiparametric MRFDG values and manual TAC-based MRFDG estimates using PMOD’s PKIN module. N = 10, corresponding to 110 correlation points. B Bland–Altman analysis of %Difference (100*(Multiparametric MRFDG − TAC-based MRFDG)/average) vs average
Correlations
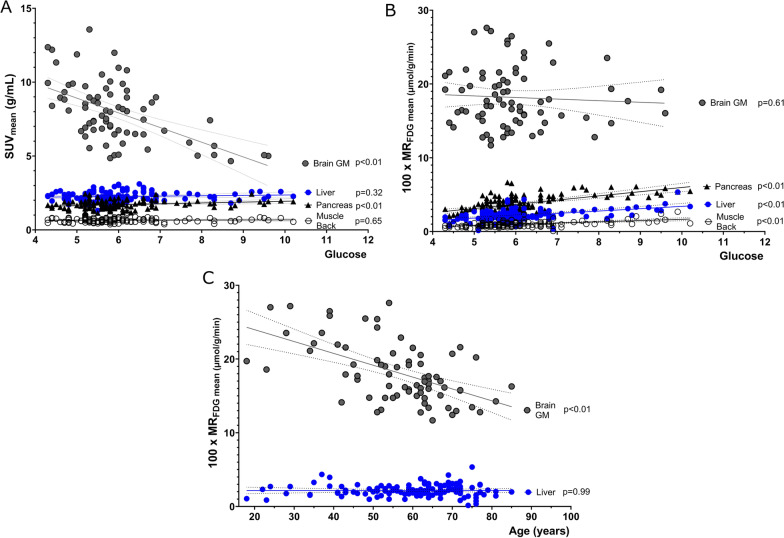
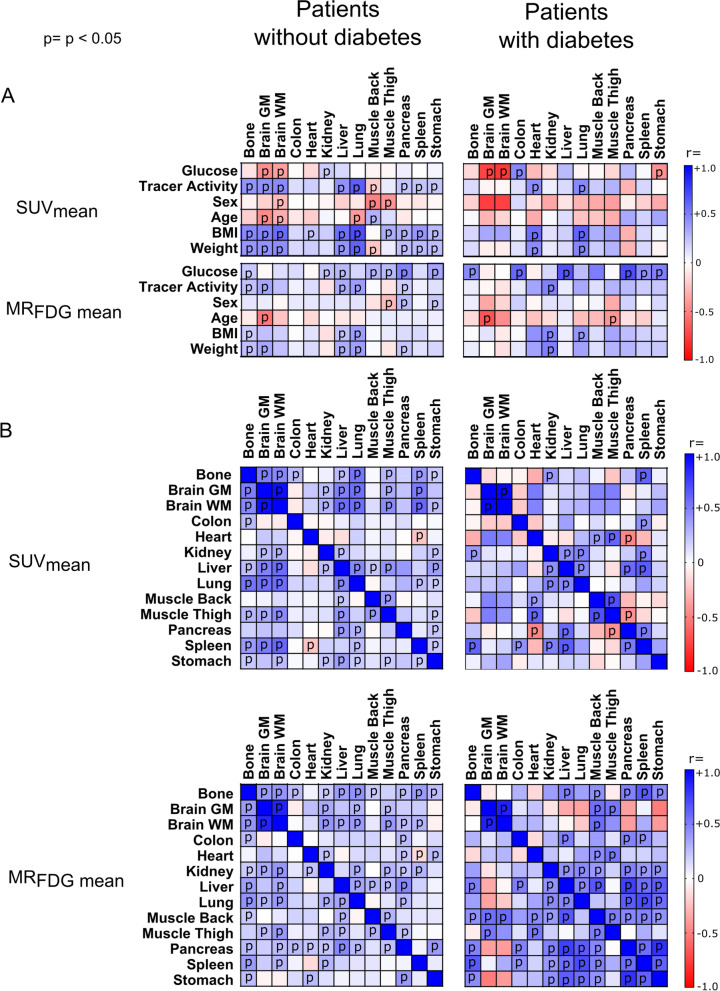
The brain and heart were the only organs where MRFDG was not directly correlated with blood glucose (Fig. 6B). By contrast, brain SUV values were negatively correlated with blood glucose (Fig. 6A). Both cardiac and cerebral MRFDG significantly decreased with increasing age as previously reported in numerous studies (Fig. 6C). Explorative correlations of SUV and MRFDG with patient glucose levels, age, injected tracer dose and patient weight/BMI are presented in Fig. 7.
Fig. 6.
Correlation of A SUVmean and B MRFDG mean with glucose for brain grey matter, pancreas, liver, and skeletal paravertebral muscle. C Correlation of MRFDG mean and age. Plotted are the VOIs for brain grey matter and liver tissue
Fig. 7.
Pearson’s correlation analysis of patients with and without diabetes, of SUVmean and MRFDG mean with glucose levels, tracer activity, sex, age, BMI, and weight (A); as well as between paired sets of organs (B). Cells labelled “p” showed statistically significant correlation (p < 0.05) without correction for multiple comparisons. The full correlation results can be found in the Additional file 1: figure S1 and S2, table S4A, S4B, S5A and S5B
Discussion
In daily clinical practice, static PET image evaluation consists fundamentally of a visual analysis that relies on good image quality and adequate contrast between target and background areas. Semiquantitative measurements of SUV, sometimes reported as a SUV ratio normalized to the blood or liver background, are often used to complement the visual information and are of particular relevance in clinical follow-up and response evaluation.
However, the use of SUV as a reference measurement has a several methodological limitations which can inhibit the correct interpretation of FDG PET/CT images. This is particularly relevant in treatment response evaluation studies as any delay in the scan acquisition time will result in different measurements [2]. Furthermore, these measurements are also dependent on factors such as hardware calibration, imaging protocols and patient physical characteristics [3, 16–18], meaning that it has so far proved impossible to reach international agreement on the use of FDG uptake levels to define pathology. Nevertheless, decades of optimization have been applied to conventional static SUV images to achieve the current standard observed in departments worldwide.
Contrasting this, multiparametric images from D-WB PET are novel in clinical PET and have yet to be optimized and standardized to the same extent [4]. Compared to SUV images, parametric images have different organ signal and lesion-to-background patterns [8, 9]. Thus, nuclear medicine clinicians therefore must update their reporting practices and measurements. The main purpose of this study was therefore to facilitate easier parametric image reading by reporting extensive parametric normal values of non-diseased tissue and organs.
It should be noted that the multiparametric images analysed in this study were produced by direct reconstruction from PET raw data [19], which lead to parametric images that are clearly less noisy than parametric images produced by the traditional indirect image-based approach [4]. Thus, the multiparametric images were of good visual quality and the distribution plots of mean organ and tissue MRFDG were almost similar to that of conventional SUV values reflecting that there was little inter-individual variation in the overall presentation of organs and tissues in the parametric images. A narrow range of mean MRFDG in background tissue and organs naturally enhances the detection of patterns of pathology and thus the clinical value of the parametric images. MRFDG in pathological lesions was also uniformly greater than in background tissue as seen in Table 3.
However, a few caveats should be mentioned. First, although great care was observed during delineation of organ and tissue structures for the quantitative analysis, some of the resulting VOIs were drawn based on poor target-to-background radioactivity and difficult CT morphology. This was particularly evident for the pancreas and other abdominal organs. Second, the length of the acquisition protocol makes D-WB PET/CT images prone to motion artefacts, which profoundly impacts quantitative measurements such as MRFDG. Bulk motion tends to result in blurry images with underestimated SUV values in static PET imaging, whereas it may dramatically affect MRFDG and DVFDG values in areas adjoining FDG avid structures. The effect can cause artefacts at the edges of organs and around lesions. Third, overlapping signal from neighbouring structures proved to be impossible to avoid in all tissues. For example, the area of the pancreas is almost impossible to correctly individualize due to poor background radioactivity and high tissue density in the upper abdomen. It is expected that the values obtained from this region includes a degree of spill-in from neighbouring structures.
MRFDG and DVFDG values of the multiparametric images could be accurately reproduced by a traditional TAC-based Patlak analysis using PMOD PKIN, and a strong correlation between parameter estimates from the two methods was observed. However, a 3% bias towards greater MRFDG estimates was present when calculated with the TAC-based Patlak analysis, which could be a consequence of the different noise propagation in the two methods: the multiparametric images are reconstructed directly from PET raw data with well-modelled Poisson noise [15], whereas post-reconstruction dynamic PET data are used for the TAC-based analysis. We found, however, that the relative difference between multiparametric images and manually calculated MRFDG was constant, and it is therefore our opinion that multiparametric MRFDG values can be used with as much confidence as manually calculated values. The reproducibility of multiparametric images could be assessed in future test–retest studies.
Excluding the brain and the heart, surprisingly little data have been published on the TACs of FDG in normal tissues and tumours with the exception of some dual-time point studies [20–25]. With D-WB PET, TACs of tracer in all tissues within the entire body during the 60-min scan are available as opposed to the regular static PET scans. D-WB thus allows clinicians to perform a multi-time point type of analysis within the normal scan duration and for any area of the body. The TACs presented in this paper are in line with previous observations of mostly increasing FDG uptake in the heart and brain, and mostly decreasing uptake in the liver, kidneys, and the spleen. As such, these TACs are confirmatory, but they also attest to the ease with which TACs of pathologies ranging from inflammatory diseases to malignancies can be obtained.
A clear physiologic interpretation of parameters obtained from full kinetic modelling requires high-quality dynamic PET data and a physiologically reasonable model. Traditionally, kinetic modelling was limited to single organs that were analysed using dedicated organ- and tracer-specific models and input functions. D-WB FDG imaging has the great advantage of covering the larger axial field of view needed for multi-organ applications and metastatic disease evaluation in oncology, but in its current form the same model is applied to all tissues, which complicates reading of the multiparametric images. The Patlak model assumes an irreversible FDG uptake (trapped in the form of FDG-6-phosphate), and the model should be applied to D-WB data only after the blood pool and reversible compartments (free FDG in blood and tissue) reach a steady-state. These assumptions, which are clearly not met by all tissues, should be taken into account when interpreting multi-parametric images: liver and kidneys are examples of tissues that are better described by reversible kinetic models. However, it should be noted that WB parametric images provide MRFDG and DVFDG values for all tissues regardless of whether these tissues are characterized by irreversible FDG uptake. The resulting MRFDG and DVFDG values may therefore not always reflect an actual physiological estimate of metabolism.
Consequently, interpretation of multiparametric images requires insight into normal glucose metabolism in all organs and tissues. For example, glucose metabolism and consequently FDG uptake in the myocardium varies significantly between patients depending on, e.g. levels of insulin (high calorie versus low-calorie diets), length of fasting or previous exercise[26–28]. The large variability observed in cardiac MRFDG values is therefore to be expected. Another particular case is the brain. For example, it is well known that SUV values in the brain decrease in patients with unregulated elevated blood sugar [18, 29]. This reduction in SUV (shown in Fig. 6A) occurs due to the competitive nature of glucose metabolism in the brain tissue and does not correspond to real metabolic changes in the brain, since brain glucose metabolism is not substrate driven. MRFDG represents the actual metabolic rate of glucose in brain tissue, and the above correlations are therefore not present in the multiparametric images. Also of interest, and as demonstrated previously [30], the metabolic rate of glucose consumption in the brain was inversely correlated with age (Fig. 6C). Therefore, in patients with unregulated blood glucose levels, the use of multiparametric images could be advantageous over traditional SUV semi-quantification. Easy access to cross-tabulated MRFDG in all tissues and organs also allows for hypothesis generating visualizations of organ cross-talk and the impact of, e.g. diabetes on glucose homeostasis (see Fig. 7).
Some limitations to the study and general use of D-WB should be noted. First, our study population consisted of patients referred for a FDG PET scan due to suspicion of pathology. Even though pathology was not always present, and no area of interest was drawn over areas visibly affected by disease, the measured normal values are not those of a healthy control population. However, our reported normal values were in line with results previously published in healthy volunteers for skeletal muscle [31, 32], the liver [33], the fasted heart [34, 35] and the brain [36, 37]. Second, current 70-min D-WB parametric protocols are laborious and occupy what is in all PET departments a finite amount of scanner time, which limits the everyday usability of this technique. However, by implementing population-based input functions, we expect that the length of the D-WB parametric imaging protocols can be reduced to 20 min, which allows a reasonable clinical throughput of two D-WB examinations per hour.
The study of whole-body metabolism is on path to make a resurgence in the nuclear medicine world. Total body PET scanners facilitate easier acquisition of whole body tracer kinetics and allow for more advanced modelling of tracer kinetics and organ interactions[38–41]. Upcoming parametric PET research could result in large libraries of human metabolic data, initially mostly based on FDG. Using FDG reference values and assisted by AI computing, clinicians may be able to interpret medical imaging in a way that was not possible before. For example, results of individual scans may be compared with values from a reference library of metabolic data in order to, e.g. modify individual treatment plans and dosages. However, this field of research should not be restricted to total body scanners, since even limited axial field of view PET scanners using automated whole-body parametric reconstruction algorithms can contribute valuable and reproducible data.
Conclusion
The automated D-WB FDG scan protocol provides high-quality multiparametric images and organ MRFDG values with little inter-subject variation and in agreement with manual TAC-based and literature values. The technique therefore facilitates multiparametric image reading, more accurate clinical reports and simpler acquisition of quantitative estimates of whole-body tissue glucose metabolism. Whole-body multi-parametric imaging can play an influential role in the future enhanced interpretation of PET images such as comparisons of individual patient data to multi-centre reference libraries of whole-body parametric images of biological relevant parameters and studies of interaction between organs.
Supplementary Information
Additional file 1. Table 1. VOI delineation methodology. Table 2A. Normal values of males and females in the population without diabetes (N = 100). Table 2B. Normal values of males and females in the population with diabetes (N = 26). Table 3. Lumped Constant. Figure 1. Pearson’s correlation results for patients without diabetes. Table 4A. Patients without diabetes P values results for SUVmean. Table 4B. Patients without diabetes P values results for MRFDG mean. Figure 2. Pearson’s correlation results for patients with diabetes. Table 5A. Patients with diabetes P values results for SUVmean. Table 5B. Patients with diabetes P values results for MRFDG mean.
Acknowledgements
We would like to acknowledge the hard work of the technologists that participated in this study.
Abbreviations
- AI
Artificial intelligence
- BMI
Body mass index
- DM
Diabetes mellitus
- DVFDG
Distribution volume of FDG
- D-WB
Dynamic whole-body
- FDG
18F-fluorodeoxyglucose
- FDG-6-P
FDG-6-phosphate
- IDIF
Image-derived input function
- MRFDG
Metabolic rate of FDG uptake
- PET/CT
Positron emission tomography/computer tomography
- SUV
Standard uptake value
- TACs
Time-activity curves
Authors' contributions
AHD processed data, analysed all images, delineated regions of interest, performed statistics and study semi-quantifications (except for the brain areas), and wrote the final version of the manuscript. AH performed the semi-quantitative analysis of brain areas. AHD, OLM and LCG conceived the study. All authors read and approved the final manuscript.
Funding
This project was partly funded by Siemens Healthineers. LCG and OLM are supported by an unrestricted grant from The Novo Nordisk Foundation (NNF19OC0055100). The participating researchers are paid by their institutions and have no affiliation with the study's financial sponsors.
Availability of data and materials
Within the restrictions applied by the EU GDPR, all data are available from the authors upon reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the local ethics committee in Region Midtjylland, Denmark (1-10-72-188-19). All methods were carried out in accordance with relevant guidelines and regulations. Informed consent was obtained from each patient before inclusion into the study.
Consent for publication
Consent for publication was obtained from each participant before inclusion into the study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lars C. Gormsen and Ole L. Munk contributed equally to this work
References
- 1.Boellaard R, Delgado-Bolton R, Oyen WJ, Giammarile F, Tatsch K, Eschner W, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging. 2015;42:328–54. doi: 10.1007/s00259-014-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keyes JW., Jr SUV: standard uptake or silly useless value? J Nucl Med. 1995;36:1836–1839. [PubMed] [Google Scholar]
- 3.Boellaard R, Krak NC, Hoekstra OS, Lammertsma AA. Effects of noise, image resolution, and ROI definition on the accuracy of standard uptake values: a simulation study. J Nucl Med. 2004;45:1519–1527. [PubMed] [Google Scholar]
- 4.Rahmim A, Lodge MA, Karakatsanis NA, Panin VY, Zhou Y, McMillan A, et al. Dynamic whole-body PET imaging: principles, potentials and applications. Eur J Nucl Med Mol Imaging. 2019;46:501–518. doi: 10.1007/s00259-018-4153-6. [DOI] [PubMed] [Google Scholar]
- 5.Dimitrakopoulou-Strauss A, Pan L, Sachpekidis C. Kinetic modeling and parametric imaging with dynamic PET for oncological applications: general considerations, current clinical applications, and future perspectives. Eur J Nucl Med Mol Imaging. 2021;48:21–39. doi: 10.1007/s00259-020-04843-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patlak C, Blasberg R. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. Generalizations. J Cereb Blood Flow Metab. 1985;5:584–590. doi: 10.1038/jcbfm.1985.87. [DOI] [PubMed] [Google Scholar]
- 7.Patlak C, Blasberg R, Fenstermacher J. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab. 1983;3:1–7. doi: 10.1038/jcbfm.1983.1. [DOI] [PubMed] [Google Scholar]
- 8.Dias AH, Pedersen MF, Danielsen H, Munk OL, Gormsen LC. Clinical feasibility and impact of fully automated multiparametric PET imaging using direct Patlak reconstruction: evaluation of 103 dynamic whole-body 18 F-FDG PET/CT scans. Eur J Nucl Med Mol Imaging. 2020 doi: 10.1007/s00259-020-05007-2. [DOI] [PubMed] [Google Scholar]
- 9.Fahrni G, Karakatsanis N, Di Domenicantonio G, Garibotto V, Zaidi H. Does whole-body Patlak (18)F-FDG PET imaging improve lesion detectability in clinical oncology? Eur Radiol. 2019;29:4812–4821. doi: 10.1007/s00330-018-5966-1. [DOI] [PubMed] [Google Scholar]
- 10.Tao Y, Peng Z, Krishnan A, Zhou XS. Robust learning-based parsing and annotation of medical radiographs. IEEE Trans Med Imaging. 2011;30:338–350. doi: 10.1109/tmi.2010.2077740. [DOI] [PubMed] [Google Scholar]
- 11.Dias AH, Pigg D, Smith AM, Shah V, Gormsen LC, Munk OL. Clinical validation of a population-based input function for dynamic whole-body 18F-FDG multiparametric PET imaging using a standard injector. In: 34th Annual Congress of the European Association of nuclear medicine: European journal of nuclear medicine and molecular imaging, Suppl. 1. 2021. p. 198–199.
- 12.Wu H, Bergsneider M, Glenn T, Yeh E, Hovda D, Phelps M, et al. Measurement of the global lumped constant for 2-deoxy-2-[18F]fluoro-D-glucose in normal human brain using [15O]water and 2-deoxy-2-[18F]fluoro-D-glucose positron emission tomography imaging. A method with validation based on multiple methodologies. Mol Imaging Biol. 2003;1:1. doi: 10.1016/s1536-1632(02)00122-1. [DOI] [PubMed] [Google Scholar]
- 13.Graham M, Muzi M, Spence A, O'Sullivan F, Lewellen T, Link J, et al. The FDG lumped constant in normal human brain. J Nucl Med. 2002;43:1157–1166. [PubMed] [Google Scholar]
- 14.Laffon E, Marthan R. Is Patlak y-intercept a relevant metrics? Eur J Nucl Med Mol Imaging. 2021 doi: 10.1007/s00259-020-04954-0. [DOI] [PubMed] [Google Scholar]
- 15.Reader A, Verhaeghe J. 4D image reconstruction for emission tomography. Phys Med Biol. 2014 doi: 10.1088/0031-9155/59/22/R371. [DOI] [PubMed] [Google Scholar]
- 16.Thie JA. Understanding the standardized uptake value, its methods, and implications for usage. J Nucl Med. 2004;45:1431–1434. [PubMed] [Google Scholar]
- 17.Huang SC. Anatomy of SUV. Standardized uptake value. Nucl Med Biol. 2000;27:643–646. doi: 10.1016/s0969-8051(00)00155-4. [DOI] [PubMed] [Google Scholar]
- 18.Eskian M, Alavi A, Khorasanizadeh M, Viglianti B, Jacobsson H, Barwick T, et al. Effect of blood glucose level on standardized uptake value (SUV) in 18 F- FDG PET-scan: a systematic review and meta-analysis of 20,807 individual SUV measurements. Eur J Nucl Med Mol Imaging. 2019;46:224–237. doi: 10.1007/s00259-018-4194-x. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Qi J. Acceleration of the direct reconstruction of linear parametric images using nested algorithms. Phys Med Biol. 2010;55:1505–1517. doi: 10.1088/0031-9155/55/5/016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wangerin K, Muzi M, Peterson L, Linden H, Novakova A, O'Sullivan F, et al. Effect of 18 F-FDG uptake time on lesion detectability in PET imaging of early stage breast cancer. Tomography. 2015;1:53–60. doi: 10.18383/j.tom.2015.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasdemir B, Güzel Y, Komek H, Can C. Evaluation of dual time-point fluorodeoxyglucose PET/computed tomography imaging in gastric cancer. Nucl Med Commun. 2020;41:1322–1327. doi: 10.1097/MNM.0000000000001290. [DOI] [PubMed] [Google Scholar]
- 22.Pang L, Bo X, Wang J, Wang C, Wang Y, Liu G, et al. Role of dual-time point 18 F-FDG PET/CT imaging in the primary diagnosis and staging of hilar cholangiocarcinoma. Abdom Radiol. 2021;46:4138–4147. doi: 10.1007/s00261-021-03071-2. [DOI] [PubMed] [Google Scholar]
- 23.Kurland B, Muzi M, Peterson L, Doot R, Wangerin K, Mankoff D, et al. Multicenter clinical trials using 18F-FDG PET to measure early response to oncologic therapy: effects of injection-to-acquisition time variability on required sample size. J Nucl Med. 2016;57:226–230. doi: 10.2967/jnumed.115.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang J, Moon J, Kim H, Lee M, Lim C, Park S, et al. Prognostic value of metabolic parameters measured by pretreatment dual-time-point 18F-fluorodeoxyglucose positron emission tomography/computed tomography in patients with intrahepatic or perihilar cholangiocarcinoma: a STROBE study. Medicine. 2021 doi: 10.1097/MD.0000000000026015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grisanti F, Zulueta J, Rosales J, Morales M, Sancho L, Lozano M, et al. Diagnostic accuracy of visual analysis versus dual time-point imaging with 18 F-FDG PET/CT for the characterization of indeterminate pulmonary nodules with low uptake. Rev Esp Med Nucl Imagen Mol. 2021;40:155–160. doi: 10.1016/j.remn.2020.03.019. [DOI] [PubMed] [Google Scholar]
- 26.de Groot M, Meeuwis A, Kok P, Corstens F, Oyen W. Influence of blood glucose level, age and fasting period on non-pathological FDG uptake in heart and gut. Eur J Nucl Med Mol Imaging. 2005 doi: 10.1007/s00259-004-1670-2. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Hsu C, Wu Y, Chen H, Chiu N. Effect of fasting duration on myocardial fluorodeoxyglucose uptake in diabetic and nondiabetic patients. Nucl Med Commun. 2021 doi: 10.1097/MNM.0000000000001339. [DOI] [PubMed] [Google Scholar]
- 28.Kang J, Lee M, Kim Y. Associations of physiologic myocardial 18 F-FDG uptake with fasting duration, HbA1c, and regular exercise. Ann Nucl Med. 2021 doi: 10.1007/s12149-020-01551-x. [DOI] [PubMed] [Google Scholar]
- 29.Sarikaya I, Sarikaya A, Sharma P. Assessing the effect of various blood glucose levels on 18 F-FDG activity in the brain, liver, and blood pool. J Nucl Med Technol. 2019;47:313–318. doi: 10.2967/jnmt.119.226969. [DOI] [PubMed] [Google Scholar]
- 30.Kakimoto A, Ito S, Okada H, Nishizawa S, Minoshima S, Ouchi Y. Age-related sex-specific changes in brain metabolism and morphology. J Nucl Med. 2016;57:221–225. doi: 10.2967/jnumed.115.166439. [DOI] [PubMed] [Google Scholar]
- 31.Gheysens O, Postnov A, Deroose C, Vandermeulen C, de Hoon J, Declercq R, et al. Quantification, variability, and reproducibility of basal skeletal muscle glucose uptake in healthy humans using 18F-FDG PET/CT. J Nucl Med. 2015;56:1520–1526. doi: 10.2967/jnumed.115.159715. [DOI] [PubMed] [Google Scholar]
- 32.Reinhardt M, Beu M, Vosberg H, Herzog H, Hübinger A, Reinauer H, et al. Quantification of glucose transport and phosphorylation in human skeletal muscle using FDG PET. J Nucl Med. 1999;40:977–985. [PubMed] [Google Scholar]
- 33.Choi Y, Hawkins R, Huang S, Brunken R, Hoh C, Messa C, et al. Evaluation of the effect of glucose ingestion and kinetic model configurations of FDG in the normal liver. J Nucl Med. 1994;35:818–823. [PubMed] [Google Scholar]
- 34.Zuo Y, López J, Smith T, Foster C, Carson R, Badawi R, et al. Multiparametric cardiac 18 F-FDG PET in humans: pilot comparison of FDG delivery rate with 82 Rb myocardial blood flow. Phys Med Biol. 2021 doi: 10.1088/1361-6560/ac15a6. [DOI] [PubMed] [Google Scholar]
- 35.Kemppainen J, Fujimoto T, Kalliokoski K, Viljanen T, Nuutila P, Knuuti J. Myocardial and skeletal muscle glucose uptake during exercise in humans. J Physiol. 2002;542:403–412. doi: 10.1113/jphysiol.2002.018135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huisman M, van Golen L, Hoetjes N, Greuter H, Schober P, Ijzerman R, et al. Cerebral blood flow and glucose metabolism in healthy volunteers measured using a high-resolution PET scanner. EJNMMI Res. 2012 doi: 10.1186/2191-219X-2-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heiss W, Habedank B, Klein J, Herholz K, Wienhard K, Lenox M, et al. Metabolic rates in small brain nuclei determined by high-resolution PET. J Nucl Med. 2004;45:1811–1815. [PubMed] [Google Scholar]
- 38.Feng T, Zhao Y, Shi H, Li H, Zhang X, Wang G, et al. Total-body quantitative parametric imaging of early kinetics of 18 F-FDG. J Nucl Med. 2021 doi: 10.2967/jnumed.119.238113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nadig V, Herrmann K, Mottaghy F, Schulz V. Hybrid total-body pet scanners-current status and future perspectives. Eur J Nucl Med Mol Imaging. 2021 doi: 10.1007/s00259-021-05536-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surti S, Pantel A, Karp J. Total Body PET: Why, How, What for? IEEE Trans Radiat Plasma Med Sci. 2020 doi: 10.1109/trpms.2020.2985403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Li E, Cherry S, Wang G. Total-Body PET Kinetic Modeling and Potential Opportunities Using Deep Learning. PET Clin. 2021 doi: 10.1016/j.cpet.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table 1. VOI delineation methodology. Table 2A. Normal values of males and females in the population without diabetes (N = 100). Table 2B. Normal values of males and females in the population with diabetes (N = 26). Table 3. Lumped Constant. Figure 1. Pearson’s correlation results for patients without diabetes. Table 4A. Patients without diabetes P values results for SUVmean. Table 4B. Patients without diabetes P values results for MRFDG mean. Figure 2. Pearson’s correlation results for patients with diabetes. Table 5A. Patients with diabetes P values results for SUVmean. Table 5B. Patients with diabetes P values results for MRFDG mean.
Data Availability Statement
Within the restrictions applied by the EU GDPR, all data are available from the authors upon reasonable request.