Abstract
Ribosome biogenesis is an essential, energy demanding process whose deregulation has been implicated in cancer, aging, and neurodegeneration. Ribosome biogenesis is therefore under surveillance of pathways including the p53 tumor suppressor. Here, we first performed a high-content siRNA-based screen of 175 human ribosome biogenesis factors, searching for impact on p53. Knock-down of 4 and 35 of these proteins in U2OS cells reduced and increased p53 abundance, respectively, including p53 accumulation after depletion of BYSL, DDX56, and WDR75, the effects of which were validated in several models. Using complementary approaches including subcellular fractionation, we demonstrate that endogenous human WDR75 is a nucleolar protein and immunofluorescence analysis of ectopic GFP-tagged WDR75 shows relocation to nucleolar caps under chemically induced nucleolar stress, along with several canonical nucleolar proteins. Mechanistically, we show that WDR75 is required for pre-rRNA transcription, through supporting the maintenance of physiological levels of RPA194, a key subunit of the RNA polymerase I. Furthermore, WDR75 depletion activated the RPL5/RPL11-dependent p53 stabilization checkpoint, ultimately leading to impaired proliferation and cellular senescence. These findings reveal a crucial positive role of WDR75 in ribosome biogenesis and provide a resource of human ribosomal factors the malfunction of which affects p53.
Subject terms: Tumour-suppressor proteins, Cancer
Introduction
Mammalian ribosomes are complex cellular machines composed of four ribosomal RNA (rRNA) and ~80 distinct ribosomal proteins (RPs) [1, 2]. Ribosomes translate the genetic information contained in mRNAs to synthesize proteins, thereby playing an essential role in the execution of gene expression programs that regulate fundamental biological processes, including cell growth, cell division, and differentiation [3, 4]. Ribosome biogenesis is initiated in the nucleolus where the 47S rRNA precursor containing the sequences for 18S, 5.8 and 28S rRNAs is transcribed by a 14-subunit enzyme RNA polymerase I (Pol I) and several Pol I-associated transcription factors, including selectivity factor 1, transcription initiation factor I A and upstream binding factor [5]. The 47S rRNA is co-transcriptionally assembled into the 90S processome with early binding RPs and a large number of ribosome biogenesis factors (RBFs) [1, 2]. During the maturation of the 90S processome into pre-40S and pre-60S ribosomal subunits, pre-rRNA is chemically modified and the mature rRNA released through processing [6]. The 5S rRNA is transcribed by Pol III in the nucleoplasm and following assembly with RPL5 (uL18) and RPL11 (uL5) (See ref. [7], for the new nomenclature of RPs) forms the 5S ribonucleoprotein particle (5S RNP), also known as the RPL5/RPL11/5S rRNA complex, that is incorporated into the pre-60S ribosomal subunit under normal growth conditions, forming the central protuberance of the mature 60S ribosomal subunits [8, 9]. The remaining RPs assemble into pre 40S and pre 60S ribosomal subunits in a step-wise manner in the nucleoplasm and the cytoplasm [1, 2]. The complete process of ribosome biogenesis involves ~200 RBFs, which transiently associate with the developing pre-ribosomes at specific assembly stages [6, 10–12]. Given the tremendous investment of cell’s resources and energy into ribosome biogenesis, it is not surprising that ribosome biogenesis is highly orchestrated by several signaling pathways that sense and respond to the availability of nutrients, intracellular energy levels, growth factors, mitogens, and various types of cellular stresses [12, 13]. As ribosome biogenesis is essential for the regulation of numerous cellular processes and quantitative and qualitative alterations in this process contribute to the pathogenesis of diverse diseases, most notably ribosomopathies and cancer, it was originally hypothesized that mechanisms had evolved to sense the fidelity of this process [12–17]. Early studies demonstrated that perturbations of ribosome biogenesis in mammalian cell culture and in vivo models are sensed by a p53 signaling pathway independently of DNA damage [18–23]. It has been subsequently demonstrated that upon impairment of ribosome biogenesis the nascent 5S RNP is redirected from assembling into ribosomes to bind the human homolog of mouse double minute 2 (HDM2), thereby inhibiting HDM2-mediated ubiquitination and degradation of the p53 protein [19, 24–29]. This checkpoint response is also known as the Impaired Ribosome Biogenesis Checkpoint (IRBC) [2]. This model might help explain how defects in the biogenesis of the 60S ribosome could activate p53 in the 5S RNP-dependent manner [19, 20]. However, it cannot easily be reconciled with the observations that the depletion of RPs of the 40S subunit, that abolishes the assembly of the 40S ribosomal subunit but not the 60S ribosomal subunit can also trigger the 5S RNP-dependent p53 activation [19–21]. Recent evidence demonstrated that the 5S RNP-dependent activation of p53 upon impairment of ribosome biogenesis plays a role in tumor suppression by triggering apoptosis or senescence as well as mediating the anticancer effects of ribosome biogenesis inhibitors [30–33]. Thus, it is of a paramount importance to identify the key factors involved, and characterize the molecular mechanisms underlying the regulation of this pathway.
In this study, we systematically depleted a large number of RBFs (known at the time when our study was initiated) that participate in distinct stages of ribosome biogenesis, including 76 RPs of the 60S or 40S ribosomal subunits to gain insights into the lesions in ribosome biogenesis that trigger the 5S RNP-dependent p53 activation. Our study therefore complements the screening efforts based on other readouts, that helped extend our view of the proteins involved in nucleolar biology and ribosome function [11, 12, 34]. Consistent with the previously published study by the Lafontaine group who focused on RPs [9], we reveal that the depletion of the majority of distinct RPs components of the 60S ribosome subunit trigger strong p53 activation, in contrast to the depletion of the 40S subunit RPs (our present results). As described below, we also identified a large number of RBFs whose depletion leads to upregulation of p53 protein abundance, including WDR75, BYSL, and DDX56. Based on our functional studies, we now also describe a mechanism of WDR75-dependent regulation of Pol I-mediated transcription, in addition to the previously suggested role of WDR75 in pre-rRNA processing within the 90S processome [12, 35]. Last but not least, we suggest that defects in both of these processes upon WDR75 knockdown likely contribute to the 5S RNP-dependent p53 activation and cell fate decision leading to cellular senescence.
Materials and methods
Cell lines
Human osteosarcoma U2OS, adenocarcinoma HeLa, normal foreskin fibroblasts BJ and diploid retinal pigment epithelium RPE1 cells were grown in DMEM (Biosera, LM-D1110/500) supplemented with 10% fetal bovine serum (Gibco, 10270106) and penicillin/streptomycin (Sigma-Aldrich, P4333). While all the above cell types have wild-type p53, the p53 protein in the HeLa cell line is strongly repressed by overexpression of the E6 oncogene from HPV16. All four cell types were purchased from ATCC, and were regularly tested for mycoplasma contamination. U2OS cells stably expressing GFP-tagged WDR75 were obtained by transfection of pCMV6-AC-WDR75-GFP plasmid (Origene, RG207339) and antibiotic selection.
Chemicals
In some experiments, cells were treated with indicated concentrations of the following drugs known to inhibit Pol I-mediated transcription: 5 nM Actinomycin D (Sigma-Aldrich, A1410) and 0.5 μM BMH-21 (Selleckchem, S7718).
Antibodies
β-Actin (Santa Cruz Biotechnology, sc-47778)
BYSL (Abcam, ab194961)
DDX18 (Sigma, HPA041056)
DDX56 (Abcam, ab115158)
Fibrillarin (Abcam, ab5821)
Fibrillarin (Invitrogen, MA3-16771)
HDM2 (Abcam, ab16895)
Histone H3 phosphorylated S10 (Millipore, 06-570)
Lamin B1 (Santa Cruz Biotechnology, sc-6217)
Nucleolin (Abcam, ab70493)
p21 (Cell Signaling, #2947)
p53 (Santa Cruz Biotechnology, sc-6243)
PARP1 (Cell Signaling, #9532)
RPA135 (Santa Cruz Biotechnology, sc-293272)
RPA194 (Santa Cruz Biotechnology, sc-48385)
RPL5 (Abcam, ab157099)
RPL11 (our own antibody [36])
RPL23 (our own antibody [19])
SMC1 (Abcam, ab9262)
α-Tubulin (Santa Cruz Biotechnology, sc-8035)
tGFP (Origene, TA150041)
UTP15 (Sigma, HPA044697)
WDR43 (Bethyl, A302-478A)
WDR75 (Abcam, ab192922)
WDR75 (Novus, NBP1-82296)
siRNA-based high-content microscopy screening
All siRNAs used in this RBF library were obtained from Ambion as Silencer Select reagents and used at a final concentration of 5 nM. As negative controls, we used non-targeting siRNAs Silencer Select Negative control #1 (Ambion, 4390846) and Silencer Select Negative control #2 (Ambion, 4390846), both of which gave comparable results to mock-transfection, the latter including the transfection reagent mix without any siRNA. For siRNA transfections in 96-well plates, 25 μl of 40 nM siRNA, diluted in DEPC-Treated Water (Ambion, AM9922), were mixed and incubated for 15 min with 25 μl of HiPerFect transfection reagent (Qiagen, 301705), diluted 1:32 in Opti-MEM medium (Gibco, 31985070), in V-bottom 96-well plates (Kisker, G060). Liquid handling was carried out with a multi-well pipetting device Liquidator 96 (Mettler-Toledo, 17010335). The transfection mix was transferred into 96-well imaging plates (Greiner, 655090) into which 150 μl of a cell suspension containing 9000 U2OS cells in culture medium was added. After 2 days of cultivation, cells were fixed, p53 protein was detected using indirect immunofluorescence and Olympus ScanR Acquisition system. Images were analyzed with the Olympus ScanR Image Analysis Software, a dynamic background correction was applied, nuclei segmentation was performed using an integrated intensity-based object detection module using the DAPI signal. P53 fluorescence intensities within segmented nuclei were quantified and are depicted as p53 fold change relative to Silencer Select negative controls.
RNA interference experiments
All siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen, 13778075) according to the manufacturer’s instructions. All siRNAs listed below were obtained from Ambion as Silencer Select reagents. Unless specified otherwise, siRNAs were used at a final concentration of 14 nM and experiments were performed 72 h after transfection. A 1:1:1 mix of siWDR75 #1, #2, and #3 was used if not specified otherwise.
siBYSL #1 (s2133, 5′-AGGUGGUUGUGGACCCUGAtt-3′)
siBYSL #2 (s2134, 5′-CCAGGAUUUUUGCCUCUAAtt-3′)
siCON (negative control #1, AM4635, 5′-AGUACUGCUUACGAUACGGTT-3′)
siDDX18 #1 (s16982, 5′-GCAUAAAAGUAUCAGACCAtt-3′)
siDDX18 #2 (s16893, 5′-GCAAUGCAGUCUUCCAAUUtt-3′)
siDDX56 #1 (s29253, 5′-GCUUCAAGCACAAAGGAAAtt-3′)
siDDX56 #2 (s29254, 5′-CAGGCAUAGUCUUAACCUUtt-3′)
siP53 (s605, 5′-GUAAUCUACUGGGACGGAAtt-3′)
siRPL5 (s56733, 5′-CAGUUCUCUCAAUACAUAAtt-3′)
siRPL11 (s12169, 5′-CAACUUCUCAGAUACUGGAtt-3′)
siRPL23 #1 (s17871, 5′-GCAGGAGUCAUAGUGAACAtt-3′)
siRPL23 #2 (s17872, 5′-CAAUAAAGGCGAGAUGAAAtt-3′)
siUTP15 #1 (s38548, 5′-GAUACUCCCAAGAACCUAUtt-3′)
siUTP15 #2 (s38549, 5′-GGAGGACAAUUGCUAGUAUtt-3′)
siWDR43 #1 (s229928, 5′-GCCCUUUGAUGGAAUUACAtt-3′)
siWDR43 #2 (s229930, 5′-GGUUUUUGCUCAGACAAAAtt-3′)
siWDR75 #1 (s38531, 5′-GGAUGAAAAACUAAACGAAtt-3′)
siWDR75 #2 (s38532, 5′-GAUUGAUCCAAGAACUAAAtt-3′)
siWDR75 #3 (s38530, 5′-CAGCUAGCAAAGAUGGUUAtt-3′)
siWDR75 3’UTR (ADPAC9P, 5′-GAGGAUCCUUGGACUUUGUtt-3′)
Immunofluorescence
Immunofluorescence staining was performed as previously described [37]. Shortly, cells grown on 12 mm wide glass coverslips (Assistent, 41001112) were washed twice in PBS, fixed for 15 min at RT with 4% formaldehyde, washed in PBS, permeabilized for 5 min with 0.2% Triton X-100 in PBS, washed in PBS and incubated with primary antibodies for 60 min at RT. After the washing step, the coverslips were incubated with goat anti-rabbit Alexa Fluor 488 or goat anti-mouse Alexa Fluor 568 secondary antibodies (Invitrogen, A11034 and A11004) for 60 min at RT, washed with PBS, and mounted using Vectashield mounting medium with DAPI (Vector Laboratories, H-1200). For detection of replicating cells, cells were incubated with 10 mM EdU 30 min before fixation and EdU detection was performed using Click-iT EdU Alexa Fluor 594 imaging kit (Invitrogen, C10639) according to manufacturer’s recommendations. For detection of nascent RNA, cells were incubated with 1 mM EU 30 min before fixation and EU detection was performed using Click-iT RNA imaging kit (Invitrogen, C10329) according to manufacturer’s recommendations.
Microscope image acquisition
Microscope images were acquired using the CellObserver spinning disc confocal microscopic system (Zeiss) equipped with CSU-X1 confocal scanner unit (Yokogawa), Evolve 512 EMCCD camera (Photometrics) and 100 x NA 1,4 plan apochromat objective (Zeiss).
Quantitative microscopy-based cytometry of the immunofluorescence-stained samples was performed using an automatic inverted fluorescence microscope BX71 (Olympus) using ScanR acquisition software (Olympus) and analyzed with ScanR analysis software (Olympus).
Immunoblotting
For immunoblotting, cells were grown in 60 mm cell culture dishes and whole cell extracts were obtained by lysis in Laemmli sample buffer (50 mM Tris–HCl (pH 6.8), 100 mM DTT, 2.0% SDS, 0.1% bromophenol blue, 10% glycerol) and analyzed by SDS–polyacrylamide gel electrophoresis following standard procedures. Primary antibodies were incubated over night at 4 °C in TBS-Tween 20 containing 5% powder milk. Secondary HRP-coupled antibodies (GE Healthcare, NA931 and NA934) were incubated at room temperature for 1 h. Chemiluminescence was detected with an ChemiDoc XRS + imaging system (BioRad).
Immunoprecipitation
Immunoprecipitation was performed as previously described [38]. Briefly, cells were washed three times in PBS and lysed in TNE buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.5% NP-40) supplemented with cOmplete and PhosSTOP tablets (Roche, 04 693 159 001 and 04 906 837 001). After 30 min incubation on ice, lysates were cleared by centrifugation (20,000 RCF, 10 min, 4 °C). Appropriate antibodies were pre-conjugated to Dynabeads M-280 sheep anti-rabbit IgG (Invitrogen, 11203D) at 4 °C for 1 h and cleared lysates incubated with beads and antibodies at 4 °C for 2 h. Immunoglobulin-antigen complexes were washed extensively by TNE buffer before elution in 20 µl 2 x Laemmli sample buffer. Protein interactions were detected by immunoblot as described above.
qRT-PCR
Total RNA was extracted using RNeasy mini kit (Qiagen) following manufacturer’s instructions, cDNA was generated using the RevertAid H Minus reverse transcriptase (Thermo Scientific, EP0451) and deoxynucleotide triphosphates (Promega, U120A, U121A, U122A, U123A) and qPCR was performed using the SYBR Green I nucleic acid gel stain (Invitrogen, S7563) in a LightCycler Nano instrument (Roche). Triplicate treatment samples and two technical replicates per sample were analyzed. Relative quantity was calculated using the ΔΔCt method and ACTB or GAPDH mRNA as internal normalizers. Primer sequences for 47S rRNA processing [33] and p21 [39] were described previously.
Cell cycle analysis
Cells were fixed in 70% ethanol, stained with propidium iodide, and examined using FACS Verse instrument (BD Biosciences). Cell cycle distribution was analyzed by FACSuite software (BD Biosciences)
DNA combing
Cells were labeled with 25 μM CldU (Sigma, I7125) for 20 min, washed and labeled with 250 μM IdU (Sigma-Aldrich, C6891) for 20 min. DNA replication was stopped by ice-cold PBS. Cells were collected and DNA was extracted using FiberPrep kit (Genomic Vision, EXT-001) following manufacturer’s instructions. Extracted DNA was combed on vinylsilane coated CombiCoverslips (Genomic Vision, COV-002-RUO) with a speed of 0.3 mm/s. Coverslips were dehydrated at 60 °C for 4 h and stored at −20 °C. Next, DNA was denatured with buffer D (0.5 M NaOH, 1 M NaCl) 8 min at RT and dehydrated with 70%, 90%, and 100% ethanol washes (1 min each), air-dried in dark and blocked using ADM buffer (10% FBS in DMEM) 30 min at RT. Coverslips were incubated with primary antibodies, mouse anti-BrdU (1:10, BD Biosciences, BD347580) and rat anti-BrdU (1:50, Abcam, ab6326) for 2 h at RT in ADM buffer. After four washes with PBS, cover glasses were incubated with secondary antibodies goat anti-mouse Alexa Fluor 488 (1:100) and goat anti-rat A549 (1:100) for 1 h at RT in ADM buffer. After four washes with PBS, cover glasses were air-dried and mounted using Vectashield (Vector Laboratories, H-1000). Images of DNA fibers were acquired using CellObserver spinning disc confocal microscopic system (Zeiss), length of labeled DNA was analyzed using ImageJ software.
Senescence-associated β-galactosidase assay
Cells were transfected with siRNAs and next day plated on glass coverslips. After 6 days from transfection, cells were fixed and stained for β-galactosidase activity using Senescence β-Galactosidase staining kit (Cell Signaling, #9860 S) following manufacturer’s instructions.
Cell fractionation
Cytoplasm, nuclei, and nucleoli, respectively, were prepared from 10 × 106 cells as previously described [40]. Briefly, cells were washed with PBS, resuspended in buffer A (10 mM HEPES-KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT), homogenized ten times using a tight pestle and centrifuged at 100 RCF for 5 min at 4 °C. Supernatant was retained as cytoplasmic fraction. Nuclear pellet was resuspended in solution S1 (0.25 M sucrose, 10 mM MgCl2), layed over solution S2 (0.35 M sucrose, 0.5 mM MgCl2) and centrifuged at 600 RCF for 5 min at 4 °C. Clean pelleted nuclei were resuspended in solution S2 and sonicated 10 × 10 s bursts using a Misonix 3000 sonicator. Sonicated sample was layered over solution S3 (0.88 M sucrose, 0.5 mM MgCl2) and centrifuged at 1150 RCF for 10 min at 4 °C. Pellet and supernatant contained nucleoli and nucleoplasmic fraction, respectively. To obtain highly purified nucleoli, the pellet was washed with solution S2 and centrifuged at 600 RCF for 5 min at 4 °C. Isolated nucleoli were resuspended in Laemmli buffer and analyzed by western blotting.
Results
siRNA screen for p53 regulators
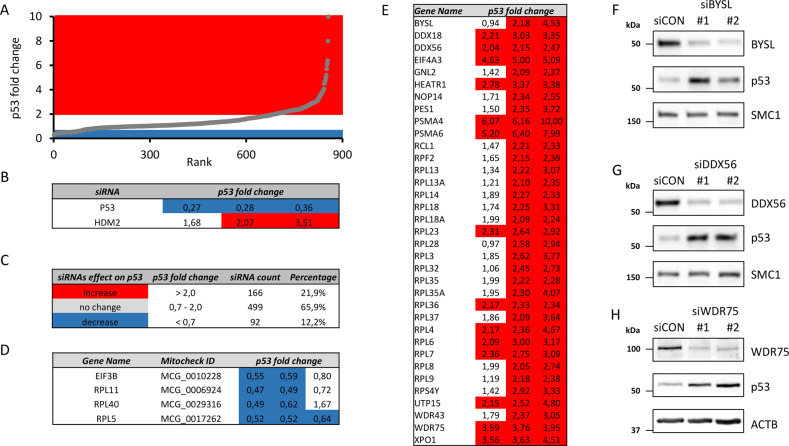
To gain a systematic overview of the relationship between the RBFs and p53, we assessed how depletion of the individual human RBFs affects the steady-state level of p53. To this end, we designed a comprehensive, customized siRNA library covering all 175 human RBFs known at the time, including 76 RPs of large and small ribosomal subunits, 6 additional proteins involved in the pathogenesis of ribosomopathies, 75 proteins involved in ribosome biogenesis based on the screen performed by the Kutay group [11] and additional 18 proteins selected based on published literature. The list of all targeted genes is presented in Supplementary Table 1. Each gene in our RBF siRNA library was targeted by three independent siRNAs positioned in separate wells in 96-well plates. Human osteosarcoma cell line U2OS, wild type for p53, was transfected with siRNAs in such 96-well plate format, and the abundance of p53 protein was assessed as a read-out, 2 days after transfection, using indirect immunofluorescence and average p53 staining intensity for each siRNA quantified by Olympus ScanR system. The results of our screen are presented in Supplementary Table 2. The changes in p53 level ranged from 0.27 to 10 fold, compared with parallel negative controls (Fig. 1A). siRNAs against HDM2 and p53 were included in the library as additional controls resulting in enhanced and diminished p53 abundance, respectively. Specifically, we observed 1.68–3.51-fold increase, and 0.27–0.36-fold decrease in p53 levels for siRNAs targeting HDM2 and p53, respectively (Fig. 1B). As primary hits that trigger a decrease or increase of p53 levels, we considered siRNAs with fold change of p53 staining intensity below 0.7 and above 2.0, respectively. About 22% (166/757) of all analyzed siRNAs resulted in at least two-fold increased p53 levels. Most of siRNAs 499/757 did not affect significantly p53 levels and 12% of siRNAs (92/757) decreased p53 levels (Fig. 1C). To apply more stringent criteria, we considered as candidate hits only those genes whose knock-down by at least two out of three siRNAs showed the suprathreshold effect on p53 protein level. After applying these criteria for hit selection, we obtained 4 and 35 genes (Fig. 1D, E), whose knockdown decreases and increases p53 protein levels, respectively. We found the known p53 regulators RPL5, RPL11 [28, 29], and RPL40 (eL40) [41] among hits whose depletion decreased p53 abundance. Further hit in this category included EIF3B, a component of the eukaryotic initiation factor 3 complex that initiates translation of subset of mRNAs involved in cell growth control processes, differentiation, and apoptosis via the mRNA 5′ untranslated region [42]. Given such function, it was therefore not surprising that EIF3B knockdown also reduced p53 protein level. Among the 35 genes whose knock-down increased the p53 levels were 19 RPs (19/76, 25%). Notably, nearly all of these are part of the large ribosomal subunit (18 of 19), with the only exception being RPS4Y (Fig. 1E). The remaining 16 hits in this category included 7 small subunit RBFs (BYSL, HEATR1, NOP14, RCL1, UTP15, WDR43, WDR75), 6 large subunit RBFs (DDX18, DDX56, GNL2, PES1, RPF2, XPO1) 2 proteasome components (PSMA4 and PSMA6) and eukaryotic initiation factor EIF4A3.
Fig. 1. siRNA screen for involvement of ribosome biogenesis factors in p53 homeostasis.
A Scatter plot of p53 fold changes derived from the screen in U2OS cells. Plotted values show both the area of reduced (blue, <0.7) and increased (red, >2.0) p53 fold changes for all siRNAs in the RBFs library ranked according their p53 fold changes. B p53 fold changes for siRNAs targeting positive controls P53 and HDM2. C siRNAs according their effect on p53 protein homeostasis. D List of candidate hits that decrease p53 protein level. E List of candidate hits that increase p53 protein level. F–H U2OS cells were transfected with indicated siRNAs, cultivated for 3 days and cell lysates analyzed by immunoblotting with indicated antibodies.
In order to further validate our screening dataset, we selected the following primary hits: Bystin (BYSL) a nucleolar protein involved in 18S rRNA processing [43], nucleolar ATP-dependent RNA helicase DDX56 [44], RP RPL23 (uL14) [45], U3 snoRNA-associated protein 15 homolog (UTP15) [46], WD-repeat containing protein WDR43 [47], ATP-dependent RNA helicase DDX18 [48] and human protein WDR75 previously implicated in pre-rRNA synthesis and early nucleolar pre-rRNA processing [12, 49]. We used independent sets of siRNAs and assessed p53 levels after knockdowns of the above listed 7 proteins by immunoblotting. In all cases, the validation siRNAs effectively reduced respective protein levels and resulted in increased p53 protein levels. (Fig. 1F, G, H and Supplementary Fig. 1, Supplementary Table 3). Our validation immunoblotting data were therefore consistent with the indirect immunofluorescence-based results from the siRNA screen.
WDR75 is a nucleolar protein
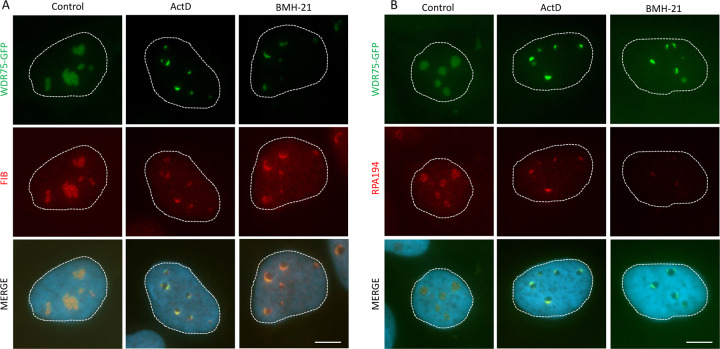
Next, we focused on the so-far functionally poorly characterized protein WDR75 and assessed its subcellular localization in cultured human cells, arguing that if WDR75 indeed played a role in ribosome biogenesis, it could be localized, at least in part, in nucleoli. Unfortunately, due to cross-reactive nature and suboptimal performance of available antibodies for indirect immunofluorescence, we were unable to reliably localize endogenous WDR75 by immunostaining. To bypass this technical obstacle, we generated U2OS cell lines expressing ectopic WDR75 fused to GFP. Under unperturbed growth conditions, such GFP-tagged WDR75 was indeed localized predominantly to nucleoli (Fig. 2A, B). Nucleolar localization of WDR75-GFP was validated by co-staining with 2 nucleolar proteins—fibrillarin (FIB) and RPA194, the catalytic subunit of RNA Pol I. To further support this important notion, the nucleolar localization of endogenous human WDR75 in both U2OS and U2OS WDR75-GFP cells was also validated through an independent approach, namely by subcellular fractionation followed by immunoblotting (Supplementary Fig. 2). Interestingly, after induction of ribosome biogenesis stress either by treating cells with Actinomycin D or a BMH-21, a specific inhibitor of RNA Pol I [50], WDR75-GFP became redistributed to nucleolar periphery forming small structures resembling nucleolar caps. Indeed, under the experimentally induced ribotoxic conditions the observed WDR75-GFP formed caps that largely colocalized with two established markers of nucleolar caps, FIB and RPA194 (Fig. 2A, B).
Fig. 2. WDR75 localizes to nucleoli and forms nucleolar caps during ribosomal stress.
U2OS cells expressing WDR75-GFP were treated for 4 h with 5 nM ActD or 0.5 μM BMH-21 and immunostained for fibrillarin (A) or RPA194 (B). Bars, 5 μM.
WDR75 knockdown activates p53 and p21, blocking proliferation
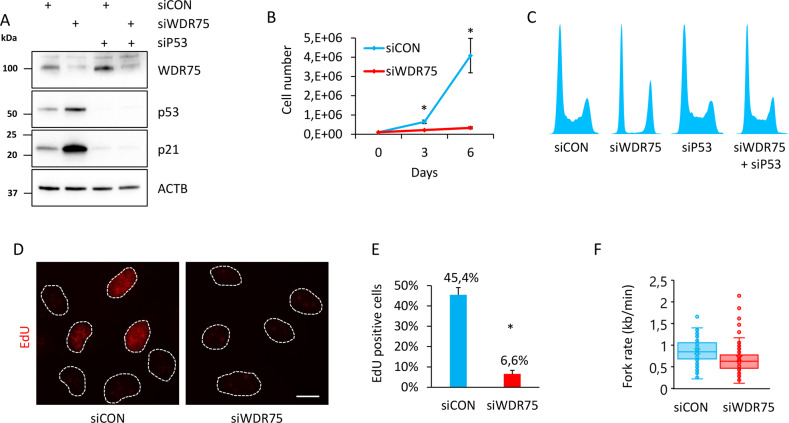
To test whether the stabilized p53 induced by WDR75 downregulation plays any functional role in controlling cell fate, we depleted WDR75 in human U2OS cells by siRNA, and assessed cell proliferation. First, our immunoblotting analysis revealed that WDR75 downregulation resulted in stabilized p53 protein and also induced the key cell-cycle inhibitory transcriptional target of p53, the CDK inhibitor p21 (Fig. 3A). Indeed, increased expression of p21 after WDR75 knockdown was p53-dependent, since concomitant downregulation of both WDR75 and p53 led to a decrease of p21 protein abundance, particularly compared to the scenario of a single knockdown of WDR75. The effect of WDR75 depletion-triggered induction of p53 and p21 was recapitulated in human diploid foreskin fibroblast strain BJ and in diploid retinal pigment epithelium RPE1 cells (Supplementary Fig. 3A), overall confirming that this cellular response to WDR75 depletion is not restricted to the cancerous U2OS cells. Notably, siRNA targeting 3’UTR of the WDR75 gene also efficiently reduced WDR75 protein level and led to induction of p53 in U2OS cells. This model system was then employed to test, and confirm, the ability of ectopically expressed WDR75-GFP (which is resistant to such siRNA targeting the 3′UTR) to functionally rescue the p53-triggering phenotype of WDR75 depletion (Supplementary Fig. 3B). These results confirmed the specificity of the observed p53-inducing effect, thereby excluding any off-target effects of the original set of siRNAs against WDR75. Consistently with the increased protein level of p21 after WDR75 knockdown, we also observed induction of p21 mRNA by quantitative RT-PCR (Supplementary Fig 3C). In order to assess impact of WDR75 status on cell cycle progression, we compared proliferation rates of WDR75-depleted and control cells. Growth rate of U2OS cells deficient in WDR75 was significantly reduced as documented by total cell counts at day 3 and 6 after plating (Fig. 3B). Further cell cycle analyses showed that WDR75 knockdown led to altered cell cycle progression, documented by an almost complete absence of cells in S phase and enhanced subpopulations of cells in G1 and G2 (Fig. 3C). Importantly, co-depletion of WDR75 and p53 completely restored normal cell cycle profile, suggesting that the observed changes in cell cycle distribution are p53-dependent. Furthermore, we used a specific mitotic marker—Ser 10-phosphorylated histone H3 (H3 pS10) and showed an almost complete absence of mitotic cells after WDR75 knockdown (Supplementary Fig. 3D). Interestingly, co-depletion of WDR75 and p53 largely rescued also the accumulation of cells positive for H3 pS10 (Supplementary Fig. 3E). In contrast to U2OS, downregulation of WDR75 in Hela cells, harboring the p53 whose function is impaired by the HPV-encoded E6 oncoprotein [51, 52], did not affect the percentage of cells positive for H3 pS10 (Supplementary Fig. 3F), an outcome that further supported the link between WDR75 status and p53 response. We also confirmed the reduced fraction of cells with ongoing S phase upon WDR75 knockdown, by monitoring incorporation of 5-ethynyl-2´-deoxyuridine (EdU) (Fig. 3D, E). Moreover, the speed of individual replication forks in WDR75-depleted cells was significantly reduced (Fig. 3F). We did not observe any induction of DNA damage after WDR75 knockdown, since there was no increase in phosphorylated histone H2Ax on S139, a commonly used surrogate marker of DNA damage response (Supplementary Fig. 3G). Overall, these data indicated that WDR75 knockdown leads to activation of p53 that leads to cell cycle arrest and thus impairs cellular proliferation.
Fig. 3. WDR75 knockdown activates p53 and p21 and impairs proliferation.
A U2OS cells were transfected with indicated siRNAs, cultivated for 3 days and cell lysates analyzed by immunoblotting with indicated antibodies. B U2OS cells were transfected with indicated siRNAs, next day 100,000 cells plated into six-well plates, incubated for indicated additional number of days and counted. Data shown as mean ± SD of three independent experiments (n = 3), each based on triplicate wells. Significance was determined by two-tailed t test: *P < 0.05. C U2OS cells were transfected with indicated siRNAs, cultivated for 3 days and cell cycle profiles analyzed by FACS. D U2OS cells were transfected with indicated siRNAs, cultivated for 3 days, treated with 10 EdU for 30 min and stained for incorporated EdU by click chemistry. Bar, 10 μM. E Quantification of EdU positive cells from D. Data shown as mean ± SD of three independent experiments (n = 3). Significance was determined by two-tailed t test: *P < 0.05. F U2OS cells were transfected with indicated siRNAs, incubated for 3 days, labeled with CldU/IdU and fork rate determined by DNA combing. Fork rates based on IdU track length are shown as box plot. Scored forks: 208 and 113 for siCON and siWDR75, respectively. Data shown is representative of two independent experiments.
WDR75 impacts pre-rRNA transcription by regulating RPA194 levels
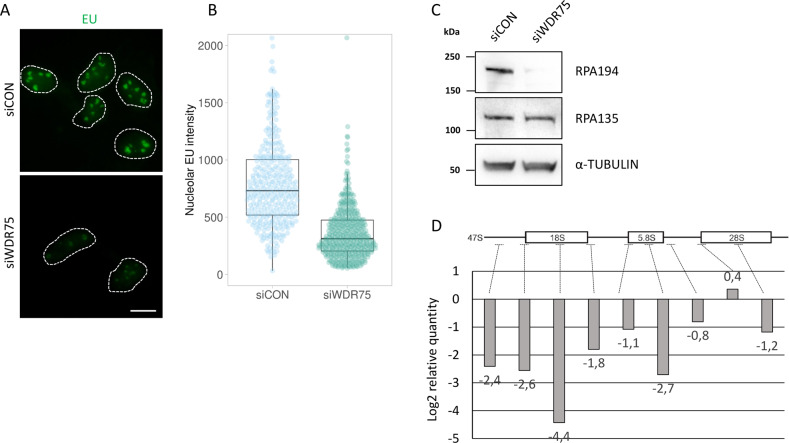
Given the nucleolar localization of WDR75, and the activation of the p53-p21 axis without the apparent occurrence of DNA damage, we argued that WDR75-depletion may cause endogenous ribosome biogenesis stress. To address this possibility, we next asked whether production of RNA is also impaired in cells deficient in WDR75. To this end, we employed a method based on incorporation of 5-ethynyl uridine (EU) into the nascent RNA. We observed reduction of nascent RNA production after WDR75 knockdown (Fig. 4A, B) and reduced protein levels of RPA194, a subunit of RNA Pol I (Fig. 4C). Notably, the observed destabilizing effect of WDR75 depletion on RPA194 was at least partly selective, not shared by RPA135, another protein component of the RNA Pol I complex (Fig. 4C). To explore this mechanistic aspect further, we assessed the impact of WDR75 knockdown on levels of 47S rRNA, using a set of primers targeting mature 18S, 5.8S, and 28S rRNAs, respectively, along with six primers specific for regions present exclusively in the 47S precursor rRNA [33, 53]. Both precursor-specific and mature regions showed a decrease in rRNA synthesis in the WDR75-depleted cells (Fig. 4D), suggesting that WDR75 is required for rRNA transcription.
Fig. 4. WDR75 impacts pre-rRNA transcription by regulating RPA194 levels.
A U2OS cells were transfected with indicated siRNAs, cultivated for 3 days, treated with 1 mM EU for 30 min and stained for EU incorporated to nascent RNAs by click chemistry. Bar, 10 μM. B Quantification of EU signal in nucleoli of cells from A. Nucleolar EU intensities are shown as box plot. Quantified nuclei: 334 and 425 for siCON and siWDR75, respectively. Data shown are representative of two independent experiments. C U2OS cells were transfected with indicated siRNAs, cultivated for 3 days and cell lysates analyzed by immunoblotting with indicated antibodies. D U2OS cells were transfected with control and WDR75-targeting siRNAs, cultivated for 3 days and RT-qPCR analysis performed. Data are expressed as relative quantity of siWDR75 treated cells compared to control cells. Data shown as mean of three independent experiments (n = 3).
WDR75 deficiency triggers the IRBC pathway and induces cellular senescence
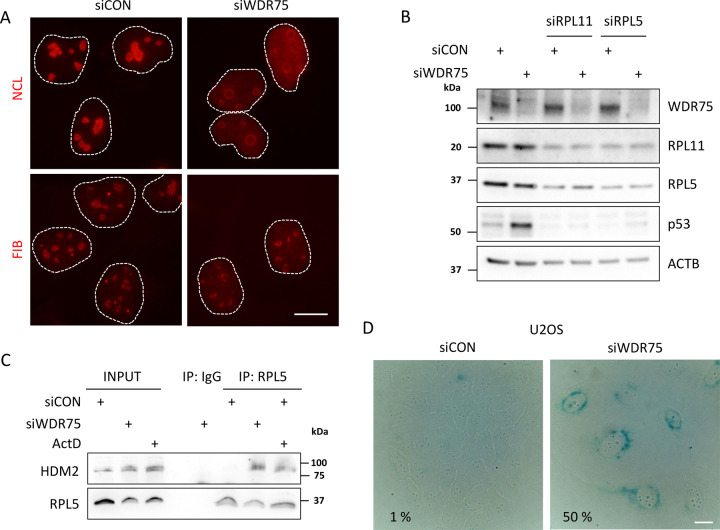
Deregulation of rRNA transcription may evoke ribosome biogenesis stress, a condition that commonly leads to induction of the IRBC pathway and p53 stabilization, largely reflecting the induced interaction between the RPL5/RPL11/5S rRNA complex and HDM2 [28, 29]. To assess this possible scenario, we first tested whether WDR75 depletion leads to ribosome biogenesis stress by analyzing localizations of nucleolar markers nucleolin and fibrillarin. Under ribosome biogenesis stress, nucleolin, which is mainly located at the granular component inside nucleoli under normal conditions, translocates to the nucleoplasm, while fibrillarin, normally located in the dense fibrillary component of the nucleoli, is known to translocate to nucleolar caps under ribosomal stress. Indeed, both these nucleolar markers changed their localization upon WDR75 knockdown. Nucleolin redistributed to nucleoplasm and fibrillarin formed nucleolar caps, respectively (Fig. 5A). Redistribution of nucleolin to nucleoplasm after WDR75 knockdown by siRNA targeting the 3′UTR of WDR75 gene was partially rescued in U2OS cells ectopically expressing GFP-WDR75 (Supplementary Fig. 4A). Together, our data so far are consistent with the notion that WDR75 knockdown induces ribosomal stress. To examine whether the p53 stabilization/activation observed in our experiments after depletion of WDR75 reflects the above mentioned RPL5/RPL11/5S rRNA pathway, with the ensuing inhibition of HDM2-mediated turnover of p53, we co-depleted WDR75 together with RPL5 or RPL11, respectively. Stabilization of p53 after WDR75 knockdown was largely abolished when knocking down either RPL5 or RPL11 (Fig. 5B). In contrast to p53 transcriptional target HDM2, RPL5 or RPL11 protein level was not affected by WDR75 knockdown (Supplementary Fig. 4B). Moreover, HDM2 was co-precipitated with RPL5 in WDR75-depleted cells or Actinomycin D-treated cells, in contrast to control cells (Fig. 5C), indicating that p53 stabilization after WDR75 knockdown reflects the activated ribosome biogenesis stress-response checkpoint. While assessing what is the long-term phenotypic impact of WDR75 knock-down, we noted that WDR75-depleted cells became enlarged, showing flatten morphology, overall resembling senescent cells. We therefore assayed cells after WDR75 downregulation for senescence-associated β-galactosidase activity as a marker of ongoing cellular senescence. Both human U2OS and RPE1 cells were found positive for senescence-associated β-galactosidase activity 6 days after WDR75 knockdown (Fig. 5D and Supplementary Fig. 4C), indicating that WDR75 knockdown indeed induces cellular senescence.
Fig. 5. WDR75 deficiency triggers the IRBC pathway and induces cellular senescence.
A U2OS cells were transfected with indicated siRNAs, cultivated for 3 days and immunostained for nucleolin (NCL), fibrillarin (FIB) and p53. Bar, 10 μM. B U2OS cells were transfected with indicated siRNAs, cultivated for 3 days and cell lysates analyzed by immunoblotting with indicated antibodies. C U2OS cells were transfected with control or WDR75 siRNAs. Cell lysates were immunoprecipitated 72 h after transfection with control (IgG) or RPL5 antibodies and immunoblotted with indicated antibodies. D U2OS cells were transfected with indicated siRNAs, cultivated for 6 days and stained for senescence-associated β-galactosidase activity. Numbers indicate percentage of cells positive for senescence-associated β-galactosidase activity. Bar, 10 μM.
Discussion
The results presented in this article can contribute to better understanding of ribosomal biogenesis and induction of IRBC in two ways. First, we provide data from our high-content siRNA-based microscopy screen to identify RPs and RBFs with impact on p53, and we present these results as an information resource to serve the global research community. Second, we functionally characterize human WDR75 as an RBF and regulator of RPA194 levels, critical for p53 homeostasis.
Our screen-derived dataset complements, validates, and robustly extends (by more than doubling the number of target genes in the examined library) results from another, smaller-scale siRNA screen based on knock-down of 80 RPs, and searching for an impact on nucleolar structure, pre-rRNA processing and p53 protein level [9]. The fact that most of our RP hits among the factors that were included in both screens scored also in the study by Nicolas et al. provides a solid validation of our screening approach (please see Supplementary Table 4 for direct comparison of RP hits). From a functional perspective, it is worth noting that these shared hits identified in both screens correspond to late-assembling structures on the ribosomal subunits [9]. Furthermore, our siRNA library encompassed not only the 76 RPs (as in the screen by Nicolas et al.) but also additional 99 targets including a spectrum of RBFs and genes mutated in human ribosomopathies. We identified additional 16 RBFs whose depletion-triggered p53 protein elevation. Interestingly, aberrations of some of the hits in this category, including the nucleolar protein NOP14, RNA helicases DDX18, DDX56 and EIF4A3, and PES1 have been linked to cancer [54–58].
Mechanistically, we focused on better understanding of WDR75, so-far a functionally poorly characterized protein that scored among RBFs whose depletion induced p53 in our screen. Human WDR75 is the structural and functional homologue of the yeast U3 small nucleolar RNA-associated protein 17 (Utp17), sharing 16% sequence identity. Unlike Utp17, however, WDR75 contains 13 WD40 repeats (WDR), short structural motifs of ~40 amino acids. WDRs are β-propeller domains that act as protein interaction scaffolds in multiprotein complexes [59]. Indeed, the yeast Utp17 had been isolated as a component of the large ribonucleoprotein called the small subunit (SSU) processome and shown to be required for 18S rRNA biogenesis [35, 60]. Our data on human WDR75, consistently with the report by Prieto and McStay [49], showed predominantly nucleolar localization of GFP-tagged ectopically expressed WDR75. Notably, we further corroborated this finding by biochemical subcellular fractionation experiments, demonstrating prominent nucleolar localization of also the endogenous WDR75 human protein. Moreover, we showed that after experimental induction of ribosomal stress in human cells WDR75 redistributed together with fibrillarin and the catalytic subunit of RNA Pol I to nucleolar caps, suggesting an active role of WDR75 in response to ribosomal stress.
From the broader perspective of cell pathophysiology and cell fate decisions, our present study provides the first evidence that WDR75 is required to prevent cells entering the state of ribosomal biogenesis stress, a condition that leads to activation of the cellular IRBC checkpoint response. We showed that absence of WDR75 caused nucleolar disintegration, and at the molecular response level, triggered association of RPL5 with HDM2 with the ensuing RPL5/RPL11 (IRBC)-dependent stabilization and functional activation of p53, accompanied by induced expression of p53´s transcriptional target, the CDK inhibitor p21. This sequence of molecular events, resulted in p53-dependent cell proliferation blockade and ultimately triggered the state of cellular senescence. In mechanistic terms, we propose that the observed cell cycle inhibition and senescence may reflect cooperative impacts of at least the following two complementary roles of p21. First, the ‘canonical’ direct CDK-inhibitory impact of p21 on the G1/S-promoting CDK2 and G2/M-promoting CDK1 kinases; a scenario consistent with the accumulation of WDR75-depleted cells in G1 and G2 phases, and fewer cells in S phase in our experiments. The other contributing function of p21 in this context likely reflects the negative impact of p21 on the speed of DNA replication fork progression, a less-established role recently discovered by us (Maya-Mendoza et al. [61]), and later shown by us to operate via direct interaction of p21, and thereby interference with, the replication factor PCNA [62]. While the direct CDK-inhibitory role may predominantly account for the observed cell cycle effects upon WDR75 knock-down, the latter p21-mediated mechanism also contributes, by slowing down the replication fork speed in the smaller yet detectable fraction of S-phase cells. Indeed, such dual contribution is also consistent with the reduced speed of replication forks upon WDR75 depletion that we report here, monitored directly by the DNA single-molecule analysis of DNA fibers. Last but not least, we suggest that the observed induction of the p53-activating cell cycle checkpoint was not caused by the otherwise commonly seen mechanism due to DNA damage-induced phosphorylation of p53, since WDR75 depletion did not lead to detectable DNA damage signaling in our experiments.
Furthermore, we report here that human WDR75 plays a positive role in pre-rRNA synthesis. Mechanistically, WDR75 colocalized with the RNA Pol I subunit RPA194 both under unperturbed growth conditions and upon inhibition of transcription by Actinomycin D. We suggest that the positive effect of WDR75 on pre-rRNA transcription is mediated via RPA194 stabilization, since WDR75 knockdown also led to reduced abundance of the RPA194 protein. The mechanistic selectivity of the WDR75-RPA194 interplay is further apparent from the fact that unlike RPA194, the RPA135 component of the RNA Pol I remained unaffected at the protein level after depletion of WDR75.
In terms of impact on our current view of ribosome biogenesis and its role in major human pathologies, recent research has demonstrated association between abnormalities of ribosome biogenesis and increased risk of cancer [63]. In this regard, while human WDR75 has not been fully explored to date, aberrantly high levels of WDR75 mRNA were identified as an unfavorable prognostic marker for renal and liver cancer [64]. Taken together, we hope that our present results, both the overall screening dataset, and the functional studies of WDR75 in the context of ribosome biogenesis, may inspire further studies in this important yet still rather under-developed area of biomedical research, including involvement in pathogenesis and potential vulnerabilities of cancer, with implications for future innovative treatments in oncology.
Supplementary information
Author contributions
PM, SV, and JB designed experiments, which were performed mostly by PM. KC performed western blot analysis. SB performed cell fractionation experiments. PM, SV, and JB discussed and interpreted the results. PM, SV, and JB wrote the manuscript, which was approved by all authors.
Funding
This project was supported by the Czech Science Foundation (grant no. 20-03457Y), the European Regional Development Fund—Project ENOCH (No. CZ.02.1.01/0.0/0.0/16_019/0000868), the MEYS CR (Large RI Project LM2018129 - Czech-BioImaging), the Danish Cancer Society (R204-A12617-B153), the Novo Nordisk Foundation (0060590), the Danish Council for Independent Research (DFF-7016-00313), the Lundbeck Foundation (R266-2017-4289), the Swedish Research Council (VR-MH 2014-46602-117891-30), the Swedish Cancer Society (Cancerfonden): #170176, the Croatian Science Foundation (no. 2079) and the Croatian Scientific Center of Excellence for Reproductive and Regenerative Medicine (KK.01.1.1.01.0008).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by M. Oren
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pavel Moudry, Email: pavel.moudry@upol.cz.
Jiri Bartek, Email: jb@cancer.dk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00882-0.
References
- 1.Bohnsack KE, Bohnsack MT. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 2019;38:e100278. doi: 10.15252/embj.2018100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelletier J, Thomas G, Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18:51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- 3.Noller HF. Evolution of protein synthesis from an RNA world. Cold Spring Harb Perspect Biol. 2012;4:a003681. doi: 10.1101/cshperspect.a003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gentilella A, Kozma SC, Thomas G. A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim Biophys Acta. 2015;1849:812–20. doi: 10.1016/j.bbagrm.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodfellow SJ, Zomerdijk JC. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell Biochem. 2013;61:211–36. doi: 10.1007/978-94-007-4525-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woolford JL, Jr., Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–81. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, et al. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–92. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolas E, Parisot P, Pinto-Monteiro C, de Walque R, De Vleeschouwer C, Lafontaine DL. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat Commun. 2016;7:11390. doi: 10.1038/ncomms11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badertscher L, Wild T, Montellese C, Alexander LT, Bammert L, Sarazova M, et al. Genome-wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Rep. 2015;13:2879–91. doi: 10.1016/j.celrep.2015.11.061. [DOI] [PubMed] [Google Scholar]
- 11.Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, et al. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell. 2013;51:539–51. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S. The relationship between the nucleolus and cancer: current evidence and emerging paradigms. Semin Cancer Biol. 2016;37-38:36–50. doi: 10.1016/j.semcancer.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–75. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 15.Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–7. [DOI] [PubMed]
- 16.Aspesi A, Ellis SR. Rare ribosomopathies: insights into mechanisms of cancer. Nat Rev Cancer. 2019;19:228–38. doi: 10.1038/s41568-019-0105-0. [DOI] [PubMed] [Google Scholar]
- 17.Lindstrom MS, Jurada D, Bursac S, Orsolic I, Bartek J, Volarevic S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene. 2018;37:2351–66. doi: 10.1038/s41388-017-0121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246–55. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bursac S, Brdovcak MC, Pfannkuchen M, Orsolic I, Golomb L, Zhu Y, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci USA. 2012;109:20467–72. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fumagalli S, Ivanenkov VV, Teng T, Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26:1028–40. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panic L, Tamarut S, Sticker-Jantscheff M, Barkic M, Solter D, Uzelac M, et al. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol. 2006;26:8880–91. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barkic M, Crnomarkovic S, Grabusic K, Bogetic I, Panic L, Tamarut S, et al. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol Cell Biol. 2009;29:2489–504. doi: 10.1128/MCB.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–33. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohrum MAE, Ludwig RL, Kubbutat MHG, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–87. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–12. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–12. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 28.Donati G, Peddigari S, Mercer CA, Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4:87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237–47. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orsolic I, Bursac S, Jurada D, Drmic Hofman I, Dembic Z, Bartek J, et al. Cancer-associated mutations in the ribosomal protein L5 gene dysregulate the HDM2/p53-mediated ribosome biogenesis checkpoint. Oncogene. 2020;39:3443–57. doi: 10.1038/s41388-020-1231-6. [DOI] [PubMed] [Google Scholar]
- 31.Bursac S, Prodan Y, Pullen N, Bartek J, Volarevic S. Dysregulated ribosome biogenesis reveals therapeutic liabilities in cancer. Trends Cancer. 2021;7:57–76. doi: 10.1016/j.trecan.2020.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espinoza JA, Zisi A, Kanellis DC, Carreras-Puigvert J, Henriksson M, Huhn D, et al. The antimalarial drug amodiaquine stabilizes p53 through ribosome biogenesis stress, independently of its autophagy-inhibitory activity. Cell Death Differ. 2019;27:773–89. doi: 10.1038/s41418-019-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farley-Barnes KI, McCann KL, Ogawa LM, Merkel J, Surovtseva YV, Baserga SJ. Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep. 2018;22:1923–34. doi: 10.1016/j.celrep.2018.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernstein KA, Baserga SJ. The small subunit processome is required for cell cycle progression at G1. Mol Biol Cell. 2004;15:5038–46. doi: 10.1091/mbc.E04-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005;19:3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frankum J, Moudry P, Brough R, Hodny Z, Ashworth A, Bartek J, et al. Complementary genetic screens identify the E3 ubiquitin ligase CBLC, as a modifier of PARP inhibitor sensitivity. Oncotarget. 2015;6:10746–58. doi: 10.18632/oncotarget.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozdian T, Holub D, Maceckova Z, Varanasi L, Rylova G, Rehulka J, et al. Proteomic profiling reveals DNA damage, nucleolar and ribosomal stress are the main responses to oxaliplatin treatment in cancer cells. J Proteomics. 2017;162:73–85. doi: 10.1016/j.jprot.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Evangelou K, Bartkova J, Kotsinas A, Pateras IS, Liontos M, Velimezi G, et al. The DNA damage checkpoint precedes activation of ARF in response to escalating oncogenic stress during tumorigenesis. Cell Death Differ. 2013;20:1485–1497. doi: 10.1038/cdd.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, et al. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Q, Hou Z, Zuo S, Zhou X, Feng Y, Sun Y, et al. LUCAT1 promotes colorectal cancer tumorigenesis by targeting the ribosomal protein L40-MDM2-p53 pathway through binding with UBA52. Cancer Sci. 2019;110:1194–1207. doi: 10.1111/cas.13951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee AS, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyoshi M, Okajima T, Matsuda T, Fukuda MN, Nadano D. Bystin in human cancer cells: intracellular localization and function in ribosome biogenesis. Biochem J. 2007;404:373–381. doi: 10.1042/BJ20061597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zirwes RF, Eilbracht J, Kneissel S, Schmidt-Zachmann MS. A novel helicase-type protein in the nucleolus: protein NOH61. Mol Biol Cell. 2000;11:1153–1167. doi: 10.1091/mbc.11.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F, Hamanaka RB, Bobrovnikova-Marjon E, Gordan JD, Dai MS, Lu H, et al. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. J Biol Chem. 2006;281:30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
- 46.Sato M, Araki N, Kumeta M, Takeyasu K, Taguchi Y, Asai T, et al. Interaction, mobility, and phosphorylation of human orthologues of WD repeat-containing components of the yeast SSU processome t-UTP sub-complex. Biochem Cell Biol. 2013;91:466–475. doi: 10.1139/bcb-2013-0062. [DOI] [PubMed] [Google Scholar]
- 47.Wada K, Sato M, Araki N, Kumeta M, Hirai Y, Takeyasu K, et al. Dynamics of WD-repeat containing proteins in SSU processome components. Biochem Cell Biol. 2014;92:191–199. doi: 10.1139/bcb-2014-0007. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Wu Z, Lu JY, Huang B, Zhou H, Xie W, et al. DEAD-Box Helicase 18 Counteracts PRC2 to Safeguard Ribosomal DNA in Pluripotency Regulation. Cell Rep. 2020;30:81–97 e87. doi: 10.1016/j.celrep.2019.12.021. [DOI] [PubMed] [Google Scholar]
- 49.Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007;21:2041–2054. doi: 10.1101/gad.436707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peltonen K, Colis L, Liu H, Trivedi R, Moubarek MS, Moore HM, et al. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell. 2014;25:77–90. doi: 10.1016/j.ccr.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus type-16 and type-18 promotes the degradation of P53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 52.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 53.Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou B, Wu Q, Chen G, Zhang TP, Zhao YP. NOP14 promotes proliferation and metastasis of pancreatic cancer cells. Cancer Lett. 2012;322:195–203. doi: 10.1016/j.canlet.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Payne EM, Bolli N, Rhodes J, Abdel-Wahab OI, Levine R, Hedvat CV, et al. Ddx18 is essential for cell-cycle progression in zebrafish hematopoietic cells and is mutated in human AML. Blood. 2011;118:903–915. doi: 10.1182/blood-2010-11-318022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kouyama Y, Masuda T, Fujii A, Ogawa Y, Sato K, Tobo T, et al. Oncogenic splicing abnormalities induced by DEAD-Box Helicase 56 amplification in colorectal cancer. Cancer Sci. 2019;110:3132–3144. doi: 10.1111/cas.14163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu YB, Liao LY, Jiang R, Xu M, Xu LW, Chen GG, et al. PES1 promotes the occurrence and development of papillary thyroid cancer by upregulating the ERalpha/ERbeta protein ratio. Sci Rep. 2019;9:1032. doi: 10.1038/s41598-018-37648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19:73. doi: 10.1186/s12943-020-01183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schapira M, Tyers M, Torrent M, Arrowsmith CH. WD40 repeat domain proteins: a novel target class? Nat Rev Drug Discov. 2017;16:773–786. doi: 10.1038/nrd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maya-Mendoza A, Moudry P, Merchut-Maya JM, Lee M, Strauss R, Bartek J, et al. High speed of fork progression induces DNA replication stress and genomic instability. Nature. 2018;559:279–84. doi: 10.1038/s41586-018-0261-5. [DOI] [PubMed] [Google Scholar]
- 62.Merchut-Maya JM, Bartek J, Maya-Mendoza A. Regulation of replication fork speed: Mechanisms and impact on genomic stability. DNA Repair. 2019;81:102654. doi: 10.1016/j.dnarep.2019.102654. [DOI] [PubMed] [Google Scholar]
- 63.Bursac S, Brdovcak MC, Donati G, Volarevic S. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim Biophys Acta. 2014;1842:817–830. doi: 10.1016/j.bbadis.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 64.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.