Abstract
Background: Malignant melanoma is a common malignant tumor and one of the tumors with the fastest growing incidence. The effect of microRNAs on the biological processing of malignant melanoma cells also have been reported. This study explores the ability of miR-498 to regulate the progression of malignant melanoma cells. Methods: The expression of miR-498 was detected by RT-qPCR. The proliferation, invasion, and migration of malignant melanoma cells were measured by cell counting kit-8, clone formation, and transwell assays. Flow cytometry assay detected the percentage of apoptotic cells. Western blot was used to detect the expression of markers related to epithelial–mesenchymal transition. The correction of miR-498 and UBE2T was explored by dual-luciferase assay and Western blot. Results: Overexpression of miR-498 inhibited the proliferation, invasion, migration, and induced cell apoptosis of M14 and A375 cells. In addition, the expression of epithelial–mesenchymal transition-related factors was altered by the overexpression of miR-498. miR-498 can directly target UBE2T 3'-UTR and inhibit UBE2T protein expression. The overexpression of UBE2T reversed the inhibitory effects of miR-498 on the progression of malignant melanoma cells. Furthermore, UBE2T mRNA was significantly highly expressed in malignant melanoma tissues. The high expression of UBE2T was associated with the poor overall survival rate of malignant melanoma patients. Conclusions: Altogether, our findings demonstrated that miR-498 significantly inhibited the proliferation, invasion, migration, and induced apoptosis of malignant melanoma cells and confirmed that miR-498 regulated malignant melanoma cell progression by targeting UBE2T.
Keywords: malignant melanoma, miR-498, UBE2T, EMT pathway
Introduction
Malignant melanoma (MM) is a common malignant tumor that occurs on the skin, mucous membranes, and pigmented membranes. MM is also one of the fastest-growing malignancies, with an annual growth rate of 3% to 5%. 1 At present, systemic chemotherapy is mainly used to treat MM in clinical practice. 2 Despite some effects, the 10-year survival rate of patients is still less than 10%. 3
MicroRNA is a short-chain uncoded RNA. 4 Its main role is to regulate the expression of target gene mRNA, which is closely related to the occurrence and development of various diseases. miR-498 is located at 19q13.41 and plays a role in many types of solid tumors.5-7 In addition, recent studies have reported the effect of microRNAs on the biological processing of MM cells, such as miR-148b, miR-33a, miR-17-92, and miR-211.8-11 The role of miR-498 in MM has not been studied.
Ubiquitin-conjugating enzyme E2T (UBE2T) is located on human chromosome 1q32.1 and encodes a total of 197 amino acids. UBE2T can mediate the ubiquitination of proteins such as FANCD2 and FANCI, and then participate in the repair of DNA damage. 12 In recent years, it has been reported that UBE2T can participate in the occurrence and development of a variety of tumors, including breast cancer, lung cancer, prostate cancer, and bladder cancer.12-14 In this study, we analyzed the function of miR-498 in MM cells.
Materials and Methods
Cell Lines and Cell Treatment
Human epidermal normal melanocytes (HEM) and MM cell lines (A375, M21, M14, and uacc62) were obtained from the Cell Bank of the Chinese Academy of Sciences. All cells were cultured in RPMI-1640 (Hyclone, GE Healthcare Life Sciences) supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific Inc.), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Sigma-Aldrich; Merck KGaA) in a humidified incubator with 5% CO2 at 37 °C. miR-498 cDNA sequence was inserted into pCMV-MIR plasmid to produce pCMV-miR-498 plasmid, which was transfected into M14 and A375 cells using Lipofectamine 2000 (miR-498 group). pCMV-MIR empty plasmid was used as negative control (NC). Similarly, M14 and A375 cells were transfected with pcDNA3.1-UBE-2T plasmid.
Quantitative Real-Time PCR Analysis
Total RNA was extracted from transfected cells using Trizol, and cDNA was synthesized using the Revert Aid First Strand cDNA Synthesis Kit (CWBIO). The expression of miR-498 was detected using ABI Prism 7300 sequence detector (Applied Biosystems) and SYBR Green reagent. The relative expression of miR-498 was calculated using the 2−ΔΔct method and normalized to U6 expression. The primers were as follows: miR-498 primer, 5′-AAGCCAGGGGGCGTTT-3′ (upstream) and 5′-GAACATGTCTGCGTATCTC-3′ (downstream); U6 primer, 5′-CTCGCTTCGGCAGCACA-3′ (upstream) and 5′-AACGCTTCACGAATTTGCGT-3′ (downstream).
Cell Counting Kit-8 Assay
Briefly, 1 × 103 cells were seeded into each well of a 96-well plate. After every 24 h of incubation, the old medium was removed, and the fresh medium containing 10 μL cell counting kit-8 (CCK8) solution (Solarbio Science & Technology) was added. After 2 h of incubation at 37 °C, the OD value of each well at 450 nm was detected with a microplate reader.
Colony Formation Assay
Approximately 3 × 102 transfected cells were planted in a 6 cm dish containing 5 mL medium and maintained at 37 °C in 5% CO2. When the number of cells in a colony exceeds 50 under the microscope, the clones were fixed and stained with 4% paraformaldehyde solution and 0.1% crystal violet. Finally, the clones were photographed and counted.
Transwell Assay
The invasion of M14 and A375 cells was determined using Matrigel-coated transwell chambers (BD Biosciences), and migration was determined using transwell chambers that were not coated with Matrigel. Briefly, 2 × 104 cells with serum-free DMEM medium were transferred to the upper chamber, while the complete medium was added to the lower chamber. After 24 h of incubation, the invasive and migrated cells were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet. Finally, the cells were photographed and counted under a microscope.
Apoptosis Detecting Assay
Apoptosis of cells was determined using Annexin V-FITC Apoptosis Detection kit I (Thermo Fisher Scientific) according to the manufacturer's instructions. Briefly, after 48 h of incubation, the cells were collected and mixed with 10 µL Annexin V-FITC and 5 µL propidium iodide at room temperature in the dark for 5 min. Apoptosis of cells was measured using flow cytometry.
Western Blot
Cells were lysed with RIPA buffer, and the lysate was centrifuged at 12 000 g for 10 min at 4 °C to extract the protein. Proteins were separated using SDS-PAGE, and transferred to a PVDF membrane. Then, the membrane was incubated with antibodies against Bcl-2 (1:1000, ab32124, Abcam), Bax (1:1000, ab32503, Abcam), cleaved caspase3 (1:1000, ab32351, Abcam), E-cad (1:1000, 20874-1-AP, Proteintech), N-cad (1:1000, 22018-1-AP, Proteintech), Vimentin (1:1000, ab92547, Abcam), UBE2T (1:1000, 10105-2-AP, Proteintech), and GAPDH (1:3000, ab8245, Abcam) overnight. Finally, the membrane was incubated with the horseradish peroxidase-conjugated secondary antibody.
Target Gene Prediction
TargetScan was used to predict the targets of miR-498. According to the predicted location in the 3'-UTR region, context++ score, target transcript size and subcellular location, site counts, and research status, target genes were selected for follow-up experiments.
Dual-Luciferase Assay
The pmirGLO-UBE2T-3'-UTR wild type (wt) or mutant (mut) reporter plasmid and miR-498 were co-transfected into cells using Lipofectamine 2000. After 48 h of transfection, cells were lysed, and the luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase activity was used for standardization.
Statistical Analysis
Data were obtained from 3 independent experiments and analyzed using SPSS 18.0. Unpaired 2-tailed Student t test was used for comparison between 2 groups, and ANOVA followed by Tukey's post hoc test was used for comparison between multiple groups. Kaplan-Meier survival analysis was used to evaluate the prognosis. The log-rank test was used to compare the survival curves. Differences with P values less than .05 were considered statistically significant.
Results
miR-498 Inhibits Malignant Melanoma Cell Proliferation
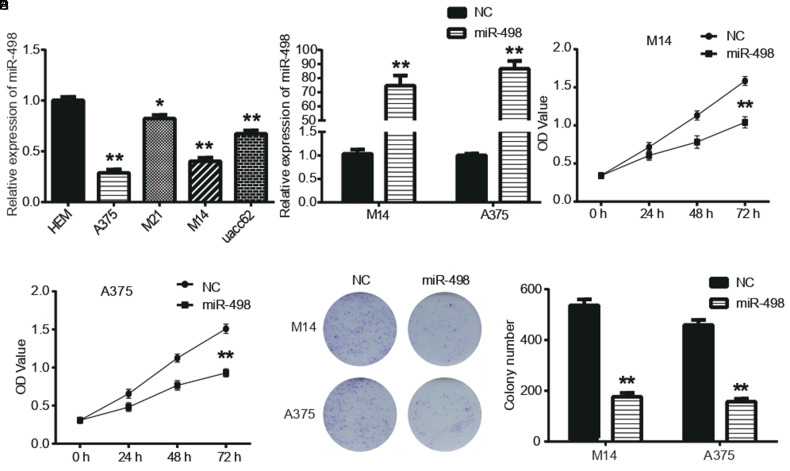
First, miR-498 expression levels in 4 human MM cell lines (A375, M21, M14, and uacc62) and HEM cells were analyzed by quantitative real-time PCR analysis. As shown in Figure 1A, miR-498 expression was significantly downregulated in MM cell lines compared to HEM cells (P < .05). We further explored the function of miR-498 in M14 and A375 cells. As shown in Figure 1B, miR-498 overexpression plasmid significantly the miR-498 levels in M14 and A375 cells (P < .05). Moreover, the results in Figure 1C and D showed that the OD values of M14 and A375 cells exogenously expressed miR-498 were significantly reduced (P < .05). The clone formation assay also showed similar results in Figure 1E that the clone formation ability of M14 and A375 cells transfected with miR-498 was significantly decreased compared with NC (P < .05).
Figure 1.
miR-498 inhibited malignant melanoma (MM) cell proliferation. (A) Quantitative real-time PCR analysis (qRT-PCR) assay detected relative expression of miR-498 in 4 human MM cell lines. (B) Relative expression levels of miR-498 in M14 and A375 cells transfected with NC or miR-498 mimics were detected by qRT-PCR assay. (C-E) Cell counting kit-8 and clone formation assays measured cell proliferation in M14 and A375 cells. *P < .05, ** P < .01 compared with NC.
miR-498 Inhibits Malignant Melanoma Cell Invasion and Migration and Induces Cell Apoptosis
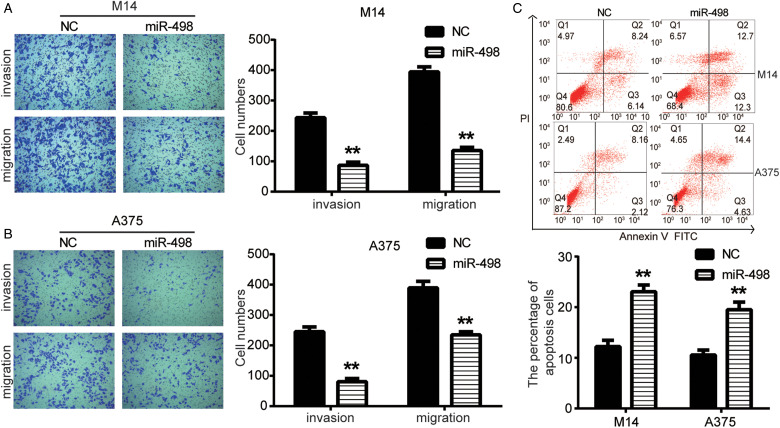
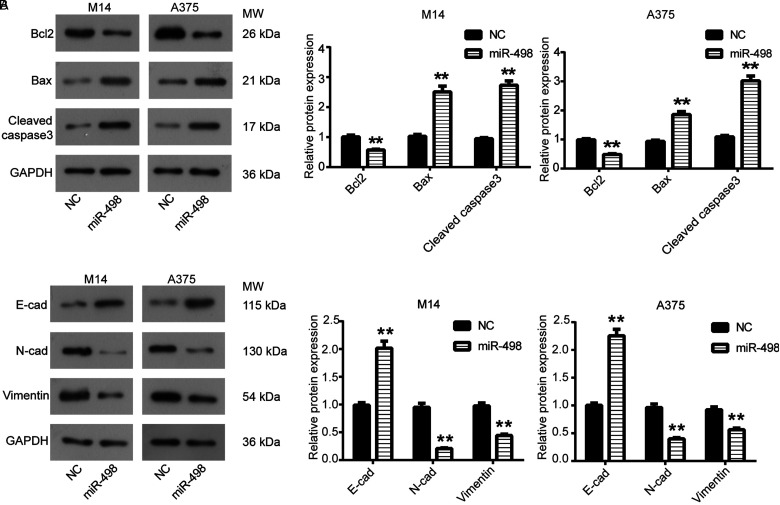
The effect of miR-498 on MM cell invasion and migration was detected using transwell assay. As shown in Figure 2A, the number of invaded and migrated M14 cells in miR-498 group was significantly reduced compared with the NC group (P < .05). Then as shown in Figure 2B, transwell assay also showed similar results that the number of invaded and migrated A375 cells in miR-498 group was significantly declined in comparison to NC group (P < .05). The apoptosis of M14 and A375 cells transfected with miR-498 was detected by flow cytometry assay. As shown in Figure 2C, the percentage of apoptotic cells was significantly increased from 14.38% or 10.28% to 25.00% or 19.03% in M14 and A375 cells, respectively, in miR-498 group compared to NC (P < .05). Western blot was further used to detect cell apoptosis. As shown in Figure 3A, the expression of antiapoptotic protein Bcl2 in miR-498 group was strongly decreased compared with the NC group, while the expression of proapoptotic proteins Bax, cleaved caspase3 was significantly increased (P < .05).
Figure 2.
miR-498 inhibited malignant melanoma (MM) cell invasion and migration and induces cell apoptosis. (A and B) Transwell assay detected MM cell invasion and migration. (C) Flow cytometry assay results showed that the percentage of apoptotic cells was significantly increased in M14 and A375 cells in miR-498 group compared to NC. **P < .01 compared with NC.
Figure 3.
miR-498 regulated epithelial–mesenchymal transition (EMT) of malignant melanoma (MM) cells. (A) Expression levels of Bcl-2, Bax and cleaved caspase3 in M14 and A375 cells were detected by Western blot assay. (B) Western blot detected the expression levels of E-cad, N-cad and Vimentin in M14 and A375 cells. **P < .01 compared with NC.
miR-498 Regulates Epithelial–Mesenchymal Transition of Malignant Melanoma Cells
The expression of epithelial–mesenchymal transition (EMT)-related proteins was detected using Western blot. We observed significant upregulation of E-cad in miR-498 group in M14 and A375 cells in comparison to NC group (P < .05), while N-cad and Vimentin were significantly downregulated in miR-498 group (Figure 3B), which suggested that miR-498 regulated EMT process in MM cells.
UBE2T Is a Novel Target Gene of miR-498 and Overexpression of UBE2T Reverses the Inhibitory Effects of miR-498 on Malignant Melanoma Cell Progression
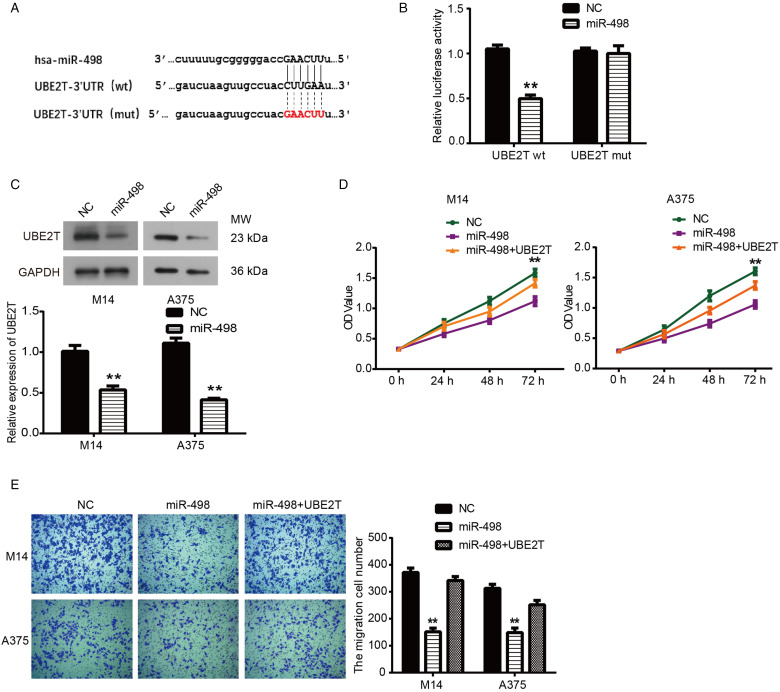
UBE2T was predicted to be the direct target of miR-498 using TargetScan (Figure 4A). In order to further determine whether UBE2T was a direct target of miR-498, we performed dual-luciferase assay and Western blot. As shown in Figure 4B, miR-498 significantly inhibited the luciferase activity of the UBE2T 3′-UTR wild-type reporter genes (P < .05). Then, the Western blot results in Figure 4C confirmed that UBE2T expression was significantly decreased in M14 and A375 cells transfected with miR-498 (P < .05). Subsequently, CCK8 and transwell assays were used to identify whether miR-498 regulated the proliferation and migration of MM cells by targeting UBE2T. The result showed in Figure 4D and E that UBE2T reversed the effect of miR-498 on inhibiting tumor growth and migration of M14 and A375 cells (P < .05). Together, these results indicated that miR-498 could inhibit tumor cell growth, invasion, and migration by directly targeting UBE2T in human MM.
Figure 4.
Overexpression of UBE2T reversed the inhibitory effects of miR-498 on malignant melanoma (MM) cell progression. (A) miR-498 binding sites in the 3′-UTR of the UBE2T gene. (B) Results of luciferase-reporter assays performed. (C) Expression levels of UBE2T in M14 and A375 cells were detected by Western blot assay. (D) Cell counting kit-8 assay detected cell proliferation in M14 and A375 cells. (E) Transwell assay detected MM cell migration. **P < .01 compared with NC.
Higher Level of UBE2T Is Associated With Poor Prognosis of Malignant Melanoma
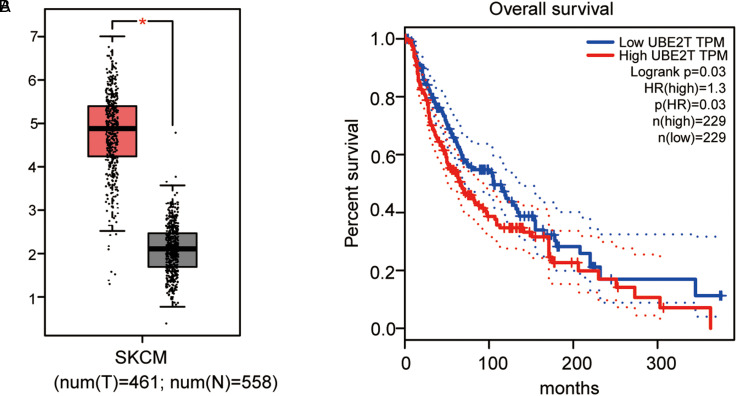
Finally, we used the TCGA database to analyze the expression pattern of UBE2T in skin cutaneous melanoma (SKCM) and its correlation with patient prognosis. As shown in Figure 5A, the expression of UBE2T in SKCM (n = 558) was significantly higher than that in normal samples (n = 461) (P < .05). In addition, compared with SKCM patients with low UBE2T expression, the survival rate of SKCM patients with high UBE2T expression was significantly lower (Figure 5B, P < .05).
Figure 5.
Higher level of UBE2T was associated with poor prognosis in malignant melanoma (MM) cells. (A) The boxplot of mRNA expression of UBE2T in skin cutaneous melanoma (SKCM). The red and gray boxes represent MM and normal tissues, respectively. (B) The survival percentage of SKCM patients with high or low UBE2T expression.
Discussion
Existing studies believe that miR-498 is a tumor suppressor miRNA, which can regulate a variety of tumor suppressor genes or proteins related to the regulation of tumor growth, invasion, and metastasis, thereby regulating the occurrence and development of various tumors.5,15,16 As shown in a previous study, miR-498 expression in colon cancer tissues is reduced, and miR-498 can inhibit the proliferation of colon cancer cells. 7 It has been reported that miR-498 can inhibit the proliferation and migration of triple-negative breast cancer cells by downregulating the BRCA1 expression. 5 Consistent with the above findings, we found that miR-498 inhibited the proliferation, invasion, and migration of MM cells, suggesting that miR-498 was a tumor suppressor miRNA in MM.
Previous studies have found that UBE2T is highly expressed in a variety of tumors tissues, and its expression level is significantly related to patient survival. 17 In breast cancer, UBE2T can not only be used as a molecular marker for predicting the prognosis of breast cancer patients but also promote the occurrence and development of tumors by participating in the ubiquitination degradation of breast cancer susceptibility protein 1. In gastric cancer, interference with the expression of UBE2T not only causes cell cycle arrest and apoptosis of gastric cancer cells, thereby inhibiting the growth of tumor cells, but also inhibits the migration and invasion of tumor cells through EMT. Clinically, the high expression of UBE2T often indicates a poor prognosis for patients with gastric cancer. In addition, the expression level of UBE2T in HCC tissues is significantly higher than that in adjacent tissues. The high expression of UBE2T in HCC is related to the intrahepatic metastasis of tumors and can promote the migration and invasion of HCC cells. 18 In this study, we found that the mRNA expression of UBE2T in SKCM tissues was significantly higher than that in normal tissues by searching on GEPIA. We explored the relationship between miR-498 and UBE2T, and first found that the overexpression of UBE2T reversed the inhibitory effects of miR-498 on the progression of MM cells.
Conclusion
In summary, miR-498 can inhibit the proliferation, invasion, migration, and EMT of M14 and A375 cells by targeting UBE2T, providing new ideas for the treatment of MM.
Abbreviations
- CCK8
cell counting kit-8
- EMT
epithelial–mesenchymal transition
- HEM
Human epidermal normal melanocytes
- MM
Malignant melanoma
- NC
negative control
- SKCM
skin cutaneous melanoma
Footnotes
Authors’ Note: The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. Our research is not on human tissue/samples, and not need Ethics/Review board approval. Furthermore, we have checked this with our review board and received their exemption.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by Natural Science Foundation of China, Grant/Award Number: 81971088; Natural Science Foundation of Shandong Province, Grant/Award Numbers: ZR2017MH074, ZR2018MC008; China Postdoctoral Science Foundation, Grant/Award Numbers: 2014M550370, 2015T80740; Academic promotion program of Shandong First Medical University, Grant/Award Number: 2019QL024; Innovation Project of Shandong Academy of Medical Sciences.
ORCID iDs: Wen Cao https://orcid.org/0000-0001-9784-563X
Shu-Hong Huang https://orcid.org/0000-0002-7415-2352
References
- 1.Gkalpakiotis S, Arenberger P, Fridman M, et al. Long-term therapy with ustekinumab for psoriasis in a patient with a history of malignant melanoma. Dermatol Ther. 2017;30(6):e12522. [DOI] [PubMed] [Google Scholar]
- 2.Matsutani T, Onda M, Miyashita M, et al. Primary malignant melanoma of the esophagus treated by esophagectomy and systemic chemotherapy. Dis Esophagus. 2001;14(3-4):241-244. [DOI] [PubMed] [Google Scholar]
- 3.Winkler JK, Buderbakhaya K, Dimitrakopouloustrauss A. Malignant melanoma current status. Radiologe. 2017;77(2):356. [Google Scholar]
- 4.Caramuta S, Egyházi S, Rodolfo M, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. Journal of Investigative Dermatology. 2010;130(8):2062–2070. [DOI] [PubMed] [Google Scholar]
- 5.Matamala N, Vargas MT, González-Cámpora R, et al. MicroRNA deregulation in triple negative breast cancer reveals a role of miR-498 in regulating BRCA1 expression. Oncotarget 2016;7(15):20068–20079. 10.18632/oncotarget.7705(15):20068-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong J, Liu R, Wang X, et al. Low miR-498 expression levels are associated with poor prognosis in ovarian cancer. Eur Rev Med Pharmacol Sci. 2016;19(24):4762. [PubMed] [Google Scholar]
- 7.Gopalan V, Smith RA, Lam AKY. Downregulation of microRNA-498 in colorectal cancers and its cellular effects. Exp Cell Res. 2015;330(2):423-428. [DOI] [PubMed] [Google Scholar]
- 8.Chen LP, Zhang NN, Ren XQ, et al. miR-103/miR-195/miR-15b regulate SALL4 and inhibit proliferation and migration in glioma. Molecules. 2018;23(11):2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian F, Wei H, Tian H, et al. miR-33a is downregulated in melanoma cells and modulates cell proliferation by targeting PCTAIRE1. Oncology Letters. 2016;11(4):2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D, Zhao J, Kong W, et al. Abstract 3054: microRNA expression profile in cutaneous melanoma: miR-17-92 cluster is a progression and prognostic marker. Cancer Res. 2010;70(8 Supplement):3054. [Google Scholar]
- 11.Bell RE, Khaled M, Netanely D, et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J Invest Dermatol. 2014;134(2):441-451. [DOI] [PubMed] [Google Scholar]
- 12.Wen M, Kwon Y, Wang Y, et al. Elevated expression of UBE2T exhibits oncogenic properties in human prostate cancer. Oncotarget. 2015;6(28):25226–25239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Xiang P, Pan Q, et al. Ubiquitin-conjugating enzyme E2T is an independent prognostic factor and promotes gastric cancer progression. Tumor Biol. 2016;37(9):11723-11732. [DOI] [PubMed] [Google Scholar]
- 14.Gong Y, Peng D, Ning X, et al. UBE2T Silencing suppresses proliferation and induces cell cycle arrest and apoptosis in bladder cancer cells. Oncology Letters. 2016(12(6):4485–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kogo R, How C, Chaudary N, et al. The microRNA-218∼survivin axis regulates migration, invasion, and lymph node metastasis in cervical cancer. Oncotarget. 2014;6(2):1090-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu K, Li C, Zheng X, et al. Prognostic significance of miR-218 in human hepatocellular carcinoma and its role in cell growth. Oncol Rep. 2014;32(4):1571-1577. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Peña J, Corrales-Sánchez V, Amir E, et al. Ubiquitin-conjugating enzyme E2T (UBE2T) and denticleless protein homolog (DTL) are linked to poor outcome in breast and lung cancers. Sci Rep. 2012;586(20):3761–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan YH, Yang M, Liu L-p, et al. UBE2S Enhances the ubiquitination of p53 and exerts oncogenic activities in hepatocellular carcinoma. Biochem Biophys Res Commun. 2016;7(1):293–307. [DOI] [PubMed] [Google Scholar]