Abstract
Breast cancer is one of the most fatal diseases leading to the death of several women across the world. But early diagnosis of breast cancer can help to reduce the mortality rate. So an efficient multi-task learning approach is proposed in this work for the automatic segmentation and classification of breast tumors from ultrasound images. The proposed learning approach consists of an encoder, decoder, and bridge blocks for segmentation and a dense branch for the classification of tumors. For efficient classification, multi-scale features from different levels of the network are used. Experimental results show that the proposed approach is able to enhance the accuracy and recall of segmentation by , , and classification by , , respectively than the methods available in the literature.
Keywords: multi-task learning, breast cancer, segmentation, classification, malignant, benign
Introduction
Breast cancer is the deadliest type of cancer that is found in women and is the leading cause of death in young women across the world. According to a study performed by the International Agency for Research on Cancer in 2018, 1 approximately 2.1 million new breast cancer cases and 0.6 million new deaths were reported worldwide. In 2020, the number of breast cancer cases exceeded lung cancer, making it more prevalent than other forms of cancer. 2 Earlier diagnosis and treatment of breast cancer can reduce the mortality rate, increase the survival chances, and quality of life of the patient.3 –5 Digital mammography is the primary imaging examination for the diagnosis of breast cancer. But exposure to ionizing radiation multiple times can increase the risk for breast cancer. 6
Ultrasound Imaging (UI) is an alternative procedure for mammography for the diagnosis of breast cancer. UI is more safer, faster, cheaper, and reproducible compared to that of digital mammography.7 –9 UI has reported higher detection rate of breast cancer in women with dense breasts than mammography. 10 But due to the high sensitivity of the ultrasound instruments, these UI images are susceptible to the influence of the environment and other tissues of the human body, resulting in large amounts of speckle noise which makes the diagnosis procedure difficult for medical professionals. Thus, efficient computerized systems for assisting medical professionals in diagnosing breast cancer, specifying surgical plans, and treatment is very essential. But designing such systems for UI based diagnosis is a complex and a challenging task because of non-uniform tumor boundary, variations in tumor size and shape, and low signal-to-noise ratio in ultrasound images.
Classification and segmentation are the two primary tasks of a computerized system in medical imaging. Malignant and benign tumors have different shape characteristics. For example, malignant tumors have speculated and irregular shapes, and benign tumors have oval, round, and smooth boundaries. 11 These shape characteristics are used in clinical diagnosis by radiologists in categorizing the tumor as benign and malignant. These properties are very useful for tumor classification and segmentation. Hence training a single network to solve two tasks (segmentation and classification of breast tumor) through feature sharing is a promising direction to explore.
In this paper, a novel multi-task learning approach is proposed to jointly perform the segmentation and classification of breast tumors in ultrasound images in an end to end model. The proposed model consists of an encoder-decoder network for segmentation and a multi-scale feature network branch for classification of a tumor. In this work, the U-Net architecture is modified by adding an residual module for building the encoder-decoder network. The classification and segmentation tasks share the common features extracted by the encoding block. To overcome the problem of varying tumor characteristics, we use multi-scale features extracted from different convolutional layers from the encoder block for tumor classification. The rest of the paper is organized as follows, Section 2 describes the existing literature, Section 3 describes the materials and methods used, Section 4 describes the results reported, and Section 5 concludes the proposed work.
Related Works
Ultrasound Image Segmentation
In previous studies, there were several methods proposed for the segmentation of ultrasound images using conventional image-processing techniques like active contour,12,13 region growing,14,15 and watershed transform.16 –18 Gomez et al. 18 proposed a robust method to segment various objects in contrast enhanced ultrasound images with closed contours using marker controlled watershed transformations. Kozegar et al. 15 proposed a two stage segmentation method, where the mass boundary is estimated using an adaptive region growing algorithm in the first stage. In the second stage, this estimation is used as an initial contour by a geometric edge based model for further refinement. The performance of this model is heavily dependent on the initial seed selection.
Over the years, there were many Convolutional Neural Network (CNN) based models designed for ultrasound image segmentation.19 –22 Xing et al. 19 developed a semi-pixel-wise cycle model using Generative Adversarial Network (GAN) and CNN for tumor segmentation. The anatomy based image segmentation is also very important in ultrasound image analysis to reduce the false positives in image segmentation. Lei et al. 23 used boundary regularization based encoder-decoder network to segment the breast anatomy in ultrasound images. Authors in Lei et al. 24 used self-co-attention mechanism to further improve the segmentation results of breast anatomy. Kumar et al. 25 designed a method named Multi-UNet based on the popular U-Net architecture for the segmentation of masses from ultrasound images. In study, 26 custom designed attention blocks were added to the existing U-Net for tumor segmentation. In this work, the salient feature maps are integrated with the deep learning attention UNet for better segmentation.
Ultrasound Image Classification
Traditional methods for ultrasound image classification rely on manual extraction of posterior acoustic features, echo patterns, lesion boundary, margin, orientation, shape, and texture features. Moon et al. 27 used texture, descriptor, and morphological features for the classification of the tumor as malignant or benign. Gómez Flores et al. 28 enhanced the diagnostic accuracy of tumor in ultrasound imaging by analyzing the distinct morphological and textual features. In Gomez et al. 29 authors used watershed transformation technique to segment the tumor area in the ultrasound images. From these segmented images 22 morphological features were computed and minimum-redundancy-maximal-relevance-criteria was used to rank these features. Later in the study, n-dimensional feature subsets have been created using ranked feature space and fisher discriminant analysis classifier. Uniyal et al. 30 proposed a novel approach, where tumor malignancy maps are generated based on an estimation of cancer likelihood from ultrasound radio-frequency time series for the classification of malignant tumors. The performance of the above discussed methods is largely dependent on the accuracy of the manually extracted features.
Deep learning based methods can overcome the limitations of traditional methods with their powerful feature extraction capability. Zeimarin et al. 31 reported enhanced performance in tumor classification while using a custom CNN model with regularization technique. More than the custom-built CNN, several architectures which are trained on large dataset, can easily be fine tuned for classification of tumors. This methodology is known as transfer learning. Transfer learning methods have shown satisfactory improvements in performance for the classification of breast tumors. 32 In study, 33 the performance of transfer learning of pre-trained models namely SD300 + ZFNet, YOLO, and VGG16 was analyzed, out of which SD300 + ZFNet reported higher performance than the other two models. Author in Hijab et al. 34 proposed a transfer learning model, which was built by fine-tuning the pre-trained model VGG16. To overcome the problem of overfitting, image augmentation techniques were been used, and the model reported to have an accuracy of 97%. Research 35 proposed a transfer learning model that uses the VGG19 pre-trained model as the base model. In this work, the ultrasound images are converted to RGB representation before training the model. Study 36 proposed a transfer learning model using pre-trained InceptionV3 for the classification of breast lesions. In study, 37 the authors performed a comparative analysis of performance reported by the transfer learning of InceptionV3, Xception, and ResNet50 to classify tumors from ultrasound datasets. In Han et al., 38 a transfer learning approach based on GoogleNet is proposed to diagnose breast cancer from ultrasound images. In this work, histogram equalization and image cropping methods are used to augment the training images. Tanaka et al. 39 employed the ensemble of VGG19 and ResNet152 for the diagnosis of malignant and benign tumors.
Joint segmentation and classification (Multi-task learning)
Multi-task learning is an approach that focuses on solving two or more different tasks parallelly at the same time. In medical imaging, multi-task learning is applied to perform segmentation and classification tasks parallelly on the same image. For ultrasound images, Wang et al. 40 modified the structure of U-Net by adding a classification branch for classification and segmentation of bone surfaces. Xie et al. 41 proposed a dual stage multi-task approach using pre-trained models for the segmentation and classification of tumors from breast ultrasound images. In this approach, ResNet is used for the extraction and the classification of candidate regions in the first stage and in the second stage a modified Mask R-CNN is used for tumor segmentation.
Materials and Methods
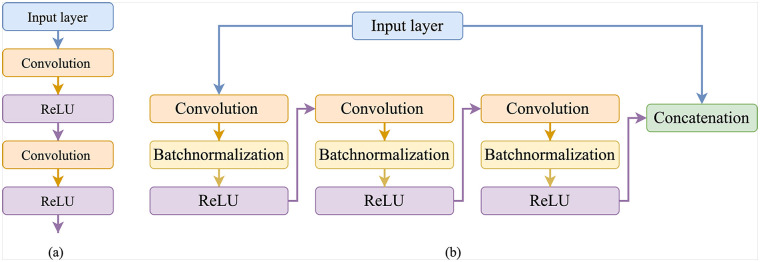
In our work, the residual learning approach was used to ease the training of the deep neural networks and to overcome the problem of degradation. 42 The architecture of the proposed residual module is different from Diakogiannis et al. 43 , Zhang et al. 44 in terms of number of layers. this proposed work used three convolutions, three batch-normalization, three ReLU, and one concatenation layer. The residual network consists of a stack of residual units, where each unit consists of a convolutional layer, batch normalization layer, and ReLU activation. Figure 1 shows the difference between the neural layers module in standard UNet and our proposed residual module.
Figure 1.
Difference between the neural layers module in standard UNet and the proposed residual module in the this work: (a) Neural layers used in U-Net and (b) proposed residual module.
As shown in Figure 1(b), shortcut connections or skip connections are the ones that skip one or more layers in the neural network. The proposed residual unit shown in Figure 1(b) can be represented by equation (1).
| (1) |
In equation (1), represents a batch-normalization layer, represents a convolutional layer with filters, represents the input, and represents the output of the residual block.
Residual-U-Net
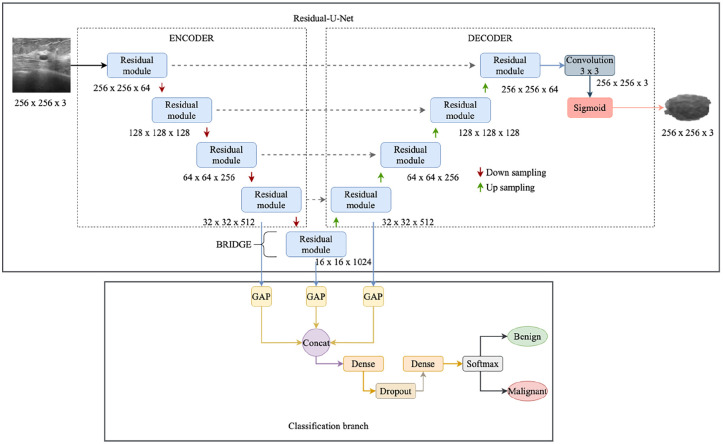
The proposed multi-task learning approach for segmentation and classification is shown in Figure 2. The proposed network is built by combining the strengths of residual and U-Net architectures. This network combination has two advantages: (1) The residual unit eases the learning of the network. (2) The skip connection between the high and low levels in the residual unit will help for the propagation of information without degradation. The proposed multi-task learning approach can perform segmentation and classification simultaneously.
Figure 2.
The proposed multi-task learning approach for segmentation and classification.
The Residual-U-Net proposed in this work is a 9 level architecture consisting of 3 parts namely encoder, decoder, and a bridge. The encoder converts the input image into compact representation and the decoder recovers this representation into pixel wise classification. Bridge acts as a connection between encoder and decoder parts. All these parts are built using residual units shown in Figure 1(b), each block consists of convolutional layers with filters, 3 batch normalization layers, 3 ReLU activation layers, and an identity mapping. The encoding block consists of 4 residual units, specifically the encoding block uses four downsampling operations after each residual unit to extract high level semantic information. In each encoding residual unit, a stride of 2 is applied to the first convolutional layer of the unit to down sample the feature map by its half instead of using a pool operation to preserve positional information. Correspondingly, the decoder path consists of 4 residual units and between each unit, there is a concatenation of the feature map from the corresponding encoding path with the upsampled feature map from the previous unit. After the last encoding unit, there is a convolutional layer and a sigmoid activation layer to project the desired segmented image.
For classifying the tumor as benign or malignant, the feature map from the last layer of the encoder block is taken and passed through a single dense layer for classification. 40 Whereas in our classification branch, the extracted features from the last block of encoder, bridge, and the first block of decoder is used for classification (Figure 2). The branch extracts and concatenates the feature maps from stages 4, 5, and 6. Since these features have dimensions that cannot be concatenated directly, Global Average Pooling (GAP) is connected to the end of each block to make them suitable for concatenation. The GAP layers are employed to enable multi-scale feature concatenation. Even fully connected (FC) layers and Global Max Pooling (GMP) layers perform the same operation, but they have a few disadvantages. FC layers increase the training time and the number of parameters. Whereas the GMP uses max-vocel to represent the whole features and neglects a lot of useful spatial information.
Then these features are concatenated and are then passed to a series of dense, dropout layers for classification. The first dense layer that receives the fused features consists of 256 units and is activated with ReLU function and the last dense layer contains 2 units and is activated with softmax function to predict the class of the input ultrasound image as malignant or benign. In comparison to Wang et al., 40 the proposed classification branch has two dense layers. Between these two dense layers, a dropout layer is added with dropout rate to 0.5 to prevent overfitting and this layer acts as regularization to the network.
Loss Function
During segmentation tasks, the variance between the background and foreground may result in segmentation bias. To solve this issue, a segmentation loss based on the Dice coefficient is used in this work. This was defined as
| (2) |
In the above equation, represents the predicted segmentation result, represents the actual segmented result, and represents the segmentation loss.
In medical imaging, class imbalancing is the most challenging problem, making the model more biased to one class if it was not resolved. In this work, the considered BUSI datasets consist of more malignant instances than benign, this may cause the model to have more bias to malignant cases. To deal with this imbalancing problem, the weighted focal loss 45 is used for the classification task and is shown in equation (3).
| (3) |
In the above equation, is the predicted classification output, is the actual class, is the focussing parameter, and is set as 2 as it has shown optimum results in Lin et al. 45 , Zunair and Hamza, 46 Azhar et al. 47 , and represent the weights assigned to malignant and benign classes. These weights are shown in equation (4).
| (4) |
In equation (4), and represent the number of malignant and benign tumors. Hence the global loss function for the proposed work is the sum of segmentation and classification loss. This is shown in equation (5).
| (5) |
Dataset
The proposed multi-task model is evaluated using the benchmark ultrasound dataset BUSI. 48 This dataset consists of a total of 780 2D breast ultrasound images collected from 600 women aging between 25 and 75. Among the 780 images, 133 are benign masses, 437 are normal cases, and 210 are malignant masses. Even though these images look like one-channeled grayscale images, but they are three-channeled images. BUSI also contains corresponding masks for all the images. The main idea behind tumor segmentation besides solo classification is to track the tumor changes and to assess the seriousness of the tumor. Since normal ultrasound images don’t have any tumor mass for segmentation, they are not used in this work.
Experimental Setting
To show a comparison between our proposed networks and the existing networks, the same experimental set-up is followed, and only benign and malignant cases are considered for training. The images are enhanced using the Generated Histogram Equalization method before feeding them into the network. It is done to increase the intensity difference between the tumor and the background. Data augmentation is done, and the images are augmented using horizontal flip, rotation by 30°, and vertical flip. Initially, the hyper-parameter values are kept same as reported in the existing work.45 –47 However, the hyper-parameters tuning is done to yield the optimum performance based on extensive evaluation. TensorFlow is used as the deep learning framework. All the experiments in this work are carried out in Google Colab. The proposed model is compiled using Adam optimizer and the proposed loss function, for validation the standard 5-fold cross validation protocol is considered. For each fold, the model is trained for 500 epochs with batch size of 16 and the learning rate is set to 0.0001 which further decreases by one tenth after every 20 epochs. Before feeding into the network, the images are enhanced by Multi peak Generated Histogram Equalization (GHE) method. 49 This method increases the intensity difference between the tumor and the background. During training, the training images are augmented by flipping horizontally, flipping vertically, and rotating by 30°. The transfer learning models are pre-trained on ImageNet and to make them suitable for our work, they were fine-tuned with the BUSI dataset.
Evaluation Metrics
In this work, eight popular metrics namely Jaccard Similarity Index (JSI), Dice Coefficient (DC), accuracy (ACC), Precision (PRE), Recall (REC), specificity (SPE), F1-score (F1), and Area under ROC curve (AUC) are used for the quantitative evaluation of the proposed model. These metrics are explained below.
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
In equations (6) and (7), and are the ground truth and segmentation results. In equations (8)–(12), True Positives ( ) represents the number of images that are correctly classified as malignant, True Negative ( ) represents the number of images that are correctly classified as benign, and False Positive ( ) and False Negative ( ) represents the number images that are wrongly classified as benign and malignant. Among these metrics, , , , , and are used for evaluating the segmentation task and , , , , , are used for evaluating the classification task.
Results
The results reported by the proposed multi-task learning method for segmentation and classification during each fold are shown in Tables 1 and 2.
Table 1.
Segmentation Results Reported by the Proposed Model.
| Exp | JSI | DC | ACC | REC | PRE |
|---|---|---|---|---|---|
| 1 | 84.37 | 83.45 | 88.03 | 84.96 | 86.77 |
| 2 | 83.80 | 84.02 | 87.80 | 84.90 | 86.56 |
| 3 | 85.06 | 85.90 | 88.35 | 85.93 | 85.60 |
| 4 | 83.65 | 83.72 | 87.55 | 86.29 | 85.36 |
| 5 | 86.74 | 87.00 | 88.67 | 86.04 | 86.38 |
| Mean | 84.72 | 84.81 | 88.08 | 85.62 | 86.13 |
Table 2.
Classification Results Reported by the Proposed Model.
| Exp | ACC | PRE | REC | SPE | F1 | AUC |
|---|---|---|---|---|---|---|
| 1 | 97.53 | 97.6 | 98.65 | 93.67 | 98.14 | 0.98 |
| 2 | 98.44 | 98.53 | 99.32 | 95.42 | 98.93 | 1.00 |
| 3 | 97.60 | 98.05 | 98.16 | 92.57 | 98.10 | 0.99 |
| 4 | 97.69 | 98.14 | 99.24 | 94.63 | 98.68 | 0.97 |
| 5 | 98.07 | 98.29 | 98.60 | 96.96 | 98.44 | 1.00 |
| Mean | 97.86 | 98.12 | 98.79 | 94.65 | 98.45 | 0.99 |
Comparison With Current STATE-of-the-ART Methods
The performance of the proposed model is compared with other existing methods50 –54 for tumor segmentation and classification. For a fair comparison, all these methods are downloaded from their public implementations and are retrained using the BUSI dataset. During retraining, optimum hyper-parameters are chosen based on extensive experiments. These experiments were conducted by varying the batch size, optimizer and the learning rate. The hyper-parameters for which the existing methods reported optimum performance is shown in Table 3.
Table 3.
Hyper-Parameters for Which the Existing Segmentation Methods Reported Optimum Performance.
For segmentation, we compare our work with pre-trained DeeplabV3+, 50 UNet++, 51 UNet, 52 feature pyramid network (FPN), 53 and also with attention based methods like DAF. 54 This comparison is shown in Table 4.
Table 4.
Comparison With Segmentation Models From the Literature.
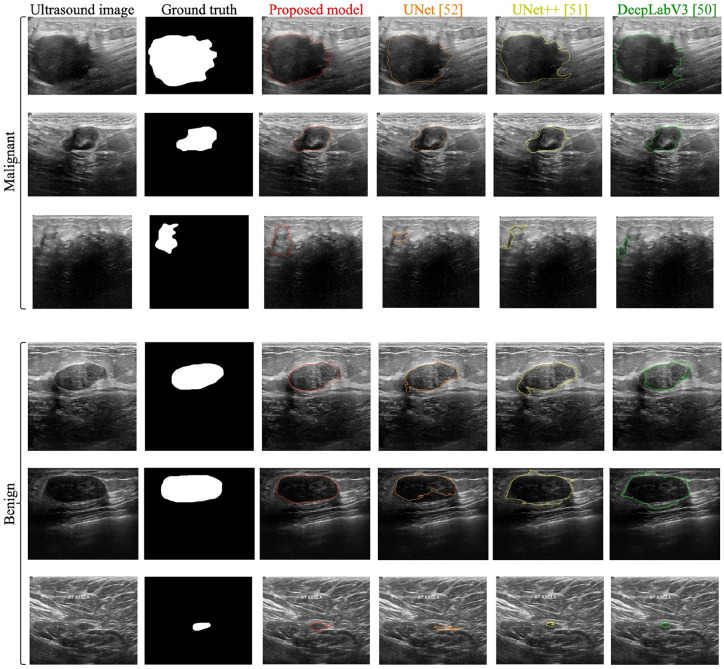
The segmentation performance reported by the proposed model and the other segmentation methods on sample images is shown in Figure 3. When normal images are passed to the network, the segmentation branch outputs the entire image without segmenting any region.
Figure 3.
Comparison of segmentation performance reported by the proposed model with other segmentation methods.
For classification, the proposed model is compared with traditional feature based classification methods like, 27 and with transfer learning methods like.30,34 For a fair comparison, these existing methods are retrained using the same data that is used by the proposed model. During retraining, the hyper-parameters were changed for the transfer learning methods30,34 for getting the optimum performance on the BUSI dataset. The hyper-parameters used for these methods are shown in Table 5.
Table 5.
Hyper-Parameters for Which the Existing Classification Methods Reported Optimum Performance.
These parameters were chosen by performing extensive experiments by changing the optimizer, batch size, and the learning rate. This comparison is shown in Table 6.
Table 6.
Comparison With Classification Models From the Literature.
The proposed model outperformed the models proposed in the literature.
Along with the comparison with the existing literature, the classification performance of the proposed model is also compared with existing transfer learning models namely ResNet50, VGG19, DenseNet201, InceptionV3, MobileNet, and InceptionResNetV2. For this work, the number of units in the last dense layer of these models is changed (n_units = 2) to make these models suitable for tumor classification. This comparison is shown in Table 7.
Table 7.
Comparison of Classification Results With Pre-Trained Models.
| Model | ACC | PRE | REC | SPE | F1 | AUC |
|---|---|---|---|---|---|---|
| ResNet50 | 86.00 | 86.66 | 82.85 | 83.33 | 89.23 | 0.83 |
| VGG19 | 86.00 | 83.33 | 89.25 | 80.90 | 86.55 | 0.87 |
| DenseNet201 | 86.00 | 86.66 | 86.66 | 85.00 | 85.66 | 0.86 |
| InceptionV3 | 88.00 | 90.00 | 86.77 | 89.00 | 88.36 | 0.88 |
| MobileNet | 86.00 | 83.33 | 86.70 | 80.90 | 86.55 | 0.87 |
| InceptionResNetV2 | 84.00 | 83.33 | 86.55 | 80.47 | 84.91 | 0.84 |
| Proposed model | 97.86 | 98.12 | 98.79 | 94.65 | 98.45 | 0.99 |
When the three classes namely Benign, Malignant, and Normal are considered the proposed model reported an classification accuracy of 97.5%, precision of 99%, recall of 98.12%, specificity of 93.59%, F1-score of 99.32%, and AUC of 0.983.
Discussion
Ultrasound imaging is extensively used for the diagnosis of breast cancer as it is safe, fast, and has better reproducibility.7 –9 But those images are susceptible to speckle noise and make the diagnosis difficult for medical professionals. So, designing an efficient system for the UI is an important task. The varying tumor characteristics like shape, size, unclear tumor boundaries, and less signal to noise ratio makes the tumor segmentation and classification task a more challenging one. In this paper, a multi-task learning approach is proposed for the efficient segmentation and classification of breast tumors from ultrasound images.
In this work, the features from multiple layers of the proposed Residual-U-Net are concatenated using GAP and used for the classification of tumors. This multi scale feature concatenation helps the classification method to overcome the problems with varying tumor characteristics and compromising UI environments. As shown in Table 6, the proposed classification method reported higher performance than single classification models. An example of a complex benign and malignant tumor is shown in the last row Figure 3, which was misclassified by methods based on UNet, 52 UNet++, 51 and DeepLabV3, 50 but correctly segmented and classified by the proposed model.
The proposed Residual-U-Net reported better segmentation performance than single segmentation models, as shown in Table 4. Besides accuracy, recall is also the most crucial metric in medical informatics, as it explains the number of correctly classified/segmented malignant tumors in this work. Among the existing related research works, methods proposed in Hijab et al. 34 , Wang et al. 54 reported higher performance in terms of accuracy and recall on segmentation and classification tasks. But the proposed multi-task approach is able to enhance the recall and accuracy of the segmentation and classification tasks by , and , , respectively. Among the five-folds, the least recall that was reported for segmentation and classification were (during fold 2) and (during fold 1) which were also higher than the best performing methods that were proposed in Hijab et al. 34 , Chen et al. 50 , Lin et al. 53 from the existing literature. This clearly shows the robustness and efficiency of the proposed method in breast cancer diagnosis from the ultrasound images.
As of now, the current limitation of the reported work is an inclusion of the Breast Imaging Reporting and Data System (BIRADS) 55 information. BIRADS is a standardized system of reporting breast cancer risk and is mainly used in mammogram, breast ultrasound, and breast magnetic resonance imaging (MRI) reports. BIRADS is used to place abnormal finding into categories that ranges between 0 and 6 (Table 8). A BIRADS score is a part of the breast imaging reports and helps in quantifying how concerning the finding is. A higher number indicates a higher risk. A change in BIRADS score from test to test helps in clearly detecting a difference between the results.
Table 8.
BIRADS Classification Categories.
| Category | Model | Details |
|---|---|---|
| 0 | Incomplete test | A possible abnormality seen by radiologists but not clear, so an additional test required. |
| 1 | Negative | Indicates a negative test and no significant abnormality to report. |
| 2 | Benign (non-cancerous) finding | Indicates a normal result and no indication of cancer. Though there can be presence of some benign cysts or masses to include in report. It ensures that the benign finding is not reported as suspicious. This finding is included in the mammogram report to help in comparing future mammograms. |
| 3 | Probably benign finding—follow-up in a short time frame is suggested | Indicates a very high chance (greater than 98%) for the findings being benign (not cancer) and these findings are not expected to change over time. However, it is not proven to be benign, it is beneficial to see if the area in question changes overtime. |
| 4 | Suspicious abnormality—biopsy should be considered | Indicates suspicious findings. The finding in this category indicates a wide range of suspicious levels as below: 4A: Finding with a low likelihood of being cancer (more than 2% but no more than 10%). 4B: Finding with a moderate likelihood of being cancer (more than 10% but no more than 50%). 4Cb Finding with a high likelihood of being cancer (more than 50% but less than 95%), but not as high as Category 5. |
| 5 | Highly suggestive of malignancy—appropriate action should be taken | High chance (at least 95%) of being cancer and biopsy is very strongly recommended. |
| 6 | Known biopsy-proven malignancy—appropriate action should be taken | This category is only used for findings on a mammogram that have already been shown to be cancer by a previous biopsy. Mammograms may be used in this way to see how well the cancer is responding to treatment. |
The literature indicates that it is relatively easy to classify BIRADS 3 and 5, whereas BIRADS 4 is difficult to classify. As a result, the lack of BIRADS 4 lesions in the used dataset can skew the result more favorably. Since the dataset used in this study is not broken into BIRADS categories, there can be chances of a skewed results. Other studies using BUSI dataset have got their images annotated manually for the BIRADS descriptors and categories with the help of a radiologist. We don’t have the resources to do the BIRADS classification as if now. In future study, we will include the information and additionally test our proposed methodology on other datasets that have BIRADS classification.
Conclusion
In this paper, a multi-task learning approach is proposed for the segmentation and classification of tumors in breast ultrasound images. In this work, the multi-task model is built by modifying the U-Net architecture by using residual units and by adding a classification branch to the network. Multi-scale features are extracted from different layers of the proposed Residual-U-Net for efficient classification of the tumor. Experimental results show that the proposed model is more efficient than the existing segmentation and classification methods in terms of accuracy and recall. This model enhanced the recall of breast image segmentation and classification tasks by and , respectively. This shows that the model is more safer as it has reported less number of false negatives than the existing models.
In future work, we will study the different networks to extract more discriminative information and will evaluate the proposed model on a larger dataset with more complex samples for understanding the robustness of the model.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Pratheepan Yogarajah  https://orcid.org/0000-0002-4586-7228
https://orcid.org/0000-0002-4586-7228
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [DOI] [PubMed] [Google Scholar]
- 2. IARC. IARC Biennial Report 2020-2021. Lyon, France: International Agency for Research on Cancer. Available from: https://publications.iarc.fr/607
- 3. Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin. 2018;68:31-54. [DOI] [PubMed] [Google Scholar]
- 4. Anitha J, Peter JD. Mammogram segmentation using maximal cell strength updation in cellular automata. Med Biol Eng Comput. 2015;53:737-49. [DOI] [PubMed] [Google Scholar]
- 5. Berg WA, Zhang Z, Lehrer D, Jong RA, Pisano ED, Barr RG, et al. Detection of breast cancer with addition of annual screening ultrasound or a single screening MRI to mammography in women with elevated breast cancer risk. JAMA. 2012;307:1394-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Skaane P, Bandos AI, Niklason LT, Sebuødegård S, Østerås BH, Gullien R, et al. Digital mammography versus digital mammography plus tomosynthesis in breast cancer screening: the Oslo Tomosynthesis Screening Trial. Radiology. 2019;291:23-30. [DOI] [PubMed] [Google Scholar]
- 7. Zanotel M, Bednarova I, Londero V, Linda A, Lorenzon M, Girometti R, et al. Automated breast ultrasound: basic principles and emerging clinical applications. Radiol Med. 2018;123:1-12. [DOI] [PubMed] [Google Scholar]
- 8. Giger ML, Inciardi MF, Edwards A, Papaioannou J, Drukker K, Jiang Y, et al. Automated breast ultrasound in breast cancer screening of women with dense breasts: reader study of mammography-negative and mammography-positive cancers. Am J Roentgenol. 2016;206:1341-50. [DOI] [PubMed] [Google Scholar]
- 9. Vourtsis A. Three-dimensional automated breast ultrasound: technical aspects and first results. Diagn Interven Imaging. 2019;100:579-92. [DOI] [PubMed] [Google Scholar]
- 10. Le EPV, Wang Y, Huang Y, Hickman S, Gilbert FJ. Artificial intelligence in breast imaging. Clin Radiol. 2019;74:357-66. [DOI] [PubMed] [Google Scholar]
- 11. Yang W, Zhang S, Chen Y, Li W, Chen Y. Measuring shape complexity of breast lesions on ultrasound images. In: Medical Imaging 2008: Ultrasonic Imaging and Signal Processing; 2008, vol. 6920, San Diego, California, United States, International Society for Optics and Photonics. pp. 169-78. [Google Scholar]
- 12. Lee CY, Chang TF, Chou YH, Yang KC. Fully automated lesion segmentation and visualization in automated whole breast ultrasound (ABUS) images. Quant Imaging Med Surg. 2020;10:568-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galiska M, Ogiego W, Wijata A, Juszczyk J, Czajkowska J. Breast cancer segmentation method in ultrasound images. In: Conference on Innovations in Biomedical Engineering; 2017, Penang, Malaysia, pp. 23-31. [Google Scholar]
- 14. Cheng JZ, Chou YH, Huang CS, Chang YC, Tiu CM, Chen KW, et al. Computer-aided US diagnosis of breast lesions by using cell-based contour grouping. Radiology. 2010;255:746-54. [DOI] [PubMed] [Google Scholar]
- 15. Kozegar E, Soryani M, Behnam H, Salamati M, Tan T. Mass segmentation in automated 3-D breast ultrasound using adaptive region growing and supervised edge-based deformable model. IEEE Trans Med Imaging. 2018;37:918-28. [DOI] [PubMed] [Google Scholar]
- 16. Cheng JZ, Chou YH, Huang CS, Chang YC, Tiu CM, Yeh FC, et al. ACCOMP: augmented cell competition algorithm for breast lesion demarcation in sonography. Med Phys. 2010;37:6240-52. [DOI] [PubMed] [Google Scholar]
- 17. Chen CM, Chou YH, Chen CS, Cheng JZ, Ou YF, Yeh FC, et al. Cell-competition algorithm: a new segmentation algorithm for multiple objects with irregular boundaries in ultrasound images. Ultrasound Med Biol. 2005;31:1647-64. [DOI] [PubMed] [Google Scholar]
- 18. Gómez W, Leija L, Alvarenga AV, Infantosi AF, Pereira WC. Computerized lesion segmentation of breast ultrasound based on marker-controlled watershed transformation. Med Phys. 2010;37:82-95. [DOI] [PubMed] [Google Scholar]
- 19. Xing J, Li Z, Wang B, Qi Y, Yu B, Zanjani FG, et al. Lesion segmentation in ultrasound using semi-pixel-wise cycle generative adversarial nets. IEEE/ACM Trans Comput Biol Bioinform. 2021;18:2555-65. [DOI] [PubMed] [Google Scholar]
- 20. Wang N, Bian C, Wang Y, Xu M, Qin C, Yang X, et al. Densely deep supervised networks with threshold loss for cancer detection in automated breast ultrasound. In: International Conference On Medical Image Computing and Computer-Assisted Intervention; 2018, Granada, Spain, pp. 641-48. [Google Scholar]
- 21. Cao X, Chen H, Li Y, Peng Y, Wang S, Cheng L. Uncertainty aware temporal-ensembling model for semi-supervised abus mass segmentation. IEEE Trans Med Imaging. 2021;40:431-43. [DOI] [PubMed] [Google Scholar]
- 22. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60-88. [DOI] [PubMed] [Google Scholar]
- 23. Lei B, Huang S, Li R, Bian C, Li H, Chou YH, et al. Segmentation of breast anatomy for automated whole breast ultrasound images with boundary regularized convolutional encoder–decoder network. Neurocomputing. 2018;321:178-86. [Google Scholar]
- 24. Lei B, Huang S, Li H, Li R, Bian C, Chou YH, et al. Self-co-attention neural network for anatomy segmentation in whole breast ultrasound. Med Image Anal. 2020;64:101753. [DOI] [PubMed] [Google Scholar]
- 25. Kumar V, Webb JM, Gregory A, Denis M, Meixner DD, Bayat M, et al. Automated and real-time segmentation of suspicious breast masses using convolutional neural network. PLoS One. 2018;13(5):e0195816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vakanski A, Xian M, Freer PE. Attention-enriched deep learning model for breast tumor segmentation in ultrasound images. Ultrasound Med Biol. 2020;46(10):2819-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moon WK, Shen YW, Huang CS, Chiang LR, Chang RF. Computer-aided diagnosis for the classification of breast masses in automated whole breast ultrasound images. Ultrasound Med Biol. 2011;37:539-48. [DOI] [PubMed] [Google Scholar]
- 28. Gómez Flores W, Pereira WCDA, Infantosi AFC. Improving classification performance of breast lesions on ultrasonography. Pattern Recognit. 2015;48:1125-36. [Google Scholar]
- 29. Gomez W, Rodriguez A, Pereira W, Infantosi A. Feature selection and classifier performance in computer-aided diagnosis for breast ultrasound. In: 10th International Conference and Expo on Emerging Technologies for a Smarter World (CEWIT); 2013, Melville, October, pp. 1-5. [Google Scholar]
- 30. Uniyal N, Eskandari H, Abolmaesumi P, Sojoudi S, Gordon P, Warren L, et al. Ultrasound RF time series for classification of breast lesions. IEEE Trans Med Imaging. 2015;34:652-61. [DOI] [PubMed] [Google Scholar]
- 31. Zeimarani B, Costa MGF, Nurani NZ, Bianco SR, De Albuquerque Pereira WC, Filho CFFC. Breast lesion classification in ultrasound images using deep convolutional neural network. IEEE Access. 2020;8:133349-59. [Google Scholar]
- 32. Ayana G, Dese K, Choe SW. Transfer learning in breast cancer diagnoses via ultrasound imaging. Cancers. 2021;13:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cao Z, Duan L, Yang G, Yue T, Chen Q, Fu H, et al. Breast tumor detection in ultrasound images using deep learning. In: International Workshop on Patch-Based Techniques in Medical Imaging; 2017, Quebec City, Canada, pp. 121-8. [Google Scholar]
- 34. Hijab A, Rushdi M, Gomaa M, Eldeib A. Breast cancer classification in ultrasound images using transfer learning. In: 2019 Fifth International Conference on Advances in Biomedical Engineering (ICABME); 2019, Tripoli, Lebanon, pp. 1-4. [Google Scholar]
- 35. Byra M, Galperin M, Ojeda-Fournier H, Olson L, O’Boyle M, Comstock C, et al. Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion. Med Phys. 2019;46:746-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y, Choi EJ, Choi Y, Zhang H, Jin GY, Ko SB. Breast cancer classification in automated breast ultrasound using multiview convolutional neural network with transfer learning. Ultrasound Med Biol. 2020;46:1119-32. [DOI] [PubMed] [Google Scholar]
- 37. Xiao T, Liu L, Li K, Qin W, Yu S, Li Z. Comparison of transferred deep neural networks in ultrasonic breast masses discrimination. Biomed Res Int. 2018;2018:4605191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Han S, Kang HK, Jeong JY, Park MH, Kim W, Bang WC, et al. A deep learning framework for supporting the classification of breast lesions in ultrasound images. Phys Med Biol. 2017;62(19):7714-28. [DOI] [PubMed] [Google Scholar]
- 39. Tanaka H, Chiu SW, Watanabe T, Kaoku S, Yamaguchi T. Computer-aided diagnosis system for breast ultrasound images using deep learning. Phys Med Biol. 2019;64(23):235013. [DOI] [PubMed] [Google Scholar]
- 40. Wang P, Patel V, Hacihaliloglu I. Simultaneous segmentation and classification of bone surfaces from ultrasound using a multi-feature guided CNN. In: International Conference on Medical Image Computing and Computer-assisted Intervention; 2018, Granada, Spain, pp. 134-42. [Google Scholar]
- 41. Xie X, Shi F, Niu J, Tang X. Breast ultrasound image classification and segmentation using convolutional neural networks. In: Pacific Rim Conference On Multimedia; 2018, Hefei, China, pp. 200-11. [Google Scholar]
- 42. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016, Las Vegas, NV, USA, pp. 770-8. [Google Scholar]
- 43. Diakogiannis FI, Waldner F, Caccetta P, Wu C. ResUNet-a: a deep learning framework for semantic segmentation of remotely sensed data. ISPRS J Photogramm Remote Sens. 2020;162:94-114. [Google Scholar]
- 44. Zhang Z, Liu Q, Wang Y. Road extraction by deep residual u-net. IEEE Geosci Remote Sens Lett. 2018;15(5):749-53. [Google Scholar]
- 45. Lin T, Goyal P, Girshick R, He K, Dollr P. Focal loss for dense object detection. In: Proceedings of the IEEE International Conference on Computer Vision; 2017, Venice, Italy, pp. 2980-8. [Google Scholar]
- 46. Zunair H, Hamza AB. Patch efficient convolutional network for multi-organ nuclei segmentation and classification. [Google Scholar]
- 47. Azhar K, Murtaza F, Yousaf MH, Habib HA. Computer vision based detection and localization of potholes in asphalt pavement images. In: 2016 IEEE Canadian Conference on Electrical and Computer Engineering (CCECE); 2016, Vancouver, British Columbia, Canada, pp. 1-5. [Google Scholar]
- 48. Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data Brief. 2020;28:104863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cheng HD, Shi XJ. A simple and effective histogram equalization approach to image enhancement. Digit Signal Process. 2004;14(2):158-70. [Google Scholar]
- 50. Chen L, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European Conference on Computer Vision (ECCV); 2018, Munich, Germany pp. 801-18. [Google Scholar]
- 51. Zhou Z, Siddiquee RMM, Tajbakhsh N, Liang J. Unet++: A nested u-net architecture for medical image segmentation. In: Maier-Hein L, Syeda-Mahmood T, Taylor Z, Lu Z, Stoyanov D, Madabhushi A, Tavares JMRS, Nascimento JC, Moradi M, Martel A, Papa JP, Conjeti S, Belagiannis V, Greenspan H, Carneiro G, Bradley A. (eds) Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. 2018, vol. 11045 LNCS, pp. 3-11. Springer Verlag. 10.1007/978-3-030-00889-5_1 [DOI] [PMC free article] [PubMed]
- 52. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2015, Munich, Germany, pp. 234-41. [Google Scholar]
- 53. Lin T, Dollr P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017, Honolulu, USA, pp. 2117-25. [Google Scholar]
- 54. Wang Y, Deng Z, Hu X, Zhu L, Yang X, Xu X, et al. Deep attentional features for prostate segmentation in ultrasound. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2018, Granada, Spain, pp.523-30. [Google Scholar]
- 55. American College of Radiology. Breast imaging reporting and data system, Breast imaging atlas. 2003. forth edition. Reston, VA: American College of Radiology. [Google Scholar]