Dear Editor,
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants is still a pandemic raging across the world (Supplementary information, Fig. S1a, b). A new variant, Omicron, was first detected in South Africa and got dominant in many regions.1 Omicron has been classified as a variant of concern by the World Health Organization (WHO), whose spike carried more than 30 mutations (Supplementary information, Fig. S1c).1,2
To make a comprehensive evaluation of the susceptibility of Omicron, we summarized plasma samples from 19 convalescent individuals infected with the wild-type (WT) virus, and measured their neutralizing activities against the WT, Beta, Delta, Mu, C.1.2, and Omicron (Supplementary information, Figs. S1d and S2). The Omicron showed more serious resistance to neutralizing antibodies (nAbs) than other variants including Beta and Mu (45.6-fold, 9.6-fold, and 15.4-fold, respectively, compared with that against the WT), the latter two of which largely escaped the antibody neutralization prior to the Omicron pandemic.3,4 More seriously, some plasma (3/19) lost their neutralizing activities against Omicron (Supplementary information, Fig. S1e).
To study the mechanism of antibody escape, we analyzed 12 published nAbs binding to the receptor binding domain (RBD) of SARS-CoV-2 with clear structural information.5,6 We used these nAbs to mimic the polyclonal antibodies in plasma to explore what kind of nAbs were mostly affected by the mutations. The Omicron decreased or abolished the neutralization and binding affinity of nearly all tested nAbs across Class 1–4 (Supplementary information, Figs. S1f, g, S3, and S4), explaining why Omicron has the most serious antibody evasion. The structural analysis showed that some key mutations were located in or near the footprint of nAbs (Supplementary information, Fig. S5a). In consistence with previous studies,1,2,7 K417N and Q493R mainly affect the recognition of Class 1 nAbs to Omicron RBD. Mutations at E484 usually result in complete insensitivity of Class 2 nAbs. For REGN10987, a Class 3 antibody, G446S may diminish the binding of this class of nAbs. The Omicron was the first variant to escape Class 4 nAbs. The structural analysis of H014 showed that S371L may mediate the resistance to this class of nAbs (Supplementary information, Fig. S5b–e). Fortunately, some minority of existing nAbs are still effective to Omicron.1,2,7 S3097 retains effective neutralizing activity and strong binding affinity against Omicron (Supplementary information, Fig. S1f, g). However, broadly nAbs (bnAbs) are rare and a head-to-head comparison of the structural characterization for the novel bnAb against SARS-CoV-2 variants is lacking.
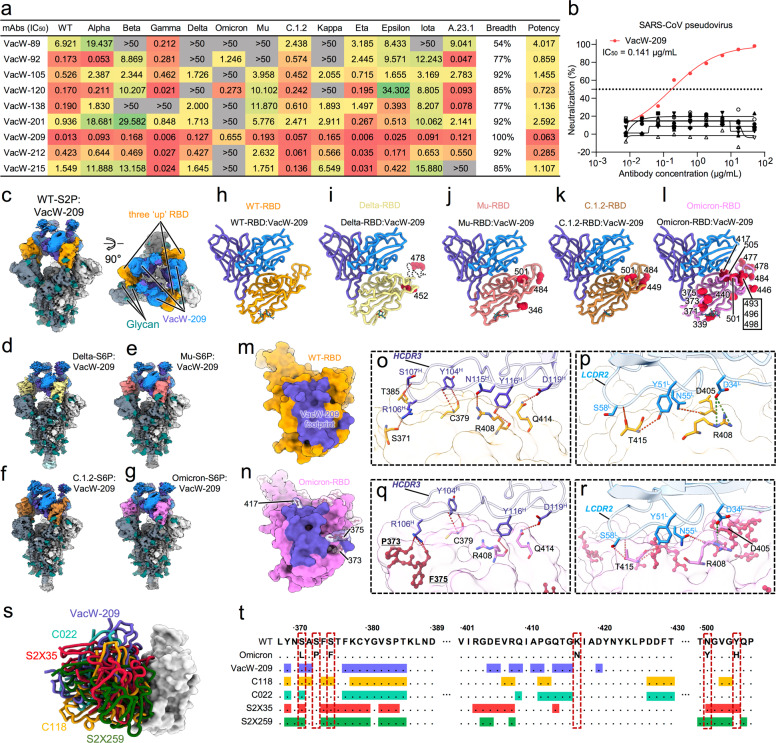
In this study, we explored bnAbs against Omicron, especially those binding to distinct epitopes away from S309. Previously, we identified nine monoclonal nAbs from individuals immunized with the SARS-CoV-2 inactivated vaccine and measured their cross-neutralizing activities against Kappa and Delta.6 Here, we further evaluated the neutralization of these nAbs against other important variants including Alpha, Beta, Gamma, Mu, Omicron, etc (Fig. 1a; Supplementary information, Fig. S6). Their neutralizing breadths ranged from 54% (7/13) to 100% (13/13) in the tested SARS-CoV-2 pseudoviruses, and a few of the nAbs (3/9) could still neutralize Omicron. VacW-92 and VacW-120 binding to overlapped epitopes with S3096 effectively neutralized Omicron (50% inhibitory concentration (IC50) = 1.246 μg/mL and IC50 = 0.273 μg/mL, respectively). VacW-209 could neutralize all tested variants with a high potency (geometric IC50 = 0.063 μg/mL). Meanwhile, the binding affinities of VacW-209 to Mu, C.1.2, and Omicron RBDs may contribute to its broadly neutralizing activity (Supplementary information, Fig. S7).
Fig. 1. The neutralizing activities and cryo-EM structures of VacW-209 against WT SARS-CoV-2 and variants.
a The neutralization of 9 nAbs against 13 tested pseudoviruses. The potency is highlighted in different colors. Red: high, yellow: moderate, green: weak, gray: non-neutralizing. The neutralizing activities of 9 nAbs to WT, Delta, and Kappa have been reported,6 which are re-tested here for comparison with other variants. b The neutralization of 9 nAbs against SARS-CoV. VacW-209 is marked in red and the other mAbs are marked in black. The data represented in (a, b) are means of at least two independent experiments. c–g Cryo-EM density maps of VacW-209 in complex with spike proteins of WT-S2P (c), Delta-S6P (d), Mu-S6P (e), C.1.2-S6P (f), and Omicron-S6P (g). h–l Cryo-EM corresponding atomic models of local refinement of VacW-209 in complex with WT-RBD (h), Delta-RBD (i), Mu-RBD (j), C.1.2-RBD (k), and Omicron-RBD (l). Models are represented as cartoon and key mutations on RBD are highlighted as red balls. m, n The binding footprints of VacW-209 (colored in purple) on WT-RBD (orange surface representation) (m) and Omicron-RBD (pink surface representation) (n). The mutated residues are rendered as red sticks with transparent surface representation on Omicron RBD and those involved in VacW-209 interactions are labeled. o, p Interaction details between WT-RBD and VacW-209 heavy chain (o) and light chain (p). q, r Interaction details between Omicron-RBD and VacW-209 heavy chain (q) and light chain (r). Hydrogen bonds and salt-bridges are labeled as red and dark green dotted lines respectively. s VacW-209-like nAbs and their binding modes on RBD. VacW-209, C118, C022, S2X35, and S2X259 are shown as sticks and colored in blue, orange, cyan, red, and green, respectively. t RBD sequence of SARS-CoV-2 WT and Omicron variant with highlighted footprints of VacW-209, C118, C022, S2X35, and S2X259. Amino acids substitutions revealed on Omicron variant are boxed.
SARS-CoV is closely related to SARS-CoV-2 sharing 80% of amino acid sequence identity in their spike proteins.8 Therefore, we detected the cross-reaction of VacW-209 to SARS-CoV, which displayed both highly neutralizing activity (IC50 = 0.141 μg/mL) and binding affinity (KD = 0.540 nM) (Fig. 1b; Supplementary information, Fig. S7). VacW-209 strongly competed with angiotensin-converting enzyme 2 (ACE2) for binding to RBD (Supplementary information, Fig. S8a). We also measured the competition of VacW-209 with 15 typical nAbs, revealing that VacW-209 bound to an epitope overlapped with those of Class 1 and Class 4 nAbs (Supplementary information, Fig. S8b). It was found that VacW-209 did not compete with two approved nAb drugs (REGN10987 and S309), suggesting that it could be used independently or in combination with these nAbs. We evaluated the neutralization of VacW-209 + REGN10987 and VacW-209 + S309 against WT, Beta, Delta, and Omicron (Supplementary information, Fig. S8c, d). REGN10987 completely lost its neutralization against Omicron, but could be effectively rescued through the combination with VacW-209. Considering that S309 and VacW-209 are both potent nAbs against all identified SARS-CoV-2 variants and SARS-CoV, the combination will open up the way against virus escape in the future.
The Mu and C.1.2 variants were identified post the global pandemic of Delta. Mutations in the spike proteins of Delta, Mu, and C.1.2 caused different resistances to the neutralization by polyclonal plasma and monoclonal nAbs.9,10 To define the structural basis of the broadly neutralizing activity of VacW-209, we resolved the cryo-electron microscopy (cryo-EM) structures of the antigen-binding fragment (Fab) of VacW-209 complexed with the spike proteins of SARS-CoV-2 WT, Delta, Mu, C.1.2, or Omicron. Five cryo-EM structures of immune complexes at 2.98–3.45 Å revealed nearly identical binding modes of VacW-209 (Fig. 1c–g, Supplementary information, Figs. S9–S13 and Table S1). Three VacW-209 Fabs bind to a completely opened spike with three “up” RBDs. We then performed the focus refinement of regions of Fab-bound RBDs of these five structures (Fig. 1h–l). High-resolution structures revealed that the binding epitope of VacW-209 completely evaded the key RBD mutations in Delta, Mu, C.1.2, and rarely overlapped with mutations in Omicron. The footprints of VacW-209 on WT-RBD and Omicron-RBD are slightly different, and three mutations in Omicron (K417N, S373P, and S375F) are involved in the nAb–RBD interaction (Fig. 1m, n).
We next analyzed the interaction details of VacW-209 binding to WT and Omicron spikes and revealed that VacW-209 mainly used its long heavy loop at complementarity determining region (HCDR) 3 to mediate spike recognition. In general, longer HCDR3 may mediate the recognition of nAbs to target some conserved epitopes, which often reside at strucutually deeper or cryptic regions in viral antigens.11,12 The identification of VacW-209 with long HCDR3 partly demonstrated the ability of SARS-CoV-2 inactivated vaccine to induce bnAbs against variants. Besides, the light chain CDR (LCDR) 2 and D34 from LCDR1 are also involved in nAb–RBD interactions (Fig. 1o–r). For WT-RBD, residues 371, 379, 408, 414, and 415 form an interaction network to VacW-209 containing 11 hydrogen bonds and 2 salt bridges (Fig. 1o, p). The heavy chain R106 (R106H) inserts its long side chain into the pocket formed by RBD aa. 371–385, which contains three key mutations of Omicron (S371L/S373P/S375F) (Supplementary information, Fig. S14a–c). Although VacW-209 showed a decreased neutralization against Omicron (Fig. 1a), our structural analysis showed that the mutations surrounding aa. 371–385 loop seemed not to obviously affect the binding of VacW-209 since the S373P and S375F build three new hydrogen bonds with R106H (Fig. 1q). Other Omicron mutations are not involved in the hydrogen bond interactions, and there are a total of 12 hydrogen bonds and 1 salt bridges formed (Fig. 1q, r), which are comparable to that in WT. We further found that the binding of VacW-209 to Omicron RBD need a slight conformational change of 371–385 loop (Supplementary information, Fig. S14d, e), which may partly account for the reduced neutralization of VacW-209 against Omicron.
Finally, we compared the binding mode of VacW-209 to several nAbs of Class 1–4 and defined a new binding mode of VacW-209 which bind to an epitope between Class 1 and Class 4, yet not overlapping with that of Class 2 or Class 3 (Supplementary information, Fig. S15a). Despite some minor differences in details, the binding of VacW-209 to RBDs of WT, Delta, Mu, C.1.2, and Omicron are all mediated by the long HCDR3 (in particular R106, Y116, and D119), LCDR2 (in particular Y51, N55, and S58), and LCDR1 residue D34 (Supplementary information, Fig. S15b–f). We also explored the potential binding sites of VacW-209 on other variant RBDs as well as SARS-CoV RBD based on the binding characterization revealed in the WT-S2P:VacW-209 (Supplementary information, Fig. S15g–k). The epitope of VacW-209, which is mainly comprised of aa. 376–385 and 405–416, nearly excludes all of above RBD mutations and is highly conserved between SARS-CoV-2 and SARS-CoV with only three amino acid substitutions (A372T, P384A, and E406D) between SARS-CoV-2 and SARS-CoV (Supplementary information, Fig. S15l).
The similar binding mode of VacW-209 was also found in some previously reported nAbs including C118, C022, S2X35, and S2X25913–15 (Fig. 1s). Available structural information revealed that the aforementioned four nAbs and VacW-209 shared lots of epitope residues located in conserved RBD aa. 376–385 and 405–416, while with diverse coverage of key mutations of Omicron (Fig. 1t; Supplementary information, Fig. S16a–e). Of these mutations, S371L, S373P, S375F, K417N, N501Y, and Y505H were structurally close to or involved in the binding epitopes of VacW-209-like nAbs. In the head-to-head comparison, although C022 and S2X35 showed significant reductions of neutralization against Omicron, these VacW-209-like nAbs generally maintained effectively neutralizing and binding activities to various SARS-CoV-2 variants and even SARS-CoV (Supplementary information, Fig. S16f, g and S17). The molecular mechanism underlying why these similar nAbs display diverse neutralizing activities need to be elucidated in the future.
In conclusion, VacW-209 identifies a highly conserved epitope on the RBDs among SARS-CoV-2 variants overlapping with the ACE2-binding site, which explains its potent neutralization. VacW-209 could strongly compete with Class 4 nAbs, indicating the potential cross-neutralization against sarbecoviruses. These VacW-209-like nAbs shared a similar antibody response to both SARS-CoV-2 and SARS-CoV, highlighting a key target for the universal vaccine design. As this binding epitope is highly conversed in different variants, vaccine design pursuant to this epitope feature may induce more bnAbs and benefit for the development of broad-spectrum COVID-19 vaccines. VacW-209, alone or in combination with S309, could also be used as countermeasure against SARS-CoV-2 variants including Omicron and even other forthcoming sarbecoviruses in the future.
Supplementary information
Acknowledgements
This study was supported by the National Science Fund for Distinguished Young Scholars (82025022), the National Natural Science Foundation of China (92169204, 82002140, 82171752, 82101861, 81991491), the National Key Plan for Scientific R&D of China (2021YFC0864500), the Guangdong Basic and Applied Basic Research Foundation (2021B1515020034, 2021B1212030010, 2019A1515011197, 2021A1515011009, 2020A1515110656), the Shenzhen Science and Technology Program (RCYX20200714114700046), and the Science and Technology Innovation Committee of Shenzhen Municipality (JSGG20200207155251653, JSGG20200807171401008, KQTD20200909113758004, JCYJ20190809115617365, JSGG20210901145200002).
Author contributions
Z.Z., N.X., S.L., and B.J. conceived and designed the study. B.J., Q.Z., H.G., Q.F., T.L., S. Song, and H.S. performed all experiments and analyzed data together with S. Shen, X.Z., W.X., L.C., and B.Z. Z.Z., N.X., S.L., B.J., and Q.Z. participated in discussion of the results and wrote the paper. All authors read and approved this version of paper.
Data availability
Structure coordinate has been deposited in the Protein Data Bank under accession code 7WP5 (Omicron-S6P:VacW-209). The corresponding density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-32674 (Omicron-S6P:VacW-209) and EMD-32669 (Omicron-S6P:VacW-209, local refinement).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Bin Ju, Qingbing Zheng, Huimin Guo, Qing Fan, Tingting Li, Shuo Song, Hui Sun.
Contributor Information
Bin Ju, Email: jubin2013@163.com.
Shaowei Li, Email: shaowei@xmu.edu.cn.
Ningshao Xia, Email: nsxia@xmu.edu.cn.
Zheng Zhang, Email: zhangzheng1975@aliyun.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00638-6.
References
- 1.Liu, L. et al. Nature10.1038/s41586-021-04388-0 (2021).
- 2.Cameroni, E. et al. Nature10.1038/s41586-021-04386-2 (2021).
- 3.Lucas C, et al. Nature. 2021;600:523–529. doi: 10.1038/s41586-021-04085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uriu K, et al. N. Engl. J. Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes CO, et al. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng L, et al. Cell Discov. 2021;7:112. doi: 10.1038/s41421-021-00347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, Y. et al. Nature10.1038/s41586-021-04385-3 (2021).
- 8.Pinto D, et al. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 9.Tada T, et al. Cell Rep. 2022;38:110237. doi: 10.1016/j.celrep.2021.110237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, et al. Cell. 2021;184:4220–4236. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekiert DC, et al. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker LM, et al. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starr TN, et al. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tortorici MA, et al. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jette CA, et al. Cell Rep. 2021;36:109760. doi: 10.1016/j.celrep.2021.109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure coordinate has been deposited in the Protein Data Bank under accession code 7WP5 (Omicron-S6P:VacW-209). The corresponding density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-32674 (Omicron-S6P:VacW-209) and EMD-32669 (Omicron-S6P:VacW-209, local refinement).