Abstract
S-layer proteins (SLPs), which are present in the external layer of certain strains of lactic acid bacteria isolated from the intestinal tract, are known to recognize and bind to specific proteins and glycan structures and contribute to adsorption to the host intestinal mucosa. The binding properties of certain SLPs are considered to exert a competitive inhibitory effect on infection because similar properties are involved in the infection mechanisms of several viruses. However, little is known regarding whether SLPs directly inhibit viral infection. In the present study, we investigated the effect of an SLP of the Lactobacillus crispatus KT-11 strain, a probiotic strain isolated from a healthy human infant, on human rotavirus infection. The impact of KT-11 lithium chloride extract (KT-11 LE), which contains SLP, on the infection of the P[4] genotype human rotavirus strain DS-1 was evaluated by monitoring the amplification of viral protein 6 (VP6) expression in human intestinal epithelial Caco-2 cells by quantitative reverse transcription-polymerase chain reaction assay after infection. KT-11 LE showed a significant suppressive effect on DS-1 infection in a dose-dependent manner with pre-infection treatment, whereas post-infection treatment was not effective. A 45 KDa protein isolated from KT-11 LE was investigated for homology using the BLAST database and was found to be a novel SLP. KT-11 SLP concentrate (KT-11 SLP) significantly inhibited the proliferative process of the DS-1 strain but not that of the P[8] genotype human rotavirus strain Wa. KT-11 SLP exerted significant inhibitory effect on DS-1 infection by pre-infection treatment even after digestion with gastric juice up to 2 h. Our results provided crucial evidence that SLPs from certain Lactobacillus strains can inhibit human rotavirus infection of intestinal epithelial cells.
Keywords: Lactobacillus crispatus, S-layer protein, lithium chloride extract, rotavirus, infection
Introduction
Lactic acid bacteria, including the genus Lactobacillus play a crucial role in the production of fermented dairy products such as cheese, yoghurt, and fermented milk. In recent years, there has been considerable focus on the action of the S-layer protein (SLP) of lactic acid bacteria as an antiviral component. SLPs are proteinaceous subunits that exist as the outermost components of the cell walls of several genera and species of bacteria and archaea. SLPs in Lactobacillus strains isolated from the intestine are considered to function as adhesion molecules to the host by recognizing extracellular proteins and carbohydrates (Hynönen and Palva, 2013). Certain SLPs are anticipated to have potential inhibitory effects on viral infections due to their immunomodulatory properties and competitive binding to target molecules of infection. Furthermore, the SLPs of several Lactobacillus species have been reported to inhibit viral infection by activating the antiviral immune system (Gao et al., 2016). In addition, the SLP from L. acidophilus ATCC 4356 has been reported to specifically block Junin virus, H9N2 avian influenza virus, alphavirus, and flavivirus infections by competitively binding to dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN) (Martínez et al., 2012; Prado Acosta et al., 2019). However, it is unclear whether the SLP is efficacious in preventing other viruses.
Rotaviruses are the leading cause of severe diarrhea in younger children worldwide, causing an estimated 453,000 deaths each year (Tate et al., 2012). A systematic analysis conducted in 2015 showed that rotavirus infections accounted for 29.3% of all diarrheal deaths in children younger than 5 years (Troeger et al., 2018). The World Health Organization recommended rotavirus vaccination in all national immunization programs in 2009; however, the total cost of vaccination remains high, especially in low- and middle-income countries (Pecenka et al., 2018). Therefore, there is an urgent need to identify and characterize effective natural materials that can be employed routinely to prevent rotavirus infections.
In the present study, we focused on the antiviral effects of the SLP of Lactobacillus crispatus strain KT-11, a probiotic strain isolated from a healthy infant (Tobita et al., 2009, 2010, 2018). Lithium chloride extract (LE) containing SLP (Johnson et al., 2013) was prepared from the KT-11 strain and evaluated for its inhibitory effect on human rotavirus infection in human intestinal epithelial cells.
Materials and Methods
Lactic Acid Bacteria
The KT-11 strain (GenBank accession number: AP025162, FERM BP-11332) was provided as a stock culture from KITII Co., Ltd. (Tokyo, Japan). KT-11 was inoculated in DeMan–Rogosa–Sharpe (MRS) broth, cultivated for 24 h at 37°C, collected by centrifugation, washed thrice with sterile water, and lyophilized.
Virus
Human rotavirus strains DS-1 (VR-2550) and Wa (VR-2018) were purchased from the American Type Culture Collection (Manassas, VA, United States). The viruses were treated with trypsin for 1 h before use, propagated in MA104 cells, and harvested after two freeze–thaw cycles.
Cells
The human colon carcinoma cell line Caco-2 was purchased from the Riken BioResource Research Center (Ibaraki, Japan). Cells were cultured in monolayer cultures using complete high-glucose Eagle’s minimal essential medium (EMEM; FUJIFILM Wako Pure Chemical, Osaka, Japan) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, United States), 100 U/mL penicillin G, and 100 μg/mL streptomycin (Sigma-Aldrich). The cells were passaged using TrypLE Express (Thermo Fisher Scientific, Waltham, MA, United States) and cultured under a humidified atmosphere of 5% CO2/95% air at 37°C.
Preparation of KT-11 Lithium Chloride Extract and S-Layer Protein
The KT-11 strain grown in MRS medium was collected by centrifugation (8,000 × g) at 4°C for 5 min. The pellet was washed by centrifugation (8,000 × g) thrice with cold distilled water. Bacterial cell pellets were suspended in 100 mL of 5 M lithium chloride and incubated in a shaker at 37°C for 2 h. Thereafter, the suspension was centrifuged (8,000 × g) at 4°C for 5 min, and the supernatant was filtered through a membrane filter with 0.20 μm pore diameter (Merck Millipore, Darmstadt, Germany). The filtrate was then dialyzed against distilled water at 4°C for 24 h. The dialyzate was centrifuged at 14,000 × g at 4°C for 5 min, and the precipitate was lyophilized to produce KT-11 lithium chloride extract (KT-11 LE) powder. KT-11 LE was obtained at a yield of 7.8%, determined on the basis of wet weight. KT-11 SLP concentrate (KT-11 SLP) was prepared from KT-11 LE using the method described by Johnson et al. (2013). Briefly, KT-11 LE was mixed with 1 M lithium chloride at 37°C for 24 h. The insoluble fraction was collected by centrifugation and lyophilized. KT-11 LE and KT-11 SLP was dissolved in sterile distilled water for subsequent study.
Rotavirus Infection of Target Cells
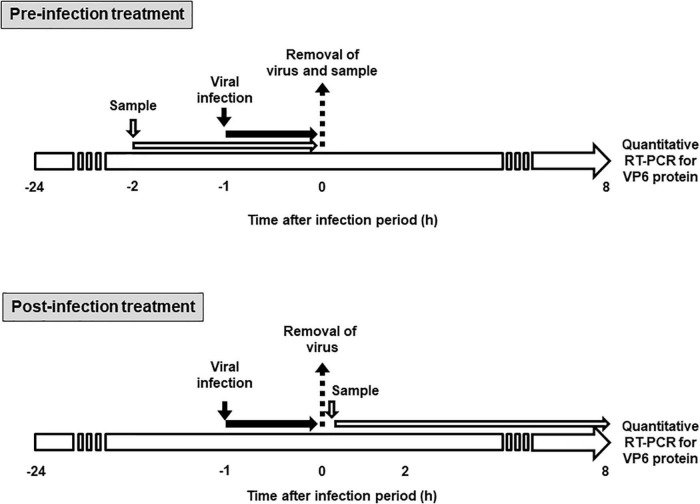
The effect of KT-11 strain and its components on rotavirus infectivity was assessed using the method shown in Figure 1. In pre-infection treatment, Caco-2 cells were seeded at 2.0 × 105 cells/mL/well in 24-well plates (Thermo Fisher Scientific, Waltham, MA, United States) and cultured at 37°C. After 24 h, the culture supernatant was discarded, and the cells were incubated with the KT-11 LE or KT-11 SLP in 200 μL of serum-free EMEM for 1 h, following which either the Wa or the DS-1 strain was added and incubated for another 1 h. The culture supernatant was removed, the cells were washed with serum-free EMEM and further cultured in complete EMEM under a humidified atmosphere of 5% CO2/95% air at 37°C. After 24 h, the cells were collected for quantitative reverse transcription-polymerase chain reaction (RT-PCR).
FIGURE 1.
Schematic diagram for sample treatment in rotavirus infection study. In pre-infection treatment, Caco-2 cells were treated with KT-11 LE or KT-11 SLP for 1 h, then infected with the virus for 1 h, and the amplification of VP6 mRNA after 8 h was analyzed by quantitative RT-PCR. In post-infection treatment, Caco-2 cells were infected with the virus for 1 h and then treated with the KT-11 LE or KT-11 SLP for 8 h, and the amplification of VP6 mRNA was analyzed by quantitative RT-PCR.
In post-infection treatment, Caco-2 cells were seeded at 2.0 × 105 cells/mL/well in 24-well plates (Thermo Fisher Scientific) and cultured at 37°C. After 24 h, the culture supernatant was discarded, and Wa or DS-1 strain was added and incubated for another 1 h. Thereafter, the culture supernatant containing uninfected virus particles was removed, the cells were washed with serum-free EMEM and further cultured in complete EMEM supplemented with the KT-11 LE or KT-11 SLP under a humidified atmosphere of 5% CO2/95% air at 37°C. After 24 h, the cells were collected for quantitative RT-PCR.
Quantitative Reverse Transcription-Polymerase Chain Reaction Assay
Total RNA was extracted from rotavirus-infected Caco-2 cells using TRI reagent (Sigma-Aldrich) according to the manufacturer’s protocol. The extracted RNA (1 μg) was reverse transcribed in a thermal cycler (PTC-200; MJ Research, Waltham, MA, United States) with 1 mM of each dNTP, 2.5 units/μL M-MLV reverse transcriptase (Thermo Fisher Scientific), and 10 pmol/μL of oligo(dT)18 primers at 42°C for 50 min. Quantitative RT-PCR of the resulting cDNA was performed using 0.5 μg cDNA with TB Green Premix Ex Taq II (Takara Bio, Shiga, Japan) and 10 pmol/μL primers. The primer sequences are shown in Table 1. The PCR comprised one cycle of preheating (95°C, 10 min) and 40 cycles of denaturation (95°C, 10 s) and primer annealing and extension (55°C, 30 s) using the Eco Real Time PCR System (Illumina, San Diego, CA, United States). Results were analyzed with the ΔΔCt method using the Eco system software (Illumina). The amount of PCR products was normalized to the expression level of GAPDH.
TABLE 1.
Primer sequences used in this study.
| Target | Sequence |
Product size (bp) | Accession number | |
| Forward | Reverse | |||
| DS-1 VP6* | 5′-CCATCAATGCACCAGCCAAC-3′ | 5′-GCCGTCGTTAATACTCTGCG-3′ | 92 | EF583028.1 |
| Wa VP6* | 5′-GTGAATCAGTGCTTGCGGAC-3′ | 5′-ATGCCTGGTGGAAATACCGG-3′ | 103 | K02086.1 |
| Human GAPDH* | 5′-AACGGATTTGGTCGTATTGG-3′ | 5′-AATGAAGGGGTCATTGATGG-3′ | 90 | BT006893.1 |
* VP6, viral protein 6; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Identification of Protein
The identification of the expressed 45 kDa protein was based on the observed molecular weights of the proteins according to Antikainen et al. (2002). Briefly, the target band of the 45 kDa protein in 12% SDS-PAGE gel was digested by trypsin, followed by analysis with liquid chromatography-tandem mass spectrometry (LC-MS/MS). The generated data were subjected to a search using the Mascot (Matrix Science, London, United Kingdom) algorithm. Protein identification was carried out using all the SLP sequences of KT-11. Homology searches were performed using the BLAST program (available at1). Sequences of eight Lactobacillus SLP sequences were aligned using CLUSTALW2. The GenBank accession numbers for the SLP sequences were as follows: AAB58734 for CbsA of L. crispatus JCM 5810; AFB69875 for SlpA of L. crispatus K313; ABI49163 for SlpA of L. crispatus ZJ001: CAA61560 for SlpA of L. acidophilus JCM 4356; YP_193101 for SlpA from L. acidophilus NCFM; ATO53009 for the SLP of L. amylovorus JCM1126T; and CAB46985 for the SLP of L. helveticus JCM1120T. Both amino acid sequencing and peptide mapping were performed by Japan Proteomics Co., Ltd. (Miyagi, Japan).
Enzymatic Digestion
Simulated digestive juice was prepared as gastric juice containing 1.6% pepsin (Sigma-Aldrich) in 0.1 M KCl/HCl (pH 2.0) and intestinal juice containing 0.1% pancreatin (Sigma-Aldrich) in PBS (pH 7.2). KT-11 LE and KT-11 SLP were treated with gastric juice or intestinal juice for the indicated time periods at 37°C. The sensitivity of KT-11 SLP to digestive enzyme treatment was evaluated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) using a discontinuous buffer system with 12.5% separating gel and 5% stacking gel. Protein bands were visualized by staining the gels with Quick-CBB (FUJIFILM Wako Pure Chemical).
To determine the inhibitory activity of SLP after gastric digestion, SLP was dissolved in gastric juice and incubated at 37°C. After treatment for 0, 0.5, 1, 2, and 3 h, the SLP solution was neutralized with NaOH to adjust pH to 7.2, and was subjected to the pre-infection treatment assay.
Statistical Analysis
The data were statistically analyzed using one-way analysis of variance (ANOVA) with Tukey–Kramer multiple comparison tests using Excel 2019 (Microsoft, Redmond, WA, United States) with the add-in software Statcel4 (OMS Publishing, Tokyo, Japan). Results with a p-value of less than 0.05 were considered statistically significant.
Results
Effect of KT-11 Lithium Chloride Extract on the Infection of DS-1 Strain in Caco-2 Cells
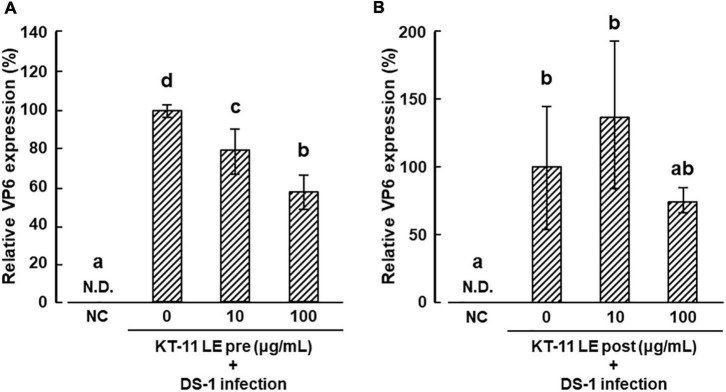
The effect of KT-11 LE on the infection of DS-1 human rotavirus in Caco-2 cells is shown in Figures 2A,B. In the pre-infection treatment, KT-11 LE significantly suppressed the amplification of VP6 mRNA in a dose-dependent manner (Figure 2A). In contrast, in the post-infection treatment, no significant suppression of DS-1 strain infection was observed with KT-11 LE at all tested concentrations (Figure 2B).
FIGURE 2.
Effect of KT-11 LE against rotavirus infection in Caco-2 cells. (A) Caco-2 cells were treated with KT-11 LE at the indicated concentration 1 h prior to DS-1 infection. The expression of VP6 in Caco-2 cells 24 h after the infection was evaluated. The results are shown as means with standard deviations relative to KT-11 LE-untreated control (n = 3). N.D. denotes not detected. Different letters denote a significant difference at p < 0.05. (B) Caco-2 cells were treated with KT-11 LE at the indicated concentration after DS-1 infection. The expression of VP6 in Caco-2 cells 24 h after the infection was evaluated. The results are shown as means with standard deviations relative to KT-11 LE-untreated and viral-infected control (n = 3). NC represents the condition of viral-uninfected (and also KT-11 LE-untreated) condition. N.D. denotes not detected.
Identification of the 45 kDa Protein
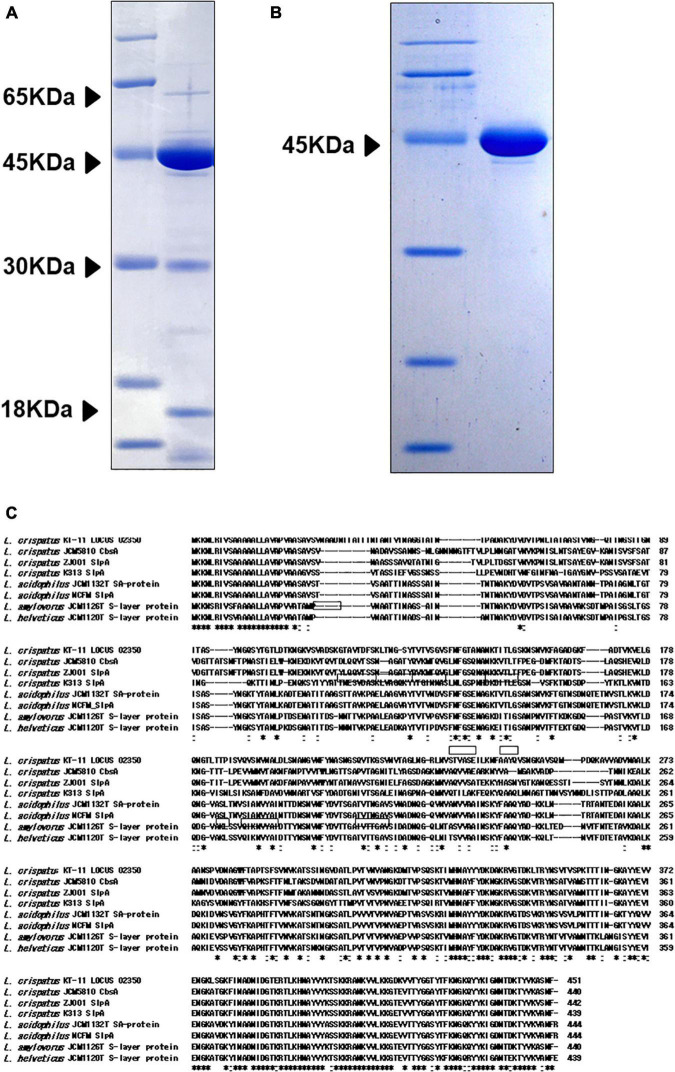
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of KT-11 LE revealed the presence of 18, 30, 45, and 65 kDa proteins as its major proteinaceous components (Figure 3A). Among them, the 45 kDa protein was concentrated as an insoluble fraction in 1 M lithium chloride (Figure 3B).
FIGURE 3.
SDS-PAGE pattern and amino acid sequence of the 45 kDa protein. SDS-PAGE patterns of (A) KT-11 LE and concentrated 45 kDa protein (B). (C) Comparison of the revealed amino acid sequences of the 45 kDa protein and SLPs from other Lactobacillus strains. The framed areas show the amino acid sequences revealed by protein identification analysis using NanoLC-MS/MS. Hyphens indicate deletions or insertions of amino acids. “*” denotes that the residues are identical in all sequences in the alignment, and “:” denotes that conserved substitutions are observed.
The nine partial amino acid sequences of the 45 kDa peptide identified by LC-MS/MS were GTAVTDFSK, GSVNVTAGLNGR, LNVSTVASEILK, NFAAYQVSNGK, AVVADVNAALK, YNSVTVSPK, AYYEVVENGK, FINADNIDGTER, and VVTYGGTYTFK. Mascot search of these results revealed that all the sequences were included in the translated product of the SLP gene (LOCUS_02350) of KT-11 (Figure 3C). Comparing this amino acid sequence with SLPs from other Lactobacillus strains revealed that the 45 kDa protein was a novel SLP with significant homology in the C-terminal region and notable difference in the N-terminal and middle regions.
Effect of KT-11 S-Layer Protein on DS-1 and Wa Strain Infection in Caco-2 Cells
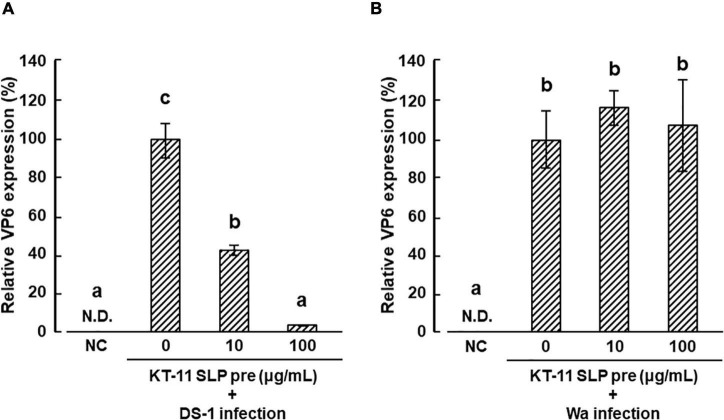
The effect of KT-11 SLP on the infection of DS-1 strain in Caco-2 cells is shown in Figure 4A. DS-1 infection was significantly suppressed by pre-infection treatment with KT-11 SLP in a concentration-dependent manner. Conversely, KT-11 SLP did not suppress the infection of the Wa strain even after pre-infection treatment at 100 μg/mL (Figure 4B).
FIGURE 4.
Effect of KT-11 SLP against rotavirus infection in Caco-2 cells. Caco-2 cells were treated with KT-11 SLP at the indicated concentrations 1 h prior to DS-1 (A) or Wa (B) strain infection. The expression of VP6 in Caco-2 cells 24 h after infection was evaluated. The results are shown as means with standard deviations relative to KT-11 SLP-untreated and viral-infected control (n = 3). NC represents the condition of viral-uninfected (and also KT-11 SLP-untreated) condition. N.D. denotes not detected. Different letters denote a significant difference at p < 0.05.
Digestive Juice Tolerance of the Protein Component of KT-11 Lithium Chloride Extract
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis patterns of KT-11 LE and the KT-11 SLP after exposure to simulated digestive juice are shown in Figures 5A,B. The 45 kDa protein band remained visible up to 3 h after the addition of simulated gastric juice, though it gradually disappeared with prolonged incubation. Only a slight decrease in the intensity of the 45 kDa protein band was observed till 6 h after the addition of simulated intestinal juice, while a more substantial decrease was observed after 24 h. Conversely, the bands of 18, 30, and 65 kDa proteins in KT-11 LE completely disappeared following the 24-h treatment with intestinal juice.
FIGURE 5.
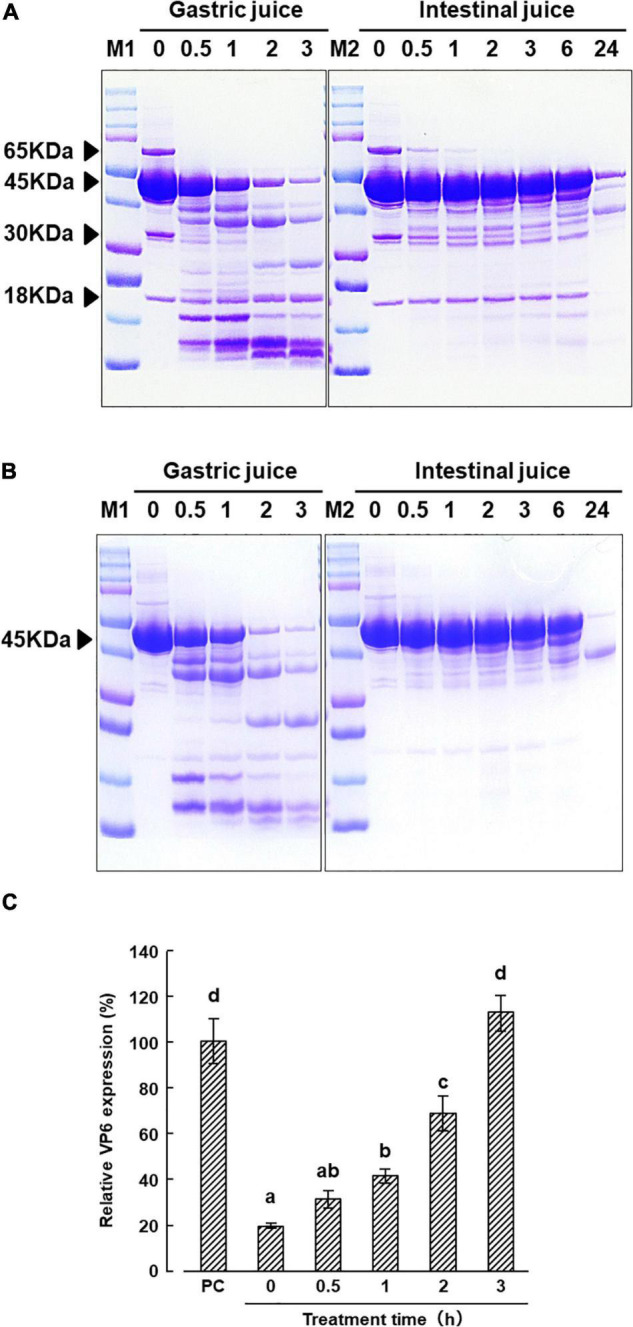
Effect of digestive juice treatment on KT-11 LE and KT-11 SLP. SDS-PAGE patterns of (A) KT-11 LE and fractionated LE, (B) KT-11 SLP after treatment with simulated gastric juice (1.6% pepsin in 0.1 M KCl/HCl, pH 2.0) and intestinal juice (0.1% pancreatin in PBS, pH 7.2) for the indicated time periods. Lane 1 (M1) and lane 7 (M2): molecular weight marker, lanes 2–6: digestion pattern by gastric juice for the indicated time periods (h), lanes 8–14: digestion pattern by intestinal juice for the indicated time periods (h). (C) Inhibitory effect on DS-1 infection in pre-infection treatment with KT-11 SLP digested with gastric juice for the indicated times periods. The results are shown as means with standard deviations relative to KT-11 SLP-untreated viral-infected control (n = 3). PC represents the condition of viral-infected and KT-11 SLP-untreated condition. Different letters denote a significant difference at p < 0.01.
From 0 to 2 h after treatment with gastric juice, the inhibitory effect of KT-11 SLP on DS-1 strain infection in the pre-infection treatment decreased in a treatment time dependent manner. Significant inhibition was observed even after 2 h, but the activity was completely lost after 3 h digestion of KT-11 SLP with gastric juice (Figure 5C).
Discussion
In the present study, we found that KT-11 LE inhibited infection of the intestinal cells by the human rotavirus DS-1 strain and confirmed that the 45 kDa protein, identified by SDS-PAGE, was one of the active components of KT-11 LE. Although SLPs have also been reported from other strains of Lactobacillus, the binding targets of most SLPs remain unelucidated. Sillanpää et al. (2000) and Sun et al. (2013) reported that CbsA from L. crispatus JCM5810 and SlpB from L. crispatus K313 bind to type I collagen and type IV collagen, respectively. Konstantinov et al. (2008) and the above-mentioned study by Martínez et al. (2012) showed that the binding target of SlpA from L. acidophilus NCFM and SLP from L. acidophilus ATCC 4356 is DC-SIGN. The inhibition of DS-1 infection only after pre-infection treatment but not post-infection treatment with KT-11 LE (Figure 2) implied that there may be an underlying mechanism of inhibition by competitive binding to the target molecules. The 45 kDa protein identified herein as KT-11 SLP may bind to the same target molecule of these known SLPs from closely related Lactobacillus species because it is homologous to their amino acid sequence (Figure 3C); however, neither target molecule has been reported as a receptor for human rotavirus.
The entry of rotavirus into cells is a complex multistep process, in which different domains of rotavirus surface proteins interact with cell-surface molecules that function as receptors for adhesion and entry (López and Arias, 2004, 2006). Among them, several carbohydrates, such as terminal sialic acids (Isa et al., 2006; Haselhorst et al., 2009) and histo-blood group antigens (HBGA) (Tan and Jiang, 2014; Jiang et al., 2017), have been reported to be involved in rotavirus attachment to target cells. As shown in Figure 4A, KT-11 SLP significantly inhibited the infection of the DS-1 strain in Caco-2 cells in a dose-dependent manner. The initial interactions of human rotavirus strains with host cells is dependent on the VP4 genotype (Ciarlet et al., 2002). According to the classification based on the molecular properties of VP4 (P-types), the DS-1 strain is classified as the P[4] genotype (Matthijnssens et al., 2008). Increasing evidence indicates that the P[4] genotype rotaviruses, including the DS-1 strain, use H-type 1 and Lewis-b antigens for infection (Huang et al., 2012; Ramani et al., 2016). H-type 1 and Lewis-b antigens have been reported in Caco-2 cells (Amano and Oshima, 1999; Murakami et al., 2013). Our results suggest that the possibility of that DS-1 infection is inhibited by competitive binding of KT-11 SLP to these antigens. However, contrary to our expectations, infection of the Wa strain, another dominant P[8] subtype reported using the same H-type antigen and Lewis-b for infection (Huang et al., 2012; Ramani et al., 2016), was not inhibited in the presence of KT-11 SLP (Figure 4B).
Although it is unclear why KT-11 SLP failed to inhibit infection of the Wa strain, the simplest possibility is that KT-11 SLP binds to a molecule used only by the DS-1 strain. However, we could not name a specific candidate molecule to explain this hypothesis; the A-type antigen reported by Böhm et al. (2015) is a possible candidate molecule, but is not expressed in Caco-2 cells derived from O-type patients. Another possible reason is that KT-11 SLP binds to common targets of DS-1 and Wa strains, such as H-type 1 and Lewis-b antigens, but may not exert its inhibitory effect due to the presence of ganglioside GM1, which has been reported to bind only to Wa strains (Ramani et al., 2016). Recently, Laucirica et al. (2017) reported that DS-1 strain infection was significantly inhibited by 3′-sialyl lactose and 6′-sialyl lactose, whereas the Wa strain was inhibited by 2′-fucosyl lactose, implying that compounds containing sialic acid may be deeply involved in the infection of DS-1 strain. In support of the involvement of sialic acid in the inhibitory effect of KT-11 SLP on human rotaviruses, we found that KT-11 SLP agglutinated sheep erythrocytes, the aggregation of which by viruses targeting sialic acid is widely known (Supplementary Figure 1). Furthermore, in preliminary studies of this study, we have confirmed the significant diarrhea-preventive effect of oral administration of KT-11 LE in an in vivo diarrheal model caused by the monkey rotavirus SA-11 virus known for sialic acid-dependent binding (Supplementary Figure 2), as well as the effect of KT-11 SLP on A similar inhibitory effect has been observed in an in vitro infection system of SA-11 strains (Supplementary Figure 3). Although the target molecule of the hemagglutinating reaction and inhibition of SA-11 infection by KT-11 SLP is not clear at present, based on the above results, we predict that DS-1 strain may be more dependent on binding to sialic acid-related structures than that of Wa strain, which may have led to the difference in inhibitory effect in this experiment. Further studies are needed to elucidate the inhibitory effect of KT-11 SLP on human rotavirus infection.
In conclusion, the present study demonstrated that KT-11 LE and its component, a novel SLP, inhibited the infection of the human rotavirus DS-1 strain. As KT-11 SLP showed some degree of resistance to digestion with gastric juice (Figure 5), it may work to prevent rotavirus infection in the intestine by withstanding digestion, particularly in infants with low stomach pH and limited action of pepsin (Nguyen et al., 2015; Neal-Kluever et al., 2019). Rotavirus infection is a serious problem in infants with a compromised immune system. Although further experimental studies are needed to fully understand the mechanism by which SLPs prevent rotavirus infection in vivo, the obtained findings suggested that SLPs derived from specific strains of lactic acid bacteria may be used as drugs or food additives to prevent or ameliorate conditions caused by rotavirus, especially in infants and young children.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
TK: conception and design, drafting the work or revising it critically for important intellectual content, and writing the manuscript. TK, IS, and YT: acquisition of data. TK, KT, MT, and IW: interpretation of results. All authors contributed to the article and approved the submitted version.
Conflict of Interest
TK, MT, and IW were employed by KITII Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
- SLP
S-layer protein
- LE
lithium chloride extract
- MRS
DeMan–Rogosa–Sharpe
- EMEM
Eagle’s minimal essential medium
- FBS
fetal bovine serum
- RT-PCR
reverse transcription-polymerase chain reaction
- VP6
viral protein 6
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- ANOVA
analysis of variance
- DC-SIGN
dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin
- HBGA
human blood group antigen.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.783879/full#supplementary-material
References
- Amano J., Oshima M. (1999). Expression of the H type 1 blood group antigen during enterocytic differentiation of Caco-2 cells. J. Biol. Chem. 274 21209–21216. 10.1074/jbc.274.30.21209 [DOI] [PubMed] [Google Scholar]
- Antikainen J., Anton L., Sillanpää J., Korhonen T. K. (2002). Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol. Microbiol. 46 381–394. 10.1046/j.1365-2958.2002.03180.x [DOI] [PubMed] [Google Scholar]
- Böhm R., Fleming F. E., Maggioni A., Dang V. T., Holloway G., Coulson B. S., et al. (2015). Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat. Commun. 6:5907. 10.1038/ncomms6907 [DOI] [PubMed] [Google Scholar]
- Ciarlet M., Ludert J. E., Iturriza-Gómara M., Liprandi F., Gray J. J., Desselberger U., et al. (2002). Initial interaction of rotavirus strains with N-acetylneuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J. Virol. 76 4087–4095. 10.1128/JVI.76.8.4087-4095.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Huang L., Zhu L., Mou C., Hodrddu Q., Yu Q. (2016). Inhibition of H9N2 Virus invasion into dendritic cells by the S-layer protein from L. acidophilus ATCC 4356. Front. Cell. Infect. Microbiol. 6:137. 10.3389/fcimb.2016.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselhorst T., Fleming F. E., Dyason J. C., Hartnell R. D., Yu X., Holloway G., et al. (2009). Sialic acid dependence in rotavirus host cell invasion. Nat. Chem. Biol. 5 91–93. 10.1038/nchembio.134 [DOI] [PubMed] [Google Scholar]
- Huang P., Xia M., Tan M., Zhong W., Wei C., Wang L., et al. (2012). Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol. 86 4833–4843. 10.1128/JVI.05507-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynönen U., Palva A. (2013). Lactobacillus surface layer proteins: structure, function and applications. Appl. Microbiol. Biotechnol. 97 5225–5243. 10.1007/s00253-013-4962-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isa P., Arias C. F., López S. (2006). Role of sialic acids in rotavirus infection. Glycoconj. J. 23 27–37. 10.1007/s10719-006-5435-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Liu Y., Tan M. (2017). Histo-blood group antigens as receptors for rotavirus, new understanding on rotavirus epidemiology and vaccine strategy. Emerg. Microbes Infect. 6:e22. 10.1038/emi.2017.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B., Selle K., O’Flaherty S., Goh Y. J., Klaenhammer T. (2013). Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology 159 2269–2282. 10.1099/mic.0.070755-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinov S. R., Smidt H., de Vos W. M., Bruijns S. C., Singh S. K., Valence F., et al. (2008). S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U.S.A. 105 19474–19479. 10.1073/pnas.0810305105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laucirica D. R., Triantis V., Schoemaker R., Estes M. K., Ramani S. (2017). Milk oligosaccharides inhibit human rotavirus infectivity in MA104 cells. J. Nutr. 147 1709–1714. 10.3945/jn.116.246090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López S., Arias C. F. (2004). Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol. 12 271–278. 10.1016/j.tim.2004.04.003 [DOI] [PubMed] [Google Scholar]
- López S., Arias C. F. (2006). Early steps in rotavirus cell entry. Curr. Top. Microbiol. Immunol. 309 39–66. 10.1007/3-540-30773-7_2 [DOI] [PubMed] [Google Scholar]
- Martínez M. G., Prado Acosta M., Candurra N. A., Ruzal S. M. (2012). S-layer proteins of Lactobacillus acidophilus inhibits JUNV infection. Biochem. Biophys. Res. Commun. 422 590–595. 10.1016/j.bbrc.2012.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Ciarlet M., Rahman M., Attoui H., Bányai K., Estes M. K., et al. (2008). Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 153 1621–1629. 10.1007/s00705-008-0155-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Kurihara C., Oka T., Shimoike T., Fujii Y., Takai-Todaka R., et al. (2013). Norovirus binding to intestinal epithelial cells is independent of histo-blood group antigens. PLoS One 8:e66534. 10.1371/journal.pone.0066534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal-Kluever A., Fisher J., Grylack L., Kakiuchi-Kiyota S., Halpern W. (2019). Physiology of the neonatal gastrointestinal system relevant to the disposition of orally administered medications. Drug Metab. Dispos. 47 296–313. 10.1124/dmd.118.084418 [DOI] [PubMed] [Google Scholar]
- Nguyen T. T. P., Bhandari B., Cichero J., Prakash S. (2015). A comprehensive review on in vitro digestion of infant formula. Food Res. Int. 76 373–386. 10.1016/j.foodres.2015.07.016 [DOI] [PubMed] [Google Scholar]
- Pecenka C., Debellut F., Bar-Zeev N., Anwari P., Nonvignon J., Shamsuzzaman M., et al. (2018). Re-evaluating the cost and cost-effectiveness of rotavirus vaccination in Bangladesh, Ghana, and Malawi: a comparison of three rotavirus vaccines. Vaccine 36 7472–7478. 10.1016/j.vaccine.2018.10.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado Acosta M., Geoghegan E. M., Lepenies B., Ruzal S., Kielian M., Guadalupe Martinez M. (2019). Surface (S) layer proteins of Lactobacillus acidophilus block virus infection via DC-SIGN interaction. Front. Microbiol. 2019:810. 10.3389/fmicb.2019.00810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani S., Hu L., Venkataram Prasad B. V., Estes M. K. (2016). Diversity in rotavirus-host glycan interactions: a “sweet” spectrum. Cell. Mol. Gastroenterol. Hepatol. 2 263–273. 10.1016/j.jcmgh.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpää J., Martínez B., Antikainen J., Toba T., Kalkkinen N., Tankka S., et al. (2000). Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 182 6440–6450. 10.1128/JB.182.22.6440-6450.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Kong J., Hu S., Kong W., Lu W., Liu W. (2013). Characterization of a S-layer protein from Lactobacillus crispatus K313 and the domains responsible for binding to cell wall and adherence to collagen. Appl. Microbiol. Biotechnol. 97 1941–1952. 10.1007/s00253-012-4044-x [DOI] [PubMed] [Google Scholar]
- Tan M., Jiang X. (2014). Histo-blood group antigens: a common niche for norovirus and rotavirus. Expert Rev. Mol. Med. 16:e5. 10.1017/erm.2014.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. E., Burton A. H., Boschi-Pinto C., Steele A. D., Duque J., Parashar U. D., et al. (2012). 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect. Dis. 12 136–141. 10.1016/S1473-3099(11)70253-5 [DOI] [PubMed] [Google Scholar]
- Tobita K., Watanabe I., Tomokiyo M., Saito M. (2018). Effects of heat-treated Lactobacillus crispatus KT-11 strain consumption on improvement of oral cavity environment: a randomised double-blind clinical trial. Benef. Microbes 9 585–592. 10.3920/BM2017.0137 [DOI] [PubMed] [Google Scholar]
- Tobita K., Yanaka H., Otani H. (2009). Heat-treated Lactobacillus crispatus KT strains reduce allergic symptoms in mice. J. Agric. Food Chem. 57 5586–5590. 10.1021/jf900703q [DOI] [PubMed] [Google Scholar]
- Tobita K., Yanaka H., Otani H. (2010). Anti-allergic effects of Lactobacillus crispatus KT-11 strain on ovalbumin-sensitized BALB/c mice. Anim. Sci. J. 81 699–705. 10.1111/j.1740-0929.2010.00795.x [DOI] [PubMed] [Google Scholar]
- Troeger C., Khalil I. A., Rao P. C., Cao S., Blacker B. F., Ahmed T., et al. (2018). Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. JAMA Pediatr. 172 958–965. 10.1001/jamapediatrics.2018.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.