ABSTRACT
Aims/Introduction
Metformin is associated with the risk of gastrointestinal complications, and probiotic Bifidobacterium bifidum G9‐1 (BBG9‐1) can improve the symptoms of diarrhea. This study aimed to clarify the effects of probiotic BBG9‐1 on the gastrointestinal symptoms of type 2 diabetes mellitus patients using metformin.
Materials and methods
In this open‐label single‐arm exploratory study, 40 patients (mean age 64.0 ± 9.4 years) were given probiotic BBG9‐1 for 10 weeks. Changes in the gastrointestinal symptom rating scale total score, which was the primary end‐point, gastrointestinal symptom rating scale subscale scores, glycated hemoglobin levels and gut microbiota after the administration of probiotic BBG9‐1 were evaluated by the Student's t‐test.
Results
The gastrointestinal symptom rating scale total score significantly improved (from 2.02 ± 0.51 to 1.59 ± 0.43, change, −0.43 ± 0.49, P < 0.001). Furthermore, all gastrointestinal symptom rating scale subscale scores, including diarrhea (from 2.32 ± 1.14 to 1.89 ± 0.99, change, −0.42 ± 0.95, P = 0.007) and constipation (from 3.00 ± 1.16 to 2.20 ± 1.07, change, −0.80 ± 1.19, P < 0.001), scores also significantly improved. However, the glycated hemoglobin levels did not change (from 7.0 ± 0.7 to 7.0 ± 0.6%, change, 0.0 ± 0.4, P = 0.91). The relative abundance of the genus Sutterella decreased by the use of probiotic BBG9‐1 (from 0.011 ± 0.009 to 0.008 ± 0.006, change, −0.003 ± 0.006, P = 0.002).
Conclusions
Type 2 diabetes mellitus patients treated with metformin showed significant improvement in all gastrointestinal symptom rating scores after using probiotic BBG9‐1 without changing the glucose control. This study showed the potential usefulness of probiotic BBG9‐1 for improving gastrointestinal symptoms, including constipation and diarrhea, in type 2 diabetes mellitus patients treated with metformin.
Keywords: Constipation, Diarrhea, Probiotics
Probiotic Bifidobacterium bifidum G9‐1 (BBG9‐1) improved gastrointestinal symptoms, including diarrhea and constipation, without changing glucose control in type 2 diabetes mellitus patients treated with metformin. The relative abundance of the genus Sutterella decreased by the use of probiotic BBG9‐1.

INTRODUCTION
The number of patients with type 2 diabetes is increasing worldwide 1 , with the number of patients in Japan exceeding 10 million 2 . Microvascular and macrovascular complications are well‐known complications of this disease 1 . Furthermore, various gastrointestinal diseases, such as reflux esophagitis, constipation and diarrhea, are also present in patients with type 2 diabetes mellitus 3 , 4 . Previous studies have shown that 75% of patients with diabetes have gastrointestinal complications, of which 10–60% have constipation and 20% have diarrhea 4 , 5 . Moreover, the use of metformin is associated with an increased risk of gastrointestinal complications 6 , 7 . Thus, many patients with type 2 diabetes discontinue metformin use, despite its hypoglycemic and protective effects against cardiovascular events 8 .
Recently, the relationship between gut microbiota and type 2 diabetes mellitus has become clearer 4 , 9 , 10 , 11 . Many dietary supplements have been marketed to improve dysbiosis 12 , 13 . However, there is little evidence of the effectiveness of these diets and supplements on the gut microbiota. Probiotic bifidobacteria have been reported to improve the symptoms of constipation and diarrhea in individuals without diabetes 14 , 15 , 16 . Furthermore, a recent animal model study showed that the probiotic Bifidobacterium bifidum G9‐1 (BBG9‐1) improved the symptoms of diarrhea caused by metformin 17 . Based on these findings, we hypothesized that probiotic bifidobacteria could improve gastrointestinal complications, especially constipation and diarrhea, in type 2 diabetes patients treated with metformin; however, there have been no previous studies examining this. Therefore, we carried out the present study to investigate the effects of the probiotic BBG9‐1 on the gastrointestinal complications in type 2 diabetes patients treated with metformin 18 .
MATERIALS AND METHODS
Study design, ethics approval and consent to participate
This study was an open‐label, single‐arm, exploratory study (Effect of Probiotics, Bifidobacteria, on Gastrointestinal Symptoms in Patients with Type 2 Diabetes Mellitus; Open‐label, Single‐Arm, Exploratory Research [Big STAR study]). The detailed protocol of the Big STAR study has been published previously 18 .
The present study was registered with the Japan Registry of Clinical Trials (jRCTs051190109), and was approved by the ethics committee of the Kyoto Prefectural University of Medicine (CRB5180001). This study was carried out in accordance with the Declaration of Helsinki, and written informed consent was obtained from all participants.
Between 8 April 2020 and 19 August 2020, we registered the study participants at Kyoto Prefectural University of Medicine (Kyoto, Japan).
Study participants
The inclusion criteria for this study were as follows: (i) symptoms of constipation or diarrhea; (ii) gastrointestinal symptom rating scale (GSRS) subscale score (diarrhea or constipation) ≥3; (iii) type 2 diabetes without diabetic polyneuropathy; (iv) taking metformin and less than four antidiabetic agents; (v) no use of antibiotics within 12 weeks before consenting to participate in the study; (vi) no new diet therapy interventions within 12 weeks before consenting to participate; (vii) no changes in concomitant medications within 12 weeks before consenting to participate; (viii) age ≥20 years and <75 years at the time of providing consent; and (ix) provision of written informed consent.
The exclusion criteria of this study were as follows: (i) average weekly defecation frequency of <1 or ≥42 times in the month before consenting to participate in the study; (ii) structural colon diseases diagnosed by colonoscopy in the 5 years before consenting to participate; (iii) celiac disease or inflammatory bowel diseases; (iv) glycated hemoglobin (HbA1c) ≥9% at the time of providing consent; (v) history of newly myocardial infarction, cerebral infarction or stroke within 12 weeks before consenting to participate; (vi) severe liver dysfunction; (vii) severe renal dysfunction; (viii) having active malignant neoplasm; (ix) history of bifidobacterial allergy; (x) use of any other medications or supplements that affect intestinal function; (xi) use of glucagon‐like peptide‐1 receptor agonists or medications that have a high likelihood of causing gastrointestinal symptoms; (xii) routine consumption of foods, supplements, or pharmaceutical agents including bifidobacteria; and (xiii) other conditions that were deemed not appropriate to participants by the investigator or researcher.
Intervention
The outline of enrollment and follow‐up visits is shown in Figure 1. Briefly, (i) the participants were selected based on the aforementioned inclusion/exclusion criteria, and provisional registration was carried out after written informed consent was obtained; (ii) observation 0 (pre‐observation): within 6 weeks of obtaining written informed consent from the participants, a pre‐observation survey was carried out, and registration was initiated; (iii) observation 1 (before treatment observation): within 8 weeks of obtaining written informed consent, baseline examination was carried out, and probiotic BBG9‐1 oral administration (Biofermin® tablets, containing 12 mg of bifidobacterial, 6 tablets 3 times per day) was started; (iv) observation 2 (10 weeks ± 2 weeks after probiotic BBG9‐1 administration; after treatment observation): the examinations, which were the same as the baseline examinations, were carried out, and probiotic BBG9‐1 administration was stopped; (v) observation 3 (12 weeks ± 2 weeks after probiotic BBG9‐1 administration; after treatment observation): a survey was carried out using the GSRS and Bristol Stool Scale. As previous studies showed that the probiotic taken internally have not been detected within a week or two weeks 19 , 20 . During the time from observation 1 to observation 3, all participants were asked to complete the Bristol Stool Scale and feces questionnaire condition daily.
Figure 1.
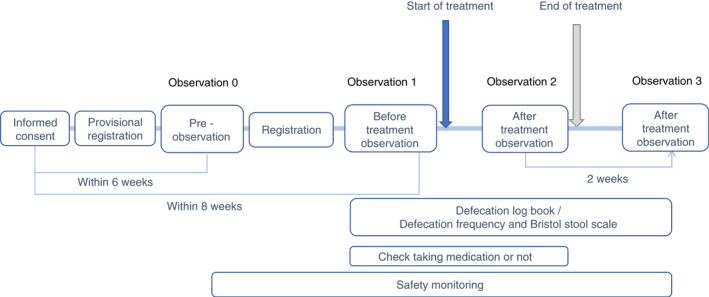
Outline of enrollment and follow‐up visits.
Data collection
Table S1 shows the protocol of data sampling schedule.
Baseline characteristics of study participants, sex, age, duration of diabetes, height, smoking, alcohol, medications for diabetes, hypertension, dyslipidemia and concomitant disease were gathered. Body mass index was calculated as weight in kilograms divided by height in meters squared. Venous blood sampling was carried out to collect the HbA1c level and fasting plasma glucose data.
GSRS was used to evaluate the gastrointestinal symptoms 21 . The GSRS assesses five symptom scores, including acid reflux, abdominal pain, constipation, diarrhea and indigestion, and a total score, using 15 disease‐specific instruments. The GSRS is rated on a 7‐point scale, with 1 representing no troublesome symptoms and 7 representing very troublesome symptoms. The Bristol Stool Scale, which classifies stool into seven categories, was used to evaluate the consistency and shape 18 of the stool: (i) nut‐like; (ii) lumpy sausage; (iii) sausage with cracks; (iv) smooth snake; (v) soft blobs; (vi) fluffy pieces; and (vii) watery. The Bristol Stool Scale score for each observation was the mean of the scores recorded 5 days before and after the start date of each observation.
The safety evaluation assessed the occurrence of adverse events, including various clinical laboratory abnormalities and drug side‐effects, by physicians at observation points.
Gut microbiota analysis
Collection of fecal sample and analysis of gut microbiota composition were carried out using a previously described method 22 . Fecal samples were collected in a guanidine thiocyanate solution (feces collection kit; Techno Suruga Laboratory, Shizuoka, Japan).
Isolation of genomic deoxyribonucleic acid (DNA) was carried out by the Nucleospin Microbial DNA kit (Macherey‐Nagel, Düren, Germany) according to the manufacturer's instructions. The distillation of extracted DNA was carried out by Agencourt AMPure XP (Beckman Coulter, Brea, CA, USA).
Analysis of DNA was carried out by 16S ribosomal ribonucleic acid metagenomic sequencing using the MiSeq platform (Illumina, San Diego, CA, USA) at the Biomedical Center at Takara Bio (Shiga, Japan). To obtain sequence libraries, two‐step polymerase chain reaction was carried out for purification of DNA samples. For the first polymerase chain reaction, amplification was carried out using 16S (V3–V4). Metagenomic library construction kit for NGS (Takara Bio Inc., Shiga, Japan) with primer pairs 341F (5′‐TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG‐3′) and 806R (5′‐GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACHVGGGTWTCTAAT‐3′) corresponding to the V3–V4 region of the 16S ribosomal ribonucleic acid gene. For the second polymerase chain reaction, adding the index sequences for the Illumina sequencer with a barcode sequence was carried out using the Nextera XT index kit (Illumina, San Diego, CA, USA). The prepared libraries were sequenced for 250 paired‐end bases using the MiSeq Reagent v3 kit and MiSeq (Illumina) at the Biomedical Center at Takara Bio.
Filtering of quality and chimeric variant were carried out using the DADA2 plugin of Quantitative Insights into Microbial Ecology 2 23 . Operational taxonomic unit numbers were constructed by clustering with a 99% identity threshold. Using VSEARCH with ≥99% identity, the representative reads for each operational taxonomic unit were designated to the 16S ribosomal ribonucleic acid gene database 24 . Comparison of each taxon in the gut microbiome was carried out at the phylum and genus levels. Evaluation of alpha diversity was carried out by the Shannon index, observed operational taxonomic units number, Chao1 index and Faith's phylogenetic diversity. Evaluation of beta diversity was carried out by computing the weighted and unweighted UniFrac distances between samples. Reducing the dimension of the distance matrix and comparing the differences in the overall bacterial gut microbiome structure was carried out by principal coordinates analysis.
End‐points
The primary end‐point of the present study was the change in the GSRS total score from observation 1 to observation 2.
Secondary end‐points were as follows: (i) change in the GSRS subscale scores, Bristol Stool Scale score, HbA1c level, fasting plasma glucose and gut microbiota from observation 1 to observation 2; (ii) change in the GSRS scores from observation 1 to observation 3; (iii) correlation between the changes in the GSRS total score from observation 1 to observation 2 and changes in other parameters from observation 1 to observation 2; and (iv) changes in the GSRS total score or HbA1c level from observation 1 to observation 2 among the subgroups: (a) age ≥65 years or not, (b) sex, (c) body mass index ≥25 kg/m2 or not, and (d) gastrointestinal symptoms, constipation and diarrhea, or none. Furthermore, as an ad hoc analysis, (v) changes in the GSRS total score or HbA1c level from observation 1 to observation 2 among the subgroup of metformin ≥1,000 mg or not.
Sample size
This was an exploratory study. The feasibility of the target number of cases to be enrolled was based on the number of cases in previous studies 25 , 26 .
Statistical analysis
A full analysis set (FAS) was used for the primary and secondary end‐points and safety analysis, and a per‐protocol set (PPS) was carried out for the primary end‐point. SAS version 9.4 (SAS Institute, Cary, NC, USA) was used for Windows 10, except the changes in GSRS total score or HbA1c level of metformin ≥1,000 mg or not. A P‐value of <5% was considered statistically significant.
The FAS group consisted of all study populations enrolled in the present study, excluding individuals who did not meet the eligibility criteria or individuals who were enrolled after the number of registrants reached the specified number. The PPS group included participants other than those excluded from the FAS group, who did not meet the eligibility criteria, were administered prohibited drugs or had medication adherence of <75%.
Categorical variables are expressed as the number of cases, and continuous variables are expressed as the mean ± standard deviation.
The change in each variable was calculated as observation 2 or observation 3 data and observation 1 data. The primary and secondary end‐points, except for the Bristol Stool Scale score, were evaluated using Student's t‐test. Changes in the Bristol Stool Scale scores were evaluated using the Wilcoxon signed‐rank test. Correlations were evaluated using Spearman's rank correlation coefficient.
In addition, as an ad hoc analysis, to compare the difference of GSRS total scores among observations 1, 2 and 3, the mixed‐effects model for repeated measures with time as fixed effects were carried out, and an unstructured covariate was used to model the covariance of within‐subject variability. The changes in GSRS total score or HbA1c level from observation 1 to observation 2 among the subgroup of metformin ≥1,000 mg or not were evaluated by Student's t‐test using JMP version 13.2.1 (SAS Institute Inc., Cary, NC, USA) for statistical analyses.
As the Bristol Stool Scale score of 4 was known as an ideal stool, we checked the proportion of Bristol Stool Scale score ≥3.5 to <4.5, for each observation point and evaluated the difference by the McNemar test, as an ad hoc analysis.
Regarding β‐diversity, based on the Unifrac distance matrix, a Monte Carlo two‐sample t‐test was carried out.
For the safety analysis, all adverse events were listed.
RESULTS
Participants
The inclusion and exclusion criteria of this study are shown in Figure 2. Among the 42 patients provisionally registered, two patients were excluded (one patient did not meet the eligibility criteria and one patient was enrolled after the number of registrants reached the specified number). In addition, among the 40 patients in the FAS group, four patients were excluded; thus, 36 patients were included in the PPS group.
Figure 2.

Inclusion and exclusion flow. FAS, full analysis set; PPS, per‐protocol set.
The baseline characteristics of the study participants in the FAS group are shown in Table 1. Mean age, body mass index and duration of diabetes were 64.0 ± 9.4 years, 25.0 ± 3.9 kg/m2 and 11.8 ± 8.2 years, respectively. In addition, the mean metformin dose was 875 ± 392 mg.
Table 1.
Baseline characteristics of study participants of full analysis set group
| n | 40 |
|---|---|
| Age (years) | 64.0 ± 9.4 |
| Sex (men/women) | 26/14 |
| Duration of diabetes (years) | 11.8 ± 8.2 |
| Height (cm) | 163.5 ± 8.7 |
| Body weight, kg (n = 38) | 67.6 ± 12.7 |
| Body mass index, kg/m2 (n = 38) | 25.0 ± 3.9 |
| Smoking (never/past/current smoker) | 20/14/6 |
| Alcohol consumption (no/yes) | 21/19 |
| Retinopathy, no/yes (n = 35) | 29/6 |
| Nephropathy, normo/micro/macroalbuminuria (n = 39) | 23/13/3 |
| Metformin dosage (mg) | 875 ± 392 |
| Sulfonylurea (no/yes) | 30/10 |
| Thiazolidine (no/yes) | 40/0 |
| α‐Glucosidase inhibitor (no/yes) | 37/3 |
| Dipeptidyl peptidase‐4 inhibitor (no/yes) | 16/24 |
| Sodium glucose cotransporter‐2 inhibitor (no/yes) | 25/15 |
| Insulin (no/yes) | 36/4 |
| Antihypertension medication (no/yes) | 21/19 |
| Dyslipidemia medication (no/yes) | 26/14 |
Change in the GSRS total score using probiotic BBG9‐1
The changes in the GSRS scores in the FAS group are shown in Table 2. The GSRS total score from observation 1 to observation 2, which was the primary end‐point of the present study, significantly improved by the use of probiotic BBG9‐1 (from 2.02 ± 0.51 to 1.59 ± 0.43, change, −0.43 ± 0.49, P < 0.001). The GSRS total score from observation 1 to observation 2 in the PPS group (n = 36) also significantly improved by the use of probiotic BBG9‐1 (from 2.02 ± 0.52 to 1.58 ± 0.45, change, −0.44 ± 0.50, P < 0.001). The results of the mixed‐effects model for repeated measures are shown in Table S2. The results of the test among observations 1, 2 and 3 were significant, and the results were almost the same as the results in Table 2.
Table 2.
Change in gastrointestinal symptom rating scale scores among full analysis set group
| GSRS scores | Observation 1 n = 40 | Observation 2 n = 40 | Change between observation 1 and observation 2 | P | Observation 3 n = 39 | Change between observation 1 and observation 3 | P |
|---|---|---|---|---|---|---|---|
| Total score | 2.02 ± 0.51 | 1.59 ± 0.43 | −0.43 ± 0.49 | <0.001 | 1.76 ± 0.48 | − 0.25 ± 0.50 | 0.003 |
| Acid reflux score | 1.61 ± 0.80 | 1.20 ± 0.42 | −0.41 ± 0.80 | 0.002 | 1.38 ± 0.48 | −0.19 ± 0.75 | 0.12 |
| Abdominal pain score | 1.32 ± 0.51 | 1.11 ± 0.25 | −0.22 ± 0.47 | 0.006 | 1.21 ± 0.36 | −0.10 ± 0.58 | 0.28 |
| Indigestion score | 1.86 ± 0.72 | 1.55 ± 0.59 | −0.31 ± 0.71 | 0.009 | 1.90 ± 0.63 | 0.03 ± 0.69 | 0.77 |
| Diarrhea score | 2.32 ± 1.14 | 1.89 ± 0.99 | −0.42 ± 0.95 | 0.007 | 1.99 ± 1.04 | −0.32 ± 0.92 | 0.034 |
| Constipation score | 3.00 ± 1.16 | 2.20 ± 1.07 | −0.80 ± 1.19 | <0.001 | 2.32 ± 1.15 | −0.68 ± 1.13 | <0.001 |
Student's t‐tests were carried out to assess statistical significance. GSRS, gastrointestinal symptom rating scale.
Changes in the GSRS subscale scores, Bristol Stool Scale score and glycemic control using probiotic BBG9‐1
The GSRS subscale scores, including diarrhea score (from 2.32 ± 1.14 to 1.89 ± 0.99, change, −0.42 ± 0.95, P = 0.007) and constipation score (from 3.00 ± 1.16 to 2.20 ± 1.07, change, −0.80 ± 1.19, P < 0.001), from observation 1 to observation 2, also significantly improved by the use of probiotic BBG9‐1. Furthermore, the improvements in the total score (change, −0.25 ± 0.50, P = 0.003), diarrhea score (change, −0.32 ± 0.92, P = 0.034) and constipation score (change, −0.68 ± 1.13, P < 0.001) were sustained at least until observation 3, 2 weeks after probiotic BBG9‐1 discontinuation.
The Bristol Stool Scale score did not change (change from observation 1 to observation 2: 0.06 ± 0.57, P = 0.44; change from observation 1 to observation 3: 0.16 ± 0.69, P = 0.14; Figure 3). In contrast, according to the ad hoc analysis, the proportions of the Bristol Stool Scale score ≥3.5 to <4.5, which is an ideal fecal condition, tended to be increased by probiotic BBG9‐1 discontinuation (47.4% in observation 1, 60.5% in observation 2 [P = 0.23], and 64.9% in observation 3 [P = 0.06]; Figure 3).
Figure 3.
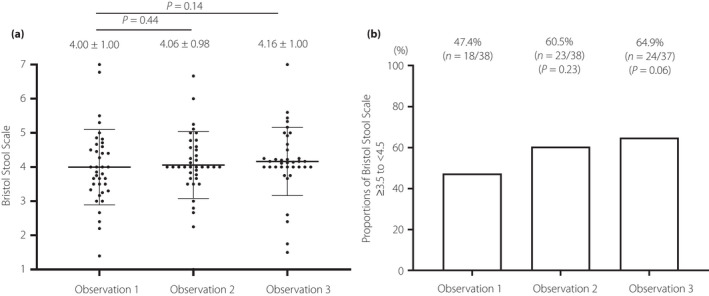
Change in Bristol Stool Scale score before and after the use of the probiotic Bifidobacterium bifidum G9‐1. (a) The Bristol Stool Scale scores of observation 1 (n = 38), observation 2 (n = 38) and observation 3 (n = 37) are shown. Change in Bristol Stool Scale score was evaluated using Wilcoxon signed‐rank test. (b) The differences in the proportions of Bristol Stool Scale scores ≥3.5 to <4.5 were evaluated by the McNemar test.
The HbA1c and fasting plasma glucose levels were not changed by the use of the probiotic BBG9‐1 (Table 3).
Table 3.
Change in glycated hemoglobin and fasting plasma glucose among full analysis set group
| Observation 1 | Observation 2 | Change | P | |
|---|---|---|---|---|
| HbA1c (%) |
7.0 ± 0.7 (n = 40) |
7.0 ± 0.6 (n = 40) |
0.0 ± 0.4 (n = 40) |
0.91 |
| HbA1c (mmol/mol) |
53.1 ± 7.5 (n = 40) |
53.2 ± 6.8 (n = 40) |
0.1 ± 4.4 (n = 40) |
0.91 |
| Fasting plasma glucose (mg/dL) |
130.9 ± 21.8 (n = 25) |
136.5 ± 24.6 (n = 28) |
4.4 ± 28.3 (n = 19) |
0.50 |
Student's t‐tests were carried out to assess statistical significance. HbA1c, glycated hemoglobin.
Changes in the gut microbiota
The α‐diversity did not show a significant difference before and after the use of probiotic BBG9‐1 (Figure 4). In addition, the β‐diversity did not show a significant difference before and after the use of probiotic BBG9‐1 (Figure 5).
Figure 4.
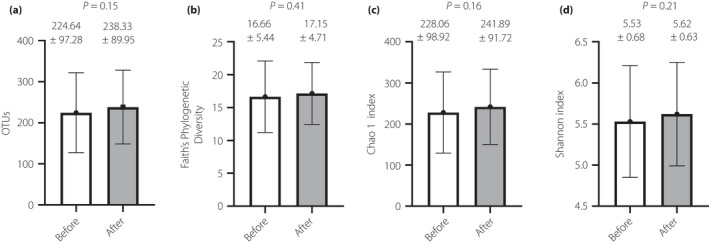
The α‐diversity comparisons between before and after use of the probiotic Bifidobacterium bifidum G9‐1. (a) Observed operational taxonomic units (OTUs) number, (b) Faith's Phylogenetic Diversity, (c) Chao1 index and (d) Shannon index. The analyses were carried out among a full analysis set group (n = 40). The differences between the groups were evaluated using Student's t‐test, with no significant differences being found.
Figure 5.

Principal coordinates analysis of the gut microbiota fecal diversity between before and after use of probiotic Bifidobacterium bifidum G9‐1. (a) Unweighted UniFrac metrics and (b) weighted Unifrac metrics. The analyses were carried out among a full analysis set group (n = 40). Based on the Unifrac distance matrix, a Monte Carlo two‐sample t‐test was carried out. There was no significant difference between the mean unweighted UniFrac distances for before‐before and that for before‐after (P = 1.00) and that for after‐after and that for before‐after (P = 0.589). There was no significant difference between the mean weighted UniFrac distances for before‐before and that for before‐after (P = 0.06) and that for after‐after and that for before‐after (P = 0.669). Blue circle represents before use of probiotic Bifidobacterium bifidum G9‐1 and red circle represents after use of probiotic Bifidobacterium bifidum G9‐1.
Changes in the phylum levels are shown in Figure 6. With the use of probiotic BBG9‐1, the relative abundance of phylum Firmicutes increased (from 0.57 ± 0.12 to 0.61 ± 0.11, P = 0.013) and phylum Bacteroidetes decreased (from 0.26 ± 0.10 to 0.22 ± 0.09, P = 0.045).
Figure 6.
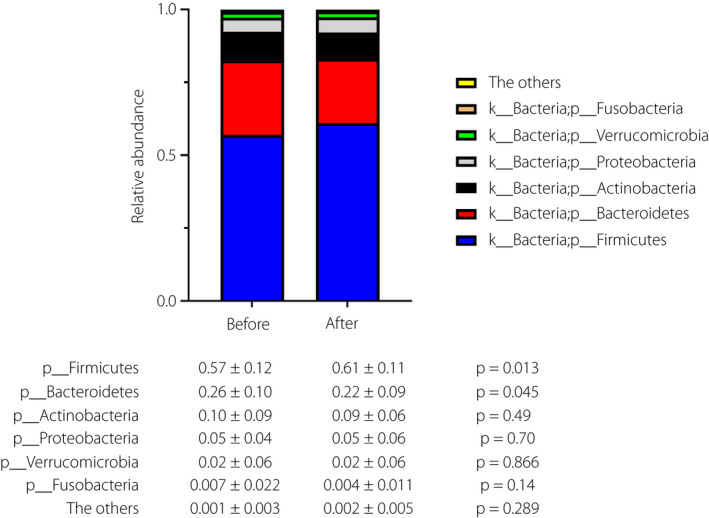
Alternation of relative abundance in phyla before and after use of probiotic Bifidobacterium bifidum G9‐1. The analyses were carried out among a full analysis set group (n = 40). The differences between the groups were evaluated using Student's t‐test.
Changes in the top 30 gut microbiota genera are shown in Table 4. The relative abundance of the genus Sutterella decreased by the use of the probiotic BBG9‐1 (from 0.011 ± 0.009 to 0.008 ± 0.006, P = 0.002).
Table 4.
Alternation of relative abundance in genera among full analysis set group
| Genera | Observation 1 n = 40 | Observation 2 n = 40 | Change | P |
|---|---|---|---|---|
| g__Bacteroides | 0.170 ± 0.103 | 0.149 ± 0.071 | −0.021 ± 0.081 | 0.11 |
| g__Bifidobacterium | 0.067 ± 0.084 | 0.056 ± 0.056 | −0.011 ± 0.054 | 0.20 |
| g__Blautia | 0.047 ± 0.030 | 0.041 ± 0.023 | −0.005 ± 0.026 | 0.25 |
| g__Ruminococcus | 0.044 ± 0.041 | 0.049 ± 0.056 | 0.006 ± 0.050 | 0.48 |
| f__Lachnospiraceae; g__[Ruminococcus] | 0.042 ± 0.059 | 0.043 ± 0.071 | 0.001 ± 0.031 | 0.81 |
| g__Faecalibacterium | 0.041 ± 0.046 | 0.050 ± 0.057 | 0.009 ± 0.035 | 0.10 |
| Unclassified f__Lachnospiraceae | 0.039 ± 0.027 | 0.040 ± 0.034 | 0.002 ± 0.03 | 0.77 |
| g__Roseburia | 0.034 ± 0.041 | 0.038 ± 0.046 | 0.004 ± 0.032 | 0.43 |
| g__Prevotella | 0.033 ± 0.072 | 0.026 ± 0.062 | −0.007 ± 0.055 | 0.43 |
| g__Coprococcus | 0.029 ± 0.027 | 0.025 ± 0.022 | −0.004 ± 0.016 | 0.17 |
| g__Megamonas | 0.028 ± 0.074 | 0.034 ± 0.070 | 0.006 ± 0.045 | 0.38 |
| g__Parabacteroides | 0.027 ± 0.029 | 0.022 ± 0.021 | −0.005 ± 0.021 | 0.14 |
| g__Collinsella | 0.026 ± 0.026 | 0.030 ± 0.028 | 0.004 ± 0.023 | 0.33 |
| g__Oscillospira | 0.026 ± 0.021 | 0.030 ± 0.022 | 0.004 ± 0.017 | 0.12 |
| Unclassified f__Enterobacteriaceae | 0.025 ± 0.032 | 0.025 ± 0.045 | −0.000 ± 0.0497 | 0.97 |
| g__Streptococcus | 0.024 ± 0.041 | 0.030 ± 0.044 | 0.006 ± 0.041 | 0.35 |
| g__Megasphaera | 0.023 ± 0.034 | 0.020 ± 0.030 | −0.003 ± 0.021 | 0.43 |
| g__Gemmiger | 0.022 ± 0.028 | 0.025 ± 0.025 | 0.002 ± 0.016 | 0.35 |
| g__Akkermansia | 0.020 ± 0.058 | 0.020 ± 0.059 | 0.001 ± 0.019 | 0.87 |
| g__Lactobacillus | 0.018 ± 0.054 | 0.018 ± 0.037 | −0.001 ± 0.039 | 0.92 |
| Unclassified f__Lachnospiraceae | 0.018 ± 0.019 | 0.020 ± 0.021 | 0.002 ± 0.019 | 0.46 |
| g__Phascolarctobacterium | 0.017 ± 0.021 | 0.015 ± 0.013 | −0.002 ± 0.016 | 0.50 |
| Unclassified f__Ruminococcaceae | 0.015 ± 0.015 | 0.017 ± 0.022 | 0.002 ± 0.022 | 0.62 |
| g__Dorea | 0.014 ± 0.009 | 0.016 ± 0.014 | 0.002 ± 0.014 | 0.34 |
| g__Veillonella | 0.012 ± 0.026 | 0.014 ± 0.029 | 0.001 ± 0.021 | 0.67 |
| g__Clostridium | 0.012 ± 0.024 | 0.011 ± 0.015 | −0.001 ± 0.027 | 0.83 |
| g__Alistipes | 0.012 ± 0.024 | 0.010 ± 0.023 | −0.002 ± 0.014 | 0.48 |
| g__Sutterella | 0.011 ± 0.009 | 0.008 ± 0.006 | −0.003 ± 0.006 | 0.002 |
| g__[Eubacterium] | 0.010 ± 0.017 | 0.007 ± 0.011 | −0.002 ± 0.012 | 0.24 |
| Unclassified f__Ruminococcaceae | 0.007 ± 0.010 | 0.009 ± 0.017 | 0.003 ± 0.012 | 0.15 |
The top 30 gut microbial genera are listed. Student's t‐tests were carried out to assess statistical significance.
Correlation between the changes in the GSRS total score and other parameters
The correlation between the changes in the GSRS total score and other parameters is shown in Table 5. Changes in the HbA1c and fasting plasma glucose levels tended to be correlated with changes in the GSRS total score, although this did not reach statistical significance.
Table 5.
Correlation between change in gastrointestinal symptom rating scale total score and change in other parameters among full analysis set group
| n | r (95% CI) | P | |
|---|---|---|---|
| Bristol stool scale | 38 | −0.08 (−0.39, 0.25) | 0.64 |
| HbA1c (%) | 40 | 0.29 (−0.03, 0.55) | 0.07 |
| Fasting plasma glucose (mg/dL) | 19 | 0.42 (−0.05, 0.73) | 0.07 |
| Body mass index (kg/m2) | 36 | 0.13 (−0.21, 0.44) | 0.44 |
| OTUs | 40 | 0.20 (−0.13, 0.48) | 0.23 |
| Faith's Phylogenetic Diversity | 40 | 0.10 (−0.22, 0.39) | 0.56 |
| Chao1 index | 40 | 0.21 (−0.11, 0.48) | 0.20 |
| Shannon index | 40 | 0.22 (−0.11, 0.49) | 0.18 |
Correlations were evaluated using Spearman's rank correlation coefficient. GSRS, gastrointestinal symptom rating scale; HbA1c, glycated hemoglobin; OTUs, operational taxonomic units.
Subgroup analysis
The results of the subgroup analyses are shown in Table 6. The GSRS total score from observation 1 to observation 2 was significantly improved by the use of probiotic BBG9‐1 among all subgroups. In addition, the HbA1c level improved from observation 1 to observation 2 among women (−1.7 ± 2.7%, P = 0.033). Among the participants taking metformin <1,000 mg (n = 19), the GSRS total score (−0.24 ± 0.44, P = 0.170) and HbA1c did not changed (0.1 ± 0.4, P = 0.707). Among the participants taking metformin ≥1,000 mg (n = 21), the GSRS total score was significantly improved by the use of probiotic BBG9‐1 (−0.61 ± 0.47, P < 0.001), whereas HbA1c did not changed (−0.1 ± 0.4, P = 0.787).
Table 6.
Subgroup analyses of change in gastrointestinal symptom rating scale total score and glycated hemoglobin among full analysis set group
| Change from observation 1 to observation 2 | P | ||
|---|---|---|---|
| Age <65 years | GSRS total score (n = 17) | −0.52 ± 0.46 | <0.001 |
| HbA1c (%) (n = 17) | 0.0 ± 0.4 | 0.73 | |
| Age ≥65 years | GSRS total score (n = 23) | −0.37 ± 0.51 | 0.002 |
| HbA1c (%) (n = 23) | 0.0 ± 0.4 | 0.88 | |
| Men | GSRS total score (n = 26) | −0.42 ± 0.51 | <0.001 |
| HbA1c (%) (n = 26) | 0.1 ± 0.4 | 0.28 | |
| Women | GSRS total score (n = 14) | −0.45 ± 0.46 | 0.003 |
| HbA1c (%) (n = 14) | −1.7 ± 2.7 | 0.033 | |
| BMI <25 kg/m2 | GSRS total score (n = 25) | −0.36 ± 0.51 | 0.002 |
| HbA1c (%) (n = 25) | 0.1 ± 0.4 | 0.29 | |
| BMI ≥25 kg/m2 | GSRS total score (n = 13) | −0.52 ± 0.48 | 0.002 |
| HbA1c (%) (n = 13) | −0.1 ± 0.4 | 0.17 | |
| Constipation symptoms (−) | GSRS total score (n = 18) | −0.39 ± 0.34 | <0.001 |
| HbA1c (%) (n = 18) | 0.0 ± 0.4 | 0.96 | |
| Constipation symptoms (+) | GSRS total score (n = 22) | −0.47 ± 0.59 | 0.001 |
| HbA1c (%) (n = 22) | 0.0 ± 0.4 | 0.191 | |
| Diarrhea symptoms (−) | GSRS total score (n = 29) | −0.39 ± 0.47 | <0.001 |
| HbA1c (%) (n = 29) | 0.0 ± 0.4 | 0.77 | |
| Diarrhea symptoms (+) | GSRS total score (n = 11) | −0.56 ± 0.53 | 0.006 |
| HbA1c (%) (n = 11) | 0.0 ± 0.5 | 0.85 |
Student's t‐tests were carried out to assess statistical significance. BMI, body mass index; GSRS, gastrointestinal symptom rating scale; HbA1c, glycated hemoglobin.
Safety analysis
No serious adverse events occurred in the present study. No specific adverse events occurred, except for pharyngitis in one patient and constipation in three patients.
DISCUSSION
The present study investigated the effect of the probiotic BBG9‐1 on gastrointestinal symptoms in type 2 diabetes patients using metformin, and found that the use of probiotic BBG9‐1 improved the GSRS total score. In addition, the use of probiotic BBG9‐1 improved all the GSRS subscale scores, such as reflux syndrome, abdominal pain, constipation syndrome, diarrhea and indigestion syndrome scores. Furthermore, the improvement of constipation syndrome score or diarrhea syndrome score continued for at least 2 weeks after discontinuation of the medication. Finally, no serious adverse events occurred due to the use of the probiotic BBG9‐1 in the present study.
Many patients with type 2 diabetes have been reported to experience various gastrointestinal diseases, such as constipation and diarrhea 3 , 4 , 5 . Furthermore, metformin use is reported to be associated with an increased risk of gastrointestinal complications 6 . We previously reported that new use of metformin increased the GSRS scores of diarrhea (from 2.9 ± 1.3 to 3.3 ± 1.7) and constipation (from 5.0 ± 2.2 to 5.9 ± 2.2) in patients with type 2 diabetes 7 . The findings of the present study showed that the use of the probiotic BBG9‐1 improved gastrointestinal complications, especially constipation and diarrhea, in type 2 diabetes patients treated with metformin, and that it decreased the GSRS scores of diarrhea and constipation, which is almost the same amount as the increase by metformin usage. Thus, there is a possibility that patients who have stopped using metformin due to abdominal symptoms might be able to continue to use metformin by using the probiotic BBG9‐1.
The possible mechanism of the improvement of gastrointestinal symptoms by using the probiotic BBG9‐1 is as follows. A recent animal model study showed that the probiotic BBG9‐1 improves the symptoms of diarrhea caused by metformin by changing the gut microbiota and fecal conditions 17 . In this study, the proportions of Bristol Stool Scale score ≥3.5 to <4.5, which is an ideal fecal condition, tended to be increased by probiotic BBG9‐1 discontinuation, although it did not reach statistical significance. In addition, the relative abundance of the genus Sutterella decreased by the use of the probiotic BBG9‐1 in this study. It has been reported that the relative abundance of the genus Sutterella, which are known to be associated with abdominal symptoms, was higher in patients with inflammatory bowel disease and Crohn's disease 27 , 28 than in those without. In addition, the relative abundance of the genus Sutterella in people with autism spectrum disorders, which is closely associated with gastrointestinal symptoms of constipation and diarrhea 29 , is reported to be higher than that in individuals without autism spectrum disorders 30 . It has also been reported that Sutterella species are involved in pro‐inflammatory activity 31 . Furthermore, bile acids, glucagon‐like peptide‐1, histamine and serotonin have been suggested to be involved in metformin‐induced gastrointestinal disorders 32 . Unfortunately, however, we did not have the data of bile acids, glucagon‐like peptide‐1, histamine and serotonin in the present study. In contrast, previous studies showed that there is a difference in gut microbiota between metformin‐treated and non‐treated groups 33 , 34 , 35 .
Furthermore, we recently showed that an increase of the genus Blautia was associated with diarrhea in patients with new use of metformin 7 . In the present study, genus Blautia tended to decrease with the use of BBG9‐1, although it did not reach statistical significance. In addition, a recent study with an animal model showed that BBG9‐1 was associated with improvement of metformin‐induced diarrhea through changing of gut microbiota 17 . Therefore, there is a possibility that the reduction of genus Sutterella and genus Blautia by the use of the probiotic BBG9‐1 might have contributed to the improvement of abdominal symptoms, although the principal coordinates analysis plot and relative abundance ratio plot did not show any significant changes before and after the use of the probiotic BBG9‐1.
In the present study, HbA1c levels did not change with the use of probiotic BBG9‐1, which was the same as in a previous animal study 17 . Furthermore, in the subgroup analysis, HbA1c levels were improved in women. It is possible that the improvement in abdominal symptoms is related to the improvement in HbA1c levels 36 , 37 . In fact, in the present study, there was a correlation between changes in HbA1c levels and changes in the GSRS score, although it did not reach statistical significance. Thus, further studies are required to investigate the association and mechanism of improving abdominal symptoms and glycemic control.
The limitations of the present study should be noted. First, the study was limited by its open‐label and single‐arm design. Furthermore, because of the exploratory nature of the study, the sample size was also small. Further double‐blinded randomized placebo‐controlled trials are desirable to clarify the results of this study. Second, the primary end‐point, the GSRS score, is a subjective survey measure using a questionnaire, it would be desirable to validate this study with additional objective measures. Third, we did not evaluate dietary habits or consider the effect of diet on the gut microbiota.
In the present study, type 2 diabetes patients treated with metformin showed significant improvement in the GSRS scores using the probiotic BBG9‐1 without serious side‐effects or changing glucose control. Furthermore, the improvement of constipation syndrome score or diarrhea syndrome score continued for at least 2 weeks after discontinuation of the medication. This study showed the potential usefulness of the probiotic BBG9‐1 for gastrointestinal symptoms, including constipation and diarrhea, in type 2 diabetes mellitus patients treated with metformin.
DISCLOSURE
Potential conflict of interest was shown in Appendix S1.
Registration: This study was registered with the Japan Registry of Clinical Trials (jRCTs051190109; 19 February 2020).
Informed consent: Written informed consent was obtained from all participants.
Approval date of registry and the registration no. of the study/trial: N/A.
Animal studies: N/A.
Supporting information
Table S1 | Schedule of data collection.
Table S2 | Differences of gastrointestinal symptom rating scale scores among observation 1, observation 2 and observation 3 among full analysis set group.
Appendix S1 | Potential conflict of interest.
ACKNOWLEDGMENTS
The authors thank all the clinical staff for their assistance with the execution of the clinical trial, and Soiken Inc. for their technical assistance in the launch and execution of the trial. The authors thank Editage (www.editage.com) for English language editing. This study, including the article processing charge, was funded by Biofermin Pharmaceutical. Co. Ltd. No drugs were donated or funded by the sponsor. No funding bodies had any role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
J Diabetes Investig.2022; 13: 489–500
Clinical Trial Registry
Japan Registry of Clinical Trials
jRCTs051190109
Contributor Information
Shinnosuke Hata, Email: hatashin@koto.kpu-m.ac.jp.
Yoshitaka Hashimoto, Email: y-hashi@koto.kpu-m.ac.jp.
REFERENCES
- 1. Chatterjee S, Khunti K, Davies MJ. Type 2 diabetes. Lancet 2017; 389: 2239–2251. [DOI] [PubMed] [Google Scholar]
- 2. The Ministry of Health, Labour, and Welfare . The National Health and Nutrition Examination Survey. 2016. (in Japanese) Available from: http://www.mhlw.go.jp/stf/houdou/0000177189.html Accessed May 1, 2021.
- 3. Zawada A, Moszak M, Skrzypczak D, et al. Gastrointestinal complications in patients with diabetes mellitus. Adv Clin Exp Med 2018; 2: 567–572. [DOI] [PubMed] [Google Scholar]
- 4. Brunkwall L, Orho‐Melander M. The gut microbiome as a target for prevention and treatment of hyperglycaemia in type 2 diabetes: from current human evidence to future possibilities. Diabetologia 2017; 60: 943–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maisey A. A practical approach to gastrointestinal complications of diabetes. Diabetes Ther 2016; 7: 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Piper MS, Saad RJ. Diabetes mellitus and the colon. Curr Treat Options Gastroenterol 2017; 15: 460–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakajima H, Takewaki F, Hashimoto Y, et al. The effects of metformin on the gut microbiota of patients with type 2 diabetes: a two‐center, quasi‐experimental study. Life (Basel) 2020; 10: 195. 10.3390/life10090195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanchez‐Rangel E, Inzucchi SE. Metformin: clinical use in type 2 diabetes. Diabetologia 2017; 60: 1586–1593. [DOI] [PubMed] [Google Scholar]
- 9. Qin J, Li Y, Cai Z, et al. A metagenome‐wide association study of gut microbiota in type 2 diabetes. Nature 2012; 490: 55–60. [DOI] [PubMed] [Google Scholar]
- 10. Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013; 498: 99–103. [DOI] [PubMed] [Google Scholar]
- 11. Blandino G, Inturri R, Lazzara F, et al. Impact of gut microbiota on diabetes mellitus. Diabetes Metab 2016; 42: 303–315. [DOI] [PubMed] [Google Scholar]
- 12. Hills R, Pontefract B, Mishcon H, et al. Gut microbiome: profound implications for diet and disease. Nutrients 2019; 11: 1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh RK, Chang H‐W, Yan DI, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med 2017; 15: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Culpepper T, Christman MC, Nieves C Jr, et al. Bifidobacterium bifidum R0071 decreases stress‐associated diarrhoea‐related symptoms and self‐reported stress: a secondary analysis of a randomised trial. Benef Microbes 2016; 7: 327–336. [DOI] [PubMed] [Google Scholar]
- 15. Ibarra A, Latreille‐Barbier M, Donazzolo Y, et al. Effects of 28‐day Bifidobacterium animalis subsp. lactis HN019 supplementation on colonic transit time and gastrointestinal symptoms in adults with functional constipation: a double‐blind, randomized, placebo‐controlled, and dose‐ranging trial. Gut Microbes 2018; 9: 236–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agrawal A, Houghton LA, Morris J, et al. Clinical trial: the effects of a fermented milk product containing Bifidobacterium lactis DN‐173 010 on abdominal distension and gastrointestinal transit in irritable bowel syndrome with constipation. Aliment Pharmacol Ther 2009; 29: 104–114. [DOI] [PubMed] [Google Scholar]
- 17. Makizaki Y, Maeda A, Yamamoto M, et al. Bifidobacterium bifidum G9–1 ameliorates soft feces induced by metformin without affecting its antihyperglycemic action. Biosci Microbiota Food Health 2020; 39: 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashimoto Y, Nakajima H, Hata S, et al. Effect of probiotics, Bifidobacterium bifidum G9–1, on gastrointestinal symptoms in patients with type 2 diabetes mellitus: study protocol for open‐label, single‐arm, exploratory research trial (Big STAR study). J Clin Biochem Nutr 2020; 67: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishizuka A, Tomizuka K, Aoki R, et al. Effects of administration of Bifidobacterium animalis subsp. lactis GCL2505 on defecation frequency and bifidobacterial microbiota composition in humans. J Biosci Bioeng 2012; 113: 587–591. [DOI] [PubMed] [Google Scholar]
- 20. Cox AJ, Makino H, Cripps AW, et al. Recovery of Lactobacillus casei strain Shirota (LcS) from faeces with 14 days of fermented milk supplementation in healthy Australian adults. Asia Pac J Clin Nutr 2019; 28: 734–739. [DOI] [PubMed] [Google Scholar]
- 21. Dimenäs E, Glise H, Hallerbäck B, et al. Well‐being and gastrointestinal symptoms among patients referred to endoscopy owing to suspected duodenal ulcer. Scand J Gastroenterol 1995; 30: 1046–1052. [DOI] [PubMed] [Google Scholar]
- 22. Hashimoto Y, Hamaguchi M, Kaji A, et al. Intake of sucrose affects gut dysbiosis in patients with type 2 diabetes. J Diabetes Investig 2020; 11: 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019; 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: High‐resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalman DS, Schwartz HI, Alvarez P, et al. A prospective, randomized, double‐blind, placebo‐controlled parallel‐group dual site trial to evaluate the effects of a Bacillus coagulans‐based product on functional intestinal gas symptoms. BMC Gastroenterol 2009; 9: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Francavilla R, Piccolo M, Francavilla A, et al. Clinical and microbiological effect of a multispecies probiotic supplementation in celiac patients with persistent IBS‐type symptoms: a randomized, double‐blind, placebo‐controlled, multicenter trial. J Clin Gastroenterol 2019; 53: e117–e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santoru ML, Piras C, Murgia A, et al. Cross sectional evaluation of the gut‐microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep 2017; 7: 9523. [published correction appears in Sci Rep. 2018 Mar 19;8(1):4993] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mangin Irène, Bonnet R, Seksik P, et al. Molecular inventory of faecal microflora in patients with Crohn's disease. FEMS Microbiol Ecol 2004; 50: 25–36. [DOI] [PubMed] [Google Scholar]
- 29. Peters B, Williams KC, Gorrindo P, et al. Rigid‐compulsive behaviors are associated with mixed bowel symptoms in autism spectrum disorder. J Autism Dev Disord 2014; 44: 1425–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benach JL, Li E, McGovern MM. A microbial association with autism. MBio 2012; 3: e00019–e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hiippala K, Kainulainen V, Kalliomäki M, et al. Mucosal prevalence and interactions with the epithelium indicate commensalism of Sutterella spp. Front Microbiol 2016; 7: 1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCreight LJ, Stage TB, Connelly P, et al. Pharmacokinetics of metformin in patients with gastrointestinal intolerance. Diabetes Obes Metab 2018; 20: 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015; 528: 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu H, Esteve E, Tremaroli V, et al. Metformin alters the gut microbiome of individuals with treatment‐naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 2017; 23: 850–858. [DOI] [PubMed] [Google Scholar]
- 35. Hung W‐W, Peng PO, Tsai Y‐C, et al. Gut microbiota compositions and metabolic functions in type 2 diabetes differ with glycemic durability to metformin monotherapy. Diabetes Res Clin Pract 2021; 174: 108731. [DOI] [PubMed] [Google Scholar]
- 36. Quan C, Talley NJ, Jones MP, et al. Gastrointestinal symptoms and glycemic control in diabetes mellitus: a longitudinal population study. Eur J Gastroenterol Hepatol 2008; 20: 888–897. [DOI] [PubMed] [Google Scholar]
- 37. Leeds JS, Hadjivassiliou M, Tesfaye S, et al. Lower gastrointestinal symptoms are associated with worse glycemic control and quality of life in type 1 diabetes mellitus. BMJ Open Diabetes Res Care 2018; 6: e000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 | Schedule of data collection.
Table S2 | Differences of gastrointestinal symptom rating scale scores among observation 1, observation 2 and observation 3 among full analysis set group.
Appendix S1 | Potential conflict of interest.


