Abstract
Reference genes are the key to study gene expression patterns using quantitative real-time PCR (qRT-PCR). No studies on the reference genes of Athetis dissimilis, an important agricultural pest, have been reported. In order to determine the reference genes for qRT-PCR normalization in A. dissimilis under different conditions, 10 candidate genes [18S ribosomal protein (18S), 28S ribosomal protein (28S), arginine kinase (AK), elongation factor 1 alpha (EF1-α), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein L32 (RPL32), ribosomal protein L40 (RPL40), alpha-tubulin (α-TUB), beta-actin (β-ACT), and beta-tubulin (β-TUB)] of A. dissimilis were selected to evaluate their stability as reference genes under different biotic and abiotic conditions by using five tools, geNorm, NormFinder, BestKeeper, ΔCt, and RefFinder. Furthermore, CSP1 and superoxide dismutase (SOD) were used as target genes to validate the candidate reference genes. The results showed that different reference genes were needed under different experimental conditions, among which, EF-1α, RPL40, and 18S are most suitable reference genes for studying genes related development stages of A. dissimilis, RPL40 and α-TUB for larval tissues, α-TUB and 28S for adult tissues, EF-1α and β-ACT for insecticidal treatments, β-ACT and 28S for temperature treatments, EF-1α and β-ACT for starvation treatments, RPL40 and 18S for dietary treatments, and 18S, 28S, and α-TUB for all the samples. These results provide suitable reference genes for studying gene expression in A. dissimilis under different experimental conditions, and also lay the foundation for further research into the function of related genes in A. dissimilis.
Keywords: reference gene, Athetis dissimilis, quantitative RT-PCR, gene expression stability, biotic and abiotic conditions
Introduction
In molecular biological research, gene expression analyses provide the information concerning gene regulatory mechanisms and functions associated with different biological processes (Zhao et al., 2018). Although there are many methods for evaluating gene expression profiles, quantitative real-time PCR (qRT-PCR) has been the most commonly used tool over the past several decades owing to its convenience, rapidity, specificity, and high sensitivity (Bustin et al., 2005; Huggett et al., 2005; Derveaux et al., 2010). For qRT-PCR studies, it is essential to select an appropriate reference gene to normalize qRT-PCR data in order to minimize the influence of RNA quality, reverse transcription efficiency, and PCR reaction conditions on the data. Thus, it is important to evaluate the validity of the candidate reference gene before qRT-PCR tests (Radonic et al., 2004; Huggett et al., 2005; Fleige and Pfaffl, 2006; Guenin et al., 2009; Xie et al., 2021).
An ideal reference gene should have steady expression levels under all the experimental conditions (Brym et al., 2013; Janska et al., 2013). However, the expression profiles of the widely used reference genes are not always constant under different experimental conditions (Selvey et al., 2001; Glare et al., 2002; Radonic et al., 2004). Numerous studies have demonstrated that even the same reference gene exhibited different expression levels under different experimental conditions, such as organismal developmental stages (Sun et al., 2009; Nakamura et al., 2016), tissues (Huis et al., 2010), cells (Nelissen et al., 2010), and temperatures (Mahanty et al., 2017). Therefore, it is crucial to identify suitable reference genes and evaluate their expression stability in certain target species under specific experimental conditions before the qRT-PCR data normalization of target gene expression levels using the reference genes (Guo et al., 2016; Wan et al., 2017; Renard et al., 2018).
For some model organisms or important economic insects, suitable reference genes for qRT-PCR have been identified and validated under various biotic or abiotic conditions (Fu et al., 2013; Yang et al., 2014; Zhang et al., 2015), but no appropriate reference genes have been identified and validated in Athetis dissimilis, an important insect pest widely distributed across many Asian countries (Dong et al., 2016; Sun et al., 2016; Guo et al., 2017; Che et al., 2019; Liu et al., 2019). The larvae of A. dissimilis bore into the seedling roots of maize, wheat, soybean, peanut, and other crop plants, causing the plants wilting and even death. Since it was first reported in Shandong Province, China in 2012, the damage of A. dissimilis has spread to Hebei, Henan, Shanxi, and other provinces. The development, mating behavior, and reproduction behavior of A. dissimilis, as well as its resistance to pesticide, are all regulated by related genes, and suitable reference genes are crucial to verify the gene expression profiles in A. dissimilis under different abiotic and biotic conditions. In addition, analysis of gene expression patterns under different conditions can provide valuable information regarding gene function and contribute to identify the important genes that may participate in these physiological and biological processes in A. dissimilis. However, the selection of reference genes of A. dissimilis in previous studies were all based on experience (Dong et al., 2016; Liu et al., 2019), a comprehensive study on the suitability of reference genes for qRT-PCR data normalization under different conditions is lacking.
Here, to identify suitable reference genes for the molecular study of A. dissimilis, 10 commonly used housekeeping genes, 18S ribosomal protein (18S), 28S ribosomal protein (28S), arginine kinase (AK), elongation factor 1 alpha (EF1-α), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein L32 (RPL32), ribosomal protein L40 (RPL40), alpha-tubulin (α-TUB), beta-actin (β-ACT), and beta-tubulin (β-TUB), were selected as the candidate reference genes (Ma et al., 2016; Yan et al., 2016; Gao et al., 2017; Hu et al., 2018; Yin et al., 2020), and their expression stabilities were analyzed under different biotic and abiotic conditions. Finally, a ranking of the stable reference genes was recommended for the corresponding experimental conditions. This work will provide suitable normalization genes for future gene expression studies and functional genomics research on A. dissimilis and its related species.
Materials and Methods
Insects
The larvae of A. dissimilis were provided by the Cotton Pest Research Group, Institute of Plant Protection, Henan Academy of Agricultural Sciences, Zhengzhou, China, and then maintained in the laboratory at 25 ± 1°C and 70 ± 5% relative humidity under a 14-h day:10-h night cycle (Guo et al., 2017). Larvae were reared on an artificial diet, and adults were provided with a 10% (w/w) honey solution.
Candidate Reference Gene Clones and qRT-PCR Primer Design
The β-ACT, RPL32, RPL40, EF1-α, α-TUB, β-TUB, 18S, 28S, and AK genes in A. dissimilis were cloned as described below using the primers listed in Supplementary Table S1, and the sequence of GAPDH (GenBank accession no. KT361883.1) was downloaded from NCBI. Total RNAs from fourth-instar larvae of A. dissimilis were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) in accordance with the manufacturer’s protocol. The purity and integrity of the total RNA were evaluated using a NanoVue spectrophotometer (GE Healthcare, United States). Then, genomic DNA in the RNA samples was removed using DNase I (TaKaRa, Japan), and the first-strand cDNA was synthesized using a PrimeScript first Strand cDNA Synthesis Kit (TaKaRa) in accordance with the manufacturer’s protocols. The PCR amplification conditions for these genes included a pre-denaturing step at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 52°C for 40 s, and 72°C for 2 min, followed by a final extension at 72°C for 10 min. Afterward, the PCR products were gel purified using a DNA gel extraction kit (Tiangen Biotech, China), sub-cloned into the pMD19-T vector (TaKaRa), and then transformed into Escherichia coli DH5α cells. The positive clones were confirmed by PCR and sequenced (Supplementary Figure S1). These sequences have been submitted to NCBI (Table 1).
Table 1.
Primer sequences and details of the candidate genes used in quantitative real-time PCR (qRT-PCR).
| Gene name | Accession no. | Sequence (5–3′) | Amplicon size (bp) | Primer efficiency (%) | Regression coefficient (R2) | Linear regression |
|---|---|---|---|---|---|---|
| GAPDH | KT361883.1 | F:CAAGATGGCTTCCTCGTA | 161 | 96.74 | 0.9996 | y = 3.4025x + 14.823 |
| R:GCACCACCCTCTAAATGA | ||||||
| β-ACT | MT880275.1 | F:GTATGGAATCTTGCGGTATC | 76 | 92.97 | 0.9996 | y = 3.5028x + 13.025 |
| R:AGGTCCTTACGGATGTCA | ||||||
| RPL32 | MT883789.1 | F:CCATCAATCGGATCGCTAT | 178 | 95.74 | 0.9958 | y = 3.4283x + 15.441 |
| R:ATTGTGGACCAGGACCTT | ||||||
| RPL40 | MT883790.1 | F:CAAGCGAAACTGGCGTAA | 96 | 91.43 | 0.9989 | y = 3.5461x + 14.565 |
| R:TTTGAACCGTAACCGATG | ||||||
| EF1-α | MT883788.1 | F:GCTGATTGTGGGAGTGAA | 144 | 90.37 | 0.9985 | y = 3.5767x + 13.088 |
| R:CCAGAAATGGGTACGAAA | ||||||
| α-TUB | MT880274.1 | F:GACTCCTTCAACACCTTCTT | 98 | 90.04 | 0.9981 | y = 3.5863x + 13.786 |
| R:CGGACCTCATCAACTACAG | ||||||
| β-TUB | MT883787.1 | F:CTCAACATCCAGAACAAGAAC | 168 | 88.65 | 0.9971 | y = −3.6278x + 13.901 |
| R:GGTGAACTGCTCCGAGAT | ||||||
| 18S | MT889641.1 | F:TGTCTGCCTTATCAACTTTC | 113 | 89.32 | 0.9999 | y = −3.6076x + 3.4139 |
| R:GATGTGGTAGCCGTTTCTCA | ||||||
| 28S | MT889642.1 | F:CCCGACGCAAGTCAACG | 96 | 85.02 | 0.9999 | y = −3.7423x + 10.850 |
| R:GCACAGTCCGAGACAGCAC | ||||||
| AK | MT880273.1 | F:CGATGACCACTTCCTGTT | 153 | 86.28 | 0.9970 | y = 3.7014x + 14.800 |
| R:ATGGAGATAAGACGGAGATG | ||||||
| CSP1 | KT357395.1 | F:TCTGCTGGTGCCCTATAT | 183 | 104.16 | 0.9940 | y = −3.2262x + 21.469 |
| R:TTCGTGGTTGATGAGATGG | ||||||
| SOD | MT883791.1 | F:ATTGAGGCGGATGTTGCT | 151 | 85.41 | 0.9991 | y = −3.7295x + 17.319 |
| R:TCTTGCTGAGGTTGTGGC |
Using the obtained sequences of these selected candidate reference genes, gene-specific primers used in the qRT-PCR experiments were designed with Primer Premier 5 software (Premier Biosoft, www.premierbiosoft.com). The primer design parameters were as follows: amplicon size, 76–178 bp; melting temperature, 52°C–62°C; primer length, 17–21 bp; and GC content, 40%–60% (Table 1). All the primers used in this study were synthesized by Sangon Biotechnology Co., Ltd. (Shanghai, China).
Collection of Insect Samples Under Different Biotic or Abiotic Conditions
All the insect samples collected included three biological replicates. After collection, the samples were flash-frozen in liquid nitrogen and then stored at −80°C for later total RNA extractions. Except for the temperature treatment, the growth environments of all other treatments were the same as in the section “Insects.” All the insects subjected to the treatments were given an adequate food supply except for starvation treatment.
Biotic Conditions
Developmental Stages
Samples of A. dissimilis at the egg stage (400 eggs), larval stage, including first- (100 individuals), second- (50 individuals), third- (20 individuals), fourth- (10 individuals), and fifth- (10 individuals) instar larvae, pupal stage (10 male and 10 female pupae, first day after pupation), and adult stage (10 male and 10 female moths, first day after emergence) were collected.
Tissues
The larval tissues, including the head, salivary gland, midgut, Malpighian tubule, fat body, hemolymph, and epidermis, were dissected from fourth-instar larvae of A. dissimilis under a dissecting microscope using a dissection needle and tweezer. Adult tissues, such as the antenna, head, thorax, abdomen, leg, and wing, were dissected from 10 males and 10 females (second day after emergence). After dissection, each tissue was washed three times with PBS solution (140 mM NaCl, 2.70 mM KCl, 10 mM Na2HPO4 and 1.80 mM KH2PO4, and pH 7.40), and then, pooled in the 1.5-ml RNase-free centrifuge tubes. For each sample, tissues of 20 individuals were dissected and collected.
Abiotic Conditions
Diets
Fourth-instar larvae of A. dissimilis were fed with Chinese cabbage leaf, maize seedlings, wheat seedlings, or artificial diet for 2 days, and then 10 whole individuals per treatment were collected.
Insecticide-Induced Stress
Two insecticides, chlorantraniliprole and lambda-cyhalothrin, that are widely used in the control of A. dissimilis, were chosen for this experiment. First, the LC50 values of the two insecticides to the fourth-instar larvae were measured using the artificial feed mixture method. Each insecticide was diluted with acetone to obtain different concentrations, and then, 10 μl of the diluent in various concentrations was dropped evenly on a piece of artificial diet (0.80 cm × 0.80 cm × 0.50 cm). After the solvent evaporated, the fourth-instar larvae that had been starved for 6 h were individually placed in a clean glass tube and fed the insecticide-treated artificial diet, with acetone as the control. Each treatment was repeated three times, with 24 larvae per replicate. After 48 h, the larval mortality rates of the different treatments were recorded to calculate the LC50 (Supplementary Table S2). Subsequently, another batch of fourth-instar larvae that had been starved for 6 h were treated with the LC50 value of each insecticide, and after 6, 12, 24, and 48 h, the surviving insects were collected, and each insect sample included 10 individuals.
Temperature
Fourth-instar larvae were placed in climatic chambers maintained at 4°C, 27°C, and 40°C. After 2, 6, and 12 h, 10 whole individuals per temperature treatment were collected.
Starvation
Fourth-instar larvae were collected after being starved for 12 h and 24 h. In total, 10 whole individuals were included per sample.
Total RNA Extraction, cDNA Synthesis, and qRT-PCR
The total RNA extraction and cDNA synthesis of each sample were performed as described in the section “Candidate Reference Gene Clones and qRT-PCR Primer Design.” qRT-PCR was performed using an ABI Q3 Real-time PCR System (Applied Biosystems, United States) with SYBR Green SuperReal PreMix Plus RT-PCR Kit (Tiangen Biotech, China) in a final volume of 20 μl, which contained 1 μl of cDNA, 0.6 μl of each primer (10 μM/L) as listed in Table 1, 10.0 μl of 2× SuperReal PreMix Plus, 0.4 μl of 50× ROX Reference Dye, and 7.4 μl of RNase-free ddH2O. The qRT-PCR conditions were as follows: 95°C for 15 min, followed by 40 cycles of 95°C for 10 s and 60°C for 32 s. The specificity levels of the primers used here were confirmed by melting curve analyses and 1.5% agarose gel electrophoresis. The melting curves were generated by measuring fluorescence through the dissociation temperature of the PCR product using a temperature transition rate of 0.1°C/s for all the reactions. For each gene, three biological samples were performed, with each sample measured in triplicate. To obtain the amplification efficiency (E), where E = [10(1/−slope) − 1] × 100%, and the correlation coefficient of each primer pair, 10-fold dilution series of cDNAs (1:1, 1:10, 1:100, 1:1,000, and 1:10,000) were used as templates to construct a standard curve (Pfaffl, 2001; Pfaffl et al., 2004).
Gene Expression Stability Analysis
The expression stabilities of the 10 selected candidate reference genes were evaluated using the comparative Δ cycle threshold (Ct) method and three commonly used software programs (geNorm version 3.5, NormFinder version 0.953, and BestKeeper version 1) in all the insect samples under different biotic and abiotic conditions (Vandesompele et al., 2002; Andersen et al., 2004; Pfaffl et al., 2004; Silver et al., 2006; Xie et al., 2012). Finally, a comprehensive tool, RefFinder,1 was used to rank the stability order of the selected reference genes (Xie et al., 2012). The detailed calculation method of each statistical algorithm was the same as described previously (Vandesompele et al., 2002; Andersen et al., 2004; Pfaffl et al., 2004; Silver et al., 2006; Xie et al., 2012).
Validation of the Selected Reference Gene
To validate the stability of the selected reference genes, the mRNA expression levels of a gene encoding a binding chemosensory protein (CSP1, GenBank accession no. KT357395.1) and the antioxidant enzyme gene encoding superoxide dismutase (SOD, GenBank accession no. MT883791.1) were examined in this study. The CSP1 transcript levels were assessed at different developmental stages and in different larval tissues (head, fat body, and midgut) of A. dissimilis with gene-specific primers listed in Table 1. For the different developmental stages, the expression profiles of CSP1 were estimated using EF1-α, RPL40, 18S (the three most stable reference genes), and GAPDH (the least stable reference gene) that were recommend by RefFinder as the reference genes. For the different larval tissues, the expression profiles of CSP1 were estimated using RPL40, α-TUB, RPL32 (the three most stable reference genes), and β-ACT (the least stable reference gene) as the reference genes. The SOD expression levels were determined in fourth-instar larvae of A. dissimilis exposed independently to two insecticides and four different diets using specific primers listed in Table 1. For the insecticide treatments, the expression levels of SOD were evaluated using EF1-α, β-ACT, GAPDH (the three most stable reference genes), and RPL32 (the least stable reference gene) as the reference genes. In contrast, the expression profiles of SOD in A. dissimilis feeding on different diets were evaluated using RPL40, 18S, RPL32 (the three most stable reference genes), and GAPDH (the least stable reference gene) as the reference genes. The qRT-PCR was performed as described in the section “Total RNA Extraction, cDNA Synthesis, and qRT-PCR,” and the relative expression levels of the two target genes were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2000). The statistical differences in the target gene expression levels among different treatments were analyzed by a one-way ANOVA with Tukey’s HSD multiple comparisons.
Results
Validation of Primer Specificity and qRT-PCR Amplification Efficiency
To validate the primer specificity of the 10 candidate reference genes and two target genes in the qRT-PCR experiment, 1.5% agarose gel electrophoresis and melting curve analyses were performed. The agarose gel electrophoresis of all the PCR products showed single bands of the expected sizes, and no nonspecific amplicons or primer dimmers were observed (Supplementary Figure S2). Additionally, all the melting curves showed single dissociation peaks (Supplementary Figure S3), which confirmed that the primers designed for the selected reference genes were highly specific and could be used for further qRT-PCR analyses. The amplification efficiency levels of all the primer pairs ranged from 85.02% (28S) to 104.16% (CSP1), and the correlation coefficients were all greater than 0.990 (p < 0.01), ranging from 0.994 (CSP1) to 0.9999 (18S and 28S; Table 1), which reflected their stability and specificity.
Transcriptional Profiles of the Candidate Reference Genes
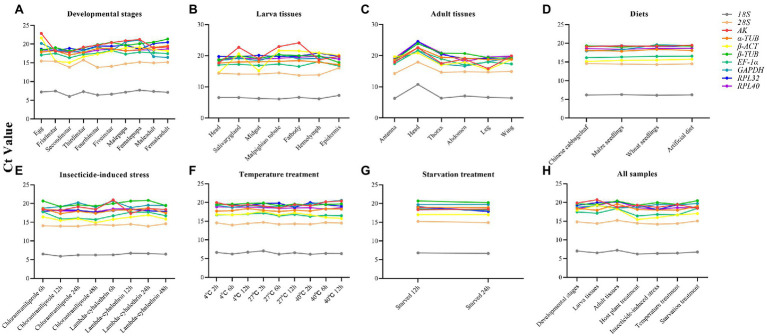
The expression profiles of the candidate reference genes in different insect samples were evaluated by comparing the Ct values. The Ct values of these reference genes varied widely, ranging from 6.03 (18S) to 24.57 (RPL40; Supplementary Table S3), for all the experimental conditions (Figure 1). The 18S gene showed the greatest expression levels, with the lowest Ct values, whereas the AK gene showed the lowest levels. The expression levels of each gene under different conditions revealed that most of these genes remained stable under different conditions, but some genes varied widely, such as AK in larval tissues (Figure 1).
Figure 1.
Variation in the reference genes expression using the distribution of Ct values in line charts. (A) Developmental stages; (B) Larvae tissues; (C) Adult tissues; (D) Diets; (E) Insecticides; (F) Temperature; (G) Starvation; (H) All samples.
Expression Stability of Candidate Reference Genes Under Biotic Conditions
Developmental Stages
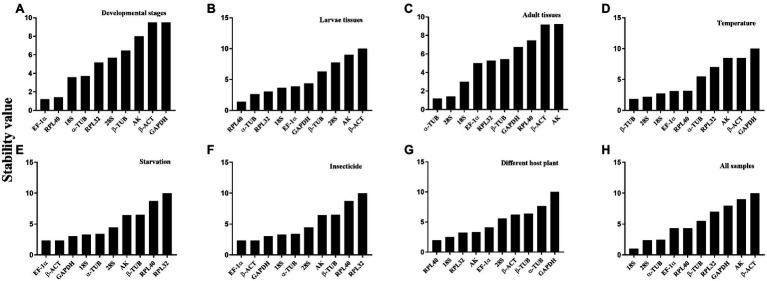
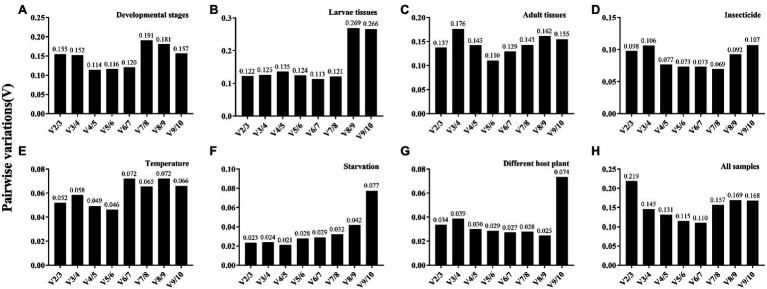
The overall stability ranking orders recommended by geNorm, NormFinder, and ΔCt methods were similar, showing EF1-α and RPL40 as the two most stable genes, and GAPDH and β-ACT as the least stable genes. The ranking order determined by BestKeeper showed GAPDH and RPL32 being the most stable genes and AK and β-ACT being the least stable genes (Table 2). The stability analysis of RefFinder ranked the genes from most stable to least stable as follows: EF1-α > RPL40 > 18S > α-TUB > RPL32 > 28S > β-TUB > AK > β-ACT > GAPDH (Figure 2A). The geNorm analysis showed that the pairwise variation values of V4/5, V5/6, and V6/7 were all less than 0.15 (Figure 3A). On the basis of cost and convenience, according to the geNorm manual, we selected three as the optimal number of reference genes. Consequently, the combination of EF1-α, RPL40, and 18S, the three most stable genes, was recommended as being suitable for normalizing qRT-PCR data at different developmental stages of A. dissimilis (Table 3).
Table 2.
Expression stability of the candidate reference genes in A. dissimilis under different biotic conditions.
| Biotic condition | Reference gene | geNorm | Normfinder | Bestkeeper | ΔCt | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | ||
| Developmental stages | 18S | 0.66 | 5 | 0.32 | 3 | 0.74 | 8 | 0.52 | 3 |
| 28S | 0.73 | 6 | 0.53 | 5 | 0.50 | 3 | 0.71 | 5 | |
| AK | 0.95 | 7 | 0.81 | 8 | 0.79 | 9 | 1.54 | 8 | |
| EF1-α | 0.44 | 1 | 0.31 | 1 | 0.64 | 7 | 0.51 | 2 | |
| GAPDH | 1.25 | 9 | 1.07 | 9 | 0.48 | 2 | 1.58 | 9 | |
| RPL32 | 0.58 | 3 | 0.59 | 6 | 0.33 | 1 | 0.76 | 6 | |
| RPL40 | 0.44 | 1 | 0.32 | 2 | 0.56 | 5 | 0.29 | 1 | |
| α-TUB | 0.49 | 2 | 0.42 | 4 | 0.62 | 6 | 0.71 | 4 | |
| β-ACT | 1.12 | 8 | 1.08 | 10 | 0.84 | 10 | 1.93 | 10 | |
| β-TUB | 0.61 | 4 | 0.64 | 7 | 0.54 | 4 | 1.00 | 7 | |
| Larval tissues | 18S | 0.58 | 4 | 0.54 | 6 | 0.07 | 2 | 0.39 | 1 |
| 28S | 0.80 | 7 | 0.77 | 8 | 0.05 | 1 | 0.78 | 7 | |
| AK | 1.17 | 8 | 1.52 | 9 | 0.79 | 9 | 2.51 | 9 | |
| EF1-α | 0.40 | 2 | 0.51 | 5 | 0.16 | 4 | 0.52 | 5 | |
| GAPDH | 0.66 | 5 | 0.24 | 2 | 0.75 | 8 | 0.72 | 6 | |
| RPL32 | 0.37 | 1 | 0.54 | 7 | 0.16 | 3 | 0.43 | 4 | |
| RPL40 | 0.37 | 1 | 0.21 | 1 | 0.59 | 7 | 0.40 | 2 | |
| α-TUB | 0.47 | 3 | 0.38 | 3 | 0.29 | 5 | 0.42 | 3 | |
| β-ACT | 1.50 | 9 | 1.83 | 10 | 0.97 | 10 | 3.02 | 10 | |
| β-TUB | 0.72 | 6 | 0.44 | 4 | 0.55 | 6 | 0.92 | 8 | |
| Adult tissues | 18S | 0.41 | 2 | 0.30 | 3 | 0.97 | 7 | 1.74 | 3 |
| 28S | 0.35 | 1 | 0.29 | 2 | 0.98 | 10 | 1.34 | 1 | |
| AK | 1.05 | 8 | 0.90 | 9 | 0.86 | 3 | 2.31 | 10 | |
| EF1-α | 0.66 | 4 | 0.49 | 5 | 0.91 | 5 | 1.75 | 5 | |
| GAPDH | 0.90 | 7 | 0.67 | 8 | 0.85 | 2 | 1.74 | 4 | |
| RPL32 | 0.70 | 5 | 0.46 | 4 | 0.97 | 8 | 2.19 | 8 | |
| RPL40 | 0.78 | 6 | 0.66 | 7 | 0.93 | 6 | 2.26 | 9 | |
| α-TUB | 0.35 | 1 | 0.19 | 1 | 0.98 | 9 | 1.52 | 2 | |
| β-ACT | 1.18 | 9 | 1.06 | 10 | 0.70 | 1 | 1.92 | 7 | |
| β-TUB | 0.58 | 3 | 0.61 | 6 | 0.90 | 4 | 1.90 | 6 | |
Figure 2.
Expression stability ranking order of the candidate reference genes for A. dissimilis at different conditions according to ReFinder. A lower-ranking value of the gene denotes it has more stable expression stability. (A) Developmental stages; (B) Larvae tissues; (C) Adult tissues; (D) Temperature; (E) Starvation; (F) Insecticides; (G) Diets; (H) All samples.
Figure 3.
Pairwise variation (V) analysis of the candidate reference genes in A. dissimilis under different conditions. Pairwise variation analysis is performed by geNorm to determine the optimal number of reference genes for normalization in different conditions. Each pairwise variation value is compared with 0.15, and when Vn/Vn+1 below 0.15, the optimal number of reference genes is n. (A) Developmental stages; (B) Larvae tissues; (C) Adult tissues; (D) Insecticides; (E) Temperature; (F) Starvation; (G) Diets; (H) All samples.
Table 3.
Suitable reference genes recommended for A. dissimilis at different experimental conditions.
| Factors | Experimental conditions | Recommended reference genes | ||
|---|---|---|---|---|
| Biotic factors | Developmental stages | EF1-α | RPL40 | 18S |
| Larval tissues | RPL40 | α-TUB | ||
| Adult tissues | α-TUB | 28S | ||
| Abiotic factors | Diets | RPL40 | 18S | |
| Insecticide treatment | EF1-α | β-ACT | ||
| Temperature treatment | β-TUB | 28S | ||
| Starvation treatment | EF1-α | β-ACT | ||
| All samples | 18S | 28S | α-TUB | |
Larval Tissues
For different larval tissues, all four programs identified β-ACT and AK as the least stable genes. RPL32 and RPL40 were identified as the most stable genes by geNorm. NormFinder, BestKeeper, and the ΔCt method identified RPL40, 28S, and 18S as the most stable gene, respectively (Table 2). RefFinder ranked the candidate reference genes from high to low stability as follows: RPL40 > α-TUB > RPL32 > 18S > EF1-α > GAPDH > β-TUB > 28S > AK > β-ACT (Figure 2B). The pairwise variation showed that two reference genes were suitable for normalizing gene expression in different larval tissues of A. dissimilis because the value of V2/3 was less than 0.15 (Figure 3B; Table 3).
Adult Tissues
In the adult tissue assessment, 28S, α-TUB, and 18S were identified as the top three stable genes by all the programs, except for BestKeeper, which identified the three best-suited genes as β-ACT, GAPDH, and AK. The least stable genes were identified by NormFinder and geNorm as AK and β-ACT, by BestKeeper as 28S and α-TUB, and by the ΔCt method as AK and RPL40 (Table 2). RefFinder ranked the stability order as follows: α-TUB > 28S > 18S > EF1-α > RPL32 > β-TUB > GAPDH > RPL40 > β-ACT > AK (Figure 2C). The pairwise variation value of V2/3 in the geNorm analysis was less than 0.15 (Figure 3C). Thus, the combination of α-TUB and 28S was suitable for normalizing qRT-PCR data in adult tissue samples of A. dissimilis (Table 3).
Dietary Treatments
Both geNorm and the ΔCt method ranked 18S, RPL32, and 28S as the top three reference genes for A. dissimili receiving different dietary treatments. NormFinder placed RPL40 as the most stable gene, whereas BestKeeper identified 28S as the most stable genes. In addition, GAPDH was identified as the least stable gene by all the methods, except BestKeeper, which identified EF1-α as the least stable gene (Table 2). The ranking of the most stable genes by the RefFinder analysis was as follows: RPL40 > 18S > RPL32 > AK > EF1-α > 28S > β-ACT > β-TUB > α-TUB > GAPDH (Figure 2G), and the pairwise variation analysis showed that the value of V2/3 was less than 0.15 (Figure 3G). Thus, RPL40 and 18S are appropriate to normalize the gene expression profiles in A. dissimili larvae fed on different diets (Table 3).
Expression Stability of Candidate Reference Genes Under Abiotic Conditions
Insecticide Treatments
The stability analyses performed by geNorm, NormFinder, and ΔCt algorithms identified AK, GAPDH, and EF1-α as the least stable genes, although their rank orders were different. The geNorm analysis inferred that α-TUB and β-TUB were the most stable genes, whereas NormFinder, BestKeeper, and the ΔCt method identified RPL40, 28S, and RPL32 as the most stable gene, respectively (Table 4). The RefFinder software ranked the expression stability of the reference genes as follows: EF1-α > β-ACT > GAPDH > 18S > α-TUB > 28S > AK > β-TUB > RPL40 > RPL32 (Figure 2D). The pairwise variation analysis showed that the value of V2/3 was less than 0.15 (Figure 3D). Thus, the two reference genes EF1-α and β-ACT are sufficient to normalize gene expression under these experimental conditions (Table 3).
Table 4.
Expression stability of the candidate reference genes in A. dissimilis under different abiotic conditions.
| Abiotic condition | Reference gene | geNorm | Normfinder | Bestkeeper | ΔCt | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | ||
| Diets | 18S | 0.07 | 1 | 0.13 | 8 | 0.77 | 3 | 0.07 | 1 |
| 28S | 0.09 | 2 | 0.16 | 9 | 0.66 | 1 | 0.12 | 3 | |
| AK | 0.13 | 3 | 0.06 | 3 | 0.92 | 9 | 0.16 | 5 | |
| EF1-α | 0.16 | 5 | 0.03 | 2 | 0.99 | 10 | 0.19 | 7 | |
| GAPDH | 0.31 | 9 | 0.51 | 10 | 0.76 | 2 | 0.80 | 10 | |
| RPL32 | 0.07 | 1 | 0.13 | 7 | 0.80 | 5 | 0.12 | 2 | |
| RPL40 | 0.14 | 4 | 0.01 | 1 | 0.88 | 8 | 0.12 | 4 | |
| α-TUB | 0.20 | 8 | 0.13 | 6 | 0.87 | 7 | 0.21 | 8 | |
| β-ACT | 0.17 | 6 | 0.09 | 4 | 0.79 | 4 | 0.26 | 9 | |
| β-TUB | 0.19 | 7 | 0.12 | 5 | 0.85 | 6 | 0.18 | 6 | |
| Insecticide | 18S | 0.37 | 3 | 0.14 | 2 | 0.82 | 7 | 0.26 | 2 |
| 28S | 0.46 | 6 | 0.34 | 7 | 0.15 | 1 | 0.27 | 3 | |
| AK | 0.69 | 9 | 0.74 | 10 | 0.32 | 2 | 1.04 | 10 | |
| EF1-α | 0.50 | 7 | 0.47 | 8 | 0.84 | 8 | 0.86 | 9 | |
| GAPDH | 0.59 | 8 | 0.48 | 9 | 0.38 | 3 | 0.68 | 8 | |
| RPL32 | 0.43 | 5 | 0.24 | 4 | 0.46 | 4 | 0.25 | 1 | |
| RPL40 | 0.39 | 4 | 0.11 | 1 | 0.72 | 5 | 0.31 | 4 | |
| α-TUB | 0.21 | 1 | 0.20 | 3 | 0.91 | 10 | 0.57 | 5 | |
| β-ACT | 0.28 | 2 | 0.28 | 5 | 0.82 | 6 | 0.62 | 6 | |
| β-TUB | 0.21 | 1 | 0.31 | 6 | 0.88 | 9 | 0.67 | 7 | |
| Temperature | 18S | 0.13 | 1 | 0.12 | 3 | 0.89 | 10 | 0.30 | 6 |
| 28S | 0.20 | 3 | 0.03 | 1 | 0.79 | 7 | 0.24 | 3 | |
| AK | 0.45 | 8 | 0.40 | 8 | 0.46 | 4 | 0.60 | 9 | |
| EF1-α | 0.13 | 1 | 0.15 | 6 | 0.83 | 8 | 0.28 | 5 | |
| GAPDH | 0.51 | 9 | 0.45 | 10 | 0.28 | 2 | 0.63 | 10 | |
| RPL32 | 0.33 | 6 | 0.35 | 7 | −0.05 | 1 | 0.41 | 7 | |
| RPL40 | 0.23 | 4 | 0.13 | 4 | 0.42 | 3 | 0.18 | 1 | |
| α-TUB | 0.25 | 5 | 0.14 | 5 | 0.68 | 6 | 0.28 | 4 | |
| β-ACT | 0.39 | 7 | 0.44 | 9 | 0.46 | 5 | 0.58 | 8 | |
| β-TUB | 0.16 | 2 | 0.04 | 2 | 0.89 | 9 | 0.22 | 2 | |
| Starvation | 18S | 0.08 | 3 | 0.03 | 3 | 0.84 | 5 | 0.11 | 5 |
| 28S | 0.14 | 6 | 0.03 | 2 | 0.91 | 7 | 0.23 | 7 | |
| AK | 0.11 | 5 | 0.24 | 8 | −0.16 | 1 | 0.11 | 4 | |
| EF1-α | 0.09 | 4 | 0.01 | 1 | 0.94 | 9 | 0.14 | 6 | |
| GAPDH | 0.03 | 1 | 0.16 | 7 | 0.35 | 2 | 0.03 | 2 | |
| RPL32 | 0.33 | 9 | 0.54 | 10 | 0.92 | 8 | 0.91 | 10 | |
| RPL40 | 0.22 | 8 | 0.19 | 8 | 0.86 | 6 | 0.47 | 9 | |
| α-TUB | 0.06 | 2 | 0.09 | 5 | 0.83 | 4 | 0.06 | 3 | |
| β-ACT | 0.03 | 1 | 0.14 | 6 | 0.38 | 3 | 0.01 | 1 | |
| β-TUB | 0.17 | 7 | 0.04 | 4 | 0.97 | 10 | 0.32 | 8 | |
Temperature Treatment
The top three least stable genes determined by geNorm, NormFinder, and the ΔCt method were GAPDH, AK, and β-ACT, whereas the determination by BestKeeper was 18S, β-TUB, and EF1-α. GeNorm identified 18S and EF1-α as the most stable genes. NormFinder, BestKeeper, and the ΔCt method identified 28S, RPL32, and RPL40 as the most stable gene, respectively (Table 4). The stability ranking of the reference genes from the most stable to least stable gene by the RefFinder analysis was as follows: β-TUB > 28S > 18S > EF1-α > RPL40 > α-TUB > RPL32 > AK > β-ACT > GAPDH (Figure 2E). The pairwise variation values were all less than 0.15 (Figure 3E). Thus, β-TUB and 28S are sufficient to normalize qRT-PCR data from the temperature-treated samples (Table 3).
Starvation Treatment
The top three ranked reference genes as determined by geNorm and the ΔCt method for insect samples after starvation were β-ACT, GAPDH, and α-TUB. NormFinder identified EF1-α, 28S, and 18S as the top three suitable reference genes, whereas BestKeeper identified AK, GAPDH, and β-ACT. The least stable gene identified by geNorm, NormFinder, and the ΔCT method was RPL32, whereas β-TUB was identified by BestKeeper (Table 4). The reference gene stability ranking, from most to least stable, as determined by the RefFinder analysis was as follows: EF1-α > β-ACT > GAPDH > 18S > α-TUB > 28S > AK > β-TUB > RPL40 > RPL32 (Figure 2F). The pairwise variation analysis showed that all the values were less than 0.15 (Figure 3F). Therefore, EF1-α and β-ACT are appropriate for normalizing gene expression data collected under starved conditions (Table 3).
Ranking of Athetis dissimilis Candidate Reference Genes Across All the Samples
For all the samples, the stability ranking results determined by geNorm were similar to those obtained by the ΔCt method, which identified 28S, 18S, and α-TUB as the three most stable genes. NormFinder identified 18S, α-TUB, and RPL40 as the three most stable genes, whereas BestKeeper selected GAPDH, AK, and β-ACT as the most appropriate candidate genes (Table 5). The RefFinder analysis ranked the candidate reference genes from the most stable to the least stable as follows: 18S > 28S > α-TUB > EF1-α > RPL40 > β-TUB > RPL32 > GAPDH > AK > β-ACT (Figure 2H). The geNorm analysis determined that the pairwise variation values of V3/4 were less than 0.15 (Figure 3H). Thus, 18S, 28S, and α-TUB are recommended as the most stable reference genes for normalizing qRT-PCR data from all the samples (Table 3).
Table 5.
Expression stability of the candidate reference genes in A. dissimilis under all samples.
| Reference gene | geNorm | Normfinder | Bestkeeper | ΔCt | ||||
|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | |
| 18S | 0.37 | 1 | 0.30 | 1 | 0.85 | 10 | 0.75 | 2 |
| 28S | 0.37 | 1 | 0.43 | 4 | 0.71 | 4 | 0.70 | 1 |
| AK | 1.08 | 8 | 0.96 | 9 | 0.61 | 2 | 1.62 | 9 |
| EF1-α | 0.62 | 3 | 0.44 | 5 | 0.78 | 8 | 0.98 | 4 |
| GAPDH | 0.91 | 7 | 0.79 | 8 | 0.57 | 1 | 1.11 | 8 |
| RPL32 | 0.77 | 6 | 0.52 | 7 | 0.75 | 5 | 1.09 | 7 |
| RPL40 | 0.72 | 5 | 0.42 | 3 | 0.79 | 9 | 1.05 | 6 |
| α-TUB | 0.58 | 2 | 0.38 | 2 | 0.77 | 7 | 0.86 | 3 |
| β-ACT | 1.23 | 9 | 1.15 | 10 | 0.66 | 3 | 2.00 | 10 |
| β-TUB | 0.67 | 4 | 0.51 | 6 | 0.76 | 6 | 0.97 | 5 |
Target Gene Validation Using the Selected Reference Genes
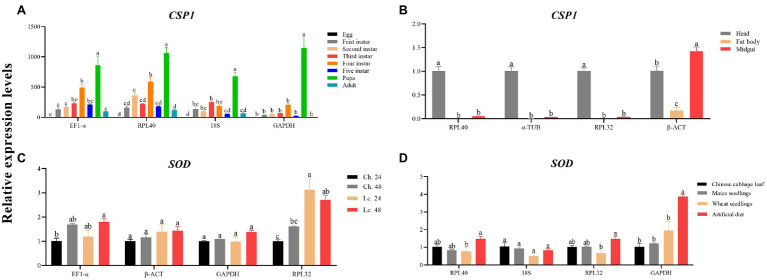
The transcript levels of CSP1 and SOD were assessed under various experimental conditions to verify the performance levels of the selected reference genes. For developmental stages, the qRT-PCR results using either EF1-α and RPL40 or 18S as normalizers were more consistent than those using GAPHD as the normalizer (Figure 4A). The same results were achieved using larval tissues (Figure 4B). Furthermore, for the insecticide treatments, the expression profile of SOD clearly exhibited differences when RPL32 was used in the normalization (Figure 4C). Moreover, for different dietary treatments, the results were much different using RPL40 and 18S or RPL32 as reference gene(s) compared with using GAPDH (Figure 4D).
Figure 4.
Validation of the candidate reference genes suitable for A. dissimilis. (A) CSP1 relative expression levels of A. dissimilis at different developmental stages. (B) CSP1 relative expression levels of A. dissimilis in three different larval tissues. (C) SOD relative expression levels of A. dissimilis treated with different insecticides (Ch.: chlorantraniliprole, Lc.: lambda-cyhalothrin) for various times. (D) SOD relative expression levels of A. dissimilis feeding on different diets. Values are means ± SEM. Different letters indicate significant differences (p < 0.05, one-way ANOVA followed by Tukey’s HSD multiple comparison).
Discussion
Quantitative real-time PCR is an important method in gene expression research that uses reference genes as standards to calibrate the expression levels of target genes. Therefore, it is essential to select the appropriate reference genes. At present, reference genes have been screened in a variety of insects (Zhang et al., 2015; Yin et al., 2020; Xie et al., 2021), but stable reference genes for A. dissimilis under various conditions remain undetermined. Recently, the transcriptomes of different tissues of A. dissimilis were sequenced (Dong et al., 2016; Sun et al., 2016; Liu et al., 2019), indicating that more genes can be studied in depth. Therefore, it is urgent to identify reference genes that are stably expressed under different experimental conditions.
In this study, four commonly used tools, geNorm, NormFinder, BestKeeper, and ΔCt, were used to analyze the expression stability levels of 10 candidate reference genes (18S, 28S, AK, EF1-α, GAPDH, RPL32, RPL40, α-TUB, β-ACT, and β-TUB) under different biotic and abiotic conditions. Because the four tools may determine different gene stability levels owing to the use of different algorithms, the web-based analysis tool RefFinder, which integrates all four major calculation programs, was used to evaluate and screen the optimal reference genes.
The optimal number of reference genes can be determined through the geNorm analysis by using the paired mutation value (Vn/n + 1; Vandesompele et al., 2002). The results showed that EF1-α, RPL40, and 18S are suitable for the normalization of data from different developmental stages, RPL40 and α-TUB for larval tissues, α-TUB and 28S for adult tissues, RPL40 and 18S for dietary treatments, EF1-α and β-ACT for insecticide treatments, β-TUB and 28S for temperature treatments, EF1-α and β-ACT for starvation treatments, and 18S, 28S, and α-TUB for all the samples.
Elongation factor 1 alpha was identified as the most stable reference gene of A. dissimilis in different developmental stages and insecticide treatments. In translation, EF1-α encodes a protein that catalyzes the GTP-dependent binding of aminoacyl tRNA to the acceptor site of the ribosome (Ponton et al., 2011). Consequently, it is one of the most abundant proteins in cells and is highly conserved among species (Xie et al., 2021). EF1-α is regarded as a suitable reference gene in other organisms under different conditions, such as Spodoptera litura at different temperatures (Lepidoptera: Noctuidae; Lu et al., 2013), Locusta migratoria at different developmental stages (Orthoptera: Acrididae; Yang et al., 2014), Frankliniella occidentalis (Thysanoptera: Thripidae; Zheng et al., 2014), and Hippodamia convergens (Coleoptera: Coccinellidae; Pan et al., 2015) under different developmental stages and temperature treatments, and Sesamia inferens in different tissues and developmental stages (Lepidoptera: Noctuidae; Sun et al., 2015). Additionally, EF1-α is the first in the reference ranking provided by the ICG website (Sang et al., 2018).
Here, RPL40 was considered as the most stable reference gene for dietary treatments and larval tissues. The protein encoded by RPL40 forms the structure of the ribosome and plays very important roles in cell life activities. Many genes in the RPL family are used as reference genes for stable expression in insects, such as Solenopsis invicta (Hymenoptera, Formicidae; Cheng et al., 2013), Anastrepha obliqua (Diptera, Tephritidae; Nakamura et al., 2016), and Cimex lectularius (Hemiptera, Cimicidae; Mamidala et al., 2011). On the ICG website, RPL was ranked as the fifth most stable reference gene.
Alpha-tubulin and β-TUB are the most stable reference genes expressed in the adult tissues and temperature treatments of A. dissimilis. TUB belongs to the structural gene family of eukaryotes, helping to form the basic components of microtubules and skeletons, and it regulates cell division, shape, movement, and intracellular activities (Nielsen et al., 2010). It has also been used as reference gene in many organisms (Huis et al., 2010; Kozera and Rapacz, 2013). In the reference ranking provided by the ICG website, the TUB family ranked sixth. TUB is also used as the best reference gene in insects under many conditions, such as the geographic populations of Nilaparvata lugens (Hemiptera: Delphacidae; Miao et al., 2014), developmental stages and temperature treatments of Sogatella furcifera (Hemiptera: Delphacidae; An et al., 2016), and temperature treatments of Bemisia tabaci (Dai et al., 2017).
Before this study, GAPDH was used as the reference gene when studying the gene expression of adult A. dissimilis (Sun et al., 2016; Liu et al., 2019). However, it is not an appropriate choice, and this study provides a basis for selecting appropriate reference genes.
RefFinder recommended 18S, 28S, and α-TUB as the most stable reference genes for A. dissimilis for all the tested samples. Ribosomal RNAs (rRNAs), including 18S rRNA and 28S rRNA, mainly participate in protein synthesis and are highly expressed in all biological cells. Because the RNA polymerase that synthesizes rRNA is different from the RNA polymerase that synthesizes mRNA, and the regulation of rRNA synthesis is not related to the mRNA level, rRNA has been regarded as an ideal reference gene (Bustin, 2000), such as in different tissues of Rhodnius prolixus (Hemiptera, Reduviidae; Paim et al., 2012) and in different body parts of N. lugens (Hemiptera: Delphacidae; Miao et al., 2014). 18S ranked eighth in the reference ranking provided by the ICG website. Although 18S is identified as the most stable reference gene under all simples in A. dissimilis, the expression levels of 18S were much higher than the target genes SOD and CSP1 used for verification, the high expression may mask our correct understanding of the actual expression of target genes, and the same goes for the 28S. Therefore, in future research, we should consider this issue and further consider the selection of reference genes according to the expression of our target genes.
An unstable reference gene is insufficient to normalize the gene expression data or may generate the wrong interpretation. According to the results of the present research, the three most stable genes and one least stable gene were used for the normalization of the expression levels of CSP1 and SOD in developmental stages, larval tissues, insecticide treatments, and dietary treatments of A. dissimilis to validate their stability. The result shows that unstable reference gene is insufficient to normalize the gene expression data or may generate the wrong interpretation Therefore, the selection and validation of the best reference genes are crucial to determine the accuracy of the expression patterns of different genes in A. dissimilis. This will benefit to the future studies on gene functions in A. dissimilis and other insects and will facilitate the generation of more reliable and accurate data on gene expression in A. dissimilis.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
JT, XG, and MZ conceived and designed the research. JT and SD conducted the experiments. JT and SS analyzed the data. JT, GL, and MZ wrote the article. All authors have read and agreed to the published version of the article.
Funding
This research was supported by the National Key Research and Development Program of China (Nos. 2018YFD0200600 and 2017YFD0201700) and the National Natural Science Foundation of China (No. 31801735).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.842195/full#supplementary-material
References
- An X. K., Hou M. L., Liu Y. D. (2016). Reference gene selection and evaluation for gene expression studies using qRT-PCR in the white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae). J. Econ. Entomol. 109, 879–886. doi: 10.1093/jee/tov333, PMID: [DOI] [PubMed] [Google Scholar]
- Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496, PMID: [DOI] [PubMed] [Google Scholar]
- Brym P., Rusc A., Kaminski S. (2013). Evaluation of reference genes for qRT-PCR gene expression studies in whole blood samples from healthy and leukemia-virus infected cattle. Vet. Immunol. Immunopathol. 153, 302–307. doi: 10.1016/j.vetimm.2013.03.004, PMID: [DOI] [PubMed] [Google Scholar]
- Bustin S. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25, 169–193. doi: 10.1677/jme.0.0250169, PMID: [DOI] [PubMed] [Google Scholar]
- Bustin S., Benes V., Nolan T., Pfaffl M. W. (2005). Quantitative real-time RT-PCR--a perspective. J. Mol. Endocrinol. 34, 597–601. doi: 10.1677/jme.1.01755, PMID: [DOI] [PubMed] [Google Scholar]
- Che Z., Tian Y., Yang J., Liu S., Jiang J., Hu M., et al. (2019). Screening of insecticidal activity of podophyllotoxin analogues against Athetis dissimilis. Nat. Prod. Commun. 14, 117–120. doi: 10.1177/1934578X1901400131 [DOI] [Google Scholar]
- Cheng D., Zhang Z., He X., Liang G. (2013). Validation of reference genes in Solenopsis invicta in different developmental stages, castes and tissues. PLoS One 8:e57718. doi: 10.1371/journal.pone.0057718, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai T. M., Lu Z. C., Liu W. X., Wan F. H. (2017). Selection and validation of reference genes for qRT-PCR analysis during biological invasions: the thermal adaptability of Bemisia tabaci MED. PLoS One 12:e0173821. doi: 10.1371/journal.pone.0173821, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derveaux S., Vandesompele J., Hellemans J. (2010). How to do successful gene expression analysis using real-time PCR. Methods 50, 227–230. doi: 10.1016/j.ymeth.2009.11.001, PMID: [DOI] [PubMed] [Google Scholar]
- Dong J., Song Y., Li W., Shi J., Wang Z. (2016). Identification of putative chemosensory receptor genes from the Athetis dissimilis antennal transcriptome. PLoS One 11:e0147768. doi: 10.1371/journal.pone.0147768, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige S., Pfaffl M. W. (2006). RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 27, 126–139. doi: 10.1016/j.mam.2005.12.003, PMID: [DOI] [PubMed] [Google Scholar]
- Fu W., Xie W., Zhang Z., Wang S., Wu Q., Liu Y., et al. (2013). Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 9, 792–802. doi: 10.7150/ijbs.5862, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. K., Zhang S., Luo J. Y., Wang C. Y., Lu L. M., Zhang L. J., et al. (2017). Identification and validation of reference genes for gene expression analysis in Aphidius gifuensis (hymenoptera: Aphidiidae). PLoS One 12:e0188477. doi: 10.1371/journal.pone.0188477, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glare E. M., Divjak M., Bailey M. J., Walters E. H. (2002). β-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax 57, 765–770. doi: 10.1136/thorax.57.9.765, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenin S., Mauriat M., Pelloux J., Van Wuytswinkel O., Bellini C., Gutierrez L. (2009). Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 60, 487–493. doi: 10.1093/jxb/ern305, PMID: [DOI] [PubMed] [Google Scholar]
- Guo H., Jiang L., Xia Q. (2016). Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol. Gen. Genomics. 291, 999–1004. doi: 10.1007/s00438-015-1125-4, PMID: [DOI] [PubMed] [Google Scholar]
- Guo T. T., Li L. L., Men X. Y., Lu Z. B., Chen H., Wang Z. Y., et al. (2017). Impact of temperature on the growth and development of Athetis dissimilis (Lepidoptera: Noctuidae). J. Econ. Entomol. 110, 274–281. doi: 10.1093/jee/tow229, PMID: [DOI] [PubMed] [Google Scholar]
- Hu Y., Fu H., Qiao H., Sun S., Zhang W., Jin S., et al. (2018). Validation and evaluation of reference genes for quantitative real-time PCR in Macrobrachium nipponense. Int. J. Mol. Sci. 19:2258. doi: 10.3390/ijms19082258, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S., Zumla A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6, 279–284. doi: 10.1038/sj.gene.6364190, PMID: [DOI] [PubMed] [Google Scholar]
- Huis R., Hawkins S., Neutelings G. (2010). Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 10:71. doi: 10.1186/1471-2229-10-71, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janska A., Hodek J., Svoboda P., Zamecnik J., Prasil I. T., Vlasakova E., et al. (2013). The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Mol. Gen. Genomics. 288, 639–649. doi: 10.1007/s00438-013-0774-4, PMID: [DOI] [PubMed] [Google Scholar]
- Kozera B., Rapacz M. (2013). Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406. doi: 10.1007/s13353-013-0173-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. L., Sun S. J., Khuhro S. A., Elzaki M. E. A., Yan Q., Dong S. L. (2019). Functional characterization of pheromone receptors in the moth Athetis dissimilis (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 158, 69–76. doi: 10.1016/j.pestbp.2019.04.011, PMID: [DOI] [PubMed] [Google Scholar]
- Livak K., Schmittgen T. (2000). Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ct method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262, PMID: [DOI] [PubMed] [Google Scholar]
- Lu Y., Yuan M., Gao X., Kang T., Zhan S., Wan H., et al. (2013). Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS One 8:e68059. doi: 10.1371/journal.pone.0068059, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K. S., Li F., Liang P. Z., Chen X. W., Liu Y., Gao X. W. (2016). Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae). J. Insect Sci. 16:17. doi: 10.1093/jisesa/iew003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty A., Purohit G. K., Mohanty S., Nayak N. R., Mohanty B. P. (2017). Suitable reference gene for quantitative real-time PCR analysis of gene expression in gonadal tissues of minnow Puntius sophore under high-temperature stress. BMC Genomics 18:617. doi: 10.1186/s12864-017-3974-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidala P., Rajarapu S. P., Jones S. C., Mittapalli O. (2011). Identification and validation of reference genes for quantitative real-time polymerase chain reaction in Cimex lectularius. J. Med. Entomol. 48, 947–951. doi: 10.1603/ME10262, PMID: [DOI] [PubMed] [Google Scholar]
- Miao Y., Lu Y., Zhu X., Wan H., Shakeel M., Zhan S., et al. (2014). Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One 9:e86503. doi: 10.1371/journal.pone.0086503, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A. M., Chahad-Ehlers S., Lima A. L., Taniguti C. H., Sobrinho I., Jr., Torres F. R., et al. (2016). Reference genes for accessing differential expression among developmental stages and analysis of differential expression of OBP genes in Anastrepha obliqua. Sci. Rep. 6:17480. doi: 10.1038/srep17480, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen K., Smeets K., Mulder M., Hendriks J. J., Ameloot M. (2010). Selection of reference genes for gene expression studies in rat oligodendrocytes using quantitative real time PCR. J. Neurosci. Methods 187, 78–83. doi: 10.1016/j.jneumeth.2009.12.018, PMID: [DOI] [PubMed] [Google Scholar]
- Nielsen M. G., Gadagkar S. R., Gutzwiller L. (2010). Tubulin evolution in insects: gene duplication and subfunctionalization provide specialized isoforms in a functionally constrained gene family. BMC Evol. Biol. 10:113. doi: 10.1186/1471-2148-10-113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paim R. M., Pereira M. H., Di Ponzio R., Rodrigues J. O., Guarneri A. A., Gontijo N. F., et al. (2012). Validation of reference genes for expression analysis in the salivary gland and the intestine of Rhodnius prolixus (Hemiptera, Reduviidae) under different experimental conditions by quantitative real-time PCR. BMC. Res. Notes 5:128. doi: 10.1186/1756-0500-5-128, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Yang X., Siegfried B. D., Zhou X. (2015). A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS One 10:e0125868. doi: 10.1371/journal.pone.0125868, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47, PMID: [DOI] [PubMed] [Google Scholar]
- Ponton F., Chapuis M. P., Pernice M., Sword G. A., Simpson S. J. (2011). Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57, 840–850. doi: 10.1016/j.jinsphys.2011.03.014, PMID: [DOI] [PubMed] [Google Scholar]
- Radonic A., Thulke S., Mackay I. M., Landt O., Siegert W., Nitsche A. (2004). Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862. doi: 10.1016/j.bbrc.2003.11.177, PMID: [DOI] [PubMed] [Google Scholar]
- Renard M., Vanhauwaert S., Vanhomwegen M., Rihani A., Vandamme N., Goossens S., et al. (2018). Expressed repetitive elements are broadly applicable reference targets for normalization of reverse transcription-qPCR data in mice. Sci. Rep. 8:7642. doi: 10.1038/s41598-018-25389-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang J., Wang Z., Li M., Cao J., Niu G., Xia L., et al. (2018). ICG: a wiki-driven knowledgebase of internal control genes for RT-qPCR normalization. Nucleic Acids Res. 46, D121–D126. doi: 10.1093/nar/gkx875, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvey S., Thompson E. W., Matthaei K., Lea R. A., Irving M. G., Griffiths L. R. (2001). Beta-actin--an unsuitable internal control for RT-PCR. Mol. Cell. Probes 15, 307–311. doi: 10.1006/mcpr.2001.0376, PMID: [DOI] [PubMed] [Google Scholar]
- Silver N., Best S., Jiang J., Thein S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. doi: 10.1186/1471-2199-7-33, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Lu M. X., Tang X. T., Du Y. Z. (2015). Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens (Lepidoptera: Noctuidae). PLoS One 10:e0115979. doi: 10.1371/journal.pone.0115979, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. F., Meng Y. P., Cui G. M., Cao Q. F., Li J., Liang A. H. (2009). Selection of housekeeping genes for gene expression studies on the development of fruit bearing shoots in Chinese jujube (Ziziphus jujube mill.). Mol. Biol. Rep. 36, 2183–2190. doi: 10.1007/s11033-008-9433-y, PMID: [DOI] [PubMed] [Google Scholar]
- Sun H., Song Y., Du J., Wang X., Cheng Z. (2016). Identification and tissue distribution of chemosensory protein and odorant binding protein genes in Athetis dissimilis (Lepidoptera: Noctuidae). Appl. Entomol. Zool. 51, 409–420. doi: 10.1007/s13355-016-0413-8 [DOI] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034. doi: 10.1186/gb-2002-3-7-research0034, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q., Chen S., Shan Z., Yang Z., Chen L., Zhang C., et al. (2017). Stability evaluation of reference genes for gene expression analysis by RT-qPCR in soybean under different conditions. PLoS One 12:e0189405. doi: 10.1371/journal.pone.0189405, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Liu T., Khashaveh A., Yi C., Liu X., Zhang Y. (2021). Identification and evaluation of suitable reference genes for RT-qPCR analysis in Hippodamia variegata (Coleoptera: Coccinellidae) under different biotic and abiotic conditions. Front. Physiol. 12:669510. doi: 10.3389/fphys.2021.669510, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Xiao P., Chen D., Xu L., Zhang B. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. doi: 10.1007/s11103-012-9885-2, PMID: [DOI] [PubMed] [Google Scholar]
- Yan Z., Gao J., Lv X., Yang W., Wen S., Tong H., et al. (2016). Quantitative evaluation and selection of reference genes for quantitative RT-PCR in mouse acute pancreatitis. Biomed. Res. Int. 2016:8367063. doi: 10.1155/2016/8367063, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Li Z., Cao J., Zhang S., Zhang H., Wu X., et al. (2014). Selection and assessment of reference genes for quantitative PCR normalization in migratory locust Locusta migratoria (Orthoptera: Acrididae). PLoS One 9:e98164. doi: 10.1371/journal.pone.0098164, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Sun L., Zhang Q., Cao C. (2020). Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: Erebidae) using qRT-PCR. Gene 749:144712. doi: 10.1016/j.gene.2020.144712, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang S., An S., Li Z., Wu F., Yang Q., Liu Y., et al. (2015). Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555, 393–402. doi: 10.1016/j.gene.2014.11.038, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao X., Fu J., Jiang L., Zhang W., Shao Y., Jin C., et al. (2018). Transcriptome-based identification of the optimal reference genes as internal controls for quantitative RT-PCR in razor clam (Sinonovacula constricta). Genes Genomics 40, 603–613. doi: 10.1007/s13258-018-0661-9, PMID: [DOI] [PubMed] [Google Scholar]
- Zheng Y. T., Li H. B., Lu M. X., Du Y. Z. (2014). Evaluation and validation of reference genes for qRT-PCR normalization in Frankliniella occidentalis (Thysanoptera: Thripidae). PLoS One 9:e111369. doi: 10.1371/journal.pone.0111369, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.