Abstract
The main oncologic events in pleomorphic adenoma (PA) are the translocations of Pleomorphic adenoma gene 1 (PLAG1) on chromosome 8q12 and High-mobility group AT-hook 2 (HMGA2) on chromosome 12q14.3 with various fusion partners. These translocations result in the transcriptional up-regulation of PLAG1 and HMGA2 proteins. We carried out a preliminary evaluation of PLAG1 translocation by fluorescence in-situ hybridization (FISH), immunohistochemistry (IHC) and HMGA2 IHC on twenty-five archived formalin-fixed paraffin-embedded tissues of PAs and its clinicopathologic features. Only eight cases were successfully hybridized and 50% of the interpretable cases were considered positive for PLAG1 translocation. PLAG1 IHC was only positive in 2 (8%) of the 25 cases stained, including one of the positive PLAG1 translocation cases. HMGA2 IHC was positive in 12 (48%) of the 25 cases stained including 2 (50%) of the 4 cases identified with PLAG1 translocation by FISH, 3 (75%) of the 4 cases negative for PLAG1 translocation by FISH and 7 (41%) of the 17 cases with failed hybridization. Overall, 15 (60%) of the 25 PA cases demonstrated PLAG1 and/or HMGA2 alterations confirmed either by FISH or IHC. In conclusion, PLAG1 and HMGA2 alterations were confirmed either by FISH or IHC in this cohort and HMGA2 alteration is a common event in PAs of salivary gland.
Keywords: Pleomorphic adenoma gene 1, high-mobility group AT-hook 2, fluorescence In-Situ hybridization, immunohistochemistry, salivary gland tumor
Introduction
Pleomorphic adenoma (PA) is the most common salivary gland neoplasm, representing ~60% of all salivary gland tumors [1]. The most common location for PA is the parotid gland, followed by the palate and the submandibular salivary gland. Histologically, PA is composed of admixed epithelial/ductal cells and myoepithelial cells in a myxoid/chondroid stroma. Because of cytological and architectural variations in PA, distinguishing PA from other salivary gland tumors can be challenging. Thus, molecular and immunohistochemical ancillary studies are sometimes necessary to distinguish PAs from other salivary gland neoplasms, especially in situations involving a limited biopsy specimen.
Several salivary gland neoplasms are known to have recurrent translocations (Table 1) [2-12]. Over half of all PAs harbor Pleomorphic adenoma gene 1 (PLAG1) translocation with various fusion partners including CTNNB1, LIFR, TCEA1, FGFR1, and CHCHD7 [10,13-16]. PLAG1 is a zinc finger transcription factor and proto-oncogene on chromosome 8q12 [17]. The High-mobility group AT-hook 2 (HMGA2) gene on chromosome 12q14.3 is the second most common recurrent cytogenetic alteration in PA. HMGA2 translocation with various fusion partners such as FHIT, NFIB, WIF1 and TMTC2 have been identified in PA [18-21]. PLAG1/HMGA2 translocations are specific to PA and carcinoma ex-PA, and they have not been identified in any other salivary gland tumors [22-26].
Table 1.
| Salivary gland tumor | Gene rearrangement/translocation |
|---|---|
| Acinic cell carcinoma | NR4A3 |
| HTN3-MSANTD3 | |
| Adenoid cystic carcinoma | MYB/MYBL1-NFIB |
| Hyalinizing clear cell carcinoma | EWSR1 |
| Intraductal carcinoma | NCOA4/TRIM27-RET |
| Mucoepidermoid carcinoma | CRTC1/CRTC3-MAML2 |
| Microsecretory adenocarcinoma | MEF2C-SS18 |
| Polymorphous adenocarcinoma/cribriform adenocarcinoma of minor salivary gland | PRKD1 |
| Pleomorphic adenoma | PLAG1 |
| HMGA2 | |
| Secretory carcinoma | ETV6 |
Translocations of PLAG1 and HMGA2 result in the transcriptional up-regulation of PLAG1 and HMGA2 oncoproteins, and the resulting overexpression of the proteins can be detected by immunohistochemistry (IHC) [24,27]. To our knowledge, no detailed molecular or immunophenotypic features of PA have been described in the Nigerian population. Thus, the aim of this study is to evaluate the prevalence and characteristics of PLAG1/HMGA2 alterations, and their association with histologic and clinical features of PA, using fluorescence in-situ hybridization (FISH) and IHC in a relatively large cohort of PAs in the Nigerian population.
Material and methods
The pathology files of the Departments of Oral and Maxillofacial Surgery and Oral Pathology, and Morbid Anatomy and Forensic Medicine of Obafemi Awolowo University Teaching Hospitals Complex and Department of Morbid Anatomy and Histopathology, LAUTECH Teaching Hospital, Nigeria, were searched for the diagnosis of PA of the salivary gland from the years 2011 to 2019. The cases were reviewed in order to identify the most predominant stromal and epithelial components, as well as the most predominant myoepithelial cell morphology. The following clinical information was also retrieved from each case file: age at diagnosis, gender, anatomic site, size, and recurrence. FISH for PLAG1 translocation and IHC for PLAG1 and HMGA2 were performed on all cases retrieved, at the University of Pittsburgh, Pennsylvania.
Inclusion and exclusion criteria for cases
Inclusion criteria
Included cases were: (1) cases with histopathologic diagnosis of salivary gland pleomorphic adenoma; (2) cases with available archived tissue for molecular and immunophenotypic evaluation.
Exclusion criteria
Excluded cases were: (1) cases of salivary gland tumors without a histopathologic diagnosis of pleomorphic adenoma; (2) cases with histopathologic diagnosis of pleomorphic adenoma of non-salivary gland origin; (3) cases with histopathologic diagnosis of salivary gland pleomorphic adenoma without available archival tissue for molecular and immunophenotypic evaluation.
Immunohistochemistry for PLAG1 and HMGA2
IHC for PLAG1 (1:60, clone 3B7; Abnova) and HMGA2 (1:100, rabbit polyclonal; GeneTex, USA) were performed on 4-μm-thick sections of formalin-fixed paraffin-embedded (FFPE) tissue. All immunostains were done on a Leica Bond III (Leica, Buffalo Grove, IL) automated stainer platform. Prior to immunohistochemical staining, heat-based antigen retrieval employing a high pH buffer (Leica, ER2) was performed on all slides for 40 mins. Antibody incubation for 15 mins. As a secondary system, a polymeric detection kit (DAB Refine) was used. The cut-off for a positive PLAG1 IHC and HMGA2 IHC was nuclear immunostaining with any intensity noted in 5% of tumor cells.
FISH for PLAG1
Formalin-fixed, paraffin-embedded tissues were serially sectioned at 4 μm intervals. An H&E stained section from each specimen was prepared and reviewed by a pathologist to identify and mark the tumor area for further analysis. Sections were deparaffinized in xylene twice for 10 minutes each, immersed twice in 100% ethanol, and then pretreated (1× sodium chloride, sodium citrate). Slides were digested in pepsin solution (0.75 mg/mL in 1N HCl) followed by drying. FISH was then performed using the Dual-color breakapart FISH Probe, PLAG1 (8q12.1) Dual Color breakapart FISH Probe labeled with 5’orange and 3’green (Abbott Molecular Inc., Des Plaines, IL, USA). The slides and probes were co-denatured in at 75°C for 5 minutes before hybridization. Slides were incubated overnight at 37°C in a humidified chamber. Post-hybridization washes were performed in 2× SSC/0.3% IGEPAL for 2 minutes at 72°C. Slides were air-dried in the dark and counterstained with 4’,6-diamidino-2-phenylindole (DAPI). Analysis was performed using a Leica Biosystems (CytoVision FISH Capture and Analysis Workstation, Buffalo Grove, IL, USA). Only individual and well-delineated cells were counted; overlapping cells were excluded from the analysis. At least 60 cells were scored from the PLAG1 slide. Normal cells without translocation show two Orange/Green fusion signals in juxtaposition for either probe set. Cells with the translocation show one Orange/Green fusion and one Orange and one Green signal separately (representing a translocation of PLAG1). The cut-off for a positive PLAG1 translocation was 9.93% of translocated cells in 60-102 cells evaluated.
Statistical analysis
The Statistical Package for the Social Sciences (SPSS), version 20 (SPSS, Inc., Chicago, III) was used to analyze the data. Descriptive statistics, for scores and percentages were for categorical data, and means with range were used for continuous variables when appropriate.
Ethical oversight
This study was reviewed and ethically approved by the Ethics and Research Committee of Obafemi Awolowo University Teaching Hospitals Complex, Obafemi Awolowo University, Ile-Ife, Nigeria (Protocol No: ERC/2019/06/06).
Results
Clinical characteristics
A total of 25 PAs were retrieved. Patient demographics are presented in Table 2. There were 19 (76%) females and 6 (24%) males, with an age range of 12 to 66 years old (mean, 43 years). The tumor sites were the parotid gland (17 cases), submandibular gland (6 cases), palate (1 case), and upper lip (1 case). Of the 25 cases, 18 reported tumor dimensions, which ranged from 2.3 to 7 cm (mean, 5.3 cm). Tumor recurrence was reported in 2 cases.
Table 2.
Clinical, molecular and immunophenotypic features of pleomorphic adenoma
| Case No. | Age | Sex | Location | PLAG1 FISH (% of cells translocated in 60-102 cells evaluated) | PLAG1 IHC | HMGA2 IHC |
|---|---|---|---|---|---|---|
| 1 | 12 | F | Parotid | + (60/64, 93.7%) | + | + |
| 2 | 23 | F | Parotid | + (56/64, 87.5%) | - | - |
| 3 | 43 | F | Parotid | + (55/60, 91.7%) | - | - |
| 4 | 66 | F | Parotid | + (43/60, 71.7%) | - | + |
| 5 | 66 | M | Parotid | - (2/62, 3.2%)* | - | + |
| 6 | 48 | F | Parotid | - (0/62, 0%) | - | + |
| 7 | 30 | M | Submandibular | - (0/102, 0%) | + | - |
| 8 | 64 | F | Submandibular | - (0/61, 0%) | - | + |
| 9 | 33 | M | Parotid | Hybridization failed x2 | - | - |
| 10 | 45 | F | Parotid | Hybridization failed x2 | - | - |
| 11 | 66 | F | Parotid | Hybridization failed x2 | - | + |
| 12 | 53 | F | Submandibular | Hybridization failed x2 | - | + |
| 13 | 42 | M | Parotid | Hybridization failed x2 | - | + |
| 14 | 60 | F | Parotid | Hybridization failed x2 | - | - |
| 15 | 65 | F | Parotid | Hybridization failed x2 | - | - |
| 16 | 56 | M | Parotid | Hybridization failed x2 | - | + |
| 17 | 42 | F | Parotid | Hybridization failed x2 | - | - |
| 18 | 39 | M | Submandibular | Hybridization failed x2 | - | - |
| 19 | 38 | F | Parotid | Hybridization failed x2 | - | + |
| 20 | 40 | F | Parotid | Hybridization failed x2 | - | - |
| 21 | 46 | F | Submandibular | Hybridization failed x2 | - | - |
| 22 | 17 | F | Upper lip | Hybridization failed x2 | - | - |
| 23 | 42 | F | Parotid | Hybridization failed x2 | - | - |
| 24 | 13 | F | Palate | Hybridization failed x2 | - | + |
| 25 | 28 | F | Submandibular | Hybridization failed x2 | - | + |
- The cutoff for positivity is translocation in 9.93% of examined cells, IHC - Immunohistochemistry, (+) - positive, (-) - negative.
Morphologic, molecular and immunophenotypic characteristics
The majority (18/25, 72%) of the cases exhibited myxoid stroma as the most predominant stromal component, followed by chondromyxoid stroma in 12% of cases. The stroma of 2 cases had tyrosine crystals, and 2 other cases had squamous differentiation (Cases 3, 7, 16 and 25) (Figure 1A & 1B). All cases exhibited ducts as the epithelial component, and a majority (18/25, 72%) of the cases exhibited plasmacytoid morphology as the most predominant myoepithelial cells, followed by spindled morphology in 12% of cases.
Figure 1.

Histopathology of pleomorphic adenoma: (A) = 56-yr old male, PLAG1 IHC negative and HMGA2 IHC positive pleomorphic adenoma of the parotid gland exhibiting tryosine crystals in the stroma (Case 16) (haematoxylin and eosin, original magnification ×200), (B) = 28-yr old female, PLAG1 IHC negative and HMGA2 IHC positive pleomorphic adenoma of the submandibular gland exhibiting squamous differentiation in the stroma (Case 25) (haematoxylin and eosin, original magnification ×200).
Of the 25 cases, PLAG1 translocations were hybridizable using FISH in only 8 cases (Cases 1-8). PLAG1 translocations were detected in only 5 cases, with a range of 3.2%-93.7% of translocated cells out of 60-102 cells examined. Based on the cut-off of 9.93%, 4 (50%) of the 8 interpretable cases were considered positive (Figure 2A and 2B; Table 2). The remaining 17 cases failed to hybridize after two attempts. PLAG1 IHC was only positive in 2 (8%) of the 25 cases stained, with weak intensity (Table 2). One of the positive PLAG1 IHC cases was determined to be negative for PLAG1 translocation by FISH (Case 7). HMGA2 IHC was positive in 12 (48%) of the 25 cases stained (Figure 3A-D; Table 2), including 3 cases with weak intensity.
Figure 2.

Fluorescence in situ hybridization for PLAG1 in a 12-yr old female with pleomorphic adenoma of the parotid gland (Case 1) (A) and in a 66-yr old female with pleomorphic adenoma of the parotid gland (Case 4) (B), respectively. FISH for PLAG1 shows one pair of split signals indicative of PLAG1 rearrangement with a balanced translocation pattern.
Figure 3.
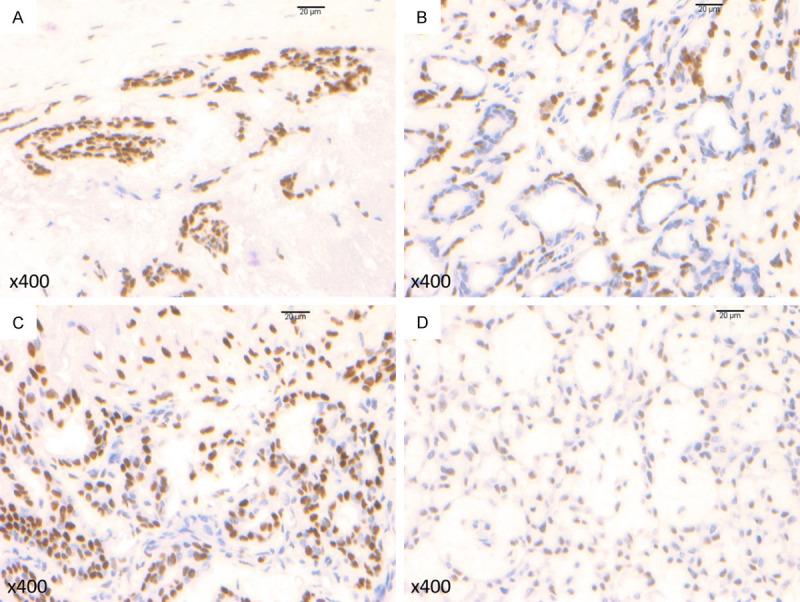
Immunohistochemistry for HMGA2 in pleomorphic adenomas: (A) photomicrograph shows diffuse HMGA2 expression with strong intensity (Case 6) (original magnification ×400), (B) photomicrograph shows diffuse HMGA2 expression with strong intensity (Case 16) (original magnification ×400), (C) photomicrograph shows diffuse HMGA2 expression with strong intensity (Case 19) (original magnification ×400), (D) photomicrograph shows diffuse HMGA2 expression with moderate intensity (Case 24) (original magnification ×400).
The 5 cases identified with PLAG1 alterations, either by FISH or IHC presented with plasmacytoid morphology as the most predominant myoepithelial cells. Nine (75%) of the 12 cases identified with HMGA2 translocation by IHC presented with plasmacytoid morphology as the most predominant myoepithelial cells. The remaining 3 cases presented with spindle, clear, and epithelioid morphology as the most predominant myoepithelial cells, respectively. Two (50%) of the 4 cases identified with PLAG1 translocation by FISH expressed HMGA2, while 3 (75%) of the 4 cases that were found to be negative for PLAG1 translocation by FISH expressed HMGA2. Seven (41%) of the 17 cases with failed hybridization for PLAG1 translocation expressed HMGA2 (Table 3). Overall, 15 (60%) of the 25 PA cases demonstrated PLAG1 and/or HMGA2 translocations either by FISH or IHC.
Table 3.
Summary of PLAG1/HMGA2 IHC in relationship to PLAG1 translocation identified by FISH in pleomorphic adenoma
| PLAG1 IHC + | PLAG1 IHC - | HMGA2 IHC + | HMGA2 IHC - | |
|---|---|---|---|---|
| PLAG1 FISH + | 1 | 3 | 2 | 2 |
| PLAG1 FISH - | 1 | 3 | 3 | 1 |
| PLAG1 FISH failed | 0 | 17 | 7 | 11 |
FISH, for the detection of PLAG1 translocation was more sensitive than PLAG1 IHC (50% VS. 12%). The sensitivity and specificity of PLAG1 IHC in predicting PLAG1 translocation in pleomorphic adenoma were 25% for each.
Discussion
PLAG1 translocation has been identified as the most prevalent oncologic event in PA, affecting 50-60% of the tumors [28] and its activation is a crucial event in other neoplasms such as AML, lipoblastoma, and hepatoblastoma [29-31]. The second most common recurrent oncologic event in PA involves translocation of HMGA2 [11], which also plays a crucial role in the oncogenic activation of different types of benign mesenchymal tumors, such as hamartoma of the breast and lung, fibroadenoma of the breast, lipoma, uterine leiomyoma, angiomyxoma, and endometrial polyps [32].
The translocations of PLAG1 and HMGA2 result in the transcriptional up-regulation of PLAG1 and HMGA2 oncoproteins, which are overexpressed in the tumor and can be detected by IHC [24,27]. In our study, we evaluated for PLAG1 translocation using FISH and IHC, and alteration in HMGA2 by IHC in a cohort of PAs from Nigerian patients. Our report of 50% prevalence of PLAG1 translocation using FISH in PAs is in concordance with other published studies [28]. We identified a higher proportion (48%) of PAs expressing HMGA2 compared to the study by Mito et al. (34%) [26], suggesting that alterations in HMGA2 in PA and carcinoma ex-PA are more frequent than previously recognized. Persson et al. identified HMGA2 alteration in 62.5% of PAs and carcinomas ex-PA by utilizing array comparative genomic hybridization and reverse transcription-polymerase chain reaction (RT-PCR) [20]. These studies and ours show that the rate of HMGA2 alteration in PA may be higher than previously reported.
Our observation of HMGA2 expression in two PLAG1 translocated-positive PAs is similar to Mito et al., who found dual expression of PLAG1 and HMGA2 immunostaining in 8 (14.5%) out of 55 PAs [26]. A possible mechanism for this is that PLAG1 may constitute a common downstream target that drives tumorigenesis in HMGA2-rearranged PA [26].
Katabi et al. evaluated for alteration in PLAG1 using both FISH and IHC, and reported a higher sensitivity of IHC in identifying PLAG1 alteration in PA than the use of FISH (96% vs. 33%) [22]. However, our report shows the reverse: FISH was more sensitive than IHC (50% vs. 8%) in identifying PLAG1 alterations in PA. Other studies evaluating PLAG1 IHC in PA have reported high (62-100%) sensitivity of the immunostain in PA [22]. It is thought that the overexpression of PLAG1 oncoprotein may be poor in PA undergoing advanced progression or malignant transformation, resulting in negative PLAG1 IHC [22,33].
Northern blot analysis, RT-PCR, and FISH are the main analytical techniques employed to detect PLAG1 translocation in PA [13,14,22,24]. FISH analysis can predict the potential for fusion transcript and it utilizes DNA probes specific to a particular DNA. It can be carried out on formalin-fixed paraffin-embedded tissue (FFPET), as obtained in this retrospective study, which employed FISH on archival FFPET. Only 8 of the 25 cases we analyzed for PLAG1 translocation using FISH were interpretable, suggesting the importance of proper fixation and storage of FFPET for molecular analysis. Degradation of DNA from improper fixation and storage leads to failure of FISH analysis, as we stated in our previous study [34]. Additionally, FISH analyses will not detect intra-chromosomal rearrangements of genes in proximity to fusion partners. For example, PLAG1 rearrangement in PA with PLAG1-FGFR1 fusion and HMGA2 rearrangement in PA with HMGA2-WIF1 fusion will not be detected by FISH because of the close proximity of the fusion genes [15,20].
The translocations of PLAG1 and HMGA2 have only been described in PA and carcinoma ex-PA, making them specific to these tumors [22-26], unlike PLAG1 IHC, which has been expressed in non-PA tumors such as de-novo myoepithelial carcinoma, basal cell adenocarcinoma, epithelial-myoepithelial carcinoma, mucoepidermoid carcinoma and adenoid cystic carcinoma [22,24,35]. As such, PLAG1 IHC expression should be used with caution when making a diagnosis of PA and carcinoma ex-PA, whereas, HMGA2 IHC has been reported to be a specific immunohistochemical marker for PA and carcinoma ex-PA [26]. Nonetheless, PLAG1 translocation is helpful in making a diagnosis of PA and carcinoma ex-PA of the salivary gland in challenging cases or in a limited biopsy. Detection of PLAG1 alteration by FISH or IHC has been shown to be useful in discriminating carcinoma ex-PA from its de-novo carcinoma counterpart [23,36,37].
Recently, Asahina et al. reported that the predominant subtype of salivary gland PAs with PLAG1 translocation demonstrated myoepithelial cells with plasmacytoid morphology compared to those without PLAG1 translocation [38]. Our study corroborated the same predominance in the 5 cases with PLAG1 alteration (FISH or IHC verified) and 75% of PA with HMGA2 expression. We can therefore suggest that PLAG1 and HMGA2 alterations may be associated with specific histologic features such as plasmacytoid morphology of the myoepithelial cells.
The limitation of this study is that we did not evaluate for alteration in HMGA2 using FISH, which could have corroborated our IHC results. In conclusion, we report the first series of PLAG1 and HMGA2 alteration-positive PAs by FISH and IHC in Nigerian (African) patients. The mainstay of management still remains surgical resection, which is associated with recurrence or malignant transformation in incomplete resections and/or tumor seeding. Patients from this population may benefit from PLAG1/HMGA2 targeted anti-neoplastic therapy. HMGA2 alteration is a common event and FISH for the detection of PLAG1 translocation was more sensitive than PLAG1 IHC in this cohort of salivary gland PAs.
Disclosure of conflict of interest
None.
References
- 1.Bell D, Bullerdiek J, Gnepp DR, Schwartz MR, Stenman G, Triantafyllou A. Pleomorphic adenoma. In: El-Naggar A, Chan JKC, Grandis JR, Takata T, Slootweg PJ, editors. World Health Organization classification of head and neck tumours. Lyon: IARC Press; 2017. pp. 185–186. [Google Scholar]
- 2.Haller F, Bieg M, Will R, Korner C, Weichenhan D, Bott A, Ishaque N, Lutsik P, Moskalev EA, Mueller SK, Bahr M, Woerner A, Kaiser B, Scherl C, Haderlein M, Kleinheinz K, Fietkau R, Iro H, Eils R, Hartmann A, Plass C, Wiemann S, Agaimy A. Enhancer hijacking activates oncogenic transcription factor NR4A3 in acinic cell carcinomas of the salivary glands. Nat Commun. 2019;10:368. doi: 10.1038/s41467-018-08069-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barasch N, Gong X, Kwei KA, Varma S, Biscocho J, Qu K, Xiao N, Lipsick JS, Pelham RJ, West RB, Pollack JR. Recurrent rearrangements of the Myb/SANT-like DNA-binding domain containing 3 gene (MSANTD3) in salivary gland acinic cell carcinoma. PLoS One. 2017;12:e0171265. doi: 10.1371/journal.pone.0171265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonescu CR, Katabi N, Zhang L, Sung YS, Seethala RR, Jordan RC, Perez-Ordonez B, Have C, Asa SL, Leong IT, Bradley G, Klieb H, Weinreb I. EWSR1-ATF1 fusion is a novel and consistent finding in hyalinizing clear-cell carcinoma of salivary gland. Genes Chromosomes Cancer. 2011;50:559–570. doi: 10.1002/gcc.20881. [DOI] [PubMed] [Google Scholar]
- 6.Skalova A, Vanecek T, Uro-Coste E, Bishop JA, Weinreb I, Thompson LDR, de Sanctis S, Schiavo-Lena M, Laco J, Badoual C, Santana Conceicao T, Ptakova N, Baneckova M, Miesbauerova M, Michal M. Molecular profiling of salivary gland intraductal carcinoma revealed a subset of tumors harboring NCOA4-RET and novel TRIM27-RET fusions: a report of 17 cases. Am J Surg Pathol. 2018;42:1445–1455. doi: 10.1097/PAS.0000000000001133. [DOI] [PubMed] [Google Scholar]
- 7.Tonon G, Modi S, Wu L, Kubo A, Coxon AB, Komiya T, O’Neil K, Stover K, El-Naggar A, Griffin JD, Kirsch IR, Kaye FJ. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a notch signaling pathway. Nat Genet. 2003;33:208–213. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]
- 8.Bishop JA, Weinreb I, Swanson D, Westra WH, Qureshi HS, Sciubba J, MacMillan C, Rooper LM, Dickson BC. Microsecretory adenocarcinoma: a novel salivary gland tumor characterized by a recurrent MEF2C-SS18 fusion. Am J Surg Pathol. 2019;43:1023–1032. doi: 10.1097/PAS.0000000000001273. [DOI] [PubMed] [Google Scholar]
- 9.Weinreb I, Zhang L, Tirunagari LM, Sung YS, Chen CL, Perez-Ordonez B, Clarke BA, Skalova A, Chiosea SI, Seethala RR, Waggott D, Boutros PC, How C, Liu FF, Irish JC, Goldstein DP, Gilbert R, Ud Din N, Assaad A, Hornick JL, Thompson LD, Antonescu CR. Novel PRKD gene rearrangements and variant fusions in cribriform adenocarcinoma of salivary gland origin. Genes Chromosomes Cancer. 2014;53:845–856. doi: 10.1002/gcc.22195. [DOI] [PubMed] [Google Scholar]
- 10.Kas K, Voz ML, Roijer E, Astrom AK, Meyen E, Stenman G, Van de Ven WJ. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet. 1997;15:170–174. doi: 10.1038/ng0297-170. [DOI] [PubMed] [Google Scholar]
- 11.Bullerdiek J, Wobst G, Meyer-Bolte K, Chilla R, Haubrich J, Thode B, Bartnitzke S. Cytogenetic subtyping of 220 salivary gland pleomorphic adenomas: correlation to occurrence, histological subtype, and in vitro cellular behavior. Cancer Genet Cytogenet. 1993;65:27–31. doi: 10.1016/0165-4608(93)90054-p. [DOI] [PubMed] [Google Scholar]
- 12.Skalova A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, Starek I, Geierova M, Simpson RH, Passador-Santos F, Ryska A, Leivo I, Kinkor Z, Michal M. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- 13.Voz ML, Astrom AK, Kas K, Mark J, Stenman G, Van de Ven WJ. The recurrent translocation t(5;8)(p13;q12) in pleomorphic adenomas results in upregulation of PLAG1 gene expression under control of the LIFR promoter. Oncogene. 1998;16:1409–1416. doi: 10.1038/sj.onc.1201660. [DOI] [PubMed] [Google Scholar]
- 14.Astrom AK, Voz ML, Kas K, Roijer E, Wedell B, Mandahl N, Van de Ven W, Mark J, Stenman G. Conserved mechanism of PLAG1 activation in salivary gland tumors with and without chromosome 8q12 abnormalities: identification of SII as a new fusion partner gene. Cancer Res. 1999;59:918–923. [PubMed] [Google Scholar]
- 15.Persson F, Winnes M, Andren Y, Wedell B, Dahlenfors R, Asp J, Mark J, Enlund F, Stenman G. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene. 2008;27:3072–3080. doi: 10.1038/sj.onc.1210961. [DOI] [PubMed] [Google Scholar]
- 16.Asp J, Persson F, Kost-Alimova M, Stenman G. CHCHD7-PLAG1 and TCEA1-PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromosomes Cancer. 2006;45:820–828. doi: 10.1002/gcc.20346. [DOI] [PubMed] [Google Scholar]
- 17.Kas K, Voz ML, Hensen K, Meyen E, Van de Ven WJ. Transcriptional activation capacity of the novel PLAG family of zinc finger proteins. J Biol Chem. 1998;273:23026–23032. doi: 10.1074/jbc.273.36.23026. [DOI] [PubMed] [Google Scholar]
- 18.Geurts JM, Schoenmakers EF, Roijer E, Stenman G, Van de Ven WJ. Expression of reciprocal hybrid transcripts of HMGIC and FHIT in a pleomorphic adenoma of the parotid gland. Cancer Res. 1997;57:13–17. [PubMed] [Google Scholar]
- 19.Geurts JM, Schoenmakers EF, Roijer E, Astrom AK, Stenman G, van de Ven WJ. Identification of NFIB as recurrent translocation partner gene of HMGIC in pleomorphic adenomas. Oncogene. 1998;16:865–872. doi: 10.1038/sj.onc.1201609. [DOI] [PubMed] [Google Scholar]
- 20.Persson F, Andren Y, Winnes M, Wedell B, Nordkvist A, Gudnadottir G, Dahlenfors R, Sjogren H, Mark J, Stenman G. High-resolution genomic profiling of adenomas and carcinomas of the salivary glands reveals amplification, rearrangement, and fusion of HMGA2. Genes Chromosomes Cancer. 2009;48:69–82. doi: 10.1002/gcc.20619. [DOI] [PubMed] [Google Scholar]
- 21.Wasserman JK, Dickson BC, Smith A, Swanson D, Purgina BM, Weinreb I. Metastasizing pleomorphic adenoma: recurrent PLAG1/HMGA2 rearrangements and identification of a novel HMGA2-TMTC2 fusion. Am J Surg Pathol. 2019;43:1145–1151. doi: 10.1097/PAS.0000000000001280. [DOI] [PubMed] [Google Scholar]
- 22.Katabi N, Xu B, Jungbluth AA, Zhang L, Shao SY, Lane J, Ghossein R, Antonescu CR. PLAG1 immunohistochemistry is a sensitive marker for pleomorphic adenoma: a comparative study with PLAG1 genetic abnormalities. Histopathology. 2018;72:285–293. doi: 10.1111/his.13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katabi N, Ghossein R, Ho A, Dogan S, Zhang L, Sung YS, Antonescu CR. Consistent PLAG1 and HMGA2 abnormalities distinguish carcinoma ex-pleomorphic adenoma from its de novo counterparts. Hum Pathol. 2015;46:26–33. doi: 10.1016/j.humpath.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuyama A, Hisaoka M, Nagao Y, Hashimoto H. Aberrant PLAG1 expression in pleomorphic adenomas of the salivary gland: a molecular genetic and immunohistochemical study. Virchows Arch. 2011;458:583–592. doi: 10.1007/s00428-011-1063-4. [DOI] [PubMed] [Google Scholar]
- 25.Martins C, Fonseca I, Roque L, Pereira T, Ribeiro C, Bullerdiek J, Soares J. PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol. 2005;18:1048–1055. doi: 10.1038/modpathol.3800386. [DOI] [PubMed] [Google Scholar]
- 26.Mito JK, Jo VY, Chiosea SI, Dal Cin P, Krane JF. HMGA2 is a specific immunohistochemical marker for pleomorphic adenoma and carcinoma ex-pleomorphic adenoma. Histopathology. 2017;71:511–521. doi: 10.1111/his.13246. [DOI] [PubMed] [Google Scholar]
- 27.Tessari MA, Gostissa M, Altamura S, Sgarra R, Rustighi A, Salvagno C, Caretti G, Imbriano C, Mantovani R, Del Sal G, Giancotti V, Manfioletti G. Transcriptional activation of the cyclin A gene by the architectural transcription factor HMGA2. Mol Cell Biol. 2003;23:9104–9116. doi: 10.1128/MCB.23.24.9104-9116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenman G. Fusion oncogenes in salivary gland tumors: molecular and clinical consequences. Head Neck Pathol. 2013;7(Suppl 1):S12–19. doi: 10.1007/s12105-013-0462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrette SF, Kuo YH, Hensen K, Barjesteh van Waalwijk van Doorn-Khosrovani S, Perrat PN, Van de Ven WJ, Delwel R, Castilla LH. Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb-MYH11. Blood. 2005;105:2900–2907. doi: 10.1182/blood-2004-09-3630. [DOI] [PubMed] [Google Scholar]
- 30.Astrom A, D’Amore ES, Sainati L, Panarello C, Morerio C, Mark J, Stenman G. Evidence of involvement of the PLAG1 gene in lipoblastomas. Int J Oncol. 2000;16:1107–1110. doi: 10.3892/ijo.16.6.1107. [DOI] [PubMed] [Google Scholar]
- 31.Zatkova A, Rouillard JM, Hartmann W, Lamb BJ, Kuick R, Eckart M, von Schweinitz D, Koch A, Fonatsch C, Pietsch T, Hanash SM, Wimmer K. Amplification and overexpression of the IGF2 regulator PLAG1 in hepatoblastoma. Genes Chromosomes Cancer. 2004;39:126–137. doi: 10.1002/gcc.10307. [DOI] [PubMed] [Google Scholar]
- 32.Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- 33.de Brito BS, Giovanelli N, Egal ES, Sanchez-Romero C, Nascimento JS, Martins AS, Tincani AJ, Del Negro A, Gondak RO, Almeida OP, Kowalski LP, Altemani A, Mariano FV. Loss of expression of plag1 in malignant transformation from pleomorphic adenoma to carcinoma ex pleomorphic adenoma. Hum Pathol. 2016;57:152–159. doi: 10.1016/j.humpath.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Owosho AA, Adesina OM, Odujoko O, Soyele OO, Komolafe A, Bauer R, Holte K, Summersgill KF. MYB-NFIB translocation by FISH in adenoid cystic carcinoma of the head and neck in nigerian patients: a preliminary report. Head Neck Pathol. 2021;15:433–437. doi: 10.1007/s12105-020-01214-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avadhani V, Cohen C, Siddiqui MT. PLAG1: an immunohistochemical marker with limited utility in separating pleomorphic adenoma from other basaloid salivary gland tumors. Acta Cytol. 2016;60:240–245. doi: 10.1159/000447622. [DOI] [PubMed] [Google Scholar]
- 36.Bahrami A, Dalton JD, Shivakumar B, Krane JF. PLAG1 alteration in carcinoma ex pleomorphic adenoma: immunohistochemical and fluorescence in situ hybridization studies of 22 cases. Head Neck Pathol. 2012;6:328–335. doi: 10.1007/s12105-012-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiosea SI, Thompson LD, Weinreb I, Bauman JE, Mahaffey AM, Miller C, Ferris RL, Gooding WE. Subsets of salivary duct carcinoma defined by morphologic evidence of pleomorphic adenoma, PLAG1 or HMGA2 rearrangements, and common genetic alterations. Cancer. 2016;122:3136–3144. doi: 10.1002/cncr.30179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asahina M, Saito T, Hayashi T, Fukumura Y, Mitani K, Yao T. Clinicopathological effect of PLAG1 fusion genes in pleomorphic adenoma and carcinoma ex pleomorphic adenoma with special emphasis on histological features. Histopathology. 2019;74:514–525. doi: 10.1111/his.13759. [DOI] [PubMed] [Google Scholar]


