Abstract
MicroRNAs (miRNAs) are highly conserved, non-coding transcripts that regulate gene expression in various ways. Evidence suggests that miRNAs may be a contributory factor in neurodegeneration, including Alzheimer’s disease (AD), Parkinson’s disease (PD), and triplet repeat disorders. In order to further understand the potential roles of miRNAs in the pathogenesis of AD, we analyzed Down syndrome (DS), a special model of AD, by using a TaqMan microRNA array and found that miRNA let-7c was up-regulated in both DS and AD. ELISA assay showed that let-7c reduced the expression level of Aβ significantly. Real-time quantitative-polymerase chain reaction (RT-qPCR) was conducted to reveal that the expression level of let-7c increased dramatically in DS cells, patients with DS and mice with AD compared with normal ones respectively. Additionally, western blotting illustrated that let-7c suppressed the expression of Aβ by inducing BACE2 to cut C99 and increase the content of C83/80. BACE2 expression was inhibited by let-7c and luciferase reporter gene assay revealed that let-7c increased the activity of wild-type BACE2 promoter but not 3’UTR. Furthermore, promoter analysis of BACE2 confirmed that let-7c could bind to BACE2 in the sequence between -1368 and -1347. In addition, immunoblotting assay demonstrated that let-7c induced BACE2 expression by RNAa. To the best of our knowledge, our study revealed for the first time that let-7c up-regulated BACE2 expression and decreased Aβ production.
Keywords: β-amyloid, Alzheimer’s disease, BACE2, Let-7c, RNA activation
Introduction
Alzheimer’s disease (AD) is the most common form of dementia. It is a progressive, ultimately fatal and neurodegenerative disease with three main pathological features: β-amyloid (Aβ) plaques, neurofibrillary tangles and neuron death [1-4]. Aβ is derived from β-amyloid precursor protein (APP) which is cleaved by β- and γ-secretase [5]. β-site APP-cleaving enzyme 1 (BACE1) is a β-secretase that cleaves APP at the β-site in vivo [6]. β-site APP-cleaving enzyme 2 (BACE2), a homologue of BACE1, cleaves APP at the θ-site and reduces the production of Aβ [7,8]. Gene mutants of APP occurring in various disease, including presenilin 1, presenilin 2 and Down syndrome (DS), may induce AD, but the complicated mechanism of AD remains unclear.
MircroRNAs (miRNAs) are a class of small, single-stranded, non-coding RNAs involved in the post-transcriptional regulation of gene expression. Many studies reveal that miRNAs participate in almost all life processes, including ontogenesis, hematopoiesis, organofaction, apoptosis, cell proliferation, and even tumorigenesis [9]. miRNAs are known to influence mRNA stability and repress protein synthesis through base-pairing with the 3’-untranslated region (3’UTR) of mRNA. Place et al. reported that miR-373 induced gene expression and identified a putative target site of miR-373 in the promoter of E-cadherin [10].
An AGO2-associated miRNA is able to bind to complementary regions of the targeted mRNA, which leads to either AGO2-mediated endonuclease cleavage of the mRNA or reduction in its translation efficiency [4]. RNA interference (RNAi) is a common negative regulation of miRNA.
Huang et al. confirmed that endogenous miRNA mediated RNA activation (RNAa), which played an important role in normal and tumor cell proliferation [11]. Similar to RNAi, RNAa depends on AGO2 to exert its roles while other AGO members including AGO1, AGO3 and AGO4 have little effects on saRNA triggering RNAa [12,13]. This is because the trigger of saRNA requires the AGO2 domain of Piwi to cut the insignificant strand of the saRNA double chain and releases it from the AGO2. However, not all RNAa is dependent on AGO2. For example, RNAa mediated by miRNA mainly recruits AGO1 [11,14]. Turchinovich and Burwinkel confirmed that there was no significant bias in association with a particular AGO for most miRNAs. However, several miRNAs clearly present a preference for association with a specific AGO protein [15].
In order to analyze the molecular mechanism of miRNAs in the pathology of AD, we studied the expression profile of miRNAs in the DS model cell line, and our findings revealed that let-7c was highly expressed in DS and AD cells. In addition, Let-7c apparently reduced Aβ production by targeting the BACE2 promoter and activated its expression in an unusual way.
Materials and methods
Animals and patients
Eight cases of patients with DS and five cases of normal controls were granted by UBC Townsend Family Laboratories. Transgenic AD mice (B6/JNju-Tg (APPswe, PSEN1dE9)/Nju) and wild-type mice (C57BL/6JNju) were bought from Nanjing University’s Model Animal Research Center. All procedures to animals have been permitted by Cheeloo College of Medicine, Shandong University Animal Care and Use Committee (Approval No. 21104) and were in accordance with the Guide for the Care and Use of Laboratory Animals issued by the National Institutes of Health. Mice were anesthetized by intraperitoneal injection of pentobarbital sodium (50 mg/kg) then they were killed by cervical dislocation. Tissues were extracted and stored in liquid nitrogen immediately. The brain samples were provided in cDNA format by UBC Townsend Family Laboratories. Besides, all the clinical assays obeyed the ethic committee of Cheeloo College of Medicine, Shandong University.
Cells culture and reagents
DS cell line MB1478 (47, XX, +21) and control cell line UMB115 (46, XX) were obtained from Maryland University’s brain library. 2EB2 (expressing APP and BACE1 stably) and 20E2 (expressing APP stably) were constructed as previously described [8]. HEK293 cells were cryopreserved by our laboratory. All cell lines were cultured at 37°C in an incubator containing 5% CO2 [7]. The cells were transfected with plasmids using Lipofectamine 2000 (11668-027, Invitrogen) according to the manufacturer’s instructions.
Plasmids construction
The expression vectors of Swedish mutant APP695 (named pZ-APPsw) [16], C99 (named pAPP-C99) [8], C99 tagged flag (named pAPP-C99Flag) [8] and C83 (named pAPP-C83) [8], luciferase reporter vectors of APP promoter (named prhβAPPluc) [17], Kv2.1 tagged flag (named pKv2.1-Flag) [18], Bace1 promoter (named pB1P-A) [19] and four truncated BACE2 promoters (named pB2Luc-A, pB2Luc-B, pB2Luc-C and pB2Luc-D) [7] were constructed as previously described. Sense oligonucleotide F1 and anti-sense oligonucleotide R1 were annealed and inserted into pSuper cloning plasmid (VEC-PBS-0002, OligoEngine) to generate pmiR-let-7c, Sense oligonucleotide F2 and anti-sense oligonucleotide R2 were annealed and inserted into pSuper cloning plasmid to generate pmiR-99a, sense oligonucleotide F3 and anti-sense oligonucleotide R3 were annealed and inserted into pSuper cloning plasmid to generate pmiR-155. Sense oligonucleotide F4 and anti-sense oligonucleotide R4 were annealed to generate vector pshAGO1 for inhibiting Argonaute 1, sense oligonucleotide F5 and anti-sense oligonucleotide R5 generated vector pshAGO2 for inhibiting Argonaute 2, sense oligonucleotide F6 and anti-sense oligonucleotide R6 generated vector pshBace2 for inhibiting Bace2, sense oligonucleotide F7 and anti-sense oligonucleotide R7 generated vector pshNCT for inhibiting Nicastrin. Forward primer F8 and reverse primer R8 amplified BACE1 3’-UTR into pmirGLO vector (E1330, Promega) to generate luciferase reporter pBACE1-3UTRluc, forward primer F9 and reverse primer R9 amplified BACE2 3’-UTR into pmirGLO vector to generate luciferase reporter pBACE2-3UTRluc (Supplementary Table 1).
Dual luciferase assay
Luciferase activity was measured following a protocol supplied by the Dual-Luciferase® Reporter Assay System (E1910, Promega), as described previously [7].
Low-density microarray and PCR
Total RNA was isolated from the cells using TRIzol® Reagent (15596018, Invitrogen) with glycogen at a final concentration of 100 ug/ml (AM9510, Invitrogen). Human Multiplex RT Set Pools 1-8 (4384791, ABI) and TaqMan® MicroRNA Reverse Transcription Kit (4366596, ABI) were used to synthesize the first-stand cDNA from an equal amount of the RNA sample following the manufacturer’s instructions. TaqMan® 2 Universal PCR Master Mix (4324018, ABI) and Taqman Low-Density Array (TLDA) Human miRNA Panel (4384792, ABI) were employed according to the manufacturer’s instructions to test the different expression of miRNAs of the DS or control cell lines. AmpliTaq Gold® 360 Master Mix (4398881, ABI) and TaqMan® 2 Universal PCR Master Mix (4324018, ABI) were used to detect the expression of miRNAs and genes by RT-PCR and RT-qPCR, respectively (Supplementary Table 2).
Aβ 40 ELISA assay
The quantity of Aβ in the cell culture medium was measured using the Aβ 40 ELISA kit (KHB3482, Invitrogen) according to the ELISA Technical Guide.
Immunoblotting
For immunoblot analysis, the cells were harvested and lysed by sonication in a RIPA buffer (150 mM NaCL, 50 MmTris-HCL, 1% TritonX-100, 2% SDS and 1% sodium deoxycholate) in the presence of a protease inhibitor cocktail (04693116001, Roche). Protein quantification was performed using the Bio-Rad Dc protein assay kit (Bio-Rad, Richmond, CA, USA). Whole-cell lysates were separated on 12% glycine SDS-PAGE gel. The primary antibodies used in this part were mouse anti-flag monoclonal antibody M2 (F1804, Sigma-Aldrich) and rabbit anti-amyloid precursor protein C-Terminal antibody C20 (A8717, Sigma-Aldrich). Mouse anti-β-actin monoclonal antibody (A1978, Sigma-Aldrich) was used to detect the protein level of β-actin as a loading control.
MTT assay
Plating cells in 96-well plate at 10,000 cells per well for 48 hours. Following the MTT Cell Proliferation Assay (30-1010K, ATCC) instructions, 10 µl MTT reagent was added into each well and incubated for two to four hours until a purple precipitate was visible. 100 µl detergent reagent was then added for two hours at room temperature in the dark. Next, the absorbance was recorded at 570 nm.
Karyotyping of DS cell lines
5 ug/ml of colchicine was added to the cell culture for three hours, and the cells were harvested and swollen in 0.075% KCl for 30 minutes, then fixed in freshly-made fixative (three parts methanol to one part glacial acetic acid). The cell suspension was dropped onto slides, and the slides were trypsinized with 0.25% trypsin (59429C, Sigma) for one minute. Giemsa solution (32884, Sigma) was used to stain the slides for 10 minutes. The cell karyotype was then analyzed under a microscope.
Data analysis
All experiments were repeated more than three times. For Western blotting (WB), the representative blots are shown in the figures. Gray values of images were analyzed by ImageJ 1.46r software. Values are given as means ± SD. An independent-samples t-test was used to compare means of two groups, one-way analysis of variance (AVOVA) was employed for comparing means among three or more groups followed by Dunnett and Bonferroni post hoc test for the comparison between two groups. The statistical significance was determined with a P value threshold of <0.05. All experimental data were analyzed by GraphPad Prism 5 software.
Results
Let-7c reduces generation of Aβ
MiRNAs are an important regulator of gene expression, and may relate to the pathology of AD. We used a natural model of AD, also known as Down syndrome, to study the mechanisms of miRNAs for AD. The karyotype of DS cell line MB1478 was 47, XX, +21 (Supplementary Figure 1A). A Taqman low-density array (TLDA) human miRNA panel was performed to test the expression of miRNAs in the DS and control cell lines. The results showed that the expression of 32 miRNAs was increased and the expression of 10 miRNAs was decreased with statistical significance in the DS cell line, including let-7c, miR-99a and miR-155, which are all located in chromosome 21 (Figure 1A).
Figure 1.
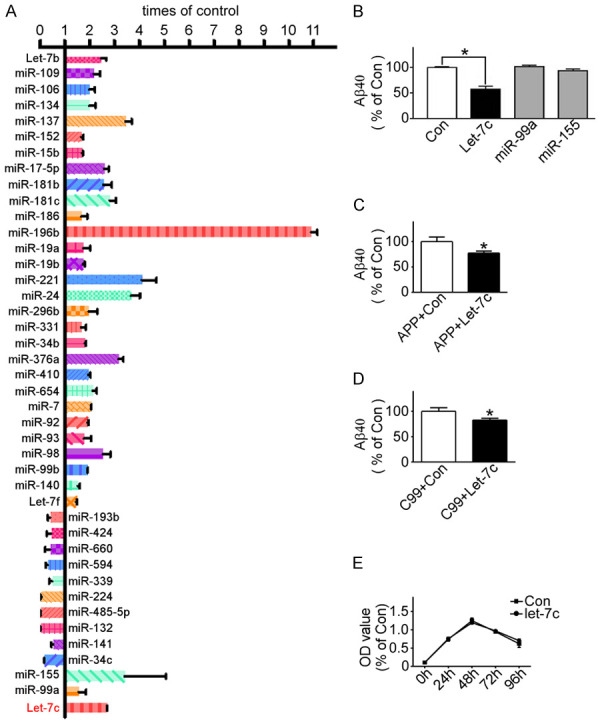
Let-7c increased in DS cell line and reduced generation of Aβ. A. Low-density Taqman array performed to identify panels of miRNA expression in DS cell line MB1478 and normal cell line UMB115; B. 20E2 cells transfected with pmiR-let-7c, pmiR-99a, pmiR-155 or control, and Aβ quantity detected by ELISA Assay. *P<0.05, let-7c compared to the control group; C and D. pZ-APPsw and pAPP-C99 transfected into HEK293 cells with pmiR-let-7c or not, respectively, and Aβ quantity detected by ELISA Assay. *P<0.05, compared to the control group; E. HEK293 cells transfected with pmiR-let-7c or not, respectively, and cell proliferation detected by MTT assay. Values represent means ± SD, n=3.
The neuropathological features of AD are neuritic plaques and neurofibrillary tangles. Neuritic plaques induced by Aβ especially by Aβ42, have strong neurotoxicity. Cleavage of APP by BACE1 produces sAPPβ and CTFβ (C99) respectively, and C99 is subsequently cleaved by γ-secretase to generate Aβ and c-terminal fragment (CTFγ). 20E2 cells that HEK293 cells stably transfected with Swedish mutant APP695 [20] were transfected with pmiR-let-7c, pmiR-99a, pmiR-155 or control to confirm whether let-7c, miR-99a and miR-155 could affect the generation of Aβ. An Aβ ELISA assay showed that let-7c but not miR-99a or miR-155 markedly reduced the expression level of Aβ to 59.11%±3.519 (P<0.05) (Figure 1B). Let-7c also reduced the load of Aβ in HEK293 cells transfected with pZ-APPsw or pAPP-C99 to 77.57%±3.678 (P<0.05) or 82.73%±3.721 (P<0.05) respectively (Figure 1C, 1D). To verify that the decrease of Aβ was caused by let-7c but not by cell death, the cell proliferation was detected. MTT assay illustrated let-7c had no effect on the proliferation of HEK293 cells (Figure 1E). These data demonstrated that the reduction of Aβ probably resulted from the effect of let-7c on the secretase-cutting of C99, not APP.
Let-7c expression panel
A semi-quantitative polymerase chain reaction (RT-PCR) showed that pre-let-7c in the DS cells increased to 136.2%±4.103 (P<0.05) compared with the normal cells (Figure 2A). This was further confirmed by a real-time quantitative-PC (RT-qPCR) of mature let-7c (P<0.05) (Figure 2B). 21-nucleotide let-7 RNA, which was first found in C.elegans, regulated developmental timing and played a vital role in brain development. Total RNAs were extracted from the brains of 8 DS patients and AD mice. Firstly, the gene-type of transgenic AD mice (APPsw/PS1AE9) was confirmed (Supplementary Figure 1B). Let-7c in the 8 DS patients was up-regulated markedly to 186.4%±37.83 (P<0.05) compared with the 5 normal patients by RT-qPCR (Figure 2C). Consistently, the load of let-7c in the brains of AD mice was increased by about 15 times (P<0.05) more than that in wild-type mice by RT-qPCR (Figure 2D).
Figure 2.
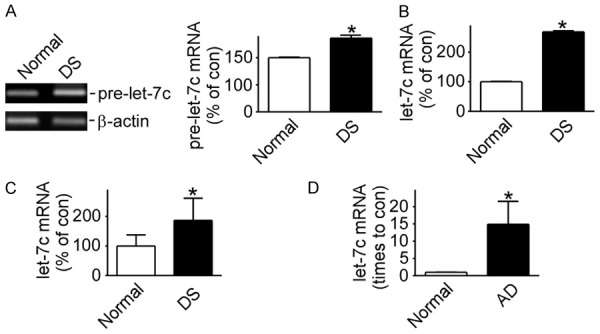
Let-7c expression panels in DS cell lines, DS patients and AD mice. A. pre-let-7c expression detected by RT-PCR in DS cell line MB1478 and normal cell line UMB115. *P<0.05, compared to the normal group; B. Mature let-7c expression detected by RT-qPCR in cell lines MB1478 and UMB115. *P<0.05, compared to the normal group; C and D. let-7c expression detected by RT-qPCR in DS patients (Patients =8, Normal =5) and AD mice (Models =12, Normal =8), respectively. *P<0.05, compared to the normal group. Values represent means ± SD, n=3.
Let-7c reduces Aβ by reducing C99 and increasing C83/80
There are two pathways for APP cleavage: amyloidogenic and non-amyloidogenic. Aβ derives from the amyloidogenic pathway. In the non-amyloidogenic pathway, cleavage of APP by α-secretase generates sAPPα and CTFα (C83) then C83 is subsequently cleaved by γ-secretase to generate P3 and CTFγ, preventing the generation of Aβ. To further examine the effect of let-7c on the cutting secretase of Aβ, the expression of C99 and C83 was measured in 2EB2 that HEK293 cells stably transfected with both the Swedish mutant APP695 and BACE1 [21]. The result showed that let-7c reduced the C99 protein to 72.54%±0.026 (P<0.05) while increased C83 to 123.2%±0.178 (P<0.05) in 2EB2 (Figure 3A). Also, let-7c markedly reduced C99 to 77.16%±0.932 (P<0.05) while increased C83 to 124.8%±3.460 (P<0.05) in HEK293 cells transfected with the pAPP-C99Flag (Figure 3B). These results demonstrated that let-7c affected secretase of C99, such as α-secretase BACE2 [8], but not BACE1. This is consistent with Aβ ELISA assay results.
Figure 3.
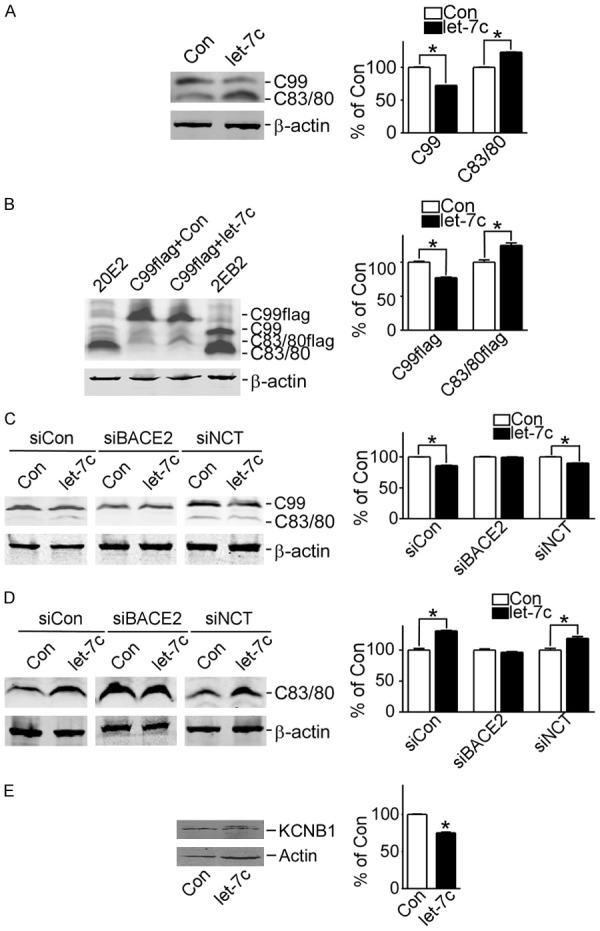
let-7c reduces Aβ production by inducing BACE2 to cut C99. A. 2EB2 cells transfected with pmiR-let-7c or not, and C99 and C83/C80 protein detected by C20 antibody. *P<0.05, let-7c compared to the control group; B. pAPP-C99Flag transfected into HEK293 cells with pmiR-let-7c or not, and C99 and C83/C80 protein detected by C20 antibody. *P<0.05, let-7c compared to the control group; C. pAPP-C99, pAPP-C99 and pshBACE2, pAPP-C99 and pshNCT transfected into HEK293 cells with pmiR-let-7c or not, and C99 protein detected by C20 antibody. *P<0.05, let-7c compared to the control group; D. pAPP-C83, pAPP-C83 and pshBACE2, pAPP-C83 and pshNCT transfected into HEK293 cells with pmiR-let-7c or not, and C83 protein detected by C20 antibody. *P<0.05, let-7c compared to the control group; E. HEK293 cells transfected with pKv2.1-Flag and pmiR-let-7c or not, and KCNB1 protein detected by M2 antibody. *P<0.05, compared to the control group. Values represent means ± SD, n=3.
Further confirmation revealed that the expression of C99 exhibited no difference in HEK293 cells whose BACE2 was disturbed using pshBace2 (Figure 3C, columns 3 and 4) while C99 was decreased by let-7c in NCT-disturbed cells (P<0.05) (Figure 3C, columns 5 and 6). Consistently, the protein level of C83 i exhibited no difference in BACE2-disturbed cells transfected with both pAPP-C83 and pmiR-let-7c but was increased in NCT-disturbed cells (P<0.05) (Figure 3D). Another substrate of BACE2, potassium voltage-gated channel Shab-related subfamily member 1, also known as KCNB1 or Kv2.1 [22], may have been reduced by let-7c in HEK293 cells transfected with pmiR-let-7c (P<0.05) (Figure 3E). These results indicated that let-7c reduced Aβ production by inducing BACE2 to cut C99 and increase C83/80.
Let-7c increases BACE2 expression by targeting its promoter
To confirm whether let-7c affects BACE2, total RNA was extracted from cells transfected with or without pmiR-let-7c. RT-qPCR assays showed that let-7c increased BACE2 mRNA expression to 212.0%±1.788 in cells with pmiR-let-7c compared with the control cells (P<0.05) (Figure 4A). A dual-luciferase reporter assay system was executed to detect the effects of let-7c on the promoter and mRNA-3’UTR of BACE2. Let-7c increased BACE2 promoter activity to 168.4%±14.080 (P<0.05), with no effect on mRNA-3’UTR (Figure 4B, 4C). The effects of let-7c on the promoter and mRNA-3’UTR of BACE1 in addition to the promoter of APP were also examined, but let-7c had no effect on them (Figure 4D-F).
Figure 4.
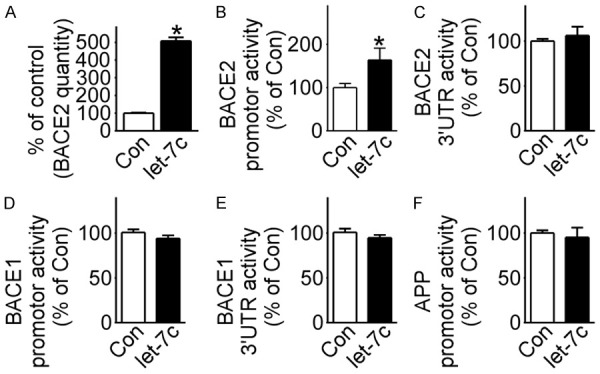
Let-7c targeted BACE2 promoter. A. HEK293 cells transfected with pmiR-let-7c or not, and BACE2 expression detected by RT-qPCR. *P<0.05, compared to the control group; B-F. pB2Luc-A, pBACE2-3UTRluc, pB1P-A, pBACE1-3UTRluc and prhβAPPluc transfected into HEK293 cells with pmiR-let-7c or not, respectively, and dual luciferase assay performed 48 hours after transfection. *P<0.05, compared to the control group. Values represent means ± SD, n=3.
Let-7c induces BACE2 expression through RNAa
To further investigate which region of the BACE2 promoter is targeted by let-7c, three deleted constructs containing different lengths of BACE2 promoter-pB2Luc-B (-446 to +278), pB2Luc-C (-371 to +278) and pB2Luc-D (-200 to +278)-while pB2Luc-A containing the whole promoter (-1580 to +278) were used (Figure 5A). A dual-luciferase reporter assay showed that, compared with the control group, let-7c increased the activity of pB2Luc-A (P<0.05), but not of pB2Luc-B, pB2Luc-C or pB2Luc-D (Figure 5B). The region targeted by let-7c is located between sequences -1580 and -446 of the BACE2 promoter. According to homologous sequence analysis, the targeted region lies between sequences -1368 and -1347 (Figure 5C).
Figure 5.
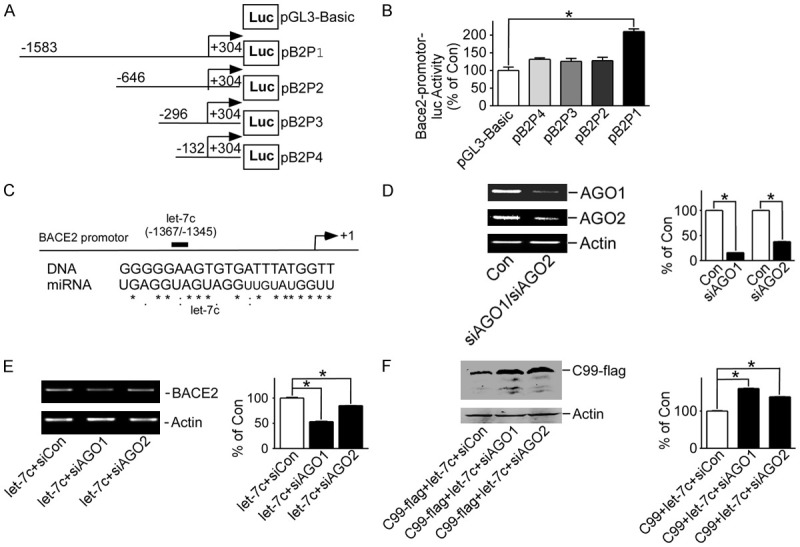
Let-7c induces BACE2 expression through RNAa. A. Schematic diagram of the structure of BACE2 promoter and three deleted constructs; B. pB2Luc-A, pB2Luc-B, pB2Luc-C and pB2Luc-D transfected into HEK293 cells with pmiR-let-7c or not, respectively, and dual luciferase assay performed 48 hours after transfection. *P<0.05, compared to the pGL3-Basic group; C. Sequence alignment of BACE2 and let-7c, with sequences of BACE2 and let-7c aligned using ClustalW2 software; D. HEK293 cells transfected with pshAGO1 or pshAGO2, and AGO1 or AGO2 expression detected by RT-PCR. *P<0.05, compared to the control group; E. HEK293 cells transfected with pshAGO1 or pshAGO2, and BACE2 expression detected by RT-PCR. *P<0.05, compared to the si-control group; F. pAPP-C99Flag and pmiR-let-7c transfected into HEK293 cells with pshAGO1 or pshAGO2, and C99 protein detected by M2 antibody. *P<0.05, compared to the si-control group. Values represent means ± SD, n=3.
RNAa was firstly reported ten years ago [23]. Since then, many types of RNAa have been found [24,25]. RNAa requires the Argonaute (AGO) protein. AGO1 and AGO2 were disturbed using pshAGO1 and pshAGO2, respectively (P<0.05) (Figure 5D). BACE2 mRNA expression increased by let-7c was decreased to 53.25%±0.071 (P<0.05) in AGO1-disturbed cells, and to 84.83%±0.017 (P<0.05) in AGO2-disturbed cells (Figure 5E). C99 down-regulated by let-7c was increased to 160.9%±0.051 (P<0.05) in AGO1-disturbed cells and to 138.2%±0.050 (P<0.05) in AGO2-disturbed cells (Figure 5F).
Discussion
AD is the leading neurodegenerative disease while its pathogenesis remains to be uncovered [26]. The discovery of miRNAs was a great accomplishment in the field of molecular biology in the 21th century. One specific miRNA can bind to multiple mRNAs and can be targeted or regulated by various miRNAs. miRNAs and mRNAs are able to form complicated gene regulatory network, among which genes may be expressed in a spatially and temporally specific manner. Disorder and imbalance in the regulation of miRNAs often cause diseases. However, little is known about the role of miRNAs in AD. In this study, we for the first time revealed that miRNA let-7c was down-regulated in DS patients and AD mice. Moreover, let-7c significantly increased the expression level of BACE2 by targeting its promotor in the manner of RNAa. Given the up-regulation of BACE2, let-7c remarkably reduced the production of Aβ by reducing C99 and increasing C83/80 (Figure 6).
Figure 6.
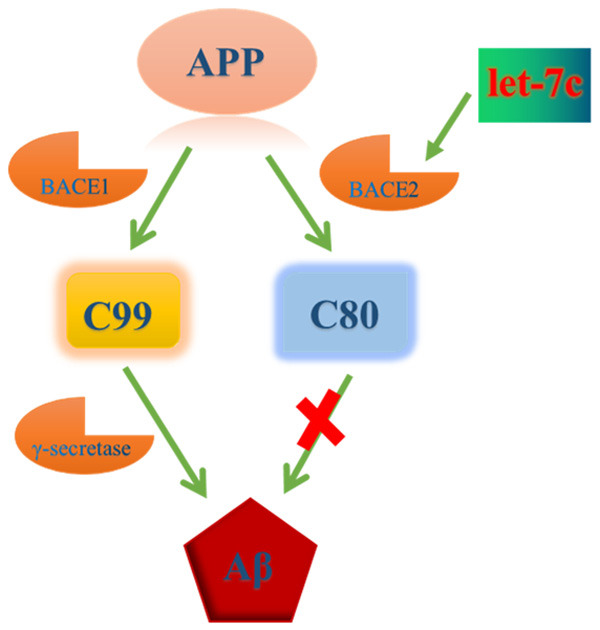
Schematic diagram of miRNA let-7c inducing BACE2 expression and inhibiting Aβ production.
AD is a progressive neurodegenerative disorder and the most common form of dementia worldwide. Several studies using profiling techniques have revealed that various miRNAs were dysregulated in AD human brain tissues. Lukiw used small-scale profiling to elicit the changes of miRNAs in AD [27]. Since then, several groups, including the Lukiw Laboratory, have performed large-scale genome-wide studies to demonstrate that miRNA expression patterns are altered not only in the AD brain but also in blood and cerebrospinal fluid [28-30]. DS is caused by the triplication of human chromosome 21 (HSA21), causing deficits in cognitive function and neurodegeneration of cholinergic basal forebrain neurons, a pathological hallmark of AD [32]. Therefore, DS is a natural disease model of AD. A small-scale profiling study of the DS cell model revealed 42 miRNAs with statistically significant differences, including let-7c, which is located at chromosome 21. Let-7c, belonging to the let-7 family and one of the earliest microRNAs, has a broad range of functions in plants and animals. Nevertheless, the roles of let-7c in patients with AD remain unclear. In this investigation, we found the expression level of let-7c, miR-99a and miR-155 was decreased in DS cell line dramatically. In addition, let-7c was also up-regulated in DS patients and AD mice.
According to previous reporters, Aβ deposition is involved in patients diagnosed with AD and contributes to the learning in addition to memory deficiency [33,34], indicating the vital effects of Aβ plaque on AD. Interestingly, we then found let-7c but not miR-99a or miR-155 down-regulated the expression of Awith AD and contributes to the learning in addition to memory uced by cell death. Based on these findings, let-7c may be related to benefits for an organism with AD even though abnormal expression of various miRNAs is usually regarded as a detrimental factor in the progression of AD [35,36]. Subsequently, we discovered that let-7c decreased the protein level of C99 while increased the level of C83/80 in NCT-disturbed cells but not in BACE2-disturbed cells, indicating that let-7c reduces Aβ production by inducing BACE2 to cut C99 and increase C83/80, which is in line accordance with previous study about the formation of Aβ [8].
It is well-known that one strand of the miRNA duplex is loaded onto an AGO2 protein in RNAi-induced silencing complex (RISC) [4,37,38]. Li et al. first reported and named the RNAa phenomenon, originally observing the positive regulation of gene transcription and epigenetic inheritance by small RNAs targeting the gene promoter and directing the RNA-Argonaute pathway [23]. Many subsequent studies have reported similar results [11,24,25,39,40]. Consistently, our experiments revealed that disturbing AGO1 or AGO2 inhibited the effect of let-7c, increased BACE2 mRNA expression and C99 cutting by BACE2, which confirmed that let-7c triggered RNAa to exert its functions. Like the 3’UTR sequence of mRNA, the promoter sequence also contains a target site of miRNA. MiRNA may positively or negatively regulate the transcriptional activity of the promoter. Analysis of the promoter-reporter system as well as blast sequence was conducted and the results indicated that the BACE2 promoter had a let-7c target site. In addition, truncation promoter of BACE2 further verified that the let-7c could bind to BACE2 in the sequence between -1368 and -1347.
There existed some limitations in our present investigation. We only uncovered that miRNA let-7c played vital roles in reduction of Aβ but we did not confirm whether these effects are associated with neuron damage in vitro and in vivo. Furthermore, more explorations should be employed to verify the clinical functions of let-7c.
In conclusion, this study is the first to report that let-7c induces BACE2 expression and inhibits Aβ production through RNAa and compensate for other adverse factors in the AD pathological progress, which provides a potential target for treatment of AD, although further studies are needed to explore the effects of let-7c on BACE2 RNAa.
Acknowledgements
This work was supported by Project of medical and health technology development program in Shandong province (2015WS0295) to Heng Liu; by Shenzhen strategic emerging industry development special funds (JCYJ20150430160921948) to Xiulian Sun; by NSFC (81701058), Shandong Academy of Sciences (ZR2017PH027), and China Postdoctoral Science Foundation (2017M612288) to Tan Wang.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold KH, Mistl C, Rothacher S, Ledermann B, Burki K, Frey P, Paganetti PA, Waridel C, Calhoun ME, Jucker M, Probst A, Staufenbiel M, Sommer B. Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc Natl Acad Sci U S A. 1997;94:13287–13292. doi: 10.1073/pnas.94.24.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cruz L, Urbanc B, Buldyrev SV, Christie R, Gomez-Isla T, Havlin S, McNamara M, Stanley HE, Hyman BT. Aggregation and disaggregation of senile plaques in Alzheimer disease. Proc Natl Acad Sci U S A. 1997;94:7612–7616. doi: 10.1073/pnas.94.14.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuo YM, Kokjohn TA, Beach TG, Sue LI, Brune D, Lopez JC, Kalback WM, Abramowski D, Sturchler-Pierrat C, Staufenbiel M, Roher AE. Comparative analysis of amyloid-beta chemical structure and amyloid plaque morphology of transgenic mouse and Alzheimer’s disease brains. J Biol Chem. 2001;276:12991–12998. doi: 10.1074/jbc.M007859200. [DOI] [PubMed] [Google Scholar]
- 4.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mori H, Takio K, Ogawara M, Selkoe DJ. Mass spectrometry of purified amyloid beta protein in Alzheimer’s disease. J Biol Chem. 1992;267:17082–17086. [PubMed] [Google Scholar]
- 6.Dislich B, Lichtenthaler SF. The membrane-bound aspartyl protease BACE1: molecular and functional properties in Alzheimer’s disease and beyond. Front Physiol. 2012;3:8. doi: 10.3389/fphys.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun X, Wang Y, Qing H, Christensen MA, Liu Y, Zhou W, Tong Y, Xiao C, Huang Y, Zhang S, Liu X, Song W. Distinct transcriptional regulation and function of the human BACE2 and BACE1 genes. FASEB J. 2005;19:739–749. doi: 10.1096/fj.04-3426com. [DOI] [PubMed] [Google Scholar]
- 8.Sun X, He G, Song W. BACE2, as a novel APP theta-secretase, is not responsible for the pathogenesis of Alzheimer’s disease in down syndrome. FASEB J. 2006;20:1369–1376. doi: 10.1096/fj.05-5632com. [DOI] [PubMed] [Google Scholar]
- 9.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 10.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC. Upregulation of cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695–1707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang V, Li LC. miRNA goes nuclear. RNA Biol. 2012;9:269–273. doi: 10.4161/rna.19354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012;9:1066–1075. doi: 10.4161/rna.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X, He G, Qing H, Zhou W, Dobie F, Cai F, Staufenbiel M, Huang LE, Song W. Hypoxia facilitates Alzheimer’s disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci U S A. 2006;103:18727–18732. doi: 10.1073/pnas.0606298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song WH, Lahiri DK. Molecular cloning of the promoter of the gene encoding the Rhesus monkey beta-amyloid precursor protein: structural characterization and a comparative study with other species. Gene. 1998;217:151–164. doi: 10.1016/s0378-1119(98)00337-0. [DOI] [PubMed] [Google Scholar]
- 18.Liu FC, Zhang Y, Liang ZL, Sun QW, Liu H, Zhao J, Xu JW, Zheng JF, Yun Y, Yu X, Song WH, Sun XL. Cleavage of potassium channel Kv2.1 by BACE2 reduces neuronal apoptosis. Mol Psychiatry. 2018;23:1542–1554. doi: 10.1038/s41380-018-0060-2. [DOI] [PubMed] [Google Scholar]
- 19.Christensen MA, Zhou W, Qing H, Lehman A, Philipsen S, Song W. Transcriptional regulation of BACE1, the beta-amyloid precursor protein beta-secretase, by Sp1. Mol Cell Biol. 2004;24:865–874. doi: 10.1128/MCB.24.2.865-874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qing H, Zhou W, Christensen MA, Sun X, Tong Y, Song W. Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 2004;18:1571–1573. doi: 10.1096/fj.04-1994fje. [DOI] [PubMed] [Google Scholar]
- 21.Deng Y, Wang Z, Wang R, Zhang X, Zhang S, Wu Y, Staufenbiel M, Cai F, Song W. Amyloid-beta protein (Abeta) Glu11 is the major beta-secretase site of beta-site amyloid-beta precursor protein-cleaving enzyme 1(BACE1), and shifting the cleavage site to Abeta Asp1 contributes to Alzheimer pathogenesis. Eur J Neurosci. 2013;37:1962–1969. doi: 10.1111/ejn.12235. [DOI] [PubMed] [Google Scholar]
- 22.Liu F, Zhang Y, Liang Z, Sun Q, Liu H, Zhao J, Xu J, Zheng J, Yun Y, Yu X, Song W, Sun X. Cleavage of potassium channel Kv2.1 by BACE2 reduces neuronal apoptosis. Mol Psychiatry. 2018;23:1542–1554. doi: 10.1038/s41380-018-0060-2. [DOI] [PubMed] [Google Scholar]
- 23.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF, Li LC. RNAa is conserved in mammalian cells. PLoS One. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo D, Barry L, Lin SS, Huang V, Li LC. RNAa in action: from the exception to the norm. RNA Biol. 2014;11:1221–1225. doi: 10.4161/15476286.2014.972853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain G, Stuendl A, Rao P, Berulava T, Pena Centeno T, Kaurani L, Burkhardt S, Delalle I, Kornhuber J, Hull M, Maier W, Peters O, Esselmann H, Schulte C, Deuschle C, Synofzik M, Wiltfang J, Mollenhauer B, Maetzler W, Schneider A, Fischer A. A combined miRNA-piRNA signature to detect Alzheimer’s disease. Transl Psychiatry. 2019;9:250. doi: 10.1038/s41398-019-0579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 28.Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiko T, Nakagawa K, Tsuduki T, Furukawa K, Arai H, Miyazawa T. MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J Alzheimers Dis. 2014;39:253–259. doi: 10.3233/JAD-130932. [DOI] [PubMed] [Google Scholar]
- 30.Tan L, Yu JT, Tan MS, Liu QY, Wang HF, Zhang W, Jiang T, Tan L. Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer’s disease. J Alzheimers Dis. 2014;40:1017–1027. doi: 10.3233/JAD-132144. [DOI] [PubMed] [Google Scholar]
- 31.Femminella GD, Ferrara N, Rengo G. The emerging role of microRNAs in Alzheimer’s disease. Front Physiol. 2015;6:40. doi: 10.3389/fphys.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alldred MJ, Lee SH, Petkova E, Ginsberg SD. Expression profile analysis of hippocampal CA1 pyramidal neurons in aged Ts65Dn mice, a model of down syndrome (DS) and Alzheimer’s disease (AD) Brain Struct Funct. 2015;220:2983–2996. doi: 10.1007/s00429-014-0839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGowan E, Eriksen J, Hutton M. A decade of modeling Alzheimer’s disease in transgenic mice. Trends Genet. 2006;22:281–289. doi: 10.1016/j.tig.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Gotz J, Gotz NN. Animal models for Alzheimer’s disease and frontotemporal dementia: a perspective. ASN Neuro. 2009;1:e00019. doi: 10.1042/AN20090042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kao YC, Wang IF, Tsai KJ. miRNA-34c overexpression causes dendritic loss and memory decline. Int J Mol Sci. 2018;19:2323. doi: 10.3390/ijms19082323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Liu D, Huang HZ, Wang ZH, Hou TY, Yang X, Pang P, Wei N, Zhou YF, Dupras MJ, Calon F, Wang YT, Man HY, Chen JG, Wang JZ, Hebert SS, Lu Y, Zhu LQ. A novel MicroRNA-124/PTPN1 signal pathway mediates synaptic and memory deficits in Alzheimer’s disease. Biol Psychiatry. 2018;83:395–405. doi: 10.1016/j.biopsych.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 37.Mallory AC, Elmayan T, Vaucheret H. MicroRNA maturation and action--the expanding roles of ARGONAUTEs. Curr Opin Plant Biol. 2008;11:560–566. doi: 10.1016/j.pbi.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 39.Meng X, Jiang Q, Chang N, Wang X, Liu C, Xiong J, Cao H, Liang Z. Small activating RNA binds to the genomic target site in a seed-region-dependent manner. Nucleic Acids Res. 2016;44:2274–2282. doi: 10.1093/nar/gkw076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Portnoy V, Lin SH, Li KH, Burlingame A, Hu ZH, Li H, Li LC. saRNA-guided Ago2 targets the RITA complex to promoters to stimulate transcription. Cell Res. 2016;26:320–335. doi: 10.1038/cr.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


