Abstract
Objectives: MUC16, a mucin marker with a high mutation probability, is closely related to the occurrence, development, response to treatment, and prognosis of melanoma. As melanoma has high immunogenicity, immunotherapy has become a routine treatment. Tumor mutation burden (TMB) is the most common indicator for determining appropriate immunotherapy. The relationship between the mutation and expression of MUC16 and the prognosis, TMB, level of immune infiltration, and drug sensitivity in melanoma was investigated in this study. Methods: Melanoma data were downloaded from the Cancer Genome Atlas and the International Cancer Genome Consortium database, and the “GenVisR” package was used to visualize the gene mutation types and frequencies. Intersections of the top 30 genes with the highest mutation frequencies were determined. Thereafter, we investigated the effects of MUC16 mutations on overall survival (OS) and TMB of melanoma patients by multivariate Cox regression and multivariate logistic analyses. Related pathways that were enriched by MUC16 and BRAF were investigated using gene-set enrichment analysis and gene-set variation analysis. The CIBERSORT calculation method was used to analyze the proportion of tumor-infiltrating immune subsets. The relationship between MUC16 expression and drug sensitivity was also discussed. Results: Twenty-two genes with high mutation frequencies were identified in both datasets. MUC16 and ADGRV1 mutations were associated with higher TMB and good clinical prognosis (P<0.05). Multivariate Cox regression analysis showed that age, clinical stage, and MUC16 mutations were independent prognostic factors affecting OS of melanoma patients. Multivariate logistic analysis showed that gender and MUC16 mutations were independent prognostic factors affecting the TMB. MUC16 mutations and high-expression groups were primarily enriched in immune-related pathways. Furthermore, T-cell CD4 memory activation and T-cell CD8 were positively correlated with MUC16 expression and activated dendritic cells were significantly enriched in the MUC16 mutant group. Abnormal MUC16 expression may be related to abnormal methylation and drug resistance. Conclusion: MUC16 was found to have a higher mutation frequency in melanoma patients, which is associated with a higher TMB. The mutation and/or expression of MUC16 may affect immune-related pathways and tumor-infiltrating immune cell subsets, which may improve the prognosis for melanoma patients.
Keywords: Cutaneous melanoma, MUC16, tumor mutation burden, immune, prognosis
Introduction
A skin cutaneous melanoma (SKCM) is a malignant tumor that originates from skin melanocytes and is potentially fatal. The incidence rate of melanoma is increasing, whereas the incidence rate of various tumors is decreasing [1]. The high malignancy of SKCM implies that its mortality accounts for 75% of the total skin cancer mortality [2]. According to a survey report by the American Joint Commission on Cancer in 2018, there are approximately 91,270 new cases and 9,320 deaths related to SKCM every year in the United States [3], and by 2020, these numbers will be 100,350 and 6,850, respectively [4]. This means that the number of new cases each year will increase, but the number of deaths will decrease slightly. Globally, there are approximately 200,000 new SKCM cases each year; however, the incidence of SKCM in the Asian population is lower than that in European/Caucasian populations. However, as the population of China is large, there are still approximately 20,000 new cases in China each year, and the mortality rate is higher than that in Western countries [5]. Thus, melanoma presents a serious threat to the health of Chinese people. When compared with those for other common malignancies, the standardized diagnosis and treatment methods for melanoma are not as advanced.
Mucoprotein (MUP) is a glycoprotein that is primarily composed of mucopolysaccharides, which not only plays an important role in intercellular signal transduction but is also closely related to intercellular adhesion and immune response [6]. The expression of MUP is associated with various cancers, and its role in tumors has received increasing attention in recent years. Studies have reported that MUP can accelerate cell metastasis and diffusion by reducing the adhesion between tumor cells and by enabling tumor cells to regulate the immune system, and thus escape it [7,8]. MUP16 (formerly known as CA12-5), is a member of the mucin family and has a high mutation frequency in many cancers, including gastric cancer, colorectal cancer, and non-small cell lung cancer, and these mutations are often associated with patient survival [9,10]. Tumor mutation burden (TMB) is defined as the total number of somatic gene coding errors, base substitutions, gene insertions, or deletion errors detected per million bases, which is the mutation density of genes [11]. A higher mutation load indicates that the tumor has a more prominent personality and will be targeted by the tumor immunity as it is more likely to be recognized by the immune system. Theoretically, the higher the tumor mutation load, the more effective the immunotherapy treatment will be, as was confirmed by Zhang et al. By analyzing the correlation between MUC16 mutations and immune checkpoint inhibitor responses in different solid tumors, MUC16 mutations appeared to be related to the response of the immune checkpoint inhibitors (ICIs) in solid tumors and the genomic factors related to improved prognosis. It has also been suggested that MUC16 could be utilized as a marker to guide immunotherapy responses [9].
In this study, we aimed to explore the associations among somatic cell mutations, TMB, and the prognosis of SKCM patients to determine the relationship between gene mutations and immune responses. Specifically, we downloaded the data of American SKCM patients from the Cancer Genome Atlas (TCGA) database and Australia SKCM patients from the International Cancer Genome Consortium (ICGC) database; the intersection of mutation genes in the two cohorts was used to analyze the TMB and prognosis of the patients. On this basis, this study further explored the immune response, the functional enrichment of the mutant genes, and the relationship between the expression level of the mutant genes and methylation and drug sensitivity.
Methods
Data
Data for 472 patients with SKCM were downloaded for this investigation from the TCGA website (http://portal.gdc.cancer.gov/projects), including transcriptome data, clinical information, and somatic mutation data. In addition, the somatic mutation data for 198 SKCM patients in Australia were downloaded from the ICGC official website (https://dcc.icgc.org).
Identification of mutant genes
In this study, MAF files were obtained from the TCGA database using the VARSCAN method to detect somatic mutations, and they were then used for mutation frequency analysis. The mutation data in the ICGC database (TSV files) were annotated according to the HG19 reference genome. Finally, the “GenVisR” package was used to visualize the frequency and type of the gene mutations in the two datasets, and the “Venn” package was used to determine the intersection of the mutant genes in the two datasets, and only the intersection genes were analyzed further.
Calculation and prognostic analysis of TMB
TMB is the total number of mutated bases per million bases. In this study, we only calculated the number of mutations that caused amino acid changes. We extracted somatic mutation information using a Perl script and corrected the TMB value for each sample by dividing the total number of mutations into the total exon length (38 Mb) [12]. Then, we combined the patient’s TMB information, gene mutation information, and clinical information using R software and visualized the relationship between the mutated genes and the TMB using the “ggboxplot” package. Finally, we divided the patients into wild-type and mutant-type groups based on whether a gene mutation occurred. K-M survival curves were drawn to compare the survival differences between the two groups, and the genes with the most significant P values (MUP16 in this study) were selected for subsequent analysis.
Independent prognostic analysis of MUP16
Using other clinical information (such as age and gender) as independent variables and the OS as a dependent variable, univariate and multivariate Cox regression analyses were conducted to explore whether MUC16 mutations could be independent of other clinical variables. On this basis, a multivariate logistic analysis was conducted to explore the independent factors affecting TMB.
Molecular characterization of MUP16
To explore the functions and pathways that were changed after gene mutation, Gene Set Enrichment Analysis (GSEA) software V4.0 was used to analyze the wild-type and mutated MUP16 patients. This study also used this method to explore the influence of the changes in the expression level of MUP16 on the related pathways in SKCM patients. Specifically, the patients were divided into two groups with high and low expression using the median expression of MUP16, and then the “GSVA” package was used to find the path most related to MUP16 and visualize it.
Relationship between MUP16 and immune cell infiltration
To evaluate the relative abundances of different MUP16 statuses and immune cell infiltration, we downloaded the “CIBERSORT” package and the gene characteristic text containing 22 types of immune cells to transform the transcriptome matrix into the matrix of the immune cell content. Only 232 tumor samples with a P<0.05 were analyzed by quality filtering, and the results were further visualized using the “corrplot” package.
In addition, we further analyzed the relationship between MUP16 and immune cell infiltration using different grouping methods. Specifically, the patients were divided into wild-type and mutant-type groups according to whether the MUP16 was mutated, and differentially expressed immune cells were obtained by differential analysis. Second, we used correlation analysis to obtain the immune cells related to MUP16 expression.
Relationship between MUP16 and methylation and drug sensitivity
The human disease methylation database (DiseaseMeth, http://bioinfo.hrbmu.edu.cn/diseasemeth/) is an interactive database designed to provide normal and abnormal DNA methylation statuses and other relevant information for human diseases (especially various cancers) [13]. We used this database to explore and visualize the differences in MUP16 methylation between normal and tumor samples of SKCM.
We downloaded the data of different cancer cell lines from the NCI-60 database (https://discover.nci.nih.gov/cellminer/home.do) and used the Pearson correlation test to explore the relationship between MUP16 expression and drug sensitivity. Only 263 FDA-approved drugs or drugs in clinical trials were included in the analysis.
Statistical analysis
All statistical analyses were performed using SPSS 22.0 (Chicago, IL, USA) and R 3.6.1 (https://www.r-project.org/), and statistical images were processed using Adobe Illustrator CC 2018. The Kaplan-Meier survival analysis was used to draw the survival curve of the relationship between gene mutation and prognosis. The Wilcoxon test was used to calculate the differentially expressed immune cells of patients with wild-type and mutant variations, and Spearman correlation analysis was used to obtain the immune cells related to the expression of MUC16. The correlation between MUC16 expression and drug sensitivity was determined using Pearson’s correlation coefficient. All data were statistically significant with P<0.05, and the definition of “*” was P<0.05, “**” was P<0.01, and “***” was P<0.001.
Results
Gene mutation in melanoma
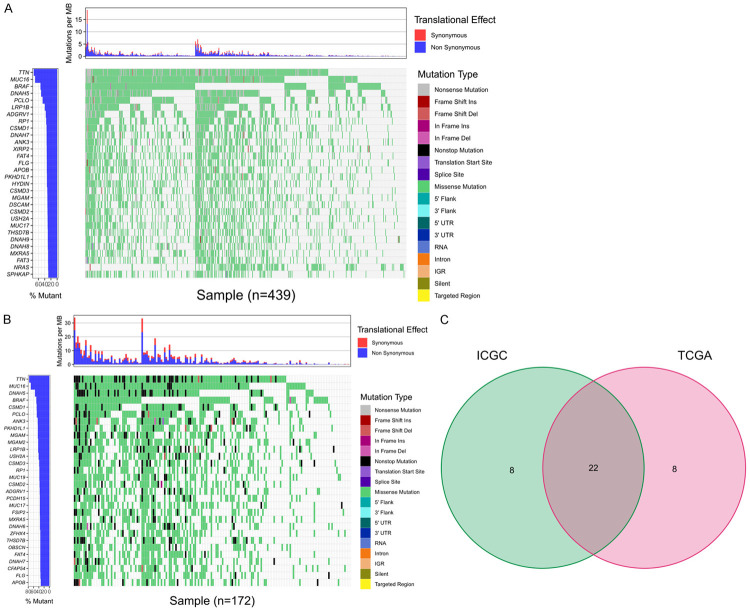
The detailed mutation information for the top 30 genes with the highest mutation frequencies from each sample cohort is presented in a waterfall diagram. In the TCGA cohort, TTN (75.6%), MUC16 (71.1%), BRAF (53.8%), DNAH5 (51.7%), and PCLO (46.9%) were the most frequently mutated genes (Figure 1A). The top five genes with the highest mutation frequency in the ICGC cohort were TTN (81.4%), MUC16 (75.6%), DNAH5 (58.7%), BRAF (54.1%), and CSMD1 (53.5%) (Figure 1B).
Figure 1.
Analysis of the somatic mutation spectrum in melanoma samples. Waterfall plot for the top 30 genes with the highest mutation frequencies in TCGA (A) and ICGC (B). Different colors are used to distinguish the different mutation types. Venn plot (C) showing the intersection of the mutated genes in the TCGA and ICGC cohorts.
We then intersected the top 30 genes with the highest mutation frequencies in the two cohorts and identified 22 overlapping genes: TTN, MUC16, DNAH5, BRAF, CSMD1, PCLO, MGAM, ANK3, LRP1B, PKHD1L1, USH2A, RP1, CSMD3, CSMD2, ADGRV1, MUC17, DNAH7, MXRA5, APOB, FAT4, FLG, and THSD7B.
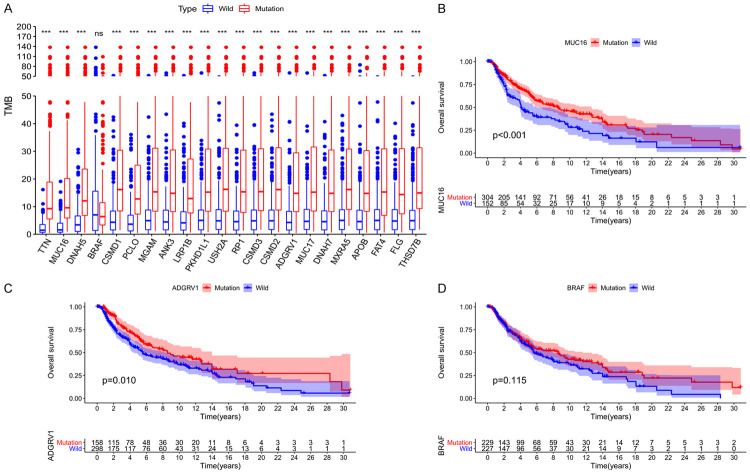
Mutations associated with TMB and survival prognosis
After calculating the TMB value of each sample, we divided the patients into wild-type and mutant-type groups according to the status of the intersection genes, and then compared the TMB values of the two groups. The results showed that, except for BRAF, the other 21 genes had higher TMB after mutation (Figure 2A). On this basis, we performed K-M analysis of the 21 genes mentioned above, and the results showed that only MUC16 and AdGRV1 mutations were associated with improved prognosis, while BRAF mutations did not affect patient OS (Figure 2B-D). Considering the important role of BRAF in the occurrence, development, and treatment of melanomas, both BRAF and the gene with the most significant P value (MUC16) were selected for subsequent analysis.
Figure 2.
Relationship between gene mutation and tumor mutation burden and clinical prognosis. Tumor mutation burden after gene mutation almost increased to different degrees (A); The OS time for patients with MUC16 (B) and ADGRV1 (C) mutations was significantly longer than that for wild type patients, and BRAF (D) mutations did not affect the OS rate of patients.
Effects of MUC16 mutations on OS and TMB
Univariate Cox regression analysis showed that age, clinical stage, and MUC16 mutations had an impact on the survival time of melanoma patients (Figure 3A). Subsequent multivariate Cox regression analysis showed that age (hazard ratio: 1.807, 95% confidence interval: 1.332-2.453, P<0.001), clinical stage (hazard ratio: 1.586, 95% confidence interval: 1.186-2.121, P=0.002), and MUC16 mutations (hazard ratio: 0.552, 95% confidence interval: 0.403-0.754, P<0.001) were all independent prognostic factors affecting OS in patients with melanoma (Figure 3B).
Figure 3.
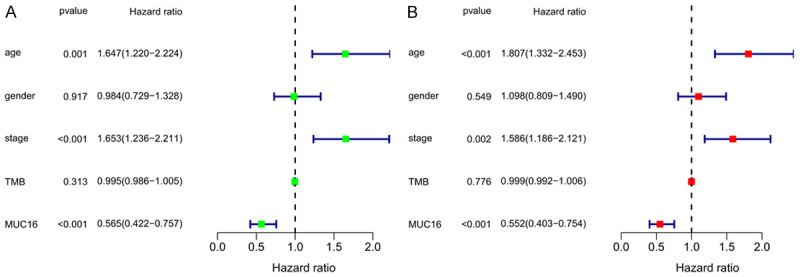
Univariate (A) and multivariate (B) COX regression analyses were used to explore the factors influencing the overall survival rate of melanoma patients.
Multivariate logistic analysis showed that gender (odds ratio: 1.932, 95% confidence interval: 1.199-3.113, P=0.007) and MUC16 mutations (odds ratio: 13.762, 95% confidence interval: 7.800-24.282, P=1.421E-19) were independent influencing factors of the TMB value (Table 1).
Table 1.
Multivariate logistic analysis results
| OR | 95% CI of OR | P value | ||
|---|---|---|---|---|
|
| ||||
| Lower limit | Upper limit | |||
| Age | 1.230 | 0.767 | 1.974 | 0.390 |
| Gender | 1.932 | 1.199 | 3.113 | 0.007 |
| Stage | 0.630 | 0.397 | 1.000 | 0.050 |
| Fustatu | 0.906 | 0.568 | 1.443 | 0.677 |
| MUC16 mutant | 13.762 | 7.800 | 24.282 | 1.421E-19 |
OR: odds ratio. CI: Confidence interval.
Functional enrichment analysis of MUC16 and BRAF
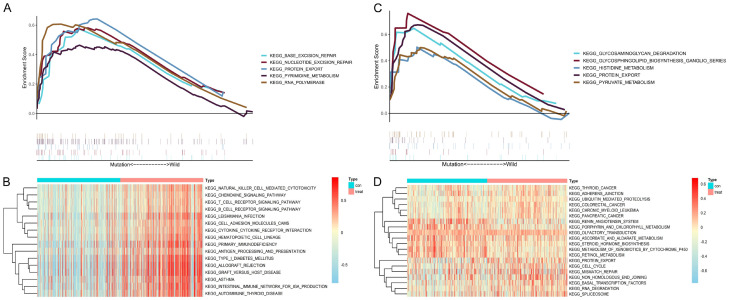
We further studied the potential function and pathway of MUC16 and BRAF in SKCM by conducting GSEA and gene set variation analysis (GSVA) on the TCGA cohort (Figure 4A-D). The GSEA results showed that BASE_EXCISION_REPAIR, NUCLEOTIDE_EXCISION_REPAIR, PROTEIN_EXPORT, PYRIMIDINE_METABOLISM, and RNA_POLYMERASE were significantly enriched in the MUC16 mutant samples. In the BRAF mutant samples, the significantly enriched pathways were GLYCOSAMINOGLYCAN_DEGRADATION, GLYCOSPHINGOLIPID_BIOSYNTHESIS_GANGLIO_SERIES, HISTIDINE_METABOLISM, PROTEIN_EXPORT, and PYRUVATE_METABOLISM.
Figure 4.
Gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) of MUC16 (A, B) and BRAF (C, D) in the TCGA dataset.
Notably, GSVA showed that the MUC16 high-expression group had significant enrichment in immune-related pathways, while the BRAF high-expression group was enriched in tumor-related pathways.
Correlation analysis of MUC16 and immune cells
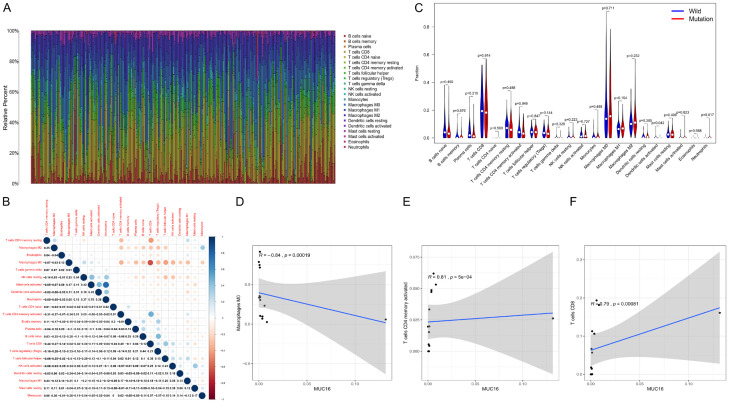
We further used the CIBERSORT algorithm to evaluate the proportion and correlation of immune cell infiltration in the SKCM tumor microenvironment. We constructed 22 immune cell maps and analyzed the correlation between the immune cells (Figure 5A, 5B). The results showed that the mast cells were activated, the neutrophils were the immune cells with the strongest positive correlation (r=0.78), and the T cells CD8 and macrophages M2 had the strongest negative correlation (r=-0.64).
Figure 5.
Relationship between MUC16 and tumor infiltrating immune cells. The CIBERSORT algorithm was used to calculate the proportions of the 22 types of immune cells in each of the skin melanoma samples (A) and the correlation matrix of the immune cells (B), with blue indicating a positive correlation and red indicating a negative correlation. Immune cells differentially expressed by the MUC16 mutation and wild type groups (C). The three types of immune cells associated with MUC16 expression (D-F).
In addition, we observed the enrichment of dendritic cells (DCs) that were activated in the MUC16 mutant group (Figure 5C). Correlation analysis showed that there were three types of immune cells associated with MUP16 expression. Two of them were positively correlated with the expression of MUC16, namely CD4 memory activated T cells and T cells CD8. M0 macrophages were negatively correlated with MUC16 expression (Figure 5D-F). These results demonstrated that the mutation status and expression level of MUC16 could affect the immune activity of SKCM patients.
Relationship between MUC16 and methylation and drug sensitivity
We explored the relationship between the expression level of MUC16 and its methylation status using the DiseaseMeth 2.0 database, and the results showed that the average methylation level of MUC16 in the SKCM tumor tissue was significantly decreased (Figure 6A).
Figure 6.
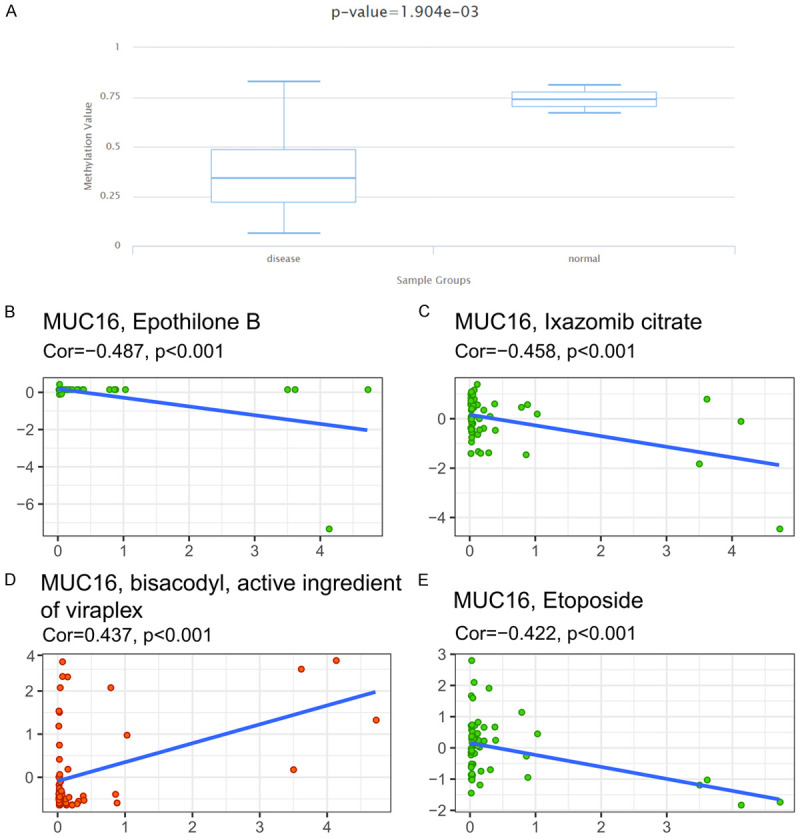
Methylation analysis of MUC16 expression and its relationship with drug sensitivity. Methylation levels in cutaneous melanoma tumors and paracancerous tissues were detected using DiseaseMeth 2.0 (A). Using NCI-60 cell line data, Pearson correlation test was used to analyze the relationship between MUC16 expression and drug sensitivity (B-E).
At the same time, we explored the relationship between the expression level of MUC16 and drug sensitivity, and only the top four drugs with the most significant P-values are shown. Among them, the sensitivity of one drug, bisacodyl, an active ingredient of viraplex, was positively correlated with the expression of MUC16. The sensitivity of the remaining three drugs, namely, epothilone B, ixazomib citrate, and etoposide, was negatively correlated with the expression of MUC16 (Figure 6B-E).
Discussion
MUP is a high molecular weight O-glycoprotein that is primarily expressed on the apical surface of epithelial cells and plays a complex role in the protection of epithelial cells and in carcinogenesis. Abnormal MUP overexpression in tumor cells can regulate various signal transduction pathways and can ultimately promote tumor cells to develop into more aggressive phenotypes [14]. Current studies have reported that MUC16 is one of the three most frequently mutated genes in tumors. It is overexpressed in different types of cancers, such as pancreatic, breast, and lung cancers, and is closely related to disease prognosis [15-18]. By analyzing gastric cancer data from the TCGA database, Li et al. found that MUC16 mutations were significantly related to patients’ OS and response to treatment, and this was further verified by using an external data set [10]. This indicates that it is feasible for clinical researchers to explore the functions of target genes and identify therapeutic targets by using gene database analysis. However, there are few studies on MUC16 expression in melanoma. Current melanoma diagnosis and treatment methods require improvement, and this highlights the need to further explore the potential functions and roles of MUC16 in melanoma.
In this study, TCGA and ICGC cohorts were analyzed, and MUC16 was confirmed to be one of the most mutable genes in melanoma. We found that MUC16 mutations were significantly associated with higher TMB and improved prognosis and were independent factors affecting patient OS. TMB is regarded as a biomarker of immunotherapy and has been used to identify patients who may benefit from immunotherapy for many cancer types [19,20]. Previous clinical trials have shown that patients with high TMB can benefit more from ICIS in patients with melanoma and non-small cell lung cancer [21,22]. As one of the most immunogenic tumors, melanoma has an ideal response to immunotherapy [23]. The above viewpoint was also confirmed by Wang et al., who demonstrated that patients with melanoma with MUC16 mutations have increased expression of immune checkpoints (e.g., PD-L1, PD-1, and CTLA-4). Additionally, the response rate to treatment in patients with melanoma with MUC16 mutations was significantly higher than that in patients without MUC16 mutations. However, these results were only observed in male patients with melanoma [24].
Consequently, melanoma has been used to promote immunotherapy use for solid tumors. Clinical trials in recent years also further confirmed the effectiveness of ICIS in patients with advanced melanoma; the 5-year OS rates of pembrolizumab, nivolumab, and nivolumab combined with ipilimumab were 34%, 44%, and 52%, respectively [25,26]. In addition, Kang et al. reached similar conclusions via an analysis of melanoma data from the TCGA database, which revealed that TMB was positively correlated with the prognosis of patients and was associated with a lower pathological stage [27]. On this basis, we have speculated that the development of drugs targeting MUC-16 could improve the prognosis of melanoma patients by increasing their TMB and consequently improving the effects of immunotherapy.
We further analyzed the influence of the MUC16 mutation and its expression on the related molecular mechanisms and pathways in melanoma patients. GSEA and GSVA showed that the mutation and upregulation of MUC16 expression increased the signaling pathways of mutation repair and immune responses. By considering the relationship between MUC16 and immune responses and immunotherapy in previous literature, we analyzed tumor-infiltrating immune cells in patients with melanoma. The results showed that more DCs were activated in the MUC16 mutant samples. Combined with the better prognosis of patients with MUC16 mutations, the results of this study support previous findings that DCs promote immune activation in the tumor microenvironment and play an important role in immune response [28-30].
This study also found that in melanoma patients, both the T cells CD4 memory activated and T cells CD8 were positively correlated with the expression of MUC16. Recent studies have also reported that to slow down the growth of tumors and prolong the survival of patients, the efficacy of adoptive immune cell therapy can be improved by infiltrating CD4+ T lymphocytes and CD8+ T lymphocytes into tumor tissues. This has been confirmed in patients with solid tumors such as melanoma, breast cancer, and lung cancer [31,32]. In addition, anti-tumor research on infiltrating T immune cells showed that the tumor tissue infiltration activated CD4+ T lymphocytes, CD8+ T lymphocytes, natural killer cells, and other immune active cells in vitro for a period of time after separation; reinjection of these cells into patients’ bodies effectively killed tumor cells and had curative effects on esophageal cancer, colorectal cancer, lung cancer, and other tumors [33-35]. Therefore, after the analysis of the mutation and expression of MUC16, it was suggested that MUC16 may interfere with the maintenance and regulation of the immune activity of the melanoma patients in both direct and indirect ways and could thus be utilized as a new therapeutic and research target for immunotherapy.
With the development of new gene research, epigenetics has been found to play an increasingly important role in tumorigenesis, and DNA methylation is an important epigenetic form for tumorigenesis [36]. Studies have confirmed that abnormal methylation is closely related to abnormal gene expression and carcinogenesis [37,38]. Therefore, the methylation level of melanoma patients was explored in this study, and it was found that MUC16 was hypomethylated in melanoma tumor tissues. However, owing to the small number of adjacent samples in the database, we were unable to compare the expression differences of MUC16 in tumors and adjacent tissues. However, based on the central law of gene expression and the possible demethylation of the MUC16 promoter region in melanoma tumor tissues, we speculated that the expression of MUC16 would be significantly upregulated in tumor tissues, but this requires further investigation [39].
Considering the in-depth research on melanoma and the continuous progress of biotechnology in recent years, targeted therapies and immunotherapies have achieved remarkable results [40]. Although melanoma patients do not respond well to chemotherapy, it is still an important treatment for patients with wild-type BRAF and those who are resistant to targeted therapies [41]. Therefore, we also explored the relationship between MUC16 expression and drug resistance in tumor cells. The results showed that with the increase in MUC16 expression, cells showed increased resistance to chemotherapeutic drugs, such as epothilone B, ixazomib citrate, and etoposide. Epothilone B has a mechanism of action similar to that of paclitaxel, but it has better anticancer activity; thus, it is expected to be a more effective anticancer drug than paclitaxel and has been approved by the Food and Drug Administration as a single or combination drug for the treatment of advanced breast cancer [42,43]. Ixazomib is an oral, highly selective proteasome inhibitor recently approved by the FDA and the European Medicine Agency for use in combination with lenalidomide and dexamethasone for the treatment of multiple myeloma that has previously received at least one line of treatment [44,45]. Etoposide is a cell cycle-specific antitumor drug that is widely used in the treatment of various cancers, including non-small cell lung cancer, breast cancer, and bladder cancer [46-48]. These data suggest that MUC16 may play a role in the sensitivity or resistance of tumor cells to drug therapy and may serve as a therapeutic target to overcome drug resistance or increase drug sensitivity.
This study has some limitations. First, data for Chinese melanoma patients were not included in the data for the two cohorts; thus, we were unable to verify whether the impact of the MUC16 mutation and changes in its expression in Chinese patients are consistent with the findings of this study. Second, further studies are required to explore the relationship between MUC16 mutations and high TMB, as well as immune cell infiltration.
Conclusions
Our study shows that MUC16 has a high mutation frequency in melanoma patients and that this mutation is associated with a higher TMB and improved prognosis; furthermore, the mutation and expression of MUC16 affects immune-related pathways in melanoma patients. Together with the results for tumor immune cell infiltration and drug resistance, the findings of this study suggest that MUC16 could form the basis for the development or improvement of existing immunotherapy regimens. Therefore, we believe that MUC16 is a potential therapeutic target, but further studies are required to confirm this.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (NSFC) [No. 81874393] and Beijing Hospital Project (BJ-2021-184).
Disclosure of conflict of interest
None.
References
- 1.MacKie RM, Hauschild A, Eggermont AM. Epidemiology of invasive cutaneous melanoma. Ann Oncol. 2009;20(Suppl 6):vi1–vi7. doi: 10.1093/annonc/mdp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook J. Surgical margins for resection of primary cutaneous melanoma. Clin Dermatol. 2004;22:228–233. doi: 10.1016/j.clindermatol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 5.Guo J, Qin S, Liang J, Lin T, Si L, Chen X, Chi Z, Cui C, Du N, Fan Y, Gu K, Li F, Li J, Li Y, Liang H, Liu J, Lu M, Lu A, Nan K, Niu X, Pan H, Ren G, Ren X, Shu Y, Song X, Tao M, Wang B, Wei W, Wu D, Wu L, Wu A, Xu X, Zhang J, Zhang X, Zhang Y, Zhu H Chinese Society of Clinical Oncology (CSCO) Melanoma Panel. Chinese guidelines on the diagnosis and treatment of melanoma (2015 edition) Ann Transl Med. 2015;3:322. doi: 10.3978/j.issn.2305-5839.2015.12.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niv Y, Ho SB, Fass R, Rokkas T. Mucin expression in the esophageal malignant and pre-malignant states: a systematic review and meta-analysis. J Clin Gastroenterol. 2018;52:91–96. doi: 10.1097/MCG.0000000000000863. [DOI] [PubMed] [Google Scholar]
- 7.Jung YS, Wang W, Jun S, Zhang J, Srivastava M, Kim MJ, Lien EM, Shang J, Chen J, McCrea PD, Zhang S, Park JI. Deregulation of CRAD-controlled cytoskeleton initiates mucinous colorectal cancer via β-catenin. Nat Cell Biol. 2018;20:1303–1314. doi: 10.1038/s41556-018-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Han X, Shi Y. Association of MUC16 mutation with response to immune checkpoint inhibitors in solid tumors. JAMA Netw Open. 2020;3:e2013201. doi: 10.1001/jamanetworkopen.2020.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Pasche B, Zhang W, Chen K. Association of MUC16 mutation with tumor mutation load and outcomes in patients with gastric cancer. JAMA Oncol. 2018;4:1691–1698. doi: 10.1001/jamaoncol.2018.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, Peters S. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, Huang F, He Y, Sun J, Tabori U, Kennedy M, Lieber DS, Roels S, White J, Otto GA, Ross JS, Garraway L, Miller VA, Stephens PJ, Frampton GM. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Y, Wei Y, Gu Y, Zhang S, Lyu J, Zhang B, Chen C, Zhu J, Wang Y, Liu H, Zhang Y. DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res. 2017;45:D888–D895. doi: 10.1093/nar/gkw1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi S, Kumar S, Choudhury A, Ponnusamy MP, Batra SK. Altered mucins (MUC) trafficking in benign and malignant conditions. Oncotarget. 2014;5:7272–7284. doi: 10.18632/oncotarget.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haridas D, Chakraborty S, Ponnusamy MP, Lakshmanan I, Rachagani S, Cruz E, Kumar S, Das S, Lele SM, Anderson JM, Wittel UA, Hollingsworth MA, Batra SK. Pathobiological implications of MUC16 expression in pancreatic cancer. PLoS One. 2011;6:e26839. doi: 10.1371/journal.pone.0026839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakshmanan I, Salfity S, Seshacharyulu P, Rachagani S, Thomas A, Das S, Majhi PD, Nimmakayala RK, Vengoji R, Lele SM, Ponnusamy MP, Batra SK, Ganti AK. MUC16 regulates TSPYL5 for lung cancer cell growth and chemoresistance by suppressing p53. Clin Cancer Res. 2017;23:3906–3917. doi: 10.1158/1078-0432.CCR-16-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakshmanan I, Ponnusamy MP, Das S, Chakraborty S, Haridas D, Mukhopadhyay P, Lele SM, Batra SK. MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene. 2012;31:805–817. doi: 10.1038/onc.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim N, Hong Y, Kwon D, Yoon S. Somatic mutaome profile in human cancer tissues. Genomics Inform. 2013;11:239–244. doi: 10.5808/GI.2013.11.4.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, RIbas A, Lo RS. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan TA, Wolchok JD, Snyder A. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2015;373:1984. doi: 10.1056/NEJMc1508163. [DOI] [PubMed] [Google Scholar]
- 22.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu TE, Badin F, Ready N, Hiltermann TJN, Nair S, Juergens R, Peters S, Minenza E, Wrangle JM, Rodriguez-Abreu D, Borghaei H, Blumenschein GR Jr, Villaruz LC, Havel L, Krejci J, Jaime JC, Chang H, Geese WJ, Bhagavatheeswaran P, Chen AC, Socinski MA CheckMate 026 Investigatiors. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marzagalli M, Ebelt ND, Manuel ER. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin Cancer Biol. 2019;59:236–250. doi: 10.1016/j.semcancer.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Yang Y, Yang M, Li X, Chen K. High mutation load, immune-activated microenvironment, favorable outcome, and better immunotherapeutic efficacy in melanoma patients harboring MUC16/CA125 mutations. Aging (Albany NY) 2020;12:10827–10843. doi: 10.18632/aging.103296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, Ferrucci PF, Smylie M, Hogg D, Hill A, Márquez-Rodas I, Haanen J, Guidoboni M, Maio M, Schöffski P, Carlino MS, Lebbé C, McArthur G, Ascierto PA, Daniels GA, Long GV, Bastholt L, Rizzo JI, Balogh A, Moshyk A, Hodi S, Wolchok JD. Five-year survival with combined Nivolumab and Ipilimumab in advanced melanoma. N Engl J Med. 2019;381:1535–1546. doi: 10.1056/NEJMoa1910836. [DOI] [PubMed] [Google Scholar]
- 26.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph R, Weber JS, Dronca R, Mitchell TC, Patnaik A, Zarour HM, Joshua AM, Zhao Q, Jensen E, Ahsan S, Ibrahim N, Ribas A. Five-year survival outcomes for patients with advanced melanoma treated with pembrolizumab in KEYNOTE-001. Ann Oncol. 2019;30:582–588. doi: 10.1093/annonc/mdz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang K, Xie F, Mao J, Bai Y, Wang X. Significance of tumor mutation burden in immune infiltration and prognosis in cutaneous melanoma. Front Oncol. 2020;10:573141. doi: 10.3389/fonc.2020.573141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janco JMT, Lamichhane P, Karyampudi L, Knutson KL. Tumor-infiltrating dendritic cells in cancer pathogenesis. J Immunol. 2015;194:2985–2991. doi: 10.4049/jimmunol.1403134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner A, de Mingo Pulido Á, Ruffell B. Dendritic cells and their role in immunotherapy. Front Immunol. 2020;11:924. doi: 10.3389/fimmu.2020.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mani V, Bromley SK, Äijö T, Mora-Buch R, Carrizosa E, Warner RD, Hamze M, Sen DR, Chasse AY, Lorant A, Griffith JW, Rahimi RA, McEntee CP, Jeffrey KL, Marangoni F, Travis MA, Lacy-Hulbert A, Luster AD, Mempel TR. Migratory DCs activate TGF-β to precondition naïve CD8(+) T cells for tissue-resident memory fate. Science. 2019;366:eaav5728. doi: 10.1126/science.aav5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldman SA, Assadipour Y, Kriley I, Goff SL, Rosenberg SA. Adoptive cell therapy--tumor-infiltrating lymphocytes, T-cell receptors, and chimeric antigen receptors. Semin Oncol. 2015;42:626–639. doi: 10.1053/j.seminoncol.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora S, Velichinskii R, Lesh RW, Ali U, Kubiak M, Bansal P, Borghaei H, Edelman MJ, Boumber Y. Existing and emerging biomarkers for immune checkpoint immunotherapy in solid tumors. Adv Ther. 2019;36:2638–2678. doi: 10.1007/s12325-019-01051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Tang Y, Huang L, Yu Q, Hu G, Zou Y, Yuan X. A high number of stromal tumor-infiltrating lymphocytes is a favorable independent prognostic factor in M0 (stages I-III) esophageal squamous cell carcinoma. Dis Esophagus. 2017;30:1–7. doi: 10.1111/dote.12518. [DOI] [PubMed] [Google Scholar]
- 34.Maus MV, June CH. Making better chimeric antigen receptors for adoptive T-cell therapy. Clin Cancer Res. 2016;22:1875–1884. doi: 10.1158/1078-0432.CCR-15-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochi T. Development of innovative T-cell immunotherapy for hematological malignancies. Rinsho Ketsueki. 2019;60:824–833. doi: 10.11406/rinketsu.60.824. [DOI] [PubMed] [Google Scholar]
- 36.Weisenberger DJ. Characterizing DNA methylation alterations from the cancer genome atlas. J Clin Invest. 2014;124:17–23. doi: 10.1172/JCI69740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iida T, Iwanami A, Sanosaka T, Koyama J, Miyoshi H, Nagoshi N, Kashiwagi R, Toyama Y, Matsumoto M, Nakamura M, Okano H. Whole-genome DNA methylation analyses revealed epigenetic instability in tumorigenic human iPS cell-derived neural stem/progenitor cells. Stem Cells. 2017;35:1316–1327. doi: 10.1002/stem.2581. [DOI] [PubMed] [Google Scholar]
- 38.Tian Z, Meng L, Long X, Diao T, Hu M, Wang M, Liu M, Wang J. DNA methylation-based classification and identification of bladder cancer prognosis-associated subgroups. Cancer Cell Int. 2020;20:255. doi: 10.1186/s12935-020-01345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng L, Tian Z, Long X, Diao T, Hu M, Wang M, Zhang W, Zhang Y, Wang J, He Y. Caspase 4 overexpression as a prognostic marker in clear cell renal cell carcinoma: a study based on the cancer genome atlas data mining. Front Genet. 2021;11:600248. doi: 10.3389/fgene.2020.600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robert C, Marabelle A, Herrscher H, Caramella C, Rouby P, Fizazi K, Besse B. Immunotherapy discontinuation - how, and when? Data from melanoma as a paradigm. Nat Rev Clin Oncol. 2020;17:707–715. doi: 10.1038/s41571-020-0399-6. [DOI] [PubMed] [Google Scholar]
- 41.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14:463–482. doi: 10.1038/nrclinonc.2017.43. [DOI] [PubMed] [Google Scholar]
- 42.Altmann KH. Epothilone B and its analogs - a new family of anticancer agents. Mini Rev Med Chem. 2003;3:149–158. doi: 10.2174/1389557033405269. [DOI] [PubMed] [Google Scholar]
- 43.Zajdel A, Wilczok A, Jelonek K, Kaps A, Musiał-Kulik M, Kasperczyk J. Cytotoxic effect of targeted biodegradable epothilone B and rapamycin co-loaded nanocarriers on breast cancer cells. J Biomed Mater Res A. 2021;109:1693–1700. doi: 10.1002/jbm.a.37164. [DOI] [PubMed] [Google Scholar]
- 44.Salvini M, Troia R, Giudice D, Pautasso C, Boccadoro M, Larocca A. Pharmacokinetic drug evaluation of ixazomib citrate for the treatment of multiple myeloma. Expert Opin Drug Metab Toxicol. 2018;14:91–99. doi: 10.1080/17425255.2018.1417388. [DOI] [PubMed] [Google Scholar]
- 45.Offidani M, Corvatta L, Caraffa P, Gentili S, Maracci L, Leoni P. An evidence-based review of ixazomib citrate and its potential in the treatment of newly diagnosed multiple myeloma. Onco Targets Ther. 2014;7:1793–1800. doi: 10.2147/OTT.S49187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamoto H, Watanabe K, Kunikane H, Yokoyama A, Kudoh S, Asakawa T, Shibata T, Kunitoh H, Tamura T, Saijo N. Randomised phase III trial of carboplatin plus etoposide vs split doses of cisplatin plus etoposide in elderly or poor-risk patients with extensive disease small-cell lung cancer: JCOG 9702. Br J Cancer. 2007;97:162–169. doi: 10.1038/sj.bjc.6603810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segar JM, Reed D, Stopeck A, Livingston RB, Chalasani P. A phase II study of Irinotecan and Etoposide as treatment for refractory metastatic breast cancer. Oncologist. 2019;24:1512–e1267. doi: 10.1634/theoncologist.2019-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loehrer PJ Sr. Etoposide therapy for testicular cancer. Cancer. 1991;67(Suppl):220–224. doi: 10.1002/1097-0142(19910101)67:1+<220::aid-cncr2820671303>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]