Abstract
BACKGROUND & AIMS:
Thirty percent to 90% of hepatocytes contain whole-genome duplications, but little is known about the fates or functions of these polyploid cells or how they affect development of liver disease. We investigated the effects of continuous proliferative pressure, observed in chronically damaged liver tissues, on polyploid cells.
METHODS:
We studied Rosa-rtTa mice (controls) and Rosa-rtTa;TRE-short hairpin RNA mice, which have reversible knockdown of anillin, actin binding protein (ANLN). Transient administration of doxycycline increases the frequency and degree of hepatocyte polyploidy without permanently altering levels of ANLN. Mice were then given diethylnitrosamine and carbon tetrachloride (CCl4) to induce mutations, chronic liver damage, and carcinogenesis. We performed partial hepatectomies to test liver regeneration and then RNA-sequencing analyses to identify changes in gene expression. Lineage tracing was used to rule out repopulation from non-hepatocyte sources. We imaged dividing hepatocytes to estimate the frequency of mitotic errors during regeneration. We also performed whole-exome sequencing of 54 liver nodules from patients with cirrhosis to quantify aneuploidy, a possible outcome of polyploid cell divisions.
RESULTS:
Liver tissues from control mice given CCl4 had significant increases in ploidy compared with livers from uninjured mice. Mice with knockdown of ANLN had hepatocyte ploidy above physiologic levels and developed significantly fewer liver tumors after administration of diethylnitrosamine and CCl4 compared with control mice. Increased hepatocyte polyploidy was not associated with altered regenerative capacity or tissue fitness, changes in gene expression, or more mitotic errors. Based on lineage-tracing experiments, non-hepatocytes did not contribute to liver regeneration in mice with increased polyploidy. Despite an equivalent rate of mitosis in hepatocytes of differing ploidies, we found no lagging chromosomes or micronuclei in mitotic polyploid cells. In nodules of human cirrhotic liver tissue, there was no evidence of chromosome-level copy number variations.
CONCLUSIONS:
Mice with increased polyploid hepatocytes develop fewer liver tumors following chronic liver damage. Remarkably, polyploid hepatocytes maintain the ability to regenerate liver tissues during chronic damage without generating mitotic errors, and aneuploidy is not commonly observed in cirrhotic livers. Strategies to increase numbers of polypoid hepatocytes might be effective in preventing liver cancer.
Keywords: HCC, Cell Division, DEN, Mouse Model
Graphical Abstract
Polyploidy refers to cells with balanced duplications of all chromosomes. Although polyploidy is commonly found in plants and fish, it is also present in mammalian organs including the heart, skeletal muscle, bone marrow, brain, and liver.1–7 Up to 90% of mouse and 40% of human hepatocytes are polyploid.8,9 Hepatocytes are diploid at birth, but the proportion of polyploid cells in-creases with age. In rodents, ploidy increases rapidly around weaning and gradually increases throughout adult-hood.10 During development, polyploid hepatocytes are mainly generated through cytokinesis failure,11 which has been associated with fluctuations in insulin/Akt signaling during weaning.12 Afterward, “unscheduled” hepatic poly-ploidization can be achieved by endoreplication, or genome duplication without mitosis.7 Polyploidy can enable selective adaptation to environmental stresses in plants and yeast13,14; however, the functional roles of hepatic poly-ploidy are still obscure.
Previously, we showed that polyploidy could protect against liver carcinogenesis after acute mutagen exposure.15 Those findings suggested relevance to environmental toxins such as aflatoxin, but it is not clear if high levels of polyploidy might influence healing and carcinogenesis in the setting of chronic injury. Such injuries and their associated proliferative demands are part and parcel of most clinically relevant liver diseases. The distinction between acute and chronic injuries is important to understand because although polyploidy might have evolved to adapt to acute environmental poisons, it might be maladaptive in the context of chronic overnutrition, nonalcoholic steatohepatitis, hepatitis viruses, alcohol, or drug-induced liver injuries.
There are reasons to expect that polyploids may not adapt well to the proliferative demands of chronic liver injury. Indeed, polyploidy is often thought to be pathologic given its appearance in disease states such as nonalcoholic steatohepatitis.16,17 Wilkinson et al18 showed that polyploid cells proliferate, but less efficiently than diploids, indicating that this may be one way that polyploids suppress tumor development. Also, Wang et al19 showed that a candidate stem cell population (peri-central AXIN2+ cells) that repopulates the liver, is more likely to be composed of diploid hepatocytes. Based on these findings, it is possible that polyploid cells and predominantly polyploid tissues might fail to regenerate properly, thereby exacerbating cirrhosis progression and promoting a cancerous microenvironment.
Moreover, enforced cell division in polyploids could present unique challenges for tumor suppression. We have shown that polyploid hepatocytes are less likely to develop cancer after a single dose of mutagen, in part because the additional tumor suppressor gene (TSG) copies made it more difficult to achieve loss of heterozygosity (LOH), but most of those data were generated in a nonproliferative setting.15 Enforced division could lead to reduced genome copies through a process called “reductive division.” These have been shown to occur in vitro,20 but the frequency and mitotic mechanisms by which these divisions might occur are not clear in vivo. Reductive divisions could in theory present another path to LOH of TSGs, given the possibility that mutated TSGs might independently assort into daughter cells.
Polyploid cell divisions, which involve centrosome duplication and multiplolar spindle formation,21,22 have also been shown to lead to aneuploidy, particularly in vitro. In vivo, wild-type (WT) livers (predominantly polyploid) vs. E2f7/E2f8 double knockout (KO) livers (predominantly diploid23,24), result in more detectable aneuploid daughter clones after chronic injury, as shown by Wilkinson et al.25 Ongoing chromosomal instability (CIN) could promote tumorigenesis via increased incidence of LOH or other mechanisms by generating permissive oncogenic karyotypes. On the other hand, aneuploid clones also could be beneficial and promote adaptation to injury.26 However, other groups have found much lower levels of aneuploidy in mouse and human livers.22,27,28 Although the functional impact of aneuploidy is known to be highly tissue and cancer-context dependent, it still would be important to know if polyploidy ultimately leads to an accumulation of aneuploidy in the human liver. The extent of aneuploidy could then be evaluated as a potential factor that influences cancer or cirrhosis phenotypes.
We tested these hypotheses about polyploid cell divisions in vivo using a variety of persistent chemical injuries to drive multiple rounds of proliferation. Remarkably, livers in which >97% of cells were poly-ploid were highly tolerant of chronic injury and protected from cancer. We tested if tumor protection was associated with gene expression changes, but RNA-sequencing (RNA-seq) showed no differences between ploidy states. Moreover, diploid and polyploids showed no differences in tissue fitness and damage sustained. In summary, polyploid hepatocytes are able to divide and regenerate while being buffered from tumor suppressor LOH and tumorigenesis. These results clarify the important functionality of polyploid cells within the liver, both in health and disease.
Methods
Mice
All mice were handled in accordance with the guidelines of the Institutional Animal Care and Use Committee at University of Texas Southwestern (UTSW). TG-shAnln and E2f8 KO mice were previously reported.15 Tp53fl/fl and Rosa-LSL-tdTomato mice were from Jackson Laboratories (Bar Harbor, ME). WT C3H/HeJ mice were used because this is a strain that is prone to liver cancer development. All experiments were done in an age- and sex-controlled fashion unless otherwise noted.
Partial Hepatectomy
Seventy percent of the liver was surgically resected as previously described.29
Chemical Injury Experiments
Diethylnitrosamine (DEN) (Sigma) was diluted in saline and injected intraperitoneally 1 time at P15 at a dose of 75 μg/g. Carbon tetrachloride (CCl4; Sigma) is diluted 1:10 in corn oil (Sigma), and administered biweekly intraperitoneally at a dose of 0.5 mL/kg of mouse30; 5-diethoxycarbonyl-1,4-dihydrocollidine (DDC; Sigma) was dosed at 0.1% in the diet (TestDiet).
Virus Experiments
A total of 100 μL of AAV-TBG-Cre (University of Pennsylvania Vector Core) was retro-orbitally injected at a dose of 5 × 1010 plaque-forming units per mouse.
In Vivo Small Interfering RNA Experiments
Control (Life Technologies, #4457289), Anln small interfering RNA (siRNA) (#4457308, s87033; Life Technologies, Carlsbad, CA) or E2f8 siRNA (#4457308, s99361; Life Technologies) were packaged and delivered as previously described.15
Liver Function Tests
Blood samples (~50 μL) were collected retro-orbitally in heparinized tubes. Tests were performed by the UTSW Molecular Genetics Core.
Histology, Immunohistochemistry, and Immunofluorescence
Tissues were fixed in 4% paraformaldehyde for 16 to 24 hours. Fixed tissues were either embedded in paraffin or cryo-protected with 30% sucrose and frozen. Immunohistochemistry was performed as previously described.31 The following primary antibodies were used: CTNNB1 (#610154; BD Transduction Laboratories), HNF-4α (sc-8987; Santa Cruz, Dallas, TX), p-Histone H3 (#9706; Cell Signaling, Danvers, MA), Ki-67 (ab15580; Abcam, Cambridge, MA), glutamine synthetase (GS) (ab49873; Abcam), α-Tubulin-FITC (F2168; Sigma) and γ-Tubulin (T5326; Sigma). The γ-Tubulin was conjugated using the Alexa Fluor 568 Antibody Labeling Kit (#A20184; Life). For immunofluorescence, the secondary antibodies Alexa Fluor 488 goat anti-mouse immunoglobulin (Ig)G1, Alexa Fluor 488 goat anti rabbit IgG, and Alexa Fluor 594 donkey anti-rabbit IgG (Life Technologies) were used. Mitosis staining was performed a; previously described.22 Fibrosis detection was performed with Sirius red (IW-3012; IHC World). Apoptosis detection was performed with the In Situ Cell Death Detection Kit (#11684817910; Sigma). Confocal images were taken by a Zeiss (Oberkochen, Germany) LSM 780 Upright confocal/multiphoton microscope. Whole slide images were taken by a Hamamatsu (Hamamatsu City, Japan) Nanozoomer 2.0HT or a Zeiss Axioscan Z1 (UTSW Whole Brain Microscopy Facility). Other images were taken by an Olympus (Tokyo, Japan) IX83 microscope.
Primary Hepatocyte Isolation
Primary hepatocytes were isolated by 2-step collagenase perfusion as described previously.15
Flow Cytometry
For detection of ploidy populations, primary hepatocytes (2 × 106/mL) were fixed in 75% ethanol at −20°C, then incubated with 500 μL (2 × 106 mL) of propidium iodide (PI)/RNase Staining Buffer (BD Pharmingen, BD 550825) at 25°C for 15 minutes. For RNA extraction from different ploidy populations, hepatocytes were fixed in 4% paraformaldehyde for 15 minutes, then permeabilized with 0.5% Triton X-100 for 30 minutes and stained with 4’,6-diamidino-2-phenylindole (DAPI). Cells were analyzed or sorted with a FACS Aria Fusion machine (BD Biosciences, San Jose, CA).
RNA-seq
Total RNA from 4 Rosa and TG-shAnln livers before surgery and after partial hepatectomy were collected using the PureLink RNA Mini Kit (Figure 3C and D). Total RNA from sorted hepatocytes with different ploidies was collected from 4 Rosa and TG-shAnln livers using the FFPE RNA Isolation Kit (Invitrogen, Carlsbad, CA) (Supplementary Figure 4). NuGEN RNA-seq libraries were made from the purified RNA and indexed libraries were multiplexed in a single flow cell lane and underwent 75-base pair single-end sequencing on a NextSeq 500 using the High Output Kit v2 (75 cycles) at the CRI Sequencing Facility. Reads were aligned to the mouse genome version mm9 using TopHat. Differential expression analysis was performed using edgeR. For the correlation matrix in, one of the noninjured control samples was excluded because of quality control problems in the sequencing.
Figure 3.

Polyploidy does not affect liver regeneration or the ability to sustain liver damage. (A) Schema for the 70% partial hepatectomy experiment in TG-shAnln mice. Doxycycline treatment established ploidy differences. Livers were harvested 48 hours after surgery. Liver and body weight ratios before and after surgery are shown on the right (n = 6 for each group). (B) Phospho-H3 staining in Rosa and TG-shAnln livers 48 hours after hepatectomy. The percentage of positive cells was quantified by whole slide images, shown on the right (n = 6 for each group). (C) Scatter plot of RNA-seq expression data showing the gene expression in Rosa and TG-shAnln livers after hepatectomy. The plot is on a log2 transformed scale (n = 4 for each group). (D) Correlation matrix showing the correlation of Rosa and TG-shAnln livers before and after hepatectomy. One of the presurgery Rosa samples (before_Ctrl_1) was excluded for this analysis because of quality control problems in the sequencing. (E) Liver function tests performed after 6 and 12 weeks of CCl4 injections (n = 8 for Rosa and n = 11 for TG-shAnln). (F) Chronically injured livers were examined for fibrosis using Sirius red staining. Four images from each mouse were quantified (in the DEN + CCl4 experiment: n = 5, 7, 9, 9 for Rosa, TG-shAnln, siCtrl, siE2f8; n = 3 for Rosa and n = 6 for TG- shAnln in the CCl4-alone experiment).
Whole-Exome Sequencing
Fifty-four fresh-frozen cirrhotic nodules were manually dissected and isolated using a razor blade under a dissection microscope. The boundaries of the nodules were not collected to avoid contamination. Exome capture was performed using the xGen Exome Research Panel (IDT). Captured nucleotides were subjected to 150-base pair paired-end sequencing on an Illumina HiSeq platform (Admera Health).
Statistical Analysis
Sample size was determined by producing a confidence interval estimate with a specified margin of error to ensure that a test of hypothesis has a high probability of detecting a meaningful difference in the parameter. The data in most figures reflect multiple experiments performed on different days using mice derived from different litters. Variation is indicated using standard error presented as mean ± SEM. Two-tailed Student t tests (2-sample equal variance) were used to test the significance of differences between groups.
In all figures, statistical significance is displayed as *P < .05, **P < .01, ***P < .001, ****P < .0001. Image analysis was blinded.
Results
Hepatic Ploidy Increases With Chronic Injury
To determine the impact of chronic injury on hepatic ploidy, we used serial dosing of CCl4 to induce cycles of hepatocyte necrosis and regeneration. WT control (Rosa-rtta heterozygous) mice were given 12 weeks of biweekly CCl4 (Figure 1A). The liver was perfused 2 weeks after the last dose of CCl4 and ploidy distribution was analyzed by flow cytometry. Ki-67 staining showed that very few cells were actively cycling at this time point, indicating that ploidy quantification was unlikely to be confused with cell division (Figure 1B). Compared with uninjured control, chronic CCl4 caused an increase in octaploid populations with a decrease in diploid and tetraploid ones (Figure 1C). Histology showed that cell and nuclear size both enlarged (Figure 1D). Previously, it was shown that oxidative stress could promote endoreplication instead of cytokinesis failure, leading to increased mononuclear polyploid hepatocytes.16 Indeed, the percentage of binuclear cells decreased after injury, suggesting that endoreplication induced polyploidization occurred during chronic CCl4 injury (Figure 1E). These data agree with other reports showing increases in ploidy after chronic injury.16,32–34
Figure 1.
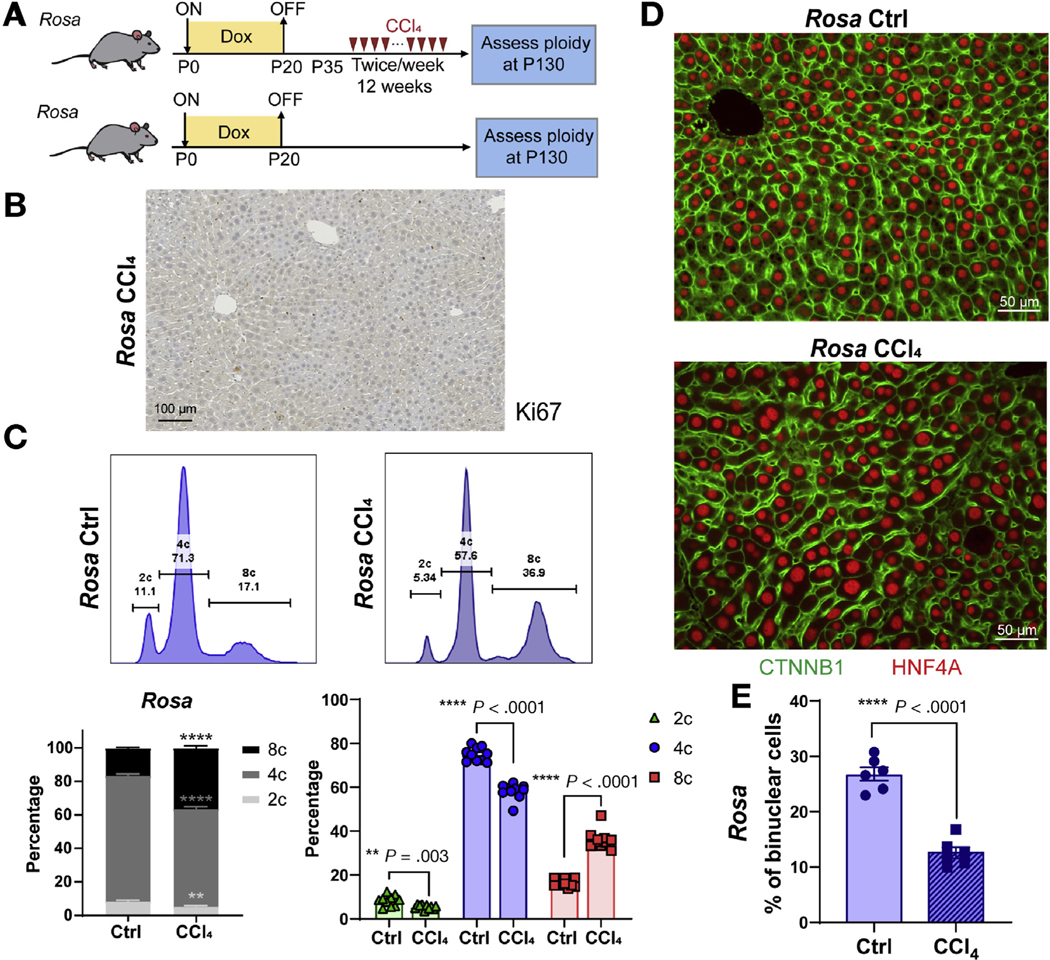
Hepatic polyploidy increases with chronic CCl4 injury. (A) Schema for the chronic CCl4 injury experiment in WT control mice. Doxycycline treatment from P0–20 in Rosa-rtta heterozygous mice served as the normal ploidy control for experiments shown in Figure 5A. Biweekly CCl4 started at P35 for 12 weeks. Hepatocytes were dissociated for ploidy analysis or livers were harvested for image analysis. (B) Immunohistochemistry Ki-67 staining of control liver collected 2 weeks after the last dose of CCl4. (C) Top: Representative cellular ploidy distribution as determined by PI staining and flow cytometry. Lower left: Average cellular ploidy distribution. Right: The percentage of each population (n = 10 mice in each group). (D) CTNNB1 (green) and HNF4A (red) -stained liver sections. (E) The percentage of binuclear hepatocytes averaged from 4 images (n = 6 mice in each group).
Polyploidy Protects From Chronic Liver Injury-Induced Hepatocellular Carcinoma Development
Chronic liver disease from any etiology can induce cellular loss, regeneration, inflammation, and fibrosis, all of which contribute to a pro-tumorigenic microenvironment. Although ploidy increased after injury, it is unclear if polyploidization has a phenotypic impact on the pathologic events leading up to cancer development. To examine the effects of changing ploidy in the chronic injury setting, we used a doxycycline-inducible short hairpin RNA mouse model to transiently knockdown Anillin (Anln) and induce ploidy as described previously.15 Control Rosa-rtTa or experimental Rosa-rtTa;TRE-shAnln (hereafter called Rosa or TG-shAnln) cohorts were given doxycycline from P0-P20. Control mice harbored 35% diploid and 5% octaploid hepatocytes, whereas super-polyploid mice had 3% and 46% (Supplementary Figure 1). Fifteen days off doxycycline allowed Anln levels to return to normal, a time at which mice were given a dose of DEN followed by biweekly CCl4 (Figure 2A). Although many hepatocellular carcinoma (HCC) models promote cancer without induction of inflammation and fibrosis, this model is routinely used to study how these processes lead to HCC development.35
Figure 2.
Polyploidy protects from chronic liver injury-induced HCC development. (A) Schema for the DEN + CCl4 chronic injury experiment. Doxycycline treatment from P0–20 established ploidy differences. At P35, mice were injected with DEN (75 μg/g), followed by biweekly CCl4 for 12 weeks starting at P38. (B) Representative gross tumor burden from (A). Arrows point to liver surface tumors. (C) Histology shows tumor nodules (circled by dashed yellow lines) from (A). (D) Quantitation of liver to body weight ratios, surface tumors, and tumor nodules from (A) to (C). (E) Schema for the dEn + CCl4 chronic injury experiment with in vivo siRNA. Scramble (siCtrl) and E2f8 siRNAs in lipid nanoparticles were injected into WT mice starting at P10. A total of 4 injections (2 intraperitoneal and 2 retro-orbital) were performed twice per week. At P35, mice were injected with DEN (75 μg/g), followed by biweekly CCl4 for 12 weeks starting at P38. (F) Representative gross tumor burden from (E). Arrows point to surface tumors. (G) Histology showing microscopic tumor nodules (circled by dashed yellow lines) from (E). (H) Liver to body weight ratios, surface tumors, and tumor nodules from (E) to (G). (I) Schema for the CCl4 chronic injury experiment in inducible short hairpin RNA mice. Doxycycline treatment from P0 to P20 established ploidy differences. Starting at P35, mice were injected with biweekly CCl4 for 20 weeks. (J) All livers from (I) are shown (n = 6 for each group).
After chronic CCl4, TG-shAnln mice showed significantly fewer nodules and no tumors as compared with controls (Figure 2B–D). To examine mice with more diploid hepatocytes, we used in vivo siRNAs to transiently knock down E2f8 to reduce ploidy (Supplementary Figure 2A and B) and challenged these mice with DEN followed by chronic CCl4 (Figure 2E). Decreasing ploidy using E2f8 knockdown was associated with greater levels of tumor formation (Figure 2F–H).
Because it was possible that polyploidization was predominantly protecting against the acute and potent mutagenic insult of DEN, we also tested chronic CCl4 in isolation. In separate experiments, we administered CCl4 for a longer period (20 weeks; see Figure 2I). Chronic CCl4, which is not thought to be strongly genotoxic, led to tumor formation in 50% of control livers but never in TG-shAnln mice (Figure 2J). Collectively, these results strongly supported the idea that the polyploidization seen in human and mouse livers is protective against cancers that arise in diseases associated with long-term proliferation.
Polyploidy Is Tumor Suppressive Even in the Tp53 Null Setting
The adaptive features of polyploidy could become maladaptive in premalignant or cancer cells. For example, polyploidy could be tumor suppressive in the Tp53 WT setting, but oncogenic in the Tp53 mutant setting, as Fujiwara et al36 had demonstrated. To test if the polyploid state could become a liability in Tp53 null settings, liver-specific Tp53 KO mice (Alb-Cre; Tp53fl/fl) were treated with transient E2f8 siRNA to reduce ploidy, then given DEN (Supplementary Figure 3A). Polyploidization was tumor suppressive even in the Tp53 null background (Supplementary Figure 3B–D). When we changed ploidy with either E2f8 or Anln siRNA while simultaneously deleting Tp53 using adeno-associated virus (Tp53fl/fl + AAV-Cre), the results were identical (Supplementary Figure 3E–I). These data suggest that polyploidization in hepatocytes is protective against cancer even in the absence of Tp53.
Polyploidy Does Not Affect Liver Regeneration or the Ability to Sustain Liver Damage
We examined mechanisms and fates of polyploid cells that potentially contribute to tumor suppression during chronic tissue repair. Previously, few differentially expressed genes were found in diploid vs. polyploid hepatocytes in uninjured settings,37 but it remained possible that when forced to regenerate, polyploids could induce a gene expression program that enables tumor suppression. We assessed gene expression changes in the midst of diploid vs. polyploid cell divisions. Using our super-polyploid model that has a much greater extent of polyploidy, we confirmed reports showing that induced polyploid livers regenerated normally after severe injuries.38 Forty-eight hours after 70% partial hepatectomy, Rosa and TG-shAnln mice had identical liver to body weight ratios and proportions of mitotic nuclei (Figure 3A and B). To ask if there were broad expression changes, we performed RNA-seq on livers before and 48 hours after partial hepatectomy. There were no differentially expressed genes between control and super-polyploid livers before resection or during regeneration (Figure 3C and D). Remarkably, these livers were transcriptomically indistinguishable even though the diploid cell fraction differed by 10-fold.
To ensure that bulk sequencing was not masking differences between cells of different ploidy levels, we also sorted and sequenced ploidy populations from control and TG-shAnln livers. We failed to identify clear transcriptional differences between ploidy states in WT or TG-shAnln hepatocytes (Supplementary Figure 4A–C). Our data agree with previous microarray analysis showing no major gene expression differences among diploid, tetraploid, and octaploid hepatocytes.37
The risk of HCC development is correlated with the pace of liver disease progression, which is influenced by hepatocyte fitness in response to insults. We asked if diploid and polyploid cells might sustain different levels of hepatotoxic damage. After a single CCl4 dose, the histology of diploid and polyploid livers showed similar levels of centrilobular necrosis and apoptosis (Supplementary Figure 5A and B). E2f8 WT and KO mice, which have predominantly diploid livers, exposed to CCl4 had similar levels of necrosis and apoptosis (Supplementary Figures 2C–F, 5A and C). Liver function tests also revealed no differences before and after 1 dose of CCl4 in both ploidy models (Supplementary Figure 5D). Even after chronic CCl4, Rosa and TG-shAnln mice showed no differences in liver function tests (Figure 3E). Importantly, there were no differences in the extent of fibrosis after either DEN + chronic CCl4 or chronic CCl4 alone (Figure 3F).
It is possible that cytochrome P450-dependent injuries such as CCl4 are uniquely unaffected by ploidy. To test if polyploidy influences other injuries, we tested the biliary toxin DDC on Rosa/TG-shAnln and E2f8 WT/KO mice (Supplementary Figure 6A). Of note, E2f8 KO mice, which have predominantly diploid livers, were used to avoid prolonged siRNA administration (Supplementary Figure 2C–F). Even with the complete ablation of E2f8 and a strong induction of diploidy, body weights, liver to body weight ratios, liver function tests, and apoptosis analysis showed no differences (Supplementary Figure 6B–D). These data demonstrated that hepatic polyploidy does not potently influence acute (partial hepatectomy) or chronic liver injury (DEN + CCl4, CCl4, or DDC).
Hepatocytes Are Still Major Contributors to Regeneration During Chronic CCl4 Injury
We hypothesized that super-polyploid livers recover from liver injuries as well as normal livers because polyploid and diploid hepatocytes have similar proliferative capacities. To challenge this, we wanted to test for contributions from rare diploid populations that might compensate for ineffectual regeneration by polyploids. Repopulation from a diploid cell source could obscure any potential negative impact associated with polyploidy. Given that >97% of the hepatocytes in super-polyploid livers are 4N or greater, 2 diploid populations have the potential to outcompete the polyploid cells: the diploids that make up 3% of the hepatocyte population or cells within the biliary lineage. Biliary epithelial cells were proposed to be a source of facultative stem cells when hepatocytes are severely impaired.39–41 To first examine this and other non-hepatocyte populations, we generated Rosa-rtta;Rosa-LSL-tdTomato and Rosa-rtta;Rosa-LSL-tdTomato;TRE-shAnln mice, gave doxycycline between P0–20, then delivered AAV-TBG-Cre, which is known to target hepatocytes only.42 Tracing unlabeled cells could identify whether or not there is a nonhepatocyte population that comes to dominate after injury.
Baseline labeling at P42 showed an even distribution of tdTomato+ hepatocytes and an absence of bile duct labeling, as would be expected for AAV-TBG-Cre. Labeled mice were subjected to biweekly CCl4 (Figure 4A). After 12 weeks, tdTomato+ cells were clustered together, but the clusters were randomly distributed and not enriched in particular zones (Figure 4B). This is important because tdTomato enrichment in pericentral zone 3 might be expected if unlabeled biliary cells near the portal tracts were trans-differentiating and expanding into zone 1. The fraction of tdTomato+ to DAPI+ cell ratios were analyzed based on whole-liver section images (Supplementary Figure 7A). Rosa and TG-shAnln mice showed similar ratios after chronic injury, suggesting that there was no substantial influx of nonlabeled cells from the portal area (Figure 4C). These data suggest that even within a super-polyploid liver, hepatocytes are still the predominant contributors to regeneration rather than a diploid population of nonhepatocytes.
Figure 4.
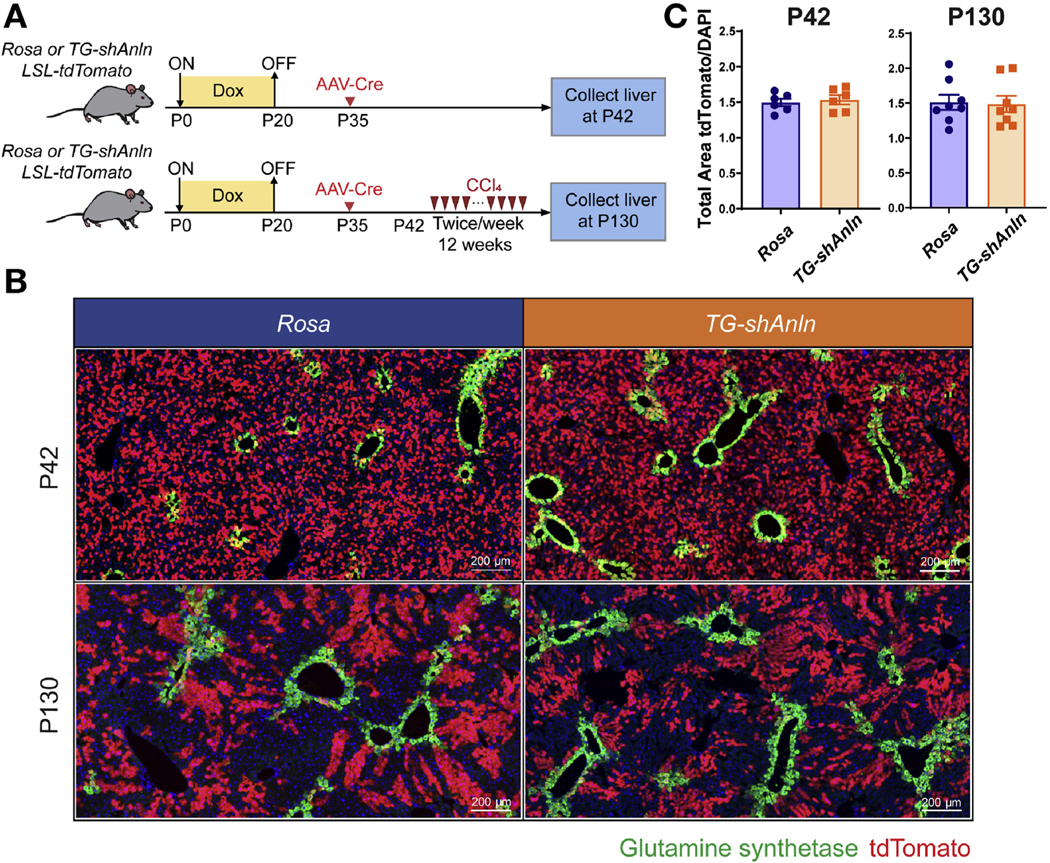
Polyploid hepatocytes are still the major cellular contributor to liver regeneration during chronic CCl4 injury. (A) Schema for the tdTomato lineage-tracing experiment. Doxycycline treatment established ploidy differences. At P35, mice were given AAV-TBG-Cre to label hepatocytes. Labeling was quantified at P42 in 6 mice. Eight mice were subjected to biweekly CCl4 starting at P42 for 12 weeks. (B) GS (green) and tdTomato (red) staining in Rosa and TG-shAnln livers at baseline (P42) and after injury (P130). (C) Quantification of the tdTomato+ to DAPI area ratios based on whole slide imaging in Supplementary Figure 4A (n = 6 for the P42 and n = 8 for the P130 group).
Polyploid Hepatocytes Divide Frequently Without Chromosome Missegregation
We then asked if diploid hepatocytes might outcompete polyploid hepatocytes by quantifying ploidy in super-polyploid mice before and after chronic injury. In Figure 1, we observed that polyploidy increased after chronic injury in WT mice, indicating that diploids likely became tetraploid, which in turn became octaploid. Here, we evaluated ploidy changes within super-polyploid mice, half of which were subjected to CCl4 and half of which were not (Figure 5A). Few cells were proliferating at the time of liver harvesting, thus making ploidy analysis more easily interpretable (Figure 5B). The super-polyploid state was maintained even after chronic injury, with no change in diploid, a small decrease in tetraploid, and an increase in octaploid frequency (Figure 5C). Even though mice with high polyploidy levels showed no evidence of increased diploids after chronic injury, we could not rule out the possibility that diploid hepatocytes were contributing to regeneration via conversion into tetraploids. However, the reduction in tetraploid frequency did suggest that tetraploid hepatocytes were also becoming octaploid as well. Consistent with a combination of polyploid hepatocyte division and endoreplication-mediated polyploidization, we saw increases in the frequency of mononuclear hepatocytes in TG-shAnln mice after chronic injury (Figure 5E).
Figure 5.
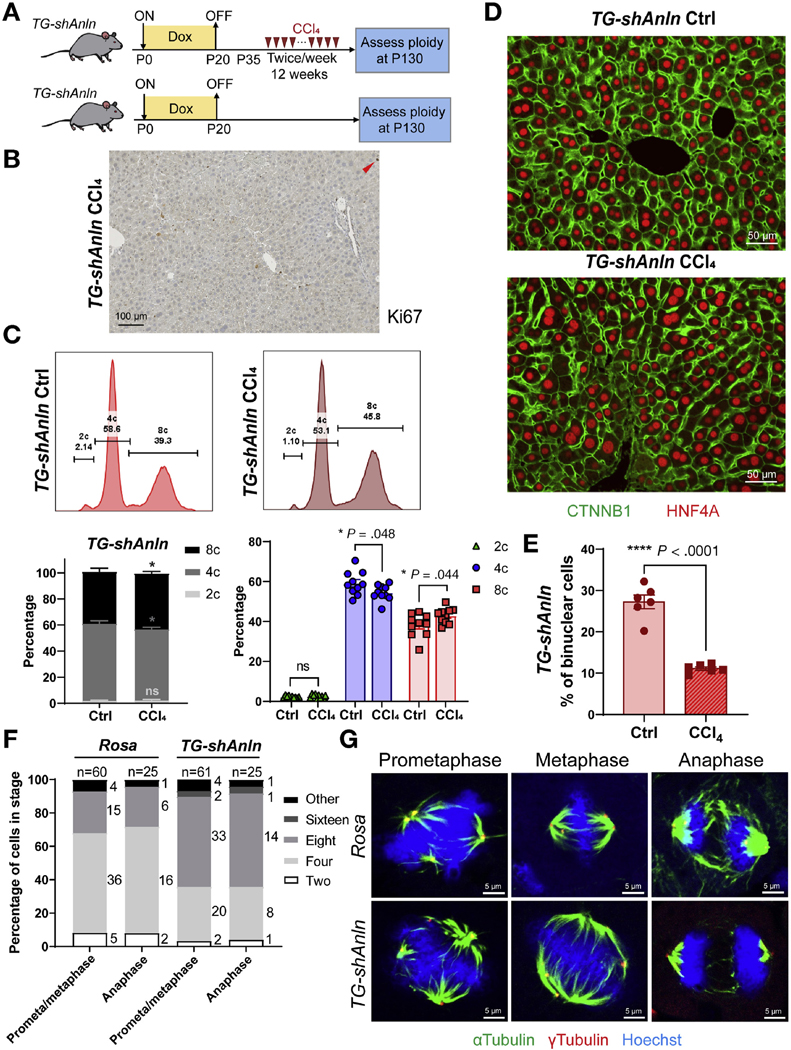
Polyploid hepatocytes in vivo readily divide and regenerate without leading to chromosome missegregation. (A) Schema for chronic CCl4 injury experiment. Doxycycline treatment established polyploidy. Twelve weeks of biweekly CCl4 started at P35. When mice were euthanized, hepatocytes were dissociated for ploidy analysis or livers were harvested for image analysis. (B) Ki-67 staining of TG-shAnln livers collected 2 weeks after the last dose of CCl4. (C) Representative cellular ploidy distribution within livers, as determined by PI staining and flow cytometry (upper). Average cellular ploidy distribution (lower left). The percentage of each population, separated from the left (right, n = 10 mice in each group were analyzed). (D) CTNNB1 (green) and HNF4A (red) stained liver sections allow ploidy analysis. (E) The percentage of binuclear hepatocytes was quantified and averaged from 4 images (n = 6 mice in each group). (F) Ploidy of hepatocytes within tissue sections of regenerating livers are estimated by centrosome number (quantified γ-tubulin foci). These hepatocytes are in prometa/metaphase and anaphase, and more than 50 and 25 mitoses in prometa/metaphase and anaphase were measured. (G) Representative images of hepatocytes within Rosa and TG-shAnln liver sections at different stages of mitosis, as stained by α-tubulin (green), γ-tubulin (red), and Hoechst (blue).
From the previously described experiments, it followed that super-polyploid livers experiencing chronic injury possessed enough hepatocyte division or cell size increases to maintain liver mass and function. Because diploids could be polyploidizing at a high rate, we needed to see if polyploid hepatocytes were actually dividing. Previously, Knouse et al22 showed that diploid and polyploid cells had similar rates of mitosis in regenerating WT livers. Likewise, we performed partial hepatectomy on control and super-polyploid mice to enforce proliferation, then used centrosome numbers to ascertain mitosis rates for cells of different ploidies. Notably, the frequency of mitotic cells with supernumerary centrosomes mirrored the original ploidy distribution (Figure 5F) and was maintained throughout regeneration (Supplementary Figure 8A). Data from livers injured with chronic CCl4 also showed frequent polyploid mitoses (Supplementary Figure 8B).
Polyploidy has been associated with multipolar spindles and high-risk cell divisions.20 We reasoned that the frequency of chromosome missegregation could increase in livers composed entirely of polyploid cells. In both Rosa and TG-shAnln livers, we observed proliferating polyploid hepatocytes with multipolar spindles (Figure 5G); however, these almost always clustered into bipolar spindles before anaphase. Despite the high frequency of polyploid cells in TG-shAnln, we did not observe any misaligned chromosomes, lagging chromosomes, or micronuclei out of 100 metaphases or anaphases analyzed (Figure 5G). These results support the idea that polyploid hepatocytes readily divide and regenerate without prevalent signs of chromosome missegregation.
Chronically Injured Heterozygous ApcMin Livers Do Not Readily Develop Apc LOH
Additional evidence for aneuploidy could come in the form of specific TSG LOH events that lead to clonal expansion. To examine whether chronic proliferation might cause LOH via reductive divisions or aneuploidy, we examined the ApcMin mouse model. Apc is a canonical 2-hit TSG in the WNT pathway that is sometimes mutated in HCC. When Apc is completely lost in hepatocytes, CTNNB1 translocates into the nucleus and activates WNT targets such as GS. In noninjured livers, only the hepatocytes that surround the central veins are GS+, but aberrant WNT activation via mutation causes ectopic GS expression in locations distant from the central veins.43 This provides a sensitive and specific marker of Apc LOH. Using WT and heterozygous ApcMin mice, we examined whether 6 weeks of biweekly CCl4 injury would trigger Apc LOH through candidate mechanisms associated with aberrant polyploid divisions. In both WT and ApcMin mice, CCl4 caused necrosis around central veins and an expected ablation of GS+ cells. However, no ectopic cells were detected in either WT or ApcMin mice (Supplementary Figure 9A and B). These data support the concept that polyploids are equipped to retain heterozygosity even after multiple rounds of cell division within a tissue where every cell is potentially at risk for LOH.
Exome Sequencing in Human Cirrhotic Nodules Did Not Reveal Aneuploidy
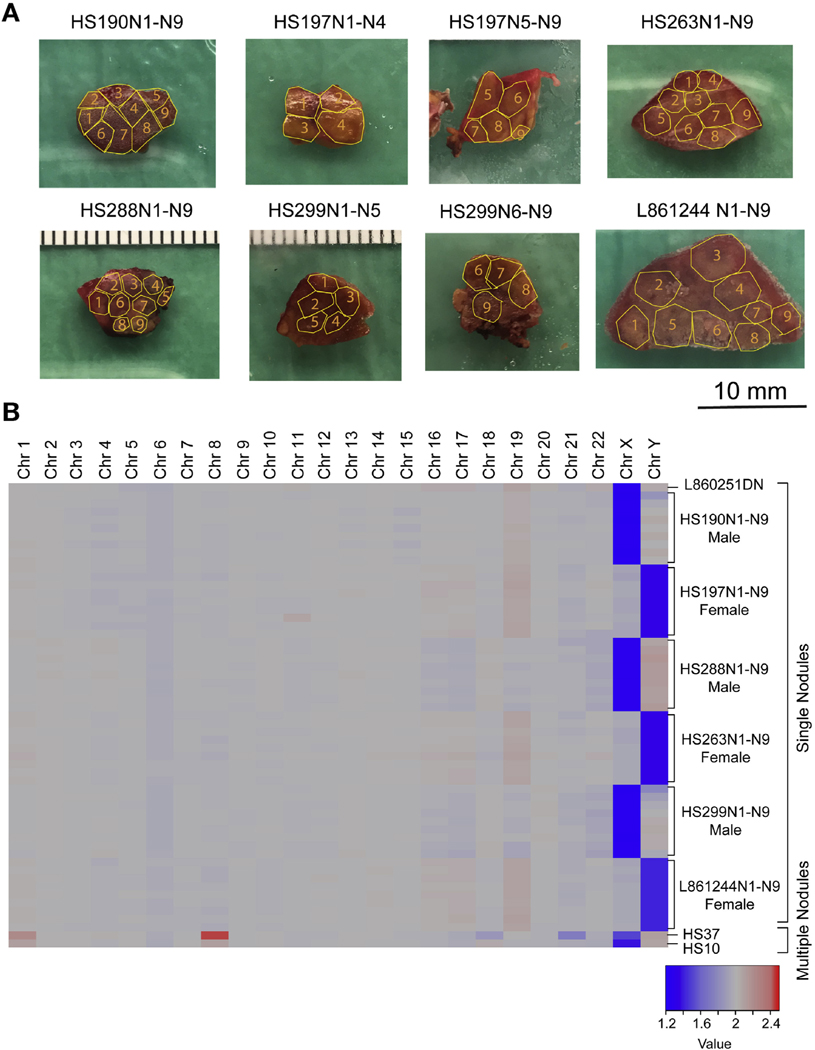
Although the inability to detect evidence of chromosomal missegregation or LOH is supporting evidence for the lack of aneuploid daughters generated from dividing polyploids, it does not directly quantitate the frequency of aneuploid clones. If aneuploidy is a common fate of polyploid cell divisions in humans, then it should be readily detected in chronically injured livers. Previous single-cell sequencing suggested low levels of aneuploidy in normal mouse livers.27 To assess the prevalence of aneuploid clones in humans, we isolated and analyzed 54 independent nodules from 6 patients with cirrhosis (Figure 6A, Supplementary Table 1). Nine nodules and a blood sample were collected from each patient and each was sequenced to a mean depth of 75x. Chromosome-level copy number variations (CNVs) were called using CNVkit.44 We were able to correctly identify the sex of each patient using X and Y chromosome copies; however, there was no evidence of whole or sub-chromosomal CNVs in somatic chromosomes. This was consistent with previous whole-exome sequencing data from Zhu et al,28 which showed that only 2 of 60 patients harbored chromosome arm CNVs in chromosomes 1 and 8 (these samples are included in Figure 6B). Similarly, a recent sequencing study also showed that aneuploidy is rare in microdissected human liver samples.45 Collectively, our data show that polyploid cell divisions are common in mice, but the generation of aneuploid clones is rare in humans, even after decades of chronic liver injury.
Figure 6.
Exome sequencing in human cirrhotic nodules did not reveal aneuploid clones. (A) Individual nodules from patients with cirrhosis were dissected within fibrotic boundaries outlined by the yellow lines. (B) Heat map of CNVs for 54 distinct nodules. L860251DN is a normal liver control. HS37 and HS10 are samples with chromosome 1 and 8 CNVs.
Discussion
Previous work showed that increasing ploidy levels in the liver could protect against tumorigenesis through reduced TSG LOH.15 Key evidence was generated after acute dosing of potent carcinogens. Polyploidy would presumably represent an advantage in the setting of catastrophic and mutagenic injuries that might occur a few times within a lifetime, but such is not the pattern of most clinically relevant liver injuries. Here, we focused on chronic injuries that demand multiple rounds of proliferation and regeneration. It is unclear whether or not increasing ploidy could become maladaptive in these chronic disease contexts. Maladaptation on the cell biological level could take the form of inability to proliferate, cellular senescence, or CIN. On the tissue level, maladaptation could manifest as inadequate regeneration, accelerated fibrosis, and/or increased cancer risk.
In agreement with previous studies, we showed that polyploid hepatocytes are able to proliferate and regenerate the liver.20 Most of the mitotic cells in super-polyploid livers exhibited supernumerary centrosomes, indicative of polyploid cell divisions.21,22 This capacity to regenerate does not necessarily mean that they are more efficient than diploid cells within competitive contexts, but does mean that they proliferate well enough to maintain the integrity of a 97% polyploid organ over months of intense tissue damage.
However, a longstanding question is whether or not the proliferation of polyploid cells gives rise to aneuploid daughters. Indeed, polyploidy in cancer cells is closely associated with CIN in part due to segregation defects. By assessing the fidelity of chromosome segregation within super-polyploid livers and CNVs in cirrhotic nodules, we found that aneuploid clones are rare in the human liver. These data are consistent with studies showing low levels of CNVs in single mouse liver cells.27 Our data have the advantage of being obtained from chronically injured human livers, where aneuploidy might be more prevalent. Still, it is possible that in chronic liver damage, aneuploid hepatocytes are readily generated but cleared before they could establish a detectable clone. In this scenario, exome sequencing may not detect a clonal CNV signal in a macroscopic nodule. A recent study performed whole-genome sequencing on microdissected liver fragments harboring 100 to 500 cells.45 Copy number analysis showed that aneuploidy was quite rare, even within these very small clones. Although single-cell sequencing would provide the strongest evidence for the existence of aneuploidy in the liver, aneuploid clones that do not grow larger than a few hundred cells is less likely to be important at the organ level.
Our data do not offer strong evidence for polyploid-associated maladaptation. Most importantly, our models support the concept that polyploid cells are more protected from carcinogenesis than diploid cells. However, we cannot assert that polyploid cells never transform or become cancer. The increased likelihood that HCCs have a diploid origin is supported by the fact that most human HCCs are diploid.15,46–49 In our experiments, super-polyploid livers are protected from both DEN + CCl4 and CCl4-induced liver cancers. These models are relevant to human HCC development because they involve cell proliferation, inflammation, and fibrosis. That said, we cannot exclude the possibility that other cancer models or genetic drivers would pose a greater risk for polyploid transformation.
It is possible that polyploidization is a compensatory mechanism for the increased cancer risk associated with regeneration. Polyploidization is closely associated with injury and regeneration in many tissues and species. For example, polyploidization serves to compensate for tissue loss during wound healing in Drosophila.50 In zebrafish, epicardial cells form a polyploid leader cell subpopulation to support tissue regeneration with enhanced capacity for surface coverage on injury.51 As shown here, the liver polyploidizes after acute or chronic injuries. Regeneration is often a response to injuries that increase cancer risk due to genotoxic stress or inflammation. Polyploidization in the setting of regeneration could represent a cell biological innovation that serves to reduce the risk of cancer associated with tissue damage. Although the human liver is less polyploid than the mouse liver, our work also suggests that induced or therapeutic polyploidization could further protect the liver from cancer while preserving the astounding regenerative capacity of this organ.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT
Many hepatocytes contain whole-genome duplications, but little is known about the fates or functions of these polyploid cells or how they affect development of liver disease.
NEW FINDINGS
Mice with increased polyploid hepatocytes develop fewer liver tumors following chronic liver damage. These polyploid hepatocytes maintain the ability to regenerate liver tissues during chronic damage without generating mitotic errors. Aneuploidy was not commonly observed in cirrhotic livers from patients
LIMITATIONS
This study was performed in mice and human tissue samples. Further studies are needed in patients.
IMPACT
Strategies to increase numbers of polypoid hepatocytes might be effective in preventing liver cancer.
Acknowledgments
We thank Helen Hobbs and Teresa Eversole for contributing human liver samples; Peter Ly and Branden Tarlow for comments on the manuscript; Cheryl Lewis and John Shelton for histopathology; the CRI Sequencing Core (Jian Xu, Xin Liu) and Admera Health (Yun Zhao) for genomics; and Nicholas Loof and the CRI Flow Core for cell sorting and analysis.
Funding
Tao Wang is supported by R03ES026397–01. Hao Zhu is supported by the Pollack Foundation, a Burroughs Wellcome Career Award for Medical Scientists, a CPRIT Individual Investigator Award (RP180268), and a Stand Up To Cancer Innovative Research Grant (SU2C-AACR-IRG 10–16). Stand Up To Cancer is a program of the Entertainment Industry Foundation and its research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C.
Abbreviations used in this paper:
- ANLN
anillin
- CCI4
carbon tetrachloride
- CIN
chromosomal instability
- CNV
copy number variation
- DDC
5-diethoxycarbonyl-1,4-dihydrocollidine
- DAPI
4’,6-diamidino-2-phenylindole
- DEN
diethylnitrosamine
- GS
glutamine synthetase
- HCC
hepatocellular carcinoma
- Ig
immunoglobulin
- KO
knockout
- LOH
loss of heterozygosity
- PI
propidium iodide
- RNA
seq
- RNA
sequencing
- siRNA
small interfering RNA
- TSG
tumor suppressor gene
- UTSw
University of Texas Southwestern
- WT
wild type
Footnotes
CRediT Authorship Contributions
Yu-Hsuan Lin, MS (Conceptualization: Equal; Investigation: Lead; Project administration: Lead; Writing - original draft: Equal). Shuyuan Zhang, PhD (Conceptualization: Equal; Investigation: Equal; Methodology: Lead; Writing - review & editing: Equal).
Min Zhu, PhD (Investigation: Supporting). Tianshi Lu, BS (Formal analysis: Lead). Kenian Chen, PhD (Formal analysis: Equal). Zhuoyu Wen, BS (Formal analysis: Supporting).
Shidan Wang, PhD (Formal analysis: Supporting). Guanghua Xiao, PhD (Formal analysis: Supporting). Danni Luo, MS (Formal analysis: Supporting). Yuemeng Jia, BS (Investigation: Supporting). Lin Li, MS (Methodology: Equal). Malcolm MacConmara, MD (Resources: Supporting). Yujin Hoshida, MD, PhD (Resources: Supporting). Amit Singal, MD (Resources: Supporting). Adam Yopp, MD (Resources: Supporting). Tao Wang, PhD (Formal analysis: Supporting). Hao Zhu, MD (Conceptualization: Lead; Funding acquisition: Lead; Supervision: Lead; Writing - original draft: Lead).
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2020.01.026.
Conflicts of interest
Publisher's Disclaimer: These authors disclose the following: At the time of publication, Hao Zhu owned Ionis Pharmaceuticals stock worth less than $5000. Hao Zhu has an active collaboration with Alnylam Pharmaceuticals and Twenty-Eight Seven Therapeutics. Hao Zhu and Shuyuan Zhang hold a patent application entitled “Inhibitory RNA-Based Therapeutics Targeting ANLN for Cancer Treatment (International Patent Application No. PCT/ US2017/034142 based on U.S. Serial No. 62/347,803).”
References
- 1.Rieseberg LH, Willis JH. Plant speciation. Science 2007; 317:910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leggatt RA, Iwama GK. Occurrence of polyploidy in the fishes. Rev Fish Biol Fish 2003;13:237–246. [Google Scholar]
- 3.Liu Z, Yue S, Chen X, et al. Regulation of cardiomyocyte polyploidy and multinucleation by CyclinG1. Circ Res 2010;106:1498–1506. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa H, Bischoff R, Holtzer H. Mitosis and intermediate-sized filaments in developing skeletal muscle. J Cell Biol 1968;38:538–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkelmann M, Pfitzer P, Schneider W. Significance of polyploidy in megakaryocytes and other cells in health and tumor disease. Klin Wochenschr 1987;65:1115–1131. [DOI] [PubMed] [Google Scholar]
- 6.Frade JM. Somatic tetraploidy in vertebrate neurons: implications in physiology and pathology. Commun Integr Biol 2010;3:201–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentric G, Desdouets C. Polyploidization in liver tissue. Am J Pathol 2014;184:322–331. [DOI] [PubMed] [Google Scholar]
- 8.Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol 2013;24:347–356. [DOI] [PubMed] [Google Scholar]
- 9.Bou-Nader M, Caruso S, Donne R, et al. Polyploidy spectrum: a new marker in HCC classification. Gut 2020; 69:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S, Lin Y-H, Tarlow B, et al. The origins and functions of hepatic polyploidy. Cell Cycle 2019; 18:1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margall-Ducos G, Celton-Morizur S, Couton D, et al. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J Cell Sci 2007;120:3633–3639. [DOI] [PubMed] [Google Scholar]
- 12.Celton-Morizur S, Merlen G, Couton D, et al. The insulin/ Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. J Clin Invest 2009;119:1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao D-Y, Dilkes B, Luo H, et al. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabi-dopsis. Science 2013;341:658–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selmecki AM, Maruvka YE, Richmond PA, et al. Poly-ploidy can drive rapid adaptation in yeast. Nature 2015; 519:349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, Zhou K, Luo X, et al. The polyploid state plays a tumor-suppressive role in the liver. Dev Cell 2018; 44:447–459.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentric G, Maillet V, Paradis V, et al. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest 2015;125:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu S-H, Duncan AW. Pathological polyploidy in liver disease. Hepatology 2015;62:968–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson PD, Delgado ER, Alencastro F, et al. The polyploid state restricts hepatocyte proliferation and liver regeneration in mice. Hepatology 2019;69:1242–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Zhao L, Fish M, et al. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature 2015;524:180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan AW, Taylor MH, Hickey RD, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 2010;467:707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faggioli F, Vezzoni P, Montagna C. Single-cell analysis of ploidy and centrosomes underscores the peculiarity of normal hepatocytes. PLoS One 2011;6:e26080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knouse KA, Lopez KE, Bachofner M, et al. Chromosome segregation fidelity in epithelia requires tissue architecture. Cell 2018;175:200–211.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H-Z, Ouseph MM, Li J, et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol 2012;14:1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandit SK, Westendorp B, Nantasanti S, et al. E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol 2012;14:1181–1191. [DOI] [PubMed] [Google Scholar]
- 25.Wilkinson PD, Alencastro F, Delgado ER, et al. Polyploid hepatocytes facilitate adaptation and regeneration to chronic liver injury. Am J Pathol 2019;189:1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duncan AW, Soto-Gutierrez A. Liver repopulation and regeneration: new approaches to old questions. Curr Opin Organ Transplant 2013;18:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knouse KA, Wu J, Whittaker CA, et al. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci U S A 2014; 111:13409–13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu M, Lu T, Jia Y, et al. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell 2019;177:608–621.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc 2008;3:1167–1170. [DOI] [PubMed] [Google Scholar]
- 30.Beer S, Komatsubara K, Bellovin DI, et al. Hepatotoxin-induced changes in the adult murine liver promote MYC-induced tumorigenesis. PLoS One 2008;3:e2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu H, Shah S, Shyh-Chang N, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet 2010; 42:626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beliaeva NN, Bonashevskaia TI, Nekrasova GI. [Poly-ploidization of hepatocytes under different regimens of rat liver exposed to CCI4]. Biull Eksp Biol Med 1980; 89:57–59; [in Russian]. [PubMed] [Google Scholar]
- 33.Vitális B. The effect of a long-term CCl4 action on the DNA content of rat liver cell nuclei. A cytophotometric study. Folia Histochem Cytochem (Krakow) 1975; 13:207–212. [PubMed] [Google Scholar]
- 34.Gerhard H, Schultze B, Maurer W. Cytological and morphometric investigation of mouse liver regeneration after CCl4 intoxication (author’s transl). Virchows Arch. B Cell Pathol 1973;14:345–359 [in German]. [PubMed] [Google Scholar]
- 35.Mu X, Español-Suñer R, Mederacke I, et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J Clin Invest 2015;125:3891–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujiwara T, Bandi M, Nitta M, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005;437:1043–1047. [DOI] [PubMed] [Google Scholar]
- 37.Lu P, Prost S, Caldwell H, et al. Microarray analysis of gene expression of mouse hepatocytes of different ploidy. Mamm Genome 2007;18:617–626. [DOI] [PubMed] [Google Scholar]
- 38.Zhang S, Nguyen LH, Zhou K, et al. Knockdown of anillin actin binding protein blocks cytokinesis in hepatocytes and reduces liver tumor development in mice without affecting regeneration. Gastroenterology 2018; 154:1421–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raven A, Lu W-Y, Man TY, et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 2017;547:350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell JO, Lu W-Y, Okabe H, et al. Hepatocyte-specific β-catenin deletion during severe liver injury provokes cholangiocytes to differentiate into hepatocytes. Hepatology 2019;69:742–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng X, Zhang X, Li W, et al. Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell 2018;23:114–122.e3. [DOI] [PubMed] [Google Scholar]
- 42.Malato Y, Naqvi S, Schürmann N, et al. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest 2011;121:4850–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benhamouche S, Decaens T, Godard C, et al. Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev Cell 2006;10:759–770. [DOI] [PubMed] [Google Scholar]
- 44.Zare F, Dow M, Monteleone N, et al. An evaluation of copy number variation detection tools for cancer using whole exome sequencing data. BMC Bioinformatics 2017;18:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brunner SF, Roberts ND, Wylie LA, et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature 2019;574:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujimoto J, Okamoto E, Yamanaka N, et al. Flow cytometric DNA analysis of hepatocellular carcinoma. Cancer 1991;67:939–944. [DOI] [PubMed] [Google Scholar]
- 47.Anti M, Marra G, Rapaccini GL, et al. DNA ploidy pattern in human chronic liver diseases and hepatic nodular lesions. Flow cytometric analysis on echo-guided needle liver biopsy. Cancer 1994;73:281–288. [DOI] [PubMed] [Google Scholar]
- 48.Nagasue N, Kohno H, Hayashi T, et al. Lack of intratumoral heterogeneity in DNA ploidy pattern of hepatocellular carcinoma. Gastroenterology 1993;105: 1449–1454. [DOI] [PubMed] [Google Scholar]
- 49.Caselitz M, Masche N, Bleck JS, et al. Increasing sensitivity of morphological diagnosis in hepatocellular carcinoma (HCC) by combination of cytological and fine-needle histological examination after ultrasound guided fine needle biopsy. Z Gastroenterol 2003; 41:559–564. [DOI] [PubMed] [Google Scholar]
- 50.Losick VP, Fox DT, Spradling AC. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr Biol 2013;23:2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao J, Wang J, Jackman CP, et al. Tension creates an endoreplication wavefront that leads regeneration of epicardial tissue. Dev Cell 2017;42:600–615.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.