Abstract
Background
Corticotropin-releasing hormone (CRH), the major secretagogue of the hypothalamic-pituitary-adrenal (HPA) axis, is intricately intertwined with the clock genes to regulate the circadian rhythm of various body functions. N6-methyladenosine (m6A) RNA methylation is involved in the regulation of circadian rhythm, yet it remains unknown whether CRH expression and m6A modification oscillate with the clock genes in chicken hypothalamus and how the circadian rhythms change under chronic stress.
Results
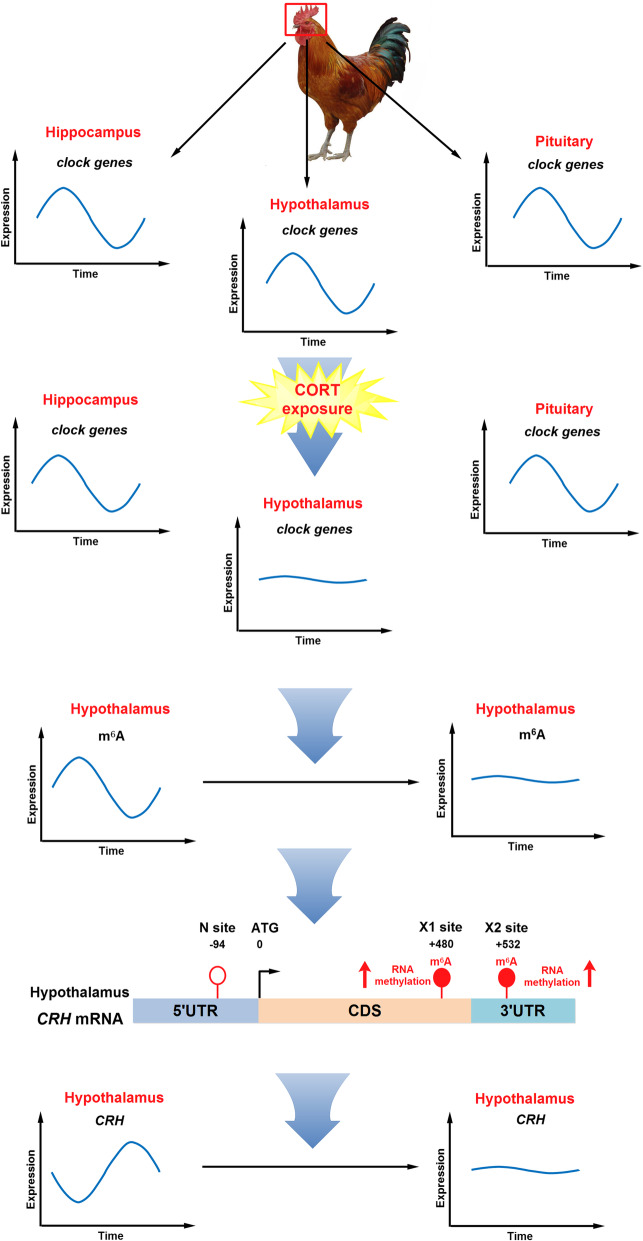
Chronic exposure to corticosterone (CORT) eliminated the diurnal patterns of plasma CORT and melatonin levels in the chicken. The circadian rhythms of clock genes in hippocampus, hypothalamus and pituitary are all disturbed to different extent in CORT-treated chickens. The most striking changes occur in hypothalamus in which the diurnal fluctuation of CRH mRNA is flattened, together with mRNA of other feeding-related neuropeptides. Interestingly, hypothalamic m6A level oscillates in an opposite pattern to CRH mRNA, with lowest m6A level after midnight (ZT18) corresponding to the peak of CRH mRNA before dawn (ZT22). CORT diminished the circadian rhythm of m6A methylation with significantly increased level at night. Further site-specific m6A analysis on 3’UTR of CRH mRNA indicates that higher m6A on 3’UTR of CRH mRNA coincides with lower CRH mRNA at night (ZT18 and ZT22).
Conclusions
Our results indicate that chronic stress disrupts the circadian rhythms of CRH expression in hypothalamus, leading to dysfunction of HPA axis in the chicken. RNA m6A modification is involved in the regulation of circadian rhythms in chicken hypothalamus under both basal and chronic stress conditions.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40104-022-00677-4.
Keywords: Chronic corticosterone exposure, Circadian rhythms, CRH, Hypothalamus, m6A
Introduction
The hypothalamus plays an important role in the regulation of hypothalamic-pituitary-adrenal (HPA) axis [1], feeding behavior [2], and circadian rhythm [3]. Corticotropin-releasing hormone (CRH) released from hypothalamus stimulates pituitary ACTH secretion to modulate the activity of HPA axis [4]. Moreover, CRH is involved in the regulation of food intake [5] via interacting with appetite inhibiting proopiomelanocortin (POMC)/cocaine amphetamine-regulated transcript (CART) neurons and the appetite-inducing neuropeptide Y (NPY) and agouti-related protein (AgRP) neurons [6]. Both the HPA axis activity and the feeding behavior exhibit diurnal patterns, which indicates complex interactive networks with the master clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus [7]. SCN can projects to the pineal gland that secretes the hormone melatonin [8]. The core molecular clock consists of a transcriptional-translational autoregulatory “loop” with a positive arm and a negative arm [9]. The clock and bmal1 genes and their protein products comprise the positive arm, while the period (PER1, PER2, and PER3) and cryptochrome (CRY1, CRY2) genes and their protein products comprise the negative arm. An early research reported that the HPA system in the chicken displays a circadian rhythm [10]. Studies in mice indicate that CRH is intricately intertwined with the clock genes to regulate the circadian rhythm of various body functions [11]. However, the circadian rhythm of CRH expression in chicken hypothalamus has not been characterized.
CRH binds to CRH receptors type 1 (CRHR1) and type 2 (CRHR2) in the pituitary, causing the production and secretion of adrenocorticotropic hormone (ACTH) [12] to regulate the stress response of the body through corticosterone (CORT) synthesis and secretion from adrenal cortex [13, 14]. CORT exerts a negative feedback regulation on CRH synthesis and secretion through its receptor, glucocorticoid receptor (GR), at different levels including hippocampus and hypothalamus [15]. Chronically elevated circulating CORT has detrimental physiological and cognitive effects [16], including HPA axis dysfunction and neuroinflammation [17], as well as depressive and anxiety-like behaviors in SD rats [18]. In addition, chronic stress causes irregular expression of circadian regulatory clock genes in mouse hippocampus [19], hypothalamus SCN [20] and pituitary [21]. However, it remains unknown how chronic CORT exposure affects the circadian rhythms of clock-related genes in the chicken brain, and how it is related to the circadian rhythm of CRH in hypothalamus.
N6-methyladenosine (m6A) is the most prevalent modification in RNAs, which plays an important role in RNA splicing, degradation, and translation [22, 23]. M6A level is finely balanced through interplay among m6A methyltransferases (“writers”, such as METTL3, METTL14 and WTAP), demethylases (“erasers”, such as fat mass and obesity-associated gene FTO and ALKBH5), and binding proteins (“readers”, such as YTHDF1, YTHDF2 and YTHDF3) [24]. Chronic stress is reported to modulate m6A modification in the brain [25]. For instance, heat exposure for 6 h increases m6A RNA methylation levels in the hypothalamus of 3-day-old chickens [26]. Yet, chronic CORT treatment reduces the m6A methylation in chicken liver [27]. Moreover, m6A methylation has been reported to have circadian rhythm [28]. Clock gene CRY1/2 knockout mice show significantly lower m6A level and lost the circadian rhythm of m6A level in RNA [28]. However, studies in the chicken are scarce. Questions remain regarding whether m6A modification in chicken hypothalamus show a circadian rhythm, whether the m6A rhythmicity, if any, is interrupted by chronic CORT exposure, and whether m6A is involved in the regulation of CRH expression in chicken hypothalamus.
Therefore, the objectives of the present study were, firstly, to elaborate the effects of chronic CORT exposure on circadian rhythms of clock-related genes in different brain areas including hippocampus, hypothalamus and pituitary; secondly, to delineate the circadian rhythms of CRH mRNA expression and m6A methylation in chicken hypothalamus, and to reveal their responses to chronic CORT exposure; and thirdly, to investigate the possible link between m6A modification and CRH mRNA expression in chicken hypothalamus.
Materials and methods
Ethics statement
The experimental protocol was approved by the Animal Ethics Committee of Nanjing Agricultural University. The project number is 31972638. The sampling procedures complied with the “Guidelines on Ethical Treatment of Experimental Animals” (2006) No.398 set by the Ministry of Science and Technology, China.
Animals and experimental design
Seventy 45-day-old male bantam chickens were purchased from Changzhou Lihua Livestock and Poultry Co., Ltd. After a three-day adaption, chickens were randomly divided into vehicle (CON) and corticosterone (CORT) group. Light regime was 12 light: 12 dark, with light on at 07:00 as zeitgeber time 0 (ZT0) and off at 19:00 as ZT12. Food and water were provided ad libitum. CORT (Sigma-Aldrich, St Louis, USA) was sonicated in saline with 0.1% Tween 80 and 0.2% DMSO until dissolved and protected from light. Chickens were injected (twice per day, 9:00–10:00 and 18:00–19:00) intraperitoneally with vehicle or CORT (4 mg/kg BW), according to previous publication [29], for 11 consecutive days. Daily food consumption and body weight were recorded every other day. By the end of the treatment, the chickens were sacrificed at the indicated time points (ZT2, ZT6, ZT10, ZT14, ZT18 and ZT22). Chickens were anesthetized with sodium pentobarbital and the brain was quickly separated from the skull. Hippocampus [30] and hypothalamus [31] were dissected as described in previous publications according to the chicken brain atlas [32]. Pituitary was removed as previously described [33]. Tissues collected were frozen immediately in liquid nitrogen and stored at − 80 °C until use.
Measurement of corticosterone and melatonin
Corticosterone concentration was determined by Enzyme Immunoassay (EIA) kit (No. ADI-900-097, Enzo, Farmingdale, NY, USA) following the manufacturer’s instructions. Serum melatonin levels were measured using Chicken MT (Melatonin) ELISA Kit (MM-34278O1, ImmunoWay Biotechnology, Plano, TX, USA) following the manufacturer’s instructions.
RNA isolation and real-time PCR
High quality total RNA was isolated from hippocampus, hypothalamus and pituitary using Trizol reagents (Invitrogen, Carlsbad, CA, USA). One microgram of RNA was reverse-transcribed according to the manufacturer’s protocol (Vazyme Biotech, Nanjing, Jiangsu, China). Four microliter cDNA was diluted (1:25) and then used for real-time PCR in a QuantStudioTM 6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Peptidylprolyl isomerase A (PPIA) was used as an internal control to normalize the technical variations. Data were analyzed using the method of 2-ΔΔCT and presented relative to the CON group. All primers (Table 1) were synthesized by Suzhou GENEWIZ Biological Technology Co., Ltd. (Suzhou, Jiangsu, China).
Table 1.
The primers sequences for RT-PCR and SELECT
| Target genes | Primer sequences (5′ to 3′) | |
|---|---|---|
| CLOCK | F: GATCACAGGGCACCTCCAATA | R: CTAGTTCTCGCCGCCTTTCT |
| B1AML1 | F: GTAGACCAGAGGGCGACAG | R: ATGAAACTGAACCAGCGACTC |
| CRY1 | F: GATGTGGCTATCCTGTAGTTCCT | R: GCTGCTGGTAGATTTGTTTCAT |
| CRY2 | F: GCACGGCTGGATAAACACT | R: AAATAAGCGGCAGGACAAA |
| PER2 | F: ATGAAACGAGCCATCCCG | R: CAGTTGTCGTGATTTTGCCTA |
| PER3 | F: CAGTGCCTTTGTTGGGTTAC | R: GATGGATTCACAAAACTGGAC |
| CRH | F: CTCCCTGGGCCTGGCTTT | R: CCTCACTTCCCGATGATT |
| CRHR1 | F: CACAGCCTTCATCCTACGCA | R: CGGAGCTTGTCGGTGGAATA |
| CRHR2 | F: TCTTTCCTGGGCTTTCACGG | R: ATTGAAGAACTCCGGGCAGG |
| NPY | F: ACTCGGCTCTGAGGCACT | R: GGTCTTCAAACCGGGATC |
| FTO | F: TCACCAAGGCGACCTCTACT | R: GCTGAACCGAGGTGAAAAGC |
| METTL3 | F: ATCCTGGAGCTGCTCAACAC | R: AGATTCGTCCGTGTGCTTGT |
| METTL14 | F: ATTCGACCAGGATGGCTGAC | R: GACTTGGGTGGTGGTGACTT |
| YTHDF1 | F: AACAACCAGCTCCGACACAT | R: GATTCTGACGTTCCTTCCGC |
| YTHDF2 | F: AAGGCCAAGGCAACAAAGTG | R: ATATGCATTGTTCGGCCGGG |
| YTHDF3 | F: CGTAATAGGGGTGTGGGCTTC | R: CACTTCCACACCAGAAGGTGA |
| PPIA | F: TTACGGGGAGAAGTTTGCCG | R: TGGTGATCTGCTTGCTCGTC |
| SELECT | ||
| CRH N site |
F: tagccagtaccgtagtgcgtgGGCGCGCAGCGCGGCCGCTG R: CCCGGTGCTGAAACGCGGCCcagaggctgagtcgctgcat |
|
| CRH X1 site |
F: tagccagtaccgtagtgcgtgTTCCCGATGATTTCCATCAG R: TTCCTGTTGCTGTGGGCTTGcagaggctgagtcgctgcat |
|
| CRH X2 site |
F: tagccagtaccgtagtgcgtgCTCTGGTGCTGACCGCGGGG R: CCCTTTGGCACGGCGCGGGGcagaggctgagtcgctgcat |
|
Analysis of mRNA m6A methylation by dot-blotting assay
Dot-blot analysis of mRNA m6A methylation was performed following a published procedure with minor modifications [34]. Briefly, total RNAs were isolated using the Trizol method and mRNAs were enriched by using GenElute™ mRNA Miniprep Kit (Sigma, Burlington, NJ, USA). The concentration and purity of mRNAs were measured by NanoDrop 2000. The mRNAs were denatured by heating at 95 °C for 5 min, followed by chilling on ice immediately. Next, the mRNA (100 ng) was spotted directly onto the positively charged nylon membrane (GE Healthcare, Pittsburgh, PA, USA) and air dried for 5 min. The membrane was then UV crosslinked in Ultraviolet Crosslinker, blocked with 5% of nonfat milk in TBST, and then incubated with anti-m6A antibody overnight at 4 °C. HRP-conjugated anti-rabbit IgG secondary antibody was added to the membrane for 2 h at room temperature with gentle shaking and then developed with enhanced chemiluminescence. Methylene blue staining was used to verify that equal amount mRNA spotted on the membrane.
Single-base elongation and ligation-based qPCR amplification method (SELECT) assay
The SELECT assay for monitoring site-specific m6A levels in the 3′UTR of CRH mRNA was performed as described previously [35]. In brief, total RNA (2 μg) was mixed with 1 μL of 100 μmol/L dNTP (NEB, Ipswich, MA, USA), 2 μL of CutSmart buffer (NEB, Ipswich, MA, USA), and 2 μL each of 400 nmol/L up and down DNA probes (Table 1). The total volume was adjusted to 17 μL with water. The DNA probes and RNA were annealed by incubating the mixture with a temperature gradient of 90 °C for 1 min, 80 °C for 1 min, 70 °C for 1 min, 60 °C for 1 min, 50 °C for 1 min, and 40 °C for 6 min. To the mixture was then added a 3 μL solution containing 0.01 U Bst 2.0 DNA polymerase, 0.5 U SplintR ligase, and 10 nmol ATP. After incubating at 40 °C for 20 min and then at 80 °C for 20 min, an aliquot (2 μL) of the reaction mixture was taken out for real-time qPCR analysis to quantify template abundance.
Statistical analysis
Data are presented as the mean ± standard error of the mean (SEM). The mRNA levels of clock-related genes and melatonin contents were analyzed using one-way analysis of variance (one-way ANOVA) with IBM SPSS Statistics 20 software (United States) to test the statistical significance of the differences among the six daily time points and confirm the daily variation (P ≤ 0.05), as the premise of cosinor analysis. To determine the circadian rhythmicity of each clock-related gene profile, the mRNA levels of clock-related genes, as well as CORT and melatonin levels were analyzed separately using MATLAB 7.0 (MathWorks Inc., USA) based on unimodal cosinor regression [y = A + (B × cos (2π(x − C)/24))]. A, B and C represent the mesor, amplitude and acrophase, respectively. The results of regression analysis were considered significant at P ≤ 0.05, which was calculated using the number of samples, R2 values and the number of predictors (mesor, amplitude and acrophase) from http://www.danielsoper.com/statcalc3/calc.aspx?i1/415 [36]. Differences of the mesor, amplitude and acrophase between CON and CORT group were tested by one-way ANOVA followed by Fisher’s least significant difference (LSD) post hoc test, considering P ≤ 0.05 to be significant.
Results
Effect of chronic CORT exposure on body weight, food intake, plasma CORT and melatonin concentration
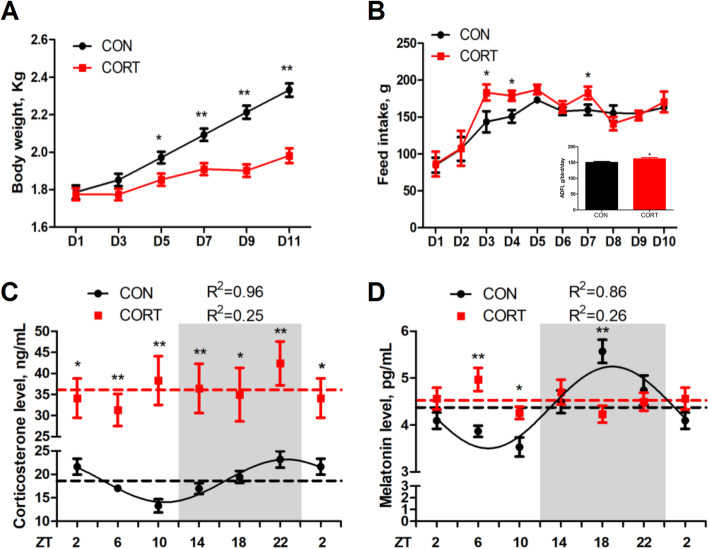
Chronic CORT exposure leads to growth retardation, with significantly lower body weight, as compared with their control counterparts, from the 5th day of CORT injection (D5) to D11 (Fig.1A). Interestingly, the feed intake was significantly increased on D3, D4 and D7, leading to significantly increased average daily feed intake (Fig.1B). Both CORT (Fig. 1C) and melatonin (Fig. 1D) levels in plasma exhibited diurnal pattern in CON group (P < 0.05, one-way ANOVA), which was eliminated in CORT group. The mesors of CORT level were significantly elevated (P < 0.01) by CORT injection, while the mesors of melatonin level did not change (Table 2).
Fig. 1.
Effect of chronic CORT exposure on body weight, food intake, plasma CORT and melatonin concentration. (A) Body weight (n = 6); (B) Feed intake (n = 6) and average daily feed intake (n = 10); (C) Plasma corticosterone content; (D) Plasma melatonin content. The curves represent the 24-hour period determined by cosinor analysis. n = 6 chickens per time point. Data from CT2 are double-plotted. R2 values represent the degree of fitting. Values are mean ± SEM, *P < 0.05, ** P < 0.01, compared with control
Table 2.
Circadian rhythm parameters of CORT and melatonin levels in plasma, as determined by cosinor analyses
| Index | Group | CORT | Melatonin |
|---|---|---|---|
| Mesor | CON | 18.62 ± 0.26 | 4.37 ± 0.12 |
| CORT | 36.10 ± 1.45** | 4.53 ± 0.10 | |
| Amplitude | CON | 3.13 ± 0.37 | 0.87 ± 0.17 |
| CORT | ND | ND | |
| Acrophase, h | CON | 23.18 ± 0.46 | 18.97 ± 0.68 |
| CORT | ND | ND |
Values are means ± SEM. **P < 0.01, compared with CON group. ND represents not determined as there was no circadian rhythm
Effect of chronic CORT exposure on the circadian rhythm of clock genes in hippocampus, hypothalamus, and pituitary
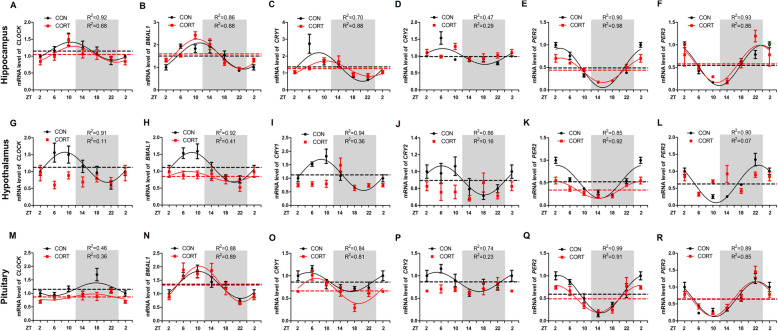
All the 6 clock genes were expressed in hippocampus (Fig. 2A-F), hypothalamus (Fig. 2G-L), and pituitary (Fig. 2M-R), in gene- and region-specific rhythmic patterns. In CON group, BMAL1, PER2 and PER3 showed more pronounced circadian pattern among 6 clock genes (P < 0.05, one-way ANOVA), regardless of the region. Among 3 brain regions, hypothalamus displayed more clearly circadian patterns for all the 6 clock genes (P < 0.05, one-way ANOVA) as shown in cosinor analysis. Chronic CORT exposure abolished or blunted the circadian rhythms of all the major clock genes in hypothalamus, while hippocampus and pituitary were less affected.
Fig. 2.
Effect of chronic CORT exposure on the circadian rhythm parameters of clock genes in chicken hippocampus, hypothalamus and pituitary. (A-F) The circadian rhythms of clock gene mRNA expression in chicken hippocampus. (A) CLOCK gene; (B) BMAL1 gene; (C) CRY1 gene; (D) CRY2 gene; (E) PER2 gene; (F) PER3 gene. (G-L) The circadian rhythms of clock gene mRNA expression in chicken hypothalamus. (G) CLOCK gene; (H) BMAL1 gene; (I) CRY1 gene; (J) CRY2 gene; (K) PER2 gene; (L) PER3 gene. (M-R) The circadian rhythms of clock gene mRNA expression in chicken pituitary. (M) CLOCK gene; (N) BMAL1 gene; (O) CRY1 gene; (P) CRY2 gene; (Q) PER2 gene; (R) PER3 gene. The relative mRNA levels of clock gene are normalized to PPIA. The data markers in the graphs indicate the clock gene mRNA expression levels, and the results are expressed as the mean ± SEM. The curves represent the 24-h period determined by cosinor analysis. n = 6 chickens per time point. Data from CT2 are double-plotted. R2 values represent the degree of fitting
Specifically, chronic CORT exposure significantly delayed (P < 0.05) the acrophase of CRY1 mRNA for 2 h (Fig. 2C and Table 3), and significantly decreased (P < 0.05) the amplitude of PER2 mRNA in hippocampus (Fig. 2E and Table 3). However, chronic CORT exposure had no impact on the rhythmicity of CLOCK (Fig. 2A), BMAL1 (Fig. 2B), CRY2 (Fig. 2D) or PER3 (Fig. 2F) mRNA expression in hippocampus (Table 3). By contrast, the circadian rhythms of CLOCK (Fig. 2G), CRY1 (Fig. 2I), CRY2 (Fig. 2J) and PER3 (Fig. 2L) mRNA in hypothalamus were lost in CORT group (Table 4). Meanwhile, the mesor and amplitude of BMAL1 (Fig. 2H) and PER2 (Fig. 2K) mRNA were significantly decreased (P < 0.05) in CORT group (Table 4). In pituitary, chronic CORT exposure significantly decreased (P < 0.05) the mesor of CLOCK (Fig. 2M) and CRY1 (Fig.2O) mRNA (Table 5). However, chronic CORT exposure had no impact on the rhythmicity of all the clock genes except CRY2 (Fig. 2P, Table 5).
Table 3.
Circadian rhythm parameters of all clock genes in hippocampus, as determined by cosinor analyses
| Index | Group | CLOCK | BMAL1 | CRY1 | CRY2 | PER2 | PER3 |
|---|---|---|---|---|---|---|---|
| Mesor | CON | 1.15 ± 0.03 | 1.50 ± 0.09 | 1.36 ± 0.18 | 0.99 ± 0.08 | 0.49 ± 0.05 | 0.54 ± 0.04 |
| CORT | 1.05 ± 0.06 | 1.60 ± 0.08 | 1.26 ± 0.06 | 1.05 ± 0.05 | 0.44 ± 0.05 | 0.58 ± 0.06 | |
| Amplitude | CON | 0.27 ± 0.04 | 0.58 ± 0.12 | 0.86 ± 0.27 | ND | 0.44 ± 0.07 | 0.45 ± 0.06 |
| CORT | 0.23 ± 0.08 | 0.65 ± 0.12 | 0.45 ± 0.27 | 0.09 ± 0.12 | 0.27 ± 0.02* | 0.41 ± 0.08 | |
| Acrophase, h | CON | 10.64 ± 0.55 | 11.14 ± 0.79 | 8.53 ± 0.28 | 5.89 ± 1.87 | 3.27 ± 0.69 | 23.80 ± 0.56 |
| CORT | 11.51 ± 1.35 | 11.43 ± 0.69 | 10.51 ± 0.70* | ND | 3.37 ± 0.32 | 23.13 ± 0.78 |
Values are means ± SEM. *P < 0.05, **P < 0.01, compared with CON group. ND represents not determined as there was no circadian rhythm
Table 4.
Circadian rhythm parameters of all clock genes in hypothalamus, as determined by cosinor analyses
| Index | Group | CLOCK | BMAL1 | CRY1 | CRY2 | PER2 | PER3 |
|---|---|---|---|---|---|---|---|
| Mesor | CON | 1.13 ± 0.05 | 1.11 ± 0.04 | 1.13 ± 0.05 | 0.90 ± 0.02 | 0.52 ± 0.06 | 0.63 ± 0.07 |
| CORT | ND | 0.84 ± 0.06* | ND | ND | 0.34 ± 0.02* | ND | |
| Amplitude | CON | 0.45 ± 0.07 | 0.44 ± 0.06 | 0.58 ± 0.07 | 0.17 ± 0.03 | 0.36 ± 0.08 | 0.55 ± 0.09 |
| CORT | ND | 0.15 ± 0.09* | ND | ND | 0.19 ± 0.03* | ND | |
| Acrophase, h | CON | 8.94 ± 0.53 | 8.43 ± 0.48 | 8.53 ± 0.43 | 6.21 ± 0.72 | 2.53 ± 0.92 | 22.88 ± 0.64 |
| CORT | ND | 7.51 ± 2.01 | ND | ND | 2.03 ± 0.63 | ND |
Values are means ± SEM. *P < 0.05, **P < 0.01, compared with CON group. ND represents not determined as there was no circadian rhythm
Table 5.
Circadian rhythm parameters of all clock genes in pituitary, as determined by cosinor analyses
| Index | Group | CLOCK | BMAL1 | CRY1 | CRY2 | PER2 | PER3 |
|---|---|---|---|---|---|---|---|
| Mesor | CON | 1.15 ± 0.09 | 1.33 ± 0.06 | 0.86 ± 0.03 | 0.86 ± 0.05 | 0.59 ± 0.02 | 0.64 ± 0.06 |
| CORT | 0.88 ± 0.05* | 1.36 ± 0.08 | 0.66 ± 0.05* | ND | 0.49 ± 0.04 | 0.65 ± 0.07 | |
| Amplitude | CON | 0.24 ± 0.13 | 0.50 ± 0.09 | 0.21 ± 0.05 | 0.21 ± 0.06 | 0.41 ± 0.02 | 0.52 ± 0.09 |
| CORT | 0.11 ± 0.08 | 0.66 ± 0.12 | 0.28 ± 0.07 | ND | 0.30 ± 0.05 | 0.51 ± 0.11 | |
| Acrophase, h | CON | 18.25 ± 2.00 | 10.34 ± 0.69 | 5.04 ± 0.84 | 4.81 ± 1.15 | 2.87 ± 0.25 | 22.53 ± 0.66 |
| CORT | 19.10 ± 2.60 | 10.57 ± 0.66 | 7.15 ± 0.84 | ND | 1.97 ± 0.68 | 21.92 ± 0.76 |
Values are means ± SEM. *P < 0.05, compared with CON group. ND represents not determined as there was no circadian rhythm
Effect of chronic CORT exposure on the circadian rhythm parameters of CRH in hypothalamus and CRH receptor genes in pituitary
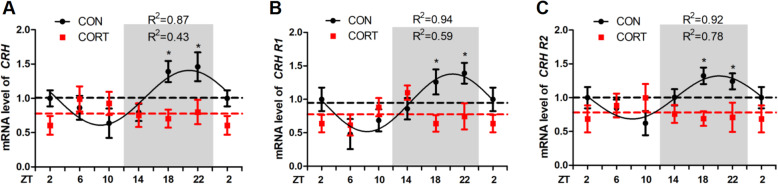
In line with the abolished rhythmicity of clock genes in hypothalamus, the circadian pattern of CRH mRNA (Fig. 3A) in hypothalamus was significantly diminished in CORT group, so was the rhythmic expression of CRHR1 (Fig. 3B) and CRHR2 (Fig. 3C) mRNA in pituitary (P < 0.05, one-way ANOVA). Chronic CORT exposure significantly decreased the mesor (P < 0.05) and amplitude (P < 0.01) of CRH mRNA in hypothalamus, as well as CRHR1 and CRHR2 mRNA in pituitary (Table 6). In general, chronic CORT exposure significantly abolished (P < 0.05) the rise of CRH (Fig. 3A), CRHR1 (Fig. 3B) and CRHR2 (Fig. 3C) mRNA expression in the dark phase after midnight at ZT18 and ZT22.
Fig. 3.
Effect of chronic CORT exposure on the circadian rhythm parameters of CRH in chicken hypothalamus and CRH receptors gene in chicken pituitary. (A) CRH gene; (B) CRHR1 gene; (C) CRHR2 gene. The relative mRNA levels of CRH and CRH receptors gene are normalized to PPIA. The data markers in the graphs indicate the CRH and CRH receptors gene mRNA expression levels, and the results are expressed as the mean ± SEM. The curves represent the 24-h period determined by cosinor analysis. n = 6 chickens per time point. Data from CT2 are double-plotted. R2 values represent the degree of fitting. *P < 0.05, compared with control
Table 6.
Circadian rhythm parameters of CRH in hypothalamus, and CRHR1, CRHR2 in pituitary, as determined by cosinor analyses
| Index | Group | CRH | CRH R1 | CRH R2 |
|---|---|---|---|---|
| Mesor | CON | 1.01 ± 0.05 | 0.95 ± 0.04 | 1.01 ± 0.03 |
| CORT | 0.78 ± 0.05* | 0.78 ± 0.05* | 0.78 ± 0.03* | |
| Amplitude | CON | 0.40 ± 0.08 | 0.43 ± 0.06 | 0.32 ± 0.05 |
| CORT | 0.13 ± 0.07** | 0.17 ± 0.07** | 0.15 ± 0.04** | |
| Acrophase, h | CON | 20.81 ± 0.65 | 20.35 ± 0.44 | 20.18 ± 0.50 |
| CORT | ND | ND | ND |
Values are means ± SEM. *P < 0.05, **P < 0.01, compared with CON group. ND represents not determined as there was no circadian rhythm
Effect of chronic CORT exposure on the circadian rhythm parameters of feeding and inflammation-related genes in hypothalamus
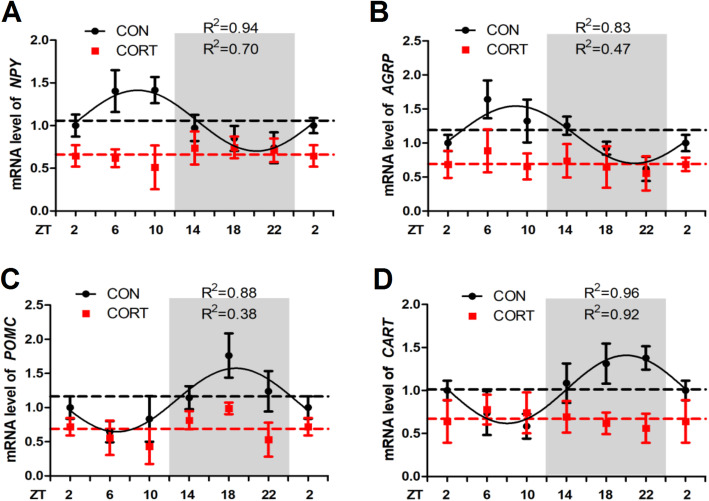
In accordance with the alterations of CRH mRNA, the diurnal patterns of hypothalamic NPY (Fig. 4A), AGRP (Fig. 4B), POMC (Fig. 4C) and CART (Fig. 4D) RNA expression were also eliminated in CORT group (P < 0.05, one-way ANOVA). The expression pattern of “the hunger genes” NPY and AGRP were opposite to that of the “the satiety genes” POMC and CART, matching the diurnal pattern of feeding behavior in the chicken. Chronic CORT exposure significantly decreased (P < 0.01) the mesor and amplitude of all the 4 feeding regulatory genes in hypothalamus (Table 7). In addition, chronic CORT exposure significantly increased (P < 0.01) the hypothalamic expression of TNF-α, IL-1β and IL-6 mRNA, among which the circadian rhythms of TNF-α and IL-6 mRNA was diminished (Additional file 1: Fig. S1).
Fig. 4.
Effect of chronic CORT exposure on the circadian rhythm parameters of feeding related genes in chicken hypothalamus. The circadian rhythms of feeding related gene mRNA expression in chicken pituitary. (A) NPY gene; (B) AGRP gene; (C) POMC gene; (D) CART gene. The relative mRNA levels of feeding related gene are normalized to PPIA. The data markers in the graphs indicate the feeding related gene mRNA expression levels, and the results are expressed as the mean ± SEM. The curves represent the 24-h period determined by cosinor analysis. n = 6 chickens per time point. Data from CT2 are double-plotted. R2 values represent the degree of fitting
Table 7.
Circadian rhythm parameters of NPY, AGRP, POMC and CART in hypothalamus, as determined by cosinor analyses
| Index | Group | NPY | AGRP | POMC | CART |
|---|---|---|---|---|---|
| Mesor | CON | 1.06 ± 0.03 | 1.12 ± 0.06 | 1.11 ± 0.06 | 1.01 ± 0.03 |
| CORT | 0.66 ± 0.02** | 0.69 ± 0.03** | 0.69 ± 0.07** | 0.67 ± 0.01** | |
| Amplitude | CON | 0.36 ± 0.05 | 0.42 ± 0.10 | 0.46 ± 0.08 | 0.40 ± 0.04 |
| CORT | 0.10 ± 0.03** | 0.10 ± 0.05** | 0.16 ± 0.10** | 0.10 ± 0.01** | |
| Acrophase, h | CON | 8.25 ± 0.44 | 8.86 ± 0.77 | 18.72 ± 0.63 | 20.03 ± 0.35 |
| CORT | ND | ND | ND | ND |
Values are means ± SEM. *P < 0.05, **P < 0.01, compared with CON group. ND represents not determined as there was no circadian rhythm
Effect of chronic CORT exposure on the circadian rhythm parameters of m6A level and m6A related genes in hypothalamus
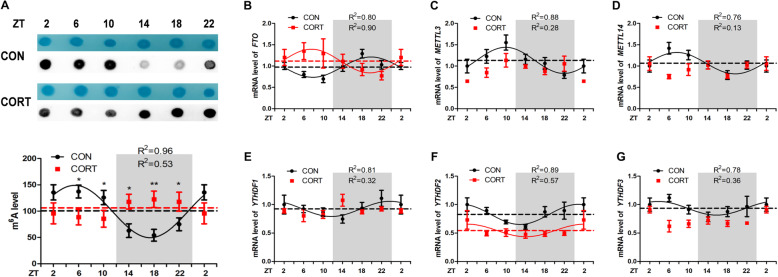
Interestingly, the global RNA m6A levels (Fig. 5A) exhibited diurnal pattern in CON group (P < 0.05, one-way ANOVA), higher m6A levels were detected in light phase. Chronic CORT exposure significantly disrupted this pattern with significantly decreased (P < 0.05) m6A levels in light phase at ZT6 and ZT10, but significantly increased (P < 0.05) m6A levels in dark phase at ZT14, ZT18 and ZT22. Meanwhile, chronic CORT exposure significantly (P < 0.01) decreased the amplitude of m6A levels and delayed the acrophase of m6A levels for 13.48 h (Table 8). Concurrently, chronic CORT exposure significantly increased (P < 0.05) the mesor of FTO (Fig. 5B) mRNA and decreased (P < 0.05) the mesor of YTHDF2 (Fig. 5F) and YTHDF3 (Fig. 5G) mRNA in hypothalamus (Table 8).
Fig. 5.
Effect of chronic CORT exposure on the circadian rhythm parameters of m6A level and m6A related genes in chicken hypothalamus. The circadian rhythms of m6A level and m6A related genes mRNA expression in chicken pituitary. (A) M6A level (n = 4); (B) FTO gene; (C) METTL3 gene; (D) METTL14 gene; (E) YTHDF1 gene; (F) YTHDF2 gene; (G) YTHDF3 gene. The relative mRNA levels of m6A related genes are normalized to PPIA, n = 6 chickens per time point. The data markers in the graphs indicate the m6A related genes mRNA expression levels, and the results are expressed as the mean ± SEM. The curves represent the 24-h period determined by cosinor analysis. Data from CT2 are double-plotted. R2 values represent the degree of fitting. *P < 0.05, compared with control
Table 8.
Circadian rhythm parameters of m6A level and m6A related genes in hypothalamus, as determined by cosinor analyses
| Index | Group | m6A | FTO | METTL3 | METTL14 | YTHDF1 | YTHDF2 | YTHDF3 |
|---|---|---|---|---|---|---|---|---|
| Mesor | CON | 99.31 ± 3.52 | 0.97 ± 0.04 | 1.13 ± 0.04 | 1.07 ± 0.05 | 0.92 ± 0.02 | 0.83 ± 0.02 | 0.94 ± 0.02 |
| CORT | 104.6 ± 1.92 | 1.12 ± 0.03* | 0.91 ± 0.09 | ND | ND | 0.54 ± 0.04* | 0.73 ± 0.05* | |
| Amplitude | CON | 49.76 ± 5.04 | 0.24 ± 0.06 | 0.31 ± 0.05 | 0.25 ± 0.07 | 0.13 ± 0.03 | 0.18 ± 0.03 | 0.12 ± 0.03 |
| CORT | 20.76 ± 2.84** | 0.28 ± 0.05 | 0.17 ± 0.07 | ND | ND | 0.11 ± 0.55 | 0.09 ± 0.06 | |
| Acrophase, h | CON | 4.31 ± 0.45 | 19.82 ± 0.83 | 9.93 ± 0.67 | 9.58 ± 0.95 | 23.73 ± 0.98 | 0.87 ± 0.75 | 3.76 ± 1.10 |
| CORT | ND | 7.56 ± 0.58** | 15.10 ± 3.43* | ND | ND | 2.12 ± 1.90 | 1.31 ± 2.87 |
Values are means ± SEM. *P < 0.05, **P < 0.01, compared with CON group. ND represents not determined as there was no circadian rhythm
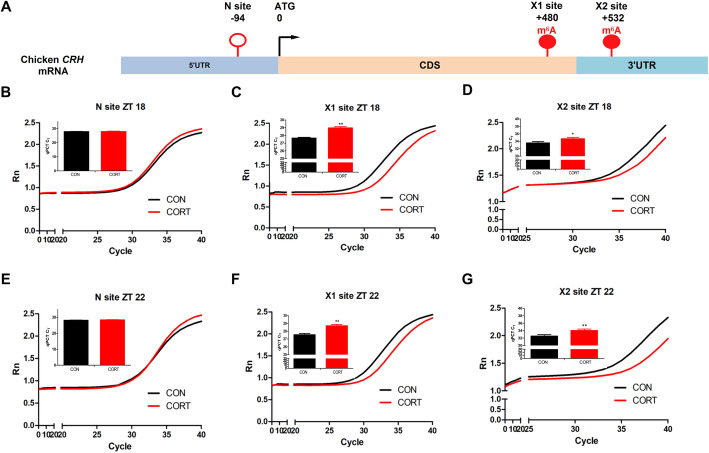
Effect of chronic CORT exposure on the site-specific m6A levels in the 3’UTR of CRH mRNA in hypothalamus
To explore the possible link between the site-specific m6A modification on CRH mRNA and CRH mRNA expression in hypothalamus, RNA samples from hypothalamus on ZT18 and ZT22 with significant changes in CRH mRNA were subjected to single-base elongation and ligation-based qPCR amplification method (SELECT) assay. Two specific m6A sites (Fig. 6A) were identified in the coding sequence (CDS) close to 3’UTR (X1) and 3’UTR (X2) of CRH mRNA, respectively, from published MeRIP-seq database [27]. N site located in the 5’UTR without consensus m6A motif was selected as a negative control. Chronic CORT exposure did not change the CT value on N site at either ZT 18 (Fig. 6B) or ZT 22 (Fig. 6E), compared with CON group. However, chronic CORT exposure significantly increased (P < 0.05) the CT value on both X1 (Fig. 6C, F) and X2 (Fig. 6D, G) at both time points (ZT18 and ZT22), which was in accordance with the significant decrease of CRH mRNA in hypothalamus at the same time points.
Fig. 6.
Effect of chronic CORT exposure on the site-specific m6A levels in the 3’UTR of CRH mRNA in chicken hypothalamus. Validation of m6A modification in CRH 3’UTR using single-base elongation and ligation-based qPCR amplification method (SELECT) when treatment with CORT in chicken hypothalamus. (A) Schematic graph of N, X1 and X2 site in CRH gene; (B) Amplification curve and qPCR CT value in CRH N site at ZT18; (C) Amplification curve and qPCR CT value in CRH X1 site at ZT18; (D) Amplification curve and qPCR CT value in CRH X2 site at ZT18; (E) Amplification curve and qPCR CT value in CRH N site at ZT22; (F) Amplification curve and qPCR CT value in CRH X1 site at ZT22; (G) Amplification curve and qPCR CT value in CRH X2 site at ZT22. Values are mean ± SEM, n = 6 chickens per time point. *P < 0.05, ** P < 0.01, compared with control
Discussion
In this study, we observed that chronic CORT exposure completely abolished the circadian rhythm of plasma melatonin levels in the chicken, indicating a disruption of the endogenous rhythmicity. The effects of CORT on plasma melatonin are biphasic, being stimulatory in the light phase when the melatonin levels are low, while inhibitory in the dark phase when the melatonin levels are high. The avian pineal gland receives circadian input through the release of norepinephrine during the day [37], and the dual effects of CORT on pineal melatonin synthesis are determined by the activation of different adrenoceptors (β or β + α1) during GR activation [38].
The circadian rhythms in birds are controlled by multiple circadian pacemakers in the central nervous system. Here we show, for the first time, the circadian expression of clock genes in chicken hippocampus, hypothalamus, and pituitary. All the 6 core clock genes show circadian rhythms in all the 3 brain areas, although the amplitude and the pattern of oscillation vary among genes and brain areas. It is noted that BMAL1 oscillates in an opposite pattern from PER2 and PER3, may be because they belong, respectively, to “negative arm” and “positive arm” of the circadian clock gene network [9]. Among 3 brain areas, hypothalamus shows more clear and significant rhythmicity and higher susceptibility to CORT treatment. This agrees with a previous publication that long-term administration of dexamethasone resulted in loss of the expression rhythms in Bmal1 and Clock genes in rat SCN [39]. The mechanisms by which chronic CORT alters the circadian gene expression in the chicken are largely unknown. It is likely that CORT directly regulates clock gene expression through GR-mediated transcriptional regulation [40]. However, as melatonin was reported to play a key role in controlling circadian behavioral responses [41] and the loss of circadian rhythm of plasma melatonin corresponded to the diminished circadian pattern of clock genes in the hypothalamus of CORT-exposed chickens in this study. We speculate that chronic CORT may indirectly affects the expression rhythm of circadian clock gene through alterations in melatonin secretion.
CRH is essential for stress adaptation by mediating HPA axis [1] and involved in the regulation of circadian rhythms [2]. Circadian variations of CRH neuron activity are driven by the SCN and likely mediate the characteristic circadian pattern of HPA axis activity [42]. Chronic unpredictable mild stress induces hyperactivity of HPA axis which is indicated by up-regulation of hypothalamic CRH mRNA expression in rats [43]. In contrast, chronic CORT exposure significantly decreased CRH expression in chicken hypothalamus during the dark phase with destroyed circadian rhythms. Many factors contribute to the disparity of the findings, including animal species (nocturnal rats vs. diurnal chickens), stress model, and the time points of the sampling.
Accordingly, genes involved in feeding regulation, including satiety genes POMC and CART and hunger genes NPY and AgRP [44], show concerted circadian expression pattern, which is in agreement with a previous report that AgRP, NPY, POMC and CART genes are expressed in a circadian rhythm in the hypothalamus [45]. The same as CRH and its receptors, the circadian rhythm of these appetite-related genes is also destroyed in chickens subjected to chronic CORT exposure. These CORT-induced alterations in hypothalamic gene expression may associated, at least partly, with the disrupted feeding behavior in the chicken.
The m6A methylation plays important roles in the regulation of neurogenesis, circadian rhythm, cognitive function, and stress responses [46]. Here, we provide the first evidence that the global m6A level in chicken hypothalamus oscillates in a day, being higher in light phase and lower in dark phase. Interestingly, the circadian rhythm pattern of diurnal chickens is opposite to that reported in nocturnal animals. This makes sense as m6A is reported to participate in many stress responses [47], and higher m6A level corresponds to higher body activity. However, in this study, chronic CORT exposure disrupted the circadian rhythms of m6A methylation levels in hypothalamus. Based on the observation that significant decrease of CRH mRNA in the dark phase corresponds to the significant increase in m6A levels at the same time points, we speculate that m6A may be involved in the post-transcriptional regulation of CRH mRNA in chicken hypothalamus. Indeed, the two predicted m6A sites X1 and X2 were both hypermethylated at detected time points (ZT18 and ZT22). Therefore, it is likely that the decrease of CRH expression was due to m6A-mediated mRNA degradation [48]. Nevertheless, a functional verification study is required to elucidate the role of m6A on these sites in CRH gene regulation in chicken hypothalamus.
Conclusion
In conclusion, our study shows that chronic CORT exposure eliminated the diurnal patterns of plasma CORT and melatonin levels in the chicken. Hypothalamus is the most susceptible brain region to CORT treatment, as almost all the genes, including clock genes, CRH, and feeding-related genes, lost their circadian rhythmicity together with the global m6A level. Higher m6A on 3’UTR of CRH mRNA coincides with lower CRH mRNA at night, indicating a possible role of m6A in the post-transcriptional regulation of CRH expression in chicken hypothalamus. These findings provide evidence of CORT-induced disruption of central circadian rhythmicity in CRH expression that leads to dysfunction of HPA axis in the chicken, and also imply a role of RNA m6A modification in the regulation of circadian rhythms in the chicken.
Supplementary Information
Additional file 1: Fig. S1. Effect of chronic CORT exposure on inflammation related genes mRNA expression in chicken hypothalamus. (A) TNF-α, IL-1β and IL-6 mRNA expression in hypothalamus, and destroyed the circadian rhythms of TNF-α and IL-6 mRNA expression (Fig. S1). The circadian rhythms of inflammation related genes and TNF-α, IL-1β and IL-6 mRNA expression in chicken hypothalamus. (A, D) TNF-α gene; (B, E) IL-1β gene; (C, F) IL-6 gene; (D) TNF-α gene. The relative mRNA levels of inflammation related genes are normalized to PPIA, n = 6 chickens per time point. The data markers in the graphs indicate the inflammation related genes mRNA expression levels, and the results are expressed as the mean ± SEM. The curves represent the 24-h period determined by cosinor analysis. Data from CT2 are double-plotted. R2 values represent the degree of fitting. **P < 0.01, compared with control.
Abbreviations
- AgRP
Agouti-related protein
- CART
Cocaine amphetamine-regulated transcript
- CORT
Corticosterone
- CRH
Corticotropin-releasing hormone
- CRHR1
CRH receptors type 1
- CRHR2
CRH receptors type 2
- Cry
Cryptochrome
- GR
Glucocorticoid receptor
- HPA
Hypothalamic-pituitary-adrenal
- m6A
N6-methyladenosine
- NPY
Neuropeptide Y
- Per
Period
- POMC
Proopiomelanocortin
- SCN
Suprachiasmatic nucleus
- SELECT
Single-base elongation and ligation-based qPCR amplification method
Authors’ contributions
YY contributed to data analysis and drafting of the manuscript. AZ, JL and WH were responsible for animal care, breeding and sampling. MZ and WC provided technical support. RZ and YJ contributed to conception, experimental design and data interpretation. RZ and DW contributed to critical revision of the manuscript. The author(s) read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31972638), the National Key Research and Development Program of China (2016YFD0500502), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_0716), and Jiangsu Collaborative Innovation Center of Meat Production and Processing, Quality, and Safety Control. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
The experimental protocol was approved by the Animal Ethics Committee of Nanjing Agricultural University. The project number is 31972638. The sampling procedures according to the “Guidelines on Ethical Treatment of Experimental Animals” (2006) No.398 set by the Ministry of Science and Technology, China.
Consent for publication
The corresponding author and all of the authors have read and approved the final submitted manuscript.
Competing interests
The authors declare no competing financial interest.
References
- 1.Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science. 1981;213(4514):1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 2.Zou H, Shi M, He F, Guan C, Lu W. Expression of corticotropin releasing hormone in olive flounder (Paralichthys olivaceus) and its transcriptional regulation by c-Fos and the methylation of promoter. Comp Biochem Physiol B Biochem Mol Biol. 2021;251:110523. doi: 10.1016/j.cbpb.2020.110523. [DOI] [PubMed] [Google Scholar]
- 3.Challet E. The circadian regulation of food intake. Nat Rev Endocrinol. 2019;15(7):393–405. doi: 10.1038/s41574-019-0210-x. [DOI] [PubMed] [Google Scholar]
- 4.Kuenzel WJ, Kang SW, Jurkevich A. The vasotocinergic system and its role in the regulation of stress in birds. Vitam Horm. 2020;113:183–216. doi: 10.1016/bs.vh.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Valassi E, Scacchi M, Cavagnini F. Neuroendocrine control of food intake. Nutr Metab Cardiovasc Dis. 2008;18(2):158–168. doi: 10.1016/j.numecd.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Van Drunen R, Eckel-Mahan K. Circadian rhythms of the hypothalamus: from function to physiology. Clocks Sleep. 2021;3(1):189–226. doi: 10.3390/clockssleep3010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez REA, Kalume F, de la Iglesia HO. Sleep timing and the circadian clock in mammals: past, present and the road ahead. Semin Cell Dev Biol. 2021. 10.1016/j.semcdb.2021.05.034. [DOI] [PMC free article] [PubMed]
- 8.Giudice A, Crispo A, Grimaldi M, Polo A, Bimonte S, Capunzo M, et al. The effect of light exposure at night (LAN) on carcinogenesis via decreased nocturnal melatonin synthesis. Molecules. 2018;23(6):1308. [DOI] [PMC free article] [PubMed]
- 9.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 10.Majsa Z, Mihaly K, Peczely P. Circadian rhythm of hypothalamo-hypophyseal-adrenal activity in the chicken. Acta Physiol Acad Sci Hung. 1976;47(2–3):101–109. [PubMed] [Google Scholar]
- 11.Muglia LJ, Jacobson L, Weninger SC, Karalis KP, Jeong K, Majzoub JA. The physiology of corticotropin-releasing hormone deficiency in mice. Peptides. 2001;22(5):725–731. doi: 10.1016/S0196-9781(01)00385-0. [DOI] [PubMed] [Google Scholar]
- 12.Herman JP, McKlveen JM, Ghosal S, Kopp B, Wulsin A, Makinson R, et al. Regulation of the hypothalamic-pituitary-adrenocortical stress response. Compr Physiol. 2016;6(2):603–21. 10.1002/cphy.c150015. [DOI] [PMC free article] [PubMed]
- 13.Darnaudery M, Maccari S. Epigenetic programming of the stress response in male and female rats by prenatal restraint stress. Brain Res Rev. 2008;57(2):571–585. doi: 10.1016/j.brainresrev.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Torres DB, Lopes A, Rodrigues AJ, Cerqueira JJ, Sousa N, Gontijo JAR, et al. Anxiety-like behavior and structural changes of the bed nucleus of the stria terminalis (BNST) in gestational protein-restricted male offspring. J Dev Orig Health Dis. 2018;9(5):536–43. 10.1017/S2040174418000399. [DOI] [PubMed]
- 15.Laryea G, Muglia L, Arnett M, Muglia LJ. Dissection of glucocorticoid receptor-mediated inhibition of the hypothalamic-pituitary-adrenal axis by gene targeting in mice. Front Neuroendocrinol. 2015;36:150–164. doi: 10.1016/j.yfrne.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolkowitz OM, Burke H, Epel ES, Reus VI. Glucocorticoids. Mood, memory, and mechanisms. Ann N Y Acad Sci. 2009;1179(1):19–40. doi: 10.1111/j.1749-6632.2009.04980.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Gao X, Wang A, Wang Y, Du Y, Li L, et al. Depression comorbid with hyperalgesia: different roles of neuroinflammation induced by chronic stress and hypercortisolism. J Affect Disord. 2019;256:117–124. doi: 10.1016/j.jad.2019.05.065. [DOI] [PubMed] [Google Scholar]
- 18.Lee B, Sur B, Shim I, Lee H, Hahm DH. Angelica gigas ameliorate depression-like symptoms in rats following chronic corticosterone injection. BMC Complement Altern Med. 2015;15(1):210. doi: 10.1186/s12906-015-0746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clement A, Pedersen MM, Stensballe A, Wiborg O, Asuni AA. Chronic stress induces NPD-like behavior in APPPS1 and WT mice with subtle differences in gene expression. Genes Brain Behav. 2021;20(8):e12766. 10.1111/gbb.12766. [DOI] [PMC free article] [PubMed]
- 20.Jiang WG, Li SX, Zhou SJ, Sun Y, Shi J, Lu L. Chronic unpredictable stress induces a reversible change of PER2 rhythm in the suprachiasmatic nucleus. Brain Res. 2011;1399:25–32. doi: 10.1016/j.brainres.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Razzoli M, Karsten C, Yoder JM, Bartolomucci A, Engeland WC. Chronic subordination stress phase advances adrenal and anterior pituitary clock gene rhythms. Am J Physiol Regul Integr Comp Physiol. 2014;307(2):R198–R205. doi: 10.1152/ajpregu.00101.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maity A, Das B. N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J. 2016;283(9):1607–1630. doi: 10.1111/febs.13614. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Fu J, Zhou Y. A review in research Progress concerning m6A methylation and Immunoregulation. Front Immunol. 2019;10:922. doi: 10.3389/fimmu.2019.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao G, Li HB, Yin Z, Flavell RA. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6(4):160003. doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chokkalla AK, Mehta SL, Vemuganti R. Epitranscriptomic modifications modulate normal and pathological functions in CNS. Transl Stroke Res. 2021. 10.1007/s12975-021-00927-z. [DOI] [PMC free article] [PubMed]
- 26.Kisliouk T, Rosenberg T, Ben-Nun O, Ruzal M, Meiri N. Early-Life m (6) A RNA demethylation by fat mass and obesity-associated protein (FTO) influences resilience or vulnerability to heat stress later in life. eNeuro. 2020;7(3). 10.1523/ENEURO.0549-19.2020. [DOI] [PMC free article] [PubMed]
- 27.Hu Y, Feng Y, Zhang L, Jia Y, Cai D, Qian SB, et al. GR-mediated FTO transactivation induces lipid accumulation in hepatocytes via demethylation of m (6) a on lipogenic mRNAs. RNA Biol. 2020;17(7):930–42. 10.1080/15476286.2020.1736868. [DOI] [PMC free article] [PubMed]
- 28.Wang CY, Yeh JK, Shie SS, Hsieh IC, Wen MS. Circadian rhythm of RNA N6-methyladenosine and the role of cryptochrome. Biochem Biophys Res Commun. 2015;465(1):88–94. doi: 10.1016/j.bbrc.2015.07.135. [DOI] [PubMed] [Google Scholar]
- 29.Luo JW, Zhou ZL, Zhang H, Ma RS, Hou JF. Bone response of broiler chickens (Gallus gallus domesticus) induced by corticosterone. Comp Biochem Physiol A Mol Integr Physiol. 2013;164(2):410–416. doi: 10.1016/j.cbpa.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Kadhim HJ, Kang SW, Kuenzel WJ. Differential and temporal expression of corticotropin releasing hormone and its receptors in the nucleus of the hippocampal commissure and paraventricular nucleus during the stress response in chickens (Gallus gallus) Brain Res. 1714;2019:1–7. doi: 10.1016/j.brainres.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Yuan L, Wang Y, Hu Y, Zhao R. In ovo leptin administration modulates glucocorticoid receptor mRNA expression specifically in the hypothalamus of broiler chickens. Neurosci Lett. 2017;638:181–188. doi: 10.1016/j.neulet.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 32.Kuenzel WJ, Masson M. A stereotaxic atlas of the brain of the chick (Gallus domesticus); 1988. Johns Hopkins University Press.2715 North Charles StreetBaltimore, Maryland 21218-4363
- 33.Sasaki F, Doshita A, Matsumoto Y, Kuwahara S, Tsukamoto Y, Ogawa K. Embryonic development of the pituitary gland in the chick. Cells Tissues Organs. 2003;173(2):65–74. doi: 10.1159/000068945. [DOI] [PubMed] [Google Scholar]
- 34.Nagarajan A, Janostiak R, Wajapeyee N. Dot blot analysis for measuring global N (6)-Methyladenosine modification of RNA. Methods Mol Biol. 1870;2019:263–271. doi: 10.1007/978-1-4939-8808-2_20. [DOI] [PubMed] [Google Scholar]
- 35.Xiao Y, Wang Y, Tang Q, Wei L, Zhang X, Jia G. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N (6) -Methyladenosine modification. Angew Chem. 2018;57(49):15995–16000. doi: 10.1002/anie.201807942. [DOI] [PubMed] [Google Scholar]
- 36.Singh D, Rani S, Kumar V. Daily expression of six clock genes in central and peripheral tissues of a night-migratory songbird: evidence for tissue-specific circadian timing. Chronobiol Int. 2013;30(10):1208–1217. doi: 10.3109/07420528.2013.810632. [DOI] [PubMed] [Google Scholar]
- 37.Cassone VM, Takahashi JS, Blaha CD, Lane RF, Menaker M. Dynamics of noradrenergic circadian input to the chicken pineal gland. Brain Res. 1986;384(2):334–341. doi: 10.1016/0006-8993(86)91169-8. [DOI] [PubMed] [Google Scholar]
- 38.Fernandes PA, Tamura EK, D'Argenio-Garcia L, Muxel SM, da Silveira Cruz-Machado S, Marcola M, et al. Dual effect of Catecholamines and corticosterone crosstalk on pineal gland melatonin synthesis. Neuroendocrinology. 2017;104(2):126–134. doi: 10.1159/000445189. [DOI] [PubMed] [Google Scholar]
- 39.Wu T, Yang L, Jiang J, Ni Y, Zhu J, Zheng X, et al. Chronic glucocorticoid treatment induced circadian clock disorder leads to lipid metabolism and gut microbiota alterations in rats. Life Sci. 2018;192:173–82. 10.1016/j.lfs.2017.11.049. [DOI] [PubMed]
- 40.Murayama Y, Yahagi N, Takeuchi Y, Aita Y, Mehrazad Saber Z, Wada N, et al. Glucocorticoid receptor suppresses gene expression of rev-erbalpha (Nr1d1) through interaction with the CLOCK complex. FEBS Lett. 2019;593(4):423–32. 10.1002/1873-3468.13328. [DOI] [PubMed]
- 41.Tosches MA, Bucher D, Vopalensky P, Arendt D. Melatonin signaling controls circadian swimming behavior in marine zooplankton. Cell. 2014;159(1):46–57. doi: 10.1016/j.cell.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholson S, Lin JH, Mahmoud S, Campbell E, Gillham B, Jones M. Diurnal variations in responsiveness of the hypothalamo-pituitary-adrenocortical axis of the rat. Neuroendocrinology. 1985;40(3):217–224. doi: 10.1159/000124078. [DOI] [PubMed] [Google Scholar]
- 43.Cai L, Li R, Tang WJ, Meng G, Hu XY, Wu TN. Antidepressant-like effect of geniposide on chronic unpredictable mild stress-induced depressive rats by regulating the hypothalamus-pituitary-adrenal axis. Eur Peuropsychopharmacol: J Eur Coll Neuropsychopharmacol. 2015;25(8):1332–1341. doi: 10.1016/j.euroneuro.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell CS, Begg DP. The regulation of food intake by insulin in the central nervous system. J Neuroendocrinol. 2021;33(4):e12952. doi: 10.1111/jne.12952. [DOI] [PubMed] [Google Scholar]
- 45.Stutz AM, Staszkiewicz J, Ptitsyn A, Argyropoulos G. Circadian expression of genes regulating food intake. Obesity. 2007;15(3):607–615. doi: 10.1038/oby.2007.564. [DOI] [PubMed] [Google Scholar]
- 46.Chokkalla AK, Mehta SL, Vemuganti R. Epitranscriptomic regulation by m (6) a RNA methylation in brain development and diseases. J Cereb Blood Flow Metab: Off J Int Soc Cereb Blood Flow Metab. 2020;40(12):2331–2349. doi: 10.1177/0271678X20960033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engel M, Eggert C, Kaplick PM, Eder M, Roh S, Tietze L, et al. The role of m (6) a/m-RNA methylation in stress response regulation. Neuron. 2018;99(2):389–403.e9. doi: 10.1016/j.neuron.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S, et al. M (6) a mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31(10):990–1006. 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Effect of chronic CORT exposure on inflammation related genes mRNA expression in chicken hypothalamus. (A) TNF-α, IL-1β and IL-6 mRNA expression in hypothalamus, and destroyed the circadian rhythms of TNF-α and IL-6 mRNA expression (Fig. S1). The circadian rhythms of inflammation related genes and TNF-α, IL-1β and IL-6 mRNA expression in chicken hypothalamus. (A, D) TNF-α gene; (B, E) IL-1β gene; (C, F) IL-6 gene; (D) TNF-α gene. The relative mRNA levels of inflammation related genes are normalized to PPIA, n = 6 chickens per time point. The data markers in the graphs indicate the inflammation related genes mRNA expression levels, and the results are expressed as the mean ± SEM. The curves represent the 24-h period determined by cosinor analysis. Data from CT2 are double-plotted. R2 values represent the degree of fitting. **P < 0.01, compared with control.
Data Availability Statement
Not applicable.