Abstract
Cryo-electron microscopy (cryo-EM) has produced a number of structural models of the SARS-CoV-2 spike, already prompting biomedical outcomes. However, these reported models and their associated electrostatic potential maps represent an unknown admixture of conformations stemming from the underlying energy landscape of the spike protein. As with any protein, some of the spike's conformational motions are expected to be biophysically relevant, but cannot be interpreted only by static models. Using experimental cryo-EM images, we present the energy landscape of the glycosylated spike protein, and identify the diversity of low-energy conformations in the vicinity of its open (so called 1RBD-up) state. The resulting atomic refinement reveal global and local molecular rearrangements that cannot be inferred from an average 1RBD-up cryo-EM model. Here we report varied degrees of “openness” in global conformations of the 1RBD-up state, not revealed in the single-model interpretations of the density maps, together with conformations that overlap with the reported models. We discover how the glycan shield contributes to the stability of these low-energy conformations. Five out of six binding sites we analyzed, including those for engaging ACE2, therapeutic mini-proteins, linoleic acid, two different kinds of antibodies, switch conformations between their known apo- and holo-conformations, even when the global spike conformation is 1RBD-up. This apo-to-holo switching is reminiscent of a conformational preequilibrium. We found only one binding site, namely that of AB-C135 remains in apo state within all the sampled free energy-minimizing models, suggesting an induced fit mechanism for the docking of this antibody to the spike.
Keywords: Cryo-EM, Molecular dynamics, Free energy landscape, Manifold machine learning, Spike protein
1. Introduction
Intensive research, primarily by cryo-Electron Microscopy (cryo-EM) techniques, has established that the spike protein plays a critical role in the process of infection by the coronaviruses (Walls et al., 2020; Lan et al., 2020). The average ‘apo’ (ligand-free) and ‘holo’ (ligand-bound) conformations assumed by the spike protein are now known to near-atomic resolution (Walls et al., 2020; Wrapp et al., 2020; Benton et al., 2020). It is recognized that the spike protein exhibits structural variability (Walls et al., 2020; Cai et al., 2020). However, access to an experimentally determined conformational path between the apo- and holo-end states would substantially elucidate the thermally accessible functionally relevant conformational motions of the spike protein (Dashti et al., 2020; Ourmazd, 2019).
As different molecular conformations populate distinct energy states, their conformational spectrum defines an energy landscape. This landscape is, in principle, multi-dimensional. Nonetheless, a two-dimensional representation described by the leading two conformational coordinates is commonly used to project the multidimensional free energy profiles on to a manifold of reduced dimensionality (Dashti et al., 2020; Ourmazd, 2019; Frank and Ourmazd, 2016). Under near-equilibrium conditions, biomolecular functions are often revealed by connecting minimum free energy conformations along these profiles (Branduardi and Faraldo-Gomez, 2013, Matsunaga et al., 2012, Medovoy et al., 2016, Meng et al., 2016, Van Der Vaart and Karplus, 2007). For example, the path underlying cell recognition of the spike protein commences along the energy landscape of the apo state transitioning to the holo state after binding the cell surface receptors, and ending at the most probable conformations on the holo state. Here, we focus on extracting a multi-model representation of only the apo-like spike conformations from a cryo-EM dataset (EMD-21375), and draw inferences on the plausible binding mechanisms of these conformations. The apo ensemble models reveal spike-ligand interactions that are obscure to a single structure interpretation, reported as PDB: 6VSB, or even microsecond-long brute force molecular dynamics (MD) simulations of this apo-state model, therefore, stressing the importance of data-guided modelling.
The Receptor Binding Domain (RBD) of the apo spike protein exists primarily in two conformations. These “down” and “up” states occur with nearly equal probability (Walls et al., 2020; Wrapp et al., 2020). In the down state, the ACE2 binding pocket is closed, rendering membrane fusion essentially unfeasible. The nature of the pathway between the down and the up states of the RBD, and the transition rate between a hidden and an accessible ACE2 binding pocket are of central importance for understanding SARS-CoV2's ability to hide vulnerable epitopes from the host's immune system (Lan et al., 2020). Several studies have employed MD simulations to investigate the transitions between these states, which uses static cryo-EM structures to characterize each endpoint (Casalino et al., 2020; Gur et al., 2020; Fallon et al., 2020; Moreira et al., 2020; Zimmerman et al., 2021; Pavlova et al., 2021a, 2021b; Acharya et al., 2021). However, a single cryo-EM map represents an ensemble of thermally accessible conformations, and not just one structure (Ourmazd, 2019; Shekhar et al., 2021; Giraldo-Barreto et al., 2021). Therefore, MD simulations that are starting from the best map-fitted model, and attempting to encompass the up-to-down transition, are expected to overcome their initial-model bias and capture at least the endpoint ensembles accurately. Enhanced sampling simulations overcome such conformational bias (Singharoy et al., 2016) but are computationally cumbersome. Yet in the absence of any benchmark, it is difficult to judge how well do the MD or enhanced sampling simulations perform towards visiting all plausible conformations underlying a reconstructed 3D density. So, complementing the body of available studies on spike conformations (Casalino et al., 2020; Gur et al., 2020; Fallon et al., 2020; Moreira et al., 2020; Zimmerman et al., 2021; Pavlova et al., 2021a, 2021b; Acharya et al., 2021), here, we seek a joint experimental-computational measure of conformational diversity of the endpoint ‘up-only ensemble’, and whether such diversity can be accurately modeled by brute force MD simulations.
By combining cryo-EM data analysis and all-atom MD simulations, we determine a collection of low-energy conformations pertaining to the up state of the RBD. Our results show that in this apo-state, the spike protein assumes a heterogeneous ensemble of nearly isoenergetic conformations. The structural difference between an ensemble model of the up vs. the down conformations is amplified relative those seen between the average up and down models in standard cryo-EM analyses (Walls et al., 2020; Wrapp et al., 2020). The local movements have a dramatic effect on key binding sites, offering fresh insights into how molecular recognition occurs at the spike's ACE2, linoleic acid, antibodies-binding domains.
Second-scale all-atom MD simulations have been performed using a distributed computing platform (Zimmerman et al., 2021) and offer new insights on the accessibility of cryptic epitopes in the up vs. down states of the RBD. Recently, a coarse-grained model of the entire SARS-CoV-2 virion has been produced (Yu et al., 2021). This work not only provides insights into the coupled behavior of the virion's structural proteins, but also outlines a methodology for incorporating statistics from all-atom MD simulations to increase the fidelity of large-scale coarse-grained models. Now, our integrative modelling reveals unique conformations in RBD binding sites not seen in the reported apo-up structures as well as microsecond-long MD simulations. Our approach also offers a protocol for future work on characterizing the most probable up to down transitions of the RBD, by simply reprocessing the existing cryo-EM datasets.
2. Results
We determine an energy landscape of the spike protein in a reduced space derived from experimental cryo-EM snapshots, and reconstruct 3D density of low-energy conformations on this landscape in the vicinity of the up state. MD simulations are then employed to interpret the density information in all-atoms detail. Finally, we present the global and local conformations of the up-state of the apo spike protein, focusing on the conformational heterogeneity of key binding pockets.
3. Conformational coordinates, energy landscape, and molecular rearrangements
We use geometric machine-learning (ManifoldEM) (Dashti et al., 2021) to extract the manifold of conformational motions from cryo-EM images. This manifold is spanned by a set of orthogonal conformational coordinates (CC) (Dashti et al., 2020). To determine the conformational changes along each coordinate, we compile a 3D movie of the density maps along each of those coordinates. (Details of the density movie generation scheme is provided in Methods). Fifty density maps were extracted along each of the two conformational coordinates. The nominal resolution of these maps varies from 3.2 to 4.4 Å. Molecular dynamics flexible fitting or MDFF was employed to construct molecular models from each of the maps (Trabuco et al., 2008, 2009), translating the density movies along CC1 and CC2 to molecular movies (Methods and Supplementary Movies 1 and 2). A string simulation with swarms of trajectories (Pan et al., 2008) was then performed using the flexibly fitted models to probe conformational transition pathways connecting the CC1 and CC2 densities (see Methods).
By construction of the conformational coordinates (Dashti et al., 2020), the motions observed along CC1 play out in the XY plane, and motions observed for CC2 evolve along the Z-axis. Movement along CC1 corresponds to a global change in the RBD's center of mass, which deforms orthogonal to the spike protein's principal axis. Similarly, motion along CC2 captures a “projectile-like” motion of the RBD parallel to the spike protein's principal axis. The functionally relevant conformational movements, however, involve a combination of CC1 and CC2 along the minimum-energy path on the conformational landscape (Van Der Vaart and Karplus, 2007). As detailed in Methods, we infer free-energy changes from the population of points on the CC1-CC2 conformational plane via the Boltzmann factor (Fig. 1A) (Dashti et al., 2020). The low energy conformations on this CC1-CC2 landscape encompasses a ‘horse-shoe’ shaped, essentially iso-energetic tube. Assuming near-equilibrium conditions, the conformational states that are remote to the horse-shoe shape energy feature are not significantly occupied. We find that comparing SASA values from MD ensembles derived by fitting a single CC (CC1 or CC2) with those concomitantly utilizing both CC1 and CC2 (i.e., along the horse-shoe locations) do not agree at the same CC value (Fig. S8). Also, the SASA values for the single CC fitting procedure generally do not span the same range seen for the minimum energy locations. This difference indicates that the coupled CC1-CC2 pathway described conformations that are distinct from 3D-densities based on single CC1 or CC2.
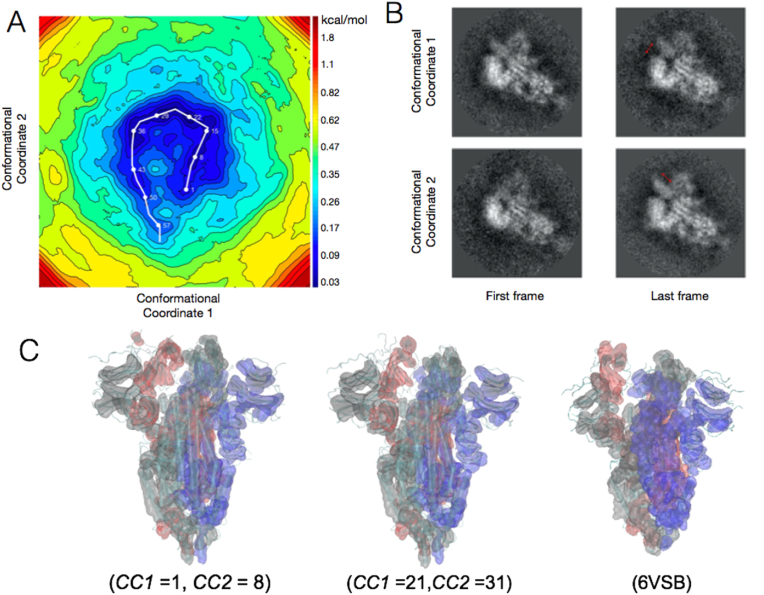
Fig. 1.
(A) The energy landscape of the SARS-CoV-2 Spike Protein spanned by two conformational coordinates. In this study, each continuous conformational degree of freedom is approximated by a spectrum of 50 states. A conformational coordinate consists of a space of 50 binned electrostatic potential maps defining conformational changes in the space of these maps. The low-energy locations are traced with a white line representing the 59 independent points on the landscape, structures for which are determined using MDFF. (B) Clustered images representing the locations traced above at CC1 = 0 and CC1 = 50, and CC1 = 0 and CC2 = 50. (C) Structural models with varied degrees of openness at locations on the free energy landscape compared to the deposited model.
Previously reported 6VSB (Wrapp et al., 2020) and 6VYB (Walls et al., 2020) densities resemble ManifoldEM models pertaining to location no. 28 (CC1 21; CC2 31) and its close vicinity on the minimum free energy structures in Fig. 1A. (See Methods for relating density maps to points on the energy landscape.) Most of the low-energy models are nearly isoenergetic, and thus populated with comparable probabilities. The pursuit of high-resolution structures, however, may preferentially select a subset of the high probability conformations present (Ourmazd, 2019). Key spike functions (e.g., antibody and receptor binding or epitope signaling) stem from the entire population of the low-energy structures. The conformational heterogeneity associated with the states in this horse-shoe shaped reaction tube highlights the range of global and local conformations often subsumed by single-model representations, which ignore the underlying conformational energy landscapes.
We already observe at the level of density changes, that the conformational spectrum along the horse-shoe profile of the apo up spike conformation can result in more open (CC1 25 → 31; CC2 28 → 32) and less open (CC1 17 → 20 and CC2 8 → 12) apo conformations (Fig. 2, Fig. 3). These conformations are as probable as those reported in the PDB-like model in location no. 28 (CC1 21; CC2 31) (Walls et al., 2020; Wrapp et al., 2020). The variation in conformations of the spike protein is only partially apparent in a comparison between the reported static apo (Walls et al., 2020; Wrapp et al., 2020), holo (Cao et al., 2020; Barnes et al., 2020; Toelzer et al., 2020), and long-time MD simulation models of the spike (Shaw, 2020). We circumvent the uncertainties in inferring function from stationary snapshots (6), by compiling density movies (Fig. 1B–C and Supplementary Movies 1 and 2) along the CC1 and CC2 dimensions to reveal a multi-model or ensemble representation of the apo spike state.
Fig. 2.
Whole RBD Solvent Accessible Surface Area (SASA) scores calculated for the up RBD (residues 320 to 510). The SASA scores for static structures are represented by red circles in the far-left panel, the non-biased equilibrium MD trajectories are represented by green circles and error bars (31) in the far-right panel, and finally each MFEP location SASA scores are represented by blue circles and error bars in the middle panel. The error bars show 1 standard deviation from the mean. The red and blue highlighted regions are used to compare the spread of SASA scores of the previously deposited static structures (red shading) with the range of SASA scores seen for all MFEP locations (blue shading). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
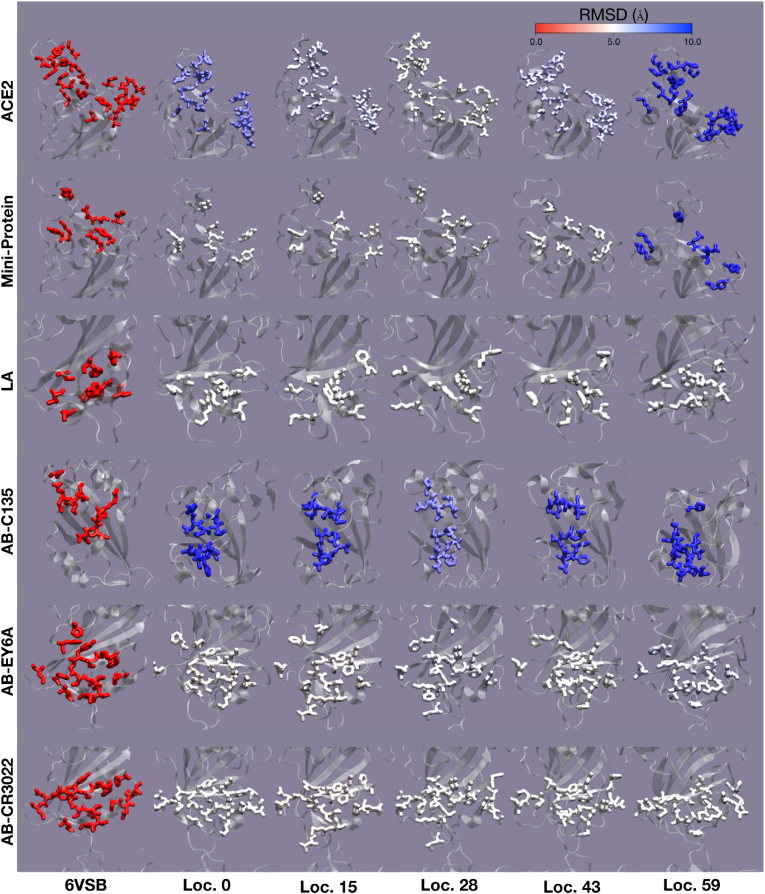
Fig. 3.
A pictorial representation of binding pocket RMSDs. The residues involved in binding are used to calculate the RMSD at minimum energy locations 0, 15, 28, 43, and 59 (see Fig. 1) and are shown in a licorice representation. The RMSD values range from 0 to 1 following the color scale at the top of the figure. The structure 6VSB is shown for comparison on the far left. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Global conformational changes along the low energy spike conformations
A total of 59 points on the horse shoe shaped low energy profile were refined using a multi-grid MDFF procedure (Vant et al., 2020a; Singharoy et al., 2019) to gauge the most probable conformational changes of the spike in an apo-only state. Shown in Supplementary Movie 3, the set of conformations reveals deviations from the static apo structures at both ends of the horseshoe-shaped tube. As expected, models stemming from the cryo-EM data-guided free energy landscape resemble previously published open state structures 6VSB and 6VYB (Walls et al., 2020; Wrapp et al., 2020), with RMSD ranging from 2.7 to 4.2 Å (Fig. S1). This agreement is especially close in the lowest-energy region of the profile (Locations: 20 to 30 in Fig. 1) and is corroborated by comparable inter-domain distances between the atomic models and those previously reported (Fig. S2). The magnitude of the sub–1 kcal/mol energy features in the vicinity of the RBD-up state in previously reported metainferencing model are also comparable to the ones found in our maifoldEM studies (Brotzakis et al., 2021). The structural attributes determined from the string simulations remain within error of the ensembles determined directly from the density data (Fig. S3 and Fig. S4), reaffirming the statistical validity of the density-guided low energy models.
Despite these similarities, Fig. 2 shows that, along the low energy ensemble of spike conformations, the RBD's Solvent Accessible Surface Area (SASA) scores span the range of values previously assigned to differences between open and closed static structures. The non-biased MD trajectories starting from the apo model span a smaller range of SASA values than those seen along the horse-shoe landscape, despite using an 800-fold longer simulation time. The up RBD's global conformation is similar to the open spike structures. However, coupling of the global conformations to local changes at key antibody or inhibitor pockets is expected to be significant. This issue is investigated in the next section.
Globally closed or in RBD-down conformation were not observed in our analysis. The distribution of RBD “all-down”, “1-up”, and “2-up” conformations varies across studies (Walls et al., 2020; Wrapp et al., 2020; Henderson et al., 2020). Typically, both the “RBD down” and “1-up” conformations have been reported to be in equilibrium, as reflected in the 2:1 to 1:1 population distribution of the 2D images (Walls et al., 2020; Wrapp et al., 2020). The lack of RBD-down conformations reflects the absence of equilibrium between the up and the down states in the picked particles used to construct the 6VSB structure. This observation supports the fact that in early SARS-CoV-2 spike protein data (Walls et al., 2020), the crown of the spike was over stabilized by adding two stabilizing proline mutations in the C-terminal S2 fusion machinery.
5. Local conformational motions at binding pockets
The starting structure can have a significant impact on the outcome of computations of ligand protein binding interactions (Robertson et al., 2019; Vant et al., 2020b). Using the energy-minimizing structures derived from experimental data, we are able to investigate the impact of the global conformational changes on individual binding sites. We choose six previously identified sites: three neutralizing antibodies (Barnes et al., 2020); linoleic acid (LA) (Toelzer et al., 2020); a computationally designed neutralizing mini-protein (Cao et al., 2020); and ACE2 (Benton et al., 2020). By comparing the ManifoldEM derived structures to apo and holo-structures, we investigate excursions from the reported models in terms of internal deformations, solvent-accessibility, and the energetics of the individual binding pockets. We also consider the impact of two important glycans N165 and N234 on stabilizing the spike protein complex.
Fig. S5 shows RMSD plots calculated using residues specific to each binding pocket. Despite the highest similarity to 6VSB at locations 20 to 30 for the global analysis, local variations in RMSD are more pronounced when the binding pockets are aligned by the residues involved in ligand-protein interactions. In Fig. 3, we investigate RMSD excursions from the static structure 6VSB aligned using the entire monomer. The pattern observed in the global RMSD analysis recurs, whereby location 28 has the lowest RMSD to 6VSB. However, each binding pocket deviates from 6VSB. The binding pockets for ACE2 and the mini-protein have high RMSD values at both ends of the horseshoe shaped energy feature, while the binding pockets for LA, AB-EY6A, and AB-CR3022 maintain similar RMSD values in all low-energy locations. The binding pocket for AB-C135 deviates strongly from 6VSB throughout the entire ensemble of models. This is discussed further below. In the RMSD space, it is difficult to determine how amenable a site is to binding. However, the LA and AB-C135 binding sites display large differences in RMSD between the apo (6VSB) and holo-structures. The LA binding pocket is distinctly more apo-like, while the converse is true for the AB-C135 binding pocket. To avoid uncertainties in interpreting the local RMSD values, we examine SASA at the local binding-pocket level.
Fig. 4 shows a clear difference in SASA values between apo and holo binding sites, allowing a direct comparison between the structures we located using ManifoldEM, and the tight vs. loose binding pocket conformations determined from the reported holo and apo models. For five of the six binding pockets, the low-energy structures along the horse shoe profile achieve solvent accessibility that spans the range of SASA seen across the apo “1-up” RBD structures (6VSB, and 6VYB), the “all-down” RBD structure (6VXX), the holo structure, and 10 μs MD ensembles (Shaw, 2020). This binding mode encompassing both holo and apo-like conformations, even prior to the ligand binding, appears to involve a conformational selection mechanism. As an example, the mini-protein binding pocket in Fig. 4, at locations 4, 47, 49, and 54, the calculated SASA scores are commensurate with the holo-structure (PDBID: 7JZU), while the rest of the locations involve more open structures (higher SASA scores). This dichotomy suggests that even without the presence of the mini-protein, the binding pocket is able to adopt a tight binding configuration amenable to spike protein neutralization. Interestingly, we observe a key interaction between the LA binding pocket of the “up” RBD and the N234 glycan of the same chain (see Fig. 5). At locations 51 to 59, the N234 glycan on chain A hydrogen bonds with residues 387 (Leu) and 388 (Asn), pulling the binding pocket open, and resulting in the “super-open” states (compared to apo 6VXX and holo 6ZB4/6ZB5) seen in Fig. 4 at the same locations. Such enhanced binding pocket opening also facilitates the binding of AB-EY6A.
Fig. 4.
Individual binding pocket Solvent Accessible Surface Area (SASA) scores calculated from only those residues involved in binding. For all six panels, the SASA scores for static structures are represented with red circles, non-biased equilibrium MD trajectories are represented with green circles and error bars (31), and finally each MFEP location SASA score is represented with blue circles and error bars. The red and blue highlighted regions are used to compare the spread of SASA scores for the previously deposited static structures (red shading) with the range of SASA scores seen for all MFEP locations (blue shading). For all binding pockets except AB-C135, the spread of SASA scores seen for all MFEP locations is comparable to those seen for the static apo and holo structures. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
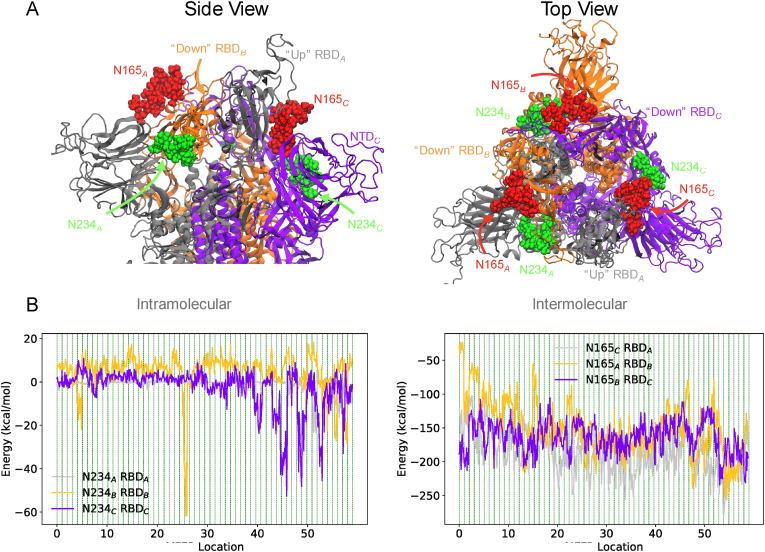
Fig. 5.
Glycan conformations and interaction energies for all chains. (A) Shows each chain of the spike protein trimer individually colored in a cartoon representation. The glycan chains are shown a red and green surface representation and are labeled accordingly. (B) Shows intramolecular or intermolecular interaction energies of the glycan and the spike protein RBD (residues 330 to 520) for each chain. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The AB-C135 binding pocket has a decidedly more closed conformation (Fig. 4). The hydrophobic residues near the AB-C135 binding pocket and at the surface of 6VSB are buried in our simulations, as indicated by the lower SASA score calculated for 6VSB (see Fig. S6). This suggests the binding mechanism involves an induced fit. The AB-C135 binding pocket is a “hidden epitope”, indicating that the closed state we find is biologically relevant (Cao et al., 2020). The local map resolution surrounding this binding pocket has a lower resolution (see Fig. S7), decreasing confidence in reported, comparatively more open conformations. Alternatively, the presence of a closed local pocket in an overall up or open spike RBD further suggests that AB-C135 binding entails an induced fit. Indeed, this pocket is found to be also closed in the recently published MD simulations (Zimmerman et al., 2021). Worth noting, locations 2 to 8, 25 to 32, 33 to 37 and 46 to 55 also exhibit less accessible ACE2 binding conformations, possessing SASA values below both the reported apo and holo structures. However, unlike AB-C135 there exists locations where the ACE2-accessible surface is comparable to both the apo and holo structures, still allowing a conformational pre-equilibrium for binding.
So far, we have observed the effect of glycosylation on the LA binding pocket. Fig. 5B shows that N165 has similar pairwise potential energies for all three intermolecular interactions between N165 and the RBD of the counterclockwise chain. The pairwise potential energies for the intramolecular interaction between N234 and the RBD of the same chain show large differences in the ability of the glycan either to stabilize or destabilize the RBD conformation. Both the N234B RBDB and N234C RBDC pairwise interactions show positive potential energies in many of the minimum energy locations due to strong van der Waals interactions. These steric interactions suggest the “down” RBD conformation is destabilized by the presence of N234, as noted by Amaro and colleagues (Casalino et al., 2020; Cao et al., 2020), albeit only for their intermolecular interactions. In contrast, the intramolecular N234A RBDA interaction is negligible in most of the locations along the horse-shoe profile, except in the 50 to 59 range of locations. In this region, RBDA is stabilized by its interactions with N234A, incidentally creating the “super-open” LA binding pocket that is only observed in the manifoldEM analysis.
6. Discussion
An equilibrium sample of biological macromolecules is inherently conformationally heterogeneous, because a range of conformational states can have significant thermally induced occupancies. Conventional cryo-EM data analysis techniques align and average snapshots with similar particle orientations. While this improves contrast in the 2D images, there is a loss of thermodynamic information. Also, static models that fit the averaged 3D reconstruction are ill-suited to describing the system's conformationally dynamical nature. Starting from such static models, even unbiased MD cannot capture all the low thermally accessible conformational states for a given system due to limited sampling times. By combining data-driven machine learning and MD computations, we have extended the conformational search around the one-up RBD state by making use of experimentally determined energy landscapes. This approach has revealed a broad spectrum of hitherto unobserved iso-energetic conformations associated with RBD binding sites. Guided by experimental data, the conformational heterogeneity observed in our modelling which totals 32 ns of MDFF simulations, and subsequent 400 ns of string simulations, is far greater than what is accessible to 10 μs of brute-force MD.
The experimentally determined horseshoe shaped energy profile shows that the simple rigid one-up RBD is inadequate to describe the complex multi-dimensional conformational motions of the Spike protein. The concerted motions of the RBD have regional effects, which entail substantial conformational heterogeneity compared with either the apo or holo structures alone. Nearly all binding pockets analyzed in this study indicate conformational selection mechanism, where specific minimum-energy locations are more or less favorable to binding. The locations with SASA values higher than those from 6VSB are interpreted as more amenable to binding, with potential therapeutic implications. The inclusion of a broad spectrum of low-energy conformations could potentially offer new drug discovery routes by considering the extensive conformational flexibility of important binding pockets. By going beyond static structures fitted to heavily averaged maps, our approach reveals the flexible nature of biological macromolecules, with possible implications for novel drugs.
7. Methods
7.1. Cryo-EM data
In this study, we used the cryo-EM images from a previous study (Wrapp et al., 2020), in which Sars-CoV-2 spike ectodomain residues 1 to 1208 were expressed based on the first reported genome sequence (Wu et al., 2020), adding two stabilizing proline mutations in the C-terminal S2 fusion machinery. Excess protein was blotted away for 6 s using grade 595 vitrobot filter paper (Ted Pella Inc.) with a force of −1 at 4 °C in 100% humidity before being plunged frozen into liquid ethane using a Vitrobot Mark IV (Thermo Fisher).
Cryo-EM grids were prepared using purified fully glycosylated spike protein. Frozen grids were imaged in a Titan Krios (Thermo Fisher) equipped with a K3 detector (Gatan). Movies were collected using Leginon at a magnification of 22,500-fold (Carragher et al., 2000), corresponding to a calibrated pixel size of 1.047 Å/pixel. A full description of sample preparation and data collection parameters can be found in (Wrapp et al., 2020). Motion correction, CTF-estimation, and non-templated particle picking were performed in Warp (Tegunov and Cramer, 2019). There were thousands of images in the 631,920 extracted particles which had artifacts e.g., harsh line/boxes. Those images were removed. The remaining 574,324 were imported to CryoSPARC and non-uniform refinement was used to get the orientation of each particle.
7.2. Geometric machine learning (ManifoldEM)
The details of our data-analytic approach are available at (Dashti et al., 2020). In brief, having assigned an orientation to each snapshot, we divide the snapshots to small orientational bins, which we call projection directions (PD). In other words, each PD includes the snapshots which are in closely similar orientations.
We select all the projection directions lying on a great circle around the orientational sphere. Then in each projection direction, we use manifold embedding (Dashti et al., 2020) to extract the conformational manifold. To this end, we use diffusion-map embedding of the snapshots in each projection direction to determine the conformational manifold. This family of dimensionality reduction techniques establishes a rigorous link between the eigenfunctions (more precisely eigenvectors) of the Laplace–Beltrami operator with respect to a Riemannian metric and the similarity between snapshots, as measured by the diffusion distance between them.
In practice, the information content of a cryo-EM snapshot depends on the defocus at which it is obtained. The effect of such defocus on the similarity measure between two otherwise identical snapshots must be eliminated from the diffusion distance. We achieve this by a double-filtering kernel that ensures a zero Euclidean distance between two snapshots differing in defocus only. Distances from this Defocus-tolerant kernel are employed to determine the eigenvectors of the Laplace–Beltrami operator to determine the conformational coordinates.
We use the two topmost conformational coordinates to describe the conformational manifold If we sort the snapshots along each of these eigenfunctions. We can compile a movie of the conformational changes along those eigenfunctions by using Non-Linear Spectral Analysis (Giannakis and Majda, 2012). Fig. 1 shows the first and last frames of the movies (Supplementary Movie 1 and 2) along with the two conformational coordinates. As the movies show, CC1 corresponds to a breathing like motion, while in CC2, the RBD moves from a down position to up.
Analysis of noisy synthetic data has shown that ManifoldEM correctly assigns the snapshots with an accuracy of 80%, with accuracy defined as the ratio of correct assignments to the total number of assigned snapshots. This error in occupation probability results in an error in the energy via the Boltzmann factor A exp –(E/kT), with A stemming from partition function. Consequently, errors in occupancy are related to errors in energy logarithmically, resulting in an error estimate of ∼0.1 kcal/mol for our approach (Dashti et al., 2020).
Integrating the information from all the PDs on a great circle, we compiled 3D conformational movies of electrostatic potential maps along each of these conformational coordinates (Supplementary Movie 1 and 2). We have measured the RMSD of MDFF models changing along the CC1 direction (setting CC2 = 0) and vice versa. We observe that changes along just the CC2 dimension are more pronounced as reflected in an RMSD of 5.5 Å between the (0,0) and (0,50) models. The same between (0,0) and (50,0) reflecting only changes along the CC1 dimension is 2.9 Å. From this analysis it can be inferred that the structural changes in most of the minimum energy conformations along the horse-shoe shaped profile are dominated by CC2.
7.3. Map preparation
We compiled 50 maps (.spi) along each conformational coordinate. The maps along each conformational coordinate were then converted from. Spi format to. mtz format with Chimera (Pettersen et al., 2004). The. mtz maps were then converted to potentials thresholding the maps at the solvent density peak by utilizing the voltools pot command, which is part of the Voltools Plugin within VMD (Humphrey et al., 1996). These potentials were used to guide the spike protein trimer's dynamics. The maps were thresholded again where the RBD density was weak to increase the magnitude of the potential map gradients for the RBD.
7.4. Molecular dynamics simulations
We used the fully glycosylated 6VSB model from the CHARMM-GUI Archive - COVID-19 Proteins Library (Woo et al., 2020). The models were stripped of all water molecules with the molecular visualization program VMD (Humphrey et al., 1996). All simulations utilized the generalized Born Implicit Solvent (GBIS) model and Charmm Force Field (Huang and Mackerell, 2013), with the molecular dynamics engine NAMD 2.14b1 (Phillips et al., 2020). The simulation parameters are provided in the supplementary NAMD input file.
7.5. Molecular dynamics flexible fitting
MDFF was used to bias simulations and fit atomic models to the extracted conformational coordinates (Trabuco et al., 2008, 2009). Noting the medium resolution of our experimental maps, only the backbone of the models was coupled to the density, with the conformations of the remainder of the system (sidechains and glycans) responding to the MD force fields. To ensure that the protein backbone conforms to the density, we constrained the protein by knowledge of its secondary structure, chirality, and cis-peptides; while the less resolved sidechains and glycans continued to refine under the chemical constraints (bonds, angles, dihedrals, and non-bonded interactions) imposed by the CHARMM36m force fields.
7.6. Conformational coordinate fitting
To obtain references for the fitting atomic models on the extracted energy landscape, atomic models were fitted to each map for both conformational coordinates. Simulations were biased with two map potentials coupling the singular threshold maps to all backbone atoms except chain A residues 320 to 520, and the doubly thresholded maps were coupled to the backbone atoms chain A and residues 320 to 520. The fitting process occurred in two steps starting from the same starting structure following the cascade-MDFF procedure (Singharoy et al., 2016). The blurred maps were created with the command voltools smooth in the VMD Voltools Plugin. The first step utilized a 2 Å Gaussian blurred potential map, while the second step used maps at their original resolution of the maps.
7.7. Flexible fitting
A movie of molecular motions was compiled along the low energy segment of the energy landscape using a so-called “multigrid” fitting procedure (Vant et al., 2020a; Singharoy et al., 2019). This procedure enables the construction of atomic models under the influence of two or more density maps. The multigrid procedure is employed within MDFF to enable the fitting of the XY-dimension of the spike protein backbone under the influence of the CC1 maps, while Z-dimensions of these atoms conform to the CC2 maps. A cascade-MDFF protocol was employed (Singharoy et al., 2016) in two steps. First, the structure was fitted to maps with a 2 Å Gaussian blur and then fitted to the original maps coming from the ManifoldEM analysis. The convergence of the fitted structures in terms of RMSD is shown in Fig. S9.
First the 6VSB model was individually fitted to all 50 maps along CC1, setting CC2 to 0, and vice-versa. This procedure created 100 single-map fitted models. To concomitantly fit two maps at any (CC1, CC2) location, that starting model is chosen from (CC1, 0) when CC1 > CC2, or from (0, CC2) if CC2 > CC1. So, to model the location (Pavlova et al., 2021b; Cao et al., 2020), the (0,30) model will be used as a starting point. Fig. S10 shows the comparison between the CC1-only, CC2-only and 2-map fitted structures in terms of radius of gyration. For each 50 locations along CC1 (setting CC2 = 0) and CC2 (setting CC1 = 0), and 59 locations along the horse-shoe profile, 200 ps of MDFF is performed. So, a total of (50 + 50 + 59) x 200 ps = 31.8 ns of flexible fitting simulations are performed.
7.8. String simulations
To corroborate the statistical relevance of the minimum energy locations derived from the multigrid fitting procedure, we choose to use the string method with swarms of trajectories (Pan et al., 2008) and monitor the deviation of the converged string method derived pathway and the ManifoldEM generated low energy models. Starting with an initial path made of M+1 images (M = 10) connecting low energy location 1 to 59 (see Fig. 1) 100 iterations of the string method were performed using 10 replica per image, 20 ps of sampling of biased MD to update the pathway and 20 ps of unbiased MD from which the drift of the pathway is calculated, totaling 400 ns of sampling. The pathway was defined in the space of 8 distance vector reaction coordinates where the distances of 4 regions of the up RBD are calculated from both endpoints of the minimum free energy pathway or MFEP.
Fig. S11 shows the convergence of the string pathway in terms of RMSD from each images starting position. The string images are well converged by iteration 80. Furthermore, we can look at the arc length of the pathway defined in the collective variable space to ensure that the string pathway has converged. From Fig. S12 we see again that the string pathway has converged by iteration 80. We also see that there is a modest 6% change in total arc length. Indicating that the ManifoldEM pathway and string method pathway are similar. Lastly, we can compare the Rg values from the ManifoldEM fitted models and the string method pathway to determine the deviation between the two pathways. Fig. S4 shows the Rg values calculated for both pathways in the XY and Z dimensions. While the deviation of the string method pathway is higher in the XY plane, the relative change remains below 15%. These simulations were repeated at room temperature and at 4 °C i.e. at a temperature prior to plunge freezing. The converged string invoked comparable arch length between the two temperatures, within 5–10% deviation, and structural features within 1–2 Å local structural difference at every image (Fig. S12).
7.9. SASA calculations
We used VMD's measure Plugin to calculate SASA values. The radius sampled around each atom selected was that atom's radius and an additional 1.4 Å. The static models used for comparison were built with VMD using the autopsf Plugin to add H at the physiological pH.
Conflict of interest
The authors declare no conflicts of interest.
Data and materials availability
The data supporting the findings of this study are available from the corresponding authors upon reasonable request.
CRediT authorship contribution statement
Ghoncheh Mashayekhi: All authors contributed to study design, extracted the manifold of conformational motions from single particle images, produced the free-energy landscape, and identified the low energy conformations. All authors provided critical input and participated in writing the manuscript. John Vant: Formal analysis, All authors contributed to study design, produced the atomic models for each conformational coordinate and along the minimum energy segment of the landscape, ran string method simulations, and analyzed the atomic models, All authors provided critical input and participated in writing the manuscript. Abhigna Polavarapu: has developed an initial glycosylated model of the spike protein. Abbas Ourmazd: proposed the study, designed the study architecture, All authors contributed to study design, All authors provided critical input and participated in writing the manuscript. Abhishek Singharoy: designed the study architecture, All authors contributed to study design.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the US National Science Foundation (NSF) under award DBI2029533. The development of underlying techniques was supported by the US Department of Energy, Office of Science, Basic Energy Sciences under award DE-SC0002164 (underlying dynamical techniques), and by the US NSF under awards STC 1231306 (underlying data analytical techniques) and 1551489 (underlying analytical models). A.S. acknowledges an NSF CAREER award MCB-1942763, and the NIH award R01GM095583. J.V. acknowledges support from the NSF Graduate Research Fellowship Grant 2020298734. We thank J. McLellan and collaborators for providing the cryo-EM data set. We are grateful to E. Seitz and S. Maji for preprocessing the data for ManifoldEM analysis. We acknowledge extensive discussions at an early stage of this work with F. Acosta, S. Maji, E. Seitz and J. Frank.
Handling editor: Glaucius Oliva
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2022.02.001.
Contributor Information
Abbas Ourmazd, Email: ourmazd@uwm.edu.
Abhishek Singharoy, Email: asinghar@asu.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Acharya A., Lynch D.L., Pavlova A., Pang Y.T., Gumbart J.C. ACE2 glycans preferentially interact with SARS-CoV-2 over SARS-CoV. Chem. Commun. 2021;57:5949. doi: 10.1039/d1cc02305e. [DOI] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., Robbiani D.F., Nussenzweig M.C., West A.P., Bjorkman P.J. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branduardi D., Faraldo-Gomez J.D. String method for calculation of minimum free-energy paths in cartesian space in freely tumbling systems. J. Chem. Theor. Comput. 2013;9:4140. doi: 10.1021/ct400469w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotzakis Z.F., Löhr T., Vendruscolo M. Determination of intermediate state structures in the opening pathway of SARS-CoV-2 spike using cryo-electron microscopy. Chem. Sci. 2021;12:9168. doi: 10.1039/d1sc00244a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Goreshnik I., Coventry B., Case J.B., Miller L., Kozodoy L., Chen R.E., Carter L., Walls A.C., Park Y.J., Strauch E.M., Stewart L., Diamond M.S., Veesler D., Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher B., Kisseberth N., Kriegman D., Milligan R.A., Potter C.S., Pulokas J., Reilein A. Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 2000;132:33. doi: 10.1006/jsbi.2000.4314. [DOI] [PubMed] [Google Scholar]
- Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., Fogarty C.A., Barros E.P., Taylor B.C., Mclellan J.S., Fadda E., Amaro R.E. ACS Cent. Sci. 2020;6:1722. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti A., Mashayekhi G., Shekhar M., Ben Hail D., Salah S., Schwander P., des Georges A., Singharoy A., Frank J., Ourmazd A. Retrieving functional pathways of biomolecules from single-particle snapshot. Nat. Commun. 2020;11:4734. doi: 10.1038/s41467-020-18403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti A., Mashayekhi G., Ourmazd A. 2021. ManifoldEM Matlab. [Google Scholar]
- Fallon L., Belfon K., Raguette L., Wang Y., Corbo C., Stepanenko D., Cuomo A., Guerra J., Budhan S., Varghese S., Rizzo R.C., Simmerling C. Free energy landscapes from SARS-CoV-2 spike glycoprotein simulations suggest that RBD opening can Be modulated via interactions in an allosteric pocket. J. Am. Chem. Soc. 2020;143:11349. doi: 10.1021/jacs.1c00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Ourmazd A. Continuous changes in structure mapped by manifold embedding of single-particle data in cryo-EM. Methods. 2016;100:61. doi: 10.1016/j.ymeth.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis D., Majda A.J. Nonlinear Laplacian spectral analysis for time series with intermittency and low-frequency variability. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2222. doi: 10.1073/pnas.1118984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Barreto J., Ortiz S., Thiede E.H., Palacio-Rodriguez K., Carpenter B., Barnett A.H., Cossio P. A Bayesian approach to extracting free-energy profiles from cryo-electron microscopy experiments. Sci. Rep. 2021;11:13657. doi: 10.1038/s41598-021-92621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur M., Taka E., Yilmaz S.Z., Kilinc C., Aktas U., Golcuk M. Conformational transition of SARS-CoV-2 spike glycoprotein between its closed and open states. J. Chem. Phys. 2020;153 doi: 10.1063/5.0011141. [DOI] [PubMed] [Google Scholar]
- Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M., Kopp M., Li D., Parks R., Hsu A.L., Borgnia M.J., Haynes B.F., Acharya P. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Nat. Struct. Mol. Biol. 2020;27:925. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Mackerell A.D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the sars-cov-2 spike receptor-binding domain bound to the ace2 receptor. Nature. 2020;581:215. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Matsunaga Y., Fujisaki H., Terada T., Furuta T., Moritsugu K., Kidera A. Minimum free energy path of ligand-induced transition in adenylate kinase. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medovoy D., Perozo E., Roux B. Multi-ion free energy landscapes underscore the microscopic mechanism of ion selectivity in the KcsA channel. Biochim. Biophys. Acta Biomembr. 2016;1858:1722. doi: 10.1016/j.bbamem.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Shukla D., Pande V.S., Roux B. Transition path theory analysis of c-Src kinase activation. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:9193. doi: 10.1073/pnas.1602790113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira R.A., Guzman H.V., Boopathi S., Baker J.L., Poma A.B. Characterization of structural and energetic differences between conformations of the SARS-CoV-2 spike protein. Materials. 2020;13:1. doi: 10.3390/ma13235362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourmazd A. Cryo-EM, XFELs and the structure conundrum in structural biology. Nat. Methods. 2019;16:941. doi: 10.1038/s41592-019-0587-4. [DOI] [PubMed] [Google Scholar]
- Pan A.C., Sezer D., Roux B. Finding transition pathways using the string method with swarms of trajectories. J. Phys. Chem. B. 2008;112:3432. doi: 10.1021/jp0777059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova A., Lynch D.L., Daidone I., Zanetti-Polzi L., Smith M.D., Chipot C., Kneller D.W., Kovalevsky A., Coates L., Golosov A.A., Dickson C.J., Velez-Vega C., Duca J.S., Vermaas J.V., Pang Y.T., Acharya A., Parks J.M., Smith J.C., Gumbart J.C. Inhibitor binding influences the protonation states of histidines in SARS-CoV-2 main protease. Chem. Sci. 2021;12:1513. doi: 10.1039/d0sc04942e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova A., Zhang Z., Acharya A., Lynch D.L., Pang Y.T., Mou Z., Parks J.M., Chipot C., Gumbart J.C. Machine learning reveals the critical interactions for SARS-CoV-2 spike protein binding to ACE2. J. Phys. Chem. Lett. 2021;12:5494. doi: 10.1021/acs.jpclett.1c01494. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips J.C., Hardy D.J., Maia J.D., Stone J.E., Ribeiro J.V., Bernardi R.C., Buch R., Fiorin G., Hénin J., Jiang W., et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020;153 doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M.J., van Zundert G.C., Borrelli K., Skiniotis G. 2019. GemSpot: A Pipeline for Robust Modeling of Ligands into CryoEM Maps. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw D.E. 2020. Molecular Dynamics Simulations Related to SARS-CoV-2. [Google Scholar]
- Shekhar M., Terashi G., Gupta C., Debussche G., Sisco N., Nguyen J., Vant J., Sarkar D., Fromme P., Van Horn W.D., Tajkhorshid E., Kihara D., Dill K., Perez A., CryoFold A. Singharoy. Determining protein structures and data-guided ensembles from cryo-EM density maps. Matter. 2021;4:3195. doi: 10.1016/j.matt.2021.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singharoy A., Teo I., McGreevy R., Stone J.E., Zhao J., Schulten K. Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps. Elife. 2016;5:1. doi: 10.7554/eLife.16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singharoy A., Maffeo C., Delgado-Magnero K., Swainsbury D.J.K., Sener M., Kleinekathofer U., Isralewitz B., Teo I., Chandler D., Vant J.W., et al. Atoms to phenotypes: molecular design principles of cellular energy metabolism. Cell. 2019;179:1098. doi: 10.1016/j.cell.2019.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D., Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods. 2019;16:1146. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toelzer C., Gupta K., Yadav S.K., Borucu U., Garzoni F., Staufer O., Capin J., Spatz J., Fitzgerald D., Berger I., Schaffitzel C. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370:725. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco L.G., Villa E., Mitra K., Frank J., Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco L.G., Villa E., Schreiner E., Harrison C.B., Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Vaart A., Karplus M. Minimum free energy pathways and free energy profiles for conformational transitions based on atomistic molecular dynamics simulation. J. Chem. Phys. 2007;126 doi: 10.1063/1.2719697. [DOI] [PubMed] [Google Scholar]
- Vant J.W., Sarkar D., Streitwieser E., Fiorin G., Skeel R., Vermaas J.V., Singharoy A. Data-guided Multi-Map variables for ensemble refinement of molecular movies. J. Chem. Phys. 2020;153:214102. doi: 10.1063/5.0022433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vant J.W., Lahey S.L.J., Jana K., Shekhar M., Sarkar D., Munk B.H., Kleinekathöfer U., Mittal S., Rowley C., Singharoy A. Flexible fitting of small molecules into electron microscopy maps using molecular dynamics simulations with neural network potentials. J. Chem. Inf. Model. 2020;60:2591. doi: 10.1021/acs.jcim.9b01167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H., Park S.J., Choi Y.K., Park T., Tanveer M., Cao Y., Kern N.R., Lee J., Yeom M.S., Croll T.I., Seok C., Im W. Developing a fully-glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B. 2020;124:7128. doi: 10.1021/acs.jpcb.0c04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Pak A.J., He P., Monje-Galvan V., Casalino L., Gaieb Z., Dommer A.C., Amaro R.E., Voth G.A. A multiscale coarse-grained model of the SARS-CoV-2 virion. Biophys. J. 2021;120:1097. doi: 10.1016/j.bpj.2020.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman M.I., Porter J.R., Ward M.D., Singh S., Vithani N., Meller A., Mallimadugula U.L., Kuhn C.E., Borowsky J.H., Wiewiora R.P., et al. SARS-CoV-2 simulations go exascale to predict dramatic spike opening and cryptic pockets across the proteome. Nat. Chem. 2021;13:651. doi: 10.1038/s41557-021-00707-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.