Abstract
SIRT6 has been in the spotlight of aging research due to progeroid phenotypes associated with SIRT6 deficiency. SIRT6 has multiple molecular functions, including DNA repair and heterochromatin regulation, which position SIRT6 as a hub regulating genome and epigenome stability. Genomic instability caused by persistent DNA damage and accumulating mutations, together with alterations in epigenetic landscape and de-repression of repetitive genetic elements emerged as mechanisms driving organismal aging. Enhanced levels of SIRT6 expression or activity provide avenues for rejuvenation strategies. This review focuses on the role of SIRT6 in the maintenance of genome and epigenome stability and its link to longevity. We propose a model where SIRT6 together with Lamins control aging and rejuvenation by maintaining epigenetic silencing of repetitive elements.
Keywords: SIRT6, longevity, DNA damage repair, epigenome, healthspan, rejuvenation
Genomic and epigenomic instability drive the aging process
The complex etiology of aging is entwined of multiple overlapping pathological processes [1]. In order to prevent pathology, each individual cell must maintain a balanced homeostasis, which relies on the maintenance of genomic stability and coordinated expression of specific sets of genes that are subject to epigenetic control. A variety of factors, such as environmental mutagens, cellular metabolism, oxidative stress or genetic predisposition result in DNA damage that poses a major threat to genome and epigenome stability [2]. Repeated cycles of DNA replication and persistent DNA damage throughout life may result in mutations, genomic re-arrangements, loss of epigenome structure and increased cell-to-cell variation in gene expression, driving aging phenotypes [3]. Chromatin organization in eukaryotic cells provides epigenetic information that dictates cell identity. However, chromatin may undergo changes with advancing age resulting in epigenetic drift (see Glossary) [4, 5]. The accumulation of epigenetic changes lead to substantial cell-to-cell heterogeneity in gene expression, which may further affect numerous signaling pathways, underlying the development of various hallmarks of aging [1].
Gene expression in defined loci relies on dynamic regulation by epigenetic factors, such as chromatin-modifying enzymes, which makes them master-regulators of the cellular homeostasis. Therefore, chromatin modifiers may be key to understanding the pathological processes comprising aging and present promising targets for longevity research. Members of the sirtuin family of proteins have been associated with aging due to their ability to influence multiple vital functions of a cell through the maintenance of genomic stability and control of gene expression [6]. SIRT6 is one of the seven mammalian homologues of the silent information regulator 2 (Sir2) and is preferentially associated with chromatin [7]. As a regulator of chromatin structure and DNA repair, SIRT6 is strategically positioned to maintain genomic and epigenomic stability, as well as influence gene expression within multiple physiological and pathological pathways determining longevity. This article reviews SIRT6 role in lifespan and healthspan regulation through the prism of its ability to control the cellular epigenetic landscape and genomic stability.
SIRT6 and lifespan in mammals
Owing to the early findings that Sir2 regulates lifespan in model organisms, SIRT6 has been investigated as a target for longevity research in mammals [8–12]. Several groups have generated SIRT6 knockout (KO) mice and reported reduced lifespan in these animals, albeit the severity of the phenotype seems to depend on the genetic background. SIRT6 KOs on a 129/SvJ background die soon after weaning and do not survive longer than 4 weeks, exhibiting a progeroid phenotype [7, 13, 14]. In animals with a mixed genetic background (129/Black Swiss/FVB), about 60% of SIRT6 KO mice also died before reaching one month of age, however, more than 75% of the female and 10% of male mice survived past 300 days [15]. SIRT6 KO in mice leads to reduced body weight, low blood glucose levels, lymphopenia and severe hypoglycemia, acute loss of subcutaneous fat, lordokyphosis and degenerative signs in bone and colonic epithelium, indicating multiple tissue homeostasis failures [7]. Studies in global SIRT6 KO and various cell type-specific SIRT6 KO models have revealed the contribution of SIRT6 deficiency to vascular aging, neurodegeneration, retinal dysfunction, stroke, muscle dystrophy, osteoporosis, obesity, fibrosis and β-cell dysfunction [16]. Even more severe phenotype is caused by SIRT6 deficiency in primates: when SIRT6 was knocked out in monkeys only females survived to birth and died within days [17]. Likewise, inactivating homozygous mutation D63H in human SIRT6 was found to cause perinatal lethality and SIRT6 deficiency resulted in sex reversal in the male fetus [18]. A recent study identified 8 mutations in SIRT6 in melanoma patients, four of which correlated with high mutation rates across the genome [19]. A strong negative correlation between SIRT6 protein expression and age has been found in human primary skin fibroblasts [20]. Reduced expression of SIRT6 was detected in the hippocampus, buccal epithelium and peripheral blood mononuclear cells (PBMC) of elderly patients [21–23]. Decreased SIRT6 level has been associated with a number of human age-related pathologies, including certain cancers, cardiovascular diseases, atherosclerosis and hypertension, liver fibrosis, Alzheimer’s disease, metabolic diseases and others [16, 24–29].
While gene knockouts often lead to various pathologies and shortened lifespan in mice, the most remarkable feature of SIRT6 is that its overexpression extends mammalian lifespan. In a recent study Roichman et al. discovered an impressive 27% increase in median lifespan for male mice and 15% – for female mice compared to wild type littermates, when SIRT6 was overexpressed under a CAG promoter [30]. Moreover, SIRT6 overexpression improved various aspects of organismal homeostasis and promoted healthspan on multiple levels [31]. One of the important aspects involving SIRT6 in longevity is its remarkable ability to influence metabolic pathways. SIRT6 deficiency impairs hepatic ability to perform β-oxidation [32]. SIRT6 overexpression promotes β-oxidation in liver, hepatic lactate and glycerol shuttling, improves nicotinamide adenine dinucleotide (NAD+)/NADH ratio, and stimulates glycerol release from adipose tissue. In addition, overexpression of SIRT6 improves the capacity to produce glucose, which decreases with age [30]. Metabolic regulation is intimately connected to the development of cancer, which becomes more prevalent with age. Interestingly, SIRT6 overexpression leads to a decrease in the incidence of neoplasms in aged mice [30]. This effect is likely to result both from improved genome stability and from SIRT6-mediated transcriptional regulation of metabolic processes, such as repression of glycolysis and suppression of Warburg effect [24, 33]. – linking epigenetics, regulation of metabolism, cancer and aging. The positive effects of SIRT6 on healthspan are not limited to metabolic regulation, as SIRT6 overexpression ameliorates endothelial cell dysfunction, improves adult neurogenesis, decreases infarct size and neurological deficit in a stroke model, protects from kidney injury and colitis, and helps maintain genomic stability in the brain [25, 29, 34–39]. Intriguingly, genetic polymorphisms in SIRT6 have been found to be associated with human longevity, however, the role these polymorphisms play in SIRT6 expression and function remains to be elucidated [40–43].
Taken together, there is strong evidence that SIRT6 deficiency shortens lifespan and leads to the development of pathologies in a variety of tissues. High levels of SIRT6 help to rescue these severe phenotypes in experimental models, implying that maintenance of stable or increased SIRT6 expression and activity throughout life could help counteract aging and positively influence healthspan.
SIRT6 structure and its enzymatic activities
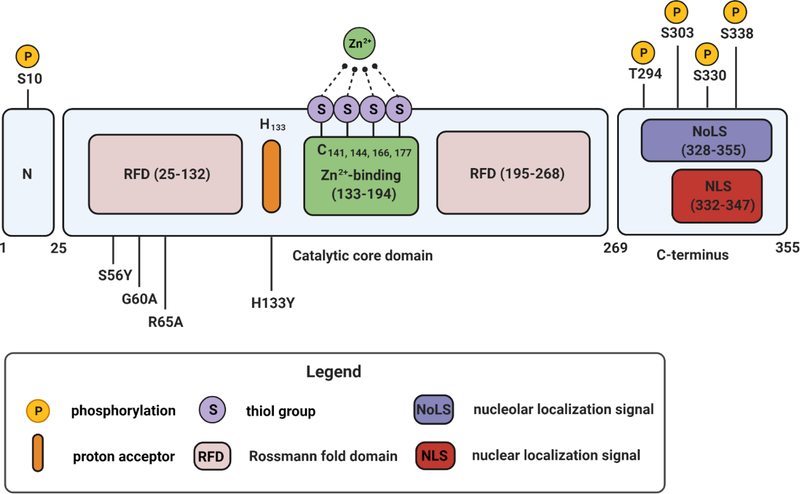
SIRT6 is predominantly associated with the chromatin fraction of a cell nucleus. SIRT6 can also be observed in the cytoplasm localized to the stress granules, where it may be an active component facilitating their assembly [44, 45]. The crystal structure of SIRT6 reveals a conserved globular catalytic core domain comprising 275 amino acids with a Rossman-fold and a Zn2+-binding domain, a less conserved N-terminus and a non-conserved and highly disordered C terminus (Fig. 1) [46]. The N-terminus, rich in positively charged residues, is critical for chromatin association and histone deacetylation in vitro, whereas the highly disordered C-terminus contains a nuclear localization signal and is responsible for the differences in the strength of SIRT6 enzymatic activity across species with various lifespans [47, 48].
Figure 1.
SIRT6 structural domains. SIRT6 protein has a length of 355 amino acids (aa) and features three main structural domains: the N-terminus (1–24 aa), the catalytic core domain (25–268) and the C-terminus (269–355 aa). The catalytic core includes an NAD+-binding Rossmann fold domain spanning the regions of 25–132 aa and 195–268 aa, a histidine in position 133 serving as proton acceptor and a Zn2+ binding domain with several cysteine residues binding Zn2+ ion (positions 141, 144, 166, 177). Several well-known mutations in the catalytic core that impair SIRT6 functions are shown: substitution of a conserved histidine H133 to tyrosine leads to a complete loss of SIRT6 catalytic activity and impaired association with chromatin, S56Y mutation causes loss of both deacetylase and mono-ADP-ribosyltransferase activities, while G60A and R65A cause a lack of mono-ADP-ribosyltransferase activity and deacetylase activity respectively [47, 60]. The C-terminus is proline-rich and structurally disordered. It includes a nuclear localization signal (NLS) and a predicted nucleolar localization signal (NoLS). Several characterized phosphorylation sites are highlighted.
The core domain of SIRT6 utilizes NAD+ to provide two main enzymatic functions that influence chromatin states – deacylation and mono-ADP-ribosylation activities [44, 49]. The most notable special case of deacylation is deacetylation and, as a nuclear sirtuin, SIRT6 has been classified as class III histone deacetylase removing acetyl from the amino group of lysines producing nicotinamide and O-acetyl-ADP-ribose as byproducts [49]. SIRT6 demonstrates only a weak deacetylation activity in vitro, however in vivo SIRT6 overexpression leads to a strong reduction in global levels of H3 acetylation. This apparent discrepancy could be resolved by at least two observations: the efficiency of deacetylation has been demonstrated to depend on complex interactions with nucleosomes and the deacetylation activity of SIRT6 can be significantly improved upon binding of fatty acids, which results in conformation changes facilitating binding acetylated H3 [50, 51]. The major histone substrates for deacetylation by SIRT6 include H3K9, H3K56 and H3K18, through which SIRT6 can directly or indirectly control the promoters of many important genes, most notably transcription factors [52–54]. Another important mechanism of gene regulation by SIRT6 occurs in intragenic regions at the level of transcription, whereby SIRT6 binding to RNA polymerase II (Pol II) stabilizes the Pol II promoter-proximal pausing and inhibits transcriptional elongation [55].
SIRT6 is also capable of removing longer fatty acyl groups, owing to the presence of the elongated hydrophobic pocket in its structure, which ensures hundreds-fold higher catalytic activity toward long-chain peptide substrates compared with acetylated substrates in vitro [56]. This feature allows it to mediate efficient deacylation of H3K9, H3K18, H3K27 and to a lower level H3K14, H3K36, H3K56 and H3K79 [57]. Apart from histone deacylation, SIRT6 has many important non-histone targets, such as tumor protein p53, histone acetyltransferase GCN5, tumor necrosis factor (TNF)-α and nuclear factor erythroid 2-related factor 2 (NRF2). The SIRT6-mediated removal of long-chain acyl groups on lysines K19 and K20 of TNF-α is particularly interesting as it has been shown to promote the secretion of this important inflammatory cytokine [58, 59]. SIRT6 also acts as ADP-ribosytransferase to mono-ADP-ribosylate nuclear proteins with the production of nicotinamide [44]. This activity of SIRT6 is less studied due to the difficulties associated with detection of mono-ADP ribosylation. SIRT6 actively ribosylates itself in vivo and in vitro, but the function of self-ribosylation is unknown [44]. The known trans- substrates of SIRT6 include poly [ADP-ribose] polymerase 1 (PARP1), (KRAB)-associated protein 1 (KAP1), BRG1-associated factor 170 (BAF170) and lysine-specific demethylase 2A (KDM2A), which mediate a wide range of cellular functions, such as DNA repair, heterochromatin relaxation and compaction, as well as antioxidant defense [60–63]. Armed with these powerful tools, SIRT6 is well equipped to influence a variety of physiological and pathological pathways related to organismal and cellular aging, most notably DNA repair and epigenome maintenance.
SIRT6 and DNA repair
SIRT6 has been implicated in DNA repair of both single-strand breaks (SSB) and double-strand breaks (DSBs). SIRT6 KO mice exhibit chromosomal abnormalities indicative of DNA repair defects [7]. The proper repair of DSBs is particularly important to counteract aging. DNA DSB repair efficiency declines with age and gets compromised during replicative senescence in mammalian cells [64]. As mammalian fibroblasts approach senescent state, they express lower SIRT6 levels compared to cells at the lower population doublings. Overexpression of SIRT6 rescues the DSB repair efficiency in senescent cells and stimulates the efficiency of both homologous recombination (HR) and non-homologous end joining (NHEJ) DSB repair pathways, as evidenced by chromosomally integrated reporter assays [60, 64].
Mammalian maximum lifespans (MLS) differ up to 100-fold between species [65] and species longevity has been shown to correlate with the efficiency of DSB repair by NHEJ and HR repair pathways [48]. Remarkably, SIRT6 was shown to be the major factor responsible for the correlation between longevity and DSB repair. SIRT6 from long-lived species has stronger deacetylation and mono-ADP-ribosylation activities and serves as a more potent activator of DSB repair. Thus, animals with the longest lifespan exhibit not only high expression of DSB repair genes, but may as well have evolved unique alleles of key aging genes allowing them to avoid, delay, or alleviate the hallmarks of aging [66, 67].
SIRT6 has been implicated in multiple stages of DSB repair. SIRT6 is rapidly recruited to the DNA DSB sites [60, 68, 69]. It has recently been reported that SIRT6 can recognize and directly bind to the damaged DNA, serving as a DNA break sensor [70, 71]. In addition, SIRT6 has been shown to anchor to γH2AX [70]. Upon DSB damage H2AX is rapidly phosphorylated by ataxia-telangiectasia mutated (ATM) kinase, which further recruits and localizes repair proteins near the DNA breaks. Interestingly, the deacetylation of SIRT6 at residue K33 by SIRT1 is required for the interaction between SIRT6 and γH2AX, revealing a synergy between the two sirtuins. Additionally, a radiation induced H2AX-interacting protein Bcl-2-associated transcription factor 1 (BCLAF1) has been reported to be a binding partner of SIRT6 [72].
A direct interaction has been shown between SIRT6 and Ku80, an abundant ring-shaped molecule that tightly associates with DSBs and initiates the assembly of the NHEJ machinery upon formation of a DSB [73, 74]. The chromatin-associated endogenous SIRT6 interacts with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and stabilizes it at the sites of DNA breaks [73]. Moreover, SIRT6 binding to Ku80 promotes phosphorylation of the DNA-PKcs at S2056, resulting in improved efficiency of NHEJ repair [74].
SIRT6 is particularly relevant under oxidative stress (OS) conditions, which may be the source of persistent DNA damage in aged tissues. Inhibitors of various stress-activated kinases were screened on human skin fibroblast cells under OS conditions, and the c-Jun N-terminal kinase (JNK) pathway has been identified as a regulator of SIRT6 ability to stimulate DSB repair via phosphorylation of SIRT6 on S10 [69]. Upon its recruitment to DSBs under OS SIRT6 mono-ADP-ribosylates PARP1 on K521, thereby stimulating its poly-ADP-ribosylase activity and enhancing DSB repair by relaxing chromatin and facilitating recruitment of DNA repair factors such as Rad51, NBS1, and 53BP1 [60, 64].
The repair of SSB DNA lesions created by endogenous alkylation, oxidation and deamination as a result of OS also deserves attention in the context of aging. A strong case has been made for the SIRT6 involvement in the SSB repair through the base-excision repair (BER) pathway supported by direct association between SIRT6 and core BER factors [7, 75, 76]. During damage sensing in BER, SIRT6 associates with the MYH/MUTYH glycosylase, heterotrimeric 9–1–1 checkpoint clamp (RAD9, RAD1 and HUS1 proteins) and apurinic/apyrimidinic endonuclease 1 (APE1), promoting their activities in response to OS [75]. Furthermore, Xu et al. showed that BER efficiency declines as a function of age in human skin fibroblasts from donors of different age and overexpression of exogenous SIRT6 rescued the decline of BER in these aged fibroblasts [20]. SIRT6 is also involved in nucleotide excision repair (NER) pathway, responsible for the removal of bulky DNA adducts induced by UV irradiation or genotoxic chemicals. The lack of SIRT6 led to the reduced NER efficiency based on the GFP plasmid reactivation assay following UV light exposure, whilst overexpression of SIRT6 promoted the NER repair [19]. SIRT6 deacetylation of DNA damage-binding protein (DDB2) has been shown, which promotes the ubiquitination of DDB2 and segregation of DDB2 from chromatin, facilitating NER signal transduction [19]. Unlike BER, defects in the NER pathway can lead to a number of human syndromes, associated with shortened lifespan, such as xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy.
DNA repair in eukaryotes requires chromatin remodeling in the areas of damaged DNA to allow the access of the DNA repair machinery to the damaged sites. SIRT6 co-operates with several other factors to shape local chromatin environment and the lack of SIRT6 has been associated with impaired chromatin remodeling and increased sensitivity to genotoxic damage. As the activator of PARP1, SIRT6 may be crucially involved in chromatin remodeling in response to DNA damage, since PARP1 rapidly PARylates chromatin and mediates initial chromatin relaxation through recruitment of chromatin remodeling complexes, such as NuRD [60, 77]. Furthermore, SIRT6 deacetylation of H3K9 is a prerequisite for PARP1 ribosylation of H3S10 during DNA repair [78]. Recently an interaction between SIRT6 and the subunit of NuRD, chromodomain helicase DNA binding protein 4 (CHD4), has been shown in response to DNA damage, leading to the displacement of heterochromatin protein (HP) 1 by CHD4 and promotion of chromatin relaxation for HR repair [79]. Furthermore, SIRT6 deacetylates H3K56 at the DSBs and recruits the chromatin remodeling factor SNF2H to the damage site, which in turn promotes local chromatin accessibility [68]. SIRT6 has also been shown to deacetylate histone acetyltransferase (HAT) GCN5 at K549, enhancing its activity, which may further lead to chromatin accommodation for repair [80]. When DNA damage occurs in the actively transcribed regions, alterations in euchromatin structure help to put a halt on local transcription in transient manner to allow the repair to proceed. Mass spectrometry followed by mutagenesis analysis showed that lysine-specific demethylase 2A (KDM2A) is mono-ADP-ribosylated by SIRT6, resulting in displacement of KDM2A from the DSB location, leading to enrichment of H3K36me2/3 marks and enhancing NHEJ repair efficiency [63].
At the final stage of DNA repair the original structure of chromatin has to be restored to prevent the accumulation of aberrantly packaged chromatin, which may promote aging of tissues. The H3K9me3-specific histone methyltransferase SUV39H1 has been shown as a binding partner of SIRT6 [81]. Methyltransferases create an initial nucleation event on the euchromatin, followed by cycles of H3K9 methylation and loading of new HP1α complexes leading to spreading of the nascent heterochromatin. It is tempting to speculate that via the interactions with SUV39H1 and possibly other chromatin factors, SIRT6 may not only regulate transcription, but also participate in restoration of the original heterochromatin structure following DNA damage.
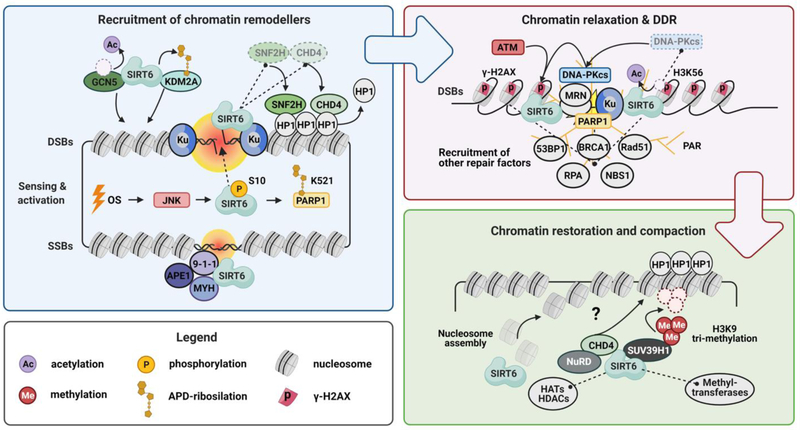
In summary, SIRT6 plays important roles regulating different stages of DNA repair. SIRT6 main functions in DNA repair involve facilitating chromatin remodeling and orderly recruitment of repair enzymes to the sites of DNA damage, acting as an upstream regulator promoting both HR and NHEJ repair pathways (Fig. 2).
Figure 2. SIRT6 at different stages of DNA repair.
Proper regulation of DNA repair is paramount for genomic stability, promoting longevity. SIRT6 is involved in the sensing of DSB through association with Ku80, 9–1-1 complex, APE1 and MYH glycosylase, as well as directly binding to DNA. Under oxidative stress (OS) SIRT6 can be phosphorylated by JNK at S10 and mono-ADP-ribosylates PARP1 at K521. PARP1 PARylates numerous proteins, promoting relaxation of chromatin. SIRT6 also promotes relaxation of chromatin via interactions with SNF2H, CHD4, GCN5 and KDM2A, which increases accessibility of the damage site for the DNA repair machinery. SIRT6 participates in DNA damage response (DDR) signaling through the regulation of DNA-PKcs, and formation of γ-H2AX foci. Furthermore, SIRT6 can interact with SUV39H1, potentially playing a role in chromatin restoration after repair.
Altering the expression or activity of DNA repair enzymes has given disappointing results and has not resulted in improved DNA repair efficiency likely due to introducing mis-coordination and imbalance between repair enzymes. SIRT6, overexpression, on the contrary, enhances multiple pathways of DNA repair due to its upstream regulatory role. The regulatory role of SIRT6 in DNA repair suggests that SIRT6 is a promising target for anti-aging interventions and enhancement of DNA genome stability.
SIRT6 and epigenome stability
Persistent DNA damage with advancing age may result in gradual changes of chromatin structure and erosion of the epigenetic landscape, which may be particularly harmful in the regions of constitutive heterochromatin. SIRT6 has been shown to co-localize with HP1β within nucleus, which potentially implicates it in the aging-related epigenetic alterations specifically in the regions of constitutive heterochromatin: at the repetitive DNA sequences, including pericentromeric and telomeric regions, retrotransposable elements and lamina-associated domains (LADs) [60].
Telomeres in actively dividing cells are subject to shortening following successive rounds of DNA replication, which may lead to genomic instability, cellular senescence and abnormal gene expression in subtelomeric regions. Cells lacking SIRT6 display telomere dysfunction and chromosomal abnormalities reminiscent of the premature aging disorder Werner syndrome, whereas overexpression of SIRT6 improves telomere integrity through H3K9 deacetylation and increased association of WRN with chromatin [82, 83]. The cell cycle-dependent regulation of H3K56 acetylation level by SIRT6 is important for the maintenance of telomeric chromatin [53, 84]. SIRT6 has also been shown to directly target the telomere repeat protein (TRF) 2, which undergoes ubiquitin-mediated proteolysis upon deacetylation by SIRT6 [85]. Another important function of telomeres is to repress nearby gene expression through telomere position effect (TPE). SIRT6 deficiency may lead to the de-repression of subtelomeric regions, rich in H3K9me3/SUV39H/HP1 environment, as shown by the increased expression of the chromosomally integrated reporter at a telomere [86]. Telomeres are also highly susceptible to oxidative damage, which accelerates telomere shortening. Upon oxidative damage the MYH foci are formed at telomeres and SIRT6 is recruited to the DNA damage site to aid in repair [76].
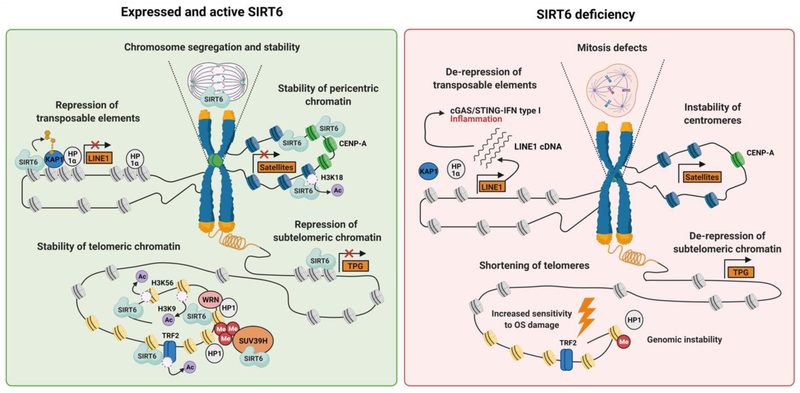
SIRT6 has also been found to co-localize with the marker of centromeres CENP-A [54]. SIRT6 deficiency is associated with aberrant expression of centromeric satellite sequences and SIRT6 is required for deacetylation of H3K18, which promotes centromeric silencing [54]. Pericentromeric heterochromatin domains have major influence on chromosome segregation and stability, and the knockdown of SIRT6 leads to an impairment of chromosome segregation, implicating SIRT6 in the regulation of mitosis (Fig. 3) [87–89].
Figure 3. SIRT6 and epigenome maintenance.
Changes in heterochromatin and de-repression of repetitive sequences pose danger for both genomic and epigenomic stability. SIRT6 deacetylates H3K9 and H3K56, TRF2, increases association of WRN with chromatin and interacts with SUV39H to promote telomere stability. SIRT6 deficiency results in telomere dysfunction, susceptibility of oxidative stress (OS) and may lead to the expression of the telomere proximal genes (TPG). At the centromeres SIRT6 deacetylates H3K18 to maintain silencing of centromeric satellite repeats and interacts and promotes proper chromosome segregation during mitosis, with the defect observed in the absence of SIRT6. SIRT6 is involved in silencing of retrotransposable elements: SIRT6 mono-ADP-ribosylates KAP1, associated with HP1α, to keep a closed conformation of chromatin at LINE1 elements, preventing retrotransposon expression. The lack of SIRT6 results in de-repression of LINE1 elements, produces genomic instability and innate immune response through accumulation of LINE1 cDNA copies and activation of cGAS-STING-IFN type I pathway.
Aging-related reorganization of heterochromatin leads to de-repression of transposable elements, genomic parasites that account for almost a half of the human genome and lead to genomic instability during aging [90]. Long interspersed elements 1 (LINE1s) are retrotransposable elements, which are particularly relevant to aging as its activity is elevated in aged somatic tissues [14, 91]. Activation of LINE1 element leads to the accumulation of LINE1 cytoplasmic DNA copies that could be detected by the DNA sensor cyclic GMP-AMP synthase (cGAS), which triggers an innate immune response through the activation of the stimulator of interferon response cGAMP interactor (STING) 1 to produce interferon (IFN) type I, promoting sterile inflammation [14]. SIRT6 KO mice show a dramatic increase in LINE1 cDNA – comparable to or higher than in old wild type mice. Conversely, overexpression of SIRT6 in mice represses LINE1 activity [14, 92]. The mechanism of LINE1 repression by SIRT6 relies on mono-ADP-ribosylation of KAP1, which is involved in the maintenance of the epigenetically stable heterochromatin [61]. Modification of KAP1 by SIRT6 at the LINE1 loci promotes KAP1 complex formation with HP1, thereby packaging the LINE1 DNA into transcriptionally silent heterochromatin [61].
In summary, SIRT6 functions to maintain heterochromatin state of a wide range of genetic elements including telomeres, centromeres and transposable elements (Fig. 3). The loss of silencing of these elements is associated with aging and age-related disease.
Rejuvenation through reprogramming involving SIRT6
Rejuvenation of the aged epigenetic landscape could be achieved through the process of reprogramming. Complete reprogramming of somatic cells is performed through the conversion of cells into induced pluripotent stem cells (iPSCs) via overexpression of the Yamanaka factors OCT4, SOX2, KLF4 and MYC (OSKM) [93]. SIRT6 deficiency decreased the reprogramming efficiency in mouse cells and SIRT6 was found to improve reprogramming efficiency in human dermal fibroblasts [94, 95]. SIRT6 activator MDL-800 improved pluripotency of murine iPSCs [96]. Moreover, when a combination of SIRT6 and Yamanaka factors was used, the NHEJ DSB repair was improved in iPSCs derived from old mice [74]. In embryonic stem cells (ESCs) SIRT6 histone deacetylation activity has been shown to play a crucial role in the stem cell fate determination through the repression of key factors of pluripotency OCT4, SOX2 and NANOG [55]. Complete reprogramming is undesirable for in vivo rejuvenation as iPSCs may be tumorigenic. As a potential solution to this problem, partial reprogramming could be employed, which has been shown to have rejuvenating effect in mouse models [97, 98]. It is tempting to speculate that the addition of SIRT6 may facilitate safer reprogramming by returning chromatin to its more youthful conformation.
SIRT6 co-operates with LMNA to maintain a youthful epigenome
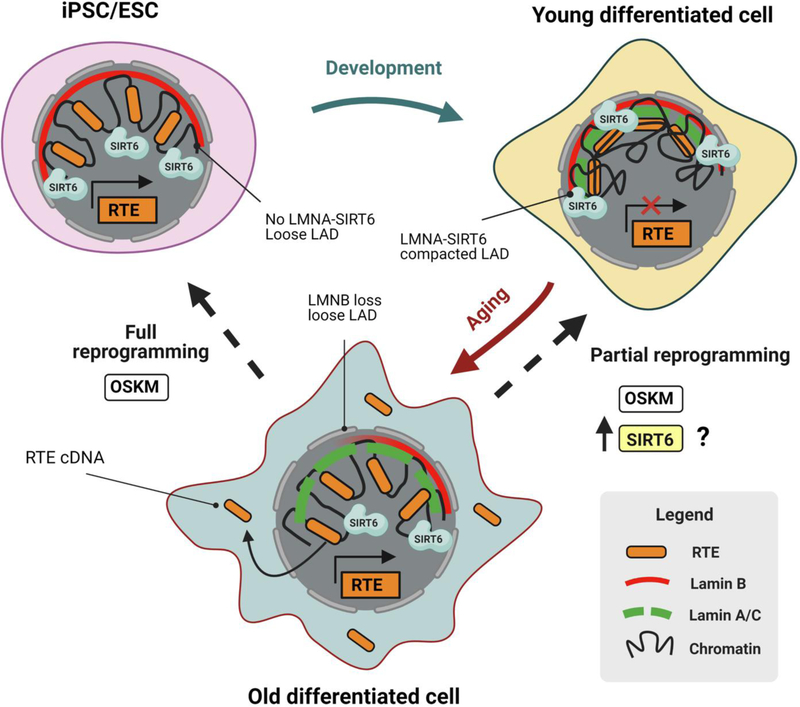
Lamin A/C (LMNA) is a nuclear scaffold protein that mediates chromatin packaging [99]. LMNA binds to the regions of LAD heterochromatin and tethers them to the nuclear periphery. LADs are enriched in retrotransposable elements, which can become de-repressed in aged cells. LMNA mutations are associated with several genetic syndromes including premature aging disease Hutchison-Gilford progeria (HGPS) [100]. Remarkably, polymorphisms in LMNA are also found in centenarians, indicating that LMNA variants may promote longevity [100]. LMNA helps to establish repressive heterochromatin at the nuclear periphery, but in the cells from HGPS patients a global loss of chromatin compartmentalization is observed [101]. LMNA is linked to epigenetic reprogramming and pluripotency. LMNA is not expressed, or expressed at very low levels, in embryonic cells and iPSCs, but is turned on with cell differentiation [102, 103]. Reprogramming of HGPS cells into iPSCs rescues their phenotype, which then reappears upon differentiation [104]. LMNA directly interacts with SIRT6 catalytic core domain via its C-terminus and promotes SIRT6 deacetylation and mono-ADP-ribosylation activities, acting as SIRT6 endogenous activator [105]. Based on the interaction between SIRT6 and LMNA and SIRT6 central role in maintaining heterochromatin organization of repetitive sequences and retrotransposable elements we propose the following model: SIRT6-LMNA axis may control epigenetic aging and the rate of epigenetic drift (Fig. 4). In a young somatic cell, SIRT6-LMNA maintains heterochromatin, which keeps retrotransposons in the packed and silent state. The LADs and retrotransposons are tethered to the nuclear periphery, thus maintaining a youthful transcriptional pattern and differentiated state of the cells. During aging, with multiple rounds of replication, transcription, and DNA repair heterochromatin unravels, leading to the de-repression of retrotransposons, accumulation of cytoplasmic retrotransposon copies and ultimately induction of sterile inflammation and functional decline. Upon full reprogramming LMNA expression is turned off and epigenome becomes permissive of retrotransposon expression. Partial reprogramming would not remove LMNA but rather “tighten up” repetitive elements into heterochromatin, thus rejuvenating the epigenome. We hypothesize that enhanced expression or activity of SIRT6, coupled to partial reprogramming or on itself, may be able to reverse these age-related changes in heterochromatin and rejuvenate the epigenome.
Figure 4. Packaging of retrotransposable elements into heterochromatin controls aging and rejuvenation.
Embryonic stem cells (ESC) and induced pluripotent cells (iPSCs) lack lamin A. Retrotransposons are open and expressed. Upon differentiation SIRT6 facilitates packaging of retrotransposons into silent lamin A-bound chromatin. During aging heterochromatin unravels, retrotransposons become expressed and drive inflammation, while SIRT6 binds to shortened telomeres and damaged DNA and becomes limiting. SIRT6 overexpression can rejuvenate chromatin structure without changing cell fate.
Modulation of SIRT6
The benefits observed in SIRT6 overexpression animal models, and pathologies associated with SIRT6 deficiency suggest activating SIRT6 as a potential rejuvenating strategy. Like other sirtuins, SIRT6 expression can be increased by calorie restriction – one of the most robust and long-known mechanisms for life extension [106]. A search for chemical SIRT6 activators revealed that SIRT6 can be activated in vitro through interactions with fatty acids, like myristoyl. Accommodation of a long-chain acyl group in the hydrophobic pocket of SIRT6 through interactions with fatty acids results in enhanced deacetylase activity of SIRT6 [51]. Taking advantage of such interactions, a screen of 432 lipid compounds based on the ability of molecules to stimulate SIRT6 deacetylation of H3K9ac peptide in vitro identified novel compounds with the ability to increase SIRT6 activity up to 48-fold [107]. Several potent synthetic activators of SIRT6 deacetylase activity have been recently identified. This includes an allosteric SIRT6 modulator MDL-800 that was shown effective in promoting SIRT6-dependent cell cycle arrest and inhibiting tumor growth in vivo, as well as promoting genomic stability by activating DNA repair pathways [96, 108, 109]. Another synthetic activator of SIRT6 deacetylation, UBCS039, has been shown to promote autophagy in a SIRT6-dependent manner [110]. In addition, a number of natural compounds have shown promise as activators of SIRT6. A polyphenol cyanidin produced a 55-fold increase in SIRT6 activity in vitro [111]. A derivative of a plant flavone quercetin could also activate the SIRT6 deacetylation activity [112]. Recently, SIRT6 transcription was found to be promoted by an alkaloid licorine, which was associated with increased NHEJ and HR repair efficiency in human fibroblasts [113]. Moreover, a polysaccharide fucoidan, isolated from brown algae has shown robust SIRT6 activation in vitro, and has been linked to multiple health benefits in humans [114–117].
Concluding remarks
Aging is associated with genomic instability and reorganization of epigenetic landscape, which are intimately linked together. SIRT6 operates at the crossroads between these processes and regulates numerous pathological pathways, working to preserve healthspan. It is possible that SIRT6 levels or activity decrease during aging, albeit to a different degree in various tissues and cells. Looking forward, a targeted restoration of SIRT6 levels or stimulation of SIRT6 activity in those cells in need could help to preserve their youthful phenotype. Many important questions remain unexplored, such as the effects of SIRT6 overexpression on DNA methylation profile and compaction of chromatin in aged tissues. The ongoing research on SIRT6 in long-living species of mammals also holds promise to discover novel, more efficient SIRT6 variants. The functions of SIRT6 in DNA repair, maintenance of heterochromatin domains, repressing transposable elements and regulation of reprogramming provide exciting opportunities for novel rejuvenation strategies to fight age-related pathologies in humans.
Outstanding questions.
Can SIRT6 overexpression reverse epigenetic aging and improve compaction of heterochromatin in aged tissues?
Can increased SIRT6 improve fidelity of DNA repair?
What are the efficient and safe SIRT6 activators?
Can SIRT6 aid in epigenetic reprogramming?
How is SIRT6 regulated in vivo and can this knowledge be harnessed to promote human longevity?
Can SIRT6 be used to prevent stem cell exhaustion – an important hallmark of aging?
Highlights.
SIRT6 extends mammalian lifespan and promotes healthspan
SIRT6 maintains genomic stability through improved efficiency of DNA repair
SIRT6 maintains heterochromatin and silences repetitive genetic elements
Regulation of SIRT6 is a potential strategy for rejuvenation
Acknowledgements
The work in authors’ laboratories is supported by grants from the US National Institutes of Health. The figures were created with BioRender.com.
Glossary
- Epigenetic clock
analysis of DNA methylation patterns that provides an estimate of chronological or biological age
- Epigenetic drift
a complex of age-related epigenetic modifications leading to aberrant expression of genetic information
- Epigenetic reprogramming
removal of epigenetic marks and change in gene transcription patterns to the state of pluripotency and further differentiation of cells into desired phenotype
- Healthspan
the part of a person’s life during which they are free from age-related disease
- Heterochromatin
compacted areas of chromatin associated with condensed, transcriptionally inactive DNA
- Lamina-associated domain (LAD)
transcriptionally silent portion of chromatin associated with nuclear lamina at the inner membrane of the nucleus
- Telomeres
sequences capping the ends of the chromosomes in eukaryotic cells
- Retrotransposable elements
selfish DNA sequences within the genome that can change their position through copy and paste mechanism, creating mutations and triggering inflammation
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Lopez-Otin C et al. (2013) The hallmarks of aging. Cell 153 (6), 1194–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbunova V and Seluanov A (2016) DNA double strand break repair, aging and the chromatin connection. Mutat Res 788, 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijg J (2021) From DNA damage to mutations: All roads lead to aging. Ageing Res Rev 68, 101316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teschendorff AE et al. (2013) Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Hum Mol Genet 22 (R1), R7–R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen P et al. (2016) Epigenetic Mechanisms of Longevity and Aging. Cell 166 (4), 822–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai S and Guarente L (2014) NAD+ and sirtuins in aging and disease. Trends Cell Biol 24 (8), 464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mostoslavsky R et al. (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124 (2), 315–29. [DOI] [PubMed] [Google Scholar]
- 8.Lin SJ et al. (2000) Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289 (5487), 2126–8. [DOI] [PubMed] [Google Scholar]
- 9.Rogina B and Helfand SL (2004) Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A 101 (45), 15998–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaeberlein M et al. (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13 (19), 2570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tissenbaum HA and Guarente L (2001) Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410 (6825), 227–30. [DOI] [PubMed] [Google Scholar]
- 12.Sinclair DA and Guarente L (1997) Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 91 (7), 1033–42. [DOI] [PubMed] [Google Scholar]
- 13.Xiao C et al. (2010) SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem 285 (47), 36776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon M et al. (2019) LINE1 Derepression in Aged Wild-Type and SIRT6-Deficient Mice Drives Inflammation. Cell Metab 29 (4), 871–885 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peshti V et al. (2017) Characterization of physiological defects in adult SIRT6−/− mice. PLoS One 12 (4), e0176371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G et al. (2021) Emerging roles of SIRT6 in human diseases and its modulators. Med Res Rev 41 (2), 1089–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W et al. (2018) SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 560 (7720), 661–665. [DOI] [PubMed] [Google Scholar]
- 18.Ferrer CM et al. (2018) An inactivating mutation in the histone deacetylase SIRT6 causes human perinatal lethality. Genes Dev 32 (5–6), 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng A et al. (2020) The deacetylase SIRT6 promotes the repair of UV-induced DNA damage by targeting DDB2. Nucleic Acids Res 48 (16), 9181–9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Z et al. (2015) SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle 14 (2), 269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pukhalskaia AE et al. (2020) Sirtuins as Possible Predictors of Aging and Alzheimer’s Disease Development: Verification in the Hippocampus and Saliva. Bull Exp Biol Med 169 (6), 821–824. [DOI] [PubMed] [Google Scholar]
- 22.Carbone A et al. (2020) Melatonin and Sirtuins in Buccal Epithelium: Potential Biomarkers of Aging and Age-Related Pathologies. Int J Mol Sci 21 (21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owczarz M et al. (2017) miR-34a and miR-9 are overexpressed and SIRT genes are downregulated in peripheral blood mononuclear cells of aging humans. Exp Biol Med (Maywood) 242 (14), 1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sebastian C et al. (2012) The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151 (6), 1185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S et al. (2016) SIRT6 protects against endothelial dysfunction and atherosclerosis in mice. Aging (Albany NY) 8 (5), 1064–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zi Y et al. (2019) Sirt6-induced autophagy restricted TREM-1-mediated pyroptosis in ox-LDL-treated endothelial cells: relevance to prognostication of patients with acute myocardial infarction. Cell Death Discov 5, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balestrieri ML et al. (2015) Sirtuin 6 expression and inflammatory activity in diabetic atherosclerotic plaques: effects of incretin treatment. Diabetes 64 (4), 1395–406. [DOI] [PubMed] [Google Scholar]
- 28.Ka SO et al. (2017) Hepatocyte-specific sirtuin 6 deletion predisposes to nonalcoholic steatohepatitis by up-regulation of Bach1, an Nrf2 repressor. FASEB J 31 (9), 3999–4010. [DOI] [PubMed] [Google Scholar]
- 29.Kaluski S et al. (2017) Neuroprotective Functions for the Histone Deacetylase SIRT6. Cell Rep 18 (13), 3052–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roichman A et al. (2021) Restoration of energy homeostasis by SIRT6 extends healthy lifespan. Nat Commun 12 (1), 3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roichman A et al. (2017) SIRT6 Overexpression Improves Various Aspects of Mouse Healthspan. J Gerontol A Biol Sci Med Sci 72 (5), 603–615. [DOI] [PubMed] [Google Scholar]
- 32.Naiman S et al. (2019) SIRT6 Promotes Hepatic Beta-Oxidation via Activation of PPARalpha. Cell Rep 29 (12), 4127–4143 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong L et al. (2010) The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140 (2), 280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okun E et al. (2017) Sirt6 alters adult hippocampal neurogenesis. PLoS One 12 (6), e0179681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liberale L et al. (2020) Endothelial SIRT6 blunts stroke size and neurological deficit by preserving blood-brain barrier integrity: a translational study. Eur Heart J 41 (16), 1575–1587. [DOI] [PubMed] [Google Scholar]
- 36.Ji L et al. (2019) Overexpression of Sirt6 promotes M2 macrophage transformation, alleviating renal injury in diabetic nephropathy. Int J Oncol 55 (1), 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu K et al. (2020) Protective Effects of SIRT6 Overexpression against DSS-Induced Colitis in Mice. Cells 9 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung ES et al. (2016) p53-dependent SIRT6 expression protects Abeta42-induced DNA damage. Sci Rep 6, 25628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein D and Toiber D (2017) DNA damage and neurodegeneration: the unusual suspect. Neural Regen Res 12 (9), 1441–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soerensen M et al. (2013) Evidence from case-control and longitudinal studies supports associations of genetic variation in APOE, CETP, and IL6 with human longevity. Age (Dordr) 35 (2), 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.TenNapel MJ et al. (2014) SIRT6 minor allele genotype is associated with >5-year decrease in lifespan in an aged cohort. PLoS One 9 (12), e115616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y et al. (2016) Association of SIRT6 Gene Polymorphisms with Human Longevity. Iran J Public Health 45 (11), 1420–1426. [PMC free article] [PubMed] [Google Scholar]
- 43.Hirvonen K et al. (2017) SIRT6 polymorphism rs117385980 is associated with longevity and healthy aging in Finnish men. BMC Med Genet 18 (1), 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liszt G et al. (2005) Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem 280 (22), 21313–20. [DOI] [PubMed] [Google Scholar]
- 45.Jedrusik-Bode M et al. (2013) The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J Cell Sci 126 (Pt 22), 5166–77. [DOI] [PubMed] [Google Scholar]
- 46.Pan PW et al. (2011) Structure and biochemical functions of SIRT6. J Biol Chem 286 (16), 14575–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tennen RI et al. (2010) Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech Ageing Dev 131 (3), 185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian X et al. (2019) SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species. Cell 177 (3), 622–638 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imai S et al. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403 (6771), 795–800. [DOI] [PubMed] [Google Scholar]
- 50.Liu WH et al. (2020) Multivalent interactions drive nucleosome binding and efficient chromatin deacetylation by SIRT6. Nat Commun 11 (1), 5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feldman JL et al. (2013) Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem 288 (43), 31350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawahara TL et al. (2009) SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 136 (1), 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Michishita E et al. (2009) Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8 (16), 2664–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tasselli L et al. (2016) SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol 23 (5), 434–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Etchegaray JP et al. (2015) The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol 17 (5), 545–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feldman JL et al. (2015) Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation. Biochemistry 54 (19), 3037–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang WW et al. (2016) A Chemical Biology Approach to Reveal Sirt6-targeted Histone H3 Sites in Nucleosomes. ACS Chem Biol 11 (7), 1973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang H et al. (2013) SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496 (7443), 110–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang H et al. (2016) Lysine fatty acylation promotes lysosomal targeting of TNF-alpha. Sci Rep 6, 24371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mao Z et al. (2011) SIRT6 promotes DNA repair under stress by activating PARP1. Science 332 (6036), 1443–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Meter M et al. (2014) SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun 5, 5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rezazadeh S et al. (2019) SIRT6 promotes transcription of a subset of NRF2 targets by mono-ADP-ribosylating BAF170. Nucleic Acids Res 47 (15), 7914–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rezazadeh S et al. (2020) SIRT6 mono-ADP ribosylates KDM2A to locally increase H3K36me2 at DNA damage sites to inhibit transcription and promote repair. Aging (Albany NY) 12 (12), 11165–11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao Z et al. (2012) Sirtuin 6 (SIRT6) rescues the decline of homologous recombination repair during replicative senescence. Proc Natl Acad Sci U S A 109 (29), 11800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorbunova V et al. (2014) Comparative genetics of longevity and cancer: insights from long-lived rodents. Nat Rev Genet 15 (8), 531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seluanov A et al. (2018) Mechanisms of cancer resistance in long-lived mammals. Nat Rev Cancer 18 (7), 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tombline G et al. (2020) Proteomics of Long-Lived Mammals. Proteomics 20 (5–6), e1800416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toiber D et al. (2013) SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell 51 (4), 454–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Meter M et al. (2016) JNK Phosphorylates SIRT6 to Stimulate DNA Double-Strand Break Repair in Response to Oxidative Stress by Recruiting PARP1 to DNA Breaks. Cell Rep 16 (10), 2641–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meng F et al. (2020) Synergy between SIRT1 and SIRT6 helps recognize DNA breaks and potentiates the DNA damage response and repair in humans and mice. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Onn L et al. (2020) SIRT6 is a DNA double-strand break sensor. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee YY et al. (2012) BCLAF1 is a radiation-induced H2AX-interacting partner involved in gammaH2AX-mediated regulation of apoptosis and DNA repair. Cell Death Dis 3, e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCord RA et al. (2009) SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 1 (1), 109–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen W et al. (2017) Sirt6 Promotes DNA End Joining in iPSCs Derived from Old Mice. Cell Rep 18 (12), 2880–2892. [DOI] [PubMed] [Google Scholar]
- 75.Hwang BJ et al. (2015) SIRT6 protein deacetylase interacts with MYH DNA glycosylase, APE1 endonuclease, and Rad9-Rad1-Hus1 checkpoint clamp. BMC Mol Biol 16, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan J et al. (2020) An ordered assembly of MYH glycosylase, SIRT6 protein deacetylase, and Rad9-Rad1-Hus1 checkpoint clamp at oxidatively damaged telomeres. Aging (Albany NY) 12 (18), 17761–17785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chou DM et al. (2010) A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A 107 (43), 18475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liszczak G et al. (2018) Acetylation blocks DNA damage-induced chromatin ADP-ribosylation. Nat Chem Biol 14 (9), 837–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hou T et al. (2020) SIRT6 coordinates with CHD4 to promote chromatin relaxation and DNA repair. Nucleic Acids Res 48 (6), 2982–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dominy JE Jr. et al. (2012) The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell 48 (6), 900–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santos-Barriopedro I et al. (2018) SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-kappaB pathway. Nat Commun 9 (1), 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michishita E et al. (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452 (7186), 492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grootaert MOJ et al. (2021) SIRT6 Protects Smooth Muscle Cells From Senescence and Reduces Atherosclerosis. Circ Res 128 (4), 474–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jia G et al. (2012) Emerging roles of SIRT6 on telomere maintenance, DNA repair, metabolism and mammalian aging. Mol Cell Biochem 364 (1–2), 345–50. [DOI] [PubMed] [Google Scholar]
- 85.Rizzo A et al. (2017) SIRT6 interacts with TRF2 and promotes its degradation in response to DNA damage. Nucleic Acids Res 45 (4), 1820–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tennen RI et al. (2011) SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun 2, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Allshire RC and Madhani HD (2018) Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol 19 (4), 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han L et al. (2015) Sirt6 depletion causes spindle defects and chromosome misalignment during meiosis of mouse oocyte. Sci Rep 5, 15366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ardestani PM and Liang F (2012) Sub-cellular localization, expression and functions of Sirt6 during the cell cycle in HeLa cells. Nucleus 3 (5), 442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maxwell PH et al. (2011) Retrotransposition is associated with genome instability during chronological aging. Proc Natl Acad Sci U S A 108 (51), 20376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Cecco M et al. (2019) L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566 (7742), 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen J et al. (2021) SIRT6 enhances telomerase activity to protect against DNA damage and senescence in hypertrophic ligamentum flavum cells from lumbar spinal stenosis patients. Aging (Albany NY) 13 (4), 6025–6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takahashi K et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (5), 861–72. [DOI] [PubMed] [Google Scholar]
- 94.Xu P et al. (2019) Sirt6 regulates efficiency of mouse somatic reprogramming and maintenance of pluripotency. Stem Cell Res Ther 10 (1), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sharma A et al. (2013) The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. J Biol Chem 288 (25), 18439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Y et al. (2020) The SIRT6 activator MDL-800 improves genomic stability and pluripotency of old murine-derived iPS cells. Aging Cell 19 (8), e13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ocampo A et al. (2016) In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell 167 (7), 1719–1733 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sarkar TJ et al. (2020) Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat Commun 11 (1), 1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burke B and Stewart CL (2013) The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol 14 (1), 13–24. [DOI] [PubMed] [Google Scholar]
- 100.Conneely KN et al. (2012) Human longevity and common variations in the LMNA gene: a meta-analysis. Aging Cell 11 (3), 475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McCord RP et al. (2013) Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res 23 (2), 260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Constantinescu D et al. (2006) Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells 24 (1), 177–85. [DOI] [PubMed] [Google Scholar]
- 103.Eckersley-Maslin MA et al. (2013) Lamin A/C is expressed in pluripotent mouse embryonic stem cells. Nucleus 4 (1), 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu GH et al. (2011) Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature 472 (7342), 221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ghosh S et al. (2015) Lamin A Is an Endogenous SIRT6 Activator and Promotes SIRT6-Mediated DNA Repair. Cell Rep 13 (7), 1396–1406. [DOI] [PubMed] [Google Scholar]
- 106.Ke Z et al. (2020) Short-term calorie restriction enhances DNA repair by non-homologous end joining in mice. NPJ Aging Mech Dis 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klein MA et al. (2020) Mechanism of activation for the sirtuin 6 protein deacylase. J Biol Chem 295 (5), 1385–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang Z et al. (2018) Identification of a cellularly active SIRT6 allosteric activator. Nat Chem Biol 14 (12), 1118–1126. [DOI] [PubMed] [Google Scholar]
- 109.Shang JL et al. (2021) MDL-800, an allosteric activator of SIRT6, suppresses proliferation and enhances EGFR-TKIs therapy in non-small cell lung cancer. Acta Pharmacol Sin 42 (1), 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iachettini S et al. (2018) Pharmacological activation of SIRT6 triggers lethal autophagy in human cancer cells. Cell Death Dis 9 (10), 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rahnasto-Rilla M et al. (2018) Natural polyphenols as sirtuin 6 modulators. Sci Rep 8 (1), 4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.You W et al. (2019) Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives. Sci Rep 9 (1), 19176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang W et al. (2021) Lycorine hydrochloride suppresses stress-induced premature cellular senescence by stabilizing the genome of human cells. Aging Cell 20 (2), e13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rahnasto-Rilla MK et al. (2017) The Identification of a SIRT6 Activator from Brown Algae Fucus distichus. Mar Drugs 15 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Negishi H et al. (2013) Supplementation of elderly Japanese men and women with fucoidan from seaweed increases immune responses to seasonal influenza vaccination. J Nutr 143 (11), 1794–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tsai HL et al. (2017) Efficacy of Low-Molecular-Weight Fucoidan as a Supplemental Therapy in Metastatic Colorectal Cancer Patients: A Double-Blind Randomized Controlled Trial. Mar Drugs 15 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Takahashi H et al. (2018) An Exploratory Study on the Anti-inflammatory Effects of Fucoidan in Relation to Quality of Life in Advanced Cancer Patients. Integr Cancer Ther 17 (2), 282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]