Abstract
Aging is a major risk factor for multiple diseases. Understanding the underlying mechanisms of aging would help to delay and prevent age-associated diseases. Short-lived model organisms have been extensively used to study the mechanisms of aging. However, these short-lived species may be missing the longevity mechanisms that are needed to extend lifespan of an already long-lived species such as humans. Unconventional long-lived animal species are an excellent resource to uncover novel mechanisms of longevity and disease-resistance. Here we review mechanisms that evolved in non-model vertebrate species to counteract age-associated diseases. Some anti-aging mechanisms are conserved across species, however, various non-model species also evolved unique mechanisms to delay aging and prevent disease. This diversity of anti-aging mechanisms has evolved due to remarkably diverse habitats and lifestyles of these species. We propose that exploring a wider range of unconventional vertebrates will provide important resource to study anti-aging mechanisms that are potentially applicable to humans.
Keywords: aging, wild vertebrate species, cancer, cardiovascular disease, inflammation, neurodegeneration
1. INTRODUCTION
Aging is defined as a persistent functional decline in fitness that occurs in most organisms over time (100; 139). In humans, the process of aging is often associated with multiple diseases, including cancer, cardiovascular disease, neurodegeneration, diabetes, osteoarthritis, and chronic inflammation (118). Traditionally, studies on the mechanisms of aging and age-associated diseases have been conducted using model animals, including nematodes (Caenorhabditis elegans), fruit flies (Drosophila melanogaster), zebrafish (Danio rerio), and mice (Mus musculus). Model animals have been used for research due to their accessibility and convenience. Their genomes are sequenced and well annotated; they have uniform genotype and phenotype due to their inbred nature; they have short lifespans and fast lifecycles. These characteristics make them a great tool for drug treatment and gene manipulation.
Extensive attempts have been made to increase mouse lifespan. Inhibiting mTOR pathway by genetically knocking out S6K gene increased mouse lifespan by 9%, while rapamycin treatment increased lifespan by 9%–14% (70; 150). Transgenic overexpression of a sirtuin protein, the Sirt6, increased lifespan in male mice by 10%–17% (84). Clearance of senescent cells (p16Ink4a-positive cells) increased only the medium but not the maximum lifespan of mice (12). These increments of lifespan, however, are not remotely comparable to the lifespan differences across species of mammals. In mammals, the maximum lifespans of different species vary from about 2 years in Mueller’s giant Sunda rats (Sundamys muelleri) (120) to 211 years in bowhead whales (Balaena mysticetus) (166). Maximum lifespan correlates positively with body mass (19), therefore larger mammals tend to be long-lived. However, there are many outliers to this rule including mouse-sized animals that are very long-lived. The naked mole rat (Heterocephalus glaber) has the maximum lifespan of over 32 years (23; 24). Multiple species of bats have maximum lifespans of up to 30–40 years (167). Remarkably, many non-model animals are also shown to be resistant to multiple age-associated diseases, which further contribute to their longevity. These facts suggest that during evolution, different species have evolved novel longevity mechanisms that are absent in short-lived model animals. Here we review the mechanisms of longevity and disease-resistance observed in a variety of unconventional model vertebrates and discuss what can be learned from them for understanding and addressing aging and age-associated diseases in humans.
2. CANCER
Cancer is one of the major age-associated diseases. In humans, although cancer can occur in young individuals, the incidence of cancer increases steeply at older age (67; 109). The epidemiological studies showing an exponential, instead of linear, increase in cancer incidence with age suggest that multiple mutations rather than a single mutation are required to develop cancer (66; 104; 136; 140). Indeed, experimental studies showed that at least two mutations (genetic hits) are required to malignantly transform mouse fibroblasts, while five mutations are required to transform human fibroblasts (93; 134). The number of hits required for malignant transformation varies across species, with larger and longer-lived animals needing a higher number of oncogenic hits (175). Additionally, in long-lived species it takes much longer for cancer to develop. Mice develop cancer in their second year of life (99), while in humans, cancer affects mostly people above 60 years of age (157). Several species of wild animals, on the other hand, are known to be extremely resistant to cancer, including naked mole rat, bind mole rat, elephant, and bowhead whale (Figure 1). Importantly, these species adopt distinct strategies to prevent cancer.
Figure 1. Distinct cancer resistance mechanisms of naked mole rat, blind mole rat, elephant, and whale.
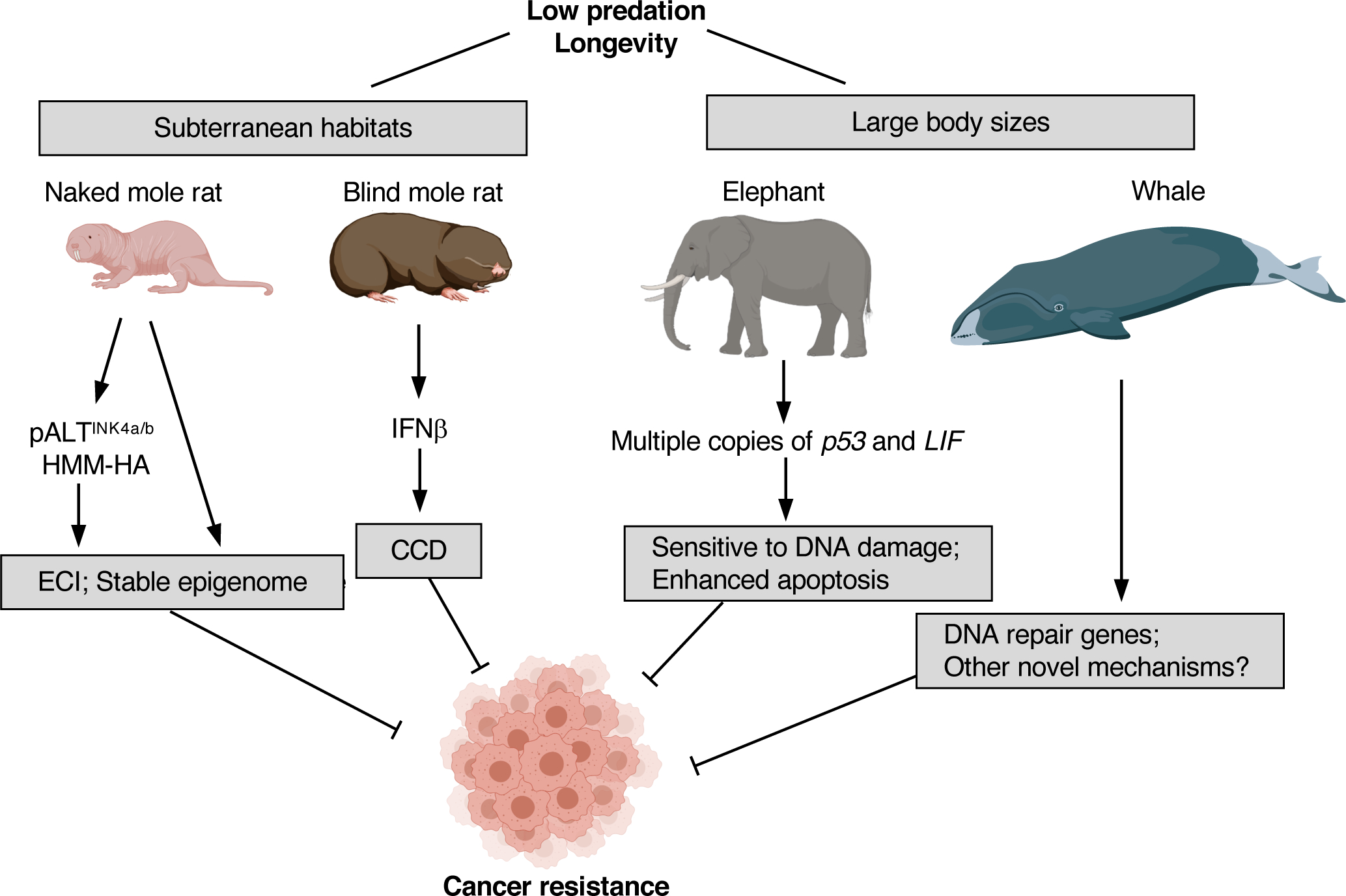
Naked mole rat cells undergo early contact inhibition (ECI) through high-molecular-mass hyaluronan (HMM-HA) and an additional INK4a/b hybrid locus (pALTINK4a/b). Blind mole rat cells undergo concerted cell death (CCD) triggered by IFNβ. Elephant has multiple copies of p53 and leukemia inhibitory factor (LIF) genes, facilitating enhanced apoptosis. Whale DNA repair genes have undergone positive selection. Other novel mechanisms for cancer resistance in whales are yet to be determined. Both naked mole rat and blind mole rat inhabit the subterranean burrows. Elephants and whales have large body size. These characteristics make them relatively protected from non-human predators, permitting the selection for longevity and cancer resistance. Part of the illustration was generated with biorender.
2.1. The naked mole rats
The naked mole rat (Heterocephalus glaber), which inhabits subterranean burrows in East Africa, is the longest-lived rodent species. A mouse-sized rodent, the naked mole rat has the maximum recorded lifespan of 37 years (23; 24). It is reported that the naked mole rats do not display aging phenotypes and age-associated diseases until very late in life, and their mortality does not increase with age, which defies the Gompertz’s law (23; 141). Most strikingly, the naked mole rats are highly resistant to cancer (98). Out of thousands of individuals that have been monitored in research laboratories and zoo facilities, only seven cases of spontaneous neoplasia and a presumptive case have been reported by three groups to date (29; 37; 172).
Some of the secrets of naked mole rat cancer resistance had been revealed by studying the behavior of naked mole rat culture cells. Primary adult naked mole rat fibroblasts are growing considerably slower in compared to mouse fibroblasts (153). Furthermore, naked mole rat fibroblasts display a unique phenotype that is termed early contact inhibition (ECI) (153). Contact inhibition is an important anti-cancer mechanism to prevent over-proliferation (2). Normal cells stop proliferating when they reach confluence and form a monolayer in the culture dishes. Cancer cells, which lost contact inhibition, continue growing to form multilayered foci. Contact inhibition is mediated by the expression of the cyclin-dependent kinase inhibitor p27kip1 (p27), which interacts with cyclin-CDK complex to induce G1 arrest (96; 127). Instead of becoming confluent, naked mole rat fibroblasts stop proliferating after loosely contacting each other, namely undergo ECI (153). Different from normal contact inhibition, ECI of naked mole rat cells is triggered by the activation of another cyclin-dependent kinase inhibitor, p16Ink4a (p16) (153). ECI adds an additional layer of control of cell proliferation to the naked mole rat. Interestingly, the gene locus of p15INK4b, p16INK4a and ARF (Alternate Reading Frame) in naked mole rat codes an additional gene, which fuses p15INK4b exon 1 and p16INK4a exons 2 and 3. This novel gene, termed pALTINK4a/b can efficiently induce cell cycle arrest (174).
In the naked mole rat, the ECI is triggered by extracellular signals mediated by a unique high-molecular-mass hyaluronan (HMM-HA) (173). Hyaluronan is a linear polysaccharide and a major component of extracellular matrix (178). The biological function of hyaluronan depends on its length. Longer molecules have anti-proliferative and anti-inflammatory effect (89; 178) whereas short molecules are pro-inflammatory and facilitate proliferation, metastasis (130; 178). The hyaluronan secreted by naked mole rat cells has the molecular weight of 6–12 MDa compared to 0.5–3 MDa in mouse and 0.5–2 MDa in human (73; 173). The high molecular weight of naked mole rat HMM-HA is achieved by two mechanisms. The naked mole rat hyaluronan synthase 2 (Has2) gene has two unique asparagine-to-serine mutations that enhance the synthesis of hyaluronan. The hyaluronan-degrading enzymes, HAases, have a lower activity in naked mole rat cells and tissues (173). Naked mole rat HMM-HA plays important roles in inhibiting cell proliferation and suppressing tumorigenesis. Naked mole rat fibroblasts are resistant to transformation by HrasG12V and SV40 Large T antigen (LT). Removing HMM-HA by either silencing Has2 or overexpressing HAase made their cells more susceptible to oncogenic transformation (173). It is known that hyaluronan mediates extracellular signals by interacting with its major receptor, CD44, on the cell surface (89; 128; 130). The naked mole rat very-high-molecular-mass hyaluronan is shown to suppress CD44 protein-protein interactions, an effect opposite to its shorter counterpart (169). As a result, naked mole rat hyaluronan uniquely regulates CD44-dependent genes and protects cells from stress in a p53-dependent manner (169).
It is proposed that naked mole rat HMM-HA has evolved as an adaptive trait to their subterranean lifestyle by providing elastic skin that facilitates squeezing in underground burrows. This trait could later had been coopted to protect naked mole rats from cancer during their long lives. The anti-cancer effect of HMM-HA may thus have been a by-product of their subterranean lifestyle.
Other mechanisms also contribute to cancer resistance of the naked mole rat. Cellular senescence is an important anti-cancer mechanism preventing damaged or premalignant cells from proliferating (30; 192). There are multiple types of cellular senescence, including replicative senescence (RS) (69; 106) induced by telomere shortening, oncogene-induced senescence (OIS) (16; 39; 155), and stress-induced premature senescence (SIPS) (180; 181) triggered by DNA damage. More recently, programmed senescence was observed during embryonic development, which plays a critical role in tissue remodeling (113; 162). Cellular senescence contributes to aging and age-associated diseases as clearance of senescent cells extends lifespan and healthspan in mice (11; 12; 165; 193). As a small-sized rodent, naked mole rat does not undergo replicative senescence due to constitutive telomerase expression (151; 154). However, all three other types of senescence have been observed in the naked mole rat (200). Compared with mouse cells, naked mole rat fibroblasts required higher dose of DNA damage to induce SIPS and are more resistant to DNA damage-induced apoptosis (200). Gene expression analysis demonstrated some unique changes of gene expression in naked mole rat senescent cells, including up-regulation of lysosomal genes, oxidative stress response, changes in extracellular matrix, and inhibition of transcription, spliceosome, and mitochondrial translation (200). These changes may contribute to stress resistance of naked mole rat cells.
It was recently shown that upon oncogene challenge, naked mole rat cells are more resistant to transcriptomic changes induced by HRASG12V. Specifically, the Ras effector pathway, the Phosphoinositide 3-kinase (PI3K) – AKT signaling pathway downstream of Ras, is inherently decreased in naked mole rat (199). This may be an evolutionary adaptation to resist oncogenic transformation in naked mole rat cells.
Another potential anti-cancer mechanism of naked mole rat is that naked mole rat cells are very resistant to induced pluripotent stem cell (iPSC) reprogramming by OSKM factors (Oct4, Sox2, Klf4 and Myc) (94; 111; 170). This characteristic may be attributable to the more stable epigenome in naked mole rat, which has higher levels of repressive H3K27 methylation marks and lower levels of activating H3K27 acetylation marks in the histone landscape (170). This more stable epigenome is likely to contribute to cancer resistance in naked mole rat as iPSC reprogramming shares similar properties with tumorigenesis (17; 164).
2.2. The blind mole rats
The blind mole rat, Spalax ehrenbergi superspecies, is a group of related subterranean rodent species that inhabit in the Middle East, East Europe, and Asia Minor (117). The blind mole rats spend their entire life in subterranean burrows and thus have been extensively studied as a model of hypoxia tolerance (117). Strikingly, it was later revealed that the blind mole rats are very long-lived and extremely cancer resistant (62). The blind mole rats are phylogenetically closer to mice and rats than to the naked mole rats (108), and their body size is similar to a rat. However, the maximum lifespan of blind mole rats in captivity reached over 21 years (117). Over thousands of blind mole rats have been captured and studied in the past 50-years, and not a single case of spontaneous cancer had been observed. Furthermore, a high-dose regimen of carcinogen DMBA/TPA, a model to induce skin cancer in mice, failed to induce cancer in blind mole rat with multiple attempts (62; 102). Therefore, blind mole rat is resistant to both spontaneous and induced tumorigenesis.
The blind mole rats adopt a distinct strategy from naked mole rats to prevent cancer. Blind mole rat fibroblasts grow faster in cell culture than the naked mole rat cells and do not undergo early contact inhibition. Instead, the cells keep proliferating for 10–15 population doublings before they enter a growth arrest. Shortly after that, the cells undergo necrotic cell death within in a few days (62). This phenotype, termed the concerted cell death (CCD), is associated with the secretion of interferon-β (IFNβ) (62). The CCD mechanism senses over-proliferation and eliminates pre-malignant cells. Genome analysis of the blind mole rat demonstrated duplications of interferon gene pathway to further support this model (48).
Interestingly, the Tp53 gene of blind mole rat carries an adaptive mutation of Arginine-to-Lysine on codon 172 (Arg-174 in human), which weakened its transactivation towards pro-apoptotic target genes (8). This mutation is believed to contribute to the hypoxia-tolerance of the blind mole rat by preventing excessive cell death induced by hypoxia. However, the weakened p53 would put the blind mole rat in risk of cancer. Therefore, it is speculated that the duplicated IFN pathway genes and the its responsiveness to cell over-proliferation act as a compensatory strategy. p53 has been shown to negatively regulate the expression of transposable elements (95; 187). Therefore, it is possible that the weak p53 in blind mole rat contributes to the derepression of transposable elements, which activates interferon through the TRAIN (transcription of repeats activates interferon) mechanism (48; 95).
2.3. Elephants
Elephants have large body sizes and a maximum lifespan of 65 years (1). The number of cell divisions throughout the lives of elephants is much larger than that of small animals, and the chance of mutation accumulation is higher, which predicts higher cancer risk. However, elephants are reported to have very low cancer incidence (1). This contradiction is referred to the Peto’s paradox (126). As reported previously, somatic cells from large animals (body mass above 5–10 kg) limit telomerase expression and undergo replicative senescence after over-proliferation (151; 152; 154). However, as implied by Peto’s paradox, additional tumor suppressive mechanisms are required for elephants to counteract the high cancer risk. In recent studies, 19 additional copies of Tp53 gene were identified in the elephant genome (1; 163). These 19 copies are believed to be pseudogenes and some of them are transcribed from transposable element derived promoters. while 2 of their transcripts are translated in elephant fibroblasts (163). As a result, the elephant cells are more sensitive to DNA damage; lower dose of DNA damage is required to induce p53-dependend apoptosis (1; 163). As there are no DNA-binding domain, tetramerization, and nuclear localization domains in these retrogenes, their products are not likely to function as transcription factors. Instead, these additional Tp53 products dimerize with the canonical p53 proteins to protect them from MDM2-dependent ubiquitination (163).
In addition to duplicated p53 genes, multiple copies of another gene, the leukemia inhibitory factor (LIF) were also discovered in the elephant genome (182). One transcript of a duplicate LIF gene (LIF6) in elephant is up-regulated by p53 in response to DNA damage, contributing to the enhanced DNA damage response in elephant. As the LIF6 gene contains a p53 responsive element, it is speculated that elephant LIF6 re-evolved into a functional target gene of p53 from a pseudogene (182). Collectively, these gene duplications provide additional tumor suppressors to mitigate cancer risk for elephants. An enhanced apoptosis in response to DNA damage helps ensure the integrity of the genome without causing problematic cell loss for the large-sized elephants.
2.4. Whales
Whales are the largest mammals on earth, with a body weight of up to 100 tons. They are also the longest-lived mammals. The maximum lifespan of the bowhead whale reaches 211 years (166), and considering both their lifespan and exceptionally large body mass, cancer incidence in whales is extremely low. Interestingly, whales did not evolve similar anti-cancer mechanism as elephants. Genomic analysis did not identify duplications of the p53 gene in whales (55; 194). Instead, several genes associated with aging and cancer were identified carrying bowhead whale specific mutations, including excision repair cross-complementing rodent repair deficiency, complementation group 1 (ERCC1), histone deacetylase 1 (HDAC1), and HDAC2 (86). ERCC1 gene is involved in DNA repair, and HDAC1 and HDAC2 are associated with age-related epigenetic modification (59; 138). Furthermore, several genes underwent duplication during evolution of bowhead whales. Proliferating cell nuclear antigen (PCNA), participating in DNA repair, is duplicated and both copies are expressed in several bowhead whale tissues. Late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 (LAMTOR1), which is involved in mTOR pathway, has an additional copy with lower but detectable expression in heart and retina (86). Several other genes associated with mitosis, cancer, and stress response had also undergone duplication, including 26S proteasome non-ATPase regulatory subunit 4 (PSMD4), ubiquitin carboxyl-terminal esterase L3 (UCHL3), cAMP-regulated phosphoprotein 19 (ARPP19), stomatin-like 2 (STOML2), heat shock factor binding protein 1 (HSBP1), spermine synthase (SMS) and suppression of tumorigenicity 13 (ST13) (86).
Transcriptome studies also demonstrated bowhead whale specific change of gene expression. Insulin signaling genes are differentially expressed in the bowhead whale, including reduced expression of growth factor receptor-bound protein 14 (Grb14) gene and elevated expression of Cited2 gene. The expression pattern of these genes mimics the effect of calorie restriction in mice, potentially contributing to the longevity of bowhead whales (149). In gray whales, several genes involved in senescence and autophagy were among the most expressed genes, including CREG1 and genes coding for CAAX box protein (179). Although the data about gray whale cancer resistance are absent, similar genes were found highly expressed in bowhead whales and minke whales (179). Meanwhile, highly expressed genes in DNA maintenance and repair, ubiquitination, and apoptosis were observed in several whale species (bowhead, gray, and minke whales) and in naked mole rats (179).
In summary, large-sized animals like elephants and whales have evolved additional tumor suppressor mechanisms to compensate for the high risk of mutations due to their large body size, but the different species adopt distinct strategies coincident with their own adaptive evolution.
3. CARDIOVASCULAR DISEASE
Cardiovascular diseases remain the number one cause of death in developed countries (137). The risk of cardiovascular diseases is associated with age, with especially high risk in populations over 65 years old (38; 79; 119). Multiple aging-related genes and pathways are associated with the cardiovascular health, including insulin/IGF-1, TOR, AMPK, LKB1, and Sirtuins (119). Several wild animals possess uniquely protective mechanisms against cardiovascular disease that evolved as an adaptation to their special lifestyle (Figure 2).
Figure 2. Cardiovascular protection mechanisms of ground squirrel, bear, whale, and giraffe.
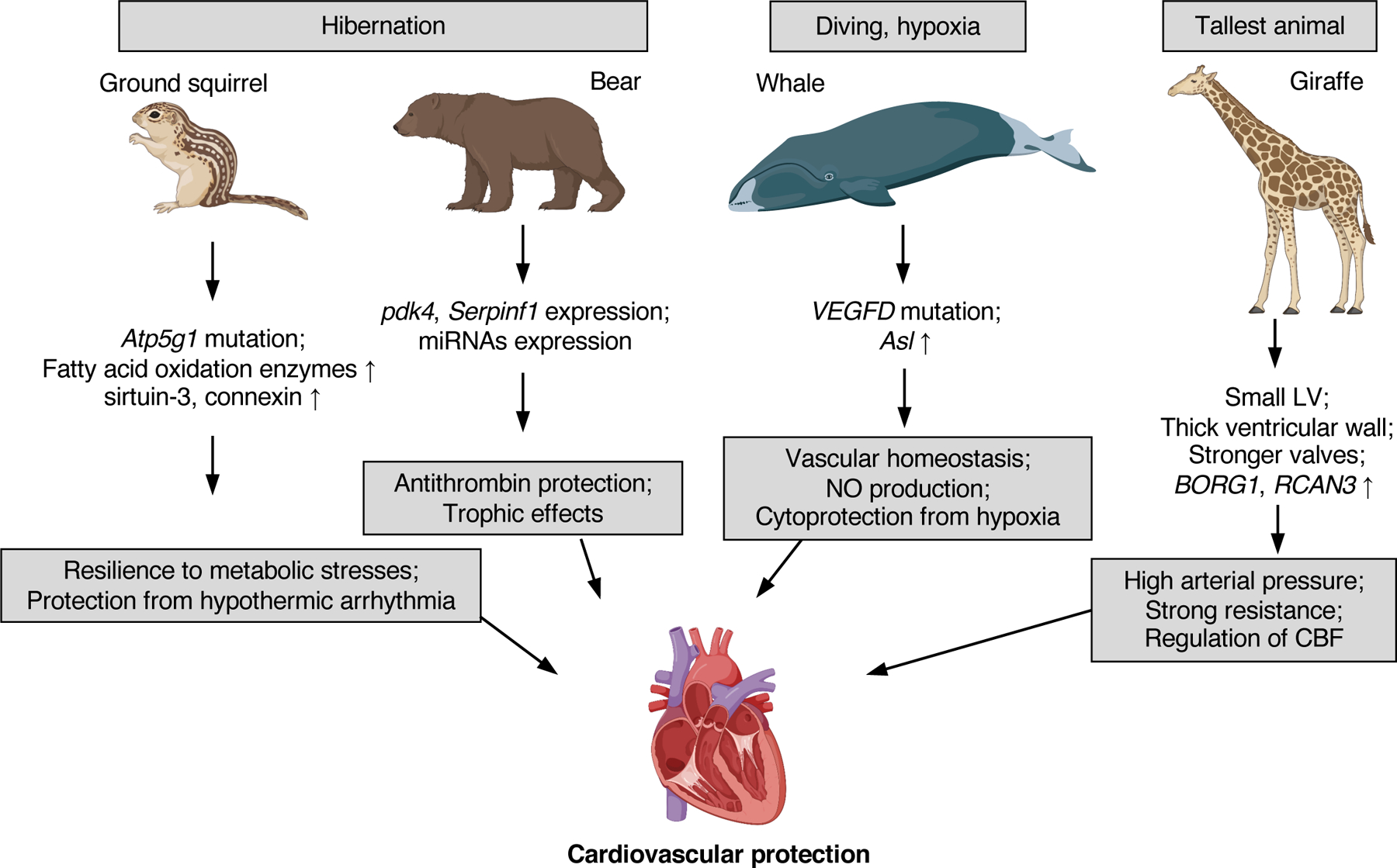
Ground squirrel and bear evolved cardioprotective mechanisms adapting to their hibernating lifestyle. These protections are achieved by altered sequences or expression of genes involved in metabolism, cell respiration, amino acid synthesis, and cell-cell communication. Whales adapt to hypoxic environment during diving in deep water and carry mutations in genes coding for VEGFD and up-regulated expression of Asl gene responsible for nitric oxide (NO) production, protecting cardiovascular function. As the tallest animal, giraffe has small left ventricular (LV) and thick ventricular wall, providing higher arterial pressure to maintain cerebral blood flow (CBF). Giraffes also developed stronger valves to resist the high blood pressure. This may be partly due to the adaptively regulated expression of genes that regulate blood pressure or cardiovascular function, including BORG1 and RCAN3.
3.1. Hibernating animals: bears and ground squirrels
One of the major cardiovascular health issues in human patients is a “hibernating myocardium”, which refers to the dysfunction of resting left ventricular (LV) due to chronically reduced coronary blood flow or repetitive ischemic episodes (40; 41; 133). Structural degeneration was observed in patients with hibernating myocardium, characterized by reduced expression of protein and mRNA, as well as disorganized contractile and cytoskeletal proteins, coupled with fibrosis and cell apoptosis (44). If blood flow to the myocardium is restored, it often takes weeks to months for contractile apparatus to reform (88). Patients with hibernating myocardium are subjected to the risk of arrhythmias (28). In the wild, many of hibernating animal species endure low metabolic rates and body temperatures, associated with limited oxygen supply to their cardiovascular system during hibernation, showing characteristics comparable to that of patients, yet they recover smoothly from hibernation to active state. Their adaptive mechanisms to the hibernating lifestyle provide important insights to human cardiovascular health (18).
Brown bears (Ursus arctos) hibernate six months every year. During hibernation, brown bear’s cardiovascular system undergoes adaptive mechanisms to the low metabolic rate, including reduced heart rate, decreased systolic and diastolic pressures of LV. These changes are not associated with cardiac atrophy (81). Similarly, in grizzly bears (Ursus arctos horribilis), LV volume:mass ratio increases during hibernation. Total left atrial emptying fraction was reduced, and atrial contraction blood flow velocities and atrial contraction ejection fraction were decreased during hibernation (115). Proteomic and transcriptomic study on hibernating grizzly bears revealed increased non-essential amino acids (NEAA) in muscle. Compared to muscle atrophy, hibernating muscles display differentially regulated genes, including Pdk4 and Serpinf1, showing trophic effects beyond hibernating animals (112). In American black bears (Ursus americanus), 24 genes were found differentially expressed during hibernation in both liver and heart (50). These genes are involved in lipid catabolism and protein biosynthesis, which enhances protein synthesis at hypothermic temperatures (50). Moreover, 15 microRNAs were also differentially expressed in the plasma of hibernating black bears. Three of these microRNAs (miR-141–3p, miR-200a-3p, and miR-200c-3p) were predicted to target SERPINC1 gene, which may contribute to antithrombin protection (49). Some changes of gene expression, including genes involved in fatty acid β oxidation, carbohydrate synthesis, lipid biosynthesis, carbohydrate catabolism, cellular respiration, and detoxification pathways, are shared between hibernating bears and small hibernators (50).
Compared to bears, the much smaller-sized hibernating animals, the ground squirrels, display much more acute hypometabolic state during hibernation. During the nonresponsive hibernating state, the body temperature of ground squirrels drops from 37°C to 3–5°C, compared to 33°C in bears (116). Cardiac output during hibernation sharply reduces in ground squirrels by 97% compared to 75% reduction in bears (116). Uniquely, the hibernation of ground squirrels is associated with periodic warming and arousals, resulting in fluctuated O2 requirements (116; 135). Well adapted to this fluctuation, ground squirrels serve as a natural model to understand the protection against ischemia/reperfusion injury in human patients.
Hibernating arctic ground squirrels (Urocitellus parryii) displayed significantly reduced plasma levels of troponin I, myocardial apoptosis, and LV contractile dysfunction (131). Most proteins involved in mitochondrial energy transduction were down-regulated in hibernating arctic ground squirrels. In contrast, fatty acid oxidation enzymes and sirtuin-3 were up-regulated (131). The up-regulation of sirtuin-3 suggests the importance of posttranslational modifications in cardiovascular protection in hibernating mammals (131). The arctic ground squirrel brain was shown to be resistant to cardiac arrest induced injury during euthermia (32). A Pro-32-Leu mutation of Atp5g1 was identified in arctic ground squirrel that contributes to resilience to metabolic stresses (159). Extremely low body temperature may cause fatal arrhythmia. Ground squirrels bypass it by regulating Ca2+ homeostasis in cardiomyocytes (43). The voltage shift of Ca2+ current (ICa) at low temperature is compensated by a higher depolarization rate and a longer duration of action potential (97; 116). To protect from hypothermic arrhythmia, connexin43 and connexin45 are up-regulated in hibernating ground squirrels, protecting against dispersion of depolarization (51).
3.2. Whales and hypoxia
In addition to their longevity and cancer resistance, whales are also well adapted to the diving lifestyle, enduring periodical insufficient oxygen supply. The hypoxia-sensitive tissues, the brain and heart, of whales tolerate low oxygen condition while diving. A unique amino acid substitution in c-fos induced growth factor (Figf) gene was identified in bowhead whales (149). The Figf gene codes for vascular endothelial growth factor D (VEGFD), which plays a role in vascular homeostasis (142). It was speculated that this mutation potentially contributes to the maintenance of vascular health in bowhead whales (149). Gene expression analysis showed a 4.4-fold higher expression of the argininosuccinate lyase (Asl) gene in whales (149). Expression of Asl gene is essential for production of nitric oxide (NO), which confers cytoprotection to tissues during hypoxia and may help preserve cardiac function in diving mammals (80; 149). However, as this differential expression analysis was based on only one bowhead whale heart and one minke whale heart (149), more thorough study is required to solidify the conclusion.
3.3. Giraffe and cerebral blood flow
Blood supply to the brain may be compromised by age-related decline of cardiovascular function through impaired cerebral blood flow (CBF) (35). Acute reduction of CBF can cause loss of consciousness. Therefore, the regulation of CBF is of great importance. Multiple factors are known determinants of CBF, including neurovascular coupling, arterial blood pressure, cardiac output (CO), and autonomic neural activity (171). Most of these factors show an age-dependent decline. For example, the CBF/CO ratio index (CCRI) is negatively correlated with age (191).
The giraffes are a perfect model to study the regulation of CBF. Giraffes are the tallest animals on earth. The positional change of giraffe head between ground level and standing upright is the largest of all animals, yet loss of consciousness never occurs as they evolved mechanisms to provide adequate blood flow (110). The rapid changes in the relative position of the brain to the heart require a well-adapted maintenance of cardiovascular homeostasis. With a normal-sized heart relative to their body size, giraffes naturally have a blood pressure twice that of humans (6; 160). Several physiological and anatomical characteristics of giraffe cardiovascular system potentially contribute to this adaptation. The giraffe heart has a small LV and thick ventricular wall, which allow the generation of high arterial pressure (160). Adapting to the high blood pressure, the aortic valve of giraffes is stronger and stiffer than that of its bovine counterpart (6). This is likely due to the higher content of collagen and elastin, which make giraffe valves more resistant to the high-pressure forces (6). Another risk is the potential high pressure in the brain when giraffes lower their heads. This is avoided by the changing blood pressures governing transcapillary exchange, precapillary vasoconstriction and low permeability of capillaries to plasma proteins (68). CBF is related with cerebral perfusion pressure (CPP). In giraffes, an increased CPP in standing upright position was observed to protect from loss of consciousness. In the head-down position, the viscous resistance in the extracranial circulation decreases more than it does in the intracranial circulation, and CBF is diverted to the jugular veins (110).
Genome sequencing compared with okapi, the closest relative of giraffe but of much shorter stature, revealed adaptive evolution of eight genes that regulate blood pressure or cardiovascular function in giraffes (3). Among these genes, BORG1 and RCAN3 are highly expressed in the heart, potentially contributing to cell shape and cardiac muscle contraction, respectively. Additionally, several metabolic genes related to cardiovascular functions showed multiple signs of adaptation (3).
4. NEURODEGENERATIVE DISEASES
4.1. Degu, a natural model for Alzheimer’s disease
Aging is the main risk factor for neurodegenerative diseases, a major type of which is the Alzheimer’s disease (AD). AD was first described as extracellular amyloid plaques and intracellular neurofibrillary tangles (NFTs) in the brain, composed of abnormally folded amyloid-β42 (Aβ42) and tau proteins (189). As an age-associated disease, AD is affecting over 40 million people worldwide (47). Despite decades of research, no curative intervention is available. Current studies use a great number of mouse models for AD. Nearly 200 transgenic rodent models for AD are available, most of which are based on mutations linked to Aβ protein misprocessing. Recent models also incorporate the mutation of tau protein (114). These mouse models mimic the familial AD but do not model the sporadic AD, which makes up to > 97% of all AD cases (114). Generation of sporadic AD models have been attempted by incorporating mutations of APOE and TREM2, mutations observed in sporadic AD, but these models did not cover many risk factors related to lifestyle, including diabetes, hypercholesterolemia, and stress (54).
The South American rodent degu (Octodon degus) has been described as a natural sporadic AD model due to its Alzheimer’s disease-like neuropathology (20; 75). Degus are highly social, thus resembling humans. Unlike other rodents, in their social interactions, degus rely largely on visual and vocal communications (22; 31; 132). They even demonstrated the ability to use tools (122). However, during aging degus experience cognitive decline, associated with memory loss (7; 36). Many degus spontaneously develop AD-like pathologies with symptoms similar to human AD patients. For example, the cognitive decline of aged degus is correlated with high level of Aβ (76). This is possibly due to a highly similar sequence of Aβ between degu and human, with only one amino acid difference (Arg13His). Other rodents like mice and rats, which do not develop AD-like pathology, have three different amino acid of Aβ peptide from human (75; 76). ApoE is the most frequently identified genetic risk factor for sporadic AD. The ApoE sequence has a higher similarity between human and degu than between human and rat (144). This may explain why degu is a good model for sporadic AD. Affected degus also share other symptoms found in human patients, including activated inflammation factors. Similar to human patients, pro-inflammatory cytokine Interleukin-6 (IL-6), TNF-α, IFNα, and granulocyte-macrophage colony-stimulating factor (GM-CSF) were found elevated in AD-like degu brains (36; 77). Therefore, degus are an excellent model to study AD and may be highly valuable for preclinical trials for therapy and interventions.
5. INFLAMMAGING AND INFLAMMATORY DISEASES
Immune response is important for protecting organisms from infection with viruses or bacteria; however, dis-regulated immune response can be damaging to the host tissues and cells. This is particularly a health concern for aged populations. Inflammation is a driver of multiple age-associated diseases, including cancer, cardiovascular disease, neurodegenerative diseases, diabetes, and osteoarthritis. This persistent and progressive increase in the proinflammatory status with aging is termed inflammaging (56). Many age-associated factors contribute to inflammaging, including cellular senescence, viral infections, microbiome bacteria, and self-products of cell damage including cytoplasmic nucleic acids (sterile inflammation) (34; 57; 158). Bat, pangolin, and naked mole rat have been shown to resist inflammatory diseases by different mechanisms (Figure 3).
Figure 3. Resistance to inflammatory diseases in bat, pangolin, and naked mole rat.
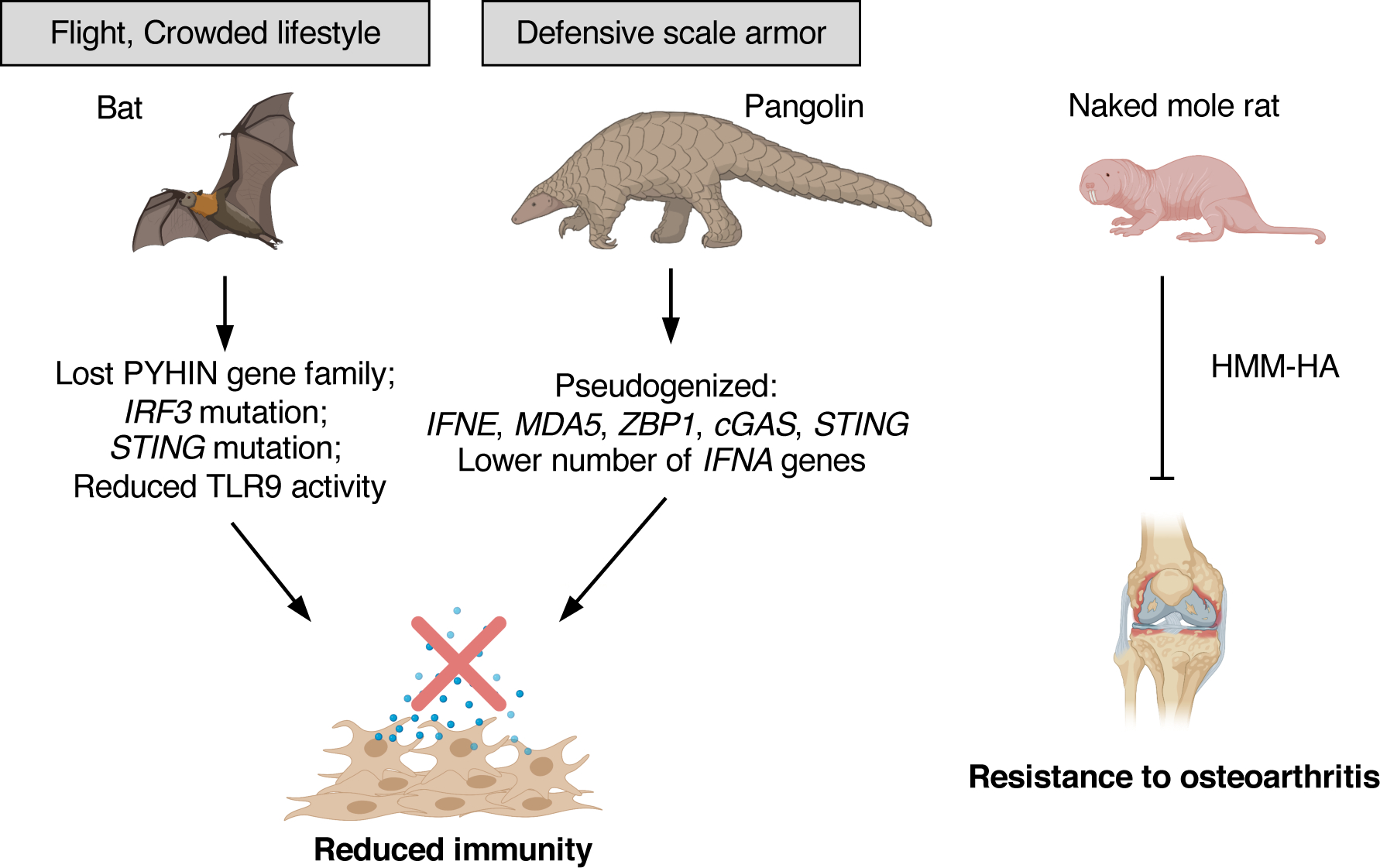
Both bats and pangolins evolved tolerance to viruses and reduced inflammatory response by mutations or pseudogenization of RNA and DNA sensors innate immune sensors. The driving forces that shape the immune responses in bats are believed to be the ability to fly and crowded lifestyle. Pangolins have defensive keratinized scale armors, which provide an additional protection from skin injuries and infection. The reduced inflammatory response in pangolins may have coevolved with the formation of scale armors. Naked mole rats are extremely resistant to osteoarthritis, possibly, through anti-inflammatory function of HMM-HA.
5.1. Dampened inflammatory response in bats and pangolins
The bats are a perfect model to study inflammatory diseases resistance and longevity. Many species of bats have extraordinarily long lifespan relative to their body size. For instance, little brown bat, Brandt’s bat, mouse-eared bat, and Indian flying fox have maximum lifespans of 30–40 years (64). Bats are also resistant to cancers; very few cancers have been identified in bats (64). Most strikingly, bats are very tolerant to different kinds of viruses, including Ebola, rabies, and coronaviruses. Often bats co-exist with viruses without displaying signs of disease or pathology.
The mechanisms of virus tolerance in bats have been extensively studied. Several studies indicate that bats have enhanced RNA sensing and dampened DNA sensing mechanisms. A novel phosphorylation site, Ser-185, was identified in IFN regulatory factor 3 (IRF3) gene in multiple bats species, which significantly improved anti-viral protection through RNA-sensing (15). Bats also constitutively express IFNα, contributing to antiviral activity and resistance to DNA damage (201).
To conquer the potentially excessive immune response, multiple immune pathways are attenuated in bats. Bats immune cells have dampened NLRP3 inflammasome (4), over-activation of which is associated with inflammatory state and age-related diseases (195). Interestingly, viral double-stranded RNA (dsRNA)-induced IFNβ in bat cells failed to activate tumor necrosis factor α (TNF-α) (14). This is due to a potential repressor on TNF promoter in bats, which is a strategy of bats to suppress excessive immune response (64). In addition, DNA sensing is remarkably dampened in bats. The PYHIN gene family, which includes cytoplasmic DNA sensors AIM2 and IFI16, are missing in 10 bat species (5; 197). The major cytoplasmic DNA sensing pathway, the cGAS-STING pathway, is dampened in bat by a mutation in STING, resulting in the missing the conserved S358 (190). The TLR9 receptor, which is activated by unmethylated CpG-containing DNA, evolved under positive selection in bats and reduced its activation (14; 46). Recently, genome-wide study on bats revealed selection and loss of NF-κB regulators and expansion of anti-viral APOBEC3 genes (78).
Bats show an enhanced autophagy in response to Australian bat lyssavirus infection (91). This acts as a compensation of the immune system to clear damaged cells; meanwhile, autophagy also contributes to longevity (143). Other pro-longevity features discovered in bats include mutations of GH and IGF1 receptors (148), positive selection on double-stranded break (DSB) repair genes (197), resistance to oxidative stress (145), higher heat-shock proteins (26), a stable microbiome composition (74), and tolerance to transposons (129).
It is believed that the dampened inflammatory response in bats may have evolved as an adaptation to flight (13; 83; 121; 197) and crowded lifestyle (64). The ability to fly is one of longevity favoring features of bats, permitting the escape from predators (9). Remarkably, dampened immune response may also serve as a longevity-promoting adaptation by reducing chronic inflammation associated with aging in other species (64).
Similar to bats, pangolins are another group of species serving as a reservoir of different viruses without getting sick. In addition to bats, pangolins were also identified as possible intermediate host of the recent SARS-CoV-2 coronavirus (92; 198). Several studies of pangolin immune system revealed mechanisms that may contribute to their viral tolerance. Genome sequencing and analysis demonstrated that the interferon epsilon (IFNE) gene was pseudogenized in all African and Asian pangolin species (27). The IFNE gene is expressed exclusively in epithelial cells and inner mucosa-protected tissues, and serves an important role in protecting skin and mucosa from infection (33; 188). Frameshift and premature stop codons were identified in IFNE gene in pangolins (27). It was proposed that this mutation is associated with the evolution of keratinized scales in pangolins, which protect the skin from injuries and infection, and possibly replace the function of IFNE (27). Other genes of IFN family are intact in pangolins, but the number of IFNA genes is lower in pangolins than in humans (27). As a result of the reduced IFN-mediated immunity, many genes involved in other immunity-related pathways have undergone positive selection (27).
Unlike bats, which have enhanced RNA-sensing and reduced DNA-sensing, pangolins seem to have dampened both RNA- and DNA-sensing pathways. The interferon-induced with helicase C domain 1 (IFIH1) gene, which codes for a cytoplasmic RNA sensor MDA5, is inactivated by frameshift and premature stop mutations in exon 1 in three pangolin species (53). Z-DNA-binding protein (ZBP1) gene, which recognizes both Z-DNA and Z-RNA, was also pseudogenized by multiple premature stop mutations (53). The loss of IFIH1 and ZBP1 in pangolins occurred shortly after their evolutionary divergence from other mammalian lineages (53). DNA sensors cGAS and its interactor STING are also inactivated by frameshift and premature stop mutations (52). It would be important to experimentally confirm these genomic findings and identify potential compensatory pathways that allow pangolins to remain healthy despite dampened immunity.
5.2. Resistance to osteoarthritis in naked mole rats
Osteoarthritis (OA) is a chronic, degenerative joint disease associated with cartilage degeneration and joint inflammation (105). Aging is the major factor causing OA. The number of OA patients is predicted to increase with the proportion of elderly increasing. Other than aging, traumatic injuries can also cause OA (45). The effects of current treatments of OA are limited (168). The long-lived naked mole rats provide a good model for better treatment of OA. The cartilage of naked mole rats was found to be stiffer than that of mouse, and the naked mole rat chondrocytes are resistant to traumatic damage (168). Using meniscal-ligamentous injury (MLI) model, naked mole rat displayed much better recovery and showed no signs of degenerative changes, suggesting that naked mole rats are extremely resistant to OA (168). Interestingly, high-molecular-mass HA (HMM-HA) is abundant in naked mole rat cartilage, suggesting a potential role of HMM-HA in resistance to OA. This discovery encourages the use of HMM-HA in OA therapies.
6. ADAPTATIONS COUNTERACTING THE HALLMARKS OF AGING
Evolutionary adaptations that slow the rate of aging may increase resistance to multiple age-related diseases. It was proposed that aging is a result of accumulation of unrepaired cellular and molecular damage (87), which may eventually cause nutrient sensing dis-regulation, genomic instability, loss of proteostasis, stem cell exhaustion, among others (118). These changes are closely correlated with aging and age-associated diseases and thus are referred to as the hallmarks of aging (100). Some wild species have evolved mechanisms that counteract the hallmarks of aging (Figure 4). Studying these non-model animals with diverse lifespans may reveal novel mechanisms critical for longevity.
Figure 4. Mechanisms targeting the hallmarks of aging.
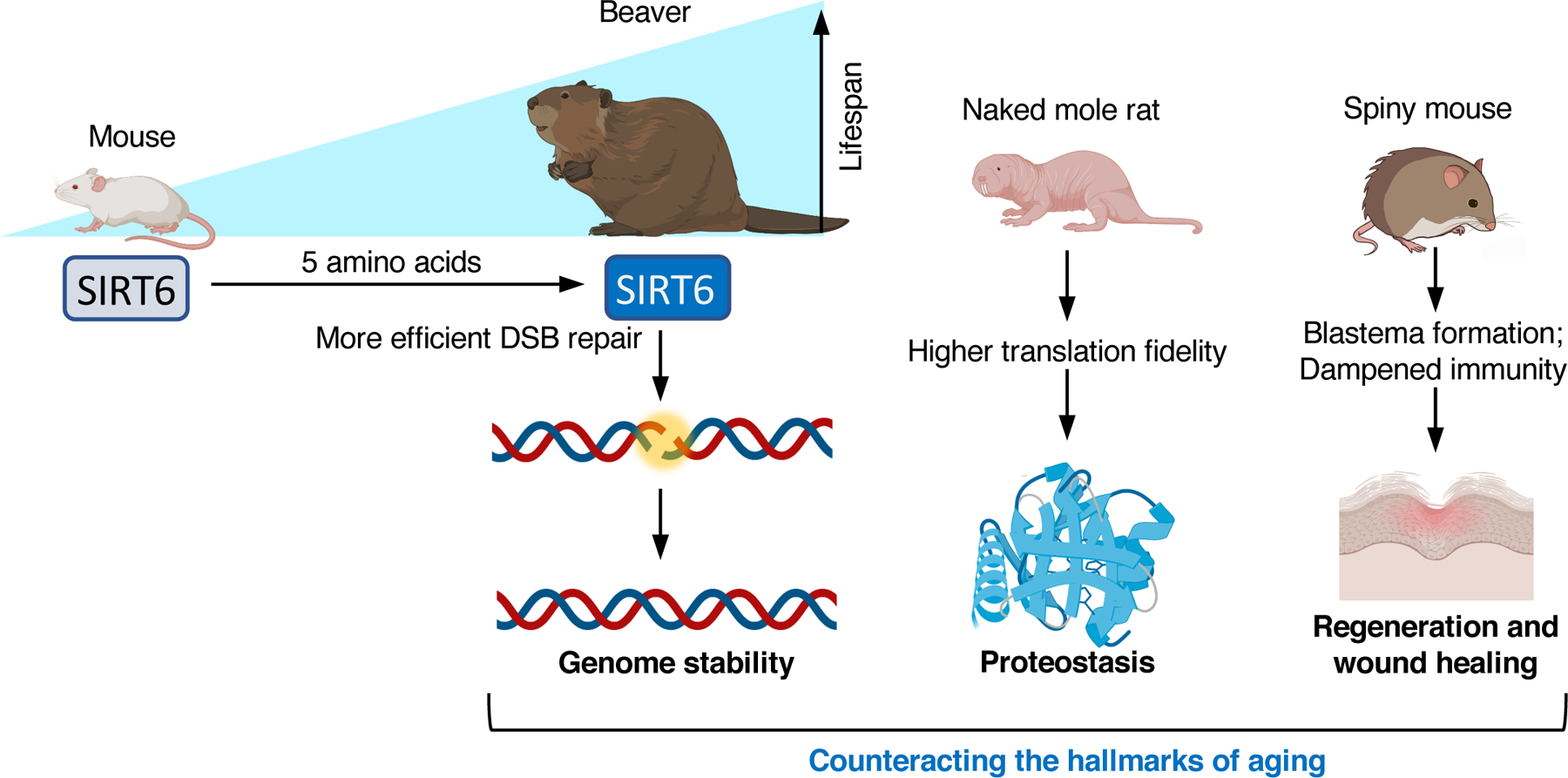
SIRT6-mediated double-stranded DNA (DSB) repair positively correlates with maximum lifespan across rodent species, with beaver having one of the highest DSB repair activities among rodents. Five amino acid substitutions in the SIRT6 gene determine the differential SIRT6 activity between beaver and mouse. Stronger DSB repair ability contributes to genome stability. Naked mole rat has higher translational fidelity, contributing to improved proteostasis. Spiny mouse has the strongest regenerative ability in mammals, due to the formation of blastema in wounded tissues, and a dampened immunity. Enhanced genome stability, proteostasis, and regenerative ability are all conserved mechanisms for longevity. These functions decline during aging and are considered aging hallmarks.
6.1. DNA repair and lifespan: from mouse to beaver
Genomic stability plays a pivotal role in longevity. DNA damage occurs every day. Without DNA repair, the accumulation of DNA damage may potentially cause mutations in critical genes like oncogenes and tumor suppressors. DNA damage has been proposed as a driving force of aging by causing genomic instability, cellular senescence, and apoptosis (185) and by destabilizing the epigenetic structure (63). Therefore, DNA repair is a key player in maintaining genomic stability. The role of DNA repair in aging is supported by multiple evidence that mutations in DNA repair genes result in premature aging phenotypes (71), however, it was harder to demonstrate that improved DNA repair would result in lifespan extension.
With comparative biology, it was possible to compare DNA repair capabilities of species with extreme lifespans differences. Comparison of transcriptomes of humans, naked mole rats, and mice, which have maximum lifespans of 120, 30, and 3 years, respectively, showed higher expression of DNA repair genes in long-lived species (humans and naked mole rats) compared with short-lived mice (101). Functional study of DNA repair was more extensively performed comparing 18 different rodent species. Skin and lung fibroblasts of all 18 rodent species were transfected with DNA repair reporter systems for two types of DNA double-stranded break (DSB) repair, homologous-recombination (HR) and non-homologous end joining (NHEJ). Both types of DSB repair showed strong positive correlation with the maximum lifespan of the species. The fibroblasts from longer-lived animals have better DSB repair than the cells from short-lived animals (176). SIRT6 is a histone deacetylase and mono-ADP-ribosylase enzyme involved in DNA repair and epigenome maintenance (103), reviewed in (25). The different DSB repair capabilities of the 18 rodent species depend largely on the different DSB repair activities of their Sirt6 gene, with the beaver having one of the most active SIRT6 and mouse having the least active SIRT6 (176). Further dissecting the SIRT6 sequences revealed five amino acids at the C-terminus that are fully responsible for the different enzymatic activities between beaver and mouse. Mouse SIRT6 carrying “beaverized” mutations resulted in more efficient DNA repair and increased lifespan, suggesting that SIRT6 activity is a determinant of lifespan (176). Therefore, protein features evolved in long-lived non-model animals provide important information for understanding aging and longevity.
6.2. High translation fidelity in the naked mole rat
The best way to address the accumulation of molecular damage, which drives aging, is to prevent it from happening in the first place. The error catastrophe theory of aging was proposed by Orgel (123–125). This theory proposed that translational fidelity plays an important role in aging. Although many in vitro studies challenged the error catastrophe theory, a theoretical study using molecular evolution simulation supported it by showing that mistranslation-induced protein misfolding acts as a strong selective pressure (42).
Using firefly luciferase reporters with various mutations that abrogate the luciferase, researchers showed that the naked mole rat cells have significantly higher translational fidelity than that of mouse (10). Coincidently, it was shown that the 28S rRNA of naked mole rat has a cleavage within the D6 region, resulting in two fragments of 2.5kb and 3kb (10). It was speculated that this cleaved 28S rRNA contributes to the higher translational fidelity of naked mole rats. Interestingly, the only other vertebrate known to have fragmented 28S rRNA is tuco-tuco, belonging to the genus of Ctenomys (107). Although phylogenetically distant from naked mole rats, Ctenomys shares similar subterranean habitats and social lifestyle (107). The maximum lifespan of Ctenomys in capacity is not determined, however, it has higher translation fidelity than many other rodents (85).
With the same luciferase reporter system, the translational fidelity of 17 rodent species with diverse lifespans was compared. The frequency of amino acid misincorporation at the first and second codon positions negatively correlated with maximum lifespan (85), suggesting that translational fidelity coevolves with longevity. As other long-lived rodent species compared in this study did not have the fragmented 28S rRNA, additional mechanisms must have played important roles in increasing the translational fidelity.
6.3. Enhanced regeneration: axolotl and spiny mouse
The ability to heal wounds and recover from injury decreases with age (58; 60; 65). Slower healing process in the elderly results in increased morbidity and mortality. Furthermore, approaches to improve regeneration would he equally valuable for younger trauma victims.
The axolotl (Ambystoma mexicanum) has been used as a model to study regeneration thanks to its remarkable capability for limb regeneration (82; 183). During limb regeneration, cartilage-derived blastema cells harbor positional identity to ensure the limb regrows properly (90). Multiple mutations of axolotl p53 were identified, which may contribute to the regulation of limb regeneration (184; 196). Interestingly, one of the mutations of axolotl p53 (Arg174Lys) was also observed in the blind mole rat (8). However, axolotl is not a mammal and some of the regenerative mechanisms discovered may be difficult to translate or may pose a cancer risk to humans.
The African spiny mice, Acomys, is the only mammal known to possess dramatically enhanced regenerative ability. Spiny mice have evolved a trait to escape from predators, termed autotomy (147). The skin of spiny mice can easily tear with very low tension. Afterwards, skin quickly regrows with no scar formation and with regenerated hair follicles (147). The remarkable regenerative ability of spiny mice also applies to ear wound healing, with complete regeneration of hair follicles, sebaceous glands, dermis and cartilage, resembling the structure of the axolotl blastema during limb regeneration (90; 147). Other than skin and ear wounds, other tissues are also highly regenerative in spiny mice, including skeletal muscle, heart, spinal cord, and kidney (146). It is speculated that the regenerative ability of spiny mice benefits from a dampened immune response following injury, expressing lower or absent inflammatory cytokines (21). Other characteristics of spiny mice fibroblasts also contribute to enhanced regeneration. Spiny mouse fibroblasts generate lower contractile forces (161) and produce more porous collagen network (147). Further studies of vertebrates with enhanced regenerative abilities will provide important avenues to promote tissue regeneration in humans.
CONCLUSIONS
Aging is a major risk factor for multiple diseases. Understanding the mechanisms of aging will significantly contribute to lifespan extension and disease prevention in humans. Nature has provided numerous wild species with remarkable diversity. Their lifespans vary up to 100-fold (166), from which novel anti-aging mechanisms can be learned. Studies on model animals, from worm to mice, revealed important conserved pathways responsible for aging, including insulin/IGF pathway, mTOR pathway, DNA repair, and Sirtuin protein family (177). Some of these pathways are modified in long-lived wild species. Examples include the mutations of GH and IGF receptors and resistance to oxidative stress in bats (145; 148), positive selection on DSB repair in whales and bats (86; 197), as well as strong DNA repair capabilities in long-lived rodents including beaver (101; 176). However, manipulating these conserved aging mechanisms in mice led to modest increases in lifespan (up to 10–15%), which cannot fully explain the large difference in lifespans between mammals. A possible reason is that naturally long-lived mammals have evolved multiple pro-longevity mechanisms, providing a synergetic effect. Combining different mechanisms learned from unconventional model animals may be a promising way to further extend lifespan. This underlies the importance of comprehensive studies on non-model animals.
It has been proposed that species living in protected environments tend to live longer, although the causal relationship is controversial (19; 186). Whales and elephants have large body sizes, protecting then from non-human predators (19); naked mole rats and blind mole rats are exposed to far fewer predators by living in subterranean burrows (61); and bats’ flight capability permits them to escape from predators (64; 72). With the low extrinsic death risk, these species have undergone selection for longevity. This prediction was further supported by the observation that arboreal mammals outlive their terrestrial counterparts (156).
Notably, although all these long-lived species seem to be selected for longevity due to protected environment, their longevity and disease-resistance mechanisms differ largely across species (Table 1). For example, naked mole rats live in subterranean burrows, where they rub their skin against underground tunnels. The HMM-HA has likely evolved to provide flexible skin in adaptation to subterranean lifestyle, while also providing protection from cancer. Likewise, dampened inflammation in bats coevolved with their flight and crowded lifestyle, while at the same time providing longevity benefit by protecting bats from inflammaging. Therefore, exploration of a broad spectrum of unconventional model animals reveals numerous anti-aging mechanisms that can be adopted to extend human healthspan and lifespan.
Table 1.
List of non-model mammals and their mechanisms and lifestyles associated with longevity and diseases.
| Age-associated disease | Species | Mechanisms | Lifestyle and ecological adaptation |
|---|---|---|---|
| Cancer | Naked mole rat | High-molecular-mass Hyaluronan (HMM-HA); Early contact inhibition; unique INK4 locus; epigenome stability | Subterranean burrows; hypoxia; protected environment |
| Blind mole rat | Interferon mediated concerted cell death | Subterranean; burrows; hypoxia; protected environment | |
| Elephant | Multiple p53 and LIF copies | Large size; lack of non-human predators | |
| Whale | Whale specific mutations in DNA repair genes | Large size; Lack of non-human predators; deep water diving; cold | |
| Cardiovascular disease | Bear | Protection and trophic effects by gene expression | Hibernation (responsive) with low metabolic rate |
| Ground squirrel | Gene mutation and expression; Protection from metabolic stresses and hypothermic arrhythmia | Hibernation (nonresponsive; periodic) with low metabolic rate | |
| Whale | Genes protecting from hypoxia | Deep water diving; cold | |
| Giraffe | Strong arterial pressure and resistance; Regulation of CBF | Tallest animal | |
| Neurodegenerative disease | Degu | AD-related genes more similar to humans | Social organization |
| Inflammatory diseases | Bat | Tolerance to viruses; reduced inflammation | Flight; crowded lifestyle |
| Pangolin | Tolerance to viruses; reduced inflammation | Defensive scale armor | |
| Naked mole rat | Resistance to osteoarthritis, possibly through HMM-HA | Longevity | |
| Counteracting hallmarks of aging | Beaver | Enhanced genome stability: High SIRT6-dependent DNA repair | Long-lived; large-sized rodent; semi-aquatic |
| Naked mole rat | Enhanced proteostasis: Higher translation fidelity | Longevity | |
| Spiny mice | Enhanced regeneration: Blastema formation; Dampened immunity | Skin autotomy and regeneration |
ACKNOWLEDGMENTS
This work was supported by grants from the US National Institutes of Health to A.S. and V.G.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, et al. 2015. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 314:1850–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abercrombie M 1979. Contact inhibition and malignancy. Nature 281:259–62 [DOI] [PubMed] [Google Scholar]
- 3.Agaba M, Ishengoma E, Miller WC, McGrath BC, Hudson CN, et al. 2016. Giraffe genome sequence reveals clues to its unique morphology and physiology. Nature communications 7:11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn M, Anderson DE, Zhang Q, Tan CW, Lim BL, et al. 2019. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat Microbiol 4:789–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn M, Cui J, Irving AT, Wang LF. 2016. Unique Loss of the PYHIN Gene Family in Bats Amongst Mammals: Implications for Inflammasome Sensing. Sci Rep 6:21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amstrup Funder J, Christian Danielsen C, Baandrup U, Martin Bibby B, Carl Andelius T, et al. 2017. How Heart Valves Evolve to Adapt to an Extreme-Pressure System: Morphologic and Biomechanical Properties of Giraffe Heart Valves. J Heart Valve Dis 26:63–71 [PubMed] [Google Scholar]
- 7.Ardiles AO, Tapia-Rojas CC, Mandal M, Alexandre F, Kirkwood A, et al. 2012. Postsynaptic dysfunction is associated with spatial and object recognition memory loss in a natural model of Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America 109:13835–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashur-Fabian O, Avivi A, Trakhtenbrot L, Adamsky K, Cohen M, et al. 2004. Evolution of p53 in hypoxia-stressed Spalax mimics human tumor mutation. Proceedings of the National Academy of Sciences of the United States of America 101:12236–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Austad SN, Fischer KE. 1991. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol 46:B47–53 [DOI] [PubMed] [Google Scholar]
- 10.Azpurua J, Ke Z, Chen IX, Zhang Q, Ermolenko DN, et al. 2013. Naked mole-rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proceedings of the National Academy of Sciences of the United States of America 110:17350–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baar MP, Brandt RMC, Putavet DA, Klein JDD, Derks KWJ, et al. 2017. Targeted Apoptosis of Senescent Cells Restores Tissue Homeostasis in Response to Chemotoxicity and Aging. Cell 169:132–47 e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, et al. 2016. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature 530:184–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee A, Baker ML, Kulcsar K, Misra V, Plowright R, Mossman K. 2020. Novel Insights Into Immune Systems of Bats. Frontiers in Immunology 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee A, Rapin N, Bollinger T, Misra V. 2017. Lack of inflammatory gene expression in bats: a unique role for a transcription repressor. Sci Rep 7:2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee A, Zhang X, Yip A, Schulz KS, Irving AT, et al. 2020. Positive Selection of a Serine Residue in Bat IRF3 Confers Enhanced Antiviral Protection. iScience 23:100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444:633–7 [DOI] [PubMed] [Google Scholar]
- 17.Ben-David U, Benvenisty N. 2011. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer 11:268–77 [DOI] [PubMed] [Google Scholar]
- 18.Berg von Linde M, Arevstrom L, Frobert O. 2015. Insights from the Den: How Hibernating Bears May Help Us Understand and Treat Human Disease. Clin Transl Sci 8:601–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blagosklonny MV. 2013. Big mice die young but large animals live longer. Aging (Albany NY) 5:227–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braidy N, Munoz P, Palacios AG, Castellano-Gonzalez G, Inestrosa NC, et al. 2012. Recent rodent models for Alzheimer’s disease: clinical implications and basic research. J Neural Transm (Vienna) 119:173–95 [DOI] [PubMed] [Google Scholar]
- 21.Brant JO, Yoon JH, Polvadore T, Barbazuk WB, Maden M. 2016. Cellular events during scar-free skin regeneration in the spiny mouse, Acomys. Wound Repair Regen 24:75–88 [DOI] [PubMed] [Google Scholar]
- 22.Braun K, Poeggel G. 2001. Recognition of Mother’s voice evokes metabolic activation in the medial prefrontal cortex and lateral thalamus of Octodon degus pups. Neuroscience 103:861–4 [DOI] [PubMed] [Google Scholar]
- 23.Buffenstein R 2008. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B 178:439–45 [DOI] [PubMed] [Google Scholar]
- 24.Buffenstein R, Jarvis JU. 2002. The naked mole rat--a new record for the oldest living rodent. Sci Aging Knowledge Environ 2002:pe7. [DOI] [PubMed] [Google Scholar]
- 25.Chang AR, Ferrer CM, Mostoslavsky R. 2020. SIRT6, a Mammalian Deacylase with Multitasking Abilities. Physiological reviews 100:145–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chionh YT, Cui J, Koh J, Mendenhall IH, Ng JHJ, et al. 2019. High basal heat-shock protein expression in bats confers resistance to cellular heat/oxidative stress. Cell stress & chaperones 24:835–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choo SW, Rayko M, Tan TK, Hari R, Komissarov A, et al. 2016. Pangolin genomes and the evolution of mammalian scales and immunity. Genome Res 26:1312–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colbert RW, Holley CT, Stone LH, Crampton M, Adabag S, et al. 2015. The Recovery of Hibernating Hearts Lies on a Spectrum: from Bears in Nature to Patients with Coronary Artery Disease. J Cardiovasc Transl Res 8:244–52 [DOI] [PubMed] [Google Scholar]
- 29.Cole JE, Steeil JC, Sarro SJ, Kerns KL, Cartoceti A. 2020. Chordoma of the sacrum of an adult naked mole-rat. J Vet Diagn Invest 32:132–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, et al. 2005. Tumour biology: senescence in premalignant tumours. Nature 436:642. [DOI] [PubMed] [Google Scholar]
- 31.Colonnello V, Iacobucci P, Fuchs T, Newberry RC, Panksepp J. 2011. Octodon degus. A useful animal model for social-affective neuroscience research: basic description of separation distress, social attachments and play. Neurosci Biobehav Rev 35:1854–63 [DOI] [PubMed] [Google Scholar]
- 32.Dave KR, Prado R, Raval AP, Drew KL, Perez-Pinzon MA. 2006. The arctic ground squirrel brain is resistant to injury from cardiac arrest during euthermia. Stroke 37:1261–5 [DOI] [PubMed] [Google Scholar]
- 33.Day SL, Ramshaw IA, Ramsay AJ, Ranasinghe C. 2008. Differential effects of the type I interferons alpha4, beta, and epsilon on antiviral activity and vaccine efficacy. J Immunol 180:7158–66 [DOI] [PubMed] [Google Scholar]
- 34.De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, et al. 2019. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566:73–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de la Torre JC. 2004. Is Alzheimer’s disease a neurodegenerative or a vascular disorder? Data, dogma, and dialectics. The Lancet. Neurology 3:184–90 [DOI] [PubMed] [Google Scholar]
- 36.Deacon RM, Altimiras FJ, Bazan-Leon EA, Pyarasani RD, Nachtigall FM, et al. 2015. Natural AD-Like Neuropathology in Octodon degus: Impaired Burrowing and Neuroinflammation. Curr Alzheimer Res 12:314–22 [DOI] [PubMed] [Google Scholar]
- 37.Delaney MA, Ward JM, Walsh TF, Chinnadurai SK, Kerns K, et al. 2016. Initial Case Reports of Cancer in Naked Mole-rats (Heterocephalus glaber). Vet Pathol 53:691–6 [DOI] [PubMed] [Google Scholar]
- 38.Dhingra R, Vasan RS. 2012. Age as a risk factor. Med Clin North Am 96:87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444:638–42 [DOI] [PubMed] [Google Scholar]
- 40.Diamond GA. 1989. Hibernating myocardium. Am Heart J 118:1361. [DOI] [PubMed] [Google Scholar]
- 41.Diamond GA, Forrester JS, deLuz PL, Wyatt HL, Swan HJ. 1978. Post-extrasystolic potentiation of ischemic myocardium by atrial stimulation. Am Heart J 95:204–9 [DOI] [PubMed] [Google Scholar]
- 42.Drummond DA, Wilke CO. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134:341–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egorov YV, Glukhov AV, Efimov IR, Rosenshtraukh LV. 2012. Hypothermia-induced spatially discordant action potential duration alternans and arrhythmogenesis in nonhibernating versus hibernating mammals. Am J Physiol Heart Circ Physiol 303:H1035–46 [DOI] [PubMed] [Google Scholar]
- 44.Elsasser A, Schlepper M, Klovekorn WP, Cai WJ, Zimmermann R, et al. 1997. Hibernating myocardium: an incomplete adaptation to ischemia. Circulation 96:2920–31 [DOI] [PubMed] [Google Scholar]
- 45.Englund M 2010. The role of biomechanics in the initiation and progression of OA of the knee. Best Pract Res Clin Rheumatol 24:39–46 [DOI] [PubMed] [Google Scholar]
- 46.Escalera-Zamudio M, Zepeda-Mendoza ML, Loza-Rubio E, Rojas-Anaya E, Mendez-Ojeda ML, et al. 2015. The evolution of bat nucleic acid-sensing Toll-like receptors. Mol Ecol 24:5899–909 [DOI] [PubMed] [Google Scholar]
- 47.Esquerda-Canals G, Montoliu-Gaya L, Guell-Bosch J, Villegas S. 2017. Mouse Models of Alzheimer’s Disease. J Alzheimers Dis 57:1171–83 [DOI] [PubMed] [Google Scholar]
- 48.Fang X, Nevo E, Han L, Levanon EY, Zhao J, et al. 2014. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nature communications 5:3966. [DOI] [PubMed] [Google Scholar]
- 49.Fazzalari A, Basadonna G, Kucukural A, Tanriverdi K, Koupenova M, et al. 2021. A Translational Model for Venous Thromboembolism: MicroRNA Expression in Hibernating Black Bears. J Surg Res 257:203–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fedorov VB, Goropashnaya AV, Toien O, Stewart NC, Chang C, et al. 2011. Modulation of gene expression in heart and liver of hibernating black bears (Ursus americanus). BMC Genomics 12:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fedorov VV, Li L, Glukhov A, Shishkina I, Aliev RR, et al. 2005. Hibernator Citellus undulatus maintains safe cardiac conduction and is protected against tachyarrhythmias during extreme hypothermia: possible role of Cx43 and Cx45 up-regulation. Heart Rhythm 2:966–75 [DOI] [PubMed] [Google Scholar]
- 52.Fischer H, Tschachler E, Eckhart L. 2020. Cytosolic DNA sensing through cGAS and STING is inactivated by gene mutations in pangolins. Apoptosis : an international journal on programmed cell death 25:474–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fischer H, Tschachler E, Eckhart L. 2020. Pangolins Lack IFIH1/MDA5, a Cytoplasmic RNA Sensor That Initiates Innate Immune Defense Upon Coronavirus Infection. Front Immunol 11:939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foidl BM, Humpel C. 2020. Can mouse models mimic sporadic Alzheimer’s disease? Neural Regen Res 15:401–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foote AD, Liu Y, Thomas GW, Vinar T, Alfoldi J, et al. 2015. Convergent evolution of the genomes of marine mammals. Nature genetics 47:272–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franceschi C, Bonafe M, Valensin S, Olivieri F, De Luca M, et al. 2000. Inflamm-aging. An evolutionary perspective on immunosenescence. Annals of the New York Academy of Sciences 908:244–54 [DOI] [PubMed] [Google Scholar]
- 57.Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. 2018. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 14:576–90 [DOI] [PubMed] [Google Scholar]
- 58.Gerstein AD, Phillips TJ, Rogers GS, Gilchrest BA. 1993. Wound healing and aging. Dermatol Clin 11:749–57 [PubMed] [Google Scholar]
- 59.Gillet LC, Scharer OD. 2006. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev 106:253–76 [DOI] [PubMed] [Google Scholar]
- 60.Goodson WH 3rd, Hunt TK. 1979. Wound healing and aging. The Journal of investigative dermatology 73:88–91 [DOI] [PubMed] [Google Scholar]
- 61.Gorbunova V, Bozzella MJ, Seluanov A. 2008. Rodents for comparative aging studies: from mice to beavers. Age 30:111–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gorbunova V, Hine C, Tian X, Ablaeva J, Gudkov AV, et al. 2012. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proceedings of the National Academy of Sciences of the United States of America 109:19392–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gorbunova V, Seluanov A. 2016. DNA double strand break repair, aging and the chromatin connection. Mutat Res 788:2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gorbunova V, Seluanov A, Kennedy BK. 2020. The World Goes Bats: Living Longer and Tolerating Viruses. Cell Metab 32:31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gosain A, DiPietro LA. 2004. Aging and wound healing. World J Surg 28:321–6 [DOI] [PubMed] [Google Scholar]
- 66.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464–8 [DOI] [PubMed] [Google Scholar]
- 67.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–74 [DOI] [PubMed] [Google Scholar]
- 68.Hargens AR, Millard RW, Pettersson K, Johansen K. 1987. Gravitational haemodynamics and oedema prevention in the giraffe. Nature 329:59–60 [DOI] [PubMed] [Google Scholar]
- 69.Harley CB, Futcher AB, Greider CW. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458–60 [DOI] [PubMed] [Google Scholar]
- 70.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, et al. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460:392–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. 2003. Aging and genome maintenance: lessons from the mouse? Science 299:1355–9 [DOI] [PubMed] [Google Scholar]
- 72.Holmes DJ, Austad SN. 1994. Fly ow, die later: life-history correlates of gliding and flying in mammals. Journal of Mammalogy 75:224–6 [Google Scholar]
- 73.Holmes MW, Bayliss MT, Muir H. 1988. Hyaluronic acid in human articular cartilage. Age-related changes in content and size. Biochem J 250:435–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hughes GM, Leech J, Puechmaille SJ, Lopez JV, Teeling EC. 2018. Is there a link between aging and microbiome diversity in exceptional mammalian longevity? PeerJ 6:e4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hurley MJ, Deacon RMJ, Beyer K, Ioannou E, Ibanez A, et al. 2018. The long-lived Octodon degus as a rodent drug discovery model for Alzheimer’s and other age-related diseases. Pharmacol Ther 188:36–44 [DOI] [PubMed] [Google Scholar]
- 76.Inestrosa NC, Reyes AE, Chacon MA, Cerpa W, Villalon A, et al. 2005. Human-like rodent amyloid-beta-peptide determines Alzheimer pathology in aged wild-type Octodon degu. Neurobiol Aging 26:1023–8 [DOI] [PubMed] [Google Scholar]
- 77.Inestrosa NC, Rios JA, Cisternas P, Tapia-Rojas C, Rivera DS, et al. 2015. Age Progression of Neuropathological Markers in the Brain of the Chilean Rodent Octodon degus, a Natural Model of Alzheimer’s Disease. Brain Pathol 25:679–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jebb D, Huang Z, Pippel M, Hughes GM, Lavrichenko K, et al. 2020. Six reference-quality genomes reveal evolution of bat adaptations. Nature 583:578–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jeck WR, Siebold AP, Sharpless NE. 2012. Review: a meta-analysis of GWAS and age-associated diseases. Aging Cell 11:727–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jensen FB. 2009. The role of nitrite in nitric oxide homeostasis: a comparative perspective. Biochimica et biophysica acta 1787:841–8 [DOI] [PubMed] [Google Scholar]
- 81.Jorgensen PG, Evans A, Kindberg J, Olsen LH, Galatius S, Frobert O. 2020. Cardiac adaptation in hibernating, free-ranging Scandinavian Brown Bears (Ursus arctos). Sci Rep 10:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Joven A, Elewa A, Simon A. 2019. Model systems for regeneration: salamanders. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kacprzyk J, Hughes GM, Palsson-McDermott EM, Quinn SR, Puechmaille SJ, et al. 2017. A potent anti-inflammatory response in bat macrophages may be linked to extended longevity and viral tolerance. Acta Chiropterol 19:219–28 [Google Scholar]
- 84.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, et al. 2012. The sirtuin SIRT6 regulates lifespan in male mice. Nature 483:218–21 [DOI] [PubMed] [Google Scholar]
- 85.Ke Z, Mallik P, Johnson AB, Luna F, Nevo E, et al. 2017. Translation fidelity coevolves with longevity. Aging Cell 16:988–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keane M, Semeiks J, Webb AE, Li YI, Quesada V, et al. 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell reports 10:112–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kirkwood TB. 2005. Understanding the odd science of aging. Cell 120:437–47 [DOI] [PubMed] [Google Scholar]
- 88.Kloner RA. 2020. Stunned and Hibernating Myocardium: Where Are We Nearly 4 Decades Later? J Am Heart Assoc 9:e015502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kothapalli D, Zhao L, Hawthorne EA, Cheng Y, Lee E, et al. 2007. Hyaluronan and CD44 antagonize mitogen-dependent cyclin D1 expression in mesenchymal cells. J Cell Biol 176:535–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, et al. 2009. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460:60–5 [DOI] [PubMed] [Google Scholar]
- 91.Laing ED, Sterling SL, Weir DL, Beauregard CR, Smith IL, et al. 2019. Enhanced Autophagy Contributes to Reduced Viral Infection in Black Flying Fox Cells. Viruses 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, et al. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583:282–5 [DOI] [PubMed] [Google Scholar]
- 93.Land H, Parada LF, Weinberg RA. 1983. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304:596–602 [DOI] [PubMed] [Google Scholar]
- 94.Lee SG, Mikhalchenko AE, Yim SH, Lobanov AV, Park JK, et al. 2017. Naked Mole Rat Induced Pluripotent Stem Cells and Their Contribution to Interspecific Chimera. Stem Cell Reports 9:1706–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, et al. 2013. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proceedings of the National Academy of Sciences of the United States of America 110:E89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Levenberg S, Yarden A, Kam Z, Geiger B. 1999. p27 is involved in N-cadherin-mediated contact inhibition of cell growth and S-phase entry. Oncogene 18:869–76 [DOI] [PubMed] [Google Scholar]
- 97.Li XC, Wei L, Zhang GQ, Bai ZL, Hu YY, et al. 2011. Ca2+ cycling in heart cells from ground squirrels: adaptive strategies for intracellular Ca2+ homeostasis. PloS one 6:e24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liang S, Mele J, Wu Y, Buffenstein R, Hornsby PJ. 2010. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber). Aging Cell 9:626–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lipman R, Galecki A, Burke DT, Miller RA. 2004. Genetic loci that influence cause of death in a heterogeneous mouse stock. J Gerontol A Biol Sci Med Sci 59:977–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153:1194–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.MacRae SL, Croken MM, Calder RB, Aliper A, Milholland B, et al. 2015. DNA repair in species with extreme lifespan differences. Aging (Albany NY) 7:1171–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Manov I, Hirsh M, Iancu TC, Malik A, Sotnichenko N, et al. 2013. Pronounced cancer resistance in a subterranean rodent, the blind mole-rat, Spalax: in vivo and in vitro evidence. BMC Biol 11:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mao Z, Hine C, Tian X, Van Meter M, Au M, et al. 2011. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332:1443–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marley AR, Nan H. 2016. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet 7:105–14 [PMC free article] [PubMed] [Google Scholar]
- 105.Martel-Pelletier J, Barr AJ, Cicuttini FM, Conaghan PG, Cooper C, et al. 2016. Osteoarthritis. Nat Rev Dis Primers 2:16072. [DOI] [PubMed] [Google Scholar]
- 106.Martens UM, Chavez EA, Poon SS, Schmoor C, Lansdorp PM. 2000. Accumulation of short telomeres in human fibroblasts prior to replicative senescence. Exp Cell Res 256:291–9 [DOI] [PubMed] [Google Scholar]
- 107.Melen GJ, Pesce CG, Rossi MS, Kornblihtt AR. 1999. Novel processing in a mammalian nuclear 28S pre-rRNA: tissue-specific elimination of an ‘intron’ bearing a hidden break site. The EMBO journal 18:3107–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, et al. 2011. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 334:521–4 [DOI] [PubMed] [Google Scholar]
- 109.Meza R, Jeon J, Moolgavkar SH, Luebeck EG. 2008. Age-specific incidence of cancer: Phases, transitions, and biological implications. Proceedings of the National Academy of Sciences of the United States of America 105:16284–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mitchell G, Bobbitt JP, Devries S. 2008. Cerebral perfusion pressure in giraffe: modelling the effects of head-raising and -lowering. J Theor Biol 252:98–108 [DOI] [PubMed] [Google Scholar]
- 111.Miyawaki S, Kawamura Y, Oiwa Y, Shimizu A, Hachiya T, et al. 2016. Tumour resistance in induced pluripotent stem cells derived from naked mole-rats. Nature communications 7:11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mugahid DA, Sengul TG, You X, Wang Y, Steil L, et al. 2019. Proteomic and Transcriptomic Changes in Hibernating Grizzly Bears Reveal Metabolic and Signaling Pathways that Protect against Muscle Atrophy. Sci Rep 9:19976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Munoz-Espin D, Canamero M, Maraver A, Gomez-Lopez G, Contreras J, et al. 2013. Programmed cell senescence during mammalian embryonic development. Cell 155:1104–18 [DOI] [PubMed] [Google Scholar]
- 114.Myers A, McGonigle P. 2019. Overview of Transgenic Mouse Models for Alzheimer’s Disease. Curr Protoc Neurosci 89:e81. [DOI] [PubMed] [Google Scholar]
- 115.Nelson OL, Robbins CT. 2010. Cardiac function adaptations in hibernating grizzly bears (Ursus arctos horribilis). J Comp Physiol B 180:465–73 [DOI] [PubMed] [Google Scholar]
- 116.Nelson OL, Robbins CT. 2015. Cardiovascular function in large to small hibernators: bears to ground squirrels. J Comp Physiol B 185:265–79 [DOI] [PubMed] [Google Scholar]
- 117.Nevo E 1999. Mosaic evolution of subterranean mammals : regression, progression, and global convergence. Oxford; New York: Oxford University Press. xxvi, 413 p. pp. [Google Scholar]
- 118.Niccoli T, Partridge L. 2012. Ageing as a risk factor for disease. Current biology : CB 22:R741–52 [DOI] [PubMed] [Google Scholar]
- 119.North BJ, Sinclair DA. 2012. The intersection between aging and cardiovascular disease. Circ Res 110:1097–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nowak RM. 1999. Walker’s mammals of the world. Baltimore: Johns Hopkins University Press [Google Scholar]
- 121.O’Shea TJ, Cryan PM, Cunningham AA, Fooks AR, Hayman DT, et al. 2014. Bat flight and zoonotic viruses. Emerg Infect Dis 20:741–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Okanoya K, Tokimoto N, Kumazawa N, Hihara S, Iriki A. 2008. Tool-use training in a species of rodent: the emergence of an optimal motor strategy and functional understanding. PloS one 3:e1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Orgel LE. 1963. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proceedings of the National Academy of Sciences of the United States of America 49:517–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Orgel LE. 1970. The maintenance of the accuracy of protein synthesis and its relevance to ageing: a correction. Proceedings of the National Academy of Sciences of the United States of America 67:1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Orgel LE. 1973. Ageing of clones of mammalian cells. Nature 243:441–5 [DOI] [PubMed] [Google Scholar]
- 126.Peto R, Roe FJ, Lee PN, Levy L, Clack J. 1975. Cancer and ageing in mice and men. Br J Cancer 32:411–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, et al. 1994. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes & development 8:9–22 [DOI] [PubMed] [Google Scholar]
- 128.Ponta H, Sherman L, Herrlich PA. 2003. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 4:33–45 [DOI] [PubMed] [Google Scholar]
- 129.Pritham EJ, Feschotte C. 2007. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proceedings of the National Academy of Sciences of the United States of America 104:1895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pure E, Assoian RK. 2009. Rheostatic signaling by CD44 and hyaluronan. Cell Signal 21:651–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Quinones QJ, Zhang Z, Ma Q, Smith MP, Soderblom E, et al. 2016. Proteomic Profiling Reveals Adaptive Responses to Surgical Myocardial Ischemia-Reperfusion in Hibernating Arctic Ground Squirrels Compared to Rats. Anesthesiology 124:1296–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Quirici V, Castro RA, Oyarzun J, Ebensperger LA. 2008. Female degus (Octodon degus) monitor their environment while foraging socially. Anim Cogn 11:441–8 [DOI] [PubMed] [Google Scholar]
- 133.Rahimtoola SH. 1989. The hibernating myocardium. Am Heart J 117:211–21 [DOI] [PubMed] [Google Scholar]
- 134.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. 2004. Species- and cell type-specific requirements for cellular transformation. Cancer Cell 6:171–83 [DOI] [PubMed] [Google Scholar]
- 135.Revsbech IG, Fago A. 2017. Regulation of blood oxygen transport in hibernating mammals. J Comp Physiol B 187:847–56 [DOI] [PubMed] [Google Scholar]
- 136.Rim SH, Seeff L, Ahmed F, King JB, Coughlin SS. 2009. Colorectal cancer incidence in the United States, 1999–2004 : an updated analysis of data from the National Program of Cancer Registries and the Surveillance, Epidemiology, and End Results Program. Cancer 115:1967–76 [DOI] [PubMed] [Google Scholar]
- 137.Ritchie H, Roser M. 2018. Causes of Death. https://ourworldindata.org/causes-of-death.
- 138.Rogina B, Helfand SL, Frankel S. 2002. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science 298:1745. [DOI] [PubMed] [Google Scholar]
- 139.Rose MR. 1991. Evolutionary biology of aging. New York: Oxford University Press. ix, 221 p. pp. [Google Scholar]
- 140.Rozhok AI, DeGregori J. 2015. Toward an evolutionary model of cancer: Considering the mechanisms that govern the fate of somatic mutations. Proceedings of the National Academy of Sciences of the United States of America 112:8914–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ruby JG, Smith M, Buffenstein R. 2018. Naked Mole-Rat mortality rates defy gompertzian laws by not increasing with age. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rutanen J, Leppanen P, Tuomisto TT, Rissanen TT, Hiltunen MO, et al. 2003. Vascular endothelial growth factor-D expression in human atherosclerotic lesions. Cardiovasc Res 59:971–9 [DOI] [PubMed] [Google Scholar]
- 143.Saha S, Panigrahi DP, Patil S, Bhutia SK. 2018. Autophagy in health and disease: A comprehensive review. Biomed Pharmacother 104:485–95 [DOI] [PubMed] [Google Scholar]
- 144.Salazar C, Valdivia G, Ardiles AO, Ewer J, Palacios AG. 2016. Genetic variants associated with neurodegenerative Alzheimer disease in natural models. Biol Res 49:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, et al. 2009. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 23:2317–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sandoval AGW, Maden M. 2020. Regeneration in the spiny mouse, Acomys, a new mammalian model. Curr Opin Genet Dev 64:31–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, Maden M. 2012. Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature 489:561–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seim I, Fang X, Xiong Z, Lobanov AV, Huang Z, et al. 2013. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nature communications 4:2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Seim I, Ma S, Zhou X, Gerashchenko MV, Lee SG, et al. 2014. The transcriptome of the bowhead whale Balaena mysticetus reveals adaptations of the longest-lived mammal. Aging (Albany NY) 6:879–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, et al. 2009. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326:140–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Seluanov A, Chen Z, Hine C, Sasahara TH, Ribeiro AA, et al. 2007. Telomerase activity coevolves with body mass not lifespan. Aging Cell 6:45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Seluanov A, Gladyshev VN, Vijg J, Gorbunova V. 2018. Mechanisms of cancer resistance in long-lived mammals. Nat Rev Cancer 18:433–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Seluanov A, Hine C, Azpurua J, Feigenson M, Bozzella M, et al. 2009. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proceedings of the National Academy of Sciences of the United States of America 106:19352–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Seluanov A, Hine C, Bozzella M, Hall A, Sasahara TH, et al. 2008. Distinct tumor suppressor mechanisms evolve in rodent species that differ in size and lifespan. Aging Cell 7:813–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593–602 [DOI] [PubMed] [Google Scholar]
- 156.Shattuck MR, Williams SA. 2010. Arboreality has allowed for the evolution of increased longevity in mammals. Proceedings of the National Academy of Sciences of the United States of America 107:4635–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Siegel RL, Miller KD, Fuchs HE, Jemal A. 2021. Cancer Statistics, 2021. CA Cancer J Clin 71:7–33 [DOI] [PubMed] [Google Scholar]
- 158.Simon M, Van Meter M, Ablaeva J, Ke Z, Gonzalez RS, et al. 2019. LINE1 Derepression in Aged Wild-Type and SIRT6-Deficient Mice Drives Inflammation. Cell Metab 29:871–85 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Singhal NS, Bai M, Lee EM, Luo S, Cook KR, Ma DK. 2020. Cytoprotection by a naturally occurring variant of ATP5G1 in Arctic ground squirrel neural progenitor cells. Elife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Smerup M, Damkjaer M, Brondum E, Baandrup UT, Kristiansen SB, et al. 2016. The thick left ventricular wall of the giraffe heart normalises wall tension, but limits stroke volume and cardiac output. J Exp Biol 219:457–63 [DOI] [PubMed] [Google Scholar]
- 161.Stewart DC, Serrano PN, Rubiano A, Yokosawa R, Sandler J, et al. 2018. Unique behavior of dermal cells from regenerative mammal, the African Spiny Mouse, in response to substrate stiffness. J Biomech 81:149–54 [DOI] [PubMed] [Google Scholar]
- 162.Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, et al. 2013. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155:1119–30 [DOI] [PubMed] [Google Scholar]
- 163.Sulak M, Fong L, Mika K, Chigurupati S, Yon L, et al. 2016. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Suva ML, Riggi N, Bernstein BE. 2013. Epigenetic reprogramming in cancer. Science 339:1567–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tacutu R, Budovsky A, Yanai H, Fraifeld VE. 2011. Molecular links between cellular senescence, longevity and age-related diseases - a systems biology perspective. Aging (Albany NY) 3:1178–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, et al. 2013. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic acids research 41:D1027–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tacutu R, Thornton D, Johnson E, Budovsky A, Barardo D, et al. 2018. Human Ageing Genomic Resources: new and updated databases. Nucleic acids research 46:D1083–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Taguchi T, Kotelsky A, Takasugi M, Chang M, Ke Z, et al. 2020. Naked mole-rats are extremely resistant to post-traumatic osteoarthritis. Aging Cell 19:e13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Takasugi M, Firsanov D, Tombline G, Ning H, Ablaeva J, et al. 2020. Naked mole-rat very-high-molecular-mass hyaluronan exhibits superior cytoprotective properties. Nature communications 11:2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tan L, Ke Z, Tombline G, Macoretta N, Hayes K, et al. 2017. Naked Mole Rat Cells Have a Stable Epigenome that Resists iPSC Reprogramming. Stem Cell Reports 9:1721–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tarumi T, Zhang R. 2018. Cerebral blood flow in normal aging adults: cardiovascular determinants, clinical implications, and aerobic fitness. J Neurochem 144:595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Taylor KR, Milone NA, Rodriguez CE. 2017. Four Cases of Spontaneous Neoplasia in the Naked Mole-Rat (Heterocephalus glaber), A Putative Cancer-Resistant Species. J Gerontol A Biol Sci Med Sci 72:38–43 [DOI] [PubMed] [Google Scholar]
- 173.Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, et al. 2013. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499:346–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tian X, Azpurua J, Ke Z, Augereau A, Zhang ZD, et al. 2015. INK4 locus of the tumor-resistant rodent, the naked mole rat, expresses a functional p15/p16 hybrid isoform. Proceedings of the National Academy of Sciences of the United States of America 112:1053–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tian X, Doerig K, Park R, Can Ran Qin A, Hwang C, et al. 2018. Evolution of telomere maintenance and tumour suppressor mechanisms across mammals. Philos Trans R Soc Lond B Biol Sci 373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tian X, Firsanov D, Zhang Z, Cheng Y, Luo L, et al. 2019. SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species. Cell 177:622–38 e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tian X, Seluanov A, Gorbunova V. 2017. Molecular Mechanisms Determining Lifespan in Short- and Long-Lived Species. Trends Endocrinol Metab 28:722–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Toole BP. 2004. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer 4:528–39 [DOI] [PubMed] [Google Scholar]
- 179.Toren D, Kulaga A, Jethva M, Rubin E, Snezhkina AV, et al. 2020. Gray whale transcriptome reveals longevity adaptations associated with DNA repair and ubiquitination. Aging Cell 19:e13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Toussaint O, Dumont P, Dierick JF, Pascal T, Frippiat C, et al. 2000. Stress-induced premature senescence. Essence of life, evolution, stress, and aging. Annals of the New York Academy of Sciences 908:85–98 [DOI] [PubMed] [Google Scholar]
- 181.Toussaint O, Medrano EE, von Zglinicki T. 2000. Cellular and molecular mechanisms of stress-induced premature senescence (SIPS) of human diploid fibroblasts and melanocytes. Experimental gerontology 35:927–45 [DOI] [PubMed] [Google Scholar]
- 182.Vazquez JM, Sulak M, Chigurupati S, Lynch VJ. 2018. A Zombie LIF Gene in Elephants Is Upregulated by TP53 to Induce Apoptosis in Response to DNA Damage. Cell reports 24:1765–76 [DOI] [PubMed] [Google Scholar]
- 183.Vieira WA, Wells KM, McCusker CD. 2020. Advancements to the Axolotl Model for Regeneration and Aging. Gerontology 66:212–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Villiard E, Brinkmann H, Moiseeva O, Mallette FA, Ferbeyre G, Roy S. 2007. Urodele p53 tolerates amino acid changes found in p53 variants linked to human cancer. BMC evolutionary biology 7:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.White RR, Vijg J. 2016. Do DNA Double-Strand Breaks Drive Aging? Mol Cell 63:729–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Williams GC. 1957. Pleiotropy, Natural Selection, and the Evolution of Senescence. Evolution; international journal of organic evolution 11:398–411 [Google Scholar]
- 187.Wylie A, Jones AE, D’Brot A, Lu WJ, Kurtz P, et al. 2016. p53 genes function to restrain mobile elements. Genes & development 30:64–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Xi Y, Day SL, Jackson RJ, Ranasinghe C. 2012. Role of novel type I interferon epsilon in viral infection and mucosal immunity. Mucosal Immunol 5:610–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xia X, Jiang Q, McDermott J, Han JJ. 2018. Aging and Alzheimer’s disease: Comparison and associations from molecular to system level. Aging Cell 17:e12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Xie J, Li Y, Shen X, Goh G, Zhu Y, et al. 2018. Dampened STING-Dependent Interferon Activation in Bats. Cell Host Microbe 23:297–301 e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xing CY, Tarumi T, Liu J, Zhang Y, Turner M, et al. 2017. Distribution of cardiac output to the brain across the adult lifespan. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism 37:2848–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xue W, Zender L, Miething C, Dickins RA, Hernando E, et al. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445:656–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yanai H, Fraifeld VE. 2018. The role of cellular senescence in aging through the prism of Koch-like criteria. Ageing Res Rev 41:18–33 [DOI] [PubMed] [Google Scholar]
- 194.Yim HS, Cho YS, Guang X, Kang SG, Jeong JY, et al. 2014. Minke whale genome and aquatic adaptation in cetaceans. Nature genetics 46:88–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Youm YH, Grant RW, McCabe LR, Albarado DC, Nguyen KY, et al. 2013. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab 18:519–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yun MH, Gates PB, Brockes JP. 2013. Regulation of p53 is critical for vertebrate limb regeneration. Proceedings of the National Academy of Sciences of the United States of America 110:17392–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, et al. 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339:456–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhang T, Wu Q, Zhang Z. 2020. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Current biology : CB 30:1346–51 e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhao J, Tian X, Zhu Y, Zhang Z, Rydkina E, et al. 2020. Reply to: Transformation of naked mole-rat cells. Nature 583:E8–E13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhao Y, Tyshkovskiy A, Munoz-Espin D, Tian X, Serrano M, et al. 2018. Naked mole rats can undergo developmental, oncogene-induced and DNA damage-induced cellular senescence. Proceedings of the National Academy of Sciences of the United States of America 115:1801–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhou P, Tachedjian M, Wynne JW, Boyd V, Cui J, et al. 2016. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proceedings of the National Academy of Sciences of the United States of America 113:2696–701 [DOI] [PMC free article] [PubMed] [Google Scholar]


