Abstract
Tuberculosis is an ancient human disease, estimated to have originated and evolved over thousands of years alongside modern human populations. Despite considerable advances in disease control, tuberculosis remains one of the world’s deadliest communicable diseases with 10 million incident cases and 1·8 million deaths in 2015 alone based on the annual WHO report, due to inadequate health service resources in less-developed regions of the world, and exacerbated by the HIV/AIDS pandemic and emergence of multidrug-resistant strains of Mycobacterium tuberculosis. Recent findings from studies of tuberculosis infection and of patients with Mendelian predisposition to severe tuberculosis have started to reveal human loci influencing tuberculosis outcomes. In this Review, we assess the current understanding of the contribution of host genetics to disease susceptibility and to drug treatment. Despite remarkable progress in technology, only a few associated genetic variants have so far been identified, strongly indicating the need for larger global studies that investigate both common and under-represented rare variants to develop new approaches to combat the disease. Pharmacogenomic discoveries are also likely to lead to more efficient drug design and development, and ultimately safer and more effective therapies for tuberculosis.
Background
Infectious diseases have ravaged mankind throughout the ages; therefore, they represent a major force in human evolution.1,2 Response to pathogen exposure is highly variable between people, and a better understanding of the biological basis of interindividual differences in infection outcomes could result in new preventive and therapeutic strategies. Exploration of the genetic underpinning of human susceptibility to infections started decades ago, but only increased in size and scope in the past few years due to newly available, more powerful approaches such as genome-wide association studies based on high-throughput genotyping and sequencing technologies.1,3,4
The potential of host genetic studies has been repeatedly shown for several infections of major public health importance. A prime example is malaria, which has exerted the strongest known selective pressure on the human genome since the start of agriculture, about 109000 years ago.5 Multiple genetic discoveries shed light on interactions between Plasmodium spp parasites and erythrocytes, including the protective effects of otherwise deleterious haemoglobin β gene variants (eg, HbS and HbC alleles) against severe disease6 or the complete protection against Plasmodium vivax conferred by a single nucleotide change in the Duffy antigen receptor for chemokines (DARC) gene, leading to no expression of this antigen receptor on red blood cells.7 Studies of chronic viral diseases also showed the ability of host genetics to uncover pathogenic mechanisms, by highlighting the role played by immune genes in shaping an individual’s response to infection. Differences were observed in immunogenetic susceptibility to HIV and hepatitis C virus: variation in adaptive immunity genes, particularly in the human leucocyte antigen class I gene family, was consistently shown as the most important determinant of HIV control,8 whereas innate immune mechanisms appeared to be more important for hepatitis C virus control, for which the strongest host contributor to spontaneous viral clearance was a variant of the interleukin 28B gene, encoding a type III interferon.9
Mycobacterium tuberculosis, the causative agent of tuberculosis, infects about a third of the world’s population. Individuals infected with M tuberculosis have a 10% lifetime risk of developing active tuberculosis. Despite remarkable advances in disease control in the past 15 years, tuberculosis ranks alongside HIV as the leading infectious cause of death worldwide: an estimated 1·8 million people died from tuberculosis in 2015, including 0·4 million resulting from tuberculosis disease in people living with HIV.10 M tuberculosis uses complex strategies for long-term survival within its host. Infection is initiated in the lungs through inhalation of a few bacteria contained within small aerosol droplets, which are engulfed by alveolar macrophages. Detection of mycobacterial pathogen-associated molecular patterns and damage-associated molecular patterns by the host cytosolic sensors within macrophages results in the induction of type I interferon responses, chemokines, and cytokines. These immune responses elicited by the host contribute substantially to bacterial containment and persistence within organised host structures in the lungs called granulomas.11 The progressive development of the granuloma lesion following infection involves a dynamic chemokine-mediated recruitment of myeloid cells—including dendritic cells, polymorphonuclear leukocytes, and myeloid-derived suppressor cells—and T and B lymphocytes, followed by the evolution of multinucleated giant cells and lipid-filled foamy macrophages encapsulated within fibrotic cuffs. Finetuning of pro-mycobacterial and antimycobacterial effects mediated by the cells within the granuloma and by their chemical mediators, while limiting bacterial growth, also suppress immune responses and provide a survival niche from which the bacteria can ultimately disseminate in immuno compromised individuals. Components of the type I interferon and inflammatory cytokine pathways are thus known to mediate host resistance and susceptibility both in animal models and human beings.
Host genetic studies of tuberculosis have been challenging for various reasons, including difficulties in phenotype definition and probable genetic heterogeneity.12 Nevertheless, recent findings from genome-wide scans and analyses of patients with Mendelian susceptibility to mycobacterial disease have started to reveal human vulnerabilities against tuberculosis. In this Review, we present a perspective on tuberculosis host genomics, based in part on the workshop “Host response to TB-HIV infection: a genomic perspective” organised by the National Institute of Health (Rockville, MD, USA, Jan 13–14, 2015), describing the latest progress and discussing the opportunities and challenges of human genetic approaches in tuberculosis research.
Early observations on the genetic basis for tuberculosis susceptibility
In Lübeck, Germany, from Dec 10, 1929, to April 30, 1930, 251 neonates were accidentally given preparations of the BCG vaccine that had been contaminated with virulent M tuberculosis.13 Of the vaccinated children, 72 died of tuberculosis and the surviving 174 children (five children died of non-tuberculosis causes) presented with a wide range of clinical presentations. A careful follow-up investigation of the accident identified variable concentrations of contamination (ie, the infectious dose) as the main source of variable clinical outcomes.13 However, even when estimated dose was considered, substantial clinical variability remained, ranging from death to very mild disease. This pronounced variability of outcome in the absence of other known environmental or societal factors might represent differences in the innate, genetically controlled ability of neonates to fight off M tuberculosis. Outbreaks of tuberculosis have been documented in members of the Navy and Marine Corps service, who are at increased risk of exposure because of close living quarters, ventilation systems aboard ships, and their travel to countries where tuberculosis is endemic. In reported instances of outbreak, although about 20% of previously unaffected sailors were found to be newly positive by tuberculin skin test (TST), results from screening of all sailors showed little transmission of M tuberculosis, despite months of potential exposure in a high-risk setting,14–16 as determined by close sleeping quarters in bunks arranged in stacks with air intake that exhausted directly overboard, and sailors in adjacent compartments connected with the patient’s. Deployments of ships spread over several weeks to months. Maximum theoretical transmission in the absence of genetic-based resistance is difficult to determine, but the closest would be intra-household transmission in susceptible individuals. For example, a simulation study indicates that the prevalence of M tuberculosis infection in household contacts in Pakistan is about 49.4%.17
Genetic basis of resistance is also substantiated by findings that previously underexposed populations are more susceptible to the disease than overexposed populations, as evidenced by the effects of European colonialism. Tuberculosis brought in by colonising Europeans has played a large part in deaths among Qu’Appelle Indians in the Saskatchewan province, Canada, in 1890, with more than half the families lost in the first three generations of the epidemic before considerable fall in death rates that could be attributed to strong selection against susceptibility genes.18 Through contact with European explorers, whalers, and missionaries, Inuit populations in Nunavut, Canada, had increasingly succumbed to many new diseases including tuberculosis.19
Tuberculosis was first introduced to Yanomami Indians in the Amazon rain forests through contact with gold miners of European descent, and since 1980 has become an epidemic even in BCG-vaccinated individuals. A study of this population showed reduced cell-mediated immunity and an increased antibody response relative to populations with extensive previous contact with M tuberculosis, again suggesting that tuberculosis exerts a selective pressure on immune responses and on human evolution.20 The protective efficacy of BCG against pulmonary tuberculosis has varied widely in different parts of the world. BCG vaccinations in Yanomami Indians did not enhance the proportion of low-grade purified protein derivative reactions, commonly observed after BCG immunisation or exposure to environmental mycobacteria in other populations, strongly suggesting an intrinsic un responsiveness of this population on first exposure to mycobacterial antigens.
Earlier studies of twins have shown that the concordance of tuberculosis was higher in monozygotic twins than in dizygotic twins under comparable environmental and social conditions,21 and these findings were further confirmed by a reanalysis in the Prophit survey.22 Studies of racially integrated nursing homes in Arkansas, USA, have shown that African Americans are twice as susceptible as people of white descent to infection with M tuberculosis, a finding that could not again be explained by environmental or social factors.23 These studies have helped to reinforce the concept of genetic susceptibility to tuberculosis.
Insight from inbred animal models of tuberculosis susceptibility
Mice and rabbits have often been used in studies to investigate genetic susceptibility to tuberculosis. Crossbreeding between resistant and susceptible inbred mice strains has shown that resistance to tuberculosis follows a complex, non-Mendelian inheritance.24 In 1981, Gros and colleagues25 found that susceptibility to infection with avirulent BCG derived from Mycobacterium bovis was determined by a host genetic locus Bcg on mouse chromosome 1. Subsequently, the gene encoding the natural resistance-associated macrophage protein 1 (Nramp1) was identified within this locus.26 Targeted disruption of Nramp1 also eliminated resistance to other infectious agents such as Leishmania donovani and Salmonella typhimurium.27 The human orthologue of mouse Nramp1 is designated NRAMP1 or SLC11A1, and is involved in susceptibility to tuberculosis. In the mouse however, Nramp1 does not affect susceptibility to virulent M tuberculosis.28,29
In a study investigating susceptibility to M tuberculosis in mice, Kramnik and colleagues29 mapped a new locus designated sst1 (susceptibility to tuberculosis 1) on mouse chromosome 1, 10–19 centimorgans distal to Nramp1. The phenotypic expression of sst1 differs from Nramp1 in that sst1 exerted a lung-specific effect. sst1-susceptible C3HeB/FeJ mice developed lung lesions characterised by extensive necrosis and uncontrolled multiplication of M tuberculosis, whereas sst1-resistant mice developed interstitial granulomas with controlled multiplication of the bacilli. Positional cloning identified the Ipr1 (intracellular pathogen resistance 1) isoform of the Ifi75 (interferon-inducible-75) gene within the sst1 locus as imparting increased susceptibility of the C3HeB/FeJ mice to M tuberculosis.30 These genetically defined mouse models provide important tools to study the pathogenesis of tuberculosis.
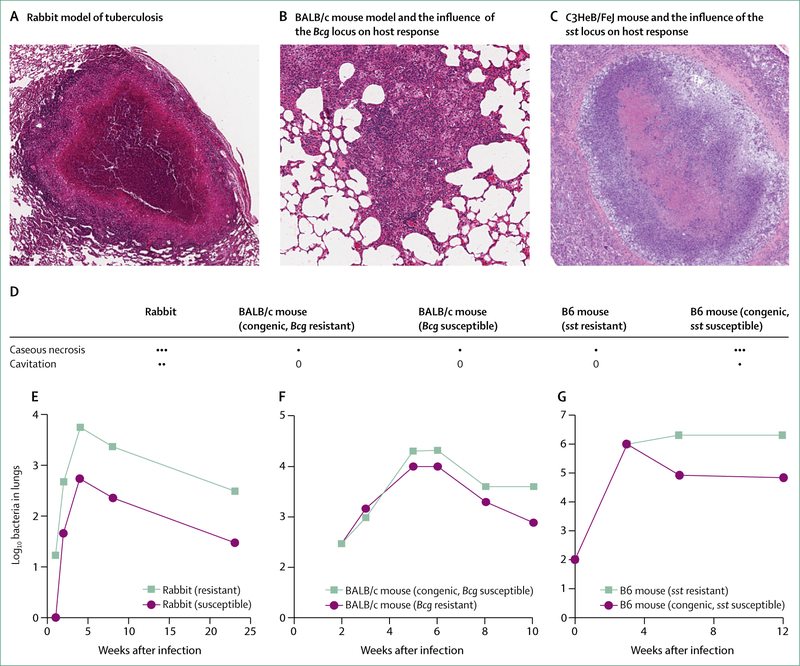
In the 1940s, Max Lurie31,32 did detailed studies over a period of 7 years on inbred families of rabbits to investigate natural resistance to tuberculosis. Lurie found that inbred strains exhibited either resistance or susceptibility to infection with virulent M bovis, and that although resistant families developed cavitary tuberculosis, susceptible families developed disseminated disease. Rabbits that were resistant lived about twice as long as susceptible ones. The inheritance pattern suggested that resistance was predominantly genetically determined. Subsequently, recognition of differential susceptibility of inbred laboratory animal strains to tuberculosis infections has enabled linkage studies and targeted gene disruptions to identify host determinants of disease susceptibility. The influence of host genetics on disease response to tuberculosis in rabbit and mouse models is indicated in figure 1.
Figure 1: Influence of host genetics on disease response to tuberculosis in rabbit and mouse models.
(A) Image captured at 40x magnification. A granuloma from a rabbit, showing central necrosis and tissue destruction. (B) Image captured at 100x magnification. A granuloma from a BALB/c mouse. (C) Image captured at 40x magnification. A necrotising granuloma from a C3HeB/FeJ mouse. Differences in pathology and host response in the C3HeB/FeJ mouse were mapped to an allele in the sst locus. (D) Pathological response to Mycobacterium tuberculosis infection in disease models compared with the pattern observed in human disease.33 (E) Pattern of bacterial burden after infection in the resistant and susceptible inbred rabbits of Max Lurie.33 (F) Pattern of bacterial burden after infection in BALB/c mice carrying a BCG-susceptible allele mapped to the Bcg locus.34 (G) Pattern of bacterial burden after infection in B6 mice compared with congenic B6 mice engineered with the sst-susceptible locus characterised in C3HeB/FeJ mice.35 ···=human disease hallmark frequently observed in model. ··=human disease hallmark sometimes observed in model. ·=human disease hallmark rarely observed in model. 0=human disease pattern not observed in model.
Infection with M bovis, a member of the M tuberculosis complex, causes tuberculosis in cattle. M bovis has a wide host range, including a number of wild ruminant animals, is infectious in human beings, and causes substantial economic hardship for farmers worldwide. Zoonotic transmission of M bovis occurs primarily through ingestion of unpasteurised milk or close contact with infected cattle. Although the global prevalence of human tuberculosis due to M bovis is low (<2%), increased prevalence has been reported in African countries, Mexico, Turkey, and India.36 In efforts to identify genotypes resistant to M bovis for the generation of resistant animal populations, Bhaladhare and colleagues37 found that single nucleotide polymorphisms (SNPs) in TLR2, a host gene important in immune sensing of pathogen-associated molecular patterns, was associated with susceptibility to M bovis in cattle. Association between TLR mutations and SNPs in TLR2 and TLR4 and increased susceptibility to Mycobacterium avium paratuberculosis has also been previously reported in cattle.38 These studies prompt additional validation in larger groups before screening and breeding of resistant animal populations can be envisaged.
Human genetics of tuberculosis
Genetic control of tuberculosis infection
There is no direct test for infection with M tuberculosis, and the latent tuberculosis infection phenotype is inferred indirectly from quantitative measurements of antimycobacterial immunity.11,39 TST is the most widely used method to test for latent infection.40 Additionally, two in-vitro blood assays measuring either the secretion of interferon γ by lymphocytes or the frequency of interferon-γ-producing blood cells in response to M tuberculosis antigens (interferon-γ release assays [IGRAs]) have been developed.41 TST and IGRAs assess different aspects of antimycobacterial immunity and are not fully concordant in predicting infection with M tuberculosis.42,43 Little or no reactivity in these tests in individuals heavily exposed to M tuberculosis is indicative of innate resistance to M tuberculosis infection. In household studies, 30–50% of contacts with heavy short-term exposure do not become infected,41 revealing substantial heterogeneity in susceptibility to infection.
Several studies have considered TST and IGRA results as quantitative traits and have shown substantial heritability for both tests following exposure to M tuberculosis. In The Gambia, a twin study estimated the heritability of TST response at 71% and IGRA response at 39%.44 The heritability of quantitative TST reactivity in children exposed to an active tuberculosis case has been estimated at 92% in Chile.45 A complex segregation analysis of TST reactivity in household contacts of tuberculosis cases in a Colombian population provided evidence for a major co-dominant gene accounting for about 65% of TST variability.46 The heritability of interferon-γ secretion has been estimated at about 43% following BCG stimulation and 58% following ESAT-6 stimulation in South Africa,47 and at between 17% and 48% following stimulation with M tuberculosis antigens, including ESAT-6, in Uganda.48,49
Few studies have aimed to identify the genetic variants underlying the detected heritability. Candidate gene association studies have focused on TST response as a binary trait, with the most interesting association found with an IL10 promoter haplotype in Ghana50 and Brazil.51 A recent genome-wide association study in a large Icelandic population identified some HLA class II variants as associated with TST positivity with weak effect (odds ratio [OR] 1·14).52 In Uganda, a genome-wide linkage analysis suggested that persistent TST negativity was linked to chromosomal regions (2)(q21;q24) and (5)(p13;q22).53 A study of TST reactivity in a sample of multiplex families from South Africa identified two major loci affecting TST responses more than 0 mm (TST1 on chromosome 11p14), and the intensity of TST-reactivity (TST2 on chromosome 5p15).54 The TST1 locus was replicated in a household contact study of French families of various ethnic origins.55 The TST1 locus might reflect innate resistance to infection with M tuberculosis whereas TST2 might account for T-cell-mediated antimycobacterial immune responses. It was subsequently discovered that a locus affecting the production of tumour necrosis factor (TNF) by blood cells in response to BCG and BCG plus interferon γ, TNF1, was genetically indistinguishable from TST1.56 This finding raises the exciting possibility that innate resistance to M tuberculosis infection might involve a TNF-mediated effector mechanism.
Finally, a linkage study identified two loci on chromosomes 8q and 3q that are controlling BCG-triggered and ESAT-6 triggered interferon-γ production, respectively, in populations of various ethnic origins living in different M tuberculosis exposure settings.57
Genetic control of severe primary tuberculosis
Many factors are known to influence the risk of progression from latent infection to clinical disease. However, as a general rule of thumb, the lifetime risk of tuberculosis in a child infected with M tuberculosis is approximately 10%.58 In a large population survey in the Netherlands, approximately two-thirds of patients who had tuberculosis developed their clinical disease within 2 years after infection,59 and it is possible that the majority of tuberculosis cases occur in this period. In highly endemic countries, this rapid onset of primary tuberculosis is particularly common in children, some of whom developing a haematogenous disseminated form (referred to in this Review as severe primary tuberculosis).13
The first molecular evidence that childhood tuberculosis might reflect a Mendelian predisposition came from the observation of severe tuberculosis in children with classic primary immunodeficiencies,60 such as chronic granulomatous disease61 or more recently complete TYK2 deficiency.62 Further progress came from the study of the syndrome of Mendelian susceptibility to mycobacterial diseases (MSMDs), which is defined by a selective vulnerability to weakly virulent non-tuberculous Mycobacterium species, such as BCG and environmental mycobacteria.63,64 Since 1996, germline mutations in seven autosomal (IFNGR1, IFNGR2, IL12B, IL12RB1, STAT1, IRF8, and ISG15) and two X-linked (NEMO and CYBB) genes have been discovered in patients with MSMDs.63,64 These defects are physiologically related, because they all result in an impairment of interferon-γ immunity. Several patients with MSMDs, particularly those with interferon-γR165,66 and interleukin-12p4067 deficiencies, have been shown to have infections due to both weakly virulent mycobacteria and M tuberculosis, raising the hypothesis that tuberculosis observed in these patients could also be attributed to a monogenic predisposition.60
Additional evidence came when several siblings of patients with MSMDs carrying the same genetic defect as the index case were found to display severe tuberculosis as their sole infectious phenotype. This finding was the case in a child with partial interferon-γR1 deficiency,68 and in a male participant from a large multiplex X-linked kindred carrying a specific mutation of CYBB impairing the interferon-?-dependent respiratory burst in macrophages.69 Several patients with tuberculosis and mutations in MSMD genes were also identified without any familial history of MSMDs.60 The most common genetic defect identified in patients with severe tuberculosis to date is complete interleukin-12Rβ1 deficiency,70 which was observed in several families.61 In a more systematic search for IL12RB1 mutations in 50 children with severe tuberculosis, two patients with complete interleukin-12Rβ1 deficiency were identified.71 Overall, these results provided proof of concept for monogenic predisposition to severe tuberculosis, and raised the possibility that severe tuberculosis could be due in a substantial proportion of children to single-gene inborn errors of immunity. This proportion has been estimated at up to 45% by theoretical calculations,72 and can be determined experimentally by next-generation sequencing approaches. These findings have paved the way for new treatments based on physiopathology. The best example is provided by patients with interleukin-12Rβ1 deficiencies, who can be treated by recombinant human interferon-? in addition to antimycobacterial drugs.73
Genetic control of pulmonary tuberculosis
Most patients with latent tuberculosis infections (90–95%) never develop clinical disease. The remaining 5–10% develop clinical tuberculosis later in life, typically pulmonary tuberculosis, due to reactivation of the original infection or, in other cases, to secondary infection.11,74 The development of pulmonary tuberculosis reflects an impairment of host resistance to M tuberculosis that might be favoured in a minority of patients by acquired immunodeficiency, such as HIV infection or anti-TNF treatment. However, in most people who do not have any overt immunodeficiency, the pathogenesis of reactivation remains elusive. There is strong evidence that the development of pulmonary tuberculosis is influenced by host genetic factors,12,75 in particular based on familial aggregation studies76 such as twin studies.21 Most classic genetic association studies investigating pulmonary tuberculosis have focused on candidate genes, and several common risk variants have been reported in immunity-related genes such as those encoding DC-SIGN, TLR1 and TLR2, vitamin D receptor, TNF, interleukin 1β, interferon γ, or some HLA class II molecules.77 However, there has been an absence of consistency and replication between most of the reported results of independent studies, partly due to underpowered studies, and strong heterogeneity across phenotype definition and epidemiological settings.77,78 One of the most convincing findings has been the identification of associated polymorphisms of the NRAMP1 gene, with an effect that is heterogeneous across populations, epidemiological settings, age, and clinical phenotypes.79–81 The importance of considering age at tuberculosis onset in host genetic analyses was further supported by the positional cloning of a major locus on chromosome 8q conferring predisposition to pulmonary tuberculosis.82 The association mapping of the linked region identified variants of the TOX gene as strongly associated with the development of early-onset pulmonary tuberculosis (before 25 years of age) in populations from Morocco and Madagascar.83 TOX encodes a nuclear factor involved in the development of T cells,84 particularly the CD4-positive T cells essential for immunity to mycobacteria.
Conversely, genome-wide association studies of pulmonary tuberculosis, considered as a single phenotype, have met with little success to date. Studies from Ghana and The Gambia,85,86 and another from Russia87 have led to the identification of only three signals that reached the genome-wide threshold for statistical significance (59×910™8). One of the two variants identified in the African genome-wide association study, rs4331426, is located in a gene desert on chromosome 18q11.286 and the other, rs2057178, is near WT1 on chromosome 11p13,85 and both had modest effect sizes (OR 1·19 and 0·77, respectively). The chromosome 11 signal was replicated in populations from Indonesia and Russia in the original study,85 and in independent investigations in South African88 and Moroccan89 populations. Attempts to replicate the chromosome 18q11.2 signal provided more conflicting results in Chinese90,91 and Moroccan89 populations. The Russian study identified a cluster of intronic ASAP1 variants in a study population of more than 159000 participants, also with a weak effect size (OR 0·84 for SNP rs4733781), with possible functional involvement in dendritic cell mobility.87 The Icelandic genome-wide association study52 also reported the association of HLA class II variants with pulmonary tuberculosis, although the precise role of these variants in tuberculosis infection or clinical tuberculosis, or both, remains to be clarified. Finally, a genome-wide association study of 581 patients with HIV (267 with active tuberculosis) from Uganda and Tanzania identified rs4921437 at chromosome 5q33, close to the IL12B gene, with a stronger protective effect size (OR 0·37) than those previously reported in HIV-negative populations.92 A striking feature of these studies is the inadequacy of replication of the pulmonary tuberculosis susceptibility factors previously detected in candidate gene analyses.4 Overall, the results from the genome-wide association studies suggest that common variants might have a little effect on individual predisposition to adult pulmonary tuberculosis, at least when considered as a single homogeneous phenotype.
Pharmacogenomics
Isoniazid
Human genetic polymorphisms affect antimicrobial pharmacokinetics and pharmacodynamics. Isoniazid is the most extensively studied anti-tuberculosis drug in this regard. It undergoes acetylation by hepatic arylamine N-acetyltransferase 2 (encoded by NAT2),93 and NAT2 loss-of-function alleles are common,93,94 with slow acetylator allele frequencies of approximately 45% with Asian, 55% with African, and 75% with European ancestry. One copy of such alleles confers an intermediate acetylator phenotype and two copies confer a slow acetylator phenotype, and increased plasma isoniazid exposure.93,94 These polymorphisms have also been associated with risk for isoniazid hepatotoxicity. In individuals prescribed isoniazid for latent tuberculosis in the USA, raised hepatic trans aminase concentrations to greater than five times the upper limit of normal were reported in approximately one case per 1000 people completing therapy,95 although risk increases with concomitant rifampicin and other factors.96 Associations between NAT2 polymorphisms and isoniazid hepatotoxicity were shown in meta-analyses involving multiple cohorts with tuberculosis, with ORs ranging from 1·93 to 4·69.97–99
Evidence for genetic associations with isoniazid hepatotoxicity beyond NAT2 is scarce. Studies have suggested associations with CYP2E1 polymorphisms,99–102 alone or with NAT2 alleles,94,97,101 but either associations have not been replicated103–105 or were seen only in east Asians.99 A meta-analysis suggested an association with GSTM1 polymorphisms,99 but not in all ancestries.99,100,106 Associations in other genes have not been replicated.106 A small study in Japan randomised patients with tuberculosis to standard isoniazid doses (5 mg/kg) versus individualised doses based on NAT2 genotype.107 Benefit was suggested, with fewer treatment failures in rapid acetylators who received higher doses than standard doses (15% vs 38%) and fewer hepatotoxicity cases in slow acetylators who received lower doses than standard doses (0% vs 78%), although failure and hepatotoxicity percentages seem excessive.
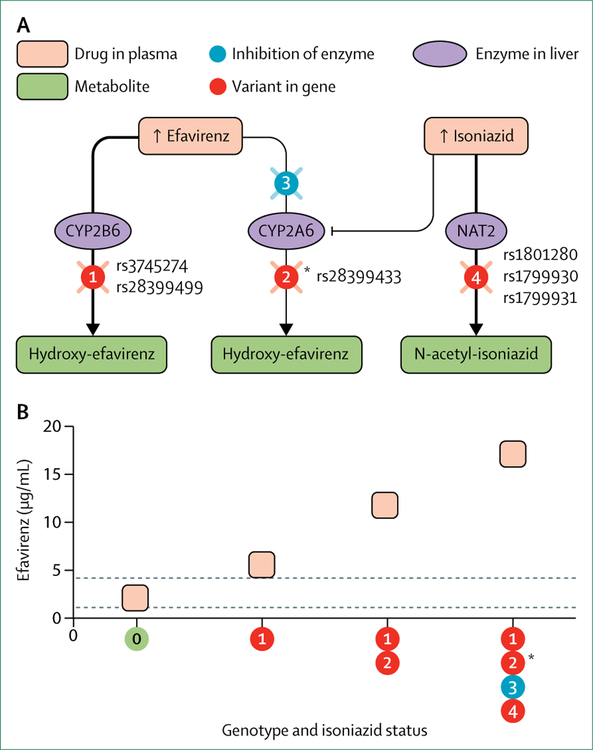
A complex drug–drug interaction involves isoniazid, the antiretroviral drug efavirenz, and several genes. Rifampicin induces hepatic cytochrome P450 (CYP) 2B6, so should decrease plasma efavirenz exposure.108 However, in some patients who are prescribed rifampicin with isoniazid, plasma efavirenz exposure paradoxically increases, particularly with CYP2B6 and NAT2 loss-of-function genotypes (figure 2).109–111 High isoniazid concentrations in NAT2 slow acetylators apparently inhibit CYP2A6, a necessary pathway for efavirenz clearance in CYP2B6 slow metabolisers.110–112 Isoniazid induced peripheral neuropathy might increase with NAT2 slow acetylator alleles.113 In five Japanese patients who had nerve biopsies for isoniazid-induced neuropathy, all were slow acetylators.114
Figure 2: Complex drug–drug interaction between isoniazid and efavirenz, and involving several pharmacogenes.
(A) Efavirenz is converted to inactive metabolites by cytochrome P450 (CYP) 2B6, with minor metabolism by CYP2A6. Individuals carrying two CYP2B6 loss-of-function alleles (rs3745274 or rs28399499, or both; circle 1) have increased plasma efavirenz exposure, which is further increased with concomitant CYP2A6 loss-of-function polymorphisms (eg, rs28399433; circle 2). When individuals with two CYP2B6 loss-of-function alleles are prescribed isoniazid, inhibition of CYP2A6 by isoniazid can also increase efavirenz exposure (circle 3), particularly with concomitant NAT2 genotypes that increase plasma isoniazid exposure (eg, rs1801280, rs1799930, and rs1799931; circle 4). (B) Depicts approximate plasma efavirenz trough concentrations by CYP2B6, CYP2A, and NAT2 genotype and isoniazid exposure. *Data are absent for CYP2A6 in isoniazid recipients with concomitant CYP2B6 and NAT2 loss-of-function genotypes, but this combination of CYP2B6, NAT2, and CYP2A6 genotypes is expected to confer even higher efavirenz concentrations.
Other anti-tuberculosis drugs
Rifampicin is a substrate for organic anion-transporting polypeptide 1B1 (coded by SLCO1B1),115 and one study associated an SLCO1B1 polymorphism (rs4149032) with rifampicin bioavailability in South Africans.116 It is not known whether immune response genes affect susceptibility to rifampicin hypersensitivity, as with other antimicrobials.117 Parenteral aminoglycosides can cause sensorineural hearing loss,118–122 and mitochondrial DNA mutations in the gene that encodes 12S ribosomal RNA confer increased risk.118,121,123,124 The mitochondrial 1555A→G risk mutation121,122,124 is present in as many as 5% of southeast Asians and approximately 0·2% of Europeans.
Linezolid is being prescribed for multidrug-resistant tuberculosis. Evidence suggests that neuropathy, myelosuppression, and hyperlactataemia with linezolid might reflect inhibition of mitochondrial protein synthesis. It is conceivable that mitochondrial mutations will affect susceptibility to these adverse events.125–130
Given the immense global burden of tuberculosis, there is opportunity for pharmacogenomic discoveries to have profound impact. Progress in this area will be facilitated by access to well phenotyped clinical datasets, including datasets that represent newer agents, with linkage to well consented DNA banks. This effect, together with advanced technologies such as high-throughput next generation sequencing, should lead to more efficient drug discovery, design, and development, and ultimately safer and more effective therapies for tuberculosis.
Future direction and perspectives
The identification of the genetic factors involved in tuberculosis susceptibility has proven difficult. As in several other human diseases, the role of common variants in pulmonary tuberculosis seems to be scarce, leading to the concept of missing heritability.131 Among the several non-mutually exclusive hypotheses that might account for this concept, an important one in tuberculosis is the oversimplification of the studied phenotypes.12 For example, tests used to assess tuberculosis infection display little concordance, and each assay captures a different aspect of the antimycobacterial response.42 In pulmonary tuberculosis, the most commonly used phenotype-defining characteristic is the presence of M tuberculosis in the sputum of patients, regardless of the other clinical, microbiological, and demographical covariables. This approach ignores the dynamic nature of pulmonary tuberculosis, and the likelihood of different stages of this process being under different genetic controls, as shown in mouse models of the BCG infection.132 The need for more homogeneous and refined tuberculosis phenotypes for genetic studies is also shown by the strong impact of age at onset of disease on the ability to detect genetic effects in clinical tuberculosis.81,83,133 Moreover, it is possible that the infectious dose has a pivotal role in tuberculosis susceptibility and that host genetic studies would be more informative if done in sporadic cases that are more likely to have received a low infectious inoculum.13
Another major aspect that has been mostly ignored so far is the possible effect of the M tuberculosis strain on latent tuberculosis infection and clinical outcomes.134,135 Evidence is accumulating that human-adapted M tuberculosis has been coevolving with modern human beings over millennia,136,137 strongly suggesting the importance of host–pathogen interaction in the different tuberculosis infection and clinical outcomes.134,138 Although some associations between human genetic variants and specific M tuberculosis lineages have been reported,139 large scale studies of interaction between human polymorphisms and pathogen variants that have been done in HIV140 are missing in tuberculosis. HIV infection has a major influence in tuberculosis infection and clinical outcomes. However, little progress has been made in the genetics of tuberculosis–HIV co-infection, with only few inconclusive association studies.139 More comprehensive studies are needed in the context of co-infection that might take advantage of extreme phenotypes such as patients with HIV infection and low CD4-positive T-cell counts who do not develop tuberculosis infection or disease despite sustained exposure. An interesting example was provided by the recent genome-wide association study92 done in participants with HIV infection that identified a locus with a strong protective effect that needs to be replicated in other samples. Likewise, the study of the genetic basis of the immune reconstitution inflammatory syndrome occurring in patients with tuberculosis–HIV co-infection after initiation of an antiretroviral treatment,141 which have similarities with reversal reactions observed in leprosy,142 will be of major interest.
Several genetic explanations might also account for the missing heritability observed in tuberculosis including genetic and allelic heterogeneity, and epistatic (gene × gene interaction) effects. Among these explanations, the contribution of rare variants to tuberculosis pathogenesis deserves attention for two main reasons: conceptually, rare variants bridge the gap between Mendelian and complex inheritance, and could account for the major loci identified through linkage studies;1,80,143 and experimentally, these variants abound in the genome can now be identified by whole-exome and whole-genome sequencing,144–146 and analysed by appropriate methods.147,148 Working on improved specified phenotypes will be pivotal for studies based on next-generation sequencing methods that typically need to be done on extreme phenotypes (eg, existence of familial cases, early age at onset, severity of the disease, or absence of comorbidities).
Transcriptomic studies, including the search for loci involved in the expression of genes (expression quantitative trait loci) in cell-specific or tissue-specific studies,149 based on RNA-sequencing, in particular,150 are also of major interest for genetic dissection of tuberculosis. A transcriptomic analysis of peripheral blood cells identified a neutrophil-driven interferon-a-inducible and interferon-β-inducible transcript signature in individuals with active pulmonary tuberculosis.151 This blood signature was validated in independent studies,152–154 and was shown to be different from that of several other infectious and pulmonary diseases,151,155 providing new insight into the most relevant pathways and candidate biomarkers for investigation in tuberculosis.156–158 A recent transcriptomic study provided evidence for a TLR and inflammasome signature in immune reconstitution inflammatory syndrome occurring in patients with tuberculosis–HIV co-infection.159 Studies done in monocyte-derived dendritic cells have also provided new insights in the identification of expression quantitative trait loci associated with M tuberculosis infection.160 Infection of dendritic cells with M tuberculosis also resulted in the up-regulation or downregulation of 155 microRNAs (miRNAs), and these changes in miRNA expression were partly under genetic control.161
The transcriptional response is strongly affected by epigenetic modifications of DNA-encoded regulatory elements in the 5'-upstream region of genes and more distal enhancers. Key molecular events are methylation and acetylation of histone proteins that result in opening of chromatin and the binding of transcriptions factors (figure 3). The epigenetic landscape of many cell types has been profiled and is available in public databases such as ENCODE. However, only one study investigated the changes in DNA methylation of human dendritic cells after infection with M tuberculosis.162M tuberculosis infection triggered the rapid demethylation of thousands of regulatory sites preferentially corresponding to distal enhancer regions (figure 3). Demethylation affected expression of adjacent genes and was associated with extensive remodelling of the epigenetic landscape of infected cells.162 A second independent line of investigation has shown a striking association between epigenetic programming of monocyte to macrophage and trained innate immunity.163–165 Collectively, these studies identify epigenetic changes as a key factor in the understanding of cellular responses to infections.
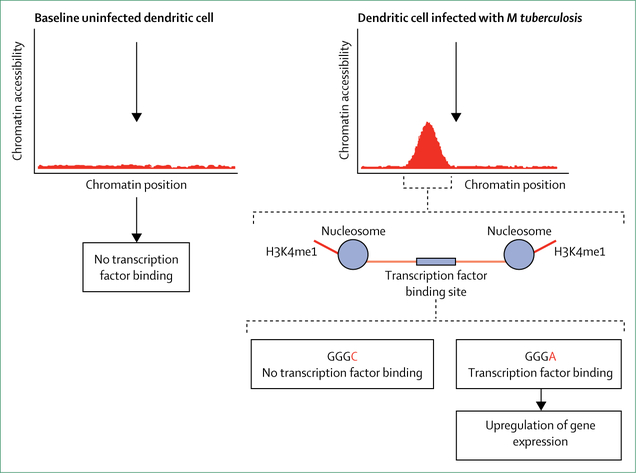
Figure 3: Interplay between epigenetic remodelling of the host cell by Mycobacterium tuberculosis and the role of germline variants in controlling host gene expression.
Opening of chromatin, a prerequisite for binding of transcription factors, is induced by histone modifications such as monomethylation at lysine 4 in histone H3 (H3K4me1). In the example shown, M tuberculosis triggered H3K4me1 histone modification that mediates chromatin accessibility in a regulatory region that carries a transcription factor binding site (GGGC or GGGA). Only the GGGA allele allows efficient binding of the transcription factor and induction of gene expression, whereas the GGGC allele does not.
Conclusion
Historical observations, heritability estimates, linkage analyses, and genome-wide scans indicate that human resistance to tuberculosis infection has a strong genetic basis, which is likely to be a consequence of evolutionary counter-response to microbial virulence. Mendelian studies have also provided the first evidence that severe tuberculosis in children could be due to single gene inborn errors of immunity. However, much is yet to be learned regarding susceptibility to disease, in particular pulmonary tuberculosis, from studies of human genetic diversity and of interactions between host and pathogen genomes. Despite remarkable progress in genotyping and sequencing technology and bioinformatic analysis, only a few associated genetic variants have been identified, strongly indicating the need for an improved definition of the phenotype, and larger studies investigating not only common variants, but also rare single nucleotide and structural variants (copy number variation, insertions, and deletions) that are under-represented on available genotyping platforms.
Although the Hapmap Project and the 1000 Genomes Project have greatly enhanced our understanding of human genetic variation globally, efforts to study African genetic diversity have been rather scarce. Therefore, synergies with the African Genome Variation Project,166 an international collaboration examining genetic diversity among 18 ethnolinguistic groups of sub-Saharan Africa where tuberculosis is endemic, become highly imperative. In this regard, a recent transcriptomic analysis of whole blood samples from independent South African and Gambian cohorts prospectively identified people at risk of developing active tuberculosis.167 In this predictive signature of 16 genes, the gene module that was predominantly represented was interferon responses, suggesting that chronic peripheral immune activation precedes onset of active disease. The tuberculosis risk signature predicted disease progression despite substantial diversity between the study cohorts, raising the possibility for targeted intervention to prevent the disease.
Recent observations in a model population of genetically diverse mice suggest that a single vaccine might not be able to protect all individuals, further emphasising the need to understand immunological variations that determine tuberculosis susceptibility and vaccine efficacy.168 Finally, whether variants associated with disease are truly causative and of any clinical relevance needs to be determined, emphasising the need to study better specified clinical and tuberculosis-relevant cellular phenotypes.
Search strategy and selection criteria.
We identified data for this Review from MEDLINE and PubMed with the search terms “host genomics AND tuberculosis”, “susceptibility to tuberculosis”, and “innate resistance to tuberculosis”. Because the Review traces the history of tuberculosis through the 20th century including the colonial period, we have included articles published in English from the database search between Jan 1, 1940, and January 31, 2017. We also identified data from our earlier work and from the references cited in these publications.
Acknowledgments
We thank all colleagues of our laboratories and workshop participants for their discussions and contributions. LA was supported in part by grants from the European Research Council (ERC-2010-AdG-268777), the French National Agency for Research (TBPAT HGEN-ANR-14-CE14-0007-01), the St Giles Foundation, the Rockefeller University, and the National Institute of Allergy and Infectious Diseases (U01AI088685, R01AI089970, U19AI111143, and R37AI095983). JF is supported by a Professorship grant from the Swiss National Science Foundation (PP00P3_133703). The work of DWH was supported in part by grants from the National Institutes of Health (R01 AI077505 and P30 AI110527). Work in the laboratory of ES is supported by a Foundation grant of the Canadian Institutes of Health Research (FDN-1433322) and a grant from the National Institutes of Health (RO1AI124349). WRB acknowledges the support of the National Institutes of Health grants (AI36973 and AI37856), and funding from the Howard Hughes Medical Institute.
Footnotes
Declaration of interests
We declare no competing interests.
For the ENCODE project see www.encodeproject.org
Contributor Information
Laurent Abel, Laboratory of Human Genetics of Infectious Diseases, INSERM U1163, Paris, France; Paris Descartes University, Imagine Institute, Paris, France; St Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York City, NY, USA.
Jacques Fellay, School of Life Sciences, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland; SIB Swiss Institute of Bioinformatics, Lausanne, Switzerland.
David W Haas, Division of Infectious Diseases, Department of Medicine, Vanderbilt University School of Medicine, Nashville, TN, USA; Department of Internal Medicine, Meharry Medical College, Nashville, TN, USA.
Erwin Schurr, McGill International TB Centre, The Research Institute of the McGill University Health Centre, Montreal, QC, Canada.
Geetha Srikrishna, Center for Tuberculosis Research, Department of Medicine Division of Infectious Diseases, Johns Hopkins School of Medicine, Baltimore, MD, USA.
Michael Urbanowski, Center for Tuberculosis Research, Department of Medicine Division of Infectious Diseases, Johns Hopkins School of Medicine, Baltimore, MD, USA.
Nimisha Chaturvedi, School of Life Sciences, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland; SIB Swiss Institute of Bioinformatics, Lausanne, Switzerland.
Sudha Srinivasan, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA.
Daniel H Johnson, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA.
William R Bishai, Center for Tuberculosis Research, Department of Medicine Division of Infectious Diseases, Johns Hopkins School of Medicine, Baltimore, MD, USA.
References
- 1.Casanova JL, Abel L. The genetic theory of infectious diseases: a brief history and selected illustrations. Annu Rev Genomics Hum Genet 2013; 14: 215–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quintana-Murci L, Clark AG. Population genetic tools for dissecting innate immunity in humans. Nat Rev Immunol 2013; 13: 280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet 2012; 13: 175–88. [DOI] [PubMed] [Google Scholar]
- 4.Abel L, Alcais A, Schurr E. The dissection of complex susceptibility to infectious disease: bacterial, viral and parasitic infections. Curr Opin Immunol 2014; 30: 72–78. [DOI] [PubMed] [Google Scholar]
- 5.Joy DA, Feng X, Mu J, et al. Early origin and recent expansion of Plasmodium falciparum. Science 2003; 300: 318–21. [DOI] [PubMed] [Google Scholar]
- 6.Malaria Genomic Epidemiology Network. Reappraisal of known malaria resistance loci in a large multicenter study. Nat Genet 2014; 46: 1197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet 1995; 10: 224–28. [DOI] [PubMed] [Google Scholar]
- 8.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science 2007; 317: 944–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 2009; 461: 798–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO. Global tuberculosis report 2015.2015. http://www.who.int/tb/publications/global_report/en/ (accessed March 25, 2016).
- 11.O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. The immune response in tuberculosis. Annu Rev Immunol 2013; 31: 475–527. [DOI] [PubMed] [Google Scholar]
- 12.Abel L, El-Baghdadi J, Bousfiha AA, Casanova JL, Schurr E. Human genetics of tuberculosis: a long and winding road. Philos Trans R Soc Lond B Biol Sci 2014; 369: 20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox G, Orlova M, Schurr E. Tuberculosis in newborns: the lessons of the “Lübeck disaster” (1929–1933). PLoS Pathog 2016; 12: e1005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navy and Marine Corps Public Health Center. Tuberculosis report: tuberculosis in the Navy and Marine Corps, 2016 report. 2016. http://www.med.navy.mil/sites/nmcphc/Documents/program-and-policy-support/NMCPHC-Tuberculosis-Report-2016.pdf (accessed Jan 6, 2017).
- 15.DiStasio AJ 2nd, Trump DH The investigation of a tuberculosis outbreak in the closed environment of a US Navy ship, 1987. Mil Med 1990; 155: 347–51. [PubMed] [Google Scholar]
- 16.Lamar JE 2nd, Malakooti MA. Tuberculosis outbreak investigation of a US Navy amphibious ship crew and the Marine expeditionary unit aboard, 1998. Mil Med 2003; 168: 523–27. [PubMed] [Google Scholar]
- 17.Akhtar S, Carpenter TE, Rathi SK. A chain-binomial model for intra-household spread of Mycobacterium tuberculosis in a low socio-economic setting in Pakistan. Epidemiol Infect 2007; 135: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lux M Perfect subjects: race, tuberculosis, and the Qu’Appelle BCG vaccine trial. Can Bull Med Hist 1998; 15: 277–95. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald N, Hebert PC, Stanbrook MB.Tuberculosis in Nunavut: a century of failure. CMAJ 2011; 183: 741–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sousa AO, Salem JI, Lee FK, et al. An epidemic of tuberculosis with a high rate of tuberculin anergy among a population previously unexposed to tuberculosis, the Yanomami Indians of the Brazilian Amazon. Proc Natl Acad Sci USA 1997; 94: 13227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kallman FJ, Reisner D. Twin studies on the significance of genetic factors in tuberculosis. Am Rev Tuberc 1942; 47: 549–74. [Google Scholar]
- 22.Comstock GW. Tuberculosis in twins: a re-analysis of the Prophit study. Am Rev Respir Dis 1978; 117: 621–24. [DOI] [PubMed] [Google Scholar]
- 23.Stead WW, Senner JW, Reddick WT, Lofgren JP. Racial differences in susceptibility to infection by Mycobacterium tuberculosis. N Engl J Med 1990; 322: 422–27. [DOI] [PubMed] [Google Scholar]
- 24.Lynch CJ, Pierce-Chase CH, Dubos R. A genetic study of susceptibility to experimental tuberculosis in mice infected with mammalian tubercle bacilli. J Exp Med 1965; 121: 1051–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gros P, Skamene E, Forget A. Genetic control of natural resistance to Mycobacterium bovis (BCG) in mice. J Immunol 1981; 127: 2417–21. [PubMed] [Google Scholar]
- 26.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell 1993; 73: 469–85. [DOI] [PubMed] [Google Scholar]
- 27.Vidal S, Tremblay ML, Govoni G, et al. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med 1995; 182: 655–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina E, North RJ. The Bcg gene (Nramp1) does not determine resistance of mice to virulent Mycobacterium tuberculosis. Ann N Y Acad Sci 1996; 797: 257–59. [DOI] [PubMed] [Google Scholar]
- 29.Kramnik I, Dietrich WF, Demant P, Bloom BR. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc Natl Acad Sci USA 2000; 97: 8560–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan H, Yan BS, Rojas M, et al. Ipr1 gene mediates innate immunity to tuberculosis. Nature 2005; 434: 767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lurie MB. Hereditary, constitution and tuberculosis. An experimental study. Am Rev Tuberc 1941; 44 (suppl 1): 1–125. [Google Scholar]
- 32.Lurie MB. Experimental epidemiology of tuberculosis: hereditary resistance to attack by tuberculosis and to the ensuing disease and the effect of the concentration of tubercle bacilli upon these two phases of resistance. J Exp Med 1944; 79: 573–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dannenberg AM Jr. Pathogenesis of human pulmonary tuberculosis: insights from the rabbit model. Washington: ASM Press, 2006. [Google Scholar]
- 34.Brown DH, Miles BA, Zwilling BS. Growth of Mycobacterium tuberculosis in BCG-resistant and -susceptible mice: establishment of latency and reactivation. Infect Immun 1995; 63: 2243–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan BS, Pichugin AV, Jobe O, et al. Progression of pulmonary tuberculosis and efficiency of bacillus Calmette–Guerin vaccination are genetically controlled via a common sst1-mediated mechanism of innate immunity. J Immunol 2007; 179: 6919–32. [DOI] [PubMed] [Google Scholar]
- 36.Olea-Popelka F, Muwonge A, Perera A, et al. Zoonotic tuberculosis in human beings caused by Mycobacterium bovis—a call for action. Lancet Infect Dis 2017; 17: e21–25 [DOI] [PubMed] [Google Scholar]
- 37.Bhaladhare A, Sharma D, Kumar A, et al. Single nucleotide polymorphisms in toll-like receptor genes and case-control association studies with bovine tuberculosis. Vet World 2016; 9: 458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mucha R, Bhide MR, Chakurkar EB, Novak M, Mikula I Sr. Toll-like receptors TLR1, TLR2 and TLR4 gene mutations and natural resistance to Mycobacterium avium subsp. paratuberculosis infection in cattle. Vet Immunol Immunopathol 2009; 128: 381–88. [DOI] [PubMed] [Google Scholar]
- 39.Ernst JD. The immunological life cycle of tuberculosis. Nat Rev Immunol 2012; 12: 581–91. [DOI] [PubMed] [Google Scholar]
- 40.Reichman LB. Tuberculin skin testing. The state of the art. Chest 1979; 76 (suppl 6): S764–70. [DOI] [PubMed] [Google Scholar]
- 41.Gallant CJ, Cobat A, Simkin L, et al. Tuberculin skin test and in vitro assays provide complementary measures of antimycobacterial immunity in children and adolescents. Chest 2010; 137: 1071–77. [DOI] [PubMed] [Google Scholar]
- 42.Pai M, Denkinger CM, Kik SV, et al. Gamma interferon release assays for detection of Mycobacterium tuberculosis infection. Clin Microbiol Rev 2014; 27: 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marks SM, Taylor Z, Qualls NL, Shrestha-Kuwahara RJ, Wilce MA, Nguyen CH. Outcomes of contact investigations of infectious tuberculosis patients. Am J Respir Crit Care Med 2000; 162: 2033–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jepson A, Fowler A, Banya W, et al. Genetic regulation of acquired immune responses to antigens of Mycobacterium tuberculosis: a study of twins in West Africa. Infect Immun 2001; 69: 3989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sepulveda RL, Heiba IM, King A, Gonzalez B, Elston RC, Sorensen RU. Evaluation of tuberculin reactivity in BCG-immunized siblings. Am J Respir Crit Care Med 1994; 149: 620–24. [DOI] [PubMed] [Google Scholar]
- 46.Cobat A, Barrera LF, Henao H, et al. Tuberculin skin test reactivity is dependent on host genetic background in Colombian tuberculosis household contacts. Clin Infect Dis 2012; 54: 968–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cobat A, Gallant CJ, Simkin L, et al. High heritability of antimycobacterial immunity in an area of hyperendemicity for tuberculosis disease. J Infect Dis 2010; 201: 15–19. [DOI] [PubMed] [Google Scholar]
- 48.Stein CM, Guwatudde D, Nakakeeto M, et al. Heritability analysis of cytokines as intermediate phenotypes of tuberculosis. J Infect Dis 2003; 187: 1679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao L, Zalwango S, Chervenak K, et al. Genetic and shared environmental influences on interferon-gamma production in response to Mycobacterium tuberculosis antigens in a Ugandan population. Am J Trop Med Hyg 2013; 89: 169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thye T, Browne EN, Chinbuah MA, et al. IL10 haplotype associated with tuberculin skin test response but not with pulmonary TB. PLoS One 2009; 4: e5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zembrzuski VM, Basta PC, Callegari-Jacques SM, et al. Cytokine genes are associated with tuberculin skin test response in a native Brazilian population. Tuberculosis 2010; 90: 44–49. [DOI] [PubMed] [Google Scholar]
- 52.Sveinbjornsson G, Gudbjartsson DF, Halldorsson BV, et al. HLA class II sequence variants influence tuberculosis risk in populations of European ancestry. Nat Genet 2016; 48: 318–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein CM, Zalwango S, Malone LL, et al. Genome scan of M. tuberculosis infection and disease in Ugandans. PLoS One 2008; 3: e4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cobat A, Gallant CJ, Simkin L, et al. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med 2009; 206: 2583–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cobat A, Poirier C, Hoal E, et al. Tuberculin skin test negativity is under tight genetic control of chromosomal region 11p14–15 in settings with different tuberculosis endemicities. J Infect Dis 2015; 211: 317–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cobat A, Hoal EG, Gallant CJ, et al. Identification of a major locus, TNF1, that controls BCG-triggered tumor necrosis factor production by leukocytes in an area hyperendemic for tuberculosis. Clin Infect Dis 2013; 57: 963–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jabot-Hanin F, Cobat A, Feinberg J, et al. Major loci on chromosomes 8q and 3q control BCG- and ESAT-6 triggered IFN-γ production, respectively, in various populations. J Infect Dis 2016; 213: 1173–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Comstock GW, Livesay VT, Woolpert SF. The prognosis of a positive tuberculin reaction in childhood and adolescence. Am J Epidemiol 1974; 99: 131–38. [DOI] [PubMed] [Google Scholar]
- 59.Borgdorff MW, Sebek M, Geskus RB, Kremer K, Kalisvaart N, van Soolingen D. The incubation period distribution of tuberculosis estimated with a molecular epidemiological approach. Int J Epidemiol 2011; 40: 964–70. [DOI] [PubMed] [Google Scholar]
- 60.Boisson-Dupuis S, Bustamante J, El-Baghdadi J, et al. Inherited and acquired immunodeficiencies underlying tuberculosis in childhood. Immunol Rev 2015; 264: 103–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conti F, Lugos Reyes SO, Galicia LB, et al. Mycobacterial disease in chronic granulomatous disease: a retrospective analysis of 71 cases. J Allergy Clin Immunol 2016; 138: 241–48. [DOI] [PubMed] [Google Scholar]
- 62.Kreins AY, Ciancanelli MJ, Okada S, et al. Human TYK2 deficiency: mycobacterial and viral infections without hyper-IgE syndrome. J Exp Med 2015; 212: 1641–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bustamante J, Boisson-Dupuis S, Abel L, Casanova JL. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-gamma immunity. Semin Immunol 2014; 26: 454–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu UI, Holland SM. Host susceptibility to non-tuberculous mycobacterial infections. Lancet Infect Dis 2015; 15: 968–80. [DOI] [PubMed] [Google Scholar]
- 65.Dorman SE, Picard C, Lammas D, et al. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet 2004; 364: 2113–21. [DOI] [PubMed] [Google Scholar]
- 66.Sasaki Y, Nomura A, Kusuhara K, et al. Genetic basis of patients with bacille Calmette-Guerin osteomyelitis in Japan: identification of dominant partial interferon-gamma receptor 1 deficiency as a predominant type. J Infect Dis 2002; 185: 706–09. [DOI] [PubMed] [Google Scholar]
- 67.Picard C, Fieschi C, Altare F, et al. Inherited interleukin-12 deficiency: IL12B genotype and clinical phenotype of 13 patients from six kindreds. Am J Hum Genet 2002; 70: 336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jouanguy E, Lamhamedi-Cherradi S, Altare F, et al. Partial interferon-gamma receptor 1 deficiency in a child with tuberculoid bacillus Calmette-Guerin infection and a sibling with clinical tuberculosis. J Clin Invest 1997; 100: 2658–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bustamante J, Arias AA, Vogt G, et al. Germline CYBB mutations that selectively affect macrophages in kindreds with X-linked predisposition to tuberculous mycobacterial disease. Nat Immunol 2011; 12: 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Beaucoudrey L, Samarina A, Bustamante J, et al. Revisiting human IL-12Rbeta1 deficiency: a survey of 141 patients from 30 countries. Medicine 2010; 89: 381–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boisson-Dupuis S, El Baghdadi J, Parvaneh N, et al. IL-12Rbeta1 deficiency in two of fifty children with severe tuberculosis from Iran, Morocco, and Turkey. PLoS One 2011; 6: e18524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alcais A, Fieschi C, Abel L, Casanova JL. Tuberculosis in children and adults: two distinct genetic diseases. J Exp Med 2005; 202: 1617–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alangari AA, Al-Zamil F, Al-Mazrou A, et al. Treatment of disseminated mycobacterial infection with high-dose IFN-gamma in a patient with IL-12Rbeta1 deficiency. Clin Dev Immunol 2011; 2011: 691956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vynnycky E, Fine PE. Lifetime risks, incubation period, and serial interval of tuberculosis. Am J Epidemiol 2000; 152: 247–63. [DOI] [PubMed] [Google Scholar]
- 75.Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol 2002; 20: 581–620. [DOI] [PubMed] [Google Scholar]
- 76.Puffer R Familial susceptibility to tuberculosis. Its importance as a public health problem. Cambridge, MA: Harvard University Press, 1944. [Google Scholar]
- 77.Azad AK, Sadee W, Schlesinger LS. Innate immune gene polymorphisms in tuberculosis. Infect Immun 2012; 80: 3343–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Möller M, de Wit E, Hoal EG. Past, present and future directions in human genetic susceptibility to tuberculosis. FEMS Immunol Med Microbiol 2010; 58: 3–26. [DOI] [PubMed] [Google Scholar]
- 79.Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med 1998; 338: 640–44. [DOI] [PubMed] [Google Scholar]
- 80.Greenwood CM, Fujiwara TM, Boothroyd LJ, et al. Linkage of tuberculosis to chromosome 2q35 loci, including NRAMP1, in a large aboriginal Canadian family. Am J Hum Genet 2000; 67: 405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Malik S, Abel L, Tooker H, et al. Alleles of the NRAMP1 gene are risk factors for pediatric tuberculosis disease. Proc Natl Acad Sci USA 2005; 102: 12183–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baghdadi JE, Orlova M, Alter A, et al. An autosomal dominant major gene confers predisposition to pulmonary tuberculosis in adults. J Exp Med 2006; 203: 1679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grant AV, El Baghdadi J, Sabri A, et al. Age-dependent association between pulmonary tuberculosis and common TOX variants in the 8q12–13 linkage region. Am J Hum Genet 2013; 92: 407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aliahmad P, Seksenyan A, Kaye J. The many roles of TOX in the immune system. Curr Opin Immunol 2012; 24: 173–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thye T, Owusu-Dabo E, Vannberg FO, et al. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat Genet 2012; 44: 257–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thye T, Vannberg FO, Wong SH, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet 2010; 42: 739–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Curtis J, Luo Y, Zenner HL, et al. Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration. Nat Genet 2015; 47: 523–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chimusa ER, Zaitlen N, Daya M, et al. Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Hum Mol Genet 2014; 23: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grant AV, Sabri A, Abid A, et al. A genome-wide association study of pulmonary tuberculosis in Morocco. Hum Genet 2016; 135: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dai Y, Zhang X, Pan H, Tang S, Shen H, Wang J. Fine mapping of genetic polymorphisms of pulmonary tuberculosis within chromosome 18q11.2 in the Chinese population: a case-control study. BMC Infect Dis 2011; 11: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang X, Tang NL, Leung CC, et al. Association of polymorphisms in the Chr18q11.2 locus with tuberculosis in Chinese population. Hum Genet 2013; 132: 691–95. [DOI] [PubMed] [Google Scholar]
- 92.Sobota RS, Stein CM, Kodaman N, et al. A locus at 5q33.3 confers resistance to tuberculosis in highly susceptible individuals. Am J Hum Genet 2016; 98: 514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang YS, Chern HD, Su WJ, et al. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology 2002; 35: 883–89. [DOI] [PubMed] [Google Scholar]
- 94.Huang YS, Chern HD, Su WJ, et al. Cytochrome P450 2E1 genotype and the susceptibility to antituberculosis drug-induced hepatitis. Hepatology 2003; 37: 924–30. [DOI] [PubMed] [Google Scholar]
- 95.Nolan CM, Goldberg SV, Buskin SE. Hepatotoxicity associated with isoniazid preventive therapy: a 7-year survey from a public health tuberculosis clinic. JAMA 1999; 281: 1014–18. [DOI] [PubMed] [Google Scholar]
- 96.Mitchell JR, Zimmerman HJ, Ishak KG, et al. Isoniazid liver injury: clinical spectrum, pathology, and probable pathogenesis. Ann Intern Med 1976; 84: 181–92. [DOI] [PubMed] [Google Scholar]
- 97.Sun F, Chen Y, Xiang Y, Zhan S. Drug-metabolising enzyme polymorphisms and predisposition to anti-tuberculosis drug-induced liver injury: a meta-analysis. Int J Tuberc Lung Dis 2008; 12: 994–1002. [PubMed] [Google Scholar]
- 98.Wang P, Xie S, Hao Q, Zhang C, Jiang B. NAT2 polymorphisms and susceptibility to anti-tuberculosis drug-induced liver injury: a meta-analysis. Int J Tuberc Lung Dis 2012; 16: 589–95. [DOI] [PubMed] [Google Scholar]
- 99.Cai Y, Yi J, Zhou C, Shen X. Pharmacogenetic study of drug-metabolising enzyme polymorphisms on the risk of anti-tuberculosis drug-induced liver injury: a meta-analysis. PLoS One 2012; 7: e47769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roy B, Ghosh SK, Sutradhar D, Sikdar N, Mazumder S, Barman S. Predisposition of antituberculosis drug induced hepatotoxicity by cytochrome P450 2E1 genotype and haplotype in pediatric patients. J Gastroenterol Hepatol 2006; 21: 784–86. [DOI] [PubMed] [Google Scholar]
- 101.Vuilleumier N, Rossier MF, Chiappe A, et al. CYP2E1 genotype and isoniazid-induced hepatotoxicity in patients treated for latent tuberculosis. Eur J Clin Pharmacol 2006; 62: 423–29. [DOI] [PubMed] [Google Scholar]
- 102.Tang S, Lv X, Zhang Y, et al. Cytochrome P450 2E1 gene polymorphisms/haplotypes and anti-tuberculosis drug-induced hepatitis in a Chinese cohort. PLoS One 2013; 8: e57526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cho H-J, Koh W-J, Ryu Y-J, et al. Genetic polymorphisms of NAT2 and CYP2E1 associated with antituberculosis drug-induced hepatotoxicity in Korean patients with pulmonary tuberculosis. Tuberculosis 2007; 87: 551–56. [DOI] [PubMed] [Google Scholar]
- 104.Kim S-H, Kim S-H, Bahn J-W, et al. Genetic polymorphisms of drug-metabolizing enzymes and anti-TB drug-induced hepatitis. Pharmacogenomics 2009; 10: 1767–79. [DOI] [PubMed] [Google Scholar]
- 105.Yamada S, Tang M, Richardson K, et al. Genetic variations of NAT2 and CYP2E1 and isoniazid hepatotoxicity in a diverse population. Pharmacogenomics 2009; 10: 1433–45. [DOI] [PubMed] [Google Scholar]
- 106.Huang YS, Su WJ, Huang YH, et al. Genetic polymorphisms of manganese superoxide dismutase, NAD(P)H: quinone oxidoreductase, glutathione S transferase M1 and T1, and the susceptibility to drug-induced liver injury. J Hepatol 2007; 47: 128–34. [DOI] [PubMed] [Google Scholar]
- 107.Azuma J, Ohno M, Kubota R, et al. NAT2 genotype guided regimen reduces isoniazid-induced liver injury and early treatment failure in the 6-month four-drug standard treatment of tuberculosis: a randomized controlled trial for pharmacogenetics-based therapy. Eur J Clin Pharmacol 2013; 69: 1091–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 2003; 306: 287–300. [DOI] [PubMed] [Google Scholar]
- 109.Kwara A, Lartey M, Sagoe KW, Court MH. Paradoxically elevated efavirenz concentrations in HIV/tuberculosis-coinfected patients with CYP2B6 516TT genotype on rifampin-containing antituberculous therapy. AIDS 2011; 25: 388–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dooley KE, Denti P, Martinson N, et al. Pharmacokinetics of efavirenz and treatment of HIV-1 among pregnant women with and without tuberculosis coinfection. J Infect Dis 2015; 211: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Luetkemeyer AF, Rosenkranz SL, Lu D, et al. Combined effect of CYP2B6 and NAT2 genotype on plasma efavirenz exposure during rifampin-based antituberculosis therapy in the STRIDE study. Clin Infect Dis 2015; 60: 1860–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.di Iulio J, Fayet A, Arab-Alameddine M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genom 2009; 19: 300–09. [DOI] [PubMed] [Google Scholar]
- 113.Goel U, Bajaj S, Gupta O, Dwivedi N, Dubey A. Isoniazid induced neuropathy in slow versus rapid acetylators: an electrophysiological study. J Assoc Physicians India 1992; 40: 671–72. [PubMed] [Google Scholar]
- 114.Yamamoto M, Sobue G, Mukoyama M, Matsuoka Y, Mitsuma T. Demonstration of slow acetylator genotype of N-acetyltransferase in isoniazid neuropathy using an archival hematoxylin and eosin section of a sural nerve biopsy specimen. J Neurol Sci 1996; 135: 51–54. [DOI] [PubMed] [Google Scholar]
- 115.Kim RB. Organic anion-transporting polypeptide (OATP) transporter family and drug disposition. Eur J Clin Invest 2003; 33 (suppl 2): 1–5. [DOI] [PubMed] [Google Scholar]
- 116.Chigutsa E, Visser ME, Swart EC, et al. The SLCO1B1 rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations: dosing implications. Antimicrob Agents Chemother 2011; 55: 4122–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aung AK, Haas DW, Hulgan T, Phillips EJ. Pharmacogenomics of antimicrobial agents. Pharmacogenomics 2014; 15: 1903–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fischel-Ghodsian N Genetic factors in aminoglycoside toxicity. Pharmacogenomics 2005; 6: 27–36. [DOI] [PubMed] [Google Scholar]
- 119.Selimoglu E Aminoglycoside-induced ototoxicity. Curr Pharm Des 2007; 13: 119–26. [DOI] [PubMed] [Google Scholar]
- 120.Guthrie O Aminoglycoside induced ototoxicity. Toxicology 2008; 249: 91–96. [DOI] [PubMed] [Google Scholar]
- 121.Guan M-X. Mitochondrial 12S rRNA mutations associated with aminoglycoside ototoxicity. Mitochondrion 2011; 11: 237–45. [DOI] [PubMed] [Google Scholar]
- 122.Xie J, Talaska AE, Schacht J. New developments in aminoglycoside therapy and ototoxicity. Hear Res 2011; 281: 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prezant TR, Agapian JV, Bohlman MC, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet 1993; 4: 289–94. [DOI] [PubMed] [Google Scholar]
- 124.Pacheu-Grau D, Gómez-Durán A, Montoya J, Ruiz-Pesini E. Influence of mtDNA genetic variation on antibiotic therapy. Pharmacogenomics 2010; 11: 1185–87. [DOI] [PubMed] [Google Scholar]
- 125.Soriano A, Miró O, Mensa J. Mitochondrial toxicity associated with linezolid. N Engl J Med 2005; 353: 2305–06. [DOI] [PubMed] [Google Scholar]
- 126.McKee E, Ferguson M, Bentley A, Marks T. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob Agents Chemother 2006; 50: 2042–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.De Vriese AS, Van Coster R, Smet J, et al. Linezolid-induced inhibition of mitochondrial protein synthesis. Clin Infect Dis 2006; 42: 1111–17. [DOI] [PubMed] [Google Scholar]
- 128.Garrabou G, Soriano A, López S, et al. Reversible inhibition of mitochondrial protein synthesis during linezolid-related hyperlactatemia. Antimicrob Agents Chemother 2007; 51: 962–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Carson J, Cerda J, Chae JH, Hirano M, Maggiore P. Severe lactic acidosis associated with linezolid use in a patient with the mitochondrial DNA A2706G polymorphism. Pharmacotherapy 2007; 27: 771–74. [DOI] [PubMed] [Google Scholar]
- 130.Pacheu-Grau D, Gómez-Durán A, Iglesias E, López-Gallardo E, Montoya J, Ruiz-Pesini E. Mitochondrial antibiograms in personalized Medicine Hum Mol Genet 2013; 22: 1132–39. [DOI] [PubMed] [Google Scholar]
- 131.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009; 461: 747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Di Pietrantonio T, Hernandez C, Girard M, et al. Strain-specific differences in the genetic control of two closely related mycobacteria. PLoS Pathog 2010; 6: e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Alcais A, Quintana-Murci L, Thaler DS, Schurr E, Abel L, Casanova JL. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann N Y Acad Sci 2010; 1214: 18–33. [DOI] [PubMed] [Google Scholar]
- 134.Brites D, Gagneux S. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol Rev 2015; 264: 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nicol MP, Wilkinson RJ. The clinical consequences of strain diversity in Mycobacterium tuberculosis. Trans R Soc Trop Med Hyg 2008; 102: 955–65. [DOI] [PubMed] [Google Scholar]
- 136.Comas I, Chakravartti J, Small PM, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 2010; 42: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Comas I, Coscolla M, Luo T, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 2013; 45: 1176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gagneux S Host–pathogen coevolution in human tuberculosis. Philos Trans R Soc Lond B Biol Sci 2012; 367: 850–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Raghavan S, Alagarasu K, Selvaraj P. Immunogenetics of HIV and HIV associated tuberculosis. Tuberculosis 2012; 92: 18–30. [DOI] [PubMed] [Google Scholar]
- 140.Bartha I, Carlson JM, Brumme CJ, et al. A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control. Elife 2013; 2: e01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Walker NF, Scriven J, Meintjes G, Wilkinson RJ. Immune reconstitution inflammatory syndrome in HIV-infected patients. HIV AIDS 2015; 7: 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Orlova M, Cobat A, Huong NT, et al. Gene set signature of reversal reaction type I in leprosy patients. PLoS Genet 2013; 9: e1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Abel L, Casanova JL. Genetic predisposition to clinical tuberculosis: bridging the gap between simple and complex inheritance. Am J Hum Genet 2000; 67: 274–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 2011; 12: 745–55. [DOI] [PubMed] [Google Scholar]
- 145.Belkadi A, Bolze A, Itan Y, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA 2015; 112: 5473–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 2010; 11: 415–25. [DOI] [PubMed] [Google Scholar]
- 147.Kiezun A, Garimella K, Do R, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet 2012; 44: 623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet 2014; 95: 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gregersen PK. Cell type-specific eQTLs in the human immune system. Nat Genet 2012; 44: 478–80. [DOI] [PubMed] [Google Scholar]
- 150.Pickrell JK, Marioni JC, Pai AA, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 2010; 464: 768–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466: 973–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cliff JM, Lee JS, Constantinou N, et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis 2013; 207: 18–29. [DOI] [PubMed] [Google Scholar]
- 153.Maertzdorf J, Repsilber D, Parida SK, et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun 2011; 12: 15–22. [DOI] [PubMed] [Google Scholar]
- 154.Ottenhoff TH, Dass RH, Yang N, et al. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One 2012; 7: e45839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bloom CI, Graham CM, Berry MP, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One 2013; 8: e70630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cliff JM, Kaufmann SH, McShane H, van Helden P, O’Garra A. The human immune response to tuberculosis and its treatment: a view from the blood. Immunol Rev 2015; 264: 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Blankley S, Berry MP, Graham CM, Bloom CI, Lipman M, O’Garra A. The application of transcriptional blood signatures to enhance our understanding of the host response to infection: the example of tuberculosis. Philos Trans R Soc Lond B Biol Sci 2014; 369: 20130427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Maertzdorf J, Kaufmann SH, Weiner J 3rd. Toward a unified biosignature for tuberculosis. Cold Spring Harb Perspect Med 2015; 5: a018531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lai RP, Meintjes G, Wilkinson KA, et al. HIV-tuberculosis-associated immune reconstitution inflammatory syndrome is characterized by toll-like receptor and inflammasome signalling. Nat Commun 2015; 6: 8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci USA 2012; 109: 1204–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Siddle KJ, Deschamps M, Tailleux L, et al. A genomic portrait of the genetic architecture and regulatory impact of microRNA expression in response to infection. Genome Res 2014; 24: 850–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pacis A, Tailleux L, Morin AM, et al. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res 2015; 25: 1801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen L, Kostadima M, Martens JH, et al. Transcriptional diversity during lineage commitment of human blood progenitors. Science 2014; 345: 1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kleinnijenhuis J, Quintin J, Preijers F, et al. Bacille Calmette–Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci USA 2012; 109: 17537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Saeed S, Quintin J, Kerstens HH, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014; 345: 1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gurdasani D, Carstensen T, Tekola-Ayele F, et al. The African Genome Variation Project shapes medical genetics in Africa. Nature 2015; 517: 327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zak DE, Penn-Nicholson A, Scriba TJ, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 2016; 387: 2312–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Smith CM, Proulx MK, Olive AJ, et al. Tuberculosis susceptibility and vaccine protection are independently controlled by host genotype. MBio 2016; 7: e01516. [DOI] [PMC free article] [PubMed] [Google Scholar]