Abstract
Type I interferons (IFN-I) are essential to establish antiviral innate immunity. Unanchored (or free) polyubiquitin (poly-Ub) has been shown to regulate IFN-I responses. However, few unanchored poly-Ub interactors are known. To identify factors regulated by unanchored poly-Ub in a physiological setting, we developed an approach to isolate unanchored poly-Ub from lung tissue. We identified the RNA helicase DHX16 as a potential pattern recognition receptor (PRR). Silencing of DHX16 in cells and in vivo diminished IFN-I responses against influenza virus. These effects extended to members of other virus families, including Zika and SARS-CoV-2. DHX16-dependent IFN-I production requires RIG-I and unanchored K48-poly-Ub synthesized by the E3-Ub ligase TRIM6. DHX16 recognizes a signal in influenza RNA segments that undergo splicing and requires its RNA helicase motif for direct, high-affinity interactions with specific viral RNAs. Our study establishes DHX16 as a PRR that partners with RIG-I for optimal activation of antiviral immunity requiring unanchored poly-Ub.
Keywords: innate immunity, unanchored ubiquitin, tripartite motif (TRIM) protein, RIG-I, DHX16, TRIM6, influenza A virus, type I interferon, splicing, SARS-CoV-2
Graphical abstract
Virus recognition triggers antiviral innate immunity. Hage et al. identify the RNA helicase DHX16 as a pattern recognition receptor. DHX16 recognizes splicing signals contained in influenza A vRNA leading to efficient RIG-I-mediated antiviral responses, which are regulated by TRIM6 and unanchored polyubiquitin. DHX16 senses infection by other viruses including SARS-CoV-2.
Introduction
The innate immune system is the first line of defense against invading microbes. It relies on the identification of pathogen-associated molecular patterns (PAMPs) via host sensors known as pattern recognition receptors (PRRs). Diverse PRRs amplify innate immune signaling in response to specific stimuli. The RIG-I-like receptor (RLR) family of PRRs containing RIG-I, MDA5, and LGP2 is central for recognition of viral RNA (vRNA) (Loo and Gale, 2011). Upon virus infection, RIG-I recognizes short, double-stranded RNA (dsRNA) with a 5′-triphosphate (Hornung et al., 2006; Schlee et al., 2009) allowing for a conformational change, releasing its two caspase activation and recruitment domains (CARDs) to enable downstream signaling via the adaptor protein MAVS (Chow et al., 2018; Liu and Gack, 2020; Loo and Gale, 2011). Ubiquitination of the RIG-I CARDs is indispensable for its activation and was originally tied to an E3-Ub ligase from the tripartite motif (TRIM) family called TRIM25 (Gack et al., 2007). Additional factors activated downstream of MAVS include the IKK-related kinases (TBK1 and IKKε), which phosphorylate IRF3 and IRF7, permitting their relocalization to the nucleus to induce type I interferon (IFN-I) (Rehwinkel and Gack, 2020). IFN-I signals in an autocrine or paracrine manner to promote STAT1, STAT2, and IRF9 assembly into the ISGF3 complex that facilitates the expression of several hundred IFN-stimulated genes (ISGs) that collectively invoke an antiviral state (Akira et al., 2006; Schoggins and Rice, 2011).
Innate immunity is tightly regulated to respond to threats while also avoiding damage. Post-translational modification (PTM) of proteins is a well-characterized method of immune regulation. Ubiquitin (Ub) is a 76 amino acid protein that functions as a PTM through covalent (anchored) or non-covalent (unanchored) interactions (Ebner et al., 2017; Hage and Rajsbaum, 2019). Unanchored Ub is unique as it is not covalently conjugated to substrates, allowing for a free C-terminal Gly residue. The significance of these Ub chains has recently been appreciated as a three-dimensional platform for protein interactions and signaling (Rajsbaum et al., 2014b; Xia et al., 2009; Zeng et al., 2010). Unanchored K63-linked poly-Ub (K63-poly-Ub) promotes the activation of TAK1, RIG-I, and MDA5 for optimal production of IFN-I and pro-inflammatory cytokines while unanchored K48-linked poly-Ub (K48-poly-Ub) enhances IFN-I signaling by stimulating IKKε (Jiang et al., 2012; Pertel et al., 2011; Rajsbaum et al., 2014b; Xia et al., 2009; Zeng et al., 2010). To identify additional factors involved in innate immunity that require unanchored Ub for their function, we used a novel approach to isolate unanchored Ub chains from in vivo samples. Employing this method, we identified the RNA helicase DHX16 as a novel unanchored Ub-interacting factor and a potential PRR that partners with RIG-I for antiviral IFN-I production and ISG expression.
DHX16 is a DEAH-box RNA helicase in the SF2 superfamily (Abdelhaleem et al., 2003; Fairman-Williams et al., 2010; Tanner and Linder, 2001). DHX16 has only been described as a pre-mRNA splicing factor that helps form a catalytically active spliceosome (Gencheva et al., 2010a; Kim and Lin, 1996; Teigelkamp et al., 1994). Mutations in DHX16 that diminish splicing result in the nuclear accumulation of unspliced transcripts including genes involved in neurodevelopment (Gencheva et al., 2010b; Paine et al., 2019). Although DHX16's involvement in pre-mRNA splicing is recognized, it is unknown if DHX16 plays a role in innate immunity and whether it is regulated by unanchored Ub.
The role of unanchored Ub has been controversial. However, several investigations have identified important functions for covalent and non-covalently linked Ub in RIG-I modulation and highlight the importance of TRIM E3-Ub ligases in innate immunity (Giraldo et al., 2020b; Hage and Rajsbaum, 2019; Rajsbaum et al., 2014a; Sun et al., 2016; van Tol et al., 2017, 2020; Versteeg et al., 2014). While several signaling partners have been described contributing to these RIG-I regulatory mechanisms, the exact benefit RIG-I gains from these interactions remains unclear. In this study, we identify DHX16 as a novel unanchored Ub-interacting factor and promoter of IFN-I production. DHX16 recognizes influenza A virus (IAV) vRNA segments 7 and 8 that encode for M1-M2 and NS1-NEP upon splicing (Dubois et al., 2014). DHX16 also preferentially recognizes the ORF1a region of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2 ). This leads to recruitment of the E3-Ub ligase TRIM6 that transfers unanchored K48-poly-Ub to DHX16 promoting RIG-I-dependent IFN-I production. This work identifies an unrecognized role for DHX16 in antiviral innate immunity and reveals unanchored Ub as a regulator to promote efficient activation of IFN-I production.
Results
DHX16 is an unanchored Ub-interacting factor
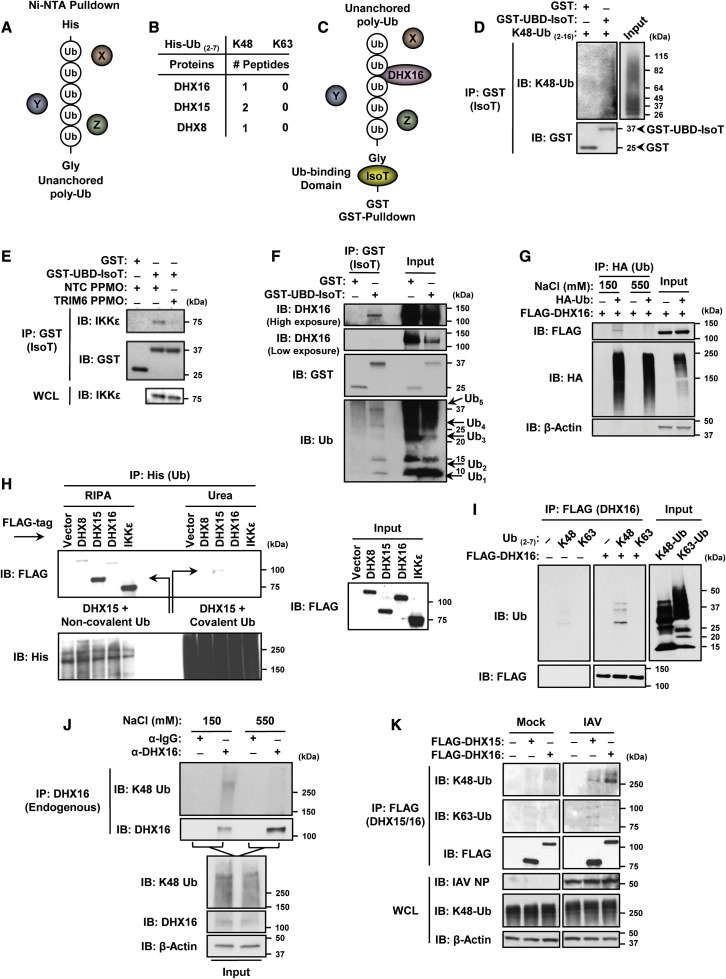
The importance of unanchored Ub in innate immunity has been shown only for a limited number of unanchored Ub interactors. We set out to investigate what additional cellular factors interact with unanchored Ub and whether unanchored Ub plays a broader role in innate immunity. To identify unanchored Ub-interacting factors from murine lungs, which we expect would represent interactions in close physiological conditions, we used His-tagged recombinant poly-Ub chains as bait. These chains were mixed with lung tissue lysates and protein complexes were isolated with Ni-NTA beads (Figure 1 A). Sample elutions were subjected to mass spectrometry (MS) analysis. We identified three DEAH-box RNA helicases (DHX8, DHX15, and DHX16) that interacted with K48-, but not K63-poly-Ub in lung tissues (Figure 1B).
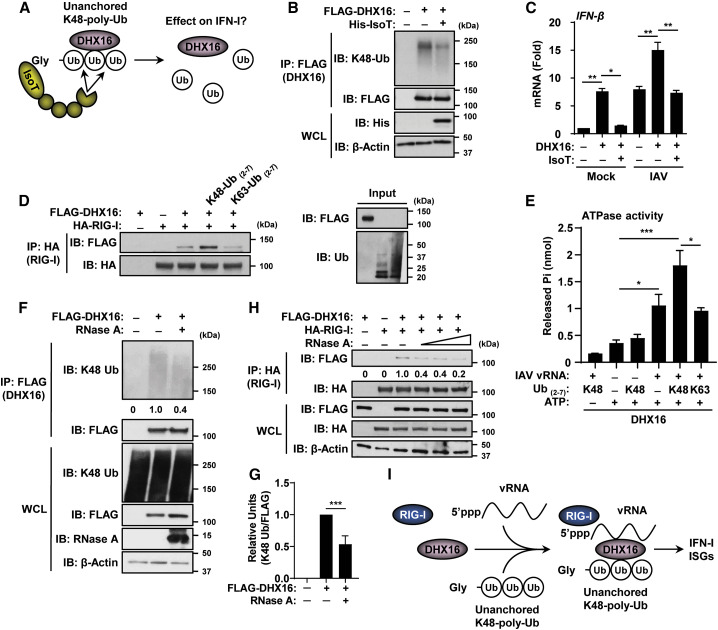
Figure 1.
DHX16 interacts with unanchored poly-Ub chains
(A) Unanchored Ub enrichment from murine lung tissue.
(B) DEAH-box RNA helicases identified from murine lungs as unanchored Ub-interacting factors (MS).
(C) Unanchored Ub enrichment utilizing GST-UBD-IsoT.
(D) Interaction between GST-UBD-IsoT and recombinant K48-poly-Ub chains (2-16 Ub/chain) in an in vitro Ub-binding assay (coIP).
(E) GST-UBD-IsoT was mixed with lung lysates from mice treated with NTC or TRIM6 PPMO followed by GST pulldown (lung samples used in Rajsbaum et al., 2014b) (coIP).
(F) Interaction between endogenous DHX16 and GST-UBD-IsoT from HEK293Ts (coIP).
(G) DHX16 interacts non-covalently with poly-Ub after normal (150 mM NaCl), but not high salt (550 mM NaCl) washes (denaturing coIP).
(H) DHX16 interacts non-covalently with poly-Ub after RIPA, but not urea washes. Arrows indicate DHX15 interactions with either non-covalent or covalent poly-Ub (denaturing coIP).
(I) DHX16 interacts with K48-poly-Ub in an in vitro Ub binding assay (coIP).
(J) Endogenous DHX16 interacts non-covalently with K48-poly-Ub after normal, but not high salt washes (denaturing coIP).
(K) DHX16 interacts with K48-poly-Ub in HEK293Ts infected with IAV for 12 hrs (PR8 MOI = 5) (coIP).
Data are representative of 2–3 independent experiments.
See also Figure S1.
To confirm that the identified helicases interact with unanchored Ub, we devised a novel co-immunoprecipitation (coIP) approach that would enrich for endogenous unanchored Ub using the Ub-specific protease isopeptidase T (IsoT, also known as USP5) (Figure 1C). IsoT interacts with unanchored Ub through its Ub-binding domain (UBD) that recognizes the exposed C-terminal Gly residue and not covalently attached Ub (Reyes-Turcu et al., 2006). IsoT was used previously to demonstrate the unanchored nature of poly-Ub (Rajsbaum et al., 2014b; Zeng et al., 2010). A GST-tagged variant of IsoT that retains its UBD while lacking the catalytic domain was used to capture endogenous unanchored Ub without degrading it (Figure 1C). As a proof of principle, the GST-UBD-IsoT bound unanchored poly-Ub in vitro while a GST control did not (Figure 1D). To validate whether this approach can be used to isolate unanchored Ub-interacting factors from mouse tissues, GST-UBD-IsoT was used to detect a known unanchored Ub-interacting factor (IKKε) (Rajsbaum et al., 2014b) from murine lung lysates. GST-UBD-IsoT co-purified IKKε from control samples, but not from lungs that were previously treated with peptide-conjugated phosphorodiamidate morpholino oligomers (PPMOs) for TRIM6 knockdown, which is required for the synthesis of K48-poly-Ub that interacts with IKKε (Rajsbaum et al., 2014b) (Figure 1E). Using this approach, endogenous DHX16 was successfully co-purified from cells by GST-UBD-IsoT (Figure 1F), confirming the interaction identified by MS. Unanchored Ub chains corresponding to as many as five Ub molecules were detected (Figure 1F). To ensure that GST-UBD-IsoT interacts with DHX16 through unanchored poly-Ub, and not via a direct interaction with DHX16, we performed a coIP with a high salt stringency wash, in which endogenous unanchored poly-Ub interacting with DHX16 was first removed before the addition of GST-UBD-IsoT (Figure S1A). GST-UBD-IsoT only interacted with DHX16 in the standard wash that retained poly-Ub, but not after the high salt wash that removed poly-Ub interacting with DHX16 (Figure S1B). Similarly, the increased ionic strength of the high salt wash decoupled overexpressed HA-Ub that co-purified with DHX16 (Figure 1G).
To further confirm non-covalent interactions between the identified helicases and unanchored Ub, we performed a side-by-side non-denaturing and denaturing coIP (Figure 1H). Although all three helicases interact with Ub after non-denaturing RIPA washes, these associations were lost after denaturing urea washes similarly to an IKKε control (Figure 1H). A weak band for DHX15 was detected after urea washes, suggesting some covalent ubiquitination on DHX15 may occur. An in vitro binding assay using purified proteins further demonstrated that DHX16 interacted directly with recombinant K48-, but not K63-poly-Ub (Figure 1I). The non-covalent interaction between DHX16 and K48-poly-Ub was also demonstrated by a coIP of endogenous proteins using the high salt wash, which removed all endogenous K48-poly-Ub from DHX16 (Figure 1J). Finally, IAV infection enhanced the association between overexpressed DHX16 and endogenous K48-poly-Ub, while DHX15 appeared to interact with both K63- and K48-poly-Ub (Figure 1K). Taken together, these results identify DHX16 as an unanchored K48-poly-Ub-interacting factor.
DHX16 is required for optimal ISG expression
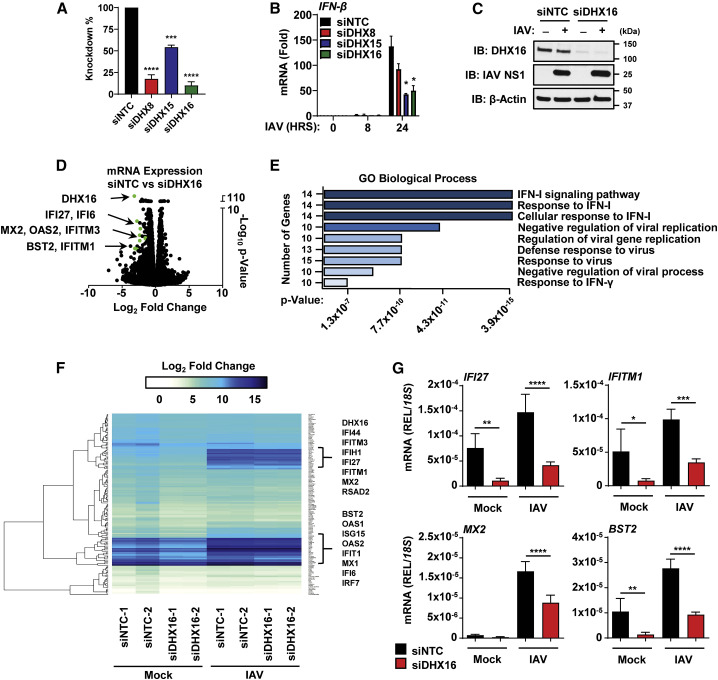
To determine if the identified helicases play a role in innate immunity, we performed gene silencing of DHX8/15/16 with a pool of small interfering RNAs (siRNA) and examined the IFN-I response to IAV infection (Figures 2A and 2B). While DHX8 knockdown did not have a statistically significant effect on IFN-β mRNA, both DHX15 and DHX16 showed significant reduction in IFN-β induction (Figure 2B). DHX15 has been characterized as a sensor for RNA viruses (Lu et al., 2014; Mosallanejad et al., 2014; Pattabhi et al., 2019), while the only known roles for DHX16 have been related to RNA metabolism and pre-mRNA splicing (Gencheva et al., 2010a, 2010b). It is unknown whether DHX16 is involved in innate immunity, or if it requires unanchored Ub, prompting us to focus our investigation on DHX16. To reduce the possibility that our observations are due to off-target effects of the siRNA pool, we tested two additional independent siRNAs for DHX16. Both siRNAs significantly reduced DHX16 protein expression and inhibited IAV-induced IFN-β and IFITM1 mRNA (Figures S1C and S1D).
Figure 2.
DHX16 is required for efficient induction of ISGs
(A–C) A549s treated with siNTC or siDHX8/15/16 followed by IAV infection for 24 h (PR8 MOI = 0.1). (A) Silencing efficiency of siRNAs and (B) IFN-β expression (qRT-PCR). (C) Silencing efficiency of DHX16 protein (immunoblot).
(D–F) Next-generation sequencing (NGS) analysis of A549s treated with siRNA and followed by IAV infection as described in (A). (D) Volcano plot of ISGs significantly downregulated in siDHX16 versus siNTC mock samples. (E) Enriched GO terms and pathways (Enrichr) of downregulated transcripts. (F) Heatmap of transcriptomic changes.
(G) Validation of selected ISGs identified in NGS (qRT-PCR).
Data are expressed as means (n = 3) ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 (Student's t test or one-way ANOVA with Dunnett's or Sidak's multiple comparisons). Data are representative of 2–3 independent experiments.
See also Figure S1.
To determine if DHX16 plays a role in innate immunity, using an unbiased approach, we performed gene silencing of DHX16 followed by a genome-wide expression analysis using next-generation sequencing (NGS). DHX16 expression at the protein and mRNA level was reduced by over 90% after knockdown (Figures 2A and 2C). Surprisingly, in mock-infected cells, the largest changes in gene expression between knockdown and control cells were found in ISGs (Figure 2D). Gene ontology (GO) enrichment analysis (Chen et al., 2013; Kuleshov et al., 2016) showed that downregulated genes were enriched in pathways involved in the IFN-I response and antiviral defenses (Figure 2E). Upregulated genes were enriched in pathways for extracellular organization (Figure S1E). Upon IAV infection, induction of ISG expression was significantly reduced in siDHX16 cells compared with siNTC (Figures 2F and 2G). Strikingly, knockdown of DHX16 adversely impacted transcript expression levels of several ISGs relevant to the formation of an antiviral state against IAV in both mock and IAV-infected samples, suggesting DHX16 is involved in driving antiviral innate immunity (Figure 2G).
These data were surprising because of the previously reported role for DHX16 in splicing, and we did not expect DHX16 function to have a bias in regulating expression of innate immune genes. To rule out whether DHX16 promotes ISG induction by regulating splicing of a factor involved in IFN-I pathways, we performed mixture of isoforms (MISO) analysis, which quantifies alternative splicing events from the NGS data and identifies differentially regulated exons across samples (Katz et al., 2010). This analysis revealed that when comparing siDHX16 to siNTC cells, a majority of the alternative splicing events were skipped exons, indicating that a dysregulation of splicing does occur (Figure S1F). However, when looking at specific gene splicing events, we did not find significant changes in splicing of well-established genes involved in the IFN-I response (Figure S1G), suggesting DHX16's function in innate immunity is independent of its role as a splicing factor. Collectively, these data suggest that DHX16 is essential for efficient ISG expression.
DHX16 is involved in antiviral IFN-I production
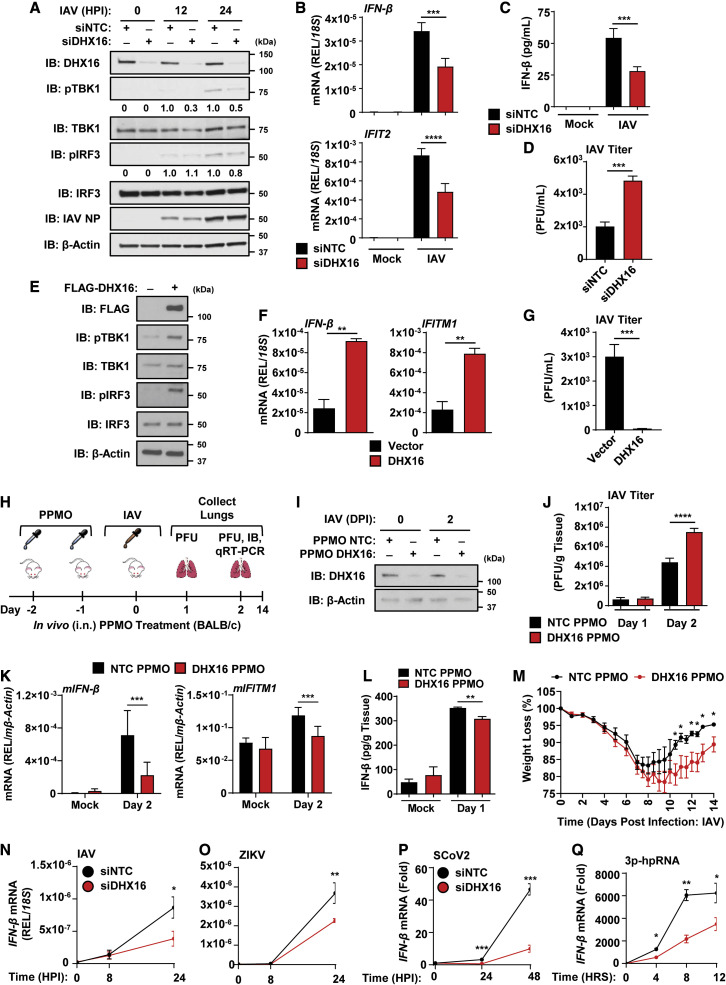
Since DHX16 knockdown affects expression of ISGs, we examined whether DHX16 plays a role downstream of the IFN-I receptor (Figure S2A). DHX16 transcript levels were not increased in response to IFN-β (Figure S2B), indicating DHX16 is not an ISG under these conditions. The induction of ISGs upon treatment with IFN-β was not significantly different between siNTC and siDHX16 cells (Figure S2C), suggesting that DHX16 does not play a role in the IFN-I signaling pathway. However, knockdown of DHX16 led to a reduction in phosphorylation of IRF3 and TBK1 during IAV infection (Figure 3 A), which are required for IFN-I production. Decreases in the active forms for these signaling factors correlated with reduced IFN-β and IFIT2 mRNA (Figure 3B) as well as a reduction of secreted IFN-β protein (Figure 3C). In line with these data, the loss in IFN-I production resulted in a significant increase of IAV titers in siDHX16 cells compared with siNTC controls (Figure 3D). Additionally, overexpression of DHX16 significantly enhanced phosphorylation of both IRF3 and TBK1 (Figure 3E), which correlated with increased expression of IFN-β and ISG transcripts (Figures 3F and S3A). Overexpression of DHX16 also significantly suppressed IAV replication (Figure 3G).
Figure 3.
DHX16 enhances activation of the IFN-I production pathway
(A–D) A549s treated with siNTC or siDHX16 followed by IAV infection for 24 h (PR8 MOI = 0.1). (A) Phosphorylation of TBK1 and IRF3 (immunoblot). (B) IFN-β and IFIT2 expression (qRT-PCR). (C) IFN-β protein in supernatants (ELISA). (D) IAV viral titers (plaque assay).
(E–G) A549s transfected with empty vector or DHX16 followed by IAV infection for 24 h (PR8 MOI = 0.1). (E) Phosphorylation of TBK1 and IRF3 (immunoblot). (F) IFN-β and IFITM1 expression (qRT-PCR). (G) IAV viral titers (plaque assay).
(H–M) PPMO-mediated in vivo silencing of DHX16 in BALB/c mice followed by IAV infection (PR8, 1000 PFU) (n = 3–4/group). (H) Schematics of experiment. (I) Validation of PPMO-mediated silencing in murine lung tissue (immunoblot). (J) IAV viral titers (plaque assay). (K) mIFN-β and mIFITM1 expression (qRT-PCR). (L) IFN-β protein in lung tissue (ELISA) (n = 5–8/group). (M) Weight loss from NTC or DHX16 PPMO-treated mice infected with IAV (PR8, 100 PFU) (n = 5/group).
(N–Q) IFN-β expression from siNTC or siDHX16 treated A549s infected with IAV (PR8 WT MOI = 0.1) (N) or ZIKV (FSS MOI = 0.1) (O), Calu-3s infected with SARS-CoV-2 (icSARS-CoV-2-mNG MOI = 1) (P) or A549s transfected with 3p-hpRNA (100 ng/mL) (Q) (qRT-PCR).
Data are expressed as means (n = 3) ± SD or ± SEM as in (M). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 (Student's t test or one-way ANOVA with Tukey's multiple comparisons). Data are representative of 2–3 independent experiments.
See also Figures S2 and S3.
To demonstrate the biological relevance of DHX16, and since DHX16 KO mice are embryonically lethal, DHX16 expression was silenced in vivo using PPMOs as we previously established (Abes et al., 2006; Rajsbaum, 2017; Rajsbaum et al., 2014b). We first validated the DHX16 PPMO knockdown in mouse embryonic fibroblasts (MEFs), which showed reduced levels of DHX16 protein (Figure S3B). BALB/c mice were administered control or DHX16 PPMOs prior to IAV challenge (Figure 3H). DHX16 PPMOs silenced murine DHX16 protein expression in lungs resulting in significantly enhanced IAV titers in infected animals, reduced mIFN-β and mIFITM1 mRNA, and a small, but statistically significant, reduction in IFN-β protein compared with control PPMO-treated mice (Figures 3I–3L). DHX16 knockdown also caused a delay in weight recovery following IAV infection (Figure 3M). The in vivo effects observed during DHX16 knockdown were consistent across different mouse strains as C57BL/6 mice also showed significantly higher viral titers for DHX16 PPMO-treated animals (Figure S3C). These data implicate a role for DHX16 in antiviral immunity in vivo.
We then tested whether DHX16 was involved in establishing a general antiviral state. The consequence of DHX16 knockdown was assessed during infection with different RNA viruses. Expression of IFN-β was significantly reduced in siDHX16 cells infected with either WT IAV (negative-sense, Orthomyxoviridae family) or a recombinant mutant incapable of effectively antagonizing IFN-I production (NS1 R38A/K41A) (Figures 3N and S3D). DHX16 silencing impaired IFN-β expression upon infections with Zika virus (ZIKV; positive-sense, Flaviviridae family) (Figure 3O) and SARS-CoV-2 (positive-sense, Coronaviridae family) (Figure 3P), but not Sendai virus (SeV; negative-sense, Paramyxoviridae family) (Figure S3E). Consistent with this, DHX16 knockdown resulted in higher SARS-CoV-2 titers (Figure S3F). These results suggest that DHX16 may play a broad role in innate immunity against multiple viruses; however specific patterns or signals may be present in different virus families that allow DHX16 to trigger an IFN-I response.
DHX16 promotes efficient antiviral IFN-I production through an interaction with RIG-I
Since DHX16 triggers an IFN-I response to multiple viruses, we asked whether DHX16 can recognize PAMPs known to stimulate specific PRRs. IFN-β was significantly reduced in siDHX16 cells in response to 3p-hpRNA, an in vitro transcribed hairpin RNA of NEP from segment 8 of IAV known to be specifically recognized by RIG-I (Liu et al., 2015; Rehwinkel et al., 2010) (Figure 3Q). In contrast, Poly(I:C) (dsRNA mimic stimulating MDA5 and RIG-I), Poly(dA:dT) (dsDNA mimic stimulating cGAS), or purified IAV vRNA (non-replicative) induced similar levels of IFN-β mRNA in siNTC and siDHX16 cells (Figures S3G–S3I). Furthermore, knockdown of DHX16 led to reduced phosphorylation of IRF3 and TBK1 after stimulation with 3p-hpRNA (Figures S3J and S3K), suggesting the involvement of the RIG-I pathway.
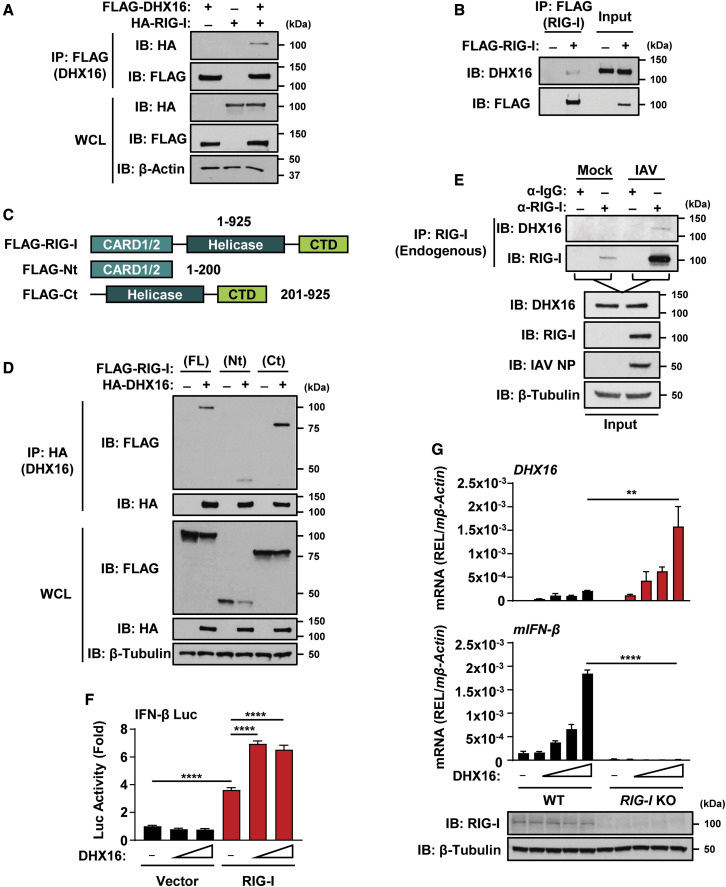
Since knockdown and overexpression of DHX16 altered TBK1 phosphorylation (Figures 3A, 3E, and S3J), DHX16 may play a role in IFN-I production at the level of TBK1 or upstream. Considering DHX16 is an RNA-binding factor, we hypothesized that it may act as a PRR for RNA viruses. We tested whether DHX16 can interact with the adaptor protein MAVS, which is known to link other PRRs to downstream signaling. Indeed, DHX16 and MAVS interacted when co-expressed (Figure S3L). Since DHX16 is involved in IFN-I induction upon stimulation with a RIG-I-specific ligand, we asked whether DHX16 cooperates with RIG-I. A coIP and reciprocal coIP showed that RIG-I is a binding partner of DHX16 (Figures 4A and S3M). Furthermore, overexpressed RIG-I interacted with endogenous DHX16 (Figure 4B). To identify the regions of RIG-I that are important for interacting with DHX16, we used deletion mutants of RIG-I encoding either the N- or C-terminal domains (Figure 4C). The C-terminal domain of RIG-I interacted with DHX16 in a similar or stronger degree to full-length RIG-I (Figure 4D). DHX16-RIG-I binding was further substantiated as endogenous DHX16 interacted with endogenous RIG-I only during IAV infection (Figure 4E). Functionally, induction of IFN-I by DHX16 required RIG-I as overexpression of DHX16 enhanced an IFN-β luciferase reporter only in the presence of RIG-I (Figure 4F). Furthermore, overexpression of DHX16 resulted in enhanced mIFN-β transcript levels in WT, but not RIG-I KO MEFs (Figure 4G). Together, these results identify DHX16 as a factor required for optimal RIG-I-mediated IFN-I production in response to specific viral PAMPs.
Figure 4.
DHX16 interacts with RIG-I to enhance IFN-I production
(A) DHX16 interacts with RIG-I following co-expression in HEK293Ts (coIP).
(B) Endogenous DHX16 interacts with ectopic RIG-I in HEK293Ts (coIP).
(C) Diagram of RIG-I truncation mutants.
(D) Interaction between DHX16 and RIG-I domain truncations following co-expression in HEK293Ts (coIP).
(E) Endogenous DHX16 and RIG-I interact in A549s infected with IAV for 24 h (PR8 MOI = 1) (coIP).
(F) IFN-β luciferase reporter activity in HEK293Ts following DHX16 and RIG-I co-expression.
(G) DHX16 and mIFN-β expression from WT or RIG-I KO MEFs following transfection of DHX16 (qRT-PCR and immunoblot).
Data are expressed as means (n = 3) ± SD. ∗∗p < 0.01; ∗∗∗∗p < 0.0001 (one-way ANOVA with Tukey's multiple comparisons). Data are representative of 2–3 independent experiments.
See also Figure S3.
Unanchored Ub chains and RNA are critical for optimal DHX16-mediated IFN-I production
To examine the role for unanchored Ub in the DHX16-RIG-I-IFN-I signaling axis, the fully catalytic form of IsoT, capable of specifically cleaving unanchored Ub chains, was used in a de-ubiquitination assay (Figure 5 A). Overexpression of IsoT reduced the levels of unanchored K48-poly-Ub interacting with DHX16 (Figure 5B). Increases in IFN-β observed upon overexpression of DHX16 were significantly reduced in the presence of IsoT under mock and IAV-infected conditions establishing a dependence on unanchored Ub (Figure 5C). Since recent investigations proposed that unanchored Ub is an important signaling mediator (Pertel et al., 2011; Rajsbaum et al., 2014b; Xia et al., 2009; Zeng et al., 2010), we questioned whether these free Ub chains could modulate DHX16 interactions with RIG-I. To address this, purified DHX16, RIG-I, and unanchored Ub chains were mixed in vitro. DHX16 and RIG-I interacted in a cell-free system, and this was further enhanced by unanchored K48-poly-Ub chains. In contrast, unanchored K63-poly-Ub chains did not increase DHX16-RIG-I binding (Figure 5D). Collectively, these results suggest that unanchored K48-poly-Ub chains promote interactions between RIG-I and DHX16 necessary for optimal downstream signaling events.
Figure 5.
DHX16 interacts with unanchored Ub and RNA to promote IFN-I production
(A) The IsoT de-ubiquitination assay.
(B) De-coupling of unanchored K48-poly-Ub from DHX16 following co-expression of IsoT in HEK293Ts (coIP).
(C) IFN-β expression following co-expression of DHX16 and IsoT in HEK293Ts infected with IAV for 24 h (PR8 MOI = 0.1) (qRT-PCR).
(D) In vitro interactions between DHX16 and RIG-I ± recombinant poly-Ub chains (coIP).
(E) Effects of recombinant poly-Ub (1 μg) and purified IAV vRNA (100 ng) on DHX16 ATPase activity in an in vitro ATPase assay.
(F) Interaction between DHX16 and unanchored K48-poly-Ub following co-expression in HEK293Ts in the presence of RNase A (100 μg/mL) (coIP).
(G) Immunoblot quantification of K48-Ub in (F) (densitometry).
(H) Interactions between DHX16 and RIG-I following co-expression in HEK293Ts in the presence of RNase A (0–100 μg/mL) (coIP).
(I) DHX16-RIG-I complex formation mediated through vRNA and unanchored poly-Ub.
Data are expressed as means (n = 3) ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 (one-way ANOVA with Tukey's multiple comparisons). Data are representative of 2–3 independent experiments.
We then tested whether unanchored Ub or vRNA altered the ability of DHX16 to hydrolyze ATP in an in vitro ATPase assay (Figure 5E). Unanchored K48-poly-Ub did not enhance DHX16's ATPase activity over basal levels, while purified IAV vRNA alone significantly increased DHX16's ATPase activity (Figure 5E). This increase could be enhanced further when IAV vRNA and unanchored K48-poly-Ub were added concurrently, whereas addition of unanchored K63-poly-Ub did not have an effect (Figure 5E). To address the possibility of a required DHX16-RNA interaction for unanchored K48-poly-Ub binding, lysates from cells expressing DHX16 were treated with RNase A to cleave ssRNA bound to DHX16 (Figure 5F). RNase A treatment reduced the amount of unanchored K48-poly-Ub co-purifying with DHX16 (Figures 5F and 5G). Since unanchored K48-poly-Ub improved DHX16-RIG-I interactions (Figure 5D), and since both RIG-I and DHX16 are known to interact with RNA, we reasoned that DHX16 and RIG-I form a complex via RNA. Treatment with RNase A prior to immunoprecipitation of lysates co-expressing DHX16 and RIG-I reduced the amount of DHX16 bound to RIG-I by as much as 80% (Figure 5H). Collectively, these data suggest that vRNA and unanchored K48-poly-Ub promote the formation of a RIG-I-DHX16 complex that is necessary for optimal IFN-I induction during virus infection (Figure 5I).
TRIM6 synthesizes unanchored K48-poly-Ub that interacts with DHX16 and regulates its activity
Since the E3-Ub ligase TRIM6 was previously found to promote IFN-I by synthesizing unanchored K48-poly-Ub (Rajsbaum et al., 2014b; van Tol et al., 2020), we hypothesized that TRIM6 may also be responsible for the unanchored K48-poly-Ub interacting with DHX16. TRIM6 and DHX16 interacted in a coIP and reciprocal coIP (Figures 6A and S4A), and the C-terminal SPRY domain of TRIM6 was responsible for this binding (Figures 6B and 6C). TRIM6 is a known cytoplasmic factor, and DHX16 is reported to localize mostly in the nucleus (Bharaj et al., 2016, 2017; Gencheva et al., 2010a; Rajsbaum et al., 2014b; Reymond et al., 2001; Zang et al., 2014). We examined whether these factors relocalize for interaction by confocal microscopy. A549s overexpressing either TRIM6 or DHX16 displayed each factor in their known locations, while co-expression resulted in a drastic redistribution of DHX16 from the nucleus to the cytoplasm where it colocalized with TRIM6 (Figures 6D and 6E). Notably, A549s maintain a sizable proportion of endogenous DHX16 in the cytoplasm (Figure S4B).
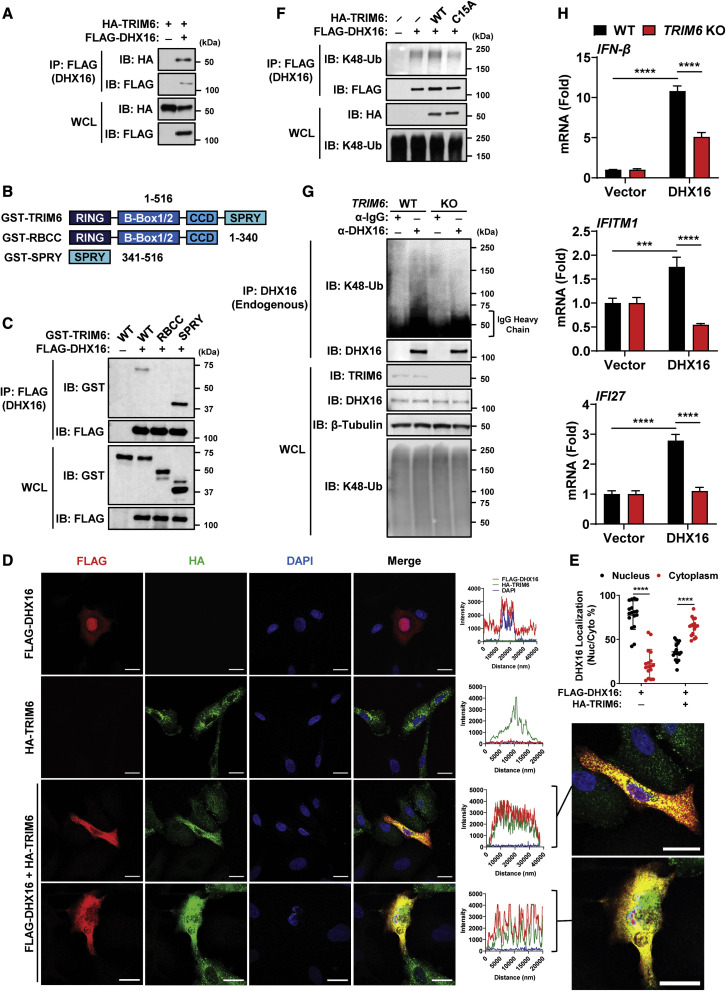
Figure 6.
DHX16 interacts with the E3-Ub ligase TRIM6
(A) Interaction between DHX16 and TRIM6 following co-expression in HEK293Ts (coIP).
(B) Diagram of TRIM6 deletion mutants.
(C) Interaction between DHX16 and the SPRY domain of TRIM6 following co-expression in HEK293Ts (coIP).
(D) Co-localization between DHX16 and TRIM6 following co-expression in A549s. Scale bar, 20 μm (confocal microscopy).
(E) Nuclear/cytoplasmic distribution of DHX16 following TRIM6 co-expression in A549s (n = 17).
(F) Interaction between DHX16 and K48-poly-Ub following co-expression with either WT or C15A TRIM6 in HEK293Ts (coIP).
(G) Interaction between endogenous DHX16 and K48-poly-Ub in WT or TRIM6 KO A549s (coIP).
(H) IFN-β, IFITM1, and IFI27 expression from WT or TRIM6 KO A549s transfected with DHX16 for 24 h (qRT-PCR).
Data are expressed as means (n = 3) ± SD. ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 (one-way or two-way ANOVA with Tukey's multiple comparisons). Data are representative of 2–3 independent experiments.
See also Figures S4 and S5.
We tested whether TRIM6 promotes interactions between unanchored K48-poly-Ub and DHX16 (Figure S4C). The association of endogenous K48-poly-Ub chains with DHX16 was enhanced in the presence of TRIM6, but not with a catalytically inactive form (TRIM6-C15A) (Bharaj et al., 2017; Rajsbaum et al., 2014b), in a coIP assay (Figure 6F). The unanchored nature of these TRIM6-dependent chains was confirmed in a coIP in which high salt washes removed K48-poly-Ub from DHX16 (Figure S4D). Interactions between endogenous K48-poly-Ub and endogenous DHX16 were detected in WT A549s but were significantly reduced in TRIM6 KO A549s (Figure 6G). Furthermore, TRIM6 was required for efficient DHX16-mediated IFN-I responses as overexpression of DHX16 significantly increased IFN-β and ISG mRNA in WT, but not TRIM6 KO A549s (Figure 6H).
We then asked whether DHX16, RIG-I, and TRIM6 interact in a complex. DHX16, RIG-I, and TRIM6 form a complex upon overexpression (Figure S4E) or in a cell-free system with purified proteins (Figure S4F). Additionally, endogenous DHX16, RIG-I, and TRIM6 form a complex during IAV infection (Figure S4G). Overexpression of all three factors resulted in relocalization of DHX16 from the nucleus to the cytoplasm, where it colocalized with both RIG-I and TRIM6 in distinct cytoplasmic puncta (Figure S5A). IAV infection promotes a small, but statistically significant, relocalization of endogenous DHX16 from the nucleus to the cytoplasm where it co-localizes with RIG-I (Figures S5B and S5C). The presence of a steady-state level of cytoplasmic DHX16, combined with further relocalization during infection, is consistent with an interaction between DHX16, RIG-I, and TRIM6 in the cytoplasm. Together, these data identify TRIM6 as a binding partner of DHX16 that promotes a complex with RIG-I. It also suggests that the unanchored K48-poly-Ub synthesized by TRIM6 is an important modulator of DHX16-mediated IFN-I.
DHX16 requires an intact motif III for RNA-binding and IFN-I production
To understand what aspects of DHX16 are required for its IFN-I response, we generated mutations to alter functional activities within the eight conserved motifs of the helicase domain (Figures 7A and S6A). Mapping of DHX16 identified a need for the HELICc domain to interact with K48-poly-Ub (Figure S6B). To narrow down the region required for K48-poly-Ub, the HELICc domain was truncated to remove motif IV (FLTG) or motif VI (QRAGRAGR) (Figure S6C). K48-poly-Ub associated with WT and Δ-VI-HELICc, but it had reduced affinity for Δ-IV-HELICc (Figure S6D). The individual DHX16 domains were made to determine the regions required for binding. The DEXDc, HELICc, and DUF domains were each sufficient for interacting with RIG-I and TRIM6 (Figures S6E and S6F). Expression of the HA2 domain was undetectable, and we could not conclude whether this domain was involved in interactions. Consistent with the interactions between HELICc and Ub, overexpression of the HELICc domain alone induced phosphorylation of IRF3 and TBK1 and enhanced expression of IFN-β mRNA, albeit to lower efficiency compared with WT DHX16 (Figures 7B and 7C). Interestingly, the S552L and DEXDc mutants did not induce phosphorylation of IRF3 and TBK1, IFN-β mRNA, or expression of IRF7 protein (an ISG), while the G724N mutant had reduced induction compared with WT DHX16 (Figures 7B and 7C). Overexpression of the S552L mutant failed to enhance basal or 3p-hpRNA-stimulated IFN-β mRNA levels compared with WT DHX16 (Figure 7D). Mutations at S552L and G724N are analogous to mutations S378L and G551N found in Prp2, the yeast homolog of DHX16, which reduce ATPase activity (Edwalds-Gilbert et al., 2000; Gencheva et al., 2010a; Plumpton et al., 1994; Zang et al., 2014). ATP hydrolysis has been shown to be an important determinant for the function of other helicases in innate immunity, or entirely dispensable (Lu et al., 2014; Mosallanejad et al., 2014; Pattabhi et al., 2019; Yoo et al., 2014). We investigated whether the loss in IFN-β mRNA for the S552L and G724N mutants was attributed to an altered ATPase activity. In the presence of a Poly U30 RNA substrate known to stimulate the ATPase activity of Prp2 (Plumpton et al., 1994; Warkocki et al., 2015), WT and G724N DHX16 hydrolyzed ATP, while S552L and constructs bearing mutations in the ATPase motifs (K428A and D520A) did not (Figure S6G). Importantly, while mutations K428A and D520A abolished ATPase activity, they did not impact IFN-β levels, suggesting that ATPase activity is dispensable for DHX16-mediated IFN-I production (Figure 7B). The requirement of both RIG-I and unanchored K48-poly-Ub led us to consider whether these mutations alter the ability of DHX16 to interact with its partners. All mutant forms of DHX16 co-purified with RIG-I and endogenous unanchored K48-poly-Ub, indicating the loss in IFN-I was not due to a reduced affinity for RIG-I or Ub (Figure S6H).
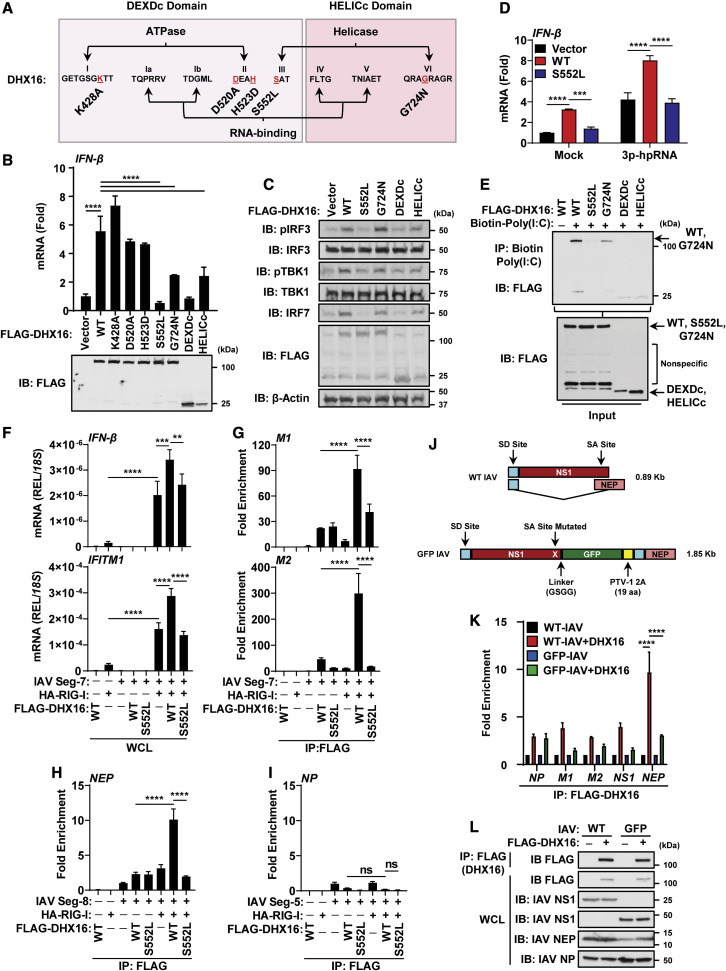
Figure 7.
DHX16 requires an intact motif III for RNA-binding and IFN-I production
(A) Diagram of DHX16 point mutations.
(B) IFN-β expression from A549s transfected with DHX16 (WT or mutants) for 24 h (qRT-PCR and immunoblot).
(C) IFN-I production pathway activation from A549s transfected with DHX16 (WT or mutants) for 24 h (immunoblot).
(D) IFN-β expression from A549s transfected with DHX16 (WT or S552L) and 3p-hpRNA (100 ng/mL) (qRT-PCR).
(E) In vitro RNA-binding ability of DHX16 (WT or mutants) (coIP).
(F–I) HEK293Ts transfected with DHX16 (WT or S552L) and segment 7, 8, or 5 of IAV ± RIG-I for 24 h (RNA-IP). (F) IFN-β and IFITM1 expression from RNA-IP (qRT-PCR). (G–I) Interaction between DHX16 (WT or S552L) and segment 7 (G), 8 (H), or 5 (I) of IAV ± RIG-I (fold enrichment over segment 7, 8, or 5 alone) (RNA-IP).
(J) Diagram of segment 8 from WT and GFP-NS1 IAV PR8 (modified from Manicassamy et al., 2010). WT PR8 expresses NS1/NEP proteins by mRNA splicing (top). GFP-NS1 PR8 expresses both NS1-GFP and NEP proteins from a single mRNA via a ribosome start/stop at the porcine teschovirus-1 2A (PTV-1 2A) site (bottom). Splice donor/acceptor sites (SD/SA).
(K) Interaction between DHX16 and IAV vRNAs from cells infected with WT or GFP-NS1 PR8 for 24 h (PR8 MOI = 0.1) (fold enrichment over IAV alone) (RNA-IP).
(L) Input protein expression from RNA-IP using IAV (WT or GFP-NS1 PR8) (immunoblot).
Data are expressed as means (n = 3) ± SD. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 (Student's t test or one-way ANOVA with Tukey's multiple comparisons). Data are representative of 2–3 independent experiments.
See also Figures S6 and S7.
RNA unwinding needs the binding energy of ATP, but it does not require its hydrolysis since conversion of ATP to ADP benefits DExD/H-box release from RNA promoting enzyme recycling for the next reaction step to proceed (Chen et al., 2008; Linder and Jankowsky, 2011; Liu et al., 2008). Once engaged with RNA, DExD/H-box helicases either catalyze strand separation of RNA duplexes or act as a nucleation point for the assembly of a larger ribonucleoprotein (RNP) complex (Linder and Jankowsky, 2011). While motifs I and II are defined as NTP-binding motifs, motifs III and VI convert the energy from NTP binding and hydrolysis into RNA unwinding and RNP remodeling through movement of the RecA1 and RecA2 domains (Bernstein et al., 2006; Tanner and Linder, 2001). An intact helicase domain, particularly motif III, is paramount for proper function as motif III employs the binding energy of ATP to orchestrate movement between motifs I, II, and VI that span the two RecA domains to generate a high-affinity ssRNA binding site (Banroques et al., 2010). To define the relevance of the S552L and G724N mutations, we asked whether these mutants had reduced binding to RNA. Purified WT DHX16 bound a biotinylated form of Poly(I:C), while the S552L mutant lost any detectable interaction. Furthermore, the G724N mutant displayed a weakened affinity for dsRNA compared with WT DHX16 (Figure 7E). Interestingly, both the DEXDc and HELICc domains could bind RNA probably due to each domain retaining RNA-binding motifs and possibly because both domains are free of the proposed inhibitory gating mechanism of the C-terminal domain (Schmitt et al., 2018; Tauchert et al., 2017). This would also explain the ability of the HELICc domain to interact with K48-poly-Ub and induce downstream signaling.
We then set out to identify the PAMP that is recognized by DHX16, and how RIG-I is involved to trigger IFN-I. Since the S552L mutant of DHX16 cannot bind RNA (Figure 7E) and has also been shown to be deficient in splicing (Gencheva et al., 2010a), we hypothesized that DHX16 may recognize a splicing signal in vRNAs. The IAV genome is comprised of eight RNA segments, two of which (segments 7 and 8) undergo splicing to produce the M1 and M2 (segment 7) and the NS1 and NEP (segment 8) products (Dubois et al., 2014). To determine whether DHX16 enhances IFN-I production through recognition of a specific IAV segment, we performed RNA-immunoprecipitation (RNA-IP). DHX16 (WT or S552L) and vectors expressing the full-length IAV segments 5, 7, and 8 were expressed in cells, and equal levels of protein were confirmed by immunoblot (Figure S6K). Although segments 7 and 8 of IAV can undergo splicing, segment 5 does not (Dubois et al., 2014). To test the contribution of RIG-I, the experiment was performed in the presence or absence of overexpressed RIG-I. Although WT DHX16 was able to bind both spliced forms of IAV segment 7 (M1 and M2) and segment 8 (NEP), these interactions were significantly increased when RIG-I was co-expressed (Figures 7G and 7H). Among the IAV segments tested, DHX16 displayed affinity for the M1 and M2 products of segment 7 and the NEP product of segment 8, but not for segment 5 (NP) (Figures 7G–7I), suggesting a splicing signal may be involved for recognition. In support of this, the S552L mutant failed to interact with any of the vRNA forms tested (Figures 7G and 7H). Strikingly, the ability of DHX16 to interact with specific IAV segments correlated with the expression levels of IFN-β and IFITM1 mRNA. When RIG-I alone was overexpressed, IFN-β and IFITM1 levels were enhanced when provided with IAV segment 7, and this was amplified in the presence of WT, but not S552L, DHX16 (Figure 7F). Of note, transfection of segment 8, which encodes the well-established IFN-I antagonist NS1, completely blocked IFN-β as expected (Figures S6I and S6J).
To examine whether DHX16's ability to recognize IAV vRNAs involved splicing, an RNA-IP was performed from IAV-infected cells using either the WT PR8 strain of IAV, capable of splicing its segment 8 into both the NS1 and NEP mRNAs, or a recombinant GFP-NS1 reporter IAV that cannot splice segment 8 and instead translates NS1 and NEP as a single polyprotein later separated via a 2A autoproteolytic cleavage site (Manicassamy et al., 2010) (Figure 7J). DHX16 showed an affinity for NEP mRNA from WT PR8, but this was significantly reduced for the GFP-NS1 virus, implicating the involvement of a splicing signal for recognition (Figure 7K). Equal levels of DHX16 and viral proteins are shown as control (Figure 7L). To rule out the possibility that DHX16 affects splicing of vRNAs, which could result in spliced forms recognized by RIG-I, we examined the RNA and protein abundance of spliced IAV transcripts upon DHX16 silencing. DHX16 knockdown did not alter vRNA ratios or protein expression for any of the spliced IAV genes (Figures S6L and S6M). Finally, since DHX16 is involved in optimal IFN-I induction upon infection with SARS-CoV-2, and because SARS-CoV-2 is not known to undergo splicing, we asked whether DHX16 interacts with a specific region of SARS-CoV-2 vRNA. SARS-CoV-2-infected cells subjected to RNA-IP (Figure S7A) showed WT DHX16, but not S552L, enriched for the ORF1a/ab region (ORF1a alone or ORF1ab) of SARS-CoV-2 vRNA (Figures S7B–S7E). Taken together, DHX16 can recognize specific vRNA signals via its conserved motif III to promote an efficient antiviral IFN-I response.
Discussion
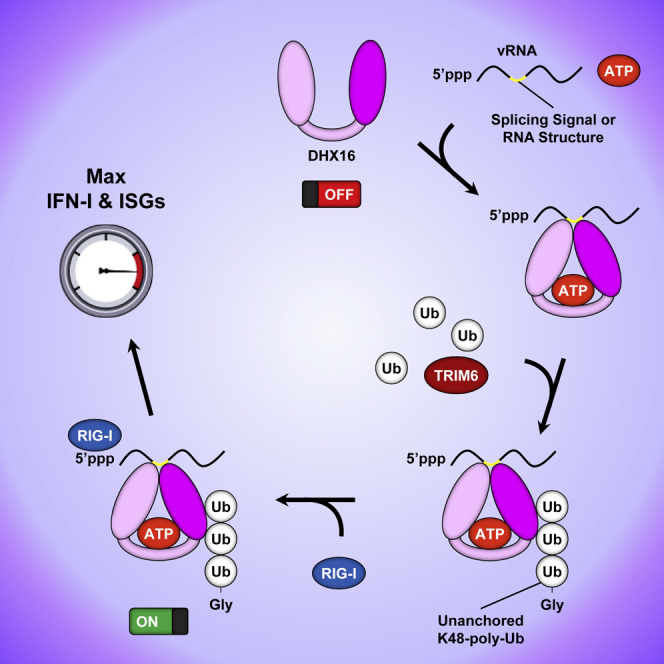
In this study, we identified the DEAH-box helicase DHX16 as required for optimal IFN-I production through an interaction with RIG-I, and this is regulated by unanchored K48-poly-Ub chains synthesized by the E3-Ub ligase TRIM6 (Figure S7F). These findings revealed a role for unanchored Ub as an activator of innate immune signaling that mirrors previous observations by ourselves and others (Jiang et al., 2012; Rajsbaum et al., 2014b; Xia et al., 2009; Zeng et al., 2010). Our data and these previous studies indicate that both unanchored K48- and K63-poly-Ub activate different signaling pathways for a full and efficient IFN-I response, but mechanistically they are needed at different steps (K63 chains promote RIG-I binding to MAVS, whereas K48 chains promote DHX16 binding to RIG-I). This suggests unanchored Ub may function as a broader signaling platform warranting further investigation. Our study also identifies TRIM6 as the responsible E3 ligase for the unanchored K48-poly-Ub interacting with DHX16. This information echoes earlier findings revealing a role for TRIM6 and unanchored K48-poly-Ub in the IFN-I response (Giraldo et al., 2020a).
These findings discern a mechanism for how DHX16 engages RIG-I to trigger IFN-I. DHX16's ability to interact with RIG-I in vitro required unanchored K48-poly-Ub and the presence of ssRNA (Figure S7F). The RNAs that activate DHX16 contain a 5′-triphosphate and a splicing signal, but they do not appear to be dsRNA. DHX16 is important for optimal IFN-I induction against ZIKV and SARS-CoV-2, indicating DHX16 may provide additional PAMP-sensing specificity for RIG-I. RIG-I recognizes flaviviruses (Chazal et al., 2018) and has been implicated in sensing coronaviruses (Chang et al., 2020; Chen et al., 2020; Hu et al., 2017; Yamada et al., 2021). Although evidence indicates SARS-CoV-2 induces IFN-I primarily through MDA5 in Calu-3 cells (Rebendenne et al., 2021; Yin et al., 2021), a role for RIG-I may apply depending on cell types and conditions. It is possible RIG-I recognizes SARS-CoV-2 in addition to MDA5 (Yamada et al., 2021), but the effects could be obscured by viral antagonism as some SARS-CoV-2 proteins inhibit IFN-I upon RIG-I stimulation (Chen et al., 2020; Xia et al., 2020).
DExD/H-box proteins have eight highly conserved motifs across their two RecA domains. Mutations in these motifs are detrimental to IFN-I responses (Yoo et al., 2014; Zhu et al., 2018). Our data indicate that a motif III mutant of DHX16 failed to recognize vRNA and was unable to potentiate an IFN-I response. A similar SAT-to-LAT motif III mutation in another SF2 helicase member, DDX3, resulted in a loss to both ATPase and helicase activities (Yedavalli et al., 2004). Motif III mutations cannot translate the binding energy of ATP's γ phosphate into a high-affinity RNA-binding site and movement of the RecA domains. This negatively impacts the RNA-dependent hydrolysis of ATP by failing to constrain the catalytic core enough to hydrolyze the β,γ phosphoanhydride bond (Banroques et al., 2010; Tanner and Linder, 2001). Motif III maintains an essential role in DExD/H-box helicases for their involvement in RNA metabolism and their non-canonical functions. Since the DHX16 helicase motif is essential for recognition of vRNAs, and since mutations within this motif result in reduced splicing activity (Gencheva et al., 2010a), we propose a model in which DHX16 couples its recognition of splicing signals with IFN-I responses. In this model, RIG-I recognizes a 5′-triphosphate, while DHX16 provides a high-affinity interaction with specific vRNAs (Figure S7G). DHX16's ability to aid RIG-I may be attributed to a unique signal or sequence in vRNAs as DHX16 targets IAV vRNAs that can be spliced. Recognition of vRNA by DHX16 required a functional motif III, suggesting that the RNA-clamp mechanism used by these helicases to engage host RNA also applies to foreign genetic material.
Recent reports have identified roles for DExD/H-box helicases in innate immunity as co-receptors to RLRs (DHX15, DHX29, DHX36, and DDX60) (Miyashita et al., 2011; Pattabhi et al., 2019; Sugimoto et al., 2014; Yoo et al., 2014; Zhu et al., 2018) and other pathway components like MAVS, STING, NLRP3, and TLRs (DHX9, DHX33, DHX36, DDX3, DDX1, DDX21, and DDX41) (Liu et al., 2014; Mitoma et al., 2013; Zhang et al., 2011a, 2011b, 2011c). This involvement of so many members from the same protein superfamily suggests DExD/H-box helicases at-large are at the forefront of combating pathogens in addition to their roles in RNA metabolism (Fullam and Schroder, 2013). A logical question that arises is why do RLRs like RIG-I rely on numerous factors to fully induce IFN-I? One explanation could be that since RIG-I itself is an ISG with low basal expression, there exists a need to rapidly induce RLR-mediated signaling. This need for a prompt, robust response may be satisfied by DExD/H-box helicases by enhancement or maintenance of PAMP recognition for PRRs (Pattabhi et al., 2019; Zhu et al., 2018), or the recruitment or redistribution of PRRs to signaling platforms (Mitoma et al., 2013; Yoo et al., 2014; Zhang et al., 2011a). Another explanation is that these helicases further identify PAMPs based on sequence signals in addition to RIG-I's recognition of 5′-triphosphate RNA and short dsRNA (Hornung et al., 2006; Schlee et al., 2009). Our study identifies segments 7 and 8 from IAV as specific vRNAs recognized by DHX16, suggesting DHX16's ability to identify PAMPs may be related to splicing. Intriguingly, ZIKV and SARS-CoV-2 are non-segmented, positive-sense RNA viruses encoding one full-length polyprotein and are not known to produce spliced vRNAs. However, DHX16 can recognize Poly U sequences (Plumpton et al., 1994), and it has been proposed that SARS-CoV-2 vRNA can be sensed by host PRRs via Poly U tails (Hackbart et al., 2020). DHX16 might also identify a structure within their genomes (Alvarez et al., 2005; Chazal et al., 2018; Tavares et al., 2020). Furthermore, coronavirus transcription is known to produce subgenomic mRNAs (Dufour et al., 2011; Gribble et al., 2021; Makino et al., 1986; Sola et al., 2015), which could provide DHX16 an unknown PAMP.
Collectively, our study provides evidence for DHX16 as a novel unanchored Ub-interacting factor necessary for optimal RIG-I-mediated IFN-I production and ISG expression in response to viral challenge. To our knowledge, DHX16 is the first example of a DEAH-box helicase requiring unanchored Ub and recognition of splicing signals to trigger an efficient antiviral response. These findings provide new insights into the involvement of DExD/H-box helicases in the immune system and may guide strategies for intervening in antiviral innate immunity via activation of non-RLR helicases against a broad spectrum of pathogens.
Limitations of the study
For the in vivo relevance of DHX16, we observed significant differences upon DHX16 knockdown with PPMOs; however, in some cases the differences are small. The use of PPMOs in vivo does present technical limitations, which could include tissue specificity of DHX16 expression, differences in immune cell infiltration, or whether the effects of DHX16 knockdown in specific cell types could be limited due to the efficiency in which the PPMO can be delivered and internalized. In vivo, our study lacks insight into the contributions of DHX16 in immune cells that produce relevant cytokines, which may be needed to appreciate the observed delay in weight gain during the recovery phase of IAV infection. Future studies may include immunostaining of tissues to assess lung injury after infection between NTC and DHX16 PPMO treatments. The inability to obtain a DHX16 KO animal limited our study to gene knockdown, which may have off-target effects. However, the PPMO and siRNAs tested recognize different sequences in human and murine DHX16 and obtained consistent phenotypes reducing this possibility; the PPMO targets the exon 2-intron 2 junction of murine DHX16 pre-mRNA, while siDHX16 #1 and #2 target nucleotides #2601-2615 and #761-779 in human DHX16 pre-mRNA, respectively.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-DHX16 | Bethyl | Cat #A301-537A; RRID:AB_1039995 |
| anti-TRIM6 N-term | Sigma-Aldrich | Cat #SAB1306751 |
| anti-K48-linked Ubiquitin | Millipore | Cat #05-1307; RRID:AB_1587578 |
| anti-K63-linked Ubiquitin | Millipore | Cat #05-1313; RRID:AB_1587585 |
| anti-Ubiquitin | Enzo Life Sciences | Cat #BML-PW0930-0100; RRID:AB_11181462 |
| anti-FLAG | Sigma-Aldrich | Cat #F7425; RRID:AB_439687 |
| anti-FLAG | Cell Signaling Technology | Cat #3768S; RRID:AB_1642183 |
| anti-HA | Sigma-Aldrich | Cat #H6908; RRID:AB_260070 |
| anti-HA | Cell Signaling Technology | Cat #2350S; RRID:AB_491023 |
| anti-GST | Bethyl | Cat #A190-122A; RRID:AB_67419 |
| anti-GST | Cell Signaling Technology | Cat #3445S; RRID:AB_1281290 |
| anti-IAV | GeneTex | Cat #GTX40388; RRID:AB_424056 |
| anti-DAPI | Thermo Fisher Scientific | Cat #D1306; RRID:AB_2629482 |
| anti-His | Sigma-Aldrich | Cat #H1029; RRID:AB_260015 |
| anti-Lamin A/C | Santa Cruz Biotechnology | Cat #sc-376248; RRID:AB_10991536 |
| anti-RIG-I 1C3 | Adolfo Garcia-Sastre | N/A |
| anti-pIRF3 (S386) | Abcam | Cat #ab76493; RRID:AB_1523836 |
| anti-IRF3 | Abnova | Cat #H00003661-B01; RRID:AB_1114076 |
| anti-pTBK1 (S172) | Abcam | Cat #3300-1; RRID:AB_10635157 |
| anti-TBK1 | Novus | Cat #NB100-56705; RRID:AB_838420 |
| anti-IRF7 | Cell Signaling Technology | Cat #4920; RRID:AB_2127551 |
| anti-IFITM1 | Thermo Fisher Scientific | Cat #PA5-20989; RRID:AB_11156379 |
| anti-RNase A | Thermo Fisher Scientific | Cat #PA5-78151; RRID:AB_2736524 |
| anti-IAV NP | Thermo Fisher Scientific | Cat #PA1-41071; RRID:AB_1076160 |
| anti-IAV M1 | GeneTex | Cat #GTX125928; RRID:AB_11171466 |
| anti-IAV M2 | Thermo Fisher Scientific | Cat #MA1-082; RRID:AB_325043 |
| anti-IAV NS1 | Adolfo Garcia-Sastre | Solórzano et al., 2005 |
| anti-IAV NEP | Thermo Fisher Scientific | Cat #PA5-32234; RRID:AB_2549707 |
| anti-SARS-CoV N | Shinji Makino | N/A |
| anti-β-Tubulin | Sigma-Aldrich | Cat #T8328; RRID:AB_1844090 |
| anti-β-Actin | Abcam | Cat #ab8227; RRID:AB_2305186 |
| anti-β-Actin | GeneTex | Cat #GTX629630; RRID:AB_2728646 |
| Alexa Fluor 555 | Thermo Fisher Scientific | Cat #A-31570; RRID:AB_2536180 |
| Alexa Fluor 633 | Thermo Fisher Scientific | Cat #A-21050; RRID:AB_141431 |
| Donkey Anti-Rabbit IgG, HRP Conjugated | GE Healthcare | Cat #NA934; RRID:AB_772206 |
| Sheep Anti-Mouse IgG, HRP Conjugated | GE Healthcare | Cat #NA931; RRID:AB_772210 |
| Bacterial and virus strains | ||
| E. coli DH5α | Invitrogen | Cat #18265017 |
| Influenza A virus (A/Puerto Rico/8/1934 (PR8) WT) | Adolfo García-Sastre | Rajsbaum et al., 2014b |
| Influenza A virus (A/Puerto Rico/8/1934 (PR8) NS1 R38A/K41A) | Adolfo García-Sastre | Donelan et al., 2003 |
| Influenza A virus (A/Puerto Rico/8/1934 (PR8) GFP-NS1) | Adolfo García-Sastre | Manicassamy et al., 2010 |
| Sendai virus (Cantell strain) | Charles River Laboratories | Cat #10100774 |
| Zika virus (ZIKV/Human/Cambodia/FSS13025/2010) | Pei-Yong Shi | Shan et al., 2016 |
| SARS-CoV-2 virus (USA-WA1/2020) | World Reference Center of Emerging Viruses and Arboviruses (WRCEVA) | N/A |
| SARS-CoV-2 virus (icSARS-CoV-2-mNG) | Pei-Yong Shi | Xie et al., 2020 |
| Chemicals, peptides, and recombinant proteins | ||
| TransIT-LT1 | Mirus Bio | Cat #MIR 2305 |
| Lipofectamine 2000 | Invitrogen | Cat #11668019 |
| Lipofectamine 3000 | Invitrogen | Cat #L3000015 |
| Lipofectamine RNAiMAX | Invitrogen | Cat #13778150 |
| Poly(I:C) HMW | InvivoGen | Cat #tlrl-pic-5 |
| Poly(dA:dT) | InvivoGen | Cat #tlrl-patn-1 |
| 3p-hpRNA | InvivoGen | Cat #tlrl-hprna |
| Biotin-labeled Poly(I:C) HMW | InvivoGen | Cat #tlrl-picb |
| IFN-β 1a | PBL Assay Science | Cat #11410-2 |
| N-ethylmaleimide | Sigma-Aldrich | Cat #E3876-25G |
| Iodoacetamide | Sigma-Aldrich | Cat #I1149-25G |
| FLAG peptide | Sigma-Aldrich | Cat #F3290-4MG |
| HA peptide | Thermo Scientific | Cat #26184 |
| L-Glutathione reduced | Sigma-Aldrich | Cat #G4251-25G |
| anti-FLAG M2 EZview agarose beads | Sigma-Aldrich | Cat #F2426 |
| anti-HA EZview agarose beads | Sigma-Aldrich | Cat #E6779-5X1ML |
| anti-GST EZview agarose beads | Sigma-Aldrich | Cat #E6406-1ML |
| Ni-NTA agarose beads | Qiagen | Cat #30210 |
| Protein A/G agarose beads | Pierce | Cat #20421 |
| High Capacity NeutrAvidin agarose beads | Thermo Scientific | Cat #29202 |
| TPCK-treated trypsin | Sigma-Aldrich | Cat #4370285 |
| ATP | Thermo Scientific | Cat # R0441 |
| Protease inhibitor cocktail | Roche | Cat #11873580001 |
| ECL chemiluminescence reagent | Pierce | Cat #32106 |
| SuperSignal West Femto chemiluminescence reagent | Pierce | Cat #34095 |
| BIOMOL Green reagent | Enzo Life Sciences | Cat #BML-AK111-0250 |
| 2-16 Ub linkage K48-poly-Ub chains | Enzo Life Sciences | Cat #BML-UW0670-0100 |
| 2-7 Ub linkage K48-poly-Ub chains | Boston Biochem | Cat #UC-230-100 |
| 2-7 Ub linkage K63-poly-Ub chains | Boston Biochem | Cat #UC-330-100 |
| GST-UBD-IsoT | UBPBio | Cat #I1400 |
| Critical commercial assays | ||
| Trans-Blot Turbo transfer system | Bio-Rad | Cat #1704150 |
| Direct-zol RNA Miniprep Kit | Zymo Research | Cat #R2053 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat #4368813 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat #1725125 |
| Dual-Luciferase reporter assay system | Promega | Cat #E1960 |
| VeriKine human IFN-β ELISA kit | PBL Assay Science | Cat #41410-2 |
| VeriKine mouse IFN-β ELISA kit | PBL Assay Science | Cat #42400-2 |
| NE-PER nuclear and cytoplasmic extraction kit | Thermo Scientific | Cat #78833 |
| QIAamp Viral RNA Mini Kit | Qiagen | Cat #52904 |
| Deposited data | ||
| Mass spectrometry proteomics data | This paper | PRIDE: PXD031365 |
| RNA-Seq data | This paper | GEO: GSE168255 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | CRL-11268 |
| HEK293T-hACE2 | Benhur Lee | Oguntuyo et al., 2021 |
| A549 (WT, TRIM6-/-) | ATCC and Ricardo Rajsbaum | CCL-185 and Bharaj et al., 2017 |
| MDCK | ATCC | CCL-34 |
| MEF (WT, RIG-I-/-) | Adolfo García-Sastre | Loo et al., 2008 |
| Calu-3 2B4 | Vineet D. Menachery | Lokugamage et al., 2020 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | The Jackson Laboratory | Cat #000664 |
| Mouse: BALBc | The Jackson Laboratory | Cat #000651 |
| Oligonucleotides | ||
| Primers for cloning DHX16 constructs | This paper | Table S1 |
| Primers for qRT-PCR used in this study | This paper and OriGene | Table S2 |
| Non-targeting siRNA control | Horizon (Dharmacon) | D-001810-10-05 |
| DHX8 siRNA | Horizon (Dharmacon) | L-010506-00-0005 |
| DHX15 siRNA | Horizon (Dharmacon) | L-011250-01-0005 |
| DHX16 siRNA | Horizon (Dharmacon) | L-011477-00-0005 |
| DHX16 siRNA | Horizon (Dharmacon) | J-011477-05-0002 |
| DHX16 siRNA | Horizon (Dharmacon) | J-011477-08-0002 |
| Non-targeting PPMO control | Hong M. Moulton | P7-NC705 |
| DHX16 PPMO | Hong M. Moulton | Dhx16-e2i2 |
| Recombinant DNA | ||
| Plasmid: pCAGGS | Ricardo Rajsbaum | Rajsbaum et al., 2014b |
| Plasmid: FLAG-DHX16 | OriGene | Cat #RC202912 |
| Plasmid: HA-RIG-I | Adolfo García-Sastre | Nistal-Villán et al., 2010 |
| Plasmid: FLAG-RIG-I | Adolfo García-Sastre | Nistal-Villán et al., 2010 |
| Plasmid: FLAG-RIG-I (2CARD: N-Terminus) | Adolfo García-Sastre | Nistal-Villán et al., 2010 |
| Plasmid: FLAG-RIG-I (C-Terminus) | Adolfo García-Sastre | Nistal-Villán et al., 2010 |
| Plasmid: HA-MAVS | Adolfo García-Sastre | Nistal-Villán et al., 2010 |
| Plasmid: HA-TRIM6 | Ricardo Rajsbaum | Rajsbaum et al., 2014b |
| Plasmid: HA-TRIM6 (C15A) | Ricardo Rajsbaum | Rajsbaum et al., 2014b |
| Plasmid: GST-TRIM6 | Ricardo Rajsbaum | Rajsbaum et al., 2014b |
| Plasmid: GST-TRIM6 (RBCC) | Ricardo Rajsbaum | Rajsbaum et al., 2014b |
| Plasmid: GST-TRIM6 (SPRY) | Ricardo Rajsbaum | Rajsbaum et al., 2014b |
| Plasmid: FLAG-DHX15 | OriGene | Cat #RC205914 |
| Plasmid: FLAG-DHX8 | OriGene | Cat #RC208403 |
| Plasmid: FLAG-IKKε | Adolfo García-Sastre | Rajsbaum et al., 2014b |
| IFN-β Luciferase | Adolfo García-Sastre | Yoneyama et al., 1996 |
| REN-Luc/pRL-TK | Promega | Cat #E2241 |
| Plasmid: His-IsoT | Ricardo Rajsbaum | Rajsbaum et al., 2014b |
| Plasmid: HA-Ub | Ricardo Rajsbaum | Rajsbaum et al., 2014b |
| Plasmid: His-Ub | Ricardo Rajsbaum | Rajsbaum et al., 2014b |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad Software, LLC | https://www.graphpad.com/scientific-software/prism/ |
| ZEN lite Software v2.5 | Carl Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html |
| FIJI (ImageJ) v1.53m | NIH | https://imagej.net/Fiji |
| Other | ||
| Cytation 5 Multi-Mode Reader | BioTek | N/A |
| Zeiss LSM 880 with Airyscan | Carl Zeiss | N/A |
| CFX384 Touch Real-Time PCR Detection System | Bio-Rad | Cat #1855484 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ricardo Rajsbaum (rirajsba@utmb.edu).
Materials availability
Plasmids generated in this study are available upon request from the lead contact.
Experimental model and subject details
Cell lines
HEK293T (CRL-11268), A549 (CCL-185), and MDCK (CCL-34) cell lines were purchased from ATCC. Calu-3 2B4 cells were kindly provided by Vineet D. Menachery (The University of Texas Medical Branch at Galveston) (Lokugamage et al., 2020). HEK293T-hACE2 cells were kindly provided by Benhur Lee (Icahn School of Medicine at Mount Sinai) (Oguntuyo et al., 2021). WT and RIG-I -/- MEFs were kindly provided by Adolfo García-Sastre (Icahn School of Medicine at Mount Sinai) (Loo et al., 2008). TRIM6 -/- A549s were generated as previously described (Bharaj et al., 2017). All cells were maintained in Dulbecco’s Modified Eagle's Medium (DMEM) (Corning) supplemented with 10% v/v fetal bovine serum (FBS) (HyClone) and 1% v/v penicillin-streptomycin (Corning). Cells used for transient transfections were plated in (DMEM) supplemented with 10% v/v (FBS) lacking 1% v/v penicillin-streptomycin.
Viruses
Viruses used in this study were handled under biosafety level 2 (BSL-2), BSL-3, and BSL-4 conditions at UTMB in accordance with institutional biosafety approvals. Influenza A/Puerto Rico/8/1934 (PR8) WT, NS1 R38A/K41A, and GFP-NS1 viruses were kindly provided by Adolfo García-Sastre (Icahn School of Medicine at Mount Sinai) (Donelan et al., 2003; Manicassamy et al., 2010) and were grown in MDCK cells as previously described (Eisfeld et al., 2014). Sendai virus (Cantell strain) was obtained from Charles River Laboratories. SARS-CoV-2 (USA-WA1/2020) was kindly provided by The World Reference Center of Emerging Viruses and Arboviruses (WRCEVA) (The University of Texas Medical Branch at Galveston). Zika virus (ZIKV/Human/Cambodia/FSS13025/2010) and SARS-CoV-2 (icSARS-CoV-2-mNG) produced from infectious cDNA clones were kindly provided by Pei-Yong Shi (The University of Texas Medical Branch at Galveston) (Shan et al., 2016; Xie et al., 2020).
Mice
All animal experiments were carried out in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines and have been approved by the IACUC of the University of Texas Medical Branch at Galveston. Eight- to 12-week-old BALB/c or C57BL/6 female mice (The Jackson Laboratory) were maintained under specific-pathogen-free conditions in the Animal Resource Center (ARC) facility at UTMB. Animal experiments involving infectious virus were performed under animal biosafety level 2 (ABSL-2) conditions at UTMB in accordance with institutional biosafety approvals.
Method details
Plasmids
The HA-TRIM6 plasmid and mutants were generated as previously described (Rajsbaum et al., 2014b). The FLAG-RIG-I, HA-MAVS, and reporter plasmids expressing firefly luciferase under the control of the IFN-β promoter were kindly provided by Adolfo García-Sastre (Icahn School of Medicine at Mount Sinai) (Nistal-Villán et al., 2010; Yoneyama et al., 1996). The reporter plasmid carrying the Renilla luciferase gene (REN-Luc/pRL-TK) was purchased from Promega. DHX16 mutants were generated by site-directed mutagenesis PCR and cloned in-frame into the pCAGGS or pCMV6-Entry (OriGene) expression plasmids using primers listed in Table S1. All sequences were confirmed by sequencing analysis at the UTMB molecular genomics core facility.
Transfections and stimulations
Transient transfections of DNA were performed with TransIT-LT1 (Mirus Bio), Lipofectamine 2000, and Lipofectamine 3000 (Invitrogen) according to the manufacture’s guidelines. Media was exchanged 4 to 6 hrs after plasmid transfection with Lipofectamine reagents. Poly(I:C) HMW, Poly(dA:dT), 3p-hpRNA (InvivoGen), and purified IAV vRNA stimulations were performed with Lipofectamine 2000. IFN stimulations were performed with human IFN-β 1a (PBL Assay Science).
Cell lysis and co-immunoprecipitation
Cells were harvested in RIPA lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% (v/v) IGEPAL CA-630, 0.5% (w/v) sodium deoxycholate, 0.1% (v/v) SDS, protease inhibitor cocktail (Roche), 5 mM N-ethylmaleimide (Sigma), and 5 mM iodoacetamide (Sigma) as deubiquitinase inhibitors. Cell lysates were clarified by centrifugation at 21,000 x RCF for 20 min at 4°C. 10% of the clarified lysate was added to 2X Laemmli buffer containing 2-Mercaptoethanol, heated for 10 min at 95°C, and stored at −20°C as the whole-cell lysate (WCL). The remaining lysate was subjected to immunoprecipitation with anti-FLAG M2, anti-HA, or anti-GST EZview Red agarose beads (Sigma), Ni-NTA agarose beads (Qiagen), or protein A/G agarose beads (Pierce). For endogenous proteins, lysates were subjected to immunoprecipitation with 1 μg of primary antibody (4–16 μg in the case of GST-UBD-IsoT: UBPBio) overnight at 4°C followed by incubation with the appropriate capture beads overnight at 4°C on a rotating platform. Beads were washed seven times in RIPA buffer (150 or 550 mM NaCl) and the bound proteins were eluted by heating samples in 2X Laemmli buffer containing 2-Mercaptoethanol for 10 min at 95°C.
Denaturing co-immunoprecipitation
Cell lysis, WCL collection, and co-immunoprecipitation were performed as above except the lysate was subjected to immunoprecipitation with nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen) overnight at 4°C on a rotating platform. Beads were washed seven times with either RIPA buffer containing 50 mM imidazole to remove excess non-specific binding to the beads (non-denaturing wash) or with a denaturing buffer (50 mM Tris-HCl pH 8.0, 6 M urea, 350 mM NaCl, 0.5% (v/v) IGEPAL CA-630, 40 mM imidazole). The bound proteins were eluted on a rotating platform at 4°C for 30 min in denaturing elution buffer (50 mM Tris-HCl pH 8.0, 300 mM imidazole).
RNA-immunoprecipitation
Cells were harvested in RIP lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM KCl, 5 mM EDTA, 0.5 mM DTT, 0.5% (v/v) IGEPAL CA-630, protease inhibitor cocktail (Roche), 5 mM N-ethylmaleimide (Sigma), and 5 mM iodoacetamide (Sigma) as deubiquitinase inhibitors. Co-immunoprecipitation was performed as above and 10% WCL was collected for RNA and protein inputs. Beads were washed seven times with RIP buffer and 10% was collected for RNA and protein inputs. The remaining beads were resuspended in TRIzol RNA extraction reagent to isolate bound RNAs.
Immunoblot analysis and antibodies
Cell lysates were resolved on 7.5 or 4–15% Mini-PROTEAN and Criterion TGX SDS-PAGE gels and then transferred to polyvinylidene difluoride (PVDF) membranes using a Trans-Blot Turbo transfer system (Bio-Rad). Membranes were blocked with 5% (w/v) non-fat dry milk in TBST (TBS with 0.1% (v/v) Tween-20) for 1 hr, and then probed with the indicated primary antibody in 3% (w/v) BSA in TBST at 4°C overnight. Following overnight incubation, membranes were probed with the following secondary antibodies in 5% (w/v) non-fat dry milk in TBST for 1 hr at room temperature: anti-rabbit or anti-mouse IgG-HRP conjugated antibody from sheep (both 1:10,000 NA934 and NA931 GE Healthcare). Proteins were visualized using ECL or SuperSignal West Femto chemiluminescence reagents (Pierce) and detected by autoradiography. The following primary antibodies were used: anti-DHX16 (1:2000 A301-537A Bethyl), anti-TRIM6 N-term (1:1000 SAB1306751 Sigma), anti-K48-linked Ubiquitin (1:1000 05-1307 Millipore), anti-K63-linked Ubiquitin (1:1000 05-1313 Millipore), anti-Ubiquitin (1:1000 P4D1 Enzo), anti-FLAG (1:2000 F7425 Sigma), anti-HA (1:2000 H6908 Sigma), anti-His (1:2000 H1029 Sigma), anti-Lamin A/C (1:1000 sc-376248 SCBT), anti-RIG-I 1C3 (1:1000 kindly provided by Adolfo Garcia-Sastre: Icahn School of Medicine at Mount Sinai), anti-pIRF3 (S386) (1:1000 ab76493 Abcam), anti-IRF3 (1:1000 H00003661-B01 Abnova), anti-pTBK1 (S172) (1:1000 3300-1 Abcam), anti-TBK1 (1:1000 NB100-56705 Novus), anti-IRF7 (1:1000 4920S CST), anti-IFITM1 (1:1000 PA5-20989 Thermo Fisher Scientific), anti-GST (1:2000 A190-122A Bethyl), anti-RNase A (1:1000 PA5-78151 Thermo Fisher Scientific), anti-IAV NP (1:1000 PA1-41071 Thermo Fisher Scientific), anti-IAV M1 (1:1000 GTX125928 GeneTex), anti-IAV M2 (1:1000 MA1-082 Thermo Fisher Scientific), anti-IAV NS1 (1:3000 kindly provided by Adolfo Garcia-Sastre: Icahn School of Medicine at Mount Sinai (Solórzano et al., 2005)), anti-IAV-NEP (1:1000 PA5-32234 Thermo Fisher Scientific), anti-SCoV N (1:2000 kindly provided by Shinji Makino: The University of Texas Medical Branch at Galveston), anti-β-Tubulin (1:2000 T8328 AA2 Sigma), and anti-β-Actin (1:1000 ab8227 Abcam and 1:10000 GTX629630 GeneTex).
Plaque assay
Influenza viral titers were determined by plaque assay on MDCK cells as previously described (Manicassamy et al., 2010). Briefly, confluent monolayers of A549 cells were washed twice with DPBS, and virus diluted in DPBS with 0.4% (w/v) bovine serum albumin (BSA) was incubated on cells at 37°C for 1 hr. Inoculations were removed, cells were washed once with DPBS, overlaid with DMEM containing 0.4% (w/v) BSA, 0.1 μg/mL TPCK-treated trypsin, and incubated at 37°C. Supernatants were collected for plaque assay at the indicated time points. For plaque assays, confluent monolayers of MDCK cells were washed twice with DPBS and virus diluted in DPBS with 0.4% (w/v) BSA was incubated on cells at 37°C for 1 hr. Infectious were removed, cells were washed once with DPBS and replaced with MEM containing 0.6% oxoid agar, 1 μg/mL TPCK-treated trypsin and incubated at 37°C for 2 days. Cells were fixed in 3.7% (v/v) paraformaldehyde in DPBS for 1 hr at room temperature, and stained with 1% (w/v) crystal violet, 20% (v/v) methanol in DPBS for 20 min at room temperature. Standard plaque assays were used for SCoV2 as previously described (Lokugamage et al., 2020).
Confocal microscopy
A549 cells seeded onto coverslips in 24-well plates (100,000 cells per well) were transfected with indicated plasmids using Lipofectamine 3000. 24 hrs post transfection, cells were washed once with DPBS and fixed for 10 min at room temperature with 4% (v/v) paraformaldehyde in DPBS. Cells were washed three times with DPBS-T (DPBS with 0.1% (v/v) Tween-20) and permeabilized for 10 min with 0.5% (v/v) IGEPAL CA-630 in DPBS. Cells were washed three times with DPBS-T and blocked for 30 min with 1% (w/v) BSA in DPBS-T. Cells were incubated with primary antibodies (anti-DHX16 1:200 A301-537A Bethyl, anti-RIG-I 1C3 1:200 kindly provided by Adolfo Garcia-Sastre: Icahn School of Medicine at Mount Sinai, FLAG 1:200 3768S CST, HA 1:800 2350S CST, GST 1:200 3445S CST, IAV 1:50 GTX40388 GeneTex) diluted in blocking buffer overnight at 4°C in a humidified container. Cells were washed three times with DPBS-T and incubated with secondary antibodies Alexa Fluor 555 and 633 (1:500 A31570 and A21050, respectively Thermo Fisher Scientific) and counterstained with DAPI (1:2000 D1306 Thermo Fisher Scientific) diluted in blocking buffer for 1 hr at room temperature in the dark. Cells were washed three times with DPBS-T and coverslips were mounted onto microscope slides using FluorSave mounting media (Millipore). Images were acquired using a Zeiss LSM 880 with Airyscan (Carl Zeiss) at 63× magnification at the UTMB Optical Microscopy Core.
Quantitative reverse-transcription-PCR (qRT-PCR)
Total RNA was isolated using the Direct-zol RNA Miniprep Kit (Zymo Research) and subjected to on column DNase digestion (Zymo Research). Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time qPCR was performed in 384-well plates in triplicate using iTaq Universal SYBR Green Supermix and a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Gene expression was normalized to either human 18S or murine β-Actin by the comparative CT method (ΔΔCT). The human and mouse primer sequences used are listed in Table S2.
IFN-β luciferase reporter assay
HEK293T cells were seeded into 24-well plates (50,000 cells/well) and were transfected with 30 ng of IFN-β reporter plasmid together with 10 ng of Renilla luciferase, 0.2–0.5 ng of FLAG-DHX16 and 1 ng of HA-RIG-I plasmid. Empty pCAGGS plasmid was used to bring the total amount of plasmid to 100 ng/well. Cells were lysed 24 hrs later and IFN-β promoter activity was measured using the Dual-Luciferase reporter assay system (Promega) on a Cytation 5 Multi-Mode Reader (BioTek) according to the manufacturer's instructions. Values were normalized to Renilla, and the fold induction was calculated as the ratio of samples transfected with stimulating plasmid versus samples transfected with empty plasmid.
vRNA purification
vRNAs were isolated from virus stocks using the QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer's instructions.
Protein purification
HEK293T cells seeded in 10 cm dishes (2.5 x 106 cells/dish) were transfected with plasmids of interest using TransIT-LT1. Cells were harvested 24 hrs later for co-immunoprecipitation using either anti-FLAG M2, anti-HA, or anti-GST EZview Red agarose beads (Sigma). Beads were washed seven times with RIPA buffer and twice with peptide elution buffer lacking peptide (10 mM Tris pH 7.4, 150 mM NaCl). Immunoprecipitated proteins were eluted with peptide buffer containing 300 μg/mL FLAG peptide (Sigma) for 30 min at 4°C, 1 mg/mL HA peptide (Thermo Scientific) for 15 min at 22°C, or 10 mM L-Glutathione reduced (Sigma) for 15 min at 22°C.
ATPase assay
100 ng of IAV vRNA or 2 μM of Poly U30 RNA was mixed with 5 μL of purified DHX16 (WT or mutant) in the presence or absence of unanchored poly-Ub (1 μg) in a total of 20 μL ATPase reaction buffer (20 mM Tris-HCl pH 8.0, 1.5 mM MgCl2, 1.5 mM DTT). Reactions were incubated at 37°C for 15 min before 1 mM ATP (Thermo Scientific) was added to the reactions and incubated at 37°C for an additional 30 min. Finally, 5 μL of reaction mixture was diluted with 45 μL ATPase reaction buffer and 100 μL BIOMOL Green reagent (Enzo Life Sciences). ATPase activity was examined by measuring sample absorbance at 620 nm using a Cytation 5 Multi-Mode Reader (BioTek).
In vitro RNA-binding assay
500 ng of biotin-labeled Poly(I:C) (HMW) (InvivoGen) was incubated with High Capacity NeutrAvidin Agarose (Thermo Scientific) in NT2 buffer (50 mM Tris-HCL pH 7.4, 150 mM NaCl, 1 mM MgCl2, and 0.05% (v/v) IGEPAL CA-630) overnight at 4°C on a rotating platform. RNA-bound beads were washed seven times in NT2 buffer and incubated further with purified DHX16 proteins in NT2 buffer overnight at 4°C on a rotating platform. Bound complexes were washed seven times in NT2 buffer and eluted by heating samples in 2X Laemmli buffer containing 2-Mercaptoethanol for 10 min at 95°C.
In vitro Ub-binding assay
Purified proteins were incubated with 1 μg of 2–7 Ub linkage K48- or K63-poly-Ub chains (Boston Biochem) in NT2 buffer overnight at 4°C on a rotating platform followed by immunoprecipitation. Bound complexes were washed seven times in NT2 buffer and eluted.
RNase a susceptibility assay
Cell lysis, WCL collection, and co-immunoprecipitation were performed with RIPA buffer (lacking N-ethylmaleimide and iodoacetamide) as above except the lysate was first subjected to treatment with RNase A (Sigma) for 15 min at 23°C on a rotating platform.
IFN-β ELISA
Secreted IFN-β was measured using the VeriKine human or mouse IFN-β enzyme-linked immunosorbent assay (ELISA) kit (PBL Assay Science) according to the manufacturer's instructions.
Cellular fractionation
Nuclear and cytoplasmic compartments were separated using the NE-PER nuclear and cytoplasmic extraction kit (Thermo Scientific) according to the manufacturer's instructions.
DHX16 knockdown in vitro using siRNA
Transient knockdown of endogenous DHX8, DHX15, and DHX16 in A549 cells, seeded in 24-well plates (30,000 cells per well), was achieved by transfection of ON-TARGETplus Non-targeting Control Pool (D-001810-10-05 Dharmacon), SMARTpool: ON-TARGETplus DHX8 siRNA (L-010506-00-0005 Dharmacon), DHX15 siRNA (L-011250-01-0005 Dharmacon), or DHX16 siRNA (L-011477-00-0005, J-011477-05-0002, J-011477-08-0002 Dharmacon) for a final concentration of 20 nM siRNA. Delivery of siRNA was achieved with Lipofectamine RNAiMAX (Invitrogen) according to the manufacture's guidelines.
DHX16 knockdown in vivo using PPMOs
The peptide-conjugated phosphorodiamidate morpholino oligomers (PPMO) were produced by previously published methods (Abes et al., 2006). The DHX16-targeting PPMO sequence (Dhx16-e2i2) was: 5′-TCCCTCTCCTTCCTTCTGACTTACC-3′, and was designed to specifically target the exon 2-intron 2 junction of DHX16 pre-mRNA in an effort to interfere with splicing and frameshift the downstream sequence. This is likely to bring a premature termination codon in-frame and trigger nonsense-mediated decay of the transcript. The non-targeting PPMO control sequence (P7-NC705) was: 5′-CCTCTTACCTCAGTTACAATTTATA-3′, which has no perfect homology to murine or influenza virus sequences. C57BL/6 or BALB/c mice (The Jackson Laboratory) were anesthetized with 5% isoflurane, followed by i.n. administration with 100 μg of PPMO in 40 μL of DPBS. After 24 hrs, PPMO treatment was repeated. At 48 hrs after the first PPMO administration, mice were infected i.n. with 100-1000 PFUs of influenza virus A/Puerto Rico/8/1934 virus (PR8) diluted in DPBS to a volume of 40 μL. Mice were euthanized and the lungs were collected and homogenization for subsequent assays at 0, 24, and 48 hrs post infection.
Mass spectrometry data processing
To prepare samples for LC-MS/MS analysis, samples were denatured and reduced in 2 M urea, 10 mM NH4HCO3, and 2 mM DTT for 30 min at 60°C with constant shaking, alkylated in the dark with 2 mM iodoacetamide for 45 min at 23°C and digested overnight at 37°C with 80 ng trypsin (Promega). Following digestion, peptides were acidified with formic acid and desalted using C18 ZipTips (Millipore) according to the manufacturer's specifications. Samples were resuspended in 4% formic acid, 2% acetonitrile solution. Digested peptide mixtures were analyzed on a Thermo Scientific Velos Pro dual linear ion trap mass spectrometer equipped with a Proxeon Easy-nLC HPLC with a pre-column (2 cm × 100 um I.D. packed with ReproSil Pur C18 AQ 5 um particles) and an analytical column (10 cm × 75 um I.D. packed with ReproSil Pur C18 AQ 3 um particles). A gradient was delivered by the HPLC from 5% to 30% ACN in 0.1% formic acid over one hr. The Velos Pro collected one full scan followed by 20 collision-induced dissociation MS/MS scans of the 20 most intense peaks from the full scan. Dynamic exclusion was enabled for 30 sec with a repeat count of 1. Data were searched against a database containing SwissProt Human protein sequences (downloaded March 21, 2018), concatenated to a decoy database where each sequence was randomized in order to estimate the false positive rate. The searches considered a precursor mass tolerance of 1 Da and fragment ion tolerances of 0.8 da, and considered variable modifications for protein N-terminal acetylation, protein N-terminal acetylation and oxidation, glutamine to pyroglutamate conversion for peptide N-terminal glutamine residues, protein N-terminal methionine loss, protein N-terminal acetylation and methionine loss, and methionine oxidation, and constant modification for carbamidomethyl cysteine. The resulting raw data were matched to protein sequences by the Protein Prospector algorithm. Prospector data were filtered using a maximum protein expectation value of 0.01 and a maximum peptide expectation value of 0.05. The proteins identified by the His-Ub pulldown are listed in Table S3.
RNA-seq data processing
A549 (siNTC and siDHX16 knockdown) cells were infected at MOI=1, and RNA was isolated at 24 hrs post infection. RNA quality was assessed by visualization of 18S and 28S RNA bands using an Agilent 2100 Bioanalyzer (Agilent Technologies); the electropherograms were used to calculate the 28S/18S ratio and the RNA integrity number. Poly(A)+ RNA was enriched from total RNA (1 μg) using oligo(dT)-attached magnetic beads. First- and second-strand synthesis, adaptor ligation, and amplification of the library were performed using the Illumina TruSeq RNA sample preparation kit as recommended by the manufacturer (Illumina). Library quality was evaluated using an Agilent DNA-1000 chip on an Agilent 2100 Bioanalyzer. Quantification of library DNA templates was performed using qRT-PCR and a known-size reference standard. Cluster formation of the library DNA templates was performed using TruSeq PE cluster kit v3 (Illumina) and the Illumina cBot workstation under conditions recommended by the manufacturer. Paired-end 50-base sequencing by synthesis was performed using TruSeq SBS kit v3 (Illumina) on an Illumina HiSeq 1500 instrument using protocols defined by the manufacturer. The alignment of NGS sequence reads was performed using the Spliced Transcript Alignment to a Reference (STAR) program, version 2.5.1b, using default parameters (Dobin et al., 2013). We used the human hg38 assembly as a reference with the UCSC gene annotation file, both downloaded from the Illumina iGenomes website. The –quantMode GeneCounts option of STAR provided read counts per gene, which were input into the DESeq2 (version 1.12.1) (Love et al., 2014) differential expression analysis program to determine expression levels and differentially expressed genes. Alternative splicing events were measured with MISO (Mixture of Isoforms) software, version 0.5.3, following the software authors' recommendations. The filter events utility script was used to filter the differential splicing events with parameters --delta-psi 0.1 –bayes-factor 10 (Katz et al., 2010).
Quantification and statistical analysis
All data were presented as means ± SD or SEM as indicated and analyzed using GraphPad PRISM software (version 9 GraphPad Software). Protein band intensity and nuclear/cytoplasmic distribution was quantified using ImageJ software (version 1.53m NIH). Confocal vector profiles were analyzed using Zen lite software (version 2.5 Carl Zeiss). Student's paired t test, one-way ANOVA, or two-way ANOVA with multiple comparisons were used. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Acknowledgments
This work was supported by NIH/NIAID grants R01AI134907, R01AI166668, R01AI155466, P01AI150585, R21AI126012, R21AI132479, and UTMB/IHII awarded to R.R. R01AI134907, UL1TR001439, and awards from Sealy & Smith, Kleberg, John S. Dunn, Amon G. Carter, Gilson Longenbaugh, and Summerfield Robert foundations awarded to P.-Y.S. T32AI007526 awarded to A.H. F31AI152422 and T32AI060549 awarded to S.v.T. K12HD052023 awarded to M.I.G. P50AI150476, U19AI135972, R01AI143292, P01AI063302, and awards from DARPA #HR0011-19-2-0020, F. Hoffmann-La Roche, Vir Biotechnology, and QCRG donors to N.J.K. U19AI135972 and CRIPT/CEIRR contract #75N93019R00028 to A.G.-S. We thank Maxim Ivannikov (UTMB Imaging Core) for assistance with confocal microscopy and Linsey Yeager for editing of the manuscript.
Author contributions
A.H. performed all aspects of this study. P.B., R.R., S.v.T., M.I.G., M.G.-O., K.M.V., A.N.W., and L.A.-A. performed experiments. X.X., B.L., P.-Y.S., A.N.F., and A.G.-S. provided critical reagents and technical advice. S.G.W. performed and analyzed NGS and MISO. H.M.M. designed and synthesized PPMOs. J.R.J. and N.J.K. performed and analyzed MS. A.H. and R.R. organized the study and prepared the manuscript. All authors read the manuscript and provided comments.
Declaration of interests
The García-Sastre laboratory received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7 Hills Pharma, PharmaMar, ImmunityBio, Accurius, nanoComposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, and Merck. The Krogan laboratory received research support from Vir Biotechnology and F. Hoffmann-La Roche. All declarations of interest are outside of the reported work. All remaining authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
Published: March 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110434.
Supplemental information
Data and code availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031365. Transcriptomic data generated during this study have been deposited to the NCBI Gene Expression Omnibus (GEO) database under GEO: GSE168255. All datasets are publically available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.
References
- Abdelhaleem M., Maltais L., Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics. 2003;81:618–622. doi: 10.1016/s0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]
- Abes S., Moulton H.M., Clair P., Prevot P., Youngblood D.S., Wu R.P., Iversen P.L., Lebleu B. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J. Control. Release. 2006;116:304–313. doi: 10.1016/j.jconrel.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., Ezcurra A.L.D.L., Fucito S., Gamarnik A.V. Role of RNA structures present at the 3′ UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Banroques J., Doere M., Dreyfus M., Linder P., Tanner N.K. Motif III in superfamily 2 “helicases” helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J. Mol. Biol. 2010;396:949–966. doi: 10.1016/j.jmb.2009.12.025. [DOI] [PubMed] [Google Scholar]
- Bernstein K.A., Granneman S., Lee A.V., Manickam S., Baserga S.J. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases involved in large ribosomal subunit biogenesis. Mol. Cell. Biol. 2006;26:1195–1208. doi: 10.1128/MCB.26.4.1195-1208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharaj P., Atkins C., Luthra P., Giraldo M.I., Dawes B.E., Miorin L., Johnson J.R., Krogan N.J., Basler C.F., Freiberg A.N., Rajsbaum R. The host E3-ubiquitin ligase TRIM6 ubiquitinates the ebola virus VP35 protein and promotes virus replication. J. Virol. 2017;91:e00833. doi: 10.1128/jvi.00833-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharaj P., Wang Y.E., Dawes B.E., Yun T.E., Park A., Yen B., Basler C.F., Freiberg A.N., Lee B., Rajsbaum R. The matrix protein of nipah virus targets the E3-ubiquitin ligase TRIM6 to inhibit the IKKepsilon kinase-mediated type-I IFN antiviral response. PLoS Pathog. 2016;12:e1005880. doi: 10.1371/journal.ppat.1005880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-Y., Liu H.M., Chang M.-F., Chang S.C. Middle East respiratory syndrome coronavirus nucleocapsid protein suppresses type I and type III interferon induction by targeting RIG-I signaling. J. Virol. 2020;94:e00099. doi: 10.1128/JVI.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazal M., Beauclair G., Gracias S., Najburg V., Simon-Lorière E., Tangy F., Komarova A.V., Jouvenet N. RIG-I recognizes the 5′ region of dengue and Zika virus genomes. Cell Rep. 2018;24:320–328. doi: 10.1016/j.celrep.2018.06.047. [DOI] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Xiao F., Hu D., Ge W., Tian M., Wang W., Pan P., Wu K., Wu J. SARS-CoV-2 nucleocapsid protein interacts with RIG-I and represses RIG-mediated IFN-β production. Viruses. 2020;13:47. doi: 10.3390/v13010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Potratz J.P., Tijerina P., Del Campo M., Lambowitz A.M., Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc. Natl. Acad. Sci. U S A. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K.T., Gale M., Jr., Loo Y.M. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 2018;36:667–694. doi: 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelan N.R., Basler C.F., García-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 2003;77:13257–13266. doi: 10.1128/jvi.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J., Terrier O., Rosa-Calatrava M. Influenza viruses and mRNA splicing: doing more with less. mBio. 2014;5:e00070. doi: 10.1128/mBio.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D., Mateos-Gomez P.A., Enjuanes L., Gallego J., Sola I. Structure and functional relevance of a transcription-regulating sequence involved in coronavirus discontinuous RNA synthesis. J. Virol. 2011;85:4963–4973. doi: 10.1128/JVI.02317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner P., Versteeg G.A., Ikeda F. Ubiquitin enzymes in the regulation of immune responses. Crit. Rev. Biochem. Mol. Biol. 2017;52:425–460. doi: 10.1080/10409238.2017.1325829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G., Kim D.H., Kim S.H., Tseng Y.H., Yu Y., Lin R.J. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DExD/H-box proteins. RNA. 2000;6:1106–1119. doi: 10.1017/s1355838200992483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld A.J., Neumann G., Kawaoka Y. Influenza A virus isolation, culture and identification. Nat. Protoc. 2014;9:2663–2681. doi: 10.1038/nprot.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams M.E., Guenther U.-P., Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullam A., Schroder M. DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. Biochim. Biophys. Acta. 2013;1829:854–865. doi: 10.1016/j.bbagrm.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J.U. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gencheva M., Kato M., Newo A.N., Lin R.J. Contribution of DEAH-box protein DHX16 in human pre-mRNA splicing. Biochem. J. 2010;429:25–32. doi: 10.1042/bj20100266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gencheva M., Lin T.Y., Wu X., Yang L., Richard C., Jones M., Lin S.B., Lin R.J. Nuclear retention of unspliced pre-mRNAs by mutant DHX16/hPRP2, a spliceosomal DEAH-box protein. J. Biol. Chem. 2010;285:35624–35632. doi: 10.1074/jbc.M110.122309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo M.I., Hage A., van Tol S., Rajsbaum R. TRIM proteins in host defense and viral pathogenesis. Curr. Clin. Microbiol. Rep. 2020:1–14. doi: 10.1007/s40588-020-00150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo M.I., Xia H., Aguilera-Aguirre L., Hage A., van Tol S., Shan C., Xie X., Sturdevant G.L., Robertson S.J., McNally K.L., et al. Envelope protein ubiquitination drives entry and pathogenesis of Zika virus. Nature. 2020;585:414–419. doi: 10.1038/s41586-020-2457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble J., Stevens L.J., Agostini M.L., Anderson-Daniels J., Chappell J.D., Lu X., Pruijssers A.J., Routh A.L., Denison M.R. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021;17:e1009226. doi: 10.1371/journal.ppat.1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbart M., Deng X., Baker S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. U S A. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage A., Rajsbaum R. To TRIM or not to TRIM: the balance of host-virus interactions mediated by the ubiquitin system. J. Gen. Virol. 2019;100:1641–1662. doi: 10.1099/jgv.0.001341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.-K., Schlee M. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017;91:e02143. doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Kinch L.N., Brautigam C.A., Chen X., Du F., Grishin N.V., Chen Z.J. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin R.J. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 1996;16:6810–6819. doi: 10.1128/mcb.16.12.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Jankowsky E. From unwinding to clamping — the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Liu F., Putnam A., Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Gack M.U. Distinct and orchestrated functions of RNA sensors in innate immunity. Immunity. 2020;53:26–42. doi: 10.1016/j.immuni.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Park H.-S., Pyo H.-M., Liu Q., Zhou Y. Influenza A virus panhandle structure is directly involved in RIG-I activation and interferon induction. J. Virol. 2015;89:6067–6079. doi: 10.1128/JVI.00232-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Lu N., Yuan B., Weng L., Wang F., Liu Y.J., Zhang Z. The interaction between the helicase DHX33 and IPS-1 as a novel pathway to sense double-stranded RNA and RNA viruses in myeloid dendritic cells. Cell Mol. Immunol. 2014;11:49–57. doi: 10.1038/cmi.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020;94:e01410–e01420. doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.-M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., García-Sastre A., Katze M.G., Gale M., Jr. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.-M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Lu N., Weng L., Yuan B., Liu Y.J., Zhang Z. DHX15 senses double-stranded RNA in myeloid dendritic cells. J. Immunol. 2014;193:1364–1372. doi: 10.4049/jimmunol.1303322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Keck J.G., Stohlman S.A., Lai M.M. High-frequency RNA recombination of murine coronaviruses. J. Virol. 1986;57:729–737. doi: 10.1128/JVI.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B., Manicassamy S., Belicha-Villanueva A., Pisanelli G., Pulendran B., García-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. U S A. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitoma H., Hanabuchi S., Kim T., Bao M., Zhang Z., Sugimoto N., Liu Y.J. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita M., Oshiumi H., Matsumoto M., Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell. Biol. 2011;31:3802. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosallanejad K., Sekine Y., Ishikura-Kinoshita S., Kumagai K., Nagano T., Matsuzawa A., Takeda K., Naguro I., Ichijo H. The DEAH-box RNA helicase DHX15 activates NF-kappaB and MAPK signaling downstream of MAVS during antiviral responses. Sci. Signal. 2014;7:ra40. doi: 10.1126/scisignal.2004841. [DOI] [PubMed] [Google Scholar]
- Nistal-Villán E., Gack M.U., Martínez-Delgado G., Maharaj N.P., Inn K.-S., Yang H., Wang R., Aggarwal A.K., Jung J.U., García-Sastre A. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J. Biol. Chem. 2010;285:20252–20261. doi: 10.1074/jbc.M109.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguntuyo K.Y., Stevens C.S., Hung C.T., Ikegame S., Acklin J.A., Kowdle S.S., Carmichael J.C., Chiu H.P., Azarm K.D., Haas G.D., et al. Quantifying absolute neutralization titers against SARS-CoV-2 by a standardized virus neutralization assay allows for cross-cohort comparisons of COVID-19 sera. mBio. 2021;12:e02492. doi: 10.1128/mBio.02492-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine I., Posey J.E., Grochowski C.M., Jhangiani S.N., Rosenheck S., Kleyner R., Marmorale T., Yoon M., Wang K., Robison R., et al. Paralog studies augment gene discovery: DDX and DHX genes. Am. J. Hum. Genet. 2019;105:302–316. doi: 10.1016/j.ajhg.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabhi S., Knoll M.L., Gale M., Jr., Loo Y.M. DHX15 is a coreceptor for RLR signaling that promotes antiviral defense against RNA virus infection. J. Interferon Cytokine Res. 2019;39:331–346. doi: 10.1089/jir.2018.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel T., Hausmann S., Morger D., Zuger S., Guerra J., Lascano J., Reinhard C., Santoni F.A., Uchil P.D., Chatel L., et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumpton M., McGarvey M., Beggs J.D. A dominant negative mutation in the conserved RNA helicase motif ‘SAT' causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994;13:879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R. Intranasal delivery of peptide-morpholinos to knockdown influenza host factors in mice. Methods Mol. Biol. 2017;1565:191–199. doi: 10.1007/978-1-4939-6817-6_16. [DOI] [PubMed] [Google Scholar]
- Rajsbaum R., Garcia-Sastre A., Versteeg G.A. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J. Mol. Biol. 2014;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R., Versteeg G.A., Schmid S., Maestre A.M., Belicha-Villanueva A., Martinez-Romero C., Patel J.R., Morrison J., Pisanelli G., Miorin L., et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKepsilon kinase-mediated antiviral response. Immunity. 2014;40:880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebendenne A., Valadão A.L.C., Tauziet M., Maarifi G., Bonaventure B., McKellar J., Planès R., Nisole S., Arnaud-Arnould M., Moncorgé O., Goujon C. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J. Virol. 2021;95:e02415–e02420. doi: 10.1128/JVI.02415-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020;34:680–692. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Tan C.P., Goubau D., Schulz O., Pichlmair A., Bier K., Robb N., Vreede F., Barclay W., Fodor E., Reis e Sousa C. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Horton J.R., Mullally J.E., Heroux A., Cheng X., Wilkinson K.D. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., et al. Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A., Hamann F., Neumann P., Ficner R. Crystal structure of the spliceosomal DEAH-box ATPase Prp2. Acta Crystallogr. D. 2018;74:643–654. doi: 10.1107/s2059798318006356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C., Xie X., Muruato A.E., Rossi S.L., Roundy C.M., Azar S.R., Yang Y., Tesh R.B., Bourne N., Barrett A.D., et al. An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe. 2016;19:891–900. doi: 10.1016/j.chom.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solórzano A., Webby R.J., Lager K.M., Janke B.H., García-Sastre A., Richt J.A. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 2005;79:7535–7543. doi: 10.1128/JVI.79.12.7535-7543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N., Mitoma H., Kim T., Hanabuchi S., Liu Y.J. Helicase proteins DHX29 and RIG-I cosense cytosolic nucleic acids in the human airway system. Proc. Natl. Acad. Sci. U S A. 2014;111:7747–7752. doi: 10.1073/pnas.1400139111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Xian H., Tian S., Sun T., Qin Y., Zhang S., Cui J. A hierarchical mechanism of RIG-I ubiquitination provides sensitivity, robustness and synergy in antiviral immune responses. Sci. Rep. 2016;6:29263. doi: 10.1038/srep29263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N.K., Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/S1097-2765(01)00329-X. [DOI] [PubMed] [Google Scholar]
- Tauchert M.J., Fourmann J.B., Lührmann R., Ficner R. Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. Elife. 2017;6:e21510. doi: 10.7554/eLife.21510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares R.d.C.A., Mahadeshwar G., Wan H., Huston N.C., Pyle A.M. The global and local distribution of RNA structure throughout the SARS-CoV-2 genome. J. Virol. 2020;95:e02190. doi: 10.1128/JVI.02190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S., McGarvey M., Plumpton M., Beggs J.D. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 1994;13:888–897. doi: 10.1002/j.1460-2075.1994.tb06332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tol S., Atkins C., Bharaj P., Johnson K.N., Hage A., Freiberg A.N., Rajsbaum R. VAMP8 contributes to the TRIM6-mediated type I interferon antiviral response during west nile virus infection. J. Virol. 2020;94:e01454. doi: 10.1128/jvi.01454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tol S., Hage A., Giraldo M.I., Bharaj P., Rajsbaum R. The TRIMendous role of TRIMs in virus-host interactions. Vaccines. 2017;5:23. doi: 10.3390/vaccines5030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., Benke S., Garcia-Sastre A., Rajsbaum R. InTRIMsic immunity: positive and negative regulation of immune signaling by tripartite motif proteins. Cytokine Growth Factor Rev. 2014;25:563–576. doi: 10.1016/j.cytogfr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z., Schneider C., Mozaffari-Jovin S., Schmitzova J., Hobartner C., Fabrizio P., Luhrmann R. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 2015;29:94–107. doi: 10.1101/gad.253070.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., Menachery V.D., Rajsbaum R., Shi P.-Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z.P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., Chen Z.J. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Sato S., Sotoyama Y., Orba Y., Sawa H., Yamauchi H., Sasaki M., Takaoka A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021;22:820–828. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- Yedavalli V.S.R.K., Neuveut C., Chi Y.-H., Kleiman L., Jeang K.-T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Yin X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., De Jesus P.D., et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34:108628. doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Suhara W., Fukuhara Y., Sato M., Ozato K., Fujita T. Autocrine amplification of type I interferon gene expression mediated by interferon stimulated gene factor 3 (ISGF3) J. Biochem. 1996;120:160–169. doi: 10.1093/oxfordjournals.jbchem.a021379. [DOI] [PubMed] [Google Scholar]
- Yoo J.S., Takahasi K., Ng C.S., Ouda R., Onomoto K., Yoneyama M., Lai J.C., Lattmann S., Nagamine Y., Matsui T., et al. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLoS Pathog. 2014;10:e1004012. doi: 10.1371/journal.ppat.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang S., Lin T.Y., Chen X., Gencheva M., Newo A.N., Yang L., Rossi D., Hu J., Lin S.B., Huang A., Lin R.J. GPKOW is essential for pre-mRNA splicing in vitro and suppresses splicing defect caused by dominant-negative DHX16 mutation in vivo. Biosci. Rep. 2014;34:e00163. doi: 10.1042/bsr20140142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z.J. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Kim T., Bao M., Facchinetti V., Jung S.Y., Ghaffari A.A., Qin J., Cheng G., Liu Y.J. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yuan B., Lu N., Facchinetti V., Liu Y.J. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J. Immunol. 2011;187:4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Tan P., Li Y., Lin M., Li C., Mao J., Cui J., Zhao W., Wang H.Y., Wang R.F. DHX29 functions as an RNA co-sensor for MDA5-mediated EMCV-specific antiviral immunity. PLoS Pathog. 2018;14:e1006886. doi: 10.1371/journal.ppat.1006886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031365. Transcriptomic data generated during this study have been deposited to the NCBI Gene Expression Omnibus (GEO) database under GEO: GSE168255. All datasets are publically available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.