Abstract
Background
The oncogene epidermal growth factor receptor variant III (EGFRvIII) is expressed in approximately one-third of all glioblastomas (GBMs). So far it is not clear if EGFRvIII expression induces replication stress in GBM cells, which might serve as a therapeutical target.
Methods
Isogenetic EGFRvIII− and EGFRvIII+ cell lines with endogenous EGFRvIII expression were used. Markers of oncogenic and replication stress such as γH2AX, RPA, 53BP1, ATR, and CHK1 were analyzed using western blot, immunofluorescence, and flow cytometry. The DNA fiber assay was performed to analyze replication, transcription was measured by incorporation of EU, and genomic instability was investigated by micronuclei and CGH-Array analysis. Immunohistochemistry staining was used to detect replication stress markers and R-loops in human GBM samples.
Results
EGFRvIII+ cells exhibit an activated replication stress response, increased spontaneous DNA damage, elevated levels of single-stranded DNA, and reduced DNA replication velocity, which are all indicative characteristics of replication stress. Furthermore, we show here that EGFRvIII expression is linked to increased genomic instability. EGFRvIII-expressing cells display elevated RNA synthesis and R-loop formation, which could also be confirmed in EGFRvIII-positive GBM patient samples. Targeting replication stress by irinotecan resulted in increased sensitivity of EGFRvIII+ cells.
Conclusion
This study demonstrates that EGFRvIII expression is associated with increased replication stress, R-loop accumulation, and genomic instability. This might contribute to intratumoral heterogeneity but may also be exploited for individualized therapy approaches.
Keywords: EGFRvIII, genomic instability, glioblastoma, irinotecan sensitivity, replication stress, R-loops
Key Points.
EGFRvIII expression is associated with increased replication stress in GBM cells and tumors.
EGFRvIII-expressing cells display elevated R-loop levels and genomic instability.
EGFRvIII expression is associated with increased irinotecan sensitivity.
Importance of the Study.
Our data show that EGFRvIII drives the oncogenic phenotype by increasing transcription activity and subsequent R-loop formation. This, in turn, results in replication stress and genomic instability. Targeting EGFRvIII-induced replication stress by inhibition of topoisomerase I revealed a significantly higher sensitivity of EGFRvIII+ cells compared to EGFRvIII− cells. We show here that increased transcription is associated with EGFRvIII-induced replication stress, providing a molecular link between upregulation of the transcription machinery and genomic instability in EGFRvIII+ GBM. These results give new insights into EGFRvIII+ GBM and highlight a molecular rationale for the application of therapeutics targeting replication stress with the potential to improve the outcome of EGFRvIII+ GBM patients. Furthermore, these results might also be of clinical relevance for GBM in general, since increased RNA synthesis, R-loop formation, and replication stress might also occur independently of EGFRvIII status, and may therefore be a rational therapeutic target in other GBM subtypes.
Glioblastoma (GBM) is the most common malignant primary brain tumor in adults with an extremely poor prognosis.1–3 GBMs are characterized by the frequent expression of a truncated epidermal growth factor receptor (EGFR), the EGFR variant III (EGFRvIII), which can be detected in approximately one-third of all GBMs.4,5 EGFRvIII lacks the exons 2–7 leading to a constitutively active receptor that can trigger downstream signaling including the MAPK, PI3K/AKT, and STAT pathways.4,6–8 Recently, we have demonstrated increased p38 signaling in EGFRvIII+ cells, which was accompanied by an elevated expression of DNA mismatch repair (MMR) proteins. This upregulation in MMR was linked to an increased temozolomide (TMZ) sensitivity of EGFRvIII-expressing and O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylated GBM.9 Both the activation of p38 signaling and the upregulation of MMR indicate oncogene-induced stress in EGFRvIII-expressing cells, which might also include replication-associated stress (RS). Recently, Carruthers et al.10 identified RS to be upregulated in radioresistant GBM stem-like cells (GSCs). GSCs displayed DNA double-strand breaks (DSBs) and increased RNA:DNA hybrids (R-loops). Targeting of RS using ATR and PARP inhibitors conferred GSC-specific cytotoxicity and a complete abrogation of GSC radiation resistance in vitro. These data demonstrate that the identification of RS can serve as a specific target with significant clinical potential. While oncogenes, such as RAS, MYC, or CYCLIN E have been shown to induce RS, EGFRvIII has not causally linked to RS so far.11–13 However, such a link would further unravel the biological relevance of EGFRvIII, thereby helping to develop new specific therapeutic strategies for EGFRvIII-positive GBM. The aim of this study was to clarify whether EGFRvIII expression is associated with RS and which biological processes might contribute to these alterations. We demonstrate that endogenous EGFRvIII expression causes hallmarks of oncogene-induced RS, such as replication fork slowing, spontaneous DNA damage, and elevated levels of single-stranded DNA (ssDNA). Furthermore, we show that EGFRvIII-expressing cells display an upregulation of transcriptional activity leading to increased R-loop formation in vitro and also in EGFRvIII+ areas of GBM patient samples, making the cells susceptible to DNA damage induction. Since it has been shown that topoisomerase I depletion leads to increased R-loop formation and therefore to DSB, targeting of EGFRvIII-induced RS by inhibition of topoisomerase I with irinotecan revealed a significantly higher sensitivity of EGFRvIII+ cells compared to EGFRvIII− cells. In summary, we show here that endogenous EGFRvIII expression is associated with elevated RS and R-loop accumulation resulting in increased genomic instability. This study identifies RS as an EGFRvIII-specific target and might provide a molecular rationale for the application of alternative treatment regimens when standard therapy is not effective in EGFRvIII-positive GBM patients.
Materials and Methods
Reagents
Irinotecan (Selleckchem) was dissolved in DMSO (Sigma-Aldrich) and stored at −20°C. 5-Ethynyluridin (Axxora) was dissolved in distilled water and stored at −20°C.
Cell Culture
The human isogenetic EGFRvIII− and EGFRvIII+ GBM sub-cell lines DKMGvIII−/+ and BS153vIII−/+ were generated, authenticated, and cultivated as described previously.9,14
GBM Patient Samples
Human tumor material was used in accordance with all local and national ethics guidelines.
Cell Proliferation
To analyze proliferation of EGFRvIII− and EGFRvIII+ cells, 1 × 105 cells were seeded. The cell number was determined for up to 8 days.
Cell Survival
The ability for self-renewal (clonogenicity) was analyzed by the colony-forming assay as described previously.9 In brief, cells were seeded and treated 24 h later with irinotecan (0.1–10 µM) for 48 h or 36 h according to their doubling time (DKMGvIII−/+ & BS153vIII− for 48 h; BS153vIII+ cells for 36 h). After treatment, the medium was replaced by AmnioMax C-100 containing 10% FCS and C-100 supplement (Life Technologies). Cells were grown until the colonies of all treatment arms had reached equal colony size.
Western Blot
Proteins from whole-cell extracts were detected by WB according to standard protocols. The Odyssey CLx Infrared Imaging System (LI-COR Biosciences) was used for signal detection and quantification. Primary antibodies: EGFR (1:1000, rabbit, Cell Signaling Technology, #2232); pEGFR (1:1000, rabbit, Cell Signaling Technology, #4407); β-Actin (1:40000, mouse, Sigma-Aldrich, #A-2228); ATM (1:1000, rabbit, Cell Signaling Technology, #2873); pATM (1:1000, rabbit, GeneTex, GTX61739); ATR (1:1000, mouse, Santa Cruz, #SC-515173); pATR (1:1000, rabbit, Cell Signaling Technology, #58014); Chk1 (1:1000, mouse, Cell Signaling Technology, #2360); pChk1 (1:1000, rabbit, US Biological, #C4200-05); Chk2 (1:1000, mouse, BD Transduction Laboratories, #2360); pChk2 (1:1000, rabbit, Cell Signaling Technology, #2661); RPA (1:1000, mouse, Santa Cruz, SC-56770); pRPA (1:1000, rabbit, Boster, #02067); RNaseH (1:1000, rabbit, Abcam, #ab229078). All primary antibodies were diluted in 5% bovine serum albumin (BSA) in PBS supplemented with 0.2 % Tween. Secondary anti-mouse and anti-rabbit antibodies were purchased from LI-COR Biosciences.
Cell Cycle
For cell cycle analysis, cells were harvested, fixed with 70% ethanol, and stored at −20°C. PI staining was performed as previously described.9
Flow Cytometry
Detection of EGFRvIII
.—EGFRvIII expression was determined as described previously.9
Detection of DNA damage
.—Cells were harvested, fixed with 4% paraformaldehyde (PFA) in PBS for 10 min, and permeabilized (PBS with 0.2% Triton X-100) before blocking overnight (1% BSA/PBS with 0.2% Triton X-100). Cells were then incubated (1 h; room temperature [RT]) with an anti-γH2AX antibody (1:3000, mouse, clone JBW301, Millipore, #05-636) in blocking solution, washed 3 times (PBS with 0.1% Tween20) before incubation (1 h; RT) with anti-mouse DyLight488 (1:1000; Jackson Immunoresearch, #35502). After an additional washing (3× PBS with 0.1% Tween20), DNA counterstaining was performed using FxCycle Far Red (Molecular Probes) plus 300 ng/ml RNAse A and 0.2 % Triton X-100 for 30 min at RT in the dark. Flow cytometric analysis was performed using a MaqsQuant Analyzer (Miltenyi Biotec). Quantification of γH2AX signal was performed using FlowLogic Software.
To analyze chromatin-bound RPA, cells were harvested, pre-extracted (0.2% Triton X-100 + DTT), fixed with 4% PFA in PBS for 10 min, and permeabilized (PBS with 0.2% Triton X-100) before blocking overnight (1% BSA/PBS with 0.2% Triton X-100). Cells were then incubated (1 h; RT) with an anti-RPA antibody (1:1000, Santa Cruz, SC-56770) in blocking solution, washed 3 times (PBS with 0.1% Tween20) before incubation (1 h; RT) with anti-mouse DyLight488 (1:1000, Jackson Immunoresearch).
5-Ethynyl-uridine incorporation assay
.—Cells were incubated with 0.5 mM 5-Ethynyl-Uridin (EU) for up to 1 h. Subsequently, cells were fixed with 70% ice-cold ethanol and permeabilized (1% BSA/PBS with 0.2% Triton X-100). Afterward, Click-iT reaction was performed (Baseclick, Tutzing). After a washing step (3 times with 1% BSA/PBS with 0.2% Triton X-100), DNA counterstaining was performed using FxCycle Far Red (Molecular Probes) in 1% BSA/PBS/0.2% Triton-X for 15 min at RT in the dark. Flow cytometric analysis was performed using a FACS Canto with FACS Diva Software (Becton Dickinson).
DNA Fiber Assay
The DNA fiber assay was performed as described in the work of Parplys et al.15 In brief, cells were pulse-labeled with 25 µM CldU (Sigma-Aldrich) and 250 µM IdU (Sigma-Aldrich) for 20 min 3 days after seeding. Cells were harvested with ice-cold PBS, and fiber spreads were prepared. Samples were incubated in 2.5 M HCl for 90 min followed by incubation in blocking buffer (2% BSA/PBS with 0.1% Tween) for 1 h. Fiber spreads were stained with rat anti-BrdU antibody (1:1000, Abcam, abcam#6326) to detect CldU, followed by mouse anti-BrdU (1:5000, mouse, BD Bioscience, #BD347580) to detect IdU. As secondary antibodies, goat anti-rat AlexaFluor555 (1:500, Invitrogen, #A-21434) and goat anti-mouse AlexaFluor488 (1:500, Invitrogen, #A110011) antibodies were used. Fiber tracts were analyzed using a confocal fluorescence microscope (Zeiss Axioplan 2; 630-fold magnification). Pictures were analyzed using ImageJ software. For fork speed analysis, replication fork speeds of CldU and IdU were measured and values were converted into kilobases (1 µM = 2.59 kb). In each independent experiment, at least 150 individual fibers were analyzed to calculate the fork speed. For the determination of fiber structures at least 400 individual fibers were analyzed.15
Immunohistochemistry
Five-micrometer sections of paraffin-embedded GBM specimens were dewaxed using standard histologic procedures. Heat-induced antigen retrieval for the detection of RPA (1:100; rabbit, Santa Cruz, SC-56770) and γH2AX (1:350, mouse clone JBW301, Millipore, #05-636) was carried out by boiling slides in sodium citrate buffer, pH 6.8, for 1 h, followed by a 20 min cooldown step. For R-loop detection, the S9.6 antibody (1:10 000, mouse, Millipore, #MABE1095) was used. Here, heat-induced antigen retrieval was carried out by boiling slides in 1 mM EDTA, pH 8.0, for 1 h, followed by a 20 min cooldown step. To confirm S9.6 specificity, slides were incubated with RNAseH (5 Units, M0297, New England Biolabs) for 30 min at 37°C. For all specimens, endogenous peroxidase was inactivated by blocking the samples with Bloxxal (Vector Laboratories). Specimens were blocked with Casein (Vector Laboratories) blocking solution. Primary antibodies were incubated overnight at 4°C. Specimens were then incubated with secondary antibodies (ImmPRESS reagent kit anti-rabbit IgG, Vector Laboratories, MP7401; ImmPRESS reagent kit anti-mouse IgG, Vector Laboratories) for 1 h. Bound secondary antibodies were detected by ImmPact DAB substrate (Vector Laboratories). EGFRvIII staining (1:250, mouse, Absolute antibody, #Ab00184-1.4) was performed on a Ventana System using standard protocols Nuclei were counterstained with hematoxylin. Substrate reaction was assessed under the microscope and stopped early upon first evidence of color development, to detect differential expression in EGFRvIII− and EGFRvIII+ areas.
Immunofluorescence
Immunofluorescence staining was performed as described previously.9 In brief, cells were fixed (4% PFA), permeabilized (PBS with 0.2% Triton X-100), and blocked (1% BSA/PBS with 0.2% Triton X-100), and DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; QBiogene). A confocal fluorescence microscope (Zeiss Axioplan 2; 630-fold magnification) was used for the analysis while the same exposure time was used to analyze EGFRvIII− and EGFRvIII+ cells for each experiment.
The micronuclei were assessed based on DAPI staining of untreated cells.
To analyze γH2AX, an anti-γH2AX (1:400, anti-mouse, clone JBW301, Millipore, #05-636) antibody and secondary antibody (goat-anti mouse ALEXA fluor 594, 1:1000, Life Technologies, #A32742) were used. At least 100 intact nuclei were randomly selected per independent experiment and counted with ImageJ.
To analyze 53BP1-foci in G1-, S-, and G2 phase, cells were incubated for 30 min with EdU (5-ethynyl-2′-deoxyuridine, baseclick GmbH) before fixation. Click-iT reaction was performed according to the manufacturer’s instructions. Cells were stained with the anti-53BP1 (1:750, mouse, Merck, #MAB3802) and geminin antibody (1:500, rabbit, Proteintech, #10802-I-AP) and the respective secondary antibodies (goat-anti-mouse Dylight 649; goat-anti-rabbit ALEXA fluor 594) at RT for 60 min.
For S9.6 staining cells were pre-extracted (0.2% Triton/PBS + dithiothreitol) for 10 min on ice before fixation. Cells were stained with anti-RNA/DNA-hybrid antibody (S9.6; 1:100, mouse, Millipore, #MABE1095) overnight. The goat-anti-mouse ALEXA fluor 594 antibody was used as a secondary antibody. At least 50 intact nuclei were randomly selected and analyzed with ImageJ. For quantification of nuclear S9.6 intensity, ImageJ was used to generate nuclear masks based on DAPI staining, and mean S9.6 fluorescence intensities per pixel were quantified per nucleus.
Genomic Copy Number Analysis
Array-based comparative genomic hybridization (CGH) was conducted using high-resolution oligonucleotide-based SurePrint G3 Human 60k CGH microarrays (AMADID 21924, Agilent Technologies), and raw data were preprocessed, analyzed, and visualized as described in the work of Wilke et al.16 For each cell line, a genome instability score (GIS) was calculated as follows: (2 × (number of regions with complete loss) + number of regions with simple loss + number of regions with simple gain + 2 × (number of regions with high-level amplification)/total number of copy number regions).
Isolation of Chromatin-Bound Proteins
The Subcellular Protein Fractionation Kit (Thermo Scientific) was used according to the manufacturer’s instructions. For WB analysis, at least 7 µg of protein were loaded on the gel.
Statistical Analysis
Experiments were repeated at least 3 times if not otherwise indicated. The data are presented as mean values (±SEM). Prism software (GraphPad Prism 5, GraphPad Software, Inc.) was used for analyzing and graphing the data. P values were calculated using Student’s t-tests (*P < .05; **P < .01; ***P < .001; ns: nonsignificant).
Results
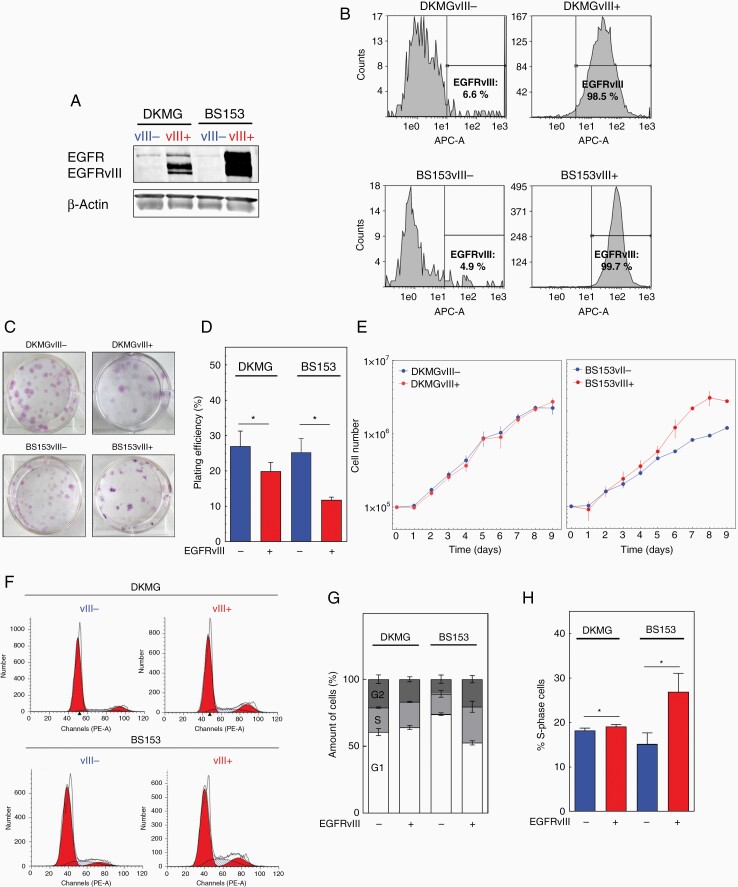
The aim of this study was to analyze the impact of EGFRvIII expression on RS and genomic instability. Therefore, we used 2 pairs of isogenic GBM cell lines with (EGFRvIII+) and without (EGFRvIII−) endogenous and stable EGFRvIII expression which originated from parental EGFR gene-amplified and EGFRvIII-positive DKMG and BS153 cell lines as described previously.2 As demonstrated in Figure 1, both EGFRvIII+ sub-cell lines (DKMGvIII+ and BS153vIII+) were tested positive for EGFRvIII by WB (Figure 1A) with more than 98% of these cells expressing EGFRvIII as quantified by flow cytometry (Figure 1B). In contrast, the EGFRvIII− counterparts (DKMGvIII− and BS153vIII−) showed no clear EGFRvIII expression in WB (Figure 1A) with less than 10% of the cells being EGFRvIII-positive (Figure 1B). We first analyzed the ability of self-renewal and observed a reduced capability of both EGFRvIII+ sub-cell lines to form colonies (Figure 1C and D). Although we detected no significant differences in proliferation (Figure 1E), we detected an increase in the fraction of S-phase cells especially for the BS153vIII+ cells (Figure 1F–H) which might be attributed to a prolonged S phase due to problems during replication, such as RS.
Figure 1.
EGFRvIII-/+ model system. (A) Detection of EGFR and EGFRvIII by Western blot analysis. β-Actin served as loading control. (B) EGFRvIII expression was detected by flow cytometry using an EGFRvIII-specific antibody (L8A4). (C) Colony formation, representative images. (D) Plating efficiency (average number of colonies per well divided by the number of seeded cells. (E) Proliferation. (F–H) Cell cycle distribution measured by flow cytometry. (F) Representative DNA content profiles, (G) cell cycle distribution and (H) quantification of S-phase cells (n = 3; mean with SEM; P values are obtained by two-tailed Student’s t-test. * P < .05).
EGFRvIII-Expressing Cells Exhibit Increased RS
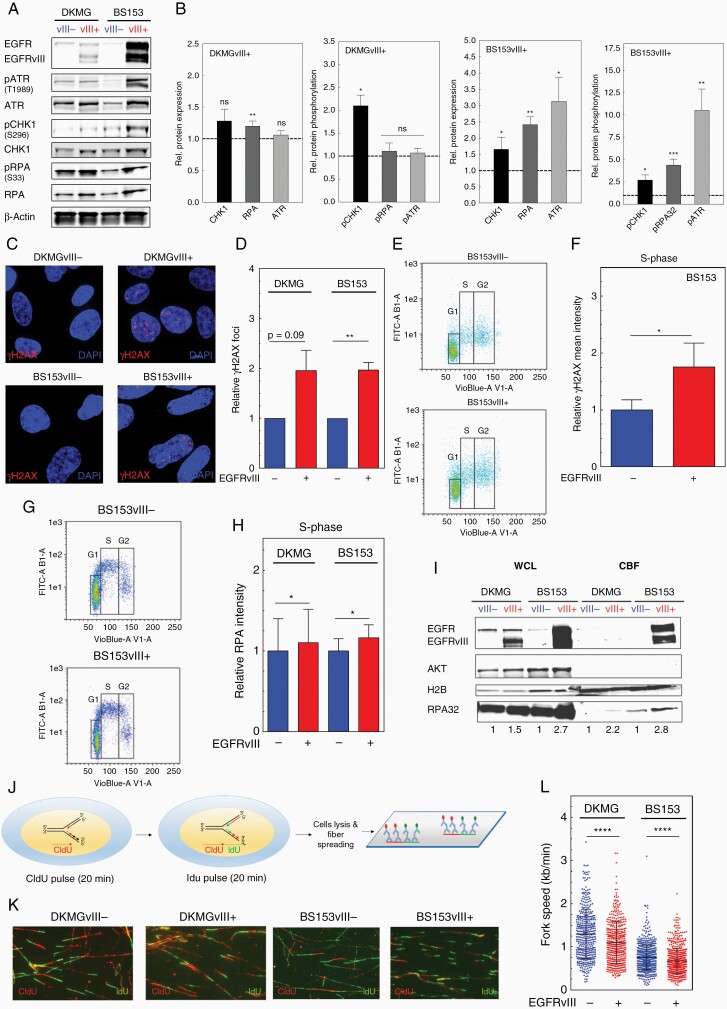
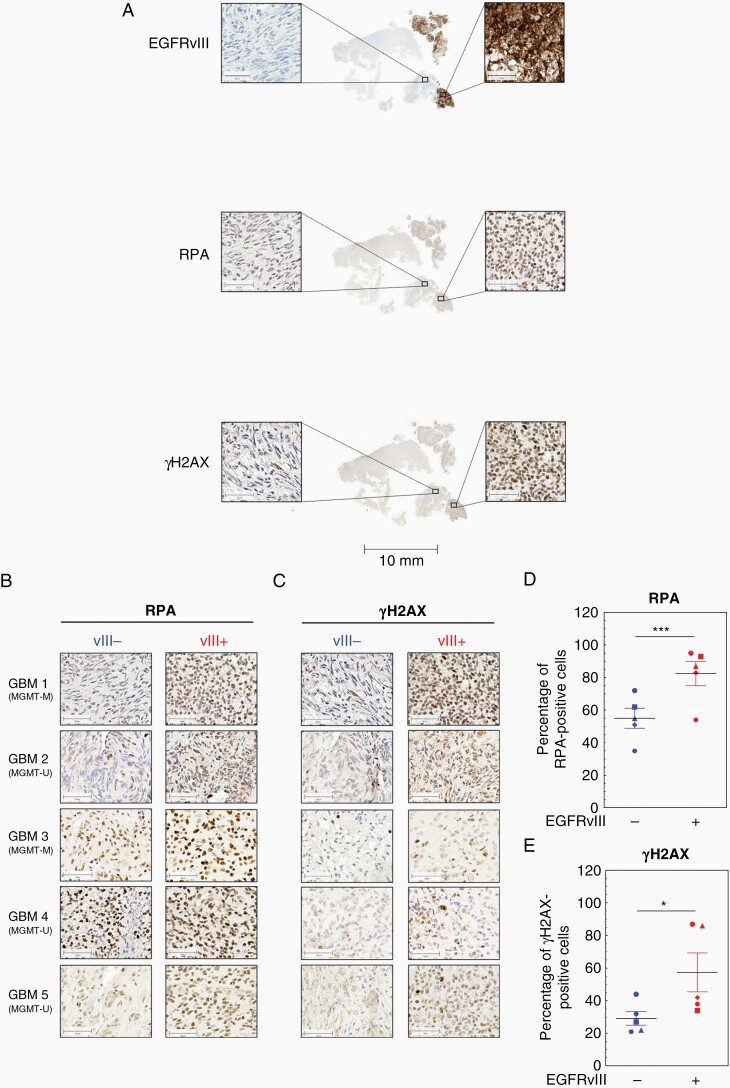
To analyze RS more specifically, we assessed the expression and phosphorylation of different marker proteins such as ATR, CHK1, RPA, and histone H2AX.17 We detected a significant increase in CHK1, RPA, and ATR expression in DKMGvIII+ and BS153vII+ cells by WB analysis. Furthermore, these proteins showed increased activation respectively phosphorylation hinting toward an activated RS response in EGFRvIII-expressing cells (Figure 2A and B). A siRNA-mediated downregulation of EGFRvIII in BS153vIII+ cells resulted in a decrease in RPA and CHK1 proportion of overall γH2AX-foci (H2AX phosphorylated at serine 139) indicating endogenous DNA damage respectively DSB in these cells (Supplementary Figure 1). Especially in S phase where replication takes place, an increased presence of γH2AX-foci indicates RS (Figure 2C and D). Therefore, we quantified γH2AX intensity in S- (Figure 2E and F) and S/G2 phase via flow cytometry (Supplementary Figure 2). The analysis revealed a nearly 2-fold increase of γH2AX intensity in BS153vIII+ cells arguing for the presence of replication-associated DNA damage. When measuring chromatin-bound RPA as a marker for ssDNA, we detected a moderate increase in S- (Figure 2G and H) and S/G2-phase cells of both EGFRvIII+ sub-cell lines (Supplementary Figure 3). To confirm this, we analyzed RPA in isolated chromatin-bound fractions via WB and detected increased amounts of chromatin-bound RPA in EGFRvIII+ cells (Figure 2I). We also observed chromatin-bound EGFRvIII in EGFRvIII+ cells, which is in line with the observations of Fan et al.6 Taking all these data together, we demonstrate that EGFRvIII+ GBM cells suffer from increased RS compared to their EGFRvIII− counterparts in cell culture. To directly analyze replication processes, we used the DNA fiber assay (Figure 2J). This allows measurement of specific replication structures and DNA replication velocities, confirming RS on a molecular level (Figure 2H).17,18 We observed a significantly slower fork progression in both EGFRvIII+ sub-cell lines (Figure 2K and L). Besides, we detected a tendency toward a higher percentage of stalled replication forks (Supplementary Figure 4A) and an in increase in first and second pulse origins (Supplementary Figure 4B) in EGFRvIII+ cells. To test if this phenotype is also detectable in human tumors in situ, we measured RPA and γH2AX in samples from EGFRvIII-positive GBM showing heterogeneous EGFRvIII expression using immunohistochemistry. An analysis of 5 GBM samples unveiled an increased presence of RPA and γH2AX in the nuclei of cells in the EGFRvIII+ areas compared to the EGFRvIII− areas (Figure 3A–C; Supplementary Figure 5). The quantification of 3 randomly chosen EGFRvIII− and EGFRvIII+ areas per patient showed a significantly higher expression of RPA and H2AX phosphorylation in the EGFRvIII+ areas, thereby confirming EGFRvIII-linked RS in human GBM (Figure 3D and E). Importantly, this seems to be MGMT status independent, since 2 GBM showed MGMT promoter methylation while 3 did not (Figure 3B).
Figure 2.
EGFRvIII expression is associated with replication stress in GBM cell lines. (A) Western blot analysis of RS response markers using specific antibodies, equal numbers of lysed cells were analyzed. β-Actin served as loading control. (B) For quantification the relative expression and phosphorylation values of EGFRvIII+ cells were normalized to the relative values of EGFRvIII− cells (n > 3; mean with SEM; P values are obtained by Wilcoxon Signed Rank test. *P < .05, **P < .01, ***P < .001, ns: not significant). (C and D) Detection of γH2AX foci by immunofluorescence. (C) Representative images (red: γH2AX foci; blue: nuclei (DAPI)). (D) Quantification of γH2AX foci (n = 3; mean with SEM; P values are obtained by Mann Whitney test, *P < .05, **P < .01, ns: not significant). (E and F) Analysis of relative γH2AX mean intensitiy in S-phase cells by flow cytometry. (E) Representative dot plot. DNA was stained with DAPI for cell cycle phase gating. (F) Quantification of γH2AX intensity in S-phase (n = 4; mean with SEM; P values are obtained by Mann Whitney test. *P < .05). (G and H) Analysis of average chromatin-bound RPA (mean intensity) in S-phase cells by flow cytometry. (G) Representative dot plots. (H) Quantification (n = 4; mean with SEM; P values are obtained by Mann Whitney test, *P < .05). (I) Western blot analyzing the level of chromatin-bound RPA32 in chromatin-bound fractions (CBF). Equal protein concentrations of CBF samples were loaded. Whole cell lysate (WCL) samples served as a control for fractionation efficiency. (J–L) DNA fiber assay. (J) Scheme: cells were incubated sequentially with CIdU (red) then IdU (green), followed by lysis and fiber spreading. (K) Representative immunofluorescent images of DNA fibers. (L) Fork speed in kb/min. The dot plot summarizes the quantification of IdU and CIdU incorporation rates (n = 3; mean with SEM; P values are obtained by two-tailed Student’s t-test. ***P < .001).
Figure 3.
EGFRvIII expression is associated with replication stress in human GBM cell samples. Immunohistochemical detection of EGFRvIII, RPA, and γH2AX in human GBM samples with heterogeneous EGFRvIII expression. (A) Representative sample (GBM1). Scale bars: Overview, 10 mm; zoom, 50 µm. Detection of (B) RPA and (C) γH2AX in 5 GBM patient samples. EGFRvIII− areas are depicted on the left, EGFRvIII+ on the right (scale bars: 50 µm). Quantification of percentage of (D) RPA-positive cells and (E) γH2AX-positive cells in the EGFRvIII− and EGFRvIII+ areas (n = 5; mean with SEM; P values are obtained by one-tailed Student’s t-test, *P < .01, ***P < .001). For quantification, 3 fields of view from the EGFRvIII− area and EGFRvIII+ area, respectively, per specimen have been evaluated.
EGFRvIII Expression Is Associated With Chromosomal Instability
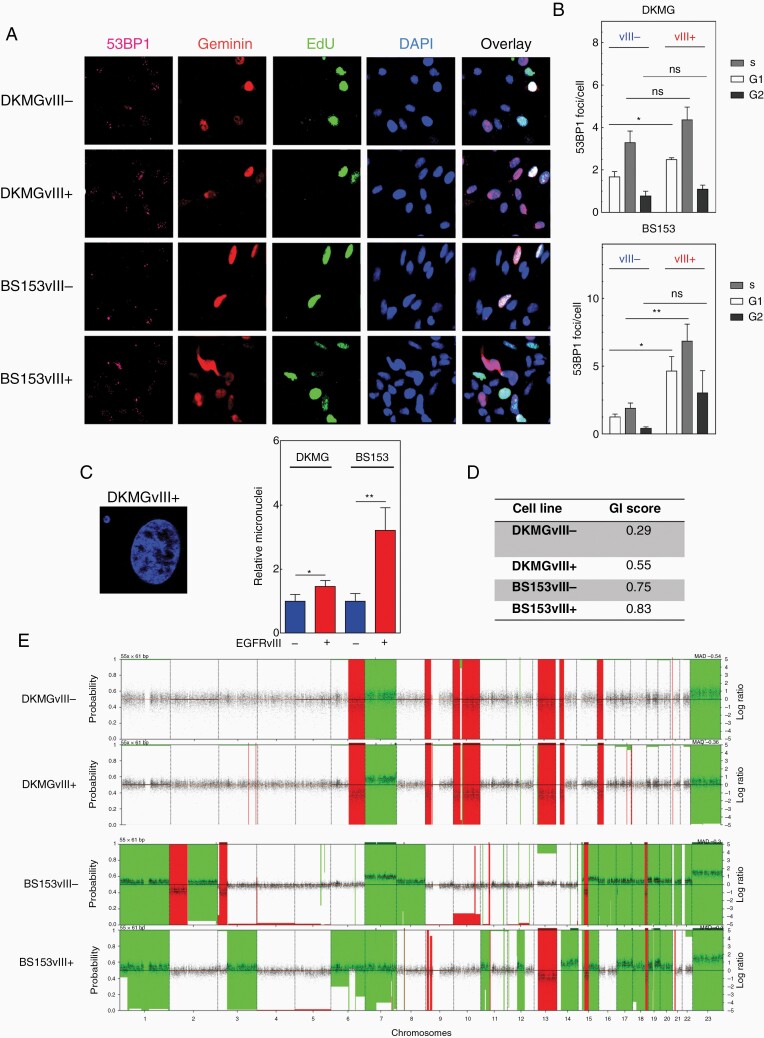
To further analyze the consequences of EGFRvIII-induced RS, we quantified 53BP1-foci and micronuclei formation, both being markers for DNA DSB and a mitotic entry with under-replicated DNA.19,20 Indeed, we observed significantly elevated numbers of 53BP1-foci in both EGFRvIII+ sub-cell lines also in the G1 phase which argues for under-replicated DNA. Besides, EGFRvIII+ cells also displayed increased 53BP1-foci in S phase which might be a consequence of EGFRvIII-induced replication stalling and collapse (Supplementary Figure 4A and B). In line with this, we observed a higher activation of ATM in EGFRvIII+ cells, which is normally activated in response to DSB (Supplementary Figure 6). When the GIS was calculated, both EGFRvIII+ cells exhibited higher genomic instability (DKMG: 0.55, BS153: 0.83) compared to EGFRvIII− cells (DKMG: 0.29, BS153 0.75, Figure 4D). The GIS was calculated on the basis of the genomic copy number alterations detected in the 4 cell lines that are visualized in Figure 4E. Taken together, these data demonstrate increased DNA damage and genomic instability as a consequence of a pronounced RS in EGFRvIII+ cells.
Figure 4.
EGFRvIII expression is associated with DNA double-strand breaks and genomic instability. (A and B) Detection of 53BP1-foci in G1-, S- and G2-phase cells by immunofluorescence. (A) Representative image (pink: 53BP1 foci). For discrimination of different cell cycle phases cells were labeld with EdU (green) before fixation and co-stained with anti-geminin antibodies (red). Cell nuclei were counterstained with DAPI (blue). (B) Quantification of 53BP-foci in G1-, S- and G2-phase cells 72 h after seeding (n = 3). (C) Detection of micronuclei by DAPI. Left: representative image; right: quantification (n = 3). (D and E) Array-based CGH-analysis with (D) genomic instability score (E) CGH-profile.
EGFRvIII-Expressing Cells Display Increased Transcription Activity and R-loop Formation
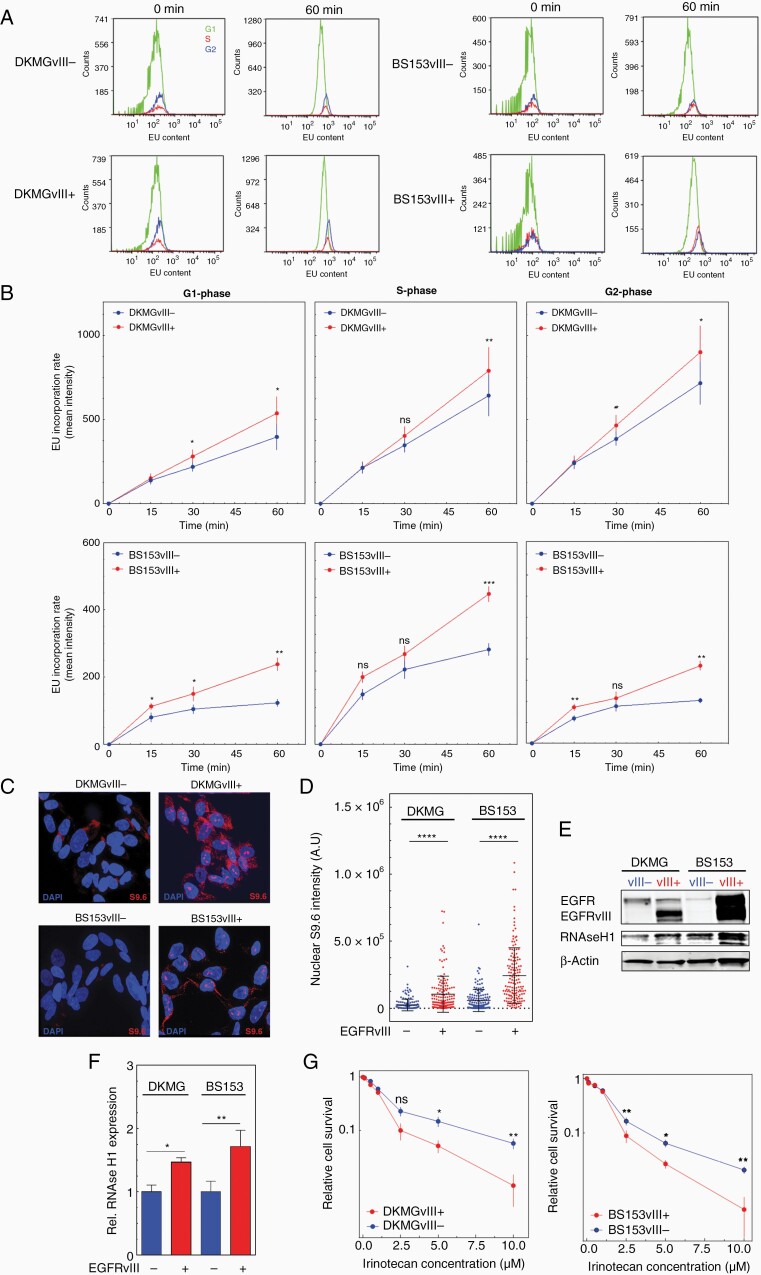
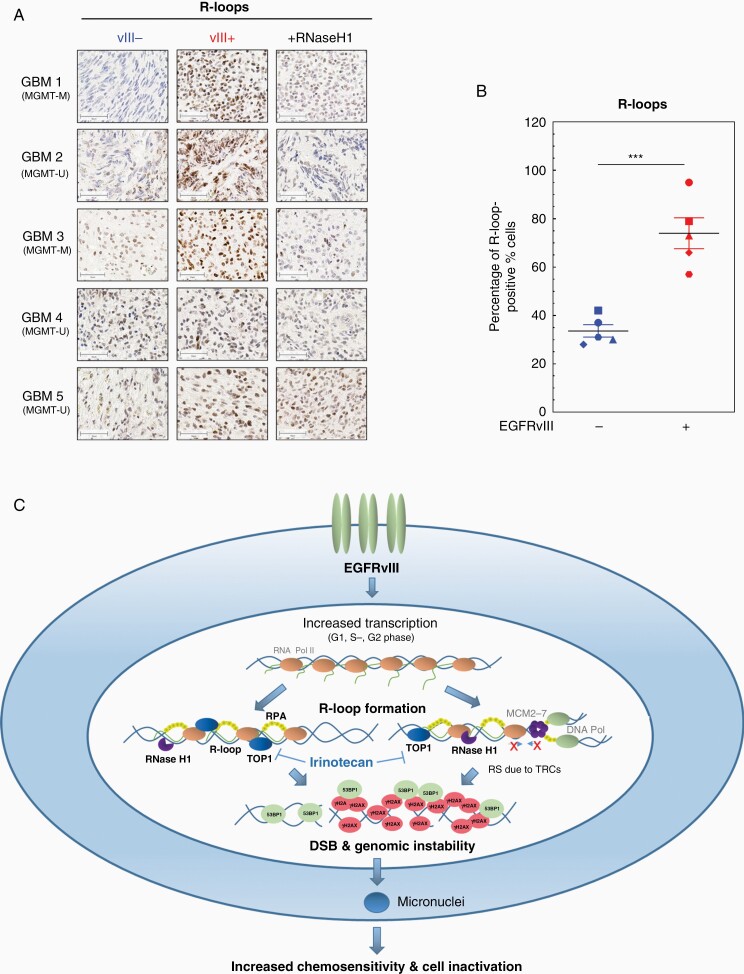
As a potential cause of increased RS, we investigated transcription activity by quantifying EU incorporation into nascent RNA. We detected an elevated RNA synthesis in both EGFRvIII+ cell lines. After 1 h, the difference was more than 1.5-fold for DKMG cells and more than 2-fold in BS153 cells (Supplementary Figure 7). As shown in Figure 5A and B, we observed significantly higher EU-incorporation rates for all cell cycle phases after 60 min (Figure 5A and B). These data indicate that increased transcription might be a cause for RS since it can lead to transcription replication conflicts (TRCs) during S phase.11 To prove this, we next analyzed the formation of R-loops via immunofluorescence. R-loops are 3 stranded RNA/DNA hybrids that can accumulate in all cell cycle phases, but can especially be stabilized due to TRCs in S phase. R-loops can promote fork stalling, but can also be a direct source of DNA damage.11,12,21,22 Using the S9.6 antibody we observed a clear upregulation of R-loops in the nuclei of all EGFRvIII+ cells, indicating that increased R-loop formation in all cell cycle phases (Figure 5C). The quantification of the nuclear signal revealed a pronounced and highly significant difference in R-loop formation between EGFRvIII− and EGFRvIII+ cells. Both EGFRvIII+ cell lines showed a 4-fold increase in nuclear S9.6 staining intensity (Figure 5D), supporting the hypothesis that EGFRvIII-induced RS is induced by TRC. In line with this, we observed higher levels of RNase H1 in EGFRvIII+ cells, which is responsible for the dissolving of R-loops (Figure 5E and F). Like RNase H1, TOP1 is of importance for the cells’ ability to deal with R-loops. Since it has been shown that tumor cells showing high levels of R-loops are highly sensitive to inhibitors of TOP1, we investigated whether R-loops could potentially be exploited for therapy.22–24 Since TOP1 inhibition by irinotecan can be used for the treatment of GBM, we tested the sensitivity of EGFRvIII+/− cells toward irinotecan with concentrations up to 10 µM. We observed that both EGFRvIII+ cell lines were more sensitive toward irinotecan, which was already pronounced at concentrations of 2.5 µM (Figure 5G). Taken together, these data show that EGFRvIII expression is associated with increased transcription and R-loop formation indicating that TRCs are the cause for the increased RS in EGFRvIII-positive GBM cells. To test if TRCs might also be a cause for increased RS in situ, we analyzed R-loops in EGFRvIII-positive GBM patient samples and detected a significantly stronger R-loop intensity in EGFRvIII+ areas (Figure 6A and B, Supplementary Figure 8). The specificity of staining was validated by pretreatment with RNase H1, which resulted in a significant loss of the R-loop signal. Therefore, we propose a model, in which EGFRvIII expression causes RS and genomic instability via the upregulation of transcription which in turn leads to the accumulation of R-loops and thereby sensitivity to TOP1 inhibition (Figure 6C).
Figure 5.
EGFRvIII expression is associated with increased transcription activity and R-loop formation. (A and B) Transcriptional activity in G1-, S- and G2-cells measured by EU incorporation and flow cytometry. (A) Representative histograms showing kinetics over 60 min of EU treatment. (B) Quantification of EU incorporation rate in G1-, S- and G2-phase cells at different time points (n = 3; mean with SEM; P values are obtained by two-tailed Student’s t-test. *P < .05, **P < .01). (C, D) Analysis of R-loops S9.6 immunostaining. (C) Representative picture (red: S9.6 signal; blue: DAPI) and (D) quantification of nuclear S9.6 intensity (n = 3; mean with SEM; P values are obtained by two-tailed Student’s t-test. *P < .0001; at least 50 nuclei were analyzed per independent experiment). (E) Western blot analysis of RNAse H1 expression. Samples were normalized to cell number. Detection of β-actin served as loading control. (F) Quanification of RNAse H1 expression intensitiy values of EGFRvIII+ cells. The values were normalized to EGFRvIII− cells (n = 3; mean with SEM; P values are obtained by two-tailed Student’s t-test. *P < .05, **P < .01). (G) Cell survival after irinotecan treatment assessed by colony-forming assay (n = 3; mean with SEM; P values are obtained by two-tailed Sudent’s t-test. *P < .05, **P < .01).
Figure 6.
Increased R-loop accumulation in EGFRvIII-expressing areas in human GBM samples and model for EGFRvIII-induced replication stress. (A and B) Immunohistochemical detection of R-loops in GBM patient samples. (A) Representative pictures for GBM samples 1–5. As a control, samples were incubated with RNase H1 before immunohistochemical staining with S9.6 antibody. EGFRvIII− areas are depicted on the left, EGFRvIII+ areas are displayed in the middle column. EGFRvIII+ areas, which were treated as a control with RNase H1, are shown on the right (scale bars, 50 µm). (B) Quantification of percentage of R-loop-positive cells (n = 5; mean with SEM; P values are obtained by one-tailed Student’s t-test. ***P < .001). (C) Model of how EGRFvIII drives RS: EGFRvIII expression increases transcriptional activity in all cell cycle phases and thereby promotes accumulation of R-loops, which cause DNA damage and genomic instability. R-loop formation during S phase can promote TRCs and can therefore lead to replication fork slowing, stalling, and finally DSB. R-loops in G1 and G2 cells can either be processed by nucleases leading directly to DSB or will form secondary DSB during replication. Both processes result in the activation of the RS response and DDR. If not properly repaired EGFRvIII-induced DSB lead to genomic instability and micronuclei formation and with that cell inactivation.
Discussion
Here we demonstrate that EGFRvIII expression is associated with oncogenic stress, elevated transcription, and R-loop formation as well as aberrant RS and increased genomic instability. These findings give important new insights into the biology of EGFRvIII+ tumors and might have therapeutic implications, since they unveil a potential Achilles heel of EGFRvIII-positive GBM as highlighted by the increased sensitivity of EGFRvIII+ cells toward the TOP1 inhibitor irinotecan (Figure 4G).
To our knowledge, this is the first study showing increased RS and R-loop accumulation in GBM cell cultures with endogenous EGFRvIII expression. Moreover, we could validate our in vitro data also in situ, using primary human GBM samples. Here, the heterogeneous expression of EGFRvIII allowed us to directly compare EGFRvIII+ and EGFRvIII− areas of the same tumor and to demonstrate increased RS as well as R-loop formation in untreated GBM samples, underscoring the physiological and clinical relevance of our results. So far EGFRvIII expression has not been directly linked to RS, and mechanistic studies are lacking. However, Nitta et al.25 reported that U87MG artificially expressing EGFRvIII displayed increased levels of reactive oxygen species and γH2AX levels, while Li et al.26 showed that EGFRvIII expression and homozygous deletion of PTEN in mice neural precursor cells drive genomic instability; both supporting the data presented here.
In contrast to Nitta et al.,25 we observed no clear increase in reactive oxygen species (data not shown). However, our data suggest that EGFRvIII-associated RS may be induced by increased transcriptional activity and subsequent R-loop formation. This is in line with the observation that the transcription machinery is frequently deregulated in cancer cells expressing oncogenes. For example, it has been shown that oncogenic HRAS activates ERK1/2 signaling, which leads to an upregulation of the transcription rate. This in turn leads to R-loop accumulation and an increase of TRCs during S phase.11,12 In line with that we have recently shown that EGFRvIII expression is associated with elevated activation of ERK1/2 and other mitogen-activated protein kinases (MAPKs)-1 leading us to hypothesize that EGFRvIII might increase transcription activity via MAPKs.9 After activation MAPKs translocate to the nucleus where they regulate the expression of multiple genes. MAPKs phosphorylate transcription factors like ATF2 and Jun proteins, which are bound to their response elements within the promoter region of early response genes, such as c-jun and c-fos. MAPKs regulate the activity of the newly synthesized proteins c-Jun and c-Fos forming AP-1, which can bind the promoter region of the oncogene c-MYC.27 So we might speculate that EGFRvIII+ cells express higher levels of c-MYC than EGFRvIII− cells contributing to the RS phenotype. Furthermore, EGFRvIII itself might also activate transcription, since we observed chromatin-bound EGFRvIII in EGFRvIII+ cells (Figure 2I), which might be able to form a nuclear complex with STAT3, thereby promoting transcription.6 As a consequence of increased transcription and R-loop formation, the cells suffer from RS and accumulate DNA damage which can cause genomic instability11,12,21,22 as we have also shown for EGFRvIII+ cells (Figure 4C and D). This EGFRvIII-driven genomic instability might add to GBM heterogeneity and therefore stem cell properties, which is in line with studies characterizing EGFRvIII as a driver for heterogeneity and stemness.28,29 In our study, DKMGvIII+ cells seem to suffer less from RS and endogenous DNA damage compared to BS153vIII+ cells. This might be due to the significantly higher expression of EGFRvIII in BS153vIII+ cells. Another reason might be the different p53 status. DKMG cells express p53 wildtype, whereas BS153 cells harbor a TP53 mutation.14 By regulating cell cycle arrests, p53 is able to protect cells from an accumulation of DNA damage and genomic instability. Besides this Yeo et al.30 could show that p53 maintains genomic stability by preventing interference between transcription and replication and that p53 loss impairs normal replication fork progression. However, in this context, p53 does not fully protect the DKMGvIII+ cells, since they also showed elevated RS, DNA damage, and R-loop formation. Importantly, RS is independent of MGMT status since we can detect increased RS and R-loop formation in both, MGMT promoter methylated and unmethylated GBM (see Figures 3 and 6). This suggests that TOP1 inhibition can be effective for most if not all EGFRvIII-positive GBM. This is, for example, in contrast to TMZ, since increased TMZ sensitivity of EGFRvIII+ GBM is clearly dependent on MGMT status.1 Whereas alkylating agents such as TMZ directly attack the DNA, TOP1 inhibitors act indirectly. TOP1 is important for the ability of cells to resolve both positive and negative supercoils, thereby also preventing conflicts between transcription and DNA replication, the TRCs. In cells with high basal levels of R-loops, TOP1 inhibition can lead to an increase in the levels of R-loops causing severe DNA damage. Furthermore, TOP1 inhibition leads to transcription-blocking TOP1 cleavage complexes (TOP1ccs), which can induce transcription blocks and subsequent R-loop formation.31 Since TOP1 inhibitor irinotecan is already approved for the treatment of recurrent GBM, makes it an attractive alternative therapeutic for EGFRvIII-positive tumors.32 Although EGFRvIII expression is not a general marker for treatment sensitivity since it is not associated with radiosensitivity, it might also confer sensitivity to other chemotherapeutics, which has to be tested in further studies. It also has to be investigated whether increased RNA synthesis, R-loop formation, and RS can occur in EGFRvIII-negative GBM, making our results clinically relevant for a broader range of GBM. In summary, our study has unveiled EGFRvIII expression to be associated with severe RS in GBM, offering new insights in GBM biology as well as options for a new personalized therapeutic concept.
Supplementary Material
Funding
This work was supported by the Federal Ministry of Education and Research (BMBF grant 02NUK032 to M.K., K.R., T.R.), University Cancer Center Hamburg Research Fellowship (L.B.), Wilhelm Sander-Stiftung (N.S., A.S.W.), E.W. Kuhlmann-Stiftung (M.H.C.), Werner-Otto-Stiftunfg (J.M.G), and the Fördergemeinschaft Kinderkrebszentrum Hamburg (U.S.).
Conflict of interest statement. The authors declare no conflict of interest.
Authorship statement. N.S., K.R, S.C.S., M.K.: Conceptualization of the study. N.S., M.K.: Writing, original draft. N.S., L.B., A.C.P, K.U., W.Y.M., T.R., S.C.S, U.S., C.P., K.R., M.K.: Review and editing. N.S.: Data analysis. M.K., N.S.: Supervision. J.L. flow cytometry for cell cycle analysis and EU incorporation assay. J.B: fiber assay and analysis. M.G.: siRNA experiments and Western blot analysis. J.M.G, K.H., M.C., N.S., A.W.: western blot analysis. L.O.: γH2AX immunofluorescence staining and analysis. K.H.: flow cytometry for analysis of chromatin-bound RPA and immunohistochemical staining. F.G.: 53BP1 and R-loop immunofluorescence staining. N.S., A.W.: colony formation assays. B.R.: 53BP1 immunofluorescence staining and analysis. K.U.: CGH-array analysis. U.S.: provided GBM specimens.
References
- 1. Smrdel U, Popovic M, Zwitter M, et al. Long-term survival in glioblastoma: methyl guanine methyl transferase (MGMT) promoter methylation as independent favourable prognostic factor. Radiol Oncol. 2016;50(4):394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Hegi M, Weller M. Neuro-oncology, a decade of temozolomide and beyond. Expert Rev Anticancer Ther. 2010;10(11):1675–1677. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 4. Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci. 2009;16(6):748–754. [DOI] [PubMed] [Google Scholar]
- 5. Heimberger AB, Hlatky R, Suki D, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11(4):1462–1466. [DOI] [PubMed] [Google Scholar]
- 6. Fan QW, Cheng CK, Gustafson WC, et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell. 2013;24(4):438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong AJ, Ruppert JM, Bigner SH, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA. 1992;89(7):2965–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng Q, Han L, Dong Y, et al. JAK2/STAT3 targeted therapy suppresses tumor invasion via disruption of the EGFRvIII/JAK2/STAT3 axis and associated focal adhesion in EGFRvIII-expressing glioblastoma. Neuro Oncol. 2014;16(9):1229–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Struve N, Binder ZA, Stead LF, et al. EGFRvIII upregulates DNA mismatch repair resulting in increased temozolomide sensitivity of MGMT promoter methylated glioblastoma. Oncogene. 2020;39(15):3041–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carruthers RD, Ahmed SU, Ramachandran S, et al. Replication stress drives constitutive activation of the DNA damage response and radioresistance in glioblastoma stem-like cells. Cancer Res. 2018;78(17):5060–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jones RM, Mortusewicz O, Afzal I, et al. Increased replication initiation and conflicts with transcription underlie cyclin E-induced replication stress. Oncogene. 2013;32(32):3744–3753. [DOI] [PubMed] [Google Scholar]
- 12. Kotsantis P, Silva LM, Irmscher S, et al. Increased global transcription activity as a mechanism of replication stress in cancer. Nat Commun. 2016;7:13087. doi: 10.1038/ncomms13087. PMID: 27725641; PMCID: PMC5062618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kotsantis P, Petermann E, Boulton SJ. Mechanisms of oncogene-induced replication stress: jigsaw falling into place. Cancer Discov. 2018;8(5):537–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Struve N, Riedel M, Schulte A, et al. EGFRvIII does not affect radiosensitivity with or without gefitinib treatment in glioblastoma cells. Oncotarget. 2015;6(32):33867–33877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parplys AC, Seelbach JI, Becker S, et al. High levels of RAD51 perturb DNA replication elongation and cause unscheduled origin firing due to impaired CHK1 activation. Cell Cycle. 2015;14(19):3190–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilke CM, Braselmann H, Hess J, et al. A genomic copy number signature predicts radiation exposure in post-Chernobyl breast cancer. Int J Cancer. 2018;143(6):1505–1515. [DOI] [PubMed] [Google Scholar]
- 17. Parplys AC, Petermann E, Petersen C, Dikomey E, Borgmann K. DNA damage by X-rays and their impact on replication processes. Radiother Oncol. 2012;102(3):466–471. [DOI] [PubMed] [Google Scholar]
- 18. Nieminuszczy J, Schwab RA, Niedzwiedz W. The DNA fibre technique—tracking helicases at work. Methods. 2016;108:92–98. doi: 10.1016/j.ymeth.2016.04.019. PMID: 27102626. [DOI] [PubMed] [Google Scholar]
- 19. Lukas C, Savic V, Bekker-Jensen S, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13(3): 243–253. [DOI] [PubMed] [Google Scholar]
- 20. Spies J, Lukas C, Somyajit K, et al. 53BP1 nuclear bodies enforce replication timing at under-replicated DNA to limit heritable DNA damage. Nat Cell Biol. 2019;21(4):487–497. [DOI] [PubMed] [Google Scholar]
- 21. Crossley MP, Bocek M, Cimprich KA. R-loops as cellular regulators and genomic threats. Mol Cell. 2019;73(3):398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sollier J, Stork CT, Garcia-Rubio ML, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56(6):777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parajuli S, Teasley DC, Murali B, et al. Human ribonuclease H1 resolves R-loops and thereby enables progression of the DNA replication fork. J Biol Chem. 2017;292(37):15216–15224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Promonet A, Padioleau I, Liu Y, et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat Commun. 2020;11(1):3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nitta M, Kozono D, Kennedy R, et al. Targeting EGFR induced oxidative stress by PARP1 inhibition in glioblastoma therapy. PLoS One. 2010;5(5):e10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li L, Dutra A, Pak E, et al. EGFRvIII expression and PTEN loss synergistically induce chromosomal instability and glial tumors. Neuro Oncol. 2009;11(1):9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Turjanski AG, Vaqué JP, Gutkind JS. MAP kinases and the control of nuclear events. Oncogene. 2007;26(22):3240–3253. doi: 10.1038/sj.onc.1210415. PMID: 17496919. [DOI] [PubMed] [Google Scholar]
- 28. Emlet DR, Gupta P, Holgado-Madruga M, et al. Targeting a glioblastoma cancer stem-cell population defined by EGF receptor variant III. Cancer Res. 2014;74(4):1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Godek KM, Venere M, Wu Q, et al. Chromosomal instability affects the tumorigenicity of glioblastoma tumor-initiating cells. Cancer Discov. 2016;6(5):532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yeo CQX, Alexander I, Lin Z, et al. p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 2016;15(1):132–146. [DOI] [PubMed] [Google Scholar]
- 31. Cristini A, Ricci G, Britton S, et al. Dual processing of R-loops and topoisomerase I induces transcription-dependent DNA double-strand breaks. Cell Rep. 2019;28(12):3167–3181 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vredenburgh JJ, Desjardins A, HerndonJE, 2nd, et al. Bevacizumab plus irinotecan in recurrent glioblastoma multiforme. J Clin Oncol. 2007;25(30):4722–4729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.