Abstract
Background:
Circular RNA-HIPK3 (CircHIPK3) has been shown to be aberrantly expressed in a variety of diseases, contributing to disease initiation and progression. The aim of the present study is to investigate the role of the circHIPK3 RNA/microRNA-124a interaction in the pathogenesis of rheumatoid arthritis (RA).
Methods:
This study included 79 RA patients and 30 control individuals. The patients involved were classified according to the disease activity score (DAS28) into mild (24 patients), moderate (24 patients), and severe (31 patients). Serum samples were collected to estimate the relative gene expression of circHIPK3 RNA and its target gene microRNA-124a by quantitative real time-PCR. Moreover, ELISA was used to detect the serum levels of monocyte chemoattractant protein-1 (MCP-1). Routine laboratory estimation of erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), and rheumatoid factor (RF) was also done.
Results:
In all grades of RA groups, there was a significantly substantial elevation of circHIPK3 RNA gene expression, with subsequent downregulation of miRNA-124a when compared to the control group. CircHIPK3 and microRNA-124a expression have been established to be inversely linked. Also, estimation of serum levels of MCP-1, ESR, CRP, and RF exhibited a significant increase in all grades of RA as compared to the control group.
Conclusion:
CircHIPK3 and microRNA-124a might be regarded as key players in the pathogenesis of RA. The cross-talk between them appears to be responsible for inducing joint inflammation by increasing MCP-1 production. Targeting circHIPK3 and microRNA-124a, and their downstream adaptor molecules, poses a new challenge for RA therapy.
Key Words: Circhipk3, Circular RNA, Microrna-124a, Rheumatoid arthritis
Introduction
Rheumatoid arthritis (RA) is a chronic systemic autoimmune disease marked by joint inflammation, which can lead to musculoskeletal abnormalities, decreased functioning, and significant morbidity if left untreated (1). The inflammatory milieu in rheumatoid joints promotes the activation of neutrophils, T-cells, B-cells, macrophages, osteoclasts, and synovial fibroblasts (2). Through cytokine production, these cells maintain a cross-talking process, and their activation promotes the secretion of enzymes and other chemicals that contribute to cartilage and bone tissue degradation (3,4). T-lymphocytes, in particular, play a crucial role in RA pathogenesis. They help synovial macrophages and rheumatoid arthritis synovial fibroblasts (RASFs) become activated (5).
Synovial hyperplasia is a phenotypical hallmark of RA in which synovial fibroblasts, and fibroblast-like synoviocytes (FLS), exhibit resistance to apoptosis. This promotes tumor-like cell proliferation (6,7). The proliferation of these RA resident cells is critical in the progression of RA because they generate inflammatory cytokines, chemokines, angiogenic factors, and enzymes that erode the bone matrix. They also orchestrate the local invasion of numerous cells, resulting in inflammatory reactions and joint destruction in RA patients (8,9). It has previously been established that monocyte chemoattractant protein 1 (MCP-1) is implicated in RA via stimulation of FLS proliferation and migration and may therefore contribute to severe joint destruction. As a result, targeting MCP-1 or its receptor may aid in the inhibition of RA progression (10).
Many epigenetic pathways involved in the transition of synovial fibroblasts into RASFs, with a more aggressive character, have been discovered in the previous decade. MicroRNAs (miRNAs) are the most extensively studied of these (11,12). miRNAs are a class of small non-coding RNAs that regulate gene expression post-transcriptionally. miRNAs bind to complementary sequence-messenger RNAs (mRNAs) and inhibit their translation, suppressing the synthesis of particular proteins. Altered miRNA expression has been linked to inflammatory pathway signaling, increased production of pro-inflammatory cytokines, and other autoimmunity-related events (13,14).
Many aspects of RASF activity are influenced by alterations in miRNA expression (15). The bulk of changes in miRNA levels resulted in the production of pro-inflammatory cytokines and enhanced RASF proliferation and survival, causing subsequent tissue damage. In vitro administration of miRNA-124a has been shown to inhibit inflammatory cell chemotaxis as well as reduce RASF proliferation (16-18). Several lines of evidence indicate that the transfection of precursor miRNA-124a into RA synoviocytes significantly reduced cell proliferation and arrested the cell cycle at the G1 phase (19).
Circular RNAs (circRNAs) are an emerging class of RNA species, with covalently closed loops that are intrinsically resistant to exoribonuclease (20). CircRNAs are derived from non‐canonical splicing processes and control gene expression and transcription by binding to particular miRNAs, sequestering them, and inhibiting their activity (21), in a phenomenon termed the competitive endogenous RNA hypothesis (22). CircRNAs have binding sites for target miRNA and serve as sponges, leading to sequestration of this miRNA, preventing its binding to the target mRNA and losing its capacity to suppress gene expression, resulting in an increase in mRNA (23).
According to emerging research, the circRNA 0000284 (circHIPK3), derived from the homeodomain-interacting protein kinase-3 (HIPK3), contains binding sites for nine miRNAs, resulting in the reduction in repression of their targeted mRNA. CircHIPK3, has been found to contain two binding sites for miRNA-124, preserving the capacity to regulate this miRNA (24). Herein, the purpose of this work is to investigate the role of the circHIPK3 RNA/miRNA-124a interaction in the pathogenesis of rheumatoid arthritis.
Materials and Methods
Ethical approval
This study followed the Declaration of Helsinki principals and was endorsed by the medical ethical committee of Kasr Al-Ainy hospitals. Written informed consent from all participants was obtained before sample collection.
Design and Participants
A cross-sectional study was carried out on 109 subjects of matched age and gender, who were recruited from the Rheumatology Department at Cairo University's Faculty of Medicine. All participants were recruited between February 2019 and February 2020, with disease durations ranging from 1 month to 18 years. All patients (n= 79) were diagnosed with RA based on the American College of Rheumatology (ACR) 2010/ European League against Rheumatism (EULAR) criteria for RA classification (25), meeting the following criteria:≥ 4 swelling joints and 6 tender joints, in addition to one of the following: morning stiffness duration≥ 45minutes, erythrocyte sedimentation rate (ESR)> 28mm/h, or C-reactive protein (CRP)> 10mg/L. All patients in this study underwent a comprehensive history and clinical examination, except for those with diabetes, neoplasia, inflammatory disorders, or other autoimmune diseases who were excluded from this study.
RA patients were classified according to disease activity score in 28 joints (DAS28) (26) into: mild RA (24 patients) with low disease activity, moderate RA (24 patients) with moderate disease activity, and severe RA (31 patients) with high disease activity. Healthy control volunteers (n= 30) with no history of inflammatory arthritis or current joint pain were matched to RA patients by age, gender, and ethnicity. This study was authorized by the Kasr Al-Ainy hospital's local ethical committee in compliance with the Declaration of Helsinki's ethical criteria. All eligible individuals provided informed consent.
All subjects were subjected to meticulous assessments that included questionnaires, rheumatologic physical examination, routine laboratory investigations including liver function tests, kidney function tests, hepatitis markers (Anti-HCV antibodies, HBsAg and Anti-HBc IgM), thyroid function tests, rheumatoid factor (RF), antinuclear antibodies(ANA), coagulation profile, and random blood glucose level. Three mL of peripheral venous blood samples were withdrawn from each subject by using a vacutainer equipment. In serum separator tubes, samples were collected and allowed to clot for 15 minutes before being centrifuged at 4000 X g for 10 minutes. The sera were separated and kept at -80 °C until analysis.
Estimation of gene expression of circHIPK3 RNA and miRNA-124a by QRT-PCR
Extraction of serum total RNA, including miRNA and circRNA, was performed using the miRNeasy mini kit (Direct-zol™ RNA MiniPrep Catalog No. R2050) following the manufacturer’s protocol. RNA samples were subjected to RNA quantitation and purity assessment using the NanoDrop® (ND)-1000 spectrophotometer (NanoDrop Technologies, Inc. Wilmington, USA). Reverse transcription (RT) was carried out using the COSMO cDNA synthesis Kit (Willowfort-1020500x, UK) according to the manufacturer’s guidance.
Five microliters of cDNA were used to assess the relative abundances of the mature miRNA and circRNA species using the HERA SYBR® Green qPCR kit (WF1030400X) (Willowfort, UK), and an ABI Prism7500 Sequence Detector System Applied Biosystem with software version 3.1 (StepOne™, USA). The PCR primers used were designed by Gene Runner Software (Hastings Software Inc., Hastings, NY, USA) from RNA sequences in GenBank (Table 1)
Table 1.
Primer sequences of circHIPK3 RNA, miR-124a, and 18S RNA.
| Parameter | Primer sequence | Gene bank accession number |
|---|---|---|
| circHIPK3 RNA | F: 5’ TGGAGACTGGGGGAAGATGA 3’ | NM001134.2 |
| R: 5’ CACACTAACTGGCTGAGGGG 3’ | ||
| miRNA-124a | F: 5’ GCAAAGCAACATCTTCCCTA 3’ | AF236056.1 |
| R: 5’ CCACAACAAACTTGCCCTC 3’ | ||
| 18s RNA | F: 5’ CAGCCACCCGAGATTGAGCA 3’ | JX132355.1 |
| R: 5’ TAGTAGCGACGGGCGGGTGT 3’ |
QRT-PCR analysis was performed according to the manufacturer’s reference (Applied Biosystems, USA). Data were calculated by Sequence Detection Software version 1.7 (PE Biosystems, USA). The relative expression of circHIPK3 RNA and miRNA-124a were calculated relative to the 18s RNA housekeeping gene by the comparative Ct method as stated by the manufacturer recommendations (Applied Biosystems, USA).
Estimation of serum monocyte chemoattractant protein 1 (MCP1) by ELISA
(MyBioSource, San Diego, California, USA): This was done in accordance with the manufacturer’s recommendations.
Routine laboratory investigations
ESR at the first hour using the Westergren method, as well as CRP and RF using latex slide agglutination assays.
Statistical Analysis
The IBM® SPSS software version 22 was used to code and enter the data. For quantitative variables, data were statistically described in terms of mean±standard deviation. Comparisons between groups were done using analysis of variance (ANOVA), followed by the Bonferroni post-hoc test (27). Statistically significant was defined as P-values less than 0.05.
Results
Comparisons of the demographic features in all studied groups
As shown in Table 2, regarding age and disease duration, there is no statistically significant difference between any of the diseased groups and the control group. As regards sex, the incidence of RA is most commonly occurring among females with a high statistically significant difference (p-value< 0.001). The disease activity score of 28 joints shows a highly significant difference between the moderate and the mild RA groups. Furthermore, the severe RA group shows a highly significant increase in DAS28 as compared to both the mild and moderate RA groups.
Table 2.
Demographic data among the studied groups.
| Control (N= 30) | Rheumatoid Patients (N= 79) | p-value | Sig. | ||||
|---|---|---|---|---|---|---|---|
| Low Disease Activity(N= 24) | Moderate Disease Activity (N= 24) | High Disease Activity(N= 31) | |||||
| Age (years) | 42±6.7 | 48.2±9.75 | 44.25±10.38 | 46.77±9.76 | > 0.05 | NS | |
| Duration (years) | 4.85±5.09 | 4.7±4.38 | 2.67±2.6 | > 0.05 | NS | ||
| DAS28 | 2.8±0.12 | 4.5±0.5 a | 16.5±6.6 a, b | <0.001a, b | HS | ||
| Sex | Male | 15 (50%) | 5 (6.3%) | <0.001* | HS | ||
| Female | 15 (50%) | 74 (93.7%) * | |||||
Data are expressed as mean±SD. P-value< 0.05 is considered statistically significant. HS: highly significant.
Significant difference versus the low disease activity group.
significant difference versus the moderate disease activity group.
Significant difference between males and females in the rheumatoid arthritis group.
Comparisons of inflammatory markers among the studied groups
Table 3 shows a highly significant increased level of ESR and RF in each group of rheumatoid patients (mild, moderate, and severe) as compared to the control group, but no significant difference is detected between the different grades of rheumatoid patients.
Table 3.
Inflammatory markers data among the studied groups.
| Control (N= 30) | Rheumatoid Patients N=79 | p-value | Sig. | |||
|---|---|---|---|---|---|---|
| Low Disease Activity(N= 24) | Moderate Disease Activity (N= 24) | High Disease Activity(N= 31) | ||||
| ESR (mm/hr) | 11±3.8 | 63±21.7 a | 63.8±21.12 a | 67.9±27.28 a | <0.001 | HS |
| CRP (mg/L) | 2.42±2.06 | 7.35±5.8 | 8.49±6.46 a | 9.9±8.05 a | <0.001 | HS |
| RF (IU/ml) | 10.32±3.4 | 119±90a | 143±71a | 149±81a | <0.001 | HS |
Data are expressed as mean±SD. P-value< 0.05 is considered statistically significant. HS: highly significant
significant difference versus the control group.
Regarding CRP, there are significantly increased levels in both the moderate and the severe disease activity groups compared to the control group, but no significant difference is found between the mild RA group and the control group (p-value= 0.19) or between the different grades of rheumatoid patients (p-value> 0.05).
MCP-1 levels among the studied groups
Figure 1 shows a highly significant increase in MCP-1 levels in all grades of RA in comparison with the control group. Furthermore, a highly significant difference is detected between the severe RA group as compared to both the mild and the moderate RA groups.
Fig. 1.
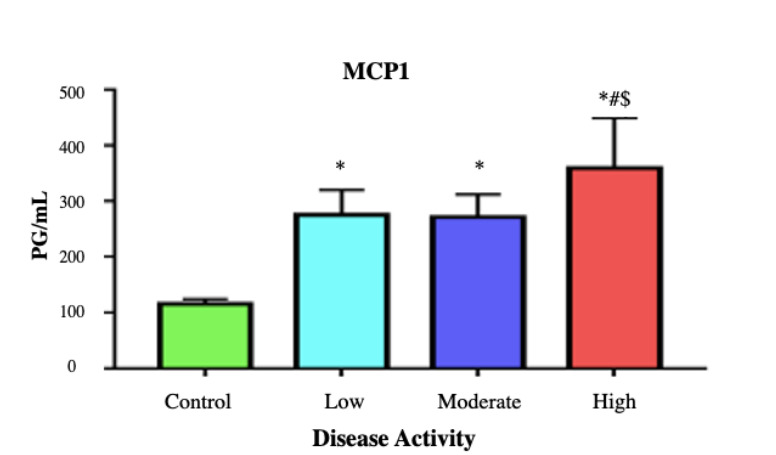
Mean±SD of MCP-1 in different studied groups. P-value< 0.001 is considered highly significant. * denotes highly significant difference versus the control group. # denotes highly significant difference versus the low disease activity group $ denotes highly significant difference versus the moderate disease activity group.
Gene expression of CircHIPK3 RNA and miRNA-124a in different studied groups
The results of CircHIPK3 RNA relative expression demonstrate a highly significant increase in all grades of RA groups compared to the control group (p< 0.001), with no significant difference among the different RA groups (p> 0.05) (Fig. 2).
Fig. 2.
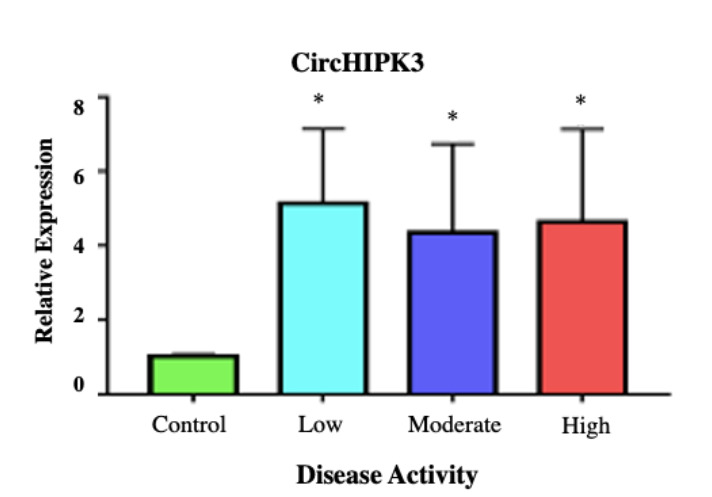
Mean±SD of the fold change of CircHIPK3 RNA expression among the studied groups. *denotes highly significant difference versus the control group (P-value< 0.001).
As for the miRNA-124a gene expression, Figure 3 shows a highly significant decrease in patients with low, moderate, and high disease activity compared to the control group (p< 0.001). In addition, a significant increase in miRNA-124a relative expression is detected in patients with moderate disease activity as compared to low disease activity group (p< 0.05). While there is no significant difference between patients with high grades of disease activity compared to both low and moderate groups (p-value> 0.05).
Fig. 3.
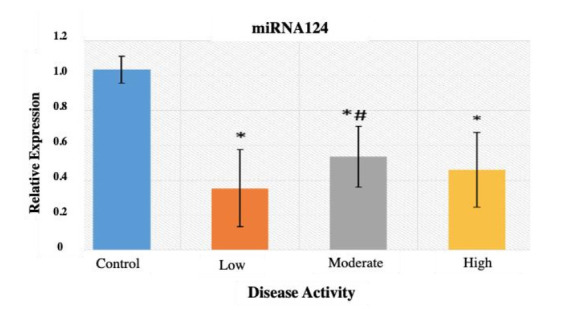
Mean±SD of the fold change of miRNA-124a expression among the studied groups. * denotes highly significant difference versus the control group (p-value< 0.001). # denotes significant difference versus the low disease activity group (p-value< 0.05).
Correlations between circHIPK3 RNA and miRNA-124a with the standard routine investigations in all studied groups
As shown in Figure 4, there is a highly significant negative correlation, with p-value< 0.001 and r= -0.767, between the circHIPK3 RNA and miRNA-124a gene expressions. On the other hand, there is a statistically significant positive correlation between circHIPK3 RNA and ESR levels (r= 0.257 & p-value= 0.022), which indicates that increased circHIPK3 RNA causes sequestration of miRNA-124a decreasing its expression, and this will be associated with increased levels of inflammatory biomarkers in RA. Moreover, there is a significant positive correlation between the RF level and the DAS28 in patients with RA (r= 0.297 & p-value= 0.008).
Fig. 4.
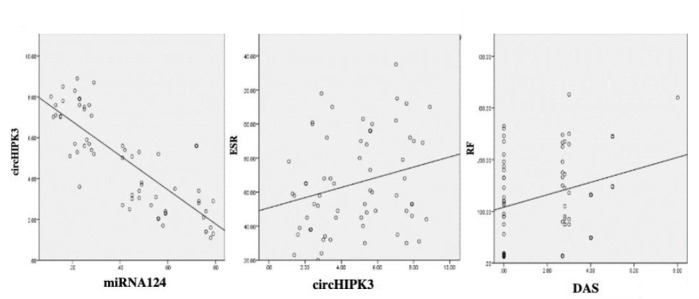
Correlations between circHIPK3 RNA and miRNA-124a with the standard routine investigations in all studied groups.
Discussion
By the virtue of targeting certain genes, a miRNA can have a variety of effects on the initiation and progression of various diseases (28). MiRNAs have the ability to control gene expression through epigenetic regulation of different genes implicated in autoimmune disorders (13). Multiple lines of evidence have revealed that circRNAs act as ubiquitous regulators of cellular and physiological processes and have been shown to be important in numerous skeletal and chondral disorders, such as RA and osteoarthritis (29, 30). Several scholars have emphasized the substantial impact of circRNAs on the regulation of gene transcription by acting as miRNA sponges, boosting the expression of downstream mRNAs (31). A comprehensive interpretation of the role of epigenetically regulated miRNAs and the miRNA-mediated epigenetic modulation of genes is critical for understanding how aberrant miRNA expression might contribute to RA, and how this knowledge can be translated into diagnostic and therapeutic tool development (32).
The current study clearly elucidated a highly significant upregulation of circHIPK3 RNA gene expression, with subsequent downregulation of miRNA-124a gene expression in all grades of RA groups compared to the control group. This interwoven relationship was evident by the strong negative correlation between circHIPK3 RNA and miRNA-124a gene expressions, emphasizing the sponging effect of circHIPK3 RNA that leads to sequestration of miRNA-124a. These findings are consistent with those of Wu et al., (33), who demonstrated that over-expression of circHIPK3 RNA in osteoarthritis cartilages could significantly promote osteoarthritis chondrocyte apoptosis by serving as a sponge for miR-124a, leading to increased expression of osteoarthritis-related genes, including the SOX8 gene, and thus accelerating osteoarthritis progression.
The redundancy of miRNA-124a's target binding sequences in the 3′-untranslated region of mRNA resulted in the regulation of a variety of components implicated in the pathogenesis of RA due to its unique characteristics. MiRNA-124a dysregulation has been shown to have a significant impact on MCP-1 mRNA, which is regarded as a critical downstream mRNA (19). In this context, our results indicated that downregulation of miRNA-124a was associated with significantly enhanced production of MCP-1 in all grades of RA as compared to the control group. Also, there was a significant difference in MCP-1 levels between the moderate RA group as compared to the mild and the severe RA groups, indicating that MCP-1 could be considered a marker of disease severity in RA patients.
Nakamachi and colleagues have previously found concordant results, suggesting that low levels of miR-124a expression during RA pathogenesis may have substantial impacts on synovial cell proliferation, leukocyte chemotaxis (MCP-1), and angiogenesis, thereby contributing to severe joint destruction. To emphasize their theory, they illustrated that co-transfection with pre-miR-124a resulted in miR-124a overexpression in RA FLS, with subsequent reduction of MCP-1 production by synoviocytes, thereby suppressing its chemotactic effects. These findings clearly imply that in RA synoviocytes, MCP-1 is a direct target of miR-124a (19).
The present study showed that enhanced secretion of MCP-1 was associated with increased disease severity, with DAS28 showing a highly significant difference between the severe and the moderate RA groups, and between the moderate and the mild RA groups as well. Furthermore, there was a highly significant increase in the inflammatory markers; ESR, CRP, and RF in all grades of RA groups compared to the control group, but no significant differences were detected between the different grades of RA. Our results go in accordance with a study performed by Abdel Fatah et al., (34), who discovered an association between blood MCP-1 levels and swollen joint count, indicating that plasma MCP-1, specifically the adapted DAS28-MCP-1, can be used to assess RA disease activity. Furthermore, they found a positive correlation between the plasma MCP-1 level and the ESR and CRP levels in RA patients.
To summarize, our research is the first, to the best of our knowledge, to assess the role of circHIPK3 RNA/miRNA-124a interaction in the pathogenesis of RA. CircHIPK3 was demonstrated to be significantly overexpressed in RA patients. Furthermore, circHIPK3 enhanced disease severity by inducing MCP-1 secretion, primarily by serving as a sponge for miRNA-124a. Further research is required to better understand how circHIPK3 and its signaling pathway might be exploited as a therapeutic target in various autoimmune diseases.
Acknowledgements
The authors extend their sincere acknowledgement to the Unit of Stem Cells and Molecular Biology, Cairo University, Egypt. All authors declare no conflict of interest.
References
- 1.He J, Wang Y, Feng M, Zhang X, Jin YB, Li X, et al. Dietary intake and risk of rheumatoid arthritis-a cross section multicenter study. Clin Rheumatol. 2016;35(12):2901–2908. doi: 10.1007/s10067-016-3383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castro-Villegas C, Pérez-Sánchez C, Escudero A, Filipescu I, Verdu M, Ruiz-Limón P, et al. Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFα. Arthritis Res Ther. 2015;17(1):49. doi: 10.1186/s13075-015-0555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh RP, Massachi I, Manickavel S, Singh S, Rao NP, Hasan S, et al. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12(12):1160–5. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Ansari-Moghaddam B, Asghar Kiani A, Sheikhian A, Birjandi M, Ahmadi SAY, Mousavi N, et al. Rheumatoid arthritis susceptibility is associated with the KIR2DS4-Full of killer-cell immunoglobulin-like receptor genes in the lur population of Iran. Rep Biochem Mol Biol. 2021;10(1):84–94. doi: 10.52547/rbmb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365(23):2205–19. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 6.Jie LG, Huang RY, Sun WF, Wei S, Chu YL, Huang QC, et al. Role of cysteine rich angiogenic inducer 61 in fibroblast like synovial cell proliferation and invasion in rheumatoid arthritis. Mol Med Rep. 2015;11(2):917–23. doi: 10.3892/mmr.2014.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang QC, Huang RY. The cyclooxygenase-2/thromboxane A2 pathway: a bridge from rheumatoid arthritis to lung cancer? Cancer Lett. 2014;354(1):28–32. doi: 10.1016/j.canlet.2014.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Huang RY, Huang QC, Burgering BM. Novel insight into the role of alpha-actinin-1 in rheumatoid arthritis. Discov Med. 2014;17:75–80. [PubMed] [Google Scholar]
- 9.Nakasa T, Nagata Y, Yamasaki Y, Ochi M. A mini-review: microRNA in arthritis. Physiol Genomics. 2011;43(10):566–70. doi: 10.1152/physiolgenomics.00142.2010. [DOI] [PubMed] [Google Scholar]
- 10.Tong X, Zeng H, Gu P, Wang K, Zhang H, Lin X. Monocyte chemoattractant protein 1 promotes the proliferation, migration and differentiation potential of fibroblast like synoviocytes via the PI3K/P38 cellular signaling pathway. Mol Med Rep. 2020;21(3):1623–1632. doi: 10.3892/mmr.2020.10969. [DOI] [PubMed] [Google Scholar]
- 11.Ospelt C, Gay S, Klein K. Epigenetics in the pathogenesis of RA. Semin Immunopathol. 2017;39(4):409–419. doi: 10.1007/s00281-017-0621-5. [DOI] [PubMed] [Google Scholar]
- 12.Chen JQ, Papp G, Szodoray P, Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016;15(12):1171–1180. doi: 10.1016/j.autrev.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Evangelatos G, Fragoulis GE, Koulouri V, Lambrou GI. MicroRNAs in rheumatoid arthritis: From pathogenesis to clinical impact. Autoimmun Rev. 2019;18(11):102391. doi: 10.1016/j.autrev.2019.102391. [DOI] [PubMed] [Google Scholar]
- 14.Shaker O, Mahfouz H, Salama A, Medhat E. Long Non-Coding HULC and miRNA-372 as Diagnostic Biomarkers in Hepatocellular Carcinoma. Rep Biochem Mol Biol. 2020;9(2):230–240. doi: 10.29252/rbmb.9.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner JD, Filer A. The role of the synovial fibroblast in rheumatoid arthritis pathogenesis. Curr Opin Rheumatol. 2015;27(2):175–82. doi: 10.1097/BOR.0000000000000148. [DOI] [PubMed] [Google Scholar]
- 16.de la Rica L, Urquiza JM, Gomez-Cabrero D, Islam AB, Lopez-Bigas N, Tegner J, et al. Identification of novel markers in rheumatoid arthritis through integrated analysis of DNA methylation and microRNA expression. J Autoimmun. 2013;41:6–16. doi: 10.1016/j.jaut.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Kawano S, Nakamachi Y. miR-124a as a key regulator of proliferation and MCP-1 secretion in synoviocytes from patients with rheumatoid arthritis. Ann Rheum Dis. 2011;70(Suppl 1):i88–91. doi: 10.1136/ard.2010.138669. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Dai L, Wu H, Zhang ZR, Wang WY, Fu J, et al. Novel anti-inflammatory target of geniposide: Inhibiting Itgbeta1/Ras-Erk1/2 signal pathway via the miRNA-124a in rheumatoid arthritis synovial fibroblasts. . Int Immunopharmacol. 2018;65(284):294. doi: 10.1016/j.intimp.2018.09.049. [DOI] [PubMed] [Google Scholar]
- 19.Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, et al., editors. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60(5):1294–304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Yu F, Wu W, Zhang Y, Chang W, Ponnusamy M, et al. Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity. Int J Biol Sci. 2017;13(12):1497–1506. doi: 10.7150/ijbs.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Rossum D, Verheijen BM, Pasterkamp RJ. Circular RNAs: Novel Regulators of Neuronal Development. Front Mol Neurosci. 2016;9:74. doi: 10.3389/fnmol.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng J, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16(1):216. doi: 10.1186/s12967-018-1582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Communications. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62(9):2569–81. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 26.Prevoo ML, van 't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL, editors. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38(1):44–8. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- 27.Chan YH. Biostatistics102: Quantitative Data – Parametric & Non-parametric Tests. Singapore Medical Journal. 2003;44(8):391–396. [PubMed] [Google Scholar]
- 28.Arif K, Elliott EK, Haupt LM, Griffiths LR. Regulatory Mechanisms of Epigenetic miRNA Relationships in Human Cancer and Potential as Therapeutic Targets. Cancers (Basel). 2020;12(10):2922. doi: 10.3390/cancers12102922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang X, Cen X, Zhang B, Liao Y, Zhu G, Liu J. Prospect of circular RNA in osteogenesis: A novel orchestrator of signaling pathways. J Cell Physiol. 2019;234(12):21450–21459. doi: 10.1002/jcp.28866. [DOI] [PubMed] [Google Scholar]
- 30.Li HZ, Lin Z, Xu XH, Lin N, Lu HD. The potential roles of circRNAs in osteoarthritis: a coming journey to find a treasure. Biosci Rep. 2018;38(5):BSR20180542. doi: 10.1042/BSR20180542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panda AC, editor. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67–79. doi: 10.1007/978-981-13-1426-1_6. [DOI] [PubMed] [Google Scholar]
- 32.Chen G, Tang W, Wang S, Long C, He X, Yang D, et al., editors. Promising diagnostic and therapeutic circRNAs for skeletal and chondral disorders. Int J Biol Sci. 2021;17(5):1428–1439. doi: 10.7150/ijbs.57887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Q, Yuan ZH, Ma XB, Tang XH. Low expression of CircRNA HIPK3 promotes osteoarthritis chondrocyte apoptosis by serving as a sponge of miR-124 to regulate SOX8. Eur Rev Med Pharmacol Sci. 2020;24(15):7937–7945. doi: 10.26355/eurrev_202008_22476. [DOI] [PubMed] [Google Scholar]
- 34.Abdel Fatah W, Atef L, Mohamed A, Refaat D, Mohsen M, Abd El-Raof M, et al. Study of Serum Monocyte Chemoattractant Protein-1AS A Marker in Rheumatoid Arthritis. The Egyptian Journal of Hospital Medicine. 2014;56(1):321–332. [Google Scholar]


