Abstract
Background
Estimating the cumulative incidence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is essential for setting public health policies. We leveraged deidentified Massachusetts newborn screening specimens as an accessible, retrospective source of maternal antibodies for estimating statewide seroprevalence in a nontest-seeking population.
Methods
We analyzed 72 117 newborn specimens collected from November 2019 through December 2020, representing 337 towns and cities across Massachusetts. Seroprevalence was estimated for the Massachusetts population after correcting for imperfect test specificity and nonrepresentative sampling using Bayesian multilevel regression and poststratification.
Results
Statewide seroprevalence was estimated to be 0.03% (90% credible interval [CI], 0.00–0.11) in November 2019 and rose to 1.47% (90% CI: 1.00–2.13) by May 2020, following sustained SARS-CoV-2 transmission in the spring. Seroprevalence plateaued from May onward, reaching 2.15% (90% CI: 1.56–2.98) in December 2020. Seroprevalence varied substantially by community and was particularly associated with community percent non-Hispanic Black (β = .024; 90% CI: 0.004–0.044); i.e., a 10% increase in community percent non-Hispanic Black was associated with 27% higher odds of seropositivity. Seroprevalence estimates had good concordance with reported case counts and wastewater surveillance for most of 2020, prior to the resurgence of transmission in winter.
Conclusions
Cumulative incidence of SARS-CoV-2 protective antibody in Massachusetts was low as of December 2020, indicating that a substantial fraction of the population was still susceptible. Maternal seroprevalence data from newborn screening can inform longitudinal trends and identify cities and towns at highest risk, particularly in settings where widespread diagnostic testing is unavailable.
Keywords: newborn screening, SARS-CoV-2, seroprevalence
Measurement of maternal antibodies in specimens collected for newborn screening offers a statewide source of seroprevalence data independent of case testing. We analyzed 72 117 Massachusetts specimens collected from November 2019–December 2020 during the COVID-19 pandemic and estimated longitudinal trends.
Disease surveillance networks operate at multiple levels of government and healthcare, yet monitoring for outbreaks of new diseases remains a challenge. There are few material sources of information with limited cohort bias available to address questions about the frequency and distribution of a new infectious agent within populations. States’ newborn screening (NBS) programs are such a resource; they collect and test infant dried blood spot (DBS) specimens at centralized clinical laboratories for markers of an ever-expanding panel of treatable disorders to inform timely care. Such programs comprise high-throughput laboratory testing with community outreach, operate under state authority, and maintain secure electronic and specimen records. Importantly, NBS-DBS infant specimens contain maternal immunoglobulin G (IgG) antibodies that cross the placenta and reflect maternal exposure to infectious agents. The use of NBS to measure the seroprevalence of an emerging infectious disease was pioneered in the late 1980s for human immunodeficiency virus [1] and adopted by a majority of states [2].
This framework offers advantages for monitoring infections by new agents, such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in the absence of symptomatic disease, particularly when diagnostic testing is limited in the initial stages of the pandemic, leading to uncertainty about the true infection burden. Many critical questions about SARS-CoV-2 infection are important to answer in order to inform public health responses to the pandemic: What proportion of our population has been exposed? How is exposure geographically distributed? Are the rates of exposure increasing in some populations faster than in others? What are the characteristics of those who become infected?
Using data generated from NBS specimens, we report findings that address these questions from a survey of SARS-CoV-2 seroprevalence in childbearing women in Massachusetts.
METHODS
Study Population
Women who were residents of Massachusetts, gave birth in Massachusetts, and whose infants’ DBS specimens had been through routine newborn screening met study surveillance inclusion criteria, with the DBS specimens serving as surrogates for the women. A control group was predefined to be those DBS arriving at the newborn screening program in March 2019, and the study group was defined as those DBS arriving from 4 November 2019 through 31 December 2020. The following inclusion–exclusion criteria were applied: only 1 DBS specimen per infant and only 1 DBS specimen from each multiples’ birth was included. All specimens of infants aged >30 days at the time specimens were collected, all DBS of infants transfused within 48 hours, all specimens determined to be of “unsatisfactory” quality for NBS, and all specimens from mothers who opted out of providing data for research were excluded.
The Massachusetts Department of Public Health and the UMass Chan Medical School institutional review boards approved waivers of consent for the deidentified public health surveillance.
Antibody Testing Optimization
Human monoclonal IgG antibody cross reactive to SARS-CoV-2 was prepared from CR3022 variable genes expressed from plasmids, as described previously for other SARS-CoV-2 antibodies [3]. The receptor-binding domain (RBD) of the spike protein used as enzyme-linked immunosorbent assay (ELISA) antigen was expressed using HEK-293F cells and prepared as follows: a plasmid containing the His-tagged RBD was transiently transfected in HEK-293F cells using polyethylenimine (PEI). The expressed protein was extracted and purified with nickel-nitrilotriacetic acid (Ni-NTA) resin and stored at –80°C until use. IgG antibodies specific to the RBD protein were detected in eluates from DBS residual to the Massachusetts newborn screening program by an adaptation of a previously described ELISA designed for serum [4]. Details of the protocol can be found in the Supplementary Methods.
Cutoff Determination
To determine positive and negative interpretations, we used a plate-specific cutoff calculation using standard deviations (SDs) above the mean of the replicates’ average IgG concentration in a 2-step process. In the first step, the mean and 2 SDs were calculated for each plate. In the second step, any specimens with a concentration ≥2 SDs above the plate mean from the first step were excluded from the calculation of the second mean and SD (mean and SD of presumed negative samples). For laboratory 1 (384-well assays), any specimen with a microgram per milliliter result that was 5.3 SDs above the second cycle mean was interpreted as positive. For laboratories 2 and 3 (96-well assays), any specimen with a microgram per milliliter result that was 3.3 SDs above the second cycle mean was interpreted as positive.
Inhibition Assay
The inhibition assay was performed in the same manner as the standard IgG antibody analysis, with the following exceptions. Concentrated RBD diluted in dilution buffer was premixed with DBS eluates so that the final dilution of the DBS eluates was identical to that used in the standard assay (1:4) and the final dilution of RBD was 1 µg/mL. This mixture was incubated for 30 minutes at room temperature, and 100 µL of the inhibited mixture was added to ELISA wells in duplicate. Uninhibited wells were also tested in duplicate, replacing the concentrated RBD with diluent alone. Percent inhibition was expressed as 100 × (average OD of inhibited wells)/(average optical density of uninhibited wells).
Data Processing
Each of 3 testing laboratories determined that test results met quality control requirements. Testing laboratories reported replicate OD and IgG concentration results for all well locations from specified plates to investigators at the New England Newborn Screening Program (NENSP). Transient linkages between plated specimens, test results, and demographic data were created and available to only 2 investigators at the NENSP (A. M .C. and J. E. H.) for application of inclusion–exclusion criteria followed by calculations for interpretation of results in a restricted database. A query of the restricted database yielded a deidentified coded specimen dataset, with the limited data variables reported here. All transient linkages to any identifiers will be irretrievably destroyed upon acceptance for publication.
Population-wide Statistical Model
We used dynamic (ie, time-varying) Bayesian multilevel regression and poststratification (MRP) models to adjust seroprevalence estimates for nonrepresentative sampling by age, sex, and Massachusetts community of residence (ie, n = 351 cities or towns) and to adjust for imperfect test specificity [5–9]. The core approach of MRP is to estimate the outcome variable conditional on sociodemographic characteristics using a nonrepresentative dataset and then project these estimates onto a representative population of interest [5, 7–11]. MRP models additionally enable estimation of longitudinal seroprevalence trends at granular geographic levels, such as communities within Massachusetts. Estimation consists of 2 steps: first, we fit a time-varying Bayesian multilevel logistic regression model to predict serostatus conditional on age category and community random effects [10]. We used 2 types of time-varying functions: we primarily assessed estimates from a model grouping data by month, but we also fit a generalized additive model that modeled time continuously [8]. We then poststratified the seroprevalence estimates over time using census data to individual communities and to the statewide population. In the monthly model, we also accounted for imperfect test specificity by allowing for measurement error when collecting data from true seronegatives [9]. Additional model details can be found in the Supplementary Methods.
To identify community-level factors associated with seropositivity, we fit a logistic regression model pooling data from the last 4 months (September 2020 through December 2020, corresponding to the height of seropositivity). We included fixed effects for multiple demographic factors from the 2015–2019 five-year American Census Survey (Supplementary Table 2) [12] and random effects for age and community. Details for all model-fitting parameters and additional data sources are in the Supplementary Methods. The code used to fit the models is available on Github: https://github.com/gradlab/covid19-newborn-seroprevalence [13].
RESULTS
Data related to neonatal specimens representing all Massachusetts women who gave birth from November 2019 through December 2020 are shown in Table 1. The 72 117 women who met study criteria resided in 337 towns and cities across Massachusetts. The 1817 presumed seronegative (March 2019) specimens were also from across the state (324 towns and cities).
Table 1.
Characteristics of Newborn Screening Dried Blood Spot Specimens Included in the Survey
| Characteristic | November 2019–December 2020 | March 2019 (Presumed Seronegative) | Totals |
|---|---|---|---|
| Total specimens punched | 93 660 | 2372 | 96 105 a |
| Types of specimens not meeting inclusion criteriab | |||
| Specimens not collected for typical NBS purposesc | 3126 | 76 | 3202 |
| Specimens declared unsatisfactory for any NBS assayd | 2304 | 79 | 2383 |
| Specimens from infants transfused within 48 hours of collection | 98 | 3 | 101 |
| Specimens collected from nonneonates (aged ≥30 days) | 2685 | 56 | 2741 |
| Specimens from all but first neonate from a multiples birth | 1967 | 57 | 2024 |
| Specimens of neonates whose mother is not Massachusetts resident | 2521 | 56 | 2577 |
| Specimens that are “repeat” NBS specimens | 8116 | 219 | 8335 |
| Research exclusion requested by parent | 0 | 0 | 0 |
| Total specimens not meeting inclusion criteria | 20 817 (22%) | 546 (23%) | 21 363 (22%) |
| Punched specimens meeting inclusion criteria | 72 843 | 1826 | 74 669 |
| Specimens not meeting technical quality data | 726 (0.99%) | 9 (0.49%) | 735 (0.98%) |
| Specimens linked to quality-controlled results | 72 117 | 1817 | 73 934 |
Abbreviation: NBS, newborn screening.
Includes 73 punched specimens that were not linkable to the newborn database.
List is progressive, and specimens are only counted in each category once.
For example, collected from patient’s parent or sibling, collected from patient to monitor treatment.
Unsatisfactory includes conventional quality control issues: poor soak, scratched/abraded, improperly dried, layered/clotted, contaminated/diluted, quantity not sufficient (QNS), no blood, no demographics. Unbolded numbers are unsatisfactory samples; bolded numbers highlight the number of specimens punched, meeting demographic inclusion criteria, and finally those linked to results.
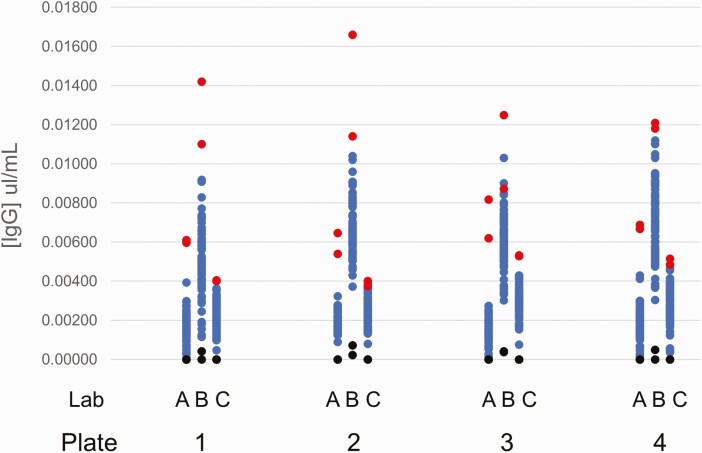
First, we measured seropositive rates from 1817 DBS, assuming that their collection significantly predated the coronavirus disease 2019 (COVID-19) pandemic. Each of the 3 testing laboratories received punches from the same 288 specimens punched across 4 plates. Figure 1 shows the individual [IgG] results of the anonymized specimens, the positive controls, and diluent controls by plate and by testing laboratory. The low positive controls showed IgG concentrations significantly above the mean of the tested specimens and all above the values for the tested specimens.
Figure 1.
IgG results of 288 anonymized specimens obtained in March 2019 tested by each of the 3 laboratories across 4 plates. Blue dots (dark gray in grayscale) are residual newborn screening dried blood spot specimens, red dots (light gray in grayscale) are positive controls, and black dots are diluent controls. Abbreviation: IgG, immunoglobulin G.
Among the 1817 presumed seronegative specimens from March 2019, 7 (0.39%) were seropositive for SARS-CoV-2. Among the 72 117 study specimens, 1261 (1.75% statewide) were seropositive; 45 seropositives were from November 2019 and December 2019. We investigated the likelihood that these early seropositive values were false positives using an inhibition assay. For comparison, of the 29 assayed seropositive specimens from July 2020, the average inhibition was 57% (95% confidence interval, 48–66; minimum, 22%). Of the available 45 specimens from November 2019 and December 2019, the average inhibition was 1.45% (95% confidence interval, 1.34–1.56), confirming that most seropositives from 2019 were false positives. Only 3 of the 45 specimens from 2019 showed notable inhibition (18%, 27%, and 36%). The difference in the positive predictive values between late 2019 and July 2020 arises because the “cutoff” to define a positive interpretation was statistically defined (using standard deviations) by results of negative specimens on a plate-by-plate basis (the 2-cycle algorithm attempts to remove potential positives when defining the final SD). This algorithm is expected (and indeed must by its design) define the highest values as “positives” even in a population where there are no true positives (eg, as in Figure 1).
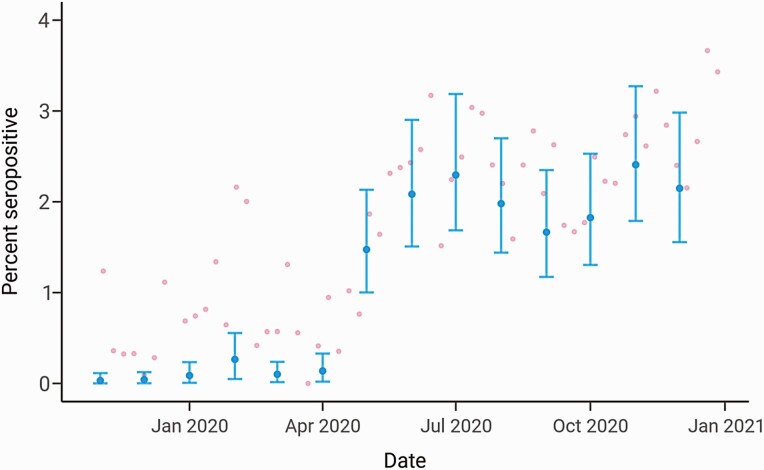
We projected the NBS seroprevalence results statewide and for Massachusetts cities and towns using MRP and additionally accounted for imperfect test specificity. MRP enables estimation of seroprevalence at granular geographic levels and addresses nonrepresentative sampling, which is a challenge for the NBS cohort since there are sampling biases by sex, age group, and geography relative to the overall Massachusetts population (Supplementary Figures 1 and 2). Statewide monthly seroprevalence in early November 2019 was estimated to be 0.03% with a 90% credible interval (CI) of 0.00%–0.11% and remained low until May 2020 (Figure 2). In May 2020, seroprevalence rose to 1.47% (90% CI: 1.00–2.13) following sustained SARS-CoV-2 transmission in the spring and plateaued at approximately 2% from July onward, reflecting decreased transmission in the summer months due to lockdowns, mask-wearing, and other factors [14, 15]. The estimate for seroprevalence for December 2020 was 2.15% (90% CI: 1.56–2.98). Statewide trends estimated from the continuous-time MRP model showed similar qualitative results (Supplementary Figure 3).
Figure 2.
Statewide longitudinal seroprevalence trend from November 2019 to December 2020 estimated using the monthly multilevel regression and poststratification model adjusting for test specificity. The mean seroprevalence estimates are indicated by the blue dots (dark gray in grayscale) with the error bars depicting 90% credible intervals; pink dots (light gray in grayscale) represent unadjusted weekly seroprevalence estimates.
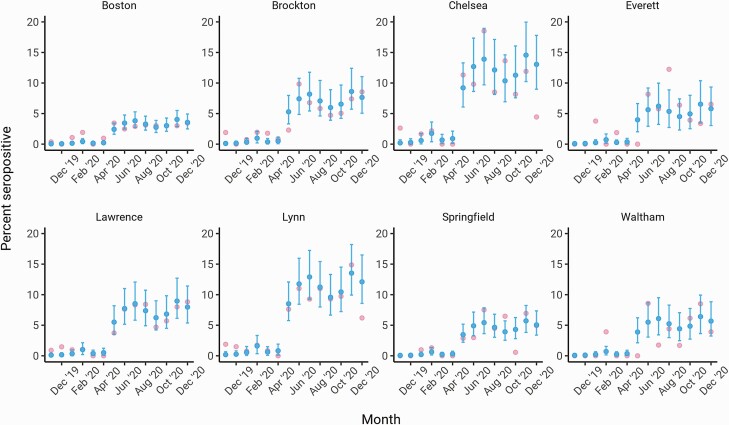
Next, we estimated longitudinal seroprevalence trends in Massachusetts cities and towns to identify geographic heterogeneities in SARS-CoV-2 infection risk. Overall seropositivity varied considerably across the state (Figure 3, Supplementary Figure 4), but cities with high seroprevalence showed similar qualitative trajectories to each other and to overall statewide trends (ie, increases from April 2020 to May 2020 followed by plateaus; Figure 3). The widths of the credible intervals also varied, reflecting uncertainty due to smaller sample sizes for some cities, such as Chelsea, compared with larger cities, such as Boston. The estimate for seroprevalence in Boston was 0.05% (90% CI: 0.00–0.17) in November 2019 and rose to 3.56% (90% CI: 2.49–4.93) at the end of 2020, which is slightly higher than the estimate for the state. Modeling time continuously yielded similar qualitative trends (Supplementary Figure 5).
Figure 3.
Longitudinal seroprevalence trends in cities and towns from November 2019 to December 2020 estimated using the monthly multilevel regression and poststratification model adjusting for test specificity. The 8 cities and towns with the highest lower 90% credible interval in December 2020 and at least 20 heel stick samples collected are shown. The mean seroprevalence estimates are indicated by the blue dots (dark gray in grayscale) with the error bars depicting the 90% credible intervals; pink dots (light gray in grayscale) represent unadjusted monthly seroprevalence estimates.
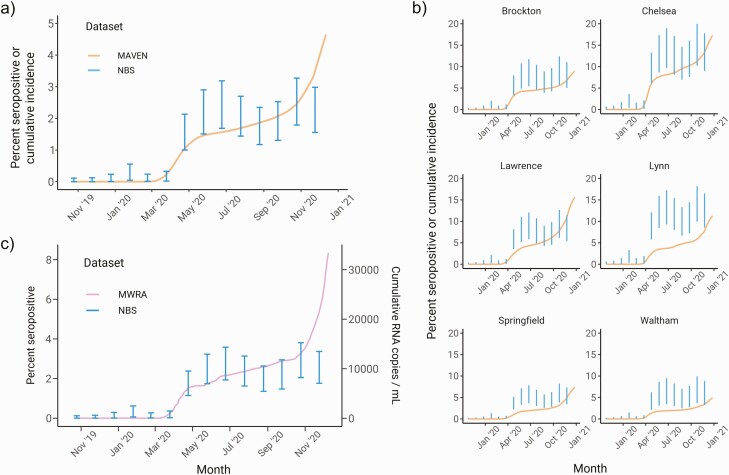
We compared the seroprevalence estimates to active case surveillance data collected by the Massachusetts Department of Public Health and wastewater surveillance conducted by the Massachusetts Water Resource Authority (MWRA) and Biobot [16, 17]. Relative to these data, which capture infected individuals shedding virus, seroprevalence levels are a lagging indicator of infection trends because of the delay from infection to antibody production. To account for this, we overlaid the statewide seroprevalence estimates with the Massachusetts Virtual Epidemiologic Network (MAVEN) and MWRA data using a 3-week lag (Figure 4). We observed good qualitative concordance between seroprevalence levels and MAVEN cumulative incidence trajectories from 2019 into 2020 but a divergence between the curves at the end of 2020. Seropositivity in the NBS cohort did not rise as sharply as cases did during the winter resurgence in transmission, both statewide (Figure 4A) and at the level of cities and towns (Figure 4B). This difference was also observed when NBS seroprevalence levels were compared with wastewater surveillance data (Figure 4C) and other serosurveys (Supplementary Table 3), indicating a deviation between the DBS data and multiple other sources of SARS-CoV-2 surveillance toward the end of the sampling timeline.
Figure 4.
Comparison of multilevel regression and poststratification seroprevalence estimates with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) surveillance data from reported cases and wastewater testing. a, Statewide longitudinal seroprevalence trend estimated from newborn screening samples (vertical bars) overlaid with cumulative incidence from MAVEN epidemiological surveillance data (smoothed line). b, Seroprevalence trends for 6 cities and towns (blue) overlaid with MAVEN cumulative incidence (smoothed line). c, Seroprevalence trend for the northern MWRA region (vertical bar) vs cumulative RNA copies per milliliter from wastewater SARS-CoV-2 testing for the same region (smoothed line). In all subpanels, the blue error bars depict the 90% credible interval for seroprevalence, and seroprevalence data are shifted backward by 3 weeks to match the timing of surveillance data. Abbreviations: MAVEN, Massachusetts Virtual Epidemiologic Network; MWRA, Massachusetts Water Resource Authority; NBS, newborn screening.
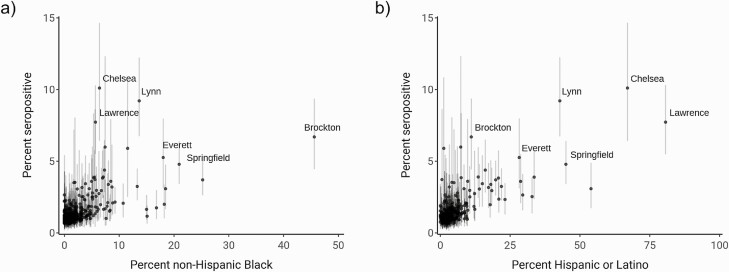
Finally, we sought to understand community-level factors associated with increased seropositivity by fitting a multivariate model that included 14 sociodemographic variables using data from the last 4 months of 2020. These variables spanned population size and density, race and ethnicity, housing, education, and socioeconomic status (Supplementary Table 2). We calculated 90% credible intervals, representing the most plausible range of values, for the 14 estimated coefficients of association between each variable and seropositivity. The percent of the community that was non-Hispanic Black was the only variable with a 90% credible interval that excluded 0 (Table 2). The mean of the posterior distribution for this variable was 0.024, indicating that a 10% increase in the percent of a community that was non-Hispanic Black was associated with 27% higher odds of seropositivity, adjusting for all other included variables. The association of percent non-Hispanic Black and percent Hispanic or Latino with increased seropositivity was driven, in part, by several cities and towns, including Brockton, Springfield, Everett, Chelsea, Lynn, and Lawrence (Figure 5). We also calculated the probability of a coefficient being greater or less than 0 for the 14 coefficients. Percent non-Hispanic Black and percent without insurance coverage had a 90% or higher probability of being greater than 0, whereas average household size, median age, and population density had a 90% or higher probability of being less than 0. These results suggest several additional sociodemographic variables that warrant further study to characterize association with seropositivity (Supplementary Figure 6).
Table 2.
Posterior Distribution Summaries for the Community-level Multivariate Association of Demographic Characteristics With Seroprevalence
| Demographic Variable | Mean | Standard Deviation | 5% | 95% | P(X > 0) | P(X < 0) |
|---|---|---|---|---|---|---|
| Percent non-Hispanic Blackb,a | 0.0238 | 0.0119 | 0.00405 | 0.0436 | 0.97 | 0.03 |
| Percent Asian | –0.00417 | 0.0167 | –0.0318 | 0.0233 | 0.4 | 0.6 |
| Average household sizea | –0.613 | 0.451 | –1.34 | 0.14 | 0.08 | 0.92 |
| Percent living in a house with more than 1 occupant per room | 0.246 | 0.217 | –0.119 | 0.585 | 0.87 | 0.13 |
| Percent born in a foreign country | 0.00932 | 0.0169 | –0.0175 | 0.037 | 0.71 | 0.29 |
| Percent Hispanic or Latino | 0.0111 | 0.00926 | –0.00413 | 0.0261 | 0.89 | 0.11 |
| Percent with a high school or higher diploma | –0.0173 | 0.0246 | –0.0567 | 0.024 | 0.23 | 0.77 |
| Percent male | 0.0301 | 0.042 | –0.041 | 0.0947 | 0.76 | 0.24 |
| Median agea | –0.0292 | 0.0223 | –0.0653 | 0.00879 | 0.1 | 0.9 |
| Median income | –8.60E-06 | 1.50E-05 | –3.37E-05 | 1.63E-05 | 0.28 | 0.72 |
| Population densitya | –3.85E-05 | 2.57E-05 | –7.89E-05 | 4.18E-06 | 0.07 | 0.93 |
| Percent under poverty threshold | –0.0298 | 0.0332 | –0.0831 | 0.0256 | 0.19 | 0.81 |
| Total population | 2.26E-07 | 6.57E-07 | –7.68E-07 | 1.36E-06 | 0.63 | 0.37 |
| Percent without insurance coveragea | 0.122 | 0.0761 | –0.00151 | 0.245 | 0.95 | 0.05 |
The
indicates either P(X >0) or P(X <0) is 90% or higher, and the
indicates that the 90% credible interval excludes 0.
Figure 5.
Comparison of percent non-Hispanic Black (left) and percent Hispanic or Latino (right) with estimated seroprevalence by community. Dots indicate mean of the posterior seroprevalence distribution, and shaded regions indicate 90% credible intervals. Communities with a lower 90% credible interval above 3% seropositivity are labeled.
DISCUSSION
Understanding the cumulative incidence of SARS-CoV-2 is essential for setting public health policies and guiding vaccine rollouts, but the cumulative incidence for states and their cities and towns is still largely unknown because of inadequate testing and a lack of representative serosurveys. In Massachusetts, we leveraged the knowledge base and repository of our NBS program for the generation of a readily accessible, retrospective, and deidentifiable source of maternal antibodies that can inform SARS-CoV-2 seroprevalence. The strengths of this approach are multifaceted. The study cohort is free of biases arising from test-seeking behaviors, test availability, and symptom presence; childbearing women were included in the study because they had given birth, not because they were thought to have been exposed to SARS-CoV-2 [18]. The specimens and demographic data are retrospectively available for public health purposes. The key idea of our study is that by using DBS as the primary data source, we can estimate longitudinal trends in seroprevalence in childbearing women and then project these results to the population at large using statistical methods.
We analyzed data from 72 117 DBS collected across Massachusetts from November 2019 through December 2020 and estimated seroprevalence using MRP, which is a Bayesian statistical method used for generalizing survey results and has been increasingly applied to epidemiological studies of SARS-CoV-2 [6, 8, 9, 19]. We found that longitudinal trends statewide and for selected cities and towns were qualitatively similar, with a rise in seropositivity in April 2020 followed by a plateau. The estimated seroprevalence levels had good concordance with cumulative incidence estimated by MAVEN case [16] data, MWRA wastewater surveillance data [17], and other serological surveys (Supplementary Table 3) until the observed resurgence of transmission toward the end of 2020. While sampling into 2021 will be necessary to fully understand the extent to which these data sources diverge, one key contributing factor could be changing behaviors of pregnant women over time. We also found evidence that community percent non-Hispanic Black was the sociodemographic variable most associated with increased seroprevalence levels, in line with other epidemiological studies [20–23]. These results underscore the importance of continuing equity and outreach initiatives for minority communities that were most affected by the initial epidemic wave of SARS-CoV-2.
Our findings are subject to several limitations and biases. As noted, selection bias could arise because fundamental risk differences between pregnant women and the general population are not accounted for in our statistical model. The direction and causes of this potential bias vary. Pregnant women could have less exposure to SARS-CoV-2 due to behavioral choices or sociodemographic characteristics but increased biological susceptibility due to immune weakening. Nonetheless, combined estimates from cohorts with clear selection biases, such as blood donors or healthy volunteers, can still meaningfully inform seroprevalence estimates in the general population [18, 24–26]. Second, misclassification bias can occur due to imperfect test specificity and sensitivity. We estimated specificity using an early prepandemic (March 2019) sample of DBS and incorporated it into the statistical model. We do not have a good estimate for test sensitivity and have not accounted analytically for waning of sensitivity over time. Accounting for imperfect test sensitivity would be expected to shift the seroprevalence estimates higher and widen the credible intervals (Supplementary Figure 7) due, in part, to the uncertainty involved in measuring sensitivity itself.
By assessing the distribution of maternal antibodies to SARS-CoV-2 statewide and over time, our study provides a strategy for the systematic evaluation and estimation of population-wide cumulative incidence of SARS-CoV-2. Our study was conducted prior to emergency use authorization of COVID-19 vaccines. Conduct of seroprevalence studies following widespread use of vaccines would require modification of the assay to detect IgG antibodies specific to gene products (eg, nucleocapsid protein) not expressed in the vaccine. Our approach, that is, leveraging an easily stored and often readily available data source, may be most useful for informing cumulative incidence estimates in areas where widespread infection testing is still unavailable or remains heavily biased [18]. Prospective use of NBS-based estimates of exposure for ongoing policy development would require the more typical same-day turnaround time of clinically based NBS programs.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We are grateful to Lisa Cavacini, PhD, Mass Biologics, UMass Chan Medical School, for providing the CR3022 monoclonal antibody and Kuang Shen, PhD, Program in Molecular Medicine, T.H. Chan School of Medicine, UMass Chan Medical School, for providing receptor-binding domain protein. We are grateful to Megan Hatch, Lori Chou, Shauna Onofrey, Melody Rush, Arthur Benjamin, Deborah Britton, and the newborn heel stick specimen processing team for laboratory and analytical support throughout the project. Finally, we thank our dear friend and mentor, Dr. George F. Grady, for his encouragement on this project.
Disclaimer. The findings, conclusions, and views expressed in this presentation are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC).
Financial support. This work was supported by the National Science Foundation Graduate Research Fellowship Program (GRFP) (DGE1745303 to K. C. M.), the National Cancer Institute Seronet Program (1U01CA261277-01 to Y. H. G.), the Morris-Singer Foundation to Y. H. G., the United States Food and Drug Administration (HHSF223201810172C to S. M. S.), the UMass Center for Clinical and Translational Science (National Institutes of Health [NIH] UL1TR001453 to K. L.), and the Centers for Disease Control and Prevention (CDC) (contract 200-2016-91779).
Potential conflicts of interest. G. A. reports support for the present manuscript from the National Institutes of Health (NIH; 3R37AI080289-11S1, R01AI146785, U19AI42790-01, U19AI135995-02, U19AI42790-01, 1U01CA260476-01, CIVIC75N93019C00052), Gates Foundation, and Musk Foundation; funding outside of the submitted work from the NIH, Gates Foundation, Musk Foundation, Ragon Institute, and Massachusetts General Hospital (MGH) Scholars Program; consulting fees from Sanofi Pasteur; payment or honoraria for lectures from Princeton, George Washington University, Johns Hopkins University, Washington University, University of Washington, Yale University, Walter Reed Medical Institute, Upstate New York, and Mount Sinai; patents for direct expression of antibodies (Novartis, PAT056060-EP-EPA), antigen-specific antibody glycosylation as a diagnostic marker of disease state (MGH21944), system and method for the multiplexed affinity purification of proteins and cells (Massachusetts Institute of Technology 16927), and a rapid Fc-enhancing screening approach (MGH in preparation, 2021; pending); served as a data and safety monitoring board (DSMB) member for the Caprisa monoclonal therapeutic trial from the University of Kwazulu Natal, South Africa; and has stock options from Systems Seromyx and Leyden Labs. C. M. B. reports grants paid to their institution to support all public health work from the CDC outside of the submitted work. K. L. reports grants or contracts to UMass Chan from NIH, Penta Foundation/Early-treated Perinatally HIV-infected Individuals: Improving Children's Actual Life with Novel Immunotherapeutic Strategies (EPIICAL), and Moderna Therapeutics (clinical trials); consulting fees from Gilead; participation on a DSMB for the Tatelo study (NIH-funded; received no compensation); participation on Duke Center for AIDS Research (CFAR) (NIH) and Boston University Clinical and Translational Institute (CTSI) (NIH); and no compensation for serving on the Management Sciences for Health Board of Directors. Y. G. reports a grant from Pfizer for unrelated research and consulting fees from GlaxoSmithKline (GSK) and Quidel for unrelated topics. All remaining authors: No reported conflicts of interest. All authors have submitted the International Committee of Medical Journal Editors (ICMJE) Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Kevin C Ma, Department of Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Jaime E Hale, New England Newborn Screening Program, UMass Chan Medical School, Worcester, Massachusetts, USA.
Yonatan H Grad, Department of Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Galit Alter, Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard University, Cambridge, Massachusetts, USA.
Katherine Luzuriaga, Program in Molecular Medicine, T.H. Chan School of Medicine at UMass Chan Medical School, Worcester, Massachusetts, USA; UMass Center for Clinical and Translational Science at UMass Chan Medical School, Worcester, Massachusetts, USA.
Roger B Eaton, New England Newborn Screening Program, UMass Chan Medical School, Worcester, Massachusetts, USA; Department of Pediatrics, T.H. Chan School of Medicine at UMass Chan Medical School, Worcester, Massachusetts, USA.
Stephanie Fischinger, Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard University, Cambridge, Massachusetts, USA.
Devinder Kaur, New England Newborn Screening Program, UMass Chan Medical School, Worcester, Massachusetts, USA; Department of Pediatrics, T.H. Chan School of Medicine at UMass Chan Medical School, Worcester, Massachusetts, USA.
Robin Brody, Program in Molecular Medicine, T.H. Chan School of Medicine at UMass Chan Medical School, Worcester, Massachusetts, USA.
Sameed M Siddiqui, Computational and Systems Biology Program, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA; Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Dylan Leach, Bureau of Infectious Disease and Laboratory Sciences, Massachusetts Department of Public Health, Boston, Massachusetts, USA.
Catherine M Brown, Bureau of Infectious Disease and Laboratory Sciences, Massachusetts Department of Public Health, Boston, Massachusetts, USA.
R Monina Klevens, Bureau of Infectious Disease and Laboratory Sciences, Massachusetts Department of Public Health, Boston, Massachusetts, USA.
Lawrence Madoff, Bureau of Infectious Disease and Laboratory Sciences, Massachusetts Department of Public Health, Boston, Massachusetts, USA.
Anne Marie Comeau, New England Newborn Screening Program, UMass Chan Medical School, Worcester, Massachusetts, USA; Department of Pediatrics, T.H. Chan School of Medicine at UMass Chan Medical School, Worcester, Massachusetts, USA.
References
- 1. Hoff R, Berardi VP, Weiblen BJ, Mahoney-Trout L, Mitchell ML, Grady GF.. Seroprevalence of human immunodeficiency virus among childbearing women. Estimation by testing samples of blood from newborns. N Engl J Med 1988; 318:525–30. [DOI] [PubMed] [Google Scholar]
- 2. Gwinn M, Pappaioanou M, George JR, et al. . Prevalence of HIV infection in childbearing women in the United States. Surveillance using newborn blood samples. JAMA 1991; 265:1704–8. [PubMed] [Google Scholar]
- 3. Ejemel M, Li Q, Hou S, et al. . A cross-reactive human IgA monoclonal antibody blocks SARS-CoV-2 spike-ACE2 interaction. Nat Commun 2020; 11:4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roy V, Fischinger S, Atyeo C, et al. . SARS-CoV-2-specific ELISA development. J Immunol Methods 2020; 484:112832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park DK, Gelman A, Bafumi J.. Bayesian multilevel estimation with poststratification: state-level estimates from national polls. Polit Anal 2004; 12:375–85. [Google Scholar]
- 6. Uyoga S, Adetifa IMO, Karanja HK, et al. . Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Kenyan blood donors. Science 2021; 371:79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang W, Rothschild D, Goel S, Gelman A.. Forecasting elections with non-representative polls. Int J Forecast 2015; 31:980–91. [Google Scholar]
- 8. Pouwels KB, House T, Pritchard E, et al. . Community prevalence of SARS-CoV-2 in England from April to November, 2020: results from the ONS coronavirus infection survey. Lancet Public Health 2021; 6:e30–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gelman A, Carpenter B.. Bayesian analysis of tests with unknown specificity and sensitivity. J R Stat Soc Series C 2020; 69:1269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gelman A, Lax J, Phillips J, Gabry J, Trangucci R.. Using multilevel regression and poststratification to estimate dynamic public opinion. 2018.. Available at http://www.stat.columbia.edu/~gelman/research/unpublished/MRT(1).pdf. Accessed 22 May 2021.
- 11. Hanretty C. An introduction to multilevel regression and post-stratification for estimating constituency opinion. Polit Stud Rev 2020; 18:630–45. [Google Scholar]
- 12.U.S. Census Bureau. Selected demographic characteristics, 2015-2019 American Community Survey 5-year estimates. U.S. Census Bureau, 2019. [Google Scholar]
- 13. Ma KC. Code associated with trends in SARS-CoV-2 seroprevalence in Massachusetts estimated from newborn screening specimens. GitHub Repository 2022. Available at: https://github.com/gradlab/covid19-newborn-seroprevalence. Accessed 04 March 2022.
- 14. Smith TP, Flaxman S, Gallinat AS, et al. . Temperature and population density influence SARS-CoV-2 transmission in the absence of nonpharmaceutical interventions. Proc Natl Acad Sci USA 2021; 118:e2019284118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flaxman S, Mishra S, Gandy A, et al. . Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature 2020; 584:257–61. [DOI] [PubMed] [Google Scholar]
- 16. Troppy S, Haney G, Cocoros N, Cranston K, DeMaria A Jr. Infectious disease surveillance in the 21st century: an integrated web-based surveillance and case management system. Public Health Rep 2014; 129:132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Massachusetts Water Resources Authority. Wastewater COVID-19 tracking. Available at: https://www.mwra.com/biobot/biobotdata.htm.
- 18. Liu F, Nguyen M, Vijayakumar P, et al. . Newborn dried blood spots for serologic surveys of COVID-19. Pediatr Infect Dis J 2020; 39:e454–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stringhini S, Wisniak A, Piumatti G, et al. . Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet 2020; 396:313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Millett GA, Honermann B, Jones A, et al. . White counties stand apart: the primacy of residential segregation in COVID-19 and HIV diagnoses. AIDS Patient Care STDS 2020; 34:417–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Millett GA, Jones AT, Benkeser D, et al. . Assessing differential impacts of COVID-19 on black communities. Ann Epidemiol 2020; 47:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bassett MT, Chen JT, Krieger N.. Variation in racial/ethnic disparities in COVID-19 mortality by age in the United States: a cross-sectional study. PLoS Med 2020; 17:e1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen JT, Krieger N.. Revealing the unequal burden of COVID-19 by income, race/ethnicity, and household crowding: US County versus zip code analyses. J Public Health Manag Pract 2021; 27 Suppl 1, COVID-19 and Public Health: Looking Back, Moving Forward:S43–S56. [DOI] [PubMed] [Google Scholar]
- 24. Jin DK, Nesbitt DJ, Yang J, et al. . Seroprevalence of anti-SARS-CoV-2 antibodies in a cohort of New York City metro blood donors using multiple SARS-CoV-2 serological assays: implications for controlling the epidemic and “Reopening.” PLoS One 2021; 16:e0250319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu R, Huang J, Duan C, et al. . Low prevalence of antibodies against SARS-CoV-2 among voluntary blood donors in Guangzhou, China. J Med Virol 2021; 93:1743–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larremore DB, Fosdick BK, Bubar KM, et al. . Estimating SARS-CoV-2 seroprevalence and epidemiological parameters with uncertainty from serological surveys. Elife 2021; 10:e64206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.