Abstract
Background
Children are less susceptible to SARS-CoV-2 infection and typically have milder illness courses than adults, but the factors underlying these age-associated differences are not well understood. The upper respiratory microbiome undergoes substantial shifts during childhood and is increasingly recognized to influence host defense against respiratory pathogens. Thus, we sought to identify upper respiratory microbiome features associated with SARS-CoV-2 infection susceptibility and illness severity.
Methods
We collected clinical data and nasopharyngeal swabs from 285 children, adolescents, and young adults (<21 years) with documented SARS-CoV-2 exposure. We used 16S ribosomal RNA gene sequencing to characterize the nasopharyngeal microbiome and evaluated for age-adjusted associations between microbiome characteristics and SARS-CoV-2 infection status and respiratory symptoms.
Results
Nasopharyngeal microbiome composition varied with age (PERMANOVA, P < .001; R2 = 0.06) and between SARS-CoV-2–infected individuals with and without respiratory symptoms (PERMANOVA, P = .002; R2 = 0.009). SARS-CoV-2–infected participants with Corynebacterium/Dolosigranulum-dominant microbiome profiles were less likely to have respiratory symptoms than infected participants with other nasopharyngeal microbiome profiles (OR: .38; 95% CI: .18–.81). Using generalized joint attributed modeling, we identified 9 bacterial taxa associated with SARS-CoV-2 infection and 6 taxa differentially abundant among SARS-CoV-2–infected participants with respiratory symptoms; the magnitude of these associations was strongly influenced by age.
Conclusions
We identified interactive relationships between age and specific nasopharyngeal microbiome features that are associated with SARS-CoV-2 infection susceptibility and symptoms in children, adolescents, and young adults. Our data suggest that the upper respiratory microbiome may be a mechanism by which age influences SARS-CoV-2 susceptibility and illness severity.
Keywords: COVID-19, pediatric microbiota, Corynebacterium, Dolosigranulum, generalized joint attribute modeling
We demonstrate that the nasopharyngeal microbiome undergoes marked shifts in composition with age during childhood and adolescence, and age-associated changes in nasopharyngeal microbiome composition are associated with SARS-CoV-2 infection and SARS-CoV-2–associated respiratory symptoms among children, adolescents, and young adults.
In contrast to most other respiratory viruses [1], children appear to be less susceptible to infection with severe acute respiratory coronavirus 2 (SARS-CoV-2), and typically have milder illness courses than adults. In a recent meta-analysis of 32 studies that included 41 640 children and adolescents and 268 945 adults, SARS-CoV-2 infection susceptibility was estimated to be 46% lower among children and adolescents relative to adults [2]. Further, a higher incidence of SARS-CoV-2 infection has been observed with increasing age, even among infants, children, and adolescents [3]. We previously demonstrated that up to one-third of SARS-CoV-2–infected children and adolescents are asymptomatic [4], and the vast majority of children who develop symptoms report mild respiratory symptoms [4, 5]. Additionally, coronavirus disease 2019 (COVID-19) hospitalization rates and mortality among children are substantially lower than among adults of all ages [6]. These data suggest that changes in host biological or immunological factors that occur with age modify susceptibility to and severity of SARS-CoV-2 infection.
Given that the upper respiratory microbiome undergoes substantial shifts in early childhood [7, 8], and is increasingly recognized to play a key role in the pathogenesis of respiratory virus infections [9, 10], we hypothesized that age-associated changes in the upper respiratory microbiome might contribute to differences in SARS-CoV-2 susceptibility and illness severity among children and adults. In this study, we used 16S ribosomal RNA (rRNA) gene amplicon sequencing to characterize the nasopharyngeal microbiomes of 285 children, adolescents, and young adults with close contact with a SARS-CoV-2–infected individual and to identify microbiome features associated with SARS-CoV-2 infection and with the presence of respiratory symptoms among SARS-CoV-2–infected individuals.
METHODS
Study Procedures
The Duke Biospecimens from RespirAtory Virus-Exposed Kids (BRAVE Kids) Study is a prospective cohort study of children, adolescents, and young adults (<21 years of age) with confirmed SARS-CoV-2 infection or close contact with an individual with confirmed SARS-CoV-2 infection, as previously described [4]. Exposure, sociodemographic, and clinical data are collected at enrollment, and we record symptoms occurring up to 14 days prior to and 28 days after study enrollment. Nasopharyngeal samples are collected with nylon flocked swabs (Copan Italia, Brescia, Italy) and placed into RNAProtect (Qiagen, Hilden, Germany) prior to storage at −80°C. Participants are classified as SARS-CoV-2 infected if the virus is detected in either a clinical or research polymerase chain reaction (PCR) assay. For the analyses presented herein, we considered SARS-CoV-2–infected individuals to have respiratory symptoms if they reported cough, rhinorrhea, nasal congestion, shortness of breath, sore throat, or anosmia at any point between 14 days prior to enrollment through 28 days after enrollment.
Processing of Nasopharyngeal Samples for 16S Ribosomal RNA Sequencing
The Duke Microbiome Core Facility extracted DNA from nasopharyngeal samples using Powersoil Pro extraction kits (Qiagen). DNA concentrations were determined using Qubit dsDNA high-sensitivity assay kits (ThermoFisher Scientific). Bacterial community composition was characterized by PCR amplification of the V4 variable region of the 16S rRNA gene [11]. Equimolar 16S rRNA PCR products were quantified and pooled prior to sequencing. Sequencing was performed by the Duke Sequencing and Genomic Technologies Core Facility on an Illumina MiSeq instrument configured for 250 base-pair paired-end sequencing. All samples were included in a single sample processing run with negative extraction and PCR controls. We analyzed raw sequences using DADA2 version 1.16 [12] and assigned taxonomy to amplicon sequence variants (ASVs) using the expanded Human Oral Microbiome Database version 15.1 [13]. We identified and removed presumed reagent contaminant ASVs (n = 35) (Supplementary Table 1) based on presence in negative control samples or negative correlation with DNA concentration using the frequency method (threshold = 0.10) implemented in the decontam R package version 1.12 [14]. We excluded samples with less than 1000 sequencing reads after quality filtering and contaminant removal. We obtained a median (interquartile range [IQR]) of 24 360 (18 117–33 371) high-quality sequencing reads from the 285 samples included in these analyses. Sequencing reads were classified into 1854 ASVs representing 202 bacterial genera from 8 phyla.
Data Analysis
We calculated nasopharyngeal microbiome alpha diversity (Shannon diversity index and number of unique ASVs) using the phyloseq R package version 1.36 [15]. We fit linear regression models to evaluate associations between patient characteristics and microbiome alpha diversity measures. The number of unique ASVs was not normally distributed and was log-transformed for these analyses. We used the microbiome R package version 1.8.0 [16] to generate centered log-ratio (CLR)–transformed sample counts to evaluate between-sample compositional differences [17]. We used k-medoids clustering and the Calinski-Harabasz index to classify samples into distinct nasopharyngeal microbiome profiles. We evaluated associations between patient characteristics and nasopharyngeal microbiome composition with permutational multivariate analysis of variance (PERMANOVA) using the adonis function within the vegan R package version 2.5.7 [18]. To evaluate associations between patient characteristics and the relative abundances of specific ASVs within the nasopharyngeal microbiome, we used generalized joint attribute modeling (GJAM) implemented in the gjam R package version 2.3.5 [19]. Analyses conducted in gjam were limited to ASVs present in at least 5% of samples. We adjusted for participant age (as a continuous variable) and assessed the significance of interaction terms in all analyses to evaluate for interactive relationships between age and the relative abundances of specific ASVs on SARS-CoV-2 infection and SARS-CoV-2–associated respiratory symptoms. Our findings in all analyses were not substantively changed when we additionally adjusted for sex and race (data not shown). All analyses were conducted in R version 4.1 [20].
RESULTS
Patient Characteristics
Two hundred eighty-five children, adolescents, and young adults were included in these analyses (Table 1). Participants were classified as SARS-CoV-2 exposed but uninfected (n = 74, 26%), SARS-CoV-2 infected without respiratory symptoms (n = 98, 34%), and SARS-CoV-2 infected with respiratory symptoms (n = 113, 40%). SARS-CoV-2–infected participants with respiratory symptoms were older than SARS-CoV-2–infected participants without respiratory symptoms (median [IQR] age: 14.1 [6.3–17.5] vs 9.3 [4.8–13.2] years; Wilcoxon rank-sum test, P = .001) and SARS-CoV-2–uninfected participants (median [IQR] age: 14.1 [6.3–17.5] vs 9.5 [5.1–15.8] years; Wilcoxon rank-sum test, P = .051). There were no significant differences in the prevalences of comorbidities or recent receipt of antibiotics or probiotics in these groups.
Table 1.
Characteristics of the Study Population
| SARS-CoV-2 Exposed, Uninfected (n = 74) | SARS-CoV-2 Infected Without Respiratory Symptoms (n = 98) | SARS-CoV-2 Infected With Respiratory Symptoms (n = 113) |
P a | ||||
|---|---|---|---|---|---|---|---|
| No. (or Median) | % (or IQR) | No. (or Median) | % (or IQR) | No. (or Median) | % (or IQR) | ||
| Age, years | 9.5 | (5.1–15.8) | 9.3 | (4.8–13.2) | 14.1 | (6.3–17.5) | .01 |
| Female sex | 37 | 50% | 53 | 54% | 59 | 52% | .87 |
| Race/ethnicity | <.0001 | ||||||
| Black or African-American | 2 | 3% | 6 | 6% | 8 | 7% | |
| Latino or Hispanic-American | 42 | 57% | 86 | 88% | 100 | 88% | |
| Non-Hispanic White | 30 | 41% | 6 | 6% | 5 | 4% | |
| Comorbiditiesb | |||||||
| Asthma | 8 | 11% | 6 | 6% | 9 | 8% | .53 |
| Obesity (BMI ≥95th percentile for age) | 20 | 27% | 24 | 24% | 40 | 35% | .19 |
| Environmental tobacco smoke in home | 11 | 15% | 9 | 9% | 14 | 12% | .51 |
| Receipt of antibiotic in prior 30 days | 1 | 1% | 2 | 2% | 3 | 3% | >.99 |
| Receipt of probiotic in prior 30 days | 3 | 4% | 0 | 0% | 1 | 1% | .09 |
Abbreviations: BMI, body mass index; IQR, interquartile range; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
P values were estimated using chi-square or Fisher’s exact tests for categorical variables and Kruskal–Wallis tests for continuous variables.
Other comorbidities included hypertension (n = 5), congenital heart disease (n = 3), chronic neurological disorder (n = 3), chronic kidney disease (n = 2), and malignancy (n = 1).
Nasopharyngeal Microbiome Diversity Differs Based on Age and SARS-CoV-2 Infection Status
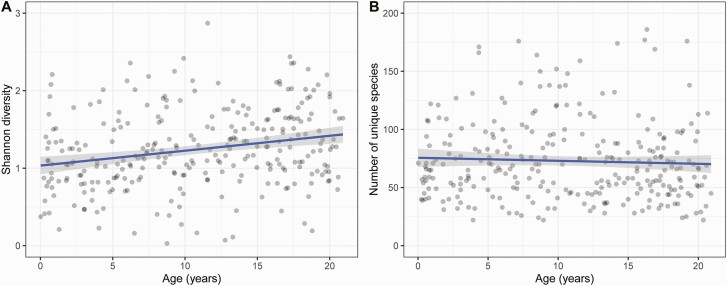
We first sought to describe changes in nasopharyngeal microbiome diversity that occur with age from infancy through early adulthood, and by SARS-CoV-2 infection and symptom status. Median (IQR) Shannon diversity and number of unique ASVs in nasopharyngeal samples were 1.49 (1.12–2.00) and 67 (48–90), respectively. Nasopharyngeal microbiome diversity, as measured by the Shannon index, increased with increasing age (Figure 1A) (analysis of covariance [ANCOVA], P < .0001), while observed richness (number of unique ASVs) was not associated with participant age (Figure 1B) (ANCOVA, P = .27). Shannon diversity was similar in nasopharyngeal samples from SARS-CoV-2–infected and uninfected participants (median [IQR]: 1.49 [1.16–2.02] vs 1.64 [1.20–2.04]; ANCOVA, P = .39); however, observed richness was higher in SARS-CoV-2–infected participants than in SARS-CoV-2–uninfected participants (median [IQR]: 69 [51–75] vs 59 [38–84]; ANCOVA, P = .01). Measures of nasopharyngeal microbiome diversity were similar in SARS-CoV-2–infected individuals with or without respiratory symptoms (data not shown).
Figure 1.
Nasopharyngeal microbiome alpha diversity by age. Shannon diversity (A) and the number of unique amplicon sequence variants (B) are shown by participant age. Each point represents an individual sample and lines correspond to the fit of the linear model between age and each alpha diversity measure. Abbreviation: ASV, amplicon sequence variant.
Nasopharyneal Microbiome Composition by Age, SARS-CoV-2 Infection, and COVID-19 Symptoms
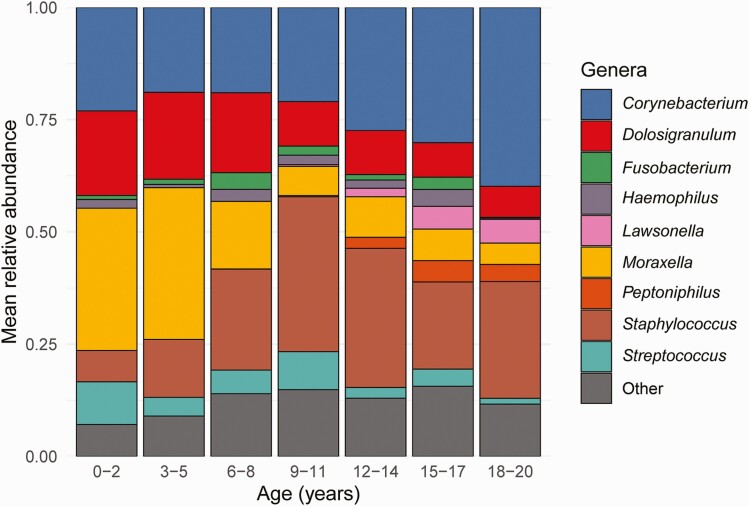
Five bacterial genera accounted for more than 80% of the sequencing reads identified in nasopharyngeal samples: Corynebacterium (26%), Staphylococcus (21%), Moraxella (15%), Dolosigranulum (13%), and Streptococcus (5%). Nasopharyngeal microbiome composition varied with age (PERMANOVA, P < .001; R2 = 0.06); specifically, increasing age was associated with decreases in the CLR-transformed abundances of the bacterial genera Moraxella (Spearman’s rank correlation; ρ = −0.40, P < .0001) and Dolosigranulum (ρ = −0.32, P < .0001) and increases in the CLR-transformed abundances of Corynebacterium (ρ = 0.24, P < .0001) and Staphylococcus (ρ = 0.45, P < .0001) (Figure 2). Two bacterial genera—Lawsonella and Peptoniphilus—were highly prevalent in participants 12 years of age or older (78% and 69%, respectively), but were identified in only 21% and 19% of children 8 years of age or younger. Nasopharyngeal microbiome composition did not differ significantly in SARS-CoV-2–infected and SARS-CoV-2–uninfected participants (PERMANOVA, P = .10; R2 = 0.004). However, the composition of the nasopharyngeal microbiome of SARS-CoV-2–infected participants with respiratory symptoms differed from that of SARS-CoV-2–infected participants without respiratory symptoms (PERMANOVA, P = .002; R2 = 0.009).
Figure 2.
Relative abundances of highly abundant bacterial genera by age. Each bar depicts the mean relative abundances of highly abundant genera in nasopharyngeal samples from participants in a specific age category. Only the 9 most highly abundant genera within nasopharyngeal samples from the entire study population are shown. Age is shown as a categorical variable only for graphical representation; all statistical analyses included age as a continuous variable.
Associations Between Nasopharyngeal Microbiome Profile, Age, and SARS-CoV-2 Status
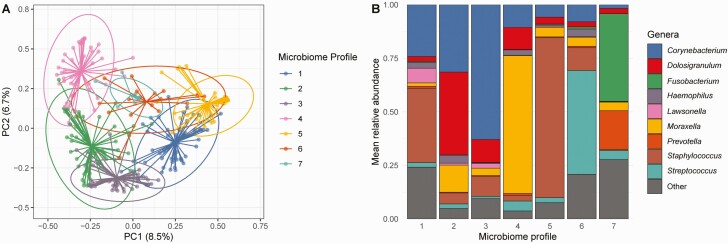
To further characterize differences in nasopharyngeal microbiome composition by age and SARS-CoV-2 status, we used unsupervised clustering to classify nasopharyngeal samples into 7 distinct microbiome profiles (Figure 3): Corynebacterium/Staphylococcus-dominant (profile 1; n = 54, 19%), Corynebacterium/Dolosigranulum-dominant (profile 2; n = 58, 20%), Corynebacterium-dominant (profile 3; n = 55, 19%), Moraxella-dominant (profile 4; n = 44, 16%), Staphylococcus-dominant (profile 5; n = 44, 15%), Streptococcus-dominant (profile 6; n = 16, 6%), and Fusobacterium-dominant (profile 7; n = 10, 4%). Participant age differed markedly by nasopharyngeal microbiome profile (Table 2) (Kruskal–Wallis test, P < .0001); however, there were no significant differences in other patient characteristics by microbiome profile. SARS-CoV-2 infection prevalence varied from 69% to 82% by microbiome profile, with the lowest prevalence observed among participants with a Moraxella-dominant microbiome profile and the highest prevalence seen among participants with a Corynebacterium-dominant microbiome profile. However, in analyses adjusting for age, there were no significant associations between nasopharyngeal microbiome profile and SARS-CoV-2 infection. Among SARS-CoV-2–infected participants, the prevalence of respiratory symptoms varied from 35% to 67% by nasopharyngeal microbiome profile, with the lowest prevalence seen among participants with a Corynebacterium/Dolosigranulum-dominant microbiome profile and the highest prevalence observed among participants with Corynebacterium/Staphylococcus-dominant or Corynebacterium-dominant microbiome profiles. SARS-CoV-2–infected individuals with a Corynebacterium/Dolosigranulum-dominant microbiome profile were less likely to have respiratory symptoms than SARS-CoV-2–infected participants with other nasopharyngeal microbiome profiles in age-adjusted analyses (logistic regression; odds ratio: .38; 95% confidence interval: .18–.81).
Figure 3.
Nasopharyngeal microbiome profiles identified by unsupervised clustering. A, Principal coordinate (PC) plot of Euclidean distances demonstrating clustering of nasopharyngeal samples by microbiome profile. Each dot corresponds to a single nasopharyngeal sample. Centroids are shown as the confluence of the lines arising from individual points from each microbiome profile. Ellipses define the regions containing 95% of all samples that can be drawn from the underlying multivariate t distribution. B, Each bar depicts the mean relative abundances of highly abundant genera in nasopharyngeal samples assigned to specific microbiome profiles. Only the 9 most highly abundant genera within nasopharyngeal samples from the entire study population are shown.
Table 2.
Characteristics of Study Participants and Nasopharyngeal Microbial Communities by Microbiome Profile
| Nasopharyngeal Microbiome Profilea | P b | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 (n = 54) | 2 (n = 58) | 3 (n = 55) | 4 (n = 48) | 5 (n = 44) | 6 (n = 16) | 7 (n = 10) | ||
| Age, median (IQR), years | 16.5 (12.9–18.1) |
7.6 (3.9–12.1) |
15.9 (10.6–18.7) |
3.8 (2.0–7.2) |
9.8 (8.3–13.2) |
7.7 (1.0–11.5) |
7.0 (6.0–13.4) |
<.0001 |
| Female sex, n (%) | 29 (53%) | 29 (50%) | 33 (60%) | 31 (65%) | 17 (39%) | 7 (44%) | 3 (30%) | .12 |
| Race/ethnicity, n (%) | ||||||||
| Black or African-American | 5 (9%) | 4 (7.0%) | 4 (7%) | 1 (2%) | 1 (2%) | 1 (6%) | 0 (0%) | .23 |
| Latino or Hispanic-American | 37 (69%) | 46 (79%) | 46 (84%) | 38 (79%) | 40 (91%) | 11 (69%) | 10 (100%) | |
| Non-Hispanic White | 12 (22%) | 8 (14%) | 5 (9%) | 9 (19%) | 3 (7%) | 4 (25%) | 0 (0%) | |
| Comorbidities, n (%) | ||||||||
| Asthma | 5 (9%) | 2 (3%) | 5 (9%) | 3 (6.3%) | 8 (18%) | 0 (0%) | 0 (0%) | .19 |
| Obesity (BMI ≥95th percentile for age) | 20 (37%) | 15 (26%) | 18 (33%) | 13 (27%) | 11 (25%) | 3 (19%) | 4 (40%) | .67 |
| Environmental tobacco smoke in home | 6 (11%) | 9 (16%) | 9 (16%) | 4 (8.3%) | 3 (7%) | 3 (19%) | 0 (0%) | .52 |
| Receipt of antibiotic in prior 30 days | 2 (4%) | 1 (2%) | 1 (2%) | 1 (2%) | 1 (2%) | 0 (0%) | 0 (0%) | .97 |
| Receipt of probiotic in prior 30 days | 3 (6%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (2%) | 0 (0%) | 0 (0%) | .18 |
| SARS-CoV-2 infection | 39 (72%) | 43 (74%) | 45 (82%) | 33 (69%) | 32 (72%) | 11 (69%) | 8 (80%) | .80 |
| With respiratory symptoms | 26 (67%) | 15 (35%) | 30 (67%) | 18 (55%) | 13 (41%) | 6 (55%) | 5 (63%) | .03 |
| Without respiratory symptoms | 13 (33%) | 28 (65%) | 15 (33%) | 15 (45%) | 19 (59%) | 5 (45%) | 3 (38%) | |
| Shannon diversity index, median (IQR) | 1.64 (1.37–1.93) | 1.15 (1.05–1.51) | 1.04 (0.91–1.35) | 0.91 (0.81–1.23) | 0.96 (0.45–1.24) | 1.5 (1.22–2.19) | 1.72 (1.37–1.97) | <.0001 |
| Number of unique ASVs, median (IQR) | 72 (45–92) | 66 (45–83) | 65 (49–82) | 58 (42–78) | 71 (53–101) | 76 (59–91) | 92 (57–106) | .16 |
Abbreviations: ASV, amplicon sequence variant; BMI, body mass index; IQR, interquartile range; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Corresponding microbiome profiles are as follows: 1 = Corynebacterium/Staphylococcus-dominant, 2 = Corynebacterium/Dolosigranulum-dominant, 3 = Corynebacterium-dominant, 4 = Moraxella-dominant, 5 = Staphylococcus-dominant, 6 = Streptococcus-dominant, 7 = Fusobacterium-dominant.
P values were estimated using Fisher’s exact tests for categorical variables and Kruskal-Wallis tests for continuous variables.
Identification of Specific Bacterial Taxa Associated With SARS-CoV-2 Infection and COVID-19 Symptoms
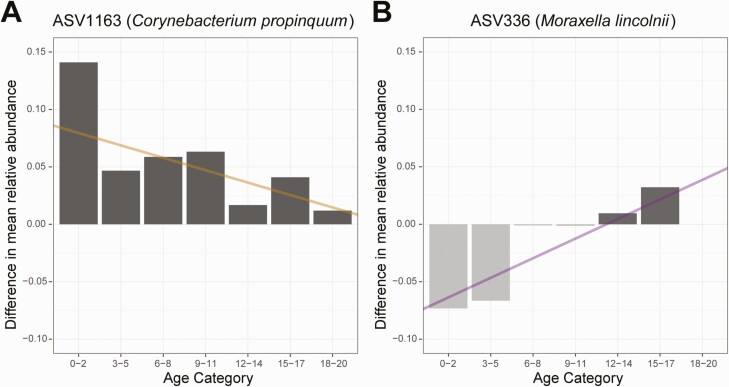
We next used GJAM to evaluate associations between specific ASVs and SARS-CoV-2 infection and SARS-CoV-2–associated respiratory symptoms. GJAM allows for the concurrent evaluation of distinct types of data derived from observations of ecological systems, where attributes of the system may be interdependent. Because we observed associations between age and nasopharyngeal microbiome composition, as well as associations between age and SARS-CoV-2 infection and respiratory symptoms, we used GJAM to separately evaluate associations between specific bacterial ASVs and SARS-CoV-2 infection and SARS-CoV-2–associated respiratory symptoms in the context of interactions of these variables with participant age. We identified 9 ASVs that were associated with SARS-CoV-2 infection (Table 3); for 8 of these ASVs, the magnitude of the association varied by participant age. For example, the relative abundance of ASV1163 (Corynebacterium propinquum) decreased with increasing participant age and was also higher among SARS-CoV-2–infected participants than uninfected participants independent of age. However, the difference in the relative abundance of ASV1163 between SARS-CoV-2–infected and uninfected participants decreased with age, such that the negative association between the relative abundance of ASV1163 and SARS-CoV-2 infection was primarily observed among young children (Figure 4A). We next used GJAM to identify ASVs associated with respiratory symptoms among participants with confirmed SARS-CoV-2 infection. We identified 6 ASVs that were differentially abundant among SARS-CoV-2–infected participants with respiratory symptoms (Table 4); for 8 of these ASVs, the magnitude of the association varied by participant age. For example, the relative abundance of ASV336 (Moraxella lincolnii) decreased with increasing age and was independently lower among SARS-CoV-2–infected participants with respiratory symptoms. However, the difference in the mean relative abundance of ASV336 between SARS-CoV-2–infected participants with and without respiratory symptoms increased with increasing age, indicating that the negative association between the relative abundance of this ASV and the presence of SARS-CoV-2–associated respiratory symptoms was observed only in younger age groups (Figure 4B).
Table 3.
Differentially Abundant Bacterial Amplicon Sequence Variants in SARS-CoV-2–Infected and Uninfected Participants in GJAM Analyses
| ASV | Bacterial Species | Age | SARS-CoV-2 Infection | Age × SARS-CoV-2 Infection | |||
|---|---|---|---|---|---|---|---|
| Estimate | (95% CI) | Estimate | (95% CI) | Estimate | (95% CI) | ||
| 306 | Moraxella nonliquefaciens | −.020 | (−.023, −.016) | −.108 | (−.158, −.055) | .005 | (.001, .009) |
| 629 | Prevotella nanceiensis | −.002 | (−.003, −.001) | −.022 | (−.037, −.006) | .002 | (.0003, .003) |
| 692 | Prevotella intermedia | −.002 | (−.003, −.0004) | −.020 | (−.038, −.001) | NS | NS |
| 712 | Prevotella melaninogenica | −.002 | (−.003, −.0009) | −.016 | (−.032, −.0006) | .001 | (.0007, .0002) |
| 1095 | Corynebacterium tuberculostearicum | .006 | (.004, .007) | .022 | (.002, .041) | −.003 | (−.004, −.001) |
| 1163 | Corynebacterium propinquum | −.004 | (−.007, −.0002) | .090 | (.040, .142) | −.004 | (−.009, −.0004) |
| 1165 | Corynebacterium propinquum | NS | NS | −.033 | (−.059, −.007) | .003 | (.001, .006) |
| 1488 | Streptococcus mitis | −.002 | (−.004, −.001) | −.202 | (−.038, −.002) | NS | NS |
| 1581 | Gemella morbillorum | −.003 | (−.004, −.001) | −.022 | (−.040, −.003) | .002 | (.0003, .003) |
ASV, amplicon sequence variant; CI, confidence interval; GJAM, generalized joint attribute modeling; NS, not significant; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Figure 4.
Interactive relationships between participant age, the relative abundances of specific bacterial ASVs in the nasopharyngeal microbiome, and SARS-CoV-2 status. A, Bar chart depicting differences in the mean relative abundance of ASV1163 (Corynebacterium propinquum) among SARS-CoV-2–infected participants relative to uninfected participants in different age categories. The line was constructed using the GJAM estimates for the association of SARS-CoV-2 infection with the relative abundance of ASV1163 (intercept) and the association of the interaction term between SARS-CoV-2 infection and age with the relative abundance of ASV1163 (slope). Higher relative abundances of ASV1163 were observed in SARS-CoV-2–infected compared with uninfected participants across all ages, but these differences were more pronounced in young children. B, Differences in mean relative abundance of ASV336 (Moraxella lincolnii) between SARS-CoV-2–infected participants with respiratory symptoms and SARS-CoV-2–infected participants without respiratory symptoms are depicted by age category. Dark (light) gray bars represent age categories in which ASV336 was more (less) abundant among SARS-CoV-2–infected participants with respiratory symptoms compared with SARS-CoV-2–infected participants without respiratory symptoms. The line was constructed using the GJAM estimates for the association of SARS-CoV-2–associated respiratory symptoms with the relative abundance of ASV336 (intercept) and the association of the interaction term between respiratory symptoms and age with the relative abundance of ASV336 (slope). The difference in the mean relative abundance of ASV336 between SARS-CoV-2–infected participants with and without respiratory symptoms differed by age, such that this ASV was less abundant in the context of SARS-CoV-2–associated respiratory symptoms among young children and more abundant in the context of SARS-CoV-2–associated respiratory symptoms in older age groups. Lines were fit using the regression coefficients generated using GJAM. Age is shown as a categorical variable only for graphical representation; all statistical analyses included age as a continuous variable. Abbreviations: ASV, amplicon sequence variant; GJAM, generalized joint attribute modeling; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Table 4.
Differentially Abundant Bacterial Amplicon Sequence Variants in SARS-CoV-2–Infected Participants With and Without Respiratory Symptoms in GJAM Analyses
| ASV | Bacterial Species | Age | SARS-CoV-2 Respiratory Symptoms | Age × SARS-CoV-2 Respiratory Symptoms | |||
|---|---|---|---|---|---|---|---|
| Estimate | (95% CI) | Estimate | (95% CI) | Estimate | (95% CI) | ||
| 336 | Moraxella lincolnii | −.006 | (−.008, −.004) | −.081 | (−.111, −.051) | .007 | (.004, .009) |
| 339 | Moraxella lincolnii | −.002 | (−.003, −.0008) | −.018 | (−.032, −.004) | .002 | (.0003, .003) |
| 692 | Prevotella intermedia | NS | NS | .024 | (.008, .039) | −.002 | (−.003, −.0002) |
| 1283 | Fusobacterium nucleatum | .003 | (.0009, .005) | .038 | (.008, .067) | −.004 | (−.007, −.002) |
| 1519 | Streptococcus pyogenes | NS | NS | .043 | (.002, .071) | NS | NS |
| 2155 | Mycoplasma lipophilum | −.001 | (−.002, −.0003) | −.014 | (−.027, −.001) | NS | NS |
Abbreviations: ASV, amplicon sequence variant; CI, confidence interval; GJAM, generalized joint attribute modeling; NS, not significant; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
DISCUSSION
We identified nasopharyngeal microbiome profiles and specific bacterial taxa associated with SARS-CoV-2 infection and the presence of respiratory symptoms in SARS-CoV-2–infected children, adolescents, and young adults. We demonstrated that nasopharyngeal microbiome diversity and composition are strongly associated with age, and that age modifies the associations between specific bacterial taxa and SARS-CoV-2 infection status and the presence of SARS-CoV-2–associated respiratory symptoms. Our findings suggest that the upper respiratory microbiome may be a previously unrecognized and potentially modifiable mechanism by which age influences SARS-CoV-2 susceptibility and respiratory symptoms.
There are accumulating data supporting a key role for the upper respiratory microbiome in the pathogenesis of respiratory virus infections. Prior studies indicate that the upper respiratory microbiome modifies susceptibility to respiratory virus infections, as has been demonstrated in studies of household influenza virus transmission [9]. The upper respiratory microbiome may also influence the symptoms of respiratory virus–infected individuals [21]. For example, among young children with respiratory syncytial virus (RSV), higher abundances of Haemophilus species in the nasopharyngeal microbiome are associated with a more exuberant host immune response [10, 22]. Nasopharyngeal microbiome profiles are also associated with inflammatory cytokine levels in nasal wash samples and the development of symptomatic infection in adults following experimental rhinovirus challenge [23]. Further, in animal models, intranasal administration of live bacterial strains directly modulates immune responses to viral infections [24, 25]. While data from clinical studies are currently lacking, this work suggests that targeted manipulation of the upper respiratory microbiome could be a promising approach to prevent or treat respiratory virus infections.
To date, studies of the upper respiratory microbiome and SARS-CoV-2 infection have primarily been conducted among cohorts of adults presenting with clinical suspicion of COVID-19. de Castilhos and colleagues [26] evaluated the oropharyngeal microbiome in 148 SARS-CoV-2–infected outpatients, 124 hospitalized patients with COVID-19, and 74 healthy adults. They did not identify any significant alterations in microbiome composition between SARS-CoV-2–infected outpatients and healthy controls but found marked dysbiosis among patients hospitalized with severe COVID-19. Mostafa and colleagues [27] reported lower nasopharyngeal microbial diversity, a lower abundance of the bacterial family Propionibacteriaceae, and a higher abundance of Corynebacterium accolens in 40 SARS-CoV-2–infected adults compared with 10 SARS-CoV-2–uninfected adults. In this study, we identified distinct bacterial ASVs associated with SARS-CoV-2 infection and SARS-CoV-2–associated respiratory symptoms. Additionally, we observed a lower prevalence of respiratory symptoms among SARS-CoV-2–infected subjects with Corynebacteriun/Dolosigranulum-dominant microbiome profiles compared with infected subjects with other microbiome profiles.
Non-diphtheriae Corynebacterium species and Dolosigranulum pigrum were previously shown to have important microbial interactions within the human nasopharynx. Corynebacterium abundance within the nasopharyngeal microbiome has been negatively associated with Streptococcus pneumoniae colonization among infants and children [28, 29]. Moreover, Corynebacterium spp. influence innate immune responses to viral infection in murine models [30, 31]. Dolosigranulum pigrum is also generally considered to play a protective role against viral and bacterial infections [32]. Islam and colleagues [33] recently demonstrated that administration of specific D. pigrum strains enhanced resistance to SARS-CoV-2 infection of cultured human respiratory epithelial cells. Further, Smith and colleagues [34] observed decreased abundance of both Corynebacterium and Dolosigranulum spp. in patients with severe COVID-19 symptoms.
Several prior studies evaluated associations between respiratory health and upper respiratory microbiome profiles. Toivonen and colleagues [35] used similar unsupervised clustering methods to create longitudinal nasopharyngeal microbiome profiles for 697 Finnish children during the first 2 years of life to evaluate associations between microbiome composition, antibiotic exposures, and later asthma development. Despite marked differences in the patient populations and sampling protocols, they identified microbiome profiles dominated by bacterial genera that were also prevalent in our cohort. Teo and colleagues [36] identified similar nasopharyngeal microbiome profiles among Australian infants during the first year of life and at 5 years of age. Notably, they found that microbiome profiles dominated by Moraxella, Streptococcus, or Haemophilus were more prevalent in samples collected during acute respiratory infections, while profiles dominated by Staphylococcus, Dolosigranulum, or Corynebacterium were more prevalent during periods of health [36]. Finally, Kelly et al [29] identified 5 nasopharyngeal microbiome profiles among 319 children less than 2 years of age in Botswana. In this study, Streptococcus-dominant or Moraxella-dominant profiles were more common among children with pneumonia or upper respiratory infection symptoms, while the majority of children without respiratory symptoms had microbiome profiles co-dominated by Corynebacterium and Dolosigranulum [29]. Taken together, these studies demonstrate that the human upper respiratory microbiome is composed largely of species from relatively few bacterial genera, and that microbiome profiles dominated by these species are observed in varied patient populations.
We observed that nasopharyngeal microbiome composition undergoes significant, age-associated shifts during infancy, childhood, and adolescence. Moreover, nearly all the associations between nasopharyngeal microbiome features and SARS-CoV-2 status that we identified in this study were modified by age, indicating that age-associated changes in the nasopharyngeal microbiome likely contribute to the susceptibility to and severity of SARS-CoV-2 infections. Much of our knowledge regarding nasopharyngeal microbiome composition is derived from studies of infants or older adults, and surprisingly little is known about how the upper respiratory microbiome changes during childhood and adolescence [7, 8, 37–39]. Our findings demonstrate that development of the nasopharyngeal microbiome continues throughout childhood and adolescence and highlight the need for future studies to identify the biological or environmental factors that contribute to the shifts in microbiome composition that occur after early childhood.
Our study had several limitations. First, nasopharyngeal samples were collected at a single time point after SARS-CoV-2 exposure; therefore, we were unable to determine if the differences in nasopharyngeal microbiome compositions observed by SARS-CoV-2 infection status preceded, or were the consequence of, SARS-CoV-2 infection. Future studies will need to use longitudinal sampling to evaluate causal relationships between upper respiratory microbiome composition and SARS-CoV-2 infection susceptibility and severity. Second, all SARS-CoV-2–infected study participants had relatively mild symptoms; thus, we were unable to identify microbiome features associated with severe COVID-19. Our use of 16S rRNA gene amplicon sequencing prevented us from evaluating other components of the upper respiratory microbiome, including viruses and fungi. Additionally, 16S rRNA gene amplicon experiments have several well-documented biases [40], although we sought to minimize these biases in our study through inclusion of all samples in a single processing run and use of appropriate negative controls. Use of metagenomic sequencing in future studies would enable improved discrimination of bacterial species and analyses of other microbial kingdoms that are components of the nasopharyngeal microbiome. Finally, residual confounding by unmeasured factors remains possible.
In conclusion, we found that age modifies the associations between specific bacterial taxa and both SARS-CoV-2 infection status and the presence of respiratory symptoms. These findings suggest that development of the nasopharyngeal microbiome during childhood and adolescence may contribute to the differences in SARS-CoV-2 susceptibility and severity observed by age. Future studies should evaluate the potential of the upper respiratory microbiome to serve as a therapeutic target for the prevention and treatment of infections caused by SARS-CoV-2 and other respiratory viruses.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the Duke University School of Medicine for use of the Microbiome Core Facility, which performed the DNA extractions and library preparations for this research, and the Duke Sequencing and Genomic Technologies Core Facility, which sequenced these libraries. We offer our sincere gratitude to the children and families who participated in this research.
Financial support. This work was supported by the Duke University School of Medicine and through grants from the Duke Microbiome Center, Children’s Miracle Network Hospitals, and the Translating Duke Health Children’s Health and Discovery Initiative. A. W. M. was supported by a grant from the National Science Foundation (DGE-1545220) and a grant from the National Institutes of Health (NIH; T32GM008555). S. M. H. was supported by an NIH training grant (T32-HD094671). M. S. K. was supported by an NIH Career Development Award (K23-AI135090). C. W. W. also reports support for this work from the NIH.
Potential conflicts of interest. M. S. K. reports advisory board fees from Adagio Therapeutics, Inc. and advisory board fees and grant funding from Merck & Co., Inc. C. W. W. reports advisory board fees from Roche Molecular Sciences, nonfinancial support from bioMérieux and Becton Dickinson, and a research collaboration with Biofire, and is co-founder of Predigen. A. W. M. reports payments for lectures from Pepperdine University. A. T. R. reports royalties for editorial work from Elsevier, honoraria for participating in a Scientific Advisory Board for Breas US, and honoraria for lecturing and development of educational materials for Vapotherm, Inc, and is the unpaid Chair for the Pediatric Section of the Society of Critical Care Medicine (SCCM). N. A. T. reports grants or contracts unrelated to this work from the Centers for Disease Control (CDC Epicenters) and the Rockefeller Foundation, and payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing, or educational events from McGraw-Hill. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data availability. The sequencing dataset supporting the conclusions of this study is available in the Sequence Read Archive (PRJNA703574). The statistical files and script used for data analyses are also publicly available (https://github.com/alexmccumber/BRAVE_Kids).
Contributor Information
Jillian H Hurst, Division of Infectious Diseases, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA; Children’s Health and Discovery Institute, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA.
Alexander W McCumber, Division of Infectious Diseases, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA.
Jhoanna N Aquino, Division of Infectious Diseases, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA.
Javier Rodriguez, Children’s Clinical Research Unit, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA.
Sarah M Heston, Division of Infectious Diseases, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA.
Debra J Lugo, Division of Infectious Diseases, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA.
Alexandre T Rotta, Division of Pediatric Critical Care Medicine, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA.
Nicholas A Turner, Division of Infectious Diseases, Department of Medicine, Duke University School of Medicine, Durham, North Carolina, USA.
Trevor S Pfeiffer, Division of Infectious Diseases, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA.
Thaddeus C Gurley, Duke Human Vaccine Institute, Duke University School of Medicine, Durham, North Carolina, USA.
M Anthony Moody, Division of Infectious Diseases, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA; Duke Human Vaccine Institute, Duke University School of Medicine, Durham, North Carolina, USA.
Thomas N Denny, Duke Human Vaccine Institute, Duke University School of Medicine, Durham, North Carolina, USA.
John F Rawls, Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA; Duke Microbiome Center, Duke University School of Medicine, Durham, North Carolina, USAand.
James S Clark, Nicholas School of the Environment, Duke University, Durham, North Carolina, USA.
Christopher W Woods, Division of Infectious Diseases, Department of Medicine, Duke University School of Medicine, Durham, North Carolina, USA; Duke Human Vaccine Institute, Duke University School of Medicine, Durham, North Carolina, USA.
Matthew S Kelly, Division of Infectious Diseases, Department of Pediatrics, Duke University School of Medicine, Durham, North Carolina, USA; Duke Microbiome Center, Duke University School of Medicine, Durham, North Carolina, USAand.
References
- 1. Lambert L, Culley FJ.. Innate immunity to respiratory infection in early life. Front Immunol 2017; 8:1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Viner RM, Mytton OT, Bonnell C, et al. Susceptibility to SARS-CoV-2 infection among children and adolescents compared with adults: a systematic review and meta-analysis. JAMA Pediatr 2021; 175:143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. COVID-19 stats: COVID-19 incidence, by age group—United States, March 1–November 14, 2020. MMWR Morb Mortal Wkly Rep 2021;69:1664. doi: 10.15585/mmwr.mm695152a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hurst JH, Heston SM, Chambers HN, et al. SARS-CoV-2 infections among children in the biospecimens from respiratory virus-exposed kids (BRAVE Kids) study. Clin Infect Dis 2020; 73:e2875-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castagnoli R, Votto M, Licari A, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children and adolescents: a systematic review. JAMA Pediatr 2020; 174:882–9. [DOI] [PubMed] [Google Scholar]
- 6. Kim L, Whitaker M, O’Halloran A, et al. Hospitalization rates and characteristics of children aged <18 years hospitalized with laboratory-confirmed COVID-19—COVID-NET, 14 states, March 1-July 25, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:1081–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mika M, Mack I, Korten I, et al. Dynamics of the nasal microbiota in infancy: a prospective cohort study. J Allergy Clin Immunol 2015; 135:905–12, e11. [DOI] [PubMed] [Google Scholar]
- 8. Biesbroek G, Tsivtsivadze E, Sanders EAM, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 2014; 190:1283–92. [DOI] [PubMed] [Google Scholar]
- 9. Tsang TK, Lee KH, Foxman B, et al. Association between the respiratory microbiome and susceptibility to influenza virus infection. Clin Infect Dis 2020; 71:1195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Steenhuijsen Piters WA, Heinonen S, Hasrat R, et al. Nasopharyngeal microbiota, host transcriptome and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016; 194:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilbert JA, Meyer F, Antonopoulos D, et al. Meeting report: the Terabase Metagenomics workshop and the vision of an Earth microbiome project. Stand Genomic Sci 2010; 3:243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP.. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13:581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Escapa IF, Huang Y, Chen T, et al. Construction of habitat-specific training sets to achieve species-level assignment in 16S rRNA gene datasets. Microbiome 2020; 8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ.. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018; 6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McMurdie PJ, Holmes S.. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lahti L, Shetty S.. Microbiome R package. 2017. Available at: https://microbiome.github.com/microbiome. Accessed 1 October 2021. [Google Scholar]
- 17. Gloor GB, Wu JR, Pawlowsky-Glahn V, Egozcue JJ.. It’s all relative: analyzing microbiome data as compositions. Ann Epidemiol 2016; 26:322–9. [DOI] [PubMed] [Google Scholar]
- 18. Oksanen J, Blanchet FG, Friendly M, et al. The vegan package. Community Ecology Package 2007; 10:719. [Google Scholar]
- 19. Clark JS, Nemergut D, Seyednasrollah B, Turner PJ, Zhang S.. Generalized joint attribute modeling for biodiversity analysis: median-zero, multivariate, multifarious data. Ecol Monogr 2017; 87:34–56. [Google Scholar]
- 20. R. Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2019. Available at: https://www.R-project.org/ Accessed 1 October 2021. [Google Scholar]
- 21. Pichon M, Lina B, Josset L.. Impact of the respiratory microbiome on host responses to respiratory viral infection. Vaccines 2017; 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ederveen TH, Ferwerda G, Ahout IM, et al. Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome 2018. ;6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allen EK, Koeppel AF, Hendley JO, Turner SD, Winther B, Sale MM.. Characterization of the nasopharyngeal microbiota in health and during rhinovirus challenge. Microbiome 2014; 2:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tomosada Y, Chiba E, Zelaya H, et al. Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC Immunol 2013; 14:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zelaya H, Tada A, Vizoso-Pinto MG, et al. Nasal priming with immunobiotic Lactobacillus rhamnosus modulates inflammation–coagulation interactions and reduces influenza virus-associated pulmonary damage. Inflamm Res 2015; 64:589–602. [DOI] [PubMed] [Google Scholar]
- 26. de Castilhos J, et al. Severe dysbiosis and specific Haemophilus and Neisseria signatures as hallmarks of the oropharyngeal microbiome in critically ill COVID-19 patients. Clin Infect Dis 2021. doi: 10.1093/cid/ciab902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mostafa HH, Zamir E, Hippchen T, et al. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect Covid-19 patients. mBio 2020; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khamash DF, Mongodin EF, White JR, et al. The association between the developing nasal microbiota of hospitalized neonates and staphylococcus aureus colonization. Open Forum Infect Dis 2019; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kelly MS, Surette MG, Smieja M, et al. The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr Infect Dis J 2017; 36:e211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanmani P, Clua P, Vizoso-Pinto MG, et al. Respiratory commensal bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae superinfection. Front Microbiol 2017; 8:1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mak N, Schiltknecht E, Ada G.. Protection of mice against influenza virus infection: enhancement of nonspecific cellular responses by Corynebacterium parvum. Cell Immunol 1983; 78:314–25. [DOI] [PubMed] [Google Scholar]
- 32. Man WH, de Steenhuijsen Piters WA, Bogaert D.. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 2017; 15:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Islam MA, Albarracin L, Melnikov V, et al. Modulates immunity against SARS-CoV-2 in respiratory epithelial cells. Pathogens 2021; 10:634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith N, Goncalves P, Charbit B, et al. Distinct systemic and mucosal immune responses during acute SARS-CoV-2 infection. Nat Immunol 2021; 22:1428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toivonen L, Schuez-Havupalo L, Karppinen S, et al. Antibiotic treatments during infancy, changes in nasal microbiota, and asthma development: population-based cohort study. Clin Infect Dis 2020; 72:1546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015; 17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bogaert D, Keijser B, Huse S, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 2011; 6: e17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hahn A, Warnken S, Pérez-Losada M, Freishtat RJ, Crandall KA.. Microbial diversity within the airway microbiome in chronic pediatric lung diseases. Infect Genet Evol 2018; 63:316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pérez-Losada M, Alamri L, Crandall KA, Freishtat RJ.. Nasopharyngeal microbiome diversity changes over time in children with asthma. PLoS One 2017; 12:e0170543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brooks JP, Edwards DJ, Harwich MD, et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol 2015; 15:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.