Abstract
Background
The increasing use of monoclonal antibodies (mAbs) to treat coronavirus disease 2019 raises questions about their impact on the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mAb-resistant variants. We assessed the impact of Casirivimab-Imdevimab on SARS-CoV-2 mutations associated with reduced mAb activity in treated patients.
Methods
We measured the nasopharyngeal (NP) viral load and sequenced the haplotypes of spike gene of 50 patients infected with the SARS-CoV-2 delta variant and treated with Casirivimab-Imdevimab using single-molecule real-time sequencing.
Results
The NP SARS-CoV-2 viral load of patients treated with Casirivimab-Imdevimab decreased from 8.13 (interquartile range [IQR], 7.06–8.59) log10 copies/mL pretreatment to 3.67 (IQR, 3.07–5.15) log10 copies/mL 7 days later (P < .001). Of the 36 patients for whom follow-up timepoints Spike sequencing were available, none of the Spike mutations that reduced mAb activity were detected.
Conclusions
Casirivimab-Imdevimab is an effective treatment for patients infected with the SARS-CoV-2 delta variant. Despite selective pressure on SARS-CoV-2 Spike quasispecies, we detected no key mutations that reduced mAb activity in our patients.
Keywords: casirivimab, COVID-19, imdevimab, quasispecies, spike protein
Patients at risk of developing severe coronavirus disease 2019 (COVID-19) can be treated early with monoclonal antibodies (mAbs) to prevent complications. Casirivimab (REGN10933) and Imdevimab (REGN10987) is a cocktail of mAbs recently developed and used to treat high-risk patients with mild to moderate COVID-19 [1, 2]. This treatment prevents the virus attaching to the human cell ACE2 receptor and entering cells by targeting the receptor-binding domain (RBD) of the virus spike protein [3, 4]. The spike protein is a homotrimeric glycoprotein whose monomers consist of 2 subunits (S1 and S2). The S1 subunit contains the RBD, including a receptor-binding motif (RBM). The RBM binds to the host ACE2 cell surface receptor, enabling the fusion of virus and host membranes via the S2 subunit [5–7].
Although the genomic diversity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been described [8–10], little is known about the impact of mAb therapy on the evolution of SARS-CoV-2 quasispecies [11]. New variants containing key changes in the RBD, the region targeted by many neutralizing mAbs, have recently appeared all around the world. These new variants can be more transmissible than the original virus, more pathogenic, and more resistant to endogenous or exogenous antibodies [12]. The B.1.617.2 (delta) variant has been the dominant SARS-CoV-2 variant in France since July 2021. It is more likely to resist mAbs therapy, particularly the Bamlanivimab and Etesevimab association, because its L452R mutation confers resistance to Bamlanivimab [13, 14].
The impact of Casirivimab-Imdevimab on SARS-CoV-2 quasispecies evolution has not been studied yet. In this study, we aimed to determine the intrahost evolution of SARS-CoV-2 Spike quasispecies by sequencing the virus in a series of nasopharyngeal (NP) samples from infected patients given Casirivimab-Imdevimab to detect mutations that reduced mAb activity.
MATERIALS AND METHODS
Subjects and Sample Collection
Nasopharyngeal samples were taken from recently infected SARS-CoV-2 patients given a single intravenous infusion of Casirivimab-Imdevimab (1200/1200 mg) at the Toulouse University Hospital between June 28 and October 29, 2021, according to the temporary authorization for use in France [15, 16]. These ambulatory patients had mild to moderate COVID-19, were treated at the early stage of infection (less than 5 days postsymptoms onset), and did not require oxygen. Nasopharyngeal samples were collected before treatment (day 0), 3 and 7 days after infusion, and then weekly until the viral load became negative or reached the 31 cycle threshold (Ct).
SARS-CoV-2 RNA Detection and Quantification
Severe acute respiratory syndrome coronavirus 2 ribonucleic acid (RNA) was extracted from NP samples with the MGI Easy Nucleic Acid Extraction kits (MGI, Shenzhen, China) and quantified using the Ct values (N-gene) obtained with the TaqPath COVID-19 reverse-transcription polymerase chain reaction (RT-PCR) assay (Thermo Fisher Scientific, Waltham, MA) and digital-droplet-RT-PCR (Bio-Rad, Hercules, CA) as previously described [11]. In brief, viral loads were derived from the Ct values using a standard curve quantified by droplet digital (dd) RT-PCR (Bio-Rad). We prepared a standard curve from a clinical strain taken from a patient’s NP at the infection acute phase. Vero cells were infected with the virus suspension (6 days). The SARS-CoV-2 RNA in cell lysates was quantified simultaneously by the RT-ddPCR and TaqPath assay.
SARS-CoV-2 RNA Sequencing
Positive NP samples with N-gene Ct values <25 were sequenced using the PacBio single-molecule real-time sequencing (SMRT) system (Pacific Biosciences, Menlo Park, CA), as previously described [11, 17]. In brief, extracted RNAs were reverse transcribed using Superscript IV VILO MasterMix (Thermo Fisher Scientific) and amplified using primers targeting the S-protein (A6-Forward, ACAAATCCAATTCAGTTGTCTTCCTATTC; A6-Reverse, TGTGTACAAAAACTGCCATATTGCA). The A6-primers target a single long amplicon named A6, encoding the S1 domain of the spike protein.
These A6-amplicon sequences were amplified in a second PCR with barcoded primers, and the resulting material was sequenced by SMRT on a Sequel IIe system sequencer, according to the manufacturer’s instructions (Pacific Biosciences) [18]. Haplotypes obtained after data processing were aligned on the reference genome (GenBank accession number NC_045512.2) [19] to detect mutations associated with reduced mAb activity: Q409E, K417E/N/R/T, D420N, N439K, N440K, K444N/Q/T, V445A, G446S/V, Y453F, L455F, N460K/S/T/Y, G476S, T478K, E484A/D/K/Q, F486V, F490S, Q493K/R, S494P, Q498R, and N501Y [20–26]. The median of the spike sequencing depth was 3085 (interquartile range [IQR], 2021–3814) reads, with a 100% coverage of the A6-amplicon.
The Complexity of the Spike Quasispecies
The S-protein region was analyzed and reads accounting for more than 0.5% of the quasispecies were retained. We used abundance (Shannon entropy), incidence (number of observed haplotypes), and functional (population nucleotide diversity π) indices to obtain a multidimensional representation of virus quasispecies complexity [27]. The Shannon entropy, normalized to the number of reads HSN, gave a measure of diversity based on haplotype frequencies. The population nucleotide diversity (π) was used to measure the average number of nucleotide differences between 2 genomes in the quasispecies. Finally, we calculated the Hill numbers, qD, to measure the complexity with variation of the weight given to rare haplotypes (q = 0, q = 1, q = 2, q=∞) [27].
Statistical Analysis
We used a D’Agostino-Pearson normality test to determine whether the viral load values were normally distributed. Matched pairs were compared using a Wilcoxon matched-pairs signed rank test. Differences between the age and the viral loads of immunocompromised and non-immunocompromised patients were compared using the Mann-Whitney U test. The frequency of men between immunocompromised and non-immunocompromised patients were compared using Fisher’s exact test. A statistically significant difference was defined as a P < .05. Statistical analyses were performed with GraphPad Prism 8.0 (GraphPad Software, San Diego, CA).
Ethical Approval
These analyses were conducted as part of the national SARS-CoV-2 surveillance effort. According to French law (CSP Art.L1121-1.1), anonymous retrospective studies do not require institutional review board approval.
Patient Consent Statement
Data were analyzed using an anonymized database. Such a protocol does not require written informed consent according to French Public Health law (CSP Art L 1121-1.1).
RESULTS
Patients’ Characteristics
The 50 SARS-CoV-2-infected patients (19 [38%] women; median age, 55 years [range, 27–88 years]) were treated with Casirivimab-Imdevimab at the Toulouse University Hospital between June 28 and October 29, 2021. The majority of them (37 of 50, 74%) were immunocompromised, 18 with solid-organ-transplants (SOTs), another 4 (8%) patients were over 80, 3 (6%) had diabetes with a high body mass index, 2 (4%) had chronic kidney disease with hemodialysis, 2 (4%) had chronic lung diseases, 1 (2%) patient had type 1 diabetes, and 1 (2%) patient had a chronic granulomatous disease (Table 1, Table 2). SARS-CoV-2 RNA was detected by RT-PCR, and sequencing detected the B.1.617.2 (delta) variant in all the patients.
Table 1.
Clinical Characteristics of the Patients Treated With Casirivimab-Imdevimab
| Immunosuppression | 37 |
| Solid organ transplanta | 18 |
| Immunosuppressive treatmentb | 19 |
| Age >80 years | 4 |
| Other medical conditions | 9 |
| Chronic kidney disease | 2 |
| Chronic lung disease | 2 |
| Obesity (BMI >30) + Diabetes | 3 |
| Type 1 diabetes | 1 |
| Chronic granulomatous disease | 1 |
Abbreviations: BMI, body mass index.
Renal transplant (n = 11), cardiac transplant (n = 5), liver transplant (n = 2).
Anti-CD20 (n = 9), chemotherapy (n = 7), antitumor necrosis factor-α (n = 2), azathioprine (n = 1).
Table 2.
Comparison Between Immunocompromised and Nonimmunocompromised Set of Individuals With Regard to Age, Sex, and Baseline NP Viral Loada
| Immunocompromised (n = 37) | Nonimmunocompromised (n = 13) | P Value | |
|---|---|---|---|
| Age (years) | 55 [39–63]b | 53 [39–83.5]b | P = .76 |
| Sex (male%) | 57 | 77 | P = .32 |
| Baseline NP viral load (log10 copies/mL) |
8.23 [7.23–8.58]b | 3.27 [2.87–4.29]b | P < .001 |
Abbreviations: IQR, interquartile range; NP, nasopharyngeal.
Age and baseline NP viral load between immunocompromised and non-immunocompromised individuals were compared using the Mann-Whitney U test, the frequency of men was compared using Fisher’s exact test.
Median [IQR].
Impact of Casirivimab-Imdevimab on Nasopharyngeal Viral Load
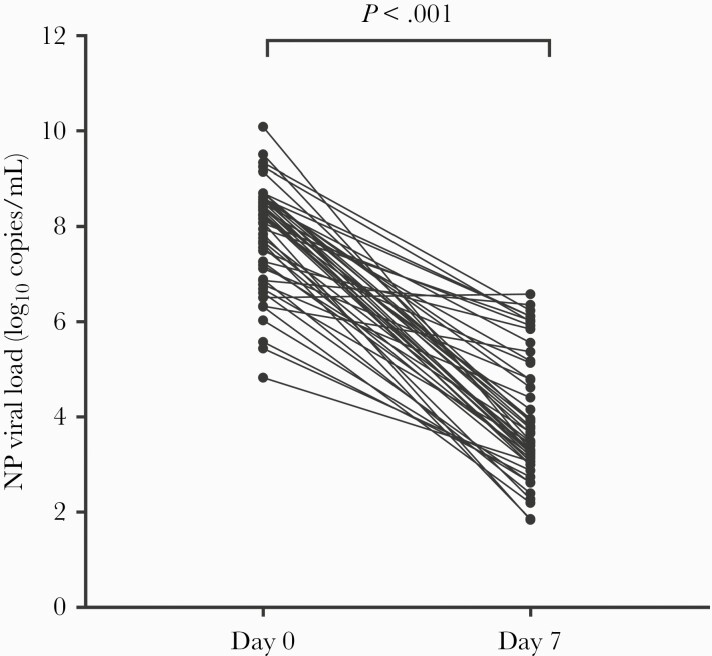
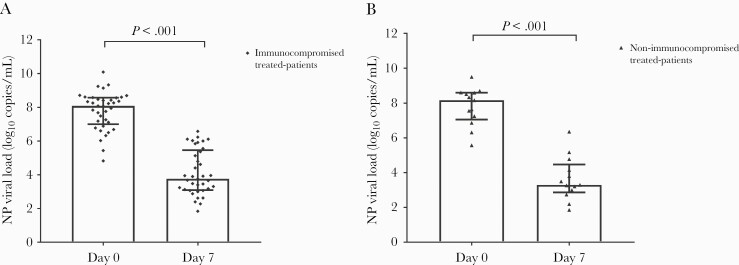
All 50 treated patients were monitored by RT-PCR with viral clearance measured over time. The NP RT-PCR N-gene values became negative or greater than 31 Ct a median of 14 days after infusion (range, 7–28 days). The median SARS-CoV-2 NP viral load decreased from 8.13 log10 copies/mL (IQR, 7.06–8.59 log10 copies/mL) on day 0 to 3.67 (IQR, 3.07–5.15 log10 copies/mL) on day 7 (P < .001) (Figure 1). The median difference in the NP viral loads before and 7 days postinfusion was 4.03 log10 copies/mL (IQR, 2.58–4.94 log10 copies/mL) (Figure 2). The NP viral load decreased by 3.99 log10 copies/mL (IQR, 2.52–4.97 log10 copies/mL) in the 37 immunocompromised patients (4.00 log10 copies/mL [IQR, 2.33–5.08 log10 copies/mL] in the 18 SOTs) and by 4.49 log10 copies/mL (IQR, 3.33–5.08 log10 copies/mL) in the other patients (P = .45). The NP viral loads of 3 of 50 (6%) patients had declined only slightly (<1 log10 copies/mL) 7 days after Casirivimab-Imdevimab infusion. The initial NP viral loads for those 3 individuals who showed slight decline in viral load on day 7 after Casirivimab-Imdevimab infusion were 6.51, 6.87, and 6.33 log10 copies/mL, respectively.
Figure 1.
Viral load trend in nasopharyngeal (NP) samples from Casirivimab-Imdevimab-treated patients: day 0 to day 7 (Wilcoxon matched-pairs signed-rank test, P < .001).
Figure 2.
Viral load trend in nasopharyngeal (NP) samples from immunocompromised (A) or non-immunocompromised (B) Casirivimab-Imdevimab-treated patients (medians + interquartile range, Wilcoxon matched-pairs signed rank test, P < .001).
Impact of Casirivimab-Imdevimab on SARS-CoV-2 Spike Quasispecies
We sequenced the spike protein of 36 (72%) patients to monitor the changes in their quasispecies. The NP viral loads from the remaining 14 patients were too low for sequencing (the viral loads of 6 patients were too low 3 days after mAbs infusion and too low in 8 patients 7 days after mAbs infusion).
Casirivimab-Imdevimab-Specific Key Mutations
None of the patients had any key mutation associated with reduced mAb activity on day 0. The SMRT sequencing indicated that the NP from 3 (8%) patients had several SARS-CoV-2 spike protein haplotypes on day 0: 2 had 2 spike haplotypes, and the third had 4 haplotypes.
The other 33 patients had only 1 spike haplotype on day 0. Nonkey Spike mutations observed on day 0 were T95I, Q628K, and Q477H.
On day 3 after Casirivimab-Imdevimab administration, new spike haplotypes were observed in NP samples from 7 patients compared to day 0, with nonkey Spike mutations L117P, T523I, A647V, and P665L.
On day 7, we found new spike haplotypes due to nonkey mutations E132K, K147R, N164S, F175S, E224Q, Q498H, and Q628K, in NP samples from 6 patients including 1 patient who showed NP viral loads slight decline (<1 log10 copies/mL) 7 days after Casirivimab-Imdevimab infusion.
Virus Quasispecies Complexity
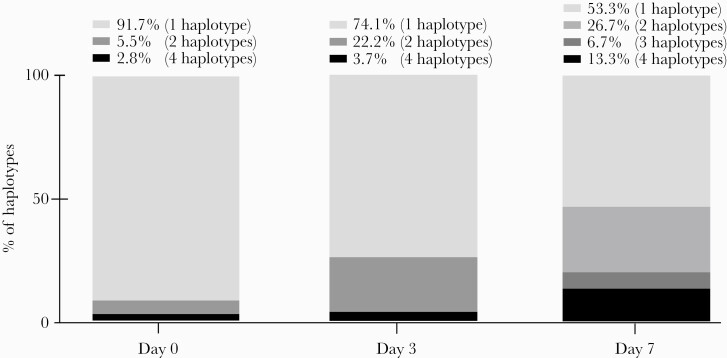
We analyzed the spike region of SARS-CoV-2 on day 0 before mAbs infusion and during follow-up to determine the complexity of the virus quasispecies: the number of spike-protein haplotypes in 16 (44%) patients varied over time (Table 3, Figure 3).
Table 3.
NP Spike Protein Haplotype Evolution in the Casirivimab-Imdevimab Treated Patients
| Number of Spike Protein Haplotypes | Patients (n = 36) | ||
|---|---|---|---|
| Day 0 | Day 3 | Day 7 | |
| 1 | 1 | ND/NC | 13 |
| 1 | 1 | 1 | 3 |
| 1 | ND/NC | 1 | 4 |
| 1 | 2 | ND | 6 |
| 1 | ND/NC | 2 | 2 |
| 1 | NC | 4 | 2 |
| 1 | 1 | 3 | 1 |
| 1 | 1 | 2 | 1 |
| 1 | 4 | ND | 1 |
| 2 | 1 | 1 | 1 |
| 2 | NC | 2 | 1 |
| 4 | 1 | ND | 1 |
Abbreviations: NC, not collected; ND, not detected; NP, nasopharyngeal.
Figure 3.
Percentages of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein haplotypes in nasopharyngeal samples from Casirivimab-Imdevimab-treated patients on days 0, 3, and 7. The number of SARS-CoV-2 spike protein haplotypes (2, 3, or 4 haplotypes) increased over time post-monoclonal antibodies infusion.
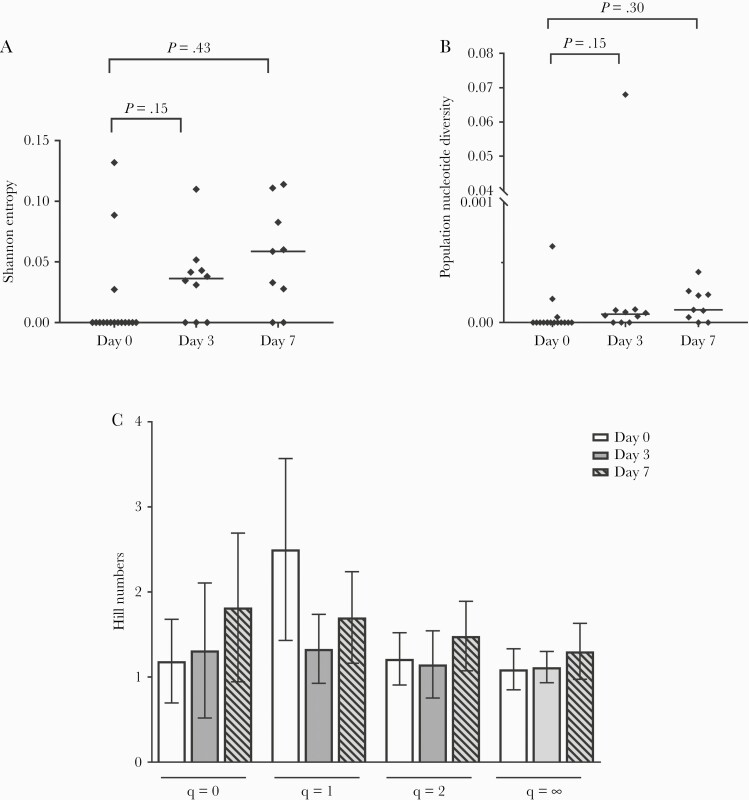
The median of the Shannon entropy, a measure of diversity based on haplotype frequencies, increased from 0.00 on day 0 to 0.036 3 days postinfusion and 0.059 7 days postinfusion (P > .05) (Figure 4A).
Figure 4.
(A) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike quasispecies Shannon entropy based on the S-protein region before (day 0) drug infusion and 3 and 7 days later. The horizontal line represents the median of the Shannon entropy for each group. (B) The SARS-CoV-2 spike quasispecies population nucleotide diversity (π) based on the S-protein region before (day 0) and 3 days and 7 days post-monoclonal antibodies (mAbs) infusion. The horizontal line represents the median of the population nucleotide diversity (π) for each group. (C) The SARS-CoV-2 spike quasispecies Hill numbers qD based on the S-protein region before mAbs (day 0) and 3 days and 7 days later. Four Hill numbers qD based on the frequency of each haplotype in the quasispecies with variations in the weight given to rare haplotypes (q = 0, q = 1, q = 2, q=∞) were calculated. Data are means + standard deviation.
The population nucleotide diversity index, a measure of the nucleotide difference between 2 sequences, increased slightly in the virus populations of 13 of 16 (81%) patients (on day 3 for 7 patients and on day 7 for 6 patients), and it decreased in 3 of 16 (19%) patients on day 7 (P > .05) (Figure 4B).
The Hill numbers, which measure virus complexity, indicated that there were slightly more SARS-CoV-2 Spike quasispecies, especially between positions 132 and 628 of the spike protein, surrounding the RBD, on day 7 postinfusion than on day 0, but the difference was not significant (Figure 4C). Although there were small changes in the Spike quasispecies after infusion of Casirivimab-Imdevimab, we detected no key mutation that reduced mAbs activity.
DISCUSSION
The use of neutralizing anti-SARS-CoV-2 mAbs raises questions about their impact on the emergence of new virus variants. This study indicates that in vivo treatment with Casirivimab-Imdevimab leads to small changes in the SARS-CoV-2 quasispecies but does not induce the emergence of mAb-resistant variants.
In the present study, the NP viral load decreased significantly (by 4.00 log10 copies/mL in the SOT patients and 4.49 log10 copies/mL in the other patients) 7 days after Casirivimab-Imdevimab infusion. This decrease was greater than the decrease observed in an historical group [11] of untreated SOT patients infected with the SARS-CoV-2 alpha variant (by 2.47 log10 copies/mL). The virus load of only 3 of 50 (6%) of the patients in the present study decreased slightly after 7 days (<1 log10 copies/mL). Among these 3 patients, 2 were immunocompromised (1 liver-transplanted-patient and 1 patient treated by anti-CD20), and 1 patient was 85 years. However, even if immune status or pharmacokinetics factors could explain a slower viral decay, their NP viral loads continued to decline and became negative after 14 to 28 days.
Ribonucleic acid viruses like SARS-CoV-2 exist as quasispecies, composed of several related genome sequences. Some haplotypes can acquire mutation and become resistant under conditions of selection such as those imposed by mAbs. We recently detected spike-protein key mutations (Q493R, Q493K, or E484K) in SARS-CoV-2 B.1.1.7 (alpha) variants in patients treated with Bamlanivimab and Etesevimab [11].
The impact of Casirivimab-Imdevimab on SARS-CoV-2 quasispecies evolution has not yet been clearly established. In vitro escape studies are inconsistent. Baum et al [23] showed that using these mAbs protected against the emergence of escape variants, whereas Starr et al [28] identified RBD mutations that enable the virus to escape Casirivimab-Imdevimab binding. A few in vivo studies have shown that this association remains potent against the B.1.617.2 variant and does not generate key mAb activity-reducing spike protein mutations [25]. It remains essential to evaluate the efficiency of mAb treatment in real life, including the pressures induced by the mAbs and by the patient himself. These mAbs are used to treat at-risk patients, whose clinical characteristics and/or treatment (immunosuppressive drugs) can exert strong selective pressure, virus persistence, and systemic disorders. Most (74%) of our patients were immunocompromised and hence likely to harbor variants with spike mutations. The 16 patients whose number of spike-protein haplotypes varied over time included 14 (88%) who were immunocompromised, including 7 SOT patients. This indicates that immunocompromised patients are more likely to have new spike variants and that using mAbs exerts selective pressure on SARS-CoV-2 quasispecies. Although 44% of the patients harbored variations in their SARS-CoV-2 quasispecies, we found no key mAb activity-reducing mutations.
A limitation of our study is the small number of samples sequenced due to the rapid decline in the SARS-CoV-2 NP viral load after mAbs infusion. This also led to a lack of statistical power in the measures of virus quasispecies complexity and diversity. However, we can show that Casivirimab-Imdevimab did not induce key mutation in a substantial number of immunocompromised patients, even if it exerted enough selective pressure to modify virus quasispecies diversity and complexity. Another limitation is the lack of a control group of untreated delta-infected patients, but we found a greater decrease in the viral load of treated SOT patients infected with this variant than in the historical group of untreated alpha-infected patients [11].
CONCLUSIONS
Finally, our results suggest that Casivirimab-Imdevimab protects against the emergence of mAbs-resistant variants. However, the Omicron variant (Pango lineage B.1.1.529), first identified in Bostwana and South Africa, could become dominant worldwide. Some recent data indicate that Casirivimab-Imdevimab does not neutralize this variant, which has many mutations, mostly in the N-terminal domain and the RBD of the spike protein [29, 30].
Acknowledgments
The English text was edited by Dr. Owen Parkes.
Author contributions. C. V. and J. I. designed the study. G. M.-B., P. D., N. K., A. D. B., and G. G. provided medical care to the participants and collected nasopharyngeal samples; C. V., P. T., and N. R. collected biological data; N. J. performed data processing; C. V. analyzed the results, prepared the figures, and performed statistical analysis; C. V. and J. I. drafted the initial version of the paper. All the authors revised the manuscript and approved the final version.
Financial support. Funding was provided by the Toulouse Institute for Infectious and Inflammatory Diseases (Infinity) (INSERM UMR1291, CNRS UMR5051, Toulouse III University) and National Agency for AIDS Research – Emerging Infectious Diseases (ANRS-MIE) (Emergen, Quasicov study).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Weinreich DM, Sivapalasingam S, Norton T, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med 2021; 384:238–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Razonable RR, Pawlowski C, O’Horo JC, et al. Casirivimab-Imdevimab treatment is associated with reduced rates of hospitalization among high-risk patients with mild to moderate coronavirus disease-19. EClinicalMedicine 2021; 40:101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020; 369:1010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tuccori M, Ferraro S, Convertino I, et al. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: clinical pipeline. mAbs 2020; 12:1854149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020; 367:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020; 581:215–20. [DOI] [PubMed] [Google Scholar]
- 7. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020; 581:221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen Z, Xiao Y, Kang L, et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin Infect Dis 2020; 71:713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li J, Du P, Yang L, et al. Two -step fitness selection for intra-host variations in SARS-CoV-2. Cell Rep 2022; 38:110205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choudhary MC, Crain CR, Qiu X, Hanage W, Li JZ.. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) sequence characteristics of coronavirus disease 2019 (COVID-19) persistence and reinfection. Clin Infect Dis 2022; 74:237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vellas C, Del Bello A, Debard A, et al. Influence of treatment with neutralizing monoclonal antibodies on the SARS-CoV-2 nasopharyngeal load and quasispecies. Clin Microbiol Infect 2022; 28:139.e5–139.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 2021; 19:409–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmann M, Hofmann-Winkler H, Krüger N, et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Reports 2021; 36:109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021; 596:276–80. [DOI] [PubMed] [Google Scholar]
- 15. French Agency for the Safety of Medicines and Health Products, Bamlanivimab/Etesevimab temporary authorisation for use. Available at: https://ansm.sante.fr/tableau-atu-rtu/bamlanivimab-700-mg-20-ml-35-mg-ml-solution-a-diluer-pour-perfusion-etesevimab-700-mg-20-ml-35-mg-ml-solution-a-diluer-pour-perfusion-en-association. Accessed 28 May 2021. [Google Scholar]
- 16. French Agency for the Safety of Medicines and Health Products, Casirivimab/Imdevimab temporary authorisation for use. Available at: https://ansm.sante.fr/tableau-atun/casirivimab-et-imdevimab-120-mg-ml-solution-a-diluer-pour-perfusion-intraveineuse-ou-solution-pour-injection-sous-cutanee. Accessed 7 March 2022. [Google Scholar]
- 17. Lhomme S, Latour J, Jeanne N, et al. Prediction of SARS-CoV-2 variant lineages using the S1-encoding region sequence obtained by PacBio single-molecule real-time sequencing. Viruses 2021; 13:2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pacifc Biosciences. Procedure & Checklist. Available at: https://www.pacb.com/wp-content/uploads/Procedure-Checklist-Preparing-SMRTbell-Libraries-using-PacBio-Barcoded-M13-Primers-for-Multiplex-SMRT-Sequencing.pdf. Accessed 7 March 2022. [Google Scholar]
- 19. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020; 579:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu C, Ginn HM, Dejnirattisai W, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021; 184:4220–36.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. US Food Drug and Administration. Fact Sheet For Health Care Providers Emergency Use Authorization (Eua) Of REGEN-COV (Casirivimab and Imdevimab). Available at: https://www.fda.gov/media/145611/download. Accessed 7 March 2022.
- 22. Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021; 593:130–5. [DOI] [PubMed] [Google Scholar]
- 23. Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020; 369:1014–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taylor PC, Adams AC, Hufford MM, de la Torre I, Winthrop K, Gottlieb RL.. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat Rev Immunol 2021; 21:382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Copin R, Baum A, Wloga E, et al. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell 2021; 184:3949–61.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim YJ, Jang US, Soh SM, Lee JY, Lee HR.. The impact on infectivity and neutralization efficiency of SARS-CoV-2 lineage B.1.351 pseudovirus. Viruses 2021; 13:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gregori J, Perales C, Rodriguez-Frias F, Esteban JI, Quer J, Domingo E.. Viral quasispecies complexity measures. Virology 2016; 493:227–37. [DOI] [PubMed] [Google Scholar]
- 28. Starr TN, Greaney AJ, Addetia A, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 2021; 371:850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022; 602:671–75. [DOI] [PubMed] [Google Scholar]
- 30. Wilhelm A, Widera M, Grikscheit K, et al. Reduced neutralization of SARS-CoV-2 omicron variant by vaccine sera and monoclonal antibodies. medRxiv 2021:2021.12.07.21267432. doi: 10.1101/2021.12.07.21267432 [DOI] [Google Scholar]