Abstract
Background
Multidrug- and methicillin-resistant staphylococci are both veterinary and public health concerns due to their zoonotic potential. Therefore, the objective of this study was to investigate patterns of antimicrobial, multidrug, and methicillin resistance among four Staphylococcus spp. commonly isolated from canine clinical specimens submitted to the Clinical Bacteriology Laboratory at the University of Tennessee College of Veterinary Medicine (UTCVM).
Methods
Results of antimicrobial susceptibility testing and mecA polymerase chain reaction (PCR) for isolates of four common Staphylococcus spp. isolates were obtained from the Bacteriology Laboratory at the UTCVM between 01/01/2006 and 12/31/2017. Cochran-Armitage trend test was used to assess temporal trends of antimicrobial resistance (AMR), multidrug resistance (MDR), and methicillin resistance. Kappa test of agreement was used to assess agreement between the results of PCR and disk diffusion tests.
Results
Most of the 7805 isolates were S. pseudintermedius (6453 isolates), followed by S. coagulans (860), S. aureus (330), and S. schleiferi (162). Among S. pseudintermedius isolates, 45.5% were MDR, and 30.8% were methicillin-resistant (MRSP). There was a significant temporal increase in MRSP (p = 0.017). Chloramphenicol resistance increased among both MRSP and methicillin-susceptible (MSSP) isolates (p < 0.0001). Among S. aureus isolates, 40.9% were MDR, 37.4% were methicillin-resistant (MRSA), and the proportion of MRSA isolates increased significantly (p = 0.0480) over time. There was an increasing temporal trend in the proportion of MDR isolates among MSSP (p = 0.0022), but a decrease among MRSP (p < 0.0001) and MRSA (p = 0.0298). S. schleiferi had the highest percentage (56.9%) of methicillin-resistant isolates. Oxacillin disk diffusion was superior to cefoxitin for the detection of mecA-mediated resistance and had almost perfect agreement with mecA PCR assay for S. pseudintermedius (95.4% agreement, kappa (κ) = 0.904; p < 0.0001), S. coagulans (95.6%, κ = 0.913; p < 0.0001) and S. schleiferi (97.7%, κ = 0.945; p < 0.0001). However, cefoxitin disk diffusion was superior to oxacillin disk diffusion and had almost perfect agreement with mecA PCR assay for S. aureus (95.3%, κ = 0.834; p < 0.0001).
Conclusions
The levels of resistance and increasing temporal trends are concerning. These findings have implications for treatment decisions and public health due to the zoonotic potential of staphylococci. Continued surveillance and use of antibiograms to guide clinical decisions will be critical.
Keywords: Antimicrobial resistance, Multidrug resistance, Methicillin resistance, Staphylococcus, MRSA, Epidemiology, Dogs, Canine, Tennessee, United States
Background
Staphylococcus bacteria are frequently implicated in opportunistic infections [1, 2]. Staphylococcus pseudintermedius is a commensal of canine skin and mucosa, and is commonly implicated in canine otitis and pyoderma [3–6]. It has also been identified as the causative agent of numerous other infections, including wound and surgical site infections, urinary tract infections, and osteomyelitis [7–10]. Carriage of Staphylococcus aureus, an important commensal of humans, has been documented in many domestic animals, and S. aureus may cause opportunistic infections in these species [11, 12]. Indeed, S. aureus has been isolated from various canine infections, although much less frequently in comparison to S. pseudintermedius [6, 7, 11, 13, 14]. Staphylococcus coagulans (formerly S. schleiferi subsp. coagulans) and coagulase-negative S. schleiferi (formerly S. schleiferi subsp. schleiferi) have been isolated from healthy dogs and those with inflammatory lesions alike, and these organisms have also been associated with recurrent pyoderma [15–19].
Numerous studies have reported that most canine Staphylococcus spp. isolates display resistance to at least one antimicrobial [6, 14, 20, 21]. Varying levels of multidrug resistance (MDR), defined as resistance to at least one drug in three or more classes of antimicrobials [22, 23], have also been detected among staphylococci isolated from companion animal specimens [14, 20, 21, 24, 25]. Moreover, methicillin resistance, which has received major attention in human medicine as the spread of hospital- and community-associated methicillin-resistant S. aureus (MRSA) challenges infection control efforts worldwide, is also increasingly being detected among canine Staphylococcus isolates, particularly S. pseudintermedius [26–31].
The occurrence of MDR and methicillin resistance among canine Staphylococcus isolates has important clinical implications. Importantly, infections with resistant isolates may require treatment with antimicrobials considered high priority for human health [32, 33]. Methicillin resistant staphylococci (MRS) express the mecA gene or its homolog, mecC, and produce an altered penicillin binding protein (PBP), which confers resistance to all β-lactam antibiotics, severely limiting treatment options [31, 34–36]. Therapeutic failures resulting from infections with resistant isolates also present serious clinical challenges to clinicians and increase the financial burdens for animal owners [37, 38]. Furthermore, evidence suggesting cross-transmission of resistant pathogens between animals and humans implies that the expansion of these organisms is also a public health concern [39–42]. S. pseudintermedius has been identified in human clinical infections, and previous reports suggest that pet owners are at risk of zoonotic transmission [43, 44]. In addition, while S. aureus is mainly a pathogen of humans, pets exposed to methicillin resistant S. aureus (MRSA) infections by their owners may play a role in perpetuating MRSA colonization in the household [45]. The potential for exposure to antimicrobial or multidrug resistant Staphylococcus isolates among those that are immune-compromised is of particular concern [46].
The existence of AMR, MDR, and methicillin resistance among canine Staphylococcus isolates, coupled with the public health implications of zoonotic transmission of these organisms, highlights the importance of monitoring antimicrobial susceptibility patterns. Temporal changes in antimicrobial resistance patterns among canine staphylococcal isolates imply that continued surveillance is essential to identify patterns, trends, and newly emerging resistance [14, 47]. Geographically relevant and timely epidemiologic monitoring is useful for the development of evidence-based antimicrobial use guidelines to inform veterinarians of current best practices. It is also important that surveillance data regarding regional AMR patterns for patients from a broad range of clinical settings, rather than specialty practices alone, are available to practitioners to enable informed antimicrobial selection when empirical therapy is necessary. Therefore, the objective of this study was to investigate patterns of antimicrobial, multidrug, and methicillin resistance among four clinically important Staphylococcus species (S. pseudintermedius, S. aureus, S. coagulans, and S. schleiferi) isolated from canine specimens submitted to the Clinical Bacteriology and Mycology Laboratory at the University of Tennessee College of Veterinary Medicine by both referring veterinarians and teaching hospital personnel between 2006 and 2017.
Methodology
Study design and data source
This descriptive retrospective study used laboratory records of canine clinical specimens processed at the Clinical Bacteriology Laboratory at the University of Tennessee College of Veterinary Medicine (UTCVM) between January 1, 2006 and December 31, 2017. Records for a total of 7810 canine clinical specimens positive for S. pseudintermedius, S. aureus, S. coagulans, or S. schleiferi were available for analysis. The data were assessed for duplicate entries and five were identified and removed, leaving 7805 unique isolates for subsequent analyses. The following data were extracted from the laboratory records: patient identification, medical record number, specimen collection site, Staphylococcus species isolated, results of antimicrobial susceptibility tests and mecA polymerase chain reaction (PCR) assay. Hospital patient records for the study period were also obtained from the UTCVM veterinary teaching hospital and matched to the laboratory records. The following data were extracted from the hospital records: medical record number, patient type (hospital patient specimen vs. submission by referring veterinarian), age, sex, species, and breed.
Bacterial isolation, antimicrobial susceptibility testing, and PCR assay
Bacterial isolation and antimicrobial susceptibility tests were performed according to standard practice using previously described methods [47]. Briefly, Staphylococcus spp. isolates were identified with a biochemical identification process, using the following tests: tube coagulase, phenol red broth with lactose and with trehalose, and the Voges-Proskauer test [48]. The diagnostic laboratory followed Clinical and Laboratory Standards Institute (CLSI) standards for antibiotic susceptibility testing [49–53]. Antimicrobial susceptibility tests for the majority of bacterial isolates were performed using the disk diffusion method. However, some were tested using a microbroth dilution method using an automated susceptibility testing system [47, 54]. Antimicrobial agents from the following classes were included in the standard antimicrobial susceptibility panel for Staphylococcus spp.: β-lactams (ampicillin, amoxicillin/clavulanic acid, cefoxitin, cefpodoxime, cephalothin, oxacillin and penicillin), fluoroquinolones (marbofloxacin), folate inhibitors (trimethoprim/sulfamethoxazole [TMS]), lincosamides (clindamycin), macrolides (erythromycin), phenicols (chloramphenicol), and tetracyclines (tetracycline). Reporting of susceptibility to β-lactam antibiotics in the present study was limited to ampicillin, oxacillin, and cefoxitin. Ampicillin (rather than penicillin) was used to represent susceptibility to penicillinase-labile penicillins in this study because the results of ampicillin testing were available for a larger number of Staphylococcus spp. isolates. For isolates deemed to be resistant to multiple drug classes, susceptibility tests for the following agents were performed in addition to the standard panel: amikacin, doxycycline, minocycline, and rifampin. Amikacin susceptibility tests were also performed for all cutaneous isolates, while gentamicin was used for otic and urinary isolates.
Oxacillin zone sizes of ≤17 mm (resistant) and ≥ 18 mm (susceptible) were used as breakpoints for classification of S. pseudintermedius, S. coagulans, and S. schleiferi [55, 56]. In addition, interpretive criteria recommended for coagulase-negative Staphylococcus spp. for cefoxitin were followed for S. pseudintermedius, S. coagulans, and S. schleiferi [49–53]. Otherwise, the laboratory followed CLSI standards in place during the year of specimen submission, and isolates were classified as susceptible, intermediate, or resistant based on these criteria [49–53]. During 2006, a real-time PCR assay was used for the detection of the mecA gene for all Staphylococcus spp. isolates, with a cycle threshold value of ≤30 being considered positive [47, 57]. During all remaining years of the study period, conventional PCR was employed for this purpose, using previously described methods [55]. After 2006, mecA PCR was performed for all staphylococcal isolates resistant to oxacillin or cefoxitin, as well as for 100 oxacillin-susceptible S. pseudintermedius isolates annually.
Data management
Data management and statistical analysis were performed using SAS Version 9.4 [58]. Medical record numbers were used to join patient data extracted from hospital records to the laboratory records. Patient age was categorized into the following groups: < 3 years of age, 3–6 years, 6–9 years, 9–12 years, and ≥ 12 years. Patient sex categories included: intact male, castrated male, intact female, and spayed female. Patient breed was placed into one of the following categories based on American Kennel Club (AKC) designations: herding, hound, toy, non-sporting, sporting, terrier, and working breed groups [59]. Patients with multiple breeds were coded as “mixed breed,” and those with a listed breed not recognized by the AKC were coded as “other”. Specimen collection sites were categorized as ear, skin, urine or bladder, joint or bone, mucosa, and “other”.
Results of antimicrobial susceptibility tests were excluded from analysis if differing interpretations were obtained upon repeated testing. Thus, results for the following agents were excluded from analysis: amikacin (2 isolates), oxacillin (6 isolates), cefoxitin (2 isolates), and cefpodoxime (2 isolates). Results of mecA PCR assay were excluded from analysis for 14 isolates for which repeated test results had differing interpretations. Therefore, results of mecA PCR assay were included for a total of 4152 isolates. All antimicrobial susceptibility test results were re-classified into two groups, susceptible and resistant, with the latter encompassing all non-susceptible isolates (those classified as “non-susceptible,” “intermediate,” or “resistant”) [23].
The antimicrobial agents used for susceptibility testing were categorized into the appropriate drug classes. Isolates that were non-susceptible to at least one agent in one or more antimicrobial classes, excluding intrinsic resistance, were classified as AMR [22, 23]. Isolates that were non-susceptible to one or more agents in 3 or more antimicrobial classes, excluding intrinsic resistance, were classified as MDR [22, 23]. Isolates were classified as methicillin-resistant based upon results of phenotypic susceptibility testing, confirmed with positive PCR results for the mecA gene.
Statistical analysis
The Kolmogorov-Smirnov test was used to assess for normality of distribution of patient age, which was found to be non-normally distributed and hence median and interquartile range (IQR) were computed. The following categorical variables were assessed: age group, patient breed category, patient sex, patient type, and specimen collection site. The Cochran-Armitage trend test was used to assess for significant temporal trends in AMR, MDR, and methicillin resistance, using a two-sided p-value with a cutoff of ≤0.05. This analysis was performed for S. pseudintermedius, S. aureus, S. coagulans, and S. schleiferi isolates. In addition, separate analyses were conducted for methicillin-resistant and methicillin-susceptible S. pseudintermedius (MRSP and MSSP) and S. aureus (MRSA and MSSA) isolates. To facilitate comparison between methicillin-resistant and methicillin-susceptible isolates, temporal trends in antimicrobial and multidrug resistance were displayed in graphs, which were generated using R version 4.1.1 [60]. Chi-square tests (or Fisher’s exact tests where appropriate due to sample size) were used to assess for significant differences in levels of antimicrobial and multidrug resistance between methicillin-resistant and methicillin-susceptible isolates. Kappa tests of agreement were used to assess agreement between the results of mecA PCR assay and oxacillin as well as cefoxitin susceptibility tests.
Results
Summary statistics
A total of 7805 canine clinical specimens with single isolates that met the inclusion criteria for the study were assessed. The most common species was S. pseudintermedius, with 6453 isolates, followed by S. coagulans (860 isolates), S. aureus (330 isolates), and S. schleiferi (162 isolates).
Most Staphylococcus spp. isolates included in the study were from the skin (56.2%), followed by ear (16.8%), urine or bladder (13.3%), joint or bone (4.5%), and mucosa (1.6%) (Table 1). Patient age ranged from 2 days to 18 years, with a median of 7 years and IQR of 4 to 10 years. The largest percentage of isolates were from spayed females (46%), followed by neutered males (38.5%), intact males (10.5%), and intact females (5.1%). Sporting breeds had the highest proportion (20.2%) of isolates, followed by mixed breeds (19.3%), toy (15.2%), working (11.1%), non-sporting (9.5%), terrier (9.2%), herding (8.4%), and hound breeds (6.9%). The majority of Staphylococcus spp. isolates were obtained from specimens submitted by referring veterinarians (59.1%), while 40.9% were from hospital patients.
Table 1.
Distribution of Staphylococcus spp. isolated from canine specimens submitted to a veterinary diagnostic laboratory in Tennessee, USA (2006–2017)
| Variable | Category | Number | Percentage | p-value* |
|---|---|---|---|---|
| Specimen Site | < 0.0001 | |||
| Ear | 1310/7804 | 16.8 | ||
| Joint/bone | 354/7804 | 4.5 | ||
| Mucosa | 122/7804 | 1.6 | ||
| Skin | 4387/7804 | 56.2 | ||
| Urine/bladder | 1041/7804 | 13.3 | ||
| Other | 590/7804 | 7.6 | ||
| Age | < 0.0001 | |||
| < 3 years | 639/4785 | 13.4 | ||
| 3 ⎯ < 6 years | 1046/4785 | 21.9 | ||
| 6 ⎯ < 9 years | 1309/4785 | 27.4 | ||
| 9 ⎯ < 12 years | 1171/4785 | 24.5 | ||
| ≥ 12 years | 620/4785 | 13.0 | ||
| Sex | 0.0098 | |||
| Female | 305/5989 | 5.1 | ||
| Female spayed | 2752/5989 | 46.0 | ||
| Male | 629/5989 | 10.5 | ||
| Male castrated | 2303/5989 | 38.5 | ||
| Breed | < 0.0001 | |||
| Herding | 492/5864 | 8.4 | ||
| Hound | 404/5864 | 6.9 | ||
| Mixed | 1129/5864 | 19.3 | ||
| Non-sporting | 559/5864 | 9.5 | ||
| Sporting | 1186/5864 | 20.2 | ||
| Terrier | 540/5864 | 9.2 | ||
| Toy | 890/5864 | 15.2 | ||
| Working | 649/5864 | 11.1 | ||
| Other | 15/5864 | 0.3 | ||
| Patient Type | < 0.0001 | |||
| Clinical | 3149/7705 | 40.9 | ||
| Referral | 4556/7705 | 59.1 |
*p-value is for Chi-square test
Antimicrobial and multidrug resistance patterns
S. pseudintermedius
The majority of S. pseudintermedius isolates were resistant to ampicillin (84.3%) (Table 2). Substantial proportions of S. pseudintermedius isolates exhibited resistance to gentamicin (27.0%), TMS (44.4%), clindamycin (39.1%), erythromycin (38.8%), and tetracycline (47.9%), while overall resistance to chloramphenicol was low (5.4%). Approximately one-third were methicillin-resistant (30.8%), and 45.5% were MDR. There was a statistically significant (p = 0.0170) increasing temporal trend in the proportion of methicillin-resistant isolates. Among the subsets of isolates tested for susceptibility to amikacin (cutaneous and MDR isolates) and rifampin (MDR isolates), few exhibited resistance to amikacin (20/2558) or rifampin (10/1026), and no significant temporal trends were observed for these agents.
Table 2.
Distribution of antimicrobial and multidrug resistance among S. pseudintermedius isolated from canine specimens submitted to a veterinary diagnostic laboratory in Tennessee, USA (2006–2017)
| 2006–2008 | 2009–2011 | 2012–2014 | 2015–2017 | Total | |
|---|---|---|---|---|---|
| Aminoglycosides | |||||
| Gentamicin | 19.3 (242/1256) | 31.7 (482/1523) | 29.2 (297/1016) | 27.4 (241/881) | 27.0 (1262/4676)*** |
| β-lactams | |||||
| Ampicillin | 84.7 (1020/1204) | 83.9 (1277/1523) | 84.5 (1566/1854) | 84.2 (1499/1781) | 84.3 (5362/6362) |
| Cefoxitin | 21.7 (272/1251) | 19.9 (301/1510) | 19.0 (353/1855) | 22.4 (396/1768) | 20.7 (1322/6384) |
| Oxacillin | 32.6 (411/1259) | 36.9 (559/1514) | 34.3 (638/1861) | 34.8 (622/1790) | 34.7 (2230/6424) |
| Fluoroquinolones | |||||
| Marbofloxacin | 31 (385/1244) | 30.1 (453/1504) | 28.3 (523/1848) | 29.1 (412/1417) | 29.5 (1773/6013) |
| Folate inhibitors | |||||
| TMSa | 42.1 (529/1258) | 42.7 (649/1521) | 42.5 (791/1863) | 49.5 (894/1805) | 44.4 (2863/6447)*** |
| Lincosamides | |||||
| Clindamycin | 38.9 (489/1256) | 40.3 (613/1522) | 39.1 (727/1859) | 38.2 (688/1801) | 39.1 (2517/6438) |
| Macrolides | |||||
| Erythromycin | 38.4 (483/1257) | 39.7 (604/1522) | 39.1 (726/1858) | 38.0 (685/1802) | 38.8 (2498/6439) |
| Phenicols | |||||
| Chloramphenicol | 2.5 (31/1258) | 3.3 (50/1523) | 5.8 (107/1862) | 8.7 (157/1805) | 5.4 (345/6448)*** |
| Tetracyclines | |||||
| Tetracycline | 52.4 (659/1257) | 50.1 (762/1521) | 45.9 (855/1862) | 44.8 (797/1780) | 47.9 (3073/6420)*** |
| AMRb | 87.1 (1096/1259) | 86.8 (1322/1523) | 86.5 (1611/1863) | 86.7 (1567/1808) | 86.7 (5596/6453) |
| MDRc | 44.2 (557/1259) | 45.8 (697/1523) | 45.1 (840/1863) | 46.4 (839/1808) | 45.5 (2933/6453) |
| MRSPd | 26.1 (300/1148) | 32.5 (479/1473) | 31.7 (585/1844) | 31.5 (544/1730) | 30.8 (1908/6195)* |
aTrimethoprim/sulfamethoxazole
bAntimicrobial resistance
cMultidrug resistance
dMethicillin-resistant S. pseudintermedius, based on oxacillin disk diffusion testing confirmed with positive mecA PCR
*p < 0.05, **p < 0.01, ***p < 0.0001 for Cochran-Armitage trend test
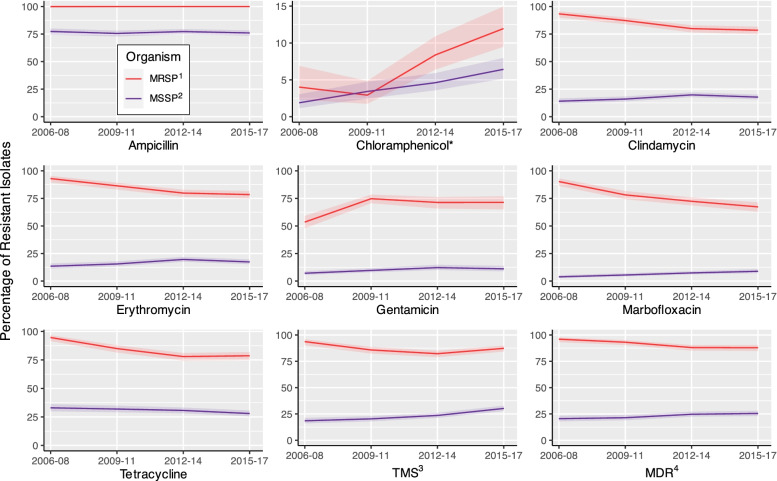
The MRSP isolates consistently had higher overall levels of antimicrobial and multidrug resistance than the MSSP isolates (Fig. 1). Statistically significant (p < 0.0001) differences were noted for ampicillin, chloramphenicol, clindamycin, erythromycin, gentamicin, marbofloxacin, TMS, and MDR. Agents with significant temporal increases among both MRSP and MSSP included gentamicin (pMRSP = 0.0001, pMSSP = 0.0019) and chloramphenicol (p < 0.0001), while tetracycline resistance decreased for both MRSP and MSSP (pMRSP < 0.0001, pMSSP = 0.0111). In contrast, MDR decreased significantly among MRSP isolates (p < 0.0001) but increased among MSSP isolates (p = 0.0022), and a similar pattern was observed for several individual antimicrobial agents: marbofloxacin (p < 0.0001), TMS (pMRSP = 0.0212, pMSSP < 0.0001), clindamycin (pMRSP < 0.0001, pMSSP = 0.007), and erythromycin (pMRSP < 0.0001, pMSSP = 0.0042).
Fig. 1.
Percentage and 95% confidence intervals of antimicrobial and multidrug resistance among methicillin-resistant and methicillin-susceptible S. pseudintermedius isolated from canine specimens submitted to a veterinary diagnostic laboratory in Tennessee, USA (2006–2017). 1Methicillin-resistant S. pseudintermedius, 2Methicillin-susceptible S. pseudintermedius, 3Trimethoprim/sulfamethoxazole, 4Multidrug resistance. *Indicates that upper limit of y-axis has been reduced to improve visual comparison
-
b)
S. aureus
Resistance to ampicillin was observed among the majority (82.6%) of S. aureus isolates (Table 3). Low levels of resistance were observed for TMS (4.2%), chloramphenicol (2.4%), gentamicin (1.1%), and tetracycline (6.4%), while resistance to clindamycin and erythromycin were relatively high (35.4 and 47.3%, respectively). The overall percentage of MRSA isolates was 37.4%, and increased significantly during the study period (p = 0.0480), from 21.6 to 40%. In addition, 40.9% of S. aureus isolates were MDR. Among the subsets of isolates tested for susceptibility to amikacin and rifampin, several exhibited resistance to amikacin (12/78), but none were resistant to rifampin (0/21). No significant temporal trends were observed for these agents.
Table 3.
Distribution of antimicrobial and multidrug resistance among S. aureus isolated from canine specimens submitted to a veterinary diagnostic laboratory in Tennessee, USA (2006–2017)
| Antimicrobials | 2006–2008 | 2009–2011 | 2012–2014 | 2015–2017 | Total |
|---|---|---|---|---|---|
| Aminoglycosides | |||||
| Gentamicin | 2.5 (2/79) | 1.2 (1/86) | 0 (0/63) | 0 (0/40) | 1.1 (3/268) |
| β-lactams | |||||
| Ampicillin | 83.6 (61/73) | 91.9 (79/86) | 74.2 (69/93) | 81.4 (57/70) | 82.6 (266/322) |
| Cefoxitin | 25.6 (20/78) | 49.4 (42/85) | 42.4 (39/92) | 45.7 (32/70) | 40.9 (133/325)* |
| Oxacillin | 27.5 (22/80) | 52.3 (45/86) | 40.2 (37/92) | 40 (28/70) | 40.2 (132/328) |
| Fluoroquinolones | |||||
| Marbofloxacin | 23.4 (18/77) | 43.5 (37/85) | 28.6 (26/91) | 40.7 (22/54) | 33.6 (103/307) |
| Folate inhibitors | |||||
| TMSa | 3.8 (3/80) | 3.5 (3/86) | 2.2 (2/93) | 8.5 (6/71) | 4.2 (14/330) |
| Lincosamides | |||||
| Clindamycin | 33.8 (27/80) | 44.2 (38/86) | 33.3 (31/93) | 29.0 (20/69) | 35.4 (116/328) |
| Macrolides | |||||
| Erythromycin | 33.8 (27/80) | 55.8 (48/86) | 48.4 (45/93) | 50.7 (36/71) | 47.3 (156/330) |
| Phenicols | |||||
| Chloramphenicol | 3.8 (3/80) | 2.3 (2/86) | 2.2 (2/93) | 1.4 (1/71) | 2.4 (8/330) |
| Tetracyclines | |||||
| Tetracycline | 5.0 (4/80) | 7.0 (6/86) | 5.4 (5/93) | 8.7 (6/69) | 6.4 (21/328) |
| AMRb | 93.8 (75/80) | 95.4 (82/86) | 89.3 (83/93) | 93.0 (66/71) | 92.7 (306/330) |
| MDRc | 35.0 (28/80) | 53.5 (46/86) | 35.5 (33/93) | 39.4 (28/71) | 40.9 (135/330) |
| MRSAd | 21.6 (16/74) | 45.8 (38/83) | 40.7 (37/91) | 40.0 (26/65) | 37.4 (117/313)* |
aTrimethoprim/sulfamethoxazole
bAntimicrobial resistance
cMultidrug resistance
dMethicillin-resistant S. aureus, based on cefoxitin disk diffusion testing confirmed with positive mecA PCR
*p < 0.05, **p < 0.01, ***p < 0.0001 for Cochran-Armitage trend test
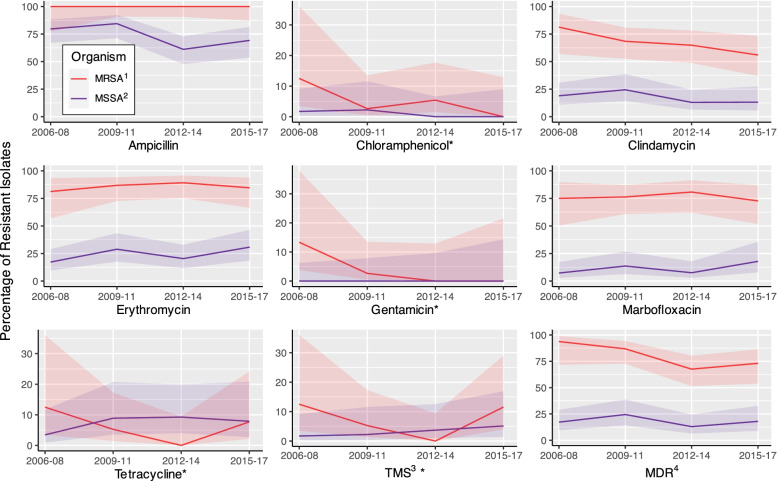
A significantly (p < 0.0001) higher proportion of MRSA isolates were MDR compared to MSSA (Fig. 2). Similarly, significant differences between MRSA and MSSA were identified for ampicillin (p < 0.0001), clindamycin (p < 0.0001), erythromycin (p < 0.0001), gentamicin (p = 0.0475), and marbofloxacin (p < 0.0001). There were significant decreases in the proportion of MRSA isolates resistant to gentamicin (p = 0.0400) and the proportion of MSSA isolates resistant to ampicillin (p = 0.0485) during the study period. In addition, the percentage of MDR MRSA isolates decreased significantly (p = 0.0298), from 93.8 to 73.1%.
Fig. 2.
Percentage and 95% confidence intervals of antimicrobial and multidrug resistance among methicillin-resistant and methicillin-susceptible S. aureus isolated from canine specimens submitted to a veterinary diagnostic laboratory in Tennessee, USA (2006–2017). 1Methicillin-resistant S. aureus, 2Methicillin-susceptible S. aureus, 3Trimethoprim/sulfamethoxazole, 4Multidrug resistance. *Indicates that upper limit of y-axis has been reduced to improve visual comparison
-
iii)
S. coagulans
Ampicillin (40.9%) resistance among S. coagulans isolates was comparatively lower than that of S. pseudintermedius and S. aureus (Table 4). Resistance to gentamicin (36.2%) and marbofloxacin (37.7%) were relatively common among S. coagulans isolates, while resistance to TMS, clindamycin, erythromycin, and tetracycline were rarely observed, and none exhibited resistance to chloramphenicol. Among the subsets of isolates tested for susceptibility to amikacin (232 isolates) and rifampin (28 isolates), none exhibited resistance.
Table 4.
Distribution of antimicrobial and multidrug resistance among S. coagulans isolated from canine specimens submitted to a veterinary diagnostic laboratory in Tennessee, USA (2006–2017)
| Antimicrobial | 2006–2008 | 2009–2011 | 2012–2014 | 2015–2017 | Total |
|---|---|---|---|---|---|
| Aminoglycosides | |||||
| Gentamicin | 35.5 (67/194) | 45.6 (93/204) | 33.1 (48/145) | 26.3 (30/114) | 36.2 (238/657) |
| β-lactams | |||||
| Ampicillin | 40.1 (75/187) | 44.1 (90/204) | 41.5 (107/258) | 37.6 (76/202) | 40.9 (348/851) |
| Cefoxitin | 21.7 (42/194) | 26.1 (53/203) | 20.6 (53/257) | 18.7 (38/203) | 21.7 (186/857) |
| Oxacillin | 35.6 (69/194) | 41.2 (84/204) | 40.9 (105/257) | 31.0 (63/203) | 37.4 (321/858) |
| Fluoroquinolones | |||||
| Marbofloxacin | 30.9 (59/191) | 44 (88/200) | 41.1 (106/258) | 33.3 (51/153) | 37.7 (304/802) |
| Folate inhibitors | |||||
| TMSa | 0.52 (1/194) | 0.49 (1/204) | 2.33 (6/257) | 0.98 (2/204) | 1.2 (10/859) |
| Lincosamides | |||||
| Clindamycin | 4.6 (9/194) | 3.4 (7/204) | 2.3 (6/256) | 3.4 (7/204) | 3.4 (29/858) |
| Macrolides | |||||
| Erythromycin | 4.1 (8/194) | 2.9 (6/204) | 2.7 (7/258) | 2.9 (6/204) | 3.1 (27/860) |
| Phenicols | |||||
| Chloramphenicol | 0 (0/194) | 0 (0/204) | 0 (0/257) | 0 (0/204) | 0 (0/859) |
| Tetracyclines | |||||
| Tetracycline | 1.6 (3/194) | 2.0 (4/204) | 3.1 (8/258) | 3.0 (6/203) | 2.4 (21/859) |
| AMRb | 56.7 (110/194) | 64.2 (131/204) | 57.0 (147/258) | 50.0 (102/204) | 57.0 (490/860) |
| MDRc | 21.7 (42/194) | 30.9 (63/204) | 16.7 (43/258) | 11.8 (24/204) | 20.0 (172/860)** |
| MRSCd | 25.2 (42/167) | 35.2 (70/199) | 39.2 (100/255) | 25.8 (49/190) | 32.2 (261/811) |
aTrimethoprim/sulfamethoxazole
bAntimicrobial resistance
cMultidrug resistance
dMethicillin-resistant S. coagulans, based on oxacillin disk diffusion testing confirmed with positive mecA PCR
*p < 0.05, **p < 0.01, ***p < 0.0001 for Cochran-Armitage trend test
The percentages of AMR (57%) and MDR (20%) S. coagulans isolates were much lower than those for S. pseudintermedius and S. aureus. Furthermore, MDR showed a significant (p = 0.0004) decreasing temporal trend, with the percentage of resistant isolates changing from 21.7% at the beginning of the study period to 11.8% at the end. On the other hand, overall methicillin resistance was similar to S. pseudintermedius and S. aureus (32.2%), but did not exhibit any significant temporal trends.
-
iv)
S. schleiferi
Similar to findings for S. coagulans, S. schleiferi isolates also exhibited a lower level of resistance to ampicillin (56.5%) than S. pseudintermedius and S. aureus (Table 5). However, S. schleiferi isolates had the highest level of methicillin resistance of all species in this study (56.9%). Approximately half of the S. schleiferi isolates showed resistance to gentamicin (45.5%) and marbofloxacin (48%), while resistance to other drugs, including TMS (2.5%), clindamycin (8.6%), erythromycin (8.6%), and tetracycline (6.8%) was much less common. However, resistance to tetracycline did increase significantly during the course of the study period (p = 0.0217). Among the subsets of isolates tested for susceptibility to amikacin (56 isolates) and rifampin (7 isolates), resistance to amikacin was observed in a single isolate, and none were resistant to rifampin.
Table 5.
Distribution of antimicrobial and multidrug resistance among S. schleiferi isolated from canine specimens submitted to a veterinary diagnostic laboratory in Tennessee, USA (2006–2017)
| Antimicrobial | 2006–2008 | 2009–2011 | 2012–2014 | 2015–2017 | Total |
|---|---|---|---|---|---|
| Aminoglycosides | |||||
| Gentamicin | 41.03 (16/39) | 48.3 (14/29) | 50.0 (11/22) | 45.5 (10/22) | 45.5 (51/112) |
| β-lactams | |||||
| Ampicillin | 60.5 (23/38) | 55.2 (16/29) | 45.0 (18/40) | 63.0 (34/54) | 56.5 (91/161) |
| Cefoxitin | 20.5 (8/39) | 17.2 (5/29) | 32.5 (13/40) | 30.2 (16/53) | 26.1 (42/161) |
| Oxacillin | 66.7 (26/39) | 65.5 (19/29) | 52.5 (21/40) | 57.4 (31/54) | 59.9 (97/162) |
| Fluoroquinolones | |||||
| Marbofloxacin | 39.5 (15/38) | 51.7 (15/29) | 51.3 (20/39) | 50 (23/46) | 48.0 (73/152) |
| Folate inhibitors | |||||
| TMSa | 0 (0/39) | 3.5 (1/29) | 2.5 (1/40) | 3.7 (2/54) | 2.5 (4/162) |
| Lincosamides | |||||
| Clindamycin | 10.3 (4/39) | 6.9 (2/29) | 0 (0/40) | 14.8 (8/54) | 8.6 (14/162) |
| Macrolides | |||||
| Erythromycin | 10.3 (4/39) | 6.9 (2/29) | 0 (0/40) | 14.8 (8/54) | 8.6 (14/162) |
| Phenicols | |||||
| Chloramphenicol | 0 (0/39) | 3.5 (1/29) | 0 (0/40) | 0 (0/54) | 0.6 (1/162) |
| Tetracyclines | |||||
| Tetracycline | 0 (0/39) | 6.9 (2/29) | 5 (2/40) | 13.0 (7/54) | 6.8 (11/162)* |
| AMRb | 84.6 (33/39) | 82.8 (24/29) | 72.5 (29/40) | 72.2 (39/54) | 77.2 (125/162) |
| MDRc | 28.2 (11/39) | 34.5 (10/29) | 20.0 (8/40) | 27.8 (15/54) | 27.2 (44/162) |
| MRSSd | 61.8 (21/34) | 64.3 (18/28) | 48.7 (19/39) | 55.8 (29/52) | 56.9 (87/153) |
aTrimethoprim/sulfamethoxazole
bAntimicrobial resistance
cMultidrug resistance
dMethicillin-resistant S. schleiferi, based on oxacillin disk diffusion testing confirmed with positive mecA PCR
*p < 0.05, **p < 0.01, ***p < 0.0001 for Cochran-Armitage trend test
The majority of S. schleiferi isolates were resistant to at least one antimicrobial (77.2%), but MDR was again comparatively lower than the other Staphylococcus species (27.2%). Methicillin resistance fluctuated but did not exhibit any significant temporal trends.
Methicillin resistance and mecA gene detection
S. pseudintermedius
A total of 97.4% of the cefoxitin-resistant S. pseudintermedius isolates (based on disk diffusion test) were positive for the mecA gene (based on PCR assay), while only 62.1% of cefoxitin-susceptible isolates were negative for mecA (Table 6). Overall, there was 74.9% agreement between cefoxitin disk diffusion testing and mecA PCR assay. The kappa test of agreement indicated moderate agreement (kappa = 0.522; p < 0.0001) between the two methods. In contrast, oxacillin disk diffusion test had almost perfect agreement with mecA PCR (95.4% agreement; κ = 0.904; p < 0.0001).
-
b)
S. aureus
Table 6.
Comparison of the results of mecA PCR and cefoxitin/oxacillin resistance among Staphylococcus spp. isolated from canine specimens submitted to a veterinary diagnostic laboratory in Tennessee, USA (2006–2017)
| Organism | Drug | Interpretation | mecA positive | mecA negative | Percent | κa | 95% CIb | p-value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Percent | Number | Percent | Number | agreement | |||||||
| S. pseudintermedius | Cefoxitin | Resistant | 97.4 | 1160/1191 | 2.6 | 31/1191 | 74.9% | 0.522 | 0.497 | 0.548 | < 0.0001 |
| Susceptible | 37.9 | 797/2104 | 62.1 | 1307/2104 | |||||||
| Oxacillin | Resistant | 95.4 | 1908/2001 | 4.7 | 93/2001 | 95.4% | 0.904 | 0.889 | 0.919 | < 0.0001 | |
| Susceptible | 4.6 | 60/1311 | 95.4 | 1251/1311 | |||||||
| S. aureus | Cefoxitin | Resistant | 96.7 | 117/121 | 3.3 | 4/121 | 95.3% | 0.834 | 0.714 | 0.953 | < 0.0001 |
| Susceptible | 12.0 | 3/25 | 88.0 | 22/25 | |||||||
| Oxacillin | Resistant | 95.9 | 117/122 | 4.1 | 5/122 | 94.0% | 0.787 | 0.653 | 0.920 | < 0.0001 | |
| Susceptible | 16.0 | 4/25 | 84.0 | 21/25 | |||||||
| S. coagulans | Cefoxitin | Resistant | 94.4 | 151/160 | 5.6 | 9/160 | 76.3% | 0.525 | 0.460 | 0.590 | < 0.0001 |
| Susceptible | 31.1 | 121/389 | 68.9 | 268/389 | |||||||
| Oxacillin | Resistant | 95.3 | 261/274 | 4.7 | 13/274 | 95.6% | 0.913 | 0.878 | 0.947 | < 0.0001 | |
| Susceptible | 4.0 | 11/275 | 96.0 | 264/275 | |||||||
| S. schleiferi | Cefoxitin | Resistant | 97.4 | 37/38 | 2.6 | 1/38 | 59.1% | 0.291 | 0.182 | 0.400 | < 0.0001 |
| Susceptible | 57.3 | 51/89 | 42.7 | 38/89 | |||||||
| Oxacillin | Resistant | 98.9 | 87/88 | 1.1 | 1/88 | 97.7% | 0.945 | 0.884 | 1.000 | < 0.0001 | |
| Susceptible | 5.0 | 2/40 | 95.0 | 38/40 | |||||||
aKappa statistic for test of agreement
bConfidence interval
Compared to S. pseudintermedius, S. aureus isolates had higher overall agreement (almost perfect) between the results of cefoxitin disk diffusion and mecA PCR (95.3% agreement; κ = 0.834; p < 0.0001). There was also substantial agreement between oxacillin disk diffusion and mecA PCR assay (94.0% agreement; κ = 0.787; p < 0.0001).
-
iii)
S. coagulans and S. schleiferi
The findings for S. coagulans and S. schleiferi isolates were similar to those for S. pseudintermedius, with oxacillin disk diffusion being more consistent with the results of mecA PCR assay than cefoxitin disk diffusion. Cefoxitin disk diffusion tests had moderate agreement with mecA PCR for S. coagulans isolates (76.3% agreement; κ = 0.525; p < 0.0001), and fair agreement for S. schleiferi isolates (59.1% agreement; κ = 0.291; p < 0.0001). In contrast, the agreement between oxacillin disk diffusion tests and mecA PCR was almost perfect for both S. coagulans (95.6% agreement; κ = 0.913; p < 0.0001) and S. schleiferi (97.7% agreement; κ = 0.945; p < 0.0001).
Discussion
This study assessed patterns of antimicrobial, multidrug and methicillin resistance among Staphylococcus isolates from canine clinical specimens submitted to the UTCVM Bacteriology Laboratory between 2006 and 2017. The majority of these staphylococci were isolated from skin, ear and mucosal specimens, reflecting their importance as causative agents of pyoderma and otitis [3–6], which are frequently encountered in clinical practice. Surveillance of antimicrobial susceptibility patterns among canine Staphylococcus isolates is therefore valuable for guiding evidence-based treatment decisions, and useful for identifying trends that may reflect selection pressure.
Patterns of antimicrobial and multidrug resistance
S. pseudintermedius
The majority of S. pseudintermedius isolates were resistant to ampicillin (84.3%) and therefore most were classified as AMR, consistent with previous reports [28, 47, 61, 62]. Indeed, resistance to penicillinase-labile penicillins has become widespread over the past several decades [63], limiting their usefulness for the treatment of staphylococcal infections in dogs.
Relatively high levels of MRSP were observed throughout the study period (30.8% overall), with a mild but statistically significant temporal increase. This follows a steep increase in mecA-mediated resistance among isolates submitted to the UTCVM bacteriology laboratory between 2001 and 2007 [55]. These findings are not unprecedented, as mecA-mediated resistance has emerged among S. pseudintermedius in North America and Europe [64, 65], and MRSP has been detected around the world [66–71]. However, the level of methicillin resistance among isolates from canine specimens in the current study was higher than previously observed in other locations within the U.S., such as Kentucky [14] and Michigan [72]. These regional differences imply that geographically relevant AMR surveillance provides valuable information for veterinary practitioners.
In addition to the substantial level of methicillin resistance, almost half of the S. pseudintermedius isolates in the current study were MDR. Though a temporal decline in the proportion of MDR MRSP isolates was observed, the vast majority of MRSP (90.5%) were MDR. A limited number of antimicrobials are available to effectively treat MDR and MRSP infections; these isolates are often resistant to drug classes used in veterinary medicine such as tetracyclines, fluoroquinolones, macrolides, trimethoprim-sulfonamides, and lincosamides [73]. Considerable levels of resistance to the above agents (exceeding 75%) were observed among MRSP isolates in the current study.
As a result of such limitations in available treatment options, infections with MDR and/or MRSP isolates may necessitate the use of antimicrobials such as rifampin or chloramphenicol, despite the potential for adverse effects [73–76]. Resistance to rifampin was only assessed among a subset of MDR isolates in the current study, and was infrequently observed. On the other hand, it is concerning that a statistically significant increase in resistance to chloramphenicol occurred among both MRSP and MSSP isolates during the study period, with the highest levels among MRSP (from 4.0 to 12.0%). This finding is particularly concerning given its utility for the treatment of MRSP infections. A similar but accelerated trend was reported among canine isolates collected at the Texas A&M Veterinary Teaching Hospital, likely due to selection pressure from the increased use of chloramphenicol in the face of MDR/MRSP infections [77]. However, while that study also reported an increase in resistance to amikacin [77], another potential treatment for MDR or MRSP infections [73], resistance to amikacin was rare among MDR and cutaneous isolates in the current study.
In addition to the clinical challenge associated with the treatment of MDR and MRSP infections in veterinary patients, the zoonotic potential of S. pseudintermedius raises concerns from a public health perspective. Human infections with S. pseudintermedius are typically associated with companion animal contact [43, 78–82], and have included MRSP infections [83, 84]. Given the potential for rapid selection of resistant organisms, ongoing surveillance is warranted to ensure prompt recognition of clinically relevant patterns of AMR and MDR. Indeed, while higher levels of resistance to non-β-lactam antibiotics among MRSP compared to MSSP are expected [63], the temporal increase in MDR MSSP isolates observed in the current study was an interesting, albeit concerning finding. This trend appears to have been driven by rising levels of resistance to fluoroquinolones, folate inhibitors, lincosamides, and macrolides, all of which exhibited decreases for MRSP isolates.
-
b)
S. aureus
S. aureus was less commonly isolated in comparison to S. pseudintermedius in this study, as dogs are not the primary reservoir for this organism [85]. In the current study, 37.4% of S. aureus isolates were methicillin-resistant, and MRSA increased substantially during the study period (from 21.6 to 40%). Varying levels of methicillin resistance have been reported among canine clinical S. aureus isolates in other recent studies, ranging from 12.8% in an Australian study [86], to 62.7% of isolates from wound infections in a German study [87].
S. aureus strains in companion animals tend to mirror those circulating in humans [87, 88], and canine colonization and infections with S. aureus often result from reverse zoonosis [45, 89–91]. However, pets may serve as reservoirs once exposed, and MRSA carriage in humans may be difficult to eliminate if household dogs harboring the organism are not treated [45, 90]. The susceptibility patterns of S. aureus differed somewhat from S. pseudintermedius, which may reflect antimicrobial use patterns, particularly if humans are a major source of S. aureus colonization in this population of dogs. For example, resistance to gentamicin, tetracycline, and TMS were much less common among S. aureus, and a temporal trend was not observed for chloramphenicol. Furthermore, the proportion of MDR isolates did not increase among MSSA. However, a more rapid increase in methicillin resistance was observed among S. aureus isolates compared to S. pseudintermedius. Further investigation is warranted to understand the epidemiology of S. aureus in this patient population and identify factors which may have contributed to the observed patterns, including the pronounced increase in MRSA.
-
iii)
S. coagulans and S. schleiferi
While S. coagulans and S. schleiferi (formerly S. schleiferi subsp. coagulans and S. schleiferi subsp. schleiferi [15]) have received increased attention for their roles as human and animal pathogens in recent years, there are fewer published reports of AMR patterns for these organisms in comparison to S. pseudintermedius and S. aureus. The percentage of methicillin resistant isolates was the most notable difference between S. coagulans and S. schleiferi (32.2 and 56.9%, respectively), a finding which has previously been reported [19]. In fact, S. schleiferi isolates had the highest proportion of methicillin resistant isolates in the current study, consistent with previous reports [19, 92]. In addition to being isolated from healthy dogs and first-time infections, S. coagulans and S. schleiferi may be particularly important in recurrent infections such as otitis and pyoderma [17, 18, 93–95]. Prior exposure to β-lactam antibiotics, commonly used for empirical treatment of skin infections, is a reported risk factor for oxacillin (methicillin) resistance among S. coagulans and S. schleiferi isolates from canine specimens [19].
Despite the considerable proportion of methicillin-resistant S. schleiferi isolates, several other antimicrobial classes appear to remain as potential treatment options. In general, similar patterns of resistance to non-β-lactam antibiotics were observed between S. coagulans and S. schleiferi. Resistance to TMS, clindamycin, erythromycin, chloramphenicol and tetracycline were relatively uncommon, while higher levels of resistance to gentamicin and marbofloxacin were observed, in agreement with the findings of Kunder and colleagues [92]. While the percentages of MDR S. coagulans and S. schleiferi (20 and 27.2%) were much lower than the other Staphylococcus spp., they were also comparable with the findings of the above study (21%) [92]. Encouragingly, a significant temporal decline in MDR occurred among S. coagulans, despite the finding that few such trends were observed for individual agents or drug classes. Further research is warranted to investigate how this finding relates to patterns of antimicrobial use in the region, and to elucidate the role of S. coagulans and S. schleiferi as pathogens and commensal organisms of dogs.
Methicillin resistance and mecA gene detection
The findings of the current study are consistent with previous reports, which have identified oxacillin disk diffusion as a superior method for detecting mecA-mediated methicillin resistance in S. pseudintermedius, S. coagulans, and S. schleiferi when compared to cefoxitin [55, 56, 96, 97]. This is reflected in the current CLSI standards [98]. While there was almost perfect agreement between oxacillin disk diffusion and PCR, a small proportion of oxacillin-susceptible S. pseudintermedius, S. coagulans and S. schleiferi isolates were mecA-positive. Detection of the mecA gene in oxacillin-susceptible isolates may reflect the effects of regulatory elements on mecA gene expression and subsequent PBP2a production [31, 67, 99]. On the other hand, a small percentage (< 5%) of oxacillin-resistant S. pseudintermedius, S. coagulans and S. schleiferi isolates were mecA-negative, in contrast with the findings of several studies that reported mecA gene detection in all phenotypically resistant isolates [31, 55, 96]. One study, however, identified a single canine S. pseudintermedius isolate speculated to have an alternate, non-mecA-mediated mechanism of oxacillin resistance. The isolate in question had a small zone size and low minimum inhibitory concentration (MIC) for oxacillin, but neither the mecA nor mecC genes were detected [97].
While MIC values may be useful when interpreting discordant results, these were not available for most isolates in the current study. Among the S. pseudintermedius isolates in question, 19 had recorded MIC values; 13 had zone sizes at or near the interpretive breakpoint but would be considered susceptible based on their MIC (≤0.25 μg/mL) [52]. For isolates that did not have borderline zone sizes and/or MIC values, the possibility of an alternate mechanism of oxacillin resistance cannot be excluded based on the information available. Nonetheless, results of the current study support the usefulness of oxacillin disk diffusion for detecting mecA-mediated resistance in S. pseudintermedius, S. coagulans, and S. schleiferi.
For S. aureus, cefoxitin was superior to oxacillin disk diffusion for detecting mecA-mediated resistance, consistent with CLSI recommendations [52, 98]. While there was almost perfect agreement between the two tests, discordant results were obtained for seven S. aureus isolates. In order to facilitate comparison between studies, CLSI-recommended interpretive breakpoints in place at the time of sample submission were used in the current study. The recommended cutoff values for cefoxitin changed during the study period, from ≤19 mm (resistant), ≥20 mm (susceptible) to ≤21 mm (resistant), ≥22 mm (susceptible) [49, 50]. Under current CLSI standards, one of the three mecA-positive, cefoxitin-susceptible isolates would have been re-classified as cefoxitin-resistant, resulting in agreement between the two tests.
Further investigation to determine the mechanism of phenotypic methicillin resistance for the mecA-negative Staphylococcus spp. isolates was beyond the scope of the present study. However, the identification of this pattern among several Staphylococcus spp. isolates in this study suggests that PCR assay for both the mecA and mecC genes may be warranted in future research.
Study strengths and limitations
The identification of antimicrobial resistance patterns and trends using a large sample of canine patients in this study are useful for informing evidence-based treatment decisions. In addition, clinical specimens were obtained from submissions by referring veterinarians in addition to hospital patients, representing a broad range of patients from both primary practice and specialty services. Antimicrobial susceptibility data were reported at the level of individual drugs, allowing for a detailed assessment of these patterns.
However, this study is not without limitations. For instance, while Staphylococcus intermedius group (SIG) species cannot be reliably distinguished by biochemical identification processes, virtually all canine SIG isolates are S. pseudintermedius; therefore, as recommended, SIG isolates were reported as S. pseudintermedius [100, 101]. S. coagulans and S. schleiferi were differentiated based upon tube coagulase testing, which may be inconsistent with genotypic testing [102], and therefore some of these isolates may have been misclassified. In addition, patient medical history, including current or prior antimicrobial therapy, was not available, precluding our ability to identify drivers of the observed patterns of AMR and MDR. Finally, the use of specimens submitted for culture may present some selection bias, as these are unlikely to be from first-time infections, and may have been submitted due to therapeutic failure. However, the majority of specimens were from submissions by referring veterinarians, and samples were not solely obtained from patients of specialty services, resulting in a broader picture of AMR patterns across a variety of clinical settings. Despite the above limitations, the findings of the current study provide valuable insights to patterns and trends of AMR in the region, and are useful for guiding both clinical decisions and directions for future research.
Conclusion
There is evidence of substantial levels of MDR among S. pseudintermedius and S. aureus isolates from canine specimens, and increasing temporal trends in MRSP and MRSA. Unfortunately, this suggests that few agents remain broadly effective against these organisms. Furthermore, the emergence of chloramphenicol resistance observed in the current study is concerning. Future studies will investigate potential drivers of the observed antimicrobial susceptibility patterns. The observed levels of MDR and methicillin-resistance highlight the importance of antibiograms to guide treatment decisions. The temporal increase in canine methicillin-resistant staphylococci observed in this study has practical implications for human health. These findings highlight the need for continued surveillance of antimicrobial resistance patterns among staphylococci, and the importance of judicious antimicrobial use practices.
Acknowledgements
We thank the UTCVM COE for providing research funds to support Nick Millis. The UTCVM Hospital Operations and Bacteriology laboratory are gratefully acknowledged for providing access to the study data. We also thank the UTCVM computer operations for helping with data extraction.
Abbreviations
- AKC
American Kennel Club
- AMR
Antimicrobial Resistance
- COE
Centers of Excellence
- CLSI
Clinical and Laboratory Standards Institute
- IQR
interquartile range
- MDR
Multidrug Resistance
- MIC
minimum inhibitory concentration
- MRS
Methicillin resistant staphylococci
- MRSA
methicillin-resistant Staphylococcus aureus
- MRSC
methicillin-resistant Staphylococcus coagulans
- MRSP
methicillin-resistant Staphylococcus pseudintermedius
- MRSS
methicillin-resistant Staphylococcus schleiferi
- PBP
penicillin binding protein
- PCR
Polymerase Chain Reaction
- SIG
Staphylococcus intermedius group
- TMS
trimethoprim/sulfamethoxazole
- UTCVM
University of Tennessee College of Veterinary Medicine
- UTVTH
University of Tennessee Veterinary Teaching Hospital
Authors’ contributions
Conceived research idea: AO. Designed the study: AO, JL, NM. Performed the experiments: RDJ, BJ, SAK. Performed statistical analysis: AO, JL, NM. Wrote draft, edited and approved final version of manuscript: AO, JL, NM, RDJ, BJ, SK.
Funding
This study was funded by the University of Tennessee College of Veterinary Medicine (UTCVM) Centers of Excellence (COE). The funder had no input on any part of the study.
Availability of data and materials
The dataset used and/or analyzed during the current study is available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The University of Tennessee institutional animal care and use committee (IACUC) indicated that the study did not use animals or animal tissues and therefore did not require IACUC oversight.
Consent for publication
This is a retrospective study that used records of dogs that are not identified and hence it was no possible to obtain consent for publication.
Competing interests
The authors declare that they do not have competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zee YC, Hirsh DC. Veterinary microbiology: EBSCOhost. Malden: John Wiley and Sons; 1999. [Google Scholar]
- 2.Foster T. Staphylococcus. In: Baron S, editor. Medical Microbiology. 4. Galveston: University of Texas Medical Branch at Galveston; 1996. pp. 309–320. [Google Scholar]
- 3.Bannoehr J, Guardabassi L. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet Dermatol. 2012;23:253–e52. doi: 10.1111/j.1365-3164.2012.01046.x. [DOI] [PubMed] [Google Scholar]
- 4.Penna B, Varges R, Medeiros L, Martins GM, Martins RR, Lilenbaum W. Species distribution and antimicrobial susceptibility of staphylococci isolated from canine otitis externa. Vet Dermatol. 2010;21:292–296. doi: 10.1111/j.1365-3164.2009.00842.x. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami T, Shibata S, Murayama N, Nagata M, Nishifuji K, Iwasaki T, et al. Antimicrobial susceptibility and methicillin resistance in Staphylococcus pseudintermedius and Staphylococcus schleiferi subsp. coagulans isolated from dogs with pyoderma in Japan. J Vet Med Sci. 2010;72:1615–1619. doi: 10.1292/jvms.10-0172. [DOI] [PubMed] [Google Scholar]
- 6.Hauschild T, Wójcik A. Species distribution and properties of staphylococci from canine dermatitis. Res Vet Sci. 2007;82:1–6. doi: 10.1016/j.rvsc.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Qekwana DN, Oguttu JW, Sithole F, Odoi A. Burden and predictors of Staphylococcus aureus and S. pseudintermedius infections among dogs presented at an academic veterinary hospital in South Africa (2007–2012) PeerJ. 2017;5:e3198. doi: 10.7717/peerj.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grönthal T, Eklund M, Thomson K, Piiparinen H, Sironen T, Rantala M. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. J Antimicrob Chemother. 2017;72:dkw559. doi: 10.1093/jac/dkw559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viñes J, Cuscó A, Francino O. Hybrid Assembly from a Pathogenic Methicillin- and Multidrug-Resistant Staphylococcus pseudintermedius Strain Isolated from a Case of Canine Otitis in Spain. Microbiol Resour Announc. 2020;9(1):e01121–19. 10.1128/MRA.01121-19. [DOI] [PMC free article] [PubMed]
- 10.Cabassu J, Moissonnier R. Surgical treatment of a vertebral fracture associated with a haematogenous osteomyelitis in a dog. Vet Comp Orthop Traumatol. 2007;20:227–230. doi: 10.1160/VCOT-06-11-0089. [DOI] [PubMed] [Google Scholar]
- 11.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peton V, Le Loir Y. Staphylococcus aureus in veterinary medicine. Infect Genet Evol. 2014;21:602–615. doi: 10.1016/j.meegid.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Faires MC, Traverse M, Tater KC, Pearl DL, Weese JS. Methicillin-resistant and -susceptible Staphylococcus aureus infections in dogs. Emerg Infect Dis. 2010;16:69–75. doi: 10.3201/eid1601.081758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conner JG, Smith J, Erol E, Locke S, Phillips E, Carter CN, et al. Temporal trends and predictors of antimicrobial resistance among Staphylococcus spp isolated from canine specimens submitted to a diagnostic laboratory. Plos One. 2018;13:e0200719. doi: 10.1371/journal.pone.0200719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madhaiyan M, Wirth JS, Saravanan VS. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcu. Int J Syst Evol Microbiol. 2020;70:5926–5936. doi: 10.1099/IJSEM.0.004498/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- 16.Zdovc I, Ocepek M, Pirš T, Krt B, Pinter L. Microbiological features of Staphylococcus schleiferi subsp. coagulans, isolated from dogs and possible misidentification with other canine coagulase-positive staphylococci. J Vet Med Ser B Infect Dis Vet Public Heal. 2004;51:449–454. doi: 10.1111/j.1439-0450.2004.00792.x. [DOI] [PubMed] [Google Scholar]
- 17.Frank LA, Kania SA, Hnilica KA, Wilkes RP, Bemis DA. Isolation of Staphylococcus schleiferi from dogs with pyoderma. J Am Vet Med Assoc. 2003;222:451–454. doi: 10.2460/javma.2003.222.451. [DOI] [PubMed] [Google Scholar]
- 18.May ER, Hnilica KA, Frank LA, Jones RD, Bemis DA. Isolation of Staphylococcus schleiferi from healthy dogs and dogs with otitis, pyoderma, or both. J Am Vet Med Assoc. 2005;227:928–931. doi: 10.2460/javma.2005.227.928. [DOI] [PubMed] [Google Scholar]
- 19.Cain CL, Morris DO, Rankin SC. Clinical characterization of Staphylococcus schleiferi infections and identification of risk factors for acquisition of oxacillin-resistant strains in dogs: 225 cases (2003-2009) J Am Vet Med Assoc. 2011;239:1566–1573. doi: 10.2460/javma.239.12.1566. [DOI] [PubMed] [Google Scholar]
- 20.Huerta B, Maldonado A, Ginel PJ, Tarradas C, Gómez-Gascón L, Astorga RJ, et al. Risk factors associated with the antimicrobial resistance of staphylococci in canine pyoderma. Vet Microbiol. 2011;150:302–308. doi: 10.1016/j.vetmic.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Qekwana DN, Oguttu JW, Sithole F, Odoi A. Patterns and predictors of antimicrobial resistance among Staphylococcus spp from canine clinical cases presented at a veterinary academic hospital in South Africa. BMC Vet Res. 2017;13:116. doi: 10.1186/s12917-017-1034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sweeney MT, Lubbers BV, Schwarz S, Watts JL. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J Antimicrob Chemother. 2018;73:1460–1463. doi: 10.1093/jac/dky043. [DOI] [PubMed] [Google Scholar]
- 23.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 24.Gandolfi-Decristophoris P, Regula G, Petrini O, Zinsstag J, Schelling E. Prevalence and risk factors for carriage of multi-drug resistant staphylococci in healthy cats and dogs. J Vet Sci. 2013;14:449–456. doi: 10.4142/jvs.2013.14.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penna B, Varges R, Martins R, Martins G, Lilenbaum W. In vitro antimicrobial resistance of staphylococci isolated from canine urinary tract infection. Can Vet J. 2010;51:738–742. [PMC free article] [PubMed] [Google Scholar]
- 26.Nienhoff U, Kadlec K, Chaberny IF, Verspohl J, Gerlach GF, Kreienbrock L, et al. Methicillin-resistant Staphylococcus pseudintermedius among dogs admitted to a small animal hospital. Vet Microbiol. 2011;150:191–197. doi: 10.1016/j.vetmic.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Hanselman BA, Kruth S, Weese JS. Methicillin-resistant staphylococcal colonization in dogs entering a veterinary teaching hospital. Vet Microbiol. 2008;126:277–281. doi: 10.1016/j.vetmic.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 28.Zur G, Gurevich B, Elad D. Prior antimicrobial use as a risk factor for resistance in selected Staphylococcus pseudintermedius isolates from the skin and ears of dogs. Vet Dermatol. 2016;27:468–e125. doi: 10.1111/vde.12382. [DOI] [PubMed] [Google Scholar]
- 29.Duquette RA, Nuttall TJ. Methicillin-resistant Staphylococcus aureus in dogs and cats: an emerging problem? J Small Anim Pract. 2004;45(12):591–7. 10.1111/j.1748-5827.2004.tb00180.x. [DOI] [PubMed]
- 30.Menandro ML, Dotto G, Mondin A, Martini M, Ceglie L, Pasotto D. Prevalence and characterization of methicillin-resistant Staphylococcus pseudintermedius from symptomatic companion animals in northern Italy: clonal diversity and novel sequence types. Comp Immunol Microbiol Infect Dis. 2019;66. 10.1016/j.cimid.2019.101331. [DOI] [PubMed]
- 31.Kania SA, Williamson NL, Frank LA, Wilkes RP, Jones RD, Bemis DA. Methicillin resistance of staphylococci isolated from the skin of dogs with pyoderma. Am J Vet Res. 2004;65:1265–1268. doi: 10.2460/ajvr.2004.65.1265. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization . Critically important antimicrobials for human medicine, 6th revision. Geneva: World Health Organization; 2019. [Google Scholar]
- 33.Foglia Manzillo V, Paola Nocera F, Gizzarelli M, Oliva G. A successful vancomycin treatment of multidrug-resistant MRSA-associated canine pyoderma. J Dermatologic Res Ther. 2015;1:19–25. [Google Scholar]
- 34.Gortel K, Campbell KL, Kakoma I, Whittem T, Schaeffer DJ, Weisiger RM. Methicillin resistance among staphylococci isolated from dogs. Am J Vet Res. 1999;60:1526–1530. [PubMed] [Google Scholar]
- 35.Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker K, Ballhausen B, Köck R, Kriegeskorte A. Methicillin resistance in Staphylococcus isolates: the “mec alphabet” with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. Int J Med Microbiol. 2014;304:794–804. doi: 10.1016/j.ijmm.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Cohn LA, Middleton JR. A veterinary perspective on methicillin-resistant staphylococci. J Vet Emerg Crit Care. 2010;20:31–45. doi: 10.1111/j.1476-4431.2009.00497.x. [DOI] [PubMed] [Google Scholar]
- 38.Grönthal T, Moodley A, Nykäsenoja S, Junnila J, Guardabassi L, Thomson K, Rantala M. Large outbreak caused by methicillin resistant Staphylococcus pseudintermedius ST71 in a Finnish Veterinary Teaching Hospital--from outbreak control to outbreak prevention. PLoS One. 2014;9(10):e110084. 10.1371/journal.pone.0110084. [DOI] [PMC free article] [PubMed]
- 39.Kmieciak W, Szewczyk EM. Are zoonotic Staphylococcus pseudintermedius strains a growing threat for humans? Folia Microbiol (Praha) 2018;63:743–747. doi: 10.1007/s12223-018-0615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youn J-H, Yoon JW, Koo HC, Lim S-K, Park YH. Prevalence and antimicrogram of Staphylococcus intermedius group isolates from veterinary staff, companion animals, and the environment in veterinary hospitals in Korea. J Vet Diagn Investig. 2011;23:268–274. doi: 10.1177/104063871102300211. [DOI] [PubMed] [Google Scholar]
- 41.Gómez-Sanz E, Torres C, Lozano C, Zarazaga M. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comp Immunol Microbiol Infect Dis. 2013;36:83–94. doi: 10.1016/j.cimid.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Faires MC, Tater KC, Weese JS. An investigation of methicillin-resistant Staphylococcus aureus colonization in people and pets in the same household with an infected person or infected pet. J AM Vet Med Assoc. 2009:540–3. [DOI] [PubMed]
- 43.Ference EH, Danielian A, Kim HW, Yoo F, Kuan EC, Suh JD. Zoonotic Staphylococcus pseudintermedius sinonasal infections: risk factors and resistance patterns. Int Forum Allergy Rhinol. 2019;9:724–729. doi: 10.1002/alr.22329. [DOI] [PubMed] [Google Scholar]
- 44.Somayaji R, Priyantha MAR, Rubin JE, Church D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn Microbiol Infect Dis. 2016;85:471–476. doi: 10.1016/j.diagmicrobio.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Mork RL, Hogan PG, Muenks CE, Boyle MG, Thompson RM, Sullivan ML, et al. Longitudinal, strain-specific Staphylococcus aureus introduction and transmission events in households of children with community-associated meticillin-resistant S aureus skin and soft tissue infection: a prospective cohort study. Lancet Infect Dis. 2020;20:188–198. doi: 10.1016/S1473-3099(19)30570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blondeau LD, Rubin JE, Deneer H, Kanthan R, Morrison B, Sanche S, et al. Persistent infection with Staphylococcus pseudintermedius in an adult oncology patient with transmission from a family dog. J Chemother. 2020;32:151–155. doi: 10.1080/1120009X.2020.1735142. [DOI] [PubMed] [Google Scholar]
- 47.Jones RD, Kania SA, Rohrbach BW, Frank LA, Bemis DA. Prevalence of oxacillin- and multidrug-resistant staphylococci in clinical samples from dogs: 1,772 samples (2001–2005) 2007. [DOI] [PubMed] [Google Scholar]
- 48.Jorgensen JH, Carroll KC, Pfaller MA. Manual of clinical microbiology. 11. Hoboken: John Wiley & Sons, Inc.; 2015. [Google Scholar]
- 49.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, vol. 16: CLSI Suppl M100; 2006. Wayne, PA, USA
- 50.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, vol. 20: CLSI Suppl M100; 2010. Wayne, PA, USA.
- 51.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, vol. 22: CLSI Suppl M100; 2012. Wayne, PA, USA.
- 52.CLSI, editor. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. 4. Wayne: CLSI supplement VET08; 2018. [Google Scholar]
- 53.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard. 3 2008. [Google Scholar]
- 54.BioMerieux. Vitek AutoMicrobicSystem (AMS). Durham, NC, USA.
- 55.Bemis DA, Jones RD, Frank LA, Kania SA. Evaluation of susceptibility test breakpoints used to predict mecA-mediated resistance in Staphylococcus pseudintermedius isolated from dogs. J Vet Diagn Investig. 2009;21:53–58. doi: 10.1177/104063870902100108. [DOI] [PubMed] [Google Scholar]
- 56.Schissler JR, Hillier A, Daniels JB, Cole LK, Gebreyes WA. Evaluation of clinical laboratory standards institute interpretive criteria for methicillin-resistant Staphylococcus pseudintermedius isolated from dogs. J Vet Diagn Investig. 2009;21:684–688. doi: 10.1177/104063870902100514. [DOI] [PubMed] [Google Scholar]
- 57.Rosato AE, Craig WA, Archer GL. Quantitation of mecA transcription in oxacillin-resistant Staphylococcus aureus clinical isolates. J Bacteriol. 2003;185:3446–3452. doi: 10.1128/JB.185.11.3446-3452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.SAS Institute . SAS 9.4. 2016. [Google Scholar]
- 59.American Kennel Club . List of Breeds by Group. 2020. [Google Scholar]
- 60.R Core Team . R: A language and environment for statistical computing. 2020. [Google Scholar]
- 61.Bardiau M, Yamazaki K, Ote I, Misawa N, Mainil JG. Characterization of methicillin-resistant Staphylococcus pseudintermedius isolated from dogs and cats. Microbiol Immunol. 2013;57:496–501. doi: 10.1111/1348-0421.12059. [DOI] [PubMed] [Google Scholar]
- 62.Windahl U, Holst BS, Nyman A, Grönlund U, Bengtsson B. Characterisation of bacterial growth and antimicrobial susceptibility patterns in canine urinary tract infections. BMC Vet Res. 2014;10:1–10. doi: 10.1186/s12917-014-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moodley A, Damborg P, Nielsen SS. Antimicrobial resistance in methicillin susceptible and methicillin resistant Staphylococcus pseudintermedius of canine origin: literature review from 1980 to 2013. Vet Microbiol. 2014;171:337–341. doi: 10.1016/j.vetmic.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 64.Ruscher C, Lübke-Becker A, Semmler T, Wleklinski CG, Paasch A, Šoba A, et al. Widespread rapid emergence of a distinct methicillin- and multidrug-resistant Staphylococcus pseudintermedius (MRSP) genetic lineage in Europe. Vet Microbiol. 2010;144:340–346. doi: 10.1016/j.vetmic.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 65.Perreten V, Kadlec K, Schwarz S, Andersson UG, Finn M, Greko C, et al. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother. 2010;65:1145–1154. doi: 10.1093/jac/dkq078. [DOI] [PubMed] [Google Scholar]
- 66.Dziva F, Wint C, Auguste T, Heeraman C, Dacon C, Yu P, et al. First identification of methicillin-resistant Staphylococcus pseudintermedius strains among coagulase-positive staphylococci isolated from dogs with otitis externa in Trinidad. West Indies Infect Ecol Epidemiol. 2015;5:29170. doi: 10.3402/iee.v5.29170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gagetti P, Wattam AR, Giacoboni G, De Paulis A, Bertona E, Corso A, et al. Identification and molecular epidemiology of methicillin resistant Staphylococcus pseudintermedius strains isolated from canine clinical samples in Argentina. BMC Vet Res. 2019;15. 10.1186/s12917-019-1990-x. [DOI] [PMC free article] [PubMed]
- 68.Tabatabaei S, Najafifar A, Askari Badouei M, Zahraei Salehi T, Ashrafi Tamai I, Khaksar E, et al. Genetic characterisation of methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in pets and veterinary personnel in Iran: new insights into emerging methicillin-resistant S. pseudintermedius (MRSP) J Glob Antimicrob Resist. 2019;16:6–10. doi: 10.1016/j.jgar.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Yang J, Logue CM, Liu K, Cao X, Zhang W, et al. Methicillin-resistant Staphylococcus pseudintermedius isolated from canine pyoderma in North China. J Appl Microbiol. 2012;112:623–630. doi: 10.1111/j.1365-2672.2012.05233.x. [DOI] [PubMed] [Google Scholar]
- 70.Kadlec K, Weiß S, Wendlandt S, Schwarz S, Tonpitak W. Characterization of canine and feline methicillin-resistant Staphylococcus pseudintermedius (MRSP) from Thailand. Vet Microbiol. 2016;194:93–97. doi: 10.1016/j.vetmic.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 71.Bell AG, Coombs GW, Cater B, Douglass C. First report of a mecA-positive multidrug-resistant Staphylococcus pseudintermedius isolated from a dog in New Zealand. N Z Vet J. 2016;64:253–256. doi: 10.1080/00480169.2016.1146171. [DOI] [PubMed] [Google Scholar]
- 72.Detwiler A, Bloom P, Petersen A, Rosser EJ. Multi-drug and methicillin resistance of staphylococci from canine patients at a veterinary teaching hospital (2006-2011) Vet Q. 2013;33:60–67. doi: 10.1080/01652176.2013.799792. [DOI] [PubMed] [Google Scholar]
- 73.Papich MG. Selection of antibiotics for meticillin-resistant Staphylococcus pseudintermedius: time to revisit some old drugs? Vet Dermatol. 2012;23:352–e64. doi: 10.1111/j.1365-3164.2011.01030.x. [DOI] [PubMed] [Google Scholar]
- 74.Short J, Zabel S, Cook C, Schmeitzel L. Adverse events associated with chloramphenicol use in dogs: a retrospective study (2007-2013) Vet Rec. 2014;175:537. doi: 10.1136/vr.102687. [DOI] [PubMed] [Google Scholar]
- 75.Bryan J, Frank LA, Rohrbach BW, Burgette LJ, Cain CL, Bemis DA. Treatment outcome of dogs with meticillin-resistant and meticillin-susceptible Staphylococcus pseudintermedius pyoderma. Vet Dermatol. 2012;23:361–e65. doi: 10.1111/j.1365-3164.2012.01034.x. [DOI] [PubMed] [Google Scholar]
- 76.De Lucia M, Bardagi M, Fabbri E, Ferreira D, Ferrer L, Scarampella F, et al. Rifampicin treatment of canine pyoderma due to multidrug-resistant meticillin-resistant staphylococci: a retrospective study of 32 cases. Vet Dermatol. 2017;28:171–e36. doi: 10.1111/vde.12404. [DOI] [PubMed] [Google Scholar]
- 77.Gold RM, Cohen ND, Lawhon SD. Amikacin resistance in Staphylococcus pseudintermedius isolated from dogs. J Clin Microbiol. 2014;52:3641–3646. doi: 10.1128/JCM.01253-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Darlow CA, Paidakakos N, Sikander M, Atkins B. A spinal infection with Staphylococcus pseudintermedius. BMJ Case Rep. 2017;2017. 10.1136/bcr-2017-221260. [DOI] [PMC free article] [PubMed]
- 79.Dahbour L, Gibbs J, Coletta C, Hummell J, Al-Sarie M, Kahlon NP, et al. Peritoneal Dialysis zoonotic bacterial peritonitis with Staphylococcus pseudintermedius. Case RepNephrol Dial. 2020;10:65–70. doi: 10.1159/000508126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robb AR, Wright ED, Foster AME, Walker R, Malone C. Skin infection caused by a novel strain of Staphylococcus pseudintermedius in a Siberian husky dog owner. JMM Case Rep. 2017;4. 10.1099/jmmcr.0.005087. [DOI] [PMC free article] [PubMed]
- 81.Riegel P, Jesel-Morel L, Laventie B, Boisset S, Vandenesch F, Prévost G. Coagulase-positive Staphylococcus pseudintermedius from animals causing human endocarditis. Int J Med Microbiol. 2011;301:237–239. doi: 10.1016/j.ijmm.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Nomoto H, Kutsuna S, Nakamura K, Nakamoto T, Shimomura A, Hirakawa T, et al. Totally implantable venous access port infection caused by Staphylococcus pseudintermedius: possible transmission from a companion dog to a human. J Infect Chemother. 2020;26:1305–1308. doi: 10.1016/j.jiac.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 83.Gagetti P, Errecalde L, Wattam AR, De Belder D, Ojeda Saavedra M, Corso A, et al. Characterization of the first mecA-positive multidrug-resistant Staphylococcus pseudintermedius isolated from an Argentinian patient. Microb Drug Resist. 2020;26:717–721. doi: 10.1089/mdr.2019.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Savini V, Barbarini D, Polakowska K, Gherardi G, Bialecka A, Kasprowicz A, et al. Methicillin-resistant staphylococcus pseudintermedius infection in a bone marrow transplant recipient. J Clin Microbiol. 2013;51:1636–1638. doi: 10.1128/JCM.03310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weese JS, van Duijkeren E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet Microbiol. 2010;140:418–429. doi: 10.1016/j.vetmic.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 86.Saputra S, Jordan D, Worthing KA, Norris JM, Wong HS, Abraham R, et al. Antimicrobial resistance in coagulase-positive staphylococci isolated from companion animals in Australia: a one year study. Plos One. 2017;12:e0176379. doi: 10.1371/journal.pone.0176379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vincze S, Stamm I, Kopp PA, Hermes J, Adlhoch C, Semmler T, et al. Alarming proportions of methicillin-resistant Staphylococcus aureus (MRSA) in wound samples from companion animals, Germany 2010–2012. Plos One. 2014;9:e85656. doi: 10.1371/journal.pone.0085656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haenni M, Châtre P, Dupieux-Chabert C, Métayer V, Bes M, Madec JY, et al. Molecular epidemiology of methicillin-resistant staphylococcus aureus in horses, cats, and dogs over a 5-year period in France. Front Microbiol. 2017;8. 10.3389/fmicb.2017.02493. [DOI] [PMC free article] [PubMed]
- 89.Rutland BE, Weese JS, Bolin C, Au J, Malani AN. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg Infect Dis. 2009;15:1328–1330. doi: 10.3201/eid1508.081635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van Duijkeren E, Wolfhagen MJHM, Box ATA, Heck MEOC, Wannet WJB, Fluit AC. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg Infect Dis. 2004;10:2235–2237. doi: 10.3201/eid1012.040387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sasaki T, Tsubakishita S, Tanaka Y, Ohtsuka M, Hongo I, Fukata T, et al. Population genetic structures of Staphylococcus aureus isolates from cats and dogs in Japan. J Clin Microbiol. 2012;50:2152–2155. doi: 10.1128/JCM.06739-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kunder DA, Cain CL, O’Shea K, Cole SD, Rankin SC. Genotypic relatedness and antimicrobial resistance of Staphylococcus schleiferi in clinical samples from dogs in different geographic regions of the United States. Vet Dermatol. 2015;26. 10.1111/vde.12254. [DOI] [PubMed]
- 93.Lee GY, Lee HH, Hwang SY, Hong J, Lyoo KS, Yang SJ. Carriage of Staphylococcus schleiferi from canine otitis externa: antimicrobial resistance profiles and virulence factors associated with skin infection. J Vet Sci. 2019;20. 10.4142/jvs.2019.20.e6. [DOI] [PMC free article] [PubMed]
- 94.May ER, Kinyon JM, Noxon JO. Nasal carriage of Staphylococcus schleiferi from healthy dogs and dogs with otitis, pyoderma or both. Vet Microbiol. 2012;160:443–448. doi: 10.1016/j.vetmic.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 95.Vanni M, Tognetti R, Pretti C, Crema F, Soldani G, Meucci V, et al. Antimicrobial susceptibility of Staphylococcus intermedius and Staphylococcus schleiferi isolated from dogs. Res Vet Sci. 2009;87:192–195. doi: 10.1016/j.rvsc.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 96.Huse HK, Miller SA, Chandrasekaran S, Hindler JA, Lawhon SD, Bemis DA, et al. Evaluation of oxacillin and Cefoxitin disk diffusion and MIC breakpoints established by the clinical and laboratory standards Institute for Detection of mecA-mediated oxacillin resistance in Staphylococcus schleiferi. J Clin Microbiol. 2018;56. 10.1128/JCM.01653-17. [DOI] [PMC free article] [PubMed]
- 97.Wu MT, Burnham CAD, Westblade LF, Bard JD, Lawhon SD, Wallace MA, et al. Evaluation of oxacillin and Cefoxitin disk and MIC breakpoints for prediction of methicillin resistance in human and veterinary isolates of Staphylococcus intermedius group. J Clin Microbiol. 2016;54:535–542. doi: 10.1128/JCM.02864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.CLSI, editor. Performance Standards for Antimicrobial Susceptibility Testing. 30. Wayne: CLSI supplement M100; 2020. [Google Scholar]
- 99.Black CC, Eberlein LC, Solyman SM, Wilkes RP, Hartmann FA, Rohrbach BW, et al. The role of mecA and blaZ regulatory elements in mecA expression by regional clones of methicillin-resistant Staphylococcus pseudintermedius. Vet Microbiol. 2011;151:345–353. doi: 10.1016/j.vetmic.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 100.Sasaki T, Kikuchi K, Tanaka Y, Takahashi N, Kamata S, Hiramatsu K. Reclassification of phenotypically identified Staphylococcus intermedius strains. J Clin Microbiol. 2007;45:2770–2778. doi: 10.1128/JCM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Devriese LA, Hermans K, Baele M, Haesebrouck F. Staphylococcus pseudintermedius versus Staphylococcus intermedius. Vet Microbiol. 2009;133:206–207. doi: 10.1016/j.vetmic.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 102.Cain CL, Morris DO, O’Shea K, Rankin SC. Genotypic relatedness and phenotypic characterization of Staphylococcus schleiferi subspecies in clinical samples from dogs. Am J Vet Res. 2011;72:96–102. doi: 10.2460/ajvr.72.1.96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset used and/or analyzed during the current study is available from the corresponding author on reasonable request.