ABSTRACT
The immune system in the large intestine is separated from commensal microbes and comparatively rare enteric pathogens by a monolayer of diverse epithelial cells overlaid with a compact and adherent inner mucus layer and a looser outer mucus layer. Microorganisms, collectively referred to as the mucus-associated (MA) microbiota, physically inhabit this mucus barrier, resulting in a dynamic and incessant dialog to maintain both spatial segregation and immune tolerance. Recent major findings reveal novel features of the crosstalk between the immune system and mucus-associated bacteria in health and disease, as well as disease-related peripheral immune signatures indicative of host responses to these organisms. In this brief review, we integrate these novel observations into our overall understanding of host-microbiota mutualism at the colonic mucosal border and speculate on the significance of this emerging knowledge for our understanding of the prevention, development, and progression of chronic intestinal inflammation.
KEYWORDS: Colon mucus layer, mucus-associated bacteria, anti-commensal IgA, anti-commensal IgG, T-dependent, T-independent, flagellin, lachnospiraceae
Introduction
Symbiosis between the host and the intestinal microbiota is dictated by the physical location of these organisms along the GI tract, including their proximity to the gut epithelium. The mammalian host has evolved multiple mechanisms that actively limit the direct interaction of gut commensals with host epithelial cells and immune cells in the underlying lamina propria. These include the epithelium-derived immunomodulatory and anti-microbial peptides, and the secretion of dimers from broadly reactive immunoglobulin A (IgA), that can help to sequester microbes away from the single-cell epithelial layer.1,2 In addition, specialized epithelial cells known as goblet cells (GC), which represent approximately 10–30% of human colonic epithelial cells, produce the secreted mucin, mucin-2 (Muc2) which forms a dense hydrogel or mucus layer that adheres to the apical surface of colonic epithelial cells. Continuous foraging of the outer region of the mucus layer by mucolytic bacteria helps to produce a loose outer mucus layer that serves as a permanent habitat for commensals including non-mucolytic opportunists such as Escherichia coli (E. coli).3 Previously, the term loosely adherent is used to refer to those organisms that forage the luminal edge of the outer mucus layer and, in so doing, supports their own existence while defending against pathogen colonization. Some commensals are also capable of occupying the dense inner mucus layer directly overlaying the epithelium. Thus, the gut microbiota is often thought of as luminal versus mucus-associated with the mucus-associated further divided into inner and outer mucus communities. Metagenomic profiling of almost 400 human gut microbiomes found that almost 90% of the organisms encode enzymes capable of cleaving or catabolizing mucin O-glycan monosaccharides suggesting that, in theory, most commensal organisms are capable of surviving in close association with the mucus layer.4 This is consistent with the fact that mucus-associated and luminal organisms are not phylogenetically distinct and taxonomic representation is virtually identical as both communities constantly admix.5 Notably, however, for the same species, there are functional differences reflected in the transcriptional profiles of bacterial cells of the same species recovered from the lumen, mucus layer, or physically associated with the gut epithelium.6,7 Thus, the term mucus-associated is a spatio-temporal description that also encompasses distinct functional and metabolic activities and immune system interactions that enable persistence in that niche. The relative proximity of these mucus-associated organisms to the epithelial border at any point in time arguably increases the likelihood of crosstalk with the intestinal immune system, from epithelial cells to the immune cells in the underlying lamina propria. Accordingly, the chronic diseases in the gastrointestinal tract collectively referred to as inflammatory bowel disease (IBD) are characterized by reproducible changes in the abundance of specific bacteria known to inhabit the mucus layer. Moreover, seroreactivity of antigens derived from these ‘border-dwellers’ is actively utilized in IBD diagnostics. Here, we discuss recent findings at the intersection of the gut mucosal barrier, the mucus-associated microbiota, and the intestinal immune system in homeostasis and in IBD. We posit that recent notable advances implore future exploration of host–microbiota interactions in the intestines that involve functional analyses of communities isolated directly from the mucus layer, elucidation of the immune cascades that are central to the persistence of these organisms in this locale, and determination of the possible connection(s) between these mutualistic interactions and the robust disease-specific anti-commensal immunoreactivity detectable systemically in patients with IBD.
Production of the colonic mucus layer
Unlike the small intestine, which features a single unattached mucus layer, the colonic mucus comprises a dense, adherent inner mucus layer and a looser, more hydrated outer layer (Figure 1A). Membrane and secreted mucins appear to have been involved in the evolutionary compromise underlying host-microbiota mutualism, particularly in the densely populated large intestine. Mucins are integral to the persistence of commensals and the resistance to colonization by pathogenic competitors. Accordingly, despite the utilization of mucus for nutritional purposes, one microbiota adaptation that helps to limit the requirement for gut mucins as a food source is the collective expression of multiple enzymes, not encoded by mammals, that can catalyze the fermentation of complex dietary plant polysaccharides into short-chain fatty acids.8 Secreted mucins are produced by goblet cells (GCs) and are released apically into the intestinal lumen via a process regulated by the Forkhead box protein O1 (Foxo1).9 These mucins form a dense disulfide-bonded hydrogel – the mucus layer – overlying the epithelial cells. In the colon, the mucus layer is highly dynamic with the main mucus protein components displaying a turnover rate approximately twice that of epithelial cells in the same region of the intestine independent of the microbiota.10
Figure 1.
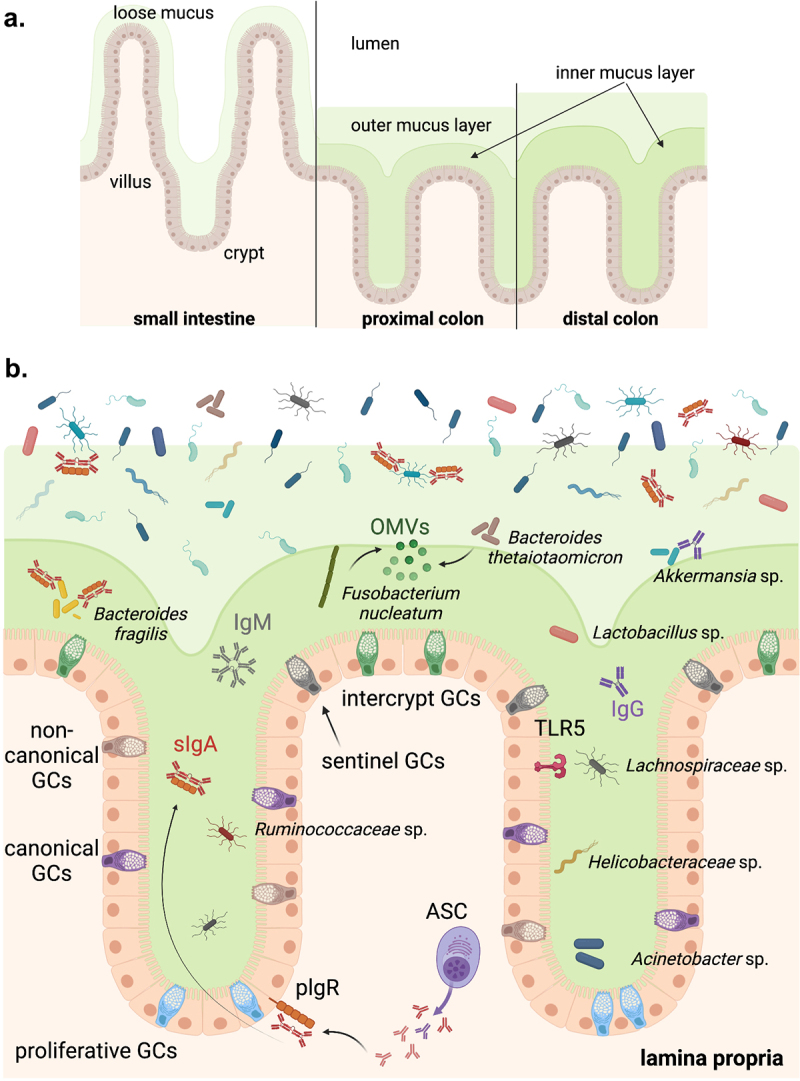
Host-microbiota spatial dynamics at the gut epithelial border. (A) The small intestinal epithelium is covered with a loose mucus layer, whereas a dense inner mucus layer and a looser outer mucus layer overlay the colonic epithelium. Both the density and thickness of the inner mucus layer increases from the proximal to the distal colon. (B) The colonic epithelial layer includes multiple subtypes of mucus-secreting goblet cells (GC). Crypt GCs include canonical and non-canonical GCs, as well as proliferative GCs that locate at the base of the crypts, whereas intercrypt GCs are found on the surface epithelium. TLR5, the receptor for bacterial flagellin, is constitutively and uniquely expressed on the luminal surface of colonic epithelial cells in the proximal colon. Microbial density is significantly higher in the outer compared to the inner mucus layer and the crypts. Bacteria including Bacteroides thetaiotaomicron and Fusobacterium nucleatum can bind mucin and release outer membrane vesicles (OMVs) which can lead to immune activation. Flagellated bacteria belonging to the Lachnospiraceae, Ruminococcaceae, and Helicobacteraceae families can colonize the crypts of the proximal colon. In addition to the physical barriers of the epithelial and mucus layers, lamina propria plasma cells produce anti-commensal immunoglobulins (Ig) including IgM, IgA, and IgG which help to regulate bacterial activation of the immune system. The most widely studied, IgA is transported into the lumen via the polymeric Ig receptor (PIGR) and helps to sequester bacteria and bacterial antigens away from the gut epithelium, but also facilitates colonization by bacteria such as Bacteroides fragilis.
GCs are highly specialized epithelial cells that, in addition to mucus production and secretion, are also essential for microbial antigen translocation, and the establishment of immune homeostasis during early life.11 An elegant study by Nystrom et al. demonstrated that based on their transcriptomic and proteomic profiles, intestinal GCs can be segregated into three major subsets.12 Proliferative GCs are found in the lower regions of intestinal crypts and give rise to two additional GC subsets – canonical GCs featuring an expression profile resembling that of mature GCs and non-canonical GCs with a transcriptional signature more closely overlapping than that of non-GC epithelial lineages (Figure 1B). Canonical and non-canonical GCs lining the intestinal crypts (collectively referred to as crypt GCs) secrete a thick, highly impenetrable mucus plume that fills and overlays the crypts, helping to protect the associated epithelial renewal machinery from bacterial invasion. Non-canonical crypt GC also give rise to sentinel GC that protect the crypt openings.13 In contrast, the terminal population of the canonical GC trajectory are the fully differentiated GCs of the surface epithelium or that open into the lumen and are collectively referred to as intercrypt goblet cells (icGCs). The icGCs produce a mucus layer that, despite being more permeable than the crypt mucus, helps to protect the surface epithelium from bacterial invasion. In both mice and humans, disruption of this intercrypt mucus results in microbiota-driven colonic inflammation.12
Major constituents of the colonic mucus layer
The primary component of the secreted colonic mucus layer is Muc2, which is heavily glycosylated. In fact, approximately 80% of the total molecular weight of mucins is derived from O-glycan carbohydrates.14 The non-redundant role of Muc2 in the structure and integrity of the colonic mucus barrier is evident based on the severe pathology that occurs in the distal colons of Muc2-deficient mice.15–18 Relative to that of the distal colon, the inner layer of the proximal colon displays a slightly reduced density and correspondingly enhanced permeability,19 which likely has important ramifications for host-microbiota crosstalk in the proximal colon. In addition to the secreted mucus layer, transmembrane mucins are tethered to the apical surface of gut epithelial cells by a single transmembrane domain and perform structural and anti-microbial functions (reviewed in 20). Like Muc2, the extracellular domains of these mucins are heavily glycosylated and can be released from the cell upon interacting with bacteria, representing an additional mechanism for restricting microbial access to epithelial cells.
Determination of mucus-associated proteins can be challenging since mucus recovered from tissue in experimental or clinical settings can potentially contain substances in transit through the intestinal lumen or derived from dead epithelial or other cells.10 An earlier study identified over 1000 proteins that were considered mucus associated, among them, several immune related proteins including anti-microbial peptides and various components of immunoglobulins.21 A recent, more refined analysis of sigmoid colon biopsies from healthy individuals and ulcerative colitis patients proposed a core mucus proteome of 29 proteins.22 These include Muc2, chloride channel regulator, calcium-activated-1 (Clca1), and zymogen granulae protein 16 (Zg16), as well as IgA and proteins associated with its dimerization and transport, J-chain (IGJ) and polymeric IgA receptor (PIGR), respectively. The proteome also includes the Fc fragment of IgG binding protein (FCGBP), the abundance and distribution of which is similar to that of MUC2. FCGBP is expressed in mucus secreting cells in the intestines and can bind covalently to MUC2 and other mucus proteins in both humans and mice via cleaved von Willebrand D domains.21,23 In mice, expression of FCGBP is sparse in the mucus layers of the stomach and small intestine and the most robust and consistent expression has been observed in the proximal colon followed by the distal colon.24 The name derives from studies demonstrating that plasma IgG from healthy individuals or Crohn’s disease (CD) patients bound via the Fc region to a high molecular weight protein detectable in the mucus granules of goblet cells and mucus layer of the intestines.25,26 This suggests a potential role for FCGBP in IgG-dependent regulation of mucus-associated microbes, particularly in the large intestine, and is also consistent with the presence of detectable levels of anti-commensal IgG in colonic mucus of mice.27 However, contrary to earlier work, a recent study using purified proteins in vitro did not detect any interaction of mouse FCGBP with IgG.28 Therefore, the functions of FCGBP in the mucus layer, including the existence and significance of any immune-related function, remain undefined. To date, there is no reported connection between FCGBP and IBD; however, reduced FCGBP has been associated with the onset and progression of colorectal cancer,29–33 which can develop secondary to chronic gut inflammation. However, the mechanism underlying this possible relationship also remains to be elucidated.
Isolation and visualization of mucus-associated bacteria
Most microbiota analyses continue to be performed on fecal bacteria, often due to practical reasons. Relative to mucus-associated samples, fecal samples are easier to collect and enable longitudinal analyses of the microbiota of a single individual. Biopsies of the gut wall can be collected from patients undergoing endoscopy and from experimental mice, but the limited amount of biological material available using this approach presents obvious challenges with the interpretation of the findings. Various technical approaches have been employed to collect and/or analyze mucus-associated bacteria from animal models. These include relatively crude methods such as scraping or vacuum-based suctioning of the mucosa following removal of luminal contents and any visible debris.6,7,27 These methods enable the collection of virtually all mucus-associated organisms from the entire mucus layer but are less than ideal for distinguishing inner mucus from outer mucus contents. Since these are terminal procedures, longitudinal analyses obviously cannot be conducted. However, samples that can be collected from any region of the intestine desired and under anaerobic conditions allow for preservation, culture, and isolation of viable, highly oxygen-sensitive organisms for culture and downstream functional analyses including transplants into axenic animals.
Different fractions of luminal microbes in the small versus large intestine are actively coated by IgA or IgG antibodies. This feature has been used extensively to enrich antibody-bound bacteria using flow cytometry before conducting genomic sequencing (IgA-Seq or IgG-Seq)34 or functional assays including gnotobiotic transplants. The organisms enriched via these methods include bacteria known to penetrate the mucus layer,2 but the point at which they become coated with these antibodies remains unclear, and therefore the utility of this approach for collecting and distinguishing mucus-associated bacteria is unknown. Importantly, whether and how the proximity of commensals to the colonic epithelium impacts homeostatic antibody responses are not very well understood. However, considering that certain commensals can utilize similar mechanisms as pathogens to gain entry into the mucus, it is possible to elicit highly specific responses to non-surface protein antigens generated after microbial engulfment and processing and presentation of lymphocyte receptor epitopes by antigen-presenting cells.
Several technical advances have enabled the visualization of microbes in whole tissue sections that preserve the host-microbiota spatial relationship.35,36 These approaches utilize fixed tissue and therefore do not allow for culture and isolation of viable organisms. A commonly utilized approach to examining the spatial segregation of gut bacteria and the mucus layers is bacterial fluorescence in situ hybridization (FISH). With this technique, undisturbed colonic tissues, still containing the luminal contents, are treated with specialized fixatives that preserve the mucus layer in its native form prior to being embedded in paraffin. Transverse tissue sections are then labeled using fluorescent probes that recognize conserved or species-specific bacterial DNA sequences.37–39 Although the mucus and microbiota can be effectively visualized, the common fixatives do not efficiently preserve the host tissue for immunostaining analysis. A recently described approach utilizing a specialized fixative formulation was demonstrated to overcome this concern.40
A more refined technique that has been utilized to study mucus-associated communities of both humans and rodents involves laser-capture microdissection (LCM) of defined areas of healthy tissue or disease lesions.41–43 This approach enables the precise collection of low biomass microbial samples from defined areas of the inner or outer mucus layer or even the epithelium. LCM requires equipment and technical expertise not readily accessible to all investigators, and given the spatial biological diversity along the length of the intestine, samples collected in this way can only be interpreted as representative of the specific area from which they were sourced.
Studying the interaction of the host and the mucus-associated microbiota
Mechanistic analysis of the functions of specific microbes often necessitates in vivo experiments that prove Koch’s postulate that the specific phenotype be reproduced when a pure culture of the organism is inoculated in a susceptible host. In the case of commensals, this generally requires that all other organisms be eliminated from the experimental system. The increased availability of gnotobiotic facilities has allowed for experiments whereby transplantation of pure cultures of known mucus-associated bacteria into axenic or ‘germ-free’ animals enable direct demonstration of specific functions in vivo. Though informative when successful, such experiments present important challenges regarding their execution and interpretation.44 Many mucus-associated bacteria are strict anaerobes, and colonization can be limited by the oxygen tension of the germ-free intestines. In addition, germ-free mice feature thinner mucus layers and under-developed immune systems, both of which are critical to the interactions of the specific microbes with the host under normal circumstances. Finally, goblet cell-associated antigen passages (GAPS) in the proximal colon, the closure of which is regulated by microbial density prior to weaning,45 are likely to be accessible in adult germ-free mice, potentially allowing for aberrant transfer of microbial antigens.
An increasingly prevalent approach to ex vivo study of the interaction of the host with the mucus-associated microbiota involves the use of colonic stem cells to generate epithelial monolayers that recapitulate the lineage diversity of the gut epithelium, including mucus-secreting goblet cells.46–48 This eliminates the need for cell lines, which may not always be representative of gut epithelial cell biology. More importantly, this approach allows for the differentiation of stem cells from specific regions of the intestine into mature lineages that recapitulate the transcriptional profile of their in vivo counterparts. In recent years, various highly innovative systems have been engineered to reproduce the environmental conditions that enable survival of oxygen-consuming host cells without compromising the viability of highly oxygen-sensitive microbes.49,50 The merger of such reductionist approaches should facilitate mechanistic discovery going forward, though with the caveat that the mammalian physiology that directly impacts gut function cannot be fully recapitulated in vitro.
Mechanisms of interaction between bacteria and mucus
A well-characterized example of a non-pathogenic interaction between host epithelial cells and commensals occurs in the mouse terminal ileum where Candidatus arthromitus, commonly known as segmented filamentous bacteria (SFB), breach the mucus layer and bind directly to epithelial cells.51 Indeed, the entire small intestine, including the ileum, is overlaid by a single, looser, and unattached mucus layer, which may potentially make it more permissive to invasion by SFB than the mucus layer of the large intestine. This might explain the absence, to date, of direct evidence of this type of invasion and interaction by non-pathogenic organisms anywhere else along the GI tract, including the colon.
Despite the relatively more robust mucus barrier, the colon mucus harbors many more microbes than the small intestine and certain microbes even gain access to the dense inner mucus layer. The diversity of organisms in the outer mucus layer is highest in regions closest to the epithelium,41 further increasing the likelihood of inner layer colonization and a consequent increase in the number of potential antigenic hits on the immune system. The probability of mucus colonization in the large bowel is further enhanced by the combination of slower transit times, increased availability of host-inaccessible carbohydrates left over from small bowel digestion, and the widespread capability to bind to and/or forage mucin O-glycans. Moreover, colonic crypts of both mice and humans harbor a diverse microbiota comprised both aerobic and anaerobic organisms that can also be found in the mucus layers.52,53 In addition to Bacteroidetes and Firmicutes, this crypt-specific core microbiota (CSCM) involves large proportions of Proteobacteria, especially in the murine cecum and proximal colon. The growth and expansion of Proteobacteria is enhanced by nutrients released from apoptotic epithelial cells,54 demonstrating that host-derived factors also facilitate microbial habitation of these privileged mucus-protected sites within the intestines.
Mucins can also serve as an anchor for bacterial colonization of the GI tract. Several taxa, including multiple species of Lactobacillus express adhesins that mediate binding to mucin glycans55 to facilitate colonization of and motility within, the mucus layer. An estimated 30% of gut commensals express type IV pili (T4P) – long and thin-surface appendages that facilitate bacterial adhesion but are also involved in motility, DNA exchange, and protein uptake.56 Fusobacterium nucleatum binds to colonic Muc2 and secretes outer membrane vesicles (OMVs) that permeate the mucus layer and can stimulate the gut epithelium.57 The genome of Bacteroides thetaiotaomicron (B. theta) consists of multiple polysaccharide utilization loci but also releases OMVs into the mucus layer that reach the immune system in the lamina propria in a sulfatase-dependent manner58 (Figure 1B).
A large fraction of the overall gut microbiota can directly target specific monosaccharides in secreted mucin O-glycans which can serve as a source of carbon and nitrogen for commensal organisms. One of the first and best described of these mucin-degraders is the aptly named Akkermansia muciniphila.59 It has become increasingly clear that the vast majority of commensals are capable of either cleaving or catabolizing at least one of the major mucin O-glycan monosaccharides – L-fucose (Fuc), D-galactose (Gal), N-acetyl-D-galactosamine (GalNAc), N-acetyl-D-glucosamine (GlcNAc), and N-acetyl-neuraminic acid (Neu5Ac).4,60 Some organisms, referred to as mucin generalists, and including several species of Muribaculaceae and Lachnospiraceae, can forage all five monosaccharides.61 Whether generalists or single monosaccharide foragers, the distinct preferences offer competitive and functional advantages within the gut microenvironment. For example, a recently described consortium of 5-sialic acid and N-acetylglucosamine utilizers can prevent colonization by the nosocomial pathogen Clostridium difficile.61
The inner mucus layer of the human and mouse colon has consistently been shown to be impermeable to bacteria-sized beads,38,62 demonstrating the reduced likelihood of passive encroachment of the gut epithelium by bacteria. However, several species of known mucus-associated bacteria encode multiple motility genes that aid their active movement into and through the mucus layer. This includes genes that encode flagellin monomers, proteins involved in flagellar biosynthesis, and the dozens of proteins that comprise the rotary motor. It also includes adhesin genes that enable binding to mucus but also potentially facilitate gliding motion along the mucus scaffold.63 Flagella are positioned on the bacterial cell in two major ways that can directly impact the motility mechanism(s) each can employ. In specific rod-shaped cells, one or more flagella can be positioned at or near one or both ends in what is known as a ‘polar’ or subterminal flagellin arrangement, respectively.63,64 Other organisms have flagellar filaments positioned along the length of the cell body in what is known as a ‘peritrichous’ distribution.63 Several mucus-associated bacteria, particularly Lachnospiraceae, express multiple distinct flagellins that exhibit distinct functional properties in vivo.65 The significance of this diversity is the subject of continued investigation but likely represents additional adaptations to help evade host control mechanisms designed to limit motility.
Establishing immune homeostasis with the mucus-associated microbiota
Innate sensing of bacterial flagellin
The existence of bacteria in such proximity to the epithelium triggers immunoregulatory programs that prevent activation of the immune system. Flagellated bacteria belonging to the families Lachnospiraceae and Ruminococcaceae are enriched in transverse folds of the mouse proximal colon.43 Accordingly, Toll-like receptor 5 (TLR5), the receptor for flagellin is constitutively expressed by epithelial cells in the proximal colon (Figure 1B) as well as Paneth cells in the small intestine.66 In neonates, TLR5 is expressed on epithelial cells in both the small and large intestines and regulates colonization by flagellated bacteria with important ramifications for microbiota composition into adulthood.66,67 After weaning, expression in the small intestine is downregulated in all but Paneth cells.66 Organoids generated in vitro from proximal colonic crypt stem cells also stably express TLR5 arguing that neither microbial nor immune signals are necessary to induce TLR5 expression.66 In epithelial cells, the NOD-like receptor (NLR) family caspase recruitment domain-containing protein 4 (NLRC4) can also be activated downstream of flagellin-sensing by NLR family apoptosis inhibitory proteins (NAIPs) in response to intracellular replication of pathogenic bacteria, such as Salmonella typhimurium. This results in the death of epithelial cells and expulsion into the gut lumen.68 However, this pathway appears dispensable for flagellin-induced gene transcription in the colonic epithelium.66 Instead, the ability to detect and respond to commensal-derived flagellin via TLR5 throughout life appears to be an intrinsic feature of epithelial cells and part of the genetic program of epithelial cell precursors and the mature enterocytes into which they differentiate. This is consistent with the non-redundant role of TLR5 as an intermediary in the generation of anti-flagellin antibody responses that regulate the composition and motility of the colonic microbiota.69 Considering the diversity of flagellins expressed even in phylogenetically related mucus-associated bacteria, evasion of TLR5-mediated sensing by epithelial cells is likely. Thus, in addition to epithelial cells, flagellin can also potently activate the NAIP/NLRC4 pathway in mouse and human macrophages.70–73 NAIP/NLRC4 in colonic macrophages likely represent a mechanism for regulating responses to such flagellated bacteria or flagellin antigens that cross the epithelial barrier.
Anti-commensal antibodies in control of microbial colonization and activity
The presence of immunoglobulins in the intestinal lumen provides an added layer of defense against epithelial invasion by pathogenic and commensal microbes. Intestinal IgM, as well as class-switched IgA and IgG antibodies, figure prominently in host–microbiota interactions from birth and extend throughout life. IgA is by far the most abundant immunoglobulin isotype present in the intestines and is involved in both exclusion of, and colonization by, mucus-associated commensals (Figure 1B). Bacteroides fragilis has evolved mechanisms for utilizing IgA to facilitate its engraftment within the intestines,74,75 and members of the phylum Firmicutes, especially those in the Lachnospiraceae family, are significantly decreased in patients with selective IgA deficiency.76 The protective role of intestinal IgG antibodies has also been widely studied but predominantly in the context of enteric infection.77 However, several recent studies (discussed below) offer compelling evidence of the involvement of anti-commensal IgG in promoting homeostasis in both the neonatal and adult stages of life.
IgA and IgG antibodies can be generated by either T-dependent or T-independent mechanisms, meaning, with or without the help of CD4 T cells, respectively. In the latter case, this occurs most commonly via direct stimulation of B cells via innate pattern-recognition receptors. T-independent anti-commensal antibodies generally target conserved carbohydrate moieties present in diverse bacteria 78 and, because of this, are sometimes referred to as being polyreactive. It should be noted that in this context, the term polyreactive relates to the diversity of microbes that can be recognized based on expression of common moieties, and not that these antibodies can bind a multiplicity of distinct antigens. In contrast, T-dependent antibodies target-specific protein epitopes either expressed externally or displayed by professional antigen-presenting cells after engulfment and degradation of microbes or microbial components. The predominant antibody isotype produced in the mammalian intestine and thus targeting gut bacteria is IgA. Rodents express a single isoform of IgA making it impossible to determine whether an IgA molecule from a wild-type mouse was produced in a T-dependent or T-independent manner. Therefore, in experimental settings, the distinction is based on whether the response can be induced when T-dependent antibody production is missing or impaired, for example, in T cell-deficient mice. Humans, on the other hand, produce two IgA subtypes, with IgA1 believed to be predominantly T-dependent and IgA2, T-independent.79,80 In contrast, both humans and mice express four distinct subtypes of IgG (IgG1-4 in humans, and IgG1, IgG2a/c, IgG2b, and IgG3 in mice). In both species, the generation of IgG1 is largely T-dependent, whereas IgG2 in humans and IgG3 in mice are generated in a T-independent manner and mainly target bacterial carbohydrate epitopes.81
The role of T-independent anti-commensal antibodies in establishing and maintaining microbial tolerance throughout life has been extensively demonstrated, especially in recent years. These include maternally derived IgA and IgG antibodies that limit immunoreactivity to the microbiota in early life,82 IgA antibodies that actively sequester the luminal bacteria in the small and large intestines,1 as well as anti-murein lipoprotein (MLP) IgG antibodies that target Gram-negative commensals and cross-react with MLP-expressing pathogens to limit systemic infection.83 Interestingly, polyreactive IgA produced by colonic plasma cells did not seem to target mucus-associated bacteria including firmicutes in the colon, further supporting the notion that at least some mucus-associated microbes necessitate high-affinity T-dependent antibody-mediated control.84
T-dependent antibody responses are known to be evoked by enteric pathogens – which also invade the mucus layer to gain access to the epithelium – and are both proportional to the threat sensed by the host and often necessary for effective clearance of the pathogen. However, the role of T-dependent antibodies in controlling commensal organisms is less well defined, with few examples of such interactions reported. IgA coating of luminal bacteria in both the small and large intestines was largely independent of T cells, except for SFB and Mucispirillum, both of which penetrate the mucus layer. Interestingly, the seemingly T-dependent coating of both organisms was also independent of somatic hypermutation Tfh cells,1 the latter of which can be directly stimulated to aid in the production of anti-commensal IgA.85 These studies provide evidence of the role of T-dependent IgA in control of a subset of gut microbes that can penetrate the mucus layer. Interestingly, total IgA-coated organisms isolated from IBD patient feces include at least a subset of organisms that efficiently colonize the colonic mucus layer and increase susceptibility of colonized mice to experimental colitis.2 The IgA-coated bacteria, which represent a minority of the total luminal community, are coated by both IgA1 and IgA286 suggesting potential cooperation between T-dependent and T-independent responses in luminal sequestration.
T-dependent IgG also appears to be involved in the regulation of host-microbiota crosstalk at the mucosal border as Akkermansia muciniphila, which can mount appreciable effector cell responses under specific conditions, also induces a robust T-dependent IgG1 response in mice under homeostatic conditions.87 Furthermore, mice with defective production of T-dependent antibodies (ICOSL-deficient mice) harbor reduced IgA and IgG antibodies specific to mucosal antigens, including antigens derived from multiple isolates of known mucus-associated bacteria27 (Figure 2). The potential colitogenicity of the microbiota appears to increase in the absence of this type of antibody-mediated control as ICOSL-deficiency predisposes to the development of severe, rapid onset colitis when immune regulation is genetically or transiently perturbed.27
Figure 2.
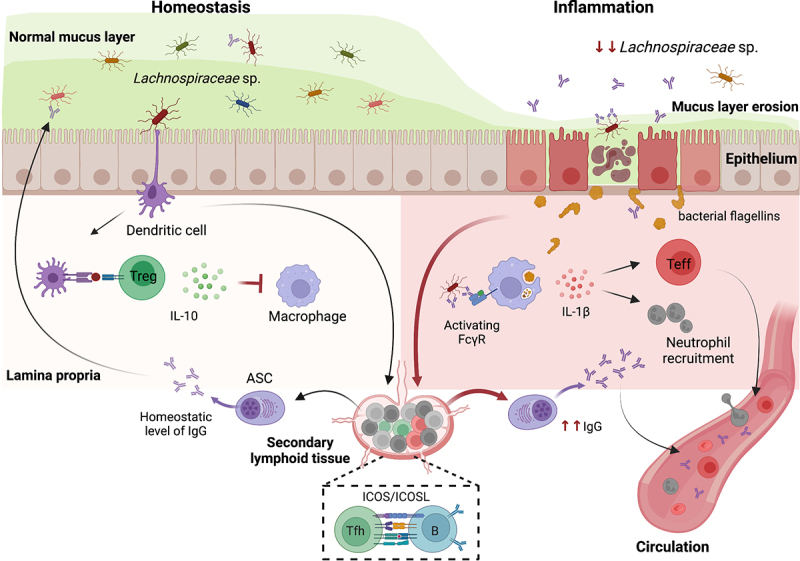
Examples of the crosstalk between mucus-associated bacteria and the intestinal immune system in health and disease. Under homeostatic conditions, the peaceful existence in the mucus layer of bacteria, including members of the Lachnospiraceae and Ruminococcaceae families, is aided by coordinated antigen collection by lamina propria myeloid cells including dendritic cells and macrophages. Intestinal dendritic cells promote the differentiation and/or maintenance of IL-10-producing Treg cells. IL-10 inhibits commensal-dependent activation of lamina propria macrophages, maintain the macrophages in a quiescent state. In addition, the presentation of commensal-derived antigens by B cells in the gut-draining lymph nodes induces differentiation into IgG+ antibody-secreting cells (ASCs) with the aid of ICOS-ICOSL-dependent T-follicular helper (Tfh) cells. This T-dependent ASC-derived IgG crosses the epithelium into the mucus layer and helps regulate the resident microbial communities. In contrast, the inflammatory state is characterized by an erosion of the mucus layer resulting in increased availability and opsonization of commensal antigens. This enables dysregulated differentiation and expansion of pro-inflammatory/effector CD4 T cells as well as hyperproduction of IgG antibodies to antigens including Lachnospiraceae flagellins. Excessive activation of intestinal macrophages via the interaction of IgG immune complexes with activating Fc gamma receptors (FcγR) induces a pro-inflammatory phenotype including secretion of cytokines such as IL-1β which can promote neutrophil recruitment. In CD, the chronic inflammatory state is also characterized by an overall reduction in the abundance of Lachnospiraceae bacteria and a breach in the so-called ‘mucosal firewall’ resulting in effector CD4 T cells and anti-commensal IgG antibodies entering the systemic circulation.
Microbial induction of T cell-mediated immune regulation
From the earliest stages of life, mucosal antigens are actively involved in promoting the development and expansion of a robust arsenal of regulatory T cells (Treg cells) that provide beneficial suppression of anti-commensal immunity well into adulthood. Mucus-producing goblet cells serve as conduits for Treg-inducing mucosal antigens during a critical window during the postnatal period.45 Although the specific antigens driving this differentiation and expansion have not been clearly defined, the time interval of postnatal days 11–20 in mice is associated with the rapid and organized acquisition of diverse organisms via maternal and potentially environmental transfer. Further, the transfer of these antigens via goblet cells suggests they derive from organisms that intimately interact with the mucus layer from the earliest stages of colonization. Experiments in which specific microbes were transferred into adolescent or adult germ-free mice have demonstrated the potential of many different mucus-associated bacteria or bacterial consortia leading to induction and/or accumulation of intestinal regulatory T cells. These organisms include Bacteroides fragilis,88 Helicobacter spp.,89,90 the Lachnospiraceae family member Roseburia intestinalis,65 and a consortium including several species of Lachnospiraceae.91
A key T cell-derived mediator of gut immune homeostasis is the immunoregulatory cytokine interleukin-10 (IL-10) which, in the intestines, is necessary to actively limit macrophage activation in response to commensal-derived triggers.92 Mucus-associated bacteria, including Bacteroides fragilis, B. acidifaciens, Akkermansia muciniphila, and Bifidobacterium spp., either induce T cell expression of IL-10 or require IL-10 to limit their activation of the immune system.93–96 It is likely that all these organisms utilize distinct mechanisms to trigger IL-10 production, but the activation of this pathway by such phylogenetically distinct organisms suggest that CD4 T cell production of IL-10 is also a dominant mechanism for tolerance of specifically mucus-associated bacteria. Expression of IL-10 by CD4 T cells increases significantly in the colonic lamina propria of mice in which production of T-dependent antibodies is impaired. Accordingly, simultaneous ablation of T-dependent antibodies and CD4 T cell-derived IL-10 in either early life or adulthood, results in rapid onset of colonic inflammation.27 These findings, coupled with the fact that Akkermansia muciniphila induces both IgG1 and IL-10, support the theory that specific mucus-associated bacteria trigger T-dependent antibodies and CD4 T cell-derived IL-10, and that synergy between both pathways may be critical for preventing pro-inflammatory responses to these microbes. Interestingly, prior to the onset of colitis that typically occurs in IL-10-deficient mice, the inner mucus layer was found to be significantly thicker but more permeable to bacteria-sized beads.62 This suggests a potential role for IL-10 might IL-10 in controlling mucus production and/or mucus layer integrity, further regulating access to the epithelium by mucus-associated bacteria.
Mucus-associated bacteria and gastrointestinal disease
Due to the non-redundant protective function of the gut mucus layer, factors that either directly or indirectly promote erosion or penetration of this layer can induce significant disturbances in microbiota composition, creating favorable environments for bacterial invasion and onset of gut inflammation. For example, the absence of fiber in the diet deprives commensals of a reliable source of nutrients, which results in excessive foraging of the mucus layer. This leads to microbiota dysbiosis, which favors outgrowth of pathogenic organisms and enhances susceptibility to colitis.97 Dietary emulsifiers, common components of processed foods, also promote bacterial encroachment of the colonic epithelium, which favors the development of metabolic syndrome and susceptibility to, or severity of, experimental colitis.98,99 It is reasonable to speculate that these observations are due to the direct effects of emulsifiers on the structure and function of mucus. However, current experimental evidence suggests that emulsifiers license pathobionts such as adherent-invasive E. coli (AIEC) for mucus penetration and epithelial activation by stimulating upregulation of virulence and motility factors.100,101 Interestingly, under these conditions, mucin expression by colonic GCs is upregulated in response to AIEC.100 Such enhanced mucus secretion is a typical response to epithelial encroachment and is seen in the presence of enteric infections by helminths, protozoan parasites, and pathogenic bacteria.102 This response can further disrupt the mucosal ecosystem and further exacerbate pathogen-induced dysbiosis leading to inflammation.
Pathobionts and IBD
The etiology of IBD is believed to involve both the overgrowth of harmful opportunists or pathobionts and the loss of beneficial bacteria. The possible relationship between CD and mucus is still somewhat controversial, whereas it is established and widely accepted that UC patients display a thinner mucus layer, reduced GCs, and defective mucus production and secretion. In the quest to understand the role of specific bacteria in promoting gastrointestinal disorders including IBD, correlative as well as mechanistic associations have been made between specific mucus-associated bacteria and chronic gut inflammation. In mice colonized with the Gram-negative mucus-associated pathobiont Mucispirillum schaedleri, combined deficiency of IBD-susceptibility gene nucleotide-binding oligomerization domain-containing protein 2 (Nod2) and the cytochrome b-245 beta-chain subunit of phagocyte NADPH oxidase develop spontaneous colitis after weaning as the protection afforded by maternal antibodies begins to wane.103 The disease is characterized by an expansion of M. schaedleri due to impaired recruitment of luminal neutrophils necessary to clear the bacteria. This illustrates how a defect in innate immune-mediated regulation can enable mucus-associated bacterial overgrowth, which overwhelms the host's adaptive immunity to promote colonic inflammation.
Ruminococcus gnavus (R. gnavus) is also significantly increased in both CD and UC, sometimes transiently reaching a relative abundance of greater than 50% of patient fecal bacteria and dominated by strains capable of surviving the oxidative stressors characteristic of the inflamed gut.43,104,105 Therefore, like AIEC,106 the expansion of R. gnavus in IBD is possibly the response of an opportunist to local conditions. Notably, however, patient isolates of R. gnavus can be distinguished by the presence or absence of a thick polysaccharide capsule that masks the cell wall components. Strains expressing this capsule are largely inert and favor tolerogenic responses in vivo. In contrast, unencapsulated strains enhance the severity of inflammation,107,108 supporting the involvement of R. gnavus as a microbial driver of IBD.
Inverse correlations of mucus-associated bacteria and colonic inflammation
Associations have also been made between low abundance, albeit in the feces, of known mucus-associated bacteria and disease activity of severity in IBD.109 Reduced fecal and mucosal abundance of Faecalibacterium prausnitzii has been observed in the feces of patients with CD and UC.110–112 The relative abundance of members of the families Ruminococcaceae and Lachnospiraceae, including Roseburia hominis are also negatively correlated with disease activity in CD and UC.113–116 Lachnospiraceae were among the most decreased bacterial families in the ileocecal biopsies in treatment-naïve Crohn’s disease patients,117 suggesting that the elimination or suppression of this family of organisms potentially contributes to disease onset. In support of this theory, transplantation of a randomly selected consortium of 23 Lachnospiraceae organisms enriched from the mouse feces, cecal contents, and cecal tissue was able to repair the dysbiosis observed in NLRP12 inflammasome-deficient mice and limit the severity of the colitis that ensued when the mice were subsequently administered dextran sulfate sodium (DSS).118
Anti-Lachnospiraceae immune responses in IBD
The total levels of fecal IgG are non-specifically elevated and bind luminal microbes in both UC and CD.119 However, immunoreactivity of antigens derived specifically from members of the Lachnospiraceae family are strongly implicated in IBD. Specifically, reactivity of Lachnospiraceae flagellin antigens continues to be utilized as a diagnostic tool in IBD and IgG seroreactivity for CBir1, a flagellin antigen isolated from cecal Lachnospiraceae, is significantly increased in patients with CD, but not in UC patients or healthy controls. This reactivity is present in CD cohorts collected from different locations and in different age groups, making it an effective biomarker for CD diagnosis and prognosis.120–122 Alexander et al. recently expanded the spectrum of flagellin antigens that are targeted by CD serum IgG to an array of human and mouse Lachnospiraceae isolates and observed that approximately 30% of CD patients displayed an elevated serum IgG response to more than 10 different flagellins. This multi-flagellin reactivity was highly correlated with heightened responses to CBir1 flagellin, but not with reactivities to flagellins from Salmonella dublin (S. dublin) or E. coli. In further support of a highly targeted response to specific commensal-derived antigenic epitopes, these patients concomitantly displayed increased flagellin-specific CD4+ effector/memory cells in the circulation 123 (Figure 2). Within this extremely diverse family, many of the organisms can express multiple flagellins, adding an additional layer of molecular complexity that likely impacts recognition by the host immune system. At a minimum, the observation that the targeted deletion of Lachnospiraceae flagellin-specific CD4 T cells in the circulation can prevent experimental colitis in mice 124 makes this axis a potentially viable target for intervention in CD.
Despite these robust IgG responses, it is still unknown whether the antibodies detectable in the circulation of CD patients were induced by disseminated bacteria, produced by circulating B cells that originate in the gut, or produced in the gut before entering the circulation. Thus, it remains to be determined whether these well-established anti-Lachnospiraceae responses are a cause or consequence of disease. IgG, acting via activating Fc gamma receptors (FcγRs) on gut-resident macrophages, potently drives inflammation in UC.125,126 Accordingly, a single-nucleotide polymorphism in FCGR2A that reduces the affinity of this receptor for IgG is protective in UC127,128 supporting the notion that massive IgG response to known mucus-associated organisms in CD could indeed be a contributor to disease induction and/or progression (Figure 2). On the other hand, a similar magnitude of Lachnospiraceae flagellin-specific serum IgG response was observed in healthy children at 12–24 months of age. This response likely originated in the infant gut and was not passively acquired since comparable responses were not detected in mothers or unrelated adult control subjects.129 This period in humans also coincides with the introduction of solid food and a turbulent period of microbiota diversification and expansion that drives the immunological “weaning reaction” documented in mice.130 Interestingly, the heightened response eventually receded to the level of healthy adults by 7 years of age.129 Thus, an alternative possibility is that the increased IgG antibodies targeting Lachnospiraceae flagellins found in adult CD patients result from an over-exuberant recall response toward mucus-associated commensal antigens triggered or enhanced by the co-occurrence of other disease-promoting events.
Conclusions and future perspectives
The colon mucus layer represents an ecosystem that is vital to the host-microbial relationships essential to mammalian life. There is still much to be learned regarding the host adaptations that facilitate this co-existence and whether and how those responses are related to the etiology and/or progression of chronic intestinal inflammation. Our understanding will improve following investigations into the nature of the physical and biochemical interactions that define the mutualism between mucus-associated microbes and the host, the potential involvement of such organisms in the initiation and/or perpetuation of gut inflammation, and the potential utility of mucus-associated microbes or microbial products toward therapeutic ends. Several emerging technical and technological advances point to an exciting future of discovery regarding host-microbiota mutualism at the colonic mucosal border. For example, continued refinement of technical approaches for (1) aseptic collection of microbes directly from the mucus layers of the colon, and (2) efficient characterization of the gene expression patterns at specific points should enable determination of the microbial molecular pathways that promote existence of specific organisms in this locale, including direct and indirect communication with the immune system. Additional improvements in, and availability of, approaches for high-throughput culture and isolation of distinct organisms from these delicate biological samples will lead to mechanistic studies at the level of bacterial strains. The differential manipulability of specific organisms implicated in gastrointestinal disease remains a persistent challenge. Genetic modifications of certain Protobacteria and Bacteroides have provided important insight into the potential functions of organisms including E. coli, and B. theta, and B. fragilis. However, other organisms, many of which still have not been cultured in a laboratory, are less genetically tractable and for now, this limits our ability to study their function. Recent innovative advances offer hope for precise genetic manipulation of individual organisms or communities of varying size and diversity.131–134 Many techniques deployed to date have predominantly engineered the bacteria in vitro, but others have demonstrated successful in situ modification of microbes recovered from the lumen of recipient animals.135,136 The latter approaches raise the possibility of whole-microbiome engineering in vivo for therapeutic purposes, although novel or modified approaches may be necessary to specifically target consequential organisms residing deep within the colonic mucus layer.
Acknowledgments
We would like to thank L. Wayne Duck for critical reading of the review. We apologize to our colleagues whose work we were unable to include due to space limitations. Figures were generated using BioRender.com.
Funding Statement
This work was supported by the Crohn’s and Colitis Foundation [Research Initiatives Award]; Kenneth Rainin Foundation [Synergy Award]; National Institutes of Health [DK118386]; National Institutes of Health [AI162736];Takeda Pharmaceuticals U.S.A.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Bunker JJ, Flynn T, Koval J, Shaw D, Meisel M, McDonald B, Ishizuka I, Dent A, Wilson P, Jabri B, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43:541–19. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palm NW, De zoete M, Cullen T, Barry N, Stefanowski J, Hao L, Degnan P, Hu J, Peter I, Zhang W, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS.. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect Immun. 2004;72:1666–1676. doi: 10.1128/IAI.72.3.1666-1676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravcheev DA, Thiele I. Comparative genomic analysis of the human gut microbiome reveals a broad distribution of metabolic pathways for the degradation of host-synthetized mucin glycans and utilization of mucin-derived monosaccharides. Front Genet. 2017;8:111. doi: 10.3389/fgene.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riva A, Kuzyk O, Forsberg E, Siuzdak G, Pfann C, Herbold C, Daims H, Loy A, Warth B, Berry D, et al. A fiber-deprived diet disturbs the fine-scale spatial architecture of the murine colon microbiome. Nat Commun. 2019;10:4366. doi: 10.1038/s41467-019-12413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donaldson GP, Chou W-C, Manson AL, Rogov P, Abeel T, Bochicchio J, Ciulla D, Melnikov A, Ernst PB, Chu H, et al. Spatially distinct physiology of bacteroides fragilis within the proximal colon of gnotobiotic mice. Nat Microbiol. 2020;5:746–756. doi: 10.1038/s41564-020-0683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, and Rupp S, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. 2015;6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Luo J, Li J, Kim G, Chen ES, Xiao S, Snapper SB, Bao B, An D, Blumberg RS, et al. Foxo1 controls gut homeostasis and commensalism by regulating mucus secretion. J Exp Med. 2021;218. doi: 10.1084/jem.20210324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arike L, Seiman A, van der Post S, Rodriguez Piñeiro AM, Ermund A, Schütte A, Bäckhed F, Johansson MEV, Hansson GC. Protein turnover in epithelial cells and mucus along the gastrointestinal tract is coordinated by the spatial location and microbiota. Cell Rep. 2020;30:1077–1087 e1073. doi: 10.1016/j.celrep.2019.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knoop KA, Newberry RD. Goblet cells: multifaceted players in immunity at mucosal surfaces. Mucosal Immunol. 2018;11:1551–1557. doi: 10.1038/s41385-018-0039-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nystrom EEL, Martinez-Abad B, Arike L, Birchenough GMH, Nonnecke EB, Castillo PA, Svensson F, Bevins CL, Hansson GC, Johansson MEV, et al. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science. 2021;372. doi: 10.1126/science.abb1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birchenough GM, Nystrom EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535–1542. doi: 10.1126/science.aaf7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gendler SJ, Spicer AP. Epithelial mucin genes. Annu Rev Physiol. 1995;57:607–634. doi: 10.1146/annurev.ph.57.030195.003135. [DOI] [PubMed] [Google Scholar]
- 15.Wenzel UA, Magnusson MK, Rydström A, Jonstrand C, Hengst J, Johansson ME, Velcich A, Öhman L, Strid H, Sjövall H, et al. Spontaneous colitis in Muc2-deficient mice reflects clinical and cellular features of active ulcerative colitis. PLoS One. 2014;9:e100217. doi: 10.1371/journal.pone.0100217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burger-van Paassen N, van der Sluis M, Bouma J, Korteland-van Male AM, Lu P, Van Seuningen I, Boehm G, van Goudoever JB, Renes IB. Colitis development during the suckling-weaning transition in mucin Muc2-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2011;301:G667–678. doi: 10.1152/ajpgi.00199.2010. [DOI] [PubMed] [Google Scholar]
- 17.Van der Sluis M, De Koning BAE, De Bruijn ACJM, Velcich A, Meijerink JPP, Van Goudoever JB, Büller HA, Dekker J, Van Seuningen I, Renes IB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K, Augenlicht L, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 19.Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 2013;305:G341–347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelaseyed T, Hansson GC. Membrane mucins of the intestine at a glance. J Cell Sci. 2020:133. doi: 10.1242/jcs.240929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson ME, Thomsson KA, Hansson GC. Proteomic analyses of the two mucus layers of the colon barrier reveal that their main component, the Muc2 mucin, is strongly bound to the Fcgbp protein. J Proteome Res. 2009;8:3549–3557. doi: 10.1021/pr9002504. [DOI] [PubMed] [Google Scholar]
- 22.van der Post S, Jabbar KS, Birchenough G, Arike L, Akhtar N, Sjovall H, Johansson MEV, Hansson GC. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut. 2019;68:2142–2151. doi: 10.1136/gutjnl-2018-317571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada N, Iijima S, Kobayashi K, Yoshida T, Brown WR, Hibi T, Oshima A, Morikawa M. Human IgGFc binding protein (FcgammaBP) in colonic epithelial cells exhibits mucin-like structure. J Biol Chem. 1997;272:15232–15241. doi: 10.1074/jbc.272.24.15232. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Pineiro AM, Bergström JH, Ermund A, Gustafsson JK, Schütte A, Johansson MEV, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am J Physiol Gastrointest Liver Physiol. 2013;305:G348–356. doi: 10.1152/ajpgi.00047.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi K, Hamada Y, Blaser MJ, Brown WR. The molecular configuration and ultrastructural locations of an IgG Fc binding site in human colonic epithelium. J Immunol. 1991;146:68–74. [PubMed] [Google Scholar]
- 26.Kobayashi K, Blaser MJ, Brown WR. Identification of a unique IgG Fc binding site in human intestinal epithelium. J Immunol. 1989;143:2567–2574. [PubMed] [Google Scholar]
- 27.Landuyt AE, Klocke BJ, Duck LW, Kemp KM, Muir RQ, Jennings MS, Blum SI, Tse HM, Lee G, Morrow CD, et al. ICOS ligand and IL-10 synergize to promote host-microbiota mutualism. Proc Natl Acad Sci U S A. 2021;118:e2018278118. doi: 10.1073/pnas.2018278118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrencrona E, van der Post S, Gallego P, Recktenwald CV, Rodriguez-Pineiro AM, Garcia-Bonete M-J, Trillo-Muyo S, Bäckström M, Hansson GC, Johansson MEV, et al. The IgGFc-binding protein FCGBP is secreted with all GDPH sequences cleaved but maintained by interfragment disulfide bonds. J Biol Chem. 2021;297:100871. doi: 10.1016/j.jbc.2021.100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhuang Q, Shen A, Liu L, Wu M, Shen Z, Liu H, Cheng Y, Lin X, Wu X, Lin W, et al. Prognostic and immunological roles of Fc fragment of IgG binding protein in colorectal cancer. Oncol Lett. 2021;22:526. doi: 10.3892/ol.2021.12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan Z, Zhao Z, Hu H, Zhu Y, Zhang W, Tang Q, Huang R, Gao F, Zou C, Wang G, et al. IgG Fc Binding Protein (FCGBP) is down-regulated in metastatic lesions and predicts survival in metastatic colorectal cancer patients. Onco Targets Ther. 2021;14:967–977. doi: 10.2147/OTT.S285171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma R, Jing C, Zhang Y, Cao H, Liu S, Wang Z, Chen D, Zhang J, Wu Y, Wu J, et al. The somatic mutation landscape of Chinese Colorectal Cancer. J Cancer. 2020;11:1038–1046. doi: 10.7150/jca.37017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W, Shi J, Zhou Y, Liu T, Zhan F, Zhang K, Liu N. Integrating proteomics and transcriptomics for the identification of potential targets in early colorectal cancer. Int J Oncol. 2019;55:439–450. doi: 10.3892/ijo.2019.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi C, Hong L, Cheng Z, Yin Q. Identification of metastasis-associated genes in colorectal cancer using metaDE and survival analysis. Oncol Lett. 2016;11:568–574. doi: 10.3892/ol.2015.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosen CE, Palm NW. Functional classification of the gut microbiota: the key to cracking the microbiota composition code: functional classifications of the gut microbiota reveal previously hidden contributions of indigenous gut bacteria to human health and disease. Bioessays. 2017;39:1700032. doi: 10.1002/bies.201700032. [DOI] [PubMed] [Google Scholar]
- 35.Mondragón-Palomino O, Poceviciute R, Lignell A, Griffiths JA, Takko H, Ismagilov RF. 2021. 3D imaging for the quantification of spatial patterns in microbiota of the intestinal mucosa. doi: 10.1101/2021.10.07.463215 https://www.biorxiv.org/content/10.1101/2021.10.07.463215v1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tropini C, Earle KA, Huang KC, Sonnenburg JL. The gut microbiome: connecting spatial organization to function. Cell Host Microbe. 2017;21:433–442. doi: 10.1016/j.chom.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavelle A, Lennon G, O’Sullivan O, Docherty N, Balfe A, Maguire A, Mulcahy HE, Doherty G, O’Donoghue D, Hyland J, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. 2015;64:1553–1561. doi: 10.1136/gutjnl-2014-307873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson ME, Hansson GC. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Mol Biol. 2012;842:229–235. doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- 39.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macedonia MC, Drewes JL, Markham NO, Simmons AJ, Roland JT, Vega PN, Scurrah CR, Coffey RJ, Shrubsole MJ, Sears CL, et al. Clinically adaptable polymer enables simultaneous spatial analysis of colonic tissues and biofilms. NPJ Biofilms Microbiomes. 2020;6:33. doi: 10.1038/s41522-020-00143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan K, Carey-Ewend K, Vaishnava S. Spatial analysis of gut microbiome reveals a distinct ecological niche associated with the mucus layer. Gut Microbes. 2021;13:1874815. doi: 10.1080/19490976.2021.1874815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chassaing B, Gewirtz AT. Identification of inner mucus-associated bacteria by laser capture microdissection. Cell Mol Gastroenterol Hepatol. 2019;7:157–160. doi: 10.1016/j.jcmgh.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter J, Armet AM, Finlay BB, Shanahan F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell. 2020;180:221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 45.Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S, Kim D, Hsieh C-S, Hogan SP, Elson CO, et al. Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol. 2017;2. doi: 10.1126/sciimmunol.aao1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sontheimer-Phelps A, Chou DB, Tovaglieri A, Ferrante TC, Duckworth T, Fadel C, Frismantas V, Sutherland AD, Jalili-Firoozinezhad S, Kasendra M, et al. Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cell Mol Gastroenterol Hepatol. 2020;9:507–526. doi: 10.1016/j.jcmgh.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 48.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Huang Y-J, Yoon JY, Kemmitt J, Wright C, Schneider K, Sphabmixay P, Hernandez-Gordillo V, Holcomb SJ, and Bhushan B, et al. Primary human colonic mucosal barrier crosstalk with super oxygen-sensitive Faecalibacterium prausnitzii in continuous culture. Med (NY) . 2021;2:74–98 e79. doi: 10.1016/j.medj.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Etienne-Mesmin L, Chassaing B, Desvaux M, De Paepe K, Gresse R, Sauvaitre T, Forano E, de Wiele TV, Schüller S, Juge N, et al. Experimental models to study intestinal microbes-mucus interactions in health and disease. FEMS Microbiol Rev. 2019;43:457–489. doi: 10.1093/femsre/fuz013. [DOI] [PubMed] [Google Scholar]
- 51.Ladinsky MS, Araujo LP, Zhang X, Veltri J, Galan-Diez M, Soualhi S, Lee C, Irie K, Pinker EY, Narushima S, et al. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science. 2019;363. doi: 10.1126/science.aat4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saffarian A, Mulet C, Regnault B, Amiot A, Tran-Van-Nhieu J, Ravel J, Sobhani I, Sansonetti PJ, and Pédron T, Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. mBio. 2019;10. doi: 10.1128/mBio.01315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pedron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, Sansonetti PJ. A crypt-specific core microbiota resides in the mouse colon. mBio. 2012:3. doi: 10.1128/mBio.00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson CJ, Medina CB, Barron BJ, Karvelyte L, Aaes TL, Lambertz I, Perry JSA, Mehrotra P, Gonçalves A, Lemeire K, et al. Microbes exploit death-induced nutrient release by gut epithelial cells. Nature. 2021;596:262–267. doi: 10.1038/s41586-021-03785-9. [DOI] [PubMed] [Google Scholar]
- 55.Muscariello L, De Siena B, Marasco R. Lactobacillus cell surface proteins involved in interaction with mucus and extracellular matrix components. Curr Microbiol. 2020;77:3831–3841. doi: 10.1007/s00284-020-02243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ligthart K, Belzer C, de Vos WM, Tytgat HLP. Bridging bacteria and the gut: functional aspects of type IV pili. Trends Microbiol. 2020;28:340–348. doi: 10.1016/j.tim.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Engevik MA, Danhof HA, Ruan W, Engevik AC, Chang-Graham AL, Engevik KA, Shi Z, Zhao Y, Brand CK, Krystofiak ES, et al. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio. 2021;12. doi: 10.1128/mBio.02706-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hickey CA, Kuhn K, Donermeyer D, Porter N, Jin C, Cameron E, Jung H, Kaiko G, Wegorzewska M, Malvin N, et al. Colitogenic bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe. 2015;17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 60.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pereira FC, Wasmund K, Cobankovic I, Jehmlich N, Herbold CW, Lee KS, Sziranyi B, Vesely C, Decker T, and Stocker, R, et al. Rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile colonization. Nat Commun. 2020;11:5104. doi: 10.1038/s41467-020-18928-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johansson ME, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjövall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wadhwa N, Berg HC. Bacterial motility: machinery and mechanisms. Nat Rev Microbiol. 2021. doi: 10.1038/s41579-021-00626-4. [DOI] [PubMed] [Google Scholar]
- 64.Duncan SH, Aminov RI, Scott KP, Louis P, Stanton TB, Flint HJ . Proposal of roseburia faecis sp. nov., roseburia hominis sp. nov. and roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol. 2006;56:2437–2441. doi: 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- 65.Patterson AM, Mulder IE, Travis AJ, Lan A, Cerf-Bensussan N, Gaboriau-Routhiau V, Garden K, Logan E, Delday MI, Coutts AGP, et al. Human gut symbiont roseburia hominis promotes and regulates innate immunity. Front Immunol. 2017;8:1166. doi: 10.3389/fimmu.2017.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price AE, Shamardani K, Lugo KA, Deguine J, Roberts AW, Lee BL, Barton GM. A map of toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity. 2018;49:560–575 e566. doi: 10.1016/j.immuni.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fulde M, Sommer F, Chassaing B, van Vorst K, Dupont A, Hensel M, Basic M, Klopfleisch R, Rosenstiel P, Bleich A, et al. Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature. 2018;560:489–493. doi: 10.1038/s41586-018-0395-5. [DOI] [PubMed] [Google Scholar]
- 68.Sellin ME, Muller AA, Felmy B, Dolowschiak T, Diard M, Tardivel A, Maslowski KM, and Hardt WD . Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 2014;16:237–248. doi: 10.1016/j.chom.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 69.Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reyes Ruiz VM, Ramirez J, Naseer N, Palacio NM, Siddarthan IJ, Yan BM, Boyer MA, Pensinger DA, Sauer JD, Shin S, et al . Broad detection of bacterial type III secretion system and flagellin proteins by the human NAIP/NLRC4 inflammasome. Proc Natl Acad Sci U S A. 2017;114:13242–13247. doi: 10.1073/pnas.1710433114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kortmann J, Brubaker SW, Monack DM. Cutting edge: inflammasome activation in primary human macrophages is dependent on flagellin. J Immunol. 2015;195:815–819. doi: 10.4049/jimmunol.1403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao Y, Yang J, Shi J, Gong Y-N, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 73.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou W-C, Conner ME, Earl AM, Knight R, Bjorkman PJ, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018;360:795–800. doi: 10.1126/science.aaq0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL, Foster KR. Host selection of microbiota via differential adhesion. Cell Host Microbe. 2016;19:550–559. doi: 10.1016/j.chom.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 76.Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, Autaa G, Gouas D, Almeida M, Lepage P, et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. 2018;10. doi: 10.1126/scitranslmed.aan1217. [DOI] [PubMed] [Google Scholar]
- 77.Chen K, Magri G, Grasset EK, Cerutti A. Rethinking mucosal antibody responses: igM, IgG and IgD join IgA. Nat Rev Immunol. 2020;20:427–441. doi: 10.1038/s41577-019-0261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mathias A, Corthesy B. Recognition of gram-positive intestinal bacteria by hybridoma- and colostrum-derived secretory immunoglobulin A is mediated by carbohydrates. J Biol Chem. 2011;286:17239–17247. doi: 10.1074/jbc.M110.209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine April. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 80.Ladjeva I, Peterman JH, Mestecky J. IgA subclasses of human colostral antibodies specific for microbial and food antigens. Clin Exp Immunol. 1989;78:85–90. [PMC free article] [PubMed] [Google Scholar]
- 81.Castro-Dopico T, Clatworthy MR. IgG and Fcgamma receptors in intestinal immunity and inflammation. Front Immunol. 2019;10:805. doi: 10.3389/fimmu.2019.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koch MA, Reiner G, Lugo K, Kreuk LM, Stanbery A, Ansaldo E, Seher T, Ludington W, Barton G. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell. 2016;165:827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, Núñez G. Gut microbiota-induced immunoglobulin g controls systemic infection by symbiotic bacteria and pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M, Jabri B, Antonopoulos DA, Wilson PC, Bendelac A, et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 2017;358. doi: 10.1126/science.aan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kubinak JL, Petersen C, Stephens W, Soto R, Bake E, O’Connell R, Round J. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 2015;17:153–163. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sterlin D, Fadlallah J, Adams O, Fieschi C, Parizot C, Dorgham K, Rajkumar A, Autaa G, El-Kafsi H,Charuel, JL, et al. Human IgA binds a diverse array of commensal bacteria. J Exp Med. 2020;217. doi: 10.1084/jem.20181635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, Brown EM, Graham DB, Xavier RJ, Moon JJ, et al. Akkermansia muciniphil induces intestinal adaptive immune responses during homeostasis. Science. 2019;364:1179–1184. doi: 10.1126/science.aaw7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, Littman DR, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 2018;554:373–377. doi: 10.1038/nature25500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chai JN, Peng Y, Rengarajan S, Solomon BDAi, TLShen Z, Perry JSA, Knoop KA, Tanoue T, Narushima S, et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol. 2017;2. doi: 10.1126/sciimmunol.aal5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 92.Zigmond E, Bernshtein B, Friedlander G, Walker C, Yona S, Kim K-W, Brenner O, Krauthgamer R, Varol C, Müller W, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 2014;40:720–733. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 93.Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wild B, Wanek W, Richter A, Rauch I, Decker T, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci U S A. 2013;110:4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, O’Connell Motherway M, Shanahan F, Nally K, Dougan G, et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A. 2012;109:2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, and Florin TH . Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 97.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–1353 e1321. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rousta E, Oka A, Liu B, Herzog J, Bhatt AP, Wang J, Habibi Najafi MB, Sartor RB. The emulsifier carboxymethylcellulose induces more aggressive colitis in humanized mice with inflammatory bowel disease microbiota than polysorbate-80. Nutrients. 2021;13:3565. doi: 10.3390/nu13103565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Viennois E, Bretin A, Dubé PE, Maue AC, Dauriat CJG, Barnich N, Gewirtz AT, Chassaing B. Dietary emulsifiers directly impact adherent-invasive E. coli gene expression to drive chronic intestinal inflammation. Cell Rep. 2020;33:108229. doi: 10.1016/j.celrep.2020.108229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chassaing B, Van de Wiele T, De Bodt J, Marzorati M, Gewirtz AT. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut. 2017;66:1414–1427. doi: 10.1136/gutjnl-2016-313099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grondin JA, Kwon YH, Far PM, Haq S, Khan WI. Mucins in intestinal mucosal defense and inflammation: learning from clinical and experimental studies. Front Immunol. 2020;11:2054. doi: 10.3389/fimmu.2020.02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Caruso R, Mathes T, Martens EC, Kamada N, Nusrat A, Inohara N, Núñez G. A specific gene-microbe interaction drives the development of Crohn’s disease-like colitis in mice. Sci Immunol. 2019:4. doi: 10.1126/sciimmunol.aaw4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:103. doi: 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser A-L, Barnich N, Bringer M-A, Swidsinski A, Beaugerie L, Colombel J-F, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 107.Henke MT, Brown EM, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J. Capsular polysaccharide correlates with immune response to the human gut microbe Ruminococcus gnavus. Proc Natl Acad Sci U S A. 2021;118:e2007595118. doi: 10.1073/pnas.2007595118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci U S A. 2019;116:12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pittayanon R, Lau JT, Leontiadis GI, Tse F, Yuan Y, Surette M, Moayyedi P. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology. 2020;158:930–946 e931. doi: 10.1053/j.gastro.2019.11.294. [DOI] [PubMed] [Google Scholar]
- 110.Altomare A, Putignani L, Del Chierico F, Cocca S, Angeletti S, Ciccozzi M, Tripiciano C, Dalla Piccola B, Cicala M, Guarino MPL, et al. Gut mucosal-associated microbiota better discloses inflammatory bowel disease differential patterns than faecal microbiota. Dig Liver Dis. 2019;51:648–656. doi: 10.1016/j.dld.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 111.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 112.Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 113.Tilg H, Danese S. Roseburia hominis: a novel guilty player in ulcerative colitis pathogenesis? Gut. 2014;63:1204–1205. doi: 10.1136/gutjnl-2013-305799. [DOI] [PubMed] [Google Scholar]
- 114.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, et al. A decrease of the butyrate-producing species roseburia hominis and faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 115.Hedin CR, McCarthy NE, Louis P, Farquharson FM, McCartney S, Taylor K, Prescott NJ, Murrells T, Stagg AJ, Whelan K, et al. Altered intestinal microbiota and blood T cell phenotype are shared by patients with Crohn’s disease and their unaffected siblings. Gut. 2014;63:1578–1586. doi: 10.1136/gutjnl-2013-306226. [DOI] [PubMed] [Google Scholar]
- 116.Duck LW, Walter MR, Novak J, Kelly D, Tomasi M, Cong Y, Elson CO. Isolation of flagellated bacteria implicated in Crohn’s disease. Inflamm Bowel Dis. 2007;13:1191–1201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- 117.Gevers D, Kugathasan S, Denson L, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song S, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen L, Wilson JE, Koenigsknecht MJ, Chou W-C, Montgomery SA, Truax AD, Brickey WJ, Packey CD, Maharshak N, Matsushima GK, et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol. 2017;18:541–551. doi: 10.1038/ni.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rengarajan S, Vivio EE, Parkes M, Peterson DA, Roberson EDO, Newberry RD, Ciorba MA, Hsieh C-S. Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes. 2020;11:405–420. doi: 10.1080/19490976.2019.1626683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee SH, Turpin W, Espin-Garcia O, Raygoza Garay JA, Smith MI, Leibovitzh H, Goethel A, Turner D, Mack D, Deslandres C, et al. Anti-Microbial antibody response is associated with future onset of crohn’s disease independent of biomarkers of altered gut barrier function, subclinical inflammation, and genetic risk. Gastroenterology. 2021;161:1540–1551. doi: 10.1053/j.gastro.2021.07.009. [DOI] [PubMed] [Google Scholar]
- 121.Torres J, Petralia F, Sato T, Wang P, Telesco SE, Choung RS, Strauss R, Li X-J, Laird RM, Gutierrez RL, et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology. 2020;159:96–104. doi: 10.1053/j.gastro.2020.03.007. [DOI] [PubMed] [Google Scholar]
- 122.Steiner CA, Berinstein JA, Louissaint J, Higgins PDR, Spence JR, Shannon C, Lu C, Stidham RW, Fletcher JG, Bruining DH, et al. Biomarkers for the prediction and diagnosis of fibrostenosing crohn’s disease: a systematic review. Clin Gastroenterol Hepatol. 2021. doi: 10.1016/j.cgh.2021.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alexander KL, Zhao Q, Reif M, Rosenberg AF, Mannon PJ, Duck LW, Elson CO. Human microbiota flagellins drive adaptive immune responses in crohn’s disease. Gastroenterology. 2021;161:522–535 e526. doi: 10.1053/j.gastro.2021.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao Q, Duck LW, Huang F, Alexander KL, Maynard CL, Mannon PJ, Elson CO. CD4 + T cell activation and concomitant mTOR metabolic inhibition can ablate microbiota-specific memory cells and prevent colitis. Sci Immunol. 2020:5. doi: 10.1126/sciimmunol.abc6373. [DOI] [PubMed] [Google Scholar]
- 125.Castro-Dopico T, Dennison TW, Ferdinand JR, Mathews RJ, Fleming A, Clift D, Stewart BJ, Jing C, Strongili K, Labzin LI, et al. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity. 2019;50:1099–1114 e1010. doi: 10.1016/j.immuni.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Uo M, Hisamatsu T, Miyoshi J, Kaito D, Yoneno K, Kitazume MT, Mori M, Sugita A, Koganei K, Matsuoka K, et al. Mucosal CXCR4 + IgG plasma cells contribute to the pathogenesis of human ulcerative colitis through FcγR-mediated CD14 macrophage activation. Gut. 2013;62:1734–1744. doi: 10.1136/gutjnl-2012-303063. [DOI] [PubMed] [Google Scholar]
- 127.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Willcocks LC, Smith KG, Clatworthy MR. Low-affinity Fcgamma receptors, autoimmunity and infection. Expert Rev Mol Med. 2009;11:e24. doi: 10.1017/S1462399409001161. [DOI] [PubMed] [Google Scholar]
- 129.Christmann BS, Abrahamsson TR, Bernstein CN, Duck LW, Mannon PJ, Berg G, Björkstén B, Jenmalm MC, Elson CO. Human seroreactivity to gut microbiota antigens. J Allergy Clin Immunol. 2015;136:1378–1386 e1371-1375. doi: 10.1016/j.jaci.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Déjardin F, Sparwasser T, Bérard M, Cerf-Bensussan N, Eberl G, et al. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity. 2019;50:1276–1288 e1275. doi: 10.1016/j.immuni.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 131.Rubin BE, Diamond S, Cress BF, Crits-Christoph A, Lou YC, Borges AL, Shivram H, He C, Xu M, Zhou Z, et al. Species- and site-specific genome editing in complex bacterial communities. Nat Microbiol. 2022;7:34–47. doi: 10.1038/s41564-021-01014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jin WB, Li -T-T, Huo D, Qu S, Li XV, Arifuzzaman M, Lima SF, Shi H-Q, Wang A, Putzel GG, et al. Genetic manipulation of gut microbes enables single-gene interrogation in a complex microbiome. Cell. 2022;185:547–562.e22. doi: 10.1016/j.cell.2021.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guo CJ, Allen BM, Hiam KJ, Dodd D, Van Treuren W, Higginbottom S, Nagashima K, Fischer CR, Sonnenburg JL, Spitzer MH, et al. Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science. 2019;366. doi: 10.1126/science.aav1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brophy JAN, Triassi AJ, Adams BL, Renberg RL, Stratis-Cullum DN, Grossman AD, Voigt CA. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat Microbiol. 2018;3:1043–1053. doi: 10.1038/s41564-018-0216-5. [DOI] [PubMed] [Google Scholar]
- 135.Farzadfard F, Gharaei N, Citorik RJ, Lu TK. Efficient retroelement-mediated DNA writing in bacteria. Cell Syst. 2021;12:860–872 e865. doi: 10.1016/j.cels.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 136.Hsu BB, Plant IN, Lyon L, Anastassacos FM, Way JC, Silver PA. In situ reprogramming of gut bacteria by oral delivery. Nat Commun. 2020;11:5030. doi: 10.1038/s41467-020-18614-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


