ABSTRACT
Although providing long-lasting immunity, smallpox vaccination was associated with local and systemic reactions and rarely with severe complications, including progressive vaccinia and postvaccinia encephalitis. As the Dryvax smallpox vaccine consists of a population of variants, we investigated a particularly pathogenic isolate called clone 3 (CL3). Virus replication was monitored by inserting the gene encoding firefly luciferase (Luc) into the genomes of CL3 and ACAM2000, the second-generation smallpox vaccine derived from a less virulent clone. Greater luminescence occurred following intranasal or intraperitoneal inoculation of mice with CL3-Luc than ACAM2000-Luc. Previous genome sequencing of CL3 and ACAM2000 revealed numerous differences that could affect pathogenicity. We focused on a 4.2-kbp segment, containing several open reading frames, in CL3 that is absent from ACAM2000 and determined that lower virulence of the latter was associated with a truncation of the interferon α/β (IFN-α/β) decoy receptor. Truncation of the decoy receptor in CL3-Luc and repair of the truncated version in ACAM2000-Luc decreased and increased virulence, respectively. Blockade of the mouse type 1 IFN receptor increased the virulence of ACAM2000-Luc to that of CL3-Luc, consistent with the role of IFN in attenuating the former. The severities of disease following intracranial inoculation of immunocompetent mice and intraperitoneal inoculation of T cell-depleted mice were also greater in viruses expressing the full-length decoy receptor. Previous evidence for the low affinity of a similarly truncated decoy receptor for IFN and the presence of a full-length decoy receptor in virus isolated from a patient with progressive vaccinia support our findings.
KEYWORDS: interferons, smallpox, smallpox vaccine, vaccine complications, vaccinia virus, viral pathogenesis
INTRODUCTION
In 1798, the British physician Edward Jenner described a classic experiment in which pustular material derived from a cowpox lesion on the hand of a dairy worker was administered by skin scratch to a boy, who was then shown to be resistant to inoculation (variolation) with infectious smallpox material (1). We can infer that Jenner’s inoculum contained an orthopoxvirus, although whether it was cowpox virus, horsepox virus, or vaccinia virus (VACV) is speculative (2). During the next century, smallpox vaccines were propagated arm-to-arm in humans or by serial passage of the vaccine in the skin of inoculated animals, and material from different sources, including patients with smallpox, were often mixed. Historical records notwithstanding, in the 20th century the vaccines from all parts of the world were comprised of strains of VACV. In the United States, the Wyeth Laboratory was licensed to manufacture the Dryvax smallpox vaccine in 1944. The original source of VACV used for Dryvax has been thought to be the New York City Board of Health strain, although alternative or multiple origins have been suggested (3, 4), and, like other first-generation smallpox vaccines propagated in animals, Dryvax did not undergo cloning steps. While Dryvax was considered safer than several other smallpox vaccines, complications, including generalized vaccinia, ocular vaccinia, and eczema vaccinatum, occurred infrequently, and fatal progressive vaccinia associated with immunodeficiency, postvaccinial encephalitis, and fetal vaccinia during early pregnancy happened rarely (5, 6). Myopericarditis, ischemic cardiac disease, and dilated cardiomyopathy were more recently recognized complications of smallpox vaccination (7).
To produce a safer second-generation smallpox vaccine, virus from six individual plaques derived from vials of Dryvax were clonally purified on human MRC-5 cells (8). As the clones had been picked by plaque isolation after terminal dilution, they should represent the majority population. The cloned viruses varied in their virulence when tested for lesion formation in rabbit skin and neurovirulence in suckling mice. Clone 2 (CL2) was selected, based on low virulence and good immunogenicity, for development of ACAM1000 and ACAM2000 vaccines, which differ only in the cells used for their propagation (9). (ACAM2000 is referred to as ACAM here for brevity). The genome of clone 3 (CL3), one of the three virulent clones, had 625 mutational differences, including 572 single nucleotide polymorphisms, of which 290 represent nonsynonymous codons and 53 insertions-deletions of various sizes compared to ACAM (10). The genomes of the other four isolates, two of which were also more pathogenic than CL2, were not sequenced. The large number of sequence differences between ACAM and CL3 indicated that Dryvax is comprised of a population of genetically varied viruses. This conclusion was supported and extended by independent isolation and sequencing studies with Dryvax clones (3, 11). Similarly, other vaccines, including Lister (12) and TianTan (13), were found to be heterogeneous.
The present study was undertaken to determine differences in the genomes of ACAM and CL3 that contribute to the increased virulence of the latter. This information could help explain variations in the side effects of different smallpox vaccines, inform development of future VACV vectors, and increase understanding of orthopoxvirus pathogenesis.
RESULTS
Development of a small-animal model to analyze the virulence of CL3.
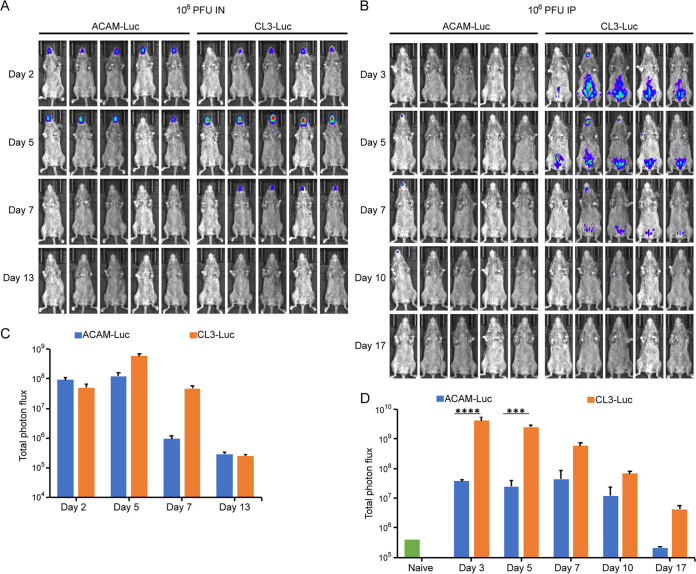
The conclusion that CL3 is more virulent than CL2, which was used for the ACAM vaccine, was based on the size and appearance of skin lesions in rabbits following intradermal inoculation and neurovirulence after intracerebral injection of suckling mice (14). Even though statistically significant, suckling mice injected with CL2 or CL3 had average survival time differences of less than two days. To develop an animal model more suitable for genetic studies, we adapted protocols successfully used to demonstrate virulence differences between strains of monkeypox virus and other orthopoxviruses (15, 16). Those studies used recombinant viruses with Luc expressed from an intergenic site that did not affect virulence to enable quantification of virus replication noninvasively in susceptible Castaneous (CAST) mice. For the present study, CL3-Luc and ACAM-Luc were constructed from CL3 and ACAM. CAST mice were inoculated intranasally (i.n.) or intraperitoneally (i.p.) with 106 PFU of CL3-Luc or ACAM-Luc. On successive days, mice were injected i.p. with d-luciferin and bioluminescence was measured. Nonsaturating exposure times were used to prepare images, and the total photon flux was measured for quantitation. Following i.n. administration, luminescence was detected in the head region for both viruses on day 2 and started declining by day 7 (Fig. 1A). On days 5 and 7, the total photon flux was higher for CL3-Luc than for ACAM-Luc (P = 0.0079), as shown in Fig. 1C. Following i.p. administration, bioluminescence remained largely in the abdominal region, although some was detected in the head (Fig. 1B). The possibility that virus was excreted following i.p. inoculation and mice were secondarily infected to account for luminescence in the nasal/mouth region was considered but not investigated. The photon flux of CL3-Luc was greater than that of ACAM-Luc on all days, and the differences were highly significant on days 3 and 5 (Fig. 1D). Luminescence started declining again on day 7. The i.p. route was chosen in preference to i.n. because of the consistently higher luminescence of CL3 compared to ACAM. Mice inoculated with 106 PFU of ACAM i.p. generally survive infection, although in some experiments one or more inoculated with CL3 succumbed.
FIG 1.
Development of an animal model for studies of virulence properties of ACAM2000 and CL3. (A and B) CAST mice (n = 5) were infected with 106 PFU of ACAM-Luc or CL3-Luc i.n. (A) or i.p. (B). The progress of infection was measured by injecting luciferin and measuring luminescence. Ventral-view images of mice are shown from the indicated days postinfection using the same exposure time and binning factor. Red, blue, and purple denote intensity of luminescence from high to low. (C and D) Total photon flux (photons per second per square centimeter per steradian) of the entire mouse was calculated and shown for each group. Bars indicate arithmetic means ± standard deviations (SD). ****, P < 0.0001; ***, P = 0.0003. Significance was calculated using the Mann-Whitney test.
A 4.5-kbp DNA segment of the CL3 genome is responsible for enhanced virulence.
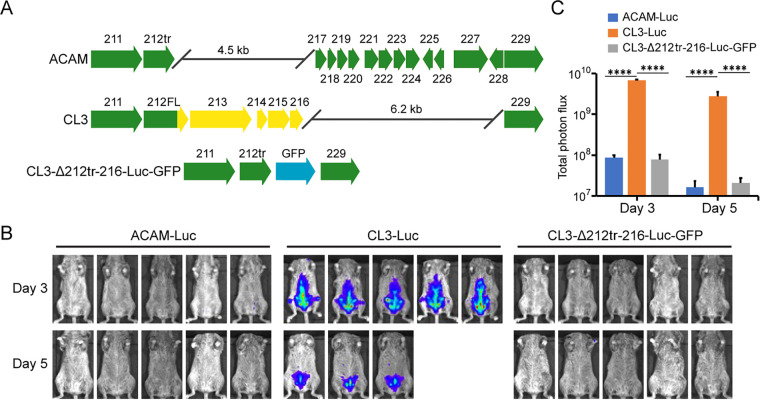
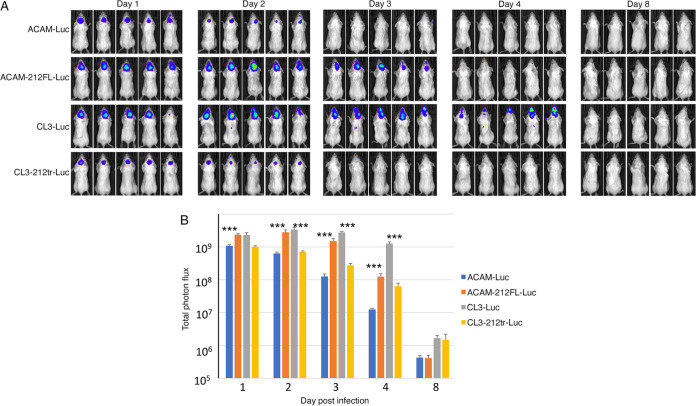
Having an animal model that quantitatively distinguishes CL3 and ACAM replication, we next sought to determine the genetic basis for virulence differences. Although numerous genomic alterations were noted between CL3 and ACAM, it had not been determined which contributed to the variation in pathogenicity. The most prominent dissimilarities are near the right ends of the genomes (Fig. 2A). Relative to other strains of VACV, there is a 4.5-kbp deletion in ACAM and a 6.2-kbp deletion in CL3 (10). Because the deletion in CL3 was within the right inverted terminal repeat, the open reading frames (ORFs) were preserved at the left end of the genome, making this difference unlikely to be responsible for enhanced virulence. In contrast, the deletion in ACAM resulted in the truncation of the single copy of ORF 212, which is 256 codons in ACAM and 353 codons in CL3, and complete loss of ORFs 213 to 216 (Fig. 2A). ORF 212 encodes a secreted interferon (IFN-α/β) decoy receptor, ORF 213 encodes an uncharacterized ankyrin repeat protein, and ORFs 214 to 216 are short and probably nonfunctional. We focused on the 4.5-kbp sequence difference between the two viruses and replaced the DNA encompassing the C-terminal segment of ORF 212 through 216 of CL3-Luc with green fluorescent protein (GFP) regulated by a VACV promoter, which facilitated isolation of the recombinant virus, named CL3Δ212tr-216-Luc-GFP (Fig. 2A). CAST mice were then inoculated i.p. with viruses from this panel. Two of the mice inoculated with CL3-Luc were moribund by day 5 and not imaged on that day. Luminescence on days 3 and 5 was diminished in mice infected with the CL3 deletion mutant CL3Δ212tr-216-Luc-GFP, and the differences in photon flux between CL3-Luc versus CL3Δ212tr-216-Luc-GFP and ACAM2000-Luc were highly significant (Fig. 2B and C). The bioluminescence of ACAM-Luc and CL3Δ212tr-216-Luc-GFP were virtually identical, indicating that one or more genetic changes in this region accounted for virulence differences.
FIG 2.
Deletion of ORFs 212tr to 216 impaired the virulence of CL3. (A) Major ORF differences between ACAM and CL3 are shown. Arrows indicate relative lengths of ORFs and the direction of transcription. ORFs are colored green for ACAM and ORFs, and parts of ORFs of CL3 that are unique are colored yellow. Note that C12 of ACAM, depicted as a green arrow, is truncated and called 212tr, whereas C12 of CL3, depicted as a green arrow with a yellow arrowhead, is full length and called C12FL. The diagram of CL3-Δ212tr-216-Luc-GFP shows a truncation of C12 and replacement of ORFs 213 to 216 by GFP regulated by the VACV P11 promoter, in blue. (B) CAST mice (n = 5) were infected i.p. with 106 PFU of ACAM-Luc, CL3-Luc, or CL3-Δ212tr-216-Luc-GFP. The progress of infection was measured by injecting d-luciferin and measuring luminescence. Ventral-view images of mice are shown for day 3 and day 5 after infection. (C) Total photon flux of the body of each mouse was calculated and shown by group. Bars indicate arithmetic means ± SD. ****, P < 0.0001. Significance was calculated using Fishers exact test.
Truncation of ORF 212, encoding the IFN-α/β decoy receptor, decreases virulence of CL3.
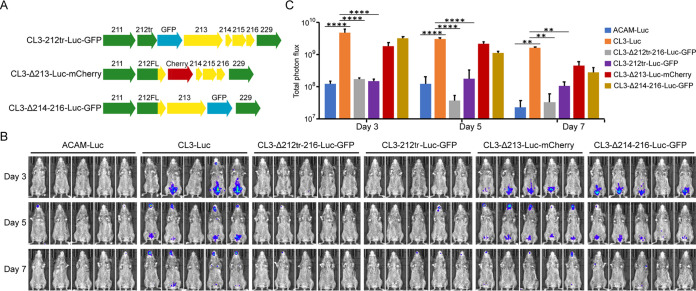
To identify the individual ORF or ORFs responsible for enhanced virulence, three new recombinant viruses with either a C-terminal truncation of ORF 212 (CL3-212tr-Luc-GFP), deletion of ORF 213 (CL3Δ213-Luc-mCherry), or deletion of ORFs 214 to 216 (CL3Δ214-216-Luc-GFP) were constructed from CL3-Luc (Fig. 3A). As indicated by their names, each virus also expressed either GFP or mCherry to facilitate isolation of the recombinant viruses. When inoculated into CAST mice, bioluminescence was strongly diminished by truncation of ORF 212 but not significantly by deletion of ORF 213 or ORFs 214 to 216 (Fig. 3B). The decrease in photon flux due to truncation of ORF 212 was highly significant, indicating that the 97 amino acids at the C terminus of ORF 212 contribute to virulence (Fig. 3C). Truncation of IFN-α/β decoy receptor is due to a stop codon between the second and third Ig domains, which has been shown to decrease the affinity for IFN-α by about 80-fold without preventing secretion and cell binding (17, 18).
FIG 3.
ORF 212 is required for virulence of CL3. (A) The top diagram shows CL3-212tr-Luc GFP, with a truncation of ORF 212, followed by the separate GFP ORF regulated by the P11 promoter depicted, in blue. The lower two diagrams depict CL3-Δ213-Luc-mCherry and CL3-Δ214-216-Luc-GFP, with ORF 213 replaced with mCherry in red or ORFs 214 to 216 replaced with GFP in blue, each regulated by the P11 promoter. Yellow and green arrows indicate CL3 ORFs that were unaltered, as in Fig. 2A. (B) CAST mice (n = 5) were infected i.p. with 106 PFU of ACAM-Luc, CL3-Luc, or CL3 deletion viruses. The progress of infection was monitored by measuring luminescence on the indicated days, as described in the legend to Fig. 1. (C) Plots of average total photon flux of animals for each group. Bars indicate arithmetic means ± SD. ****, P < 0.0001; **, P < 0.01. Significance was calculated using Fishers exact test.
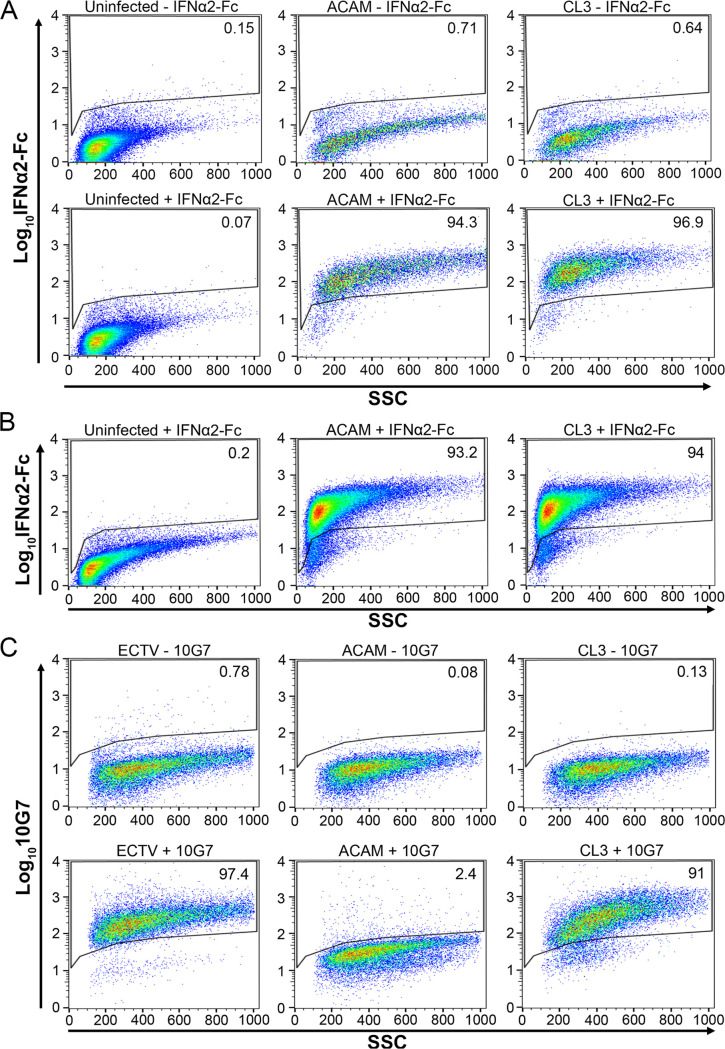
To confirm expression of the full-length and truncated IFN-α/β decoy receptors encoded by CL3 and ACAM, respectively, unpermeabilized infected cells were incubated with human IFN-α2 protein tagged with an Fc peptide (IFN-α2-Fc) followed by anti-human Fc-allophycocyanin (APC) monoclonal antibody (MAb) and analyzed by flow cytometry. Labeling of ACAM and CL3 were similar (Fig. 4A).
FIG 4.
Detection of cell associated and soluble truncated and full-length IFN-αβ decoy receptors. (A) Uninfected HeLa cells and HeLa cells infected for 16 to 18 h with ACAM or CL3 were incubated without (-) or with (+) IFN-α-2Fc followed by anti-Fc-APC, and fluorescence was detected in a flow cytometer. A total of 10,000 to 70,000 events were collected for each sample. The percentages of cells above the gate are indicated. (B) The medium from HeLa cells that were uninfected or infected with ACAM or CL3 for 16 to 18 h was collected and clarified by centrifugation. Uninfected HeLa cells were incubated with the medium followed by IFN-α-2Fc and anti-Fc-APC. The percentages of cells above the gate are indicated. (C) HeLa cells were infected with ectromelia (ECTV), ACAM, or CL3 for 24 h. The lower panels show staining with MAb 10G7 (+) followed by APC-conjugated anti-mouse secondary antibody. The upper panels are controls in which the 10G7 MAb was omitted (-) and staining was just with the secondary antibody. The percentages of cells above the gate are indicated.
Alcami et al. (17, 18) showed that secreted full-length or truncated VACV IFN-α/β decoy receptor can bind to cells, although the membrane receptor was not identified. Similarly, in our study, soluble forms of the full-length and truncated IFN-α/β decoy receptors were obtained from the clarified medium of infected cells and incubated with uninfected cells. Association of IFN-α/β decoy receptor made by ACAM-Luc and CL3-Luc with the surface of uninfected cells was demonstrated by labeling the latter with IFN-α2-Fc, followed by anti-human Fc-APC MAb, and analysis by flow cytometry (Fig. 4B). We concluded that the biological effect of the truncation of the decoy receptor is due to the previously shown decrease in affinity for IFN rather than low expression by ACAM.
In contrast to the similar binding of IFN-α2-Fc, a MAb to the homologous ectromelia virus IFN-α/β decoy receptor (19) was found to bind avidly to cells infected with CL3 but not to cells infected with ACAM, distinguishing the full-length and truncated receptors (Fig. 4C). This result suggests that the C terminus of the decoy receptor is the site of binding of the MAb, although this had not previously been investigated (19).
Repair of ORF 212, encoding the IFN-α/β decoy receptor, enhances virulence of ACAM.
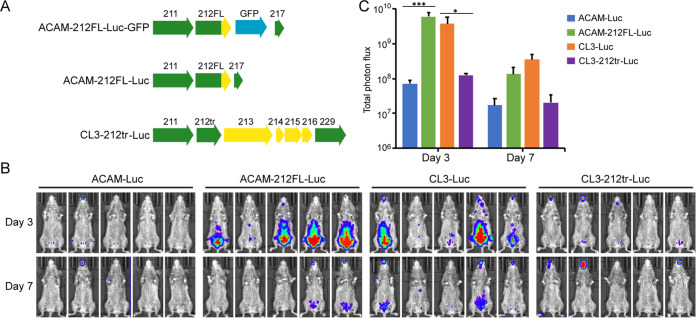
The experiments described above showed that the full-length IFN-α/β decoy receptor was necessary for the virulence of CL3. However, since there are numerous sequence differences between ACAM and CL3, it was important to carry out the reciprocal experiment to determine whether extension of the truncated ORF 212 was sufficient to enhance virulence of ACAM. Recombinant ACAM-212FL-Luc-GFP containing the full-length (FL) 212 ORF was constructed, and the GFP ORF was then deleted to produce ACAM-212FL-Luc (Fig. 5A). We also removed the GFP ORF from CL3-212tr-Luc-GFP to produce CL3-212tr-Luc to ensure that expression of GFP was not altering results (Fig. 5A). On day 3 following i.p. inoculation, the bioluminescence of CAST mice receiving ACAM-212FL-Luc expressing the full-length decoy receptor was like that of CL3-Luc and was significantly higher than that of mice receiving ACAM-Luc expressing the truncated decoy receptor (Fig. 5B and C). Bioluminescence of mice receiving CL3-Luc was also higher than that of mice receiving CL3-212tr-Luc (Fig. 5B and C). Thus, truncation of ORF 212 reduces virulence of CL3 and extension of ORF 212 increases the virulence of ACAM.
FIG 5.
Repair of ORF 212 in ACAM increases virulence. (A) The top diagram shows ACAM-212FL-Luc-GFP, which was constructed by adding the 3′ end of full-length ORF 212 and the adjacent GFP ORF under the p11 promoter into ACAM-Luc. The GFP ORF was removed to generate ACAM-212FL-Luc (middle diagram) and CL3-212tr-Luc (bottom diagram). Color coding is the same as that in Fig. 1A and 3A. (B) CAST mice (n = 5) were infected i.p. with 106 PFU of the indicated viruses. Virus spread was determined from bioluminescence as described in Fig. 1, and ventral images are shown. (C) Total photon flux of the entire mouse was calculated and plotted for each group. Bars indicate arithmetic means ± SD. ***, P < 0.001; *, P = 0.03. Significance was calculated using Fishers exact test.
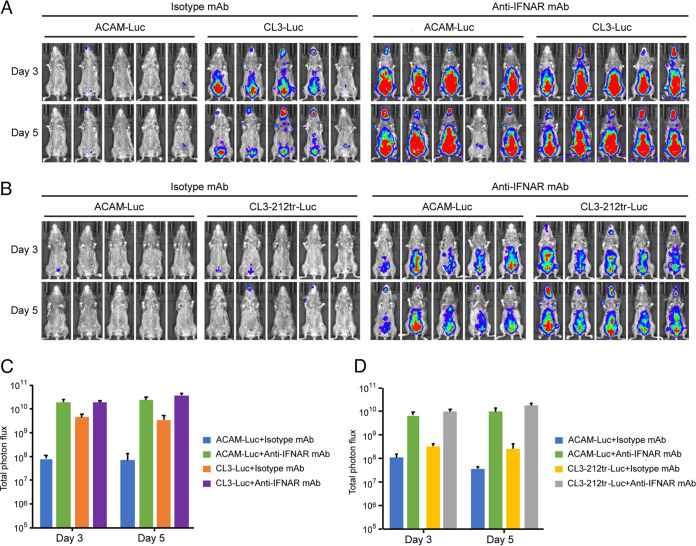
Enhanced virulence of ACAM-Luc in CAST mice treated with blocking antibody to IFNAR1.
The VACV IFN-α/β decoy receptor competes with the mouse type 1 IFN receptor (IFNAR1) for binding IFN and reduces IFN-mediated signaling (17, 20). If the difference in virulence between CL3 and ACAM is due to the low affinity of the truncated IFN-α/β decoy receptor for IFN in vivo, then the virulence of the two viruses should be similar in IFNAR1-inactivated mice. An anti-IFNAR1 blocking MAb (21) was administered to uninfected mice, and splenocytes were isolated and stained with anti-IFNAR1 conjugated to phycoerythrin (PE) to detect unblocked receptors. In parallel, mice were injected with an isotype MAb. Staining was reduced by more than 96% in the anti-IFNAR1 MAb-treated mice, as determined by flow cytometry, indicating a high level of blocking (see Fig. S1 in the supplemental material). Having demonstrated blocking in control experiments, mice were treated before and after infection with anti-IFNAR MAb and bioluminescence determined. In mice infected with ACAM-Luc, luminescence increased by more than 2 logs relative to the isotype MAb, whereas the luminescence of CL3-Luc was increased by much less, although to a similar final value. When the IFN-α/β decoy receptor of CL3-Luc was truncated, the increase in virulence due to anti-IFNAR MAb was like that of ACAM-Luc (Fig. 6B and D). Thus, blockade of the IFN receptor had a much greater effect on ACAM-Luc and CL3-C12tr-Luc with truncated decoy receptors than CL3-Luc with a full-length decoy receptor.
FIG 6.
Virulence of ACAM is enhanced by blocking the mouse IFNAR. (A and B) CAST mice (n = 5) were injected i.p. with 500 μg of anti-mouse IFNAR MAb or isotype MAb as a control 1 day before infection and 250 μg on the day of infection and days 2 and 4 after infection. The mice were infected i.p. with 106 PFU of the indicated viruses. Luminescence on days 3 and 5 are shown. (C and D) Plots of mean total photon flux of animals for each group in panels A and B. Bars indicate arithmetic means ± SD.
Blockade of IFNAR1 by IFN-α2-Fc. CAST mice were injected i.p. with 500 μg of anti-mouse IFNAR MAb or anti-mouse IgG1 isotype MAb as described in the legend to Fig. 6, except that the mice were not infected. Splenocytes were isolated one day after the third injection and stained with anti-IFNAR1 or anti-IgG isotype conjugated to phycoerthyrin (PE). Fluorescence was determined by flow cytometry, and the numbers within the panels refer to the percentage of cells labeled by IFNAR1-PE. Approximately 30,000 total events were collected for each sample. Download FIG S1, TIF file, 0.2 MB (250.8KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Enhanced neurovirulence correlated with expression of full-length IFN-α/β decoy receptor.
The original characterization of the clones isolated from Dryvax included intracranial (i.c.) inoculation of 3- to 5-day-old ICR mice (8). In developing our protocol, 6-week-old BALB/c mice were injected i.c. with 104, 105, or 106 PFU of ACAM-Luc or CL3-Luc. Bioluminescence was localized in the head region, and maximal photon flux was proportional to dose (Fig. 7A and B). For all doses of CL3-Luc, the photon flux increased from day 1 to day 3 and then declined by day 6, and the mice survived. Except for the lowest dose of ACAM-Luc, the photon flux was greatest on day 1. The difference between ACAM-Luc and CL3-Luc was highly significant on day 3 for all doses.
FIG 7.
Greater neurovirulence of CL3-Luc than ACAM-Luc. (A) BALB/c mice (n = 5) were injected i.c. with 104, 105, or 106 PFU of ACAM-Luc or CL3-Luc. Bioluminescence was measured as described for Fig. 1, except dorsal images are shown. (B) Total photon flux of animals shown in panel A. Bars indicate arithmetic means ± SD. ****, P < 0.0001; ***, P = 0.0004. Significance was calculated using Fishers exact test.
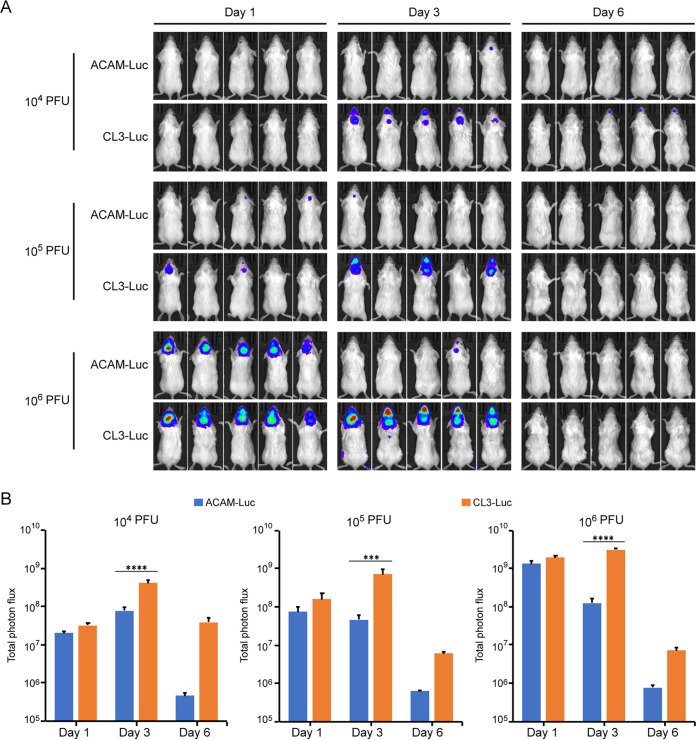
In a follow-up experiment, expression of the full-length IFN-α/β decoy receptor was shown to be associated with neurovirulence. Mice were injected i.c. with 106 PFU of ACAM-Luc, ACAM-212FL-Luc, CL3-Luc, or CL3-212tr-Luc. As in the previous experiment, bioluminescence was significantly higher in mice injected i.c. with CL3-Luc than ACAM-Luc (Fig. 8A and B). Furthermore, the level of bioluminescence was significantly higher in mice injected with ACAM-212FL-Luc than with ACAM-Luc and lower in mice injected with CL3-212tr-Luc than with CL3-Luc. Thus, viruses with a full-length decoy receptor were more neurovirulent than viruses with the truncated form.
FIG 8.
Attenuation of neurovirulence of CL3-Luc by truncation of ORF 212. (A) BALB/c mice were injected i.c. with 106 PFU of the indicated viruses. Virus replication was determined by bioluminescence as described for Fig. 7. (B) Total photon flux of animals shown in panel A. Bars indicate arithmetic means ± SD. ***, P < 0.001. Significance was calculated using Mann-Whitney test. This experiment was repeated with essentially the same result.
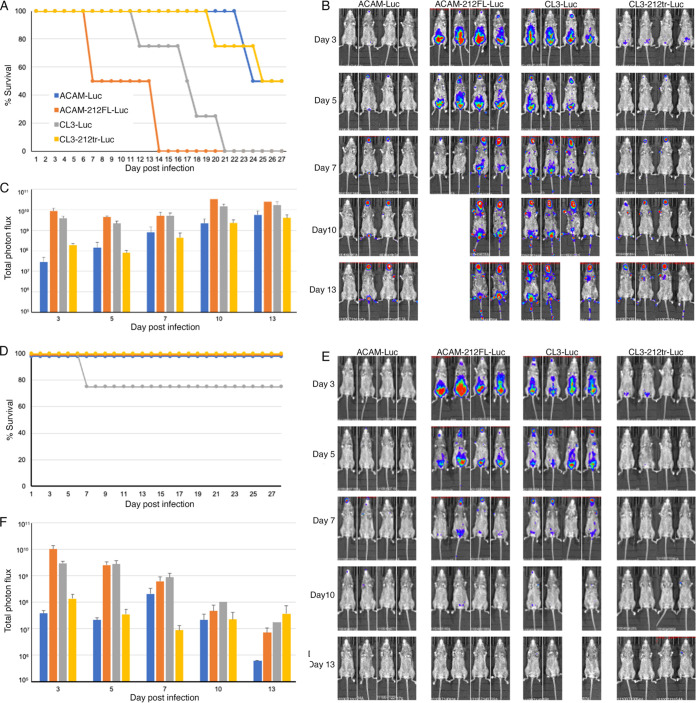
Shortened survival and enhanced virus spread in T cell-depleted mice correlated with expression of full-length IFN-α/β decoy receptor.
Defects in cellular immunity were correlated with progressive vaccinia following vaccination with Dryvax. We investigated the effects of CD4/CD8 T cell depletion of CAST mice on virus spread. Depletion was carried out using specific antibodies before and after virus challenge, as previously described (22), and control mice were injected with an isotype antibody. Depletion of CD4+ and CD8+ T cells was 98% or more, as determined by flow cytometry (Fig. S2). The T cell-depleted mice infected with the two viruses, ACAM-212FL-Luc and CL3-Luc, containing full-length IFN-α/β decoy receptor, all died by day 21, whereas the T cell-depleted mice infected with ACAM or CL3-212tr-Luc, with truncated IFN-α/β decoy receptor, survived longer (Fig. 9A). In contrast, the mice that received the isotype MAb all survived, except for one infected with CL3 (Fig. 9D). In the T cell-depleted mice, the difference in times to death for the mice inoculated with full-length versus truncated decoy receptor, namely, between ACAM-Luc and both ACAM-212FL-Luc and CL3-Luc (P < 0.01) and between CL3-Luc and CL3-212tr-Luc (P = 0.028), were significant, but the times to death were not significant between the two viruses with full-length decoy receptor CL3-Luc and ACAM-212FL-Luc (Fig. 9A). For all viruses, luminescence continually increased after infection of T cell-depleted mice (Fig. 9B and C), whereas luminescence decreased after the first week in the control mice (Fig. 9E and F), presumably due to T cell activation. The higher luminescence in the mice infected with CL3-Luc and ACAM-212FL-Luc correlated with earlier time to death in T cell-depleted mice (Fig. 9C).
FIG 9.
Enhanced virus spread and decreased survival of T cell-depleted mice infected with viruses expressing the full-length IFN-α/β decoy receptor. CAST mice were treated with anti-CD4+ and anti-CD8+ antibodies (A, B, and C) or isotype antibody (D, E, and F) and infected i.p. with 106 PFU of ACAM-Luc, ACAM-212FL-Luc, CL3-Luc, or CL3-212tr Luc. Survival was determined daily (A and D), and mice were injected with d-luciferin and imaged on the indicated days (B, C, E, and F). In the T cell-depleted mice, the differences in times to death between ACAM-Luc and both ACAM-212FL-Luc and CL3-Luc (P < 0.01) and between CL3-Luc and CL3-212tr-Luc (P = 0.028) were significant, as determined by the Mantel-Cox test.
Depletion of CD4+ and CD8+ T cells. Anti-CD4 and anti-CD8 MAb or anti-KLH isotype MAb were injected i.p. into CAST mice on days 3 and 4 prior to infection as described for Fig. 6. At 24 h before infection, a sample of whole blood was collected and lymphocytes were isolated, incubated with anti-CD4-PE and anti-CD8-PerCP, and analyzed by flow cytometry as shown here. Approximately 28,000 to 40,000 total events were captured. Depletion was estimated to be >98%, and similar results were obtained by repeat analysis of whole blood at 11 days after infection (not shown). Download FIG S2, TIF file, 0.7 MB (754.8KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
The genome sequence of pathogenic CL3 compared to the clone used to derive the ACAM second-generation smallpox vaccine revealed 290 nonsynonymous codon differences and 53 insertions-deletions of various sizes (10). However, which of these changes is responsible for the difference in pathogenicity was not determined. In the present study, we constructed recombinant CL3 and ACAM that express Luc and used mouse models to compare their virulence. Greater bioluminescence, a measure of virus replication, was found in mice inoculated i.n. or i.p. with CL3-Luc than ACAM-Luc. By constructing additional recombinant viruses, we determined that full-length ORF 212 in CL3, which is truncated in ACAM, contributes significantly to virulence in the mouse model. Indeed, the virulence of ACAM with 97 codons added to the C terminus of the truncated ORF 212 was like that of CL3, and the virulence of CL3 with truncation of ORF 212 was like that of ACAM. These results are explained by the role of the IFN α/β decoy receptor encoded by ORF 212 and the decreased affinity for IFN-α of a version that is truncated between the second and third Ig domains (17), although the effect of truncation on virulence had not previously been determined. The importance of IFN in diminishing the virulence of ACAM in mice is consistent with the enhancement of replication by anti-IFNAR MAb, which brought the virulence up to that of CL3. Although ORF 213, encoding an ankyrin repeat protein with an unknown role, is present in CL3 but not ACAM, deletion of the gene from CL3 did not reduce virulence either in our study or as reported previously (3).
The serious side effects of smallpox vaccines include encephalitis and progressive vaccinia in individuals with defects in cellular immunity. Using an i.c. injection model, we confirmed the greater neurovirulence of CL3-Luc compared to that of ACAM-Luc and demonstrated that the virulence was associated with expression of the full-length IFN-α/β decoy receptor. Mice that were depleted of CD4+ and CD8+ T cells were more susceptible to CL3 and ACAM-212FL, in which the ORF expressing the IFN-α/β decoy receptor was repaired, than to ACAM and CL3-212tr, in which the receptor was truncated.
ORFs encoding full-length IFN-α/β decoy receptor homologs predominate in vaccine and laboratory strains of VACV as well as in most other orthopoxviruses. Lister, an exception with a deletion, and Dryvax, with variants that have a truncation, had less severe local and systemic reactions and fewer side effects in humans than other first-generation smallpox vaccines, whereas Copenhagen, Tashkent, and CVA smallpox vaccines, which encode a full-length IFN-α/β decoy receptor, were considered more pathogenic (23, 24). Additionally, a comparison of the pathogenicity of smallpox vaccine strains in mice and rabbits classified TianTan (Temple of Heaven) and Copenhagen, both of which encode full-length decoy receptors, as having higher pathogenicity than Lister and Dryvax (25). Thus, the full-length IFN-α/β decoy receptor correlates with adverse reactions in smallpox vaccines. Laboratory strains of VACV such as WR, IHD-W, and rabbitpox, encoding full-length IFN-α/β decoy receptors, are also notably pathogenic in animal models, and deletion of the ORF attenuates the WR strain in mice (18). VACV strains, including some with full-length decoy receptor homologs, have been used for development of oncolytic therapy, where greater pathogenicity might be advantageous (26). ORF 213, which we showed is not responsible for virulence of CL3, is truncated or absent from all VACV strains except for rabbitpox and horsepox and does not correlate with smallpox vaccines that have high incidence of adverse reactions. With regard to any concerns, genetic manipulation of viruses was carried out with viral genes present in the original Dryvax stock, and CL3 and ACAM-212FL are both considerably less pathogenic in CAST mice than common laboratory strains of VACV and cowpox virus (16).
Further support for the idea that clones encoding the full-length IFN-α/β decoy receptor are responsible for the rare but severe side effects that were associated with Dryvax comes from a patient with a smallpox vaccine-related complication (27). This individual was diagnosed with progressive vaccinia at the Duke University Medical Center and treated with VACV immunoglobulin. Genome sequencing of VACV-Duke, isolated from a lesion, confirmed an origin from Dryvax and suggested clonality. Like CL3, ORF 212 in VACV-DUKE was full length, but unlike CL3, ORF 213 was truncated (3). Regrettably, isolates from other patients that suffered smallpox vaccine complications are not available. However, the finding that two single nucleotide polymorphisms associated with adverse reactions to the related Aventis-Pasteur smallpox vaccine mapped to the interferon regulatory factor 1 gene also implicates the importance of IFN (28). Finally, we point out that the VACV IFN-α/β decoy receptor has a lower affinity for mouse IFN-α than human IFN-α and inhibits human but not mouse IFN-α (17), suggesting that the difference in virulence of CL3 and ACAM would be greater in humans than mice.
MATERIALS AND METHODS
Ethics statement.
All experiments and procedures using mice were approved under protocol LVD29E by the NIAID Animal Care and Use Committee according to standards set forth in the NIH guidelines, Animal Welfare Act, and U.S. federal law. Euthanasia was carried out using carbon dioxide inhalation in accordance with the American Veterinary Medical Association guidelines for euthanasia of animals (2013 Report of the AVMA Panel of Euthanasia). Procedures using VACV were carried out in a biosafety level 2 (BSL-2) laboratory by personnel that received smallpox vaccine as required by NIH.
Mice.
Female 6- to 7-week-old CAST/EiJ mice were purchased from Jackson Laboratories (Bar Harbor, ME), and female 5-week-old BALB/cAnNTac mice were from Taconic Biotechnology (Germantown, NY). Mice were housed in small, ventilated microisolator cages in an animal BSL-2 pathogen-free environment.
Cells.
African green monkey BS-C-1 cells (ATCC CCL-26) were propagated at 37°C and 5% CO2 in modified Eagle minimal essential medium (EMEM) (Quality Biologicals, Inc., Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO), 2 mM l-glutamine, 100 U of penicillin, and 100 μg of streptomycin per mL (Quality Biologicals, Inc.) and used for replication and titration of viruses.
Viruses.
ACAM2000 and CL3 were obtained from the CDC. ACAM-Luc and CL3-Luc expressing Luc regulated by a synthetic early-late promoter were generated by inserting the cassette between the F12 and F13 genes of ACAM2000 (GenBank accession no. AY313847) and CL3 (GenBank accession no. AY313848), respectively, and identifying plaques containing virus that expresses Luc. Additional modified viruses were constructed by homologous recombination using the fluorescent reporter genes encoding GFP or mCherry, regulated by the P11 late promoter for selection. Recombinant viruses CL3-Δ213-Luc-mCherry and CL3-Δ214–216-Luc-GFP were constructed by deleting the ORFs from CL3-Luc. To generate CL3-212tr-Luc-GFP, ORF 212 was truncated by replacing the last 294 bp with the last 30 bp present in ACAM, followed by the P11 VACV promoter-driven GFP ORF. CL3-Δ212tr–216-Luc-GFP was constructed by truncation of ORF 212 as described for vCL3-212tr-Luc GFP and deletion of ORF 213 to ORF 216. To generate ACAM-212FL-Luc-GFP, the DNA segment containing the last 294 bp from CL3 along with P11-GFP was inserted into ACAM-Luc. Homologous recombination was used to excise GFP to generate ACAM2000-212FL-Luc and CL3-212tr-Luc.
Viruses were grown in BS-C-1 cells. Unmodified CL3 and ACAM were purified by sedimentation through a sucrose cushion. All recombinant viruses expressing Luc were purified by centrifugation through a 36% sucrose cushion, followed by banding in a 24% to 40% sucrose gradient, and used for infections after determination of infectivity by plaque assay in BS-C-1 cells (29).
Infection of mice.
Six- to 7-week-old CAST/EiJ mice were inoculated i.p. with 100 μL of virus or i.n. with 10 μL of virus. For i.c. viral administration, 5-week-old BALB/cAnNTac mice were anesthetized with isoflurane and injected with 30 μL of virus using a 27-gauge needle into the right frontal cerebral hemisphere (30). A small portion of virus was saved and titrated on BS-C-1 cells to verify the challenge dose.
Bioluminescence imaging.
Bioluminescence imaging of live animals was performed with an IVIS Lumina LT series III system (Perkin Elmer, Waltham, MA) as previously described (16). Infected mice were anesthetized with isoflurane, and Xenolight d-luciferin substrate (Perkin Elmer) was injected i.p. (150 μg/g body weight) 8 to 10 min prior to live imaging. Animals remained under isoflurane sedation for the duration of each imaging session, which occurred at various times postinfection until death or total virus clearance was achieved. Luminescent exposures were collected for 1 to 60 s with small or medium binning factors and various f-stop settings. Living Image Software (Perkin Elmer) was used for acquisition and analysis. For photon flux, a single region of interest was drawn around the entire body, and light emission was measured in photons per second per square centimeter per steradian.
In vivo IFNAR blockade.
CAST/EiJ mice were injected i.p. with 500 μg of anti-IFNAR MAb (clone MAR1-5A3; BioXCell, Lebanon, NH) or anti-mouse IgG1 isotype (clone MOPC-21; BioXCell) on days −1 and 0 prior to infection and 250 μg of anti-IFNAR MAb or isotype control MAb i.p. on days 2 and 4 after infection.
T cell depletion.
Depletion of T cells was carried out as previously described (22) using MAbs against CD4 (clone GK1.5), CD8 (clone YTS 169.4), or KLH (isotype control, clone LTF-2) (Bio X Cell). Cast mice were injected i.p. with 0.2 mg of both anti-CD4 and anti-CD8 MAb or 0.4 mg of anti-KLH MAb in 200 μl of phosphate-buffered saline (PBS) on days 3 and 4 before virus challenge. The efficacy of depletion was tested by incubating splenocytes at 24 h before infection or whole blood 11 days after infection with anti-CD4-PE and anti-CD8-peridinin chlorophyll protein (PerCP) (BioLegend, San Diego, CA) and analyzing by flow cytometry.
Detection of truncated and full-length IFN-αβ decoy receptors.
HeLa cells were uninfected or infected with 3 PFU/cell ACAM2000 or CL3 for 2 h at 37°C, 5% CO2. After 2 h, virus was removed, and monolayers were washed and incubated with EMEM supplemented with 2% FBS. After 16 h at 37°C, 5% CO2, the liquid overlay was removed and transferred to a sterile 1.5-mL tube for further processing, as indicated below. Infected monolayers were washed with cold Dulbecco’s phosphate-buffered saline (DPBS) with 0.5 mM EDTA. Cells were scraped in fresh cold DPBS plus 0.5 mM EDTA, transferred to a new 1.5-mL tube, and centrifuged at 4°C, 180 × g for 10 min. Cell pellets were resuspended in Fc block (clone 2.4G2; gift of Jack Bennick) and incubated at 4°C for 20 min. After blocking, cells were spun at 4°C, 180 × g for 10 min, and pellets were washed two times in cold binding buffer (RPMI 1640 medium supplemented with 1% bovine serum albumin and 20 mM HEPES). Cells were then incubated with 11 nM IFN-α2-Fc (Sino Biologicals, Wayne, PA) diluted in cold binding buffer for 3 h on ice. After incubation with IFN-α2-Fc, cells were washed two times in cold binding buffer and then incubated with anti-human Fc-APC (BioLegend, San Diego, CA) for 1 h on ice. Prior to analysis using a FACSCalibur (BD Biosciences, San Jose, CA), cells were washed two times in cold DPBS plus 0.5 mM EDTA and fixed in DPBS containing 2% paraformaldehyde.
Medium from uninfected HeLa cells or HeLa cells infected with ACAM2000 or CL3 was centrifuged at 20,000 × g for 30 min at 4°C. Clarified supernatants were incubated with fresh HeLa cells for 3 h on ice, after which the cells were washed in cold binding buffer, scraped into a fresh portion of cold binding buffer, and centrifuged at 4°C, 180 × g for 10 min. Cell pellets were then incubated with 11 nM IFN-α2-Fc for 3 h on ice and processed as described above.
Statistical analysis.
P values were calculated as indicated in the figure legends with Mann-Whitney test, Fisher’s exact test, or Mantel-Cox test using Prism (GraphPad).
Data availability.
Data and materials will be made available upon request.
ACKNOWLEDGMENTS
We thank Subbian Satheshkumar Panayampalli for providing ACAM2000, Dryvax, and CL3, Luis Sigal for MAb 10G7, Andrea S. Weisberg for modifying figures, and the technical staff of the NIAID Comparative Medical Branch for expert animal care.
This research was supported by the Intramural Research Program of NIAID, NIH.
Footnotes
This article is a direct contribution from Bernard Moss, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Stuart Isaacs, University of Pennsylvania, and Gunasegaran Karupiah, University of Tasmania.
Contributor Information
Bernard Moss, Email: bmoss@nih.gov.
Barry R. Bloom, Harvard School of Public Health
REFERENCES
- 1.Jenner E. 1798. An inquiry into the causes and effects of the variolae vaccinae, a disease discovered in some of the western countries of England, particularly near Gloucestershire, and known by the name of the cow pox, London, p 213–240. In Camac CNB (ed), Classics of medicine and surgery, 1959. Dover, New York, NY. [Google Scholar]
- 2.Baxby D. 1977. The origins of vaccinia virus. J Infect Dis 136:453–455. doi: 10.1093/infdis/136.3.453. [DOI] [PubMed] [Google Scholar]
- 3.Qin L, Favis N, Famulski J, Evans DH. 2015. Evolution of and evolutionary relationships between extant vaccinia virus strains. J Virol 89:1809–1824. doi: 10.1128/JVI.02797-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damaso CR. 2018. Revisiting Jenner's mysteries, the role of the Beaugency lymph in the evolutionary path of ancient smallpox vaccines. Lancet Infect Dis 18:e55–e63. doi: 10.1016/S1473-3099(17)30445-0. [DOI] [PubMed] [Google Scholar]
- 5.Lane JM, Ruben FL, Neff JM, Millar JD. 1969. Complications of smallpox vaccination, 1968. National surveillance in the United States. N Engl J Med 281:1201–1208. doi: 10.1056/NEJM196911272812201. [DOI] [PubMed] [Google Scholar]
- 6.Aragon TJ, Ulrich S, Fernyak S, Rutherford GW. 2003. Risks of serious complications and death from smallpox vaccination: a systematic review of the United States experience. BMC Public Health 3:26–1968. doi: 10.1186/1471-2458-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poland GA, Grabenstein JD, Neff JM. 2005. The US smallpox vaccination program: a review of a large modern era smallpox vaccination implementation program. Vaccine 23:2078–2081. doi: 10.1016/j.vaccine.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Weltzin R, Liu J, Pugachev KV, Myers GA, Coughlin B, Blum PS, Nichols R, Johnson C, Cruz J, Kennedy JS, Ennis FA, Monath TP. 2003. Clonal vaccinia virus grown in cell culture as a new smallpox vaccine. Nat Med 9:1125–1130. doi: 10.1038/nm916. [DOI] [PubMed] [Google Scholar]
- 9.Weltzin R, Drabik G, Liu YX, Georgakopoulos K, Pougatcheva S, Vellom D, Monath TP. 2003. Equivalence of ACAM1000 and ACAM2000 smallpox vaccines in virulence and immunogenicity. FASEB J 17:C26–C27. [Google Scholar]
- 10.Osborne JD, Da Silva M, Frace AM, Sammons SA, Olsen-Rasmussen M, Upton C, Buller RM, Chen N, Feng Z, Roper RL, Liu J, Pougatcheva S, Chen W, Wohlhueter RM, Esposito JJ. 2007. Genomic differences of Vaccinia virus clones from Dryvax smallpox vaccine: the Dryvax-like ACAM2000 and the mouse neurovirulent Clone-3. Vaccine 25:8807–8832. doi: 10.1016/j.vaccine.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 11.Qin L, Upton C, Hazes B, Evans DH. 2011. Genomic analysis of the vaccinia virus strain variants found in Dryvax vaccine. J Virol 85:13049–13060. doi: 10.1128/JVI.05779-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morikawa S, Sakiyama T, Hasegawa H, Saijo M, Maeda A, Kurane I, Maeno G, Kimura J, Hirama C, Yoshida T, Asahi-Ozaki Y, Sata T, Kurata T, Kojima A. 2005. An attenuated LC16m8 smallpox vaccine: analysis of full-genome sequence and induction of immune protection. J Virol 79:11873–11891. doi: 10.1128/JVI.79.18.11873-11891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin L, Liang M, Evans DH. 2013. Genomic analysis of vaccinia virus strain TianTan provides new into the evolution and evolutionary relationships between Orthopoxviruses. Virology 442:59–66. doi: 10.1016/j.virol.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 14.Monath TP, Caldwell JR, Mundt W, Fusco J, Johnson CS, Buller M, Jian L, Gardner B, Downing G, Blum PS, Kemp T, Nichols R, Weltzin R. 2004. ACAM2000 clonal Vero cell culture vaccinia virus (New York City Board of Health strain)–a second-generation smallpox vaccine for biological defense. Int J Infect Dis 8:31–44. doi: 10.1016/j.ijid.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Americo JL, Moss B, Earl PL. 2010. Identification of wild-derived inbred mouse strains highly susceptible to monkeypox virus infection for use as small animal models. J Virol 84:8172–8180. doi: 10.1128/JVI.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Americo JL, Sood CL, Cotter CA, Vogel JL, Kristie TM, Moss B, Earl PL. 2014. Susceptibility of the wild-derived inbred CAST/Ei mouse to infection by orthopoxviruses analyzed by live bioluminescence imaging. Virology 449:120–132. doi: 10.1016/j.virol.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Symons JA, Alcami A, Smith GL. 1995. Vaccinia virus encodes a soluble type 1 interferon receptor of novel structure and broad species specificity. Cell 81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 18.Alcami A, Symons JA, Smith GL. 2000. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J Virol 74:11230–11239. doi: 10.1128/jvi.74.23.11230-11239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu RH, Rubio D, Roscoe F, Krouse TE, Truckenmiller ME, Norbury CC, Hudson PN, Damon IK, Alcami A, Sigal LJ. 2012. Antibody inhibition of a viral type 1 interferon decoy receptor cures a viral disease by restoring interferon signaling in the liver. PLoS Pathog 8:e1002475. doi: 10.1371/journal.ppat.1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM. 1995. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem 270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- 21.Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, Aronow BJ, Karp CL, Brooks DG. 2013. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earl PL, Americo JL, Moss B. 2017. Insufficient innate immunity contributes to the susceptibility of the castaneous mouse to orthopoxvirus infection. J Virol 91:e01042-17. doi: 10.1128/JVI.01042-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polak MF, Beunders BJ, Van Der Werff AR, Sanders EW, Van Klaveren J, Brans LM. 1963. A comparative study of clinical reaction observed after application of several smallpox vaccines in primary vaccination of young adults. Bull World Health Organ 29:311–322. [PMC free article] [PubMed] [Google Scholar]
- 24.Fenner F, Henderson DA, Arita I, Jezek Z, Ladnyi ID. 1988. Smallpox and its eradication, 1st ed. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 25.Marrenikova SS, Svet-Moldavskaya IA, Maltseva NN. 1969. On laboratory markers of reactogenicity of smallpox vaccines. Voprosy Virusologii 14:579–584. [PubMed] [Google Scholar]
- 26.Guo ZS, Lu BF, Guo ZB, Giehl E, Feist M, Dai EY, Liu WL, Storkus WJ, He YK, Liu ZQ, Bartlett DL. 2019. Vaccinia virus-mediated cancer immunotherapy: cancer vaccines and oncolytics. J Immunother Cancer 7:6. doi: 10.1186/s40425-018-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Chen N, Feng Z, Buller RM, Osborne J, Harms T, Damon I, Upton C, Esteban DJ. 2006. Genomic sequence and analysis of a vaccinia virus isolate from a patient with a smallpox vaccine-related complication. Virol J 3:88. doi: 10.1186/1743-422X-3-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reif DM, McKinney BA, Motsinger AA, Chanock SJ, Edwards KM, Rock MT, Moore JH, Crowe JE. 2008. Genetic basis for adverse events after smallpox vaccination. J Infect Dis 198:16–22. doi: 10.1086/588670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyatt LS, Earl PL, Moss B. 2017. Generation of recombinant vaccinia viruses. Curr Protoc Mol Biol 117:16–18. doi: 10.1002/cpps.33. [DOI] [PubMed] [Google Scholar]
- 30.Israely T, Paran N, Erez N, Cherry L, Tamir H, Achdout H, Politi B, Israeli O, Zaide G, Cohen-Gihon I, Vitner EB, Lustig S, Melamed S. 2019. Differential response following infection of mouse CNS with virulent and attenuated vaccinia virus strains. Vaccines 7:19. doi: 10.3390/vaccines7010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blockade of IFNAR1 by IFN-α2-Fc. CAST mice were injected i.p. with 500 μg of anti-mouse IFNAR MAb or anti-mouse IgG1 isotype MAb as described in the legend to Fig. 6, except that the mice were not infected. Splenocytes were isolated one day after the third injection and stained with anti-IFNAR1 or anti-IgG isotype conjugated to phycoerthyrin (PE). Fluorescence was determined by flow cytometry, and the numbers within the panels refer to the percentage of cells labeled by IFNAR1-PE. Approximately 30,000 total events were collected for each sample. Download FIG S1, TIF file, 0.2 MB (250.8KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Depletion of CD4+ and CD8+ T cells. Anti-CD4 and anti-CD8 MAb or anti-KLH isotype MAb were injected i.p. into CAST mice on days 3 and 4 prior to infection as described for Fig. 6. At 24 h before infection, a sample of whole blood was collected and lymphocytes were isolated, incubated with anti-CD4-PE and anti-CD8-PerCP, and analyzed by flow cytometry as shown here. Approximately 28,000 to 40,000 total events were captured. Depletion was estimated to be >98%, and similar results were obtained by repeat analysis of whole blood at 11 days after infection (not shown). Download FIG S2, TIF file, 0.7 MB (754.8KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Data and materials will be made available upon request.