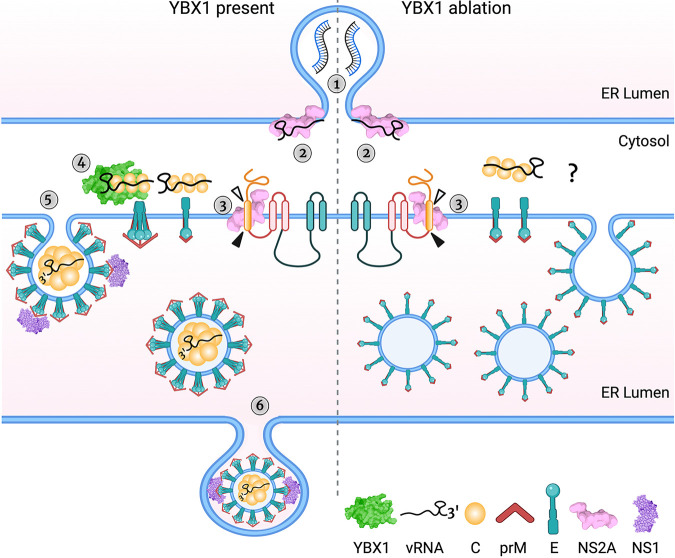
FIG 8.
YBX1 is required for DENV assembly: a model. The schematic shows DENV assembly and egress steps in WT (left of the dashed line) and YBX1 KO cells (right of the dashed line) (1) There is no difference in vRNA accumulation in WT and YBX1 KO cells and we interpret this to indicate normal DENV vRNA replication, which occurs in ER invaginations in close proximity to the assembly sites. (2) NS2A binds to vRNA and recruits it to the assembly sites in both WT and KO cells. (3) Processing of capsid-prM junctions allows the formation of prM-E dimers, the dimerization of capsid proteins and its association with vRNA. (4) YBX1 binds to the nucleocapsid enabling the interaction of capsid dimers with prM-E trimers. We posit that this interaction is disrupted in YBX1 KO cells. (5) The viral membrane proteins drive the budding of assembled particles into the ER lumen. In YBX1 KO cells, budded particles are empty-looking and with a rough surface. We propose that these anomalous particles arise from an assembly defect affecting the organization of E on the host membrane, likely due to the lack of interaction between this protein with the nucleocapsid, which is mediated by YBX1. (6) Assembled particles are secreted via the secretory pathway, in a process likely mediated by NS1 and other sorting mechanism involving prM-E interacting proteins. In the anomalous particles found in YBX1 KO cells, the secretion motifs are lacking or hidden, and thus particles accumulate in the ER lumen. Created with BioRender.com.