ABSTRACT
The highly transmissible severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 253 million people, claiming ∼5.1 million lives to date. Although mandatory quarantines, lockdowns, and vaccinations help curb viral transmission, there is a pressing need for cost-effective systems to mitigate the viral spread. Here, we present a generic strategy for capturing SARS-CoV-2 through functionalized cellulose materials. Specifically, we developed a bifunctional fusion protein consisting of a cellulose-binding domain and a nanobody (Nb) targeting the receptor-binding domain of SARS-CoV-2. The immobilization of the fusion proteins on cellulose substrates enhanced the capture efficiency of Nbs against SARS-CoV-2 pseudoviruses of the wild type and the D614G variant, the latter of which has been shown to confer higher infectivity. Furthermore, the fusion protein was integrated into a customizable chromatography with highly porous cellulose to capture viruses from complex fluids in a continuous fashion. By capturing and containing viruses through the Nb-functionalized cellulose, our work may find utilities in virus sampling and filtration through the development of paper-based diagnostics, environmental tracking of viral spread, and reducing the viral load from infected individuals.
IMPORTANCE The ongoing efforts to address the COVID-19 pandemic center around the development of diagnostics, preventative measures, and therapeutic strategies. In comparison to existing work, we have provided a complementary strategy to capture SARS-CoV-2 by functionalized cellulose materials through paper-based diagnostics as well as virus filtration in perishable samples. Specifically, we developed a bifunctional fusion protein consisting of both a cellulose-binding domain and a nanobody specific for the receptor-binding domain of SARS-CoV-2. As a proof of concept, the fusion protein-coated cellulose substrates exhibited enhanced capture efficiency against SARS-CoV-2 pseudovirus of both the wild type and the D614G variant, the latter of which has been shown to confer higher infectivity. Furthermore, the fusion protein was integrated into a customizable chromatography for binding viruses from complex biological fluids in a highly continuous and cost-effective manner. Such antigen-specific capture can potentially immobilize viruses of interest for viral detection and removal, which contrasts with the common size- or affinity-based filtration devices that bind a broad range of bacteria, viruses, fungi, and cytokines present in blood (https://clinicaltrials.gov/ct2/show/NCT04413955). Additionally, since our work focuses on capturing and concentrating viruses from surfaces and fluids as a means to improve detection, it can serve as an “add-on” technology to complement existing viral detection methods, many of which have been largely focusing on improving intrinsic sensitivities.
KEYWORDS: COVID-19, SARS-CoV-2, nanobody, cellulose, cellulose binding protein, cellulose binding domain
INTRODUCTION
Since the first documented coronavirus disease 2019 (COVID-19) case at the end of 2019 (1), the highly contagious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in at least 253 million positive cases and 5.1 million deaths (∼2.02% fatality rate) in 223 countries and territories (as of 12 November 2021 according to https://covid19.who.int/). To contain the spread of SARS-CoV-2, nonpharmaceutical interventions were originally deployed, including wearing masks, handwashing, and routine testing and contact tracing, as well as public measures, such as city lockdowns, travel restrictions, and social distancing. However, the long-term adherence to these preventative measures has led to severe societal and economic burdens (2). Meanwhile, the administration of SARS-CoV-2 vaccines has helped to reduce infection and death rates worldwide (3). Nevertheless, due to the constant emergence of SARS-CoV-2 variants, COVID-19 poses a continued threat to the public (4), which necessitates complementary technologies to existing diagnosis platforms, as well as reducing viral load from COVID-19 patients (5, 6; https://clinicaltrials.gov/ct2/show/NCT04413955).
SARS-CoV-2 is a single-stranded RNA virus that consists of four major structural proteins: the spike (S), the membrane (M), the envelope (E), and the nucleocapsid (N) (7). Viral infections rely upon cellular entry to utilize the host’s machinery for replicating viral copies that are then released by the host. The S protein facilitates the attachment of the virus to the host’s cellular receptors and promotes the fusion between host and viral membranes (8). In particular, the S protein contains the receptor-binding domain (RBD), which binds to the extracellular domain of the host receptor angiotensin-converting enzyme 2 (ACE2) for viral entry (9–12). Recent work demonstrated that SARS-CoV-2 targets the same functional host receptor, ACE2, as SARS-CoV. However, the effectiveness of SARS-CoV-2’s RBD is ∼10- to 20-fold higher than that of SARS-CoV in ACE2 binding. Due to the key roles of S protein or the subdomain RBD in the entry of SARS-CoV-2 into host cells, the S protein or RBD has been extensively explored as a pivotal target for the development of antiviral antibodies, among which nanobodies (Nbs) represent a unique class in these efforts. Nbs are single-domain nano-size antibodies, which are derived from variable fragments of Camelidae (including camels and llamas) heavy-chain-only antibodies (13, 14). Nbs offer a variety of advantages over other antibodies for diagnostic development: (i) nanometer size, (ii) high-affinity and specificity, (iii) deep penetration in tissues, (iv) low immunogenicity, and (v) easy scalability for mass production (14, 15). Due to these benefits, to date, several high-affinity neutralizing Nbs directed against the S protein and RBD have been identified, among which Ty1 has shown nanomolar binding affinity and effective neutralization after sequential steps of alpaca immunization and phage display (16–20).
In this study, we repurposed a recently developed Nb, Ty1, to capture SARS-CoV-2 from complex fluids by immobilizing cellulose materials with functionally oriented Nbs. Such a strategy could have implications in virus sampling from the environment and infected individuals for the development of paper-based diagnostics (21), as well as virus filtration from blood in an antigen-specific manner (5, 6). Specifically, we designed a bifunctional fusion protein that comprises a cellulose-binding domain (CBD) and Nb for cellulose immobilization and SARS-CoV-2 capture, respectively (22, 23). Nbs are generally easier to manufacture (e.g., Escherichia coli fermentation) than conventional human immunoglobin (IgG)-based antibodies, the latter of which require mammalian cell hosts for production (24). Additionally, the use of CBD fusion proteins has been demonstrated for the biofunctionalization of cellulose substrates in various applications, including protein purification (25, 26), textile manufacturing (27), and immunoassay development (28–31). Notably, the CBD can facilitate the absorption of CBD-containing fusion proteins to cellulose in molar quantities, which allows for an excess amount of immobilized proteins relative to the soluble target (28, 32). As a result, our fusion technology is highly cost-effective and scalable to overcome various challenges posed by the pandemic, including but not limited to disrupted supply chains, restricted deployment to remote areas, and mass production. As a proof of concept, we performed an immunoassay on cellulose-based filter paper for the detection of SARS-CoV-2’s RBD using our bifunctional proteins. Furthermore, we developed a customized cellulose-based affinity chromatography to remove SARS-CoV-2 viral particles from biological fluids, which may have utilities in sampling viruses of low concentration from the environment to track viral spread (33), as well as reducing the viral load from COVID-19 patients through extracorporeal filtration (5, 6; https://clinicaltrials.gov/ct2/show/NCT04413955). Given the modularity of our bifunctional protein platform and the ease of rapidly identifying target-specific Nbs through immunization and directed evolution, our work can potentially provide a framework to address other emerging infectious diseases by similar approaches.
RESULTS
Genetic fusion of an RBD-specific nanobody with a cellulose-binding domain.
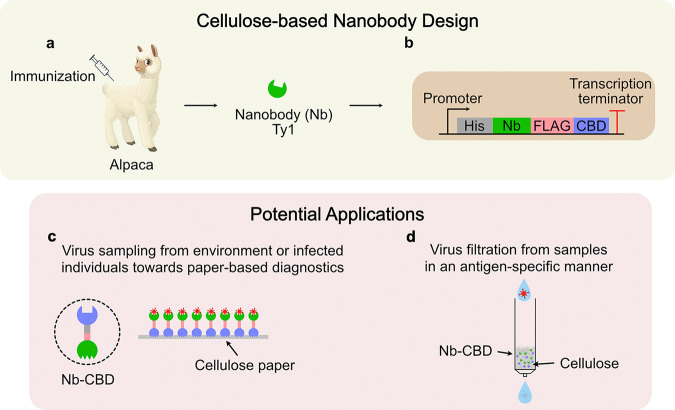
Our overall scheme for low-cost capture and detection of SARS-CoV-2 capitalizes on generating a bifunctional protein through the genetic fusion between the high-affinity Nb Ty1 targeting the RBD of SARS-CoV-2 and the cellulose-binding domain (CBD) (Fig. 1). SARS-CoV-2 can be transmitted via airborne particles or through directly contacting contaminated surfaces (34). Therefore, considering that cellulose is prevalent in many materials, such as paper towels and the inner coating of face masks, we immobilized the fusion proteins to the surface of cellulose materials, such as filter paper, to enable sampling and immobilization of SARS-CoV-2 from the environment or specimens of infected individuals (Fig. 1c). Importantly, due to the specific interaction between the CBD and cellulose, we reason that the bifunctional Nb-CBD can be immobilized in a defined orientation that favors the interaction between the Nb of interest and the antigen, compared to random immobilization. On the other hand, viral transmission and contamination through blood products present a concern during the pandemic. Because blood products are susceptible to heat and chemical denaturation, it is desirable to devise a strategy that reduces and eliminates the viral load in blood products while maintaining the blood’s bioactivities. To assess whether our strategy can address these challenges, we integrated the bifunctional fusion protein Nb-CBD into a customized cellulose affinity purification column to allow for a SARS-CoV-2-specific filtration in a continuous manner (Fig. 1d).
FIG 1.
Development of a bifunctional fusion protein to enable cellulose immobilization and subsequent detection and capture of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). (a) An alpaca-derived high-affinity nanobody (Nb), Ty1, for the receptor-binding domain (RBD) of SARS-CoV-2 was genetically fused with (b) a cellulose-binding domain (CBD) isolated from Clostridium thermocellum. (c) As SARS-CoV-2 is transmitted through surface contact, CBD fusion proteins or CBD-containing E. coli cell lysate was immobilized on the surface of cellulose materials, such as cellulose paper, for viral enrichment toward the development of paper-based diagnostics. (d) We also customized Nb-dependent regenerated amorphous cellulose (RAC) materials to specifically deplete the viral load of SARS-CoV-2 from virus-containing samples.
Production and purification of the bifunctional protein Nb-CBD.
Compared to human IgG, Nbs can be produced in E. coli with high yield and purity. Therefore, DNA encoding the fusion protein Nb-CBD was first cloned in a standard expression vector for recombinant protein production in E. coli. Because the antigen-binding site of Nbs is closed to the N terminus, we placed the CBD at the C terminus of Nb to circumvent potential steric hindrance. Meanwhile, a short 6×His tag was attached to the N terminus for metal affinity purification, while a FLAG epitope (DYKDDDDK) was inserted between Nb and the CBD, simultaneously serving as a hydrophilic flexible linkage and a tag for immunostaining (Fig. 1a and b). The yield of fusion proteins was estimated to be ∼50 mg/L of bacterial culture in a shake flask mode and was purified to high homogeneity, as evidenced by denaturing SDS-PAGE and size exclusion chromatography (see Fig. S1a and S1b in the supplemental material).
The fusion protein is functionally active in cellulose binding and RBD detection.
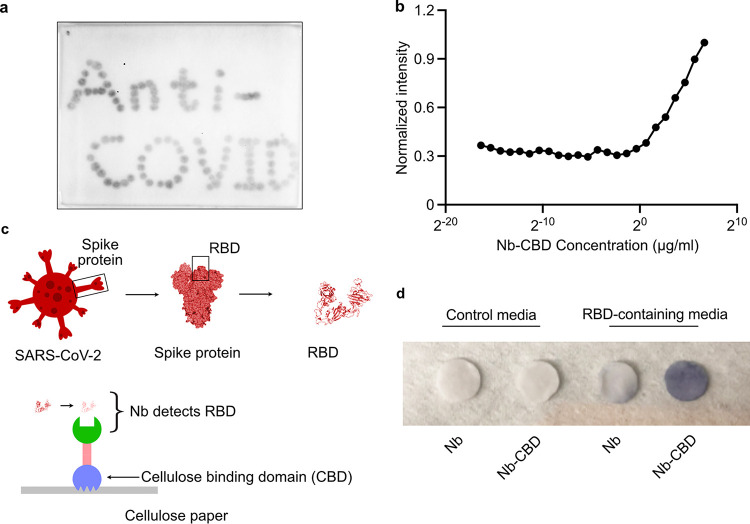
Next, we sought to evaluate whether the fusion proteins were capable of cellulose binding and Nb-specific target recognition. To this end, we first spotted purified fusion proteins onto the surface of cellulose paper (Fig. 2a). Upon air drying, the paper was stained with a rat antibody against the FLAG epitope in the fusion protein, followed by an anti-rat secondary antibody conjugated with horseradish peroxidase (HRP). A dark precipitate was visualized after incubation with an HRP substrate, 3,3′-diaminobenzidine (DAB). To quantify the binding efficiency of the fusion protein to the cellulose paper, we immobilized serially diluted fusion proteins within a defined area on the Whatman filter paper followed by immunostaining with an anti-FLAG antibody. Indeed, the extent of protein immobilization correlated with the staining in a certain concentration range (Fig. 2b; Fig. S1c), from which we estimated that a surface area of 1 mm2 can be saturated by 500 ng (∼0.02 nmol) of Nb-CBD proteins in the Whatman filter paper.
FIG 2.
The fusion protein maintains its activities in binding cellulose and the RBD of SARS-CoV-2. (a) Detection of immobilized Nb (Ty1)-CBD on a cellulose paper. Nb-CBD was first spotted onto a piece of cellulose paper. Upon air drying, the paper was incubated with a rat antibody against the FLAG epitope (DYKDDDDK), followed by an anti-rat secondary antibody conjugated with HRP. The dark precipitate “Anti-COVID” was visualized after incubation with 3,3′-diaminobenzidine (DAB). (b) Quantification of maximal protein absorption on Whatman filter paper. Ten microliters of serially diluted fusion protein solutions was applied to the filter paper, followed by immunoblotting with anti-FLAG directly on the filter paper. Based on the normalized unit intensity quantified by ImageJ, protein abundance increased with concentration. We estimate that 500 ng of Nb-CBD binds to 1 mm2 of cellulose paper at saturation status. (c) Schematic of an immunoassay to evaluate the function of the fusion protein. Nb-CBD fusion proteins were immobilized on cellulose paper and then submerged in culture medium containing RBD-Fc (∼100 ng/mL) as a proxy for actual SARS-CoV-2. The capture capability was confirmed by anti-human Fc-HRP and the DAB substrate. The structure of the RBD was adapted from PDB ID no. 6ZXN. (d) Testing the capture capability of protein-coated cellulose paper discs in RBD-containing medium. Representative discs were prepared by a 6-mm biopsy punch and then coated with E. coli lysates containing the indicated recombinant fusion proteins. The functionalized discs were incubated with RBD-containing or control (no RBD) medium. The intensity of dark staining was strongest from the combination of Nb-CBD-coated disc and RBD-containing medium (∼100 ng/mL).
Having validated the high binding capacity of purified Nb-CBD to filter paper, we speculated that because the CBD itself can act as a natural affinity ligand to cellulose, we could directly immobilize E. coli cell lysate containing the CBD fusion proteins on filter paper, followed by extensive washing to remove nonspecific proteins. This approach circumvents the need to purify the desired proteins beforehand, which is labor-intensive and impractical when it comes to the large-scale manufacturing of functionalized cellulose materials (Fig. 2c). To this end, we first incubated E. coli cell lysate with filter discs and removed nonspecific proteins via washing. Then we subjected the functionalized filter discs to cell culture medium containing secreted recombinant RBD as a proxy for actual SARS-CoV-2. As shown in Fig. 2d, Nb-CBD-coated filter paper was able to capture SARS-CoV-2’s RBD, as evidenced by the intense dark staining reflecting the detection of RBD. Of note, filter discs precoated with Nb alone exhibited light staining, likely due to nonspecific but weak absorption of Nb to cellulose. In comparison, in the control medium without the RBD, neither Nb- nor Nb-CBD-coated discs displayed the dark staining. Our findings here indicate that the CBD promoted the immobilization of Nb-CBD on cellulose substrates, while Nb remained able to specifically recognize the target.
Immobilization of Nb-CBD on filter paper increases the capture efficiency of Nb against SARS-CoV-2 pseudovirus.
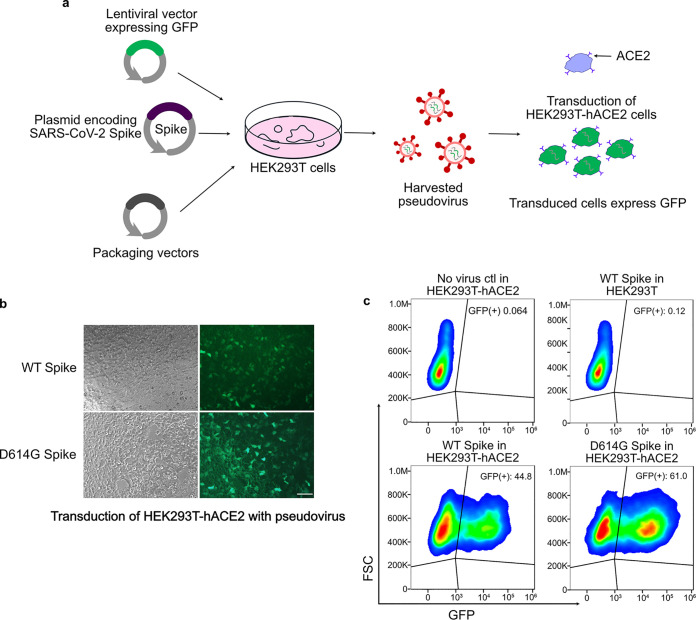
The stoichiometry and kinetics of a target-binding interaction can be favorably influenced by three general approaches: (i) increasing the molar abundance and the soluble antigen concentration, (ii) improving the binding interaction affinity under relevant assay conditions, and (iii) raising the capturing reagents’ (e.g., antibodies) abundance and concentration through surface immobilization according to the law of mass action (35, 36). Since it is not practical to raise the concentration of antigens or the affinity of already optimized antibodies, here we sought to explore the third strategy of increasing the surface densities of Nb-CBD via immobilization on cellulose paper. To do so, we evaluated the capability of Nb-CBD fusion proteins in capturing SARS-CoV-2 mimics (referred to as pseudovirus in this work). One of the “gold standards” for SARS-CoV-2-related studies is to use nonreplicative lentivirus pseudotyped with the S protein derived from SARS-CoV-2 in conjunction with mammalian cells engineered to express human ACE2 (hACE2) (37) (Fig. 3a). Since emerging SARS-CoV-2 variants with higher transmission rates bear mutations in the S protein, one advantage of the pseudovirus system is that it can rapidly evaluate intervention approaches against different spike variants. Using this system, we compared the original wild-type (WT) S protein to the D614G mutant, in which the 614th aspartate is converted to glycine in the S protein of SASR-CoV-2. Notably, epidemiology and molecular biology studies have demonstrated that the D614G mutant confers higher transmission and worse symptoms in humans (38). Therefore, it is of particular interest to assess our fusion protein strategy in the context of both the WT and the D614G variant. As shown in Fig. 3b and c, after HEK293T-hACE2 cells were transduced with WT or D614G pseudotyped lentivirus carrying a green fluorescence protein (GFP) reporter, ∼50% of cells were GFP positive, with D614G pseudovirus exhibiting a higher transduction efficacy than that of the WT. These findings agreed with the increased infectivity by the D614G mutation (39, 40). In comparison, transduction of the parental HEK293T cell line (lacking hACE2 expression) with the same SAR-CoV-2 pseudovirus did not result in GFP expression, which validated an ACE2-dependent infection by SARS-CoV-2 (41).
FIG 3.
Generation of wild-type and D614G pseudoviruses for functional assays by Nb (Ty1)-CBD-functionalized cellulose. (a) Schematic overview of the pseudovirus production. HEK293T cells were transfected with a lentiviral vector expressing a green fluorescent protein (GFP), a plasmid encoding SARS-CoV-2 spike, and packaging vectors. The transfected cells produced lentiviral particles pseudotyped with the S protein of SARS-CoV-2, and the pseudovirus can transduce HEK293T expressing human angiotensin-converting enzyme 2 (hACE2) to express GFP. (b) Microscope images showing that the HEK293T-hACE2 cells expressed GFP after transduction with lentivirus pseudotyped with the wild-type (WT) SARS-CoV-2 spike protein or the D614G variant. Scale bar, 100 μm. (c) Representative flow cytometric analysis evaluating the transduction efficiency of SARS-CoV-2 WT and D614G pseudoviruses compared with two negative-control groups: HEK293T-hACE2 without any transduction and HEK293T transduced with SARS-CoV-2 WT pseudotyped lentivirus. Results are representative of three independent experiments.
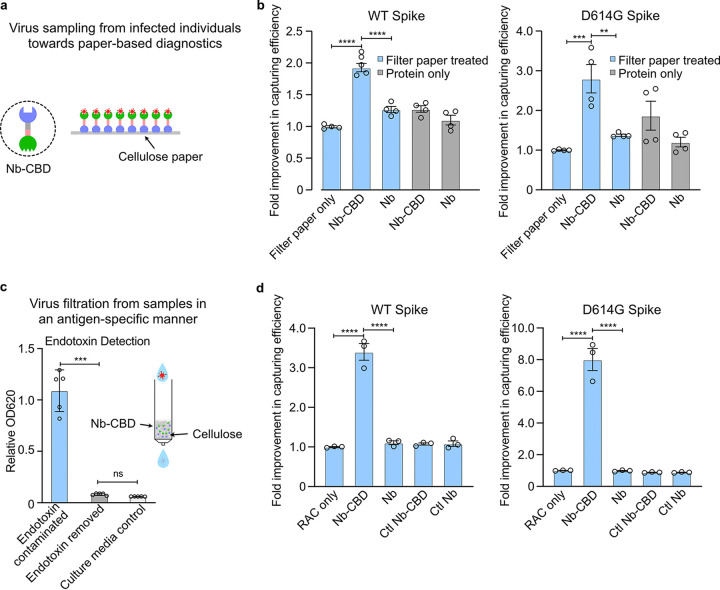
It is worth noting that the levels of SARS-CoV-2 in COVID-19 patients range from 104 to 109 copies/mL, depending on the type of bodily fluids and degree of the symptoms (42, 43). Meanwhile, we calculated the titers of WT or D614G pseudotyped lentivirus and estimated that ∼105 viral particle particles/mL were present in the culture medium. Therefore, to demonstrate the capability of capturing SARS-CoV-2 at the lower end of the viral titer range for SARS-CoV-2-containing fluids, we further diluted the medium to contain approximately ∼104 pseudovirus copies/mL and quantified the viral capture efficacy of Nb-CBD-immobilized cellulose paper (Fig. 4a). In addition, filter paper alone or filter paper coated with Nb but lacking the CBD module served as a negative control. The capture efficiency of Nb-CBD-immobilized filter paper was calculated by dividing the titer of medium treated with Nb-CBD-immobilized filter paper or other control groups by the initial viral titer (i.e., without any treatment). Indeed, using medium containing WT or D614G pseudoviruses (Fig. 4b), Nb-CBD-immobilized filter paper resulted in an ∼2-fold increase of the capture efficacy compared to that of filter paper only and an ∼1.5-fold enhancement over filter paper precoated with Nb alone. Moreover, filter paper precoated with Nb-CBD or Nb displayed an ∼1.65-fold improvement in binding pseudovirus from the medium over free proteins (Nb-CBD or Nb) at equal concentrations, which indicated that the surface immobilization itself can facilitate target recognition. Notably, a similar strategy has been proposed for the detection of SARS-CoV-2-specific antibody through cellulose filter paper immobilized with fusion proteins (22, 23), in which the nucleocapsid (N) protein of SARS-CoV-2 was linked with the CBD to capture N protein-specific antibodies. In comparison, the present study directly captured the viral particles by linking CBD with an Nb specific for the S protein of SARS-CoV-2, which can facilitate subsequent investigation of SARS-CoV-2, such as monitoring the viral load and spread in the environment.
FIG 4.
SARS-CoV-2 pseudovirus capture by Nb (Ty1)-CBD-immobilized cellulose in two different formats. (a) Schematic of increasing surface densities of Nb-CBD through protein immobilization on cellulose materials for SARS-CoV-2 neutralization. (b) Increased neutralization efficacy of pseudovirus through protein immobilization on cellulose paper over free proteins. After incubation of the pseudovirus with 200 μL of 10 μg/mL fusion protein Nb-CBD or Nb (negative control) immobilized on cellulose paper or free protein with equal concentrations, the titers of wild-type (WT) and D614G pseudoviruses were quantified by transducing HEK293T-hACE2 cells with the remaining viruses in the supernatant. Fold changes from each treatment group were normalized to that of filter paper only. (c) An in vitro cellular assay to confirm the complete removal of bacterial toxins from the Nb-CBD-functionalized RAC column. Bacterial toxins were removed in a single step with 0.1% Triton X-114. Flowthrough factions from Nb-CBD-functionalized RAC were added to RAW-Blue cells, an engineered murine macrophage cell line that can detect trace levels of endotoxin and other bacterial toxins. The secretion of a reporter protein, phosphatase, can indicate the presence of bacterial toxins as innate immune agonists. It was found that direct flowthrough fractions were indeed contaminated with bacterial toxins, denoted by “Endotoxin contaminated,” while flowthrough fractions from Triton X-114-treated RAC (denoted by “Endotoxin removed”) did not have detectable levels of toxins compared to the culture medium control. A schematic of an Nb-CBD-functionalized RAC column is shown in the inset. (d) Capture efficacy of Nb-CBD-functionalized RAC. The flowthrough samples from functionalized RAC columns were used to transduce HEK293T-hACE2 cells to quantify viral titers for WT and D614G SARS-CoV-2 pseudoviruses, respectively. Fold changes from each treatment group were normalized to that of RAC only. Graphs are expressed as mean ± SEM (n = 4) in panel b and as mean ± SEM (n = 3) in panel d. Statistical analysis was performed by one-way analysis of variance (ANOVA) according to the following scale: **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Integration of the bifunctional protein with an amorphous cellulose column further enhanced the capture efficiency.
Having validated the increased capture efficiency of fusion proteins immobilized on filter paper, we further sought to enhance the capture efficiency for SARS-CoV-2 by incorporating the fusion proteins into regenerated amorphous cellulose (RAC). Since RAC has been shown to exhibit a higher surface area per unit of mass than filter paper (44), RAC may improve the rate and the degree of target capture by increasing the immobilization densities of Nb-CBD on cellulose (22, 23). To demonstrate the ease of preparing the Nb-CBD-functionalized cellulose column, we directly perfused total E. coli lysate containing the fusion proteins through a plastic column packed with ∼0.1 mL (∼50 mg dry weight) of RAC. As shown in Fig. S1d, only Nb-CBD but not Nb alone was able to specifically bind RAC after total proteins were extracted from protein-coated RAC columns, which were pretreated with cell lysate containing Nb-CBD or Nb. While using total cell lysate instead of purified proteins could greatly reduce the time and cost of the column preparation, the possible contamination with bacterial toxins can pose a challenge to certain downstream applications. Interestingly, we found that an additional washing step using 0.1% Triton X-114 (TX-114) was able to completely eliminate bacterial toxins that are immunostimulatory in macrophage cells (Fig. 4c).
Having demonstrated the simplicity and the lack of bacterial toxin contamination in preparing the functionalized RAC columns, the culture medium containing WT or D614 pseudoviruses was passed through the functionalized column by gravity, and viral titers were determined for different flowthrough samples. Compared to RAC alone, Nb-CBD-immobilized columns increased the capture efficiency for WT and D614G pseudoviruses by ∼3.5 times and ∼8 times, respectively. In contrast, RAC columns carrying an irrelevant Nb (caffeine specific) fused with CBD or irrelevant Nb alone failed to further enhance the degree of the capture efficiency compared to that of RAC alone (Fig. 4d). Taken together, we demonstrated that the Nb-CBD fusion protein can be integrated into an RAC column to markedly increase the filtration efficiency of SARS-CoV-2 pseudovirus in a highly specific and continuous fashion.
DISCUSSION
During the pandemic, the affordability and deployability associated with the supply chain represent increasingly essential but often neglected aspects of outbreak response, especially in developing countries. To this end, we developed a simple yet versatile technology to immobilize SARS-CoV-2 on cost-effective cellulose substrates, which were functionalized by Nbs to achieve antigen-specific capture. While cellulose represents the most abundant and commonly used biopolymers, Nbs have been gaining increasing attention due to their stability and ease of production from bacteria. In addition to minimizing the cost of reagents, we further demonstrated that the Nb-functionalized cellulose substrate can be easily prepared by first incubating cell lysate containing the fusion proteins and then washing off the unbound nonspecific proteins. Thanks to these attributes, our approach holds promise as a highly adaptable add-on technology to existing cellulose-based diagnostic tests and filtration systems.
In addition to immobilizing virus on cellulose-based diagnostic platforms, sampling and concentrating viral particles (e.g., SARS-CoV-2) from surfaces and fluids can greatly facilitate downstream detection. Recently, the Centers for Disease Control and Prevention called for the development of a wastewater surveillance sampling strategy to monitor viral threats in the community. It has been suggested that in most cases, untreated wastewater will likely require concentration prior to RNA extraction, and the number of infections needed to detect the virus in wastewater without concentration is difficult to determine (33). Having recognized this pressing need, we introduced a generic strategy to enrich viral particles of low concentrations from surfaces and fluids. To do so, we were inspired by how our immune system exploits antibodies to capture viruses from the blood. Because our work focuses on capturing and concentrating viruses from surfaces and fluids as a means to improve detection, we reasoned that it can serve as an “add-on” technology to complement existing viral detection methods, many of which have been largely focusing on increasing the sensitivities of various diagnostic platforms.
Regarding the clinical relevance of our work, for individuals infected with SARS-CoV-2, it has been postulated that the clearance of SARS-CoV-2 virus from COVID-19 patients may provide a more opportunistic environment for the immune system to clear the virus and establish lasting immunity. For this reason, there is an ongoing clinical trial that explores a broad-spectrum sorbent hemoperfusion filter for removing virus from the blood in patients (5, 6). While such a filter was designed to bind a broad range of bacteria, viruses, fungi, and cytokines present in blood, our approach is more pathogen or antigen dependent due to the fact that specific Nbs can be immobilized on the surface of cellulose.
While our study only focused on Ty1, a recently developed Nb against the spike protein of SARS-CoV-2 (16), the conceptual framework can be easily adapted to target other emerging viruses by substitution for the Nb module with other target-specific Nbs. Moreover, since many other SARS-CoV-2-specific Nbs have been identified through immunization and phage display to target different epitopes for SARS-CoV-2, future work may investigate a combination of CBD fusion proteins comprising different Nbs in a multivalent manner to enhance capture efficiency (45). Despite the promises demonstrated in this study, one limitation is that only pseudovirus-containing culture media were used to characterize the fusion proteins for proof of concept. Therefore, it is necessary to further evaluate our approach in the near future in real specimens, such as actual wastewater contaminated with SARS-CoV-2 or blood from COVID-19 patients.
MATERIALS AND METHODS
Reagents and chemicals.
Tween 20, Triton X-100 (TX-100), and Triton X-114 (TX-114) were obtained from Sigma-Aldrich (St. Louis, MO). Strep-tag and Strep-Tactin XT were purchased from IBA Lifesciences (Gottingen, Germany). A detergent-compatible (DC) protein assay kit was bought from Bio-Rad Laboratories (Hercules, CA). Anti-FLAG epitope (DYKDDDDK; catalog no. 637301) and horseradish peroxidase (HRP)-conjugated donkey anti-human IgG antibody (catalog no. 410902) were purchased from Biolegend (San Diego, CA). The secondary antibody anti-rat IgG-HRP (catalog no. 7077) was bought from Cell Signaling Technology (CST [Danvers, MA]). All other reagents and chemicals, including nickel-nitrilotriacetic acid (Ni-NTA) agarose and the Pierce Rapid Gold BCA (bicinchoninic acid) protein assay kit, were purchased from Fisher Scientific International, Inc., (Hampton, NH) and were of the highest purity or analytical grade commercially available.
Cell lines.
HEK293T cells expressing human angiotensin I-converting enzyme 2 (HEK293T-hACE2) were kindly provided by Jesse Bloom (Fred Hutchinson Cancer Research Center, Seattle, WA, USA) (46). The Lenti-X 293T cell line was purchased from TaKaRa Bio USA, Inc. (San Jose, CA). RAW-Blue cells were purchased from Invivogen (San Diego, CA, USA). These cell lines were maintained in complete Dulbecco’s modified Eagle’s medium (DMEM) (Corning, Corning, NY) supplemented with 10% fetal bovine serum (FBS) (Corning) and 100 U/mL penicillin-streptomycin (Corning) at 37°C in a humidified incubator with 5% CO2. Cells at passages 2 to 10 were used for the experiments.
Plasmid construction, protein expression, and purification.
Nb (Ty1) variants, including Nb-CBD and control protein Nb without CBD module, were cloned into the pSH200 vector (a generous gift from Xiling Shen at Duke University), containing a 6×His tag, between BamHI and XbaI sites. Both plasmids were validated by sequencing before expression. Nb-CBD and Nb were expressed and produced in the same manner previously described (44, 47). Produced protein was sequentially purified by affinity chromatography using Ni-NTA agarose beads and fast protein liquid chromatography (FPLC) (NGC Quest 10 chromatography system; Bio-Rad, Hercules, CA). Protein fractions detected at λ = 280 nm were collected. Collected protein fractions were quantified by a detergent-compatible (DC) protein assay according to the manufacturer’s instructions, and purities were verified by SDS-PAGE. Validated protein was aliquoted and kept at −80°C with 50% glycerol at all times until future use.
Cellulose paper-based immunoblotting.
To validate the binding ability of Nb-CBD, purified Nb-CBD was spotted onto cellulose paper and dried at room temperature for ∼2 min. Cellulose paper with dried Nb-CBD was incubated with 5 mL of 5% nonfat milk in Tris-buffered saline (TBS) for 30 min to block nonspecific binding sites. After blocking, the cellulose paper was then incubated with anti-FLAG epitope (DYKDDDDK), which was diluted at 1:2,000 in 3 mL TBS plus 5% nonfat milk overnight at 4°C. The paper was washed three times with 5 mL 1× TBS containing 0.05% Tween 20 (TBST), with 15 min per wash cycle. The paper was incubated with HRP-conjugated anti-rat IgG (1:2,000) for 1 h at room temperature. After being washed with 1× TBS with 5 mL 0.05% Tween 20, premixed Pierce 3,3′-diaminobenzidine (DAB) substrate was directly added onto the cellulose paper. The reaction was terminated with water after dark spots appeared.
Pseudovirus production.
HDM-SARS2-Spike-delta21 and HDM-SARS2-Spike-del21-614G encoding the SARS-CoV-2 spike protein with a 21-amino-acid C-terminal deletion for lentiviral pseudotyping were purchased from Addgene (Watertown, MA) under Addgene serial no. 155130 and 158762, respectively. Both plasmids were isolated following the manufacturer’s instructions and quantified through a Nanodrop spectrophotometer. Twenty-four hours prior to transfection, Lenti-X 293T cells in the logarithmic growth phase were trypsinized, and the cell density was adjusted to 1.0 × 106 cells/mL with complete DMEM. The cells were reseeded into 10-cm cell culture dishes to reach 70% confluence on the day of transfection. The plasmid mixture was prepared according to Tables 1 and 2. After the plasmid mixture was inverted 5 to 8 times and kept at room temperature for 30 min, the plasmid mixture was gently added to the Lenti-X 293T cells (46). After 48 and 72 h of transfection, the pseudovirus was collected and centrifuged at 1,000 × g at 4°C for 20 min to remove the debris. The cell culture medium was replaced with fresh complete DMEM once the pseudovirus was collected. Aliquots of the harvested virus were stored at 4°C for immediate use or frozen at −80°C for future use.
TABLE 1.
Compositions of the plasmid mixtures for generation of WT or D614G mutant SARS-CoV-2 pseudovirus
| Composition | Amt |
|---|---|
| HDM-SARS-2-Spike-delta21 (or D614G) | 2.5 μg |
| pFuw-Ubc-GFP | 10 μg |
| psPAX2 | 7.5 μg |
| TransIT-X2 2000 | 30 μL |
| Opti-MEM | 1,000 μL |
TABLE 2.
Compositions of the plasmid mixture for generation of lentivirus as a positive control
| Composition | Amt |
|---|---|
| pMD2.G | 2.5 μg |
| pFuw-Ubc-GFP | 10 μg |
| psPAX2 | 7.5 μg |
| TransIT-X2 2000 | 30 μL |
| Opti-MEM | 1,000 μL |
Quantification of viral titers by flow cytometry.
HEK293T-hACE2 cells were seeded 24 h before the pseudovirus transduction assay in 96-well plates (2 × 104 cells/well in a volume of 100 μL complete DMEM). On the coculture day, the medium was removed and 200 μL of prewarmed pseudovirus was added to the cells. Polybrene (Sigma-Aldrich) was added into cultured HEK293T-hACE2 cells to a final concentration of 8 μg/mL, to facilitate lentiviral infection by minimizing charge repulsion between virus and cells. After transduction for 48 h, cells were collected through trypsinization and transferred to a 96-well V-bottom plate with complete culture DMEM. Cells were pelleted at 300 × g for 3 min and washed twice with phosphate-buffered saline (PBS: 137 mM NaCl, 10 mM phosphate, 2.7 mM KCl [pH 7.4]). After the final wash, the cells were resuspended in 200 μL fluorescence-activated cell sorter (FACS) buffer (5% FBS, 2 mM EDTA, 0.1% sodium azide in PBS) for flow cytometric analysis in an Attune NxT flow cytometer (Thermo Fisher), and data were analyzed by FlowJo (Franklin Lakes, NJ). Using a well that has 1 to 20% GFP-positive cells, the titer was calculated according to the following formula: titer = [(F × Cn)/V] × DF. F represents the frequency of GFP-positive cells determined by flow cytometry, Cn represents the total number of target cells infected, V represents the volume of the inoculum, and DF represents the virus dilution factor. For all experiments, triplicate samples were analyzed, data are representative of two or more experiments, and the standard error of the mean (SEM) is shown.
Preparation of the RAC column.
Regenerated amorphous cellulose (RAC) was produced by RAC-based affinity protein purification as previously described (44). The RAC column was first equilibrated with 1× PBS, and proteins of interest were added to the RAC column until the column was saturated by quantifying the amount of fusion protein in flowthrough fractions and comparing it to the original supernatant via SDS-PAGE. The saturated RAC was washed with 10 column volumes of 1× PBS followed by loading with 400 μL complete DMEM. Four hundred microliters of pseudovirus was flowed through the saturated RAC and collected for future use. To functionalize the RAC column preparation with E. coli lysate, 1× PBS-equilibrated RAC was applied to the column, followed 3 times by equilibration with 2 mL 3× PBS. A 0.5-mL sample of E. coli lysate was gently filtered through the RAC column. Two milliliters of ice-cold 3× PBS with 0.1% (vol/vol) TX-114 was applied through the E. coli lysate-treated RAC column three times at 4°C. After the RAC column was drained, 2 mL ice-cold 3× PBS was added to the RAC column to remove the TX-114. Two milliliters of DMEM was flowed through the RAC column for later endotoxin detection.
Preparation of Nb-CBD-functionalized cellulose paper.
For the capture capability assay, cellulose filter paper discs were fitted into a 96-well plate or 1.5-mL microcentrifuge tube followed by blocking with 1% bovine serum albumin (BSA) in 1× TBS for 1 h. After aspiration of the blocking buffer, 50 to 100 μL purified proteins at concentrations of 100, 10, and 1 μg/mL in 1× TBS or 400 μL protein lysate of interest was applied directly to the coated cellulose paper. After 1 h of incubation at room temperature with slow shaking, purified proteins or lysates were removed from 96-well plates or the 1.5-mL microcentrifuge tube, followed by washing 3 times with 1× TBS, with 10 min between washes. Cellulose paper was blocked with 5% nonfat milk in TBS at room temperature for 15 to 30 min. Fifty to 100 μL medium with spike protein expression was applied to the cellulose paper, and the paper was incubated at room temperature for 1 h. The treated cellulose paper was washed with TBST 4 times for 5 to 10 min each. The washed cellulose paper was incubated with HRP-conjugated donkey anti-human IgG antibody diluted in 1× TBS supplemented with 5% milk with antibody dilution of 1:2,000. After 1 h, the cellulose paper was washed with TBST 4 times for 5 to 10 each. Premixed DAB substrate was directly added onto the cellulose paper after the TBST was removed. The reaction was terminated with distilled water until the color development.
For the preparation of the cellulose filter paper discs to evaluate the filtration efficiency against the pseudovirus, cellulose filter paper discs were fitted into a 96- or 48-well plate followed by blocking with 1% BSA in 1× TBS for 1 h. After aspiration of the blocking buffer, 50 to 100 μL of 10 μg/mL purified fusion proteins in 1× TBS was applied directly to the coated cellulose paper. After 1 h of incubation at room temperature with slow shaking, diluted purified proteins were aspirated from 96- or 48-well plates, followed by washing 2 times with 1× TBS, with 10 min between washes. One hundred to 400 μL of pseudoviruses was added into each well with cellulose paper. After 1 h of incubation, the cellulose paper-treated pseudovirus was gently transferred to transfect HEK293T-hACE2 cells with 80% confluence, followed by addition of Polybrene to a final concentration of 8 μg/mL. Twenty-four hours postransduction, HEK293T-hACE2 cells were split into a ratio of 1:3. Forty-eight hours posttransfection, cells were collected at 300 × g for 3 min, followed by resuspension in 200 μL FACS buffer. Samples were loaded for Attune flow cytometry followed by analysis through FlowJo. After gating of the HEK293T-hACE2 cells without pseudovirus (negative control), with less than 2% GFP positive, the transduction efficiency for the experimental group was calculated as GFP+/(GFP+ + GFP−). For all experiments, triplicate samples were analyzed, data are representative of two or more experiments, and the standard error of the mean (SEM) is shown.
Endotoxin detection.
For endotoxin detection in flowthrough from Nb-CBD-coated cellulose, RAW-Blue cells were seeded in 96-well plates at 3 × 105 cells mL−1 in 100 μL DMEM supplemented with 10% heat-inactivated FBS and 100 U/mL penicillin-streptomycin per well. After 24 h of incubation, the collected DMEM that flowed through extensively washed Nb-CBD lysate-coated cellulose was added to RAW-Blue cells, and the cells were incubated overnight. After incubation, 20 μL of the conditioned cell supernatant was added to 180 μL QUANTI-Blue solution per well of a 96-well plate. The plate was incubated at 37°C until a visible color difference was observed. Interferon (IFN)-secreted embryonic alkaline phosphatase activity was then determined with a spectrophotometer by the absorbance at 620 nm.
Statistical analysis.
Statistical significance was evaluated using a one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test using GraphPad PRISM (San Diego, CA, USA). P values of <0.05 were considered statistically significant. Statistical significance of interest is indicated in all figures according to the following scale: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. All graphs are expressed as means ± SEM.
ACKNOWLEDGMENTS
This work was supported by the Northeastern University COVID19 Rapid Seed Grant (J.L.) and the Peer Reviewed Medical Research Program from the Department of Defense's Congressionally Directed Medical Research Programs (W81XWH-19-1-0041) (J.L.). We extend our appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia (J.L. and A.A.A.-D.), for funding this research work.
We would like to express our gratitude to Sara Rouhanifard at Northeastern University’s Department of Bioengineering for generously sharing her lab’s fluorescent microscope and Ke Zhang at Northeastern University’s Department of Chemistry for sharing his lab’s equipment.
J.L. designed the study. X.S., S.Y., Y.N., M.G., and M.Y. performed the experiments. J.L., X.S., S.Y., A.A.A.-D., S.B., Y.N., and M.Y. analyzed the data. J.L., X.S., and A.A.A.-D. wrote the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Jiahe Li, Email: jiah.li@northeastern.edu.
Robert M. Kelly, North Carolina State University
REFERENCES
- 1.Liu Y-C, Kuo R-L, Shih S-R. 2020. COVID-19: the first documented coronavirus pandemic in history. Biomed J 43:328–333. 10.1016/j.bj.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai S, Ruktanonchai NW, Zhou L, Prosper O, Luo W, Floyd JR, Wesolowski A, Santillana M, Zhang C, Du X, Yu H, Tatem AJ. 2020. Effect of non-pharmaceutical interventions to contain COVID-19 in China. Nature 585:410–413. 10.1038/s41586-020-2293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haghpanah F, Lin G, Levin SA, Klein E. 2021. Analysis of the potential impact of durability, timing, and transmission blocking of COVID-19 vaccine on morbidity and mortality. EClinicalMedicine 35:100863. 10.1016/j.eclinm.2021.100863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krause PR, Fleming TR, Longini IM, Peto R, Briand S, Heymann DL, Beral V, Snape MD, Rees H, Ropero A-M, Balicer RD, Cramer JP, Muñoz-Fontela C, Gruber M, Gaspar R, Singh JA, Subbarao K, Van Kerkhove MD, Swaminathan S, Ryan MJ, Henao-Restrepo A-M. 2021. SARS-CoV-2 variants and vaccines. N Engl J Med 385:179–186. 10.1056/NEJMsr2105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson SW, Oliver JD, Collen J, Bunin J, Gleeson TD, Foster BE, Simmons MP, Chen HW, Ficke JB, Brown TE, Nau MT, Cebula BR, Kielstein J, Chung KK. 2020. Treatment for severe coronavirus disease 2019 with the Seraph-100 Microbind affinity blood filter. Crit Care Explor 2:e0180. 10.1097/CCE.0000000000000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Children’s Hospital Medical Center, Cincinnati. 2021. Registry of Seraph-100 Microbind affinity blood filter for the treatment of COVID-19 under EUA. https://clinicaltrials.gov/ct2/show/NCT04413955.
- 7.Luan J, Lu Y, Jin X, Zhang L. 2020. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun 526:165–169. 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoeman D, Fielding BC. 2019. Coronavirus envelope protein: current knowledge. Virol J 16:69. 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bangaru S, Ozorowski G, Turner HL, Antanasijevic A, Huang D, Wang X, Torres JL, Diedrich JK, Tian J-H, Portnoff AD, Patel N, Massare MJ, Yates JR, Nemazee D, Paulson JC, Glenn G, Smith G, Ward AB. 2020. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science 370:1089–1094. 10.1126/science.abe1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 11.Sternberg A, Naujokat C. 2020. Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination. Life Sci 257:118056. 10.1016/j.lfs.2020.118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassanzadeh-Ghassabeh G, Devoogdt N, De Pauw P, Vincke C, Muyldermans S. 2013. Nanobodies and their potential applications. Nanomedicine (Lond) 8:1013–1026. 10.2217/nnm.13.86. [DOI] [PubMed] [Google Scholar]
- 14.Steeland S, Vandenbroucke RE, Libert C. 2016. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov Today 21:1076–1113. 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Zhao G, He L, Sun S, Qiu H, Tai W, Chen J, Li J, Chen Y, Guo Y, Wang Y, Shang J, Ji K, Fan R, Du E, Jiang S, Li F, Du L, Zhou Y. 2018. A novel nanobody targeting Middle East respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J Virol 92:e00837-18. 10.1128/JVI.00837-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanke L, Vidakovics Perez L, Sheward DJ, Das H, Schulte T, Moliner-Morro A, Corcoran M, Achour A, Karlsson Hedestam GB, Hällberg BM, Murrell B, McInerney GM. 2020. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat Commun 11:4420. 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huo J, Le Bas A, Ruza RR, Duyvesteyn HME, Mikolajek H, Malinauskas T, Tan TK, Rijal P, Dumoux M, Ward PN, Ren J, Zhou D, Harrison PJ, Weckener M, Clare DK, Vogirala VK, Radecke J, Moynié L, Zhao Y, Gilbert-Jaramillo J, Knight ML, Tree JA, Buttigieg KR, Coombes N, Elmore MJ, Carroll MW, Carrique L, Shah PNM, James W, Townsend AR, Stuart DI, Owens RJ, Naismith JH. 2020. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol 27:846–854. 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 18.Koenig P-A, Das H, Liu H, Kümmerer BM, Gohr FN, Jenster L-M, Schiffelers LDJ, Tesfamariam YM, Uchima M, Wuerth JD, Gatterdam K, Ruetalo N, Christensen MH, Fandrey CI, Normann S, Tödtmann JMP, Pritzl S, Hanke L, Boos J, Yuan M, Zhu X, Schmid-Burgk JL, Kato H, Schindler M, Wilson IA, Geyer M, Ludwig KU, Hällberg BM, Wu NC, Schmidt FI. 2021. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science 371:eabe6230. 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoof M, Faust B, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesbølle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Deshpande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Liu Y, Krogan NJ, Ralston CY, et al. 2020. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370:1473–1479. 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Xu K, Jung S, Conte A, Lieberman J, Muecksch F, Lorenzi JCC, Park S, Schmidt F, Wang Z, Huang Y, Luo Y, Nair MS, Wang P, Schulz JE, Tessarollo L, Bylund T, Chuang G-Y, Olia AS, Stephens T, Teng I-T, Tsybovsky Y, Zhou T, Munster V, Ho DD, Hatziioannou T, Bieniasz PD, Nussenzweig MC, Kwong PD, Casellas R. 2021. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature 595:278–282. 10.1038/s41586-021-03676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen PQ, Soenksen LR, Donghia NM, Angenent-Mari NM, de Puig H, Huang A, Lee R, Slomovic S, Galbersanini T, Lansberry G, Sallum HM, Zhao EM, Niemi JB, Collins JJ. 2021. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat Biotechnol 39:1366–1374. 10.1038/s41587-021-00950-3. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Hao Y, Miller EA, Tay DMY, Yee E, Kongsuphol P, Jia H, McBee M, Preiser PR, Sikes HD. 2021. Vertical flow cellulose-based assays for SARS-CoV-2 antibody detection in human serum. ACS Sens 6:1891–1898. 10.1021/acssensors.1c00235. [DOI] [PubMed] [Google Scholar]
- 23.Kim S, Yee E, Miller EA, Hao Y, Tay DMY, Sung K-J, Jia H, Johnson JM, Saeed M, Mace CR, Yüksel Yurt D, Sikes HD. 2021. Developing a SARS-CoV-2 antigen test using engineered affinity proteins. ACS Appl Mater Interfaces 13:38990–39002. 10.1021/acsami.1c08174. [DOI] [PubMed] [Google Scholar]
- 24.Harmsen MM, De Haard HJ. 2007. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol 77:13–22. 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugimoto N, Igarashi K, Samejima M. 2012. Cellulose affinity purification of fusion proteins tagged with fungal family 1 cellulose-binding domain. Protein Expr Purif 82:290–296. 10.1016/j.pep.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Tomme P, Boraston A, McLean B, Kormos J, Creagh AL, Sturch K, Gilkes NR, Haynes CA, Warren RA, Kilburn DG. 1998. Characterization and affinity applications of cellulose-binding domains. J Chromatogr B Biomed Sci Appl 715:283–296. 10.1016/s0378-4347(98)00053-x. [DOI] [PubMed] [Google Scholar]
- 27.Levy I, Shoseyov O. 2002. Cellulose-binding domains: biotechnological applications. Biotechnol Adv 20:191–213. 10.1016/s0734-9750(02)00006-x. [DOI] [PubMed] [Google Scholar]
- 28.Dai G, Hu J, Zhao X, Wang P. 2017. A colorimetric paper sensor for lactate assay using a cellulose-binding recombinant enzyme. Sens Actuators B Chem 238:138–144. 10.1016/j.snb.2016.07.008. [DOI] [Google Scholar]
- 29.Holstein CA, Chevalier A, Bennett S, Anderson CE, Keniston K, Olsen C, Li B, Bales B, Moore DR, Fu E, Baker D, Yager P. 2016. Immobilizing affinity proteins to nitrocellulose: a toolbox for paper-based assay developers. Anal Bioanal Chem 408:1335–1346. 10.1007/s00216-015-9052-0. [DOI] [PubMed] [Google Scholar]
- 30.Hussack G, Luo Y, Veldhuis L, Hall JC, Tanha J, MacKenzie R. 2009. Multivalent anchoring and oriented display of single-domain antibodies on cellulose. Sensors (Basel) 9:5351–5367. 10.3390/s90705351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H-D, Choi S-L, Kim H, Sohn JH, Lee S-G. 2013. Enzyme-linked assay of cellulose-binding domain functions from Cellulomonas fimi on multi-well microtiter plate. Biotechnol Bioproc E 18:575–580. 10.1007/s12257-013-0242-3. [DOI] [Google Scholar]
- 32.Li M, Yue Y, Zhang Z-J, Wang Z-Y, Tan T-W, Fan L-H. 2016. Site-specific and high-loading immobilization of proteins by using cohesin-dockerin and CBM-cellulose interactions. Bioconjug Chem 27:1579–1583. 10.1021/acs.bioconjchem.6b00282. [DOI] [PubMed] [Google Scholar]
- 33.Schang C, Crosbie ND, Nolan M, Poon R, Wang M, Jex A, John N, Baker L, Scales P, Schmidt J, Thorley BR, Hill K, Zamyadi A, Tseng C-W, Henry R, Kolotelo P, Langeveld J, Schilperoort R, Shi B, Einsiedel S, Thomas M, Black J, Wilson S, McCarthy DT. 2021. Passive sampling of SARS-CoV-2 for wastewater surveillance. Environ Sci Technol 55:10432–10441. 10.1021/acs.est.1c01530. [DOI] [PubMed] [Google Scholar]
- 34.Morawska L, Cao J. 2020. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int 139:105730. 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esteban Fernández de Ávila B, Watkins HM, Pingarrón JM, Plaxco KW, Palleschi G, Ricci F. 2013. Determinants of the detection limit and specificity of surface-based biosensors. Anal Chem 85:6593–6597. 10.1021/ac4012123. [DOI] [PubMed] [Google Scholar]
- 36.Miller EA, Baniya S, Osorio D, Al Maalouf YJ, Sikes HD. 2018. Paper-based diagnostics in the antigen-depletion regime: high-density immobilization of rcSso7d-cellulose-binding domain fusion proteins for efficient target capture. Biosens Bioelectron 102:456–463. 10.1016/j.bios.2017.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, Zhang L, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Fan C, Huang W, Xu M, Wang Y. 2020. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect 9:680–686. 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O’Toole Á, Southgate J, Johnson R, Jackson B, Nascimento FF, Rey SM, Nicholls SM, Colquhoun RM, da Silva Filipe A, Shepherd J, Pascall DJ, Shah R, Jesudason N, Li K, Jarrett R, Pacchiarini N, Bull M, Geidelberg L, Siveroni I, Koshy C, Wise E, Cortes N, Lynch J, Kidd S, Mori M, Fairley DJ, Curran T, McKenna JP, Adams H, Fraser C, Golubchik T, Bonsall D, Moore C, Caddy SL, Khokhar FA, Wantoch M, Reynolds N, Warne B, Maksimovic J, Spellman K, McCluggage K, John M, Beer R, Afifi S, Morgan S, et al. 2021. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184:64–75.e11. 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, Rangarajan ES, Pan A, Vanderheiden A, Suthar MS, Li W, Izard T, Rader C, Farzan M, Choe H. 2020. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun 11:6013. 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. 2020. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis 20:411–412. 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 44.Yang M, Zhu G, Korza G, Sun X, Setlow P, Li J. 2020. Engineering Bacillus subtilis as a versatile and stable platform for production of nanobodies. Appl Environ Microbiol 86:e02938-19. 10.1128/AEM.02938-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang Y, Nambulli S, Xiao Z, Liu H, Sang Z, Duprex WP, Schneidman-Duhovny D, Zhang C, Shi Y. 2020. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science 370:1479–1484. 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD. 2020. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12:513. 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun X, Ni Y, He Y, Yang M, Tani T, Kitajima S, Barbie DA, Li J. 2021. Engineering the immune adaptor protein STING as a functional carrier. Adv Therap 4:2100066. 10.1002/adtp.202100066. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download aem.02303-21-s0001.pdf, PDF file, 0.9 MB (926KB, pdf)