ABSTRACT
RNA polymerase (RNAP) is a highly conserved macromolecular machine that contributes to the flow of genetic information from genotype to phenotype. In Bacillus subtilis, mutations in the rpoB gene encoding the β-subunit of RNAP have been shown to alter a number of global phenotypes, including growth, utilization of unusual nutrient sources, sporulation, germination, and production of secondary metabolites. In addition, the spectrum of mutations in rpoB leading to rifampin resistance (Rifr) can change dramatically depending upon the environment to which B. subtilis cells or spores are exposed. Rifr rpoB mutations have historically been associated with slower growth and reduced fitness; however, these assessments of fitness were conducted on limited collections of mutants in rich laboratory media that poorly reflect natural environments typically inhabited by B. subtilis. Using a novel deep-sequencing approach in addition to traditional measurements of growth rate, lag time, and pairwise competitions, we demonstrated that the competitive advantages of specific rpoB alleles differ depending on the growth environment in which they are determined.
IMPORTANCE Microbial resistance to antibiotics is a growing threat to public health across the world. Historically, resistance to antibiotics has been associated with reduced fitness. A growing body of evidence indicates that resistance to rifampin, a frontline antibiotic used to treat mycobacterial and biofilm-associated infections, may increase fitness given an appropriate environment even in the absence of the selective antibiotic. Here, we experimentally confirm this phenomenon by directly comparing the fitness of multiple rifampin-resistant mutants of Bacillus subtilis in rich LB medium and an asparagine minimal medium. Our research demonstrates that the fitness cost of rifampin resistance can vary greatly depending upon the environment. This has important implications for understanding how microbes develop antimicrobial resistance in the absence of antibiotic selection.
KEYWORDS: Bacillus subtilis, evolution, RNA polymerase, fitness, rifampin, rifampin resistance, transcription
INTRODUCTION
Transcription in bacteria depends on a complex of proteins collectively known as RNA polymerase (RNAP), and bacterial responses to their environment are in large part due to alterations in their global pattern of transcription, i.e., the transcriptome. Because RNAP physically contacts every promoter in the bacterial genome, mutations altering RNAP can exert wide-ranging pleiotropic effects on the transcriptome, and hence, the resulting phenotype and fitness of a cell growing in a particular environment. The antibiotic rifampin (Rif) is a potent inhibitor of prokaryotic transcription initiation (1) and has a long history of use in the treatment of infectious diseases, particularly tuberculosis and leprosy caused by Mycobacterium spp., as well as biofilm-associated infections (2). In addition to its clinical importance, Rif has long been used to probe the structure and function of RNAP. Resistance to Rif (Rifr) arises from mutations in the rpoB gene, encoding the β subunit of RNAP (3). Binding of Rif to RNAP occurs at a specific site on the β subunit, the Rif-binding pocket, which physically blocks the progression of nascent transcripts through the RNA exit channel (4, 5). Mutations conferring Rifr are localized on the primary sequence of rpoB in four regions designated the N cluster and clusters I, II, and III (4, 6). The majority of Rifr mutations occur in cluster I, and elucidation of the three-dimensional structure of RNAP bound to Rif has revealed that the Rif binding pocket consists of several key amino acids within cluster I that make direct contact with Rif (4). In the Gram-positive model organism Bacillus subtilis, Rifr mutations in rpoB have been shown to affect a number of global phenotypes, including deficiencies in sporulation, competence, growth rate, and yield (7), activation of cryptic genes involved in the utilization of rare carbon sources (8), altered resistance of spores to various sporicidal stresses (9), temperature-sensitive sporulation (10), and spore germination (7).
Not all sites in cluster I of rpoB are prone to mutation at equal frequency; some bases mutate at higher frequency (“hot spots”) or lower frequency (“cold spots”). Such patterns of mutation in a gene are referred to as its mutational spectrum (11). In prior communications, we have reported several instances in which the Rifr mutational spectrum in the B. subtilis rpoB gene is changed when cells are cultivated in different physiological environments or when spores are exposed to different environmental extremes (12–17). Most recently, we reported on differences in the spectrum of Rifr mutations between populations grown in complete Miller LB medium (LB) and populations grown in Spizizen minimal medium containing l-asparagine as the sole carbon source (SMMAsn) (18). We observed that the major Rifr amino acid change in LB cultures was H482Y (a change of H to Y at position 482), while the major mutation in SMMAsn cultures was S487L (18). How does exposure of a cell or spore to a particular environment lead to an altered mutational spectrum? At least two explanations for this phenomenon can be posited, which are not mutually exclusive.
(i) One of the explanations is that under different physiological conditions, changes in the intracellular environment can cause alterations in nucleoid architecture, DNA supercoiling, and DNA conformation; such structural alterations can alter the probability of mutation at a particular base within a gene. This notion is supported by the observed differences both in DNA structure and in the spectrum of rpoB mutations between vegetative cells and spores in B. subtilis, where the mutagenic spectrum of spores more closely resembled the spectrum observed in clinical Mycobacterium tuberculosis isolates (12). Differences in the spectrum of rpoB mutations have also been observed in B. subtilis spores exposed to a number of extreme environmental conditions, including simulated Mars conditions (13), ultrahigh vacuum (19), and exposure to space in low-Earth orbit (15). It should be noted that in the examples listed above, dormant spores themselves were exposed to different environments and then allowed to germinate and grow under the same laboratory conditions; hence, differences in mutational spectrum cannot be attributed to different growth environments.
(ii) The second explanation is that growth under selective pressure produces a spectrum of mutations within a population and those mutants with the lowest fitness cost or those with a fitness advantage become more prevalent. Supporting this line of thought, Wrande et al. (20) illustrated that the logarithmic accumulation of Rifr mutations in aging Salmonella enterica and Escherichia coli colonies is due to clonal growth of Rifr mutants in localized “jackpots” within the colony. These Rifr alleles were found to have a selective advantage over their respective ancestral strains within the context of these aging colonies. Similarly, study of Rifr Staphylococcus aureus mutants isolated from biofilms revealed that Rifr mutants exhibited greater fitness when cocultured with the wild-type strain in a biofilm liquid culture (21).
It is possible that these two mechanisms work together in determining the spectra of mutations in both vegetative cells and spores, due to the integral roles that DNA bending and supercoiling play in regulating bacterial transcription and, in turn, the role that transcription plays in determining DNA architecture (reviewed in references 22, 23, and 24). This inextricable linkage between gene expression and DNA supercoiling makes these notions explaining variations in spectra of mutations difficult to disentangle.
To determine whether growth under differing selective pressures is a contributing factor in differences in the spectra of mutations, we report here the development of a competition assay utilizing next-generation-sequencing technology to quantify the fitness of multiple rpoB alleles in a mixed culture simultaneously. We compared the population frequencies of 18 rpoB alleles in mixed cultures grown in two distinct environments: LB and SMMAsn. We found significant differences in the fitness of the various Rifr mutant rpoB alleles in each medium. In LB, the wild-type allele dominated, whereas in SMMAsn, the H482N mutant became the dominant allele present in the population. These results were subsequently confirmed by pairwise competition experiments, as well as assessments of exponential growth rates and lag times.
RESULTS AND DISCUSSION
Levels of Rif resistance conferred by mutant rpoB alleles.
The natural environment often contains inhibitory or subinhibitory levels of various antibiotics; thus, natural antibiotic resistance would confer a selective advantage in such environments (reviewed in reference 25). Through our studies of Rifr over the years, we have amassed a collection of congenic B. subtilis strains, each carrying 1 of 17 different Rifr mutations in rpoB (Table 1). We measured the levels of Rifr in these strains by measuring the 50% inhibitory concentration (IC50) for Rif in LB medium by the broth dilution method (Fig. 1). As expected, the wild-type laboratory strain was very sensitive to Rif (IC50 of 0.06 ± 0.01 μg/mL). All mutant rpoB alleles that had been selected on a high concentration of Rif (50 μg/mL) (12, 13) exhibited high IC50 values of >100 μg/mL, whereas mutant rpoB alleles selected on a low concentration of Rif (5 μg/mL) (17) exhibited generally lower IC50 values, ranging from 4 to 84 μg/mL, the lowest of which was still at least 67-fold higher than that of the wild-type strain (Fig. 1). Thus, in the context of growth in a selective environment containing Rif, all the Rifr rpoB mutants would be expected to be more fit than the wild-type strain.
TABLE 1.
B. subtilis strains and plasmids used in this study
| Strain or plasmid | Genotype, phenotype, or description | Reference and/or sourcea |
|---|---|---|
| Strain | ||
| WN547 | trpC2 pheA1 cat 6His-rpoC; Cmr | 61 |
| WN624 | trpC2 amyE::spc; Spcr | 53 |
| WN758 | trpC2 pheA1 rpoB-Q469R cat 6His-rpoC; Rifr Cmr | 7 |
| WN759 | trpC2 pheA1 rpoB-H482R cat 6His-rpoC; Rifr Cmr | 7 |
| WN760 | trpC2 pheA1 rpoB-H482Y cat 6His-rpoC; Rifr Cmr | 7 |
| WN761 | trpC2 pheA1 rpoB-S487L cat 6His-rpoC; Rifr Cmr | 7 |
| WN999 | trpC2 pheA1 rpoB-S487F cat 6His-rpoC; Rifr Cmr | 13 |
| WN1000 | trpC2 pheA1 rpoB-S487Y cat 6His-rpoC; Rifr Cmr | 13 |
| WN1002 | trpC2 pheA1 rpoB-H482D cat 6His-rpoC; Rifr Cmr | 13 |
| WN1004 | trpC2 pheA1 rpoB-V135F cat 6His-rpoC; Rifr Cmr | 13 |
| WN1007 | trpC2 pheA1 rpoB-Q469K cat 6His-rpoC; Rifr Cmr | 13 |
| WN1009 | trpC2 pheA1 rpoB-H482P cat 6His-rpoC; Rifr Cmr | 13 |
| WN1011 | trpC2 pheA1 rpoB-Q469L cat 6His-rpoC; Rifr Cmr | 13 |
| WN1191 | trpC2 pheA1 rpoB-A478D cat 6His-rpoC; Rifr Cmr | This study |
| WN1261 | trpC2 amyE::neo; Neor | 62 |
| WN1624 | trpC2 pheA1 rpoB-D472V cat 6His-rpoC; Rifr Cmr | This study |
| WN1625 | trpC2 pheA1 rpoB-D472Y cat 6His-rpoC; Rifr Cmr | This study |
| WN1626 | trpC2 pheA1 rpoB-Δ(E479-H482) cat 6His-rpoC; Rifr Cmr | This study |
| WN1637 | trpC2 pheA1 rpoB-H482N cat 6His-rpoC; Rifr Cmr | This study |
| WN1647 | trpC2 pheA1 rpoB-A478V cat 6His-rpoC; Rifr Cmr | This study |
| WN1651 | trpC2 pheA1 amyE::spc cat 6His-rpoC; Cmr Spcr | This study |
| WN1653 | trpC2 pheA1 rpoB-A478D,I528V cat 6His-rpoC; Rifr Cmr; reconstructed | This study |
| WN1658 | trpC2 rpoB-A478D amyE::neo; Neor Rifr | This study |
| WN1662 | trpC2 rpoB-A478D,I528V amyE::neo; Neor Rifr | This study |
| WN1663 | Isolate from LB culture no. 3 evolved from WN1191; rpoB-A478D,I528V; Rifr | This study |
| Plasmids | ||
| pDG1730 | amyE spectinomycin integration vector | BGSC; 63 |
| pJOE8999 | CRISPR-Cas9 system | BGSC; 51 |
| pWN1630 | pJOE8999 + sgRpoB + 1409b | This study |
| pWN1633 | pWN1630 + rpoB-H482N homology template | This study |
| pWN1641 | pWN1630 + WT rpoB homology template | This study |
| pWN1645 | pWN1630 + rpoB-A478V homology template | This study |
BGSC, Bacillus Genetic Stock Center.
sgRpoB, single guide RNA for RpoB.
FIG 1.
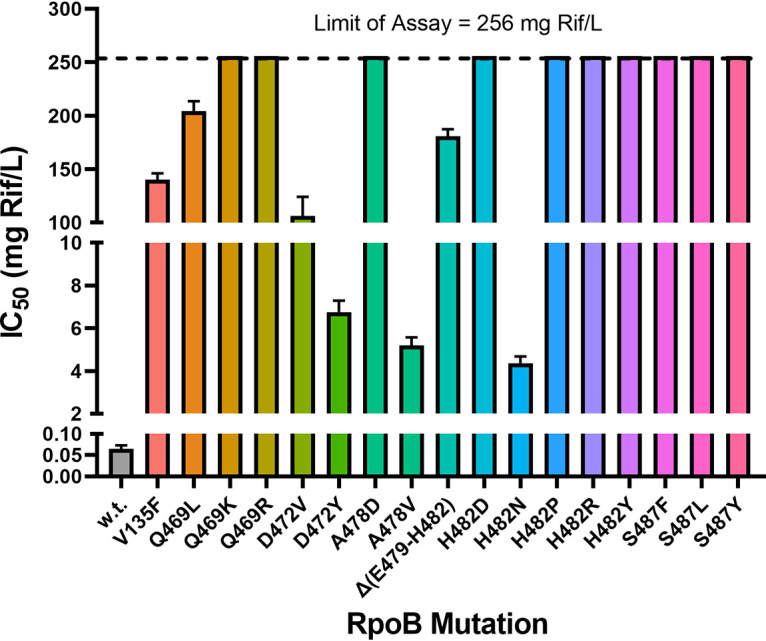
Determination of IC50 values of Rif in B. subtilis strains used in this study. Results are presented as averages ± standard deviations (n = 3). w.t., wild type.
Growth rates of rpoB mutants in LB cultures.
In the absence of Rif selection, it has been noted previously that mutations in rpoB leading to Rifr often exert a fitness cost, such as a lower growth rate as measured in standard laboratory media (26; reviewed in reference 27). To test this notion, we measured the growth rates of wild-type B. subtilis and 17 congenic rpoB mutants in individual liquid-LB shake flask cultures (Fig. 2). It was observed that 10 of the 17 rpoB mutant strains grew significantly more slowly than the wild type (Fig. 2), consistent with the notion that rpoB mutations can exert a fitness cost (26). However, 8 of the 17 rpoB mutants grew at a rate statistically indistinguishable from that of the wild-type strain (Fig. 2). The results indicate that not all Rifr mutants exhibit lowered fitness in LB under standard laboratory conditions, as measured by growth rate.
FIG 2.
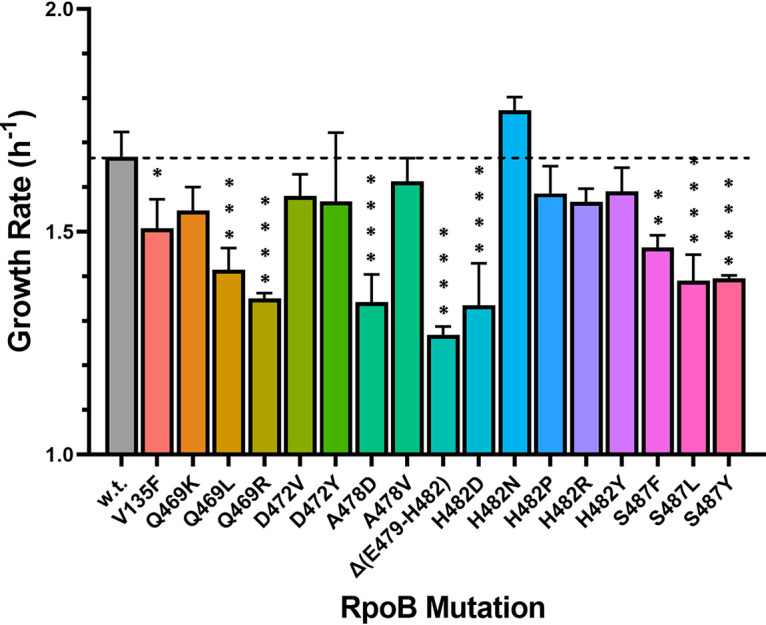
Growth rates of B. subtilis strains cultivated in liquid LB in shake flasks with aeration. Strains carrying the indicated rpoB mutations were grown as described in Materials and Methods. Results are presented as averages ± standard deviations (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (ordinary one-way ANOVA with Dunnett’s test for multiple comparison to the mean value for the wild-type strain).
Relative fitness of rpoB alleles in LB medium.
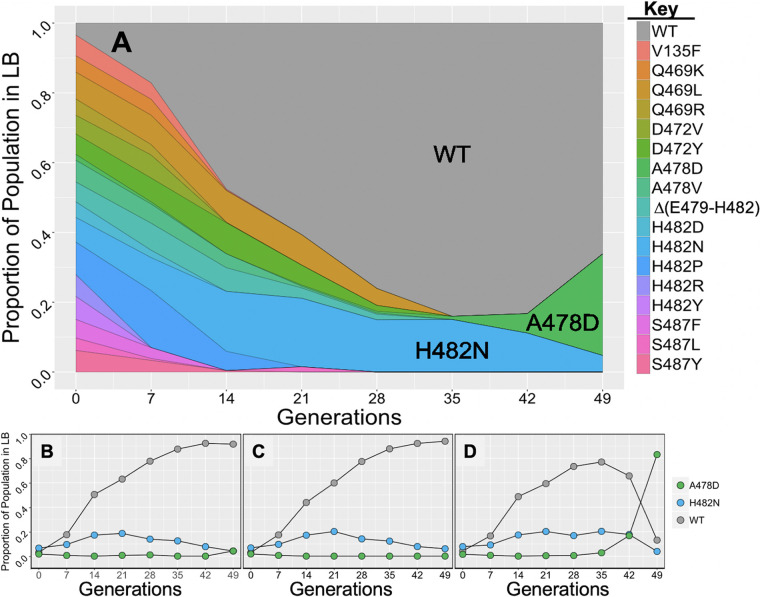
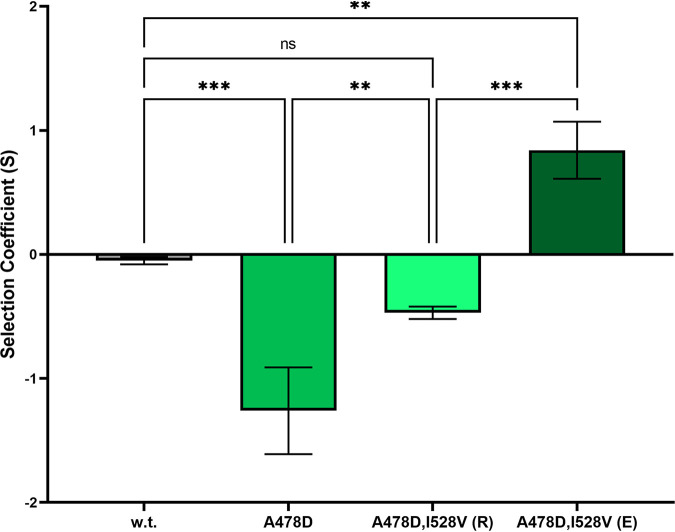
The growth rate experiments described above were performed on individual strains in monoculture, which may not reflect their true fitness in a complex competitive environment such as would likely prevail in nature. Therefore, to directly compare the fitness of rpoB alleles, we performed, in triplicate cultures, a multigenerational and multistrain competition experiment in which the wild-type strain and all 17 Rifr rpoB mutant strains were competed simultaneously in LB (Fig. 3). At daily (∼7-generation) intervals, the proportion of each rpoB allele in the mixed population was determined by next-generation sequencing of two ∼500-bp PCR amplicons spanning either the N cluster or clusters I, II, and III of the rpoB gene (Fig. 3A). At generation zero of the experiment, each rpoB allele was present at ∼5 to 6% of the total population. By generation 35, the wild-type strain came to dominate all three cultures, and the proportion of most Rifr mutant alleles had dropped below the level of detection (∼0.1%) (Fig. 3A). Two Rifr rpoB mutants persisted until the end of the experiment, the H482N and A478D variants. Although the growth rate of the H482N mutant was not significantly higher than that of the wild-type strain when grown separately in monoculture (Fig. 2), the mutant appeared to be less fit in a competitive environment (Fig. 3A). Interestingly, the A478D mutant appeared to almost disappear from the population by 14 generations, but beginning at ∼35 generations, it increased again in frequency to an average of ∼25% of the population by generation 49 (Fig. 3A). This anomalous result prompted us to more closely examine the allelic frequencies of the wild-type, H482N, and A478D strains in each of the three replicate mixed cultures. We observed that in two of the three replicate cultures, the A478D allele behaved like the majority of the Rifr rpoB mutations, becoming reduced to a very low, barely detectable frequency (Fig. 3B and C). In contrast, in the third culture, the A478D mutant initially dropped below the limit of detection (∼0.1%), but then starting at ∼generation 35, it increased dramatically in frequency to comprise over 80% of that population by generation 49 (Fig. 3D). Closer examination of the PCR amplicon spanning clusters I, II, and III from the A478D mutant arising after generation 35 in the third culture revealed the appearance of a second mutation in rpoB, I528V. Furthermore, Sanger sequencing of the rpoB gene of a Rifr isolate from this culture, strain WN1663, confirmed the presence of the I528V mutation in addition to the original A478D mutation. These observations led us to suspect that during the competition experiment in the third culture, the original A478D strain had suffered a second-site mutation in rpoB, dramatically increasing its competitive fitness. To determine if this compensatory mutation was responsible for the observed increase in relative fitness, both the ancestral (A478D) and evolved (A478D+I528V) alleles were transferred into a clean wild-type background to create strains WN1658 and WN1662, respectively. To test their competitive fitness, the four congenic strains WN1621 (wild type), WN1658 (A478D), WN1663 (evolved A478+I528V, isolated from the generation 49 culture of the experiment whose results are shown in Fig. 3D), and WN1662 (A478D+I528V, reconstructed in a wild-type background) were each competed separately against the same isogenic wild-type strain (strain WN624, carrying a spectinomycin resistance [Spcr] marker) in LB medium (Fig. 4). Pairwise competition experiments showed that strain WN1658, the A478D rpoB mutant, was dramatically less fit than the wild type (selection coefficient [S] = −1.26 ± 0.35), whereas the evolved isolate strain WN1663 (A478D+I528V) was dramatically more fit than the wild type (S = +0.84 ± 0.23) (Fig. 4). However, simple reconstruction of the A478D+I528V rpoB double mutation in an otherwise wild-type background (strain WN1662) resulted in only a modest increase in fitness (S = −0.47 ± 0.05) compared to that seen in the single A478D mutant and failed to replicate the dramatic increase in fitness seen in the evolved strain WN1663 (Fig. 4). We therefore conclude that while the second-site mutation I528V in rpoB increased competitive fitness to a certain extent, it was not sufficient to account for the dramatic increase in fitness seen in strain 1663 arising in the third multiallelic competition culture in the experiment whose results are shown in Fig. 3D. The results lead us to hypothesize that strain WN1663 had sustained genomic mutation(s) in addition to I528V that also contributed to its dramatically increased fitness. These putative additional mutations could be revealed by whole-genome sequencing, which is beyond the scope of the present study. Results from the experiment whose results are shown in Fig. 3 indicated that, when challenged in a more complex competitive environment (18 rpoB variants growing together for 49 generation in LB medium in the absence of Rif selection), all of the Rifr mutations in rpoB led to a generalized reduction in fitness. This result is somewhat at odds with the conclusions drawn from measurements of growth rates of the same strains in monoculture (Fig. 2), indicating that fitness cannot be predicted by growth rate in monoculture alone.
FIG 3.
(A) Population dynamics of wild-type (WT) and 17 rpoB mutant strains of B. subtilis in LB liquid medium grown with aeration at 37°C. Data shown are average values (n = 3). (B to D) Population dynamics of wild-type (WT), A478D, and H482N strains during the three replicate competition experiments from which the average values in panel A were derived.
FIG 4.
Results from pairwise competition experiments in LB medium. The indicated Rifr rpoB strains were competed against the appropriate congenic wild-type strain. The strains used were WN1261 (w.t.), WN1658 (A478D), WN1663 [A478D+I528V, evolved from the culture used in the experiment whose results are shown in Fig. 3D; displayed in the figure as A478D,I528V(E)], and WN1662 [A478D+I528V, reconstructed in a wild-type strain by transformation; displayed in the figure as A478D,I528V(R)]. The selection coefficients (S) (54) shown are the average values ± standard deviations from 3 independent experiments. **, P < 0.01; ***, P < 0.001 (ordinary one-way ANOVA with Tukey’s HSD test for multiple comparisons).
Relative fitness of rpoB alleles in SMMAsn medium.
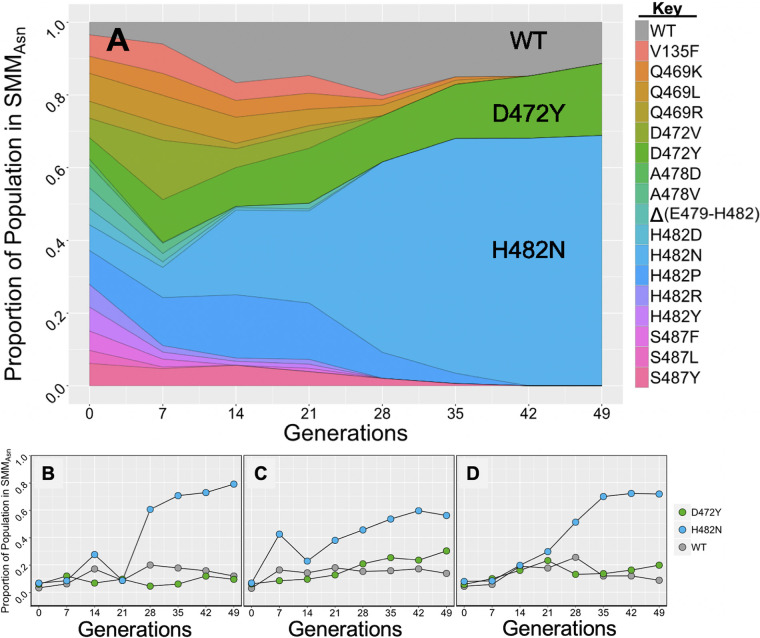
The multiallele competition experiment whose results are shown in Fig. 3 was performed in nutrient-rich LB medium. However, in nature, B. subtilis is found in soils or associated with plants and their roots. Such natural environments are either subjected to “feast or famine” conditions or are chronically oligotrophic; therefore, cells would need to utilize limited quantities of rarer, more ephemeral nutrient sources. In previous work, we performed OmniLog global metabolic profiling on wild-type B. subtilis and 11 Rifr rpoB mutants to assess how mutations in rpoB could affect metabolic capabilities (8). Indeed, we found that several environmental nutrients, normally utilized poorly by the wild-type laboratory strain of B. subtilis, were used significantly better by certain Rifr rpoB mutants. In particular, we noted that a strain carrying the S487L rpoB substitution utilized the amino acids l-alanine, l-asparagine, l-glutamate, and l-serine significantly better than the wild type (8). l-Asparagine specifically has been found to be substantially present in root exudates and plays a significant role in determining the bacterial species composition of the rhizosphere (28). Based on these observations, we were interested in measuring the relative fitness of wild-type and 17 Rifr rpoB mutant strains of B. subtilis in a more stringent selective environment. We therefore modified Spizizen minimal medium by replacing glucose with l-asparagine as the sole carbon source (SMMAsn; see Materials and Methods for details) and again performed a multiallele, multigenerational competition experiment (Fig. 5). Based on its performance in OmniLog metabolic assays (8), we reasoned that the S487L mutant would fare better in this more stringent environment than the wild type or other Rifr mutant strains.
FIG 5.
(A) Population dynamics of wild-type (WT) and 17 rpoB mutant strains of B. subtilis in SMMAsn liquid medium grown with aeration at 37°C. Data shown are average values (n = 3). (B to D) Population dynamics of wild-type (WT), D472Y, and H482N strains during the three replicate competition experiments from which the average values in panel A were derived.
In the multiallele competition experiment conducted in SMMAsn, each of the 18 strains was initially present at 5 to 6% of the total population (Fig. 5A). As seen earlier in the multiallele competition performed in LB medium (Fig. 3), most of the strains carrying various rpoB mutations dropped out of the population during the course of propagation in SMMAsn (Fig. 5A). Interestingly, in this experiment, we observed that the strain carrying the H482N rpoB mutation rose to dominate the population, gaining an average frequency of ∼70% by generation 49, followed by the strain carrying the D472Y mutation (∼20%) and then the wild-type strain (∼10%) (Fig. 5A). These results were in stark contrast to those obtained in LB medium, where the wild-type strain dominated the population, with the H482N mutant forming a minority (Fig. 3). Interestingly, the strain carrying the S487L mutation, which we predicted would become the most frequent allele, did not succeed in this medium as we had expected but, rather, dropped below the threshold of detection in the population by generation 14 of the competition (Fig. 5A). Comparison of the results presented in Fig. 3 with the results in Fig. 5 demonstrated a dramatic difference in the population dynamics and strain fitness of the same rpoB mutants depending upon whether they were competed in the rich LB or the minimal SMMAsn environment. In addition, the lack of increased competitive fitness displayed by the S487L rpoB mutant competed in SMMAsn (Fig. 5A) brought into question whether an increased utilization of a nutrient, such as l-asparagine, as measured by Omnilog assay (8), is a relevant parameter to predict fitness.
During multiallele competition in LB medium, we noted that the A478D rpoB mutant rose to prominence in one of the three replicate cultures (Fig. 3D), presumably due to a second-site mutation(s). Accordingly, we also examined the frequencies of the wild-type, D472Y, and H482N strains in each individual SMMAsn culture (Fig. 5B to D). In this case, all three cultures behaved similarly, with the H482N strain arising to dominate each of the three cultures. This observation suggested that no second-site mutation(s) improving fitness had arisen in the SMMAsn cultures during the course of the multiallele competition experiment.
Growth rates and lag times of rpoB mutations in SMMAsn cultures.
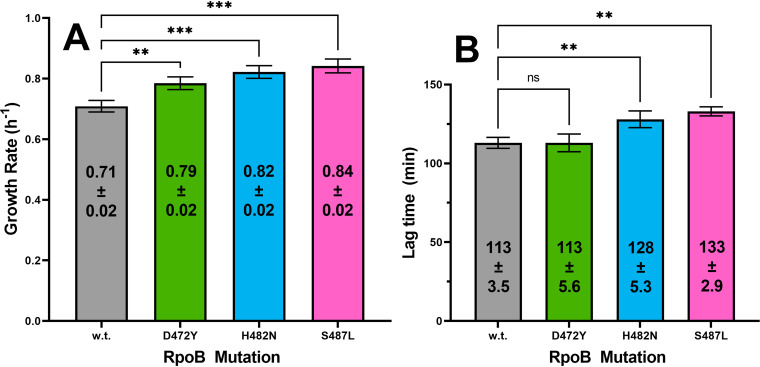
We sought to further explore the role played by growth rate in the fitness of rpoB mutants by measuring the exponential growth rates during cultivation in SMMAsn of the wild-type B. subtilis strain and congenic mutant strains harboring the D472Y, H482N, or S487L rpoB allele (Fig. 6). In individual SMMAsn monocultures, all three Rifr mutant strains exhibited a significantly increased exponential growth rate compared to the growth rate of the wild-type strain, but there were no statistical differences among the growth rates of the Rifr mutants themselves (Fig. 6A). These results were distinctly different from the relative growth rates of the same strains in LB medium, in which the rpoB mutants either grew more slowly (S487L) or at essentially the same rate (D472Y and H482N) as the wild type (Fig. 2). If the exponential growth rate were the sole determinant of fitness, it would be expected that the three Rifr mutants would have risen to dominate the multiallele competition experiment performed in SMMAsn, but this was clearly not the case (Fig. 5).
FIG 6.
Exponential growth rates (A) and lag times (B) in SMMAsn of the wild-type (w.t.) strain and congenic rpoB mutant strains carrying the D472Y, H482N, and S487L variants. Data are averages ± standard deviations (n = 3). **, P < 0.01; ***, P < 0.001 (ordinary one-way ANOVA with Dunnett’s test for multiple comparison to the mean value for the wild type).
In addition to the exponential growth rate, the ability of cells to adjust to a new environment is reflected in the lag time preceding exponential growth. In one of the few studies examining this phenomenon in rpoB mutants, it was observed that a Mycobacterium tuberculosis Rifr mutant strain carrying an H526D substitution (the equivalent of the H482D substitution in B. subtilis rpoB) suffered significantly delayed growth following resuscitation from nutrient starvation (29). We thus compared the lag time of the wild-type strain to the lag times of congenic strains carrying the D472Y, H482N, and S487L rpoB variants in SMMAsn. No significant difference was observed in the lag times between the wild type and the D472Y variant; however, both the H482N and S487L variants demonstrated significant increases in lag time relative to that of the wild type (Fig. 6B). In fact, the H482N and S487L variants were statistically indistinguishable in their growth rates (increased over that of the wild type) and lag times (longer than that of the wild type). Taken together, the results shown in Fig. 6 indicated that in SMMAsn medium, the growth rate and/or lag time of individual monocultures corresponded poorly with increased or decreased relative fitness in competition experiments.
Pairwise competitions in LB versus SMMAsn.
To further examine the discrepancies between the observed growth metrics and the results of our multiallele competition, we performed a pairwise competition for each Rifr mutant strain against the wild-type strain in both LB and SMMAsn media (Fig. 7). The H482N Rifr mutant strain showed the most dramatic fitness difference compared to the fitness of the wild type; it was less fit in LB (S = −0.047 ± 0.01) but more fit (S = +0.403 ± 0.063) in SMMAsn medium (Fig. 7). The S487L mutant was less fit than the wild type in both media, although its fitness in SMMAsn (S = −0.144 ± 0.005) was significantly improved over that in LB (S = −0.337 ± 0.01) (Fig. 7). The D472Y mutant was also less fit than the wild type in both media, but the difference in its fitness between the two media was just above the cutoff for significance (P = 0.061). Measurements of the relative fitness for each mutant rpoB allele in pairwise competition agreed with the results of the multiallelic competitions in SMMAsn.
FIG 7.
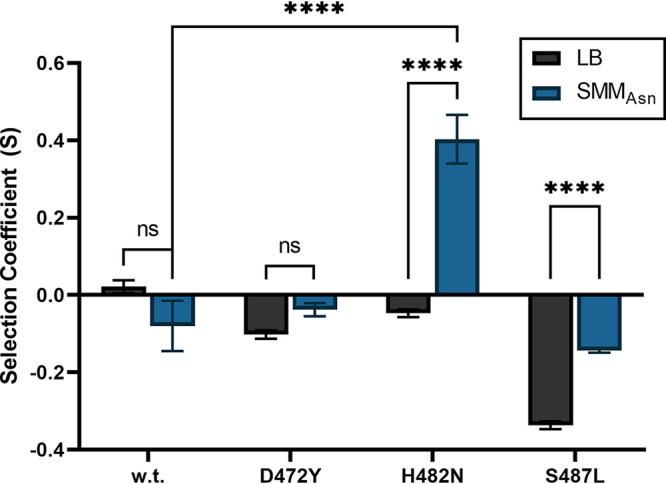
Pairwise competition experiments. Rifr strains carrying the variant D472Y (strain WN1625), H482N (strain WN1637), or S487L (strain WN761) were each competed against congenic wild-type strain WN1651 in LB or SMMAsn medium for 21 generations, and fitness coefficients (S) were calculated as described previously (54). Data are average values ± standard deviations (n = 3). ***, P < 0.001; ****, P < 0.0001; ns, not significantly different (two-way ANOVA with Tukey’s HSD test for multiple comparisons).
Previous work in our laboratory has shown differing spectra of mutations in the rpoB gene across several different environment (12, 13, 16–18). The reproducibility of these changes in the spectra of mutations indicates that the environment somehow dictates these changes. Multiple studies have found that mutations in the rpoB gene that confer rifampin resistance can provide a fitness advantage or increased growth rate in the environment from which they were isolated. Several examples illustrate the repeated occurrence of rpoB mutations in adaptive laboratory evolution of Escherichia coli, wherein these mutations significantly increased the fitness of the organism in different environments, including optimized growth on various minimal media and increased fitness at elevated temperatures (30–32).
Our multiallele, multigenerational competition experiments (Fig. 3 and 5) showed that the wild-type rpoB allele was most fit in nutrient-rich LB medium, while the H482N variant was most fit in populations cultivated in minimal SMMAsn medium. The different rpoB alleles and the corresponding changes in global gene expression associated with them likely explain the differences in relative fitness depending upon the growth environment. Numerous studies suggest changes in global gene expression to be the cause of changes in fitness observed with mutations in rpoB (7, 33–37), although one study examining the system-wide impacts of mutations in the Pseudomonas aeruginosa rpoB gene found few differences in global gene expression and attributed mutant phenotypes to increased expression of RNAP (38). Ultimately, a better understanding of the specific mechanisms behind these differences in fitness will require an examination of global gene expression of these different rpoB alleles in SMMAsn.
Which changes in gene expression are specifically responsible for the success of the H482N mutant can only be speculated upon at present, as this particular allele is new to our collection and not yet very well characterized. However, the lack of fitness of the S487L mutant in SMMAsn may be a bit easier to explain. Previous work studying the rpoB-S487L allele in B. subtilis revealed that this particular allele autoinduced the expression of genes responsible for the production of the antibiotic 3,3′-neotrehalosadiamine (NTD), which is not produced by the wild-type strain under the same conditions (39). This autoinduction of NTD production might place a significant metabolic burden on the organism and may be responsible for the lack of relative fitness observed in the rpoB-S487L allele.
The discrepancies between fitness and exponential growth rate reveal that, while fairly simple to determine, growth rate is a poor proxy for relative fitness, as a similar study has found (40). Simply measuring the exponential growth rate fails to account for differences in fitness during stationary and lag phase, in addition to potential antagonistic or cooperative interactions between the strains when competing for the same resources, such as dramatic acidification of the medium (41). Our study highlights the danger of conflating exponential growth rates or results from simplified metabolic assays (8) with relative fitness. Three Rifr rpoB variants exhibited an increased growth rate relative to the growth rate of the wild-type strain in SMMAsn (Fig. 6); however, only one of those variants (H482N) proved to be more fit than the wild-type strain in the same medium (Fig. 7). There are many potential explanations for this discrepancy that could be controlled at the transcriptional level, such as increased probability of differentiation in the stationary phase (42) or shaping of the environment by an evolved variant in a manner that preferentially benefits itself (41, 43).
Unlike other results from this study, the increased relative fitness of the evolved strain WN1663 cannot be explained solely by a compensatory mutation in rpoB (Fig. 4). Our data indicate that other changes within the B. subtilis genome must account for the observed sweep of the population by WN1663. The increase in fitness of the A478D+I528V double mutant relative to the fitness of the A478D single mutant is consistent with other studies that have reported the ability of compensatory mutations in rpoB and rpoC to restore fitness defects (44–46). However, these studies did not find increases in fitness relative to that of the wild-type allele. In summary, this study illustrates the fitness impacts of rpoB alleles in different environments, highlights the deficiencies of growth or metabolic metrics of monocultures as a sole measure of relative fitness, and demonstrates the role of compensatory mutations in rpoB to recover fitness cost.
MATERIALS AND METHODS
Bacterial strains, media, plasmids, and growth conditions.
All B. subtilis strains and plasmids used in this study are listed in Table 1. The strains were constructed by DNA-mediated transformation of competent cells with purified genomic DNA as previously described (47). The complex media used were Miller LB medium (48) and Trypticase-soy-yeast extract (TSY) medium (49). The minimal medium used was Spizizen minimal medium (50) containing the auxotrophic requirements tryptophan and phenylalanine (50 μg/mL each) and asparagine (95 mM) in place of glucose as the carbon source (SMMAsn). Media were solidified with agar (15 g/L) as needed. The antibiotics used (final concentration) were chloramphenicol (10 μg/mL), rifampin (5 μg/mL), kanamycin (5 μg/mL), neomycin (5 μg/mL), or spectinomycin (100 μg/mL). Plasmids for CRISPR-Cas9 introduction of point mutations were constructed and used as previously described (51). Site-directed mutagenesis was conducted via inverse PCR as previously described (52) to introduce the point mutation A478V into an rpoB template. All mutations were confirmed by Sanger sequencing (Genewiz LLC) of PCR amplicons.
Rif IC50 determination.
For quantification of Rifr, an overnight culture of each strain was grown in liquid TSY without antibiotics. One microliter of overnight culture was inoculated into 100 μl of liquid TSY medium without Rif or containing Rif at a 2-fold range of concentrations from 4 to 256 μg/mL. Cells were grown in 96-well microtiter plates sealed with gas-permeable membranes (Breathe-Easy; Sigma-Aldrich). Growth at 37°C was measured by the optical density at 620 nm (OD620) at 15-minute intervals in a temperature-controlled microplate absorbance spectrophotometer (model ELx808; BioTek). The resulting growth data from triplicate experiments were then used to determine the 50% inhibitory concentration (IC50).
Growth rate and lag time determinations.
Liquid cultures were propagated in 125-mL sidearm (Klett) flasks in 10 mL of LB at 37°C in a rotary shaking bath with vigorous aeration. Growth was measured at 15-minute intervals by the OD660 using a Klett-Summerson colorimeter fitted with the no. 66 (red, 660-nm) filter. Growth rates were calculated from the exponential portion of each growth curve. Lag times were determined from the point of intersection of the lag and exponential portions of each growth curve. Growth rates and lag times were determined from triplicate cultures. Data sets were tested for normality using the Shapiro-Wilk test online calculator (https://www.statskingdom.com/shapiro-wilk-test-calculator.html), and all data sets were found to be normally distributed. Statistical parameters and tests of significance (analysis of variance [ANOVA]) were computed using KaleidaGraph version 4.5.2 (Synergy Software, Reading, PA, USA).
Competition assays.
Individual overnight cultures of the wild-type strain and all congenic Rifr strains were inoculated into fresh individual 2-mL LB cultures. Upon entering stationary phase, the OD660 of each culture was measured and a volume corresponding to ∼108 cells of each strain was combined into a single mixed culture, which was then diluted 1:100 into 10 mL of either LB or SMMAsn in 125-mL Erlenmeyer flasks. Cultures were grown at 37°C in a temperature-controlled rotary shaking bath at 200 rpm. Every 24 h, cultures were diluted 1:100 into fresh medium. Under these conditions, cultures progress through ∼7 generations per day (53). For multiallelic competition experiments, every 24 h, from each culture, a 1-mL aliquot was pelleted for chromosomal DNA extraction; 0.8 mL was stored in 25% (vol/vol) glycerol at −70°C, and 0.1 mL was diluted serially 10-fold in phosphate-buffered saline (PBS), plated on LB with chloramphenicol (Cm), and incubated overnight at 37°C for total viable counts. For two-allele competition experiments, every 24 h, 0.1 mL of each culture was serially diluted and plated on two different LB plates, each supplemented with a different antibiotic. These LB plates were then incubated overnight at 37°C to determine the viable counts of each strain. Relative fitness was quantified with a previously described selection coefficient model (54).
Next-generation library preparation, sequencing, and data analysis.
From purified chromosomal DNA, two ∼500-bp amplicons of the rpoB gene, one spanning the N cluster and one spanning clusters I, II, and III, were amplified by PCR using Q5 high-fidelity DNA polymerase (New England Biolabs) and the corresponding primer pairs (Table 2). PCR products were purified (Qiagen PCR purification kit) and quantified by Qubit fluorometry, and equal amounts were combined for library preparation. Next-generation sequencing libraries were prepared using an Illumina Nextera XT DNA library preparation kit with some modifications. Due to the high levels of NaOH in the Nextera XT bead-based normalization process, the native normalization process is not compatible with the iSeq100. In place of the bead-based normalization process, normalization was performed manually by capillary electrophoresis using the Agilent Bioanalyzer 2100 expert software to calculate the molar amount of DNA fragments in each library. Equivalent amounts of DNA fragments from 24 individual libraries were pooled at a final DNA fragment concentration of 4 nM. Pooled libraries were diluted to 50 pM prior to loading into an Illumina i1 reagent box and sequenced on an Illumina iSeq100 next-generation-sequencing platform.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′–3′) | Purposed |
|---|---|---|
| I-cluster-F | CGGAGTAGGCGACACAGATG | Amplification of clusters I, II, and III for NGS |
| I-cluster-R | ATTCTGCCCGTTACCTTCCC | Amplification of clusters I, II, and III for NGS |
| N-cluster-F | TTCAGTATGGACGACACCGC | Amplification of N cluster for NGS |
| N-cluster-R | TTACGTGTGCGATCAATGCG | Amplification of N cluster for NGS |
| Q5SDM_A478V_F | AACCCGCTTGTTGAATTAACG a | Site-directed mutagenesis for rpoB-A478V |
| Q5SDM_A478V_R | CGTCTGATCCATGAATTG | Site-directed mutagenesis for rpoB-A478V |
| rpoB+1409_sgRNA+ | AAACcttgctgaattaacgcacaa b | sgRNA oligonucleotide |
| rpoB+1409_sgRNA- | TACGttgtgcgttaattcagcaag b | sgRNA oligonucleotide |
| rpoB-903F_HT | AAGGCCAACGAGGCCggcgagctttttctttgcca c | rpoB homology template |
| rpoC+181R_HT | AAGGCCTTATTGGCCtcgcttgtacttcccgcaat c | rpoB homology template |
| rpoB-24F | CGCATGATTTGAGGGG | N-cluster amplification and sequencing |
| rpoB+737R | GGCGGCTCTCCAGG | N-cluster amplification |
| rpoB+1319F | CGAATACAATTACGCCTCAGC | Cluster I, II, and III amplification and sequencing |
| rpoB+2000R | CCTGATACGTATTCCATACC | Cluster I, II, and III amplification |
Bold underlined letter represents mutation being introduced.
Uppercase letters represent bases complementary to BsaI cut site on pJOE8999 (51); lowercase letters represent wild-type rpoB sequence.
Uppercase letters represent SfiI recognition and cut site complementary to pJOE8999 (51); lowercase letters represent wild-type genomic sequence.
NGS, next-generation sequencing; sgRNA, single guide RNA.
Raw data from the iSeq100 was demultiplexed and converted to fastq files using Illumina bcl2fastq software. Quality control of reads was performed with FastQC (55). Quality reads were mapped to the Bacillus subtilis strain 168 reference genome (GenBank accession number AL009126.3) using Bowtie 2 (56). From mapped reads, LoFreq (57) was used to call single-nucleotide polymorphisms and small deletions in the rpoB gene. Preliminary calibration experiments performed with known proportions of mutant rpoB alleles established that the lower limit of detection was ∼0.1% (data not shown).
Statistical analysis.
All data sets were tested for normality using the Shapiro-Wilk method (58). Data sets passing the normality test were analyzed for differences by ANOVA with Tukey’s honestly significant difference (HSD) test (59) or Dunnett’s test for multiple comparison analysis (60). All statistical analyses were performed with the statistical graphing software Prism (GraphPad Software). Differences with a P value of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank the Department of Microbiology and Cell Science at the University of Florida for use of the iSeq100 instrument.
This work was supported by grants from the NASA Space Biology (grant number NNX14AT38G) and USDA-Hatch (grants number FLA-MCS-005500 and FLA-MCS-006066) programs to W.L.N.
Contributor Information
Wayne L. Nicholson, Email: WLN@ufl.edu.
Maia Kivisaar, University of Tartu.
REFERENCES
- 1.Wehrli W, Knüsel F, Schmid K, Staehelin M. 1968. Interaction of rifamycin with bacterial RNA polymerase. Proc Natl Acad Sci USA 61:667–673. 10.1073/pnas.61.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tupin A, Gualtieri M, Roquet-Banères F, Morichaud Z, Brodolin K, Leonetti J-P. 2010. Resistance to rifampicin: at the crossroads between ecological, genomic and medical concerns. Int J Antimicrob Agents 35:519–523. 10.1016/j.ijantimicag.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Jin DJ, Gross CA. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol 202:45–58. 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 4.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 5.Naryshkina T, Mustaev A, Darst SA, Severinov K. 2001. The beta′ subunit of Escherichia coli RNA polymerase is not required for interaction with initiating nucleotide but is necessary for interaction with rifampicin. J Biol Chem 276:13308–13313. 10.1074/jbc.M011041200. [DOI] [PubMed] [Google Scholar]
- 6.Severinov K, Soushko M, Goldfarb A, Nikiforov V. 1993. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the beta subunit of Escherichia coli RNA polymerase. J Biol Chem 268:14820–14825. 10.1016/S0021-9258(18)82407-3. [DOI] [PubMed] [Google Scholar]
- 7.Maughan H, Galeano B, Nicholson WL. 2004. Novel rpoB mutations conferring rifampin resistance on Bacillus subtilis: global effects on growth, competence, sporulation, and germination. J Bacteriol 186:2481–2486. 10.1128/JB.186.8.2481-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perkins AE, Nicholson WL. 2008. Uncovering new metabolic capabilities of Bacillus subtilis using phenotype profiling of rifampin-resistant rpoB mutants. J Bacteriol 190:807–814. 10.1128/JB.00901-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moeller R, Vlašić I, Reitz G, Nicholson WL. 2012. Role of altered rpoB alleles in Bacillus subtilis sporulation and spore resistance to heat, hydrogen peroxide, formaldehyde, and glutaraldehyde. Arch Microbiol 194:759–767. 10.1007/s00203-012-0811-4. [DOI] [PubMed] [Google Scholar]
- 10.Leighton TJ. 1973. An RNA polymerase mutation causing temperature-sensitive sporulation in Bacillus subtilis. Proc Natl Acad Sci USA 70:1179–1183. 10.1073/pnas.70.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg EC, Walker GC, Siede W. 2005. DNA repair and mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 12.Nicholson WL, Maughan H. 2002. The spectrum of spontaneous rifampin resistance mutations in the rpoB gene of Bacillus subtilis 168 spores differs from that of vegetative cells and resembles that of Mycobacterium tuberculosis. J Bacteriol 184:4936–4940. 10.1128/JB.184.17.4936-4940.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkins AE, Schuerger AC, Nicholson WL. 2008. Isolation of rpoB mutations causing rifampicin resistance in Bacillus subtilis spores exposed to simulated Martian surface conditions. Astrobiology 8:1159–1167. 10.1089/ast.2007.0224. [DOI] [PubMed] [Google Scholar]
- 14.Moeller R, Reitz G, Berger T, Okayasu R, Nicholson WL, Horneck G. 2010. Astrobiological aspects of the mutagenesis of cosmic radiation on bacterial spores. Astrobiology 10:509–521. 10.1089/ast.2009.0429. [DOI] [PubMed] [Google Scholar]
- 15.Moeller R, Reitz G, Nicholson WL, Horneck G, the PROTECT team. 2012. Mutagenesis in bacterial spores exposed to space and simulated Martian conditions: data from the EXPOSE-E spaceflight experiment PROTECT. Astrobiology 12:457–468. 10.1089/ast.2011.0739. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson WL, Park R. 2015. Anaerobic growth of Bacillus subtilis alters the spectrum of spontaneous mutations in the rpoB gene leading to rifampicin resistance. FEMS Microbiol Lett 362:fnv213. 10.1093/femsle/fnv213. [DOI] [PubMed] [Google Scholar]
- 17.Fajardo-Cavazos P, Leehan JD, Nicholson WL. 2018. Alterations in the spectrum of spontaneous rifampicin-resistance mutations in the Bacillus subtilis rpoB gene after cultivation in the human spaceflight environment. Front Microbiol 9:192. 10.3389/fmicb.2018.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leehan JD, Nicholson WL. 2021. The spectrum of spontaneous rifampin resistance mutations in the Bacillus subtilis rpoB gene depends on the growth environment. Appl Environ Microbiol 87:e01237-21. 10.1128/AEM.01237-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munakata N, Takahashi K, Hieda K, Panitz C, Horneck G. 2004. Mutagenesis of Bacillus subtilis spores exposed to simulated space environment, abstr 04-A-00898. Abstr 35th COSPAR Sci Assembly, Paris, France.
- 20.Wrande M, Roth JR, Hughes D. 2008. Accumulation of mutants in “aging” bacterial colonies is due to growth under selection, not stress-induced mutagenesis. Proc Natl Acad Sci USA 105:11863–11868. 10.1073/pnas.0804739105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maudsdotter L, Ushijima Y, Morikawa K. 2019. Fitness of spontaneous rifampicin-resistant Staphylococcus aureus isolates in a biofilm environment. Front Microbiol 10:988. 10.3389/fmicb.2019.00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee DJ, Minchin SD, Busby SJW. 2012. Activating transcription in bacteria. Annu Rev Microbiol 66:125–152. 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 23.Pruss GJ, Drlica K. 1989. DNA supercoiling and prokaryotic transcription. Cell 56:521–523. 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- 24.Dorman CJ, Dorman MJ. 2016. DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression. Biophys Rev 8:209–220. 10.1007/s12551-016-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christaki E, Marcou M, Tofarides A. 2020. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol 88:26–40. 10.1007/s00239-019-09914-3. [DOI] [PubMed] [Google Scholar]
- 26.Cohan FM, King EC, Zawadzki P. 1994. Amelioration of the deleterious pleiotropic effects of an adaptive mutation in Bacillus subtilis. Evolution 48:81–95. 10.1111/j.1558-5646.1994.tb01296.x. [DOI] [PubMed] [Google Scholar]
- 27.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 28.Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM. 2015. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol Plant Microbe Interact 28:1049–1058. 10.1094/MPMI-01-15-0016-R. [DOI] [PubMed] [Google Scholar]
- 29.Rifat D, Campodónico VL, Tao J, Miller JA, Alp A, Yao Y, Karakousis PC. 2017. In vitro and in vivo fitness costs associated with Mycobacterium tuberculosis RpoB mutation H526D. Future Microbiol 12:753–765. 10.2217/fmb-2017-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM. 2015. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol 81:17–30. 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM. 2014. Evolution of Escherichia coli to 42°C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol 31:2647–2662. 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad TM, Frazier M, Joyce AR, Cho B-K, Knight EM, Lewis NE, Landick R, Palsson BØ. 2010. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci USA 107:20500–20505. 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Utrilla J, O’Brien EJ, Chen K, McCloskey D, Cheung J, Wang H, Armenta-Medina D, Feist AM, Palsson BO. 2016. Global rebalancing of cellular resources by pleiotropic point mutations illustrates a multi-scale mechanism of adaptive evolution. Cell Syst 2:260–271. 10.1016/j.cels.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodríguez-Verdugo A, Tenaillon O, Gaut BS. 2016. First-step mutations during adaptation restore the expression of hundreds of genes. Mol Biol Evol 33:25–39. 10.1093/molbev/msv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao W, Cameron DR, Davies JK, Kostoulias X, Stepnell J, Tuck KL, Yeaman MR, Peleg AY, Stinear TP, Howden BP. 2013. The RpoB H481Y rifampicin resistance mutation and an active stringent response reduce virulence and increase resistance to innate immune responses in Staphylococcus aureus. J Infect Dis 207:929–939. 10.1093/infdis/jis772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colicchio R, Pagliuca C, Pastore G, Cicatiello AG, Pagliarulo C, Talà A, Scaglione E, Sammartino JC, Bucci C, Alifano P, Salvatore P. 2015. Fitness cost of rifampin resistance in Neisseria meningitidis: in vitro study of mechanisms associated with rpoB H553Y mutation. Antimicrob Agents Chemother 59:7637–7649. 10.1128/AAC.01746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hua X, Wang H, Wang C, Tian B, Hua Y. 2011. Global effect of an RNA polymerase β-subunit mutation on gene expression in the radiation-resistant bacterium Deinococcus radiodurans. Sci China Life Sci 54:854–862. 10.1007/s11427-011-4209-3. [DOI] [PubMed] [Google Scholar]
- 38.Qi Q, Preston GM, MacLean RC. 2014. Linking system-wide impacts of RNA polymerase mutations to the fitness cost of rifampin resistance in Pseudomonas aeruginosa. mBio 5:e01562-14. 10.1128/mBio.01562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inaoka T, Takahashi K, Yada H, Yoshida M, Ochi K. 2004. RNA polymerase mutation activates the production of a dormant antibiotic 3,3′-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J Biol Chem 279:3885–3892. 10.1074/jbc.M309925200. [DOI] [PubMed] [Google Scholar]
- 40.Concepción-Acevedo J, Weiss HN, Chaudhry WN, Levin BR. 2015. Malthusian parameters as estimators of the fitness of microbes: a cautionary tale about the low side of high throughput. PLoS One 10:e0126915. 10.1371/journal.pone.0126915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maughan H, Nicholson WL. 2011. Increased fitness and alteration of metabolic pathways during Bacillus subtilis evolution in the laboratory. Appl Environ Microbiol 77:4105–4118. 10.1128/AEM.00374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yüksel M, Power JJ, Ribbe J, Volkmann T, Maier B. 2016. Fitness trade-offs in competence differentiation of Bacillus subtilis. Front Microbiol 7:888. 10.3389/fmicb.2016.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholson WL. 2012. Increased competitive fitness of Bacillus subtilis under nonsporulating conditions via inactivation of pleiotropic regulators AlsR, SigD, and SigW. Appl Environ Microbiol 78:3500–3503. 10.1128/AEM.07742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stefan MA, Ugur FS, Garcia GA. 2018. Source of the fitness defect in rifamycin-resistant Mycobacterium tuberculosis RNA polymerase and the mechanism of compensation by mutations in the β′ subunit. Antimicrob Agents Chemother 62:e00164-18. 10.1128/AAC.00164-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Z, Zhou A, Wu J, Zhou A, Li J, Zhang S, Wu W, Karakousis PC, Yao Y-F. 2018. Transcriptional approach for decoding the mechanism of rpoC compensatory mutations for the fitness cost in rifampicin-resistant Mycobacterium tuberculosis. Front Microbiol 9:2895. 10.3389/fmicb.2018.02895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brandis G, Hughes D. 2018. Mechanisms of fitness cost reduction for rifampicin-resistant strains with deletion or duplication mutations in rpoB. Sci Rep 8:17488. 10.1038/s41598-018-36005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cutting SM, Vander Horn PB. 1990. Genetic analysis, p 27–74, In Harwood CR, Cutting SM (ed), Molecular biological methods for Bacillus. John Wiley & Sons Ltd, Chichester, England. [Google Scholar]
- 48.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Morrison MD, Fajardo-Cavazos P, Nicholson WL. 2017. Cultivation in space flight produces minimal alterations in the susceptibility of Bacillus subtilis cells to 72 different antibiotics and growth-inhibiting compounds. Appl Environ Microbiol 83:e01584-17. 10.1128/AEM.01584-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA 44:1072–1078. 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altenbuchner J. 2016. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl Environ Microbiol 82:5421–5427. 10.1128/AEM.01453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva D, Santos G, Barroca M, Collins T. 2017. Inverse PCR for point mutation introduction. Methods Mol Biol 1620:87–100. 10.1007/978-1-4939-7060-5_5. [DOI] [PubMed] [Google Scholar]
- 53.Maughan H, Callicotte V, Hancock A, Birky CW, Nicholson WL, Masel J. 2006. The population genetics of phenotypic deterioration in experimental populations of Bacillus subtilis. Evolution 60:686–695. 10.1111/j.0014-3820.2006.tb01148.x. [DOI] [PubMed] [Google Scholar]
- 54.Woods RJ, Barrick JE, Cooper TF, Shrestha U, Kauth MR, Lenski RE. 2011. Second-order selection for evolvability in a large Escherichia coli population. Science 331:1433–1436. 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data, v0.11.5. Babraham Bioinformatics. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 56.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilm A, Aw PPK, Bertrand D, Yeo GHT, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shapiro SS, Wilk MB. 1965. An analysis of variance test for normality (complete samples). Biometrika 52:591–611. 10.2307/2333709. [DOI] [Google Scholar]
- 59.Haynes W. 2013. Tukey’s test, p 2303–2304. In Dubitzky W, Wolkenhauer O, Cho K-H, Yokota H (ed), Encyclopedia of systems biology. Springer, New York, NY. 10.1007/978-1-4419-9863-7_1212. [DOI] [Google Scholar]
- 60.Dunnett CW. 1955. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50:1096–1121. 10.1080/01621459.1955.10501294. [DOI] [Google Scholar]
- 61.Qi Y, Hulett FM. 1998. PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol Microbiol 28:1187–1197. 10.1046/j.1365-2958.1998.00882.x. [DOI] [PubMed] [Google Scholar]
- 62.Waters SM, Zeigler DR, Nicholson WL. 2015. Experimental evolution of enhanced growth by Bacillus subtilis at low atmospheric pressure: genomic changes revealed by whole-genome sequencing. Appl Environ Microbiol 81:7525–7532. 10.1128/AEM.01690-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guérout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61. 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]